Significance
γδ T cells are an abundant component of the human immune system that protect against infection and cancer. Here we show that γδ T cells recognize CD1b proteins, which are known for their capacity to present lipid antigens to αβ T cells. Recognition of CD1b is dependent on the presented lipid antigen for some γδ T cells, but a lipid-independent pattern was also observed. Thus, γδ T cells show diversity in the type of protein recognized as well as differing surfaces of the protein.
Keywords: butyrophilin-like molecule, Vd1, lipid antigen, CD1b, gd T cell
Abstract
γδ T cells form an abundant part of the human cellular immune system, where they respond to tissue damage, infection, and cancer. The spectrum of known molecular targets recognized by Vδ1-expressing γδ T cells is becoming increasingly diverse. Here we describe human γδ T cells that recognize CD1b, a lipid antigen-presenting molecule, which is inducibly expressed on monocytes and dendritic cells. Using CD1b tetramers to study multiple donors, we found that many CD1b-specific γδ T cells use Vδ1. Despite their common use of Vδ1, three CD1b-specific γδ T cell receptors (TCRs) showed clear differences in the surface of CD1b recognized, the requirement for lipid antigens, and corecognition of butryophilin-like proteins. Several Vγ segments were present among the CD1b-specific TCRs, but chain swap experiments demonstrated that CD1b specificity was mediated by the Vδ1 chain. One of the CD1b-specific Vδ1+ TCRs paired with Vγ4 and shows dual reactivity to CD1b and butyrophilin-like proteins. αβ TCRs typically recognize the peptide display platform of MHC proteins. In contrast, our results demonstrate the use of rearranged receptors to mediate diverse modes of recognition across the surface of CD1b in ways that do and do not require carried lipids.
In all species with T cells, two major lineages can be distinguished based on T cell receptor (TCR) gene usage, αβ T cells and γδ T cells. In humans, the γδ T cell population comprises ∼4% of circulating T cells (1), of which the Vδ1- and Vδ2-expressing subsets are the two most abundant. Semi-invariant Vγ9Vδ2 T cells form the most prominent γδ T cell population found in the circulation, while the highly diverse Vδ1 T cells form the majority in tissues, such as intestine and spleen (2, 3).
The mechanism of antigen recognition by αβ T cells has been well established to involve TCR contact with the membrane distal surface of MHC I and II, where peptide antigens are exposed (4). Much less is known about the identities and nature of molecular targets of γδ T cells. γδ TCRs have been implicated in sensing molecular signals of microbial and nonmicrobial stress via recognition of cellular proteins, soluble proteins, or small molecules, suggesting a role for these cells in stress surveillance. The Vγ9Vδ2 population is stimulated by small phosphorylated metabolites in a butyrophilin (BTN) 2A1-, BTN3A1-, and BTN3A2-dependent way (5–7). Colonic Vγ4+ T cells respond to gut epithelial cells by binding to butyrophilin-like (BTNL) 3 and BTNL8 heterodimers through the Vγ4 chain (8–10). Human Vδ1 T cells recognize an array of proteins, including annexin A2, phycoerythrin, MR1, CD1c, and CD1d proteins (11–21). Knowing how αβ T cells depend on a large number of highly diverse V(D)J joints to recognize almost every imaginable peptide bound to major histocompatibility complex (MHC) proteins, it seems counterintuitive that γδ T cells would use the same diversity-generating mechanisms to generate millions of unique TCRs for recognition of only a handful of nonpolymorphic targets that do not present an antigen.
The CD1 family of proteins (CD1a, CD1b, CD1c, and CD1d) consists of nonpolymorphic MHC class I–like molecules encoded outside the MHC that present self- and pathogen-derived lipid antigens to T cells. CD1d is constitutively expressed on both hematopoietic and nonhematopoietic cells, while CD1a and CD1c are expressed by professional antigen-presenting cells (APCs), such as Langerhans cells (CD1a), B cells (CD1c), and dendritic cells (CD1a and CD1c) (22). Expression of CD1b in the periphery is limited to mature dendritic cells (DCs). However, CD1a, CD1b, and CD1c expression can rapidly be up-regulated on monocytes in response to cytokines (23–26). The first study describing the interaction of CD1 molecules with γδ T cells showed that Vδ1 TCRs are activated by CD1c in the absence of a foreign antigen (16). The development of CD1 tetramers has enabled the detection of γδ T cells from human peripheral blood that recognize CD1c (18, 21). In addition to CD1c, CD1d-reactive γδ T cells have also been described to recognize CD1d presenting the self-lipid sulfatide or α-galactosylceramide (17, 19, 20).
Most known CD1-specific γδ T cells express Vδ1. The biased use of Vδ1 by CD1c- and CD1d-specific γδ TCRs suggests that there could be a conserved docking mode for the interaction between Vδ1 and CD1 isoforms. Mutational analysis and crystal structures of CD1-specific γδ TCRs demonstrate a dominant role for the Vδ1 chain in binding of CD1c and CD1d, while the γ chain was either minimally involved or not involved at all (18–20). The recent discoveries that γδ TCRs interact with BTNs and BTNLs and that the interaction is mediated by the Vγ domain (5, 9, 10) point to γδ TCRs as potentially bispecific receptors and are consistent with Vδ1-dominated CD1 recognition.
The importance and function of CD1 recognition by γδ T cells is not well understood. Recognition of endogenous self-lipids might be of importance under conditions of cell stress, such as infections, cancer, and cell damage (27). γδ TCRs might sense stress through recognition of CD1 proteins that are up-regulated upon inflammatory signals and combine this with conserved binding of BTNs or BTNLs to recognize self. On the other hand, CD1 is well known to present bacterial glycolipids such as glucose monomycolate, mycolic acid, dideoxymycobactin, sulfoglycolipid, and mannosyl phosphomycoketide to αβ T cells, which is reminiscent of the presentation of pathogen-derived peptides by classical MHC molecules, the cornerstone of adaptive immunity.
The previous findings on γδ T cells that recognize CD1c and CD1d led us to the question of whether γδ T cells that recognize other CD1 molecules also exist. CD1b is, in this respect, particularly interesting because it is known to present bacterial as well as self-antigens to αβ T cells via mechanisms that are understood in great molecular detail (28–31). In addition, CD1b is the most stress regulated CD1 isoform in the sense that no human tissue other than the thymus expresses CD1b under homeostatic conditions, yet it is up-regulated under inflammatory conditions (23, 32). Therefore, we decided to specifically look for CD1b-specific γδ T cells and determine whether they recognize CD1b per se or CD1b–lipid complexes.
Results
Identification of CD1b Tetramer-Reactive γδ T Cells.
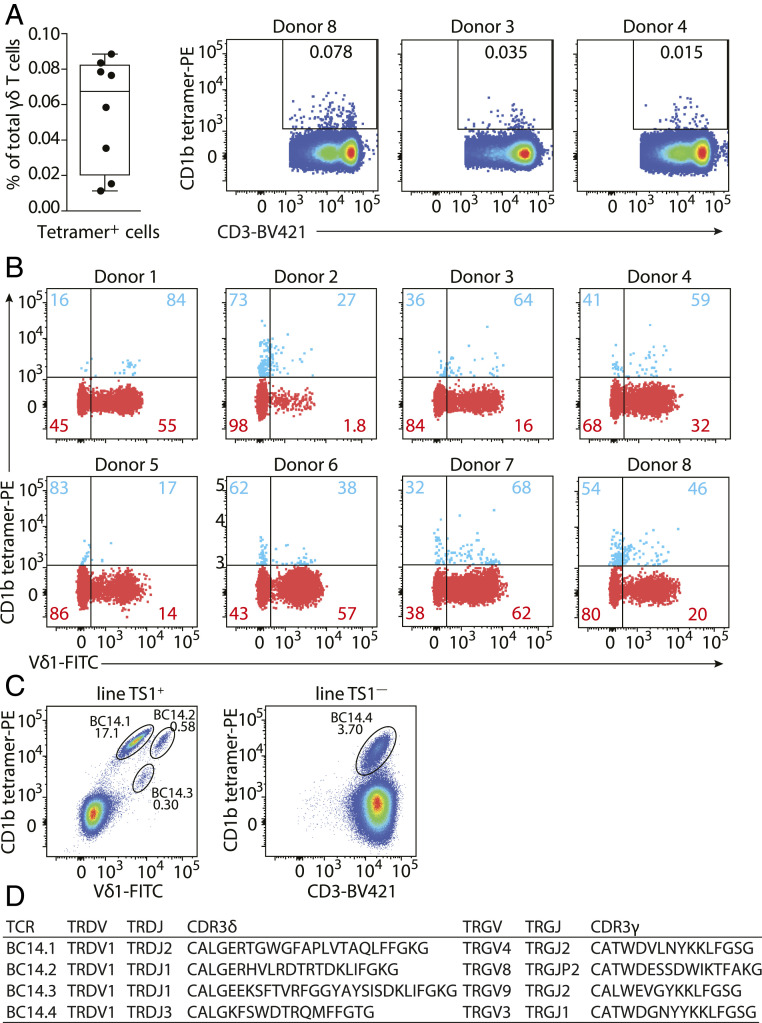
To determine if γδ T cells might recognize CD1b, we enriched γδ T cells from peripheral blood mononuclear cells (PBMCs) from eight random blood bank donors by column purification and stained them with CD1b tetramers. Tetramers were not treated with antigens and so carried a mixture of endogenous (CD1b-endo) lipids from the mammalian protein expression system. Low numbers of residual αβ T cells were excluded by gating on CD3+αβTCR− cells (SI Appendix, Fig. S1A). CD1b tetramer+ cells were detected in all donors tested (SI Appendix, Fig. S1B), with frequencies ranging from 0.011 to 0.088% of total γδ T cells, as illustrated by flow cytometry dot plots from donor 8, donor 3, and donor 4, representing frequencies at the high, intermediate, and low end of the range (Fig. 1A). Most published γδ TCRs that recognize other members of the CD1 family, CD1c and CD1d, are Vδ1+ γδ T cells (17–21), and the majority of Vδ2+ T cells recognizes targets other than CD1. Therefore, we hypothesized that CD1b-specific γδ T cells might be of the Vδ1 type. The frequency of Vδ1-utilizing TCRs among the CD1b tetramer+ γδ T cells was higher than among tetramer− γδ T cells in seven out of eight donors, as demonstrated by costaining with an anti-Vδ1 antibody (Fig. 1B), but Vδ1+ γδ T cells formed the majority of the CD1b-endo tetramer+ fraction in only four of the eight donors. Thus, CD1b-binding γδ T cells are present in the blood of healthy donors, and about half of them express Vδ1+ TCRs.
Fig. 1.
Identification of CD1b-endo-recognizing γδ T cells. (A) Percentages of CD1b-endo tetramer+ γδ T cells of total γδ T cells enriched from PBMCs by column purification (n = 8 PBMC samples) are shown. Flow cytometry dot plots show CD1b-endo tetramer staining of γδ T cells from three representative donors (gated as shown in SI Appendix, Fig. S1). (B) Overlaid flow cytometry dot plots of CD1b-endo tetramer+ (blue) and CD1b-endo tetramer− (red) γδ T cell populations from eight donors (gated as shown in SI Appendix, Fig. S1). Percentages of Vδ1− and Vδ1+ cells are shown separately for the CD1b tetramer+ and CD1b tetramer− populations. (C) Flow cytometric dot plots of line TS1+ (gated on CD3+ cells) and line TS1−. Four CD1b tetramer-positive populations can be observed: BC14.1, BC14.2, BC14.3, and BC14.4. (D) Paired TCR sequences of four human CD1b-endo-recognizing γδ TCRs determined using a multiplex PCR and Sanger sequencing-based approach.
To discover TCRs and enable functional studies of T cell response, we used flow cytometry to sort CD1b tetramer+ T cells from PBMCs. After two rounds of T cell sorting and expansion, we obtained an oligoclonal T cell line from a blood bank–derived buffy coat (BC14) in which ∼3% of the cells stained brightly with CD1b tetramer (SI Appendix, Fig. S2A). Further sorting based on the anti-Vδ1 antibody (clone TS1) generated two separate cell lines: line TS1+ (anti-Vδ1+) and line TS1−, which was CD3+ but anti-TCR αβ−, anti-Vδ1−, and anti-Vδ2− (SI Appendix, Fig. S2B). Staining of line TS1+ with CD1b tetramer and anti-Vδ1 antibody showed three distinct tetramer positive subpopulations, which we named BC14.1, BC14.2, and BC14.3, while line TS1− contained one tetramer-positive population called BC14.4 (Fig. 1C). To fully characterize the CD1b-binding γδ TCR sequences, we bulk sorted each of the four populations and used a multiplex PCR approach (Fig. 1D), which resulted in one TCR γ chain and one TCR δ chain nucleotide sequence for each population, solving four paired clonal TCRs. All CD1b tetramer-positive populations, including the BC14.4 TCR, which did not stain with the TS1 antibody, expressed Vδ1 TCRs but differed in their CDR3δ regions. The TCR γ chains were all different and used the Vγ4, Vγ8, Vγ9, or Vγ3 segments. Thus, we were able to detect CD1b tetramer-binding γδ T cells ex vivo and derive paired TCR γ and δ chain sequences of four Vδ1+ γδ T cell clones.
Primary CD1b-Endo-Recognizing γδ T Cells Show Autoreactivity.
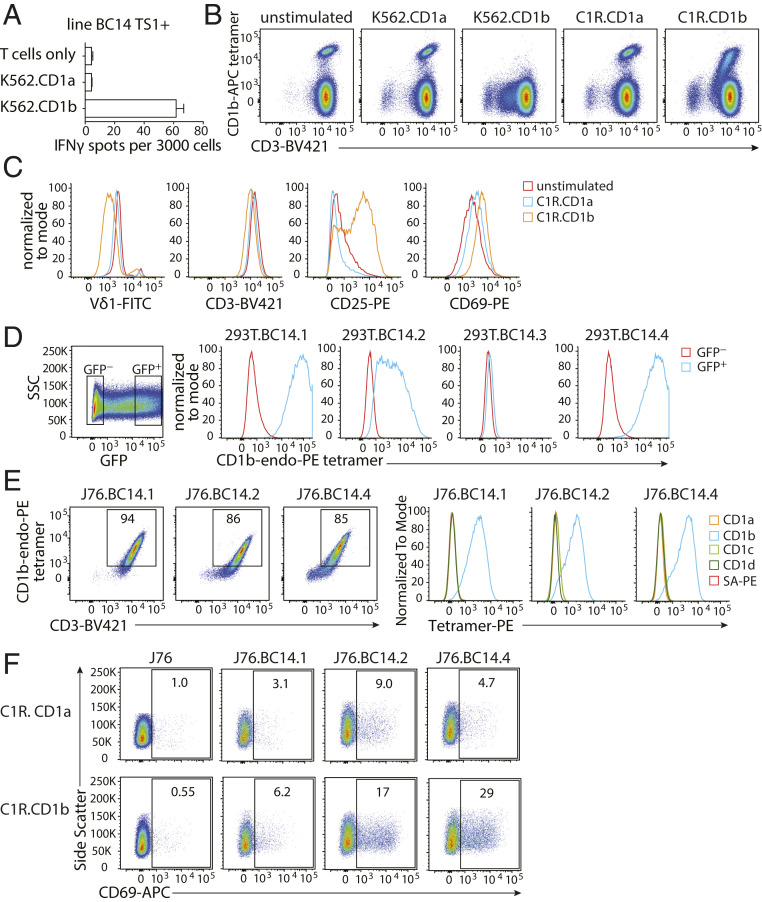
The usual ligand for tetramers of antigen-presenting molecules is the TCR. However, because γδ T cells that are functionally reactive to CD1b were unknown, we asked whether T cells sorted on CD1b tetramers could functionally recognize CD1b. Line TS1+ γδ T cells produced interferon γ (IFNγ) when stimulated with the myelogenous leukemia cell line K562 transfected with CD1b (K562.CD1b) in the absence of an added foreign lipid but not when stimulated with K562.CD1a (Fig. 2A). In addition, we tested down-regulation of the TCR and up-regulation of activation markers as an outcome of TCR activation. Stimulation of line TS1+ with K562.CD1b cells but not K562.CD1a cells induced complete down-regulation of the TCR, as seen by loss of tetramer binding and the appearance of a CD3-low population, while stimulation with C1R.CD1b showed a partial response (Fig. 2B). Tetramer+ cells cocultured with C1R.CD1b showed TCR down-regulation, as evidenced by the lower intensity of staining for Vδ1 and CD3. T cell activation was apparent based on CD25 and CD69 up-regulation (Fig. 2C). Thus, primary CD1b tetramer+ T cells show functional responses to CD1b+ cells, which is consistent with direct TCR recognition of CD1b.
Fig. 2.
Tetramer-positive γδ T cells are autoreactive to CD1b. (A) IFN-γ ELISPOT of line TS1+ stimulated with K562.CD1a or K562.CD1b cells in the absence of exogenously added antigen. Error bars represent the SEM of triplicate wells. One representative experiment of three is shown. (B) Flow cytometry dot plots of line TS1+ after coculture with CD1a- or CD1b-expressing K562 or C1R cells. One representative experiment of three is shown. (C) Flow cytometry histograms show changes in cell surface levels of Vδ1, CD3, CD25, and CD69 on CD1b tetramer+CD3+ cells after stimulation by C1R.CD1a or C1R.CD1b cells for 20 h. (D) Flow cytometry histograms show 293T cells transfected with γδ TCRs and CD3 complex proteins stained with CD1b tetramers. The mean fluorescence intensity (MFI) of the CD1b tetramer is shown for GFP-negative and GFP-positive 293T cells. (E) Flow cytometry dot plots and histograms of J76 cells stably transduced with γδ TCRs. (F) Flow cytometry dot plots of J76 cell lines after stimulation by C1R.CD1a or C1R.CD1b cells for 20 h. One representative experiment of three experiments is shown.
γδ TCR Recognition of CD1b.
To formally test if CD1b was activating the γδ T cells by binding to the TCRs we sequenced, we transiently transfected 293T cells with the γδ TCRs and the CD3 complex proteins CD3γ, CD3δ, CD3ζ, and CD3ε and stained them with CD1b-endo tetramers. Each transfection showed TCR expression on the cell surface as measured by antibodies against both CD3 and the γδ TCR (SI Appendix, Fig. S3A). Staining of the transfected 293T cells confirmed binding of CD1b-endo tetramers by three of the four TCRs (Fig. 2D). BC14.3 did not bind the CD1b-endo tetramer, which could mean that the TCR was of low affinity.
Next, we stably transduced Jurkat 76 (J76) cells (a TCR-negative T cell line, ref. 33) with the confirmed CD1b tetramer-binding TCRs to generate stable cell lines (called J76.BC14.1, J76.BC14.2, and J76.BC14.4) for further tetramer assays (SI Appendix, Fig. S3B). J76 cells normally express CD1b, so to prevent self-activation upon transduction with CD1b-reactive TCRs, we first deleted CD1b (SI Appendix, Fig. S3C). The resulting CD1b-negative, TCR-transduced lines J76.BC14.1, J76.BC14.2, and J76.BC14.4 displayed comparable staining intensity of the CD1b-endo tetramer (Fig. 2 E, Left) but did not stain with CD1a-, CD1c-, or CD1d-endo tetramers (Fig. 2 E, Right). When cocultured with C1R.CD1b cells, a fraction of each line up-regulated the activation marker CD69 (Fig. 2F), consistent with TCR-dependent signaling observed for the primary γδ T cells. Even though BC14.1 stained intensely with CD1b tetramers, it was the weakest responder in terms of CD69 up-regulation in three independent experiments. Altogether, these data prove that we have identified three Vδ1 γδ TCRs that bind CD1b directly and can initiate signaling.
Key CD1b Residues for γδ TCR Binding.
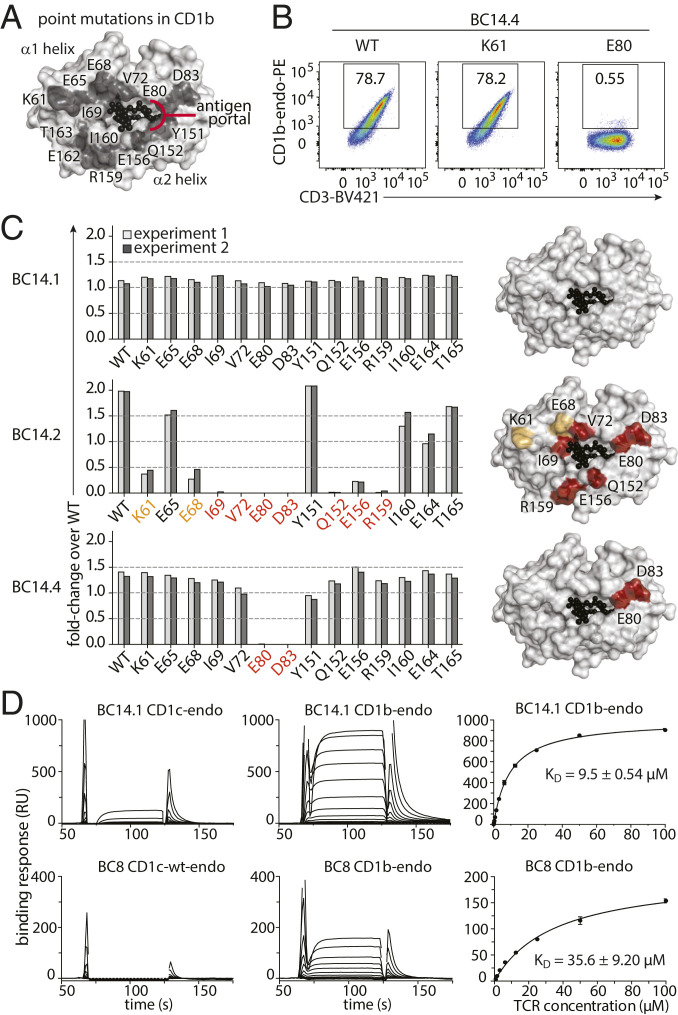
To begin to consider how γδ TCRs might bind CD1b, we first considered where four known CD1b-specific αβ TCRs map onto the antigen display platform formed by the α1 and α2 helices of CD1b. When bound to MHC proteins, peptide antigens span nearly the entire width of the display platform. In contrast, CD1b has a portal where lipid antigens emerge, whereas the remainder of the CD1b antigen display platform is a closed structure known as the roof (28–31). To determine which residues that form the CD1b antigen display platform are important for binding of γδ T cells, based on the CD1b crystal structure, we generated 14 mutant CD1b-endo tetramers and tested staining of J76.BC14.1, J76.BC14.2, and J76.BC14.4 clones. Alanine-substituted residues were surface-exposed residues in the α1 helix (K61A, E65A, E68A, I69A, V72A, E80A, D83A) or the α2 helix (Y151A, Q152A, E156A, R159A, I160A, E164A, T165A) and, as such, were considered not to impact lipid antigen display (Fig. 3A). As a reference value we used unmutated wild-type (WT) CD1b made in the same system. As a positive control we used WT CD1b from the NIH tetramer facility, which allowed us to determine a fraction of positively staining cells for each tetramer (Fig. 3B).
Fig. 3.
Differential docking modes of CD1b-restricted γδ TCRs. (A) Location of tested mutations in CD1b tetramers. (B and C) J76.BC14.1, J76.BC14.2, and J76.BC14.4 were tested for binding of CD1b-endo tetramers with the indicated point mutations by flow cytometry (representative dot plots in B; complete data set in SI Appendix, Fig. S4) summarized in bar graphs showing fold change in staining compared to wild-type CD1b-endo produced using the same platform (C, Left). Representation of the CD1b surface (white), with residues that affect tetramer binding when mutated to alanine substantially (red, greater than fourfold reduction) or moderately (orange, twofold to fourfold reduction) (C, Right). Light and dark gray bars show two independent experiments per mutant tetramer. WT: wild-type CD1b tetramer from the NIH tetramer facility. (D) Representative SPR sensorgrams (Left and Middle) and derived steady-state affinity measurements (Right) of the BC14.1 TCR (Top) and control TCR BC8 (Bottom) against endogenously loaded CD1b and CD1c. Two independent SPR experiments were conducted, each containing experimental replicates with derived steady-state (KD) values and error bars denoting SD.
Given the nonpolymorphic nature of CD1b and the conserved use of Vδ1, we hypothesized the existence of a conserved mode of CD1b recognition. However, staining patterns of CD1b tetramer-positive cells across all mutants were clearly different for each of the three Vδ1 TCRs, and the results were highly reproducible in two independent experiments (Fig. 3C and SI Appendix, Fig. S4). For BC14.1 none of the tested mutant CD1b tetramers affected tetramer binding. For BC14.4 strong effects of E80A and D83A mutations on tetramer binding were observed, but not of other residues. Binding to BC14.2 was reduced by mutations K61A, E68A, I69A, V72A, E80A, D83A, Q152A, E156A, and R159A. Every mutant CD1b tetramer stained at least one of the Jurkat 76 clones, demonstrating that none of the used mutant tetramers failed to fold or was otherwise functionally defective. Therefore, the differing patterns were encoded in the TCRs. These studies demonstrate that the footprints of these three TCRs on CD1b are markedly different from each other, ranging from an extensive area of CD1b that interacts with the BC14.2 TCR to no effect of any of the 14 mutants spread throughout the CD1b antigen display platform on binding to the BC14.1. The latter observation suggests a docking mode that is very different from known CD1b-αβ TCR interactions, which bind the antigen display platform of CD1b.
Affinity of the BC14.1 TCR toward CD1 Molecules.
In surface plasmon resonance (SPR) assays, CD1b and CD1c loaded with endogenous lipids were coupled to the sensor surface, with BC14.1 TCR used as an analyte (Fig. 3D). The steady state dissociation constant (KD) of the BC14.1 TCR bound to CD1b was ∼9.5KD ± 0.54, which represents a high-affinity interaction, and exhibited no cross-reactivity to CD1c. Notably, the BC14.1 TCR bound CD1b with a higher affinity than the BC8 TCR and represents a slightly higher affinity interaction than the low to middle micromolar interactions obtained for other MHC-like molecules such as CD1c (18), CD1d (20), and MR1 (15).
γδ TCR Specificity for Lipids Presented by CD1b.
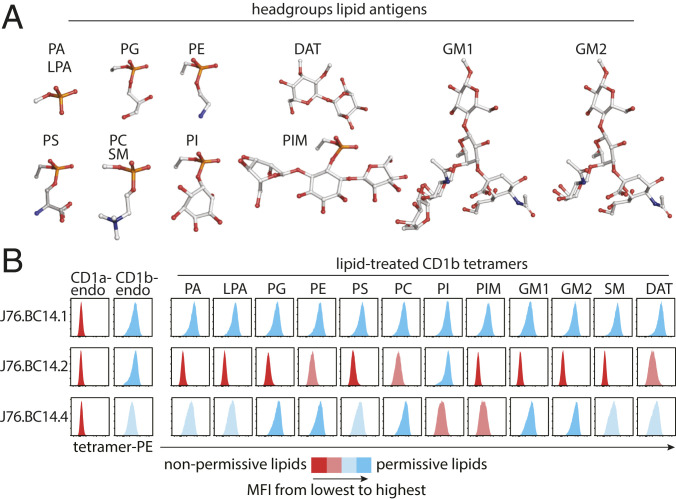
Antigens insert their alkyl chains into CD1b to position their hydrophilic head groups on the outside of CD1b for TCR contact. Some CD1-autoreactive TCRs bind on the closed roof of CD1 proteins and so are not dependent on the bound lipid (34, 35). However, large head groups on “nonpermissive ligands” tend to block these CD1–TCR interactions, whereas smaller head groups on “permissive ligands” permit CD1–TCR interactions. The BC14.2 TCR footprint on the CD1b antigen display platform surrounds the antigen portal through which antigens normally protrude to the surface of CD1b (Fig. 3C). Therefore, binding of this TCR is predicted to be affected by the type of lipid carried in the CD1b cleft. To test this, we determined the pattern of lipid reactivity of γδ TCRs by staining them with a panel of CD1b tetramers treated with common self-glycerophospholipids or sphingolipids or the bacterial lipids phosphatidylinositol dimannoside (PIM2) and diacyltrehalose (DAT). This panel included lipids with known CD1b-binding properties whose head groups ranged from small to large, containing one, two, or more carbohydrates (Fig. 4A) (29). Staining of cell lines expressing the HD1B and PG90 TCRs, two CD1b-specific αβ TCRs with known lipid reactivity patterns (27, 29, 30), was included as a positive control for tetramer loading with lysophosphatidic acid, phosphatidylglycerol (PG), and phosphatidylserine (PS) (SI Appendix, Fig. S5). Both J76.BC14.1 and J76.BC14.4 recognized CD1b treated with a broad range of lipids, including phospholipids and sphingolipids, while J76.BC14.2 strongly recognized phosphatidylinositol (PI) and, to a lesser extent, phosphatidylethanolamine (PE), phosphatidylcholine (PC), and DAT (Fig. 4B). Thus, the BC14.2 TCR is an antigen-specific TCR. BC14.4 is influenced by the loaded lipid but has much less stringent requirements for the lipid that is present in the CD1b groove, and BC14.1 shows no detectable signs of sensitivity to lipid ligands in CD1b. Unlike all other CD1-reactive TCRs that we know of, BC14.1 recognition is not influenced by 14 CD1b antigen display platform residues, nor does the lipid that is bound by CD1b contribute to the specificity of the TCR, suggesting docking at the extreme end or the sides of the α1 and α2 helices or the α3 domain of CD1b or β2M.
Fig. 4.
Antigen specificity of CD1b-restricted γδ TCRs. (A) Head groups of lipids used to treat CD1b monomers are shown as ball-and-stick plots. Red: oxygen; orange: phosphorus; white: carbon; blue: nitrogen. (B) Flow cytometry histograms of the indicated J76 lines stained with a panel of 13 CD1b tetramers and one CD1a tetramer carrying endogenous (endo) lipids or treated with the indicated lipid are shown. One representative experiment of two is shown.
Dual Specificity of the BC14.1 TCR.
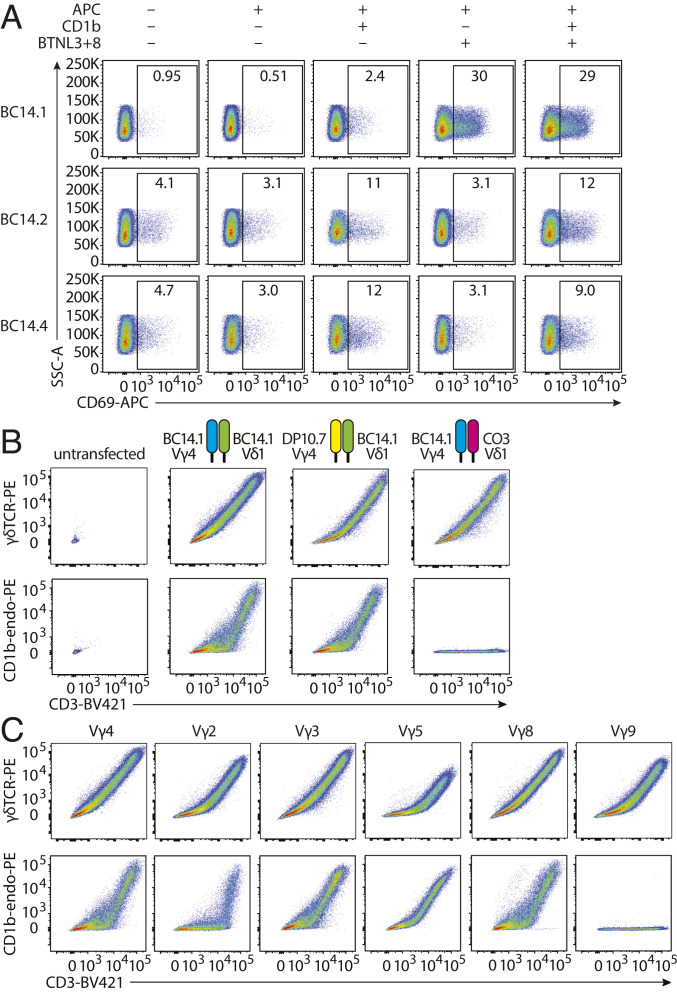
Even though we have firmly established the specificity of the BC14.1 TCR for CD1b, other Vγ4Vδ1 cells have been reported to respond to cells cotransduced with human BTNL3 and BTNL8 (8). The hypervariable region 4 (HV4) domain of Vγ4 is essential for interaction with the BTNL3 and BTNL8 heterodimer (BTNL3+8) and may act independently of the clonally rearranged parts of the TCR, such as the CDR3γ and the CDR3δ (9, 10). To investigate whether this BTNL responsiveness also applies to the BC14.1 Vγ4Vδ1 TCR, we cocultured the TCR-transduced Jurkat cell lines with K562 cells expressing BTNL3+8 (K562.EV or K562.CD1b with or without BTNL3+8, SI Appendix, Fig. S6). We observed only minor CD69 up-regulation on J76.BC14.1 cells when stimulated with K562.CD1b cells (Fig. 5A), as expected based on results with C1R.CD1b stimulation (Fig. 2F). However, when stimulated with K562.EV.BTNL3+8, J67.BC14.1 cells strongly up-regulated CD69 (Fig. 5A). The percentage of CD69 positive J76.BC14.1 cells after stimulation with K562.CD1b.BTNL3+8 was similar to the expression observed after stimulation with the K562.EV.BTNL3+8-stimulated cells. As expected based on their lack of expression of a Vγ4 chain, both J76.BC14.2 and J76.BC14.4 cell lines showed CD69 up-regulation when stimulated with K562 cells expressing CD1b, regardless of the coexpression of BTNL3+8 (Fig. 5A). Melandri et al. recently proposed dual recognition of CD1c and butyrophilins or EPCR and butyrophilins by human Vγ4+ TCRs (9), and these studies confirm this model and extend it to CD1b.
Fig. 5.
Binding of the BTNL3-BTNL8 heterodimer and CD1b by the BC14.1 TCR. (A) J76 cell lines were stimulated with K562.EV, K562.CD1b, K562.EV.BTNL3+8, or K562.CD1b.BTNL3+8 cells and analyzed for CD69 expression by flow cytometry. One representative experiment of two is shown. (B) Flow cytometry was performed on untransfected 293T cells and 293T cells transfected with the WT BC14.1 TCR or a modified version of the BC14.1 TCR consisting of the DP10.7 Vγ4 combined with the BC14.1 Vδ1 or the BC14.1 Vγ4 combined with the CO3 Vδ1. (C) Flow cytometry was performed on 293T cells transfected with either the WT BC14.1 TCR or the BC14.1 Vδ1 chain paired with a different functional Vγ chain from published CD1c or CD1d-specific γδ TCRs (SI Appendix, Table S1).
Specificity for CD1b Is Mediated by the δ Chain.
No additional effect of CD1b was observed when J76.BC14.1 was stimulated with K562 cells expressing both CD1b and BTNL3+8. This observation raised the question of whether BTNL3+8 creates maximal binding such that effects of CD1b are present but not additive or whether BTNL3+8 and CD1b compete for binding to the TCR. A first step toward answering this would be to determine which parts of the BC14.1 TCR interact with CD1b. Whereas the genome-encoded part of Vγ4 was previously demonstrated to interact with BTNL3-BTNL8, CDR3 regions almost always contribute to recognition of antigenic targets. To test whether the CDR3γ of BC14.1 was necessary for CD1b binding, we transfected 293T cells with a modified version of the BC14.1 TCR in which the native Vγ4 chain was replaced by a Vγ4 chain from the CD1d-specific γδ TCR DP10.7 (17). This TCR has a different CDR3γ, while the BC14.1 Vδ1 chain remained unchanged. Expression of modified TCRs by the 293T cells was confirmed by anti-CD3 and anti-γδ TCR staining (Fig. 5 B, Upper). We observed that when stained with CD1b-endo tetramers, the TCR using BC14.1 Vδ1 combined with a different Vγ4 chain was still able to bind the tetramer at a level comparable to the wild-type receptor, whereas nontransfected cells did not bind the tetramer (Fig. 5 B, Lower, first three plots). Thus, the CDR3γ of BC14.1 is not required for recognition of CD1b.
Replacement of the Vγ4 chain of BC14.1 with the DP10.7 TCR Vγ4 chain changes the CDR3γ but not the CDR1γ and CDR2γ loops. To assess whether the CDR1γ or CDR2γ loops or other genome-encoded parts of Vγ4 contribute to CD1b binding, we transfected and stained 293T cells with additional modified BC14.1 TCRs. These TCRs consist of the BC14.1 Vδ1 chain combined with representatives of all functional human γ chains: Vγ2, Vγ3, Vγ5, Vγ8, and Vγ9, which we selected from CD1c- and CD1d-specific γδ T cells (SI Appendix, Table S1). These modified TCRs expressed well on the surface of 293T cells (Fig. 5C). Replacing the Vγ4 chain with Vγ2, Vγ3, Vγ5, and Vγ8 did not cause loss of CD1b tetramer binding. However, replacement with a Vγ9 chain completely abrogated binding of CD1b by the TCR (Fig. 5C). Based on sequence comparisons, TRGV9 is quite distinct from the other Vγ segments, so loss of CD1b binding could not be assigned to the CDR1γ and CDR2γ loops or any other specific part of the sequence (SI Appendix, Fig. S7). When comparing TRGV2, TRGV3, TRGV5, and TRGV8, the high variability in CDR1γ and CDR2γ loop sequences contrasts with their uniform support of CD1b binding and suggests that CDR1γ and CDR2γ are not determinative of CD1b binding. However, the CDR1γ and CDR2γ loops of these TRGV genes do share some residues that are not present in TRGV9, including amino acids Y at the end of CDR1γ and YD at the beginning of CDR2γ. Therefore, involvement of the CDR1γ and CDR2γ loops cannot be completely ruled out. However, because the Vγ4 chain does not seem to play a major role in binding of CD1b, it is extremely unlikely that the binding sites of the BTNL3+BTNL8 heterodimer and CD1b overlap.
Our results also suggest that the CD1b specificity of BC14.1 is mostly determined by its Vδ1 chain. To test whether the CDR3δ or other parts of the δ chain direct binding of CD1b, we transfected 293T cells with a modified version of BC14.1 where we preserved the γ chain and combined it with a Vδ1 chain with a different CDR3δ, from the T cell line CO3. The cells expressing the altered TCR did not bind the CD1b-endo tetramer but were expressed at the surface (Fig. 5B). Thus, CD1b specificity of BC14.1 is dominated by the CDR3δ. Other parts of the TCR may contribute to CD1b binding but are not sufficient for binding without the correct CDR3δ. This situation is reminiscent of the G8 γδ TCR where CDR3δ dominates the recognition of the murine T22 molecule (36).
Discussion
Here we describe recognition of the antigen-presenting molecule CD1b by human γδ T cells. CD1b-specific γδ T cells were found at low frequency in the blood of all donors tested and were activated by stimulation with CD1b. CD1b tetramer staining of TCR-transduced cells formally ruled in γδ TCRs as CD1b ligands. These findings contribute to the understanding that members of the human CD1 protein family function as natural targets for Vδ1 T cells. Recognition of CD1b likely has functional parallels with recognition of CD1c and CD1d. However, unlike CD1c and CD1d, which are constitutively expressed in the periphery, CD1b has a unique pattern of activation-induced expression on myeloid cells (22, 26, 37). Also, CD1b has a larger antigen-binding cleft that is suitable for lipid antigens that cannot fit within CD1c and CD1d.
Further, the particular patterns of TCR-mediated recognition of CD1b and BTNLs shown here provide insights into how γδ TCRs approach and recognize their targets. The expansion of diverse, antigen-specific γδ T cell clones upon exposure to pathogens like cytomegalovirus, Epstein–Barr virus, and herpes simplex virus is a prime example of how γδ T cells take part in adaptive immune responses (38–43). On the other hand, semi-invariant γδ T cell populations that are present in large numbers in all individuals of a species, such as dendritic epidermal γδ T cells in mice and the Vγ9Vδ2 T cell population in humans, seem to play a role in innate-like tissue surveillance. These semi-invariant populations can exert rapid responses without the need for clonal expansion and differentiation (44, 45). In discussing three CD1b-reactive γδ TCRs, we emphasize the question of whether their targets are constitutively expressed molecules, consistent with a role in homeostatic survival; molecules regulated by stress or inflammation, consistent with a role in innate immunity; or complexes of antigen-presenting molecules and lipids, consistent with a role in adaptive immunity.
All known αβ T cells (28–31) dock on the CD1b antigen display platform, where they bind near or over the antigen portal. Because the Vδ1 frequency was increased among CD1b tetramer+ cells and CD1b is nonpolymorphic, Vδ1 might be part of a TCR pattern that binds CD1b with a conserved docking mode. Unexpectedly, we found a unique footprint for each of the three tested γδ TCRs. BC14.2 and BC14.4 TCR binding is influenced by treating CD1b with lipid ligands and is lost after alanine substitutions close to the antigen portal of CD1b, indicating that lipid sensing by these two Vδ1 TCRs likely occurs through TCR contact with lipid as it protrudes from the antigen portal. BC14.2 TCR binding is lost after mutation of any of nine residues located on either side of the antigen exit portal. In contrast the BC14.4 TCR requires E80 and E83, which are located on the right side of the portal. These findings suggest a footprint on the CD1b antigen display platform in the vicinity of the antigen portal. Thus, these TCRs use different approaches from CD1b, but both recognize antigens, supporting the idea that γδ TCRs recognize distinct combinations of CD1b and lipid.
Binding by the BC14.1 TCR is different, as we could not establish a footprint on the antigen display platform of CD1b with the mutant tetramers tested. After our studies were completed, a new mode of ligand binding by γδ TCRs was published, challenging the general view on how TCRs bind antigen-presenting molecules. Instead of binding the membrane-distal surface, the antigen display platform, of MR1, a γδ TCR was shown to bind the platform “down under,” contacting the α3 domain (15). It is possible that TCR recognition of antigen-presenting molecules at locations other than the top surface of the antigen display platform is more common than previously thought, and the CD1b-BC14.1 interaction might be the second example. Attempts at crystallization have not succeeded, so structural proof is lacking. It is notable that for the 12 lipid ligands tested, nearly all of which affect other TCRs, none affected BC14.1 binding to CD1b. This outcome independently suggests TCR binding at a site distant from the antigen portal, which is readily explained by the proposed down-under binding mechanism. Whatever the point of contact, the lipid-independent recognition of an immune-inducible protein like CD1b is consistent with the TCR behaving as a receptor to amplify innate inflammation, like members of the immunoglobulin-like transcript (ILT) family of receptors recognizing CD1c and CD1d (46–49).
The discovery of γδ TCR recognition of the constitutively expressed BTNL family of proteins provides a candidate explanation for the selection and maintenance of γδ TCRs (9, 10). The human TRGV4 segment in Vγ4+ TCRs mediates the recognition heterodimer of BTNL3 and BTNL8. More specifically, the evolutionally conserved hypervariable region 4 (HV4) motif of the Vγ4 chain is essential (9, 10). Recognition of BTNL3+8 heterodimers by Vγ4+ T cells is considered innate since the HV4 region is germ line encoded, and BTNLs do not present antigens. Inspired by this, we wanted to know if BC14.1, which also uses TRGV4, recognizes BTNL3+8. In addition to CD1b, Jurkat cells expressing the Vγ4+ TCR BC14.1 also recognized BTNL3+8. In fact, BTNL3+8 was a stronger stimulus for these cells than CD1b. Melandri et al. proposed that the γ chain HV4 mediates recognition of BTNLs and other parts of the TCR, including the rearranged CDR3s, contact CD1d-lipid or CD1c-lipid, and interpreted these interactions as innate and adaptive recognition modes by one TCR, respectively (9). Our findings show that recognition of CD1b is mediated by an adaptive, rearranged part of the BC14.1 TCR, but the antigen-presenting capacity of CD1b is ignored and redundant. Binding of CD1b by BC14.1 is dominated by the Vδ1 chain, leaving the Vγ4 accessible for binding of BTNL3+8. Our data raise the possibility that simultaneous engagement of CD1b and BTNL by γδ T cells might be possible.
Thus, in addition to being a bona fide antigen-presenting molecule, the mere presence of CD1b, regardless of the bound lipid, can stimulate certain γδ T cells. This strongly supports a physiologic role for regulation of surface expression of CD1b in immune responses. CD1b is up-regulated during inflammation-induced monocyte differentiation, via GM-CSF, IL-4, and IL-1β (23–25), which in turn can promote the activation of γδ T cells, such as BC14.1, that take part in innate stress surveillance.
Materials and Methods
Recombinant Proteins and Tetramers.
For lipid loading, WT CD1b monomers were obtained from the NIH tetramer facility. In a 10 mm wide glass tube, 16 μg of dry lipid were sonicated at 37 °C for 1 h in 45 μL of 0.5% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate in 50 mM sodium citrate buffer (pH 4.5) for DAT or 45 μL of 0.5% CHAPS 50 mM sodium citrate buffer (pH 7.4) for other lipids. Subsequently, CD1b monomers (10 μg) were added to the tubes and incubated overnight at 37 °C. The next day the solution for DAT-loaded monomers was neutralized to pH 7.4 by adding 5 μL 1M Tris (pH 8). Mutant monomers were generated as described and used without treatment with exogenous lipids (28). CD1b was expressed in HEK293S GnTI cells (from ATCC). Alanine mutants of the CD1b molecules were generated by site-directed mutagenesis and expressed in mammalian cells under equivalent conditions used to produce the WT CD1b molecule. Monomers were tetramerized using streptavidin-APC (Molecular Probes) or streptavidin-PE (Invitrogen). For WT CD1b-endo, mutant CD1b-endo, CD1a-endo, CD1c-endo, and CD1d-endo tetramers the monomers were diluted in Tris-buffered saline (0.2 mg/mL) and tetramerized.
Lipids.
PC (#850475; C18:1/C16:0), sphingomyelin (#860584; C18:1/C16:0), PI (#840042; mixture from bovine liver with a range of fatty acids), PS (#840032; mixture from porcine brain with a range of fatty acids), phosphatidic acid (PA, #840857; C18:1/C16:0), lyso-PA (#857130; C18:1), PE (#850757; C18:1/C16:0), and PG (#840503; C18:1/C18:0) were purchased from Avanti polar lipids. Gangliosides GM1 (G7641; mixture from bovine brain with a range of fatty acids) and GM2 (G8397; mixture from bovine brain with a range of fatty acids) were purchased from Sigma. PIM2 and DAT, previously described as DAT2a, were provided by the Bill and Melinda Gates Foundation lipid bank.
Staining Protocol.
Human PBMCs, T cell lines, and Jurkat lines were stained with tetramers at 2 μg/mL in phosphate-buffered saline (PBS) containing 1% bovine serum albumin and 0.01% sodium azide. Cells and tetramers were incubated for 10 min at room temperature, followed by the addition of antibodies and another incubation for 10 min at room temperature, followed by 20 min at 4 °C. Cells were analyzed using the BD LSRFortessa flow cytometer and FlowJo software. Antibodies that were used are as follows: CD3-BV421 (UCHT1; Biolegend), CD3-FITC (SK7; BD Bioscience), γδTCR-PE (B1; Biolegend), αβTCR-APC (IP26; Biolegend), Vδ1-FITC (TS-1; Invitrogen), Vδ2-PE (B6; Biolegend), CD25-PE (BC96; Biolegend), CD69-PE (FN50; Biolegend), CD69-APC (FN50; Biolegend), CD1b-PE (SN13/K5-1B8; Ancell), DYKDDDDK Tag-BV421 (Flag tag, L5; Biolegend), and HA-APC (16B12; Biolegend).
T Cells and T Cell Lines.
PBMCs were obtained from leukoreduction collars provided by the Brigham and Women’s Hospital Specimen Bank, as approved by the Partners Healthcare Institutional Review Board, and cultured overnight in medium containing 0.2 ng/mL IL-15 before staining. γδ T cells were enriched using the untouched TCRγ/δ + T cell isolation kit (Miltenyi Biotec). Cells were analyzed using the BD LSRFortessa flow cytometer and FlowJo software. For generation of T cell lines, total PBMCs or PBMC-derived T cells were stained with CD1b-DAT tetramer and anti-CD3. PBMCs were sorted for double positive staining of CD3 and tetramer. Expansion of sorted cells was performed by plating cells at 100 to 700 cells/well in round-bottom 96-well plates containing 2.5 × 105 irradiated allogeneic PBMCs, 5 × 104 irradiated Epstein–Barr virus transformed B cells, and 30 ng/mL anti-CD3 antibody (clone OKT3) per plate. The next day human IL-2 or a mix of human IL-2, IL-7, and IL-15 was added to the wells. After 2 wk, the sorting and expansion procedure was repeated as needed.
γδ TCR Sequencing and Transient Transfection.
TCR sequences were determined by isolating RNA from bulk sorted γδ T cell populations using the RNeasy kit (QIAGEN), followed by cDNA synthesis using the QuantiTect Reverse Transcription Kit (QIAGEN). TCRγδ transcripts were amplified using a multiplex approach (50), followed by direct Sanger sequencing of the PCR product. Full-length TCR γ and TCR δ chains separated by self-cleaving 2A peptide were purchased from GENEWIZ and cloned into a (MSCV)-IRES-GFP (pMIG) vector. All plasmids used for transfection were purified using a QIAprep Spin Miniprep Kit (QIAGEN) or NucleoBond Xtra Midi EF kit (Macherey-Nagel). HEK293T cells were cotransfected with pMIG-TCR and pMIG-CD3δγεζ using FuGENE-6 (Promega) (51). Expression of TCR and CD3 and binding of tetramer were analyzed using the BD LSRFortessa flow cytometer and FlowJo software. For replacement of TCR γ chains, full-length TCRγ gene segments were purchased from GENEWIZ and used to replace the γ chain of the BC14.1 TCR in the (MSCV)-IRES-GFP (pMIG) using EcoRI and BspEI.
Transduction of Jurkat 76 Cells with TCR and K562 Cells with BTNL.
pMIG-TCR and pMIG-CD3δγεζ were cotransfected into HEK293T cells in the presence of the retroviral packaging vectors pPAM-E and pVSV-g. Medium was replaced 16 h after transfection. Supernatant containing virus was collected at 48 h and filtered through 0.45 μm nylon mesh, followed by concentration of virus using a Mag4C-LV kit (OZ Biosciences). Concentrated virus was used to stably transduce TCR-deficient Jurkat clone 76 cells (33) to generate the BC14 T cell lines. After 5 d, cells with the highest expression of TCR, GFP, and CD3 were single-cell sorted using the BD FACSAria. K562.EV and K562.CD1b cell lines were transduced with FLAG-BTNL3 and HA-BTNL8 cloned into pCSIGPW (9). Cells with the highest expression of GFP and BTNL3 (anti-Flag staining) or BTNL8 (anti-HA staining) were single-cell sorted and expanded.
Functional T Cell Assays.
For enzyme-linked immune absorbent spot (ELISPOT) assays, cocultures of 2 × 104 APCs and 3 × 103 T cells were incubated for 16 h in a Multiscreen-IP filter plate (96 wells; Millipore) coated according to the manufacturer’s instructions (Mabtech). For coculture assays, T cell lines and Jurkat cell lines were cultured with C1R or K562 cells (1:2) overnight in round-bottom 96-well plates, and their activation status was measured by flow cytometry to detect TCR down-regulation and IL2R and CD69 up-regulation for T cells and CD69 up-regulation for Jurkat cell lines.
CRISPR/Cas9 Editing of Jurkat 76 Cells.
CRISPR/Cas9 ribonuclear protein (RNP) complexes were assembled as previously described (52). In short, 40 μM Cas9 protein (QB3 Mircolabs) was mixed with equal volumes of 40 μM modified single guide (sg)RNA targeting CD1B (Synthego; targets: rs158331053, rs158331042, rs158331007, rs158330955) and incubated at 37 °C for 15 min to form RNP complexes. Jurkat 76 cells were nucleofected with RNPs in an Amaxa 4D nucleofector (SE protocol: CL-120). Cells were immediately transferred to 24-well plates with prewarmed media and cultured. After 7 to 10 d, CD1b expression was assessed by flow cytometry, and cells were single-cell sorted based on the absence of CD1b staining. Editing of clonal cell lines was confirmed by PCR amplifying and sequencing genomic DNA around CD1b, and sequences were analyzed by Tracking of Indels by Decomposition (TIDE) analysis (tide.nki.nl).
Recombinant Proteins and Surface Plasmon Resonance.
Recombinant BC14.1 γδ TCR chimeras were cloned into the pET30a vector utilizing the α and β constant domains, expressed, refolded, and purified from Escherichia coli inclusion bodies. Inclusion bodies were resuspended in 6 M guanidine hydrochloride, 20 mM Tris⋅HCl (pH 8.0), 0.5 mM sodium-ethylenediaminetetraacetic acid, and 1 mM dithiothreitol. The TCR was refolded by rapid dilution into 3 M urea, 100 mM Tris (pH 8.5), 2 mM Na-EDTA, 400 mM l-arginine-HCl, 0.5 mM oxidized glutathione, 5 mM reduced glutathione, and 1mM phenylmethylsulfonyl fluoride. The refolding solution was then dialyzed into 20 mM Tris⋅HCl (pH 8.5). The refolded protein was purified via diethylethanolamine anion exchange and further purified by size exclusion chromatography. Soluble CD1b was expressed using HEK293 S GnTI− (American Type Culture Collection) cultured in Dulbecco's Modified Eagle Medium supplemented with 10% (vol/vol) fetal calf serum. Media containing soluble CD1b was dialyzed extensively into 20 mM Tris⋅HCl (pH 8.0) and 150 mM NaCl and purified by HisTrap nickel nitrilotriacetic acid-affinity chromatography and further purified via size exclusion chromatography (29). SPR experiments were conducted at 25 °C on a BIAcore 3000 instrument in 20 mM Tris (pH 8.0) and 150 mM NaCl supplemented with 0.5% (wt/vol) BSA. CD1 proteins were captured on a streptavidin-coated SA sensor chip (GE Healthcare), where ∼1,500 response units of endogenously loaded CD1b or CD1c were immobilized. Serial dilutions from 100 to 0 µM of BC14.1 TCR and the BC8 TCR control were injected over the immobilized CD1 proteins at a flow rate of 5 µL min−1. All experiments were performed in duplicate with an independent experimental replicate conducted again in duplicate. The sensorgram plots and kinetic parameters were calculated in Prism using a one-site Langmuir binding model.
Supplementary Material
Acknowledgments
This work is supported by the NIH (Grants AI049313, AR048632) and the Australian Research Council (Grant CE140100011). A.C.H. and P.V. were supported by Wellcome Trust Investigator Award 106292/Z/14/Z. J.R. is supported by an Australian Research Council Laureate fellowship. The CD1a and CD1b monomers were made available by the NIH Tetramer Core Facility.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010545117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Groh V.et al., Human lymphocytes bearing T cell receptor γ/δ are phenotypically diverse and evenly distributed throughout the lymphoid system. J. Exp. Med. 169, 1277–1294 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falini B.et al., Distribution of T cells bearing different forms of the T cell receptor γ/δ in normal and pathological human tissues. J. Immunol. 143, 2480–2488 (1989). [PubMed] [Google Scholar]
- 3.Deusch K.et al., A major fraction of human intraepithelial lymphocytes simultaneously expresses the γ/δ T cell receptor, the CD8 accessory molecule and preferentially uses the V δ 1 gene segment. Eur. J. Immunol. 21, 1053–1059 (1991). [DOI] [PubMed] [Google Scholar]
- 4.La Gruta N. L., Gras S., Daley S. R., Thomas P. G., Rossjohn J., Understanding the drivers of MHC restriction of T cell receptors. Nat. Rev. Immunol. 18, 467–478 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Rigau M.et al., Butyrophilin 2A1 is essential for phosphoantigen reactivity by γδ T cells. Science 367, eaay5516 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Vavassori S.et al., Butyrophilin 3A1 binds phosphorylated antigens and stimulates human γδ T cells. Nat. Immunol. 14, 908–916 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Harly C.et al., Key implication of CD277/butyrophilin-3 (BTN3A) in cellular stress sensing by a major human γδ T-cell subset. Blood 120, 2269–2279 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Marco Barros R.et al., Epithelia use butyrophilin-like molecules to shape organ-specific γδ T cell compartments. Cell 167, 203–218.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melandri D.et al., The γδTCR combines innate immunity with adaptive immunity by utilizing spatially distinct regions for agonist selection and antigen responsiveness. Nat. Immunol. 19, 1352–1365 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willcox C. R.et al., Butyrophilin-like 3 directly binds a human Vγ4+ T cell receptor using a modality distinct from clonally-restricted antigen. Immunity 51, 813–825.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willcox B. E., Willcox C. R., γδ TCR ligands: The quest to solve a 500-million-year-old mystery. Nat. Immunol. 20, 121–128 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Zeng X.et al., γδ T cells recognize a microbial encoded B cell antigen to initiate a rapid antigen-specific interleukin-17 response. Immunity 37, 524–534 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marlin R.et al., Sensing of cell stress by human γδ TCR-dependent recognition of annexin A2. Proc. Natl. Acad. Sci. U.S.A. 114, 3163–3168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willcox C. R.et al., Cytomegalovirus and tumor stress surveillance by binding of a human γδ T cell antigen receptor to endothelial protein C receptor. Nat. Immunol. 13, 872–879 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Le Nours J.et al., A class of γδ T cell receptors recognize the underside of the antigen-presenting molecule MR1. Science 366, 1522–1527 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Porcelli S.et al., Recognition of cluster of differentiation 1 antigens by human CD4-CD8-cytolytic T lymphocytes. Nature 341, 447–450 (1989). [DOI] [PubMed] [Google Scholar]
- 17.Bai L.et al., The majority of CD1d-sulfatide-specific T cells in human blood use a semiinvariant Vδ1 TCR. Eur. J. Immunol. 42, 2505–2510 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy S.et al., Molecular analysis of lipid-reactive Vdelta1 gammadelta T cells identified by CD1c tetramers. J. Immunol. 196, 1933–1942 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luoma A. M.et al., Crystal structure of Vδ1 T cell receptor in complex with CD1d-sulfatide shows MHC-like recognition of a self-lipid by human γδ T cells. Immunity 39, 1032–1042 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uldrich A. P.et al., CD1d-lipid antigen recognition by the γδ TCR. Nat. Immunol. 14, 1137–1145 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Spada F. M.et al., Self-recognition of CD1 by γ/δ T cells: Implications for innate immunity. J. Exp. Med. 191, 937–948 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dougan S. K., Kaser A., Blumberg R. S., CD1 expression on antigen-presenting cells. Curr. Top. Microbiol. Immunol. 314, 113–141 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Yakimchuk K.et al., Borrelia burgdorferi infection regulates CD1 expression in human cells and tissues via IL1-β. Eur. J. Immunol. 41, 694–705 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasinrerk W., Baumruker T., Majdic O., Knapp W., Stockinger H., CD1 molecule expression on human monocytes induced by granulocyte-macrophage colony-stimulating factor. J. Immunol. 150, 579–584 (1993). [PubMed] [Google Scholar]
- 25.Sallusto F., Lanzavecchia A., Pillars Article: Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor α. J. Exp. Med. 1994. 179:1109–1118. J. Immunol. 200, 887–896 (2018). [PubMed] [Google Scholar]
- 26.Porcelli S., Morita C. T., Brenner M. B., CD1b restricts the response of human CD4-8-T lymphocytes to a microbial antigen. Nature 360, 593–597 (1992). [DOI] [PubMed] [Google Scholar]
- 27.Van Rhijn I.et al., Human autoreactive T cells recognize CD1b and phospholipids. Proc. Natl. Acad. Sci. U.S.A. 113, 380–385 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gras S.et al., T cell receptor recognition of CD1b presenting a mycobacterial glycolipid. Nat. Commun. 7, 13257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shahine A.et al., A T-cell receptor escape channel allows broad T-cell response to CD1b and membrane phospholipids. Nat. Commun. 10, 56 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shahine A.et al., A molecular basis of human T cell receptor autoreactivity toward self-phospholipids. Sci. Immunol. 2, eaao1384 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melián A.et al., Molecular recognition of human CD1b antigen complexes: Evidence for a common pattern of interaction with α β TCRs. J. Immunol. 165, 4494–4504 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Chancellor A., Gadola S. D., Mansour S., The versatility of the CD1 lipid antigen presentation pathway. Immunology 154, 196–203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heemskerk M. H. M.et al., Redirection of antileukemic reactivity of peripheral T lymphocytes using gene transfer of minor histocompatibility antigen HA-2-specific T-cell receptor complexes expressing a conserved alpha joining region. Blood 102, 3530–3540 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Wun K. S.et al., T cell autoreactivity directed toward CD1c itself rather than toward carried self lipids. Nat. Immunol. 19, 397–406 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Birkinshaw R. W.et al., αβ T cell antigen receptor recognition of CD1a presenting self lipid ligands. Nat. Immunol. 16, 258–266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams E. J., Chien Y. H., Garcia K. C., Structure of a gammadelta T cell receptor in complex with the nonclassical MHC T22. Science 308, 227–231 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Sieling P. A.et al., CD1 expression by dendritic cells in human leprosy lesions: Correlation with effective host immunity. J. Immunol. 162, 1851–1858 (1999). [PubMed] [Google Scholar]
- 38.Davey M. S.et al., Clonal selection in the human Vδ1 T cell repertoire indicates γδ TCR-dependent adaptive immune surveillance. Nat. Commun. 8, 14760 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Couzi L.et al., Common features of gammadelta T cells and CD8(+) alphabeta T cells responding to human cytomegalovirus infection in kidney transplant recipients. J. Infect. Dis. 200, 1415–1424 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Déchanet J.et al., Implication of gammadelta T cells in the human immune response to cytomegalovirus. J. Clin. Invest. 103, 1437–1449 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farnault L.et al., Clinical evidence implicating gamma-delta T cells in EBV control following cord blood transplantation. Bone Marrow Transplant. 48, 1478–1479 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Fujishima N.et al., Skewed T cell receptor repertoire of Vdelta1(+) gammadelta T lymphocytes after human allogeneic haematopoietic stem cell transplantation and the potential role for Epstein-Barr virus-infected B cells in clonal restriction. Clin. Exp. Immunol. 149, 70–79 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pitard V.et al., Long-term expansion of effector/memory Vdelta2-gammadelta T cells is a specific blood signature of CMV infection. Blood 112, 1317–1324 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davey M. S.et al., The human Vδ2+ T-cell compartment comprises distinct innate-like Vγ9+ and adaptive Vγ9- subsets. Nat. Commun. 9, 1760 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asarnow D. M.et al., Limited diversity of γ δ antigen receptor genes of Thy-1+ dendritic epidermal cells. Cell 55, 837–847 (1988). [DOI] [PubMed] [Google Scholar]
- 46.Willcox B. E., Thomas L. M., Bjorkman P. J., Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nat. Immunol. 4, 913–919 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Shiroishi M.et al., Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc. Natl. Acad. Sci. U.S.A. 100, 8856–8861 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li D.et al., Ig-like transcript 4 inhibits lipid antigen presentation through direct CD1d interaction. J. Immunol. 182, 1033–1040 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li D.et al., A novel role of CD1c in regulating CD1d-mediated NKT cell recognition by competitive binding to Ig-like transcript 4. Int. Immunol. 24, 729–737 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Guo X.-J.et al., Rapid cloning, expression, and functional characterization of paired αβ and γδ T-cell receptor chains from single-cell analysis. Mol. Ther. Methods Clin. Dev. 3, 15054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szymczak A. L.et al., Correction of multi-gene deficiency in vivo using a single “self-cleaving” 2A peptide-based retroviral vector. Nat. Biotechnol. 22, 589–594 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Schumann K.et al., Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. U.S.A. 112, 10437–10442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.