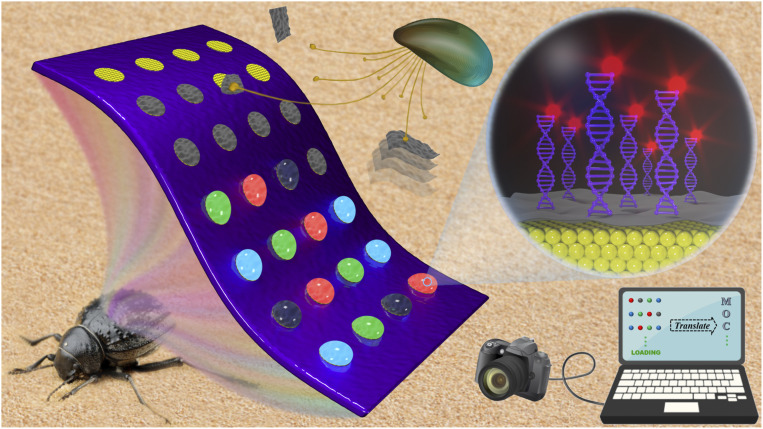
Significance
Inspired by the wettability structure of Stenocara beetle and adhesion ability of mussel, we present an innovative MXene-integrated PhC array for multichannel bioinformation coding. The bioinspired hydrophilic–hydrophobic surfaces can selectively capture colloidal droplets to form PhC arrays; while the mussel’s byssus-mimicked DA catechol units help in the immobilization of MXene nanosheets on these arrays. MXene can quench the fluorescence signal of QD-DNA probes, which can be recovered when the corresponding target is present. In this case, complex sequence information of DNA can be converted into visualized fluorescence signals to facilitate information reading. Based on this strategy, high-throughput information encoding and encryption are realized, which indicates its broad application prospect in big data storage.
Keywords: bioinspired, colloidal crystal, structural color, MXene, wettability
Abstract
Information coding strategies are becoming increasingly crucial due to the storage demand brought by the information explosion. In particular, bioinformation coding has attracted great attention for its advantages of excellent storage capacity and long lifetime. Herein, we present an innovative bioinspired MXene-integrated photonic crystal (PhC) array for multichannel bioinformation coding. PhC arrays with similar structure to Stenocara beetle’s back are utilized as the substrate, exhibiting properties of high throughput and stability. MXene nanosheets are further integrated on the PhC array’s substrate with the assistance of the adhesion capacity of mussel-inspired dopamine (DA). Benefitting from their fluorescence resonance energy transfer effect, MXene nanosheets can quench the fluorescence signals of quantum dot (QD) modified DNA probes unless the corresponding targets exist. Additionally, these black MXene nanosheets can enhance the contrast of structural color. In this case, the encrypted information can be easily read out by simply observing the fluorescence signal of DNA probes. It is demonstrated that this strategy based on bioinspired MXene-integrated PhC arrays can realize high-throughput information encoding and encryption, which opens a chapter of bioinformation coding.
With the advent of the information age, the exponential growth of data stimulated storage exploration and research. Various information coding strategies have been proposed to realize data storage, classification, and encryption in different areas, such as communication and anticounterfeiting fields. Among these strategies, bioinformation coding, such as DNA, has attracted great interest from researchers because of its large storage capacity and long storage life (1). In general, primers are first attached to the substrate for the fixation of sequences. Subsequently, four kinds of nucleotides would be linked to the primers in a specific order with the help of DNA polymerase. After sequencing, the synthesized DNA sequences are matched with decode statements under coding rules, thus realizing the bioinformation coding and decoding strategy based on DNA molecules (2–4). It is worth mentioning that DNA molecules can store 215 petabytes of information per gram, equivalent to 100,000 common hard disks. This feature greatly expands information storage capacity, and the stored information would not be destroyed by demagnetization or power outages as hard drives (5‒6). Although with great progress, the simple substrate-based DNA bioinformation coding strategies are often monotonous and are not conducive to the further expansion of their values. In addition, the reading of this coded information is usually complicated and expensive. Therefore, new bioinformation coding strategies with advanced substrates and facile readout processes are still highly anticipated.
Here, we present an MXene-integrated photonic crystal (PhC) array for multichannel bioinformation coding, as schemed in Fig. 1. PhCs are periodically structured materials with photonic bandgap (PBG) and characteristic reflection peaks (7–11). The internal periodic structure does not change when the PhC surface is modified by two-dimensional (2D) materials such as graphene oxide (12). In addition, PhCs have the characteristics of fluorescence enhancement and can be used to improve the signal-to-noise ratio. These advantages enable PhCs to serve as an excellent substrate for bioinformation coding (12–16). MXene is an emerging 2D material with extraordinary physicochemical properties, which shows great potential in the field of biosensors (17‒18). Because of the fluorescence resonance energy transfer (FRET), MXene can quench the fluorescence signal of closer DNA probes, while when the corresponding target is present, the fluorophore can be kept away from MXene to recover the fluorescence signal (19–24). Based on this case, the complex sequence information of DNA can be converted into visualized fluorescence signals, which facilitates information reading. Therefore, the integration of MXene into PhC substrate is expected to play an important role in the field of bioinformation coding.
Fig. 1.
Schematic diagram. Bioinspired MXene-integrated colloidal crystal arrays for multichannel bioinformation coding.
In this paper, inspired by the wettability structure of Stenocara beetle and adhesion ability of mussel (25–31), we developed an effective approach to realize this integration. Stenocara beetle is a kind of magical creature living in the desert. Its hydrophobic elytra have a series of hydrophilic protrusions on the surface. These bumps are used to collect water from the air to ensure the beetle’s survival. By selectively treating a hydrophobic slide substrate with hydrophilic pattern, monodispersed colloidal nanoparticles can only self-assemble in the hydrophilic region to form a Stenocara beetle’s back-inspired PhC arrays, which could be used as high-throughput PhC substrates for bioinformation coding. With the assistance of the mussel’s byssus-mimicked dopamine (DA) catechol units, the MXene nanosheets were tightly immobilized on the PhC arrays. These black MXene nanosheets could not only realize effective regulation of fluorescence signals, but also enhance the contrast of structure color (32‒33). Thus, through simply detecting the fluorescence signal of the bioreaction on the surface of MXene-integrated PhC arrays, the hidden bioinformation in the substrate could be achieved. It was demonstrated that the MXene-integrated PhC arrays are completely feasible for multichannel coding, which indicates a broad application prospect in big data storage.
Results
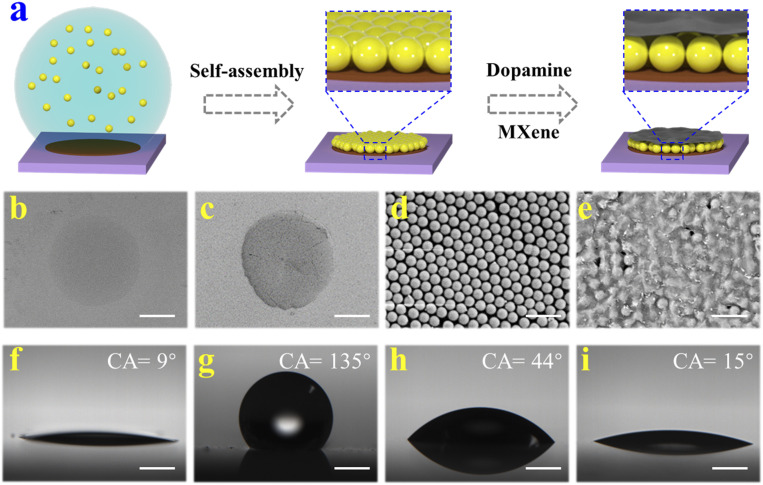
In a typical experiment, the Stenocara beetle’s back-inspired MXene-integrated PhC arrays were fabricated by a simple and effective approach, as shown in Fig. 2A. A glass slide was first treated with NaOH solution to expose the hydroxyl groups on its surface and further modified with fluorosilane to obtain the hydrophobic property (Fig. 2 F and G). Subsequently, a stainless steel mask with holes was tightly attached to the fabricated glass slide for plasma treatment. Under this condition, the unprotected portion formed a hydrophilic array while the protected area remained hydrophobic, which was similar to the construction of Stenocara beetle’s back. Because of the wettability contrast, only the hydrophilic arrays could capture the droplets (about 0.5 μL) when Tris⋅HCl buffer containing DA slightly rinsed through the substrate. After evaporation of water, the surfaces of these hydrophilic arrays were successfully coated with a DA layer (Fig. 2B and SI Appendix, Fig. S1).
Fig. 2.
(A) Schematic illustration of the preparation process of bioinspired MXene-integrated PhC arrays. (B–E) The SEM images of (B) DA layer, (C) PhC pattern, (D) the surface of PhC pattern, and (E) the surface of MXene-integrated PhC pattern. (F–I) The water contact angles (CA) of (F) glass slide, (G) hydrophobic substrate, (H) DA layer, and (I) the MXene-integrated PhC pattern. (Scale bars are 300 μm in B and C, 1 μm in D and E, and 500 μm in F–I.)
Subsequently, silica colloidal particle-contained solution was dragged on the wettability-patterned surface, and only the hydrophilic DA arrays could capture a certain volume of colloidal solution (about 0.5 μL). With the evaporation of water, silica colloidal particles gradually self-assembled into periodic and orderly PhC structure under the driving of capillary force (Fig. 2 C and D). It is worth mentioning that PhC structure could be firmly fixed on the hydrophilic array of substrate with the aid of DA layer as DA is a small-molecular analog of the adhesion protein in the mussel’s byssus which could increase the adhesion ability for anchoring function materials. Then, dispersed solution containing MXene nanosheets (more than 500 nm) and DA were dripped onto PhC arrays. Because the silica colloidal particles were tightly assembled, the gaps between the particles were just a few dozen nanometers. As a consequence, these MXene nanosheets could only deposit on the surface of PhC arrays (Fig. 2E). In addition, the existence of DA made MXene nanosheets tightly adhere to PhC patterns. Based on the above steps, a bioinspired hydrophobic substrate with hydrophilic MXene-integrated PhC arrays was successfully fabricated.
It is worth noting that the “coffee-ring” effect would occur during the self-assembly of silica colloidal particles into PhC patterns (SI Appendix, Fig. S2A). The phenomenon was caused by the faster water evaporation rate at the edges compared with that in the center of the droplets captured by PhC arrays (33–35). In this case, capillary flow was generated in the colloidal droplets, which drove the colloidal particles to the edge of the droplets and assembled into a coffee ring. Considering that the coffee-ring effect would lead to uneven deposition of colloidal crystals which might affect the recognition of PhC patterns, efforts were devoted to avoiding this phenomenon. It has been found that the PhC arrays could self-assemble at a low temperature to reduce the capillary flow, thus inhibiting the effect of the coffee ring. In addition, the introduction of ethylene glycol could enhance the Marangoni effect to counteract the influence of capillary flow, which was conductive to the formation of flat PhC patterns, as shown in SI Appendix, Fig. S2B.
In these hydrophilic PhC patterns, silica colloidal particles closely arranged into periodic ordered structure. This structure would create a PBG that allows PhC to reflect bright structural colors (Fig. 3 A and B). PhC with different sizes would produce distinct characteristic reflection peaks at different angles of incident light, which can be calculated by Bragg formula. Under normal incidence case, the wavelength of the structural color can be calculated by the following simplified formula,
| [1] |
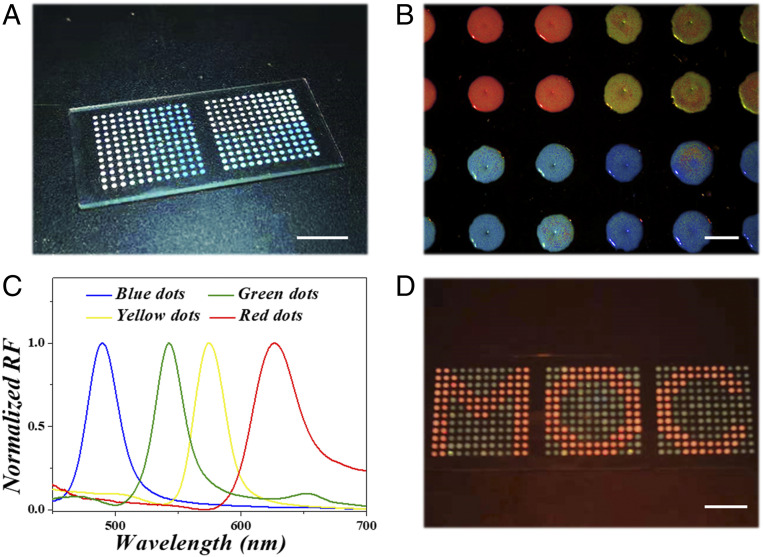
where λ refers to the wavelength of structural color, d is size of silica colloidal particles we chose in the experiment, naverage means the average refractive index of the entire PhC layer. The value of naverage can be calculated according to the refractive index of silica and the medium, which has been reported in our previous work. Therefore, the characteristic reflection peak of structural color is only determined by the size of silica colloidal particles (Fig. 3C). Because of the Stenocara beetle’s back-inspired structure, colloidal droplets would be captured and restricted in the hydrophilic region. Through using silica colloidal solutions with different particle sizes, PhC arrays with various colors could be formed on the substrate (SI Appendix, Fig. S3). By further precisely manipulating various droplets, each hydrophilic array can deposit PhCs with different reflection peaks and form special patterns together (Fig. 3D).
Fig. 3.
(A) The digital photograph of MXene-integrated PhC arrays on a hydrophobic substrate. (B) The bright structural colors of different PhC patterns and (C) their corresponding reflection (RF) spectra. (D) The PhC arrays form a special pattern “MOC.” (Scale bars are 1 cm in A and D and 1 mm in B.)
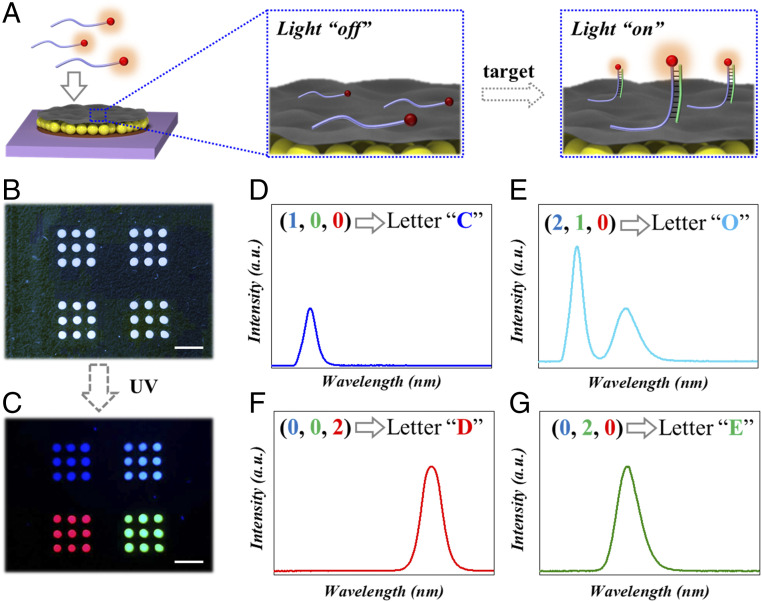
Because of the excellent physicochemical properties of MXene nanosheets, they were integrated with PhC arrays for bioinformation coding. During the process, quantum dot (QD) labeled oligonucleotides were fixed on MXene nanosheets by π-π stacking to serve as fluorescent probes. Under the effect of FRET, the fluorescence of QDs was quenched by MXene nanosheets. With the addition of the corresponding target, the QD-labeled probe end formed double-stranded DNA with it (Fig. 4A and SI Appendix, Fig. S4), while the other end of the probe kept single-stranded and ensured the attachment of QD-DNA to MXene nanosheets. In this case, the QD kept away from MXene nanosheets to recover the fluorescence signal, which increased with the quantity of added targets (SI Appendix, Figs. S5 and S6). In addition, the PhC array below MXene nanosheets could enhance the recovered fluorescence signal (SI Appendix, Fig. S7). From a macro perspective, the MXene-integrated PhC array underwent the process of fluorescence signal from “off” to “on” (Fig. 4A). Furthermore, the strategy is also applicable to other silicon-based substrates such as quartz (hard material) and polydimethylsiloxane (soft material), as shown in SI Appendix, Fig. S8. These results showed that our hydrophilic–hydrophobic composite platform provided a solid foundation for bioinformation coding.
Fig. 4.
(A) Schematic illustration of the fluorescence signal from “off” to “on” on the MXene-integrated PhC arrays. (B) Bright field and (C) fluorescent field image of an array with information “CODE.” (D–G) Fluorescence spectra corresponding to four different fluorescent patterns and their coding information. (Scale bars are 5 mm in B and C.)
QDs have been proven to possess the advantages of wide-spectrum absorption, narrow-spectrum emission, high quantum yield, and stable fluorescence signal, which results in excellent readability for bioinformation coding (36–39). Compared with PhC with single-characteristic reflection peak, the encoding ability can be greatly extended when each array is loaded with different QD-labeled probes. The corresponding spectra were obtained by scanning the fluorescence signal of each MXene-integrated PhC array (SI Appendix, Fig. S9). Assuming each array contains m kinds of QD-labeled probes and n kinds of fluorescence intensity, finally, nm different spectra could be achieved. Different spectrograms corresponded to different coding units, and as a result, the measured spectral information established a relationship with coding information. The fluorescence spectra were translated to reveal the encrypted bioinformation according to standard coding/decoding program and corresponding relation designed. For example, three different QDs (red, green, and blue) and three different fluorescence intensities (0, 1, and 2) were used for bioinformation coding, and the resulting 27 spectra represented 26 letters and 1 blank (SI Appendix, Table S1). Under this rule, seemingly random arrays could be translated into complete information, thus realizing the bioinformation coding (Fig. 4 B–G). Most importantly, the correct “key,” that was matched target sequence, must be obtained before decoding. Otherwise, the fluorescence signal of the array would be lost, and the result of translation was incorrect. By increasing the number of QD-labeled probes and analyzing different fluorescence intensities with sophisticated instruments, more coding units could be obtained for bioinformation coding. In addition, these fluorescent signals have good stability under different temperature conditions (SI Appendix, Fig. S10). These results indicated that the hydrophilic–hydrophobic composite platform had potential applications for storing and encrypting large amounts of information.
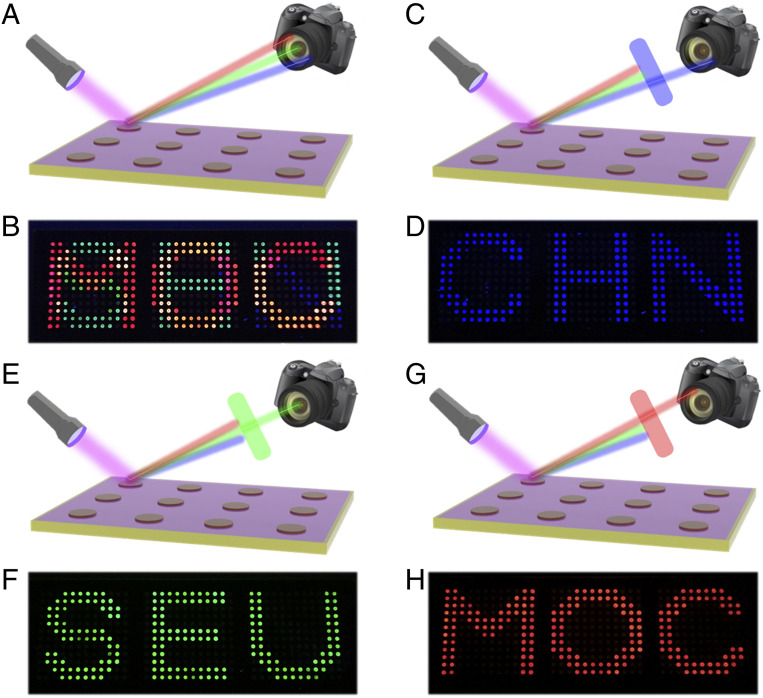
It is noteworthy that there were several QD-labeled probes on each array, and the fluorescence signals between them appeared to be superimposed. With the assistance of a fluorescence spectrometer, the fluorescence signals of different QDs could be distinguished without affecting each other (SI Appendix, Fig. S11). Here, optical filters, which are widely used in various cameras, microscopes, and telescopes, were used to select the desired band of optical signals (SI Appendix, Fig. S12 and Movie S1). Since different groups of QD-labeled probes were immobilized on each array, distinct fluorescence signals could be displayed by adding the same target. Without the assistance of optical filters, three fluorescence signals interfered with each other, and the pattern could not be identified (Fig. 5 A and B). When observed under different optical filters, the blue channel’s “CHN” pattern, the green channel’s “SEU” pattern, and the red channel’s “MOC” pattern could be distinguished respectively (Fig. 5 C–H). Therefore, the fluorescence signal could be selectively read out under different filters, which enhanced the security of this biological information.
Fig. 5.
(A and B) Schematic and digital images of the encrypted information on PhC arrays without filter. (C and D) Schematic and digital images of the decoded “CHN” pattern on PhC arrays under a blue filter. (E and F) Schematic and digital images of the decoded “SEU” pattern on PhC arrays under a green filter. (G and H) Schematic and digital images of the decoded “MOC” pattern on PhC arrays under a red filter.
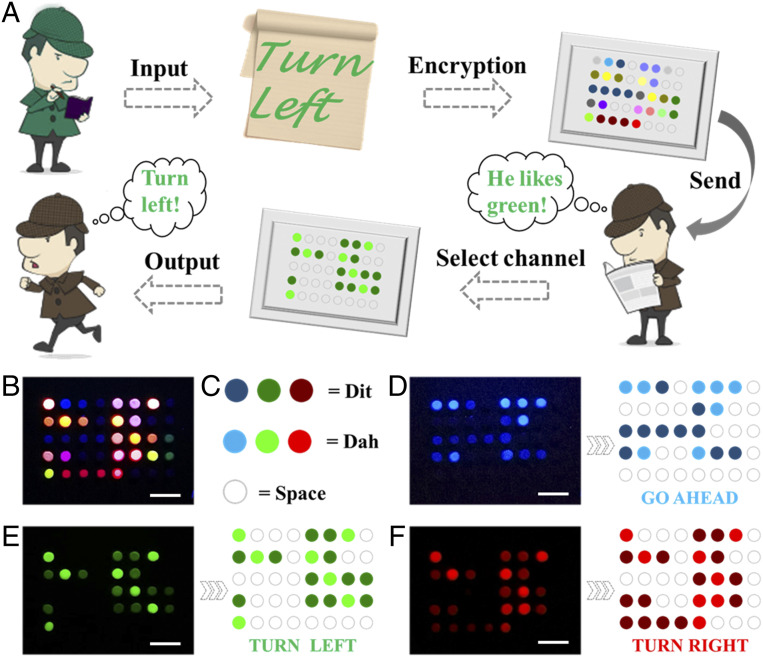
Under the action of the optical filter, only one kind of fluorescence signal corresponding to the filter can be observed. In this case, the fluorescence intensity could serve as an encoding element. Inspired by Morse code, we set each array of four dots as a coding unit, and the corresponding relationship was shown in SI Appendix, Table S2. According to the encoding rules, information contained could be easily read out. However, different fluorescence channels hid different information, which should be distinguished and observed under different optical filters. Therefore, not only the encoding rules, but also the right channels were necessary for decoding. For instance, Detective Green transcodes the real password “TURN LEFT” and the other two disguised passwords in Morse code fashion (Fig. 6A), where the true password corresponds to green fluorescence channel, and the camouflage passwords correspond to red and blue channels. Subsequently, the encrypted fluorescent array is sent to Detective Brown. Once he selects the corresponding channel, he could decode the message to obtain the correct password. Here, the weak fluorescence, strong fluorescence, and no fluorescence were defined as Dit, Dah, and space, respectively (Fig. 6C). Without separating channels, the array appears colorful and cannot be decoded (Fig. 6B). Only after separating the channels, the password “GO AHEAD,” “TURN LEFT,” and “TURN RIGHT” can be translated separately (Fig. 6 D–F).
Fig. 6.
(A) Schematic illustration of the bioinformation coding strategy. (B–F) Fluorescent patterns observed under different channels and their corresponding bioinformation, (B) invalid information, (C) the meaning of different intensities of fluorescence, (D) “GO AHEAD,” (E) “TURN LEFT,” (F) “TURN RIGHT.” (Scale bars are 5 mm in B–F.)
This bioinformation coding strategy was extremely secure because the data contained in these MXene-integrated PhC arrays were protected by many different protection strategies. First, oligonucleotide was used as the basis of the whole bioinformation coding and avoided the problem that long gene sequences were easy to break down and hard to store. Besides, the usage of base complementary pairing principle and the fluorescence signal as the coding unit made the decoding more accurate and easy to read. Second, different coding rules could be used to interpret a variety of results. This kind of algorithm encryption could develop different corresponding relations for each information coding, making the coding uncertain and greatly avoiding the possibility of being cracked. The final safeguard was the reliance on selective readability of optical filters. In the absence of an optical filter, the coded pattern looked haphazard. Different optical filters could read different information, and only the right channel could achieve the corresponding information. The combination of these three encryption methods greatly improved the security level of this bioinformation coding strategy.
Discussion
In summary, we have developed an innovative bioinspired MXene-integrated PhC array for multichannel bioinformation coding. Inspired by Stenocara beetle, a hydrophilic–hydrophobic patterned substrate was successfully prepared. Through the adhesion of DA, a PhC array was tightly fixed on the hydrophilic region of the substrate, followed by the integration of MXene nanosheets for subsequent bioinformation coding. The highly precise matching of oligonucleotides based on the complementary pairing principle ensured great safety. In addition, the conversion of coding elements into fluorescence signals overcame the complex and difficult decoding problem of traditional bioinformation coding. By applying and combining different fluorescence signals, multichannel bioinformation coding was realized, which improved throughput and security of information. It is worth mentioning that the functional elements utilized in this strategy are at the submicron level, which makes effective bioinformation coding feasible from micro to macro scale. These results showed that the proposed coding platform had great potential in data storage, transmission, and encryption, which was expected to open up a horizon in the field of bioinformation coding.
Materials and Methods
Materials.
Glass slide was obtained from Feizhou Co., Ltd. Stainless steel mask was purchased from Mirotech Technology Co., Ltd. Single-layer MXene dispersion (Ti3C2, 2 mg mL−1) was derived from Nanjing XF Nano Material Tech Co., Ltd. QD-labeled oligonucleotides were synthesized by Ruizhen Biotech Co., Ltd. Ethyl orthosilicate (TEOS), ethanol, ammonia, DA, NaOH, and (Heptadecafluoro-1,1,2,2-tetradecyl) trimethoxysilane were brought from Aladdin Reagent Co., Ltd. Silica colloidal solutions were synthesized by ourselves. Milli‐Q ultrapure water machine with ultraviolet (UV) sterilization (Millipore) was used throughout the experiment.
Preparation of SiO2 of Different Sizes.
TEOS (1.6 mL) was dissolved in anhydrous ethanol (44 mL) to get solution A, ammonia (10 mL) was dissolved in anhydrous ethanol (44 mL) to get solution B, and TEOS (2 mL) was dissolved in anhydrous ethanol (88 mL) to get solution C. Add solution A and solution B to the flask with mechanical stirring at a speed about 350 rpm. After 30 min of reaction, 8 mL ammonia was added in flask. After another 5 min of reaction, solution C was added to the system. The reaction continued for 2 h and stopped. SiO2 colloidal nanoparticles with different particle sizes could be obtained by changing the amount of TEOS in solution A.
Preparation of MXene-Integrated PhC Arrays.
Glass slides were immersed in a solution of 0.1 M NaOH for 0.5 h at room temperature. The slides were then evaporated and plated with 1% (vol/vol) (Heptadecafluoro-1,1,2,2-tetradecyl) trimethoxysilane dichloromethane solution. After hydrophobic treatment, cover the glass slides with a metal mask and put them into plasma for 15 s. Tris⋅HCl buffer (pH 8.5) containing DA (2 mg/mL) was utilized for forming adhesive layers on the glass slide. Twenty percent silica colloidal solutions (wt/wt) was then dripped on DA arrays to self-assemble into photonic crystals. MXene dispersion (2 mg/mL) containing DA was then incubated with PhC arrays at 20 °C for 0.5 h. Then the MXene-integrated PhC arrays were successfully fabricated.
Bioinformation Coding Based on Oligonucleotides.
Three kinds of 100 nM QD-labeled oligonucleotides solution were dripped on each dot of the MXene-integrated PhC array, and fluorescence signals were quenched. Then the mixed saturated solution (1 μM) of the three targets was added on the dots to restore the fluorescence.
Characterization.
The microstructures were characterized by a scanning electron microscope (SEM, Hitachi, S-300N). Fluorescence images were taken by the metallographic microscope (Olympus, BX51) equipped with a CCD camera (Olympus, DP30BW). The reflectance spectra were recorded by the same microscope equipped with a fiber-optic spectrometer (Ocean Optics, USB 2000+). Fluorescence images of cross-section were snapped by a Laser Scanning Confocal Microscope (Carl Zeiss, LSM510). The fluorescence intensity was detected by a fluorescence microscope (Olympus, CKX41) equipped with the same optic spectrometer.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grants 61927805 and 51522302), the National Natural Science Foundation of China (NSAF) Foundation of China (Grant U1530260), the Natural Science Foundation of Jiangsu Province (Grant BE2018707), the Scientific Research Foundation of Southeast University, and the Scientific Research Foundation of the Graduate School of Southeast University.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2011660117/-/DCSupplemental.
Data Availability.
All data are contained in the manuscript text and SI Appendix.
References
- 1.Bancrof C., Bowler T., Bloom B., Clelland C. T., Long-term storage of information in DNA. Science 293, 1763–1765 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Goldman N.et al., Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature 494, 77–80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Church G. M., Gao Y., Kosuri S., Next-generation digital information storage in DNA. Science 337, 1628 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Grass R. N., Heckel R., Puddu M., Paunescu D., Stark W. J., Robust chemical preservation of digital information on DNA in silica with error-correcting codes. Angew. Chem. Int. Ed. Engl. 54, 2552–2555 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Erlich Y., Zielinski D., DNA Fountain enables a robust and efficient storage architecture. Science 355, 950–954 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Li H.et al., Amplification of fluorescent contrast by photonic crystals in optical storage. Adv. Mater. 22, 1237–1241 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y. S., Zhu C., Xia Y., Inverse opal scaffolds and their biomedical applications. Adv. Mater. 29, 1701115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu W., Li Z., Yin Y., Colloidal assembly approaches to micro/nanostructures of complex morphologies. Small 14, e1801083 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Cai L.et al., Stomatocyte structural color-barcode micromotors for multiplex assays. Natl. Sci. Rev. 7, 644–651 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi J.et al., Bioinspired dynamic structural color‐based cryptographic surface. Adv. Opt. Mater. 8, 1901259 (2020). [Google Scholar]
- 11.Li Z., Yin Y., Stimuli-responsive optical nanomaterials. Adv. Mater. 31, e1807061 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Bian F.et al., Bioinspired photonic barcodes with graphene oxide encapsulation for multiplexed MicroRNA quantification. Small 14, e1803551 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Hou J., Li M., Song Y., Patterned colloidal photonic crystals. Angew. Chem. Int. Ed. Engl. 57, 2544–2553 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Bian F.et al., Colloidal crystals from microfluidics. Small 16, e1903931 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Qin M., Sun M., Hua M. T., He X. M., Bioinspired structural color sensors based on responsive soft materials. Curr. Opin. Solid State Mater. Sci. 23, 13–27 (2019). [Google Scholar]
- 16.Zhang H.et al., Immunotherapeutic silk inverse opal particles for post-surgical tumor treatment. Sci. Bull. (Beijing) 65, 380–388 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Zhou T.et al., Super-tough MXene-functionalized graphene sheets. Nat. Commun. 11, 2077 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar S., Lei Y., Alshareef N. H., Quevedo-Lopez M. A., Salama K. N., Biofunctionalized two-dimensional Ti3C2 MXenes for ultrasensitive detection of cancer biomarker. Biosens. Bioelectron. 121, 243–249 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Manzanares-Palenzuela C. L.et al., Interaction of single- and double-stranded DNA with multilayer MXene by fluorescence spectroscopy and molecular dynamics simulations. Chem. Sci. (Camb.) 10, 10010–10017 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohammadniaei M., Koyappayil A., Sun Y., Min J., Lee M. H., Gold nanoparticle/MXene for multiple and sensitive detection of oncomiRs based on synergetic signal amplification. Biosens. Bioelectron. 159, 112208 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Liu L., Wei Y., Jiao S., Zhu S., Liu X., A novel label-free strategy for the ultrasensitive miRNA-182 detection based on MoS2/Ti3C2 nanohybrids. Biosens. Bioelectron. 137, 45–51 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Bian F.et al., Molybdenum disulfide-integrated photonic barcodes for tumor markers screening. Biosens. Bioelectron. 133, 199–204 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Peng X., Zhang Y., Lu D., Guo Y., Guo S., Ultrathin Ti3C2 nanosheets based “off-on” fluorescent nanoprobe for rapid and sensitive detection of HPV infection. Sens. Actuators B Chem. 286, 222–229 (2019). [Google Scholar]
- 24.Bian F. K.et al., Binary optical barcodes for label-free multiplex detection based on molybdenum disulfide composites. Compos. Commun. 16, 136–142 (2019). [Google Scholar]
- 25.Parker A. R., Lawrence C. R., Water capture by a desert beetle. Nature 414, 33–34 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Hou J.et al., Bio-inspired photonic-crystal microchip for fluorescent ultratrace detection. Angew. Chem. Int. Ed. Engl. 53, 5791–5795 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Lee H., Dellatore S. M., Miller W. M., Messersmith P. B., Mussel-inspired surface chemistry for multifunctional coatings. Science 318, 426–430 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang S. M.et al., Simultaneous reduction and surface functionalization of graphene oxide by mussel‐inspired chemistry. Adv. Funct. Mater. 21, 108–112 (2011). [Google Scholar]
- 29.Cui W.et al., A strong integrated strength and toughness artificial nacre based on dopamine cross-linked graphene oxide. ACS Nano 8, 9511–9517 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Deng J.et al., A bioinspired medical adhesive derived from skin secretion of Andrias davidianus for wound healing. Adv. Funct. Mater. 29, 1809110 (2019). [Google Scholar]
- 31.Wu J.et al., Natural hydrogel in American lobster: A soft armor with high toughness and strength. Acta Biomater. 88, 102–110 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Li M.et al., Ultrasensitive DNA detection using photonic crystals. Angew. Chem. Int. Ed. Engl. 47, 7258–7262 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Xiao M.et al., Bio-inspired structural colors produced via self-assembly of synthetic melanin nanoparticles. ACS Nano 9, 5454–5460 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Hou J.et al., Four-dimensional screening anti-counterfeiting pattern by inkjet printed photonic crystals. Chem. Asian J. 11, 2680–2685 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Li Y.et al., Patterned photonic crystals for hiding information. J. Mater. Chem. C Mater. Opt. Electron. Devices 5, 4621–4628 (2017). [Google Scholar]
- 36.Bian F., Wang H., Sun L., Liu Y., Zhao Y., Quantum-dot-encapsulated core-shell barcode particles from droplet microfluidics. J. Mater. Chem. B Mater. Biol. Med. 6, 7257–7262 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Zhang M.et al., Ultrasmooth quantum dot micropatterns by a facile controllable liquid-transfer approach: Low-cost fabrication of high-performance QLED. J. Am. Chem. Soc. 140, 8690–8695 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Li X.et al., Continuous and controllable liquid transfer guided by a fibrous liquid bridge: Toward high-performance QLEDs. Adv. Mater. 31, e1904610 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Bian F., Sun L., Cai L., Wang Y., Zhao Y., Quantum dots from microfluidics for nanomedical application. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 11, e1567 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained in the manuscript text and SI Appendix.