Significance
This manuscript identifies a pathway of recombination which joins DNA ends on two homologous chromosomes in human cells. This pathway supports efficient recombination at targeted DSBs and nicks, at frequencies exceeding those of the spontaneous recombination events that ameliorate genetic diseases in human blood and skin cells. Recombination is further stimulated by depletion of POLQ, a factor with both DNA polymerase and helicase activities. POLQ depletion causes an especially dramatic increase in the frequency of copy-neutral loss of heterozygosity (cnLOH) events. These results have immediate practical implications for therapeutic correction of both compound heterozygous and autosomal dominant mutations, and they identify a mechanistic origin for prominent but poorly understood mutagenic signatures in cancer.
Keywords: DNA nick, double-strand break, gene editing, gene therapy, polymerase
Abstract
Interhomolog recombination (IHR) occurs spontaneously in somatic human cells at frequencies that are low but sufficient to ameliorate some genetic diseases caused by heterozygous mutations or autosomal dominant mutations. Here we demonstrate that DNA nicks or double-strand breaks (DSBs) targeted by CRISPR-Cas9 to both homologs can stimulate IHR and associated copy-neutral loss of heterozygosity (cnLOH) in human cells. The frequency of IHR is 10-fold lower at nicks than at DSBs, but cnLOH is evident in a greater fraction of recombinants. IHR at DSBs occurs predominantly via reciprocal end joining. At DSBs, depletion of POLQ caused a dramatic increase in IHR and in the fraction of recombinants exhibiting cnLOH, suggesting that POLQ promotes end joining in cis, which limits breaks available for recombination in trans. These results define conditions that may produce cnLOH as a mutagenic signature in cancer and may, conversely, promote therapeutic correction of both compound heterozygous and dominant negative mutations associated with genetic disease.
Interhomolog recombination (IHR) occurs rarely in somatic cells, but when it does it may profoundly affect genomic structure and function, especially if it results in copy-neutral loss of heterozygosity (cnLOH). In cancer, cnLOH can replace long blocks of chromosome arms, creating homozygosity that contributes to clonal expansion and tumor progression. In genetic disease, spontaneous recombination between homologs can correct autosomal dominant or compound heterozygous mutations.
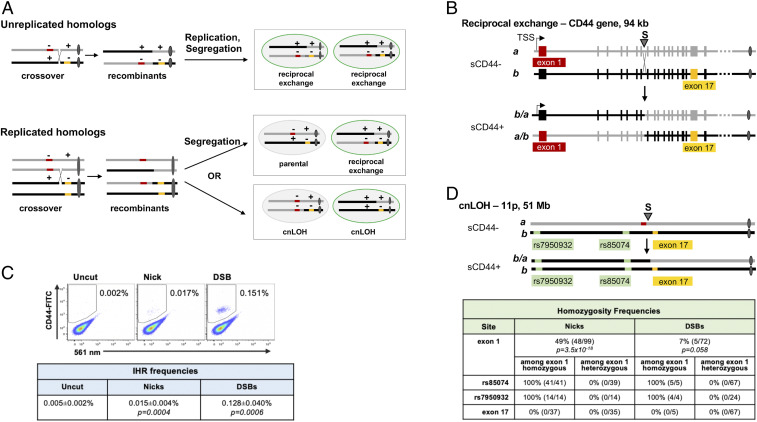
One pathway to cnLOH involves reciprocal recombination between replicated homologs in late S or G2 phase of the cell cycle, with segregation generating daughter cells that receive one unrecombined and one recombinant chromosome. The alternative segregation pattern generates one daughter cell bearing both unrecombined homologs and another bearing both recombined homologs, neither exhibiting cnLOH (Fig. 1 A, Bottom). If segregation is random, the two segregation patterns should occur with equal frequency, in which case 50% of reciprocal IHR events between replicated homologs will exhibit cnLOH.
Fig. 1.
Targeted nicks or DSBs initiate IHR and LOH. (A) The diagram shows two homologous chromosomes, distinguished by gray or black lines, with centromeres as ovals. Mutations are indicated by red or gold bars, with + and − above a homolog distinguishing wild-type and mutant sequences. Reciprocal exchange that corrects a compound heterozygous mutation (++ homolog) enables protein expression, indicated as a green cell perimeter. (A, Top) Cross-over recombination between unreplicated homologs can correct compound heterozygous mutations. (A, Bottom) Recombination between replicated homologs can correct compound heterozygous mutations and may result in cnLOH. cnLOH occurs if chromosome segregation distributes one cross-over and one non–cross-over homolog to each daughter cell (Bottom Right). cnLOH extends from the recombination site to the telomere. cnLOH can yield a corrected chromosome restoring homozygosity to the wild-type allele, as shown (Bottom Right). (B) Reciprocal exchange at the CD44 gene. (B, Top) Diagram of homologs a and b (gray and black, respectively) bearing alleles of CD44 engineered to carry mutations in exon 1 (red; homolog a) and exon 17 (gold; homolog b), showing the site targeted by gRNA S in intron 7 (arrowhead). Cells carrying these homologs are sCD44-. (B, Bottom) Diagrams of recombinants that are products of reciprocal exchange (b/a and a/b) that corrects the CD44 gene, generating sCD44+ cells. Also shown are the transcription start site and centromeres (ovals). (C) Representative flow cytometry analysis of HT1080-K1 cells in which cleavage of CD44 intron 7 was targeted by transfection of plasmids expressing gRNA S alone (no cleavage) or together with Cas9D10A (nicks) or Cas9 (DSBs). Frequencies of CD44+ cells (in gate) are indicated. (C, Bottom) Frequencies are shown as mean and SEM (n = 6). P values were determined by two-tailed t test. (D) Diagram of chromosome 11p, showing homologs a and b carrying mutations in CD44 (red, gold) as in B. Recombinant b/a, bearing a corrected CD44 gene, which has cosegregated with homolog b, resulting in cnLOH. Exon 1 and two telomeric SNPs, rs7950932 and rs85074, are homozygous in this recombinant. (D, Bottom) Frequencies of homozygosity among sCD44+ clones produced by IHR at nicks or targeted DSBs. P values were determined by Fisher’s exact test. The frequency of homozygosity at SNPs and at exon 17 is shown for a subset of clones, both heterozygous and homozygous at exon 1.
IHR has frequently been documented in studies of cancer and genetic disease, where recombination in individual somatic cells confers a proliferative or selective advantage that supports clonal expansion. IHR leading to cnLOH has been inferred in many tumor types, especially hematopoietic tumors, where cnLOH correlates with poor prognosis (1). In contrast, IHR and cnLOH can result in gene correction and amelioration of genetic disease (2–4). Beneficial outcomes of IHR have been documented most frequently in blood and skin, where corrected cells can be readily identified and characterized. Spontaneous IHR in blood cells corrects mutations that cause DOCK8 immunodeficiency syndrome (5), Bloom syndrome (6), Fanconi anemia (7), dyskeratosis congenita (8), Diamond–Blackfan anemia (9), and ataxia-pancytopenia syndrome (10). In skin cells, IHR corrects mutations that cause ichthyosis with confetti (11), loricrin keratoderma (12), and the severe skin fragility disorder epidermolysis bullosa (13). IHR also corrects mutations in seven different genes that cause congenital disorders of glycosylation affecting various tissues (14). Molecular analyses have shown that, in some patients, correction results from reciprocal IHR that occurs between two compound heterozygous mutations, while in those suffering from dyskeratosis congenita, Diamond–Blackfan anemia, ataxia-pancytopenia syndrome, loricrin keratoderma, and ichthyosis with confetti, cnLOH has occurred to eliminate the autosomal dominant mutation associated with disease.
The clear potential of IHR and cnLOH to affect disease progression in both deleterious and beneficial ways prompted us to learn more about the mechanism and regulation of these pathways. We have analyzed IHR and cnLOH at breaks targeted to an endogenous gene in a diploid human cell line, as analyses of mammalian cells (15–20) and yeast (21–23) have shown that IHR normally occurs at very low frequencies but can be stimulated by DNA damage, including targeted nuclease cleavage. Here we report that spontaneous IHR occurs infrequently in human cells (<0.01%), as anticipated by previous reports, but that IHR can be stimulated as much as 200-fold by nicks or double-strand breaks (DSBs) targeted to both homologous chromosomes. The frequency of IHR is 10-fold lower at nicks than at DSBs, but cnLOH is associated with half of all recombination events at nicks and a much lower fraction at DSBs. Both clonal analysis and deep sequencing establish that IHR at DSBs occurs predominantly by a reciprocal end-joining (EJ) pathway. IHR frequencies at DSBs were stimulated fivefold (from 0.4 to 2%) by depletion of POLQ, a DNA helicase and translesion polymerase that is key to alternative end joining (alt-EJ) and suppresses some forms of genomic instability but also promotes insertions and random chromosomal integration (24–29). This stimulation was accompanied by a striking increase in cnLOH. These results suggest that POLQ may promote end joining in cis and thereby limit the number of breaks available for end joining in trans; and that, in the absence of POLQ, DSBs may persist and become substrates for reciprocal EJ in G2 phase of the cell cycle, when subsequent segregation can result in cnLOH. The pathways documented here may contribute to cnLOH in tumors. They may also potentially be utilized to correct both compound heterozygous and autosomal dominant mutations that cause human genetic disease.
Results
Nicks and DSBs Targeted to Both Homologs Stimulate IHR.
To better understand the mechanism of IHR and cnLOH in human cells, we developed an assay that uses flow cytometry to score IHR at DNA breaks targeted to the endogenous CD44 gene. The CD44 gene is composed of 18 exons spanning 94 kb, ∼35 Mb from the telomere on chromosome 11p (Fig. 1B). It encodes a cell-surface glycoprotein that is expressed in most cells but is dispensable for proliferation in culture. IHR at the CD44 gene was assayed in human HT1080 cells, a p53-positive fibrosarcoma cell line that maintains a near-diploid karyotype in culture (30, 31). We generated an HT1080 derivative line, HT1080 K1, in which mutations inactivate exon 1 on one homolog and exon 17 on the other (homologs denoted a and b in Fig. 1 B, Top). Reciprocal IHR at a site between these mutations is predicted to restore gene function, yielding sCD44+ cells (Fig. 1 B, Bottom). If homologs recombine prior to replication, reciprocal recombination will correct the compound heterozygous mutation in all daughter cells (Fig. 1 A, Top). If replicated homologs recombine, then segregation may generate either of two distinct pairs of daughter cells (Fig. 1 A, Bottom). In one pair, both daughters exhibit cnLOH extending from the break site to the telomere, and each daughter carries one parental homolog and either a mutant (sCD44-) or corrected (sCD44+) recombinant homolog. In the other pair, neither daughter exhibits cnLOH, and one daughter carries two parental homologs (sCD44-) and the other carries two recombinant homologs (sCD44+). Thus, this assay of IHR at the endogenous CD44 gene scores events that can correct either compound heterozygous or autosomal dominant mutations.
To ask if DNA lesions can stimulate IHR and cnLOH, nicks or DSBs were targeted, by transfection with plasmids expressing Streptococcus pyogenes Cas9D10A or Cas9 and CRISPR guide RNA (gRNA) S, to a site in CD44 intron 7 that is more than 60 kb downstream of the transcription start site (TSS) and exon 1 mutation and more than 20 kb upstream of the mutation in exon 17 (Fig. 1B and SI Appendix, Fig. S1B). The frequency of sCD44+ cells was very low in control populations (0.005%), but IHR was stimulated to reach frequencies of 0.015% at nicks and 0.128% at DSBs (P < 0.001), with recombinant populations readily distinguished by flow cytometry (Fig. 1C).
IHR Is Accompanied by cnLOH.
To quantify frequencies of cnLOH among products of IHR, single sCD44+ recombinant clones were sorted by flow cytometry and expanded, generating populations that exhibited continuous stable expression of surface CD44. Homozygosity was assessed by restriction fragment-length polymorphism (RFLP) analysis of CD44 exons 1 and 17 and of two heterozygous single-nucleotide polymorphisms (SNPs) telomeric to CD44 (SI Appendix, Figs. S1 and S2). The frequency of cnLOH among sCD44+ cells approached 50% at nicks, sevenfold higher than at DSBs (48.5 vs. 6.9%; P = 1.6 × 10−9; Fig. 1D). In all tested recombinants, heterozygosity was maintained at exon 17 and zygosity of both telomeric SNPs matched that of CD44 exon 1. Thus, cnLOH extended from the break site to the telomere, as predicted for cnLOH events caused by segregation of cross-over homologs.
IHR Frequencies Are Largely Independent of Break Site.
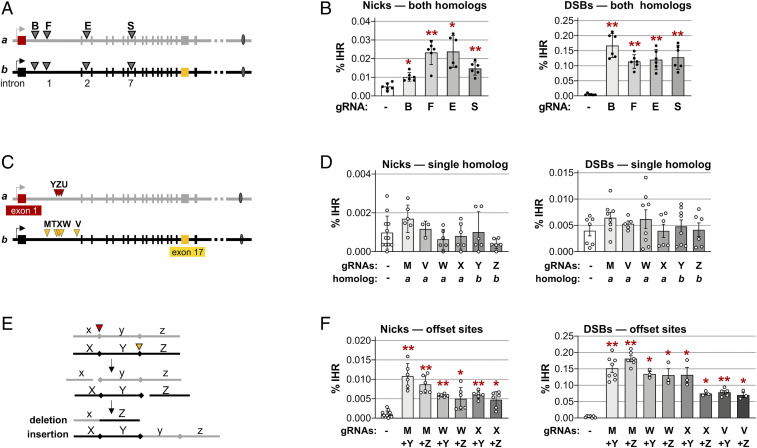
To ask if IHR frequency depends upon the location of the break site, nicks and DSBs were targeted to four different sites in the CD44 gene, by gRNAs B, F, E, and S (Fig. 2A). Frequencies of IHR ranged from 0.01 to 0.02% at nicks and 0.11 to 0.17% at DSBs, in both cases significantly above the background (0.001%) in control populations expressing only Cas9 but no gRNA (P < 3 × 10−3 and P < 7 × 10−4, respectively; Fig. 2B). Frequencies of IHR at DSBs were 10-fold higher than at nicks, but for both DSBs and nicks the variation in IHR frequencies among sites separated by nearly 50 kb was less than twofold. Thus, there was little dependence of IHR frequency upon the position of the target site within the CD44 gene.
Fig. 2.
IHR joins offset nicks and DSBs. (A) Map of sites in introns 1, 2, and 7 of the CD44 gene targeted by gRNAs B, F, E, and S. (B) Frequencies of IHR in HT1080-K1 cells in which nicks or DSBs were targeted to both homologs by transfection of plasmids expressing Cas9D10A or Cas9 and no gRNA (-) or the indicated gRNA. Control cells were transfected with expression constructs for gRNA only, not enzyme. IHR frequencies are shown as mean and SD (n = 6); asterisks indicate significant differences from the no-gRNA control (*P < 0.05 and **P < 10−3). (C) Map of sites in CD44 targeted on only a single homolog by gRNAs Y, Z, or U (homolog a; red arrowheads) or M, T, X, W, or V (homolog b; gold arrowheads) in the engineered CD44-/- gene in HT1080-K2 cells. Sequences are shown in SI Appendix, Fig. S3. (D) Frequencies of IHR in HT1080-K2 cells in which nicks or DSBs were targeted to sites on a single homolog by transfection with an expression clone for Cas9D10A or Cas9, respectively, and no gRNA (-) or the indicated gRNA. IHR frequencies are shown as mean and SD (n ≥ 3). No IHR frequencies were significantly different from the no-gRNA control. (E) Diagram of recombinants predicted to result from the joining of ends of offset DSBs. Adjacent regions x, y, and z on the upper homolog (gray) and X, Y, and Z on the lower homolog (black) are cleaved at offset sites indicated by red arrowheads on either side of the central fragment, which has caps at its boundaries. Interhomolog EJ generates one recombinant in which region y is deleted and one in which it is inserted. See also Fig. 5A. (F) Frequencies of IHR at nicks and DSBs targeted to the indicated offset sites on the two homologs in HT1080-K2 cells by transfection with an expression clone for Cas9D10A or Cas9, respectively, and no gRNA (-) or the indicated gRNA. IHR frequencies are shown as mean and SD (n ≥ 3); asterisks indicate significant differences from the no-gRNA control (*P < 0.05 and **P < 10−3).
Nicks or DSBs Targeted to a Single Homolog Do Not Stimulate IHR.
If IHR depends upon invasion of a duplex by a cleaved DNA end, as occurs in canonical homologous recombination (HR), then a DSB targeted to a single homolog is predicted to stimulate IHR. To test this, IHR frequencies were determined at nicks or DSBs targeted to intron 1 by gRNAs that recognize only a single homolog. gRNAs Y, Z, and U target homolog a within a heterologous region engineered 27.5 kb downstream of the CD44 TSS; gRNAs T, W, and X target homolog b at sites protected by the engineered heterology; and gRNAs M and V target homolog b at naturally occurring SNPs (Fig. 2C and SI Appendix, Fig. S3). Nicks or DSBs targeted to a single homolog did not stimulate IHR at frequencies significantly above background (<0.002 and <0.007%, respectively; Fig. 2D). These results suggest that canonical HR is not the predominant pathway of IHR.
IHR Requires a Break on Each Homolog.
If an EJ pathway supports IHR, then IHR will require that both homologs be targeted by breaks (Fig. 2E). We tested this by determining frequencies of IHR in cells in which combinations of gRNAs targeted nicks or DSBs to each of the two homologs at distinct positions (“offset” sites). Target sites were offset by 5 bp (gRNAs W+Y, W+Z, X+Y, and X+Z), 0.6 kb (gRNAs M+Y and M+Z), or 3.5 kb (gRNAs V+Y and V+Z) (Fig. 2C and SI Appendix, Fig. S3). The frequencies of IHR at offset nicks were in the range of 0.005 to 0.011%, and those at offset DSBs were in the range of 0.07 to 0.17% (Fig. 2F), significantly above background and comparable to the range at breaks targeted to the same site on both homologs (Fig. 2B). While there were some significant differences between IHR frequencies at different cut sites (for example, at DSBs targeted by gRNAs M+Y [0.6-kb separation] vs. gRNAs V+Y [5-bp separation]; P = 7.7 × 10−6; Fig. 2F), this could reflect different cleavage efficiencies of individual gRNAs.
DNA-Protein Kinase and POLQ Inhibit IHR at DSBs.
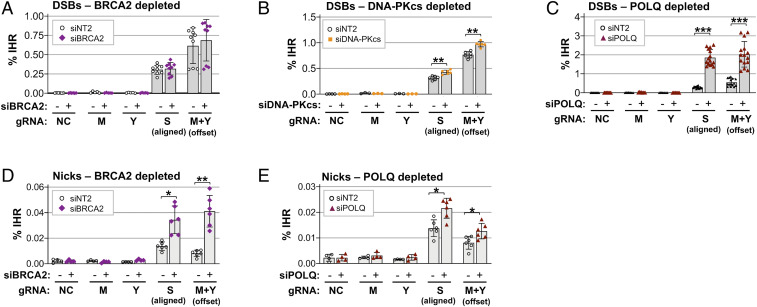
To further assess the contributions of HR and EJ to IHR, we assayed IHR frequencies in cells depleted of key factors in each pathway. BRCA2 is critical for canonical HR at DSBs, and control experiments showed that depletion of BRCA2 caused a fivefold decrease in canonical HR supported by a dsDNA plasmid donor at a DSB targeted to the CD44 gene (SI Appendix, Fig. S4A). However, depletion of BRCA2 did not affect IHR frequencies at aligned or offset DSBs (Fig. 3A). Thus, canonical HR is not the primary pathway of IHR at DSBs.
Fig. 3.
POLQ, DNA-PK, and BRCA2 can limit IHR. Frequencies of IHR at DSBs (A–C) or nicks (D and E) targeted by transfection of RNPs containing Cas9 or Cas9D10A and the indicated gRNAs (gRNA NC, noncutting control) in HT1080-K2 cells treated with siRNAs to BRCA2, DNA-PKcs, or POLQ, as indicated. IHR frequencies are shown as mean and SD (n ≥ 3); asterisks indicate significant differences from the nontargeting control siRNA, siNT2 (*P < 0.05, **P < 10−3, and ***P < 10−6).
The contribution of classical nonhomologous end joining [c-NHEJ (32, 33)] to IHR at DSBs was tested by depletion of the catalytic subunit of DNA-protein kinase (DNA-PKcs), which caused a modest but significant stimulation of IHR at both aligned and offset DSBs (P = 5 × 10−4 and 1 × 10−4; Fig. 3B). A specific role for DNA-PK in inhibition of IHR was confirmed by analyzing the effect of treatment with NU7441, a highly selective inhibitor of DNA-PK activity. Culture with NU7441 increased IHR frequencies, with twofold stimulation evident at 2 µM NU7441 (SI Appendix, Fig. S4B). These results suggest that c-NHEJ, which is regulated by DNA-PK, may compete with IHR at DSBs.
The contribution of the alt-EJ pathway was tested by depletion of POLQ, which encodes DNA Polθ, a DNA helicase and translesion polymerase that promotes repair of DSBs formed by a variety of processes and mechanisms, including replication fork stalling, CRISPR-Cas9 cleavage, and ionizing radiation, and, conversely, promotes insertions and random chromosomal integration (24, 26–29). Depletion of POLQ, using a pool of four small interfering RNAs (siRNAs), stimulated IHR frequencies fourfold or more at both aligned and offset DSBs (Fig. 3C; P = 5.3 × 10−10 and 3.4 × 10−7, respectively). The specificity of depletion for POLQ was confirmed by demonstrating that depletion with any one of the four pooled siRNAs also increased IHR frequencies (SI Appendix, Fig. S4C). We conclude that POLQ inhibits IHR at DSBs.
BRCA2 and POLQ Inhibit IHR at Nicks.
We also tested the contribution of BRCA2 and POLQ to IHR at nicks. Depletion of BRCA2 caused a 2.5-fold stimulation of IHR at aligned nicks and a 5.0-fold stimulation at offset nicks (P = 0.005 and 0.001, respectively; Fig. 3D). BRCA2 depletion also suppressed alternative homology-directed repair (alt-HDR) at nicks supported by either single-stranded DNA donors or nicked plasmid donors (34, 35). Depletion of POLQ caused a modest stimulation of IHR at both aligned and offset nicks (1.5-fold, P < 0.018; Fig. 3E). These results identify previously unappreciated roles of BRCA2 and POLQ in suppressing IHR to maintain genomic stability.
POLQ Inhibits cnLOH at DSBs.
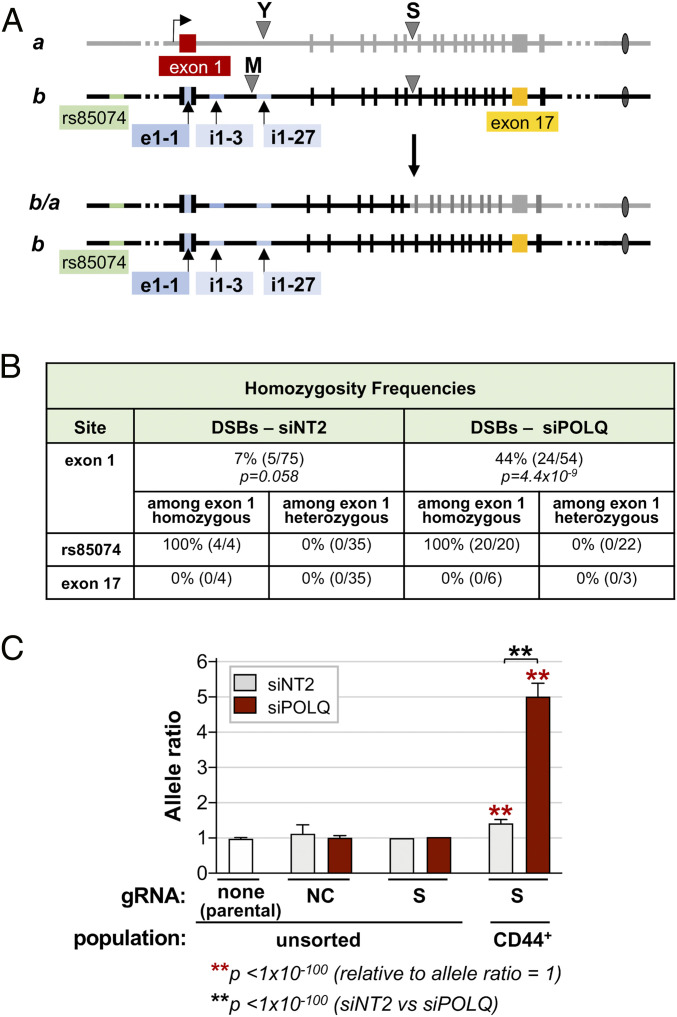
The possibility that POLQ depletion might increase frequencies of cnLOH as well as IHR was first tested by querying RFLPs in sCD44+ recombinants initiated by offset DSBs targeted by gRNAs M and Y (Fig. 4A). Homozygosity at CD44 exon 1 was evident in 6.7% (5/75) of CD44+ recombinant clones from the control population and in 44.4% (24/54) of recombinant clones from cells treated with siPOLQ, a 6.6-fold difference (P = 7.3 × 10−7; Fig. 4B). In all recombinant clones tested, heterozygosity was retained at exon 17 and the zygosity of telomeric SNP rs85074 matched that of CD44 exon 1. Thus, cnLOH extended from the cross-over to the telomere.
Fig. 4.
Depletion of POLQ stimulates cnLOH at DSBs. (A, Top) Map of the CD44 gene identifying SNPs in exon 1 (e1-1) and intron 1 (i1-3, i1-27) that were queried to determine allele ratios (sequences are in SI Appendix, Fig. S1). (A, Bottom) Map of predicted recombinants exhibiting cnLOH after a DSB targeted to intron 7 by gRNA S. (B) Frequencies of homozygosity as determined by RFLP mapping at the indicated regions in sCD44+ cells. IHR was stimulated by offset DSBs targeted by transfection of RNPs containing Cas9 and gRNAs M and Y. P values were determined by Fisher’s exact test. Frequencies of homozygosity at SNP rs85074 and at exon 17 are shown for a subset of clones, both heterozygous and homozygous, at exon 1. (C) Allele ratios of diagnostic SNPs in exon 1 and intron 1 of the CD44 gene as determined by amplicon sequencing of the indicated cell populations. Results are shown for parental cells and for cells transfected with RNPs containing Cas9 and either nonspecific gRNA NC or gRNA S, targeted to CD44 intron 7. Cells were unsorted or sorted for sCD44+ and treated with control siNT2 or siPOLQ, as indicated. An allele ratio greater than 1 within a region is characteristic of a population that has lost heterozygosity in that region. Allele ratios are shown as the mean and SD of the individual allele ratios at the SNPs in exon 1 (e1-1) and intron 1 (i1-3, i1-27). Significant differences are indicated relative to an allele ratio of 1 and between sorted siNT2 and siPOLQ samples (**P < 1 × 10−100, Fisher’s exact test).
Analysis of cnLOH was extended by using next-generation sequencing (NGS) to determine the allele ratios of cell populations at diagnostic SNPs between the cut site and the telomere, which are predicted to become homozygous following cnLOH (Fig. 4A and SI Appendix, Fig. S1). The allele ratio will equal 1.0 in a population of diploid cells in which all cells are heterozygous but will be greater than 1.0 if either of the two alternative alleles queried is homozygous. Importantly, the allele ratio will also equal 1.0 in a population in which both alternative alleles are homozygous in the same number of cells, such as in an unsorted population that has been induced to undergo IHR (Fig. 1 A, Bottom).
Cells were stimulated to carry out IHR by targeting DSBs to intron 7 with gRNA S, DNA prepared from >106 cells, and regions spanning three diagnostic SNPs were amplified and sequenced. The average of the allele ratios for these diagnostic SNPs was close to 1 in the parental population and in the unsorted cell populations. In sorted populations of recombinant sCD44+ cells, the average allele ratio was 1.4 in the siNT2-treated population and 5.0 in the siPOLQ-treated population. Both these allele ratios are significantly greater than 1 (P < 10−100; Fig. 4C). Moreover, the increase in allele ratio resulting from POLQ depletion was highly significant (P < 10−100). Thus, the results of amplicon sequencing are consistent with the IHR-associated cnLOH observed in sCD44+ clones.
IHR at DSBs Proceeds by Reciprocal EJ.
Reciprocal EJ between homologs targeted by offset DSBs will generate characteristic recombinants in which the sequence flanked by the DSBs has been deleted from one homolog and inserted into the other (Figs. 2E and 5A). Reciprocal EJ at offset DSBs will also eliminate both target cut sites, so while ends may undergo processing prior to or during joining, they will not be subject to further cleavage by Cas9 subsequent to joining. Junction sequences will therefore directly reflect the process of reciprocal EJ.
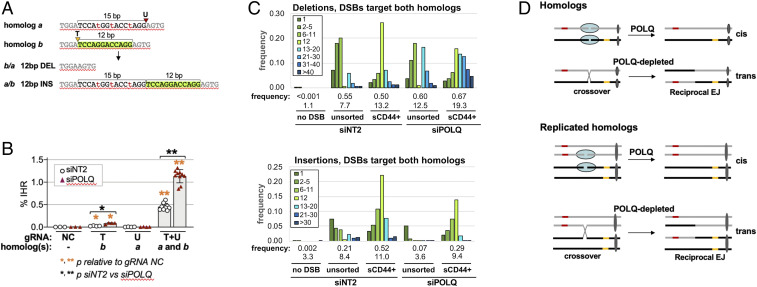
Fig. 5.
Amplicon sequencing of products of IHR at DSBs. (A) IHR at DSBs targeted to homolog a by gRNA U or to homolog b by gRNA T. (A, Top) Parental configuration. (A, Bottom) Recombinants. Lengths of deleted and inserted sequences are indicated by brackets, with the 12-bp region deleted in homolog a/b and inserted in homolog b/a highlighted in green. Arrowheads identify target sites; sequence polymorphisms are shown in lowercase red font. gRNA sequences and map are shown in SI Appendix, Fig. S3. (B) Frequencies are shown (mean and SD) of IHR at DSBs targeted to CD44 intron 1 on the indicated homolog(s) by transfection of RNPs containing Cas9 and the indicated gRNAs (NC; no DSB) in HT1080-K2 cells treated with control siNT2 or siPOLQ. Asterisks indicate significant differences (*P < 0.05 and **P < 10−6) either between gRNA NC and another gRNA or between siNT2 and siPOLQ (indicated by a bracket). (C) Frequencies of binned deletions (Top) and insertions (Bottom) in parental cells (no DSB, control) or at DSBs targeted by gRNAs T+U to homologs a and b. Cell populations were treated with siNT2 or siPOLQ, and DSBs were targeted by transfection of RNPs containing Cas9 and gRNA T+U. Total frequencies and average lengths (bp) are shown below each sample. Frequencies are calculated among total reads (both homologs). Note the 12-bp bins are nearly entirely compromised of the deletion and duplication products predicted to result from IHR. Frequencies and lengths of indels are shown below binned results for each sample (also tabulated in SI Appendix, Fig. S8). The distributions of deletion and insertion lengths were similar in two independent T+U samples, as is evident by comparing these data with a duplicate experiment shown in SI Appendix, Fig. S10. (D) Diagram showing how POLQ may promote EJ in cis, and thereby limit the availability of DSBs for EJ in trans that results in IHR.
To establish whether IHR junctions are formed by reciprocal EJ and to learn how junction structure or sequence was affected by depletion of POLQ, amplicon-based NGS was carried out on populations of cells in which DSBs were targeted by gRNAs T or U alone or by gRNAs T+U together. Reciprocal EJ between DSBs targeted by gRNAs T+U to offset sites on homologs a and b is predicted to result in deletion of a 12-bp region from one recombinant homolog and its insertion into the other (Fig. 5A). Depletion of POLQ was shown to stimulate IHR at these offset sites, as at other sites (2.5-fold, P = 4.6 × 10−9; Fig. 5B). Additional control experiments assaying positional read depth established that gRNAs T or U preferentially targeted the intended homolog (SI Appendix, Fig. S5). Amplicon-based sequencing of cell populations targeted by either gRNA T or U alone showed that POLQ depletion caused a modest increase in deletion frequency and a 7- to 10-fold decrease in the frequency of insertions larger than 1 bp accompanied by a decrease in average insertion length (SI Appendix, Figs. S6 and S7). This evidence that POLQ depletion reduced the frequency of insertions, particularly longer insertions (26, 29), provides an internal control for the effectiveness of POLQ depletion.
In populations that had undergone IHR (sCD44+), 12-bp indels diagnostic of reciprocal EJ were enriched more than 70-fold relative to unsorted cells (P < 10−100; Fig. 5C). Diagnostic 12-bp indels comprised 26% of deletions and 21% of insertions in control populations and 15% of deletions and 13% of insertions in cells depleted for POLQ (Fig. 5C). Frequencies of indels were similar at DSBs targeted to only a single homolog or to both homologs, and average insertion length was reduced in response to depletion of POLQ (SI Appendix, Figs. S7 and S8). POLQ-mediated alt-EJ uses microhomologies to rejoin DNA ends (26, 29). Fractional representation of microhomologies at deletion junctions identified differences between cut sites, but no consistent effect of POLQ depletion was evident in this limited dataset (SI Appendix, Fig. S9).
It was initially surprising that depletion of POLQ decreased the frequency of 12-bp indels in populations that had undergone IHR (sCD44+). This may reflect processing that occurs during IHR junction formation, as evident when single DSBs were rejoined in cis (SI Appendix, Figs. S6 and S7). Independent of IHR, the average deletion length was at least 1.5-fold greater in POLQ-depleted than in control cells, regardless of whether only one (SI Appendix, Fig. S7) or both (SI Appendix, Fig. S8) homologs were targeted by DSBs. Similar deletions at an IHR junction will reduce the fraction of junctions bearing diagnostic 12-bp indels. This was evident as an increase in deletions longer than 12 bp in products of IHR in POLQ-depleted sCD44+ cells (Fig. 5C and SI Appendix, Fig. S10).
These results support the view that IHR occurs by reciprocal EJ. Processing that accompanies joining of DSBs in trans may be accompanied by both insertions and deletions, but frequently occurs in the absence of either. POLQ suppresses reciprocal EJ that results in IHR while limiting the length of deletions and promoting insertions, especially longer insertions.
Discussion
We have shown that targeted nicks or DSBs can stimulate IHR that corrects compound heterozygous mutations and causes cnLOH in human cells. IHR frequencies were nearly an order of magnitude greater at DSBs than at nicks (0.17 vs. 0.02% in control cells; Fig. 2). Depletion of BRCA2 stimulated IHR at nicks to reach frequencies of 0.04% (Fig. 3D) and depletion of POLQ stimulated IHR at DSBs to reach frequencies of 2.0% (Fig. 3C), suggesting unexpected roles for BRCA2 and POLQ in protecting cells from IHR and associated cnLOH.
Our experiments assayed IHR at compound heterozygous mutations in exons 1 and 17 of the endogenous CD44 gene of HT1080 cells, a p53+ diploid human cell line derived from a fibrosarcoma. IHR was initiated at intronic sites from 0.3 to 60.3 kb downstream of the CD44 gene TSS, in experiments that used 12 different gRNAs to target breaks at both aligned and offset sites. IHR required a nick or DSB on each homolog. IHR was stimulated by breaks at sites that were aligned or offset by 5 bp and 0.6 and 3.5 kb (Fig. 2F). IHR frequencies were largely independent of the cut site within CD44 or the distance between cut sites on the two homologs (Fig. 2).
IHR has previously been analyzed at DSBs targeted to a single homolog in mouse embryonic stem cells or human B-lymphoblastoid cell lines, generally using assays that reported on both short gene conversion events and cross-overs. In those experiments, maximum frequencies documented were <0.01% and most events were not cross-overs (15, 17, 18, 20). The higher frequencies documented here probably result from targeting cleavage to both homologs.
Reciprocal EJ at DSBs May Compete with c-NHEJ and alt-EJ.
Several lines of evidence argue that IHR at DSBs proceeds predominantly via a reciprocal EJ pathway that joins DNA ends in trans, as diagrammed in Fig. 5D. Clear evidence of reciprocal exchange was provided by amplicon sequencing of junctions generated during IHR at offset DSBs, which identified a 70-fold enrichment of diagnostic indels (P < 10−100; Fig. 5C). The reciprocal EJ pathway that supports IHR is not only distinct from both c-NHEJ and alt-EJ, it is actively suppressed by both pathways. IHR was stimulated by depletion or inhibition of DNA-PK (Fig. 3B and SI Appendix, Fig. S4B) and by depletion of POLQ (Fig. 3C and SI Appendix, Fig. S4C). Frequencies of IHR at DSBs were unaffected by depletion of BRCA2 (Fig. 3A), indicating that canonical HR is unlikely to support this pathway.
Cell cycle may regulate repair pathway choice (33). cnLOH is associated with IHR that occurs in S/G2 phases of the cell cycle (Fig. 1A). POLQ depletion caused a fivefold increase in IHR at DSBs and a sixfold increase in cnLOH. The dramatic increase in cnLOH caused by POLQ depletion suggests that DSBs that would be repaired in cis in the presence of POLQ may persist in its absence, necessitating their repair late in the cell cycle. One mechanism by which POLQ may inhibit IHR and cnLOH is by promoting end joining that limits the availability of DNA ends that are potential substrates for reciprocal EJ. POLQ may do so indirectly by regulating alt-EJ or more directly by carrying out repair synthesis, as suggested by the reduced length of junctional insertions in cells depleted for POLQ (Fig. 5C and SI Appendix, Figs. S6–S8 and S10). The evidence that depletion or inhibition of DNA-PK can stimulate reciprocal EJ suggests that the c-NHEJ and reciprocal EJ pathways compete to repair DSBs. Reciprocal EJ may also compete with alt-EJ and, as a regulator of alt-EJ, POLQ may similarly function indirectly to tip the balance toward alt-EJ and away from reciprocal EJ.
The ability of POLQ to promote EJ in cis (alt-EJ) while suppressing EJ in trans, documented here in the context of IHR, echoes another of its roles in maintaining genomic stability. In activated B cells, the immunoglobulin heavy-chain locus undergoes class-switch recombination, a regulated process of recombination in which DNA breaks on a single chromosome undergo end joining that results in deletion of an intervening sequence. POLQ promotes intrachromosomal joining of ends generated during class-switch recombination, and conversely suppresses interchromosomal joining of DSBs that results in Myc/IgH translocations (24). The role of POLQ in other translocations is less clear (25, 26).
BRCA2 and POLQ Inhibit IHR at Nicks.
Approximately half of all IHR events at nicks were associated with cnLOH (Fig. 1D), raising the possibility that most or even all of these events occurred in G2 phase of the cell cycle (Fig. 1A). The DNA ends that are substrates for IHR may be generated during S phase, if replication at a nick results in a “one-ended” DSB that persists into G2 phase. BRCA2 or POLQ may limit IHR at nicks (Fig. 3 D and E) by maintaining replication fork integrity (29, 36–39).
Alternatively, the indels that occur at nicks at low but detectable frequencies (40–43) suggest that nicks may be subject to unwinding and nucleolytic resection that can create substrates for IHR. In this context, BRCA2 may inhibit the processing of unwound nicks, as suggested by its suppression of alt-HDR and mutagenic end joining (34, 35, 44), and BRCA2 may similarly protect nicks from IHR. POLQ, in contrast, may actively prevent nicks from becoming substrates for IHR by filling gaps formed upon unwinding or nucleolytic resection, analogous to a function ascribed to POLQ in base-excision repair (45).
IHR requires nicks targeted to both homologs (Fig. 2), as does a pathway that supports homologous recombination between a nicked target and a nicked dsDNA plasmid donor (34, 43, 46, 47). The use of a plasmid donor by that pathway confers length limitations, consistent with recombination via gene conversion or double cross-over. In contrast, half of the sCD44+ cells generated by IHR at nicks exhibit homozygosity from the cut site to telomeric markers (Fig. 1D). This suggests that EJ occurs at nicks via a pathway similar to the one active at DSBs.
Suppression of IHR and cnLOH Is Key to Maintaining Genomic Stability.
IHR and cnLOH are potential sources of genomic instability. The evidence that BRCA2, DNA-PK, and POLQ can all suppress IHR suggests that increased cnLOH frequencies documented in many kinds of tumors (1, 48, 49) may in some cases correlate with increased IHR frequencies resulting from dysregulation of one of these pathways. Nicks and DSBs caused by ordinary nuclear transactions may provide endogenous points of entry to these pathways. Nicks are the most common form of DNA damage, as they are generated as a consequence of inevitable damage from oxidation and ionizing radiation and in the course of DNA repair, and the frequency with which nicks occur suggests that they will sometimes be in proximity on homologous chromosomes. DSBs occur less frequently than nicks, but they may be in proximity on homologs at common fragile sites (50), transcription start sites of highly active genes (51), and programmed TOP2-dependent DSBs that mediate hormone-dependent stimulation of gene expression (52).
Gene Correction by IHR.
The same pathways that support IHR and cnLOH at targeted nicks and DSBs may potentially be harnessed for therapeutic correction of compound heterozygous and autosomal dominant mutations that contribute to genetic disease. The ability of IHR to support spontaneous correction of mutations that ameliorates more than 10 genetic diseases (2, 3, 14) provides physiological “proof of principle” for the efficacy of donor-free gene correction in vivo. Recent evidence that, in mice, adeno-associated virus-delivered CRISPR-Cas9 can ameliorate liver disease by correcting compound heterozygous mutations in the FAH and IDUA genes (53) provides experimental support for this strategy. Gene correction by IHR is donor-free, offering a clear advantage as it eliminates issues about donor design, delivery, and off-target insertions. Success will depend upon the frequency of IHR and on the selective advantage of corrected cells. The fundamental mechanistic understanding reported here will drive future optimization and application.
Materials and Methods
Cells and Cell Culture.
HT1080 fibrosarcoma cells (30) were used as a model. These cells express functional p53; propagate with a stable near-diploid karyotype, which avoids complications due to polyploidy; and have a high cloning efficiency, which facilitates molecular analysis of individual recombinants. Cells were cultured in DMEM/FBS (Dulbecco’s modified Eagle’s medium [HyClone] supplemented with 10% fetal bovine serum [Gemini Bio-Products]) and 200 U/mL penicillin, 200 mg/mL streptomycin (HyClone), and 2 mM l-glutamine (HyClone) at 37 °C, 5% CO2. The starting population of HT1080 cells (ATCC) was confirmed to be near-diploid and sCD44+.
Engineering HT1080 Cells to Report on IHR.
Mutations were engineered at DSBs or nicks targeted by transfection with the indicated plasmid constructs expressing Cas9 or Cas9D10A and gRNA, as previously described (34, 35, 44). Cells were cultured 4 to 7 d and then single cells were sorted into 96-well plates containing 100 µL conditioned medium and expanded for further analysis. Flow cytometry and genotyping were carried out as described below.
The HT1080-K1 IHR reporter line, bearing mutations in exons 1 and 17, was engineered in three steps: 1) DSBs were targeted to exon 1 by gRNA-CD44-4, single sCD44- cells were sorted and expanded, and exon 1 was analyzed by PCR with primers CD44-F2 and R2 and Sanger sequencing. A clone (HT1080-G1-C11) in which CD44 exon 1 was mutated at both alleles was used for subsequent engineering. 2) The mutation in one allele of exon 1 was corrected by alt-HDR at a nick targeted to exon 1 using gRNA-CD44-D1-2 and supported by single-stranded deoxyoligonucleotide (SSO) donor CD44-1-TOP. Single sCD44+ cells were sorted and expanded, and exon 1 was analyzed by PCR with primers CD44-F2 and R2 and Sanger sequencing. A clone (HT1080-B7) heterozygous at exon 1 (CD44+/-) was used for subsequent engineering. 3) A deletion in CD44 exon 17 was engineered by targeting a nick to this exon with gRNA-CD44T-17-1 in cells provided with SSO donor CD44T-17eng1. Resulting alt-HDR replaced 38 bp of coding sequence with a 17-bp insertion that created a diagnostic HindIII site (5′-AAGCTT-3′; SI Appendix, Fig. S1A). Single sCD44- clones were sorted and expanded, and exon 17 was amplified by PCR with primers CD44-F4 and R4 and sequenced. Clone HT1080-K1 (sCD44-) was shown to bear compound heterozygous mutations at CD44 exons 1 and 17.
We confirmed that alt-HDR at either exon 1 or exon 17 could restore expression of CD44 in HT1080-K1 cells (sCD44+) by using either gRNA-CD44-K1B and SSO donor CD44-1-TOP3 (exon 1) or gRNA-CD44T-17-2 and SSO donor CD44T-17-0 (exon 17) to correct those mutations. HT1080-K1 cells were used to determine IHR and cnLOH frequencies in the experiments shown in Figs. 1 C and D and 2B and SI Appendix, Fig. S2.
The HT1080-K2 derivative line, bearing SNPs in CD44 intron 1, was engineered to enable analysis of IHR by nicks or DSBs targeted to a single homolog or to offset sites on both homologs, and was used for analysis of IHR by offset and aligned DSBs in Figs. 2 D and F, 3, 4, and 5 and SI Appendix, Figs. S4–S8. It was generated from HT1080-K1 by alt-HDR at a nick targeted by gRNA W, using SSO repair donor CD44-intron 1-1 to create three single-base insertions. These insertions in intron 1 of homolog a created target sites for gRNAs Y and Z and prevented cleavage targeted by gRNA W or X (SI Appendix, Fig. S3B). Single cells were sorted and expanded; the target was amplified with PCR with primers CD44-F16 and CD44-R14; clones were screened for the new diagnostic KpnI site (5′-GGTACC-3′); and mutations were confirmed by Sanger sequencing.
Targeted DNA Cleavage.
Nicks or DSBs were targeted by transfection with Cas9D10A or Cas9 and gRNA expression plasmids (Figs. 1 and 2) or by transfection with Cas9D10A/gRNA or Cas9/gRNA ribonucleoproteins (RNPs; Figs. 3, 4, and 5 and SI Appendix, Figs. S4–S8). Typically, for plasmid transfections, on day 0, cells were seeded in a 24-well plate format at 5 × 104 cells per well in 0.5 mL DMEM/FBS. To transfect (day 1), each well received 100 µL of transfection mix consisting of 150 ng Cas9D10A or Cas9 expression plasmid, 75 ng gRNA expression plasmid (as a single plasmid or 37.5 ng each of two different plasmids), and 1.2 µL Lipofectamine LTX (Thermo Fisher Scientific), combined in 100 µL Opti-MEM (Thermo Fisher Scientific) reduced serum medium, and preincubated for 20 min at room temperature prior to addition to the cells. Cells were expanded into 6-well plates (day 2 or 3) and collected for analysis on day 4. The Cas9 expression construct (pCas9[wild type]-T2A-mTagBFP) was previously described (35). All gRNA expression plasmids were constructed by cloning annealed and extended oligos (IDT) into AflII-digested gRNA-cloning vector (54). The donor plasmid used for the canonical HR assay (SI Appendix, Fig. S4A) was constructed by cloning a 491-bp fragment of CD44 into pCR2.1-TOPO (Invitrogen) by TOPO TA cloning. The cloned PCR product was confirmed by Sanger sequencing with primers CD44-F2 and CD44-R2. DNA sequences of the protospacer and the protospacer adjacent motif (PAM) of all gRNAs are shown in SI Appendix, Table S1.
Cells were transfected with RNPs (CRISPR RNA, trans-activating CRISPR RNA, and Cas9D10A [IDT]; Cas9[wild type] [IDT or QB3 Berkeley]) using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the IDT protocol but at amounts corresponding to 25% of the recommended scaled amounts. Cells were plated at 1.5 × 105 cells per well in 24-well plates containing the transfection mix (day 1) and then expanded into 6-well plates (day 2 or 3) and collected for analysis on day 4.
Transfections were scaled to 48-well or 6-well plates or 6-cm plates without evident effect on IHR frequencies. Experiments routinely included triplicate transfections and were performed at least two times. Sample size (n) is indicated in the figure legends and represents a single measurement from each sample.
siRNAs and siRNA Depletion.
siRNAs used for depletion were the nontargeting siRNA siNT2 (Thermo Fisher Scientific; 4390847) and siRNAs targeting BRCA2 (Thermo Fisher Scientific; s2085; validated by the manufacturer), DNA-PKcs (Qiagen; SI02224229, SI02224236, SI02663633; all three validated by the manufacturer), and POLQ (Qiagen; SI02665215, SI00090083, SI00090076, SI00090069). When using multiple siRNAs, transfections were with equimolar pools. The effect on IHR of the pool of four siRNAs designed for POLQ depletion was confirmed by testing each siRNA individually (SI Appendix, Fig. S4B), suggesting that the effect is specific to POLQ and not a result of off-target effects.
To analyze effects of siRNA depletion on IHR frequencies (Figs. 3, 4, and 5 and SI Appendix, Figs. S4–S8), cells were seeded in 10-cm plates at 1.8 × 106 cells per plate in 11 mL DMEM/FBS and cultured for 1 to 2 h, and then 1.8 mL Opti-MEM containing 60 pmol siRNA and 22.5 µL Lipofectamine RNAiMAX (Thermo Fisher Scientific) were added to each plate; the next day, cells were transferred to smaller wells prior to transfection with Cas9D10A/gRNA or Cas9/gRNA RNPs. For 24-well plates, 1.5 × 105 siRNA-treated cells in 0.6 mL DMEM/FBS were added to wells containing 75 µL Opti-MEM, 4.5 pmol RNP, and 1.8 µL Lipofectamine RNAiMAX. Following 1 to 2 d of incubation, cells were expanded into 6-well plates. Four days after transfection, cells were collected for analysis.
Analytic Quantification of IHR Frequencies by Flow Cytometry.
For analytic quantification of sCD44+ cell frequencies by flow cytometry, cells (∼106) were washed once with phosphate-buffered saline (PBS), removed from plates in 200 µL TrypLE Select (Thermo Fisher Scientific), and mixed with an equal volume of 4% formaldehyde (2% final concentration). Fixed cells could be stored at 4 °C for up to a week. For staining, fixed cells were pelleted by centrifugation at 1,000 rpm for 5 min, the supernatant was aspirated, and cells were washed first with 1 mL ice-cold Dulbecco’s PBS (DPBS; HyClone) and then with 1 mL ice-cold DPBS containing 1% FBS (DPBSF), and then pelleted and resuspended in 300 µL of a 1:200 dilution of fluorescein isothiocyanate (FITC)-conjugated anti-CD44 antibody (clone G44-26; BD Biosciences) in DPBSF, gently mixed at 4 °C for 1 h, pelleted, and resuspended in 100 to 300 µL DPBSF for flow cytometry analysis. For flow cytometry, cells were gated for single cells and CD44-FITC was detected with a 488-nm laser on an LSRII flow cytometer (Becton Dickinson), as previously described (55). sCD44− control populations were used to gate expression of sCD44+. IHR frequencies are presented as the fraction of sCD44+ cells in a population. Statistical significance was determined in Excel by two-tailed t test.
Single-Cell Sorting, Cell Expansion, and DNA Preparation.
Single cells were sorted into 96-well plates at 7 to 10 d after transfection. Cells were washed and then removed from plates with TrypLE Select (1 mL per 10-cm plate), which was then inactivated by addition of five volumes of fresh DMEM/FBS, and cells were pelleted, washed, and stained as above. Cells were sorted using an Aria III cytometer (Becton Dickinson), deposited into 96-well plates containing 100 µL conditioned medium per well, cultured for 10 to 14 d, and then transferred to 24-well plates and cultured an additional 4 to 6 d. Medium was aspirated and cells were dissociated by adding 105 µL TrypLE Select per well. For DNA preparation, 35 µL of this cell suspension was added to 100 µL DMEM to inactivate the TrypLE Select, centrifuged, and washed once with DPBS, and genomic DNA was prepared using DirectPCR Lysis Reagent (Viagen Biotech).
RFLP Analysis of Homozygosity.
For RFLP assays of homozygosity, exon 1 of CD44 was amplified in a 30-µL reaction using the primer pair CD44-F2 and CD44-R2, Taq DNA polymerase, and ThermoPol buffer (NEB). Restriction digestion was carried out by adding 12 µL PCR product to 3 µL 1× ThermoPol buffer containing 1.75 U (0.35 µL) Tth111I (NEB; cleaves 5′-GACN↓NNGTC-3′) and then incubating at 65 °C for 90 min. Undigested and digested DNAs were resolved by 1.3% agarose gel electrophoresis. Similar analyses were performed at exon 17 (CD44-F4 and R4, HindIII or ApoI), rs85074 (rs85074-F and R, BstBI), and rs7950932 (rs7950932-F and R, BsaHI). Primer sequences are shown in SI Appendix, Table S2. Data were tabulated and Fisher’s exact tests were performed in Prism 8 (GraphPad Software; Figs. 1D and 4B).
Amplicon-Sequencing Analyses of IHR and cnLOH.
For amplicon sequencing, cell populations were recovered for further analysis at 6 to 7 d post transfection. For control cell populations, ∼1 × 106 cells were used directly for genomic DNA preparation using a Qiagen Blood and Tissue Kit. Other samples were split such that ∼1 × 106 cells were used directly for genomic DNA preparation and 3 × 106 cells were stained as above and sorted by flow cytometry to recover sCD44+ cells. At least 5,000 sCD44+ cells per sample were sorted into conditioned medium and cultured for 7 to 10 d. Cells were confirmed to be sCD44+ by flow cytometry and ∼1 × 106 cells were used for genomic DNA preparation.
To add unique molecular identifiers (UMIs), Nextera targeting primers were used to amplify 150 ng of genomic DNA (>20,000 genomes) for three cycles of PCR (SI Appendix, Table S2; IDT). Reactions were purified with 0.7 volumes AMPure XP beads (Beckman Coulter) following the manufacturer’s protocol and then used as a template for 23 cycles of PCR with Nextera indexing primers. These reactions were purified, pooled, and run on a MiSeq System with the MiSeq Reagent Kit (Illumina) at the Fred Hutchinson Cancer Research Center.
Reads were first converted to unaligned SAM files using the FastqToSam command from PICARD tools. Then, the UMI sequences were extracted and converted to “ab/ba” format, where “a” and “b” are the tag sequences from read 1 and read 2 using an in-house script. Remaining reads were then assembled to BAM files, in which reads having the same UMI were assembled to consensus sequences using single-strand consensus sequence (SSCS) assembly in FASTQ format. Then, remaining reads were aligned with the reference sequences and analyzed using two established methods: (1) SSCS read 1 and read 2 were aligned by Burrows–Wheeler Aligner MEM and realigned around INDELs by GATK. Variants were called using Samtools mpileup. INDELs were normalized using bcftools. Output of variant frequencies (single-nucleotide variants [SNVs], insertions, and deletions) at each position was generated by a Python program derived from ref. 56. (2) SSCS reads were analyzed using CRISPResso2 (57). VCF files were analyzed in Excel. Indel average size and frequency were calculated based only on those indels within a window 40 bp on either side of the DSB site, except when both alleles were analyzed cumulatively, in which case the window spanned positions 111 to 165 of both alleles. All frequencies were calculated as a fraction of the highest read depth in that sample for the allele in question or, when calculating cumulative frequencies, the sum of the highest read depths for each allele. Allele ratios were tabulated and Fisher’s exact tests were performed in Prism 8 (Fig. 4C).
For amplicons encompassing the DSB sites (Fig. 5C and SI Appendix, Figs. S5–S10), the total number of mapped, UMI collapsed reads was at least 65,000. This coverage was sufficient to identify thousands of unique indels and SNVs in unsorted samples and to thoroughly sample the unique events in sorted samples, which were bottlenecked when sorting 5,000 to 10,000 cells. For the amplicons used for allele ratio determinations (Fig. 4C), the total number of mapped, UMI collapsed reads was at least 22,000, with one exception (gRNA-NC, siNT2, i1-3; 8,535 reads). Deviations from 1:1 allele frequencies were easily observed at this level of coverage, though the i1-3 sample was an outlier among the control samples, possibly due to sampling error caused by the low read count.
CRISPResso2 (57) was used to view alignments in order to confirm that the most frequent indels, as seen in the VCF files, were not primarily a part of more complex sequence alterations. For these analyses, the fastq output from GATK of the UMI collapsed and trimmed read 1 and read 2 were used as input for the CRISPResso2 web interface (https://crispresso.pinellolab.partners.org/submission).
Supplementary Material
Acknowledgments
This work was supported by US NIH Grants R01 CA183967 and R21 CA190675. We thank our colleagues for their interest and support.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. R.D.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008073117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Gronseth C. M.et al., Prognostic significance of acquired copy-neutral loss of heterozygosity in acute myeloid leukemia. Cancer 121, 2900–2908 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Hirschhorn R., In vivo reversion to normal of inherited mutations in humans. J. Med. Genet. 40, 721–728 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim Y. H., Fisher J. M., Choate K. A., Revertant mosaicism in genodermatoses. Cell. Mol. Life Sci. 74, 2229–2238 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Revy P., Kannengiesser C., Fischer A., Somatic genetic rescue in Mendelian haematopoietic diseases. Nat. Rev. Genet. 20, 582–598 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Zhang Q., Jing H., Su H. C., Recent advances in DOCK8 immunodeficiency syndrome. J. Clin. Immunol. 36, 441–449 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis N. A.et al., Somatic intragenic recombination within the mutated locus BLM can correct the high sister-chromatid exchange phenotype of Bloom syndrome cells. Am. J. Hum. Genet. 57, 1019–1027 (1995). [PMC free article] [PubMed] [Google Scholar]
- 7.Gross M.et al., Reverse mosaicism in Fanconi anemia: Natural gene therapy via molecular self-correction. Cytogenet. Genome Res. 98, 126–135 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Jongmans M. C.et al., Revertant somatic mosaicism by mitotic recombination in dyskeratosis congenita. Am. J. Hum. Genet. 90, 426–433 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garelli E.et al., Spontaneous remission in a Diamond-Blackfan anaemia patient due to a revertant uniparental disomy ablating a de novo RPS19 mutation. Br. J. Haematol. 185, 994–998 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Chen D. H.et al., Ataxia-pancytopenia syndrome is caused by missense mutations in SAMD9L. Am. J. Hum. Genet. 98, 1146–1158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choate K. A.et al., Mitotic recombination in patients with ichthyosis causes reversion of dominant mutations in KRT10. Science 330, 94–97 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki S.et al., Somatic recombination underlies frequent revertant mosaicism in loricrin keratoderma. Life Sci. Alliance 2, e201800284 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiritsi D.et al., Mechanisms of natural gene therapy in dystrophic epidermolysis bullosa. J. Invest. Dermatol. 134, 2097–2104 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Kane M. S.et al.; NISC Comparative Sequencing Program , Mitotic intragenic recombination: A mechanism of survival for several congenital disorders of glycosylation. Am. J. Hum. Genet. 98, 339–346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moynahan M. E., Jasin M., Loss of heterozygosity induced by a chromosomal double-strand break. Proc. Natl. Acad. Sci. U.S.A. 94, 8988–8993 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quintana P. J., Neuwirth E. A., Grosovsky A. J., Interchromosomal gene conversion at an endogenous human cell locus. Genetics 158, 757–767 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stark J. M., Jasin M., Extensive loss of heterozygosity is suppressed during homologous repair of chromosomal breaks. Mol. Cell. Biol. 23, 733–743 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neuwirth E. A., Honma M., Grosovsky A. J., Interchromosomal crossover in human cells is associated with long gene conversion tracts. Mol. Cell. Biol. 27, 5261–5274 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moynahan M. E., Jasin M., Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 11, 196–207 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaRocque J. R.et al., Interhomolog recombination and loss of heterozygosity in wild-type and Bloom syndrome helicase (BLM)-deficient mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 108, 11971–11976 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Symington L. S., Rothstein R., Lisby M., Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics 198, 795–835 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin Y., Dominska M., Yim E., Petes T. D., High-resolution mapping of heteroduplex DNA formed during UV-induced and spontaneous mitotic recombination events in yeast. eLife 6, e28069 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hum Y. F., Jinks-Robertson S., DNA strand-exchange patterns associated with double-strand break-induced and spontaneous mitotic crossovers in Saccharomyces cerevisiae. PLoS Genet. 14, e1007302 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yousefzadeh M. J.et al., Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 10, e1004654 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mateos-Gomez P. A.et al., Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wyatt D. W.et al., Essential roles for polymerase θ-mediated end joining in the repair of chromosome breaks. Mol. Cell 63, 662–673 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito S., Maeda R., Adachi N., Dual loss of human POLQ and LIG4 abolishes random integration. Nat. Commun. 8, 16112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zelensky A. N., Schimmel J., Kool H., Kanaar R., Tijsterman M., Inactivation of Pol θ and C-NHEJ eliminates off-target integration of exogenous DNA. Nat. Commun. 8, 66 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brambati A., Barry R. M., Sfeir A., DNA polymerase theta (Polθ)—An error-prone polymerase necessary for genome stability. Curr. Opin. Genet. Dev. 60, 119–126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasheed S., Nelson-Rees W. A., Toth E. M., Arnstein P., Gardner M. B., Characterization of a newly derived human sarcoma cell line (HT-1080). Cancer 33, 1027–1033 (1974). [DOI] [PubMed] [Google Scholar]
- 31.Becerikli M.et al., Growth rate of late passage sarcoma cells is independent of epigenetic events but dependent on the amount of chromosomal aberrations. Exp. Cell Res. 319, 1724–1731 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Deriano L., Roth D. B., Modernizing the nonhomologous end-joining repertoire: Alternative and classical NHEJ share the stage. Annu. Rev. Genet. 47, 433–455 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Ceccaldi R., Rondinelli B., D’Andrea A. D., Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 26, 52–64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis L., Maizels N., Homology-directed repair of DNA nicks via pathways distinct from canonical double-strand break repair. Proc. Natl. Acad. Sci. U.S.A. 111, E924–E932 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis L., Maizels N., Two distinct pathways support gene correction by single-stranded donors at DNA nicks. Cell Rep. 17, 1872–1881 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ceccaldi R.et al., Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 518, 258–262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z.et al., DNA polymerase θ (POLQ) is important for repair of DNA double-strand breaks caused by fork collapse. J. Biol. Chem. 294, 3909–3919 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng W., Jasin M., BRCA2 suppresses replication stress-induced mitotic and G1 abnormalities through homologous recombination. Nat. Commun. 8, 525 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng L.et al., Mitotic CDK promotes replisome disassembly, fork breakage, and complex DNA rearrangements. Mol. Cell 73, 915–929.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ran F. A.et al., Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho S. W.et al., Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 24, 132–141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen B.et al., Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods 11, 399–402 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Hyodo T.et al., Tandem paired nicking promotes precise genome editing with scarce interference by p53. Cell Rep. 30, 1195–1207.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Maizels N., Davis L., Initiation of homologous recombination at DNA nicks. Nucleic Acids Res. 46, 6962–6973 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshimura M.et al., Vertebrate POLQ and POLbeta cooperate in base excision repair of oxidative DNA damage. Mol. Cell 24, 115–125 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen X.et al., In trans paired nicking triggers seamless genome editing without double-stranded DNA cutting. Nat. Commun. 8, 657 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakajima K.et al., Precise and efficient nucleotide substitution near genomic nick via noncanonical homology-directed repair. Genome Res. 28, 223–230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stirewalt D. L., Pogosova-Agadjanyan E. L., Tsuchiya K., Joaquin J., Meshinchi S., Copy-neutral loss of heterozygosity is prevalent and a late event in the pathogenesis of FLT3/ITD AML. Blood Cancer J. 4, e208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sinclair P. B.et al., SH2B3 inactivation through CN-LOH 12q is uniquely associated with B-cell precursor ALL with iAMP21 or other chromosome 21 gain. Leukemia 33, 1881–1894 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ozeri-Galai E., Bester A. C., Kerem B., The complex basis underlying common fragile site instability in cancer. Trends Genet. 28, 295–302 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Marnef A., Cohen S., Legube G., Transcription-coupled DNA double-strand break repair: Active genes need special care. J. Mol. Biol. 429, 1277–1288 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Puc J., Aggarwal A. K., Rosenfeld M. G., Physiological functions of programmed DNA breaks in signal-induced transcription. Nat. Rev. Mol. Cell Biol. 18, 471–476 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang D.et al., Cas9-mediated allelic exchange repairs compound heterozygous recessive mutations in mice. Nat. Biotechnol. 36, 839–842 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mali P.et al., CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–838 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis L., Zhang Y., Maizels N., Assaying repair at DNA nicks. Methods Enzymol. 601, 71–89 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kennedy S. R.et al., Detecting ultralow-frequency mutations by duplex sequencing. Nat. Protoc. 9, 2586–2606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clement K.et al., CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.