Significance
Our identification and characterization of two anion channelrhodopsins families, one from nonphotosynthetic microorganisms, shows that light-gated anion conductance is more widely spread among eukaryotic lineages than previously thought. The strongly red-shifted absorption spectra of the subset that we designate RubyACRs make them promising candidates for the long-sought inhibitory optogenetic tools producing large passive currents activated by red light, enabling deep tissue penetration. Previously only low-efficiency ion-pumping rhodopsins were available for neural inhibition at similar long wavelengths. The unusual residue composition of the retinal-binding pocket in RubyACRs expands our understanding of color tuning in rhodopsins. Finally, activation of chloride currents by energy transfer from a cytoplasmic fluorescent tag on RubyACRs introduces a potential dimension in molecular engineering of optogenetic tools.
Keywords: channelrhodopsins, optogenetics, ion channels, photobiology
Abstract
Channelrhodopsins are light-gated ion channels widely used to control neuronal firing with light (optogenetics). We report two previously unknown families of anion channelrhodopsins (ACRs), one from the heterotrophic protists labyrinthulea and the other from haptophyte algae. Four closely related labyrinthulea ACRs, named RubyACRs here, exhibit a unique retinal-binding pocket that creates spectral sensitivities with maxima at 590 to 610 nm, the most red-shifted channelrhodopsins known, long-sought for optogenetics, and more broadly the most red-shifted microbial rhodopsins thus far reported. We identified three spectral tuning residues critical for the red-shifted absorption. Photocurrents recorded from the RubyACR from Aurantiochytrium limacinum (designated AlACR1) under single-turnover excitation exhibited biphasic decay, the rate of which was only weakly voltage dependent, in contrast to that in previously characterized cryptophyte ACRs, indicating differences in channel gating mechanisms between the two ACR families. Moreover, in A. limacinum we identified three ACRs with absorption maxima at 485, 545, and 590 nm, indicating color-sensitive photosensing with blue, green, and red spectral variation of ACRs within individual species of the labyrinthulea family. We also report functional energy transfer from a cytoplasmic fluorescent protein domain to the retinal chromophore bound within RubyACRs.
Channelrhodopsins are light-gated ion channels, first found in chlorophyte (green) flagellate algae as phototaxis receptors that depolarize the cell membrane by cation conduction (1–3). The genomes of cryptophyte algae encode a related class of channelrhodopsins that are, however, strictly anion-selective (4). Both functional classes of channelrhodopsins, known as cation channelrhodopsins (CCRs) and anion channelrhodopsins (ACRs), are widely used to, respectively, activate (5) and inhibit (6) neurons with light (optogenetics). Cation conductance appears to have independently evolved also within the family of cryptophyte CCRs, referred to as “bacteriorhodopsin-like” CCRs (BCCRs) because of their sequence homology to archaeal proton-pumping rhodopsins (7).
Recently, a distinct family of ACRs was identified in environmental DNA samples of unknown organismal origin collected by the Tara Oceans project (8). This finding raised the possibility that channelrhodopsin genes were not limited to algae and were more widespread among protists. Here we report two distinct ACR families from labyrinthulea and haptophytes. Labyrinthulea (also known as labyrinthulomycetes) are a class of aquatic heterotrophic microbes. Despite the lack of flagella in vegetative stages, the peculiar lifestyles and the presence of the bothrosome in many members, the labyrinthulea are typical stramenopiles as evidenced by their heterokont zoospores (when present). Labyrinthulomycete ancestors are believed to have never been photosynthetic (9), unlike some other heterotrophic eukaryotic lineages that evolved by plastid loss. Haptophytes are flagellate algae distantly related to cryptophytes (10, 11).
Long-wavelength light better penetrates biological tissue, and therefore actuator molecules with red-shifted absorption are highly desired for optogenetic applications. A natural CCR variant with peak absorption in the orange-red spectral region has been discovered and used for photostimulation of neuronal firing (12). However, this CCR (named Chrimson) could not be converted to an anion channel by mutagenesis (13), and no natural ACRs with an absorption maximum beyond 540 nm have been found so far, despite extensive screening of homologous proteins from various cryptophyte species (14, 15). Here we report that among labyrinthulea ACRs are four that we name “RubyACRs” that exhibit maximal spectral sensitivity in the orange-red region of the spectrum, dependent on unique residues of their retinal-binding pocket. Furthermore, we also observed energy transfer between a fluorescent protein, fused as a tag to the cytoplasmic C terminus of a RubyACR, and its photoactive site retinal chromophore, an engineered antenna effect that has not been reported to photoactivate channelrhodopsins.
Results
Channelrhodopsin Sequences.
We found channelrhodopsin homologs in the genomes of 12 strains of labyrinthulea from the family Thraustochytriaceae. Most of the analyzed genomes (listed in SI Appendix, Table S1) encode three paralogs. The exceptions are the genomes of Schizochytrium aggregatum ATCC 28209, in which we found only two paralogs, and of Aplanochytrium kerguelense PBS07, in which we could not find a homolog. The Aurantiochytrium sp. KH105 genome encodes three pairs of nearly identical paralogs that may have resulted from recent gene duplications in a single genome or from two different strains/species in a mixed culture. Some other sequences from different organisms were also completely or nearly identical to each other. The mean length of the encoded polypeptides was ∼660 residues. The seven transmembrane helical (rhodopsin) domain was composed of ∼270 residues and was followed by a large cytoplasmic fragment, as in previously known algal channelrhodopsins. In the cytoplasmic fragments of some labyrinthulea homologs, signal receiver domains (accession number cl19078; also known as response regulator or CheY-like domains) were detected by bioinformatic analysis, although the E-values were very large (0.01). Very recently, such domains have also been identified in the cytoplasmic fragment of green algae CCRs and ACRs from prasinophytes and their viruses (16). Based on residue conservation analysis the authors suggest that, in contrast to the earlier known homologous domains associated with histidine kinase rhodopsins (17), these domains in channelrhodopsins do not undergo phosphorylation and either are constitutively active or have no signaling function (16).
We also found six and three channelrhodopsin homologs in the genomes of the haptophytes Phaeocystis antarctica and Phaeocystis globosa, respectively. The length of the encoded polypeptides ranges from 312 to 1,682 amino acid residues. In addition to the rhodopsin domain, no other known conserved domains were detected even in the longest sequence. Our search of the whole-genome shotgun (WGS) contigs at the National Center for Biotechnology Information (NCBI) also returned two sequences from Stramenopiles sp. TOSAG23-3. Their rhodopsin domains, however, clustered with Metagenomically discovered, Marine, Anion-conducting and Intensely Desensitizing channelrhodopsins (MerMAIDs) (8), whereas labyrinthulea and haptophyte homologs formed separate branches of the phylogenetic tree of channelrhodopsins (Fig. 1A).
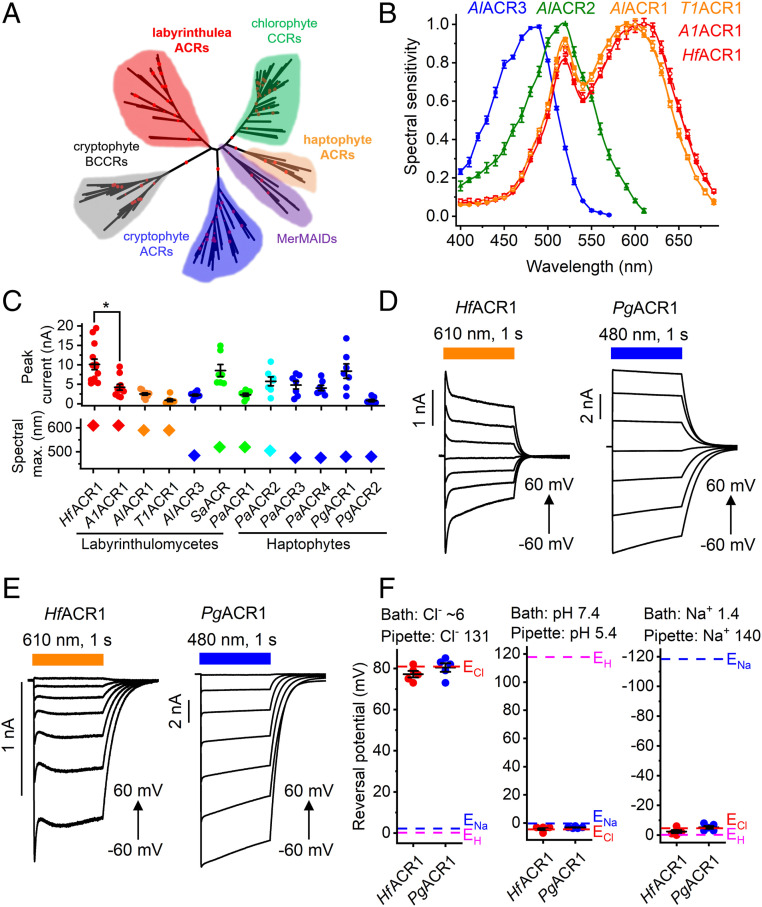
Fig. 1.
(A) A phylogenetic tree of rhodopsin domains constructed by the neighbor-joining method. The ultrafast bootstrap support values are shown by red circles (95 to 100% range). The GenBank accession numbers and source organism names of the sequences used are listed in SI Appendix, Tables S2 and S3, and in Dataset S1. (B) The action spectra of photocurrents generated by indicated ACRs. The data points are the mean values ± SEM (n = 6 to 10 scans). The spectral sensitivity was defined as the slope of current rise; see Materials and Methods for more detail. (C) The peak current amplitudes (Top) and spectral maxima (Bottom) of all functional ACRs tested in this study, except AlACR2, were generated in response to the first 1-s light pulse after seal formation. The data from individual cells are shown as circles; the lines show the mean values ± SEM (n = 7 to 13 cells). The diamonds show spectra maxima determined from the averaged spectra. (D and E) Series of photocurrents recorded using the indicated Cl− concentrations (in mM) at voltages changed from −60 to 60 mV at the amplifier output. (F) The reversal potentials (Erev) of photocurrents measured under indicated ionic conditions (in mM). The data from individual cells are shown as circles; the lines show the mean values ± SEM (n = 5 cells). The Erev values were corrected for liquid junction potentials, as described in Materials and Methods.
The GenBank accession numbers, abbreviated protein names, source organism names, and gene model/WGS sequence numbers of the identified homologs, are listed in SI Appendix, Tables S2 and S3. The first two italicized letters of the protein names are derived from the genus and species name of the source organism; if no species name was assigned to a strain, consecutive numbers were used to distinguish orthologs. When numbering paralogs, we followed a historical numbering convention and assigned the number 1 to the most red-shifted paralog, and consecutively numbered other paralogs according to the position of their spectral maxima. The numbers were assigned to untested labyrinthulea homologs by taking into account their sequence homology with tested ones.
Protein alignments of the rhodopsin domains of labyrinthulea and haptophyte channelrhodopsins are shown in SI Appendix, Figs. S1 and S2, respectively. The Schiff base Lys is found in the seventh transmembrane helix (TM7) of all homologs, as is the immediately following Gln, characteristic of most earlier known channelrhodopsins except BCCRs. The position of the proton donor in bacteriorhodopsin (Asp85) is occupied by a noncarboxylate residue in all of the sequences, as in cryptophyte ACRs and MerMAIDs. This feature distinguishes them from chlorophyte CCRs and cryptophyte BCCRs, most of which contain a carboxylate residue in this position. The second photoactive site carboxylate (Asp212 of bacteriorhodopsin) is universally conserved. Only two glutamates (Glu60 and Glu68 using Guillardia theta ACR1 [GtACR1] numbering) are conserved in TM2 of the haptophyte homologs. This is also a feature shared with cryptophyte ACRs and MerMAIDs, in contrast to chlorophyte CCRs, most of which contain four conserved glutamates in this helix. Most labyrinthulea homologs completely lack glutamates in TM2, except two sequences from Aplanochytrium stocchinoi recently identified in the Marine Microbial Eukaryote Transcriptome Sequencing Project database (16), in which the homolog of Glu60 is conserved (SI Appendix, Fig. S1). An unrooted phylogenetic tree of labyrinthulea homologs is shown in SI Appendix, Fig. S3.
Screening of ACR Homologs by Patch Clamp Electrophysiology.
We synthesized mammalian codon-adapted polynucleotides encoding the rhodopsin domains (residues 1 to 270 or 1 to 300; see Materials and Methods) of seven channelrhodopsin homologs from various labyrinthulea species, six from P. antarctica and three from P. globosa, fused them to a C-terminal enhanced fluorescent yellow protein (EYFP) tag and expressed them in human embryonic kidney (HEK293) cells. As shown below, labyrinthulea and haptophyte homologs are strictly anion-selective, so we will refer to them as “ACRs.”
All seven labyrinthulea and six haptophyte homologs generated photocurrents when probed with whole-cell patch clamping. First, we determined their action spectra by measuring the initial slope of photocurrent in the linear range of the stimulus intensity, as shown in SI Appendix, Fig. S4A. The spectra of AlACR1, AlACR2, and AlACR3 peaked at 590, 520, and 485 nm, respectively (Fig. 1B), reminiscent of the red, green, and blue color system of human vision. A1ACR1, HfACR1, and T1ACR1 sequences, although they originate from different organisms, are 76 to 80% identical to that of AlACR1 (SI Appendix, Fig. S1). In the action spectrum of T1ACR1 photocurrents, the position of the main peak was identical to that of AlACR1 (590 nm), but, in both A1ACR1 and HfACR1, it was at 610 nm (Fig. 1B). Other functional homologs exhibited maximal sensitivity to green or blue light (Fig. 1C and SI Appendix, Fig. S4 B–D).
When probed at the wavelength of their maximal sensitivity, most of the tested rhodopsins generated photocurrents in the nanoampere range. Fig. 1C shows the peak current amplitude for all functional homologs except AlACR2, the currents of which are described in a separate section below. HfACR1 generated the largest currents among labyrinthulea homologs, and PgACR1, among the haptophyte homologs. Of the two most red-shifted homologs, HfACR1 generated significantly larger photocurrents than A1ACR1 (P < 0.001, Mann–Whitney U test), although the sequences of their rhodopsin domains differ only at five positions in the range of residues 1 to 270 (SI Appendix, Fig. S5). SI Appendix, Fig. S6 shows normalized photocurrent traces recorded from all functional homologs except AlACR2. The amplitude of channel currents decreased during continuous illumination (a phenomenon known as desensitization). The amplitude of stationary current, degree of desensitization, and half-time of current decay after switching off the light varied between different homologs (SI Appendix, Fig. S7 A–C). On average, desensitization was greater in labyrinthulea homologs (89 ± 3%, mean ± SEM, n = 6 homologs) than in haptophyte homologs (41 ± 8%, mean ± SEM, n = 6 homologs). As in strongly desensitizing channelrhodopsins studied earlier (3), the peak-to-stationary current ratio in labyrinthulea ACRs reduced in a series of light pulses, whereas in haptophyte ACRs this effect was negligible (SI Appendix, Fig. S7D).
With standard bath and pipette solutions (∼156 and 131 mM Cl−, respectively; for other components, see Materials and Methods) the photocurrents reversed their direction near zero voltage, indicating passive ionic conductance (Fig. 1D). To test relative permeability for Cl−, Na+, and H+, we individually changed the concentrations of these ions in the bath. Representative photocurrent traces recorded from HfACR1 and PgACR1 with ∼6 mM Cl− in the bath at incremental voltages are shown in Fig. 1E. We then measured the current-voltage relationships and calculated the reversal potentials (Erev). Under all tested conditions, the Erev matched the Nernst equilibrium potential for Cl− (Fig. 1F), indicating that the tested channelrhodopsins were strictly permeable for anions. SI Appendix, Fig. S8A, shows a current-voltage curve for HfACR1 activated at 630 nm and tested under ionic conditions typical of mature neurons (Materials and Methods). SI Appendix, Fig. S8B, shows photocurrent traces recorded upon pulsed stimulation at different frequencies.
Color Tuning in Labyrinthulea ACRs.
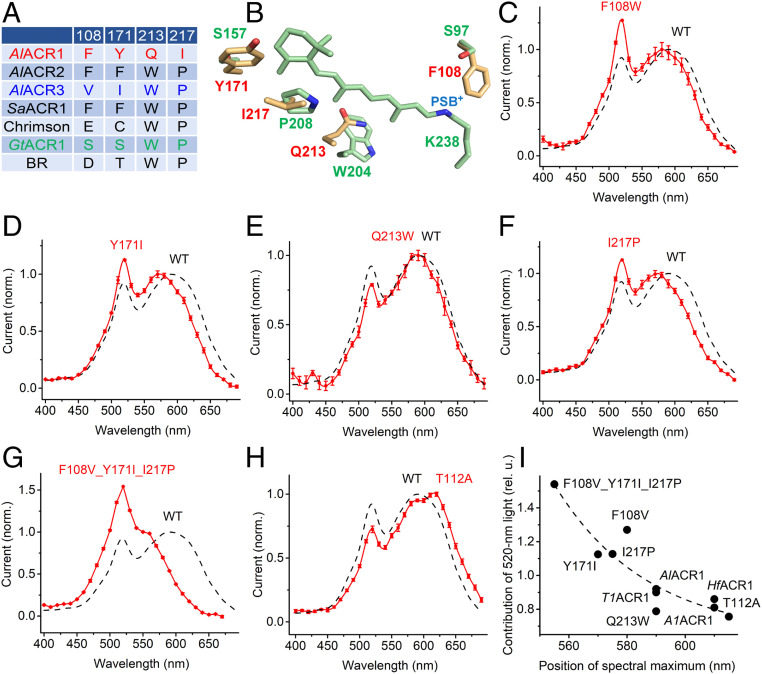
Comparison of the retinal-binding pockets of AlACR1 (spectral maximum 590 nm) and AlACR3 (487 nm) revealed four divergent positions (Fig. 2A). Most unusual were Gln213 and Ile217, which correspond, respectively, to Trp182 and Pro186 of bacteriorhodopsin and are highly conserved in the entire superfamily of microbial rhodopsins. These four unusual residues are conserved in all RubyACRs (AlACR1, A1ACR1, HfACR1, and T1ACR1) (SI Appendix, Fig. S1), but not in Chrimson (Fig. 2A), although the latter also exhibits a red-shifted absorption peak at 590 nm (12).
Fig. 2.
(A) The residues in the retinal-binding pocket of indicated proteins. The numbers correspond to the sequence of AlACR1. BR, bacteriorhodopsin. (B) A homology model of AlACR1 showing the side chains of the residues from A (yellow with red labels) and the chromophore and corresponding side chains of GtACR1 (green; PDB entry code: 6EDQ). PSB+, protonated Schiff base. (C–H) The action spectra of photocurrents generated by the indicated mutants of AlACR1 (red) and that of the wild type from Fig. 1B (black dashed line). (I) The dependence of contribution of the 520-nm peak (its ratio to the rhodopsin peak) on the position of the rhodopsin peak.
Fig. 2B shows locations of these residues according to a homology model of AlACR1. To test their possible role in color tuning, we individually replaced the residues in AlACR1 with those found in the corresponding positions in the blue-absorbing AlACR3 and measured the action spectra of photocurrents in the resultant mutants. In the AlACR1_F108W, Y171I, and Y217P mutants the position of the main peak shifted 10, 20, and 15 nm to shorter wavelengths, respectively, whereas in the Q213W mutant it remained almost unchanged (Fig. 2 C–F). The effects of the three color-tuning mutations were additive (Fig. 2G). In Chrimson, the S169A mutation caused an 18-nm red shift of the spectral maximum (18). The corresponding T112A mutation caused a similar red shift in AlACR1 (Fig. 2H) despite the differences in the retinal-binding pockets of the two wild-type proteins. SI Appendix, Fig. S9, shows photocurrent amplitudes generated by the binding-pocket mutants.
The action spectra of photocurrents generated by all four RubyACRs exhibited an additional sharp peak at 520 nm, suggesting the presence of a second chromophore (Fig. 1B). In addition to a shift of the main (rhodopsin) spectral peak, the AlACR1_F108W, T112A, Y171I, and Y217P mutations also changed the relative contribution of the 520-nm peak (its ratio to the rhodopsin peak). Notably, there was an inverse correlation between this contribution and the rhodopsin peak position (Fig. 2I) as would be expected from Förster resonance energy transfer (FRET) from the second chromophore to the retinal chromophore, because efficiency of FRET is proportional to the extent of spectral overlap between the donor and acceptor.
Previously, we have shown that incorporation of 3,4-dehydroretinal (A2 retinal) instead of native chromophore (A1 retinal) causes a red shift of the spectral sensitivity in several microbial rhodopsins expressed in HEK293 cells (19). Supplementation of AlACR1-expressing cells with A2 retinal led to a red shift of the rhodopsin peak of the photocurrent action spectrum as compared to that measured with A1 retinal, but did not shift the 520-nm peak (SI Appendix, Fig. S10A). In HfACR1 (spectral maximum with A1 retinal at 610 nm), a red shift of the rhodopsin peak was observed, and also the relative amplitude of the 520-nm peak decreased without a change in its position (SI Appendix, Fig. S10B). Both of these effects were clearly resolved in the difference (A2 retinal – A1 retinal) spectra (SI Appendix, Fig. S10C). These observations confirmed that the 520-nm peak originated from a second chromophore, as its spectral position was not affected by the type of retinal used. Energy migration from the antenna pigment to the retinal chromophore was further confirmed by the inverse correlation of the contribution of the antenna peak to the spectrum with the wavelength of the rhodopsin peak, as in the case of the color-tuning mutants (Fig. 2I).
Retinal Chromophore-EYFP Interaction.
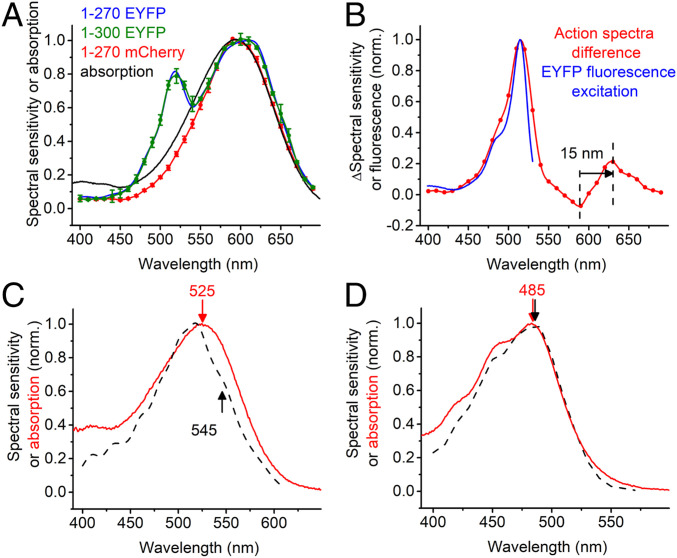
To test whether the second chromophore responsible for the 520-nm peak in the photocurrent action spectra is derived from the EYFP tag and gain more information about the spectral properties of RubyACRs, we expressed A1ACR1 and HfACR1 without fluorescent tags in Pichia and purified the encoded proteins in nondenaturing detergent. The absorption spectra of detergent-purified A1ACR1 (Fig. 3A, black line) and HfACR1 (SI Appendix, Fig. S10D, red line) lacked the 520-nm band observed in the action spectra of the corresponding EYFP fusions. When we replaced the EYFP tag (absorption maximum at 513 nm) with the mCherry tag (587 nm), the 520-nm band disappeared from the action spectrum (Fig. 3A, red line). The difference between the action spectra of photocurrents generated by A1ACR1_EYFP and A1ACR1_mCherry matched the fluorescence excitation spectrum of EYFP (Fig. 3B). The difference between the action spectra also clearly showed that the position of the rhodopsin peak was ∼15 nm shifted to shorter wavelengths by the tag replacement (the arrow in Fig. 3B). The maximum of the action spectrum of photocurrents generated by A1ACR1_mCherry was close to that of the absorption spectrum of the fluorescence tag-free protein (595 nm; Fig. 3A, red line and black line, respectively). The action spectra of photocurrents generated by A1ACR1 versions comprising amino acid residues 1 to 270 or 1 to 300 were identical within experimental error (Fig. 3A, blue and green).
Fig. 3.
(A) The action spectra of photocurrents generated by A1ACR1 fused to EYFP (amino acids 1 to 270 or 1 to 300) or mCherry and the absorption spectrum of fluorescence tag-free A1ACR1 purified from Pichia. (B) The difference between the action spectra obtained with A1ACR1_EYFP and AlACR1_mCherry and the fluorescence excitation spectrum of EYFP from FPbase (https://www.fpbase.org/protein/eyfp/). (C and D) The action spectra of photocurrents recorded by AlACR2 (C) and AlACR3 (D) EYFP fusions (black dashed lines, reproduced from Fig. 1B) and the absorption spectra of the respective purified proteins (red lines).
The absorption spectrum of the Pichia-expressed green-absorbing AlACR2 peaked at 525 nm (Fig. 3C, red line). The action spectrum of photocurrents recorded from the AlACR2_EYFP fusion exhibited a shoulder at ∼545 nm (Fig. 3C, black dashed line). Comparison with the absorption spectrum shows that this shoulder corresponds to rhodopsin absorption, whereas the main peak at 520 nm reflects energy transfer from EYFP. The absorption spectrum of the Pichia-expressed blue-absorbing AlACR3 closely matched the action spectrum of photocurrents recorded from the corresponding EYFP fusion (Fig. 3D).
Channel Gating in AlACR1.
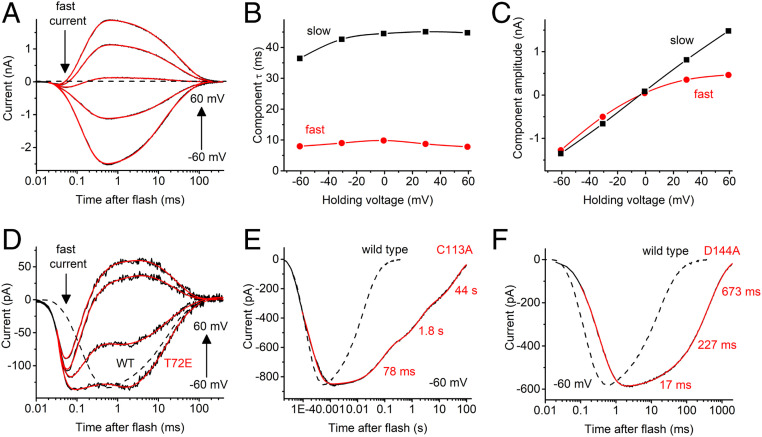
To analyze the kinetics of channel gating in AlACR1, we recorded photocurrents under single-turnover conditions using 6-ns laser flash excitation. Laser-evoked current traces could be fit with four exponentials (two for the current rise and two for the decay) (Fig. 4A). Channel opening (the time constant [τ] of which was ∼100 µs) was preceded with a fast, only weakly voltage-dependent negative peak that could be clearly resolved near the reversal voltage for channel currents (the arrow labeled fast current in Fig. 4A). Its rise τ was <20 µs, the lower limit of time resolution of our system. Such currents have previously been recorded from several other channelrhodopsins and attributed to a charge displacement associated with retinal chromophore isomerization, integrated by the recording circuit (20).
Fig. 4.
(A) A series of photocurrent traces recorded from AlACR1 upon laser flash excitation at the voltages changed in 20-mV increments from −60 to 60 mV at the amplifier output. Black lines, experimental data; red lines, multiexponential fit. (B and C) The dependence of the decay components τ (B) and amplitude (C) on the holding voltage for the series of traces shown in A. (D) A series of photocurrent traces recorded from the AlACR1_T72E mutant at incremental voltages from −60 to 60 mV. The wild-type trace at −60 mV from A is shown as a dashed line for comparison. (E and F) The current traces recorded from the indicated AlACR1 mutants at −60 mV (solid lines) as compared to the wild-type trace (dashed lines).
AlACR1 channel closing was biphasic, and both phases were faster (τ, 13 ± 1 ms and 77 ± 11 ms at −60 mV, n = 5 cells) than those in GtACR1 (21). Moreover, in AlACR1, τ of both decay phases showed only weak (if any) voltage dependence (Fig. 4B), in contrast to GtACR1 in which the fast decay was strongly accelerated, and the slow decay slowed upon depolarization (21). The amplitude of the fast decay component in AlACR1 exhibited inward, and that of the slow decay, no rectification (Fig. 4C), again in contrast to GtACR1, in which the amplitude of the fast component showed outward, and that of the slow component, inward rectification (21).
Previously, we had found that Glu68 controls the kinetics of the fast channel closing in GtACR1 (21). The corresponding position in AlACR1 is occupied by a noncarboxylate residue, Thr72 (SI Appendix, Fig. S1), which is very unusual for channelrhodopsins, in most of which a Glu is conserved. The T72E mutation significantly reduced laser-evoked channel currents from 2.7 ± 0.5 nA measured in the wild type to 76 ± 17 pA (mean ± SEM, n = 8 and 7 cells, respectively; P < 0.005, Mann–Whitney U test), which made the fast isomerization peak more obvious in the current trace (Fig. 4D). The current decay rate was little affected by this mutation. Cys128 from TM3 and Asp156 from TM4 form an interhelical hydrogen bond (the “DC gate”) in Chlamydomonas reinhardtii channelrhodopsin-2 (CrChR2) (22). The homologs of these residues in AlACR1 are Cys113 and Asp144. Alanine replacement of each of these residues slowed channel closing (Fig. 4 E and F). The effect was much more pronounced in the C113A mutant (the slowest decay phase was on the timescale of tens of seconds) than in the D144A mutant.
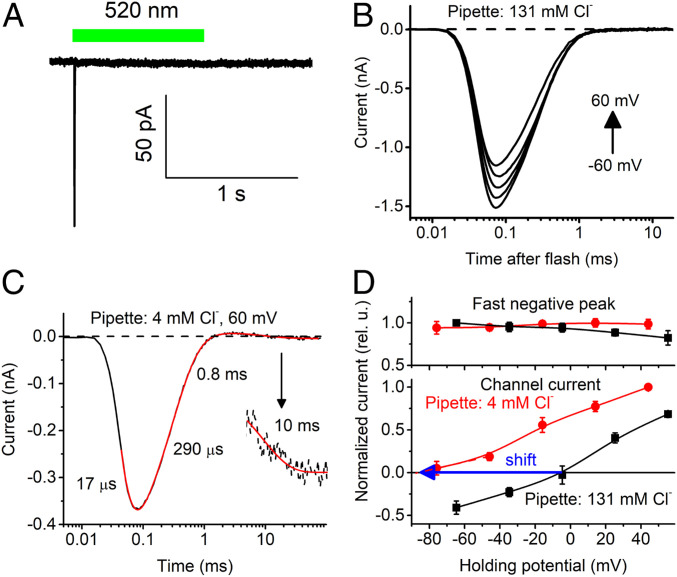
Unusual Photocurrents of AlACR2.
In contrast to all other channelrhodopsins studied in heterologous expression systems amenable to electrophysiological recording, photocurrents from AlACR2 were dominated by a fast negative signal (Fig. 5A). AlACR2’s signal peak amplitude recorded in response to a 1-s pulse of continuous light at −60 mV was 103 ± 15 pA (mean ± SEM, n = 7 cells). It showed a very weak, if any, dependence of the holding voltage (Fig. 5B), as is typical of similar fast negative currents associated with retinal chromophore isomerization in other channelrhodopsins (20). To detect a possible contribution of passive Cl− conductance in photocurrents, we reduced the Cl− concentration in the pipette to 4 mM and held the cells at +60 mV. Under these conditions, passive Cl− flux would be inward, producing an outward hyperpolarizing electrical current, opposite to the inward isomerization current. The laser flash-evoked current traces could be fit with four exponentials, the slowest of which had τ 10 ms (Fig. 5C), which is within the range of channel closing time in other ACRs. Next, we measured the voltage dependence of the amplitude of this component with 4 and 131 mM Cl− in the pipette. Fig. 5D shows that, in contrast to the peak amplitude, the reversal potential of this slowest component followed the equilibrium potential for Cl−, which confirmed that AlACR2 possessed residual passive conductance for this ion.
Fig. 5.
(A) A photocurrent trace recorded from AlACR2 upon stimulation with a continuous light pulse, the duration of which is schematically shown as the green bar. (B) A series of photocurrent traces recorded from AlACR2 upon laser flash excitation at the voltages changed from −60 to 60 mV at the amplifier output with the standard pipette solution. (C) A photocurrent trace recorded from AlACR2 at 60 mV using the pipette solution with a reduced Cl− concentration. (Inset) A portion of the trace from 5 to 100 ms vertically stretched to better resolve the channel current decay. Black lines, experimental data; red lines, multiexponential fit, dashed black line, zero current level. (D) The current-voltage dependencies of the peak current and the slowest signal component at the indicated Cl− concentrations in the pipette. The data points are the mean ± SEM values (n = 5 cells).
The action spectrum of the unusual photocurrents generated by the AlACR2_EYFP fusion exhibited a main peak at 520 nm and a minor shoulder at ∼540 nm (Fig. 1B, green). The position of the main peak was very close to that of EYFP absorption. To test the possibility of energy transfer from EYFP to the AlACR2 retinal chromophore, we measured the action spectrum after incubation of the cells with A2 retinal. A clear shoulder appeared at ∼580 nm, which can be explained by a shift of the rhodopsin peak to longer wavelengths as expected from A2 retinal binding in the chromophore pocket, whereas the peak at 520 nm remained unchanged (SI Appendix, Fig. S11), consistent with energy transfer between EYFP and AlACR2.
Discussion
We report channelrhodopsins in two eukaryotic lineages phylogenetically distant from chlorophyte and cryptophyte algae in which such proteins had so far been found. Both labyrinthulea and haptophyte channelrhodopsins exclusively conduct anions and therefore should be classified as ACRs. This expansion of the ACR family benefits elucidation of the structural requirements for anion conductance by comparative analysis of the structure–function relationships in different ACR groups and offers a possibility of using alternative scaffolds for molecular design of the physicochemical properties desirable in optogenetic tools. The most important in this respect are the four RubyACRs, closely related labyrinthulea ACRs with spectral maxima in the orange-red region. The only known inhibitory optogenetic tool with peak absorption at ∼590 nm has been the engineered halorhodopsin from Halobacterium salinarum strain Shark, referred to as “Jaws” (23). Although Jaws generates larger photocurrents than the earlier known Cl− pumps, its efficiency is limited by translocation of only one ion per captured photon across the membrane. RubyACRs generate large passive anion currents and offer highly efficient optogenetic inhibition for the long-wavelength spectral range.
Our analysis of spectral-tuning mutants and A1/A2 retinal substitution strongly indicate FRET from a fluorescent protein (EYFP) to the retinal chromophore in red-shifted rhodopsins, which may provide an additional avenue for the development of optogenetic tools. Previously, energy transfer from a light-harvesting antenna (the carotenoid salinixanthin) to the retinal chromophore was reported in xanthorhodopsin, a eubacterial proton pump (24). This phenomenon has also been demonstrated in the heterologously expressed proton-pumping rhodopsin from the cyanobacterium Gloeobacter violaceous reconstituted with the carotenoid echinenone present in this organism (25). In the crystal structure of xanthorhodopsin, the ring of salinixanthin is bound 5 Å from the retinal chromophore’s β-ionone ring (26). Salinixanthin binding depends on the presence of retinal, but does not require formation of the Schiff base linkage (27). Fluorescence spectroscopy and femtosecond transient absorption spectroscopy have shown that the energy transfer mostly occurs from the excited singlet 2 (S2) state of salinixanthin to the S1 state of the retinal chromophore (28).
The molecular system in which we observed the energy transfer in this study differs in that the two chromophores occur within two different protein domains: water-soluble EYFP and membrane-embedded rhodopsin. FRET in similar complexes between fluorescent proteins and rhodopsins has been reported (29–31). In those studies quenching of or increase in the donor fluorescence was used to monitor the state of the acceptor (a voltage reporter). Instead, our observations show “functional FRET.” The FRET efficiency depends on the inverse sixth power of the distance between the donor and the acceptor, scaled by the factor that reflects the relative orientation of the donor emission dipole moment and the acceptor absorption dipole moment. The linker is flexible, so neither the distance, nor the orientation of the fluorescent protein with respect to the channelrhodopsin domain, can accurately be predicted without crystallographic data and may be different for different rhodopsin variants. A longer linker may actually bring the fluorescent tag closer to rhodopsin and/or reorient its absorption dipole moment in a more favorable position for FRET. We did not find any difference between the action spectra of photocurrents generated by A1ACR1 versions comprising 270 and 300 amino acid residues (Fig. 3A, blue and green, respectively), but the absence of a difference cannot be considered a general rule.
The influence of retinal-binding pocket residues on the absorption spectra (color tuning) in rhodopsins has been extensively studied by experimental and computational approaches (32–35). However, the residue pattern conserved in RubyACRs has not been found in any other microbial rhodopsins, including the cation-conducting Chrimson that exhibits peak absorption at 590 nm. Of the three pocket residues that contribute to the red-shifted spectral sensitivity of RubyACRs, Phe108 (AlACR1 numbering) occupies the position of the counterion of the Schiff base (Asp85 in bacteriorhodopsin), the importance of which for color tuning is well documented (32, 36, 37). As shown in bacteriorhodopsin, a neutral residue in this position destabilizes the ground state, decreasing the energy gap between the ground and excited states as evidenced by red-shifted absorption (38). A bulky aromatic residue in the counterion position is very unusual among microbial rhodopsins. A previously known group of sequences in which phenylalanine was found at this position are schizorhodopsins from Asgardarchaeota (39) and some of their bacterial relatives, Antarctic rhodopsins (40), but they show very little homology to channelrhodopsins and act as inward proton pumps. To the best of our knowledge, a possible role of this substitution in color tuning in these proteins has not been tested.
According to our homology modeling of AlACR1, Tyr171 and Ile217 are located near the β-ionone ring, as shown overlaid with the retinal binding pocket of GtACR1 (Fig. 2B), the only ACR crystal structure available. The red-shifting effect of polar Tyr171 on the AlACR1 spectrum is likely due to stabilization of the excited state of the chromophore, which would occur if the dipole moment associated with the side chain is oriented with its negative pole toward the retinal chromophore (33). The orientation of the Tyr171 side chain needs to be resolved by atomic structure determination. Ile217 of AlACR1 corresponds to Pro219 of the sodium-pumping rhodopsin KR2. Replacement of this residue with nonpolar Gly caused a 10-nm red spectral shift in KR2, which was explained by the influence of this mutation on the dipole created by the C=O and N-H groups of the backbone (41). A similar explanation may be suggested for Ile217, but again needs structural verification and computational analysis.
The combination of the three color-tuning mutations, F108V, Y171I, and I217P, did not shift the spectral maximum of AlACR1 all of the way to that of AlACR3 (Fig. 2G), indicating that there are other residues that contribute to the spectral difference. One candidate is the residue corresponding to Leu93 in bacteriorhodopsin. This position has been shown to determine the spectral difference between green- and blue-absorbing proteorhodopsins (42). All RubyACRs have Ile in this position (Ile116 in AlACR1) as do green-absorbing AlACR2 and SaACR2, whereas the blue-absorbing AlACR3 has Met (SI Appendix, Fig. S1).
The only species in which the functional role of channelrhodopsins has been demonstrated is the green flagellate alga C. reinhardtii, in which they function as phototaxis receptors (1). All labyrinthulea and haptophyte species in which we found channelrhodopsins produce free-swimming zoospores in their life cycles (43, 44). Therefore, it is plausible that channelrhodopsins guide phototaxis also in these microorganisms. Consistent with this hypothesis is that no channelrhodopsin homologs have been found in the genome of the labyrinthulea A. kerguelense PBS07, which does not produce zoospores (44). However, two paralogs have been found in the transcriptome of A. stocchinoi (16), although there are no known flagellate stages in its life cycle (45).
Materials and Methods
Bioinformatics.
A keyword search of gene annotations was used to identify rhodopsin genes at the genome portals of Aurantiochytrium limacinum MYA-1381, P. antarctica CCMP1374, and P. globosa Pg-G of the US Department of Energy Joint Genome Institute (JGI). The three hits showing protein sequence homology to previously known algal channelrhodopsins found in the A. limacinum genome were then used for a tblastn search of the JGI genome portal for S. aggregatum ATCC 28209 and whole-genome shotgun contigs from stramenopiles at the NCBI portal.
Protein sequence alignments were created using the MUSCLE algorithm implemented in DNASTAR Lasergene MegAlign Pro software. Phylogenetic trees were analyzed with the W-IQ-TREE online tool (46) using automatic model selection, 1,000 ultrafast bootstrap replicates, and visualized with iTOL 5.5.1 (47). Homology models were obtained using the commonly used structure prediction servers I-TASSER (48) and Robetta (49, 50), a membrane protein-adapted form of the Rosetta program, and visualized with PyMol software (www.pymol.org). A comparative study of several homology modeling protocols applied to microbial rhodopsins with known crystal structures (51) found that I-TASSER and Rosetta with membrane-specific terms are overall highly accurate but with less fidelity in prediction of active site residue orientation. We emphasize that we present homology models as first approximations and that determination of the physicochemical basis of tuning responsible for the red-shift will require a more accurate atomic structure of a RubyACR, which we are pursuing by X-ray crystallography. The best templates for AlACR1 modeling were 3ug9A (hybrid channelrhodopsin C1C2), root-mean-square deviation (rmsd) 2.68 selected by the I-TASSER server, and 5zihB (Chrimson), rmsd 1.47, by the Robetta server.
Molecular Biology.
For expression in HEK293 cells, DNA polynucleotides encoding the transmembrane domains showing homology to previously known ACRs optimized for human codon usage were synthesized (GenScript) and cloned into the mammalian expression vector pcDNA3.1 (Life Technologies) in frame with an EYFP or mCherry tag. Mutants were generated using a Quikchange XL kit (Agilent Technologies) and verified by sequencing. For expression in Pichia, the opsin-encoding constructs were fused in frame with a C-terminal 8-His tag and subcloned into the pPIC9K (A1ACR1, HfACR1, and AlACR2) or pPICZα (AlACR3) vector (Invitrogen) according to the manufacturer’s instructions. Expression of the constructs for A1ACR1 from Aurantiochytrium sp. and T1ACR from Thraustochytrium sp. in HEK293 cells was poor, as judged by the tag fluorescence, but a C-terminal truncation to 270 encoded residues improved it. We also synthesized constructs encoding the entire predicted polypeptides for AlACR1 and AlACR2 from A. limacinum (696 and 635 residues, respectively), but in both cases no membrane fluorescence was observed.
HEK293 Transfection and Patch Clamp Recording.
HEK293 cells were transfected using the JetPRIME transfection reagent (Polyplus). All-trans-retinal (Sigma) was added at the final concentration of 3 µM immediately after transfection. Photocurrents were recorded 48 to 96 h after transfection in the whole-cell voltage clamp mode with an Axopatch 200B amplifier (Molecular Devices) using the 10-kHz low-pass Bessel filter. The signals were digitized with a Digidata 1440A using pClamp 10 software (both from Molecular Devices). Patch pipettes with resistances of 2 to 4 MΩ were fabricated from borosilicate glass. The standard pipette solution contained (in mM): KCl 126, MgCl2 2, CaCl2 0.5, Na-EGTA 5, and Hepes 25, pH 7.4. The standard bath solution contained (in mM): NaCl 150, CaCl2 1.8, MgCl2 1, glucose 5, and Hepes 10, pH 7.4. A 4-M KCl bridge was used in all experiments, and possible diffusion of Cl− from the bridge to the bath was minimized by frequent replacement of the bath solution with fresh buffer. For measurements of the reversal potential shifts under varied ionic conditions, Na+ was substituted for K+ in the pipette solution to minimize the number of ionic species in the system. To reduce the Cl− concentration in the bath, NaCl was replaced with Na-aspartate; to reduce the Na+ concentration, with N-methyl-d-glucamine chloride; to increase the H+ concentration, pH was adjusted with H2SO4. To mimic ionic conditions of mature neurons, the pipette solution contained (in mM): K-gluconate 135, MgCl2 2, and Hepes 20, pH 7.2, and the bath solution contained (in mM): NaCl 125, KCl 2, MgCl2 1, CaCl2 3, glucose 30, and Hepes 25, pH 7.3. The holding voltages were corrected for liquid junction potentials calculated using the Clampex built-in LJP calculator (52). Continuous light pulses were provided by a Polychrome V light source (T.I.L.L. Photonics) in combination with a mechanical shutter (Uniblitz Model LS6, Vincent Associates) with a half-opening time of 0.5 ms. The action spectra were constructed by calculation of the initial slope of photocurrent as shown in SI Appendix, Fig. S4A, corrected for the quantum density measured at each wavelength and normalized to the maximal value. Light intensity <25 µW mm−2 was used to provide linear dependence of the parameter on the quantum density. The time window for determination of the linear slope of the photocurrent rise (5 ms) was first selected for the most efficient wavelength and then used for all other wavelengths. Laser excitation was provided by a Minilite Nd:YAG laser (532 nm, pulse width 6 ns, energy 12 mJ; Continuum). The current traces were logarithmically filtered using custom software. Curve fitting was performed by Origin Pro software (OriginLab).
Expression and Purification of ACRs from Pichia.
The plasmids encoding labyrinthulea ACRs were linearized with SalI or PmeI and used to transform Pichia pastoris strain SMD1168 (his4, pep4) by electroporation according to the manufacturer’s instructions. Resistant transformants were screened on 4 mg/mL geneticin or 1 mg/mL zeocin and first cultivated on a small scale. Rhodopsin gene expression was induced by the addition of methanol. All-trans retinal (5 µM final concentration) was added simultaneously. Clones of the brightest color were selected for further experimentation. For protein purification, a starter culture was inoculated into buffered complex glycerol medium until absorbance at 600 nm (A600) reached 4 to 8, after which the cells were harvested by centrifugation and resuspended at A600 1 to 2 in buffered complex methanol medium supplemented with 5 μM all-trans retinal (Sigma Aldrich). Expression was induced by the addition of 0.5% methanol. After 24 to 30 h, the cells were harvested and disrupted in a bead beater (BioSpec Products) in buffer A (20 mM sodium phosphate, pH 7.4, 100 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 5% glycerol). After removing cell debris by low-speed centrifugation, membrane fragments were collected by ultracentrifugation, resuspended in buffer B (20 mM Hepes, pH 7.4, 300 mM NaCl, 5% glycerol) and solubilized by incubation with 1.5% dodecyl maltoside (DDM) for 1.5 h or overnight at 4 °C. Nonsolubilized material was removed by ultracentrifugation, and the supernatant was mixed with nickel-nitrilotriacetic acid or cobalt superflow agarose beads (Thermofisher) and loaded on a column. The proteins were eluted with buffer C (20 mM Hepes, pH 7.4, 300 mM NaCl, 5% glycerol, 0.02% DDM) containing 300 mM imidazole, which was removed by repetitive washing with imidazole-free buffer C using YM-10 centrifugal filters (Amicon).
Absorption Spectroscopy.
Absorption spectra of purified proteins were recorded using a Cary 4000 spectrophotometer (Varian).
Statistics.
The data are presented as mean ± SEM values. Normality and equal variances of the data were not assumed, and therefore the nonparametric two-sided Mann–Whitney U test was used for pairwise comparison of independent datasets. P values > 0.05 were considered not significant. The sample size was estimated from previous experience and published work on a similar subject, as recommended by the NIH guidelines (53).
Supplementary Material
Acknowledgments
This work was supported by NIH Grant R01GM027750 and Endowed Chair AU-0009 from the Robert A. Welch Foundation (to J.L.S.) and by the Natural Sciences and Engineering Research Council of Canada Discovery Grant RGPIN-2018-04397 (to L.S.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. P.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005981117/-/DCSupplemental.
Data Availability.
The expression construct sequences of labyrinthulea and haptophyte ACRs have been deposited in the GenBank database (their accession numbers are listed in the second column in SI Appendix, Tables S2 and S3). Polynucleotide sequence data have been deposited in the GenBank database under accession numbers MT002463–MT002481.
References
- 1.Sineshchekov O. A., Jung K.-H., Spudich J. L., Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 99, 8689–8694 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagel G.et al., Channelrhodopsin-1: A light-gated proton channel in green algae. Science 296, 2395–2398 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Nagel G.et al., Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. U.S.A. 100, 13940–13945 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Govorunova E. G., Sineshchekov O. A., Janz R., Liu X., Spudich J. L., NEUROSCIENCE. Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics. Science 349, 647–650 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyden E. S., Zhang F., Bamberg E., Nagel G., Deisseroth K., Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Mohammad F.et al., Optogenetic inhibition of behavior with anion channelrhodopsins. Nat. Methods 14, 271–274 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Sineshchekov O. A., Govorunova E. G., Li H., Spudich J. L., Bacteriorhodopsin-like channelrhodopsins: Alternative mechanism for control of cation conductance. Proc. Natl. Acad. Sci. U.S.A. 114, E9512–E9519 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oppermann J.et al., MerMAIDs: A family of metagenomically discovered marine anion-conducting and intensely desensitizing channelrhodopsins. Nat. Commun. 10, 3315 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leyland B., Leu S., Boussiba S., Are Thraustochytrids algae? Fungal Biol. 121, 835–840 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Reeb V. C.et al., Interrelationships of chromalveolates within a broadly sampled tree of photosynthetic protists. Mol. Phylogenet. Evol. 53, 202–211 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Burki F., Roger A. J., Brown M. W., Simpson A. G. B., The new tree of eukaryotes. Trends Ecol. Evol. 35, 43–55 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Klapoetke N. C.et al., Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wietek J.et al., Anion-conducting channelrhodopsins with tuned spectra and modified kinetics engineered for optogenetic manipulation of behavior. Sci. Rep. 7, 14957 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Govorunova E. G.et al., The expanding family of natural anion channelrhodopsins reveals large variations in kinetics, conductance, and spectral sensitivity. Sci. Rep. 7, 43358 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Govorunova E.G.et al., Extending the time domain of neuronal silencing with cryptophyte anion channelrhodopsins. eNeuro 5, ENEURO.0174-0118.2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rozenberg A., et al., Lateral gene transfer of anion-conducting channelrhodopsins between green algae and giant viruses. bioRxiv, 2020.2004.2015.042127 (16 April 2020). [DOI] [PubMed] [Google Scholar]
- 17.Mukherjee S., Hegemann P., Broser M., Enzymerhodopsins: Novel photoregulated catalysts for optogenetics. Curr. Opin. Struct. Biol. 57, 118–126 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Oda K.et al., Crystal structure of the red light-activated channelrhodopsin Chrimson. Nat. Commun. 9, 3949 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sineshchekov O. A., Govorunova E. G., Wang J., Spudich J. L., Enhancement of the long-wavelength sensitivity of optogenetic microbial rhodopsins by 3,4-dehydroretinal. Biochemistry 51, 4499–4506 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sineshchekov O. A., Govorunova E. G., Wang J., Li H., Spudich J. L., Intramolecular proton transfer in channelrhodopsins. Biophys. J. 104, 807–817 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sineshchekov O. A., Govorunova E. G., Li H., Spudich J. L., Gating mechanisms of a natural anion channelrhodopsin. Proc. Natl. Acad. Sci. U.S.A. 112, 14236–14241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nack M.et al., The DC gate in channelrhodopsin-2: Crucial hydrogen bonding interaction between C128 and D156. Photochem. Photobiol. Sci. 9, 194–198 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Chuong A. S.et al., Noninvasive optical inhibition with a red-shifted microbial rhodopsin. Nat. Neurosci. 17, 1123–1129 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balashov S. P., Lanyi J. K., Xanthorhodopsin: Proton pump with a carotenoid antenna. Cell. Mol. Life Sci. 64, 2323–2328 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balashov S. P.et al., Reconstitution of gloeobacter rhodopsin with echinenone: Role of the 4-keto group. Biochemistry 49, 9792–9799 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luecke H.et al., Crystallographic structure of xanthorhodopsin, the light-driven proton pump with a dual chromophore. Proc. Natl. Acad. Sci. U.S.A. 105, 16561–16565 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imasheva E. S.et al., Chromophore interaction in xanthorhodopsin: Retinal dependence of salinixanthin binding. Photochem. Photobiol. 84, 977–984 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polívka T.et al., Femtosecond carotenoid to retinal energy transfer in xanthorhodopsin. Biophys. J. 96, 2268–2277 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong Y., Wagner M. J., Zhong Li J., Schnitzer M. J., Imaging neural spiking in brain tissue using FRET-opsin protein voltage sensors. Nat. Commun. 5, 3674 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou P.et al., Bright and fast multicoloured voltage reporters via electrochromic FRET. Nat. Commun. 5, 4625 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azimi Hashemi N.et al., Rhodopsin-based voltage imaging tools for use in muscles and neurons of Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 116, 17051–17060 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujimoto K., Hayashi S., Hasegawa J. Y., Nakatsuji H., Theoretical studies on the color-tuning mechanism in retinal proteins. J. Chem. Theory Comput. 3, 605–618 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Melaccio F., Ferré N., Olivucci M., Quantum chemical modeling of rhodopsin mutants displaying switchable colors. Phys. Chem. Chem. Phys. 14, 12485–12495 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Katayama K., Sekharan S., Sudo Y., “Color tuning in retinylidene proteins” in Optogenetics: Light-Sensing Proteins and Their Applications, Yawo H., Kandori H., Koizumi A., Eds. (Springer, Tokyo, 2015), pp. 89–109. [Google Scholar]
- 35.Gozem S., Luk H. L., Schapiro I., Olivucci M., Theory and simulation of the ultrafast double-bond isomerization of biological chromophores. Chem. Rev. 117, 13502–13565 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Hufen J., Sugihara M., Buss V., How the counterion affects ground- and excited-state properties of the rhodopsin chromophore. J. Phys. Chem. B 108, 20419–20426 (2004). [Google Scholar]
- 37.Karasuyama M., Inoue K., Nakamura R., Kandori H., Takeuchi I., Understanding colour tuning rules and predicting absorption wavelengths of microbial rhodopsins by data-driven machine-learning approach. Sci. Rep. 8, 15580 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marti T., Rösselet S. J., Otto H., Heyn M. P., Khorana H. G., The retinylidene Schiff base counterion in bacteriorhodopsin. J. Biol. Chem. 266, 18674–18683 (1991). [PubMed] [Google Scholar]
- 39.Inoue K.et al., Schizorhodopsins: A novel family of rhodopsins from Asgard archaea that function as light-driven inward H+ pumps. Sci. Adv. 6, eaaz2441 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris A.et al., Mechanism of inward proton transport in an antarctic microbial rhodopsin. J. Phys. Chem. B 124, 4851–4872 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Inoue K.et al., Red-shifting mutation of light-driven sodium-pump rhodopsin. Nat. Commun. 10, 1993 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Man D.et al., Diversification and spectral tuning in marine proteorhodopsins. EMBO J. 22, 1725–1731 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peperzak L., Colijn F., Vrieling E. G., Gieskes W. W. C., Peeters J. C. H., Observations of flagellates in colonies of Phaeocystis globosa (Prymnesiophyceae): A hypothesis for their position in the life cycle. J. Plankton Res. 22, 2181–2203 (2000). [Google Scholar]
- 44.Bennett R. M., Honda D., Beakes G. W., Thines M., “Labyrinthulomycota” in Handbook of the Protists, Archibald J., Simpson A., Slamovits C., Eds. (Springer, 2017), pp. 507–542. [Google Scholar]
- 45.Moro I., Negrisolo E., Callegaro A., Andreoli C., Aplanochytrium stocchinoi: A new Labyrinthulomycota from the southern ocean (Ross Sea, Antarctica). Protist 154, 331–340 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Trifinopoulos J., Nguyen L. T., von Haeseler A., Minh B. Q., W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Letunic I., Bork P., Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang J.et al., The I-TASSER suite: Protein structure and function prediction. Nat. Methods 12, 7–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ovchinnikov S., Park H., Kim D. E., DiMaio F., Baker D., Protein structure prediction using Rosetta in CASP12. Proteins 86 (suppl. 1), 113–121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haas J.et al., Introducing “best single template” models as reference baseline for the Continuous Automated Model Evaluation (CAMEO). Proteins 87, 1378–1387 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nikolaev D. M.et al., A comparative study of modern homology modeling algorithms for rhodopsin structure prediction. ACS Omega 3, 7555–7566 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barry P. H., JPCalc, a software package for calculating liquid junction potential corrections in patch-clamp, intracellular, epithelial and bilayer measurements and for correcting junction potential measurements. J. Neurosci. Methods 51, 107–116 (1994). [DOI] [PubMed] [Google Scholar]
- 53.Dell R. B., Holleran S., Ramakrishnan R., Sample size determination. ILAR J. 43, 207–213 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The expression construct sequences of labyrinthulea and haptophyte ACRs have been deposited in the GenBank database (their accession numbers are listed in the second column in SI Appendix, Tables S2 and S3). Polynucleotide sequence data have been deposited in the GenBank database under accession numbers MT002463–MT002481.