ABSTRACT
p63 (also known as TP63) is a transcription factor of the p53 family, along with p73. Multiple isoforms of p63 have been discovered and these have diverse functions encompassing a wide array of cell biology. p63 isoforms are implicated in lineage specification, proliferative potential, differentiation, cell death and survival, DNA damage response and metabolism. Furthermore, p63 is linked to human disease states including cancer. p63 is critical to many aspects of cell signaling, and in this Cell science at a glance article and the accompanying poster, we focus on the signaling cascades regulating TAp63 and ΔNp63 isoforms and those that are regulated by TAp63 and ΔNp63, as well the role of p63 in disease.
KEY WORDS: Cancer, Development, Isoforms, p63, Stemness, Differentiation, Signal transduction
Summary: A review of the signaling cascades regulating and regulated by the different isoforms of the p53-related transcription factor p63, as well the role of p63 in disease.
Introduction
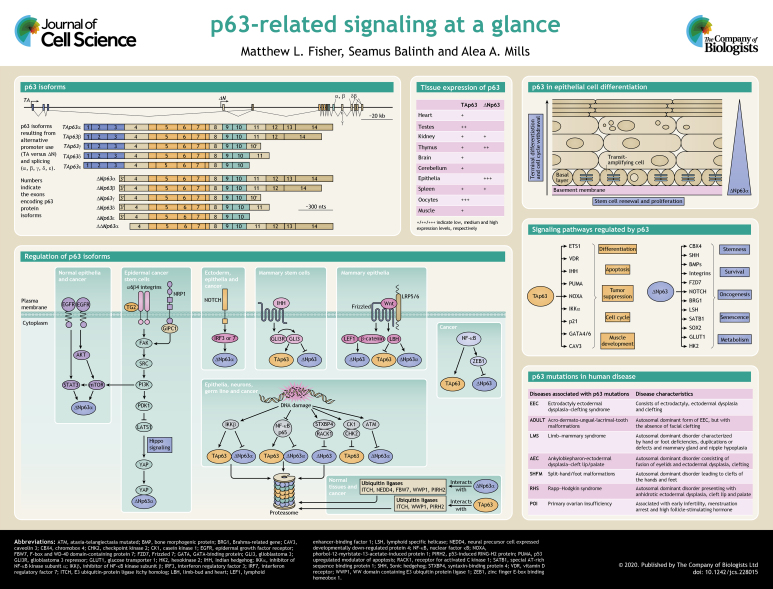
p63, also known as Trp63 (transformation related protein 63), TP63 (tumor protein 63) or AIS (amplified in squamous cell carcinoma), is a member of the p53 family of transcription factors, which also includes p73 (Botchkarev and Flores, 2014; Soares and Zhou, 2018). Despite initial ideas that p63 evolved to modulate p53 function, p63 has been shown to be the primordial family member of the p53/p63/p73 gene family (Amelio et al., 2012). Several isoforms of p63 exist, and these are linked to numerous biological functions, including development and differentiation, senescence, proliferation, stem cell maintenance, aging, and apoptosis (Galoczova et al., 2018; Yoh and Prywes, 2015; Keyes and Mills, 2006; Gonfloni et al., 2015). p63 is an important player in embryonic epidermal development and in epidermal keratinocyte morphogenesis, proliferation and differentiation, where it directly transactivates a plethora of target genes involved in cellular proliferation, differentiation and adhesion (Kouwenhoven et al., 2015; Moll and Slade, 2004; Boughner et al., 2018; van Bokhoven et al., 2011; Westfall and Pietenpol, 2004). Additionally, p63 plays a role in the modulation of the chromatin landscape in epidermal keratinocytes by directly regulating chromatin-modulating factors and by engaging and opening chromatin regions (Qu et al., 2018, Pattison et al., 2018). The existence of multiple isoforms with distinct, often opposing, functions allows p63 to have a wide array of effects on essential cellular functions (Westfall and Pietenpol, 2004) (see Box 1 and poster). Different p63 isoforms have been shown to impact various cells, tissues and diseases (Soares and Zhou, 2018). Because p63 isoforms have such a diverse array of functions, they must act on numerous pathways to carry out these functions. Furthermore, there is an equally large number of pathways that regulate p63 expression. We will focus here on some of the key signaling pathways that regulate, and are regulated by, the TAp63 and ΔNp63 isoforms to demonstrate the vast array of functions p63 isoforms can carry out.
Box 1. Structure of p63 isoforms.
The p63 gene consists of 15 exons spanning ∼270 kb (Murray-Zmijewski et al., 2006; Mills et al., 1999) (see poster) and maps to chromosome 3q27 (Murray-Zmijewski et al., 2006). It encodes two main classes of isoforms that are generated by alternative promoters. TAp63 transcripts are produced from the promoter upstream of exon 1 (Yang et al., 1998). TAp63-encoding transcripts contain three TA-specific exons (exons 1, 2 and 3) that encode an N-terminal transactivation domain, homologous to that of p53 (Murray-Zmijewski et al., 2006). An alternate promoter within the intron downstream of exon 3 results in transcripts encoding truncated ΔNp63 isoforms that lack the N-terminal transactivation domain (Sethi et al., 2015). However ΔNp63 isoforms retain the ability to induce genes via a region in exon 3′ (Helton et al., 2006). The ability of ΔNp63 to activate target genes depends on the first 26 residues and the first 14 residues for ΔNp63α and ΔNp63β, respectively (Bergholz and Xiao, 2012; Helton et al., 2006). Additionally, the PXXP and PPXY motif are required for optimal transactivation. Therefore, the 14 residues unique to the ΔNp63 isoforms and the adjacent region, comprise an activation domain for ΔNp63 isoforms (Helton et al., 2006) (see poster). A further truncated form of p63 can be produced, and is denoted the ΔΔNp63 isoform. This isoform lacks the first 26 amino acids of ΔNp63 isoforms due to an alternative translational start site in exon 4 (Rinne et al., 2008).
Alternative splicing occurring at the 3′ end of the p63 mRNA generates multiple C-terminal variants (α, β, γ, δ and ε) for both TAp63 and ΔNp63 (King et al., 2013; Mangiulli et al., 2009). At the C-terminus, the longest isoform is α, which contains exons 11–14. At the protein level, the α isoform contains a sterile alpha-motif (SAM) involved in protein–protein interactions and a transactivation inhibitory domain (TID) (Soares and Zhou, 2018). The β and δ isoforms lack exon 13, and γ does not have exons 11–14 but has a γ-specific exon 10′ (Ghioni et al., 2002). The ε isoform is generated by a premature stop codon located in exon 10 (Mangiulli et al., 2009). All isoforms contain the DNA-binding domain and oligomerization domain (OD). TAp63 and ΔNp63 isoforms are typically differentially expressed. TAp63 variants are prevalent in the heart, testis, kidney, thymus, brain and cerebellum, while ΔNp63 transcripts are highly detected in epithelia, kidney, spleen and thymus, but not the heart, liver, testis or brain (Di Como et al., 2002; Kawasaki et al., 2006; Nylander et al., 2002) (see poster).
Regulation of p63
p63 isoforms are critical to a number of aspects of cell biology, and below we discuss some of the many signaling events that can regulate p63 expression (see poster).
DNA damage
A major regulator of p63 is DNA damage. The response to DNA damage appears to be isoform and cell type specific (Moll and Slade, 2004; Yoh and Prywes, 2015). TAp63 isoforms are expressed in response to DNA damage or other stresses in epithelial tissue, neurons and the germline (Su et al., 2013). TAp63 is primarily found as a closed dimer with low activity due to the C-terminal inhibitory domain and N-terminal TA domain blocking the oligomerization domain (Deutsch et al., 2011). This differs from ΔNp63 isoforms typically found as tetramers (Russo et al., 2018). However, in response to DNA damage, TAp63α is phosphorylated by CHK2 (also known as CHEK2) and casein kinase 1 (CK1), leading first to the formation of an open dimer and then an active tetramer (Deutsch et al., 2011). Furthermore, in oocytes, TAp63 mediates apoptosis in response to DNA damage by inducing Puma and Noxa (also known as BBC3 and PMAIP1, respectively) (Kerr et al., 2012). Conversely, GSK-3β reduces TAp63 expression to protect oocytes from premature death (Wen et al., 2019). In head and neck squamous cell carcinoma (HNSCC), DNA damage triggered by chemotherapy decreases ΔNp63-mediated transcriptional repression by blocking p63-responsive elements or sequestering TAp63 into less-active heterotetramers (Melino, 2011). A different report found that, in response to cisplatin treatment, c-Abl phosphorylates the Y55, Y137 and Y308 residues of ΔNp63α; this stabilizes it by inducing its binding to Yes-associated protein (YAP; also known as YAP1), consequently regulating cell viability in HNSCC cells (Yuan et al., 2010). Additionally, c-Abl was shown to phosphorylate TAp63α on Y149, Y171 and Y289 residues, resulting in increased protein stability of TAp63α (Gonfloni et al., 2009).
The ataxia-telangiectasia mutated (ATM) kinase is also critical to the p63 response to DNA damage. ATM reduces ΔNp63α and increases p53 after irradiation (Huang et al., 2008). This impairs the proliferative capacity of epithelial stem cells with regard to p53-mediated repair of DNA damage. Furthermore, DNA damage leads to phosphorylation of TAp63γ by inhibitor of nuclear factor-κB kinase-β (IKKβ; also known as IKBKB) in response to ionizing radiation, resulting in its stabilization (MacPartlin et al., 2008).
Additionally, two mechanisms related to the NF-κB pathway have been found to mediate DNA-damage-induced ΔNp63 degradation. IKKβ is capable of binding to ΔNp63, phosphorylating it to induce degradation (Chatterjee et al., 2010). The p65 subunit of NF-κB can also bind directly to ΔNp63 in cisplatin-treated cells, leading to degradation of ΔNp63 (Sen et al., 2010).
Degradation of ΔNp63 in response to DNA damage is carried out by the proteasome. The scaffold protein syntaxin-binding protein 4 (STXBP4) binds to ΔNp63 and stabilizes it (Otaka et al., 2017). In response to DNA damage, homeodomain-interacting protein kinase 2 (HIPK2) phosphorylates ΔNp63 on T397. STXBP4 is downregulated, resulting in the destabilization of ΔNp63 through receptor of activated kinase C1 (RACK1), which facilitates the binding of ΔNp63 to ubiquitin ligases (Lazzari et al., 2011). Multiple ubiquitin ligases that bind p63 have been identified, including neuronal precursor cell-expressed developmentally downregulated 4 (NEDD4), itchy E3 ubiquitin protein ligase (ITCH), WW domain containing E3 ubiquitin protein ligase 1 (WWP1), F-box and WD-40 domain-containing protein 7 (FBW7, also known as FBXW7), putative E3 ubiquitin-protein ligase RING1B, and p53-induced RING-H2 protein (PIRH2; also known as RCHY1) (Li and Xiao, 2014). Pirh2 is also a transcriptional target of ΔNp63, establishing an auto-regulatory loop, and is required for epithelial differentiation (Jung et al., 2013). Binding of these E3 ligases in turn targets ΔNp63 for proteasomal degradation (Li and Xiao, 2014). Furthermore, feedback loops occur frequently as part of p63 regulation, as discussed further below.
Cell surface receptors
The tyrosine kinase receptor epidermal growth factor receptor (EGFR) can induce ΔNp63 expression. EGFR activation of phosphatidylinositol 3-kinase (PI3K) signaling has been shown to upregulate ΔNp63α in keratinocytes (Barbieri et al., 2003); in cancer, EGFR activation of ΔNp63α is mediated by STAT3 (Ripamonti et al., 2013). These pathways are likely linked through mammalian target of rapamycin (mTOR). PI3K activation of mTOR has been shown to lead to mTOR-dependent activation of the STAT3-p63-Jagged pathway (Ma et al., 2010). In addition to EGFR-mediated activation of ΔNp63α, the interaction of α6β4 integrin with specific proteins stimulates ΔNp63α expression, such as interaction with transglutaminase 2 (TG2; also known as TGM2) (Fisher et al., 2016). In normal epithelial tissue, α6β4 integrin is associated with stable adhesion and does not activate FAK–SRC signaling (Duperret and Ridky, 2013; Epifano and Perez-Moreno, 2012). However, in squamous cell carcinoma, α6β4 integrin associates with the actin cytoskeleton and forms new signaling complexes that allow for the activation of FAK–SRC and PI3K–PD1 signaling (Duperret and Ridky, 2013; Epifano and Perez-Moreno, 2012; Kim and Gumbiner, 2015). FAK and SRC are non-receptor tyrosine kinases that form a complex and phosphorylate various adapter proteins that promote cell motility, cell cycle progression and survival. PI3K is responsible for generating phosphoinositol lipids that regulate cell growth, proliferation, survival and cytoskeletal changes. PI3K activation results in pyruvate dehydrogenase kinase 1 (PDK1) activation, which is responsible for phosphorylating AKT family proteins, among other targets, such as large tumor suppressor kinase 1 (LATS1), a key kinase in the Hippo signaling cascade (Fisher et al., 2016; Kim and Gumbiner, 2015). LATS1 phosphorylates and inactivates YAP (Hao et al., 2008), so when LATS1 is inactivated by PDK1, YAP is free to enter the nucleus where it binds to ΔNp63a and stabilizes its expression by preventing its proteasomal degradation (Fisher et al., 2016).
In addition to TG2, neuropilin-1 (NRP-1), a transmembrane protein that acts as a co-receptor for numerous extracellular ligands, forms a complex with the scaffolding protein GAIP C-terminus interacting protein 1 (GIPC1) and α6β4 integrin to activate a downstream kinase cascade that also leads to ΔNp63α stabilization (Grun et al., 2018). NRP-1 interacts with vascular endothelial growth factor A (VEGF-A); this leads to an interaction of VEGF-A with its receptor VEGFR, resulting in enhanced angiogenesis (Klagsbrun et al., 2002). Consistent with this role, NRP-1 is often overexpressed in cancer, where it increases blood vessel formation and tumor growth (Grun et al., 2016; Jimenez-Hernandez et al., 2018; Lin et al., 2018). In squamous cell carcinoma, NRP-1 has been shown to interact with α6β4 integrin and GIPC1 independently of binding to VEGF-A. This interaction results in a suppression of Hippo signaling, relocation of YAP to the nucleus, and ultimately increased ΔNp63α expression (Grun et al., 2018).
Finally, Notch, a key regulator of maintenance, cell fate specification and differentiation, has been shown to regulate p63 in a cell-type-specific manner (Dotto, 2009; Siebel and Lendahl, 2017). In keratinocytes, mammary epithelial and ectodermal progenitor cells, Notch activation reduces p63 expression in a manner dependent on interferon-regulatory factor 3 (IRF3) and IRF7, transcriptional regulators associated with regulation of the type 1 interferon system (Nguyen et al., 2006).
Wnt/β-catenin pathway
Wnt/β-catenin signaling is a highly conserved pathway involved in the regulation of cellular proliferation, differentiation, migration, apoptosis and stem cell renewal. p63 is directly regulated by the Wnt/β-catenin pathway through binding of lymphoid enhancer binding factor 1 (Lef1) along with β-catenin between the TAp63 and ΔNp63 promoters (Ferretti et al., 2011). Additionally, there is a β-catenin-responsive element within the proximal ΔNp63 promoter (Chu et al., 2008). Wnt/β-catenin has also been shown to activate the transcriptional co-factor limb-bud and heart (LBH) (Rieger et al., 2010). LBH increases transcription of ΔNp63α in mammary epithelial cells, while downregulating TAp63α transcription, leading to enhanced replicative potential and stemness (Lindley et al., 2015).
Hedgehog signaling
The Hedgehog signaling pathway is an important, evolutionarily conserved pathway that transmits signals from the cell membrane to the nucleus. Hedgehog is involved in embryonic development and is critical for the formation of organismal polarity, as well as wound healing and maintenance of somatic stem cells and pluripotent cells. An important downstream target of Hedgehog signaling is p63 (Takebe et al., 2015). The hedgehog ligand Indian hedgehog (IHH) activates the transcription factor Gli3, which in the absence of IHH, is in a repressive form denoted Gli3R (Wang et al., 2000). The balance between the levels of Gli3 and Gli3R affects the level of p63; IHH-mediated induction of active Gli3 upregulates TAp63 expression, while reducing the usage of the ΔNp63 promoter. Additionally, shRNA-mediated disruption of Gli3 expression is sufficient to alter p63 promoter usage. This signaling likely acts on the mammary gland in a stage-specific manner, and can initiate elaboration of mammary progenitors and enhance clonigenicity of mammary stem cells to maintain mammary epithelial stasis (Li et al., 2008).
NF-κβ
In addition to the above-mentioned role of NF-κB in regulating p63 in response to DNA damage, NF-κB-mediated repression of p63 also has a role in epithelial cell differentiation. Overexpression the p65 subunit (also known as RelA) in epithelial cells leads to the induction of epithelial-mesenchymal transition (EMT) and reduced p63. This is likely due to NF-κB-mediated enhancement of zinc finger E-box binding homeobox (ZEB1) expression. ZEB1 is a transcription factor critical to the induction of EMT, that represses the ΔNp63 promoter (Chua et al., 2007). Additionally, NF-κB activates the TAp63 promoter, suggesting that NF-κB can induce a shift to TAp63-driven differentiation (Wu et al., 2010).
Major transcriptional targets of p63
p63 has been shown to be critical for many aspects of life. A major reason for this is the multitude of signaling cascades that p63 isoforms are able to regulate. Below we discuss some of these and their cellular effects.
Chromatin-modifying proteins
The polycomb group proteins (PcG) are a family of proteins involved in regulating differentiation via transcriptional repression and can form two major polycomb repressive complexes (PRCs), PRC1 or PRC2. The PcG family member chromobox protein homolog 4 (CBX4) is a transcriptional repressor required for preserving cell cycle dormancy of quiescent stem cells (Luis et al., 2011). Cbx4 is a direct downstream target of p63 and mediates its effects on epidermal cell proliferation, repressing non-epidermal lineage genes during epidermal differentiation via its canonical PRC1 function, as well as by regulating basal keratinocyte proliferation and differentiation via its SUMO E3 ligase activity (Cohen and Ezhkova, 2016; Mardaryev et al., 2016).
Another chromatin-modifying protein that is regulated by p63 is lymphoid-specific helicase (LSH; also known as HELLS) (Keyes et al., 2011), which is implicated in embryonic development and cellular senescence (Muegge, 2005). The LSH promoter contains consensus p63-binding sites, and ΔNp63α has been shown to be the predominant isoform binding the LSH promoter. ΔNp63α-driven Lsh expression is required for senescence bypass, and is induced during tumor initiation in keratinocytes and progression to squamous cell carcinoma (Keyes et al., 2011).
Additionally, p63 directly regulates the expression of the ATP-dependent chromatin remodeler BRG1 (also known as SMARCA4) in epidermal progenitor cells (Botchkarev, 2015). During epidermal morphogenesis, BRG1 binds to the epidermal differentiation complex (EDC), a keratinocyte lineage-specific gene cluster comprising over 50 genes encoding proteins involved in terminal differentiation and cornification of keratinocytes (Mardaryev et al., 2014). BRG1 binds to distinct regions of the EDC and is required for its relocation towards the nuclear interior (Mardaryev et al., 2014); this has a critical role in proper formation of stratified epithelia.
Another key chromatin-modifying protein target of p63 is Satb1. Satb1 is a global chromatin organizer and transcription factor that plays an important role in integrating higher-order chromatin architecture with gene regulation (Ding et al., 2018). Satb1 contributes to epidermal morphogenesis by establishing a tissue-specific chromatin organization and gene expression pattern in epidermal progenitor cells. p63 binds to a proximal regulatory region of the Satb1 gene, and ablation of p63 results in reduced Satb1 expression levels in the epidermis (Fessing et al., 2011). This results in failure to properly activate EDC-associated genes, impairing epidermal morphogenesis (Fessing et al., 2011).
Cell surface proteins
As mentioned above, multiple cell surface markers regulate p63 expression. Many of these markers are in turn transcriptionally activated by p63, creating feedback loops. For instance, ΔNp63α expression increases both the mRNA and protein levels of EGFR, as well as increasing EGFR activity, as measured by determining levels of phospho-EGFR (Y1086) (Holcakova et al., 2017). Furthermore, silencing of ΔNp63α in epithelial cells reduces both the total and phospho-EGFR levels, inhibiting the ability of EGF to activate EGFR (Holcakova et al., 2017). Additionally, direct transcriptional activation of the NRG1 gene – a member of the EGF family – by p63 occurs selectively in epithelial basal cells. Here, activation of NRG1 is required for activation of luminal ERBB4 and/or STAT5A expression, which leads to luminal progenitor cell maturation in breast epithelia (Forster et al., 2014).
Another group of cell surface markers regulated by p63 are integrins. TAp63γ and ΔNp63α induce expression of genes encoding integrins, as shown by their downregulation upon expression of p63-directed shRNAs (Carroll et al., 2006; Yang et al., 2011). In particular, genes encoding integrins α6, β4 and α3 are transcriptionally regulated by ΔNp63α (Carroll et al., 2006). α6β4 integrin is an essential component of hemidesmosomes, which provide stable adhesion to basal epithelial cells and the underlying basement membrane. Additionally, the PERP gene, encoding a key desmosomal protein important for desmosome stabilization, is also transactivated by ΔNp63α (Beaudry et al., 2009). Thus, ΔNp63α-induced expression of these adhesion markers increases cellular adhesion to the exogenous extracellular matrix (ECM) and also confers resistance to anoikis – a form of anchorage-dependent cell death – in breast epithelial cells (Carroll et al., 2006). This makes ΔNp63α critical for maintaining epithelial integrity, adhesion and survival.
Another cell surface marker gene that is a transcriptional target of p63 is the vitamin D receptor (VDR). ΔNp63α and TAp63γ can bind the VDR promoter, and silencing of p63 reduces VDR expression (Kommagani et al., 2006). Furthermore, a naturally occurring p63 missense mutant of TAp63γ (R279H) acts in a dominant-negative manner to inhibit TAp63γ-mediated upregulation of VDR (Kommagani et al., 2006).
Finally, ΔNp63 isoforms also induce the expression of the gene encoding the cell surface antigen CD44, which defines cancer stem cells in breast, colorectal and squamous cell carcinomas (Wang et al., 2018). Regulation of CD44 abundance by ΔNp63 could be achieved either directly or through indirect activation of the Hedgehog signaling pathway (Compagnone et al., 2017; Di Franco et al., 2016; Gatti et al., 2018). By sustaining the production of CD44, ΔNp63 contributes to the maintenance of cancer stem cell self-renewal (Gatti et al., 2018).
Proteins involved in cell metabolism
p63 isoforms are important regulators of energy metabolism. This is particularly true in squamous cell carcinomas, where the major glucose transporter GLUT1 is highly overexpressed (Hsieh et al., 2019). This high GLUT1 expression is driven by cooperation between ΔNp63 and SOX2. However, the precise mechanism in which ΔNp63 and SOX2 interact to transacvtivate GLUT1 is yet to be elucidated (Hsieh et al., 2019). Additionally, hexokinase 2 (HK2) is a direct ΔNp63α target gene in human keratinocytes. HK2 regulates mitochondrial ROS generation and catalyzes the first step of glycolysis, producing glucose-6-phosphate (Viticchiè et al., 2015). Furthermore, ΔNp63α transcriptionally regulates glutathione biogenesis, utilization and regeneration, and The Cancer Genome Atlas (TCGA) database shows that p63 amplification or overexpression upregulates the glutathione metabolism pathway in tumors (Wang et al., 2017).
Proteins involved in regulation of muscle cells
TAp63 isoforms play important roles in regulating skeletal and cardiac muscle. During myoblast differentiation, TAp63γ regulates numerous proteins involved in skeletal muscle contractility, including the components of the dystrophin-glycoprotein complex (Cav3), titin complex (Myh1, Myh2, Neb, Tnnt2 and Ttn), fast-twitch muscle fibers (Atp2a1, Myh1 and Myh2) and slow-twitch muscle fibers (Mb, Myh1 and Tnnc1) (Cefalù et al., 2015). These genes are critical to basic muscle function and growth. In addition to skeletal muscle, TAp63 is also important in cardiomyocytes (Rouleau et al., 2011). GATA transcription factors are a family of zinc finger proteins that bind the consensus DNA sequence GATA (Romano and Miccio, 2020). GATA4 and GATA6 are required for cardiogenesis, and in embryonic stem cells, TAp63 regulates GATA4 and GATA6. p63-null mice display defects in cardiac development including, alterations in cardiac gene expression, myofibrillogenesis and beating activity, which can be rescued with GATA6 expression in TAp63-deficient cells (Rouleau et al., 2011).
IKKα
The NF-κB family of transcription factors is a key regulator of immune responses, inflammation and cancer. NF-κB is quickly activated in response to various stimuli, allowing the rapid activation of target genes that encode cytokines, membrane proteins and transcription factors (Mitchell et al., 2016). Inhibitor of NF-κB kinase subunit α (IKKα) functions in the activation of NF-κB in response to a subset of tumor necrosis factor (TNF) family members. Moreover, IKKα is involved in keratinocyte differentiation, but this function is independent of its kinase activity (Israel, 2010). TAp63 is capable of driving the expression of the IKKα-encoding gene (CHUK) both directly and indirectly (Candi et al., 2006). Direct regulation of CHUK by TAp63 is through one of the three p53-consensus motifs within the CHUK promoter. In addition, TAp63 also regulates CHUK through the induction of ETS1, and GATA3, which subsequently induce CHUK expression (Candi et al., 2006).
p21
p21 (also known as CDKN1A) is a cyclin-dependent kinase (CDK) inhibitor that has an important role in cell cycle progression. p21 can arrest cell cycle progression in G1/S and G2/M transitions by inhibiting the CDK4–CDK6–cyclin-D and CDK2–cyclin-E complexes (Karimian et al., 2016). All isoforms of TAp63 transactivate the p21-encoding gene, with TAp63β showing the highest levels of p21 induction (Helton et al., 2008). The transcriptional activation of p21 by TAp63 requires both the AD and proline-rich domains of TAp63. Specifically, the PXXP motif of the proline-rich domain at residues 124–127 are critical (Helton et al., 2008). Therefore, TAp63 plays a critical role in regulating senescence and proliferation via regulation of p21 (Guo et al., 2009).
Hedgehog signaling
Much like integrin signaling, the Hedgehog pathway is involved in the regulation of p63, and, in turn, is also regulated by p63 itself. p63-mediated regulation of Hedgehog signaling contributes to stem cell maintenance through the ability of p63 to bind to the promoters of several genes encoding Hedgehog signaling pathway components (Chari et al., 2013; Melino et al., 2015; Sari et al., 2018). For instance, IHH is directly transcriptionally regulated, either positively or negatively by TAp63 and ΔNp63, respectively, and IHH is a driver of mammary progenitor maturation (Sari et al., 2018). p63 also transactivates sonic hedgehog (SHH) and GLI2 in mammary cancer progenitor cells by directly binding to their promoters (Memmi et al., 2015). These components of the Hedgehog signaling pathway are thus important mediators of the p63-driven stem cell phenotype.
Wnt
The canonical Wnt cascade has emerged as another critical regulator of p63, particularly in epithelial stem cells. In the absence of a Wnt ligand, β-catenin is constitutively phosphorylated, and degraded by the proteasome (Zhan et al., 2017). When Wnt ligands are present, a member of the Frizzled (FZD) receptor family and its co-receptor low-density-lipoprotein-related protein (LRP) are engaged, inhibiting β-catenin phosphorylation (Zhan et al., 2017). This results in β-catenin accumulation in the nucleus, where it interacts with the LEF/TCF family of DNA-binding proteins to bind to its target promoters (Zhan et al., 2017). In particular, the FZD7 receptor and WNT5B ligand are under the direct transcriptional control of ΔNp63 (Chakrabarti et al., 2014). ΔNp63 depletion in mammary progenitors results in impaired self-renewal, and FZD7 expression can rescue the impaired self-renewal of both normal and malignant mammary progenitors following ΔNp63 depletion, suggesting that FZD7 is a key downstream regulator of ΔNp63-driven stemness (Chakrabarti et al., 2014).
Notch
The Notch signaling pathway is induced in response to the engagement of Notch receptors (Notch1–4) by membrane-bound Delta or Jagged ligands, leading to Notch cleavage and release of its intracellular domain (NICD) into the cytoplasm (Siebel and Lendahl, 2017). NICD then translocates into the nucleus, where it converts the DNA-binding protein CBF1 from a repressor into a transcriptional activator (Siebel and Lendahl, 2017).
ΔNp63 affects Notch signaling by regulating the expression of multiple genes encoding Notch pathway components, including Notch1, Notch3, Jag1, Jag2 and Hes1 (Kent et al., 2011; Mezzomo et al., 2017; Ravindran and Devaraj, 2012; Yalcin-Ozuysal et al., 2010). ΔNp63α is directly involved in transactivation of Notch by binding to the Notch1 promoter; this increases mRNA and protein levels of Notch1 and leads to increased accumulation of the Notch1 intracellular domain. This in turn induces the expansion of breast cancer stem cell subpopulations (Du et al., 2010). In the mammary gland, Notch inhibits ΔNp63α to induce luminal commitment. However, ΔNp63α maintains mammary cell fate by counteracting Notch activation (Balboni et al., 2013).
Additionally, reciprocal antagonistic interactions between ΔNp63α and Notch regulate epithelial cells (Koh et al., 2015; Nguyen et al., 2006; Tadeu and Horsley, 2013). In the basal layer of the epidermis, opposing gradients of ΔNp63α and Notch activation govern the balance between self-renewing and transit-amplifying cells (Senoo, 2013). Therefore, the interplay between p63 and Notch are critical to the regulation of stem cell quiescence and progenitor commitment (Melino et al., 2015).
BMP
Bone morphogenetic proteins (BMPs) are members of the transforming growth factor β (TGF-β) superfamily of cytokines. BMP ligands activate signaling by forming complexes with receptor kinases at the cell surface. Typically, BMP signaling induces cellular quiescence and maintenance of embryonic and adult stem cells (Wu et al., 2016). In the mammary epithelium, ΔNp63α activates canonical BMP signaling by inducing the expression of Bmp7, which induces the stem cell phenotype of mammary progenitors (Balboni et al., 2013).
p63 in disease
p63 is involved in a number of disease syndromes. Germline mutations of p63 are found in humans and cause six rare autosomal dominant developmental diseases (see Box 2) (Soares and Zhou, 2018). In addition to mutations in p63 that cause disease, changes in p63 expression are associated with numerous cancers. p63 was initially hypothesized to be a tumor suppressor based on its homology to p53. However, mutation of the p63 gene in cancer is rare, indicating that p63 is not a classical tumor suppressor such as p53 (Gonfloni et al., 2015). Furthermore, spontaneous tumors are extremely rare in mouse models heterozygous for null p63 alleles (Gonfloni et al., 2015; van Bokhoven et al., 2011; Keyes et al., 2006). However, the human p63 gene maps to chromosome 3q27–28, a region frequently amplified in squamous cell carcinomas and some adenocarcinomas, with copy number of the p63 locus and p63 protein levels being significantly increased in 88% of squamous carcinomas, and in 42% of large cell carcinomas and adenocarcinomas of lung (Massion et al., 2003). The predominant isoform expressed is ΔNp63α (Botchkarev and Flores, 2014). ΔNp63α expression correlates with a poor response to cisplatin in multiple cancers. This is likely due to the number of pro-survival pathways regulated by ΔNp63α, allowing it to act as an oncogene (Botchkarev and Flores, 2014; Moll and Slade, 2004; Gonfloni et al., 2015). In contrast to ΔNp63α, exogenous expression of TAp63 isoforms, can induce cellular senescence in cultured cells and inhibit tumor formation and metastasis upon xenograft implantation in nude mice, suggesting that TAp63 is a tumor suppressor (Su et al., 2013; Guo et al., 2009).
Box 2. Germline mutations of p63.
Germline mutations are genetic variations within germ cells, resulting in the mutation being passed on to offspring. Below are known diseases that result from various mutations occurring in the p63 gene within germ cells.
Ectrodactyly ectodermal dysplasia–clefting
Ectrodactyly ectodermal dysplasia–clefting (EEC) syndrome has three major features: ectrodactyly (a split or cleft anomaly of the hands and feet), ectodermal dysplasia (developmental defects of hair, teeth, nails and sweat glands) and clefting (cleft lip with or without cleft palate). Mutations in codons 204, 227, 279, 280, and 304 of p63 lead to amino acid substitutions and are present in ∼75% of EEC patients. These mutations affect the p63 DNA-binding domain (Rinne et al., 2007; van Bokhoven and Brunner, 2002).
Acro-dermato-ungual-lacrimal-tooth malformations
Acro-dermato-ungual-lacrimal-tooth malformations (ADULT) syndrome differs from EEC syndrome owing to the absence of facial clefting (Brunner et al., 2002a,b).
Limb–mammary syndrome
Limb–mammary syndrome (LMS) consists of split-hand and/or foot malformation, mammary gland and nipple hypoplasia and isolated cleft palate (Rinne et al., 2007; van Bokhoven and Brunner, 2002).
Ankyloblepharon-ectodermal dysplasia-cleft lip/palate
Ankyloblepharon-ectodermal dysplasia-cleft lip/palate (AEC) syndrome consists of partial or complete fusion of eyelids and ectodermal dysplasia-clefting (Zhang et al., 2019).
Split-hand/foot malformations
Split-hand/foot malformations (SHFM) patients have median clefts of the hands and feet but no other features of EEC syndrome (Ianakiev et al., 2000).
Rapp–Hodgkin syndrome
Rapp–Hodgkin syndrome (RHS) patients suffer from ectodermal dysplasia, cleft lip and palate, deformed ears and genitourinary abnormalities (Holder-Espinasse et al., 2007).
Primary ovarian insufficiency
Primary ovarian insufficiency (POI) patients experience infertility associated with early arrest of menstruation and high follicle stimulating hormone (FSH) before the age of 40 (Veitia, 2020).
Conclusions
The p63 gene encodes multiple protein isoforms that serve a vast array of functions encompassing nearly all aspects of cellular signaling. Genetic studies in mice have shed much light on the roles of TAp63 versus ΔNp63 isoforms and their various functions. However, the roles of p63 in multiple aspects of cancer, including tumorigenesis, cancer progression, and metastasis as well as how they impact other diseases are still being discovered. Thus, a better understanding of the functions and interactions of the different p63 isoforms will undoubtedly prove beneficial to the design of effective therapies for cancer and beyond.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This review was made possible through the support of the Cold Spring Harbor Laboratory and Northwell Health Affiliation, the Office of the Director, National Institutes of Health under award numbers R01CA190997 and R21OD018332 (to A.A.M.) and 5F32CA225134 (to M.L.F.), and the Cancer Center Support Grant 5P30CA045508 and 1F31CA247400 (to S.B). Deposited in PMC for release after 12 months.
Cell science at a glance
A high-resolution version of the poster and individual poster panels are available for downloading at http://jcs.biologists.org/lookup/doi/10.1242/jcs.228015.supplemental
References
- Amelio I., Grespi F., Annicchiarico-Petruzzelli M. and Melino G. (2012). p63 the guardian of human reproduction. Cell Cycle 11, 4545-4551. 10.4161/cc.22819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboni A. L., Hutchinson J. A., DeCastro A. J., Cherukuri P., Liby K., Sporn M. B., Schwartz G. N., Wells W. A., Sempere L. F., Yu P. B. et al. (2013). ΔNp63α-mediated activation of bone morphogenetic protein signaling governs stem cell activity and plasticity in normal and malignant mammary epithelial cells. Cancer Res. 73, 1020-1030. 10.1158/0008-5472.CAN-12-2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri C. E., Barton C. E. and Pietenpol J. A. (2003). ΔNp63α expression is regulated by the phosphoinositide 3-kinase pathway. J. Biol. Chem. 278, 51408-51414. 10.1074/jbc.M309943200 [DOI] [PubMed] [Google Scholar]
- Beaudry V. G., Pathak N., Koster M. I. and Attardi L. D. (2009). Differential PERP regulation by TP63 mutants provides insight into AEC pathogenesis. Am. J. Med. Genet. A 149A, 1952-1957. 10.1002/ajmg.a.32760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergholz J. and Xiao Z.-X. (2012). Role of p63 in development, tumorigenesis and cancer progression. Cancer Microenviron. 5, 311-322. 10.1007/s12307-012-0116-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev V. A. (2015). Integration of the transcription factor-regulated and epigenetic mechanisms in the control of keratinocyte differentiation. J. Investig. Dermatol. Symp. Proc. 17, 30-32. 10.1038/jidsymp.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev V. A. and Flores E. R. (2014). p53/p63/p73 in the epidermis in health and disease. Cold Spring Harb. Perspect Med. 4, a015248 10.1101/cshperspect.a015248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughner J. C., van Eede M. C., Spring S., Yu L. X., Rostampour N. and Henkelman R. M. (2018). P63 expression plays a role in developmental rate, embryo size, and local morphogenesis. Dev. Dyn. 247, 779-787. 10.1002/dvdy.24622 [DOI] [PubMed] [Google Scholar]
- Brunner H. G., Hamel B. C. and van Bokhoven H. (2002a). P63 gene mutations and human developmental syndromes. Am. J. Med. Genet. 112, 284-290. 10.1002/ajmg.10778 [DOI] [PubMed] [Google Scholar]
- Brunner H. G., Hamel B. C. and van Bokhoven H. (2002b). The p63 gene in EEC and other syndromes. J. Med. Genet. 39, 377-381. 10.1136/jmg.39.6.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E., Terrinoni A., Rufini A., Chikh A., Lena A. M., Suzuki Y., Sayan B. S., Knight R. A. and Melino G. (2006). p63 is upstream of IKK alpha in epidermal development. J. Cell Sci. 119, 4617-4622. 10.1242/jcs.03265 [DOI] [PubMed] [Google Scholar]
- Carroll D. K., Carroll J. S., Leong C. O., Cheng F., Brown M., Mills A. A., Brugge J. S. and Ellisen L. W. (2006). p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 8, 551-561. 10.1038/ncb1420 [DOI] [PubMed] [Google Scholar]
- Cefalù S., Lena A. M., Vojtesek B., Musarò A., Rossi A., Melino G. and Candi E. (2015). TAp63gamma is required for the late stages of myogenesis. Cell Cycle 14, 894-901. 10.4161/15384101.2014.988021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R., Wei Y., Hwang J., Hang X., Andres Blanco M., Choudhury A., Tiede B., Romano R. A., DeCoste C., Mercatali L. et al. (2014). ΔNp63 promotes stem cell activity in mammary gland development and basal-like breast cancer by enhancing Fzd7 expression and Wnt signalling. Nat. Cell Biol. 16, 1004-1015, 1-13. 10.1038/ncb3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari N. S., Romano R. A., Koster M. I., Jaks V., Roop D., Flores E. R., Teglund S., Sinha S., Gruber W., Aberger F. et al. (2013). Interaction between the TP63 and SHH pathways is an important determinant of epidermal homeostasis. Cell Death Differ. 20, 1080-1088. 10.1038/cdd.2013.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A., Chang X., Sen T., Ravi R., Bedi A. and Sidransky D. (2010). Regulation of p53 family member isoform DeltaNp63alpha by the nuclear factor-kappaB targeting kinase IkappaB kinase beta. Cancer Res. 70, 1419-1429. 10.1158/0008-5472.CAN-09-2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu W.-K., Dai P.-M., Li H.-L. and Chen J.-K. (2008). Glycogen synthase kinase-3beta regulates DeltaNp63 gene transcription through the beta-catenin signaling pathway. J. Cell. Biochem. 105, 447-453. 10.1002/jcb.21839 [DOI] [PubMed] [Google Scholar]
- Chua H. L., Bhat-Nakshatri P., Clare S. E., Morimiya A., Badve S. and Nakshatri H. (2007). NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene 26, 711-724. 10.1038/sj.onc.1209808 [DOI] [PubMed] [Google Scholar]
- Cohen I. and Ezhkova E. (2016). Cbx4: A new guardian of p63's domain of epidermal control. J. Cell Biol. 212, 9-11. 10.1083/jcb.201512032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagnone M., Gatti V., Presutti D., Ruberti G., Fierro C., Markert E. K., Vousden K. H., Zhou H., Mauriello A., Anemone L. et al. (2017). ΔNp63-mediated regulation of hyaluronic acid metabolism and signaling supports HNSCC tumorigenesis. Proc. Natl. Acad. Sci. USA 114, 13254-13259. 10.1073/pnas.1711777114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch G. B., Zielonka E. M., Coutandin D., Weber T. A., Schäfer B., Hannewald J., Luh L. M., Durst F. G., Ibrahim M., Hoffmann J. et al. (2011). DNA damage in oocytes induces a switch of the quality control factor TAp63α from dimer to tetramer. Cell 144, 566-576. 10.1016/j.cell.2011.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como C. J., Urist M. J., Babayan I., Drobnjak M., Hedvat C. V., Teruya-Feldstein J., Pohar K., Hoos A. and Cordon-Cardo C. (2002). p63 expression profiles in human normal and tumor tissues. Clin. Cancer Res. 8, 494-501. [PubMed] [Google Scholar]
- Di Franco S., Turdo A., Benfante A., Colorito M. L., Gaggianesi M., Apuzzo T., Kandimalla R., Chinnici A., Barcaroli D., Mangiapane L. R. et al. (2016). ΔNp63 drives metastasis in breast cancer cells via PI3K/CD44v6 axis. Oncotarget 7, 54157-54173. 10.18632/oncotarget.11022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M., Pan J., Guo Z., Liu Q., Yang C. and Mao L. (2018). SATB1 is a novel molecular target for cancer therapy. Cancer Invest. 36, 28-36. 10.1080/07357907.2018.1423688 [DOI] [PubMed] [Google Scholar]
- Dotto G. P. (2009). Crosstalk of Notch with p53 and p63 in cancer growth control. Nat. Rev. Cancer 9, 587-595. 10.1038/nrc2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Li J., Wang L., Bian C., Wang Q., Liao L., Dou X., Bian X. and Zhao R. C. (2010). Overexpression of ΔNp63α induces a stem cell phenotype in MCF7 breast carcinoma cell line through the Notch pathway. Cancer Sci. 101, 2417-2424. 10.1111/j.1349-7006.2010.01700.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duperret E. K. and Ridky T. W. (2013). Focal adhesion complex proteins in epidermis and squamous cell carcinoma. Cell Cycle 12, 3272-3285. 10.4161/cc.26385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epifano C. and Perez-Moreno M. (2012). Crossroads of integrins and cadherins in epithelia and stroma remodeling. Cell Adh. Migr. 6, 261-273. 10.4161/cam.20253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E., Li B., Zewdu R., Wells V., Hebert J. M., Karner C., Anderson M. J., Williams T., Dixon J., Dixon M. J. et al. (2011). A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Dev. Cell 21, 627-641. 10.1016/j.devcel.2011.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessing M. Y., Mardaryev A. N., Gdula M. R., Sharov A. A., Sharova T. Y., Rapisarda V., Gordon K. B., Smorodchenko A. D., Poterlowicz K., Ferone G. et al. (2011). p63 regulates Satb1 to control tissue-specific chromatin remodeling during development of the epidermis. J. Cell Biol. 194, 825-839. 10.1083/jcb.201101148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher M. L., Kerr C., Adhikary G., Grun D., Xu W., Keillor J. W. and Eckert R. L. (2016). Transglutaminase interaction with α6/β4-integrin stimulates YAP1-Dependent ΔNp63α stabilization and leads to enhanced cancer stem cell survival and tumor formation. Cancer Res. 76, 7265-7276. 10.1158/0008-5472.CAN-16-2032 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Forster N., Saladi S. V., van Bragt M., Sfondouris M. E., Jones F. E., Li Z. and Ellisen L. W. (2014). Basal cell signaling by p63 controls luminal progenitor function and lactation via NRG1. Dev. Cell 28, 147-160. 10.1016/j.devcel.2013.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galoczova M., Coates P. and Vojtesek B. (2018). STAT3, stem cells, cancer stem cells and p63. Cell. Mol. Biol. Lett. 23, 12 10.1186/s11658-018-0078-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti V., Fierro C., Compagnone M., Giangrazi F., Markert E. K., Bongiorno-Borbone L., Melino G. and Peschiaroli A. (2018). DeltaNp63 regulates the expression of hyaluronic acid-related genes in breast cancer cells. Oncogenesis 7, 65 10.1038/s41389-018-0073-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghioni P., Bolognese F., Duijf P. H., van Bokhoven H., Mantovani R. and Guerrini L. (2002). Complex transcriptional effects of p63 isoforms: identification of novel activation and repression domains. Mol. Cell. Biol. 22, 8659-8668. 10.1128/MCB.22.24.8659-8668.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonfloni S., Di Tella L., Caldarola S., Cannata S. M., Klinger F. G., Di Bartolomeo C., Mattei M., Candi E., De Felici M., Melino G. et al. (2009). Inhibition of the c-Abl-TAp63 pathway protects mouse oocytes from chemotherapy-induced death. Nat. Med. 15, 1179-1185. 10.1038/nm.2033 [DOI] [PubMed] [Google Scholar]
- Gonfloni S., Caputo V. and Iannizzotto V. (2015). P63 in health and cancer. Int. J. Dev. Biol. 59, 87-93. 10.1387/ijdb.150045sg [DOI] [PubMed] [Google Scholar]
- Grun D., Adhikary G. and Eckert R. L. (2016). VEGF-A acts via neuropilin-1 to enhance epidermal cancer stem cell survival and formation of aggressive and highly vascularized tumors. Oncogene 35, 4379-4387. 10.1038/onc.2015.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun D., Adhikary G. and Eckert R. L. (2018). NRP-1 interacts with GIPC1 and α6/β4-integrins to increase YAP1/ΔNp63α-dependent epidermal cancer stem cell survival. Oncogene 37, 4711-4722. 10.1038/s41388-018-0290-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Keyes W. M., Papazoglu C., Zuber J., Li W., Lowe S. W., Vogel H. and Mills A. A. (2009). TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat. Cell Biol. 11, 1451-1457. 10.1038/ncb1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Chun A., Cheung K., Rashidi B. and Yang X. (2008). Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 283, 5496-5509. 10.1074/jbc.M709037200 [DOI] [PubMed] [Google Scholar]
- Helton E. S., Zhu J. and Chen X. (2006). The unique NH2-terminally deleted (ΔN) residues, the PXXP motif, and the PPXY motif are required for the transcriptional activity of the ΔN variant of p63. J. Biol. Chem. 281, 2533-2542. 10.1074/jbc.M507964200 [DOI] [PubMed] [Google Scholar]
- Helton E. S., Zhang J. and Chen X. (2008). The proline-rich domain in p63 is necessary for the transcriptional and apoptosis-inducing activities of TAp63. Oncogene 27, 2843-2850. 10.1038/sj.onc.1210948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcakova J., Nekulova M., Orzol P., Nenutil R., Podhorec J., Svoboda M., Dvorakova P., Pjechova M., Hernychova L., Vojtesek B. et al. (2017). ΔNp63 activates EGFR signaling to induce loss of adhesion in triple-negative basal-like breast cancer cells. Breast Cancer Res. Treat. 163, 475-484. 10.1007/s10549-017-4216-6 [DOI] [PubMed] [Google Scholar]
- Holder-Espinasse M., Martin-Coignard D., Escande F. and Manouvrier-Hanu S. (2007). A new mutation in TP63 is associated with age-related pathology. Eur. J. Hum. Genet. 15, 1115-1120. 10.1038/sj.ejhg.5201888 [DOI] [PubMed] [Google Scholar]
- Hsieh M. H., Choe J. H., Gadhvi J., Kim Y. J., Arguez M. A., Palmer M., Gerold H., Nowak C., Do H., Mazambani S. et al. (2019). p63 and SOX2 dictate glucose reliance and metabolic vulnerabilities in squamous cell carcinomas. Cell Rep 28, 1860-78.e9. 10.1016/j.celrep.2019.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Sen T., Nagpal J., Upadhyay S., Trink B., Ratovitski E. and Sidransky D. (2008). ATM kinase is a master switch for the Delta Np63 alpha phosphorylation/degradation in human head and neck squamous cell carcinoma cells upon DNA damage. Cell Cycle 7, 2846-2855. 10.4161/cc.7.18.6627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianakiev P., Kilpatrick M. W., Toudjarska I., Basel D., Beighton P. and Tsipouras P. (2000). Split-hand/split-foot malformation is caused by mutations in the p63 gene on 3q27. Am. J. Hum. Genet. 67, 59-66. 10.1086/302972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel A. (2010). The IKK complex, a central regulator of NF-kappaB activation. Cold Spring Harb. Perspect Biol. 2, a000158 10.1101/cshperspect.a000158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Hernandez L. E., Vazquez-Santillan K., Castro-Oropeza R., Martinez-Ruiz G., Munoz-Galindo L., Gonzalez-Torres C., Cortes-Gonzalez C. C., Victoria-Acosta G., Melendez-Zajgla J. and Maldonado V. (2018). NRP1-positive lung cancer cells possess tumor-initiating properties. Oncol. Rep. 39, 349-357. 10.3892/or.2017.6089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y.-S., Qian Y., Yan W. and Chen X. (2013). Pirh2 E3 ubiquitin ligase modulates keratinocyte differentiation through p63. J. Invest. Dermatol. 133, 1178-1187. 10.1038/jid.2012.466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimian A., Ahmadi Y. and Yousefi B. (2016). Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair 42, 63-71. 10.1016/j.dnarep.2016.04.008 [DOI] [PubMed] [Google Scholar]
- Kawasaki S., Tanioka H., Yamasaki K., Connon C. J. and Kinoshita S. (2006). Expression and tissue distribution of p63 isoforms in human ocular surface epithelia. Exp. Eye Res. 82, 293-299. 10.1016/j.exer.2005.07.001 [DOI] [PubMed] [Google Scholar]
- Kent S., Hutchinson J., Balboni A., Decastro A., Cherukuri P. and Direnzo J. (2011). ΔNp63α promotes cellular quiescence via induction and activation of Notch3. Cell Cycle 10, 3111-3118. 10.4161/cc.10.18.17300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr J. B., Hutt K. J., Michalak E. M., Cook M., Vandenberg C. J., Liew S. H., Bouillet P., Mills A., Scott C. L., Findlay J. K. et al. (2012). DNA damage-induced primordial follicle oocyte apoptosis and loss of fertility require TAp63-mediated induction of Puma and Noxa. Mol. Cell 48, 343-352. 10.1016/j.molcel.2012.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes W. M. and Mills A. A. (2006). p63: a new link between senescence and aging. Cell Cycle 5, 260-265. 10.4161/cc.5.3.2415 [DOI] [PubMed] [Google Scholar]
- Keyes W. M., Pecoraro M., Aranda V., Vernersson-Lindahl E., Li W., Vogel H., Guo X., Garcia E. L., Michurina T. V., Enikolopov G. et al. (2011). ΔNp63α is an oncogene that targets chromatin remodeler Lsh to drive skin stem cell proliferation and tumorigenesis. Cell Stem Cell 8, 164-176. 10.1016/j.stem.2010.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.-G. and Gumbiner B. M. (2015). Adhesion to fibronectin regulates Hippo signaling via the FAK-Src-PI3K pathway. J. Cell Biol. 210, 503-515. 10.1083/jcb.201501025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King K. E., Ha L., Camilli T. and Weinberg W. C. (2013). Delineating molecular mechanisms of squamous tissue homeostasis and neoplasia: focus on p63. J. Skin Cancer 2013, 632028 10.1155/2013/632028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klagsbrun M., Takashima S. and Mamluk R. (2002). The role of neuropilin in vascular and tumor biology. Adv. Exp. Med. Biol. 515, 33-48. 10.1007/978-1-4615-0119-0_3 [DOI] [PubMed] [Google Scholar]
- Koh L. F., Ng B. K., Bertrand J. and Thierry F. (2015). Transcriptional control of late differentiation in human keratinocytes by TAp63 and Notch. Exp. Dermatol. 24, 754-760. 10.1111/exd.12764 [DOI] [PubMed] [Google Scholar]
- Kommagani R., Caserta T. M. and Kadakia M. P. (2006). Identification of vitamin D receptor as a target of p63. Oncogene 25, 3745-3751. 10.1038/sj.onc.1209412 [DOI] [PubMed] [Google Scholar]
- Kouwenhoven E. N., van Bokhoven H. and Zhou H. (2015). Gene regulatory mechanisms orchestrated by p63 in epithelial development and related disorders. Biochim. Biophys. Acta 1849, 590-600. 10.1016/j.bbagrm.2015.03.003 [DOI] [PubMed] [Google Scholar]
- Lazzari C., Prodosmo A., Siepi F., Rinaldo C., Galli F., Gentileschi M., Bartolazzi A., Costanzo A., Sacchi A., Guerrini L. et al. (2011). HIPK2 phosphorylates ΔNp63α and promotes its degradation in response to DNA damage. Oncogene 30, 4802-4813. 10.1038/onc.2011.182 [DOI] [PubMed] [Google Scholar]
- Li C. and Xiao Z. X. (2014). Regulation of p63 protein stability via ubiquitin-proteasome pathway. Biomed. Res. Int. 2014, 175721 10.1155/2014/175721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Singh S., Cherukuri P., Li H., Yuan Z., Ellisen L. W., Wang B., Robbins D. and DiRenzo J. (2008). Reciprocal intraepithelial interactions between TP63 and hedgehog signaling regulate quiescence and activation of progenitor elaboration by mammary stem cells. Stem Cells 26, 1253-1264. 10.1634/stemcells.2007-0691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Zhang Y., Wu J., Li L., Chen N., Ni P., Song L. and Liu X. (2018). Neuropilin 1 (NRP1) is a novel tumor marker in hepatocellular carcinoma. Clin. Chim. Acta 485, 158-165. 10.1016/j.cca.2018.06.046 [DOI] [PubMed] [Google Scholar]
- Lindley L. E., Curtis K. M., Sanchez-Mejias A., Rieger M. E., Robbins D. J. and Briegel K. J. (2015). The WNT-controlled transcriptional regulator LBH is required for mammary stem cell expansion and maintenance of the basal lineage. Development 142, 893-904. 10.1242/dev.110403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis N. M., Morey L., Mejetta S., Pascual G., Janich P., Kuebler B., Cozutto L., Roma G., Nascimento E., Frye M. et al. (2011). Regulation of human epidermal stem cell proliferation and senescence requires polycomb- dependent and -independent functions of Cbx4. Cell Stem Cell 9, 233-246. 10.1016/j.stem.2011.07.013 [DOI] [PubMed] [Google Scholar]
- Ma J., Meng Y., Kwiatkowski D. J., Chen X., Peng H., Sun Q., Zha X., Wang F., Wang Y., Jing Y. et al. (2010). Mammalian target of rapamycin regulates murine and human cell differentiation through STAT3/p63/Jagged/Notch cascade. J. Clin. Invest. 120, 103-114. 10.1172/JCI37964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPartlin M., Zeng S. X. and Lu H. (2008). Phosphorylation and stabilization of TAp63γ by IκB kinase-β. J. Biol. Chem. 283, 15754-15761. 10.1074/jbc.M801394200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiulli M., Valletti A., Caratozzolo M. F., Tullo A., Sbisa E., Pesole G. and D'Erchia A. M. (2009). Identification and functional characterization of two new transcriptional variants of the human p63 gene. Nucleic Acids Res. 37, 6092-6104. 10.1093/nar/gkp674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardaryev A. N., Gdula M. R., Yarker J. L., Emelianov V. U., Poterlowicz K., Sharov A. A., Sharova T. Y., Scarpa J. A., Joffe B., Solovei I. et al. (2014). p63 and Brg1 control developmentally regulated higher-order chromatin remodelling at the epidermal differentiation complex locus in epidermal progenitor cells. Development 141, 101-111. 10.1242/dev.103200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardaryev A. N., Liu B., Rapisarda V., Poterlowicz K., Malashchuk I., Rudolf J., Sharov A. A., Jahoda C. A., Fessing M. Y., Benitah S. A. et al. (2016). Cbx4 maintains the epithelial lineage identity and cell proliferation in the developing stratified epithelium. J. Cell Biol. 212, 77-89. 10.1083/jcb.201506065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massion P. P., Taflan P. M., Jamshedur Rahman S. M., Yildiz P., Shyr Y., Edgerton M. E., Westfall M. D., Roberts J. R., Pietenpol J. A., Carbone D. P. et al. (2003). Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res. 63, 7113-7121. 10.1378/chest.125.5_suppl.102S-a [DOI] [PubMed] [Google Scholar]
- Melino G. (2011). p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ. 18, 1487-1499. 10.1038/cdd.2011.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melino G., Memmi E. M., Pelicci P. G. and Bernassola F. (2015). Maintaining epithelial stemness with p63. Sci. Signal. 8, re9 10.1126/scisignal.aaa1033 [DOI] [PubMed] [Google Scholar]
- Memmi E. M., Sanarico A. G., Giacobbe A., Peschiaroli A., Frezza V., Cicalese A., Pisati F., Tosoni D., Zhou H., Tonon G. et al. (2015). p63 Sustains self-renewal of mammary cancer stem cells through regulation of Sonic Hedgehog signaling. Proc. Natl. Acad. Sci. USA 112, 3499-3504. 10.1073/pnas.1500762112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzomo L. C., Pesce F. G., Marcal J. M., Haag T., Ferreira N. P., Lima J. F., Leaes C. G., Oliveira M. C. and da Fonte Kohek M. B. (2017). Decreased TAp63 and DeltaNp63 mRNA levels in most human pituitary adenomas are correlated with Notch3/Jagged1 relative expression. Endocr Pathol. 28, 13-21. 10.1007/s12022-016-9463-2 [DOI] [PubMed] [Google Scholar]
- Mills A. A., Zheng B., Wang X. J., Vogel H., Roop D. R. and Bradley A. (1999). p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398, 708-713. 10.1038/19531 [DOI] [PubMed] [Google Scholar]
- Mitchell S., Vargas J. and Hoffmann A. (2016). Signaling via the NFκB system. Wiley Interdiscip. Rev. Syst. Biol. Med. 8, 227-241. 10.1002/wsbm.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll U. M. and Slade N. (2004). p63 and p73: roles in development and tumor formation. Mol. Cancer Res. 2, 371-386. [PubMed] [Google Scholar]
- Muegge K. (2005). Lsh, a guardian of heterochromatin at repeat elements. Biochem. Cell Biol. 83, 548-554. 10.1139/o05-119 [DOI] [PubMed] [Google Scholar]
- Murray-Zmijewski F., Lane D. P. and Bourdon J.-C. (2006). p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 13, 962-972. 10.1038/sj.cdd.4401914 [DOI] [PubMed] [Google Scholar]
- Nguyen B. C., Lefort K., Mandinova A., Antonini D., Devgan V., Della Gatta G., Koster M. I., Zhang Z., Wang J., Tommasi di Vignano A. et al. (2006). Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 20, 1028-1042. 10.1101/gad.1406006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander K., Vojtesek B., Nenutil R., Lindgren B., Roos G., Zhanxiang W., Sjöström B., Dahlqvist A. and Coates P. J. (2002). Differential expression of p63 isoforms in normal tissues and neoplastic cells. J. Pathol. 198, 417-427. 10.1002/path.1231 [DOI] [PubMed] [Google Scholar]
- Otaka Y., Rokudai S., Kaira K., Fujieda M., Horikoshi I., Iwakawa-Kawabata R., Yoshiyama S., Yokobori T., Ohtaki Y., Shimizu K. et al. (2017). STXBP4 drives tumor growth and is associated with poor prognosis through PDGF receptor signaling in lung squamous cell carcinoma. Clin. Cancer Res. 23, 3442-3452. 10.1158/1078-0432.CCR-16-1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattison J. M., Melo S. P., Piekos S. N., Torkelson J. L., Bashkirova E., Mumbach M. R., Rajasingh C., Zhen H. H., Li L., Liaw E. et al. (2018). Retinoic acid and BMP4 cooperate with p63 to alter chromatin dynamics during surface epithelial commitment. Nat. Genet. 50, 1658-1665. 10.1038/s41588-018-0263-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J., Tanis S. E. J., Smits J. P. H., Kouwenhoven E. N., Oti M., van den Bogaard E. H., Logie C., Stunnenberg H. G., van Bokhoven H., Mulder K. W. et al. (2018). Mutant p63 affects epidermal cell identity through rewiring the enhancer landscape. Cell Rep 25, 3490-503.e4. 10.1016/j.celrep.2018.11.039 [DOI] [PubMed] [Google Scholar]
- Qu J., Yi G. and Zhou H. (2019). p63 cooperates with CTCF to modulate chromatin architecture in skin keratinocytes. Epigenetics Chromatin 12, 31 10.1186/s13072-019-0280-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran G. and Devaraj H. (2012). Aberrant expression of β-catenin and its association with ΔNp63, Notch-1, and clinicopathological factors in oral squamous cell carcinoma. Clin. Oral Investig. 16, 1275-1288. 10.1007/s00784-011-0605-0 [DOI] [PubMed] [Google Scholar]
- Rieger M. E., Sims A. H., Coats E. R., Clarke R. B. and Briegel K. J. (2010). The embryonic transcription cofactor LBH is a direct target of the Wnt signaling pathway in epithelial development and in aggressive basal subtype breast cancers. Mol. Cell. Biol. 30, 4267-4279. 10.1128/MCB.01418-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinne T., Brunner H. G. and van Bokhoven H. (2007). p63-associated disorders. Cell Cycle 6, 262-268. 10.4161/cc.6.3.3796 [DOI] [PubMed] [Google Scholar]
- Rinne T., Clements S. E., Lamme E., Duijf P. H., Bolat E., Meijer R., Scheffer H., Rosser E., Tan T. Y., McGrath J. A. et al. (2008). A novel translation re-initiation mechanism for the p63 gene revealed by amino-terminal truncating mutations in Rapp-Hodgkin/Hay-Wells-like syndromes. Hum. Mol. Genet. 17, 1968-1977. 10.1093/hmg/ddn094 [DOI] [PubMed] [Google Scholar]
- Ripamonti F., Albano L., Rossini A., Borrelli S., Fabris S., Mantovani R., Neri A., Balsari A., Magnifico A. and Tagliabue E. (2013). EGFR through STAT3 modulates ΔN63α expression to sustain tumor-initiating cell proliferation in squamous cell carcinomas. J. Cell. Physiol. 228, 871-878. 10.1002/jcp.24238 [DOI] [PubMed] [Google Scholar]
- Romano O. and Miccio A. (2020). GATA factor transcriptional activity: insights from genome-wide binding profiles. IUBMB Life 72, 10-26. 10.1002/iub.2169 [DOI] [PubMed] [Google Scholar]
- Rouleau M., Medawar A., Hamon L., Shivtiel S., Wolchinsky Z., Zhou H., De Rosa L., Candi E., de la Forest Divonne S., Mikkola M. L. et al. (2011). TAp63 is important for cardiac differentiation of embryonic stem cells and heart development. Stem Cells 29, 1672-1683. 10.1002/stem.723 [DOI] [PubMed] [Google Scholar]
- Russo C., Osterburg C., Sirico A., Antonini D., Ambrosio R., Würz J. M., Rinnenthal J., Ferniani M., Kehrloesser S., Schäfer B. et al. (2018). Protein aggregation of the p63 transcription factor underlies severe skin fragility in AEC syndrome. Proc. Natl. Acad. Sci. USA 115, E906-Ee15. 10.1073/pnas.1713773115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sari I. N., Phi L. T. H., Jun N., Wijaya Y. T., Lee S. and Kwon H. Y. (2018). Hedgehog signaling in cancer: a prospective therapeutic target for eradicating cancer stem cells. Cells 7, 208 10.3390/cells7110208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen T., Chang X., Sidransky D. and Chatterjee A. (2010). Regulation of ΔNp63α by NFκΒ. Cell Cycle 9, 4841-4847. 10.4161/cc.9.24.14093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo M. (2013). Epidermal stem cells in homeostasis and wound repair of the skin. Adv. Wound Care (New Rochelle) 2, 273-282. 10.1089/wound.2012.0372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi I., Romano R. A., Gluck C., Smalley K., Vojtesek B., Buck M. J. and Sinha S. (2015). A global analysis of the complex landscape of isoforms and regulatory networks of p63 in human cells and tissues. BMC Genomics 16, 584 10.1186/s12864-015-1793-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel C. and Lendahl U. (2017). Notch signaling in development, tissue homeostasis, and disease. Physiol. Rev. 97, 1235-1294. 10.1152/physrev.00005.2017 [DOI] [PubMed] [Google Scholar]
- Soares E. and Zhou H. (2018). Master regulatory role of p63 in epidermal development and disease. Cell. Mol. Life Sci. 75, 1179-1190. 10.1007/s00018-017-2701-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Chakravarti D. and Flores E. R. (2013). p63 steps into the limelight: crucial roles in the suppression of tumorigenesis and metastasis. Nat. Rev. Cancer 13, 136-143. 10.1038/nrc3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadeu A. M. and Horsley V. (2013). Notch signaling represses p63 expression in the developing surface ectoderm. Development 140, 3777-3786. 10.1242/dev.093948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe N., Miele L., Harris P. J., Jeong W., Bando H., Kahn M., Yang S. X. and Ivy S. P. (2015). Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat. Rev. Clin. Oncol. 12, 445-464. 10.1038/nrclinonc.2015.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bokhoven H. and Brunner H. G. (2002). Splitting p63. Am. J. Hum. Genet. 71, 1-13. 10.1086/341450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bokhoven H., Melino G., Candi E. and Declercq W. (2011). p63, a story of mice and men. J. Invest. Dermatol. 131, 1196-1207. 10.1038/jid.2011.84 [DOI] [PubMed] [Google Scholar]
- Veitia R. A. (2020). Primary ovarian insufficiency, meiosis and DNA repair. Biomed. J. 43, 115-123. 10.1016/j.bj.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viticchiè G., Agostini M., Lena A. M., Mancini M., Zhou H., Zolla L., Dinsdale D., Saintigny G., Melino G. and Candi E. (2015). p63 supports aerobic respiration through hexokinase II. Proc. Natl. Acad. Sci. USA 112, 11577-11582. 10.1073/pnas.1508871112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Fallon J. F. and Beachy P. A. (2000). Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 100, 423-434. 10.1016/S0092-8674(00)80678-9 [DOI] [PubMed] [Google Scholar]
- Wang G. X., Tu H. C., Dong Y., Skanderup A. J., Wang Y., Takeda S., Ganesan Y. T., Han S., Liu H., Hsieh J. J. et al. (2017). ΔNp63 Inhibits oxidative stress-induced cell death, including ferroptosis, and cooperates with the BCL-2 family to promote clonogenic survival. Cell Rep. 21, 2926-2939. 10.1016/j.celrep.2017.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zuo X., Xie K. and Wei D. (2018). The role of CD44 and cancer stem cells. Methods Mol. Biol. 1692, 31-42. 10.1007/978-1-4939-7401-6_3 [DOI] [PubMed] [Google Scholar]
- Wen J., Yan H., He M., Zhang T., Mu X., Wang H., Zhang H., Xia G. and Wang C. (2019). GSK-3β protects fetal oocytes from premature death via modulating TAp63 expression in mice. BMC Biol. 17, 23 10.1186/s12915-019-0641-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall M. D. and Pietenpol J. A. (2004). p63: Molecular complexity in development and cancer. Carcinogenesis 25, 857-864. 10.1093/carcin/bgh148 [DOI] [PubMed] [Google Scholar]
- Wu J., Bergholz J., Lu J., Sonenshein G. E. and Xiao Z. X. (2010). TAp63 is a transcriptional target of NF-kappaB. J. Cell. Biochem. 109, 702-710. 10.1002/jcb.22449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Chen G. and Li Y. P. (2016). TGF-beta and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 4, 16009 10.1038/boneres.2016.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin-Ozuysal O., Fiche M., Guitierrez M., Wagner K. U., Raffoul W. and Brisken C. (2010). Antagonistic roles of Notch and p63 in controlling mammary epithelial cell fates. Cell Death Differ. 17, 1600-1612. 10.1038/cdd.2010.37 [DOI] [PubMed] [Google Scholar]
- Yang A., Kaghad M., Wang Y., Gillett E., Fleming M. D., Dotsch V., Andrews N. C., Caput D. and McKeon F. (1998). p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2, 305-316. 10.1016/S1097-2765(00)80275-0 [DOI] [PubMed] [Google Scholar]
- Yang C., Hayashida T., Forster N., Li C., Shen D., Maheswaran S., Chen L., Anderson K. S., Ellisen L. W., Sgroi D. et al. (2011). The integrin alpha(v)beta(3-5) ligand MFG-E8 is a p63/p73 target gene in triple-negative breast cancers but exhibits suppressive functions in ER(+) and erbB2(+) breast cancers. Cancer Res. 71, 937-945. 10.1158/0008-5472.CAN-10-1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoh K. and Prywes R. (2015). Pathway regulation of p63, a director of epithelial cell fate. Front. Endocrinol. (Lausanne) 6, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Luong P., Hudson C., Gudmundsdottir K. and Basu S. (2010). c-Abl phosphorylation of ΔNp63α is critical for cell viability. Cell Death Dis. 1, e16 10.1038/cddis.2009.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan T., Rindtorff N. and Boutros M. (2017). Wnt signaling in cancer. Oncogene 36, 1461-1473. 10.1038/onc.2016.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Cheng R., Liang J., Lu Z., Wang Y., Li M., Yu H. and Yao Z. (2019). Ankyloblepharon-ectodermal dysplasia-clefting syndrome misdiagnosed as epidermolysis bullosa and congenital ichthyosiform erythroderma: case report and review of published work. J. Dermatol. 46, 422-425. 10.1111/1346-8138.14837 [DOI] [PubMed] [Google Scholar]