Significance
Control of gene expression and the cell cycle is critical for appropriate cell growth and timely cell division. Failure of the mechanisms regulating these processes can result in proliferative diseases. A molecular complex termed the CDK-activating kinase (CAK) impinges on both of these regulatory networks in human cells and is thus a possible drug target for treatment of cancer. Here, we use cryoelectron microscopy to describe the detailed molecular structure of the human CAK, revealing its architecture and the interactions between its regulatory elements. Additionally, we have obtained the structure of the CAK in complex with a small-molecule inhibitor.
Keywords: cryoelectron microscopy, cyclin-dependent kinase, transcription, cell cycle, phosphorylation
Abstract
The human CDK-activating kinase (CAK), a complex composed of cyclin-dependent kinase (CDK) 7, cyclin H, and MAT1, is a critical regulator of transcription initiation and the cell cycle. It acts by phosphorylating the C-terminal heptapeptide repeat domain of the RNA polymerase II (Pol II) subunit RPB1, which is an important regulatory event in transcription initiation by Pol II, and it phosphorylates the regulatory T-loop of CDKs that control cell cycle progression. Here, we have determined the three-dimensional (3D) structure of the catalytic module of human CAK, revealing the structural basis of its assembly and providing insight into CDK7 activation in this context. The unique third component of the complex, MAT1, substantially extends the interaction interface between CDK7 and cyclin H, explaining its role as a CAK assembly factor, and it forms interactions with the CDK7 T-loop, which may contribute to enhancing CAK activity. We have also determined the structure of the CAK in complex with the covalently bound inhibitor THZ1 in order to provide insight into the binding of inhibitors at the CDK7 active site and to aid in the rational design of therapeutic compounds.
Transcription initiation by eukaryotic RNA polymerase II (Pol II) depends on the formation of a preinitiation complex (PIC) on promoter DNA, which includes general transcription factors, Pol II, and Mediator (1). Like many other cellular processes, transcription and the assembly of the PIC are regulated by phosphorylation events. During PIC assembly, Mediator interacts with the C-terminal heptapeptide repeat domain of RPB1, the largest subunit of Pol II (2), also referred to as the Pol II-CTD. Phosphorylation of this region by the cyclin-dependent kinase (CDK) 7 subunit of transcription initiation factor IIH (TFIIH) disrupts its interactions with Mediator, allowing initiating Pol II to break free from the PIC and clear the promoter (3). Pol II-CTD phosphorylation and dephosphorylation are thus key events that regulate transcription of eukaryotic protein-coding genes and have been implicated in the formation of phase-separated transcriptional subcompartments (4, 5). This process is not only important for normal transcription, but also contributes to aberrantly elevated transcription levels in cancer cells. Consequently, transcription-related CDKs have been identified as promising drug targets (6), and inhibition of CDK7 by compounds such as THZ1 or SY-1365 has been shown to selectively kill cancer cells that require elevated levels of transcription (7, 8).
In addition to its conserved role in transcription, human CDK7 phosphorylates CDKs that control the cell cycle, a function that is not conserved in yeast. The activity of cell cycle CDKs is generally controlled by association with partner cyclins (9). Cyclin binding provides the bulk of CDK stimulation (10) by inducing conformational changes in the CDK that lead to extension of a regulatory loop near the active site, known as the T-loop, and to the proper arrangement of the active site for phosphoryl transfer (11). However, in addition to cyclin binding, full activation of cell cycle CDKs requires phosphorylation of the T-loop (9, 12). In animal cells, these activating phosphorylations are carried out by CDK7 (13, 14), itself a cyclin-dependent kinase whose activity depends on cyclin H (14).
In human and other metazoan cells, regulation of transcription initiation by phosphorylation of the Pol II-CTD and phosphorylation of cell cycle CDKs thus depend on a single kinase, CDK7, which functions in a complex termed the CDK-activating kinase (CAK) (14). In addition to CDK7 and cyclin H, human CAK comprises a third subunit, MAT1 (15–18). CDK7 and cyclin H form a canonical CDK-cyclin pair, while MAT1 serves as a CAK assembly factor (15–17), increases CAK activity toward target CDKs (16, 19), and tethers the CAK to the core of TFIIH when this 10-subunit complex carries out its function in transcription initiation (19–21). When TFIIH functions in DNA repair, MAT1 is dislodged from its binding sites on the TFIIH core complex, releasing the 3-subunit CAK module (22, 23). The functional unit of the human CAK complex is thus the CDK7-cyclin H-MAT1 trimer (16, 18).
While X-ray crystal structures of CDK7 and of cyclin H have been determined (24–26), structural information on the entire three-subunit CAK, which is critical for a mechanistic understanding of the role of MAT1 in CAK assembly and the regulation of its activity, has remained elusive. Here, we present cryoelectron microscopy (cryo-EM) structures of human CAK bound to either ATPγS or the inhibitor THZ1, thereby revealing the architecture of the human CAK and the structural basis for its inhibition by covalent inhibitors. Our structure of the CAK also completes the high-resolution structural analysis of human TFIIH and shows how MAT1 interlinks CDK7, XPB, and XPD, the three major functional centers in TFIIH.
Results and Discussion
Structure Determination of the Human CAK.
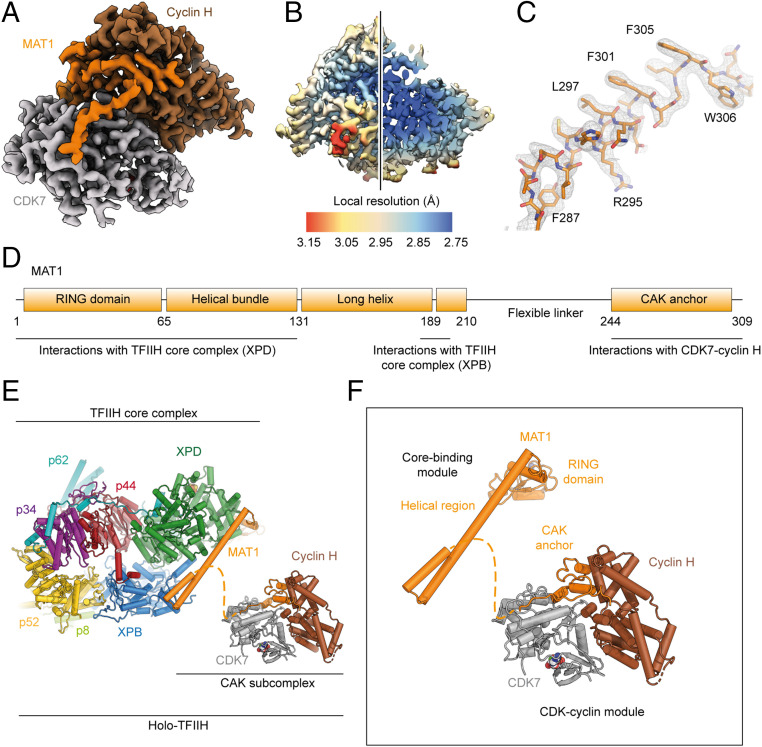
To determine the structure of the human CAK, we recombinantly coexpressed CDK7, cyclin H, and MAT1 in insect cells, purified the resulting complex by affinity and size-exclusion chromatography, and collected cryo-EM data of frozen-hydrated CAK bound to the nucleotide analog ATPγS (SI Appendix, Fig. S1) using a 200-kV instrument (27). Three-dimensional (3D) reconstruction resulted in a structure at 2.8-Å resolution (Fig. 1 A and B and SI Appendix, Fig. S2 A–C), which allowed unambiguous docking of the high-resolution crystal structures of CDK7 and cyclin H (24–26). We rebuilt the docked models according to the cryo-EM map, which showed clear density for amino acid side chains and the bound nucleotide (Fig. 1C and SI Appendix, Fig. S2 D and E). After assignment and rebuilding of the CDK7-cyclin H dimer, the remaining density allowed building of residues 244 to 308 of MAT1, consistent with biochemical data showing that a C-terminal fragment of MAT1 (residues 229 to 309) is sufficient for binding to CDK7-cyclin H and to enhance kinase activity of CDK7 (19). The reconstructed volume thus corresponds to a mass of ∼84 kDa, which is among the smallest asymmetric particles for which better than 3-Å resolution have been obtained using cryo-EM.
Fig. 1.
Structure determination of CAK and structural models of holo-TFIIH and the full CAK complex. (A) Cryo-EM map of human CAK at 2.8-Å resolution. Subunits are colored and indicated. (B) Local resolution estimation. (C) Assignment of MAT1 according to features of the cryo-EM density. (D) Schematic of domain organization and interactions of MAT1 with partners within TFIIH. (E) Depiction of holo-TFIIH, composed of the core complex (PDB ID 6NMI) and the CAK subcomplex. (F) The complete CAK subcomplex consists of a core-binding module (visualized with the TFIIH core complex) and the CDK-cyclin module (reported in this work).
Complete Structural Description of Human TFIIH.
The CAK subcomplex is an important functional unit of TFIIH and critical for its activity as a general transcription factor. Previous cryo-EM studies of human and yeast TFIIH have revealed the structure of the TFIIH core complex and residues 1 to 210 of MAT1 (28–32), but were lacking the remainder of the CAK module due to disorder (28, 29) or low local resolution (30, 31). Our structure of human CAK completes the structure of human TFIIH (Fig. 1 D–F). The combined structures show how MAT1 simultaneously interacts with both the TFIIH core complex and the CDK7-cyclin H module. Distinct and independently folding structural modules, the RING domain and helical regions near the N terminus, and the CAK anchor near the C terminus of MAT1, engage with the TFIIH core complex and the CDK-cyclin module, respectively. These regions are connected by a flexible tether of ∼35 residues that are not resolved in any of the structures reported (Fig. 1 D–F). The flexible nature of the tether and the lack of observed interactions of the CDK7-cyclin H module with other regions of MAT1 in the full-length CAK complex (Fig. 1A) explain why the CDK7-cyclin H module and parts of MAT1 bound to it were not resolved in previous structures of free TFIIH (28, 29). This structural flexibility may support the ability of the CAK module to occupy distinct positions in the Pol II-PIC depending on the presence of mediator (30, 31, 33). Our findings also underscore the role of MAT1 as a key regulator of TFIIH that physically links the DNA translocase XPB, the helicase XPD, and the kinase CDK7 (Fig. 1E), the three ATP-consuming, functional centers of the complex. MAT1 forms interactions with regulatory elements on both CDK7 (see below) and XPD (28, 32, 34), highlighting its central regulatory role.
Role of MAT1 in CAK Architecture and Stability.
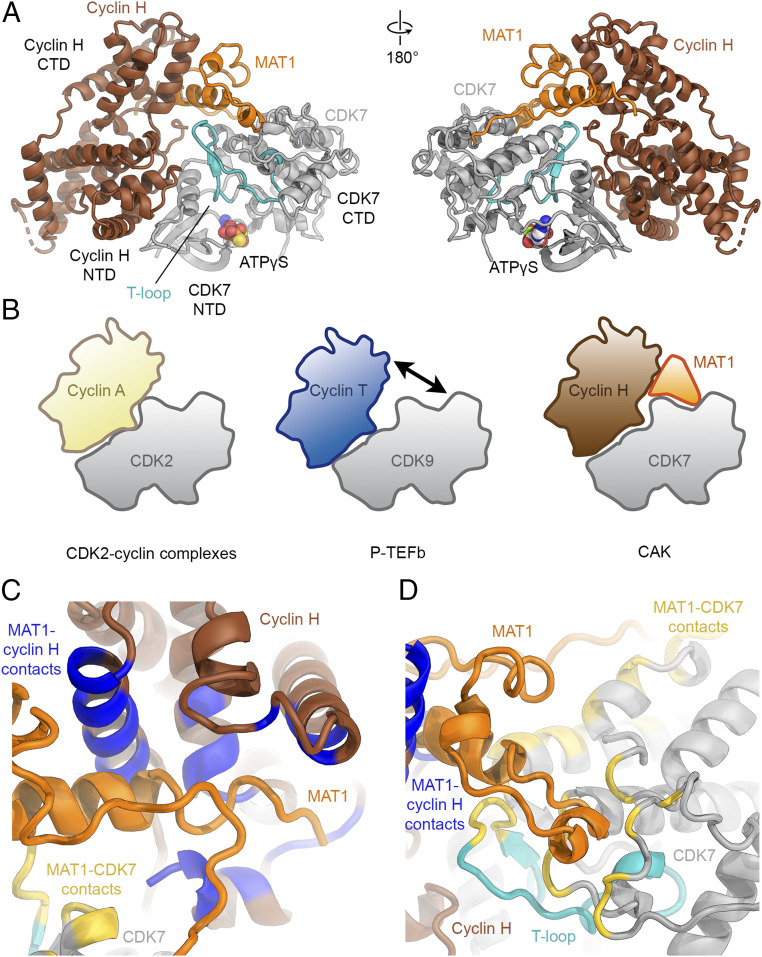
Overall, the arrangement of CDK7 and cyclin H resembles that of other CDK-cyclin complexes (reviewed in ref. 35). Interactions are mostly confined to the N-terminal domains of both CDK7 and cyclin H (Fig. 2A and SI Appendix, Fig. S3A), burying approximately 1,100 Å2 of accessible surface area. Previous analysis revealed that both the extent of the interaction surface and the geometry of CDK-cyclin complexes show marked variability (36). We find that the C-terminal lobe of CDK7 and the C-terminal cyclin fold of cyclin H are rotated away from each other compared to the structure of CDK2-cyclin A (11), resulting in a geometry that is intermediate between that of CDK2-cyclin A and that described for the transcriptional CDK9-cyclin T complex (P-TEFb) (37) (Fig. 2B and SI Appendix, Fig. S3 B and C). In the CAK, the C-terminal CAK-anchor region of MAT1 (residues 255 to 309) fills the space vacated by this rotation (Fig. 2 A and B). Most of this region of MAT1 is sandwiched between the C-terminal lobe of CDK7 and the C-terminal cyclin fold of cyclin H, thereby bridging the two subunits (Fig. 2 C and D). Indeed, MAT1 buries more surface area with CDK7 and cyclin H (>1,200 Å2 each) than CDK7 and cyclin H bury with each other. This rationalizes biochemical data that established MAT1 as a CAK assembly factor that enhances the stability of the CDK7-cyclin H interaction (15, 16), but requires revision of a previously proposed architecture of the CAK based on low-resolution cryo-EM data, which placed the MAT1 components such that the space between CDK7 and cyclin H was left unoccupied (33).
Fig. 2.
Structure of the human CAK and role of MAT1 in its architecture. (A) Atomic model of CAK shown in “front” and “back” views (CDK7 gray with the T-loop shown in teal, cyclin H brown, and MAT1 orange). (B) Schematics of the architecture of CDK2-containing complexes, transcriptional CDK complexes, and CAK. (C and D) Interactions of MAT1 with cyclin H and CDK7. Contact sites (distance <4 Å) on cyclin H are colored blue, contact sites on CDK7 are colored yellow, and the T-loop is shown in cyan.
The most prominent and well-ordered part of the MAT1 density is its C-terminal α-helix (Fig. 1C, residues 288 to 300), which spans the distance between the C-terminal lobe of CDK7 and the C-terminal cyclin fold of cyclin H and interacts with both proteins. Comparison of the CAK structure to that of CDK2 with cyclins A, B, and E (11, 12, 38, 39) shows that the C-terminal α-helix of MAT1 occupies a similar position as an extended α-helix at the N terminus of the cyclin folds in cyclin A, B, and E that contributes to the enlarged interaction surface in CDK2-cyclin complexes (SI Appendix, Fig. S3D). Both cyclin H and cyclin T within P-TEFb (37), which is more closely related to cyclin H than are the cell cycle cyclins (40), lack this extended α-helix at the N terminus of the cyclin folds, leaving room for other interactions at this site. However, it is unclear whether any associated factors bind at this site in P-TEFb or other transcriptional CDKs, and thus the cooption of this interaction site by an associated protein may be unique to CAK.
The yeast equivalent of the human CDK7-cyclin H-MAT1 complex is called TFIIK and is formed by the proteins Kin28, Ccl1, and Tfb3 (41, 42). Analysis of our structure shows that the MAT1 residues that form contacts to cyclin H and CDK7 are mostly conserved in Tfb3 (SI Appendix, Figs. S4 and S5), suggesting that the molecular contacts between MAT1/Tfb3 and the CDK-cyclin pair are likely equivalent, and confirming a conserved architecture between CAK and TFIIK, and likely across all eukaryotes.
In summary, our data establish that MAT1 forms extensive, conserved interfaces with both CDK7 and cyclin H, explaining its role as a CAK assembly factor.
Conformation and Interactions of the CDK7 T-Loop in the Human CAK.
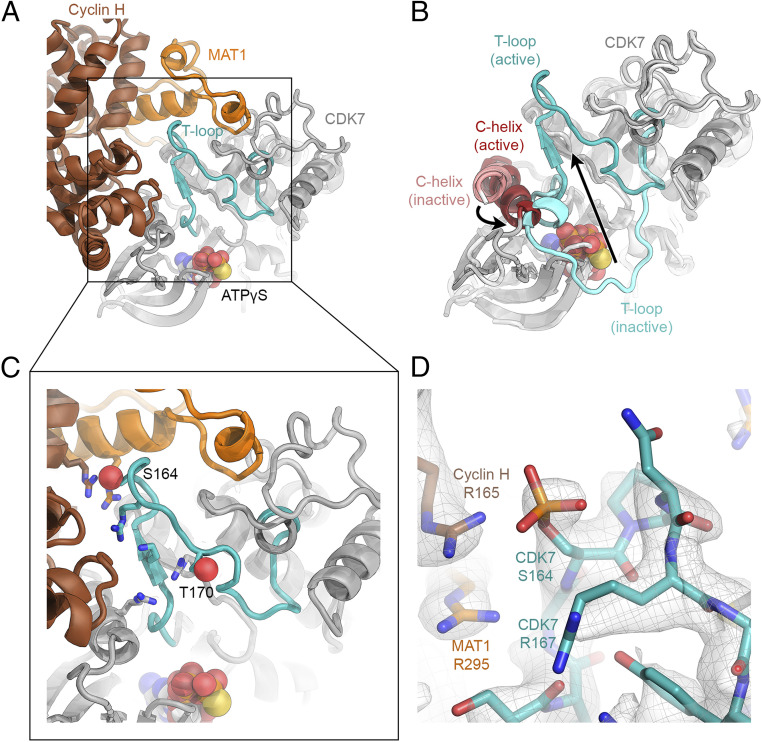
In our structure, which has clear density for ATPγS in the CDK7 active site (SI Appendix, Fig. S2D), the regulatory T-loop of CDK7 is found in an extended, active conformation, pointing away from the nucleotide-binding site and toward cyclin H (Fig. 3 A and B and SI Appendix, Fig. S6 A–D). This conformation contrasts with the structure of isolated CDK7, in which the T-loop occludes the substrate-binding site (Fig. 3B and SI Appendix, Fig. S6C). Furthermore, the presence of cyclin H pushes the C-helix (CDK7 residues 57 to 68) into position to promote assembly of the CDK7 active site (Fig. 3B), analogous to the conformational change of the C-helix described for CDK2-cyclin A complex (12). Accordingly, our purified CAK was able to phosphorylate a synthetic (YSPTSPS)3KKKK peptide, mimicking the RPB1 CTD, on Ser5 of the YSPTSPS heptapeptide repeat (SI Appendix, Fig. S6 E and F). Notably, the tip of the CDK7 T-loop closely approaches the C-terminal α-helix of MAT1 (Fig. 2D), suggesting that interactions with MAT1 promote the extended conformation of the CDK7 T-loop within the CAK complex (Fig. 3 A and C), likely contributing to the reported CAK activation by MAT1 (19).
Fig. 3.
Extended T-loop of CDK7. (A) The T-loop in the CAK-ATPγS complex is extended and points toward MAT1 and cyclin H. (B) In the crystal structure of free, inactive CDK7 (light gray), the T-loop (cyan) is folded across the active site and the C-helix is dislodged from the active site (PDB ID 1UA2) (26). Our structure is superposed (CDK7, gray; T-loop, teal; C-helix, dark red). Large conformational changes between the inactive and active states are indicated with arrows. (C) Positively charged residues near the CDK7 phosphorylation sites (S164, T170; Oγ shown as red spheres) are shown as sticks. (D) Structure of the CAK near S164; cryo-EM density shown in gray (combined surface and mesh) suggests that S164 is partially phosphorylated in our preparation of CAK. Positively charged residues surrounding the phosphate group are labeled.
Like its CDK substrates, CDK7 itself is the target of regulatory phosphorylation. Specifically, its T-loop harbors residues S164 and T170 (Fig. 3C), both of which are phosphorylation sites (16, 43, 44). In our cryo-EM maps, S164 and to a lesser degree T170 showed enlarged densities that may arise from the presence of substoichiometric levels of the phosphorylated species (Fig. 3D). These map features are consistent with mass spectrometric analysis, indicating the presence of some phosphorylated molecules in our CAK samples (SI Appendix, Fig. S7). The presence of both phosphorylated and unphosphorylated CDK7 in otherwise homogeneously stable CAK is in agreement with previous reports of MAT1-mediated assembly of CAK and with data showing that full, active complex can exist in the absence of T-loop phosphorylation (16).
Interestingly, S164 within the CDK7 T-loop is found in proximity to three arginine side chains (Fig. 3 C and D), one each from all three CAK subunits (CDK7 R167, MAT1 R295, and cyclin H R165). This patch of arginines is reminiscent of a positively charged pocket that accommodates the phosphate group of phosphorylated T160 in CDK2, an arrangement that was first visualized in the structure of phosphorylated CDK2 in complex with cyclin A (12), and which is also found in CDK7 (CDK7 residues R61, R136, and K160; Fig. 3C). The human CAK thus harbors two such positively charged pockets, the first of them formed by CDK7 alone and conserved across CDKs, the second specific to the CAK and formed by all three subunits. Phosphate binding in the latter pocket might influence complex assembly and/or the conformation of the T-loop. Indeed, we found that a phosphate group modeled into our density closely approaches cyclin H R165, suggesting formation of an intermolecular salt bridge, and the phosphate may form additional interactions with CDK7 R167 and N166 (Fig. 3D). These potential additional intermolecular interactions following phosphorylation of S164 might promote CAK stability or an extended conformation of the T-loop, both of which would likely contribute to regulation of CAK activity toward certain substrates. This idea is consistent with previous findings showing that T-loop phosphorylation can stabilize CDK7-cyclin H association in the absence of MAT1, and that an S164A mutant shows lower kinase activity toward CDK2 (16). Additionally, an S164A T170A double mutant shows reduced CAK assembly in Drosophila (45) and an equivalent mutant in Xenopus laevis CDK7 (S170A) shows impaired activation in response to cyclin H binding (43). Our structural results, which suggest formation of intermolecular contacts involving the phosphorylated T-loop, can now rationalize these biochemical data. However, our structure does not provide a clear mechanism for the inhibition of CTD-kinase activity by S164 phosphorylation (46), and it cannot easily explain why T170 phosphorylation was found to contribute more strongly to CAK assembly than S164 phosphorylation (16, 43), given that T170 is far removed from any structural elements of cyclin H or MAT1 (Fig. 3C). This interplay between T-loop phosphorylation and complex stability was also observed in the yeast Kin28-Ccl1-Tfb3 complex (42), even though yeast Kin28 lacks a secondary phosphorylation site equivalent to human S164. It is possible that T170 phosphorylation has an indirect effect by affecting CDK7 conformation, or that it stabilizes an extended T-loop conformation that enables formation of interactions involving the tip of the T-loop.
Taken together, our data suggest that T-loop contacts with MAT1 are a specific feature of the CAK, contribute to an extended state of the T-loop, and promote an active CDK7 conformation upon formation of the trimeric CAK. T-loop phosphorylation may enable further regulation of the activity of the CAK toward specific substrates or depending on the cell cycle stage (45, 46).
Structural Mechanism of Irreversible Inhibition of CDK7 by THZ1.
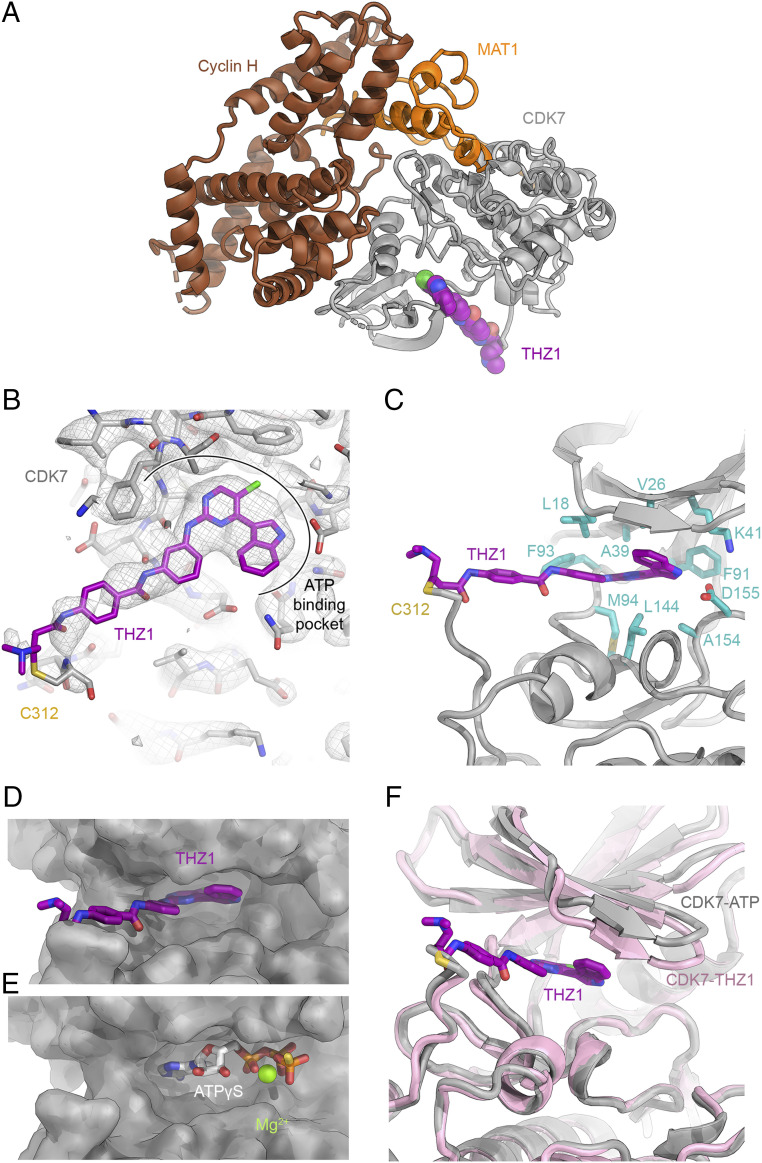
Due to its activity in transcription and cell cycle regulation, the human CAK plays an important role in regulating cell growth and cell division. Consequently, inhibition of the CAK has been found to be a promising strategy for treatment of proliferative diseases (6, 7). To obtain insight into the covalent binding of the cysteine-reactive anticancer compound THZ1 (7) to CDK7 within the CAK, we have determined the 3.3-Å resolution structure of THZ1 bound to a CDK7-cyclin H-MAT1Δ219 complex (CAK-MAT1Δ219), in which the N-terminal region of MAT1, including its RING domain, has been removed (Fig. 4A and SI Appendix, Figs. S1A and S8). Consistent with mass spectrometric analysis that shows essentially complete occupancy of our complex with THZ1 (SI Appendix, Fig. S7), we observe well-defined density for the small-molecule inhibitor in the active site of CDK7 (Fig. 4B). THZ1 occupies the mostly hydrophobic binding pocket that otherwise accommodates the base and ribose of bound ATP (Fig. 4 C–E). Less defined density extends toward the comparably poorly ordered C-terminal region of CDK7, which is only visible until residue C312. In inhibitor-bound CDK7, the nucleotide-binding pocket is slightly contracted at the site where the phosphate groups would be located in the nucleotide-bound state (Fig. 4F), likely because there are no corresponding groups in the inhibitor occupying this position.
Fig. 4.
The CAK-THZ1 complex. (A) Structure of the CAK-THZ1 complex. THZ1 is shown in purple. (B) THZ1 shown in the cryo-EM density contoured at 4.5σ. THZ1 is bound in the ATP-binding pocket of CDK7 and stretches to the covalently modified cysteine C312. (C) Molecular environment of THZ1 in the nucleotide-binding pocket of CDK7, dominated by hydrophobic residues. (D) THZ1 (purple) in the nucleotide-binding cavity of CDK7. (E) ATPγS in the nucleotide-binding cavity of CDK7. The aromatic head group of THZ1 and the base and ribose of the nucleotide occupy the same site. (F) Comparison of the conformation of CDK7 in the nucleotide-bound (gray) and THZ1-bound (pink) states.
Our observations are in overall agreement with the proposed mode of binding of THZ1 based on computational modeling (7). Our density is best defined for the aromatic groups of THZ1 that are buried inside CDK7 and suggests that the THZ1 indole ring is rotated such that it points toward the open side of the nucleotide-binding pocket, rather than more deeply into it (Fig. 4 B and C). This aspect of the conformation of THZ1 more closely resembles a recent computational model for the binding of SY-1365 to CDK7 (8) and the structure of the related compound THZ531 bound to CDK12-cyclin K (47), as compared to the originally proposed model for binding of THZ1 (7).
In addition to CDK7, THZ1 can irreversibly inhibit CDK12 and CDK13 because of the high sequence conservation of their active site pockets and because similarly to CDK7, CDK12 and CDK13 also have cysteine residues located in proximity to the active site that can be targeted for covalent modification (7, 47). The structure of CDK12-cyclin K bound to the related cysteine-reactive compound THZ531 (47) shows that the aromatic ring systems of THZ1 and THZ531 inserted into the kinase active sites exhibit very similar conformations, in agreement with the sequence similarity of the binding pockets in CDK7 and CDK12 (SI Appendix, Fig. S9 A and B). In contrast, the arms of the inhibitors spanning the distance to the covalently modified cysteines assume dramatically different conformations, both between CAK-THZ1 and CDK12-cyclin K-THZ531 as well as between the two copies of the molecule present in the asymmetric unit of the latter structure (SI Appendix, Fig. S9C).
In summary, our structure is in overall agreement with a previous computational model for the interaction of THZ1 with CDK7 at the ATP-binding pocket and, together with structural data on the related CDK12-cyclin K-THZ531 complex, informs on the interactions of different regions of the inhibitor with its target. Additionally, our structure now refines our understanding of the conformation of the inhibitor inside the CDK7 nucleotide-binding pocket.
Conclusion
The cryo-EM structure of the 84-kDa CDK-cyclin module of the human CAK reveals the molecular architecture of this trimeric CDK assembly, how the unique third subunit MAT1 contributes to the stability of the complex, and how molecular interactions within the CAK promote the extended, activated conformation of the T-loop of CDK7. We also determined the structure of the small-molecule inhibitor THZ1 at the active site of CAK. Our study shows that cryo-EM is suitable for structure determination of a small, asymmetric complex at better than 3-Å resolution, opening the way for structure-guided design and improvement of therapeutic compounds that inhibit the activity of the human CAK and other important targets of similar size.
Methods
Cloning and Expression of CAK and CAK-MAT1Δ219.
Codon-optimized genes for MAT1, cyclin H, and CDK7 (sequences were synthesized by GenScript) were amplified by PCR using primers with suitable overhangs and cloned into 438-series vectors harboring expression cassettes with baculoviral promoters and terminators (48) using the in-fusion variant (49) of SLIC cloning (50). The expression cassettes encoding His6-tagged MAT1, Strep-tagged CDK7, and tag-free cyclin H (SI Appendix, Fig. S1A) were then combined into expression constructs containing MAT1-cyclin H-CDK7 using PmeI (MssI) and SwaI (SmiI) restriction digests (FastDigest enzymes, Thermo Fisher Scientific) and subsequent in-fusion reactions. The construct for expression of MAT1Δ219-cyclin H-CDK7 (CAK-MAT1Δ219) was assembled using the same approach but using cloning primers that remove the N-terminal 219 residues of MAT1 and allow creation of an N-terminal MBP-His6 fusion construct. A construct harboring a Pol II-CTD peptide mimic fused to the CDK7 N terminus was constructed by linearization of the plasmid encoding CDK7 by PCR, followed by insertion of a gene block (synthesized by IDT DNA) encoding the peptide sequence and a linker by an in-fusion reaction. The peptide could not be visualized in any of the subsequent cryo-EM maps (see below).
The vectors encoding the expression constructs were transformed into EMBacY cells (51) for preparation of bacmids by standard methods. Bacmid DNA was prepared by isopropanol precipitation and transfected into Sf9 (Spodoptera frugiperda) insect cells using FuGENE HD transfection reagent (Promega). After two rounds of virus amplification in Sf9 cells, 1-L cultures of High5 (Trichoplusia ni) insect cells were infected with 10 mL supernatant from the previous virus amplification cultures. The cells were harvested by centrifugation after 72 h of incubation and frozen in liquid N2 for later use.
Purification of CAK.
CAK complexes were purified using Nickel affinity, Strep-Tactin affinity, and size exclusion chromatography. Cells were resuspended in lysis buffer (250 mM KCl, 40 mM Hepes-KOH pH 7.9, 5 mM MgCl2, 5 mM β-mercaptoethanol, 10 mM imidazole, and 10% [vol/vol] glycerol supplemented with protease inhibitors and DNaseI) and homogenized by sonication. The lysate was cleared using a JA-20 rotor (18,000 rpm for 30 min at 4 °C) and incubated with Ni-NTA superflow resin (Qiagen) for 30 min. The beads were washed in purification buffer (250 mM KCl, 25 mM Hepes-KOH pH 7.9, 5 mM MgCl2, 5 mM β-mercaptoethanol, and 10% [vol/vol] glycerol) supplemented with 25 mM imidazole and eluted in purification buffer with 300 mM imidazole. The eluate was incubated with Strep-Tactin superflow plus resin (Qiagen) for 30 min, washed with 10 bed volumes of purification buffer, and eluted with purification buffer supplemented with 10 mM desthiobiotin (Sigma-Aldrich). The eluted fractions were concentrated by ultrafiltration using 30-kDa molecular weight cutoff Centricon centrifugal filter units (EMD Millipore) and applied onto a Superdex 200 10/300 GL column (GE Healthcare) for size exclusion chromatography (SI Appendix, Fig. S1B). Purified fractions (SI Appendix, Fig. S1C) were pooled, concentrated to 2 mg/mL, and frozen in liquid N2 for storage at −80 °C.
Purification of CAK-MAT1Δ219 and THZ1 Complex Formation.
For studies involving THZ1 binding to CAK, the CAK-MAT1Δ219 construct was used because THZ1 induced precipitation of full-length CAK at various stages during the preparation of the complex, possibly because the zinc-coordinating cysteines in the MAT1 RING domain can engage in unwanted reactions with THZ1. CAK-MAT1Δ219 was purified as described above, with modifications to the protocol to enable binding of THZ1. Specifically, the MBP-His6 tag on MAT1 and the Strep-tag on CDK7 were cleaved using TEV (tobacco etch virus) protease (2 h at room temperature). The resulting protein was incubated with nickel beads to remove the cleaved MBP-tag and TEV protease and subsequently subjected to gel filtration to remove β-mercaptoethanol. This step was added to avoid side reactions of the sulfhydryl-reactive acrylamide group in THZ1 with the thiol on β-mercaptoethanol. After formation of the CAK-MAT1Δ219-THZ1 complex by incubation of 0.2 mg/mL CAK-MAT1Δ219 (approximately 2 μM) with 5 μM THZ1 (EMD Millipore) for 1.5 h at room temperature, the complex was concentrated and subjected to a second gel filtration step, again without β-mercaptoethanol, to remove excess THZ1 and residual DMSO (dimethyl sulfoxide), in which THZ1 had been solubilized. The complex was then concentrated, flash frozen, and stored at −80 °C.
Cryo-EM Specimen Preparation.
Cryo-EM specimens were prepared on UltrAuFoil R1.2/1.3 holey gold grids (Quantifoil microtools) that were plasma cleaned using a Tergeo plasma cleaner (PIE Scientific) for 15 to 45 s. A total of 2 mg/mL CAK was diluted 10× in 200 mM KCl, 2 mM MgCl2, 20 mM Hepes-KOH pH 7.9, 5 mM β-mercaptoethanol and incubated for 5 min with 2 mM ATPγS (lithium salt, Sigma Aldrich). A total of 4 µL of this mixture were then applied to the plasma-cleaned grids, which were blotted for 4 to 6 s using a Vitrobot Mk IV (Thermo Fisher Scientific) and vitrified by plunging into liquid ethane at liquid N2 temperature. Specimens for CAK-MAT1Δ219-THZ1 were prepared analogously, except that β-mercaptoethanol and ATPγS were omitted from the dilution buffer, and that CAK was diluted only five times from the frozen stock solution. Grids were initially screened using a Tecnai F20 cryoelectron microscope (Thermo Fisher Scientific/FEI) equipped with a Gatan US 4000 charge-coupled device camera, and promising specimens after optimization of conditions were clipped into autoloader cartridges.
Cryo-EM Data Collection.
All datasets were acquired using a Thermo Fisher Scientific Talos Arctica cryo-EM operated at 200-kV acceleration voltage according to a conventional defocus-based cryo-EM approach (27). Electron micrograph movies (example shown in SI Appendix, Fig. S1D) were recorded using a K3 camera (Gatan) operated in superresolution counting mode, with the microscope set to 72,886 × magnification (0.686 Å/physical pixel on the camera) at 35 e−/Å2 per second for a total dose of 69 e−/Å2 during a 2-s movie exposure, which was fractionated into 69 frames. The magnification was calibrated using the X-ray crystal structures of CDK7 (PDB ID 1UA2) and cyclin H (PDB ID 1KXU, 1JKW) as a reference. Defocus values ranged from approximately 0.3 μm to 2.5 μm. Data collection was controlled by SerialEM (52), using image shift for rapid image acquisition in a 3 × 3 pattern in nine holes with active beam tilt compensation enabled.
The structure of the CAK complex was obtained from two data collection sessions. One session used a specimen in which a Pol II-CTD peptide mimic had been fused to the N terminus of CDK7; however, no density for the peptide was observed and the structures of the two complexes were identical. The data were thus merged with the wild-type CAK data to obtain a higher-resolution reconstruction (SI Appendix, Fig. S2A).
Cryo-EM Data Processing.
Cryo-EM data were processed in RELION 3.1 (53) as detailed in SI Appendix, Figs. S2 and S8. Movie frames were aligned using MOTIONCOR2 (54) from within RELION 3.1 and binned two times (to 0.686 Å/pixel) during motion correction. After defocus estimation and contrast transfer function (CTF) fitting using CTFFIND4 (55) from within RELION, particles were selected using the 3D reference-based autopicking routine in RELION 3.1. To obtain the initial autopicking template, particles were selected using the Laplacian-of-Gaussian (LoG) algorithm in RELION 3.1 (53) from a preliminary dataset, which was not included in this manuscript because the dataset quality compared unfavorably to the subsequent datasets used here. To obtain an initial cryo-EM reconstruction, these particles were two-dimensional (2D) classified and aligned to an initial reference model generated from converting PDB coordinates of CDK7 and cyclin H (PDB ID 1UA2, 1KXU) to a map using University of California San Francisco (UCSF) Chimera (56). The cryo-EM map refined from these LoG-picked particles subsequently served as the autopicking template and initial reference for processing of all other datasets.
From a typical micrograph with high CAK concentration, 1,000 to 1,500 particles could be picked. To speed up calculations, given the computational cost of processing several million particle picks per dataset, particles were initially extracted at 2.744 Å/pixel in 64-pixel boxes for initial 2D and 3D classification. Initial 2D classifications were performed using fast subsets in RELION 3.1 (53) and the resulting classes were generously selected, excluding mostly false positives or ice contamination. The subsequent low-resolution refinement reached near-Nyquist frequency resolution, and 3D classification without alignment (which applies to all 3D classifications in this work) from this refinement allowed selection of a pool of high-quality particles that immediately refined to better than 4-Å resolution upon reextractions of the particles at 1.28 Å/pixel (SI Appendix, Fig. S2A). These particles were then either directly subjected to Bayesian polishing (57) in RELION 3.1 (dataset 2) or subclassified and polished (dataset 1). In subclassifications, class selection was guided mostly by inspection of the slices of the 3D volumes, and classes with the crispest features were retained. During polishing, the box size was expanded from 144 pixels to 216 pixels to account for signal delocalization by the CTF (58). In our case, the resolution gain from this procedure was minimal, possibly because the high-resolution signal is contributed mostly by very low defocus particles, which are less affected by signal delocalization. However, more particles were retained in equivalent 3D classification steps than when using smaller boxes, which appeared to benefit the map quality, and thus the larger box sizes were retained. After a 2D classification to remove particles impacted by detector defects and other artifacts revealed by polishing and box size expansion, these datasets refined to resolution of 3.1 to 3.2 Å. One more round of 3D classification and optimized CTF refinement (53, 59) yielded subsets of 67,890 and 101,016 particles, which were joined, subjected to CTF refinement again, and refined to the final 2.8-Å resolution map after removal of all particles that came from micrographs with power spectra that did not extend to 4 Å or better.
The resolution of the reconstruction, obtained by gold-standard refinement of fully independent particle subsets (60), was assessed by Fourier shell correlation (FSC) (SI Appendix, Fig. S2B) at the FSC = 0.143 cutoff (61). Additionally, the 3D reconstruction was validated using the 3D FSC validation server (62), which confirmed that the map is isotropic (SI Appendix, Fig. S2C) with a sphericity of 0.96.
The procedure for reconstruction of the CAK-THZ1 structure (SI Appendix, Fig. S8A) was essentially as described for the ATPγS-bound reconstruction, with minor modifications. Apparent larger counts of low-quality particle images necessitated two 3D classification steps at the 2.744 Å/pixel stage to eliminate these and reach 5.5-Å resolution before particle reextraction at smaller pixel size. Due to the lower overall resolution, the final pixel size was chosen to be 1.372 Å/pixel, and the corresponding enlarged box size after particle polishing was 192 pixels. The resolution of the map immediately after polishing (from 310,052 particles) was estimated at 3.2 Å, which is nominally better than the 3.3 Å estimated for the final map from 31,198 particles, obtained after two more 3D classification steps (SI Appendix, Fig. S8A). However, the latter map exhibited far better side chain density, better overall connectivity, and a lower B-factor (approximately −60 Å2 vs. −90 Å2 as automatically determined by the RELION postprocessing function) and was therefore used for interpretation, model building, and refinement.
Model Building, Refinement, and Validation.
Before model building, the 3D volumes obtained from cryo-EM map refinement were reboxed to 150 pixels (CAK reconstruction) or 134 pixels (CAK-THZ1) without changing the pixel size. The atomic model of the CAK complex was assembled by fitting the high-resolution structures of CDK7 and cyclin H (24–26) into the cryo-EM map using UCSF Chimera (56), followed by rebuilding in COOT (63). Both crystal structures showed a good initial fit to the cryo-EM map and needed only minor rebuilding, with the exception of the CDK7 T-loop, which occupies the inactive conformation in the crystal structure, and CDK7 residues 104 to 111, which were found to be register shifted by one residue. After rebuilding of CDK7 and cyclin H according to the density, a continuous stretch of remaining unassigned density was identified and assigned to MAT1 based on agreement of the density with secondary structure prediction and the pattern of large side chains near the C terminus of MAT1 (Fig. 1C). The model was then iteratively rebuilt in COOT and refined using the real space refinement program in PHENIX (64, 65). To maintain good model geometry at the given resolution, Ramachandran, rotamer, Cβ, and secondary structure restraints were used throughout.
The molecular structure and geometric restraints for THZ1 were generated using PHENIX ELBOW (66), and the CAK-THZ1 structure was initially refined using the higher-resolution CAK-ATPγS structure to provide reference restraints, before five final macrocycles of real-space refinement were carried out without reference restraints.
The structures were validated by computing the FSC between the model and the map to assess the agreement of the model and the cryo-EM density (SI Appendix, Figs. S2B and S8B), and using MOLPROBITY (67) to assess the quality of the model geometry. Validation statistics are given in SI Appendix, Table S1. Components of the refined atomic models are listed in SI Appendix, Table S2.
Mass Spectrometry.
The 10-µL aliquots of CAK and CAK-THZ1 were diluted to an approximate concentration of 10 µM and subjected to mass spectrometry analysis to evaluate the binding of THZ1 to the CAK. Intact protein samples were analyzed using a Synapt G2-Si mass spectrometer (Waters), as described (68). The deconvoluted mass spectra for the samples are shown in SI Appendix, Fig. S7. Samples for detection of phosphorylated Pol II-CTD peptide from activity assays (SI Appendix, Fig. S6F) were prepared as described below and analyzed analogously.
Activity Assay and Western Blotting.
Activity assays were performed in 25 mM Hepes-KOH pH 7.7, 150 mM KCl, 5 mM MgCl2, 5 mM β-mercaptoethanol, 5% glycerol. The 100-µl-reactions containing 500 nM CAK, 500 µM (YSPTSPS)3KKKK-biotin substrate peptide (synthesized by GenScript), and 2 mM ATP were incubated for 1, 2, 3, and 5 h at 30 °C. Assays for mass spectrometric analysis of phosphopeptides (SI Appendix, Fig. S6F) were performed under identical conditions, except that the concentration of substrate peptides was lowered to 20 µM.
Proteins and peptides were separated on 4 to 20% MiniProtean TGX SDS/PAGE gels (Bio-Rad) and transferred to nitrocellulose membranes using the TransBlot Turbo Western blotting system (Bio-Rad). The membrane was transferred into TBST (50 mM Tris⋅HCl pH 7.6, 150 mM NaCl, 0.1% Tween 20) with 5% bovine serum albumin and incubated for 2 h, followed by incubation with primary antibodies (rat α-CTD-phospho-Ser5 IgG, clone 3E8, EMD Millipore and rabbit α-CDK7 IgG, clone C-4, Santa Cruz Biotechnology) at 4 °C for 16 h. After washing with TBST, the membrane was incubated with secondary antibodies conjugated to fluorescent dyes (α-rabbit Cy5 and α-rat Cy3) for 1.5 h at room temperature. The fluorescent signal was recorded using a ChemiDoc imaging system (Bio-Rad).
Other Software Used.
Software used in this project was configured by SBGRID (69). Molecular graphics were created using UCSF Chimera (56), UCSF ChimeraX (70), and PyMol (the PyMol molecular graphics system, Schrödinger LLC). Local resolution of cryo-EM maps was computed using RELION 3.1 (53). Buried surface areas were computed using PDBePISA (71) via the PDBePISA web server interface (https://www.ebi.ac.uk/pdbe/pisa/).
Supplementary Material
Acknowledgments
We thank A. Chintangal and P. Tobias for support with computation, J. Remis and P. Grob for support with microscopy, the lab of A. Martin for help with Western blot imaging, C. Ward for discussions, and A. Killilea and M. Fischer at the University of California Berkeley cell culture facility for providing insect cell cultures. This work was funded through National Institute of General Medical Sciences grants R01-GM63072, P01-GM063210, and R35-GM127018 to E.N. The Synapt mass spectrometer was purchased with NIH support (grant 1S10OD020062-01). E.N. is a Howard Hughes Medical Institute Investigator.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009627117/-/DCSupplemental.
Accession Numbers.
Cryo-EM maps and raw electron micrograph movie datasets of CAK and CAK-MAT1Δ219-THZ1 have been deposited at the EM Data Resource with accession codes EMD-22123, EMD-22131, EMPIAR-10431, and EMPIAR-10438, respectively. The refined coordinate models have been deposited at the Protein Data Bank (PDB) with accession codes 6XBZ and 6XD3, respectively.
References
- 1.Sainsbury S., Bernecky C., Cramer P., Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 129–143 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Myers L. C.et al., The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 12, 45–54 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Søgaard T. M. M., Svejstrup J. Q., Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J. Biol. Chem. 282, 14113–14120 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Harlen K. M., Churchman L. S., The code and beyond: Transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 18, 263–273 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Jeronimo C., Bataille A. R., Robert F., The writers, readers, and functions of the RNA polymerase II C-terminal domain code. Chem. Rev. 113, 8491–8522 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Galbraith M. D., Bender H., Espinosa J. M., Therapeutic targeting of transcriptional cyclin-dependent kinases. Transcription 10, 118–136 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwiatkowski N.et al., Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 511, 616–620 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu S.et al., Discovery and characterization of SY-1365, a selective, covalent inhibitor of CDK7. Cancer Res. 79, 3479–3491 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Morgan D. O., Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13, 261–291 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Connell-Crowley L., Solomon M. J., Wei N., Harper J. W., Phosphorylation independent activation of human cyclin-dependent kinase 2 by cyclin A in vitro. Mol. Biol. Cell 4, 79–92 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeffrey P. D.et al., Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature 376, 313–320 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Russo A. A., Jeffrey P. D., Pavletich N. P., Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat. Struct. Biol. 3, 696–700 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Larochelle S., Pandur J., Fisher R. P., Salz H. K., Suter B., Cdk7 is essential for mitosis and for in vivo Cdk-activating kinase activity. Genes Dev. 12, 370–381 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher R. P., Morgan D. O., A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell 78, 713–724 (1994). [DOI] [PubMed] [Google Scholar]
- 15.Yee A.et al., Molecular cloning of CDK7-associated human MAT1, a cyclin-dependent kinase-activating kinase (CAK) assembly factor. Cancer Res. 55, 6058–6062 (1995). [PubMed] [Google Scholar]
- 16.Fisher R. P., Jin P., Chamberlin H. M., Morgan D. O., Alternative mechanisms of CAK assembly require an assembly factor or an activating kinase. Cell 83, 47–57 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Devault A.et al., MAT1 (“menage à trois”) a new RING finger protein subunit stabilizing cyclin H-cdk7 complexes in starfish and Xenopus CAK. EMBO J. 14, 5027–5036 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tassan J. P.et al., In vitro assembly of a functional human CDK7-cyclin H complex requires MAT1, a novel 36 kDa RING finger protein. EMBO J. 14, 5608–5617 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Busso D.et al., Distinct regions of MAT1 regulate cdk7 kinase and TFIIH transcription activities. J. Biol. Chem. 275, 22815–22823 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Shiekhattar R.et al., Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature 374, 283–287 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Serizawa H.et al., Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature 374, 280–282 (1995). [DOI] [PubMed] [Google Scholar]
- 22.Svejstrup J. Q.et al., Different forms of TFIIH for transcription and DNA repair: Holo-TFIIH and a nucleotide excision repairosome. Cell 80, 21–28 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Coin F.et al., Nucleotide excision repair driven by the dissociation of CAK from TFIIH. Mol. Cell 31, 9–20 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Kim K. K., Chamberlin H. M., Morgan D. O., Kim S. H., Three-dimensional structure of human cyclin H, a positive regulator of the CDK-activating kinase. Nat. Struct. Biol. 3, 849–855 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Andersen G.et al., The structure of cyclin H: Common mode of kinase activation and specific features. EMBO J. 16, 958–967 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lolli G., Lowe E. D., Brown N. R., Johnson L. N., The crystal structure of human CDK7 and its protein recognition properties. Structure 12, 2067–2079 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Herzik M. A. Jr., Wu M., Lander G. C., High-resolution structure determination of sub-100 kDa complexes using conventional cryo-EM. Nat. Commun. 10, 1032 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greber B. J., Toso D. B., Fang J., Nogales E., The complete structure of the human TFIIH core complex. eLife 8, e44771 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greber B. J.et al., The cryo-electron microscopy structure of human transcription factor IIH. Nature 549, 414–417 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schilbach S.et al., Structures of transcription pre-initiation complex with TFIIH and Mediator. Nature 551, 204–209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He Y.et al., Near-atomic resolution visualization of human transcription promoter opening. Nature 533, 359–365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kokic G.et al., Structural basis of TFIIH activation for nucleotide excision repair. Nat. Commun. 10, 2885 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan C.et al., Transcription preinitiation complex structure and dynamics provide insight into genetic diseases. Nat. Struct. Mol. Biol. 26, 397–406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peissert S.et al., In TFIIH the Arch domain of XPD is mechanistically essential for transcription and DNA repair. Nat. Commun. 11, 1667–1613 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood D. J., Endicott J. A., Structural insights into the functional diversity of the CDK-cyclin family. Open Biol. 8, 180112–180126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Echalier A., Endicott J. A., Noble M. E. M., Recent developments in cyclin-dependent kinase biochemical and structural studies. Biochim. Biophys. Acta 1804, 511–519 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Baumli S.et al., The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J. 27, 1907–1918 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown N. R.et al., Cyclin B and cyclin A confer different substrate recognition properties on CDK2. Cell Cycle 6, 1350–1359 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Honda R.et al., The structure of cyclin E1/CDK2: Implications for CDK2 activation and CDK2-independent roles. EMBO J. 24, 452–463 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma Z.et al., Phylogenetic analysis reveals the evolution and diversification of cyclins in eukaryotes. Mol. Phylogenet. Evol. 66, 1002–1010 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Feaver W. J., Svejstrup J. Q., Henry N. L., Kornberg R. D., Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell 79, 1103–1109 (1994). [DOI] [PubMed] [Google Scholar]
- 42.Keogh M.-C., Cho E.-J., Podolny V., Buratowski S., Kin28 is found within TFIIH and a Kin28-Ccl1-Tfb3 trimer complex with differential sensitivities to T-loop phosphorylation. Mol. Cell. Biol. 22, 1288–1297 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez A. M.et al., Dual phosphorylation of the T-loop in cdk7: Its role in controlling cyclin H binding and CAK activity. EMBO J. 16, 343–354 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Labbé J. C.et al., p40MO15 associates with a p36 subunit and requires both nuclear translocation and Thr176 phosphorylation to generate cdk-activating kinase activity in Xenopus oocytes. EMBO J. 13, 5155–5164 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larochelle S.et al., T-loop phosphorylation stabilizes the CDK7-cyclin H-MAT1 complex in vivo and regulates its CTD kinase activity. EMBO J. 20, 3749–3759 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akoulitchev S., Reinberg D., The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7. Genes Dev. 12, 3541–3550 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang T.et al., Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat. Chem. Biol. 12, 876–884 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gradia S. D.et al., MacroBac: New Technologies for Robust and efficient large-scale production of recombinant multiprotein complexes. Methods Enzymol. 592, 1–26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Irwin C. R., Farmer A., Willer D. O., Evans D. H., In-fusion® cloning with vaccinia virus DNA polymerase. Methods Mol. Biol. 890, 23–35 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Li M. Z., Elledge S. J., SLIC: A method for sequence- and ligation-independent cloning. Methods Mol. Biol. 852, 51–59 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Bieniossek C., Richmond T. J., Berger I., “MultiBac: Multigene baculovirus-based eukaryotic protein complex production” in Curr. Protoc. Protein Sci., (2008), Vol. Chapter 5, p. Unit 5.20. [DOI] [PubMed] [Google Scholar]
- 52.Schorb M., Haberbosch I., Hagen W. J. H., Schwab Y., Mastronarde D. N., Software tools for automated transmission electron microscopy. Nat. Methods 16, 471–477 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zivanov J.et al., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng S. Q.et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goddard T. D., Huang C. C., Ferrin T. E., Visualizing density maps with UCSF Chimera. J. Struct. Biol. 157, 281–287 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Zivanov J., Nakane T., Scheres S. H. W., A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sigworth F. J., Principles of cryo-EM single-particle image processing. Microscopy (Oxf.) 65, 57–67 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zivanov J., Nakane T., Scheres S. H. W., Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheres S. H. W., Chen S., Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosenthal P. B., Henderson R., Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Tan Y. Z.et al., Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Afonine P. V.et al., Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adams P. D.et al., PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moriarty N. W., Grosse-Kunstleve R. W., Adams P. D., Electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 65, 1074–1080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Williams C. J.et al., MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Light S. H.et al., A flavin-based extracellular electron transfer mechanism in diverse Gram-positive bacteria. Nature 562, 140–144 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morin A.et al., Collaboration gets the most out of software. eLife 2, e01456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goddard T. D.et al., UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM maps and raw electron micrograph movie datasets of CAK and CAK-MAT1Δ219-THZ1 have been deposited at the EM Data Resource with accession codes EMD-22123, EMD-22131, EMPIAR-10431, and EMPIAR-10438, respectively. The refined coordinate models have been deposited at the Protein Data Bank (PDB) with accession codes 6XBZ and 6XD3, respectively.