Significance
The yeast Saccharomyces cerevisiae has been utilized extensively as a cell factory to produce numerous products. The de novo synthesis of medium-chain fatty alcohols (MCFOHs) through conversion of free fatty acids by carboxylic acid reductase (CAR) has been studied in yeast. However, the broad substrate specificity of CAR is one of the biggest challenges to improve MCFOH production. Here, different protein engineering strategies were employed to modify the CAR enzyme toward a more specific catalytic activity on shorter chain fatty acids. For this, we created a high-throughput screening assay relying on the detoxification of the toxic substrate to select for efficient enzyme variants. This study represents an important step forward in the selective synthesis of MCFOHs in yeast.
Keywords: carboxylic acid reductase, protein engineering, high-throughput screening, medium-chain fatty alcohols, Saccharomyces cerevisiae
Abstract
Medium-chain fatty alcohols (MCFOHs, C6 to C12) are potential substitutes for fossil fuels, such as diesel and jet fuels, and have wide applications in various manufacturing processes. While today MCFOHs are mainly sourced from petrochemicals or plant oils, microbial biosynthesis represents a scalable, reliable, and sustainable alternative. Here, we aim to establish a Saccharomyces cerevisiae platform capable of selectively producing MCFOHs. This was enabled by tailoring the properties of a bacterial carboxylic acid reductase from Mycobacterium marinum (MmCAR). Extensive protein engineering, including directed evolution, structure-guided semirational design, and rational design, was implemented. MmCAR variants with enhanced activity were identified using a growth-coupled high-throughput screening assay relying on the detoxification of the enzyme’s substrate, medium-chain fatty acids (MCFAs). Detailed characterization demonstrated that both the specificity and catalytic activity of MmCAR was successfully improved and a yeast strain harboring the best MmCAR variant generated 2.8-fold more MCFOHs than the strain expressing the unmodified enzyme. Through deletion of the native MCFA exporter gene TPO1, MCFOH production was further improved, resulting in a titer of 252 mg/L for the final strain, which represents a significant improvement in MCFOH production in minimal medium by S. cerevisiae.
Concerns about climate change drive the scientific community to explore alternative ways to produce renewable biofuels (1). With straight hydrocarbon chains ranging from C6 to C12, medium-chain fatty alcohols (MCFOHs) are designated as higher alcohols in contrast to lower alcohols, such as butanol, ethanol, and methanol. MCFOHs are considered to be more suitable to substitute the traditional diesel and jet fuels due to their higher energy density, higher cetane number, and lower vapor pressure (2, 3). MCFOHs are also widely used in surfactants, cosmetics, and plasticizers (4). Currently, fatty alcohols are synthesized from petrochemical sources or produced from fatty acids (FAs) extracted from oil seeds (5). A major obstacle with sourcing from oil seeds is the competition with food production by using arable land and deforestation (6), and there is therefore interest in producing MCFOHs through microbial fermentation, allowing for the utilization of a wide range of feedstocks (7), including biomass.
In organisms, fatty alcohols are derived from FAs. The biosynthesis of fatty alcohols can proceed through either fatty aldehyde intermediates via two steps of two-electron reductions or directly from fatty acyl-CoA/ACP via a four-electron reduction (8). Fatty aldehydes can be generated from 1) activated fatty acyl-CoA/ACP by fatty acyl-CoA/ACP reductase [ACR (9) or AAR (10)] or 2) free FAs by carboxylic acid reductase (CAR) (11). MmCAR from Mycobacterium marinum has shown the highest efficiency toward formation of fatty aldehydes that can subsequently be converted to fatty alcohols in a previous study (12). CARs utilize free FAs as substrates (Fig. 1A), which are usually more abundant than the activated acyl-CoA and acyl-ACP in yeast cells (13). Moreover, thioesterases hydrolyzing acyl-CoA or acyl-ACP, thus releasing free FAs, can be used not only for controlling the chain length of synthesized FAs but also for increasing the FA synthesis flux, which is restricted by feedback inhibition mediated by acyl-CoA and acyl-ACP (14). Therefore, fatty alcohol formation based on free FAs as substrates through CAR enzymes possesses great advantages. Previous studies on engineering the FA synthesis pathway through augmenting the precursor flux and improving FA synthetase (FAS) activity in yeast have generated microbial cells producing substantial amounts of medium-chain FAs (MCFAs) (Fig. 1A), which could be utilized for the synthesis of MCFOHs (15, 16). In our previous study (17), an engineered MCFA producing FAS and MmCAR were expressed in yeast to generate aldehyde precursors for alkane synthesis. Substantial quantities of MCFOHs produced by the engineered strain ZW540 (SI Appendix, Fig. S1) showed that endogenous alcohol dehydrogenases and aldehyde reductases were able to support a strong downstream flux toward MCFOH formation. However, a promiscuous CAR enzyme reducing C6–C18 FAs is not ideal for MCFOH production, as it deprives the cell of C16–C18 long-chain FAs (LCFAs) that are essential for cell growth, and also invests NADPH and ATP in the formation of long-chain by-products (18, 19). It is therefore of great interest to narrow the substrate spectrum of the enzyme or increase its activity on MCFAs to more specifically produce MCFOHs. Protein engineering of a CAR enzyme would offer the opportunity to change its substrate specificity in the desired direction.
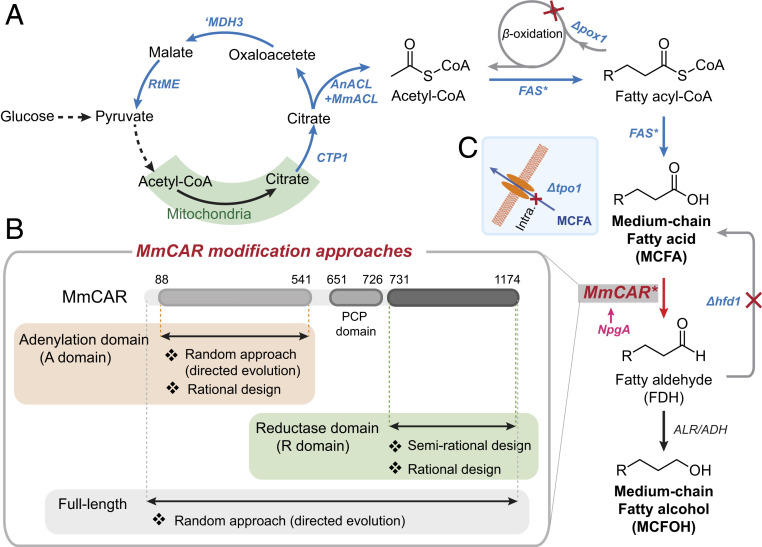
Fig. 1.
Overview of genetic manipulations in yeast enabling selective production of MCFOHs. (A) Schematic illustration of the pathway engineering strategies in background strain ZWE243. AnACL, ATP-citrate lyase from Aspergillus nidulans; Ctp1, mitochondrial citrate transporter; FAS*, engineered endogenous fatty acid synthase for MCFAs production; Hfd1, aldehyde dehydrogenase; ′Mdh3, malate dehydrogenase without peroxisomal signal; MmACL, ATP-citrate lyase from Mus musculus; MmCAR, carboxylic acid reductase from Mycobacterium marinum; NpgA, phosphopantetheinyl transferase from Aspergillus nidulans; Pox1, fatty acyl-CoA oxidase; RtME, malic enzyme from Rhodosporidium toruloides; . The genes encoding ′Mdh3, RtME, MmACL, AnACL, and Ctp1 were overexpressed to increase the acetyl-CoA supply, and POX1 and HFD1 genes were deleted to avoid reverse reactions. R represents C3–C9. (B) MmCAR was modified using diverse approaches. A domain (residues 88 to 541); PCP domain (residues 651 to 728); R domain (residues 731 to 1,174). (C) The predicted MCFA transporter gene TPO1 was deleted to prevent the secretion of MCFAs.
The CAR enzyme consists of an adenylation domain (A domain), a reductase domain (R domain), and a peptidyl carrier protein domain (PCP domain) that links the A domain and R domain (Fig. 1B). A phosphopantetheinyl transferase (PPTases) is required to activate the enzyme by covalently attaching a 4-phosphopantetheine moiety to the PCP domain (20, 21). The carboxylic group of the FA substrate is converted to the corresponding aldehyde in the presence of Mg2+, ATP, and NADPH through a process of two-electron reduction (20). The A domain exhibits a mechanism of substrate recognition and activation similar to that of the ANL superfamily of adenylating enzymes, where the α-phosphate of ATP is attacked by the carboxylate to generate an AMP-acyl phosphoester (21–23). The R domain shows homology to the short-chain dehydrogenases/reductase family whose members commonly exist as subdomains in polyketide synthases (PKSs) and nonribosomal peptide synthases (NRPSs), where they mediate the off-loading of the final products (24, 25). The R domain of CAR enzymes catalyzes the strict two-electron reduction by consuming one NADPH and releases the corresponding aldehyde (21, 26).
To change the activity or specificity of an enzyme, different engineering strategies can be chosen that highly depend on the amount of knowledge available on the protein of interest. Random approaches, referred to as directed evolution, with less information being required always rely on the availability of a high-throughput screening method for large libraries of enzyme variants (27). Smaller libraries are usually generated for rational and semirational approaches that require structural and mechanistic information (28). The revealed catalytic mechanism and the availability of the enzyme structure provided us with the opportunity to rationally engineer the CAR enzyme (19). In addition, directed evolution as a powerful tool enables the development of reaction schemes and catalytic mechanisms not found in nature (29), and could be utilized to access the untapped function of the CAR enzyme using efficient high-throughput selection. In this study, we first developed a high-throughput screening method relying on the detoxification of the toxic MCFAs that proved to be beneficial for selection of efficient MmCAR variants. We designed multiple strategies for comprehensive engineering of MmCAR, including a random approach using directed evolution, a structure-guided semirational design, and a rational approach (Fig. 1B). Measuring the in vivo biotransformation capacity as well as in vitro enzymatic activity on FAs of different chain length showed that some mutants with increased catalytic activities toward MCFAs could be generated and used for the more selective production of MCFOHs. In addition, with enhancing the endogenous MCFA precursor supply through deleting an MCFA transporter (Fig. 1C), a further increase in MCFOH production was achieved. This study demonstrates that engineering key metabolic enzymes can significantly improve the synthesis of the desired product, while reducing by-product formation.
Results
Growth-Coupled Screening Approach for MmCAR Libraries.
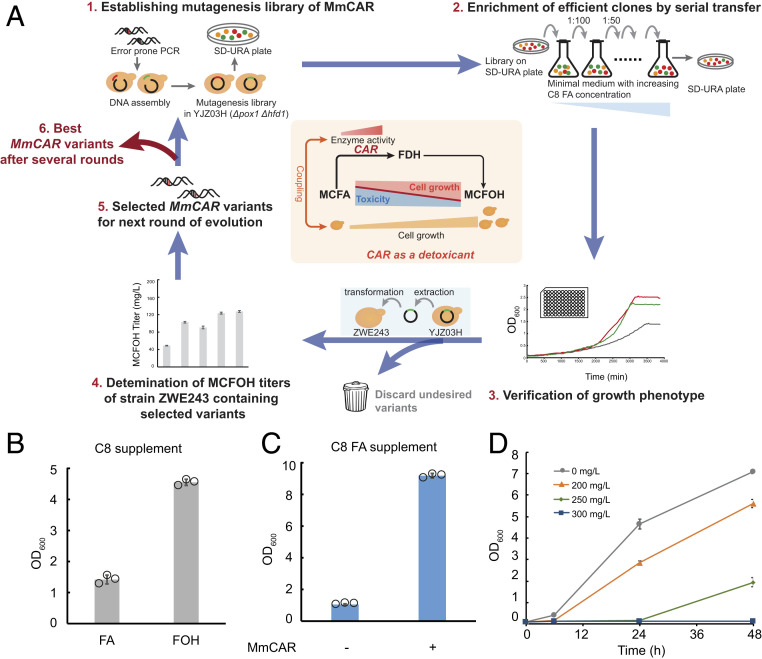
MCFAs are toxic to yeast cells, the growth of which was significantly reduced by concentrations as low as 1 mM octanoic acid and decanoic acid (30). However, the toxicity of MCFAs could be exploited for selection of efficient variants of enzymes leading to their detoxification (Fig. 2A). To investigate if MCFOH formation represents a potential detoxification pathway for MCFAs, we first evaluated the cellular toxicity of MCFAs (octanoic acid and decanoic acid) and compared it to the toxicity of the respective fatty alcohols of the same chain length. Strain YJZ03H, which is deficient in both POX1 and HFD1, was used for the toxicity test. Deletion of POX1 eliminated FA β-oxidation and the deletion of aldehyde dehydrogenase gene HFD1 was shown to be beneficial for alcohol formation (12, 17, 31). Next, 0.4 mM (∼70 mg/L) of C10 FA or fatty alcohol and 1 mM (∼150 mg/L) of C8 FA or fatty alcohol was used to evaluate their toxicity. Results showed that 70 mg/L C10 FA and C10 fatty alcohol were equally toxic to yeast cells (SI Appendix, Fig. S2), while the presence of 150 mg/L C8 fatty alcohol allowed cell growth to a much higher final OD than C8 fatty alcohol at the same concentration (Fig. 2B). Correspondingly, expression of MmCAR was beneficial for yeast cell growth at 150 mg/L C8 FA (Fig. 2C), which can be attributed to the detoxifying activity of MmCAR that converts C8 FA to the less toxic C8 fatty alcohol. A further investigation of the detoxification capacity of MmCAR showed that yeast cells containing MmCAR could hardly grow in the presence of 300 mg/L of C8 FA (Fig. 2D), indicating that ∼300 mg/L of C8 FA would be suitable for the selection process. Based on these data, we proposed a growth-coupled screening scheme for the selection of MmCAR variants exhibiting a higher activity on MCFAs (Fig. 2A).
Fig. 2.
Design of the growth-coupled screening approach. (A) Workflow of the high-throughput screening method coupled to cell growth. 1) Libraries of MmCAR variants expressed from a plasmid were established in yeast strain YJZ03H (Δpox1 Δhfd1) via error-prone PCR and homologous recombination. 2) The variants beneficial to cell growth in minimal medium with C8 FA were selected and 3) their growth phenotype was verified using a Bioscreen or Growth Profiler. 4) The variants leading to better growth were tested in the high MCFA-producing strain ZWE243, and 5) the most efficient one was used as the template for the next round of evolution. (B) Comparison of the C8 FA/alcohol toxicity to yeast cells. Strain YJZ03H was cultivated in minimal medium with 150 mg/L of C8 FA/alcohol for 72 h. The final optical density was measured at a wavelength of 600 nm (OD600). (C) Growth of yeast strain with or without MmCAR in medium with C8 FAs. Strain YJZ03H carrying an empty vector or plasmid pZW01 harboring wild-type MmCAR was cultivated in minimal medium with 150 mg/L C8 FA. The final OD600 was measured after 72 h of cultivation. (D) The effect of MmCAR expression on cell growth at different concentrations of C8 FAs. The yeast strain YJZ03H with pZW01 harboring MmCAR was cultivated in minimal medium with 0 mg/L, 200 mg/L, 250 mg/L, and 300 mg/L C8 FA, respectively. The OD600 was measured during the cultivation. The mean ± SD of three biological replicates is presented (B, C, and D).
Directed Evolution of the Full-Length MmCAR.
Phosphopantetheinylation undertaken by PPTases is required for the activity of CAR enzymes. We first evaluated the effects of PPTases on the biosynthesis of MCFOH through combined expression of the wild-type MmCAR with different PPTases from various species in the MCFA producing yeast strain ZW2071 (SI Appendix, Fig. S3). Among the tested candidates, expression of NpgA from Aspergillus nidulans resulted in the relatively higher MCFOH titers. NpgA was therefore used for all following experiments.
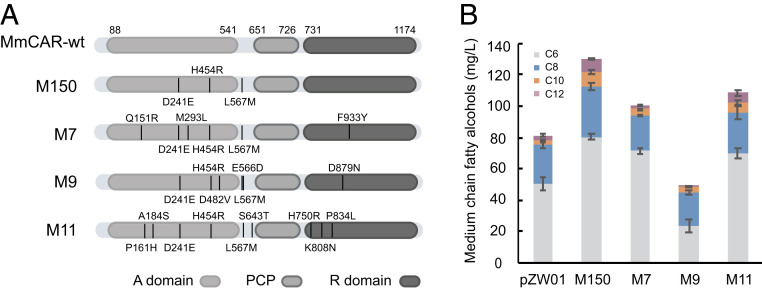
In order to improve the catalytic activity of MmCAR for MCFA conversion, directed evolution was performed by first establishing a random mutagenesis library of the full-length MmCAR. The library was cultivated at an increasing concentration of C8 FA from 290 mg/L to 330 mg/L to enrich it for improved enzyme variants. After the enrichment process, single clones were isolated and tested for improved growth on C8 FA. This led to identification of variant M150, whose expression enabled the fastest growth of yeast strain YJZ03H in medium with 330 mg/L C8 FA (SI Appendix, Fig. S4A). Sequencing of M150 revealed three amino acid changes compared to the wild-type enzyme. Two of the mutations, D241E and H454R, were located in the A domain of MmCAR, while the mutation L567M was situated between the A domain and PCP domain (Fig. 3A). When expressed in an MCFA-producing yeast strain, M150 enabled a more than 50% increase in MCFOH production compared to the wild-type MmCAR, reaching a titer of 129 mg/L (Fig. 3B). We therefore used M150 as the template for the second round of evolution, from which the efficient variants M7, M9, and M11 were selected on the basis of conferring a superior growth phenotype (SI Appendix, Fig. S4B). Although all of these three CAR mutants showed lower in vivo activities on MCFA conversion than M150, two of them still performed better than the wild-type MmCAR in terms of MCFOH production (Fig. 3B). These results clearly demonstrate that our growth-coupled screening was suitable for the selection of CAR variants with improved activity on MCFAs.
Fig. 3.
The synthesis of MCFOHs in S. cerevisiae by expressing variants derived from directed evolution of full-length MmCAR. (A) Schematic illustration of the mutated MmCARs. (B) Production of MCFOHs in S. cerevisiae by enzyme variants derived from two rounds of directed evolution based on full-length MmCAR. pZW01 represents the plasmid containing wild-type MmCAR. M150 represents the variant from the first round of evolution. M7, M9, and M11 represent the variants from the second round of evolution. Strain ZWE243 was used for evaluation, all of the cultivations were performed in minimal medium with histidine for 48 h. The mean ± SD of three biological replicates is shown.
Modification of the A Domain.
MmCAR is able to reduce diverse aliphatic FAs with chain length ranging from C6 to C18 (18). The broad substrate spectrum of MmCAR is not optimal for MCFOH production, as the generation of long-chain fatty alcohols (LCFOHs) as by-products not only reduces the overall product yield but may also lead to impaired growth as a result of competition for essential LCFAs in yeast cells. According to the catalytic mechanism revealed in a previous study (19), the volume of the active-site in the A domain could potentially be altered to tailor the substrate specificity of CAR.
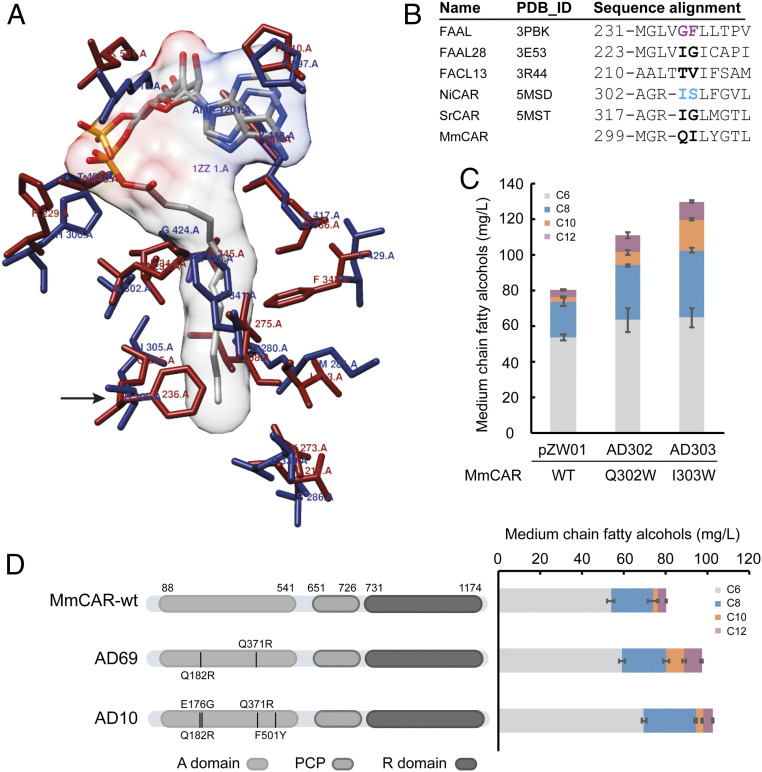
Fatty acyl-AMP ligases (FAALs) and fatty acyl-CoA ligases (FACLs) belong to a superfamily of acyl-activating enzymes that activate the carboxyl groups of their substrates to acyl-adenylates and then transfer these to other acceptor groups, thus having a function similar to the A domain of CAR (32, 33). The FAAL from Escherichia coli (PDB ID code 3PBK) containing an acyl-AMP ligand was aligned with the A domain of CAR from Nocardia iowensis (NiCAR, PDB ID code 5MSD) to assign the acyl-residing tunnel. A potentially crucial residue S306 (I303 in MmCAR) adjacent to the C11 position was identified (Fig. 4 A and B). A previous study revealed that the mutated proteins FAAL28 (I227W) and FACL13 (T214W), which possessed a bulky tryptophan residue at the position equivalent to I305 in NiCAR and Q302 in MmCAR (Fig. 4B), showed a significant decrease in the activity toward longer acyl chains and an increase in the activity on C10 or C8 FAs (32). We therefore substituted I303 and Q302 of MmCAR with a tryptophan residue. The strains expressing the MmCAR variants containing the Q302W (AD302) and I303W (AD303) mutations were able to produce 110 mg/L and 129 mg/L of MCFOHs, which was, respectively, 37% and 61% higher than those produced by the strain expressing the wild-type MmCAR (Fig. 4C). Therefore, we speculated that the bulkier tryptophan residue might cause a steric hindrance by which the catalytic pocket of MmCAR was narrowed to result in impaired binding of long-chain acyl groups.
Fig. 4.
The synthesis of MCFOHs in S. cerevisiae by MmCAR variants generated through A domain modifications. (A) Structure superposition of EcFAAL (PDB ID code 3PBK, in dark red) and the A domain of NiCAR (PDB ID code 5MSD, in dark blue). Residues surrounding the substrate binding pocket are shown in stick representation. The C12 acyl-AMP ligand (gray) is shown in surface representation. The arrow indicates the residues (F236 in EcFAAL and S306 in NiCAR) proximate to C11 of the acyl chain. (B) Structure-based sequence alignment of selected acyl-activating enzymes. Residues crucial for medium-chain carboxylic acid substrates are shown in bold. Production of MCFOHs in S. cerevisiae by mutated MmCARs derived from rational design (C) and directed evolution (D) of the A domain, respectively. The schematic illustration of variants from plasmids AD69 and AD10 is shown in d. All variants were assessed in strain ZWE243; all cultivations were performed in minimal medium with histidine for 48 h. The mean ± SD of three biological replicates is shown.
Parallel to the targeted engineering approach, we also engineered the A domain of MmCAR by random mutagenesis (residues 88 to 541) (Fig. 4D). The same workflow as in Fig. 2A was performed by using 290 to 310 mg/L of C8 FA as the enrichment reagent. The best variant AD69 enabled better cell growth (SI Appendix, Fig. S5A) and led to a higher production of MCFOHs of 96.9 mg/L, which is a 21% increase compared with that of the wild-type MmCAR (Fig. 4D). Cells expressing the AD10 variant isolated after a second round of evolution (SI Appendix, Fig. S5B) produced an approximately equal amount of MCFOHs, which was 102 mg/L (Fig. 4D). Although the total amount of MCFOHs produced by the strain carrying AD10 was not significantly improved compared to the AD69 carrying strain, more C6 and C8 fatty alcohols (around 18%) were produced by the AD10-containing strain, which might indicate that this mutant preferably reduces FAs with a shorter chain length and thereby provided the cells with an increased tolerance toward C8 FA in the medium.
Modification of the R Domain.
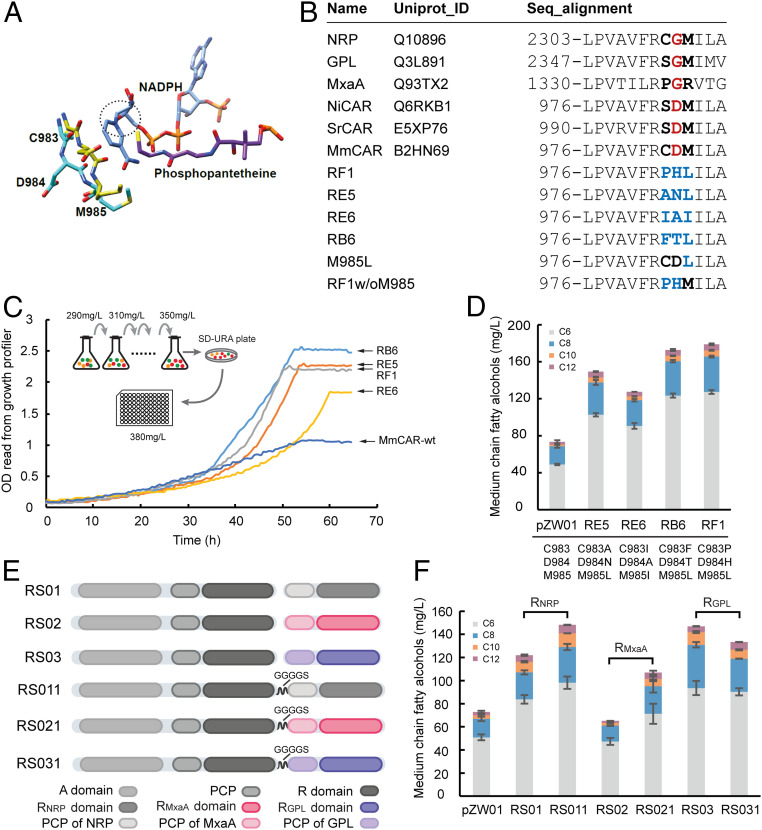
The R domain of MmCAR (residues 731 to 1,174) enables the enzyme to catalyze a strict two-electron reduction of the acyl-PCP thioester to a fatty aldehyde. This is due to the fact that the conformation adopted by residues 983 to 985 appears to be linked to the position of the smaller substrate-binding domain, which ensures that the reduction cannot proceed beyond the aldehyde product (19). However, the backbone reorientation of residues 983 to 985 was reported to be able to affect the conformational equilibrium between on- and off-states of the MmCAR R domain (34). In the active form, the D984 will be positioned pointing away from the nicotinamide and buried within the protein matrix, while in the inactive form, the D984 and the S983 carbonyl group will locate within the nicotinamide binding pocket (34) (Fig. 5A).
Fig. 5.
The synthesis of MCFOHs in S. cerevisiae by MmCAR variants generated through R domain modifications. (A) The on- and off-states of the R domain determined by the conformation change of C983–M985. In the active form (cyan), the D984 residue is positioned away from the active center (shown in a dashed ellipse), while in the inactive form (yellow), D984 and the backbone of C983–M985 occupy the pocket for the nicotinamide moiety of NADPH. (B) Partial sequence alignment of MmCAR, SrCAR, NiCAR, MxaA, GPL, and NRP. (C) Growth curves of strains expressing MmCAR variants selected from the site-directed saturation mutagenesis library. After an enrichment process in the medium with 290 to 350 mg/L C8 FA, the cell growth of selected clones in medium with 380 mg/L C8 FA was monitored by a Growth Profiler. (D and F) Production of MCFOHs in S. cerevisiae by mutated MmCAR variants derived from the site-directed mutagenesis library (D) and di-domain expression (F). All plasmids were transformed into ZWE243 for evaluation; all of the cultivations were performed for 48 h in minimal medium with histidine. The mean ± SD of three biological replicates is shown. (E) Schematic illustration of the didomain expression constructs. The PCP-R didomains from NRPS enzymes (RGPL, RNRP, and RMxaA) were expressed with MmCAR separately (RS01, RS02, and RS03) or fused with MmCAR via a GGGGS linker (RS11, RS21, and RS31).
MmCAR with an engineered R domain that could reduce free FAs via aldehydes all of the way to fatty alcohols could be beneficial for MCFOH production in yeast. In a previous study (19), the mutation D998G (equivalent to D984 in MmCAR) (Fig. 5B) in CAR from Segniliparus rugosus (SrCAR) led to the formation of alcohol products. However, the same mutation in MmCAR showed a negative effect on MCFOH production in yeast (SI Appendix, Fig. S6A). It was reported that the R1339A (equivalent to M985 in MmCAR) (Fig. 5B) mutant of the MxaA R domain from Stigmatella aurantiaca exerted significantly increased activities for the reduction of C10 acyl-PCP and C10 aldehyde (35). After introducing the mutation M985A into MmCAR, we did not observe augmented production of MCFOHs by this mutant (SI Appendix, Fig. S6B).
To obtain an engineered R domain free of conformational regulation by C983–M985 or more efficient for reduction of medium-chain substrates, we created a site-directed saturation mutagenesis library targeting residues 983 to 985. Four selected variants (RE5, RE6, RB6 and RF1) (Fig. 5C), which enabled a higher growth rate and final cell mass of yeast cells in medium with 380 mg/L C8 FA (Fig. 5C and SI Appendix, Fig. S7), were also beneficial for MCFOH production in yeast (Fig. 5D). Specifically, variant RF1 containing mutations C983P, D984H, M985L, led to the highest titer of MCFOHs of 178 mg/L, a 2.4-fold increase compared with the wild-type MmCAR (Fig. 5D). A common mutation (M985L or M985I) was found among these four MmCAR variants (Fig. 5B). Therefore, M985L and RF1 without the mutation on M985 (RF1w/o985) variants, which contained either only the M985L mutation or the other two mutations (C983P and D984H) observed in RF1, respectively, were constructed to separately test the effects of these mutations on the enzyme activity. Interestingly, we found that the M985L mutant only led to production of 95.5 mg/L MCFOHs in yeast. The RF1w/o985 variant (Fig. 5B), in contrast, exerted an even slightly stronger effect on MCFOH production than the RF1 mutant (SI Appendix, Fig. S8). These results indicated that these two mutations might play a more critical role in RF1 for MCFOH production than M985L.
The comparison based on crystal structures between MmCAR and NRPSs elucidated that they share a high similarity in the structures of the R domains (24, 35). Nevertheless, some of the R domains from NRPSs differ in their final product determination compared with CARs, as they release alcohols produced through aldehyde intermediates. Three R domains from three NRPS enzymes (RGPL, RNRP, and RMxaA) sharing a high level of identity (∼50%) with the R domain of MmCAR (SI Appendix, Fig. S9), were reported to catalyze the four-electron reduction of fatty acyl thioesters to the corresponding alcohols (24, 35). We attempted replacing the MmCAR R domain or PCP-R didomain with the respective domains from NRP, GPL, and MxaA (SI Appendix, Figs. S9 and S10) in order to enable a four-electron reduction that could benefit fatty alcohol production. Unfortunately, all of the chimeras seemed to lose their function as no MCFOHs were detected in the yeast strains expressing these constructs. Thus, in order to avoid the potential misfolding of MmCAR, the heterologous PCP-R didomains were introduced into yeast cells through two strategies: 1) Separate expression together with MmCAR or 2) fusion with MmCAR using the flexible linker GGGGS (Fig. 5E). Most of the constructs expressing an additional PCP-R didomains successfully promoted MCFOH production in yeast (Fig. 5F). Among the evaluated variants, the most prominent effect on MCFA conversion was observed for the MmCAR-RNRP fusion (RS011), which enabled production of 148 mg/L MCFOHs (Fig. 5F).
Combination of Modifications.
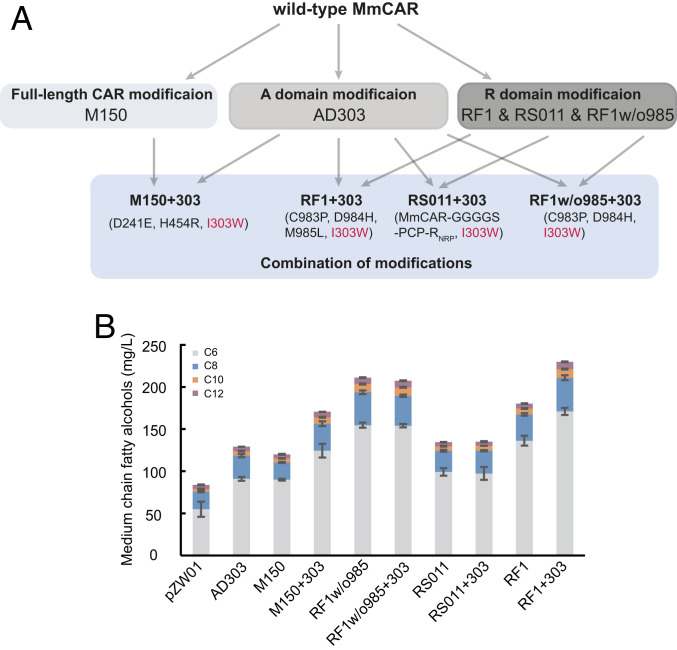
Different modifications evaluated above were combined to achieve a more efficient MmCAR variant for MCFOH production in yeast. We combined all of the other beneficial mutations with I303W to ensure a more specific substrate preference toward MCFAs. The highest production of MCFOHs was obtained in strains expressing RF1+303, which resulted in production of MCFOHs at a level of 229 mg/L, a 2.8-fold increase over that of the wild-type MmCAR (Fig. 6). Additionally, to enable a further enhancement of MCFOH production, we also incorporated two additional modification (M150 and RS011) into the two best variants RF1+303 and RF1w/o985+303, respectively. However, none of these combinations reached the MCFOHs levels of RF1+303 (SI Appendix, Fig. S11).
Fig. 6.
The production of MCFOHs in strains with combined modification in MmCAR. (A) Modifications and changed residues in each enzyme variant. (B) The production of MCFOHs was tested in the strain ZWE243 containing plasmids pZW01, M150, ADD303, M150+303, RF1w/o985, RF1w/o985+303, RS011, RS011+303, RF1, and RF1+303, respectively. All strains were cultivated for 48 h in minimal medium and the final products were quantified by gas chromatography-flame ionization detector (GC-FID); the mean ± SD of three biological replicates is presented.
Evaluation of Substrate Specificity of MmCAR Variants.
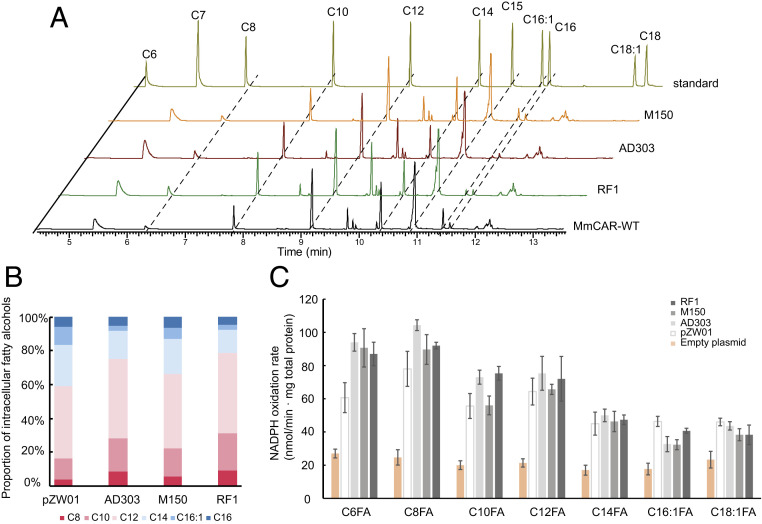
The strain ZWE243 producing MCFAs provided a platform to evaluate the variants in vivo through measurement of the production of MCFOHs. Since this strain generates both LCFAs and MCFAs, the investigation of the profile of intracellular fatty alcohols could therefore represent a complementary assay to interpret the catalytic activities of each enzyme variant. The intracellular fatty alcohols extracted from strains containing the wild-type MmCAR, AD303, M150, and RF1, respectively, were quantified. The GC spectra showed that most of the products were MCFOHs with relatively low amounts of LCFOHs detected (Fig. 7A). We subsequently quantified the intracellular fatty alcohols for each strain and calculated the proportion of each component in the total products. According to the results, the wild-type MmCAR led to a production of 41% of LCFOHs (including C14, C16:1, and C16) and 59% of MCFOHs (including C8, C10, and C12) among the intracellular fatty alcohols (Fig. 7B). Compared to the wild-type, all of the other mutant CARs resulted in a higher proportion of MCFAs, 75%, 78%, and 67% of MCFOHs in the strains expressing AD303, RF1, and M150, respectively (Fig. 7 A and B).
Fig. 7.
Characterization of the substrate specificity of mutated MmCARs. (A) GC spectra of intracellular fatty alcohols derived from strain ZWE243 with pZW01, M150, AD303, and RF1, respectively. Cultivation was performed for 72 h in minimal medium. (B) Proportion of intracellular fatty alcohols in strain ZWE243 with pZW01, M150, AD303, and RF1, respectively. (C) The NADPH oxidation rate of the crude protein extracted from strain ZWE243 containing RF1, M150, AD303, wild-type CAR and an empty plasmid, respectively, supplemented with different FAs. The samples were taken after cultivation in minimal medium for 24 h. The mean ± SD of three biological replicates is presented.
To investigate if these MmCAR variants indeed possessed altered substrate preferences, the enzyme activities on FAs of different chain length were measured in crude protein extracts. The crude protein extracted from the strain with the empty plasmid exerted background NADPH oxidation activity, which was ∼23 nmol·min−1·mg−1 total protein and comparatively constant between the different assays. Except for the reduction activity of M150 on C10 FA, which was comparable to that of wild-type MmCAR, all three mutants had improved activities toward C6–C10 FAs, but comparable or decreased activities toward C12–C18 FAs (Fig. 7C). Particularly, the I303W mutant showed a roughly twofold increase in activity on C6 FA compared with the wild-type. We therefore compared the kinetic properties between the I303W mutant and wild-type, and observed that, compared with the wild-type, the I303W mutant had a twofold higher kcat, albeit a higher Km value (SI Appendix, Fig. S12). These results could partially explain the elevated production of MCFOHs.
Further Increase in MCFOH Production through Deletion of TPO1.
The endogenously produced MCFAs were found to be inadequate for MCFOH production in strain ZWE243 containing RF1+303 (SI Appendix, Fig. S13); we thus fed the cells with the C8 FA in the medium as the precursor. However, both MCFOH production and cell growth were negatively affected when 100, 200, and 250 mg/L of C8 FAs were added into the medium, respectively (SI Appendix, Fig. S14), which might be due to the cellular toxicity caused by high concentrations of C8 FA. Moreover, a bilayer cultivation was conducted through addition of dodecane, but a large amount of the MCFA precursors were captured by the dodecane phase, leading to a low-level production of MCFOHs (SI Appendix, Fig. S15).
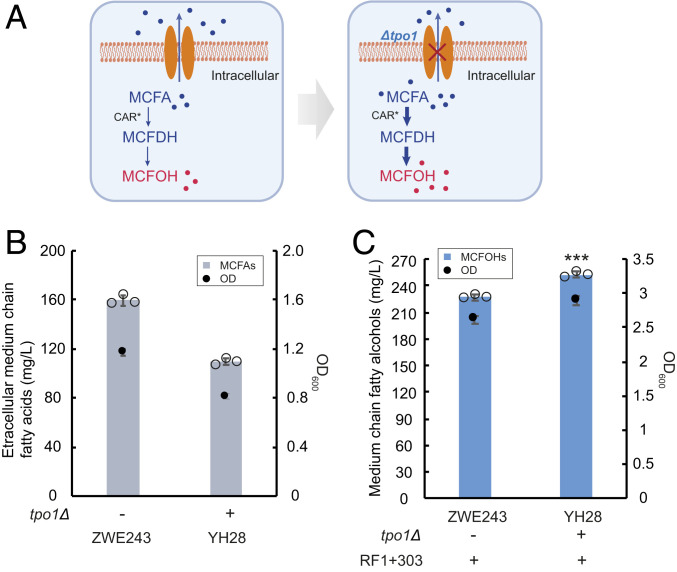
Tpo1 is a native transporter located on the plasma membrane of yeast that was suggested to contribute to the resistance to polyamines by exporting these substrates (36). Later, it was identified to confer tolerance against MCFAs as well (30, 37). In our previous work (37), an improved Tpo1 variant was able to significantly increase the production of extracellular MCFAs in yeast. We assumed that the deletion of TPO1 might augment the intracellular MCFA pool by potentially reducing the secretion of MCFAs (Fig. 8A), which would benefit MCFOH synthesis. As expected, upon introducing the TPO1 deletion, the accumulation of extracellular MCFAs in the resulting strain YH28 was significantly decreased to 109 mg/L compared with the titer of 159 mg/L generated by the parental strain (Fig. 8B). In line with this, strain YH28 showed a lower final OD, which could result from the inhibitory effect of intracellular MCFAs. Subsequently, the most efficient MmCAR variant RF1+303 was introduced into strains ZWE243 and YH28, respectively. As expected, the knockout of TPO1 led to an ∼11% improvement of the final titer of MCFOHs to 252 mg/L, which is around 3.2-fold higher than that of the original strain (ZWE243) harboring wild-type MmCAR (Fig. 8C).
Fig. 8.
The effect of TPO1 deletion on MCFOH production in S. cerevisiae. (A) Schematic illustration of the postulated effect of a TPO1 deletion. Deleting TPO1 is assumed to affect MCFA efflux from the cells. With the increased intracellular pool of MCFAs, the flux toward MCFOH production would be enhanced. MCFAs in blue dots, MCFOHs in red dots. MCFDH, medium-chain fatty aldehyde. (B) Production of extracellular MCFAs and final OD600 for strains ZWE243 and YH28. Strains were cultivated in minimal medium with histidine and uracil for 48 h. (C) Production of MCFOHs and final OD600 for strains ZWE243 and YH28 with RF1+303 expression. The strains were cultivated for 48 h in minimal medium with histidine and the final products were quantified by GC-FID. The P value was 0.0005 (***P < 0.001). Statistical analysis was conducted using a Student’s t test (one-tailed; two-sample unequal variance). The mean ± SD of three biological replicates is presented in B and C.
Discussion
Enlarging the variety of compounds produced by cell factories requires a feasible engineering approach to break the restriction imposed by the native activity of enzymes. Protein engineering has been applied as a powerful tool for fine-tuning enzyme activities (38), often through the modification of amino acid sequences that are found in nature to expand the borders of biocatalysis. Conferred by protein engineering strategies, such as incorporation of mutations and domain modifications, it was shown that the property of CARs can be improved toward desired functions (39, 40).
In this study, the MmCAR enzyme was modified to stimulate high-level production of MCFOHs. MmCAR is a relatively large enzyme of ∼130 kDa with multiple domains. Thus, we designed different engineering approaches based on the distinct domains. A high-throughput screening method coupled with cell growth was applied, which enabled an efficient selection process for beneficial enzyme variants. We identified several mutations based on directed evolution of the full-length MmCAR that potentially enhanced the catalytic activity toward MCFAs. According to the structural information obtained in a recent study (19), two of the mutated residues (D241E, H454R) from the best variant M150 are located in the core A domain (Acore, residues 1 to 510), but on the surface of the A domain, not in the active site. The third mutated residue, L567M, is situated in the mobile domain (Asub, residues 511 to 636), whose conformation changes substantially between the adenylation and thiolation states (SI Appendix, Fig. S16). Hence, this indicates that these mutations displayed indirect effects on the activity or stability of MmCAR. Although the mechanism of these effects is still unclear, the improved activity of M150 toward shorter chain length substrates, especially C6 FA rather than LCFAs (C16:1 and C18:1), was confirmed in vitro (Fig. 7C).
Additional efforts of shifting the substrate preference to MCFAs were made through establishing a random mutagenesis library based on only the A domain of MmCAR and rational design to narrow the substrate-binding pocket. Two rounds of evolution were executed, resulting in two CAR variants that benefitted the production of MCFOHs. Four mutations (E176G, Q182R, Q371R, F501Y) were identified in the most-efficient variant, AD10, after the second round of evolution. However, all of them were located on the surface of the Acore domain. Although the F517 residue is close to the adenine base of the substrate ATP, the structure analysis showed no involvement of this residue in ATP binding (19) (SI Appendix, Fig. S17). It was intriguing that the mutated residues in both M150 and AD10 tended to be on the protein surface and change the charge state, which might influence the protein stability or solubility and also indirectly increase the overall catalytic activity (41). A smaller space in the substrate-binding pocket would be beneficial for accommodating shorter-chain FA, which was demonstrated in our work by incorporating two tryptophan residues individually substituting two residues (Q302 and I303) close to the binding pocket of the A domain. The best performing mutant, AD303, enabled production of around 130 mg/L of MCFOHs, which was a 1.6-fold increase compared with the wild-type MmCAR, indicating that the rational design directly interfering with the binding pocket was potentially more efficient than the random approach in terms of altering substrate specificity in our study. Similarly, a previous study showed that the rational redesign of CAR enzymes successfully enabled the extension of their reaction scope toward amide synthesis (42).
Although successful studies on domain exchange in NRPS systems (43, 44) and CAR systems (19, 45) have been reported, replacing the native R or PCP-R domains with the ones from NRPS enzymes sharing high sequence similarity resulted in the complete loss of the activity of MmCAR (SI Appendix, Fig. S12), which might be due to an inappropriate domain boundary (SI Appendix, Fig. S7) used for domain-swapping, as shown in previous reports on PKS engineering (46). Thus, in the context of maintaining the biocatalytic activity of MmCAR, we sought to express the heterologous off-loading PCP-R domains without replacing the respective MmCAR domains. The observed positive effect on MCFOH production might be due to the extra aldehyde reductase activity from these off-loading PCP-R domains. In addition, a modification of the native R domain was conducted as well through establishing a site-directed saturation mutagenesis library toward residues 983 to 985. The mutant RF1 showed highest catalytic activity, resulting in a more than twofold increase in MCFOH production compared to the wild-type CAR (Fig. 5C). Consistently, the change in substrate preference toward MCFAs was also observed in the enzymatic assay in vitro. Although the positive effect on MCFOH production by the modified residues 983 to 985 has been proven in our study, it was difficult to investigate whether the improvement of the reduction efficiency was conferred by the final product scope being extended to fatty alcohols directly or by other mechanisms, which is due to the complex reaction environment and the presence of other endogenous reductases/dehydrogenases in vivo.
Adding additional MCFA substrates to the medium impaired cell growth, which also had a negative effect on the final MCFOH titer. Thus, we sought to increase intracellular MCFAs by deleting a potential MCFA exporter-coding gene TPO1. As intracellular accumulation is expected to increase the toxicity of MCFAs, the lower OD of TPO1-deficient strain YH28 indicated that the level of intracellular MCFAs was indeed increased. The successful improvement of MCFOH production in YH28 containing RF1+303 further indicated that the flux toward MCFOH formation was augmented due to the enhanced intracellular substrate level. The cellular toxicity was relieved by the conversion to alcohols, as a higher OD (around 3) was observed for the CAR-expressing strains (Fig. 8C). However, the impaired growth and low final biomass yield resulting from the toxicity of MCFOHs is still a major obstacle for further improvements in MCFOH production (47). Thus, strategies increasing cellular tolerance against MCFOHs and optimization of the fermentation process through decoupling the cell growth and product formation would be potential future directions in order to obtain further improvements in the production of MCFOHs.
The broad substrate scope of CAR enzymes enables their usage in the bioproduction of diverse aldehydes and alcohols. However, for the production of MCFOHs by microbial cell factories, the substrate promiscuity of CAR enzymes represents an obstacle. Through multiple protein engineering strategies, we succeeded in optimizing MmCAR to significantly enhance MCFOH production to 252 mg/L. Many efforts have been devoted to enable biosynthesis of MCFOHs in yeast cells in previous studies. Targeting a fatty acyl-CoA reductase (TaFAR) to the peroxisomes was demonstrated to be an efficient strategy and resulted in the production of MCFOHs (C10–C12) in Saccharomyces cerevisiae in a dodecane bilayer cultivation (SI Appendix, Table S1). However, the C16 fatty alcohol was still the major product (>60%) in this study, for which the substrate preference of TaFAR might be one of the reasons. Our study, on the other hand, provides new insight into MCFOH production. Considering the difference in cultivation conditions, we achieved a significant improvement in both MCFOH titer and yield in minimal medium (SI Appendix, Table S1). The catalytic activity of the engineered MmCAR was also investigated both in vitro and in vivo. The efficient reduction of MCFAs by the engineered CAR enzymes will enable the synthesis of versatile aldehyde intermediates with broad applications for further production of, for example, the corresponding alka(e)nes and FA acyl esters. Although CAR enzymes were previously modified by different strategies (39, 42, 45), this work is unique in comprehensively engineering the CAR enzyme for more selective biosynthesis of MCFOHs. In addition, our engineering strategies may inspire and promote the engineering of other complex multidomain enzymes.
Methods
Directed Evolution of Full-Length MmCAR.
The mutagenesis library of MmCAR was constructed by error-prone PCR (GeneMorph II Random Mutagenesis Kit, Agilent Technologies) with a low mutation frequency (0 to 4.5 mutations/kb) using the primers MmCARwm-F/MmCARwm-R. The product was then cotransformed with two backbone fragments of pZW01 into yeast strain YJZ03 (pox1Δ hfd1Δ). The primers used for amplification are shown in SI Appendix, Table S4. The plasmid-based libraries were established in yeast. The library sizes for the two rounds of evolution were around 6 × 107 and 4.5 × 107, respectively. All of the colonies from the SD-URA (synthetic complete medium without uracil) plate were all scratched into 20 mL minimal medium with 290 mg/L C8 FA (330 mg/L for the second round). After 24 h, the cells (1:100 diluted) were transferred into 20 mL minimal medium with 310 mg/L C8 FAs (350 mg/L for the second round) for 48 h. Then the cells were diluted 1:50, and transferred into 20 mL minimal medium with 330 mg/L C8 FA (370 mg/L for the second round) for 48 h. The enriched cells were plated on SD-URA solid medium, and around 60 single colonies were picked randomly into 96-well plate with minimal medium containing 330 mg/L C8 FAs (370 mg/L for the second round). The growth curves were monitored by a Bioscreen C MBR instrument (Growth Profiler 960 was used for the second round). The plasmids from fast-growing colonies were extracted and used to transform strain ZWE243 to determine MCFOH production. The mutant CAR with mutations D241E, H454R, L567M was used as the template for the second round of evolution. The same process was conducted as described above.
Directed Evolution of MmCAR a Domain.
The random mutagenesis library based on the A domain (residues 88 to 541) of MmCAR was established using error-prone PCR (GeneMorph II Random Mutagenesis Kit) with the low mutation frequency (0 to 4.5 mutations/kb) by primer pair CAR-A-F/CAR-A-R. The primers are listed in SI Appendix, Table S4. The wild-type MmCAR was the template for the first round of evolution and the mutant CAR with mutations Q182R; Q371R was used for the second round. The sizes of the two libraries were both around 5 × 104. The same workflow for the library enrichment was used as described above, but different concentrations of C8 FA were used, which were 290 mg/L, 300 mg/L, and 310 mg/L for the first round and 310 mg/L, 330 mg/L, and 350 mg/L for the second round. Around 60 single colonies from first round and second round were cultivated in minimal medium with histidine containing 330 mg/L and 370 mg/L C8 FAs, respectively. The growth curves were monitored by a Bioscreen C MBR instrument (Growth Profiler 960 was used for the second round).
Site-Directed Saturation Mutagenesis Library.
The primers used for establishing the site-directed saturation mutagenesis library based on residues 983 to 985 are listed in SI Appendix, Table S4. The same enrichment method was used as described above with concentrations of 290 mg/L, 310 mg/L, 330 mg/L, and 350 mg/L C8 FAs. After isolating the variants on an SD-URA plate, 80 of the colonies were picked to grow in medium with 380 mg/L of C8 FA. Based on the growth curves generated with the help of a Growth Profiler 960, the clones with the highest growth rates were selected.
Enzymatic Assays.
The yeast cells (50 OD) were harvested after 24 h by centrifuging at 4,000 × g at 4 °C for 5 min, then washed twice with PBS buffer. After discarding the supernatant, the cells were resuspended in 0.5 mL extraction buffer (50 mM Tris⋅Cl, 1 mM EDTA, 1 mM KCl, pH 7.5) containing 10 mM DTT and 1% (vol/vol) protease inhibitor. The suspensions were vortexed for 20 s ×5 (5-min intervals on ice) with 300 mg 0.2- to 0.4-mm glass beads. The supernatants of the samples were collected by centrifuging at 20,000 × g at 0 °C for 20 min. Proteins were quantified using a Pierce BCA Protein Assay Kit (ThermoFisher Scientific). The MmCAR activity assay was performed in 96-well microplates; 90-μL reaction mix containing 50 mM Tris⋅Cl (pH 7.5), 10 mM MgCl2, 1 mM NADPH, 1 mM ATP, and FAs (0.5 mM C10, C12, C14, C16:1, C18:1 FA, and 5 mM C6, C8 FA) and 10 μL of extracts (5 μg total protein) were mixed to initiate the reaction. Then the reactions were monitored at 340 nm for 30 min at room temperature with a FLUOstar Omega micropate reader (BMG Labtech). The MmCAR activity was defined as NADPH oxidation rate (1 μmol/min) of total protein.
Supplementary Material
Acknowledgments
We thank Quanli Liu, Yi Liu, Martin Engqvist, and Tao R. Yu for critical discussion; and the Chalmers Mass Spectrometry Infrastructure for assistance with metabolite analysis. This work was funded by the Novo Nordisk Foundation (Grant NNF10CC1016517), the Swedish Foundation for Strategic Research, and the Knut and Alice Wallenberg Foundation. We also thank the Energimyndigheten for support. M.W. thanks the Austrian science fund (Elise-Richter Fellowship V415-B21) for financial support. Z.Z. acknowledges support from the Fundamental Research Funds for the Central Universities [DUT20RC(3)044].
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010521117/-/DCSupplemental.
Data Availability.
All details of strains, plasmids, media, and methods can be found in SI Appendix, SI Materials and Methods. All data related to this study, including MCFOH production, cell growth in Bioscreen and Growth profiler, and enzymatic assays, can be found in Dataset S1.
References
- 1.Peralta-Yahya P. P., Zhang F., del Cardayre S. B., Keasling J. D., Microbial engineering for the production of advanced biofuels. Nature 488, 320–328 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Rajesh Kumar B., Saravanan S., Rana D., Nagendran A., A comparative analysis on combustion and emissions of some next generation higher-alcohol/diesel blends in a direct-injection diesel engine. Energy Convers. Manage. 119, 246–256 (2016). [Google Scholar]
- 3.He M., Wang M., Tang G., Fang Y., Tan T., From medium chain fatty alcohol to jet fuel: Rational integration of selective dehydration and hydro-processing. Appl. Catal. A Gen. 550, 160–167 (2018). [Google Scholar]
- 4.Pfleger B. F., Gossing M., Nielsen J., Metabolic engineering strategies for microbial synthesis of oleochemicals. Metab. Eng. 29, 1–11 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Rupilius W., Ahmad S., Palm oil and palm kernel oil as raw materials for basic oleochemicals and biodiesel. Eur. J. Lipid Sci. Technol. 109, 433–439 (2007). [Google Scholar]
- 6.Hassan S. N., Sani Y. M., Abdul Aziz A. R., Sulaiman N. M. N., Daud W. M. A. W., Biogasoline: An out-of-the-box solution to the food-for-fuel and land-use competitions. Energy Convers. Manage. 89, 349–367 (2015). [Google Scholar]
- 7.Yaguchi A., Spagnuolo M., Blenner M., Engineering yeast for utilization of alternative feedstocks. Curr. Opin. Biotechnol. 53, 122–129 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y., Nielsen J., Liu Z., Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived hydrocarbons. Biotechnol. Bioeng. 115, 2139–2147 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Reiser S., Somerville C., Isolation of mutants of Acinetobacter calcoaceticus deficient in wax ester synthesis and complementation of one mutation with a gene encoding a fatty acyl coenzyme A reductase. J. Bacteriol. 179, 2969–2975 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schirmer A., Rude M. A., Li X., Popova E., del Cardayre S. B., Microbial biosynthesis of alkanes. Science 329, 559–562 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Winkler M., Carboxylic acid reductase enzymes (CARs). Curr. Opin. Chem. Biol. 43, 23–29 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Zhou Y. J.et al., Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat. Commun. 7, 11709 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schjerling C. K.et al., Disruption of the gene encoding the acyl-CoA-binding protein (ACB1) perturbs acyl-CoA metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 271, 22514–22521 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Runguphan W., Keasling J. D., Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metab. Eng. 21, 103–113 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Zhu Z.et al., Expanding the product portfolio of fungal type I fatty acid synthases. Nat. Chem. Biol. 13, 360–362 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Gajewski J., Pavlovic R., Fischer M., Boles E., Grininger M., Engineering fungal de novo fatty acid synthesis for short chain fatty acid production. Nat. Commun. 8, 14650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Z.et al., Enabling the synthesis of medium chain alkanes and 1-alkenes in yeast. Metab. Eng. 44, 81–88 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Akhtar M. K., Turner N. J., Jones P. R., Carboxylic acid reductase is a versatile enzyme for the conversion of fatty acids into fuels and chemical commodities. Proc. Natl. Acad. Sci. U.S.A. 110, 87–92 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gahloth D.et al., Structures of carboxylic acid reductase reveal domain dynamics underlying catalysis. Nat. Chem. Biol. 13, 975–981 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venkitasubramanian P., Daniels L., Rosazza J. P. N., Reduction of carboxylic acids by Nocardia aldehyde oxidoreductase requires a phosphopantetheinylated enzyme. J. Biol. Chem. 282, 478–485 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Finnigan W.et al., Characterization of carboxylic acid reductases as enzymes in the toolbox for synthetic chemistry. ChemCatChem 9, 1005–1017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gulick A. M., Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol. 4, 811–827 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hisanaga Y.et al., Structural basis of the substrate-specific two-step catalysis of long chain fatty acyl-CoA synthetase dimer. J. Biol. Chem. 279, 31717–31726 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Chhabra A.et al., Nonprocessive [2 + 2]e- off-loading reductase domains from mycobacterial nonribosomal peptide synthetases. Proc. Natl. Acad. Sci. U.S.A. 109, 5681–5686 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manavalan B., Murugapiran S. K., Lee G., Choi S., Molecular modeling of the reductase domain to elucidate the reaction mechanism of reduction of peptidyl thioester into its corresponding alcohol in non-ribosomal peptide synthetases. BMC Struct. Biol. 10, 1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stolterfoht H.et al., Identification of key residues for enzymatic carboxylate reduction. Front. Microbiol. 9, 250 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bloom J. D.et al., Evolving strategies for enzyme engineering. Curr. Opin. Struct. Biol. 15, 447–452 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Lutz S., Beyond directed evolution—Semi-rational protein engineering and design. Curr. Opin. Biotechnol. 21, 734–743 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnold F. H., Directed evolution: Bringing new chemistry to life. Angew. Chem. Int. Ed. Engl. 57, 4143–4148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Legras J. L.et al., Activation of two different resistance mechanisms in Saccharomyces cerevisiae upon exposure to octanoic and decanoic acids. Appl. Environ. Microbiol. 76, 7526–7535 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buijs N. A., Zhou Y. J., Siewers V., Nielsen J., Long-chain alkane production by the yeast Saccharomyces cerevisiae. Biotechnol. Bioeng. 112, 1275–1279 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Goyal A., Verma P., Anandhakrishnan M., Gokhale R. S., Sankaranarayanan R., Molecular basis of the functional divergence of fatty acyl-AMP ligase biosynthetic enzymes of Mycobacterium tuberculosis. J. Mol. Biol. 416, 221–238 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Gulick A. M., Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol. 4, 811–827 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Y., “Multidimensional engineering for the production of fatty acid derivatives in Saccharomyces cerevisiae,” PhD thesis, Chalmers University of Technology, Gothenburg, Sweden (2019).
- 35.Barajas J. F.et al., Comprehensive structural and biochemical analysis of the terminal Myxalamid reductase domain for the engineered production of primary alcohols. Chem. Biol. 22, 1018–1029 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Albertsen M., Bellahn I., Krämer R., Waffenschmidt S., Localization and function of the yeast multidrug transporter Tpo1p. J. Biol. Chem. 278, 12820–12825 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Zhu Z.et al., Multidimensional engineering of Saccharomyces cerevisiae for efficient synthesis of medium-chain fatty acids. Nat. Catal. 3, 64–74 (2020). [Google Scholar]
- 38.Renata H., Wang Z. J., Arnold F. H., Expanding the enzyme universe: Accessing non-natural reactions by mechanism-guided directed evolution. Angew. Chem. Int. Ed. Engl. 54, 3351–3367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fedorchuk T. P., Khusnutdinova A. N., Flick R., Yakunin A. F., Site-directed mutagenesis and stability of the carboxylic acid reductase MAB4714 from Mycobacterium abscessus. J. Biotechnol. 303, 72–79 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Schwendenwein D.et al., Random mutagenesis-driven improvement of carboxylate reductase activity using an amino benzamidoxime-mediated high-throughput assay. Adv. Synth. Catal. 361, adsc.201900155 (2019). [Google Scholar]
- 41.Strickler S. S.et al., Protein stability and surface electrostatics: A charged relationship. Biochemistry 45, 2761–2766 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Wood A. J. L.et al., Adenylation activity of carboxylic acid reductases enables the synthesis of amides. Angew. Chem. Int. Ed. Engl. 56, 14498–14501 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Mootz H. D., Schwarzer D., Marahiel M. A., Construction of hybrid peptide synthetases by module and domain fusions. Proc. Natl. Acad. Sci. U.S.A. 97, 5848–5853 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hur G. H., Vickery C. R., Burkart M. D., Explorations of catalytic domains in non-ribosomal peptide synthetase enzymology. Nat. Prod. Rep. 29, 1074–1098 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kramer L.et al., Engineering and characterization of hybrid carboxylic acid reductases. J. Biotechnol. 304, 52–56 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Yuzawa S.et al., Comprehensive in vitro analysis of acyltransferase domain exchanges in modular polyketide synthases and its application for short-chain ketone production. ACS Synth. Biol. 6, 139–147 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Henritzi S., Fischer M., Grininger M., Oreb M., Boles E., An engineered fatty acid synthase combined with a carboxylic acid reductase enables de novo production of 1-octanol in Saccharomyces cerevisiae. Biotechnol. Biofuels 11, 150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All details of strains, plasmids, media, and methods can be found in SI Appendix, SI Materials and Methods. All data related to this study, including MCFOH production, cell growth in Bioscreen and Growth profiler, and enzymatic assays, can be found in Dataset S1.