Significance
In many plants, flowering is regulated by environmental cues, such as day length. Under flowering-inductive conditions, leaves synthesize and transmit FLOWERING LOCUS T (FT) protein to the shoot apex, where FT activates flowering. Dodder Cuscuta australis, which is a root- and leafless parasitic plant, however, very likely does not have fully functional FT genes, and it flowers only when the host plants flower. We show that host-synthesized FT protein is able to move into dodder stems, where FT physically interacts with dodder FD transcription factor, activating flowering of dodder. This specific manner of flowering allows dodder to synchronize its flowering time with that of the host plant, and this is likely a trait that is beneficial for dodder’s reproductive success.
Keywords: Cuscuta, dodder, flowering, FLOWERING LOCUS T, parasitic plant
Abstract
Many plants use environmental cues, including seasonal changes of day length (photoperiod), to control their flowering time. Under inductive conditions, FLOWERING LOCUS T (FT) protein is synthesized in leaves, and FT protein is a mobile signal, which is able to travel to the shoot apex to induce flowering. Dodders (Cuscuta, Convolvulaceae) are root- and leafless plants that parasitize a large number of autotrophic plant species with varying flowering time. Remarkably, some dodder species, e.g., Cuscuta australis, are able to synchronize their flowering with the flowering of their hosts. Detailed sequence inspection and expression analysis indicated that the FT gene in dodder C. australis very likely does not function in activating flowering. Using soybean host plants cultivated under inductive and noninductive photoperiod conditions and soybean and tobacco host plants, in which FT was overexpressed and knocked out, respectively, we show that FT-induced flowering of the host is likely required for both host and parasite flowering. Biochemical analysis revealed that host-synthesized FT signals are able to move into dodder stems, where they physically interact with a dodder FD transcription factor to activate dodder flowering. This study demonstrates that FTs can function as an important interplant flowering signal in host–dodder interactions. The unique means of flowering regulation of dodder illustrates how regressive evolution, commonly found in parasites, may facilitate the physiological synchronization of parasite and host, here allowing the C. australis parasite to time reproduction exactly with that of their hosts, likely optimizing parasite fitness.
Parasitic plants independently evolved 12 to 13 times, constituting about 1% of angiosperms (∼4,500 species) (1). Parasitic plants use a special organ, named haustorium, to attach themselves to the hosts and penetrate into host tissues, forming vascular connections. Through haustoria, parasitic plants extract water and nutrients from hosts. Some parasitic plants, termed holoparasites, do not photosynthesize, whereas hemiparasites are photosynthetically active. Facultative parasites can live alone without parasitizing host plants, while obligate parasites are not able to complete their life cycles without host plants. Most hemiparasites still have leaves, but almost all holoparasites evolved to be leafless, and some even no longer have roots. Parasitic plants have unique physiology, ecology, and evolutionary histories, and host plant–parasite systems have become exciting models for studying plant–plant interactions (2).
Dodders (Cuscuta spp., Convolvulaceae) are worldwide distributed parasitic plant (3). They are leaf- and rootless parasites with little to no photosynthetic activity and are usually considered to be holoparasitic. Dodders twine around host plants, producing numerous haustoria along dodder stems, which penetrate into host stems. Dodder haustorial phloem and xylem fuse with those of its hosts to obtain nutrients and water. Recent studies have revealed that through dodder haustoria, dodders and host plants also have extensive interplant trafficking of systemic signals, secondary metabolites, mRNAs, small RNAs, and even proteins (4–8). Kim et al. (9) demonstrated that in an Arabidopsis–dodder parasitization system, thousands of Arabidopsis and dodder mRNA species could be detected in the recipient dodder and Arabidopsis, respectively. At the haustorial interface, miRNAs from dodder are able to enter host stem tissues and silence target host mRNAs, functioning as virulence factors during parasitism (5). Using proteomic analysis, it was recently demonstrated that hundreds to more than 1,500 proteins trafficked between dodder and the host plants Arabidopsis and soybean, and hundreds of interplant mobile proteins were even detected in dodder and soybean seeds; importantly, using Arabidopsis expressing different reporter proteins, it was shown that these reporter proteins retained their activity in dodder parasites (10).
Appropriate timing of flowering is critical for successful reproduction of plants. Various environment cues, especially changes in day length (photoperiod), are perceived by leaves (11, 12). Under “inductive” conditions, such as short photoperiods for soybean (Glycine max) and rice (Oryza sativa) and long photoperiods for Arabidopsis (Arabidopsis thaliana), the synthesis of a mobile signal FLOWERING LOCUS T (FT) is triggered in leaves; FT is then transported to the shoot apical meristem, where it induces flowering by facilitating the transcription of genes functioning in flower development (13, 14).
Yet very little is known about how flowering of parasitic plants is regulated. Plant parasites are vascularly connected with hosts and there is a possibility that in some of the host–parasite systems, the host or parasite might regulate the other’s flowering by interplant transfer of signaling molecules. Holoparasites have dramatic changes in body plans, and their physiology is expected to be very different from that of autotrophic plants. Given that holoparasites are mostly leafless, how these parasites sense environmental changes and activate flowering remains unclear. Dodders parasitize a wide range of host plants that span multiple plant families with a great diversity of flowering time. More than half a century ago, Fratianne (15) reported that Cuscuta campestris flowered only when the short-day (SD) hosts, the cocklebur (Xanthium strumarium) and soybean and the long-day (LD) plants, feverfew (Matricaria parthenoides) and henbane (Hyoscyamus niger), flowered under their respective flowering-inductive conditions, but not under the noninductive photoperiods. Thus, flowering of dodder seemed to be synchronized with flowering of their hosts.
In this study, we show that Cuscuta australis does not have an autonomous flowering behavior but aligns its flowering time with that of its host. Our genetic and biochemical analyses indicate that host-produced FT proteins are able to travel to C. australis and physically interact with dodder FD transcription factor, likely leading to flowering of C. australis. This study identifies FT as an important interplant mobile flowering signal, which allows C. australis parasites to synchronize their flowering with the flowering of hosts.
Results
C. australis FT Is Very Likely Nonfunctional in Regulating Flowering.
Recent sequencing efforts of the C. australis genome revealed the loss of 25 genes, including CaELF3, CaELF4, CaFLC, CaFRI, CaSVP, CaAGL17, and CaCO (16), whose Arabidopsis homologs have been intensively studied and are known to regulate flowering time; our analysis also indicated a very similar loss of these flowering regulatory genes in C. campestris (17) (Dataset S1). Thus, dodders likely no longer possess these critical flowering regulation genes that are normally conserved in autotrophic plants.
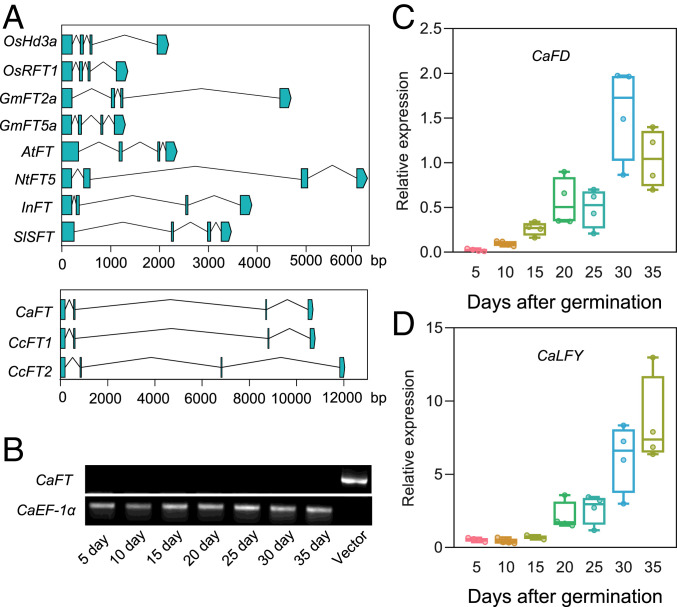
In autotrophic plants, FT is located downstream of multiple flowering pathways and plays an important role in inducing floral transition and floral development genes. Therefore, we inspected the FT genes in the C. australis and C. campestris genomes (16, 17). The FT genes in both dodder species were found to harbor a large transposon-like insertion in their second introns, compared with the second introns of FT genes in rice, soybean, Arabidopsis, tobacco (Nicotiana tabacum), morning glory (Ipomoea nil), and tomato (Solanum lycopersicum) (Fig. 1A). Next, real-time quantitative PCR (qPCR) and reverse transcription-PCR (RT-PCR) analysis were performed to detect the expression of the C. australis FT gene in samples collected from 5- to 35-d-old dodder stems with 5-d intervals. The 35-d-old dodders had started to flower; thus, these samples represent dodders from the vegetative to reproductive stage. CaFT expression was not detected in any samples (Fig. 1B and Dataset S2 A and B), even though the flowering inducers CaFD and CaLFY were highly induced in 20- to 35-d-old dodder samples (Fig. 1 C and D).
Fig. 1.
The structures of FT genes in different species and expression of CaFT, CaFD, and CaLFY at different stages of C. australis development. (A) The genomic structure of FT genes from rice (O. sativa, OsHd3a and OsRFT1), soybean (G. max, GmFT2a and GmFT5a), cultivated tobacco (N. tabacum, NtFT5), tomato (S. lycopersicum, SlSFT), morning glory (I. nil, InFT), dodder (C. australis, CaFT and C. campestris, CcFT1/2), and thale cress (A. thaliana, AtFT). Exons are shown as blue boxes and introns are shown as lines. The x axes indicate the lengths (bp) of genes. Accession nos. are as follows: OsHd3a (LOC_Os06g06320), OsRFT1 (LOC_Os06g06300), GmFT2a (Glyma.16G150700), GmFT5a (Glyma.16G044100), AtFT (AT1G65480), NtFT5 (KY306471), SlSFT (Solyc03g063100.2), InFT (INIL09g31483), CaFT (RAL41934), CcFT1 (Cc042091), and CcFT2 (Cc008736). (B–D) Relative expression levels of flowering-related genes CaFT, CaFD, and CaLFY in C. australis. Seedlings (5 d old) and stems (10 to 35 d old; note: dodder was grown on soybean Williams 82 under SD conditions, and both dodder and soybean had flowered on day 35) of C. australis were collected for qPCR or RT-PCR analysis. The relative expression levels of CaFT were analyzed by RT-PCR (B) and CaEF-1α was used as the internal control. The “vector” lane (positive control for CaFT amplification) indicates the PCR product obtained using the plasmid pJET1.2-CaFT-CDS as the template and CaFT-specific primers. PCR products were separated on an agarose gel. The relative transcript levels of CaFD (C) and CaLFY (D) were quantified by qPCR (n = 4). For C and D, the horizontal bars within boxes indicate medians. The tops and bottoms of boxes indicate upper and lower quartiles, respectively. The upper and lower whiskers represent maximum and minimum, respectively.
A construct containing C. australis CaFT full-length genomic sequence driven by a CaMV 35S promoter was transformed into Arabidopsis (SI Appendix, Fig. S1A), and both the wild-type (WT) and the transgenic lines were cultivated under SD conditions, which suppresses the expression of the Arabidopsis endogenous AtFT. These transgenic lines flowered at the same time as the WT plants (SI Appendix, Fig. S1B), and RT-PCR analysis indicated no detectable CaFT transcripts, maybe due to its very large intron, which might interfere in the splicing of CaFT pre-mRNA (SI Appendix, Fig. S1C). In addition, a construct containing an artificially synthesized C. australis CaFT coding sequence, driven by a CaMV 35S promoter, was also transformed into Arabidopsis. Similarly, under SD conditions, the flowering times of both WT and three transgenic lines were not different (SI Appendix, Fig. S1D), even though the CaFT transcripts were detected by qPCR in all three independent lines examined (SI Appendix, Fig. S1E).
From these data, we infer that CaFT protein seems to be nonfunctional, at least when being heterologously expressed in Arabidopsis. Consistent with this scenario, we aligned the deduced C. australis CaFT and C. campestris CcFT1 and CcFT2 protein sequences with the FT proteins from a few autotrophic plants (SI Appendix, Fig. S2). It was found that three amino acids (positions 10, 99, and 131 in the alignment shown in SI Appendix, Fig. S2), which are well conserved among monocotyledonous rice and the other dicotyledonous plants, had mutated in both C. australis and C. campestris. Notably, none of these three amino acids overlapped with either of the previously identified two amino acids that are important for Arabidopsis AtFT function (18). It is probable that one or more of these mutated sites may be the reason for the loss of CaFT function in flowering regulation.
C. australis Flowering Requires Host FT Expression.
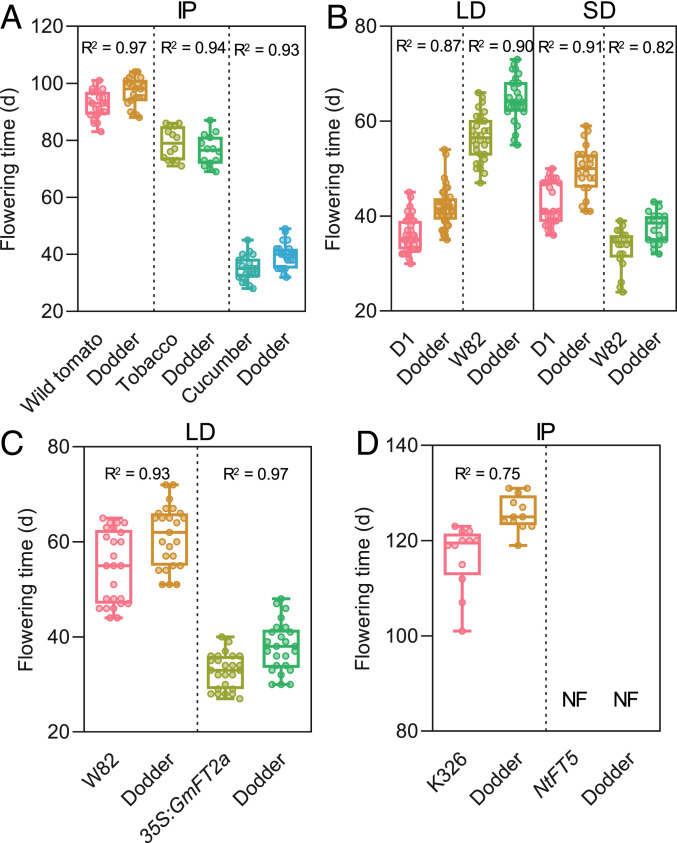
To confirm this unusual nonautonomous flowering behavior of dodder, we infested three photoperiod-insensitive species, the wild tomato (Solanum pennellii), tobacco, and cucumber (Cucumis sativus) with C. australis seedlings. These host plants flowered on days 87, 75, and 35, respectively, and their respective parasitizing C. australis flowered on days 92, 79, and 40 (Fig. 2A). Next, we used a SD plant, the soybean cultivar Williams 82 (W82) as a host. Under LD conditions, W82 soybean and the parasitizing C. australis, respectively, flowered on days 57 and 64, while under SD conditions, W82 and C. australis flowered on days 33 and 38 (Fig. 2B and SI Appendix, Fig. S3). Another soybean cultivar Dongsheng 1 (D1; bred for soybean cultivation under the LD condition of northeastern China) was also grown under LD and SD. D1 flowered moderately earlier under LD than under SD, and again the flowering times of C. australis closely synchronized with those of D1 under both conditions (Fig. 2B). Thus, these experiments indicate that C. australis flowering time is highly flexible and closely tracks that of its host plants.
Fig. 2.
The flowering times of C. australis on different hosts under different photoperiod conditions. (A) The flowering times of day-neutral wild tomato, tobacco (cv Samsun), and cucumber hostplants and their respective parasitizing dodder cultivated under intermediate photoperiod (IP) (n = 21, 21, 20, 20, 21, and 21; from Left to Right). (B) The flowering times of soybean Dongsheng 1 (D1) and Williams 82 (W82) and their parasitizing dodder under LD and SD conditions (n = 40, 40, 30, and 26 under LD and n = 26, 23, 20, and 18 under SD; from Left to Right). (C) The flowering times of WT soybean (W82) and transgenic soybean overexpressing GmFT2a (35S:GmFT2a) and their respective parasitizing dodder, under LD conditions (n = 27, 27, 27, and 27; from Left to Right). (D) The flowering times of WT tobacco (K326) and NtFT5-knockout mutants and their parasitizing dodder (n = 12, 12, 16, and 16; from Left to Right). NF, no flowering until 180 d. Each panel indicates the data from the same group of host–dodder interaction partners. The horizontal bars within boxes indicate medians. The tops and bottoms of boxes indicate upper and lower quartiles, respectively. The upper and lower whiskers represent maximum and minimum, respectively. R2 indicates correlation coefficients, which were obtained from correlation analyses using the flowering times of each pair of host–dodder interaction partners.
Given that FT is well known as a phloem mobile signal controlling flowering (13, 14), we hypothesized that under inductive conditions, FT is synthesized in host leaves and transported through the phloem to the host shoot apex, as well as through phloem fusions into the C. australis to trigger C. australis flowering. A genetic approach was used to test this hypothesis. WT and GmFT2a-overexpressing soybean plants (19) were infested with C. australis and it was found that overexpressing GmFT2a in the soybean greatly accelerated the flowering of both soybean and C. australis (Fig. 2C). Conversely, if C. australis parasitized a tobacco line whose flowering inducer NtFT5 (20) had been knocked out, the C. australis plants did not flower at all and finally died (Fig. 2D and SI Appendix, Fig. S4).
Therefore, these genetic data indicate that expression of host FT is required for C. australis flowering.
Host FT Can Travel into C. australis and Interact with C. australis FD Transcription Factor.
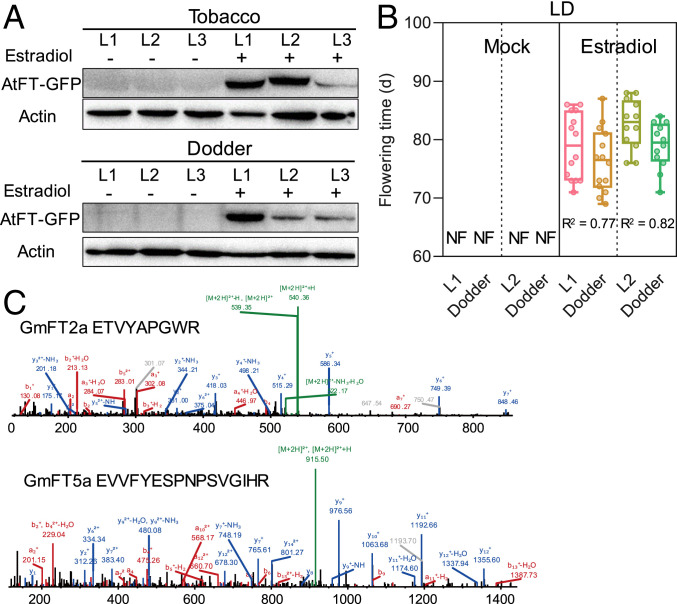
To biochemically evaluate if the FT protein can be translocated from host to dodder, we transformed the SD tobacco cultivar, Maryland Mammoth (MM) (21), with the Arabidopsis AtFT fused with GFP (green fluorescence protein) under the control of an estradiol-inducible promoter. WT MM and transformants were cultivated under LD conditions, in order to suppress the expression of the endogenous NtFTs in MM, and the leaves were treated with estradiol to activate the expression of transgene AtFT-GFP. Estradiol treatment highly activated the expression of the AtFT-GFP transgene (SI Appendix, Fig. S5A). Western blotting indicated that AtFT-GFP protein was indeed induced in the MM hosts, and importantly, AtFT-GFP was also detected in C. australis (Fig. 3A). In contrast, very little or no signals were detected in the mock-treated transgenic MM hosts (controls) and the parasitizing C. australis (Fig. 3A). Consistently, the transgenic MM treated with estradiol and its parasitizing C. australis flowered on days 81 and 78, respectively, while the control MM and its parasitizing C. australis did not flower, even on day 120, when the experiment was terminated (Fig. 3B). To exclude the possibility that AtFT-GFP mRNA could be transported from the MM hosts to C. australis and translated into protein therein, RT-PCR analysis was performed to detect AtFT-GFP transcripts in C. australis. No detectable AtFT-GFP transcripts were detected in C. australis (SI Appendix, Fig. S5B).
Fig. 3.
Translocation of FT proteins from hosts to C. australis. (A) Western blotting detection of AtFT-GFP in the transgenic tobacco and parasitizing dodder. Three lines (L1 to L3) of tobacco (cv Maryland Mammoth) plants, which had been transformed with AtFT-GFP driven by an estradiol-inducible promoter, were treated with estradiol or mock treated. After 48 h, tobacco leaves and stems of the parasitizing dodders were harvested for detection of AtFT-GFP protein. (B) The flowering times of the transgenic tobacco (L1 and L2) and parasitizing dodder. Tobacco plants were treated with estradiol or mock treated for 16 d (once every 2 d) (n = 21, 21, 15, 15, 14, 14, 12, and 12; from Left to Right). NF, no flowering until 120 d. L1 and L2 represent two independent lines. The horizontal bars within boxes indicate medians. The tops and bottoms of boxes indicate upper and lower quartiles, respectively. The upper and lower whiskers represent maximum and minimum, respectively. R2 indicates correlation coefficients, which were obtained from correlation analysis using the flowering times of each pair of host–dodder. (C) Mass spectrometry characterization of GmFT2a- and GmFT5a-derived peptides identified in dodder stem proteome. Dodders were grown on soybean Williams 82, which was cultivated under SD condition for about 20 d. Mass spectra indicate sequences ETVYAPGWR and EVVFYESPNPSVGIHR from GmFT2a and GmFT5a, respectively.
Two FT genes in soybean W82, GmFT2a and GmFT5a, are flowering inducers (22). Under the flowering-inducing SD conditions, the transcript levels of both GmFT2a and GmFT5a in W82 soybean plants attained peak values 20 d after germination, and on this day, we harvested the stem tissues of the parasitizing C. australis. Proteomic analysis of these C. australis stems revealed the presence of 327 soybean mobile proteins (Dataset S3 A and B), among which GmFT2a and GmFT5a were identified (Fig. 3C), although none of the other host mobile proteins are known to be flowering inducing.
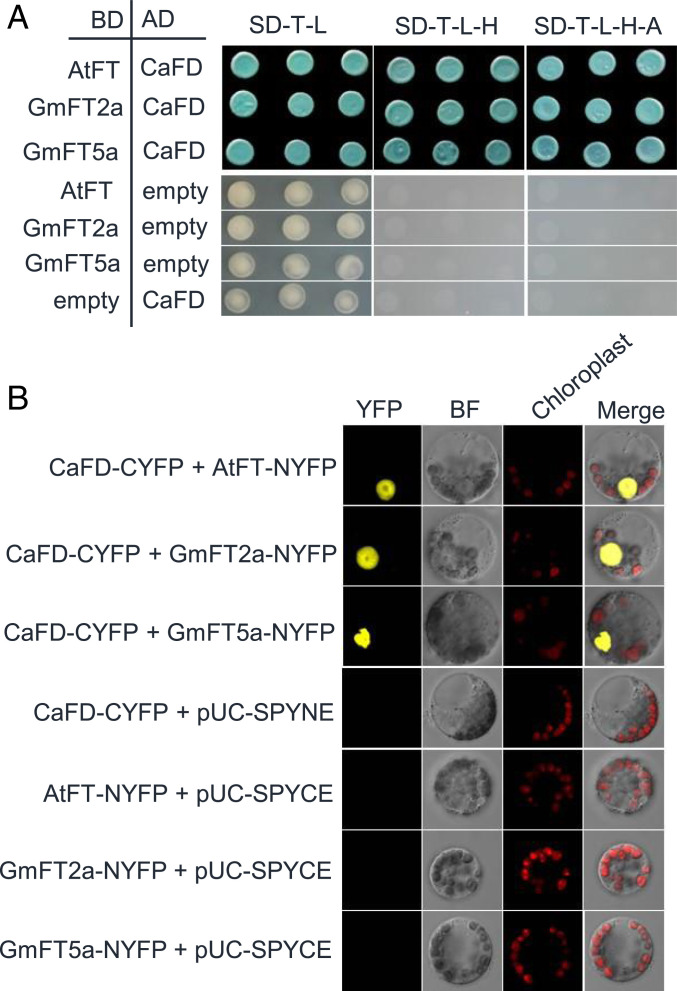
FT binds to a b-ZIP transcription factor FD to activate the transcription of downstream flowering genes (23). Next, we used yeast two-hybrid (Y2H) assays to examine whether host FT can interact with C. australis CaFD in vitro. The Arabidopsis AtFT and soybean GmFT2a and GmFT5a indeed interacted with CaFD in yeast (Fig. 4A). We also confirmed these interactions using BiFC (bimolecular fluorescence complementation) in Arabidopsis protoplasts (Fig. 4B). In addition, we found that CaFT did not interact with C. australis CaFD or Arabidoposis AtFD, further corroborating the notion that C. australis CaFT protein is nonfunctional, even if it could be expressed (SI Appendix, Fig. S6 A and B).
Fig. 4.
Protein interaction assays between C. australis CaFD and host FT proteins. (A) Y2H assay for interaction between CaFD and AtFT, GmFT2a, and GmFT5a. After cotransformation, positive clones were sequentially screened on plates without Trp (T) and Leu (L) first, on the plates without T, L, and His (H), and then on the plates without T, L, H, and adenine (A). For the control experiment of Y2H assays, pGADT7-CaFD and empty vector pGBKT7 were cotransformed into yeast AH109, and pGBKT7-GmFT2a, pGBKT7-GmFT5a, and pGBKT7-AtFT were respectively cotransformed with empty vector pGADT7 into yeast AH109. To all plates, 200 ng/mL Aureobasidin A and 20 mg/mL X-α-Gal were added. AD, GAL4 activation domain; BD, GAL4 DNA binding domain. (B) Bimolecular fluorescence complementation (BiFC) assays of the interactions between CaFD and AtFT, GmFT2a, and GmFT5a in Arabidopsis protoplasts. For the control experiment of BiFC assays, the plasmids CaFD-CYFP and pUC-SPYNE, GmFT2a-NYFP and pUC-SPYCE, GmFT5a-NYFP and pUC-SPYCE, and AtFT-NYFP and pUC-SPYCE were respectively cotransformed into Arabidopsis protoplasts. The plasmids pUC-SPYNE and pUC-SPYCE are for expression of split YFP N- and C-terminal fragments, respectively. BF, bright field; chloroplast, chloroplast autofluorescence.
From these data we infer that host FT signals are able to travel through dodder–host junctions (phloem fusion) and enter C. australis stems, where they bind to the C. australis CaFD transcription factor.
Host FT Activates Flowering-Related Genes in C. australis.
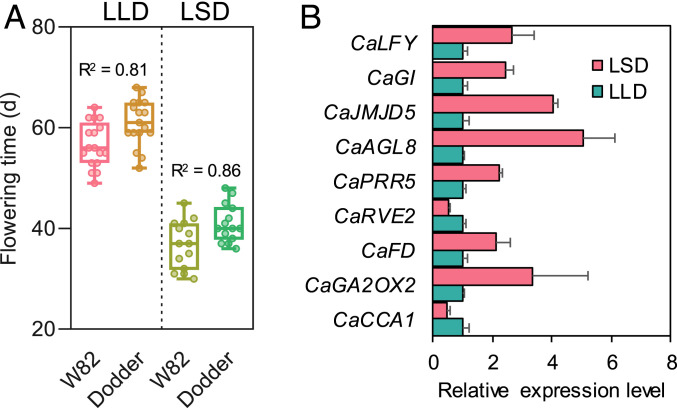
A transcriptomic approach was employed to further understand the host-induced flowering in C. australis. Fifteen-day-old W82 soybean plants, parasitized by C. australis and cultivated under the flowering suppressive LD conditions, were either retained under the LD condition (LD to LD, LLD) or transferred to SD conditions (LD to SD, LSD) to rapidly induce host flowering. Indeed, the LD-to-SD shift highly accelerated the flowering of soybean and C. australis (Fig. 5A), and this was associated with the increased expression of both GmFT2a and GmFT5a, which attained peak values 6 d in soybean leaves after the LD-to-SD shift (SI Appendix, Fig. S7 A and B). The parasitizing C. australis stems were harvested at this time for RNA sequencing (RNA-seq) analysis. In total, we identified 188 differentially expressed genes (DEGs) between the C. australis from the treatment and control groups (Dataset S4), among which, the expression of the flowering-related genes CaGA2OX2, CaFD, CaPRR5, CaAGL8, CaGI, CaLFY, and CaJMJD5 were up-regulated, while CaCCA1 and CaRVE2 were moderately down-regulated (Fig. 5B). Gene ontology (GO) analysis also indicated enrichment of the GO terms “circadian rhythm” and “flower development” (SI Appendix, Table S1). The elevated expression of CaFD, CaLFY, and CaAGL8 were confirmed by qPCR in the samples from day 6, as well as from days 8 and 10 (SI Appendix, Fig. S8). These RNA-seq data are consistent with the scenario that host FT signals activate flowering-related genes in C. australis.
Fig. 5.
LD to SD photoperiod shift-induced changes of flowering times and transcriptomes in C. australis. Soybean (Williams 82) and the parasitizing dodders were initially grown under LD. They were kept under LD (LLD) or moved to SD condition (LSD). (A) The flowering times of soybean and parasitizing dodders (n = 17, 17, 14, and 14; from Left to Right). Each panel indicates the data from the same group of host–dodder interaction partners. The horizontal bars within boxes indicate medians. The tops and bottoms of boxes indicate upper and lower quartiles, respectively. The upper and lower whiskers represent maximum and minimum, respectively. R2 indicates correlation coefficients, which were obtained from correlation analyses using the flowering times of each pair of host–dodder. (B) The relative transcript levels of nine flowering-related differentially regulated genes identified between LSD- and LLD-grown dodders, 6 d after the photoperiod shift. Data were retrieved from the RNA-seq data (Dataset S2 A and B; n = 3; error bars are SE).
Various mobile mRNAs can be translocated between scions and rootstocks in grafting systems, and similarly, host and dodders also exchange large numbers of mRNAs (9, 24, 25). Next, the above transcriptome data were used to determine whether soybean mRNAs could be transported to C. australis stems. Indeed, under LLD and LSD conditions, 245 and 112 mobile soybean mRNAs were respectively identified in C. australis stems (Dataset S5 A and C). However, transcripts from soybean FT/FT-like genes or other flowering accelerator genes (e.g., GmCO and GmFTIP) were not detected in C. australis stems (Dataset S5 B and D). Thus, it is unlikely that transport of mRNAs from host plants could have impacted flowering of C. australis.
C. campestris and Cuscuta europaea Have a Similar Host-Regulated Flowering Behavior.
By cultivating W82 soybeans under LD and SD photoperiod and by using the transgenic soybean overexpressing GmFT2a, we confirmed that C. campestris synchronized its flowering with its hosts. Moreover, another dodder species, C. europaea (subgenus Cuscuta), which is phylogenetically relatively distant from C. australis and C. campestris (both subgenus Grammica), also exhibited synchronized flowering with its hosts (SI Appendix, Figs. S9 and S10). Thus, it is possible that the common ancestor of the Cuscuta genus had evolved this host FT-dependent flowering mechanism.
Discussion
There is large-scale interplant trafficking of mRNAs, small RNAs, and proteins between dodder and hosts (5, 9, 10). However, very little is known about the physiological and ecological functions of most of these interplant mobile molecules. In this study, we reveal a highly likely mechanism underlying the unique flowering behavior of C. australis parasite and demonstrate a means of parasite–host physiological synchronization: the host’s leaf-synthesized mobile flowering inducer, FT, moves through host–C. australis junctions into C. australis to activate flowering, illustrating that FT proteins are interplant mobile signals and are physiologically and ecologically important in host–C. australis interaction. C. australis and many other dodder species have very wide host ranges, and some of these host species might use special flowering regulation mechanisms that do not require FT signals under certain circumstances. Thus, in addition to the host FT-mediated flowering regulation, dodders may have evolved other mechanisms to perceive cues other than FT from hosts to flower, especially when the hosts’ flowering does not require FT signals. Future investigations on flowering of dodders on hosts from various families and under different environmental factors, including photoperiod, temperature, biotic and abiotic stresses, and genetic and biochemical analyses are needed to gain further insight into the flowering regulation of dodders.
We found that C. europaea, which belongs to the subgenus Cuscuta and is relative distantly related to C. australis and C. campestris (both subgenus Grammica), also has a host-regulated flowering behavior. Thus, there is a possibility that this nonautonomous flowering behavior was a trait evolved in the dodder common ancestor, before the split of descendent species. Host-driven flowering behavior is likely an adaptive trait of dodder. Dodder host plants’ flowering times vary substantially. If dodder parasites flower autonomously, they could flower much earlier or later than do their hosts. Compared with dodder whose flowering time is similar to that of the host, 1) in the scenario that dodder flowers much earlier, the early-flowering dodder should grow smaller, as the vegetative growth is prematurely ended, and likely does not produce as many seeds; and 2) in the scenario that dodder flowers much later than the host, dodder may suffer from decreased nutrient levels in the host, as flowering and subsequent seed development drain the nutrients of the host, and there is even a possibility that there is a too small time window between host flowering and death, and therefore, dodder may have no chance to produce seeds. Dodder is an opportunistic parasite, and by “eavesdropping” on these host flowering signals (FT proteins and/or certain other flowering-related signals), dodder can synchronize its reproduction with that of its host, thereby allowing these heterotrophic plants to parasitize a wide range of host plants.
The evolution of dodder FT genes seems to be complex. C. australis and C. campestris genomes both have lost many flowering regulator genes (16, 17), and in this study we show that the FT in C. australis very likely had lost its function in activation of flowering during evolution, in light of three lines of evidence: 1) CaFT expression was undetectable; 2) expressing CaFT under the 35S promoter in Arabidopsis did not change Arabidopsis flowering time; and 3) CaFT protein did not interact with CaFD or AtFD. However, we cannot completely rule out the possibility that C. australis CaFT may be expressed in particular scenarios (e.g., under specific environmental conditions or in specific tissues). Moreover, the intact full-length C. australis and C. campestris FT protein sequences imply that these dodder FTs might still have certain unknown functions, as they should have accumulated many mutations if they are completely nonfunctional. Since CaFT and CcFT have novel amino acids at the well-conserved sites of the FT proteins from autotrophic plants, these dodder FTs may even have neofunctionalized to regulate certain physiological processes (but very likely not flowering). In autotrophic plants, FTs may have other functions, apart from regulating flowering: Kinoshita et al. (26) demonstrated that in Arabidopsis AtFT also controls stomatal opening. The sequences of FT genes from many more dodder species can be further studied for molecular evolution analyses, including their sequence divergence and synonymous vs. nonsynonymous changes, shedding light on the selection pressure on dodder FT genes, and importantly, expression analysis and genetic and biochemical studies are needed for understanding the actual function of dodder FT genes, if they really had neofunctionalized.
This unique flowering mechanism illustrates what has long been considered a likely adaptive advantage of gene loss and regressive evolution commonly found in obligate parasites and bacterial symbionts (27, 28), namely, by losing its own flowering regulatory network (and even leaves), the ancestral dodders could readily adjust their reproductive physiology to that of their host physiology by assimilating and responding to host-derived signals, such as FTs. The holoparasitic lifestyle has independently evolved more than 10 times and most holoparasites have no or only vestigial leaves, and some of them also have multiple host species with diverse life spans (e.g., the broomrapes Phelipanche [Orobanchaceae]). It will be interesting to explore how many times regressive evolution has convergently discovered this solution to the challenges of physiological synchronization of parasites with hosts.
Materials and Methods
Plants and Growth Conditions.
Soybean (G. max cvs Williams 82) (WT and transgenic plants) and Dongsheng 1 were cultivated under SD (8 h light, 16 h dark) and/or under LD (16 h light, 8 h dark) conditions at 18 to 25 °C. The wild tomato (S. pennellii ecotype LA0716), tobacco (N. tabacum cvs Samsun and K326), transgenic tobacco (background K326) plants, and cucumber (C. sativus cv Chinese Long) were growth in a glasshouse under a light regime of intermediate photoperiod (12 h of light, 12 h of dark) at 22 to 25 °C. Transgenic tobacco plants with a genetic background cv Maryland Mammoth were grown under LD conditions at 22 to 25 °C. A. thaliana (Col-0) were growth in a growth chamber under SD conditions at 22 °C.
For the day-length shift experiment, 15-d-old soybean plants (cv William 82), which had been infested with dodder and cultivated under LD conditions, were divided to two group: in one group, the plants remained under the LD conditions (LLD), while in the other group, the plants were transferred to SD conditions (LSD).
The dodder (C. australis, C. campestris, or C. europaea) seeds were scarified in concentrated sulfuric acid for 40 min and then thoroughly rinsed 10 times with water (29). The scarified dodder seeds were allowed to develop for 4 d on wet paper towels in a growth chamber (intermediate photoperiod; 12/12 h light/dark; 25 °C), and the 4-d-old seedlings were used for infestations. The stems of soybean, wild tomato, and cucumber plants, which were about 4, 10, and 4 d old, respectively, were infested with dodder seedlings. Leaf petioles, rather than stems, of tobacco (30 d old) were infested, since dodder seedlings have a low success rate when infesting tobacco stems. Details of experimental methods are given in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Fanjiang Kong (Guangzhou University) for providing the GmFT2a-overexpressing soybean seeds and Y2H and BiFC plasmids, Drs. Xiaolan Zhang (China Agricultural University) and Susann Wicke (University of Muenster) for cucumber and C. europaea seeds. We thank Drs. Wenhao Yan (Huazhong Agricultural University) and Jinyong Hu (Kunming Institute of Botany, Chinese Academy of Sciences [KIB, CAS]) for valuable comments. We thank Ms. Ting Yang, Drs. Jing Li, Shalan Li, and Hui Liu, and Mr. Tianyin Zheng (KIB, CAS) for technical assistance. This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB11050200), the National Science Foundation of China (31970274), and the International Partnership Program of the Chinese Academy of Sciences (151853KYSB20170025).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009445117/-/DCSupplemental.
Data Availability.
All data are available in the main text or SI Appendix, Materials and Methods. The original sequencing data of the transcriptome are available at the National Genomics Data Center (NGDC) under the BioProject PRJCA001885 (30).
References
- 1.Westwood J. H., Yoder J. I., Timko M. P., dePamphilis C. W., The evolution of parasitism in plants. Trends Plant Sci. 15, 227–235 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Clarke C. R., Timko M. P., Yoder J. I., Axtell M. J., Westwood J. H., Molecular dialog between parasitic plants and their hosts. Annu. Rev. Phytopathol. 57, 279–299 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Poulin R., The many roads to parasitism: A tale of convergence. Adv. Parasitol. 74, 1–40 (2011). [DOI] [PubMed] [Google Scholar]
- 4.David-Schwartz R., Runo S., Townsley B., Machuka J., Sinha N., Long-distance transport of mRNA via parenchyma cells and phloem across the host-parasite junction in Cuscuta. New Phytol. 179, 1133–1141 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Shahid S.et al., MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 553, 82–85 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Haupt S., Oparka K. J., Sauer N., Neumann S., Macromolecular trafficking between Nicotiana tabacum and the holoparasite Cuscuta reflexa. J. Exp. Bot. 52, 173–177 (2001). [PubMed] [Google Scholar]
- 7.Smith J. D., Woldemariam M. G., Mescher M. C., Jander G., De Moraes C. M., Glucosinolates from host plants influence growth of the parasitic plant cuscuta gronovii and its susceptibility to aphid feeding. Plant Physiol. 172, 181–197 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hettenhausen C.et al., Stem parasitic plant Cuscuta australis (dodder) transfers herbivory-induced signals among plants. Proc. Natl. Acad. Sci. U.S.A. 114, E6703–E6709 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim G., LeBlanc M. L., Wafula E. K., dePamphilis C. W., Westwood J. H., Plant science. Genomic-scale exchange of mRNA between a parasitic plant and its hosts. Science 345, 808–811 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Liu N.et al., Extensive inter-plant protein transfer between Cuscuta parasites and their host plants. Mol. Plant 13, 573–585 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Kim D. H., Doyle M. R., Sung S., Amasino R. M., Vernalization: Winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 25, 277–299 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Song Y. H., Ito S., Imaizumi T., Flowering time regulation: Photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 18, 575–583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turck F., Fornara F., Coupland G., Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59, 573–594 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Corbesier L.et al., FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Fratianne D. G., The interrelationship between the flowering of dodder and the flowering of some long and short day plants. Am. J. Bot. 52, 556–562 (1965). [Google Scholar]
- 16.Sun G.et al., Large-scale gene losses underlie the genome evolution of parasitic plant Cuscuta australis. Nat. Commun. 9, 2683 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogel A.et al., Footprints of parasitism in the genome of the parasitic flowering plant Cuscuta campestris. Nat. Commun. 9, 2515 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahn J. H.et al., A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 25, 605–614 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nan H.et al., GmFT2a and GmFT5a redundantly and differentially regulate flowering through interaction with and upregulation of the bZIP transcription factor GmFDL19 in soybean. PLoS One 9, e97669 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beinecke F. A.et al., The FT/FD-dependent initiation of flowering under long-day conditions in the day-neutral species Nicotiana tabacum originates from the facultative short-day ancestor Nicotiana tomentosiformis. Plant J. 96, 329–342 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Amasino R. M., My favourite flowering image: Maryland Mammoth tobacco. J. Exp. Bot. 64, 5817–5818 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Kong F.et al., Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 154, 1220–1231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abe M.et al., FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Thieme C. J.et al., Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants 1, 15025 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Zhang W.et al., tRNA-related sequences rrigger systemic mRNA transport in plants. Plant Cell 28, 1237–1249 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinoshita T.et al., FLOWERING LOCUS T regulates stomatal opening. Curr. Biol. 21, 1232–1238 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Ochman H., Moran N. A., Genes lost and genes found: Evolution of bacterial pathogenesis and symbiosis. Science 292, 1096–1099 (2001). [DOI] [PubMed] [Google Scholar]
- 28.McCutcheon J. P., Moran N. A., Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 10, 13–26 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Li J.et al., The parasitic plant Cuscuta australis is highly insensitive to abscisic acid-induced suppression of hypocotyl elongation and seed germination. PLoS One 10, e0135197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen G., et al., Cuscuta australis (dodder) parasite eavesdrops on the host plants’ FT signals toflower. Sequence Read Archive. https://bigd.big.ac.cn/bioproject/browse/PRJCA001885. Deposited 5 November 2019. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or SI Appendix, Materials and Methods. The original sequencing data of the transcriptome are available at the National Genomics Data Center (NGDC) under the BioProject PRJCA001885 (30).