Significance
CRISPR-Cas genome engineering has enabled scientists to modify the genome of the popular model organism Drosophila melanogaster with unprecedented ease. However, so far only the CRISPR nuclease Cas9 has been used for this task. Additional nucleases could expand the genomic target space and facilitate modification of independent cell populations in the same organism. Here we describe genome editing with Cas12a in Drosophila. Cas12a recognizes target sites orthogonal to Cas9, can use compact arrays of crRNAs for multiplexed gene editing, and conditional expression leads to tissue-specific mutagenesis. We also show that a point mutant in Cas12a substantially increases editing rates. This work significantly expands the CRISPR toolbox in Drosophila and will inform the development of similar systems in other organisms.
Keywords: Cas12a, Drosophila, CRISPR, genome engineering, Cas9
Abstract
CRISPR-Cas genome engineering has revolutionized biomedical research by enabling targeted genome modification with unprecedented ease. In the popular model organism Drosophila melanogaster, gene editing has so far relied exclusively on the prototypical CRISPR nuclease Cas9. Additional CRISPR systems could expand the genomic target space, offer additional modes of regulation, and enable the independent manipulation of genes in different cells of the same animal. Here we describe a platform for efficient Cas12a gene editing in Drosophila. We show that Cas12a from Lachnospiraceae bacterium, but not Acidaminococcus spec., can mediate robust gene editing in vivo. In combination with most CRISPR RNAs (crRNAs), LbCas12a activity is high at 29 °C, but low at 18 °C, enabling modulation of gene editing by temperature. LbCas12a can directly utilize compact crRNA arrays that are substantially easier to construct than Cas9 single-guide RNA arrays, facilitating multiplex genome engineering. Furthermore, we show that conditional expression of LbCas12a is sufficient to mediate tightly controlled gene editing in a variety of tissues, allowing detailed analysis of gene function in a multicellular organism. We also test a variant of LbCas12a with a D156R point mutation and show that it has substantially higher activity and outperforms a state-of-the-art Cas9 system in identifying essential genes. Cas12a gene editing expands the genome-engineering toolbox in Drosophila and will be a powerful method for the functional annotation of the genome. This work also presents a fully genetically encoded Cas12a system in an animal, laying out principles for the development of similar systems in other genetically tractable organisms for multiplexed conditional genome engineering.
Understanding the development of multicellular organisms and the mechanisms that govern their health and disease requires detailed knowledge of the underlying genetic loci. Research in genetically tractable model organisms has been particularly powerful for functional genome annotation as it allows experimentally introducing genetic variants and observing the resulting phenotype. Chemical, biological, and physical methods to induce mutations in the genome have been used for many decades, but the fact that these methods introduce genetic variation at random makes systematic gene discovery inefficient. As a result, functionally characterized genetic alleles have so far been described only for a minority of genes (1). RNAi constructs for gene knockdown have partially filled this gap, but are themselves limited by off-target effects and residual gene expression (2). The efficient targeted introduction of mutations at specific loci would facilitate a more systematic assessment of the functional roles of genomic elements, but methods for such genome engineering have become broadly accessible only relatively recently. This has been largely driven by the discovery that the prokaryotic CRISPR-Cas system can be easily programmed to induce DNA double-strand breaks (DSBs) at defined loci in the genome of a large variety of organisms (3–5).
In the widely used model organism Drosophila melanogaster, CRISPR-Cas genome engineering has so far almost exclusively relied on the use of the RNA-guided DNA endonuclease Cas9 from Streptococcus pyogenes (SpCas9) (6). When expressed from a stable transgenic source, SpCas9 mediates highly efficient induction of targeted DSBs in the Drosophila genome, which often lead to small insertions or deletions (Indels) at the target locus or can be harnessed for the precise introduction of novel sequences through homology-directed repair (7–10). However, the use of Cas9 for mutagenesis is limited to target sites adjacent to a cognate protospacer adjacent motif (PAM), which in the case of SpCas9 is NGG (4). Furthermore, Cas9-induced mutations are predominantly small (1 to 10 bp) deletions, which when occurring in-frame often do not disrupt gene function. In addition, a single genome-engineering system is not sufficient to independently engineer target loci in different cell types of the same organism, a prerequisite to take full advantage of an in vivo model to study complex organismal biology. Therefore, there exists a need for additional genome-engineering systems in Drosophila, which can be used instead or in parallel to Cas9.
CRISPR-Cas12a (formerly known as Cpf1) is a class 2 type V CRISPR system that has been adopted for genome engineering in several organisms (11–14). Cas12a is an RNA-guided DNA endonuclease with a number of attributes distinct from Cas9, making it an interesting candidate to complement the CRISPR toolbox also in Drosophila. First, the target space for Cas12a-mediated genome editing is nonoverlapping with that of Cas9, as it requires a TTTV PAM (11, 15). Second, Cas12a mediates a staggered DSB, which has been suggested to lead to distinct DNA repair outcomes (11, 16). Third, Cas12a only requires a short CRISPR RNA (crRNA) for gene targeting and has RNase activity to process single crRNAs from a crRNA array, a potential advantage for multiplex genome engineering (17–19). The most commonly used Cas12a enzymes originate from Acidaminococcus (AsCas12a) and Lachnospiraceae bacterium (LbCas12a) and both enzymes function with comparable efficiency in mammalian cells (11, 20). We previously assessed Cas12a performance in Drosophila, but focused exclusively on AsCas12a, and reported low activity compared to SpCas9 (21). A subsequent study in zebrafish and Xenopus demonstrated that Cas12a activity is strongly temperature-dependent and showed that AsCas12a has low activity at temperatures below 30 °C (12). This is likely to limit the utility of AsCas12a in Drosophila, which do not tolerate long-term exposure to temperatures above 30 °C. However, in fish and frogs LbCas12a was found to have higher activity at low temperatures, raising the possibility that LbCas12a might efficiently function as RNA-guided DNA endonuclease also in flies (12).
Here, we report efficient multiplexed and conditional Cas12a genome engineering in Drosophila. We show that LbCas12a, in contrast to AsCas12a, mediates robust mutagenesis of endogenous target genes. With most, but not all, crRNAs, LbCas12a activity is strongly influenced by temperature, being inactive at 18 °C and becoming active at 29 °C, and thus enabling temperature-modulated genome editing. We also show that LbCas12a can utilize compact crRNA arrays for multiplex genome engineering and demonstrate that tissue-specific expression of LbCas12a is sufficient for spatially restricted mutagenesis in vivo. Finally, we test a point mutant variant of LbCas12a (D156R) and find that it has substantially higher activity than the wild-type enzyme and outperforms a current Cas9-based system in identifying essential genes. The tools presented here significantly expand the genome-engineering toolbox in Drosophila and enable targeting of previously inaccessible genomic target sites, facilitate simultaneous mutagenesis of more than two loci and pave the way for the independent manipulation of different cell populations in the same organism.
Results and Discussion
Temperature-Controlled Cas12a Gene Editing.
In order to systematically characterize LbCas12a and AsCas12a for genome engineering applications in Drosophila, we established a toolbox for the expression of each Cas12a enzyme and its cognate crRNAs in vivo (Fig. 1 A and B). We previously described a plasmid for the expression of AsCas12a crRNAs (pCFD7) and a transgenic fly line expressing AsCas12a under the ubiquitous act5C promoter (21). We generated an equivalent expression plasmid for LbCas12a crRNAs (pCFD8) and an act5c-LbCas12a transgenic fly line, which has the transgene located at the same genomic landing site as the AsCas12a line (Fig. 1B). Plasmids pCFD7 and pCFD8 both express crRNAs from the strong, ubiquitous U6:3 promoter, but differ in the repeat-derived 5′ stem-loop of the crRNA, which is unique for each enzyme. Importantly, AsCas12a and LbCas12a require the same PAM sequence (11), enabling direct comparisons of the two enzymes using crRNAs targeting the exact same target site.
Fig. 1.
LbCas12a mediates efficient temperature-sensitive gene editing in Drosophila. (A) Schematic of the Cas12a genome engineering system. Cas12a possesses RNase activity and can cleave crRNA arrays into individual crRNAs. Upon on-target binding, Cas12a cuts dsDNA, producing DSBs with 5′ overhangs. These are often imprecisely repaired by the endogenous DNA repair machinery, resulting in mutations at the target locus. (B) Schematic of the workflow of experiments presented in C and D. Single Cas12a crRNAs are cloned into expression vectors pCFD7 (for AsCas12a) or pCFD8 (for LbCas12a). Plasmids are integrated at defined genomic locations and crossed to the respective nuclease. Offspring inheriting Cas12a and crRNA transgenes are raised at different temperatures and assessed for on-target gene editing. (C) Cas12a-mediated mutagenesis of ebony (e) at different temperatures. Images of flies expressing crRNA-e and either AsCas12a or LbCas12a and raised at the indicated temperatures are shown. Disruption of e results in dark coloration of the cuticle. AsCas12a mediates efficient mutagenesis only at 29 °C, while LbCas12a mediates gene disruption at all tested temperatures. (D) Assessment of gene-editing efficiency of 11 target sites by Amp-seq. Activity of each nuclease was tested at 18 °C, 25 °C, and 29 °C. Genomic target sites of each crRNA were PCR amplified and amplicon pools were subjected to deep sequencing. Gene-editing activity was strongly temperature-sensitive with both nucleases. LbCas12 displayed much more robust activity, mediating strong mutagenesis with 7 of 11 crRNAs at 29 °C, compared to 2 of 11 in the case of AsCas12a. Rates of modified reads being called due to sequencing errors was between 0.5% and 1.6% (SI Appendix, Fig. S1).
We first combined the two Cas12a enzymes with crRNAs transgenes targeting a characterized site in ebony (e, crRNA-e), which we previously found to be susceptible to AsCas12a-induced gene editing in the germline with low efficiency when flies were reared at 25 °C (21). Flies transgenic for act-Cas12a and U6:3-crRNA-e, or other crRNAs targeting nonessential genes (see below), are viable and fertile, demonstrating that genetically encoded Cas12a is well tolerated in Drosophila. Consistent with previous results, act-AsCas12a U6:3-crRNA-e flies at 25 °C had normal coloration of the cuticle, as did flies of the same genotype raised at the lower temperature of 18 °C, indicating little or no disruption to e (Fig. 1C). In contrast, act-AsCas12a U6:3-crRNA-e flies reared at 29 °C had dark cuticle typical of e loss-of function mutants, suggesting efficient mutagenesis of e at this higher temperature. Interestingly, flies expressing LbCas12a and crRNA-e had ebony body coloration regardless of the temperature at which they were raised, suggesting that in Drosophila LbCas12a has higher activity at low temperatures than AsCas12a (Fig. 1C).
To evaluate if such differences in the activity of the two Cas12a enzymes at different temperatures are observed more generally, we generated 10 additional pairs of crRNA transgenes, 8 targeting additional sites in e, as well as 1 crRNA targeting yellow (y) and 1 crRNA targeting sepia (se) (SI Appendix, Table S1). We then tested these in combination with their cognate Cas12a enzyme at either 18 °C, 25 °C, or 29 °C. To quantitatively assess the amount of gene editing with high sensitivity, we performed deep sequencing of PCR amplicons spanning the crRNA target sites. AsCas12a did not result in detectable gene editing at any site when flies were reared at 18 °C and only mediated medium to high editing in combination with 2 of the 11 crRNAs at 29 °C (Fig. 1D; see SI Appendix, Fig. S1 for negative control). In contrast, LbCas12a displayed much higher activity at 29 °C, resulting in efficient gene editing when combined with 7 of the 11 crRNAs (Fig. 1D). Activity of LbCas12a was markedly reduced when animals were raised at 25 °C and very low or absent at 18 °C (Fig. 1D). The exception was crRNA-e, which mediated high levels of mutagenesis at all three temperatures (Fig. 1D). Amplicon sequencing (Amp-seq) data were consistent with the level of visible phenotypes observed in adult flies (SI Appendix, Fig. S2). We also tested the temperature dependency of Cas9-mediated gene editing with seven single-guide RNA (sgRNA) targeting genes with visible phenotypes and did not observe any difference in phenotypic outcomes, suggesting that Cas9 functions with similar efficiency at temperatures between 18 °C and 29 °C (SI Appendix, Fig. S3). Together, these experiments establish that the efficiency of Cas12a genome engineering with transgenic components in Drosophila is strongly temperature dependent. LbCas12a displays significantly higher activity than AsCas12a in the temperature range tolerated by Drosophila and in combination with many, but not all, crRNAs LbCas12a can be switched from very low to high activity by increasing the temperature from 18 °C to 29 °C. LbCas12a is therefore a powerful CRISPR effector for inducible genome editing in Drosophila and we focused on this nuclease for the remainder of this study.
Targeted deep-sequencing of induced on-target edits also allowed us to profile the mutations induced by LbCas12a. Each crRNA resulted in a specific mutagenesis pattern dominated by deletions. These deletions most commonly ranged in size between 10 and 15 bp (SI Appendix, Fig. S4). These results confirm and expand previous observations by us and others that Cas12a induces mutations that are significantly larger than those induced by Cas9 (21–23). Larger deletions are often desirable as they have a higher chance to impair the function of genetic elements.
Multiplexed Gene Editing with crRNA Arrays.
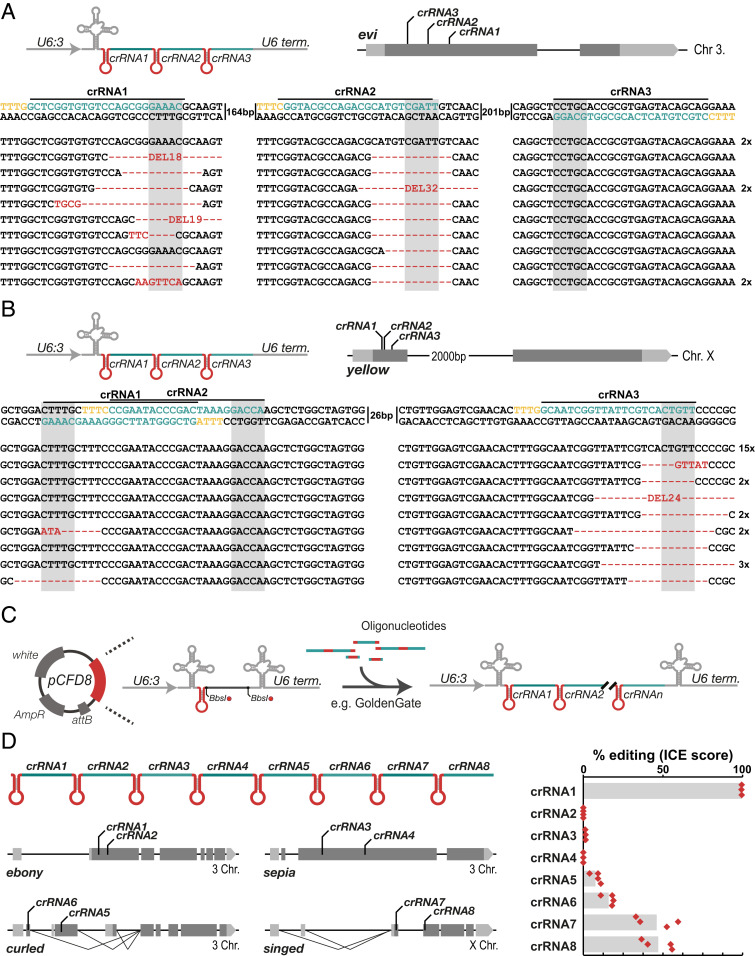
Cas12a has been shown to possess RNase activity and be able to excise single crRNAs from a multiplexed crRNA array (17). This substantially simplifies multiplex gene editing compared to the Cas9 system, which requires additional functional elements, such as tRNAs or ribozymes, to produce multiple sgRNAs from a single transcript (21, 24, 25). Furthermore, Cas12a crRNAs are significantly smaller than Cas9 sgRNAs, which facilitates the cloning of such multiplexed arrays. To test if LbCas12a is able to simultaneously edit several target sites in the Drosophila genome in combination with an array of crRNAs encoded on the same transcript, we generated transgenic flies harboring an array of three crRNAs targeting either the Wnt secretion factor evenness interrupted (evi/wls) or the pigmentation gene y. Flies transgenic for act-LbCas12a and either U6:3-crRNA-evi3x or U6:3-crRNA-y3x raised at 29 °C died at pupal stage or developed yellow pigmentation, respectively, indicating efficient gene disruption. To test if these phenotypes resulted from the activity of several crRNAs in an array, we extracted genomic DNA from act-LbCas12a pCFD8-crRNA3x animals, amplified the target locus, and cloned and sequenced individual PCR products. Several PCR amplicons contained mutations at two crRNA target sites, indicating that crRNAs encoded in an array are correctly utilized to mutate several target sites in parallel (SI Appendix, Fig. S5). Using this assay, we also frequently detected deletions either between target sites or encompassing the entire target region and not starting or ending at a crRNA target site (SI Appendix, Fig. S5). The latter type of deletions is rarely observed in CRISPR mutagenesis experiments, although similar deletions have been described to sometimes occur in Cas9-mediated gene-editing in mammalian cells (26). To test if large deletions are commonly induced by multiplex gene editing of evi and y or if they might be artificially enriched by subcloning PCR amplicons, we assayed mutations induced by the same crRNA arrays in the germline. We did not detect large deletions using this assay, indicating they are likely overrepresented when assaying mutations by cloning PCR amplicons (SI Appendix, Fig. S5). Importantly, germline editing confirmed activity by more than one crRNA per array, indicating that also in Drosophila arrays are processed into individual crRNAs and loaded into Cas12a (Fig. 2 A and B).
Fig. 2.
Multiplexed gene editing through crRNA arrays. (A) Gene editing of evi with three arrayed crRNAs. crRNAs were expressed from a U6:3 promoter and combined with nos-Gal4 UAS-LbCas12a transgenes for germline-restricted mutagenesis. Flies were raised at 29 °C and viable, indicative of efficient restriction of gene disruption to the germline. nos-Gal4 UAS-LbCas12a pCFD8-evi3x males were crossed to wild-type virgin females and genomic DNA from individual offspring was subjected to PCR amplification of the evi locus. PCR amplicons were sequenced and sequencing traces were parsed into wildtype and alternative allele sequences (Materials and Methods). The majority of animals harbor mutant evi alleles modified at the target sites of crRNA1 and crRNA2. Edited sequences are indicated in red and the crRNA target site is indicated in green, the PAM in yellow, and the window in which LbCas12a is expected to cut is shaded in gray. Indels at individual target sites are typically larger than 10 bp, further supporting data presented in SI Appendix, Fig. S4. (B) Gene editing of y with three arrayed crRNAs. The experiment was conducted as described above using a pCFD8-y3x transgene. Sequencing of PCR amplicons revealed highly efficient gene editing at the target site of the third crRNA in the array and editing with low efficiency at the target site of the first crRNA. No mutations were detected at the target site of the second crRNA. (C) Due to the compact size of Cas12a, several crRNAs can be encoded on commercially available oligonucleotides and oligos can be fused to construct larger crRNA arrays encoded in pCFD8 (see SI Appendix, Supplementary Cloning Protocol). (D) Multiplex gene targeting of four genes with two crRNAs each. Flies transgenic for a 8x crRNA array and act5C-LbCas12a were raised at 29 °C and had ebony cuticle and bristles displaying the singed phenotype. Each target locus was PCR amplified and amplicons were subjected to Sanger sequencing. Efficiency of gene editing was inferred from sequencing traces by ICE analysis (Materials and Methods). High levels of activity were detected for crRNA1 targeting e, crRNAs 7 and 8 had intermediate activity and crRNAs 5 and 6 showed low to medium activity. crRNAs 2 to 4 were inactive. crRNAs 1 (crRNA-e in Fig. 1 C and D), 2 (crRNA-e6) and 4 (crRNA-se) were in parallel also tested when expressed as single crRNAs in pCFD8 (Fig. 1D) with consistent results.
Next, we set out to test if a larger crRNA array would also be efficiently utilized by LbCas12a and could be used to edit several genes in parallel. Creation of larger crRNA arrays is significantly helped by the fact that multiple crRNAs can be encoded on commercially available oligonucleotides and oligos can be efficiently assembled into crRNA arrays in pCFD8 (Materials and Methods and Fig. 2C). We generated a pCFD8 plasmid containing eight crRNAs, targeting the genes e, se, curled (cu), and singed (sn) with two crRNAs per gene. We crossed pCFD8-crRNA-e2x-se2x-cu2x-sn2x transgenic flies to act-LbCas12a flies and incubated the flies at 29 °C. The offspring had ebony cuticle and bristles displaying the sn loss-of-function phenotype, but did not have sepia eye coloration or curled wings. To directly monitor the editing efficiency at each target site, we extracted genomic DNA, amplified the target sites, and sequenced the resulting PCR amplicons. Deconvolving the Sanger sequencing chromatograms (Materials and Methods) revealed that three target sites were edited with high efficiency, two sites with low efficiency, and editing was undetectable at three sites (Fig. 2D). Of the three inactive crRNAs, two were also tested as individual crRNAs in pCFD8 and also showed no activity in this configuration (Fig. 1D and SI Appendix, Table S1), suggesting that low activity in these cases is intrinsic to the crRNA and not caused by their presence in a larger array.
Together, these results demonstrate that in Drosophila LbCas12a can directly utilize arrays of several crRNAs to mediate multiplexed genome editing.
Tissue-Specific Cas12a Gene Editing.
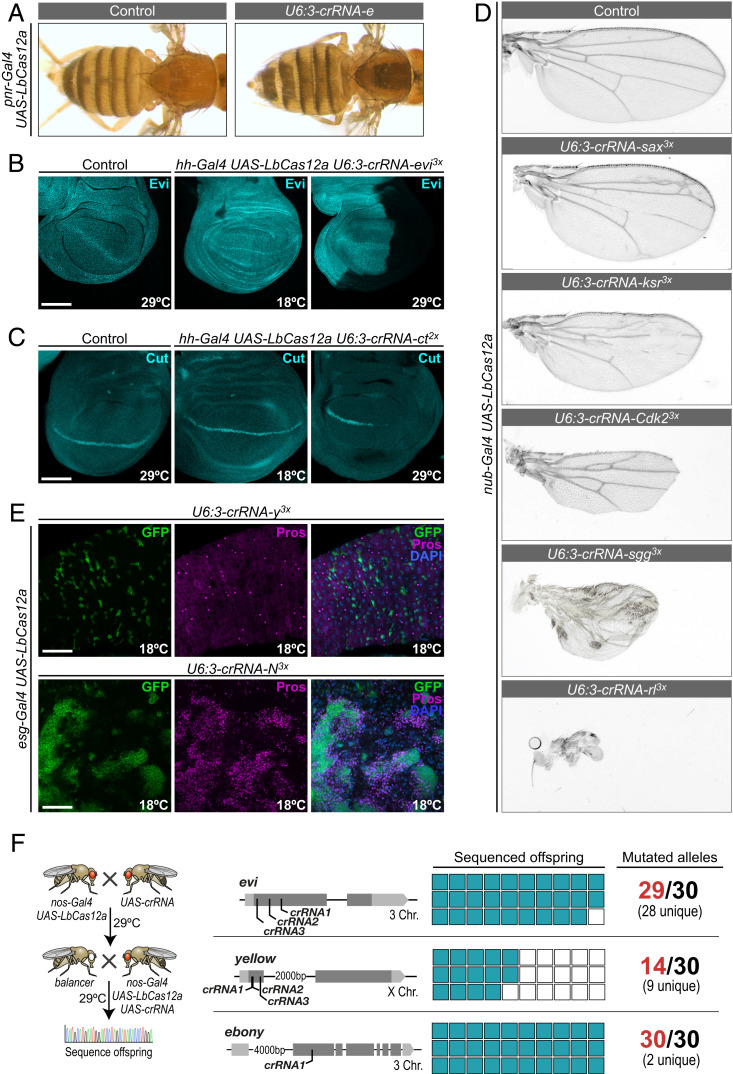
Dissection of the often-multifaceted functions of genes in multicellular organisms requires the ability to manipulate their sequence with spatial and temporal precision in vivo. We therefore explored whether LbCas12a could be utilized for tissue-specific mutagenesis in Drosophila. Conditional gene editing is typically achieved by linking the expression of CRISPR components to cis-regulatory elements with tissue-specific expression patterns. In Drosophila this is most commonly done via the binary Gal4-UAS system (27). We generated UAS-LbCas12a flies and combined them with a range of Gal4 drivers, resulting in tissue-specific LbCas12a expression (SI Appendix, Fig. S6). We then crossed them with animals transgenic for U6:3-crRNAs to induce gene editing in Gal4-expressing cells of the offspring. First, we generated pnr-Gal4 UAS-LbCas12a U6:3-crRNA-e flies, which displayed dark cuticle exclusively in the pnr-Gal4 expression domain along the dorsal midline, indicative of disruption of e exclusively in Gal4 expressing cells (Fig. 3A).
Fig. 3.
Tissue-specific gene editing with UAS-LbCas12a. (A) LbCas12a-mediated mutagenesis of e in the pnr-Gal4 expression domain along the dorsal midline leads to the development of dark pigment in the cuticle. Flies were raised at 29 °C. (B) Mutagenesis of evi in the posterior compartment of the wing imaginal disc with hh-Gal4. Evi protein is detected throughout the wing imaginal disc in control and hh-Gal4 UAS-LbCas12a U6:3-crRNA-evi3x animals raised at 18 °C, but absent from the posterior compartment in animals raised at 29 °C, demonstrating efficient, temperature-sensitive, and spatially defined gene disruption. (Scale bars, 50 μm.) (C) Mutagenesis of ct in the posterior compartment with hh-Gal4. Ct protein is detected in a stripe of cells in control and hh-Gal4 UAS-LbCas12a U6:3-crRNA-evi3x animals raised at 18 °C, but absent from the posterior compartment in animals raised at 29 °C, demonstrating efficient, temperature-controlled and spatially defined gene disruption. (Scale bars, 50 μm.) (D) Tissue-specific mutagenesis of essential genes in the wing primordium with nub-Gal4. Wings from animals expressing nub-Gal4 UAS-LbCas12a and the indicated crRNA array are shown. When combined with ubiquitous act5C-LbCas12a these crRNAs lead to lethality, but tissue-restricted expression with nub-Gal4 gives rise to viable or semiviable flies. Wings of such animals display specific and informative phenotypes affecting wing size, vein formation, bristle formation, or wing margin formation. (E) Intestinal tumorigenesis induced by conditional LbCas12a mutagenesis of N. esg-Gal4 was used to express LbCas12a in adult intestinal stem cells. esg-Gal4 UAS-LbCas12a U6:3-crRNA-N3x animals raised at 29 °C are lethal before adulthood, consistent with the known expression of esg-Gal4 in larval and pupal tissues. Reducing the culture temperature to 18 °C leads to viable adults that develop intestinal tumors characterized by expression of pros, a hallmark of tumors caused by loss of N, indicating low level mutagenesis at this temperature. Mutagenesis of y was used as a control, as y does not play a role in intestinal stem cells. (Scale bars, 50 μm.) (F) Efficient Cas12a-mediated mutagenesis in the germline to create heritable alleles. Images in A–E are representative examples of at least nine samples from two independent experiments.
Next, we attempted conditional mutagenesis in wing imaginal discs. We used hh-Gal4 UAS-LbCas12a to knockout evi selectively in the posterior compartment and analyzed the outcome by directly visualizing Evi protein. Evi is ubiquitously expressed in wild-type tissue, but was undetectable in the P-compartment of wing discs of hh-Gal4 UAS-LbCas12a U6:3-crRNA-evi3x animals raised at 29 °C (20 of 22 discs) (Fig. 3B). Wing discs of animals of the same genotype raised at 18 °C expressed Evi throughout the wing disc and displayed only weak and infrequent reductions in Evi staining in the P-compartment of 7 of 13 discs (Fig. 3B), which likely reflect infrequent gene editing and possibly interference with efficient transcription by binding of the Cas12a–crRNA complex to the target gene (13). These results were further corroborated by targeting a second gene, cut (ct), a transcription factor expressed in a stripe of cells along the dorsal-ventral boundary (Fig. 3C). Mutagenesis of ct with hh-Gal4 UAS-LbCas12a at 29 °C lead to lethality at the third-instar larval stage. We made use of the temperature sensitivity of LbCas12a and raised animals for the first 24 h at 18 °C and then shifted them to 29 °C. These animals survived until pupal stages, allowing us to assess ct expression in late third-instar discs and highlighting the advantages of a temperature-sensitive gene-editing system. In 10 of the 13 analyzed wing discs, ct expression was completely lost exclusively in the hh-Gal4 expression domain, while in the remaining 3 discs a few ct-expressing cells remained (Fig. 3C). Mutagenesis was strongly suppressed in wing discs from animals raised at 18 °C, where ct expression was unaffected in five of nine discs and only few cells had lost ct in the remaining four samples (Fig. 3C).
Together, these experiments establish UAS-LbCas12a as a powerful tool for tissue-specific gene disruption. Interestingly, in all cases mutagenesis appeared strictly restricted to the hh-Gal4 expression domain. This is in contrast to Cas9, where leaky expression of Cas9 from UAS vectors results in ectopic gene editing in combination with U6:3-driven sgRNAs (21). In addition to conditional control of gene editing by controlling Cas12a transcription, it is possible to further control mutagenesis by changing the incubation temperature from 18 °C, typically resulting in low gene-editing efficiency, to 29 °C, where Cas12a is often highly active. Of note, the morphology of wing imaginal discs expressing an active Cas12a system was sometimes abnormal, with tissue folds forming at abnormal positions (SI Appendix, Fig. S7A). This was not caused by excessive amounts of cell death in these tissues (SI Appendix, Fig. S7B) and was independent of the target of the coexpressed crRNA. In the future, careful titration of Cas12a protein levels, as we have recently demonstrated with Cas9 (8), might alleviate these effects.
We also used an additional line, nub-Gal4 UAS-LbCas12a, to restrict mutagenesis to the pouch region of the wing imaginal disc and a few other tissues and tested it with crRNA arrays targeting essential genes. Adult flies emerged from crosses with 21 of the 23 tested crRNA transgenes, supporting tight restriction of gene editing to Gal4-expressing cells. Wings from such animals typically displayed highly specific phenotypes affecting their size, veins, bristles, texture, or margin, which can give valuable clues about the function of the target gene (Fig. 3D).
Next, we analyzed Cas12a-mediated mutagenesis in an adult tissue, the Drosophila midgut. Many Gal4 drivers are active at multiple stages during development, making transgene-mediated gene editing of essential genes in adults challenging. We reasoned that the temperature-sensitivity of the Cas12a system could be used to avoid premature lethality in such experiments. We generated an esg-Gal4 UAS-LbCas12a line, which is expressed in adult intestinal stem cells, as well as various embryonic, larval, and pupal tissues, and crossed it to a crRNA array targeting Notch (N). When esg-Gal4 UAS-LbCas12a U6:3-crRNA-N animals were reared at 29 °C they died at larval to pupal stage, consistent with efficient mutagenesis of N in esg-expressing cells during early development. In contrast, when animals of the same genotype were raised at 18 °C they developed to adulthood, demonstrating effective suppression of mutagenesis at this temperature. However, 14-d-old adults frequently displayed large tumors in their midgut, suggesting some disruption of N occurred at this temperature (Fig. 3E). These tumors were formed by prospero-expressing cells with a small nucleus, the known phenotype of tumors caused by loss of N (28), and were not observed in control tissue (Fig. 3E). This demonstrates how temperature can be used to tune Cas12a gene-editing rates to allow analysis of loss-of-function phenotypes in essential genes in later stages of life. However, as LbCas12a-mediated gene editing is typically inefficient, but not absent at 18 °C, it is difficult to exactly time the induction of individual genetic perturbations by this method.
Finally, we established Cas12a-mediated mutagenesis restricted to the germline, an important application allowing the creation of heritable alleles. We generated a nos-Gal4 UAS-LbCas12a fly line and tested it with crRNA arrays targeting evi and y and a single crRNA targeting e at 29 °C. Flies expressing crRNAs targeting evi were viable and animals with crRNAs targeting e or y had normal coloration of their cuticle, indicative of very low or no Cas12a activity in somatic tissues (SI Appendix, Fig. S8). In contrast, all three crRNA arrays mediated high levels of on-target mutagenesis in the germline, as revealed by sequencing of their target sites in individual offspring (Figs. 2 A and B and 3F). Together, these data demonstrate that germline-specific expression of LbCas12a allows the creation of heritable alleles in the germline without affecting viability of the animal.
Highly Efficient Gene Disruption by Cas12a+.
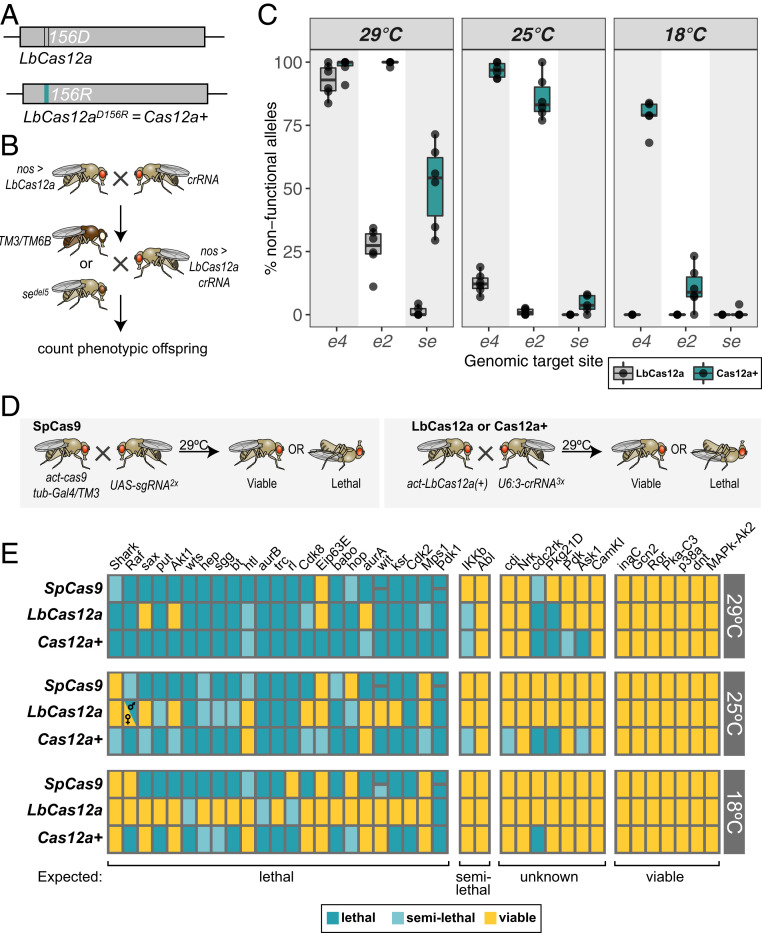
The experiments described so far establish LbCas12a as a potent genome-engineering tool in Drosophila. However, the robustness of the Cas12a system (i.e., the fraction of crRNAs mediating efficient editing of their target site) appears lower than what is typically observed with transgenic SpCas9 systems (8, 10, 29). However, this could potentially be compensated for by a larger degree of crRNA multiplexing, which is facilitated by the small size of such arrays. Furthermore, rationally designed point mutants of AsCas12a with increased activity have been described previously (30) and one such mutation, D156R, has recently been shown to also enhance the activity of LbCas12a in Arabidopsis (31). We generated a transgenic fly line expressing LbCas12aD156R under the ubiquitous act5c promoter (Fig. 4A). We then compared the activity of this variant with wild-type LbCas12a at different temperatures using three crRNAs that we had found earlier to have high, medium, and low activity with LbCas12a at 29 °C (Fig. 1D). For practical reasons, we assayed activity by genetic complementation of germline-induced alleles (Fig. 4B). Activity levels observed by this method were very similar to our earlier Amp-seq experiment, showing high, medium and low activity when LbCas12a crRNA animals were raised at 29 °C, low or no activity at 25 °C, and no activity at 18 °C (Fig. 4C). LbCas12aD156R mediated substantially higher levels of mutagenesis at all three temperatures and with all three crRNAs (Fig. 4C). For simplicity, we refer to LbCas12aD156R from now on as Cas12a+, referring to its strong increase in activity.
Fig. 4.
Highly efficient gene disruption mediated by Cas12a+. (A) Cas12a+ is a point mutant variant of LbCas12a harboring a D156R mutation. This variant is based on earlier work describing rationally designed variants of AsCas12a and has been shown to have increased activity in Arabidopsis (30, 31). (B) Scheme of the experimental workflow of the experiment shown in C. Gene-editing efficiency was measured by counting the number of nonfunctional alleles induced in the germline by testing for genetic complementation with known null alleles. Note that this method does not account for the induction of functional alleles. Levels of gene editing were in excellent agreement with levels measured by Amp-seq using the same crRNAs (Fig. 1D). (C) Cas12a+ induces much higher levels of gene editing than LbCas12a at all three target sites and at all tested temperatures. Dots represent the result of individual crosses of each condition and boxplots represent the 25% and 75% percentile and the median as a solid line. (D) Schematic of the experimental set-up of the experiment shown in E. Gene targeting by SpCas9 was performed using sgRNA transgenes of the Heidelberg CRISPR Fly Design library, which encode two sgRNAs expressed from the Gal4/UAS system (8). LbCas12a or Cas12a+ gene editing was induced with crRNA arrays encoding three crRNAs per gene encoded in pCFD8. Flies ubiquitously expressing either nuclease and the respective crRNA or sgRNAs were raised at 29 °C, 25 °C, or 18 °C and lethality was scored 5 d after the first flies eclosed. (E) Cas12a+ outperforms Cas9 and LbCas12a in identifying essential genes. Summary of the observed phenotypes is shown, with genes grouped according to the phenotype expected based on existing literature (noted below each group; see Materials and Methods). Semilethal refers to lethality with incomplete penetrance, where between 30% and 70% of animals do not survive. Two independent Cas9 sgRNA lines targeting wit and Pdk1 were available and each is represented by a small square. Targeting Raf with LbCas12a at 25 °C resulted in only female offspring, which likely reflects the fact that males have only a single copy of Raf, as it is located on the X chromosome. Differential mutagenesis of genes with limited prior knowledge has been analyzed in greater detail in SI Appendix, Fig. S9.
Next, we set out to compare the efficiency of gene disruption between LbCas12a, Cas12a+, and the current standard gene-editing system in the fly, SpCas9. A direct comparison of the activity of Cas9 and Cas12a is not possible due to their different PAM requirements and crRNA topologies. Instead, we approached this question from a practical standpoint, comparing both Cas12a systems with crRNA arrays that are easily and economically generated with a publicly available, state-of-the-art resource for Cas9-mediated gene disruption.
We generated LbCas12a crRNA arrays by pooled one-step cloning, each encoding three crRNAs targeting the same gene at independent positions. These were stably integrated into the Drosophila genome at a defined position and combined with either act-LbCas12a or act-Cas12+ through a genetic cross (Fig. 4D). We then compared the phenotypic outcome of CRISPR-Cas12a mutagenesis with experiments set up in parallel utilizing the Heidelberg CRISPR Fly Design (HD_CFD) library, a large-scale collection of transgenic Cas9 sgRNA pairs (8). Experiments were performed at 29 °C, 25 °C, and 18 °C. In total, we targeted 39 genes, each encoding a protein kinase, with either CRISPR-Cas12a or CRISPR-Cas9. Of these, 23 are described to be essential for Drosophila development, 2 have been shown to lead to lethality with incomplete penetrance (referred to as “semilethal”) when inactivated, and 7 are established to be nonessential. For the remaining seven only limited information is available about their loss-of-function phenotype (e.g., no existing validated null alleles) (Materials and Methods and Fig. 4E).
Targeting any of the seven nonessential genes with any of the three nucleases gave rise to viable offspring, indicating that gene editing with these enzymes is well tolerated in Drosophila (Fig. 4E). Furthermore, gene targeting of essential genes frequently resulted in lethality of the offspring, suggesting efficient on-target mutagenesis. Remarkably, mutagenesis with Cas12a+ at 29 °C lead to lethality in combination with all 23 crRNA constructs targeting essential genes (21 fully lethal, 2 semilethal), indicating extremely efficient gene disruption with this nuclease (Fig. 4E). Similarly, CRISPR-Cas9 mutagenesis with HD_CFD sgRNAs targeting essential genes lead to lethality or semilethality in 22 of 23 cases, highlighting that this system has very high activity as well (Fig. 4E). Gene disruption with LbCas12a was slightly less effective, but still resulted in lethality with 19 of 23 crRNA arrays targeting essential genes when animals were raised at 29 °C (Fig. 4E). With all three nucleases, recall of essential genes became less effective when animals were raised at a lower temperature. At 25 °C, the standard culture condition for Drosophila, Cas12a+, and Cas9 still correctly identified the great majority of essential genes (19 of 23 lethal or semilethal with Cas9, 21 of 23 lethal or semilethal with Cas12a+), but LbCas12a only resulted in lethality with 13 of 23 crRNA constructs (Fig. 4E). Reduction of gene-editing efficiency was much more pronounced at 18 °C, in particular with LbCas12a, where only 1 crRNA line targeting an essential gene resulted in lethality and 3 lines in semilethality, while the remaining 19 lines all give rise to viable offspring (Fig. 4E).
We also observed lethality at 29 °C in crosses with crRNA arrays targeting four genes with limited prior knowledge: cdc2rk, Pkg21D, Pdk, Ask1. Gene targeting of cdc2rk gave rise to lethality with all three nucleases (although to a lesser extend with Cas9), but phenotypes upon targeting the other three genes were inconsistent between nucleases. To explore whether such differences in phenotype are likely caused by differential on-target gene editing or might reflect off-target effects, we directly monitored mutagenesis at each target site (SI Appendix, Fig. S9). We found that editing rates in Ask1 and Pkg21D matched the observed phenotypes, with low or no editing observed with Cas9, which gave rise to viable offspring, and efficient editing with Cas12a+, which resulted in lethality (SI Appendix, Fig. S9 A and B). Differences in editing were less pronounced or not observed at target sites in cdc2rk and Pdk (SI Appendix, Fig. S9 C and D). In these cases, off-target effects could cause or contribute to phenotypic differences in these cases. Alternatively, small differences in efficiency or differential effects of the induced mutations might give rise to the relatively mild differences in phenotype.
Together, these experiments establish that Cas12a gene abrogation in combination with arrays of three crRNAs per gene is highly efficient and robust.
Conclusion
Here we present a system to perform efficient Cas12a genome engineering in a variety of tissues in D. melanogaster. It consists of transgenic fly lines expressing LbCas12a or Cas12a+ from the ubiquitous act5C promoter or under the control of the conditional Gal4/UAS system and a plasmid for the expression of one or several crRNAs. These tools substantially expand and improve the current genome-engineering toolbox in Drosophila. The PAM sites utilized by Cas12a are orthogonal to those of the SpCas9 nuclease and the availability of an efficient Cas12a system, therefore, substantially expands the genomic target space available for genome engineering. In addition, the Cas12a system greatly facilitates multiplex gene editing of several sites in parallel. crRNA arrays are easy and economical to construct and are efficiently utilized by Cas12a. Simultaneous editing of several sites in parallel can compensate for the induction of mutations that do not abrogate gene function or for inactive crRNAs, which—at least in combination with LbCas12a—are observed at significant frequency. Multiplex gene editing also allows for the concurrent editing of several genes in parallel, for example to test for functional redundancies or genetic interactions.
The multiple layers of control that can be exerted over Cas12a add further appeal to this system. First, the level of on-target editing can be influenced by the nuclease that is utilized in an experiment. We find that LbCas12a and its D156R variant Cas12a+ differ significantly in activity, with Cas12a+ outperforming the wild-type LbCas12a nuclease, as well as a current Cas9 system, in identifying essential genes. However, high mutation rates are not always desirable and in cases where spare mutagenesis would be preferred, LbCas12a is likely the better choice. Gene-editing efficiency can be further influenced by the temperature at which experiments are conducted. Both Cas12a+ and LbCas12a are sensitive to temperature and the latter enzyme is often strongly suppressed at low temperature. However, even at 18 °C the enzyme often retains a low level of activity and the presence of a minority of crRNAs that mediate efficient mutagenesis at this temperature make modulation of Cas12a activity through this parameter somewhat unpredictable. Finally, a genetically encoded Cas12a system also enables mutagenesis with spatial and temporal precision by linking Cas12a expression to specific cis-regulatory elements. Interestingly, gene editing appears to be strictly restricted to cells with high levels of Cas12a expression, which is typically not the case with Cas9, where low level “leaky” expression often mediates substantial levels of mutagenesis in undesired locations.
Future work with Cas12a will clarify further aspects of this gene-editing system. For example, it will be important to explore in more detail the potential for false-positive results through off-target cleavage or other bystander effects. Work in other systems has indicated that Cas12a requires a higher degree of complementarity between crRNA and the genomic target site than Cas9, and hence induces fewer off-target mutations (32–35). Furthermore, Cas12a has also been shown to possess indiscriminate single-stranded DNase activity and it will be important to carefully investigate whether this can lead to adverse effects in cells (36). However, the results in the present study that targeting multiple nonessential genes results in viable flies indicate that such effects are unlikely to be widespread or severe.
We envision that in the future Cas12a will often be used instead or in parallel to Cas9 for genome engineering in Drosophila. Cas12a will likely become a particularly important genome-engineering system for AT-rich loci, for applications that benefit from the possibility to control gene-editing activity by multiple means or in cases that require highly multiplexed mutagenesis. Cas9 benefits from the overall higher density of target sites across the genome and the so far better characterization of the system. Having two orthogonal genome-engineering systems available will enable scientists to independently confirm novel findings made with one technology through use of the other. In addition, both technologies can be used in combination to manipulate genes in different cells of the same animal. As such, this study substantially expands the applications for genome engineering in Drosophila. The advantages of a genetically encoded Cas12a system demonstrated here should also inform the development of CRISPR tools in other genetically tractable organisms.
Materials and Methods
Plasmid Construction.
Unless indicated otherwise, PCRs were performed with the Q5 Hot-start 2 master mix (New England Biolabs) and cloning was performed using the In-Fusion HD cloning kit (Takara Bio) or restriction/ligation-dependent cloning. Newly introduced sequences were verified by Sanger sequencing. Oligonucleotide and gBlock sequences are listed in the SI Appendix, Table S2.
act5c-LbCas12a.
Plasmid act5c-LbCas12a was constructed by replacing the Cas9 ORF in act5c-Cas9 (7) by a LbCas12a ORF. DNA encoding LbCas12a (based on ref. 11) was ordered as gBlocks from Integrated DNA Technologies (IDT). Individual gBlocks were fused by extension PCR and PCR amplicons were purified using the Qiagen PCR purification kit according to the instructions supplied by the manufacturer. Plasmid act5c-Cas9 was digested with EcoRI-HF and XhoI (New England Biolabs) and the plasmid backbone was gel-purified. The plasmid backbone and the double-stranded DNA (dsDNA) fragment encoding LbCas12a were assembled by In-Fusion cloning. The proportion of the plasmid encoding LbCas12a was sequence verified by Sanger sequencing.
act5c-LbCas12aD156R (act5c-Cas12a+).
Plasmid act5c-LbCas12aD156R was constructed from act5c-LbCas12a by amplifying the Cas12a coding sequence with primers Cas12aD156Rfwd1, Cas12aD156Rrev1 and Cas12aD156Rfwd2, Cas12aD156Rrev2. Both PCR amplicons were gel purified and cloned into the act5c-LbCas12a backbone digested with EcoRI and XhoI by In-Fusion cloning.
UAS-LbCas12a.
Plasmid UAS-LbCas12a was generated by cloning LbCas12a encoded in act5c-LbCas12a into pUASTattB (37). Plasmid pUASTattB was digested with EcoRI-HF and XbaI and gel-purified. LbCas12a was PCR amplified from act5c-LbCas12a with primers UASLbCas12afwd and UASLbCas12arev. The purified PCR amplicon was assembled with the digested plasmid backbone using In-Fusion cloning.
pCFD8.
Plasmid pCFD8 was constructed as the default cloning plasmid for one or several LbCas12a crRNAs. It was generated by replacing the Cas9 sgRNA cassette in pCFD5_w (Addgene 112645) with the LbCas12 crRNA stem loop, a 2xBbsI restriction cassette, and a downstream tRNA. The necessity of the tRNAs in pCFD8 has not been assessed, but previous work indicated that crRNAs from tRNA vectors have slightly higher efficiency (21). Plasmid pCFD5_w was digested with BbsI-HF and XbaI and the backbone was gel-purified. The LbCas12a crRNA cassette was ordered as a gBlock from IDT and assembled with the plasmid backbone using In-Fusion cloning.
Cloning of crRNAs into pCFD8.
The compact size of Cas12a crRNA arrays allows to directly encode one or several crRNAs on commercially available oligonucleotides. A step-by-step cloning protocol is available as in SI Appendix, Supplementary Cloning Protocol, and possible updates will be posted at crisprflydesign.org/. Briefly, pCFD8 was digested with BbsI-HF and gel-purified. Sequences to be introduced into pCFD8 were ordered as complementary oligos (one for the top and one for the bottom strand) and resuspended to 100 µM in ddH2O. Oligo pairs were mixed at an equimolar ratio in PCR tubes containing T4 Ligation buffer and T4 polynucleotide kinase (New England Biolabs) for 5′ phosphorylation of oligos, incubated at 37 °C for 30 min, and then heated to 95 °C and allowed to cool down to 25 °C at a rate of −3 °C/min. Annealed oligos were diluted 1:200 in ddH2O. The oligos were designed such that after annealing the dsDNA had single-stranded overhangs complementary for the BbsI sites in pCFD8. The annealed oligos were ligated into the digested pCFD8 backbone using T7 DNA ligase. Successful introduction of crRNAs was confirmed by Sanger sequencing.
Drosophila Strains and Culture.
Transgenic Drosophila strains used or generated in this study are listed in SI Appendix, Table S3. Flies were kept in incubators with 50 ± 10% humidity with a 12-h light/12-h dark cycle. Temperature during the individual experiments are indicated in the figures and were regularly checked with independent thermometers.
Transgenesis.
Transgenesis was performed with the PhiC31/attP/attB system and plasmids were inserted at landing site (P{y[+t7.7]CaryP}attP40) on the second chromosome. Microinjection of plasmids into Drosophila embryos was carried out using standard procedures. Transgenesis of crRNA plasmids was typically performed by a pooled injection protocol, as previously described (38). Briefly, individual plasmids were pooled at equimolar ratio and DNA concentration was adjusted to 250 ng/μL in dH2O. Plasmid pools were microinjected into y[1] M{vas-int.Dm}ZH-2A w[*]; (P{y[+t7.7]CaryP}attP40) embryos, raised to adulthood and individual flies crossed to P{ry[+t7.2] = hsFLP}1, y[1] w[1118]; Sp/CyO-GFP. Transgenic offspring were identified by orange eye color and individual flies crossed to P{ry[+t7.2] = hsFLP}1, y[1] w[1118]; Sp/CyO-GFP balancer flies and a stable transgenic stock was generated.
Genotyping of Transgenic crRNA Flies.
Individual transgenic flies from pooled plasmid injections were genotyped to determine which plasmid was stably integrated into their genome. If transgenic flies were male or virgin female, animals were removed from the vials once offspring were apparent and prepared for genotyping. In the case of mated transgenic females, genotyping was performed in the next generation after selecting and crossing a single male offspring, to prevent genotyping females fertilized by a male transgenic for a different construct. The crRNA transgene was amplified by PCR from genomic DNA with primers U63seqfwd2 and pCFD8genorev2 and PCR amplicons were analyzed by Sanger sequencing using primer U63seqfwd2.
Preparation of Genomic DNA.
Single flies were collected in PCR tubes containing 50 µL squishing buffer (10 mM Tris⋅HCl pH8, 1 mM EDTA, 25 mM NaCl, 200 µg/mL Proteinase K). Flies were disrupted in a Bead Ruptor (Biovendis) for 20 s at 30 Hz. Samples were then incubated for 30 min at 37 °C, followed by heat inactivation for 3 min at 95 °C. Typically, 3 µL of supernatant were used in 30-µL PCR reactions.
Quantification of Gene-Editing Efficiency.
Estimation of editing efficiency by Sanger sequencing bulk PCR amplicons.
To estimate the efficiency of gene editing in animals potentially harboring a large variety of different indels (e.g., through somatic mutagenesis with ubiquitously expressed Cas12a and crRNAs) by decomposition of Sanger-sequencing traces, the target locus was amplified from genomic DNA by PCR. Primers (SI Appendix, Table S2) were typically designed to anneal ∼300 bp 5′ or 3′ of the crRNA target site. PCR amplicons were purified with the Qiagen PCR purification kit according to the instructions supplied by the manufacturer and subjected to Sanger sequencing. Sequencing chromatograms were then analyzed by ICE (Inference of CRISPR Edits) analysis (39).
Identification of edited alleles by Sanger sequencing of PCR amplicons of germline-transmitted alleles.
Germline-transmitted alleles present a good opportunity to identify the sequence of CRISPR-induced mutations, as offspring of crosses between nos-Gal4 UAS-nuclease pCFD8-crRNA males and wild-type females carry only two alleles, the CRISPR-modified and the wild-type allele, for which the sequence is known. Genomic DNA of single animals of the offspring was extracted as described above and the target locus was amplified by PCR. PCR amplicons were purified and subjected to Sanger sequencing. Sequencing chromatograms were parsed with Poly Peak Parser (yosttools.genetics.utah.edu/PolyPeakParser/) (40).
Identification of edited alleles by Sanger sequencing of subcloned PCR amplicons.
To identify the sequence of individual CRISPR-edited alleles among a pool of many different alleles, PCR amplicons were subcloned and subjected to Sanger sequencing. Genomic DNA was extracted from act-Cas12a pCFD8-crRNA animals and the target locus was amplified by PCR, as described above. After the final elongation, 1 unit of Taq polymerase was added and the reaction was incubated for 10 min at 72 °C to add 3′ A-overhangs. PCR amplicons were then cloned into the pCR4-TOPO vector using the TOPO TA cloning Kit for Sequencing (Life Technologies) according to the instructions by the manufacturer. DNA extracted from individual colonies was subjected to Sanger sequencing using the M13fwd primer and sequencing traces were inspected in Snapgene software. We found that this method leads to a strong overrepresentation of small amplicons carrying deletions (Fig. 2 and SI Appendix, Fig. S5).
Quantification of editing efficiency by Amp-seq.
For accurate and sensitive detection of on-target mutations, PCR amplicons spanning the crRNA target site were sequenced on the Illumina MiSeq platform. Transgenic flies expressing the respective Cas12a enzyme under the ubiquitous act5C promoter were crossed to the indicated crRNA transgene and crosses were incubated at the indicated temperature (either 18 °C, 25 °C, or 29 °C). As a control act-Cas12a flies were crossed to transgenic flies expressing a Cas9 sgRNA, which was present in the same genetic background as the Cas12a crRNAs (SI Appendix, Fig. S1). Several flies of the offspring from each cross were pooled and 700 ng of genomic DNA was extracted and used as template to amplify genomic regions spanning each crRNA target site (see SI Appendix, Table S2 for primer sequences). PCR amplicons were purified with the Qiagen PCR purification kit according to the instructions supplied by the manufacturer. Amplicons were then PCR-amplified using primers containing Illumina adaptors and indices. Following purification as described above, DNA concentration of each sample was measured with the Qubit 1.0 fluorometer (Thermo Fisher Scientific). Equimolar (10 nM) dilutions of amplicons were pooled, 30% PhiX was added, and the library was sequenced on an Illumina MiSEq. (300-bp single-end reads) by the High-Throughput Sequencing Unit of the Genomics and Proteomics Core Facility (German Cancer Research Facility, DKFZ). Obtained sequences were analyzed with CRISPResso (41). Fig. 1D shows the fraction of modified reads as categorized by CRISPResso as a fraction of all reads, with the exception of samples containing crRNA e5. We found that in our crRNA-e5 transgenic fly line some animals carry an SNP at the crRNA e5 target site. act-LbCas12a crRNA-e5 flies carrying the SNP never contained indels at the target site and had wild-type coloration at 29 °C, while flies not carrying the SNP were entirely ebony. This indicates that the SNP renders the target site refractory to Cas12a cutting. We therefore excluded sequencing reads containing the SNP and analyzed the fraction of modified reads among the remaining reads.
Categorization of Essential and Nonessential Genes.
Target genes (Fig. 4) were categorized as essential or nonessential based on information available in Flybase (release FB2019_5). For each gene we manually curated the information available in the phenotype category. We did not consider information based on RNAi experiments, as these were typically performed with tissue-specific Gal4 drivers, and residual gene expression, which is the norm with this method, is expected to rescue lethality in some cases. Genes classified as “unknown” had no null alleles described in Flybase or the allele is known to also affect neighboring genes (as is the case for cdi).
Immunohistochemistry.
Immunohistochemistry of Drosophila tissue was performed using standard procedures. Briefly, larva or adult intestine were dissected in ice-cold PBS and fixed in either 80% ice-cold methanol in PBS for 1 h on ice (for staining with anti-LbCas12a antibody) or in 4% paraformaldehyde in PBS containing 0.05% Triton-X 100 for 25 min at room temperature (for staining with any of the other antibodies). Larva were washed three times in PBS containing 0.3% Triton-X 100 (PBT) and then blocked for 1 h at room temperature in PBT containing 1% heat-inactivated normal goat serum. Subsequently, larva were incubated with first antibody [mouse anti-LbCas12a (Sigma Aldrich) 1:20; mouse anti-Cut (Gary Rubin, Developmental Studies Hybridoma Bank) 1:30; rabbit anti-Evi (42) 1:800; mouse anti-Prospero (MR1A, C. Q. Doe, Developmental Studies Hybridoma Bank) 1:1,000] in PBT overnight at 4 °C. The next day, samples were washed three times in PBT for 15 min and incubated for 2 h at room temperature with secondary antibody (antibodies coupled to Alexa fluorophores, Invitrogen) diluted 1:600 in PBT containing Hoechst dye. Samples were washed three times 15 min in PBT and mounted in Vectashield (Vectorlabs). To visualize apoptotic cells, wing discs expressing the apoptosis sensor GC3Ai (43) were fixed in 4% PFA, washed in PBT containing Hoechst and mounted in Vectashield.
Image Acquisition, Processing, and Analysis.
Microscopy images were acquired with a Zeiss LSM800, Leica SP5 or SP8 confocal microscope in the sequential scanning mode. Image processing and analysis was performed with FIJI (44). Experiments were performed at least twice and more than three samples were analyzed for each experiment. Imaging of adult flies or wings was performed with a stereomicroscope equipped with a 14.2 Color Mosaic camera (Visitron Systems). If wings were analyzed, the flies were incubated for at least 4 h in 50% ethanol/50% glycerol solution and individual wings were then mounted on microscopy slides in the same solution. Lightning was kept constant during image acquisition. Contrast and brightness were modified uniformly within an experiment with Photoshop or Fiji.
Supplementary Material
Acknowledgments
We thank Laura Lange and Nora Langner for excellent technical assistance; Luisa Henkel and Siamak Redhai for helpful comments on the manuscript; and the High-Throughput Sequencing Unit of the Genomics and Proteomics Core Facility and Light Microscopy Facility at The German Cancer Research Facility for support. This work has in part been supported by grants from the European Research Council (DECODE) and the German Research Foundation (SFB1324, Z03).
Footnotes
The authors declare no competing interest.
See online for related content such as Commentaries.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004655117/-/DCSupplemental.
Data Availability.
All material reported in this study is available upon request. Plasmids pCFD8, act5c-LbCas12a, act5c-Cas12a+, and UAS-LbCas12a will become available from Addgene (Addgene plasmids 140619–140621). All study data are included in the main text and SI Appendix.
References
- 1.Kaufman T. C., A short history and description of Drosophila melanogaster classical genetics: Chromosome aberrations, forward genetic screens, and the nature of mutations. Genetics 206, 665–689 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perkins L. A.et al., The transgenic RNAi project at Harvard Medical School: Resources and validation. Genetics 201, 843–852 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knott G. J., Doudna J. A., CRISPR-Cas guides the future of genetic engineering. Science 361, 866–869 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jinek M.et al., A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll D., Genome engineering with targetable nucleases. Annu. Rev. Biochem. 83, 409–439 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Bier E., Harrison M. M., O’Connor-Giles K. M., Wildonger J., Advances in engineering the fly genome with the CRISPR-Cas system. Genetics 208, 1–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Port F., Chen H.-M., Lee T., Bullock S. L., Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 111, E2967–E2976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Port F.et al., A large-scale resource for tissue-specific CRISPR mutagenesis in Drosophila. eLife 9, e53865 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gratz S. J.et al., Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kondo S., Ueda R., Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195, 715–721 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zetsche B.et al., Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreno-Mateos M. A.et al., CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat. Commun. 8, 2024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malzahn A. A.et al., Application of CRISPR-Cas12a temperature sensitivity for improved genome editing in rice, maize, and Arabidopsis. BMC Biol. 17, 9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chow R. D.et al., In vivo profiling of metastatic double knockouts through CRISPR-Cpf1 screens. Nat. Methods 16, 405–408 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H. K.et al., In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat. Methods 14, 153–159 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Swarts D. C., Jinek M., Mechanistic insights into the cis- and trans-acting DNase activities of Cas12a. Mol. Cell 73, 589–600.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fonfara I., Richter H., Bratovič M., Le Rhun A., Charpentier E., The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Zetsche B.et al., Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol. 35, 31–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campa C. C., Weisbach N. R., Santinha A. J., Incarnato D., Platt R. J., Multiplexed genome engineering by Cas12a and CRISPR arrays encoded on single transcripts. Nat. Methods 16, 887–893 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Wang Y.et al., Systematic evaluation of CRISPR-Cas systems reveals design principles for genome editing in human cells. Genome Biol. 19, 62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Port F., Bullock S. L., Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat. Methods 13, 852–854 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D.et al., Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 34, 863–868 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Bernabé-Orts J. M.et al., Assessment of Cas12a-mediated gene editing efficiency in plants. Plant Biotechnol. J. 17, 1971–1984 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie K., Minkenberg B., Yang Y., Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. U.S.A. 112, 3570–3575 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nissim L., Perli S. D., Fridkin A., Perez-Pinera P., Lu T. K., Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol. Cell 54, 698–710 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kosicki M., Tomberg K., Bradley A., Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36, 765–771 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brand A. H., Perrimon N., Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993). [DOI] [PubMed] [Google Scholar]
- 28.Ohlstein B., Spradling A., The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470–474 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Port F., Muschalik N., Bullock S. L., Systematic evaluation of Drosophila CRISPR tools reveals safe and robust alternatives to autonomous gene drives in basic research. G3 (Bethesda) 5, 1493–1502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleinstiver B. P.et al., Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 37, 276–282 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schindele P., Puchta H., Engineering CRISPR/LbCas12a for highly efficient, temperature-tolerant plant gene editing. Plant Biotechnol. J. 18, 1118–1120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh D.et al., Real-time observation of DNA target interrogation and product release by the RNA-guided endonuclease CRISPR Cpf1 (Cas12a). Proc. Natl. Acad. Sci. U.S.A. 115, 5444–5449 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strohkendl I., Saifuddin F. A., Rybarski J. R., Finkelstein I. J., Russell R., Kinetic basis for DNA target specificity of CRISPR-Cas12a. Mol. Cell 71, 816–824.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kleinstiver B. P.et al., Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 34, 869–874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D., et al., Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 34, 863–868 (2016). Correction in: Nat Biotechnol.34, 888 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Chen J. S.et al., CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U.S.A. 104, 3312–3317 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bischof J.et al., A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development 140, 2434–2442 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Hsiau T., et al., Inference of CRISPR edits from Sanger trace data. bioRxiv:10.1101/251082 (10 August 2019). [DOI] [PubMed]
- 40.Hill J. T.et al., Poly Peak Parser: Method and software for identification of unknown indels using Sanger sequencing of PCR products. Dev. Dyn. 243, 1632–1636 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinello L.et al., Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 34, 695–697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Port F.et al., Wingless secretion promotes and requires retromer-dependent cycling of Wntless. Nat. Cell Biol. 10, 178–185 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Schott S.et al., A fluorescent toolkit for spatiotemporal tracking of apoptotic cells in living Drosophila tissues. Development 144, 3840–3846 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Schindelin J.et al., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All material reported in this study is available upon request. Plasmids pCFD8, act5c-LbCas12a, act5c-Cas12a+, and UAS-LbCas12a will become available from Addgene (Addgene plasmids 140619–140621). All study data are included in the main text and SI Appendix.