Significance
B cells serve as a key weapon against infectious diseases. They also contribute to multiple autoimmune diseases, including multiple sclerosis (MS) where depletion of B cells is a highly effective therapy. We describe a comprehensive profile of central nervous system (CNS)-specific transcriptional B cell phenotypes in MS at single-cell resolution with paired immune repertoires. We reveal a polyclonal immunoglobulin M (IgM) and IgG1 cerebrospinal fluid B cell expansion polarized toward an inflammatory, memory and plasmablast/plasma cell phenotype, with differential up-regulation of specific proinflammatory pathways. We did not find evidence that CNS B cells harbor a neurotropic virus. These data support the targeting of activated resident B cells in the CNS as a potentially effective strategy for control of treatment-resistant chronic disease.
Keywords: multiple sclerosis, neuroimmunology, B cell, immune repertoire
Abstract
Central nervous system B cells have several potential roles in multiple sclerosis (MS): secretors of proinflammatory cytokines and chemokines, presenters of autoantigens to T cells, producers of pathogenic antibodies, and reservoirs for viruses that trigger demyelination. To interrogate these roles, single-cell RNA sequencing (scRNA-Seq) was performed on paired cerebrospinal fluid (CSF) and blood from subjects with relapsing-remitting MS (RRMS; n = 12), other neurologic diseases (ONDs; n = 1), and healthy controls (HCs; n = 3). Single-cell immunoglobulin sequencing (scIg-Seq) was performed on a subset of these subjects and additional RRMS (n = 4), clinically isolated syndrome (n = 2), and OND (n = 2) subjects. Further, paired CSF and blood B cell subsets (RRMS; n = 7) were isolated using fluorescence activated cell sorting for bulk RNA sequencing (RNA-Seq). Independent analyses across technologies demonstrated that nuclear factor kappa B (NF-κB) and cholesterol biosynthesis pathways were activated, and specific cytokine and chemokine receptors were up-regulated in CSF memory B cells. Further, SMAD/TGF-β1 signaling was down-regulated in CSF plasmablasts/plasma cells. Clonally expanded, somatically hypermutated IgM+ and IgG1+ CSF B cells were associated with inflammation, blood–brain barrier breakdown, and intrathecal Ig synthesis. While we identified memory B cells and plasmablast/plasma cells with highly similar Ig heavy-chain sequences across MS subjects, similarities were also identified with ONDs and HCs. No viral transcripts, including from Epstein–Barr virus, were detected. Our findings support the hypothesis that in MS, CSF B cells are driven to an inflammatory and clonally expanded memory and plasmablast/plasma cell phenotype.
Multiple sclerosis (MS) is a common autoimmune demyelinating disease of the central nervous system (CNS), affecting ∼1 million people in the United States (1). Although T cells are important effector cells in MS, it is now clear that B cells play a central role in both the relapsing and progressive forms of the disease (2–5).
To date, microarray and bulk RNA-sequencing (RNA-Seq) studies of B cells from MS subjects have been carried out on CNS and blood samples with a focus on understanding differential expression of B cell receptor (BCR) genes in MS compared with healthy controls (HCs) (6–12). These studies have not yet been able to clearly define the transcriptome-wide profiles of CNS B cell subpopulations or compare them with their peripheral counterparts. More effective therapies against MS, especially against progressive disease, will likely require the targeting of residual CNS B cells, a heterogeneous population that may include culprit autoreactive clones as well as beneficial regulatory B cells that serve homeostatic functions. Thus, better clarifying the functional phenotypes of CNS B cell subtypes in MS may not only shed light on disease pathogenesis but also potentially provide more disease-specific and safer therapeutic targets to guide development of the next-generation of B cell therapeutics.
Similar to a recent study (13), we performed RNA-Seq at single-cell resolution of paired cerebrospinal fluid (CSF) and blood samples from relapsing-remitting MS (RRMS), other neurologic diseases (ONDs), and HCs. Additionally, we paired single-cell transcriptome data with immunoglobulin repertoire sequencing (Ig-Seq) of B cells in MS so that transcriptomic phenotypes of B cells could be further delineated based on both Ig subclass as well as the degree to which a cell is clonally expanded. While this methodology makes it possible to simultaneously obtain transcriptional phenotypes and paired Ig heavy- and light-chain sequences from a single cell, the number of genes whose messenger RNA (mRNA) transcripts can be reliably detected in each cell remains relatively small with today’s single-cell technology (∼1,000 genes). Thus, to extend our findings to include more in-depth transcriptional phenotyping, we performed bulk RNA-Seq on five classical B cell subsets defined by CD19, CD27, and IgD expression from the CSF and blood on an independent cohort of seven treatment-naïve, RRMS subjects. We further leveraged our bulk RNA-Seq dataset to assess nonhost and human endogenous retrovirus (HERV) transcripts to look for evidence of viral transcription in both blood and CSF B cells.
Materials and Methods
Single-Cell RNA-Seq Cohort.
Study cohort and inclusion criteria.
All RRMS and clinically isolated syndrome (CIS) subjects were participants in either the University of California, San Francisco (UCSF) Origins or Expression, Proteomics, Imaging, Clinical (EPIC) studies (14, 15) and were diagnosed according to the 2017 McDonald criteria (16). Subjects were not on immunomodulatory or immunosuppressive disease-modifying therapy at the time of sample collection. OND and HC participants were enrolled in a biobanking study “Immunological Studies of Neurologic Subjects” (Dataset S1A).
CSF and blood processing and single-cell sequencing.
CSF and blood were obtained during diagnostic and research procedures after subjects had provided informed consent; 20 to 30 mL of CSF was centrifuged immediately after collection at 400 × g for 15 min at 4 °C. The pellet was resuspended in ∼80 µL of residual supernatant, and lymphocytes were counted using a hemocytometer. Peripheral blood mononuclear cells (PBMCs) were isolated using cell preparation tubes as described previously (17) and resuspended in 2% fetal bovine serum. All experiments were performed immediately with freshly collected, unsorted cells. To obtain additional single-cell Ig-Seq (scIg-Seq) data for some patients, CSF or PBMCs were enriched for total B cells or for memory B cells via bead-based selection (EasySep Human Pan-B cell enrichment kit and EasySep Human Memory B cell Isolation Kit, respectively; StemCell Technologies). Sequencing libraries were prepared using 3′ or 5′ library preparation kits (10x Genomics).
Bioinformatic analysis.
Both single-cell RNA-Seq (scRNA-Seq) and scIg-Seq datasets were analyzed using the Cell Ranger (v3.0.1) count and variable diversity joining (VDJ) functions using the Ensembl GRCh38.v93 and GRCh38.v94 references, respectively. Data were analyzed using a custom bioinformatics pipeline that included Seurat (v3.1.2), the Spliced Transcripts Alignment to a Reference (STAR) algorithm (v2.5.1), SingleR (v1.1.7), and DoubletFinder (v2.0.2). The Immcantation pipeline, specifically IgBLAST (v1.4.0) and Change-O (v0.4.6), was used to assemble heavy- and light-chain contigs for each cell.
Quality control for barcode hopping.
Using a custom pipeline, overlapping reads present in more than one sample that shared the same cell barcode and unique molecular identifier (UMI) were counted and filtered using a single sample read assignment threshold percentage of 80% as previously described (18). If no sample accounted for at least 80% of the overlapping reads, the reads were removed and excluded from further analysis from both samples (code available at https://github.com/UCSF-Wilson-Lab/SingleCellVDJdecontamination).
Quality control for RNA-Seq data.
The 3′ and 5′ single-cell datasets were analyzed separately. All gene counts were aggregated together using Seurat. Only genes present in two or more cells were included. Only cells containing transcripts for 700 or more genes were included. For both the 5′ and 3′ data, maximum cutoffs of 2,500 and 4,000 genes were used, respectively. The PercentageFeatureSet function was used to calculate the percentage of mitochondrial transcript expression for each cell. Cells were omitted if they expressed at least 10% mitochondrial genes or were defined by the platelet markers PPBP and PF4. Gene counts were normalized using the R package SCTransform (19). In addition, nonregularized linear regression was used on the UMI counts per cell. Both datasets were clustered using 20 principal components in Seurat. Clusters were formed using a shared nearest neighbor graph in combination with dimensional reduction using uniform manifold approximation and projection (UMAP) (20). In order to optimize the total number of principal components, cumulative sums were iteratively calculated for each principal component to measure the percent variance accounted for with the data. A threshold of 90% variance was applied. As a result, 10 and 17 principal components were used for the 5′ and 3′ data, respectively. Both datasets were reclustered with the newly calculated principal components. Doublet detection and removal were performed using DoubletFinder (21) with expected doublet rates set based upon the 10x Genomics reference manual.
Cell type annotation and differential gene expression analysis.
Immune cell type identity was defined by performing differential gene expression (DGE) analysis for each cluster. The normalized gene expression profile for each cluster was compared with the remaining cells using a Wilcoxon rank sum test as provided in Seurat’s FindAllMarkers function (default parameters). The most up-regulated genes, with the highest positive average log fold change, were compared with a custom panel of canonical gene makers (Dataset S2) spanning several key immune cell types, including B cells, CD4+ T cells, CD8+ T cells, natural killer (NK) cells, classical monocytes, inflammatory monocytes, macrophage, plasmacytoid dendritic cells, and monocyte-derived dendritic cells. Clusters that predominantly expressed PPBP or HEMGN genes were omitted.
In addition, to improve specificity, all B cells included in our analysis were required to have reads to CD79B. All clusters annotated as B cells were extracted and annotated further with SingleR using the combined Blueprint and ENCODE reference dataset (22–24). DGE analysis for heat map generation was performed using the FindMarkers command in Seurat with the Wilcoxon test and the following parameters: P-adjusted value cutoff = 0.05 and logFC cutoff = 0.25. DGE was performed using zinbwave (v1.8.0) (default parameters) (25) and DESeq2 (v1.26.0) (Dataset S3). For the DGE and subsequent downstream analysis, only paired CSF and blood transcriptome profiles of RRMS patients were included.
Pathway analysis.
Pathway analysis was carried out on both the 5′ and 3′ scRNA-Seq datasets using the Ingenuity Pathway Analysis (IPA) toolkit (v01.12) (26) (Dataset S4). We reported all pathways that were significant at Benjamini–Hochberg P adjusted of 0.05. In addition, upstream regulator analysis and causal network analysis were carried out using the IPA toolkit to identify likely upstream regulators of genes in the dataset. The activation state of a pathway or gene was calculated based on differentially expressed genes (P adjusted = 0.05), and all pathways/causal networks/master upstream regulators with a P adjusted less than 0.05 and Z score greater than or equal to absolute value of two were reported. Upstream and master regulators with a Z score ≥2 were considered activated and ≤−2 were considered inhibited. The ToppGene suite of tools and clusterProfiler (27) were used to identify Gene Ontology (GO) terms (biological and molecular processes) enriched in the differentially expressed genes. Only the overlapping statistically significant differentially expressed genes in the 5′ and 3′ scRNA-Seq datasets were used in ToppGene and clusterProfiler.
Identification of clonally related B cells.
Assembled contigs outputted from Cell Ranger were inputted into the Immcantation pipeline for a second round of alignment to the VDJ region using IgBLAST. Contigs containing fewer than three UMIs were omitted. Only contigs that aligned in frame (both the FUNCTIONAL and IN_FRAME output fields were TRUE) and across the constant region were retained. B cells were clustered based on similarities between their IGHV (immunoglobulin heavy-chain variable region gene), IGHJ (immunoglobulin heavy-chain joining region gene), and H-CDR3 (heavy-chain Complementary Determining Region 3) amino acid sequence. The Change-O toolkit generated a length-normalized bimodal distance-to-nearest distribution of hamming distances for all cells in the dataset. A Gaussian mixture model was applied to predict an appropriate hamming distance threshold (threshold = 0.15). B cells were defined as highly similar if they shared identical V and J genes as well as a CDR3 amino acid sequence within the accepted threshold. Any cell containing a single chain was omitted. Within an individual, B cells that are highly similar based on these criteria are likely clonally related (i.e., originating from the same progenitor B cell), but that is less likely when comparing B cells with highly similar heavy-chain sequences between individuals. Clonal expansion within an individual was defined as a highly similar B cell cluster containing two or more B cells (28–30).
B Cell Subset Bulk RNA-Seq Cohort.
Study cohort and inclusion criteria.
All subjects were participants in either the Origins or EPIC studies at UCSF. Similar to patients included in the single-cell sequencing cohort, subjects had to be treatment-naïve at the time of sample collection with a clinical diagnosis of MS according to the 2017 McDonald criteria (16) (Dataset S1B). Further, each subject had to have at least 7 of 10 B cell subsets (5 CSF and 5 blood subsets total) with 50 or more cells sorted by flow cytometry.
Sample collection and flow cytometry analysis.
CSF and blood were obtained during diagnostic and research procedures following informed consent. Fresh whole CSF and blood lymphocytes were labeled with the following antibodies: CD3, CD19, CD27, CD38, CD138, and IgD. Using a Beckman MoFlo Astrios cell sorter, CSF and blood B cell subsets were sorted directly into lysis buffer as previously described (17, 31): naïve B cells (N; CD19+IgD+CD27–), unswitched memory B cells (USM; CD19+IgD+CD27+), switched memory B cells (SM; CD19+IgD–CD27+), double-negative B cells (DN; CD19+IgD–CD27–), CSF plasmablast/plasma cells (CD19+IgD–CD27hi), and blood plasmablast/plasma cells (CD19+IgD–CD27hiCD38hi) (Dataset S1C). These distinct B cell populations represent classic B cell subsets, as described previously (17, 32–34). Flow sorted cell lysates were stored at −80 °C until RNA extraction was performed.
Complementary DNA (cDNA) library preparation and next-generation sequencing.
RNA was extracted from the CSF and blood B cell subsets using the AllPrep DNA/RNA Micro kit (Qiagen), automated on the QIAcube robotics platform. RNA-Seq libraries were prepared with the Next Ultra II RNA Library Prep kit (New England Biolabs), automated on a Beckman-Coulter Biomek NXP robot. Individual cDNA libraries were barcoded with dual-indexing barcodes, pooled evenly, and sequenced on an Illumina HiSeq 4000 machine to generate 150-base pair paired-end reads.
Bioinformatic Analyses.
DGE and pathway analysis.
The bulk RNA-Seq cohort was analyzed independently of the scRNA-Seq cohort. Quality filtering of raw sequencing files was performed using the Paired-Read Iterative Contig Extension software tool kit (v1.2) (80% of sequences with a Q score > 0.98) (35). High-quality reads were aligned to the human genome (hg38) and the Genome-based Endogenous Viral Element database (36) using STAR (37). Final gene counts for the human genome (including HERV transcripts) were obtained using STAR (v2.5.3a) and samtools (v1.7), respectively (36–38). DGE and principal component analysis was conducted using DESeq2 (v2.1.14.1) using counts of all protein-coding genes expressed in greater than 10% of the samples (39) across all CSF and blood B cell subsets (Dataset S3). A false discovery rate and P-adjusted (Benjamini–Hochberg) cutoff of 0.05 were used to select the final list of differentially expressed genes. Cook’s distance, used for identification of outliers, was calculated in DESeq2 (v2.1.14.1). Genes containing an outlying count as defined by a Cook’s distance above the default threshold were flagged and not included in downstream analysis. DGE analysis was focused on memory B cells and plasmablast/plasma cells, with additional B cell subsets described in the SI Appendix. Pathway analysis was carried out using the IPA toolkit, with methods described in the single-cell cohort section.
In Results and Discussion, we highlight pathways and GO terms that were statistically significant by at least one of the single-cell (3′ or 5′) and bulk RNA-Seq methods.
Immune repertoire analysis.
A custom bioinformatics pipeline incorporating MiXCR (v2.1.9) adapted for RNA-Seq data were used to identify Ig heavy-chain (IGH) and light-chain (IGK, IGL) germline genes from raw, paired-end sequencing reads (40). BCRs using identical IGHV and IGHJ germline segments and identical or near-identical H-CDR3 regions (minimum eight-amino acid length, maximum Hamming distance of two at the amino acid level) were considered highly similar (17, 41). Each candidate BCR sequence had to have a coverage depth of two or more across the entire length of the sequence to be included in the analysis.
Virus identification.
Viral transcripts were identified from raw sequencing reads using IDseq (v2.11) as previously described (42–44). To distinguish potential viruses from laboratory reagent and skin commensal flora contaminants, a Z score was calculated for the value observed for each genus relative to water (“no template”) and uninfected CSF controls (45).
Results
Overview of Study.
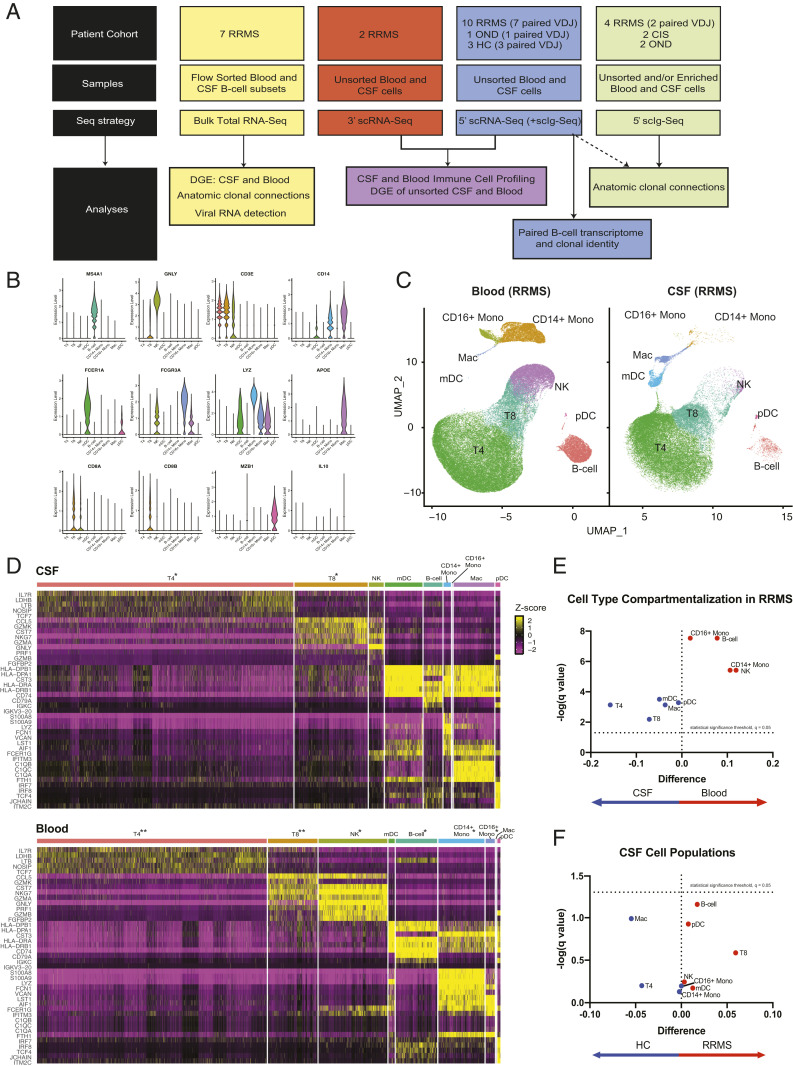
The structure of this study is outlined in Fig. 1A. Across the scRNA-Seq and scIg-Seq cohorts, 24 total subjects were enrolled (n = 16 RRMS, 3 OND, 3 HC, 2 CIS) (Table 1 and Dataset S1A). For gene expression analysis using scRNA-Seq, 12 RRMS (n = 10 from 5′ and n = 2 from 3′), 1 OND, and 3 HC were analyzed. For paired scIg-Seq, a subset of the scRNA-Seq gene expression cohort (seven RRMS, one OND, three HC) and two additional RRMS subjects were included. For analysis of B cell clonal connections, all available subjects, including those from whom matched unsorted CSF and blood samples were not available, were included (n = 14 RRMS, n = 3 OND, n = 2 CIS, and n = 3 HC). Sequencing libraries were prepared using the 10x Genomics 3′ and 5′ library preparation kits (detailed in Fig. 1A and Dataset S1A).
Fig. 1.
Study overview and the CSF immune landscape in HCs and RRMS subjects. (A) A schematic representation and overview of sample collection, processing, and bioinformatic analysis. (B) Representative violin plots of paired CSF and blood from 5′ scRNA-Seq (n = 10 RRMS). The expression of canonical genes is shown for each of the Seurat clusters generated, with manual cell type annotations inputted for each Seurat cluster. (C) The 5′ gene expression from the samples represented in B with UMAP coordinates, and in D, heat maps for CSF (Upper) and blood (Lower) are shown. The top five genes exhibiting the strongest differential expression for each of the major lineages are displayed. The following scaling factors were chosen to maintain the overall proportion of cell types found in the CSF and blood: *Cell type down sampled by 66.67% to enable visualization of low-proportion cell types; **Cell type down sampled by 90% to enable visualization of low-proportion cell types. (E) Volcano plot of difference in proportion of each major cell type in the CSF compared with the blood with false discovery Q value of 0.05 for multiple t tests represented as a dashed line on a log scale. (F) Similar volcano plot of difference in proportion of each major cell type in RRMS vs. HC.
Table 1.
Subject demographics
| Diagnosis | Age (y) | Female (%) | EDSS | CSF WBC | OCBs (% positive) | IgG index | Gd (% positive) | Months from onset |
| RRMS (n = 7, bulk) | 25–40 | 71 | 1.5–2.5 | 1–20 | 86 | 0.66–1.4 | 43 | 1–79 |
| RRMS (n = 16, single cell) | 22–54 | 69 | 0–4 | 0–13 | 81 | 0.46–1.75 | 63 | 0–166 |
| CIS (n = 2, single cell) | 35–53 | 100 | 2.5–4 | 2–11 | 0 | 0.47–0.51 | 50 | 1–3 |
| OND* (n = 3, single cell) | 21–72 | 100 | N/A | 0–4 | 100 | 0.52–0.70 | 0 | 5–90 |
| HC (n = 3, single cell) | 27–41 | 33 | N/A | 0–1 | 0 | 0.51–0.58 | N/A | N/A |
Seven treatment-naïve RRMS subjects whose age at enrollment ranged from 23 to 40 were included in the bulk RNA-Seq cohort. Twenty-four subjects (n = 16 RRMS, n = 2 CIS, n = 3 OND, n = 3 HC) (Fig. 1A and Dataset S1A have a breakdown of samples included in the RNA-Seq and Ig-Seq analyses) whose age at enrollment ranged from 21 to 72 were included in the single-cell cohort. OCB, oligoclonal band; EDSS, expanded disability status scale; WBC, white blood cell; N/A, not applicable; Gd, gadolinium.
Subjects in the OND group had diagnoses of neurosarcoidosis, atypical neuroinflammatory syndrome with uveitis, and AQP4+ neuromyelitis optica.
Seven treatment-naïve RRMS subjects were enrolled in the bulk RNA-Seq cohort (Table 1 and Dataset S1B). Flow cytometric B cell subset distributions for the CSF and blood were determined for all seven subjects in the bulk RNA-Seq cohort (Dataset S1C). Principal component analysis demonstrated that B cells primarily clustered based on their subset type and the body compartment from which they were isolated (SI Appendix, Fig. S1).
Distinct T Cells and Myeloid Cells Define the Cerebrospinal Fluid Immune Environment, with B Cells Abnormally Enriched in RRMS Compared with HCs.
We analyzed 5′ scRNA-Seq libraries from paired blood and CSF from 10 RRMS subjects. Expression of canonical immune cell markers was determined for each unsupervised Seurat cluster (Fig. 1B). To determine the relationship between cells, we plotted the data using UMAP coordinates and revealed distinct cell types (Fig. 1 C and D and SI Appendix, Fig. S2A). We found distinct differences in the myeloid and lymphoid populations in the CSF relative to blood of MS patients, consistent with previous reports (13, 46, 47). CSF macrophages, also termed microglia-like or CSF microglial cells, were present in both RRMS and HCs (SI Appendix, Fig. S2B). In patients with RRMS, comparison of CSF cell types relative to blood found no cell type with a Q value < 0.05 (Fig. 1E). In comparison with HCs, CSF B cells were 13 times more abundant in RRMS subjects (Fig. 1F and SI Appendix, Fig. S2C).
Integrated Bulk and Single-Cell Transcriptomic Analysis of B Cells in RRMS Identifies Distinct Changes in CSF Memory Cells and Plasmablast/Plasma Cells.
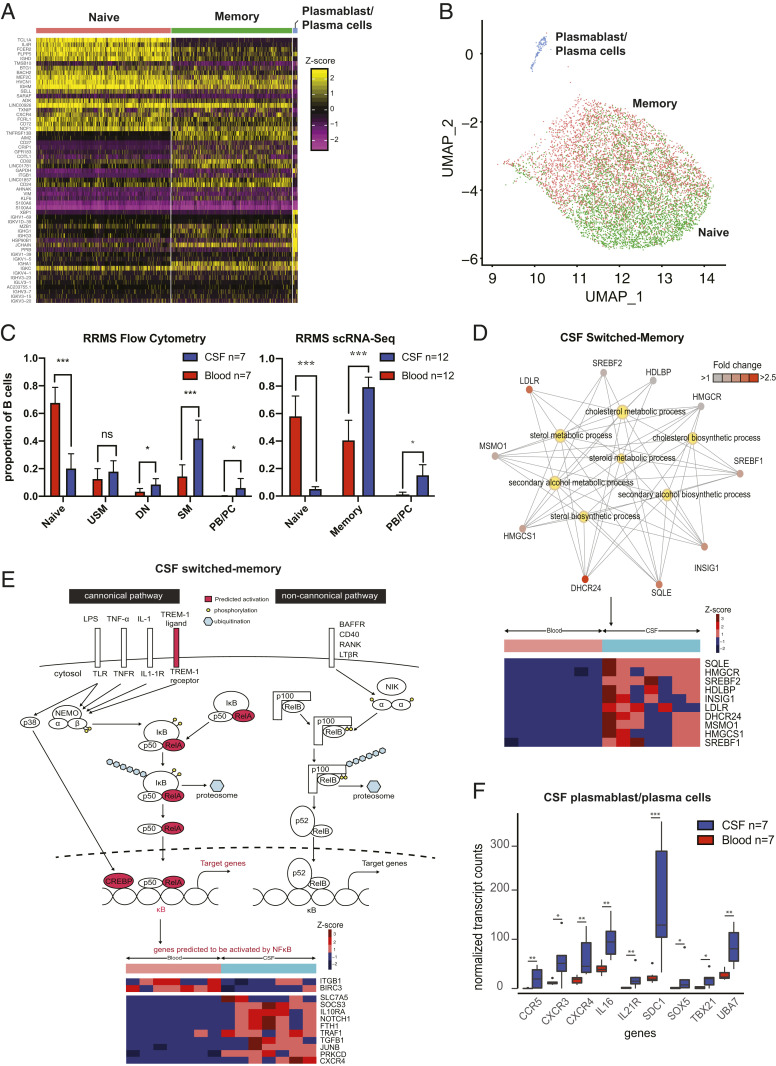
We were able to clearly identify three B cell subtypes in the 5′ and 3′ scRNA-Seq datasets: naïve, memory, and plasmablast/plasma cells (Fig. 2A), with naïve and memory B cells clustered in a transcriptional continuum in UMAP space (Fig. 2B). In both the bulk (flow cytometry) and single-cell cohorts, CSF contained higher proportions of memory B cells and plasmablast/plasma cells than in peripheral blood (Fig. 2C). Expressions of classic B cell markers were similar across bulk and scRNA-Seq (SI Appendix, Fig. S3).
Fig. 2.
Single-cell and bulk profiling of B cells in RRMS. (A) Heat map of B cells from paired RRMS CSF and blood samples from the 5′ scRNA-Seq gene expression analysis (n = 10 subjects) revealing clustering into three main groups of B cells. (B) Representation of this dataset in UMAP space extracted from and zoomed in on from the UMAP coordinates represented in Fig. 1C. Annotations generated algorithmically using SingleR with the Encode reference set. (C) Flow cytometry B cell results from 7 paired RRMS blood and CSF samples for bulk RNA-Seq and 12 paired RRMS blood and CSF samples (n = 10 from 5′ and n = 2 from 3′ scRNA-Seq) from scRNA-Seq. Statistical tests were performed using the Mann–Whitney U test; mean ± SEM is shown. (D) Network diagram of CSF switched memory cells with a focus on the cholesterol biosynthesis pathways (Upper) and corresponding heat map of differentially expressed genes (Lower) in the bulk RNA-Seq cohort. Pathway data were generated using clusterProfiler. Genes are depicted as outer nodes in shades of red (corresponding to fold enrichment), while GO terms are highlighted centrally in yellow. (E) Schematic representation of upstream NF-κB pathway members predicted to be activated (Upper), and heat map of differentially expressed downstream NF-κB pathway members genes (Lower) in CSF SM cells from the bulk RNA-Seq cohort. (F) Normalized expression of genes involved in inflammatory pathways in CSF and blood plasmablast/plasma cells. PB/PC, plasmablast/plasma cells. *P < 0.05, **P < 0.01, ***P < 0.001.
CSF Antigen-Experienced Memory B Cells Activate Nuclear Factor kappa-Light-Chain-Enhancer of Activated B Cells (NF-κB) and Proinflammatory Cytokines.
IgD–CD27+ SM.
A total of 294 genes were differentially expressed between CSF and blood SM B cells, with 166 of these up-regulated in the CSF (Dataset S3). Compared with their peripheral counterparts, CSF SM B cells up-regulated CD138 and CD83 and down-regulated CD52, CD180, CD53, CD22, and CD48 (Table 2 and Dataset S3). Additionally, IL10RA, IL2RB, TGFB1, TBX21, CXCR3, CXCR4, CCR2, and CCR5 were up-regulated by CSF SM B cells. A total of 18 upstream regulators were significant, including the transmembrane receptor TREM1 (Z = 2.2) and the NF-κB (Z = 2.7) complex, both predicted to be activated (Fig. 2E and Dataset S4A). Additionally, CEBPB, a transcriptional regulator downstream of the NF-κB pathway, was also predicted to be activated (Z = 2.2), implying activation of the canonical NF-κB pathway. Among chemokines, both CCL2 and CCL11 were predicted to be activated (Dataset S4B). By IPA, 59 networks were significant, 50 of which were predicted to be activated (Dataset S4B). CSF SM B cells showed enrichment of GO terms corresponding to an inflammatory profile, not observed in their peripheral counterparts (Dataset S4D). Similarly, clusterProfiler analysis also revealed an enrichment of GO terms related to cholesterol metabolism and biosynthesis (Fig. 2D) (Dataset S4E). An enrichment of GO terms corresponding to transcription, translation, and catabolic processes was also seen in the blood SM B cells (Dataset S4E).
Table 2.
RNA-Seq analysis findings in B cell subsets
| B cell subset | Cell surface transcripts up in CSF | Cell surface transcripts down in CSF | Transcripts up in CSF | Significant upstream regulators | Significant networks |
| N | CD82 | CD22, CD52, CD74 | IL10RA, CXCR4, RASGRP2, IL2RB | IL3, TLR9 | — |
| DN | CD3E, CD5 | CD200 | — | — | TPSD1, USP21 |
| USM | CD83 | CD52, CD180, CD27 | IL10RA, CXCR4, TGFB1 | NF-κB, TREM-1 | IL3, IL6, IL7, IL32, OSM, TSLP, CXCL8 |
| SM | CD138, CD83 | CD52, CD180, CD53, CD22, CD48 | IL10RA, CXCR3, TBX21, IL2RB, CXCR4, CCR5, CCR2, AKNA, TGFB1 | NF-κB, CCL2, CCL11 | — |
| Plasmablast/plasma cells | CD138, CD74, CD79A, CD46, BCR | — | AKNA, CCR5, IL21R, UBA7, CXCR4, TBX21, IL16, SOX5, CXCR3, PPARA | IL4* | — |
Curated overview of transcripts up- and down-regulated between CSF and blood, significant causal networks, and upstream regulators predicted to be activated/inhibited using the IPA toolkit in five B cell subsets (N, DN, USM, SM, and plasmablasts/plasma cells).
Pathway inhibited.
IgD+CD27+ USM.
A total of 228 genes were differentially expressed between CSF and blood USM B cells; 96 were up-regulated in the CSF (Dataset S3). Compared with their blood counterparts, CSF USM B cells up-regulated CD83, IL10RA, CXCR4, and TGFB1 and down-regulated CD180, CD52, and CD27 (Table 2 and Dataset S3). IPA network analysis predicted that five upstream regulators were activated including TREM-1 (Z = 2.5) and NF-κB (Z = 2.6) (Dataset S4A). Thirty-four master regulators were significant, of which 29 were predicted to be activated, including the following cytokines: IL3, IL6, IL7, IL32, OSM, TSLP, and CXCL8 (Dataset S4B). Only one GO term was enriched in CSF USM B cells: “skeletal muscle development.” GO terms related to transcription, translation, and catabolic processes were enriched in blood USM B cells (Dataset S4 D and E). One pathway, “EIF2 signaling,” was significantly inhibited in both USM and SM CSF B cell subsets (Dataset S4C).
We next compared our bulk RNA-Seq data with our scRNA-Seq data. Totals of 1,231 and 1,221 genes were differentially expressed between CSF and blood memory B cells in the 3′ and 5′ single-cell gene expression datasets, respectively (Dataset S3). Of these, 430 were differentially expressed (59 up-regulated in the CSF) across both the 3′ and 5′ datasets. In comparison with their blood counterparts, CSF memory B cells up-regulated CXCR4 and CD70 and down-regulated CD180, CD22, CD27, CD37, CD40, CD52, CD53, CD72, CD74, CD79A, and CD79B. Eight upstream regulators (three activated: PRDM1, NR4A1, and MAPK1; five inhibited: STK11, PAX5, PML, PPARA, and PPARGC1A) were significant (Dataset S4F). Seven upstream regulators including TREM1, NF-κB complex, and RELA were all predicted to be activated in the single-cell CSF memory B cell 5′ cohort, similar to the bulk RNA-Seq cohort (Dataset S4F). Seventeen causal networks (10 activated, 7 inhibited) were significant in both 3′ and 5′ datasets; 11 causal networks, including CXCL8, were found overlapping with the bulk RNA-Seq cohort (Dataset S4G). Additionally, pathway (5′ dataset) and GO (3′ and 5′ datasets) analyses showed that cholesterol biosynthesis was activated in CSF memory B cells (Dataset S4 H and I). In blood memory B cell counterparts, GO terms associated with antigen processing such as “peptide antigen binding” and “MHC class II protein complex binding” were enriched.
CSF CD27hi Plasmablast/Plasma Cells Display an Inflammatory Phenotype and Down-Regulate Genes in the TGF-β1 Pathway.
A total of 204 genes (63 up-regulated in the CSF) were differentially expressed between CSF and blood plasmablast/plasma cells (Dataset S3). Compared with their peripheral counterparts, CSF plasmablast/plasma cells up-regulated CD138, CD74, CD79A, CD46, and BCR (Table 2 and Dataset S3). Genes related to cytokines/chemokines up-regulated in CSF plasmablast/plasma cells included IL21R, UBA7, TBX21, IL16, SOX5, CXCR4, CXCR3, and CCR5 (Fig. 2F). CSF plasmablast/plasma cells enriched for inflammation GO terms including “cytokine receptor activity,” “chemokine receptor activity,” and “G protein-coupled chemoattractant receptor activity” and terms associated with cholesterol biosynthesis (Dataset S4D). Similarly, clusterProfiler analysis revealed enrichment of terms associated with fatty acid biosynthesis and inflammation (Dataset S4E). “SMAD protein import into the nucleus,” corresponding to regulation of SMAD protein signal transduction, involved in the TGF-β1 signaling pathway (BMPR1A, TOB1, JUN, SPTBN1) was down-regulated in CSF plasmablast/plasma cells (Dataset S4E). Finally, CSF plasmablast/plasma cells did not express more IL10RA than their blood counterparts (Dataset S3). Three upstream regulators were significant using IPA (Dataset S4B), including inhibition of IL4, a Th2 cytokine (Z = −2).
In comparison, 225 and 49 genes were differentially expressed between CSF and blood plasmablast/plasma cells in the 3′ and 5′ scRNA-Seq datasets, respectively (Dataset S3). Of these, only 30 genes (10 up-regulated in the CSF) were shared, with no overlaps in the pathways, predicted master regulators, or causal networks. Additionally, the three significant upstream regulators that were significant in the bulk RNA-Seq analysis were not significant in the single-cell cohort. In the 3′ gene expression dataset, similar to our findings in the bulk study, IL16 was up-regulated in the CSF plasmablast/plasma cells (Dataset S3). Interestingly, TGFB-1 was predicted to be inhibited in the 5' gene expression dataset (Dataset S4F); with a downregulation of genes in the TGF-β1 signaling pathway also observed in the CSF plasmablast/plasma cells using the clusterProfiler analysis in the bulk RNA-Seq cohort (Dataset S4E). GO analysis using only the genes differentially expressed across both datasets revealed an abundance of terms associated with iron transport protein targeting and translocation (Dataset S4I), and while GO terms associated with cholesterol biosynthesis were not enriched, a fourfold enrichment of the enzyme SQLE, necessary for the initiation of cholesterol biosynthesis (P adjusted = 0.001), was observed in the 3′ gene expression dataset. Similar to the bulk RNA-Seq cohort, IL10RA was not differentially expressed between the CSF plasmablast/plasma cells and their blood counterparts (Dataset S3).
Paired scRNA-Seq and scIg-Seq Reveals Clonally Expanded, Somatically Hypermutated IgM and IgG1 CSF B Cells Associated with Acute Inflammation and Intrathecal Ig Synthesis.
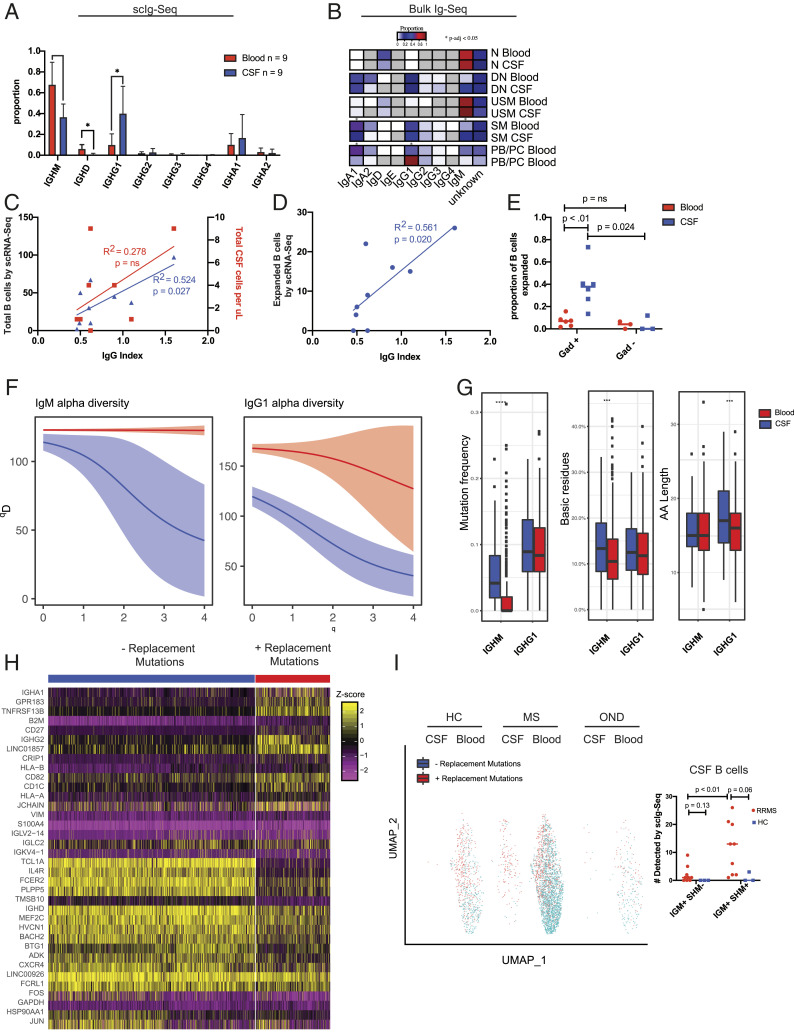
We next leveraged our 5′ scRNA-Seq libraries to perform full-length scIg-Seq analysis on paired CSF and blood from RRMS subjects (n = 9) (Fig. 1A and Dataset S1A). As expected, we found a statistically greater proportion of IgG1+ B cells between the CSF and blood and a roughly equal proportion of IgG1+ and IgM+ B cells within the CSF (Fig. 3A). In our bulk RNA-Seq dataset, we observed similar findings, with IgM-expressing cells found in the naïve and USM subsets and IgG1-expressing cells found predominantly in the plasmablast/plasma cells, SM, and DN subsets (Fig. 3B). These findings recapitulate expected changes in the isotypes of CSF B cells (48, 49).
Fig. 3.
Paired scIg-Seq in RRMS ties clonal B cell expansion to intrathecal Ig synthesis and blood–brain barrier breakdown and to the emergence of IgM+ B cells undergoing SHM in the CSF. (A) Isotype usage by scIg-Seq in the 5′ dataset from n = 9 paired CSF and blood RRMS subjects. Statistics were performed with Mann–Whitney U test; mean ± SD is shown. (B) Isotype usage by limited Ig-Seq data extracted from bulk RNA-Seq. (C) Linear correlation between the clinical CSF WBC count vs. total B cells by scRNA-Seq and the clinical IgG index. (D) Linear correlation between the number of expanded B cells detected by scRNA-Seq and the clinical IgG index. (E) Enrichment of clonally expanded CSF, but not blood, B cells in the same subjects with active gadolinium on MRI. Statistics performed with Mann–Whitney U test. (F) Alpha diversity plots from the Immcantation framework depicting restricted diversity in the CSF relative to the blood for both IgM+ and IgG1+ B cells (n = 9 RRMS subjects). There were too few B cells in the HCs to generate comparison plots. Blood is depicted in red, and CSF is depicted in blue. (G) Mutation frequency, basic residue count, and CDR3 length in the CSF and blood of RRMS subjects by scIg-Seq. Statistical test was performed using a Student’s t test. (H) Heat map of the top 20 most differentially expressed genes by the Seurat FindMarkers command in IgM+ B cells with and without replacement mutations. (I) UMAP of IgM+ B cells from scIg-Seq overlaid onto 5′ scRNA-Seq gene expression data from HC (n = 3; 2 had zero B cells with productive VDJ sequences), RRMS (n = 7), and OND (n = 1) subjects. Dot plot adjacent to UMAP with statistical testing comparing the total number of B cells detected in the CSF and blood with either IGM+ SHM or IGM− SHM. Statistical testing was performed with the Mann–Whitney U test. PB/PC, plasmablast/plasma cells; WBC, white blood cell. *P < 0.05, ***P < 0.001, ****P < 0.0001.
We next investigated whether these findings correlated with clinical laboratory testing or radiographic characteristics of the patients. We found a significant linear correlation between the clinical IgG index and the total number of CSF B cells we identified in each subject by scRNA-Seq, relative to a weaker correlation between the clinical total white blood cell count and the CSF IgG index (R2 = 0.524, P = 0.027 vs. R2 = 0.278, P = nonsignificant [ns]) (Fig. 3C). There was an even stronger correlation between the number of CSF clonally expanded B cells by scRNA-Seq and the CSF IgG index (R2 = 0.561, P = 0.02) (Fig. 3D). Further, we found a greater number of clonally expanded CSF B cells in RRMS subjects with gadolinium enhancement on MRI (P = 0.024) (Fig. 3E). This increase was unique to the CSF, as there was clonal expansion of CSF B cells relative to the blood in gadolinium-positive subjects (P < 0.01) and no difference in the proportion of clonally expanded B cells between the blood samples of gadolinium-positive and -negative subjects (P = ns) (Fig. 3E).
We next explored the B cell immune repertoire diversity in the CSF and blood. Interestingly, the B cell clonal expansion in the CSF manifested as restricted diversity in both the IgM+ and IgG1+ cells (Fig. 3F). Further comparing all of the scIg-Seq–derived IgM+ and IgG1+ RRMS B cells, we observed an increase in the replacement mutation frequency of the CSF IgM+ CDR sequences vs. their blood counterparts but no difference between blood IgG1+ cells, extending previous bulk Ig-Seq findings (Fig. 3G) (P < 0.001 and P = ns, respectively) (50). The H-CDR3 of CSF IgM+ cells contained more basic amino acid residues relative to the blood, and the CDR3 of CSF IgG1+ cells was longer relative to the blood, supportive of antigen experience in these cells (Fig. 3G) (P < 0.01).
To determine the phenotype of replacement mutation-bearing IgM+ B cells, we combined scRNA-Seq and scIg-Seq to analyze CSF and blood IgM+ B cells, categorizing them as either replacement mutation negative (i.e., germline) or replacement mutation positive (i.e., having undergone somatic hypermutation [SHM]) to identify differentially expressed genes between them. The genes that best distinguished germline and SHM IgM+ B cells are shown (Fig. 3H). Germline B cells were TCL1A+, CD27-, TNFRSF13B-, and GPR183-, supporting their identity as naïve B cells. SHM+ IgM+ B cells were TCL1A-, CD27+, GPR183+, and TNFRSF13B+, supporting their identity as memory B cells with a phenotype resembling previously described prediversified splenic marginal zone IgM+ B cells (Fig. 3H) (51). Indeed, when we superimposed IgM+ germline and SHM+ B cells onto our 5′ scRNA-Seq gene expression UMAP space, the SHM- cells clustered primarily with naïve B cells, and SHM+ cells clustered primarily with memory B cells (Figs. 2B and 3I). Of note, we also identified three IgM+ SHM+ B cells in HC CSF, suggesting that their simple presence in the CNS is not necessarily indicative of a diseased state.
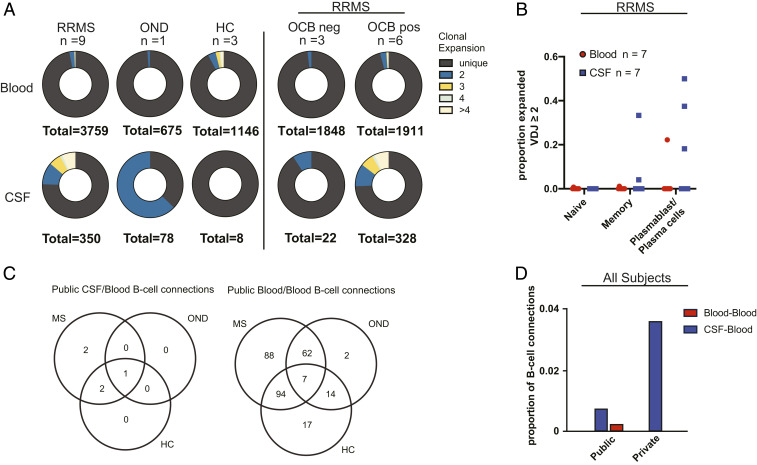
CSF, but Not Blood, B Cell Clonal Expansion Is Linked to the Presence of Unique Oligoclonal Bands in RRMS, and Some CSF B Cells in RRMS Are Highly Similar to B Cells in the Blood of Controls.
To examine whether B cell clonal expansion in RRMS is unique to the CSF, we compared the degree of clonal expansion in the blood and CSF of nine subjects with RRMS, one subject with an atypical neuroinflammatory disorder and uveitis (OND), and three HCs. Of the three HCs, two had no B cells detected in the CSF by scIg-Seq. Substantial B cell clonal expansion was observed in the CSF of patients with RRMS and OND. In contrast, clonal expansion was not detected in the CSF of the HC (Fig. 4A). As expected, we observed that CSF B cell clonal expansion was linked to the presence of unique oligoclonal bands in RRMS (Fig. 4A). We overlapped the scRNA-Seq and scIg-Seq data from seven of our RRMS subjects and identified clonal expansion primarily in the memory and plasmablast/plasma cell compartments (Fig. 4B).
Fig. 4.
(A) Donut charts of paired scIg-Seq results from blood and CSF in RRMS (n = 9), OND (n = 1), and HC (n = 3; 2 subjects had zero CSF B cells by scIg-Seq). Adjacent plots demonstrating clonal expansion in RRMS subjects with or without unique CSF oligoclonal bands (OCBs). Unique indicates a clone seen in only one cell in a subject’s sample. Clonal expansion was further divided into clone counts of two, three, four, and more than four. (B) Dot plot of cell types with clonal expansion in seven RRMS subjects in whom overlapping gene expression data were available. (C) Venn diagrams of shared B cells, determined by highly similar Ig heavy-chain sequences in the scIg-Seq cohort from an expanded set of subjects, including those without paired CSF and blood samples and with sorted B cell subsets as listed in Fig. 1A and Dataset S1 (n = 14 RRMS, n = 3 HC, n = 3 OND, n = 2 CIS). (D) Proportions of public (present in more than one subject) and private (present in only one subject) CSF B cells connected to blood (CSF/blood) and public blood B cells connected to other blood B cells (blood/blood). Of 693 total CSF B cell connections, 5 of 693 were highly similar to blood across subjects (publicly), and 25 of 693 were highly similar to blood within the same subject (privately). Of the 137,416 blood B cell connections, 284 of 137,416 were shared publicly.
Nine of 8,692 (0.1%) and 90 of 138,109 (0.06%) B cells identified exclusively across RRMS patients in the bulk and single-cell cohorts were highly similar, respectively (Fig. 4D and SI Appendix, Fig. S4). However, the presence of highly similar B cells across subjects was not specific to a given diagnosis. In the scRNA-Seq cohort, we found that 5 of 693 (0.72%) blood/CSF B cells across subjects, including in HC and OND controls, were highly similar, compared with 284 of 137,416 (0.21%) blood/blood B cells. In the scRNA-Seq cohort, we found a greater number (25 of 693, 3.61%) of B cells that were highly similar across the blood/CSF immune axis within individuals (Fig. 4D).
Lack of Epstein–Barr Virus mRNA in MS Blood and CSF B Cell Subsets.
To address the possibility of Epstein–Barr virus (EBV) transcription (or transcription of RNA from any other pathogen) in the B cells from RRMS subjects, we mined our bulk dataset for nonhuman reads. An average of 1,725,779 (interquartile range: 9,657 to 2,326,989) sequences per sample were nonhuman, high quality, and nonredundant (Dataset S1C). Of these, all sequences aligned to common environmental and reagent contaminants. No human virus transcripts, including EBV transcripts, were detected using IDseq despite detecting EBV transcripts in positive controls (i.e., RNA from the EBV-infected EHEB cell line) at total input RNA masses ranging from 10 pg to 10 ng. EBV transcripts in the positive controls ranged from 24.4 to 68.7 reads per million. Based on the limit of detection we calculated in the EHEB control, we calculate for each CSF and blood sample the approximate minimum number of EBV-infected cells we would have been able to detect at the sequencing depths we used for each sample (Dataset S1C). Additionally, no DGE in HERVs/repetitive elements was observed between individual blood and CSF B cell subsets. However, eight genes, including HERVK9-int (P adjusted = 4.68E-07) (SI Appendix, Fig. S5), were up-regulated in blood B cells compared with the CSF. The other seven genes up-regulated in the blood were LINE/L1 elements.
Discussion
While monoclonal antibody therapy against CD20-expressing B cells represents a highly effective treatment approach in MS, the roles B cells play in this complex disease are still not fully understood (52). A better understanding of B cell phenotypes in MS, particularly in the relevant body compartment (i.e., CNS), is vital to develop more effective and better targeted B cell depletion strategies. In this study, we independently characterized the transcriptional phenotypes and immune repertoires of B cell subsets from the CSF and blood of treatment-naïve RRMS subjects at both the bulk and single-cell levels. We have highlighted the potential contribution of distinct subsets of B cells to ongoing inflammation. In addition, we characterized SHM patterns with corresponding transcriptome profiles in CSF B cells in the single-cell cohort and mined the bulk B cell subset transcripts for evidence of EBV and HERV replication.
Using scRNA-Seq, we show that in MS patients, the CSF has a remarkably different cellular profile from the blood. We found very few B cells in healthy CSF, barely enough to be detected with current single-cell technology, extending previous reports using flow cytometry (53, 54). In addition, we found an expansion of CD4+ and CD8+ T cells in the CSF. The myeloid population was also distinct, with blood CD14+ and CD16+ monocytes existing in a transcriptional continuum with what we broadly termed CSF macrophages and monocyte-derived dendritic cells. The identification of these CSF macrophages by scRNA-Seq mirrors what was found in recently published scRNA-Seq studies of CSF obtained from patients with HIV-1 infection, RRMS, and anti-myelin oligodendrocyte glycoprotein antibody disorder. These investigators variously called these cells “microglia,” “microglia like,” or “monocytes” (13, 46, 47). Like the study by Esaulova and colleagues (47), we also identified these cells in the CSF of all our HCs, illustrating that their presence is not necessarily indicative of a neuroinflammatory disease process. The CSF also contained a relative dearth of NK cells compared with the blood and a relative enrichment for rare plasmacytoid dendritic cells, a cell type associated with MS relapse and activated via TLR-9 to produce large amounts of type I interferon (IFN) (55). Further subtyping of these cells was beyond the scope of our study.
Genetic variation in multiple genes in the NF-κB pathway has been identified by genome-wide association studies as a risk factor for MS (56, 57). We observed NF-κB complex activation in CSF memory (unswitched and switched) B cells. The canonical NF-κB pathway is triggered by signaling through the BCR, toll-like receptors, TREM1, and a variety of TNF receptor superfamily members, through the generation of RELA and NF-κB1 heterodimers (58, 59). TREM1, NF-κB complex, and RELA were predicted to be activated both in the bulk RNA-Seq and scRNA-Seq datasets. Additionally, in CSF SM B cells, CEBPB and ECSIT pathway members directly involved in the activation of NF-κB (58–61) were also predicted to be activated. CEBPB, the CCAAT/Enhancer Binding Protein β transcription factor, is activated by multiple inflammatory stimuli and promotes inflammation in experimental autoimmune encephalomyelitis (EAE) (60). CEBPB−/− mice are resistant to EAE with reduced lymphocyte and antigen presenting cell infiltration into the CNS. In addition, pathway and GO analyses of the bulk and single-cell datasets revealed an enrichment of terms associated with inflammation, activation, and fatty acid biosynthesis in CSF memory and plasmablast/plasma cells. Previous work indicated that inhibition of downstream biosynthesis of cholesterol promotes regulatory Th2 bias and reverses paralysis in EAE models (62, 63). While human trials did not demonstrate a clinical benefit from statins in CIS and RRMS (64–67), there are indications that statins may be beneficial in the secondary progressive form of the disease (68). Further work may identify novel therapeutic targets for MS related to the cholesterol biosynthesis pathway.
We also found evidence for a compartmentalized CSF CD27hi plasmablast/plasma cell inflammatory profile in MS. IL16, a chemoattractant whose ligand is CD4, was significantly up-regulated both in the single-cell and bulk analyses, suggesting cross-talk between CSF plasmablast/plasma cells and CD4+ T cells (69). Additionally, in the bulk RNA-Seq analyses, the CSF plasmablast/plasma cells up-regulated IL21 receptor, SOX5, UBA7, and TBX21 and inhibited IL4, suggesting these cells display an inflammatory phenotype. UBA7, an ISG15 activating enzyme, has been previously reported in systemic lupus erythematosus to represent a distinct proinflammatory subset of B cells (70). TBX21, the master transcription factor for Th-1 cell differentiation and IFN-γ gene transcription, has been recently shown to promote transmigration of B cells into the CNS (71–73). Consistent with prior MS studies, genes associated with the SMAD/TGF-β1 signaling pathway were down-regulated in CSF plasmablast/plasma cells using both single-cell and bulk analyses (74). Furthermore, TOB1, a gene involved in the SMAD/TGF-β1 signaling pathway, was down-regulated in CSF plasmablast/plasma cells (75). Low TOB1 expression, previously only reported in T cells from MS subjects and here reported in B cells, has been associated with a higher risk of MS conversion from CIS to RRMS, and its ablation worsens disease severity in EAE models (76, 77). Collectively, these findings support the conclusion that antigen-experienced CSF B cells and plasmablast/plasma cells in MS are polarized toward an inflammatory phenotype compared with their peripheral counterparts.
Previous immune repertoire studies in MS suggested that B cell maturation occurs both in the periphery and CNS with Ig class-switched B cells (IgG and IgM) providing an antigen-experienced immune axis connecting the two compartments (41, 78). In this study, we detected similar connections of highly similar B cells across the immune axis that, in a given person, are likely to be clonally related. Data from both bulk and single-cell cohorts support the finding that highly similar B cells are also shared across patients. In addition to observing B cell connections across the CSF/blood and blood/blood of subjects with RRMS, highly similar B cells were also identified between subjects in the OND group and HCs, with no significant difference in the proportion of highly similar B cells shared between MS subjects and individuals in the OND and HC groups. A recent study found that 1 to 6% of identical Ig heavy chain clones (identical IGHV, IGHJ, and H-CDR3) in the blood were shared between three HCs (79). Independent of the presence or absence of genetically similar BCRs across disease subjects and controls, more work needs to be done to characterize the antigenic specificity of expanded B cell populations in the CSF of RRMS patients and to determine whether genetically similar but not identical BCRs are indeed specific for the same antigen.
Using bulk and scIg-Seq, we found that most CSF B cells are IgM+ or IgG1+. Using diversity analysis, we showed that both IgM+ and IgG1+ B cells in RRMS subjects are less diverse in the CSF and are clonally expanded. The number of clonally expanded B cells in the CSF of MS subjects correlated with the IgG index, consistent with what was seen with proteomics studies (80). Further, we linked clonal B cell expansion to gadolinium enhancement on MRI, suggesting CSF clonal B cell expansion is heightened during active demyelination and blood–brain barrier breakdown. This extends findings from a recent study that identified a connection between the IgG index and elevated CSF neurofilament light chain, a marker of axonal damage (81). There was increased SHM in CSF IgM+ cells with increased basic residues in the H-CDR3, consistent with neoantigen exposure in the CSF. Further, IgG1+ CSF B cells had longer H-CDR3 sequences relative to their blood counterparts, again suggestive of neoantigen exposure in the CNS. Finally, we showed that while there was a greater number of CSF IgM+ cells that had undergone SHM, these cells were also present in HCs, and so, their presence in low numbers is potentially a part of normal CNS immune surveillance.
It has been hypothesized that EBV, the most consistent environmental risk factor for MS, may latently infect MS subjects’ B cells (82, 83). Some groups have identified EBV transcripts by real-time PCR, in situ hybridization, and immunohistochemistry in the CNS (84, 85), whereas others have not been able to detect EBV in these locations (86–88). We did not detect EBV transcripts (or other viral transcripts) in any of the B cell subsets using an unbiased metagenomics platform, even though EBV was detected in the latently EBV-infected control cell line EHEB at 10 pg RNA input, comparable with the amount of RNA in a typical single cell (45, 89, 90). While our findings do not rule out rare latent EBV infection with intermittent transcription, they do argue against constitutive EBV transcription in the B cells of MS subjects. Lastly, a variety of HERV transcripts were present in all of the B cell subsets we studied, but there was no difference in their expression between CSF and blood B cells. The significance of this finding is unclear without a comparable dataset generated from HC and OND cohorts.
Our study has several limitations. It is possible that transcriptional profiles are altered by sample handling, although we did use a standardized protocol and processed samples immediately upon acquisition to minimize the potential for batch effect. For the bulk RNA-Seq cohort, we sequenced sorted CSF and blood B cell subsets based on traditional CD19, IgD, and CD27 surface markers. However, this is not the only strategy for defining B cell subsets, so the particular sorting protocol we utilized is important context for appropriately interpreting the transcriptomic profiles that are derived from these sorted cell populations. To account for the bias that arises from defining cell populations based on a priori selection of a limited number of cell surface markers, we also performed scRNA-Seq on unsorted CSF and blood from an independent cohort of MS patients and defined cell identity based on scRNA-Seq profiles alone. While we highlighted pathways that were differentially regulated between CSF and blood in both the bulk and single-cell datasets, the DGE analyses for each of the cohorts were carried out independently, and the data for each set of analyses are available in Dataset S4. Future studies should take advantage of recently developed protocols that allow for the simultaneous single-cell measurements of RNA transcripts and surface proteins using DNA barcoded antibodies (cellular indexing of transcriptomes and epitopes by sequencing or CITE-Seq) (91). Finally, we cannot conclude that the CSF B cell transcriptional signatures we identified are MS specific without additional OND controls and without normative RNA-Seq profiles of HC CSF B cells, which were not feasible to obtain here given the low numbers of CSF B cells in HC CSF, as has been seen in other studies (53, 54). Nevertheless, we have used the combination of bulk RNA-Seq and scRNA-Seq, including with full-length VDJ analysis, to identify a core set of transcriptional programs in antigen-experienced CSF B cells with respect to blood B cells in patients with treatment-naïve RRMS.
In this study, we present a comprehensive profile of CNS-specific transcriptional B cell phenotypes in MS at single-cell resolution with paired immune repertoires. Further, we combine this with in-depth profiling of sorted B cell subsets. We reveal a polyclonal IgM and IgG1 CSF B cell expansion polarized toward an inflammatory, memory, and plasmablast/plasma cell phenotype, with differential up-regulation of specific proinflammatory pathways that can serve as potential therapeutic targets. Our data are consistent with a pathogenic role for CNS B cells, including evidence for active cross-talk with T cells in the CNS. We did not find evidence that CNS B cells harbor a neurotropic virus. Taken together, these data support the targeting of activated resident B cells in the CNS as a potentially effective strategy for control of treatment-resistant chronic disease.
Subject Consent.
All studies were approved by the UCSF Institutional Review Board, and written informed consent was obtained from each participant before inclusion in the study.
Supplementary Material
Acknowledgments
We express gratitude to the individuals who agreed to participate as research subjects in this study. We thank the UCSF EPIC and Origins Study Teams for valuable aid in subject recruitment and H.-Christian von Büdingen for his valuable comments on the paper. The Origins and EPIC cohorts are supported by the Valhalla Foundation. S.L.H., A.R., S.S.Z., and M.R.W. received support from NIH/National Institute of Neurological Disorders and Stroke (NINDS) Grant R01NS092835. S.L.H., A.R., and M.R.W. received support from NIH/NINDS Grant R35NS111644. R.D.S. received research support from the National Multiple Sclerosis Society and American Brain Foundation Clinician-Scientist Development Award FAN-1608-25607. A.L.G. was supported by National Multiple Sclerosis Society Clinician-Scientist Development Award Kathleen C. Moore Postdoctoral Fellowship FAN-1507-05479. S.S.Z., R.D., J.J.S., and M.R.W. received support from NIH/National Institute of Allergy and Infectious Diseases Grant R21AI142186. J.J.S. is supported by NIH/NINDS Grant K08NS107619, National Multiple Sclerosis Society Grant TA-1903-33713, and a Race to Erase MS Young Investigator Award. A.-K.P. is supported by Swiss National Science Foundation Fellowships P2SKP3_164938/1 and P300PB_177927/1 and National Multiple Sclerosis Society Kathleen C. Moore Postdoctoral Fellowship FG-1708-28871. M.R.W. received support from the Debbie and Andy Rachleff Foundation.
Footnotes
Competing interest statement: R.D.S. served on an advisory board for Sanofi Genzyme and is currently an employee of Asceneuron SA. A.J.G. reports personal fees from Inception Sciences and Mylan Pharmaceuticals and has reported serving on an end point adjudication committee for Biogen and Medimmune. He has served on trial steering committees for Novartis and serves on the Scientific Advisory Board for Bionure. R.M.B. has received personal compensation for medical legal consulting and for consulting or serving on the advisory boards of F. Hoffmann-La Roche Ltd., Sanofi-Genzyme, and Novartis. J.M.G. has received research support to University of California, San Francisco from Genentech, personal compensation for consulting for Biogen and Alexion, and personal compensation for medical legal consulting; he has also received honoraria from Dynamed Plus for editorial work. B.A.C.C. receives personal compensation for consulting from Abbvie, Biogen, EMD Serono, GeNeuro, Novartis, and Sanofi Genzyme. S.S.Z. is Deputy Editor of Neurology, Neuroimmunology and Neuroinflammation and is an Associate Editor for Frontiers in Immunology and Frontiers in Neurology. He serves on the Advisory Committee for the American Congress on Treatment and Research in Multiple Sclerosis and on the grant review committee for the National Multiple Sclerosis Society (NMSS). Previously, he has served on the Editorial Board of the Journal of Clinical Investigation, The Journal of Immunology, and The Journal of Neurological Sciences and has been a charter member of the grant review committee for the NIH Clinical Neuroimmunology and Brain Tumors. He has served as a consultant and received honoraria from Biogen-Idec, EMD-Serono, Genzyme, Novartis, Roche/Genentech, and Teva Pharmaceuticals, Inc. and has served on Data Safety Monitoring Boards for Lilly, BioMS, Teva, and Opexa Therapeutics. Currently, S.S.Z. receives research grant support from the NIH, NMSS, Weill Institute, Race to Erase MS, and the Maisin Foundation. S.L.H. currently serves on the Scientific Advisory Board of Alector, Annexon, Bionure, and Molecular Stethoscope and on the Board of Trustees of Neurona. S.L.H. also has received travel reimbursement and writing assistance from F. Hoffmann-La Roche Ltd. and Novartis for CD20-related meetings and presentations. M.R.W. received research funding from Roche/Genentech.
This article is a PNAS Direct Submission.
3A complete list of the University of California, San Francisco MS-EPIC Team can be found in SI Appendix.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008523117/-/DCSupplemental.
Data Availability.
The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus (GEO) repository under BioProject PRJNA549712 (GEO accession no. GSE133028). Additionally, gene count data for the bulk RNA-Seq cohort analyzed during this study are included in this article as part of Dataset S5.
References
- 1.Betts K. A.et al., The prevalence of hyperkalemia in the United States. Curr. Med. Res. Opin. 34, 971–978 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Kappos L.et al., Ocrelizumab in relapsing-remitting multiple sclerosis: A phase 2, randomised, placebo-controlled, multicentre trial. Lancet 378, 1779–1787 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Hauser S. L.et al.; HERMES Trial Group , B-cell depletion with rituximab in relapsing-remitting multiple sclerosis. N. Engl. J. Med. 358, 676–688 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Hauser S. L.et al.; OPERA I and OPERA II Clinical Investigators , Ocrelizumab versus interferon beta-1a in relapsing multiple sclerosis. N. Engl. J. Med. 376, 221–234 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Montalban X.et al.; ORATORIO Clinical Investigators , Ocrelizumab versus placebo in primary progressive multiple sclerosis. N. Engl. J. Med. 376, 209–220 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Nickles D.et al., Blood RNA profiling in a large cohort of multiple sclerosis patients and healthy controls. Hum. Mol. Genet. 22, 4194–4205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ottoboni L., et al., An RNA profile identifies two subsets of multiple sclerosis patients differing in disease activity. Sci. Transl. Med. 4, 153ra131–153ra131 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baranzini S. E.et al., Transcriptional analysis of multiple sclerosis brain lesions reveals a complex pattern of cytokine expression. J. Immunol. 165, 6576–6582 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Mostafavi S., Baranzini S., Oksernberg J., Mousavi P., Predictive modeling of therapy response in multiple sclerosis using gene expression data. IEEE 2006, 5519–5522 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Ratzer R.et al., Gene expression analysis of relapsing-remitting, primary progressive and secondary progressive multiple sclerosis. Mult. Scler. 19, 1841–1848 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Dutta R., Trapp B. D., Gene expression profiling in multiple sclerosis brain. Neurobiol. Dis. 45, 108–114 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raddatz B. B. R.et al., Transcriptomic meta-analysis of multiple sclerosis and its experimental models. PLoS One 9, e86643 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schafflick D.et al., Integrated single cell analysis of blood and cerebrospinal fluid leukocytes in multiple sclerosis. Nat. Commun. 11, 247 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cree B. A. C.et al.; University of California, San Francisco MS-EPIC Team , Long-term evolution of multiple sclerosis disability in the treatment era. Ann. Neurol. 80, 499–510 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cree B. A. C.et al.; University of California, San Francisco MS-EPIC Team , Silent progression in disease activity-free relapsing multiple sclerosis. Ann. Neurol. 85, 653–666 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson A. J.et al., Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 17, 162–173 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Eggers E. L.et al., Clonal relationships of CSF B cells in treatment-naive multiple sclerosis patients. JCI Insight 2, e92724 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffiths J. A., Richard A. C., Bach K., Lun A. T. L., Marioni J. C., Detection and removal of barcode swapping in single-cell RNA-seq data. Nat. Commun. 9, 2667 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hafemeister C., Satija R., Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becht E.et al., Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2018). [DOI] [PubMed] [Google Scholar]
- 21.McGinnis C. S., Murrow L. M., Gartner Z. J., DoubletFinder: Doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aran D.et al., Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernández J. M.et al.; BLUEPRINT Consortium , The BLUEPRINT data analysis portal. Cell Syst. 3, 491–495.e5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sloan C. A.et al., ENCODE data at the ENCODE portal. Nucleic Acids Res. 44, D726–D732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Risso D., Perraudeau F., Gribkova S., Dudoit S., Vert J.-P., A general and flexible method for signal extraction from single-cell RNA-seq data. Nat. Commun. 9, 284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krämer A., Green J., Pollard J. Jr., Tugendreich S., Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30, 523–530 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu G., Wang L.-G., Han Y., He Q.-Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nouri N., Kleinstein S. H., Optimized threshold inference for partitioning of clones from high-throughput B cell repertoire sequencing data. Front. Immunol. 9, 1687 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vander Heiden J. A.et al., pRESTO: A toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics 30, 1930–1932 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta N. T.et al., Change-O: A toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 31, 3356–3358 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenfield A. L.et al., Longitudinally persistent cerebrospinal fluid B cells can resist treatment in multiple sclerosis. JCI Insight 4, 126599 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Gent R.et al., Refined characterization and reference values of the pediatric T- and B-cell compartments. Clin. Immunol. 133, 95–107 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Klein U., Rajewsky K., Küppers R., Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J. Exp. Med. 188, 1679–1689 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanz I., Wei C., Lee F. E.-H., Anolik J., Phenotypic and functional heterogeneity of human memory B cells. Semin. Immunol. 20, 67–82 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruby J. G., Bellare P., Derisi J. L., PRICE: Software for the targeted assembly of components of (Meta) genomic sequence data. G3 (Bethesda) 3, 865–880 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakagawa S., Takahashi M. U., gEVE: A genome-based endogenous viral element database provides comprehensive viral protein-coding sequences in mammalian genomes. Database (Oxford) 2016, baw087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dobin A.et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H.et al., The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolotin D. A.et al., MiXCR: Software for comprehensive adaptive immunity profiling. Nat. Methods 12, 380–381 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Palanichamy A., et al., Immunoglobulin class-switched B cells form an active immune axis between CNS and periphery in multiple sclerosis. Sci. Transl. Med. 6, 248ra106–248ra106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramesh A.et al., Metagenomic next-generation sequencing of samples from pediatric febrile illness in Tororo, Uganda. PLoS One 14, e0218318 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalantar K. L., et al., IDseq—an open source cloud-based pipeline and analysis service for metagenomic pathogen detection and monitoring. bioRxiv:2020.04.07.030551 (9 April 2020). [DOI] [PMC free article] [PubMed]
- 44.Saha S.et al., Unbiased metagenomic sequencing for pediatric meningitis in Bangladesh reveals neuroinvasive chikungunya virus outbreak and other unrealized pathogens. MBio 10, e02877-e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson M. R.et al., Chronic meningitis investigated via metagenomic next-generation sequencing. JAMA Neurol. 75, 947–955 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farhadian S. F.et al., Single-cell RNA sequencing reveals microglia-like cells in cerebrospinal fluid during virologically suppressed HIV. JCI Insight 3, e121718 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Esaulova E.et al., Single-cell RNA-seq analysis of human CSF microglia and myeloid cells in neuroinflammation. Neurol. Neuroimmunol. Neuroinflamm. 7, e732 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams A. C., Mingioli E. S., McFarland H. F., Tourtellotte W. W., McFarlin D. E., Increased CSF IgM in multiple sclerosis. Neurology 28, 996–998 (1978). [DOI] [PubMed] [Google Scholar]
- 49.Losy J., Mehta P. D., Wisniewski H. M., Identification of IgG subclasses’ oligoclonal bands in multiple sclerosis CSF. Acta Neurol. Scand. 82, 4–8 (1990). [DOI] [PubMed] [Google Scholar]
- 50.Beltrán E.et al., Intrathecal somatic hypermutation of IgM in multiple sclerosis and neuroinflammation. Brain 137, 2703–2714 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Weller S.et al., Human blood IgM “memory” B cells are circulating splenic marginal zone B cells harboring a prediversified immunoglobulin repertoire. Blood 104, 3647–3654 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greenfield A. L., Hauser S. L., B-cell therapy for multiple sclerosis: Entering an era. Ann. Neurol. 83, 13–26 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Svenningsson A., Andersen O., Edsbagge M., Stemme S., Lymphocyte phenotype and subset distribution in normal cerebrospinal fluid. J. Neuroimmunol. 63, 39–46 (1995). [DOI] [PubMed] [Google Scholar]
- 54.de Graaf M. T.et al., Central memory CD4+ T cells dominate the normal cerebrospinal fluid. Cytom. Part B Clin. Cytom. 80B, 43–50 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Longhini A. L. F.et al., Plasmacytoid dendritic cells are increased in cerebrospinal fluid of untreated patients during multiple sclerosis relapse. J. Neuroinflammation 8, 2 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hussman J. P.et al., GWAS analysis implicates NF-κB-mediated induction of inflammatory T cells in multiple sclerosis. Genes Immun. 17, 305–312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Housley W. J., et al., Genetic variants associated with autoimmunity drive NFκB signaling and responses to inflammatory stimuli. Sci. Transl. Med. 7, 291ra93–291ra93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lawrence T., The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 1, a001651 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tammaro A.et al., TREM-1 and its potential ligands in non-infectious diseases: From biology to clinical perspectives. Pharmacol. Ther. 177, 81–95 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Simpson-Abelson M. R.et al., CCAAT/Enhancer-binding protein β promotes pathogenesis of EAE. Cytokine 92, 24–32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kopp E.et al., ECSIT is an evolutionarily conserved intermediate in the Toll/IL-1 signal transduction pathway. Genes Dev. 13, 2059–2071 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Youssef S.et al., The HMG-CoA reductase inhibitor, atorvastatin, promotes a Th2 bias and reverses paralysis in central nervous system autoimmune disease. Nature 420, 78–84 (2002). [DOI] [PubMed] [Google Scholar]
- 63.Greenwood J., Steinman L., Zamvil S. S., Statin therapy and autoimmune disease: From protein prenylation to immunomodulation. Nat. Rev. Immunol. 6, 358–370 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kamm C. P.et al., Atorvastatin added to interferon β for relapsing multiple sclerosis: A randomized controlled trial. J. Neurol. 259, 2401–2413 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lanzillo R.et al.; ARIANNA study group , No evidence for an effect on brain atrophy rate of atorvastatin add-on to interferon β1b therapy in relapsing-remitting multiple sclerosis (the ARIANNA study). Mult. Scler. 22, 1163–1173 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Sorensen P. S.et al.; SIMCOMBIN study investigators , Simvastatin as add-on therapy to interferon β-1a for relapsing-remitting multiple sclerosis (SIMCOMBIN study): A placebo-controlled randomised phase 4 trial. Lancet Neurol. 10, 691–701 (2011). [DOI] [PubMed] [Google Scholar]
- 67.Waubant E.et al., Randomized controlled trial of atorvastatin in clinically isolated syndrome. Neurology 78, 1171–1178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chataway J.et al., Effect of high-dose simvastatin on brain atrophy and disability in secondary progressive multiple sclerosis (MS-STAT): A randomised, placebo-controlled, phase 2 trial. Lancet 383, 2213–2221 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Skundric D. S., Cruikshank W. W., Montgomery P. C., Lisak R. P., Tse H. Y., Emerging role of IL-16 in cytokine-mediated regulation of multiple sclerosis. Cytokine 75, 234–248 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Care M. A.et al., Network analysis identifies proinflammatory plasma cell polarization for secretion of ISG15 in human autoimmunity. J. Immunol. 197, 1447–1459 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peng S. L., Szabo S. J., Glimcher L. H., T-bet regulates IgG class switching and pathogenic autoantibody production. Proc. Natl. Acad. Sci. U.S.A. 99, 5545–5550 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ji N., Sosa R. A., Forsthuber T. G., More than just a T-box: The role of T-bet as a possible biomarker and therapeutic target in autoimmune diseases. Immunotherapy 3, 435–441 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Langelaar J.et al., Induction of brain-infiltrating T-bet-expressing B cells in multiple sclerosis. Ann. Neurol. 86, 264–278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mokhtarian F.et al., Defective production of anti-inflammatory cytokine, TGF-beta by T cell lines of patients with active multiple sclerosis. J. Immunol. 152, 6003–6010 (1994). [PubMed] [Google Scholar]
- 75.Didonna A., Cekanaviciute E., Oksenberg J. R., Baranzini S. E., Immune cell-specific transcriptional profiling highlights distinct molecular pathways controlled by Tob1 upon experimental autoimmune encephalomyelitis. Sci. Rep. 6, 31603 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Corvol J.-C.et al., Abrogation of T cell quiescence characterizes patients at high risk for multiple sclerosis after the initial neurological event. Proc. Natl. Acad. Sci. U.S.A. 105, 11839–11844 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schulze-Topphoff U.et al., Tob1 plays a critical role in the activation of encephalitogenic T cells in CNS autoimmunity. J. Exp. Med. 210, 1301–1309 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stern J., et al., B cells populating the multiple sclerosis brain mature in the draining cervical lymph nodes. Sci. Transl. Med. 6, 248ra107–248ra107 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Soto C.et al., High frequency of shared clonotypes in human B cell receptor repertoires. Nature 566, 398–402 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Obermeier B.et al., Matching of oligoclonal immunoglobulin transcriptomes and proteomes of cerebrospinal fluid in multiple sclerosis. Nat. Med. 14, 688–693 (2008). [DOI] [PubMed] [Google Scholar]
- 81.Engel S.et al., Association of intrathecal pleocytosis and IgG synthesis with axonal damage in early MS. Neurol. Neuroimmunol. Neuroinflamm. 7, e679 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ascherio A., Munger K. L., Environmental risk factors for multiple sclerosis. Part I. The role of infection. Ann. Neurol. 61, 288–299 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Levin L. I., Munger K. L., O’Reilly E. J., Falk K. I., Ascherio A., Primary infection with the Epstein-Barr virus and risk of multiple sclerosis. Ann. Neurol. 67, 824–830 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aloisi F., Serafini B., Magliozzi R., Howell O. W., Reynolds R., Detection of Epstein-Barr virus and B-cell follicles in the multiple sclerosis brain: What you find depends on how and where you look. Brain 133, e157 (2010). [DOI] [PubMed] [Google Scholar]
- 85.Serafini B.et al., Dysregulated Epstein-Barr virus infection in the multiple sclerosis brain. J. Exp. Med. 204, 2899–2912 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lassmann H., Niedobitek G., Aloisi F., Middeldorp J. M.; NeuroproMiSe EBV Working Group , Epstein-Barr virus in the multiple sclerosis brain: A controversial issue—report on a focused workshop held in the Centre for Brain Research of the Medical University of Vienna, Austria. Brain 134, 2772–2786 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Willis S. N.et al., Epstein-Barr virus infection is not a characteristic feature of multiple sclerosis brain. Brain 132, 3318–3328 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sargsyan S. A.et al., Absence of Epstein-Barr virus in the brain and CSF of patients with multiple sclerosis. Neurology 74, 1127–1135 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wilson M. R.et al., A novel cause of chronic viral meningoencephalitis: Cache Valley virus. Ann. Neurol. 82, 105–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wilson M. R.et al., Diagnosing balamuthia mandrillaris encephalitis with metagenomic deep sequencing. Ann. Neurol. 78, 722–730 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stoeckius M.et al., Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus (GEO) repository under BioProject PRJNA549712 (GEO accession no. GSE133028). Additionally, gene count data for the bulk RNA-Seq cohort analyzed during this study are included in this article as part of Dataset S5.