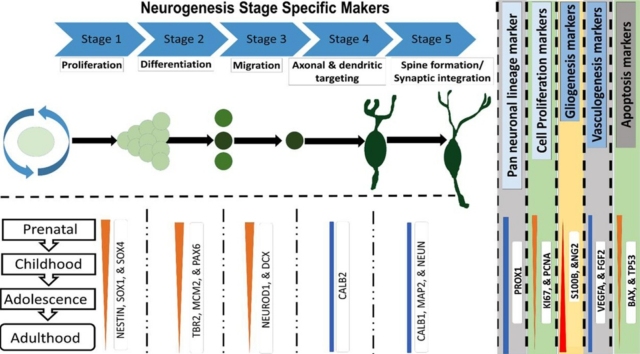
Graphical abstract
Keywords: Adult human neurogenesis, Developmental stages, Hippocampus, Signature genes, Transcriptome
Highlights
-
•
NESTIN, SOX1, and SOX4 decreased progressively from prenatal to adult age.
-
•
KI67 and TBR2 reached zero expression level at adolescence.
-
•
NEUROD1, DCX, PSA NCAM remained unchanged post-childhood.
-
•
VEGF and FGF2 did not change significantly from prenatal to adult age.
-
•
BAX and TP53 decreased progressively from prenatal to adult age.
Abstract
Purpose
Immunohistological investigations have given rise to divergent perspectives about adult hippocampal neurogenesis in humans. Therefore, this study aimed to examine whether a comprehensive transcriptomic analysis of signature markers of neurogenesis, supplemented with markers of gliogenesis, vasculogenesis, cell proliferation, and apoptosis, may help discern essential aspects of adult hippocampal neurogenesis in humans.
Materials and Methods
RNA expression data for salient marker genes of neurogenesis, gliogenesis, vasculogenesis, and apoptosis in post-mortem human hippocampal tissue [from prenatal (n = 15), child (n = 5), adolescent (n = 4), and adult (n = 6) brains] were downloaded from the Allen Human Brain Atlas database (http://www.brainspan.org/rnaseq/search/index.html). Gene expression data was categorized, median values were computed, and age group-specific differential expression was subjected to statistical analysis (significance level, α = 0.01).
Results
With the exception of the genes encoding GFAP, BLBP, SOX2, and PSA-NCAM (unchanged), and the post-mitotic late maturation markers CALB1, CALB2, MAP2, and NEUN as well as the pan-neuronal marker PROX1 which were persistently expressed throughout, expression of all other genes associated with neurogenesis was steeply and progressively downregulated between perinatal life and adulthood. Interestingly, expression of the classical proliferation marker KI67 and a progenitor cell marker TBR2 were found to have reached baseline expression levels (zero expression score) at adolescence while the expression of immature neuronal, post-mitotic early and late maturation markers remained at a constant level after childhood. In contrast, markers of gliogenesis (other than PDGFRA and Vimentin) were significantly upregulated between prenatal life and childhood. Expression of the vasculogenesis markers VEGFA and FGF2 did not differ across any of the age groups studied, whereas the expression of apoptotic markers was progressively decreased after prenatal life.
Conclusions
Our findings indicate that the progression of neurogenesis from progenitor cells is highly restricted in the human brain from childhood onwards. An alternative possibility that limited neurogenesis may be continued in adolescents and adults from a developmentally arrested pool of immature neurons needs to be examined further through experimental studies.
1. Introduction
In adult humans and other mammals, the subgranular zone (SGZ) of the dentate gyrus (DG) region of the hippocampus is a primary site of neurogenic activity (Ming and Song, 2011). Newly formed neurons from the SGZ migrate to the granular layer of DG and integrate into the existing cortical neuronal circuitries (Diana et al., 2014; Luna et al., 2019). Induced adult hippocampal neurogenesis (AHN) has been shown to improve spatial learning and memory in transgenic mouse models (Sahay et al., 2011) and to protect against neuropsychiatric disorders associated with a decline of these cognitive functions (Braun and Jessberger, 2014; Sahay et al., 2011). Apart from a few exceptions (Amrein et al., 2007; Patzke et al., 2015), neurogenesis in the hippocampus occurs over a protracted period in most mammals. Drawing upon published data, a recent analysis of hippocampal neurogenesis across the life span in commonly studied mammals, including humans, revealed a gradual decline in the rate of neuronal birth from prenatal life to adulthood (Snyder, 2019). Importantly, newborn cells in the hippocampus were found to retain unique plastic properties for long intervals and to have the potential to exert distinct functions depending on the neurodevelopmental stage at which they appeared. Snyder (2019) concluded that the continued formation of new neurons might be essential for the formation of new memories and to allow adaptive flexibility to new experiences (Snyder, 2019).
While the phenomenon of adult hippocampal neurogenesis (AHN) may be considered an accepted dogma (Andreae, 2018; Bergmann et al., 2015; Spalding et al., 2013), its extent has been challenged previously (Rakic, 1985) and more recently (Andreae, 2018; Kumar A et al., 2019). The results of recent studies in humans are disparate, with reports of complete absence of AHN in individuals aged 18+ (Cipriani et al., 2018; Dennis et al., 2016; Sorrells et al., 2018) as well as reports that AHN continues into the ninth and tenth decades of life (Boldrini et al., 2018; Moreno-Jiménez et al., 2019; Tobin et al., 2019). While some of the discrepant findings may be attributed to differences in methodology, or technical confounds (Kempermann et al., 2018), the work by Boldrini et al. (2018) stands out because it involved multiple analytical approaches (stereological cell counting, complemented by measures of vasculogenesis) and examined only samples from subjects without potentially confounding conditions before death (Kempermann et al., 2018). While these authors found persistent vasculogenesis in the neurogenic niche, they did not detect newborn neurons in the adult human hippocampus (Kempermann et al., 2018). In contrast, Moreno-Jiménez et al. (2019) who employed standardized tissue fixation, a supposed confounding factor (Kempermann et al., 2018), and introduced a unique pretreatment of brain tissue, identified thousands of immature neurons exhibiting variable degrees of maturation within the DG of neurologically healthy aged human subjects. Interestingly, in contrast to Tobin et al. (2019) who observed lifelong AHN, even in patients with mild cognitive impairments and Alzheimer’s disease, Moreno-Jiménez et al. (2019) noted that neurogenesis occurs at a drastically reduced rate in patients with Alzheimer’s disease and that overall higher AHN is associated with better cognitive status. On the other hand, in an immunohistological study, Seki et al. (2019) observed a paucity of Ki67 and doublecortin (DCX) expressing cells in the post-mortem neurogenic niche of the DG of healthy adults, despite the presence of a substantial population of PSA-NCAM expressing immature neurons. Another study that deserves mention is that of Sorrells et al. (2019), who studied neurogenesis in the para-laminar (PL) nuclei of the amygdala (involved in fear and anxiety) of adolescent and adult humans. Using a sophisticated combination of morphological, transcriptomic and ultrastructural methods, these authors found negligible evidence of immature neurons (DCX+ PSA-NCAM+) or mature excitatory (TBR1+VGLUT2+) neurons derived from dividing precursor cells in the adolescent and adult amygdala; these findings led them to suggest that, rather than the formation of new neurons per se, a developmentally arrested pool of immature neurons may give rise to late-maturing neurons during postnatal life.
Neurogenesis is a multi-step process, each step recognizable by the expression of lineage-specific protein markers (von Bohlen und Halbach, 2007). These markers have been mostly studied in varying combinations using immunohistological methods within confined age groups. However, advances in spatio-temporal transcriptomic analysis now allow assembly of differential expression patterns of neurogenesis signature markers, including information on the expression of markers of gliogenesis, vasculogenesis, cell proliferation, and apoptosis markers, allowing the development of a more comprehensive picture, especially with respect to the question of the temporal extent of AHN. In this work, we performed an in silico analysis of the developmental transcriptome (from prenatal life through to adulthood) of the human hippocampus in an attempt to help resolve the controversy surrounding the persistence of AHN in humans.
2. Materials and methods
RNA expression data for neurogenesis signature genes in post-mortem human hippocampal tissue of the prenatal (n = 15), childhood (from birth up to 3 years age, n = 5), adolescent (11–19 years, n = 4), and adulthood (20–40 years, n = 6) were downloaded from the development transcriptome database of the Allen Brain Atlas (http://www.brainspan.org/rnaseq/search/index.html).
2.1. Acquisition of data (from Allen Brain Atlas)
As per data source, fresh post-mortem brain (only specimens from neurologically healthy individuals that were free from significant genetic errors) were considered for original data retrieval. In addition, to ensure consistency between the samples and to decrease potential variation arising from ante- and post-mortem conditions, specific tissue preservation, storage, and RNA extraction and analysis criteria were followed (all described in detail at https://help.brain-map.org/display/devhumanbrain/Documentation). Briefly, RNA quality was confirmed using Bioanalyzer RNA 6000 Nano Kit or Bioanalyzer RNA 6000 Pico Kit (Agilent), RNA sequencing was performed using an Illumina Genome Analyzer II (GAIIx) instrument, and Gencode (v10 and Gencode v3c annotations) was used for RNA sequencing alignment and expression quantification. The metadata for the subjects included in this study were age, gender, ethnicity, and post-mortem interval (PMI), pH, cerebral hemisphere used for the tissue biopsy, and RNA integration number (RIN); the quality of the data are given in Table S1. All work was performed according to guidelines for the research use of human brain tissue and with prior approval by the Human Investigation Committees and Institutional Ethics Committees of each institute from which samples were obtained.
2.2. In silico data analysis
We categorized the gene expression data according to age, and computed median expression values for the neurogenesis signature genes of interest, as well as genes implicated in gliogenesis (OLIG2, Vimentin (or VIM), S100B, PDGFRA, NG2), cell proliferation (KI67 and PCNA), vasculogenesis (VEGFA and FGF2), and apoptosis (BAX and TP53); the list of genes selected for analysis in this study are shown in Table 1. Differential gene expression, corresponding to the neurogenesis maturation stages across the age groups studied was statistically analyzed, using non-parametric statistical analyses: respectively, the Kruskal-Wallis (KW) test and Mann-Whitney U (MWU) test to examine whether significant differences existed between the four different age groups as well as between any two age groups (Tables S2 and S3). In all cases, the level of significance was set to α = 0.01. The p values reported in Table S3 are after false discovery rate adjustment (FDR). Box plots, showing medians, minimum and maximum bars are depicted, allowing inference of trends of gene expression in the different age groups.
Table 1.
Developmental stage-specific expression of immuno-histological protein markers in the neurogenic niche of adult hippocampus.
| Stage 1 (Neural stem cell markers) | Stage 2 (Progenitor intermediate cell forms markers) | Stage 3 (Migration/ Immature granule cell markers) | Stage 4 (Axonal & dendritic targeting/Early maturation markers) | Stage 5 (Synaptic integration/Late maturation markers) | |
|---|---|---|---|---|---|
| NESTIN, BLBP, GFAP, SOX1, 2 & 4 | TBR2#, MCM2, PAX6 | NEUROD1, DCX*, PSA NCAM | STMN2, SEMA3C, Calretinin**, (CALB2), TUBB3*** | MAP2, Calbindin (CALB1), NEUN****#, | |
| Pan neuronal lineage marker | PROX1 | ||||
| Cell proliferation markers | KI67#, PCNA | ||||
| Gliogenesis markers | OLIG2, Vimentin#, S100B, PDGFRA, NG2 | ||||
| Vasculogenesis markers | VEGFA, FGF2 | ||||
| Apoptosis markers | BAX, TP53 | ||||
*Pan neuronal lineage marker (expressed in Stage 2–5), *Also expressed in Stage 4, **Also expressed in Stage 5, ***A postmitotic neuronal lineage marker, also expressed in stage 3 and 5, ****\Also expressed in stage 4.
#Alternative gene names:TBR2 or EOMES, KI67 or MKI67, NEUN or RBFOX3, Vimentin or VIM.
3. Results
For analyzing the developmental stage-specific expression of the neurogenesis signature, studied genes were cataegorized as Stages 1–5, based on their known chronological appearance and maximum expression (Kempermann et al., 2018; Kumar et al., 2019; von Bohlen und Halbach, 2007). Some genes which express across more than one stage are marked with a star. Additional categories were made for pan-neuronal lineage, cell proliferation, gliogenesis, vasculogenesis, and apoptosis markers (Table 1).
3.1. Neurogenesis signature markers (Stage 1–5)
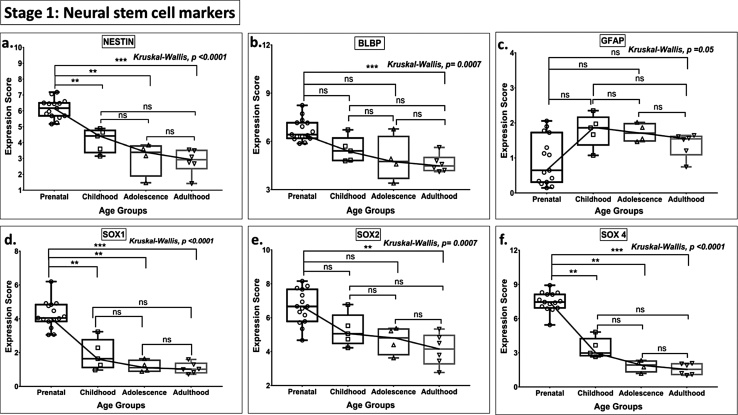
A steep downregulation of expression of neural stem cell (NSC), intermediate neural progenitor (INP), immature granule cell, and post-mitotic early and late maturation cell markers was noted between prenatal life and childhood. However, the expression of GFAP, BLBP, SOX2, PSA NCAM, NEUROD1, CALB1, CALB2, STMN2, MAP2, NEUN, and PROX1 did not differ between these two age groups (Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5). None of the stage 1–5 markers were significantly altered in postnatal age groups.
Fig. 1.
Box plot presentation (median with minimum and maximum bars showing all data points) of Neurogenesis Stage 1 - Neural stem cell markers (NESTIN, BLBP, GFAP, SOX1, SOX2, SOX4), gene expression scores in log2 of RPKM(reads per kilobase of exon model per million mapped reads).Statistical comparisons were made using the Kruskal-Wallis test followed by a post hoc Mann Whitney U test and FDR correction. Adjusted p-values for MWU comparisons represented by, **** ≤0.0001, ***≤0.001, **≤0.01, and ns for non-significant.
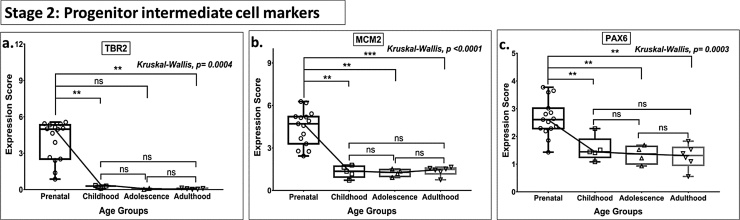
Fig. 2.
Box plot presentation (median with minimum and maximum bars showing all data points) of Neurogenesis Stage 2 - Progenitor intermediate cell markers (TBR2, MCM2, PAX6), gene expression scores in log2 of RPKM(reads per kilobase of exon model per million mapped reads). Statistical comparisons were made using the Kruskal-Wallis test followed by a post hoc Mann Whitney U test and FDR correction. Adjusted p-values for MWU comparisons represented by, **** ≤0.0001, ***≤0.001, **≤0.01, and ns for non-significant.
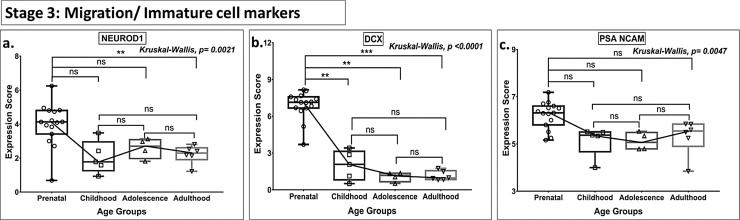
Fig. 3.
Box plot presentation (median with minimum and maximum bars showing all data points) of Neurogenesis Stage 3 - Migration/Immature cell markers (NEUROD1, DCX, PSA NCAM), gene expression scores in log2 of RPKM(reads per kilobase of exon model per million mapped reads). Statistical comparisons were made using the Kruskal-Wallis test followed by a post hoc Mann Whitney U test and FDR correction. Adjusted p-values for MWU comparisons represented by, **** ≤0.0001, ***≤0.001, **≤0.01, and ns for non-significant.
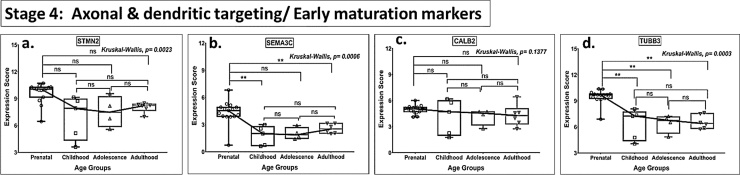
Fig. 4.
Box plot presentation (median with minimum and maximum bars showing all data points) of Neurogenesis Stage 4 - Axonal and dendritic targeting/Early maturation markers (STMN2, SEMA3C, CALB2, TUBB3), gene expression scores in log2 of RPKM(reads per kilobase of exon model per million mapped reads). Statistical comparisons were made using the Kruskal-Wallis test followed by a post hoc Mann Whitney U test and FDR correction. Adjusted p-values for MWU comparisons represented by, **** ≤0.0001, ***≤0.001, **≤0.01, and ns for non-significant.
Fig. 5.
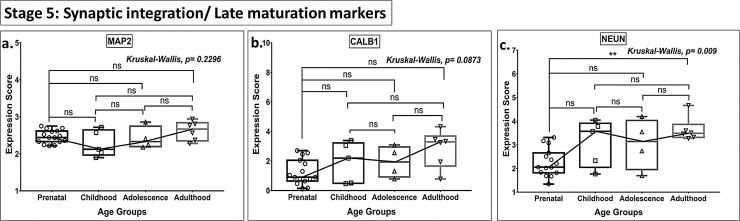
Box plot presentation (median with minimum and maximum bars showing all data points) of Neurogenesis Stage 5 - Synaptic integration/Late maturation markers (MAP2, CALB1, NEUN), gene expression scores in log2 of RPKM(reads per kilobase of exon model per million mapped reads). Statistical comparisons were made using the Kruskal-Wallis test followed by a post hoc Mann Whitney U test and FDR correction. Adjusted p-values for MWU comparisons represented by, **** ≤0.0001, ***≤0.001, **≤0.01, and ns for non-significant.
Markers of NSC (NESTIN, SOX1, SOX4 —Stage 1, Fig. 1), and INP cells (TBR2, MCM2, PAX6 — Stage 2, Fig. 2) declined progressively with age across all age groups (from prenatal to adult). As compared to all other stem cell marker genes, GFAP, BLBP, and SOX2 (Stage 1) showed no significant downregulation in postnatal expression (prenatal vs. adolescent groups). Uniquely, the marked postnatal downregulation of TBR2 continued with advancing age, reaching baseline (zero expression value) during adolescence, and remained undetectable in adult (Stage 2, Fig. 2.a).
There was no statistically significant age-related change in the expression of immature granule cell markers (NEUROD1, DCX, PSA NCAM—Stage 3, Fig. 3) and post-mitotic early maturation markers (STMN2, SEMA3C, CALB2, TUBB3, Stage 4, Fig. 4) beyond childhood. However, the expression of DCX, a marker of immature neurons, showed a gain in expression between childhood and adulthood group, when both of these age groups were compared with prenatal values (Fig. 3b).
The expression levels of post-mitotic late maturation markers (MAP2, CALB1, NEUN (Stage 5, Fig. 5)) did not differ significantly between the prenatal stage and any other ages analyzed, but interestingly, NEUN showed a gain in expression between adolescence and adulthood, when both of these age groups were compared to prenatal values (Fig. 5c).
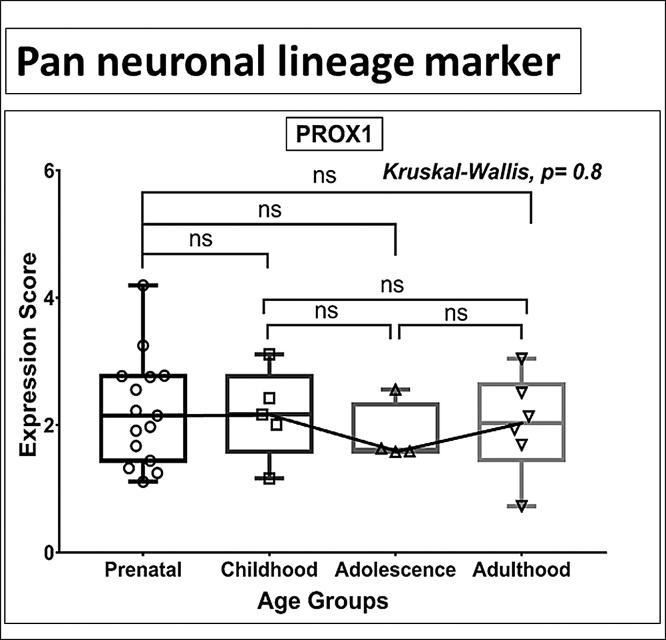
No significant differences were noted in the expression for GFAP (Stage 1), PSA-NCAM (Stage 3), PSA NCAM & CALB2 (Stage 4), and CALB1, MAP2 (Stage 5) and PROX 1 (Stage 2–5, a pan-neuronal marker) (Figs. 1c, 3 c, 4 c, 5 a-b, 6 , Table S2) in any of the age groups (all MWU comparisons).
Fig. 6.
Box plot presentation (median with minimum and maximum bars showing all data points) of the pan-neuronal lineage marker PROX1), gene expression score in log2 of RPKM(reads per kilobase of exon model per million mapped reads). Statistical comparisons were made using the Kruskal-Wallis test followed by a post hoc Mann Whitney U test and FDR correction. Adjusted p-values for MWU comparisons represented by, **** ≤0.0001, ***≤0.001, **≤0.01, and ns for non-significant.
3.2. Gliogenesis markers
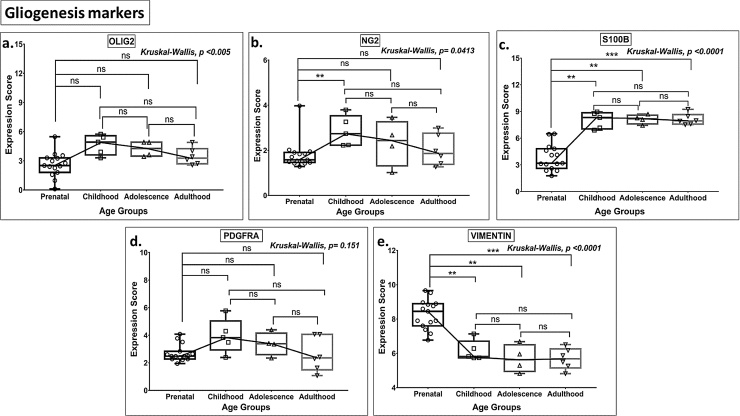
The expression of S100B and NG2 increased significantly during the prenatal to postnatal transition. PDGFRA and OLIG2 expression were unaltered although the expression of Vimentin was significantly reduced (Fig. 7, Tables S2 and S3), and none of the gliogenesis markers differed in expression levels (Fig. 7) during this period.
Fig. 7.
Box plot presentation (median with minimum and maximum bars showing all data points) of Gliogenesis markers (OLIG2, NG2, S100B, PDGFRA, Vimentin), gene expression scores in log2 of RPKM(reads per kilobase of exon model per million mapped reads). Statistical comparisons were made using the Kruskal-Wallis test followed by a post hoc Mann Whitney U test and FDR correction. Adjusted p-values for MWU comparisons represented by, **** ≤0.0001, ***≤0.001, **≤0.01, and ns for non-significant.
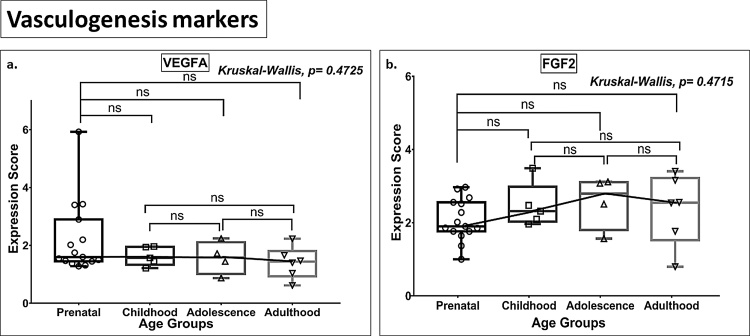
3.3. Vasculogenesis markers
The expression of the vasculogenesis markers VEGFA and FGF2 did not show significant age-dependent changes (Fig. 8a-b, Tables S2 and S3).
Fig. 8.
Box plot presentation (median with minimum and maximum bars showing all data points) of Vasculogenesis markers (VEGFA, FGF2), gene expression scores in log2 of RPKM(reads per kilobase of exon model per million mapped reads). Statistical comparisons were made using the Kruskal-Wallis test followed by a post hoc Mann Whitney U test and FDR correction. Adjusted p-values for MWU comparisons represented by, **** ≤0.0001, ***≤0.001, **≤0.01, and ns for non-significant.
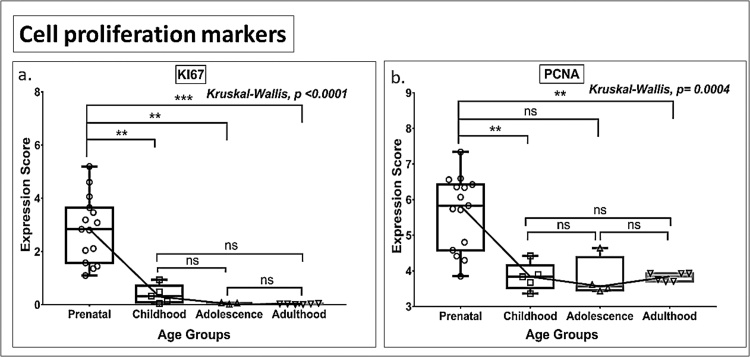
3.4. Cell proliferation markers
The cell proliferation markers KI67 and PCNA showed downward trends in expression across all age groups. KI67 expression reached the baseline (zero expression value) by adolescence. PCNA expression plateaued but remained above baseline from childhood onwards (Fig. 9a-b, Tables S2, and S3).
Fig. 9.
Box plot presentation (median with minimum and maximum bars showing all data points) of Cell proliferation markers (KI67, PCNA), gene expression scores in log2 of RPKM(reads per kilobase of exon model per million mapped reads). Statistical comparisons were made using the Kruskal-Wallis test followed by a post hoc Mann Whitney U test and FDR correction. Adjusted p-values for MWU comparisons represented by, **** ≤0.0001, ***≤0.001, **≤0.01, and ns for non-significant.
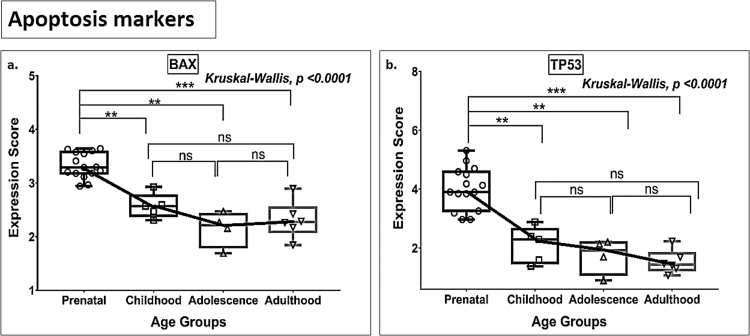
3.5. Apoptosis markers
BAX and TP53 showed significant downregulation in expression between prenatal life and childhood (Fig. 10a-b, Tables S2 and S3). Both markers displayed this downward trend, which continued through to adulthood, although no statistically significant differences were found between adjacent age groups (Fig. 10a-b, Tables S2 and S3).
Fig. 10.
Box plot presentation (median with minimum and maximum bars showing all data points) of Apoptosis markers (BAX, TP53), gene expression scores in log2 of RPKM(reads per kilobase of exon model per million mapped reads). Statistical comparisons were made using the Kruskal-Wallis test followed by a post hoc Mann Whitney U test and FDR correction. Adjusted p-values for MWU comparisons represented by, **** ≤0.0001, ***≤0.001, **≤0.01, and ns for non-significant.
4. Discussion
The study of the hippocampal developmental transcriptome provides insights into the events occurring in this brain area’s neurogenic niche. We here performed an in silico analysis of the hippocampal developmental transcriptome data made available by Allen Human Brain Atlas. Specifically, we focused on the chronologic (stage-specific) expression of signature genes involved in neurogenesis; we also examined key genes implicated in gliogenesis, vasculogenesis, cell proliferation, and apoptosis. The results of this analysis contribute to resolving the ambiguity regarding the temporal extent of AHN in humans.
Results shown in Table S2 and S3, and Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5 indicate age- and stage-related (from prenatal life through to adulthood) patterns in the expression of genes contributing to hippocampal neurogenesis. Teleologically, one would expect that, in case of continued neurogenesis, markers for all neurogenesis stages (Stage 1–5), cell proliferation, and vasculogenesis marker genes, will show significant expression across the age groups (prenatal to adult), conversely if AHN is a residual process, neurogenesis-associated genes would be expressed at increasingly lower levels along with an increase in expression of apoptotic marker genes with the advancement of age. However, our analysis revealed that whereas genes expressed by neural stem cells (NSC) and intermediate neural progenitors (INP) (Stages 1 and 2) are downregulated over time, there was no parallel reduction in the total number of immature granule cells (Stage 3) and post-mitotic forms (early mature and mature granule cells; Stages 4 and 5), i.e., the number of these cell types remain stable from prenatal/childhood to adult ages and may be interpreted to support the notion of persistent AHN. The continued expression of immature granule cell markers (Stage 3, Fig. 3) and post-mitotic early maturation markers (Stage 4, Fig. 4) during the post-childhood phase may be explained by either (a) sustained genesis of cells with a neuronal lineage that can differentiate into immature and early mature neurons, albeit to a limited extent as indicated by our data (Fig. 3-5), or (b) the operation of an as yet unidentified mechanism that serves to maintain a constant total number of maturing neurons (Sorrells et al., 2019; Seki et al., 2019). Here, the observation of detectable levels of the post-mitotic maturation marker genes NEUN, calbindin (CALB1) and MAP2 (Stage 5, Fig. 5), stable expression of the pan-neuronal lineage marker gene PROX1 across age groups (Fig. 6), as well as of TUBB3 (a neuronal lineage marker for post-mitotic Stages 3–5) (Fig. 4d) deserves mention as it adds support for the existence of lifelong neurogenesis (AHN). Unexpectedly, our analysis also showed that there is a continual downregulation of apoptosis marker genes such as BAX and TP53, albeit without detectable significant differences between adjacent age groups, indicating that cell death does not increase during aging of the human hippocampus; this finding is consistent with the data of Boldrini et al. (2018) showing that the number of mature neurons is maintained between the ages of 14 and 79 years. Together, these observations suggest that neurogenesis persists in the adult, a rate just sufficient to replenish small neuronal losses. They are further supported by the results of the present analysis which failed to find any significant age-related reductions in the expression of the key vasculogenesis genes VEGFA and FGF2 (Tables S2 and S3, Fig. 8); VEGFA and FGF2 were specifically examined in light of previous work that suggested parallel regulation of neurogenesis and vasculogenesis (Boldrini et al., 2012, 2018; Heine et al., 2005; Warner-Schmidt and Duman, 2007), allowing markers of vasculogenesis to serve as proxy markers of AHN.
On the other hand, the present analysis failed to find robust evidence for a linear progression of neurogenesis from NSC and INP cells. Specifically, key markers of NSC (NESTIN, SOX1, and SOX4), progenitors (TBR2), and cell proliferation (KI67) were observed to undergo steeper downregulation with age (Figs. 1,2,9) than that of immature neuron markers (DCX, PSA-NCAM, NEUROD1) (Fig. 3). This observation suggests a considerable mismatch between proliferation and differentiation/maturation of newly born neurons. Moreover, KI67 and TBR2 (but not PCNA and MCM2) were not being expressed (∼zero expression value) by the time of adolescence, indicating the depletion of progenitor cells by this age.
In contrast to the markers of cells in the neuronal lineage, expression patterns of the key gliogenesis markers, NG2 (oligodendrocytes), and S100B (post-mitotic astrocytes) were significantly upregulated from prenatal life to childhood, without further changes beyond the latter stage of development (Tables S2 and S3, Fig. 7a–c). These data indicate continued gliogenesis and are consistent with the above-discussed results for KI67 and other proliferation markers that point to a limited linear progression of progenitor-derived glial cells during post-childhood life.
Based on the transcriptomic patterns revealed in this in silco analysis, we conclude that a linear progression of neurogenesis (and gliogenesis) from progenitor cell pools is most likely restricted after childhood ages, and becoming absent from adolescence onwards.
The mismatch observed between proliferation (Stage 1–2) and differentiation/maturation of newly born neurons (Stage 3–5) in the results is consistent with an alternative explanation provided by Sorrells et al. (2019) and Seki et al. (2019) for the continued AHN, that newborn neurons in the adult hippocampus may be attributed to a developmentally-arrested pool of immature neurons, rather than to progenitor cells per se. In case of continued neurogenesis from developmentally protracted immature neurons, marker genes for pre-mitotic neurogenesis stages (Stage 1–2) will show a dip but those for post-mitotic stages (Stage 3–5) will keep showing significant expression with the advancement of age. However, even by this mechanism, the formation of new neurons has to be very limited, as is reflected from the expressions of immature neuronal markers in our study (Stage 3, Fig. 3). An alternative possibility that limited neurogenesis may be continued in adolescents and adults from a developmentally arrested pool of immature neurons needs to be examined further through experimental studies.
Limitations of this study are: (i) the transcriptomic analysis in the original data source was from the hippocampus (hippocampal anlage and hippocampal formation), but not the neurogenic niche of DG specifically, (ii) the dataset is unevenly distributed with relatively small sample sizes for some age groups, and (iii) many of the neurogenesis markers we studied are known to be expressed in more than one precursor cell subtype and across developmental stages, but the nature of the bulk transcriptomic analysis did not permit exact cell lineage tagging and stage-specific demarcation. The first limitation is unlikely to have affected the relative expression of the markers studied across age groups despite possible effects on their absolute expression values, and this limitation is therefore considered to have a minimal impact on the overall analysis. Our choice of non-parametric tests to compute differential expression of the genes was based on the need to overcome the limitations of homogenous distribution considering small sample sizes (second limitation).
Future studies to answer the question of whether AHN is a significant event will benefit from the combined use of immunohistological and single-cell RNA sequencing (scRNA-seq) techniques to assess lineage-specific expression of neurogenesis markers and to delineate their expression trajectories across developmental stages (Artegiani et al., 2017; Mu et al., 2019).
Author (s) contributions
AK conceived, and AK and VP designed the study. AK, VP analysed the data and wrote the first draft. AK, VP, MF, PK, CK, HS, SG revised the first draft. All authors have consented for the submission of the final draft.
Funding statement
None.
Ethics statement
Primary data used for this study were collected from the published databases of BrainSpan Atlas of the Developing Human Brain managed by Allen Brain Atlas (http://brainspan.org/), hence, an ethical clearance from Institutional Ethics Committee was precluded.
Conflict of Interest
None.
Acknowledgments
Data for this study was collected from the BrainSpan Atlas of the Developing Human Brain managed by Allen Brain Atlas. We are very thankful to Professor Osborn Almeida, Max Planck Institute of Psychiatry, Munich, Germany for language editing and providing valuable inputs for the preparation of this manuscript.
Footnotes
The findings of this study were first presented at the Annual Meeting of Society for Neuroscience (SFN) 2018, San Diego, USA.
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ibror.2020.08.003.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Amrein I., Dechmann D.K.N., Winter Y., Lipp H.-P. Absent or low rate of adult neurogenesis in the Hippocampus of bats (Chiroptera) PLoS One. 2007;2:e455. doi: 10.1371/journal.pone.0000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreae L.C. Adult neurogenesis in humans: dogma overturned, again and again? Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aat3893. [DOI] [Google Scholar]
- Artegiani B., Lyubimova A., Muraro M., van Es J.H., van Oudenaarden A., Clevers H. A single-cell RNA sequencing study reveals cellular and molecular dynamics of the hippocampal neurogenic niche. Cell Rep. 2017;21:v3271–3284. doi: 10.1016/j.celrep.2017.11.050. [DOI] [PubMed] [Google Scholar]
- Bergmann O., Spalding K.L., Frisén J. Adult neurogenesis in humans. Cold Spring Harb. Perspect. Biol. 2015;7 doi: 10.1101/cshperspect.a018994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldrini M., Hen R., Underwood M.D., Rosoklija G.B., Dwork A.J., Mann J.J., Arango V. Hippocampal angiogenesis and progenitor cell proliferation are increased with antidepressant use in major depression. Biol. Psychiatry. 2012;72:562–571. doi: 10.1016/j.biopsych.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldrini M., Fulmore C.A., Tartt A.N., Simeon L.R., Pavlova I., Poposka V., Rosoklija G.B., Stankov A., Arango V., Dwork A.J., Hen R., Mann J.J. Human hippocampal neurogenesis persists throughout aging. Cell Stem Cell. 2018;22:589–599.e5. doi: 10.1016/j.stem.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S.M.G., Jessberger S. Adult neurogenesis: mechanisms and functional significance. Development. 2014;141:1983–1986. doi: 10.1242/dev.104596. [DOI] [PubMed] [Google Scholar]
- Cipriani S., Ferrer I., Aronica E., Kovacs G.G., Verney C., Nardelli J., Khung S., Delezoide A.-L., Milenkovic I., Rasika S., Manivet P., Benifla J.-L., Deriot N., Gressens P., Adle-Biassette H. Hippocampal radial glial subtypes and their neurogenic potential in human fetuses and healthy and alzheimer’s disease adults. Cereb. Cortex. 2018;28:2458–2478. doi: 10.1093/cercor/bhy096. [DOI] [PubMed] [Google Scholar]
- Dennis C.V., Suh L.S., Rodriguez M.L., Kril J.J., Sutherland G.T. Human adult neurogenesis across the ages: an immunohistochemical study. Neuropathol. Appl. Neurobiol. 2016;42:621–638. doi: 10.1111/nan.12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diana X.Y., Marchetto M.C., Gage F.H. How to make a hippocampal dentate gyrus granule neuron. Development. 2014;141:2366–2375. doi: 10.1242/dev.096776. [DOI] [PubMed] [Google Scholar]
- Heine V.M., Zareno J., Maslam S., Joëls M., Lucassen P.J. Chronic stress in the adult dentate gyrus reduces cell proliferation near the vasculature and VEGF and Flk-1 protein expression. Eur. J. Neurosci. 2005;21:1304–1314. doi: 10.1111/j.1460-9568.2005.03951.x. [DOI] [PubMed] [Google Scholar]
- Kempermann G., Gage F.H., Aigner L., Song H., Curtis M.A., Thuret S., Kuhn H.G., Jessberger S., Frankland P.W., Cameron H.A., Gould E., Hen R., Abrous D.N., Toni N., Schinder A.F., Zhao X., Lucassen P.J., Frisén J. Human adult neurogenesis: evidence and remaining questions. Cell Stem Cell. 2018 doi: 10.1016/J.STEM.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Pareek V., Faiq M.A., Ghosh S.K.K.C. Adult neurogenesis in humans: basics, history, status quo, and relevance. Innov. Clin. Neurosci. 2019;16:30–37. [PMC free article] [PubMed] [Google Scholar]
- Luna V.M., Anacker C., Burghardt N.S., Khandaker H., Andreu V., Millette A., Leary P., Ravenelle R., Jimenez J.C., Mastrodonato A., Denny C.A. Adult-born hippocampal neurons bidirectionally modulate entorhinal inputs into the dentate gyrus. Science. 2019;364:578–583. doi: 10.1126/science.aat8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming G., Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/J.NEURON.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Jiménez E.P., Flor-García M., Terreros-Roncal J., Rábano A., Cafini F., Pallas-Bazarra N., Ávila J., Llorens-Martín M. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat. Med. 2019;25:554–560. doi: 10.1038/s41591-019-0375-9. [DOI] [PubMed] [Google Scholar]
- Mu Q., Chen Y., Wang J. Deciphering brain complexity using single-cell sequencing. GPB. 2019 doi: 10.1016/j.gpb.2018.07.007. pii: S1672-0229(19)30127-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzke N., Spocter M.A., Karlsson K.Æ., Bertelsen M.F., Haagensen M., Chawana R., Streicher S., Kaswera C., Gilissen E., Alagaili A.N., Mohammed O.B., Reep R.L., Bennett N.C., Siegel J.M., Ihunwo A.O., Manger P.R. In contrast to many other mammals, cetaceans have relatively small hippocampi that appear to lack adult neurogenesis. Brain Struct. Funct. 2015;220:361–383. doi: 10.1007/s00429-013-0660-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Limits of neurogenesis in primates. Science. 1985;227:1054–1056. doi: 10.1126/science.3975601. [DOI] [PubMed] [Google Scholar]
- Sahay A., Scobie K.N., Hill A.S., O’Carroll C.M., Kheirbek M.A., Burghardt N.S., Fenton A.A., Dranovsky A., Hen R. Increasing adult hippocampal neurogenesis is sufficient to improve pattern separation. Nature. 2011;472:466–470. doi: 10.1038/nature09817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki T., Hori T., Miyata H. Analysis of proliferating neuronal progenitors and immature neurons in the human hippocampus surgically removed from control and epileptic patients. Sci. Rep. 2019;18194 doi: 10.1038/s41598-019-54684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder J.S. Recalibrating the relevance of adult neurogenesis. Trends Neurosci. 2019;0 doi: 10.1016/j.tins.2018.12.001. [DOI] [PubMed] [Google Scholar]
- Sorrells S.F., Paredes M.F., Cebrian-Silla A., Sandoval K., Qi D., Kelley K.W., James D., Mayer S., Chang J., Auguste K.I., Chang E.F., Gutierrez A.J., Kriegstein A.R., Mathern G.W., Oldham M.C., Huang E.J., Garcia-Verdugo J.M., Yang Z., Alvarez-Buylla A. Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature. 2018;555:377–381. doi: 10.1038/nature25975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrells S.F., Paredes M.F., Velmeshev D., Herranz-Pérez V., Sandoval K., Mayer S. Immature excitatory neurons develop during adolescence in the human amygdala. Nat. Commun. 2019;10(1):2748. doi: 10.1038/s41467-019-10765-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding K.L., Bergmann O., Alkass K., Bernard S., Salehpour M., Huttner H.B., Boström E., Westerlund I., Vial C., Buchholz B.A., Possnert G., Mash D.C., Druid H., Frisén J. Dynamics of hippocampal neurogenesis in adult humans. Cell. 2013;153:1219–1227. doi: 10.1016/j.cell.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin M.K., Musaraca K., Disouky A., Shetti A., Bheri A., Honer W.G., Kim N., Dawe R.J., Bennett D.A., Arfanakis K., Lazarov O. Human hippocampal neurogenesis persists in aged adults and alzheimer’s disease patients. Cell Stem Cell. 2019 doi: 10.1016/J.STEM.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bohlen und. Halbach O. Immunohistological markers for staging neurogenesis in adult hippocampus. Cell Tissue Res. 2007;329:409–420. doi: 10.1007/s00441-007-0432-4. [DOI] [PubMed] [Google Scholar]
- Warner-Schmidt J.L., Duman R.S. VEGF is an essential mediator of the neurogenic and behavioral actions of antidepressants. Proc. Natl. Acad. Sci. U.S.A. 2007;104:4647–4652. doi: 10.1073/pnas.0610282104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.