Significance
We present a site-specific NP labeling strategy for large RNAs by transcription using the TPT3-NaM UBP system. The applicability of this labeling strategy is validated by XSI measurements on 97-nt and 719-nt RNAs with site-specific Nanogold labels. This strategy is generally applicable to any natural and artificial RNAs that can be transcribed by T7 RNA polymerase (RNAP) or covalent conjugation of RNAs with other metal NPs. The usage of a far upstream forward primer during PCR enables easy purification of RNA from DNA templates, and the nondenaturing conditions for conjugation reactions and subsequent purification steps avoids potential large RNA misfolding. This labeling strategy expands our capability to site-specifically conjugate RNAs with NPs for many applications.
Keywords: site-specific nanoparticle labeling, expanded genetic alphabet, TPT3-NaM, X-ray scattering interferometry, large RNAs
Abstract
Conjugation of RNAs with nanoparticles (NPs) is of significant importance because of numerous applications in biology and medicine, which, however, remains challenging especially for large ones. So far, the majority of RNA labeling relies on solid-phase chemical synthesis, which is generally limited to RNAs smaller than 100 nucleotides (nts). We, here, present an efficient and generally applicable labeling strategy for site-specific covalent conjugation of large RNAs with a gold nanoparticle (Nanogold) empowered by transcription of an expanded genetic alphabet containing the A-T/U and G-C natural base pairs (bps) and the TPT3-NaM unnatural base pair (UBP). We synthesize an amine-derivatized TPT3 (TPT3A), which is site specifically incorporated into a 97-nt 3′SL RNA and a 719-nt minigenomic RNA (DENV-mini) from Dengue virus serotype 2 (DENV2) by in vitro T7 transcription. The TPT3A-modified RNAs are covalently conjugated with mono-Sulfo-N-hydroxysuccinimidyl (NHS)-Nanogold NPs via an amine and NHS ester reaction and further purified under nondenaturing conditions. TPT3 modification and Nanogold labeling cause minimal structural perturbations to the RNAs by circular dichroism, small angle X-ray scattering (SAXS), and binding activity assay. We demonstrate the application of the Nanogold-RNA conjugates in large RNA structural biology by an emerging molecular ruler, X-ray scattering interferometry (XSI). The internanoparticle distance distributions in the 3′SL and DENV-mini RNAs derived from XSI measurements support the hypothetical model of flavivirus genome circularization, thus, validate the applicability of this labeling strategy. The presented strategy overcomes the size constraints in conventional RNA labeling strategies and is expected to have wide applications in large RNA structural biology and RNA nanotechnology.
Many RNAs, including the long noncoding RNAs that are arbitrarily defined as transcripts longer than 200 nts but with no or little protein coding potentials, have been found to play widespread and crucial roles in a variety of biological processes and emerge as key players in the etiology of several disease states (1). The various functions of RNAs are dictated by their propensities to form stable and complex structures (2). In light of the structure and function properties of natural RNA that can be found in nature, a large variety of artificial RNA nanostructures have been designed and constructed for diverse biotechnology applications, such as biosensing and therapeutics over the past decade (3, 4). To elucidate and manipulate the natural and artificial RNA structures, the ability to precisely incorporate either useful labels or reactive groups to specific regions within RNA molecules is required (4, 5). These labels are essential for further investigation and applications of RNAs as they enable the structural elucidation, visualization, localization, biodistribution, etc. (6).
Conjugation of NPs, particularly, metal NPs to biomolecules (protein, DNA, and RNA) is of significant importance because of numerous applications in medicine and biology, such as in sensing, imaging, diagnosis, targeted delivery, therapeutics, and structural biology (7). Due to their amenability of synthesis, unique surface, high electron density, and strong optical absorption, Nanogolds are an obvious choice in biomedical research (8). For example, Nanogolds have been used widely for labeling proteins for molecular localization by high-resolution electron microscopy (EM) (9) or as markers to directly visualize the structure and conformational changes of biomolecules by atomic force microscopy (AFM) imaging (10).
SAXS is an evolving structural technique that provides information about the shape, structure, and dynamics of biomolecules under varying solution conditions (11). It has been widely used for structural analysis of RNAs (12), DNA/RNA nanostructures (13–15), or as restraints in RNA 3D structures modeling (16). Recently, it has been found that the information content of SAXS measurements can be significantly enhanced by conjugating biomolecules or DNA nanostructures with a single or a pair of Nanogolds (17–20). SAXS measurements of biomolecules labeled with a single Nanogold can reliably identify the label positions in the low-resolution electron density maps reconstructed from SAXS data (19). SAXS measurements of biomolecules labeled with pairs of Nanogolds emerge as a new biophysical technique, XSI (20, 21). XSI is a new class of molecular ruler that can provide high-resolution label–label distance distributions ranging from 50 up to 400 Å in a model-independent fashion (17, 18), thus, can be used to quantify ensembles of macromolecule structures and be directly related to three-dimensional structures (20). Over the past few years, XSI has been fruitfully used in nucleic acids and nucleic acid/protein complexes research (22), such as probing the structure and conformational changes of DNA origami (13), DNA, or DNA-protein complexes (17, 18, 23), the conformational landscape of a complex RNA motif in response to changes in solution condition and protein binding, etc. (24, 25).
The prerequisite for successful applications of XSI is to prepare biomolecules conjugated with a single and/or a pair of Nanogolds at specific sites (20, 22). While labeling techniques have been extensively developed for proteins and DNAs (9, 17, 18, 21, 23, 26), NP conjugation to RNAs is more challenging, especially for large RNAs. So far, such RNA labelings are limited to end labeling or internal labeling through solid-phase chemical synthesis (19, 22, 24, 25). The chemical synthesis method has great capability and flexibility in generating chemical diverse RNAs but is generally limited to RNAs smaller than 100 nts. The size limits might be mitigated by combining chemical synthesis with splint-assisted enzymatic ligation where larger RNAs are assembled from smaller pieces (5, 27) or by the complementary-addressed approaches (28, 29) where custom-designed DNA probes were utilized to direct the incorporation of reactive groups into RNAs for subsequent functional coupling. All these approaches are laborious, resulting in low yield of the final products. Also, the denaturing steps to remove the DNA splints or probes during purification are likely to prompt misfolding of RNAs (27–29) especially for large ones. Alternative approaches are, therefore, highly desirable for site-specific conjugation of large RNAs with NPs under nondenaturing conditions.
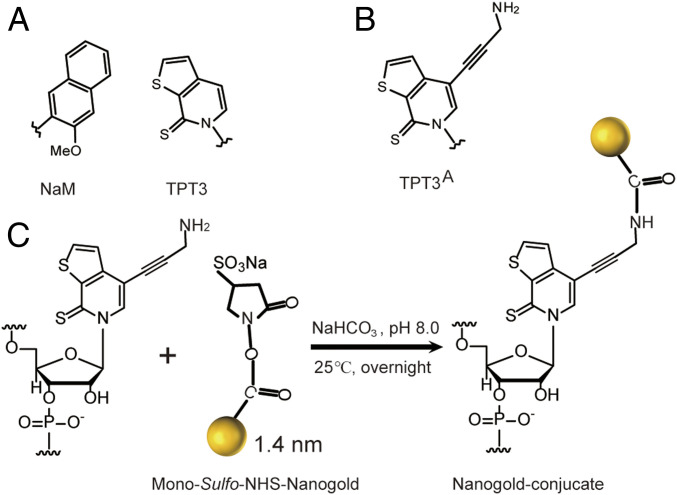
In this paper, we demonstrate an efficient and generally applicable strategy for site-specific NP labeling of large RNAs empowered by transcription of an expanded genetic alphabet containing the A-T and G-C natural base pairs and TPT3-NaM UBP. In the past decades, three groups of Benner, Hirao, and Romesberg have developed different types of UBPs that can function as a third bp in replication, transcription, and/or translation, thus, expanding the genetic alphabet (30). The expanded genetic alphabet can be used to direct the site-specific incorporation of a functionalized unnatural nts into DNAs via PCR or RNAs via in vitro transcription catalyzed by RNAPs, such as T7 (31, 32). Recently, the TPT3-NaM UBP originally developed in Romesberg’s group (Fig. 1A) was reported to exhibit natural-like efficiency and fidelity in in vitro replication and transcription (33), but its potential in directing site-specific RNA labeling has not been fully explored (34–36). We, here, synthesize a TPT3A (Fig. 1B), which is site-specifically incorporated into a 97-nt 3′SL RNA and a 719-nt DENV-mini from DENV2 (Fig. 2) by in vitro transcription with T7 RNAP. The purified unnatural RNAs are covalently conjugated with monoSulfo-NHS-Nanogolds via the highly selective amine-NHS ester reaction and purified under nondenaturing conditions (Fig. 1C). We demonstrate the application of the Nanogold-RNA conjugates in large RNA structural biology by XSI measurements. The inter-NP distance distributions in the 3′SL and DENV-mini RNAs measured by XSI support the hypothetical model of flavivirus genome circularization, thus, validate the applicability of this labeling strategy. The presented strategy overcomes the size constraints in conventional RNA labeling strategies and is expected to have wide applications in large RNA structural biology and RNA nanotechnology.
Fig. 1.
The TPT3-NaM UBP for genetic alphabet expansion. (A) The parental TPT3-NaM UBP. (B) Chemical structure of the amine-derivatized TPT3 (TPT3A). (C) Schematic for covalent conjugation of rTPT3A-modified RNAs with the mono-sulfo-NHS-Nanogold via amine-NHS ester reaction.
Fig. 2.
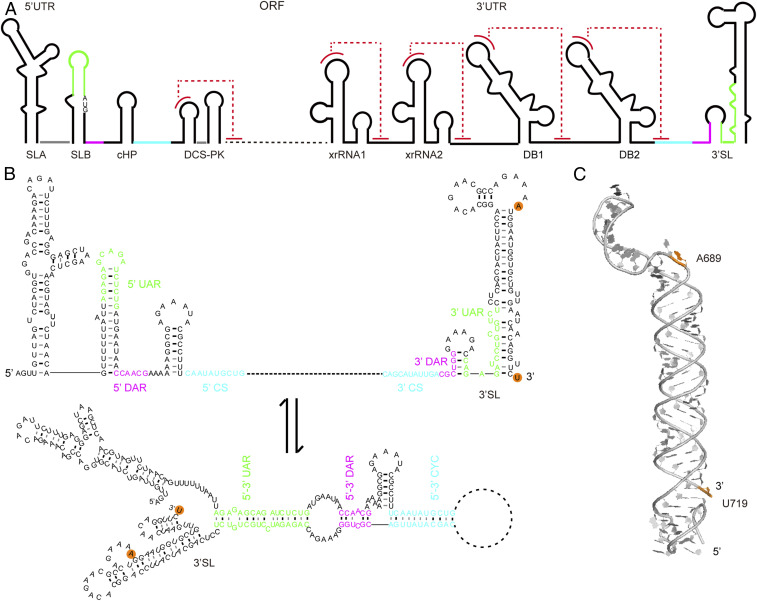
Conformational changes in the 3′SL RNA element of DENV2 upon genome cyclization. (A) The single-stranded positive sense genomic RNA of DENV2 consists of an ORF flanked with many highly structured RNA elements, such as the SLA, SLB, cHP, and DCS-PK elements located in the 5′-UTRs and the capsid protein coding regions, the xrRNA1, xrRNA2, DB1, DB2, and 3′SL elements located within the 3′ UTR. Pseudoknot formations in these elements are colored by dashed lines in red. (B) Three sets of long-range RNA–RNA interactions: 5′-3′ upstream of AUG region (UAR) (green), 5′-3′ downstream of AUG region (DAR) (magenta), and 5′-3′ cyclization sequence (CS) (cyan) mediate the linear-to-circular conformational transition of the genomic RNA upon genome cyclization, resulting in significant structural changes in RNA elements, such as the SLB and 3′SL. The modification or labeling sites corresponding to A689 and U719 in DENV-mini are indicated with orange backgrounds. (C) Atomic model of the 97-nt 3′SL element by SAXS and computation, which forms an extended conformation coaxially stacked by the sHP and the long SL (42). The distance between N9 of A689 and N1 of U719 is measured as 83.3 Å in the atomic model.
Results
Chemical Synthesis of Unnatural NTs.
To implement in vitro replication and transcription by the expanded genetic alphabet containing TPT3-NaM (Fig. 1A), the deoxyribont phosphoramidites, triphosphorylated deoxynt, and ribonts of TPT3 and NaM (dTPT3-CEP, dNaM-CEP, dTPT3TP, dNaMTP, rTPT3TP, and rNaMTP) were synthesized according to the literature procedures (31, 33, 37). Since extremely uniform 1.4 nm AuNP (Nanogolds with a single sulfo-NHS (Monosulfo-NHS) that can react with primary amines is commercially available, we synthesized amine-modified triphosphorylated ribonts of TPT3 (rTPT3ATP) (Fig. 1B) as described in the SI Appendix, SI Text. rTPT3ATP is expected to be incorporated into RNA transcripts at specific sites by expanded genetic alphabet transcription, and the TPT3A-modified RNAs are expected to allow stable covalent conjugation of the Nanogold via amine-NHS ester reaction in solution (Fig. 1C).
Selection of Labeling Sites.
Dengue virus is an important human pathogen featuring a single-stranded positive-sense RNA genome which contains an open reading frame (ORF) flanked by highly structured untranslated regions (UTRs) (Fig. 2A). During infection, the DENV genome serves as mRNA for translation, template for RNA synthesis, and substrate for encapsidation and is predicted to undergo conformational transition between the linear and the circular forms to fulfill its diverse function (38, 39). While the linear conformation is translation competent, the circular conformation is more efficiently replicated (39). The circular conformation is stabilized by long-range RNA–RNA interactions mediated by the inverted complementary sequences (5′-3′ UAR, 5′-3′ DAR, and 5′-3′ CS) at both the 5′ and the 3′ ends of the genome (Fig. 2B) (40), resulting in conformational changes in several conserved RNA structures. One example is the 3′SL structure, which harbors the 3′ UAR and 3′ DAR sequences and consists of a small hairpin (sHP) followed by a large stem loop (SL) in the linear genomic RNA, and this feature is highly conserved among all flavivirus genomes (Fig. 2B) (41). Upon genome circularization, the 3′SL RNA element is expected to open the large stem and release the last nt of the genome, thus, adopts a conformation that has a large SL ahead of a small one (Fig. 2B) (38, 39), acting as template RNA to initiate replication. A balance between circular and linear forms of the DENV genome is crucial for viral replication (39), however, the conformational changes at the tertiary level have not been studied before.
The 97-nt 3′SL RNA alone (SI Appendix, Fig. S1A) lacks the complementary sequences within the 5′ end of the genome, thus, representing its conformation in the linear genomic RNA. Recently, the 3′SL RNA alone is confirmed to have an extended rodlike structure in solution by SAXS (Fig. 2C) (42). The 719-nt DENV-mini RNA, which consists of all of the elements in 5′ UTR, 3′UTR, and part of the capsid coding sequence of the genomic RNA, is a minimal and efficient template for translation and minus strand RNA synthesis in vitro (43). The secondary structure of DENV-mini (SI Appendix, Fig. S1B) was recently probed by SHAPE probing, which verified the long-range RNA–RNA interactions between the 5′ and the 3′ terminal regions that mediates genome circularization (44). Thus, the smaller DENV-mini RNA can resemble the circular conformation of the full-length genomic RNA of DENV2, which is generally larger than 11 kb and difficult to study by XSI directly. To study the conformational changes in the 3′SL upon genome circularization by XSI, the small 3′SL RNA alone and DENV-mini RNA are chosen to represent the linear and circular genomic RNAs, respectively. Two identical sites in both 3′SL and DENV-mini corresponding to A689 and U719 (referred to as DENV-mini) are selected to be labeled with TPT3A for Nanogold conjugation. Based on the secondary structures, A689 and U719 are located at the internal loop and 3′-terminal regions, respectively, in both 3′SL and DENV-mini (SI Appendix, Fig. S1), incorporation of TPT3A at these sites is expected to avoid significant structural perturbations and allow for conformational changes to be monitored.
Preparation of DNA Templates Containing UBPs at Specific Sites.
Previously, it was reported that dNaM in the template strand of the double-stranded DNA (dsDNA) template can direct the incorporation of its partner into RNA more efficiently and selectively than dTPT3 in the template strand (45). To prepare single and/or double TPT3- or TPT3A-labeled RNAs by expanded genetic alphabet transcription, we generate dsDNA templates containing one or two dNaMs in the template strand at specific sites by PCR. Two plasmid codings for 3′SL and DENV-mini RNAs with an upstream T7 promoter, respectively, are total gene synthesized as PCR templates (SI Appendix, Table S1). To introduce the UBPs into the dsDNA templates, the UBPs are first incorporated into the single-stranded DNA (ssDNA) primers using standard solid-phase chemical synthesis with the phosphoramidites of dNaM or dTPT3 (Fig. 3A). As the two labeling sites (A689 and U719) are close to the 3′ end of 3′SL, only a pair of forward and reverse primers are needed for generating the dsDNA templates for each constructs in one PCR. In the case of preparation of dsDNA templates with two distant labeling sites, two pairs of primers can be synthesized, and an overlap extension PCR protocol can be employed (46, 47). A common forward primer pMVF along with three reverse primers containing one or two dNaMs at sites corresponding to A689 and/or U719 are initially synthesized (SI Appendix, Table S2). The forward primer pMVF is designed to target a common sequence 390 bp upstream of the T7 promoter in the plasmids (Fig. 3A), such a design results in dsDNA templates significantly larger than the RNA transcripts, enabling subsequent efficient and easy RNA purification (see below). Directed by the respective pair of the natural forward and unnatural reverse primers, PCR reactions are performed with a mixture of dNTPs of A, T, G, C, NaM, and TPT3. The PCR amplifications produce dsDNA templates containing one or two dNaMs in the template strand at sites corresponding to A689 or/and U719 for both 3′SL and DENV-mini RNAs with lengths of 504 and 1128 bps, respectively (SI Appendix, Fig. S2). Since the TPT3-NaM UBP has a natural bplike efficiency and fidelity in in vitro replication, and the dNaMs are first introduced into the reverse primers, no mutagenesis in the template strand of dsDNA templates is expected during PCR amplification. The PCR products are directly used for subsequent transcription without further purification.
Fig. 3.
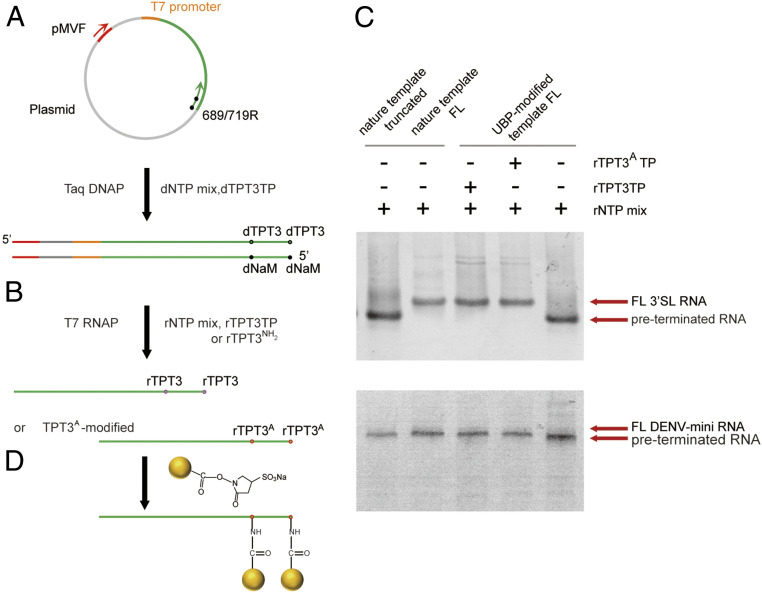
A general strategy for site-specific Nanogold labeling of large RNAs using an expanded genetic alphabet containing TPT3-NaM. (A) The UBPs were first incorporated into the reverse ssDNA primers targeting the plasmids by solid-phase chemical synthesis with the phosphoramidites of dNaM or dTPT3. Directed by an upstream forward primer and the respective reverse primers, dsDNA templates containing UBPs at specific sites were amplified by PCR. (B) rTPT3 or rTPT3A can be incorporated into RNAs by in vitro transcription with dsDNA templates containing UBPs (dNaM in the template strand) using the six-letter expanded genetic alphabet. (C) 10% (for 3′SL, top) or 6% (for DENV-mini, down) native PAGE analysis of the in vitro transcripts from natural DNA templates or UBP-modified unnatural DNA templates. The transcription of truncated (up to A688) or full-length (FL) natural DNA template (Left two) using rNTP mix resulted in one single band for 3′SL (top) and DENV-mini (down). The transcription of unnatural DNA template (Right most) in the absence of unnatural nts resulted in preterminated smeared bands. The transcription of the unnatural DNA template (Middle) in the presence of rTPT3 or rTPT3A resulted in a singe dense band with similar migration as FL RNAs. (D) MonoSulfo-NHS-Nanogold labels are coupled to TPT3A-modified RNAs via the amine-NHS ester reaction.
Site-Specific Incorporation of rTPT3 or rTPT3A into RNAs.
Although the in vivo replication and transcription of TPT3-NaM UBP in Escherichia coli, eukaryotic yeast semisynthetic organisms (48, 49), and the site-specific incorporation of three derivatives of TPT3 into RNAs have been validated recently (34–36), the ribonts of TPT3 and TPT3A have not been tested in in vitro transcription with T7 RNAP. The dsDNA templates containing dNaMs in the template strand for 3′SL and DENV-mini are used to explore site-specific incorporation of rTPT3 and/or rTPT3A into RNAs.
We first characterize the ability of T7 RNAP to incorporate rTPT3 and rTPT3A into the 97-nts 3′SL RNA (Fig. 3B). The transcription of a DNA template containing one dNaM at a site corresponding to A689 of 3′ SL RNA is first examined with the natural ribotriphosphates and either rTPT3 or rTPT3A. Under the conditions employed (SI Appendix, Table S4), virtually no full-length but preterminated product is observed in the absence of an unnatural triphosphate (Fig. 3C). The termination site is likely right before the modified site (A689) as a truncated natural transcript up to 688 migrates at the same rate as the preterminated product. These data show that rTPT3 or rTPT3A is essential for correct transcription of DNA templates containing dNaM in the template strand. On the contrary, addition of either rTPT3 or rTPT3A results in efficient production of the full-length transcription product (Fig. 3C). These data suggest that enzymatic incorporation of both rTPT3 and rTPT3A into RNA is feasible via in vitro T7 transcription. We further confirm the selective incorporation of rTPT3 or rTPT3A into the 719-nt DENV-mini RNA using its dsDNA template containing dNaM at a site corresponding to A689, and similar results are observed (Fig. 3C).
For XSI data acquisition, usually a sample quartet including the unmodified, the two orthogonal single site, the one double- modified construct, and the free Nanogold sample are prepared (20, 22). We then prepared the A689TPT3/U719TPT3 (for the unlabeled), A689TPT3A (for one single-site Nanogold labeling), U719TPT3A (for another single-site Nanogold labeling), and A689TPT3A/U719TPT3A (for double-site Nanogold labeling) modified RNAs of 3′SL and DENV-mini, respectively, by in vitro transcription in the presence of natural ribonts triphosphates mix (A, U, G, and C) and rTPT3TP or rTPT3ATP. As the dsDNA templates are much larger than the RNA transcripts, each of the RNA transcripts is well resolved from the dsDNA template in the polyacrylamide gel electrophoresis (PAGE), digestion of dsDNA templates is not needed prior to native purification. The transcription supernatants are directly applied to the Superdex 75 (for 3′SL) or 200 (for DENV-mini) gel filtration columns, the RNAs are purified from the dsDNA templates, the abortive transcripts, and the excess rNTPs by size-exclusion chromatography (SEC) (SI Appendix, Fig. S3A) and shown as a single band in native PAGE gel (SI Appendix, Fig. S3B).
Site-Specific Nanogold Labeling of RNAs.
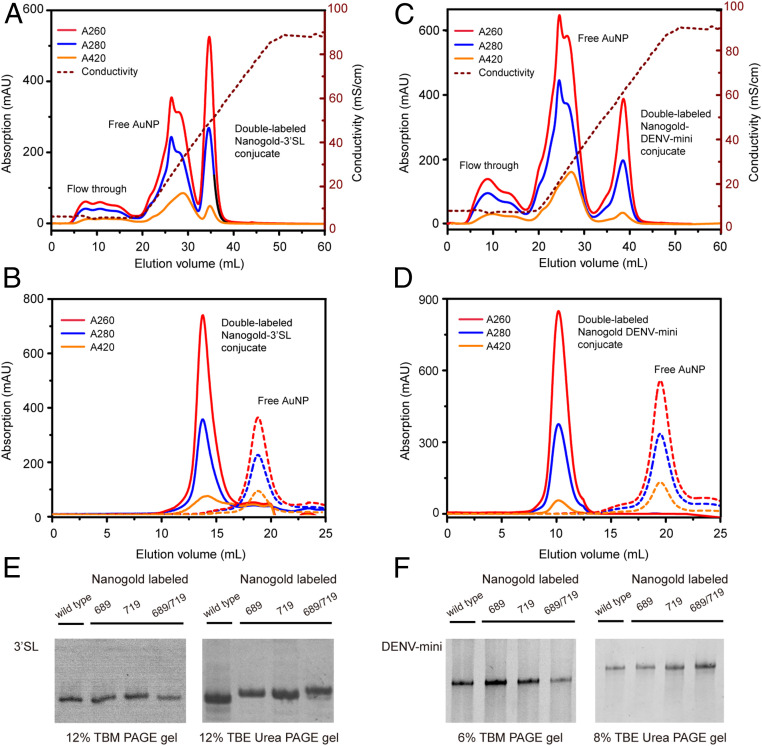
Purified unnatural 3′SL and DENV-mini RNAs containing one TPT3A at sites of 689 or 719 or two TPT3A at both sites of 689 and 719 are subjected to covalent conjugation with a two- to threefold excess of the 1.4 nm MonoSulfo-NHS-Nanogold from Nanoprobe (https://www.nanoprobes.com/), resulting in stable RNA conjugates with one site or double-labeled Nanogold (Fig. 3D). Reaction of primary amine with NHS esters at physiological pH in aqueous environment is fast and highly selective, and cross-linking by this reaction is one of the most commonly used methods in RNA labeling (5). Freshly prepared MonoSulfo-NHS-Nanogold is immediately reacted with TPT3A-modified RNA samples in order to avoid hydrolysis of the NHS ester. The pH is maintained between 7 and 9 in 0.1 M NaHCO3 since the hydrolysis of the NHS ester is pH dependent. The Nanogold-RNA conjugates can be purified from the excess free Nanogold by anion exchange chromatography using the Hitrap Q column (Fig. 4 A and B). The Hitrap Q column is preequilibriumed with three column volumes of the low salt buffer A (20 mM 4-[2-hydroxyethyl]-1-piperazineethanesulfonic acid [Hepes], 20 mM KCl, 5 mM Mg2+, 3% glycerol, and 1 mM tris[2-carboxyethyl]phosphine [TCEP], pH 7.5). During sample application, a typical elution profile shows small but broad flow-through peaks containing free Nanogold, which do not efficiently bind the Hitrap Q matrix at the low salt concentration of buffer A since the surface charge of Nanogold is low. After loading, a salt gradient up to 100% buffer B (buffer A supplemented with 0.98 M KCl) in six column volumes is applied to ensure complete removal of the free Nanogold before elution of the Nanogold-RNA conjugate. Nanogold-RNA conjugates elute depending on the overall phosphate charge per molecule. While the elution profiles are similar and well separated between peaks for the double-labeled Nanogold-3′SL conjugate (Fig. 4A) and the double-labeled Nanogold-DENV-mini RNA conjugate (Fig. 4B), the elutions start from 55% up to 70% buffer B (Fig. 4A) and from 60% up to 75% buffer B (Fig. 4B), respectively. This can be partly explained by more negative charges in the Nanogold-DENV-mini conjugate than in the Nanogold-3′SL conjugate. The Nanogold-RNA conjugates are further purified with SEC, and each elutes as a single peak (Fig. 4 C and D). The nondenaturing purification protocols for Nanogold-labeled 3′SL and DENV-mini RNAs avoid the denaturing steps used in other labeling methods (27–29), which help the RNAs preserve conformations formed during cotranscriptional folding.
Fig. 4.
Purification of Nanogold-labeled RNA samples under nondenaturing conditions. (A and B) The elution profiles of double Nanogold-labeled 3′SL RNA (A) and double Nanogold-labeled DENV-mini RNA B by anion exchange chromatography. Peaks corresponding to the flow-through, free Nanogold, the Nanogold-RNA conjugates, and the curves of the conductivity are indicated. (C and D) The elution profiles of double Nanogold-labeled 3′SL (C) and double Nanogold-labeled DENV-mini (D) RNAs by SEC. The elution profile of free Nanogold is shown in C and D for comparison. (E) Native (Left) and denaturing urea (Right) PAGE gel analysis of singly (at sites of 689 and 719, respectively) and doubly (at sites of both 689 and 719) Nanogold-labeled 3′SL RNAs. Wild type 3′SL is included for comparison. (F) Native (Left) and denaturing urea (Right) PAGE gel analysis of singly (at sites of 689 and 719, respectively) and doubly (at sites of both 689 and 719) Nanogold-labeled DENV-mini RNAs. Wild type DENV-mini RNA is included for comparison.
Purified singly and doubly labeled Nanogold-RNA conjugates are shown as a single band in both native and denaturing PAGE gels (Fig. 4 E and F). While the migration rates for singly, doubly, and unlabeled Nanogold-RNA conjugates are very similar in native PAGE gels, they differ obviously in denaturing PAGE gels, especially for that of 3′SL RNAs. The differences in migration rates in denaturing PAGE gels can be explained by the differences in the overall molecular weight (MW) of the Nanogold-RNA conjugates. Given an estimated MW of 15 kDa for Nanogold (50), the Nanogold label accounts for 31.7% and 48.2% mass of the singly and doubly labeled Nanogold-3′SL conjugate but 6% and 11.2% mass of the singly and doubly labeled Nanogold-DENV-mini conjugate, thus, the differences in migration rates are more prominent for Nanogold-labeled 3′SL than DENV-mini.
The successful conjugation of Nanogold to the TPT3A-modified RNAs can also be verified by ultraviolet–visible (UV–vis) absorbance at 420 nm. Generally, Nanogold exhibits prominent absorption at both 260 and 420 nm, and natural RNA has strong absorption at 260 nm but very weak absorption at 420 nm. Thus, the absorption peak at 420 nm for the RNA conjugates indicate stable Nanogold coupling to RNAs (Fig. 4 A–D). The Nanogold coupling efficiency can be calculated from the absorption of the RNA conjugates at both 260 and 420 nm, respectively, using the respective molar extinction coefficients of RNAs and Nanogold (26). The efficiencies of the singly and doubly labeling range from 74 to 88% (SI Appendix, Table S5).
Effects of TPT3 Modification and Nanogold Labeling on RNA Structures.
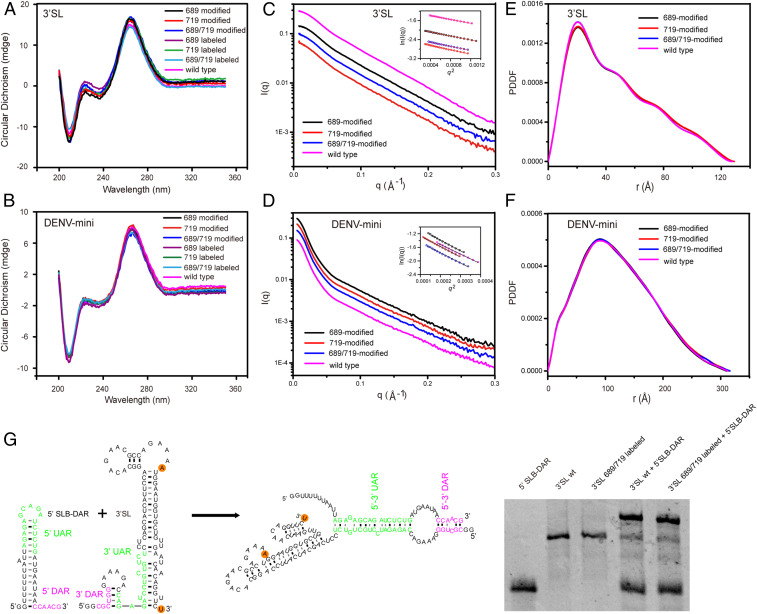
To study the effects of UBP modification and NP labeling on the structures of 3′SL and DENV-mini RNAs, we measure CD spectra of wild type, TPT3 modified, as well as Nanogold-labeled 3′SL and DENV-mini RNAs under the same solution conditions. The CD spectrum provides information on the helical structures of DNA and RNA, thus, has been widely used to study secondary structure formation in nucleic acids (51). As shown in Fig. 5 A and B, all CD spectra of 3′SL (Fig. 5A) or DENV-mini (Fig. 5B) RNAs (wild type, single- and double-TPT3-modified, and single- and double-Nanogold labeled) display the same characteristic absorption profiles. These CD data reveal that the UBP modification and Nanogold labeling have minor effects on secondary structures of the 3′SL and DENV-mini RNAs, consistent with previous results that Au nanocrystal labels do not substantially perturb RNA structures (25).
Fig. 5.
Effects of TPT3 modification and Nanogold labeling on RNA structures. (A and B) CD spectra of TPT3-modified (689 singly, 719 singly, and 689/719 doubly) and Nanogold-labeled (689 singly, 719 singly, and 689/719 doubly) 3′SL (A) and DENV-mini (B) RNAs. Wild type 3′SL (A) and DENV-mini (B) are included for comparison. (C and D) Experimental scattering profiles of native (magenta), 689-modified (black), 719-modified (red), and 689/719 double-modified (blue) RNAs for 3′SL (C) and DENV-mini (D). The Insets in C and D are the respective linear fittings of the Guinier plotting. (E and F) Overlay of the respective PDDFs of native (magenta), 689-modified (black), 719-modified (red), and 689/719 double-modified (blue) RNAs for 3′SL (E) and DENV-mini (F). (G) EMSA of 5′SLB-DAR in the absence or presence wild type or Nanogold-labeled 3′SL RNAs (Right). The secondary structures of 5′SLB-DAR, 3′SL, and the complex are also shown (Left).
We next investigate the influence of the incorporation of TPT3 on RNA overall structures by SAXS. The scattering profiles, with scattering intensity I(q) plotted against momentum transfer q, along with pair distance distribution functions (PDDFs) transformed from the scattering profiles for all of the UBP-modified RNAs and the wild-type RNAs of 3′SL and DENV-mini are shown in Fig. 5 C–F. The Guinier regions of all of the scattering profiles are linear (Fig. 5 C and D), indicating that all of the RNA samples are monodisperse and homogeneous in solution. Comparing the scattering profiles and PDDFs of the TPT3-modified RNAs with the wild-type RNAs of the 3′SL and DENV-mini, respectively, we concluded that TPT3 modification has little effects on the overall structures of the respective RNAs. The overall structural parameters, including the radius of gyration Rg calculated from the slopes of Guinier fitting Rg, the maximum dimension Dmax from PDDF functions, as well as MWs derived from volume-of-correlation (Vc) (52), are summarized in SI Appendix, Table S6, among which the MWs calculated from SAXS data are consistent with those predicted from sequences, indicating that all of the RNA constructs are monomeric in solution. The slight variations of the Rg and Dmax upon UBP modification further prove the absence of significant overall structural perturbations.
We further assess whether Nanogold labeling affects 3′SL RNA function. As shown in Fig. 2B and 3′SL harbors the 3′ UAR and 3′ DAR sequences. A RNA called 5′ short SL (SLB)-DAR that harbors the 5′ UAR and 5′ DAR is constructed and its interactions with wild type and Nanogold-labeled 3′SL RNAs are evaluated using electrophoretic mobility shift assay (EMSA). As shown in Fig. 5G, both wild type and double Nanogold-labeled 3′SL RNAs interact with SLB-DAR to form a complex, suggesting that Nanogold labeling on 3′SL RNA does not impair its folding and binding activities. Overall, circular dichroism (CD) spectroscopy, SAXS, and EMSA assays show that the TPT3 modification and Nanogold labeling minimally perturb the structure and function of the 3′SL and DENV-mini RNAs.
Distance Measurements by XSI.
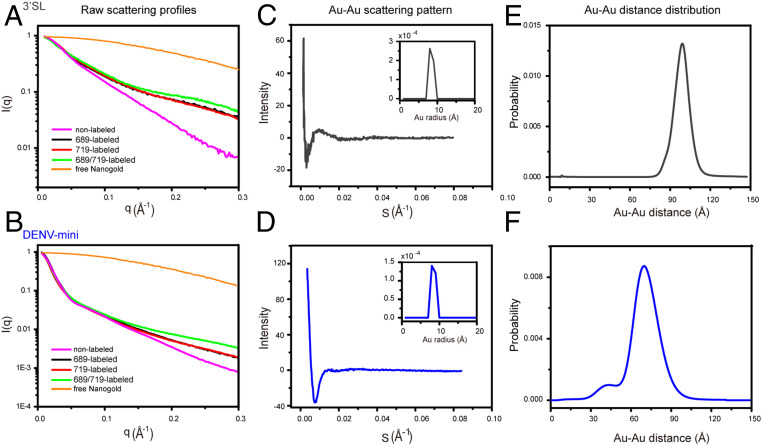
Measurements of intra- or intermolecular distances are key to dissect the structures, dynamics, and functions of biomolecules (21). To demonstrate the applications of Nanogold-conjugated RNAs in large RNA structural biology, we measured the distance distributions between sites of A689 and U719 in the 3′SL and DENV-mini RNAs by XSI. SAXS experiments are performed for the RNA sample quartets of 3′SL and DENV-mini RNAs along with the free Nanogold, respectively. The scattering profiles normalized against I0 for the free Nanogold, nonlabeled RNAs, and the two orthogonal single-labeled, doubled-labeled Nanogold-RNA conjugates of 3′SL and DENV-mini are shown in Fig. 6 A and B, respectively. In comparison with the scattering profiles of nonlabeled 3′SL, the two single-labeled and the double-labeled Nanogold-RNA conjugates show systematic increase in intensity at elevated q range from 0.15 to 0.30 Å−1 (Fig. 6A), which is consistent with stable conjugation of the RNAs with one or two Nanogolds that scatter the X rays strongly. A similar pattern is observed for the DENV-mini RNAs (Fig. 6B). The overall structural parameters derived from scattering profiles including Rg and Dmax are shown in SI Appendix, Table S6. While the Rg and Dmax of internal single-labeled (689-labeled) Nanogold-RNA conjugates are similar to that of the nonlabeled native RNA, the Rg and Dmax of the 3′-terminal single-labeled (719-labeled) and double-labeled (689/719-labeled) Nanogold-RNA conjugates are slightly larger than that of the nonlabeled native RNA, this may be due to the size of the Nanogold and the differences in the labeling sites.
Fig. 6.
Measuring Nanogold–Nanogold distance distributions from Nanogold-conjugated 3′SL and DENV-mini RNA samples by XSI. (A and B) I0-normalizzed scattering profiles of free Nanogold (orange), 689 single-labeled (black), 719 single-labeled (red), 689/719 double-labeled (green) Nanogold-RNA conjugates, nonlabeled native RNA (magenta) for 3′SL (A), and DENV-mini (B). (C and D) Scattering interference pattern of the two Nanogold labels in 3′SL (C) and DENV-mini (D). The Insets are the radius distribution of the free Nanogold used in the experiment, showing a maximum probability at 7 Å. (E and F) Probability distribution of the center-to-center distance between the two Nanogold labels in 3′SL (97.7 Å) (E) and DENV-mini (78.2 Å) (F) with variance of 52.7 and 83.3 Å2, respectively. A shoulder peak with mean distance of 40 Å was observed for DENV-mini.
The full set of scattering profiles of the RNA quartets and the free Nanogold were combined to derive Nanogold–Nanogold scattering interference profiles (Fig. 6 C and D) and the center-to-center distance distributions between the pairs of Nanogold (Fig. 6 E and F) for the 3′SL and DENV-mini RNAs, respectively, by following the previously described protocols (20, 22). Both Nanogold–Nanogold scattering interference profiles show oscillating peaks, but the oscillation pattern is more prominent for the 3′SL than the DENV-mini RNA, suggesting enhanced flexibility in the DENV-mini than in 3′SL alone, which has been confirmed to be relatively rigid (42). Transformation of the scattering interference profiles result in major distance distributions with mean values of 97.7 and 78.2 Å and variance of 52.72 and 83.3 Å2 for the 3′SL (Fig. 6 C and E) and DENV-mini (Fig. 6 D and F) RNA samples, respectively. The distance between N1 of U719 and N9 of A689 is measured as 83.3 Å in the atomic model of 3′SL derived from SAXS data (Fig. 1C). Given the radius of the Nanogold and the length of the linkers, the mean Nanogold–Nanogold distance of 97.7 Å in 3′SL RNA is reasonably consistent with the atomic model. The 20 Å decrease of the mean Nanogold–Nanogold distance between labeling sites in DENV-mini RNA supports conformational changes in 3′SL upon genome circularization mediated by long-range RNA–RNA interactions between the 5′ and the 3′ ends. The larger variance of the distance distribution in DENV-mini RNA is consistent with its enhanced flexibility, suggesting that the 3′SL element becomes more flexible and may adopt an ensemble of conformations upon genome circularization, which is further evidenced by the small shoulder peak with mean distance of 40 Å (Fig. 6F).
Discussion
In this paper, we present an efficient and generally applicable labeling strategy for site-specific covalent conjugation of large RNAs with Nanogold empowered by expanded genetic alphabet transcription. By site-specific labeling of 97-nt 3′SL and 719-nt DENV-mini RNAs with one or a pair of Nanogold labels, we measured the inter-Nanogold distance distributions between the labeling sites with XSI, which validates the applicability of this labeling strategy for large RNAs. We provide experimental evidence on tertiary conformational changes to such large RNAs caused by long-range RNA–RNA interactions, thus, supporting the hypothetical flavivirus genome circularization model. This labeling strategy overcomes the size constraints in conventional RNA-labeling strategies. In principle, it is applicable to RNAs with sizes from tens up to thousands of nts due to the high processivity of T7 RNAP. The design of a far upstream forward primer enables easy and efficient native purification of the RNA transcripts from DNA templates and excess nts. Since all of the reactions are performed in near physiological conditions, it avoids the denaturing steps in other labeling strategies and helps preserve the native conformations of large RNAs for subsequent analysis.
The presented strategy is expected to promote wide applications of Nanogold-RNA conjugates in large RNA structural biology. To our knowledge, the 97-nt 3′SL and the 719-nt DENV-mini RNAs are among the largest ones that have been site-specifically conjugated with Nanogolds and studied by XSI. As XSI can measure much broader distance distributions (ranging from 50 up to 400 Å) than other molecular rulers, such as pulsed electron paramagnetic resonance spectroscopy and Förster resonance energy transfer that provide distance information from 20 to 80 Å (18), XSI can play important roles in the structural study of large RNAs and RNA complexes, e.g., the long noncoding RNAs. For example, the XSI-derived distance distributions can be used to aid in computational 3D modeling of large RNAs (16) or integrative structural modeling of RNA-protein complexes (53). Furthermore, we envision that Nanogold-RNA conjugates can be utilized in studying the structure and dynamics of RNAs by cryo-EM, an EM-based technique called individual-particle electron tomography or high-resolution AFM (9, 54), which site-specific Nanogold-labeled RNA samples are required. Very recently, the structure and motion of the long noncoding RNA HOTAIR (2,158 nts) have been visualized by AFM, but better anatomy assignments could be achieved with AFM imaging if site-specific Nanogold-labeled RNA is prepared with the presented labeling strategy (55).
The presented strategy could also have wide applications in RNA nanotechnology (4). Recently, several fluorogenic or cell-specific RNA aptamers have been developed for live cell applications (4). Bioconjugation of RNA aptamers with NPs could combine unique and orthogonal strengths of the specific interactions of RNA nanostructures and novel material properties of NPs and open up new options for a wide range of applications (10). For example, the utility of SAXS and AFM for analysis of NP-conjugated RNA nanostructures could also be established (13). RNA aptamer-NP conjugates could be powerful diagnostic and therapeutic tools (56).
Last but not least, the presented strategy can be used for site-specific conjugation of RNAs with other metal NPs including silver, titanium oxide, zinc, and ion NPs for diverse biomedical applications (57). The expanded genetic alphabet transcription also allows for flexible chemistry for site-specific covalent conjugation. For example, alkyne- or azide-derivatized UBP (e.g., TPT3) can be synthesized and incorporated into RNAs by expanded genetic alphabet transcription for subsequent site-specific conjugation with commercially available Click Nanogold probes (https://www.nanoprobes.com/) via click chemistry (31, 32).
Materials and Methods
Materials.
The dNaM-CEP/dTPT3-CEP, dTPT3TP/dNaMTP, and rTPT3TP/rNaMTP were synthesized according to the literature procedures (31, 33, 37). The 2× golden mix of Taq DNA polymerase (with buffer and natural dNTPs mix included) was purchased from TSINGKE Biological Technology Co., Ltd. (Beijing, China). The T7 RNAP was homemade (58). Briefly, the full-length T7 RNAP with N-terminal His6-tag was expressed in E. coli BL21 by induction with isopropyl-1-thio-d-galactopyranoside. T7 RNAP was purified by nickel chromatography followed with SEC. The T7 RNAP stock (1 to 2 mg/mL) was stored at −80 °C in 20 mM 2-amino-2-hydroxymethyl-1,3-propanediol 8.0, 100 mM KCl, 10 mM dithiothreitol, and 50% vol/vol glycerol prior to use. The 1.4 nm Mono-Sulfo-NHS-Nanogold with the molar extinction coefficient at 420 nm being 155,000 M−1cm−1, was purchased from Nanoprobes (https://www.nanoprobes.com/). Plasmids encoding the 3′SL or the DENV-mini RNAs of DENV2 with an upstream T7 promoter were total-gene synthesized and sequenced by Wuxi Qinglan Biotechnology, Inc., Wuxi, China. All ssDNA primers containing natural or unnatural nts were synthesized and purified with ogliont purificationcartridge purification by TSINGKE. The DNA sequences of the plasmids and the natural and unnatural primers in this paper could be found in SI Appendix, Tables S1 and S2, respectively. All of the polyacrylamide gels were stained with GelSafe (YPR-Bio) and imaged with a Tanon 2,500 gel imaging system.
Preparation of Natural- or UBP-Modified DNA/RNAs.
With the synthesized plasmids as templates and directed by the respective pairs of primers (SI Appendix, Table S2), the dsDNA templates for the respective RNA constructs were generated by PCR reactions (SI Appendix, Table S3). Prior to large scale (2–5 mL transcription reaction mixture) RNA sample preparation, the optimal conditions for in vitro transcription were screened against Mg2+ and DNA template concentrations in a 20 µL system. The same NaM-containing dsDNA templates were used for incorporation of rTPT3 or rTPT3A into 3′SL and DENV-mini RNAs by in vitro transcription (SI Appendix, Table S4). The transcription supernatants were directly applied to Superdex 75 (for 3′SL) or 200 (for DENV-mini) gel filtration columns, and the RNAs were purified by SEC. The SEC buffer contains 20 mM Hepes pH 7.5, 100 mM KCl, and 5 mM Mg2+. RNA fractions were collected and concentrated with the 10 K Amicon Ultra centrifugal filter devices (Sigma-Aldrich) and stored at −80 °C for further use. The concentrations of native or Nanogold-labeled RNA samples were determined by UV–vis absorbance at 260 or 420 nm, respectively, on a NanoDrop 2,000 (Thermo Scientific). The molar extinction coefficients of RNAs at 260 nm were calculated from sequences with the OligoAnalyzer Tool (https://sg.idtdna.com/pages/tools/oligoanalyzer).
Site-Specific Nanogold Labeling of RNAs.
The purified single or double TPT3A-modified 3′SL and DENV-mini RNAs were buffer exchanged into 0.1 M NaHCO3, pH 8.0 with the 10 K Amicon Ultra centrifugal filter devices and subjected to Nanogold labeling. Some 1.4 nm Nanogold was dissolved with DEPC H2O following the product instruction manuals (https://www.nanoprobes.com/). The coupling reactions were initiated by mixing 8 nM single- or double-site TPT3A-modified RNA samples with 16 or 32 nM 1.4 nm Nanogold, respectively, then incubated at 25 °C overnight. To increase the solubility and coupling efficiency of Nanogold, 0.05% (vol/vol) Triton X-100, 0.1% (vol/vol) Tween 20, and 20% (vol/vol) dimethyl sulfoxide was added in the reaction mixture. Reactions were carried out at room temperature overnight. Nanogold-RNA conjugates were purified after the reactions were performed. The reaction mixtures were first injected into Hitrap Q column (GE Healthcare), preequilibrated with buffer A (20 mM Hepes, 20 mM KCl, 5 mM Mg2+, 3% glycerol, and 1 mM TCEP, pH 7.5), and, then, eluted with a salt gradient to 100% buffer B (20 mM Hepes, 1 M KCl, 5 mM Mg2+, 3% glycerol, and 1 mM TCEP, pH 7.5). Free Nanogold and Nanogold-labeled 3′SL and DENV-mini RNAs were eluted at different concentrations of buffer B, respectively. The combined fractions of Nanogold-labeled RNAs were further purified by passing through the Superdex 75 (for 3′SLs) or the Superdex 200 (for DENV-minis) gel filtration column.
CD Spectroscopy.
CD spectra of wild type, TPT3 modified, and Nanogold-labeled 3′SL or DENV-mini RNAs were measured on an Applied Photophysics Chirascan-plus spectrophotometer (Leatherhead) controlled by Pro-Data Chirascan v4 software. Spectra were recorded between 200 and 350 nm with a step resolution of 1 nm, a slit width of 0.6 nm, and an integration time of 5 s. Acquisition was performed at 25 °C using a 0.2 mm path-length cuvette with RNA concentrations being 1 and 5 μM for DENV-mini and 3′SL, respectively. The spectra were averaged over three scans and corrected by subtraction of the buffer signal.
EMSA Assay.
A total of 4 pmol wild type or double Nanogold-labeled 3′SL RNAs were mixed with 2 pmol 5′ SLB-DAR in SEC buffer. The samples were immediately loaded into 8% native polyacrylamide gels after being incubated at 37 °C for 30 min. The electrophoresis was conducted at 120 V at 4 °C for 1 h.
SAXS.
The wild type- and UBP-modified RNA samples as well as the Nanogold-labeled RNAs and free Nanogold were exchanged into final SAXS buffer (20 mM Hepes pH 7.5, 100 mM KCl, 5 mM Mg2+, 0.5 mM TCEP, and 3% [vol/vol] glycerol) using SEC columns or 10 K Amicon Ultra centrifugal filter devices (Millipore). All of the wild type- and UBP-modified RNA samples were diluted to final concentrations of 0.25–1 mg/mL, all Nanogold-labeled RNA solutions were concentrated and adjusted to 30 µM, and the free Nanogold sample was concentrated to 100 μM. The parameters for data collection and software employed for data analysis are summarized in SI Appendix, Table S7.
The data collection and processing procedures are similar as described before in ref. 42. Briefly, SAXS measurements were carried out at room temperature at the beamline 12 ID-B of the Advanced Photon Source, Argonne National Laboratory. The setups were adjusted to achieve scattering q values of 0.005 < q < 0.89 Å-1 (12ID-B), where q = (4π/λ) sin θ, and 2θ is the scattering angle. Thirty two-dimensional (2D) images were recorded for each buffer or sample solution using a flow cell with the exposure time of 1 s for wild type- and UBP-modified RNA samples or of 0.1 s for Nanogold-labeled RNA samples so as to minimize radiation damage and obtain a good signal-to-noise ratio. The 2D images were reduced to one-dimensional scattering profiles by using Matlab on site. The scattering profiles, the forward scattering intensity I(0), the radius of gyration (Rg), the PDDF p(r), as well as the maximum dimension Dmax of the RNAs were calculated using same procedures as described before in ref. 42. The Volumes-of-correlation (Vc) were calculated by using the program Scatter, and the MWs of wild type- and UBP-modified RNAs were calculated on a relative scale using the Rg/Vc power law developed by Rambo et al. (52), independent of RNA concentrations and with minimal user bias.
XSI Data Processing.
The scattering profiles of the RNA quartets for 3′SL, DENV-mini RNAs, and the free Nanogold were used to calculate the Nanogold–Nanogold scattering interference profiles from which the Nanogold–Nanogold distance distributions were inferred with the previously described protocols (20, 22) and the Matlab scripts in AuSAXSGUI (https://github.com/thomas836/AuSAXSGUI) shared by the Jan Lipfert group. Briefly, after standard SAXS data processing, the radius distribution of Nanogold was first determined from the free Nanogold scattering profile. The Nanogold–Nanogold scattering interference profiles IAu–Au(S) were then calculated as the sum of the concentration-normalized scattering signals of the double-labeled samples and nonlabeled sample minus the signals of the two single-labeled samples. Systems with two Nanogolds separated at different distances will give different interference profiles. The experimentally obtained IAu–Au(S) was then decomposed into contributions from 200 uniformly spaced Nanogold–Nanogold distances of 1–200 Å to generate the Nanogold–Nanogold distance probability distribution using a procedure that maximized the entropy.
Supplementary Material
Acknowledgments
We thank Dr. Xiaobing Zuo at the beamline 12-ID-B, Advanced Photon Source, Argonne National Laboratoryfor assistance during data collection. This work was supported by grants from the National Natural Science Foundation of China (U1832215), the Beijing Advanced Innovation Center for Structural Biology, and the Tsinghua-Peking Joint Center for Life Sciences to X.F.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. S.C.K. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005217117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Engreitz J. M., Ollikainen N., Guttman M., Long non-coding RNAs: Spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 17, 756–770 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Bernat V., Disney M. D., RNA structures as mediators of neurological diseases and as drug targets. Neuron 87, 28–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohno H., Akamine S., Saito H., RNA nanostructures and scaffolds for biotechnology applications. Curr. Opin. Biotechnol. 58, 53–61 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Jasinski D., Haque F., Binzel D. W., Guo P., Advancement of the emerging field of RNA nanotechnology. ACS Nano 11, 1142–1164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paredes E., Evans M., Das S. R., RNA labeling, conjugation and ligation. Methods 54, 251–259 (2011). [DOI] [PubMed] [Google Scholar]
- 6.George J. T., Srivatsan S. G., Posttranscriptional chemical labeling of RNA by using bioorthogonal chemistry. Methods 120, 28–38 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Tiwari P. M., Vig K., Dennis V. A., Singh S. R., Functionalized gold nanoparticles and their biomedical applications. Nanomaterials (Basel) 1, 31–63 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giljohann D. A.et al., Gold nanoparticles for biology and medicine. Angew. Chem. Int. Ed. Engl. 49, 3280–3294 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Powell R. D., Hainfeld J. F., Preparation and high-resolution microscopy of gold cluster labeled nucleic acid conjugates and nanodevices. Micron 42, 163–174 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tessmer I., Kaur P., Lin J., Wang H., Investigating bioconjugation by atomic force microscopy. J. Nanobiotechnology 11, 25 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brosey C. A., Tainer J. A., Evolving SAXS versatility: Solution X-ray scattering for macromolecular architecture, functional landscapes, and integrative structural biology. Curr. Opin. Struct. Biol. 58, 197–213 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang X., Stagno J. R., Bhandari Y. R., Zuo X., Wang Y. X., Small-angle X-ray scattering: A bridge between RNA secondary structures and three-dimensional topological structures. Curr. Opin. Struct. Biol. 30, 147–160 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker M. A. B.et al., Dimensions and global twist of single-layer DNA origami measured by small-angle X-ray scattering. ACS Nano 12, 5791–5799 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Bruetzel L. K.et al., Conformational changes and flexibility of DNA devices observed by small-angle X-ray scattering. Nano Lett. 16, 4871–4879 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Fischer S.et al., Shape and interhelical spacing of DNA origami nanostructures studied by small-angle X-ray scattering. Nano Lett. 16, 4282–4287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ponce-Salvatierra A.et al., Computational modeling of RNA 3D structure based on experimental data. Biosci. Rep. 39, BSR20180430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathew-Fenn R. S., Das R., Harbury P. A., Remeasuring the double helix. Science 322, 446–449 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathew-Fenn R. S., Das R., Silverman J. A., Walker P. A., Harbury P. A., A molecular ruler for measuring quantitative distance distributions. PLoS One 3, e3229 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zettl T.et al., Gold nanocrystal labels provide a sequence-to-3D structure map in SAXS reconstructions. Sci. Adv. 4, eaar4418 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi X., Bonilla S., Herschlag D., Harbury P., Quantifying nucleic acid ensembles with X-ray scattering interferometry. Methods Enzymol. 558, 75–97 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Zettl T.et al., Absolute intramolecular distance measurements with angstrom-resolution using anomalous small-angle X-ray scattering. Nano Lett. 16, 5353–5357 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Zettl T.et al., Recording and analyzing nucleic acid distance distributions with X-ray scattering interferometry (XSI). Curr. Protoc. Nucleic Acid Chem. 73, e54 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hura G. L.et al., DNA conformations in mismatch repair probed in solution by X-ray scattering from gold nanocrystals. Proc. Natl. Acad. Sci. U.S.A. 110, 17308–17313 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi X., Huang L., Lilley D. M., Harbury P. B., Herschlag D., The solution structural ensembles of RNA kink-turn motifs and their protein complexes. Nat. Chem. Biol. 12, 146–152 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi X., Walker P., Harbury P. B., Herschlag D., Determination of the conformational ensemble of the TAR RNA by X-ray scattering interferometry. Nucleic Acids Res. 45, e64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ackerson C. J., Powell R. D., Hainfeld J. F., Site-specific biomolecule labeling with gold clusters. Methods Enzymol. 481, 195–230 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lebars I.et al., A fully enzymatic method for site-directed spin labeling of long RNA. Nucleic Acids Res. 42, e117 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Babaylova E. S.et al., Complementary-addressed site-directed spin labeling of long natural RNAs. Nucleic Acids Res. 44, 7935–7943 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao M.et al., Site-specific dual-color labeling of long RNAs for single-molecule spectroscopy. Nucleic Acids Res. 46, e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malyshev D. A., Romesberg F. E., The expanded genetic alphabet. Angew. Chem. Int. Ed. Engl. 54, 11930–11944 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seo Y. J., Malyshev D. A., Lavergne T., Ordoukhanian P., Romesberg F. E., Site-specific labeling of DNA and RNA using an efficiently replicated and transcribed class of unnatural base pairs. J. Am. Chem. Soc. 133, 19878–19888 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Someya T., Ando A., Kimoto M., Hirao I., Site-specific labeling of RNA by combining genetic alphabet expansion transcription and copper-free click chemistry. Nucleic Acids Res. 43, 6665–6676 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L.et al., Natural-like replication of an unnatural base pair for the expansion of the genetic alphabet and biotechnology applications. J. Am. Chem. Soc. 136, 826–829 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Domnick C., Eggert F., Kath-Schorr S., Site-specific enzymatic introduction of a norbornene modified unnatural base into RNA and application in post-transcriptional labeling. Chem. Commun. (Camb.) 51, 8253–8256 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Eggert F., Kath-Schorr S., A cyclopropene-modified nucleotide for site-specific RNA labeling using genetic alphabet expansion transcription. Chem. Commun. (Camb.) 52, 7284–7287 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Domnick C.et al., EPR distance measurements on long non-coding RNAs empowered by genetic alphabet expansion transcription. Angew. Chem. Int. Ed. Engl. 59, 7891–7896 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seo Y. J., Hwang G. T., Ordoukhanian P., Romesberg F. E., Optimization of an unnatural base pair toward natural-like replication. J. Am. Chem. Soc. 131, 3246–3252 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gebhard L. G., Filomatori C. V., Gamarnik A. V., Functional RNA elements in the dengue virus genome. Viruses 3, 1739–1756 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iglesias N. G., Gamarnik A. V., Dynamic RNA structures in the dengue virus genome. RNA Biol. 8, 249–257 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Nicholson B. L., White K. A., Functional long-range RNA-RNA interactions in positive-strand RNA viruses. Nat. Rev. Microbiol. 12, 493–504 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brinton M. A., Basu M., Functions of the 3′ and 5′ genome RNA regions of members of the genus Flavivirus. Virus Res. 206, 108–119 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y.et al., Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution. EMBO Rep. 20, e47016 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.You S., Padmanabhan R., A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 274, 33714–33722 (1999). [DOI] [PubMed] [Google Scholar]
- 44.Sztuba-Solinska J.et al., Structural complexity of Dengue virus untranslated regions: cis-acting RNA motifs and pseudoknot interactions modulating functionality of the viral genome. Nucleic Acids Res. 41, 5075–5089 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seo Y. J., Matsuda S., Romesberg F. E., Transcription of an expanded genetic alphabet. J. Am. Chem. Soc. 131, 5046–5047 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lavergne T.et al., FRET characterization of complex conformational changes in a large 16S ribosomal RNA fragment site-specifically labeled using unnatural base pairs. ACS Chem. Biol. 11, 1347–1353 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R., Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 (1989). [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y.et al., A semi-synthetic organism that stores and retrieves increased genetic information. Nature 551, 644–647 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou A. X., Sheng K., Feldman A. W., Romesberg F. E., Progress toward eukaryotic semisynthetic organisms: Translation of unnatural codons. J. Am. Chem. Soc. 141, 20166–20170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hainfeld J. F., Furuya F. R., A 1.4-nm gold cluster covalently attached to antibodies improves immunolabeling. J. Histochem. Cytochem. 40, 177–184 (1992). [DOI] [PubMed] [Google Scholar]
- 51.Kypr J., Kejnovská I., Renciuk D., Vorlícková M., Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 37, 1713–1725 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rambo R. P., Tainer J. A., Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477–481 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dimitrova-Paternoga L., Jagtap P. K. A., Chen P. C., Hennig J., Integrative structural biology of protein-RNA complexes. Structure 28, 6–28 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Zhang L.et al., Three-dimensional structural dynamics and fluctuations of DNA-nanogold conjugates by individual-particle electron tomography. Nat. Commun. 7, 11083 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spokoini-Stern R.et al., Visualizing the structure and motion of the long noncoding RNA HOTAIR. RNA 26, 629–636 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jo H., Ban C., Aptamer-nanoparticle complexes as powerful diagnostic and therapeutic tools. Exp. Mol. Med. 48, e230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Azharuddin M.et al., A repertoire of biomedical applications of noble metal nanoparticles. Chem. Commun. (Camb.) 55, 6964–6996 (2019). [DOI] [PubMed] [Google Scholar]
- 58.He B.et al., Rapid mutagenesis and purification of phage RNA polymerases. Protein Expr. Purif. 9, 142–151 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.