Significance
Rational development of successful vaccines requires utilization of predictive models of vaccination. One approach for development of an HIV vaccine has been to study broadly neutralizing antibodies (bnAbs) and revert the mutations back to germline. However, there are limitations to such models. Therefore, we generated three knockin mice expressing B cell receptors (BCRs) from authentic naive VRC01-class B cells from healthy human donors (“HuGL” mice). This approach revealed that human VRC01-class naive B cell BCRs are indeed competent for antigen-specific responses in vivo. Additionally, a series of experiments shows the importance of precursor frequency and affinity on B cell responses to vaccine antigens. Overall, these HuGL mouse models validate a central tenet of the germline-targeting approach to vaccine design.
Keywords: vaccine, HIV, germline targeting, immunodominance, germinal center
Abstract
Animal models of human antigen-specific B cell receptors (BCRs) generally depend on “inferred germline” sequences, and thus their relationship to authentic naive human B cell BCR sequences and affinities is unclear. Here, BCR sequences from authentic naive human VRC01-class B cells from healthy human donors were selected for the generation of three BCR knockin mice. The BCRs span the physiological range of affinities found in humans, and use three different light chains (VK3-20, VK1-5, and VK1-33) found among subclasses of naive human VRC01-class B cells and HIV broadly neutralizing antibodies (bnAbs). The germline-targeting HIV immunogen eOD-GT8 60mer is currently in clinical trial as a candidate bnAb vaccine priming immunogen. To attempt to model human immune responses to the eOD-GT8 60mer, we tested each authentic naive human VRC01-class BCR mouse model under rare human physiological B cell precursor frequency conditions. B cells with high (HuGL18HL) or medium (HuGL17HL) affinity BCRs were primed, recruited to germinal centers, and they affinity matured, and formed memory B cells. Precursor frequency and affinity interdependently influenced responses. Taken together, these experiments utilizing authentic naive human VRC01-class BCRs validate a central tenet of germline-targeting vaccine design and extend the overall concept of the reverse vaccinology approach to vaccine development.
Vaccinology has largely been an empirical science over the past century. While this approach has produced many successful vaccines (1, 2), the lack of vaccines to pathogens like HIV has suffered from our lack of knowledge of the fundamental biology of how B cells compete in vivo following immunization (3). Nearly all vaccines are thought to work by induction of protective antibody responses (4). There is increasing optimism that effective vaccine strategies for HIV might be found. Recent efforts have focused on recurrent broadly neutralizing antibody (bnAb) specificities, such as the VRC01 class, that might be elicited by vaccines (5). The concept of reverse vaccinology (also known as [aka] reverse vaccinology 2.0) involves engineering proteins as immunogens based on knowledge of bnAbs, or protective mAbs more broadly (6, 7). Succinctly put, reverse vaccinology defines antibody-to-vaccine strategies of vaccine design. Germline-targeting vaccine design is a vaccine concept based on designing a priming immunogen specifically capable of binding B cell receptors (BCRs) of “germline” precursors of bnAbs, based on BCR sequence relationships, and thereby eliciting related B cell responses in vivo (8, 9). One apparent barrier to eliciting bnAbs is that the inferred germline (iGL) BCR precursors of most bnAbs have negligible affinity for HIV Env (5), and so are subdominant. BnAb precursors are also predicted to be quite rare (10). Competing B cells reactive to nonconserved epitopes tend to dominate the response (3) and fail to provide protection, as evidenced during natural infection by the ubiquity of strain-specific antibodies and the rarity of bnAbs (11).
In response to these challenges, germline-targeting immunogens have been engineered. First generation germline-targeting immunogens have been developed based on inferred unmutated ancestor, i.e., iGL, bnAb sequences (8, 9, 12–18). Development of candidate vaccines in that manner has two challenges that relate to BCR sequences and the human B cell repertoire. First, authentic gl-bnAb BCR sequences are almost always difficult to predict (15, 19–21). This is particularly true for CDR3 sequences, as these regions are not germline encoded and are stochastically generated during B cell development. Second, the protein engineering was frequently focused on binding to a single (or small set of) bnAb-precursor sequence match(es) that may not sufficiently relate to sequences normally found in the human B cell repertoire. Germline targeting requires the ability to target BCRs of precursors of the desired bnAb, specifically BCRs actually found in the human naive B cell repertoire. While knockin mouse models have provided substantial value in understanding the in vivo challenges that exist in maturing B cells toward broad neutralization (12, 13, 22–27), it is unclear if these models are capable of revealing all of the hurdles that authentic VRC01-class B cells will face with germline-encoded CDR3s. Second generation germline-targeting immunogens have incorporated broader sets of precursors of representative bnAbs and BCR sequences identified from next-generation sequencing (NGS) of HIV-negative donors predicted to be related to bnAb precursors (28, 29). VRC01-class germline-targeting immunogens have progressed furthest in development by these criteria. eOD-GT8 60mer is a germline-targeting HIV Env-engineered outer domain nanoparticle that has now entered human clinical trials (30).
VRC01-class bnAbs can neutralize upwards of 98% of HIV viral isolates. The VRC01 class is special in several respects (31, 32). Broad and potent, owing to its favorable angle of approach and because it mimics interactions of CD4 (engaging the CD4 binding site of HIV Env [CD4bs]), the VRC01 class uses a single VH gene, VH1-2, along with a critical short 5 amino acid (aa)-long l-CDR3 (typically QQYEF). To develop neutralizing breadth, VCR01-class Abs also require acquisition of a small l-CDR1 deletion or other accommodation to avoid a clash with the Env glycan at N276 (10, 19, 33). VRC01-class B cells undergo 19 to 40% amino acid somatic hypermutation (SHM) from germline to develop into bnAbs in HIV+ individuals (32–36). Germinal centers (GCs) are the anatomical site in which antigen-activated B cells undergo SHM, clonal competition, and Darwinian selection by T follicular helper (TFH) cells for survival (37–40). These rounds of mutation and selection produce mutated high-affinity antibodies in a process known as affinity maturation (41, 42). VRC01-class B cells will have to compete successfully in these processes in response to immunization if a germline-targeting VRC01-class bnAb vaccine is to succeed.
One way to rigorously assess the quality of a germline-targeting immunogen is to determine if it can identify bnAb precursor naive B cells in humans, via direct binding. To date, this has only been accomplished for two germline-targeting immunogen designs (22, 28, 29, 43, 44). We discovered that VRC01-class B cells (i.e., B cells that contain the VH1-2*02 [or VH1-2*04], heavy chain [HC] allele paired with a light chain [LC] with a 5 aa l-CDR3) do exist in the naive B cell repertoire of most humans, at a frequency of 1 in 300,000 B cells, and those B cells can be identified by their binding to eOD-GT8 (28, 43). VRC01-class eOD-GT8-bound human naive B cells could be grouped into subclasses, based on the LC V gene. eOD-GT8-binding VRC01-class naive human B cells of different subclasses are predicted to differ in their ability to develop into bnAbs, based on the ease with which they accommodate the HIV Env N276 glycan (43). VK1-5+ or VK1-33+ VRC01-class B cells can likely do so by point mutations within l-CDR1, whereas VK3-20+ VRC01-class B cells appear to require l-CDR1 deletions or longer H-CDR3s (19, 32, 45). Further investigation of the VRC01-class human naive B cell repertoire revealed the BCRs vary widely in affinity for eOD-GT8, H-CDR3 sequence, and H-CDR3 length (43). Importantly, it is not yet known whether subsets of these eOD-GT8-binding VRC01-class naive human B cells can respond in vivo, and it is not yet known whether they differ in their abilities to compete in vivo.
Mice do not possess a VH1-2 gene homolog, and thus cannot make VRC01-class Abs, necessitating the analysis in various types of knockin mice. eOD-GT8 60mer has shown promise as a priming immunogen in mice carrying an inferred germline BCR of the original VRC01 bnAb (VRC01gH or VRC01gHL) (13, 27, 44, 46), or the 3BNC60 bnAb HC (12), or in recombining HC knockin mice carrying the VH1-2 gene (22, 44). Given that VRC01-class precursors are rare in humans, we developed a VRC01gHL mouse B cell transfer strategy to better model human physiological precursor frequencies and affinities (27). Precursor frequency and antigen affinity were both shown to be important factors in determining B cell competitive success in GCs (27). The VRC01gHL model relied on an iGL bnAb sequence, which has limitations, both because the sequence was used in the original germline-targeting immunogen design process and because the l-CDR3 and H-CDR3 sequences were relatively unchanged from the VRC01 bnAb because these regions are not germline encoded.
We sought to develop animal models that directly reflect true naive human precursor B cells, with sequences that have not undergone germline reversion. To achieve this, we created three different BCR knockin H/L mouse models carrying authentic VRC01-class BCRs identified from isolated human naive B cells. B cells from these models were transferred into congenic recipients to achieve a human-like physiologically reasonable VRC01-class precursor frequency and tested for their ability to respond in vivo to germline targeting immunogen, to participate in GCs, and to accrue SHMs, including VRC01-class mutations.
Results
Generation of Three BCR Knockin Mouse Models with Authentic Antigen-Specific Human Naive B Cell BCR Specificity: HuGL16, HuGL17, and HuGL18.
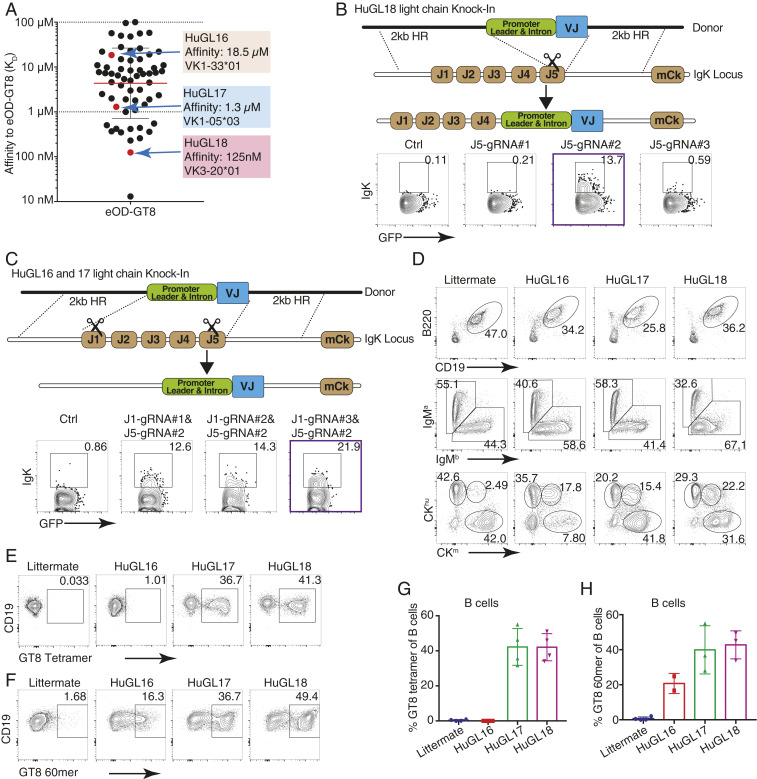
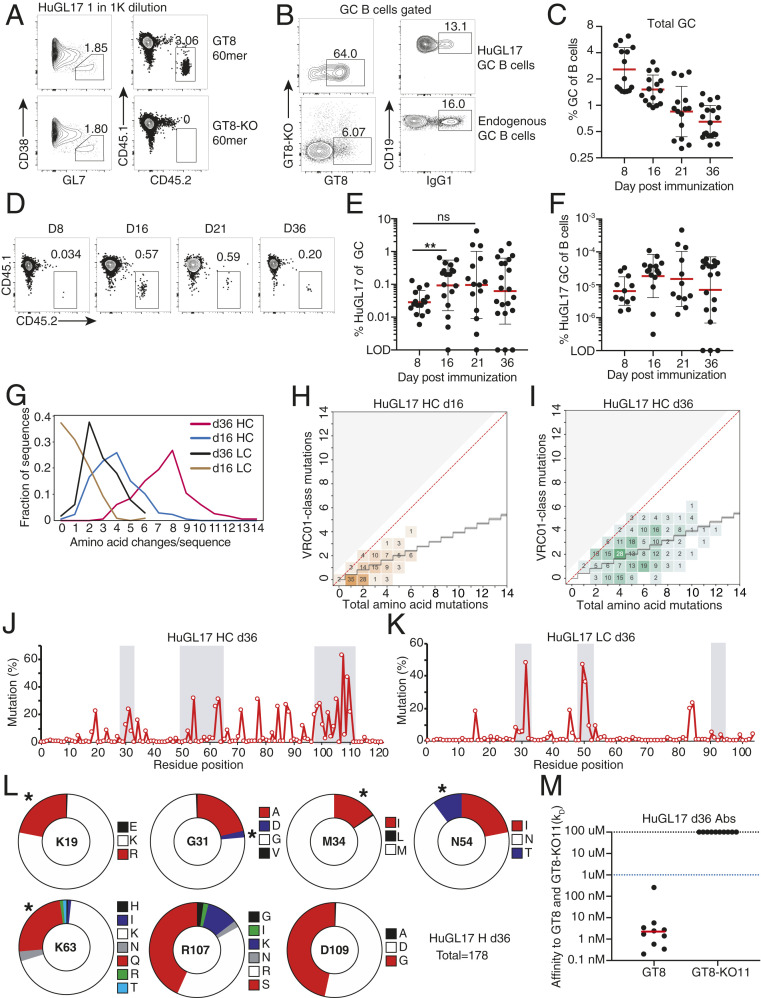
We generated mouse models carrying three different VRC01-class BCRs identical to those from human naive B cell donors. These are termed “HuGL” mice due to their expression of authentic antigen-specific “human germline” naive B cell BCR specificities. All use VH1-2*02, JH4*02, and LCs with 5 aa-long CDRL3 CQQY(E/D)X motif (underlining shows the 5 amino acid stretch referred to downstream of the conserved C) but vary in H-CDR3 length, affinity for eOD-GT8, and VK usage (Fig. 1A and SI Appendix, Table S1). HuGL16 (KD 18.5 μM) was chosen for its use of VK1-33, which is capable of Env N276 glycan-accommodating glycine mutations in l-CDR1 rather than deletions. HuGL17 (KD 1.3 μM) uses VK1-5, which should similarly be able to accommodate the N276 glycan by l-CDR1 point mutations based on similarity with bnAb lineage PCIN63 (34). HuGL17 and HuGL18 have an l-CDR3 sequence of CQQYETF, only 1 aa different from mature VRC01. HuGL18 was chosen for its high affinity (125 nM) and the use of VK3-20, a commonly used VK carrying a relatively short l-CDR1 of 7 aa. HC knockin mice were generated using embryonic stem cell targeting (13, 47, 48). Knockin allele usage for HuGL16, HuGL17, and HuGL18 was 89%, 79%, and 93%, respectively, as determined in crosses with IgHa/a wild-type strains (SI Appendix, Fig. S1A, similar to the 85% usage in VRC01gH mice) (13). LC knockins were generated directly in zygotes using CRISPR/Cas9 technology using either a single-cut strategy for HuGL18 (Fig. 1B) or a double-cut strategy for HuGL16 and HuGL17 (Fig. 1C). As is commonly seen in such models, only a fraction (∼30%) of cells in LC-only mice retained expression of the knockin allele (SI Appendix, Fig. S1B). When bred together, HuGL16 H/L, HuGL17 H/L, and HuGL18 H/L mice (“HuGL16,” “HuGL17,” and “HuGL18” hereafter) generated 20%, 40%, and 40% eOD-GT8-binding cells, respectively, as measured for HuGL17 and HuGL18 using eOD-GT8:streptavidin or, for HuGL16, by binding of eOD-GT8 60mer nanoparticles (Fig. 1 F and G). B cell numbers were somewhat lower than normal C57BL/6 B cell numbers (SI Appendix, Fig. S1), comparable to many BCR knockin models. A significant proportion of eOD-GT8-binding cells in the bone marrow, spleen, and lymph nodes of HL mice expressed only the knockin chains; however, some cells coexpressed an additional endogenous LC (Fig. 1D and SI Appendix, Fig. S1 D and E). Importantly, eOD-GT8-binding cells were present in similar percentages in spleen and lymph nodes, indicating they were likely functional, as lymph nodes are enriched in long-lived, nonanergic naive B cells (Fig. 1 E–H and SI Appendix, Fig. S1 F and G). B cells from HuGL17 and HuGL18 were able to make robust calcium flux responses to eOD-GT8 nanoparticles, whereas HuGL16 B cells failed to respond detectably under these conditions (SI Appendix, Fig. S2). Epitope-mutated negative control nanoparticles (eOD-GT8-KO 60mer) failed to elicit any response (SI Appendix, Fig. S2). These three HuGL mouse models thus generated B cells with properties predicted to allow response to eOD-GT8; however, the in vitro Ca2+ response of low-affinity HuGL16 B cells was poor.
Fig. 1.
Naive human B cell BCR VRC01-class H/L knockin mouse models HuGL16, HuGL17, and HuGL18. Figure shows the design and features of HuGL knockin mice. (A) Affinities of VRC01-class BCRs cloned from HIV– healthy human donors (43) highlighting the KD values and LC usage of HuGL16, HuGL17, and HuGL18. (B and C) HC knockins were generated using a conventional targeting strategy as described (13, 47, 48). LC knockin strategies are shown in the schematics, detailing the CRISPR-facilitated targeting strategy used for HuGL18 (B) or HuGL16 and HuGL17 (C), which involved one or two cuts in the Jκ locus, respectively. HR, arms of homology. Flow plots below each schematic show transient transfection analysis of gene targeting efficiencies in a Rag1−/− pro-B cell line using CRISPR ribonucleoproteins carrying the indicated guide RNAs and LC targeting construct (Materials and Methods). (D–H) Characterization of lymphocytes in HuGL mice. (D) Alleles were marked by breeding BCR knockins of interest (C57BL/6 background: Ighb/bCκm/m) to Igha/aCκhu/hu mice, followed by flow cytometry analysis with antibodies that distinguish C regions. (E–H) Evaluation of antigen-binding by B cells of the indicated strains using eOD-GT8 streptavidin tetramers (E) or eOD-GT8 60mer nanoparticles (F), with quantification shown in G and H. Data are representative of multiple litters per HuGL. (E–H) n = individual experiments, n = mice per group. n = 3. n = 1 to 3 mice per experiment. See also SI Appendix, Fig. S1.
HuGL18 VRC01-Class B Cells Can Be Primed and Recruited to GCs In Vivo.
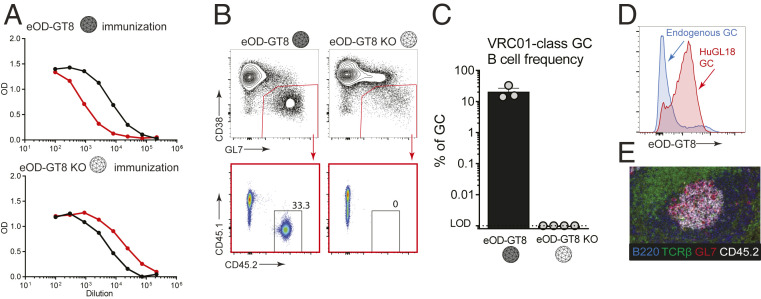
We next tested whether these B cells carrying authentic naive human VRC01-class BCR specificities could respond functionally in vivo, to confirm that the germline-targeting immunogen “bait” used to isolate VRC01-class human naive B cells cannot only bind intended BCRs ex vivo but can activate these cells in vivo. It was also of substantial interest to know whether VRC01-class HuGL B cells would be capable of participating in GCs. We first tested HuGL18 B cells. We transferred a high number of HuGL18 B cells (CD45.2+ allotype) into CD45.1+ congenic recipients, establishing a 1 in 1,000 HuGL18 B cell precursor frequency, and immunized the recipients with eOD-GT8 60mer in alum. Env CD4bs-specific IgG responses were detectable in HuGL18 recipients by day 8 (d8) postimmunization (Fig. 2A). CD4bs-specific IgG responses were not detected in mice immunized with eOD-GT8-KO 60mer (Fig. 2A). After immunization with eOD-GT8 60mer, large numbers of HuGL18 B cells were detected in GCs by flow cytometry (CD38– GL7+) (Fig. 2 B and C). As expected, these HuGL18 B cells were antigen-specific (Fig. 2D). Immunofluorescence staining of spleen histology sections revealed numerous GCs that contained HuGL18 B cells (Fig. 2E), in agreement with the flow cytometric data. Taken together, the data show that B cells expressing authentic VRC01-class human naive B cell BCRs are functional in vivo.
Fig. 2.
VRC01-class B cells from a BCR knockin mouse with an authentic naive human VRC01-class BCR specificity (HuGL18) can be primed and recruited to GCs. (A) Day 8 ELISA binding curves from CD45.1+ mice that received HuGL18 B cells (1 in 103 precursor frequency) and immunized with eOD-GT8 60mer (Top) or eOD-GT8-KO 60mer (Bottom). eOD-GT8 IgG titers are shown in black, KO IgG titers are shown in red. Responses are shown from individual mice that are representative of two mice per group. Bars indicate SEM. (B) Flow cytometric analysis of total BGC (gated as SSL/B220+/CD4−/CD38−/GL7+) and HuGL18 B cell occupancy of GCs (SSL/B220+/CD4−/CD38−/GL7+/CD45.1−/CD45.2+ cells) of mice immunized with eOD-GT8 60mer (Left), or eOD-GT8-KO 60mer (Right). Day 8 splenic B cells were analyzed. (C) Quantitation of VRC01-class B cells among BGC cells. (D) Flow cytometric analysis of antigen binding (eOD-GT8) by endogenous or HuGL18 BGC cells. Representative of five mice tested. Mice were immunized as in B. (E) Representative histological analysis of GCs. Frozen splenic sections were stained with B220 (blue), TCRβ (green), GL7 (red), and CD45.2 (white) for identification of HuGL18 B cells within GCs. SSL, singlet scatter live. n = 3 (B and C), n = 1 (A, D, and E). n = 3 to 4 mice per experiment.
HuGL18 VRC01-Class B Cells at Rare Physiological Precursor Frequencies Can Be Primed by eOD-GT8 60mer and Recruited to GCs.
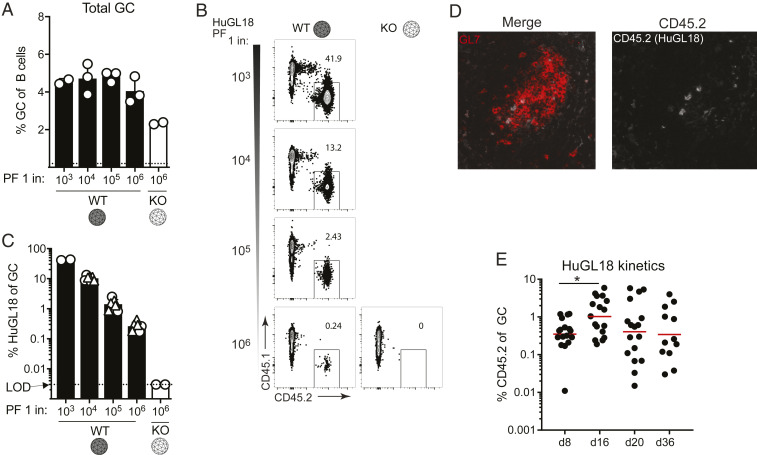
In vivo validations of germline-targeting approaches to vaccine design are key, particularly under physiologically relevant precursor frequency conditions, given that bnAb precursor B cells are rare in humans (28, 29, 43). It is not uncommon for in vivo mouse models to utilize precursor frequencies of 1 in 10 B cells to 1 in 1,000 B cells, which are far higher than the physiological range of a normal repertoire (45, 49). We therefore next assessed if HuGL18 VRC01-class B cells could be primed at physiologically relevant precursor frequencies found in the human repertoire. Human naive VRC01-class B cells with affinities better than a KD of 3 μM (the top ∼33%) are found at a precursor frequency of ∼1 in 106 B cells (43). We hypothesized that precursor frequency would be an important determinant for recruitment of authentic HuGL18 B cells to early GCs. To test the hypothesis, we transferred a range of HuGL18 B cells into congenic CD45.1+ hosts and immunized. The spleens of mice receiving HuGL18 B cell transfers were assessed for grafting efficiency, and precursor frequencies of 1 in 103 B cells to 1 in 106 B cells were confirmed (SI Appendix, Fig. S3 A–F). On day 8 postimmunization with eOD-GT8 60mer, all HuGL18 recipient animals developed similar total GC B cell (BGC) frequencies (Fig. 3A). However, the frequency of HuGL18 B cells among BGC cells was sharply dependent on the original precursor frequency. When HuGL18 B cells started from high precursor frequencies (1 in 103 to 104), HuGL18 B cells occupied a high proportion of the BGC compartment (∼10 to 40%, Fig. 3 B and C). In contrast, when precursor frequency was restricted to the physiological level found in humans (1 in 106 B cells), HuGL18 B cell frequencies among BGC postimmunization were 40- to 160-fold lower (∼0.25% of the GC compartment, Fig. 3 B and C). The HuGL18 cells localized to GCs histologically, consistent with the frequencies determined by flow cytometry (Fig. 3D). The HuGL18 B cell response was also confirmed to be CDbs-epitope specific; immunization with mutant eOD-GT8-KO 60mer did not prime HuGL18 B cells (Fig. 3 B and C). We next sought out how HuGL18 GC B cells could compete within the GC compartment over time when starting from physiologically rare precursor frequencies of 1 in 106 B cells. HuGL18 BGC cells showed outgrowth within GCs in most animals over time as GC reactions gradually waned, reaching levels as high as 5.7% of BGC cells in some animals at day 20 postimmunization (Fig. 3E and SI Appendix, Fig. S3 I and J). In summary, B cells expressing authentic VRC01-class human BCRs could be primed to proliferate and be recruited to GCs by the eOD-GT8 60mer germline-targeting antigen, even when the HuGL B cells started from rare physiologically relevant precursor frequencies.
Fig. 3.
Authentic human VRC01-class naive B cell BCRs can be primed by eOD-GT8 60mer and recruited to GCs at rare physiological precursor frequencies. (A) Frequency of splenic BGC cells in mice immunized with eOD-GT8 60mer, or the eOD-GT8-KO variant, when starting from the indicated HuGL18 B cell precursor frequency (PF). Day 8 splenocytes were analyzed and BGC cells were gated as SSL/B220+/CD4−/CD38−/GL7+ and plotted as percent of total B cells (B220+ cells). (B) Representative flow cytometric plots of HuGL18 B cells in GCs utilizing allotype mark. Cells are gated on GCs as indicated in A. (C) Quantitation of HuGL18 B cells in GCs as indicated in B. Experiments are pooled between two experiments (circles and triangles are separate experiments). A total of 25 mice were examined. (D) Representative histological analysis of individual GCs for HuGL18 B cells in individual GCs as indicated in B. Dotted blue line represents starting affinity. (E) Kinetic experiment showing HuGL18 B cell competitiveness within GCs over time. All data are shown pooled from multiple independent experiments. *P < 0.05. n = 3. n = 3 to 4 mice per experiment.
HuGL18 B Cells Develop Memory after eOD-GT8 60mer Immunization.
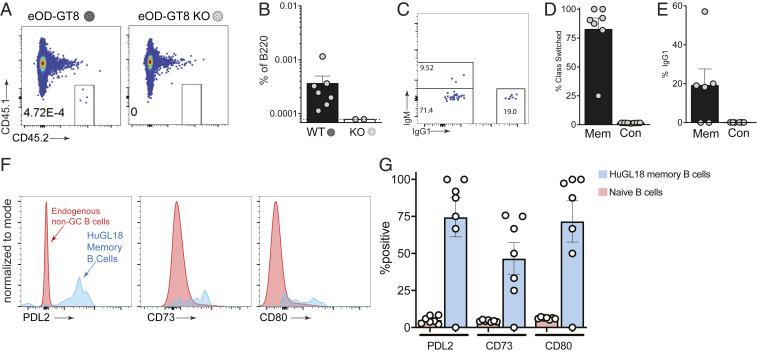
A bnAb-based HIV vaccine approach will involve multiple booster immunogens, designed to recruit VRC01-class memory B cells back to GCs and “shepherd” their affinity maturation pathway toward broad neutralization (10, 15, 25). We therefore evaluated VRC01-class memory B cell development after an eOD-GT8 60mer priming immunization, starting from physiologically rare naive VRC01-class B cell precursor frequencies. All immunized mice developed HuGL18 memory B cells by day 36 postimmunization (Fig. 4A). The frequency of memory B cells was variable among mice and reached values of 1 in 105 to 106 B cells, representing a threefold expansion on average from the naive frequency (Fig. 4B). Nearly all of the HuGL18 memory B cells were class switched (Fig. 4 C–E). Additionally, the majority of HuGL18 memory B cells expressed CD73, PD-L2, and CD80 (Fig. 4 F and G), three memory B cell markers associated with GC-derived memory B cells (50, 51). Altogether, the data show that HuGL B cells can form GC-derived memory B cells, even when starting from physiologically rare precursor frequencies.
Fig. 4.
HuGL18 B cells develop memory after eOD-GT8 60mer immunization. (A) Mice were assessed by flow cytometry for HuGL18 memory B cell formation in the spleen on day 36 postimmunization with eOD-GT8 60mer. eOD-GT8-KO 60mer immunization was done as a negative control. Precursor frequency of HuGL18 naive B cells was 1 in 106 before immunization. Memory B cells were gated as SSL/B220+/CD4−/IgD−/CD38+/GL7−/CD45.2+/CD45.1−. (B) Quantitation of memory B cells in A, as percent of B220+ B cells. (C) Flow cytometric analysis of class switched memory B cells gated in A. (D–E) Quantitation of (D) HuGL18 class switched memory B cells gated as IgM− B cells and (E) HuGL18 IgG1+ memory B cells on day 36 postimmunization. (F and G) Memory B cells phenotypes. (F) Representative flow cytometry and (G) quantitation of indicated memory B cell markers on memory HuGL18 B cells on day 36. Red histograms are total non-BGC cells (SSL/B220+/CD4−/CD38+/GL7−). n = 3. n = 4 to 7 mice per experiment. Mem, memory; Con, control.
Vk3-20+ VRC01-Class B Cells Develop SHM and Affinity Maturation after a Priming Immunization.
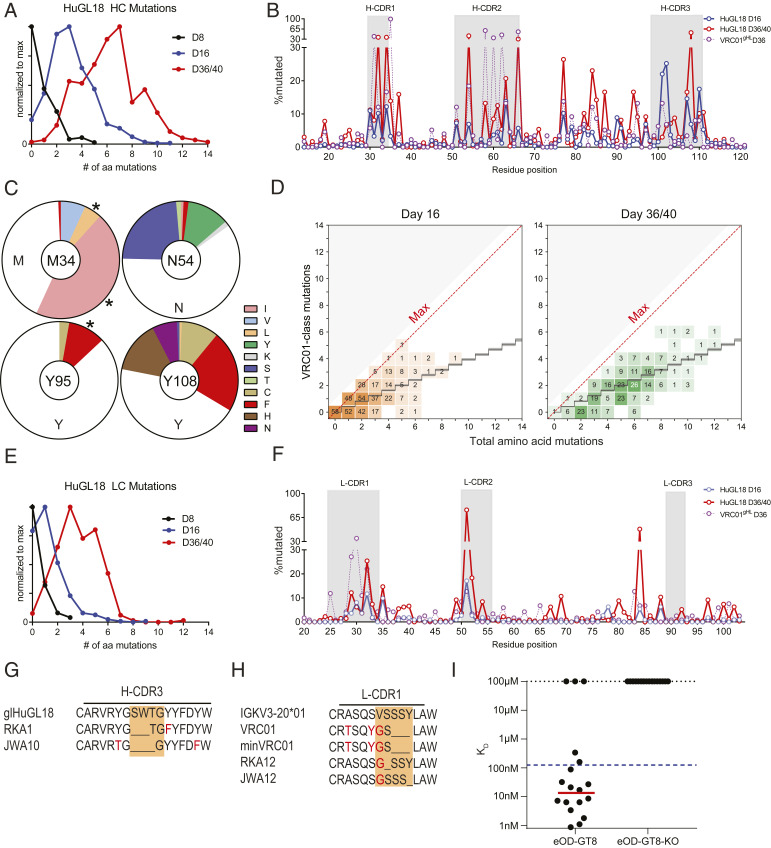
HuGL18 VH1-2*02 HCs accumulated substantial amino acid SHMs within 16 d postimmunization, with a maximum of 11 aa mutations, a median of 3 aa mutations, and with >95% of HuGL18 BGC clones containing at least 1 aa mutation (Fig. 5A and SI Appendix, Fig. S4B). Substantially more SHM accumulated in HuGL18 BGC cells by d36 (Fig. 5 A–D). SHMs in H-CDR2 were present (Fig. 5 B and C and SI Appendix, Fig. S4 A and B), consistent with the H-CDR2 dominant interaction with eOD-GT8 (28). Substantial mutations were also present in H-CDR1 and H-CDR3. SHM accrual in the H-CDR3 of HuGL18 was particularly prominent when compared to VRC01gHL (Fig. 5 B and C), illustrating a value of assessing B cells with authentic naive H-CDR3s. Nearly all VRC01-class bnAbs contain a W at the W100b (Kabat numbering) equivalent position of VRC01 in the H-CDR3, though one bnAb possesses a Y. Naive human VRC01-class B cells generally possess a Y or W at this position (43). Mutations were observed in HuGL18 BGC cells at Y107 (100b), including Y→P or Y→W (SI Appendix, Fig. S4B). VH1-2 positions known to be important for VRC01-class broad neutralization capacity were mutating, including M34L/I in H-CDR1, as well as K63Q/R and Y95F (Fig. 5 B and C and SI Appendix, Fig. S4B) (10, 34). We next determined how many VRC01-class amino acid mutations existed in each VH1-2 gene sequence and compared that to the total number of VH1-2 gene amino acid mutations to ascertain whether HuGL18 B cells were capable of maturing along a desired affinity maturation pathway in response to eOD-GT8 60mer. Encouragingly, we observed VRC01-class SHMs in HuGL18 B cells in GCs over time after a single immunization (Fig. 5D). We conducted additional VRC01-class mutational analysis with an expanded set of recently discovered VRC01-class bnAbs showing nearly identical results (SI Appendix, Fig. S4C). Statistical analysis of these VRC01-class mutations found no significant difference above observed patterns for antigen agnostic amino acid mutation accumulation in human VH1-2 memory B cells, which would indicate a specific VRC01-class mutational directionality (Materials and Methods). However, some HuGL18 clones at d16 achieved a perfect or near perfect VRC01-class mutational trajectory (Fig. 5D), while >77% of clones contained one or more VRC01-class mutations by d36.
Fig. 5.
Vk3-20+ VRC01-class B cells develop SHM and affinity maturation after a single priming immunization. Mice were generated with a HuGL18 B cell precursor frequency of 1 in 106 B cells, then immunized with eOD-GT8 60mer. Splenic IgG1+ d16 HuGL18 BGC cells were sorted from eOD-GT8 60mer immunized mice on days 8 to 40 postimmunization and HCs (A, D, and G) and LCs (E, F, and H) were assessed for amino acid SHMs. Cells were gated as SSL/B220+/CD4−/CD38−/GL7+/CD45.2+/CD45.1−/IgG1+/IgD−/CD138−. (A) Distribution of HC mutations over time. S, individual sequences. s = 134 for d8, s = 455 for d16, s = 295 for d36/40. (B) HuGL18 HC amino acid SHM distribution. For comparison, d36 VRC01gHL SHM data from animals with 1 in 106 precursor frequencies immunized with eOD-GT5 60mer (from ref. 27) are overlaid. (C) Specific HuGL18 HC amino acid mutations at d36. Asterisks (*) mark VRC01-class mutations. (D) VRC01-type mutations detected in HuGL18 HCs. A VRC01-class mutation (y axis) was any mutation observed in a representative set of VRC01-class bnAbs (12a12, 3BNC60, PGV04, PGV20, VRC-CH31, and VRC01) (13, 46). The black staircase depicted is a computational estimate of antigen-agnostic mutation accumulation in VH1-2 B cells (46). Each number within each square on the graph represents the number of antibody sequences that contained those specific numbers of mutations. Sequences were recovered from GC B cells that were single-cell sorted (one sequence per cell). The density of coloring is proportional to the stated number of sequences at each point relative to the total sequences analyzed. (E) Total LC amino acid mutations. s = 72 for d8, s = 343 for d16, s = 272 for d36/40. (F) Per residue HuGL18 LC amino acid SHM distribution. D36 VRC01gHL data are shown for comparison, as in B. (G) Alignment of two representative HuGL18 clones recovered on d36 with deletions in H-CDR3, out of seven total clones recovered with deletions in H-CDR3. (H) Alignment of two representative HuGL18 clones recovered on d16 with deletions in l-CDR1, out of 13 total clones recovered with deletions in l-CDR1. (I) Surface plasmon resonance (SPR) measured KD affinities for mAbs recovered from paired d36 HuGL18 GC B cell sequences. Dotted blue line represents affinity of HuGL18 precursor. Red line placed at geometric mean of clones with detectable affinity. Total sequences are from all experiments pooled. n = 2 for d16/d36. n = 1 for d8. n = 3 to 7 mice per experiment. See also SI Appendix, Fig. S4.
The majority of HuGL18 BGC cells at day 16 postimmunization also possessed LC mutations, with up to 9-aa SHMs detected (Fig. 5E). Minimal mutation was seen in l-CDR3 (Fig. 5F), which only differed from mature VRC01 by 1 aa, thus preserving the critical VRC01-class features of the l-CDR3. Most mutations were in l-CDR1 (Fig. 5F), consistent with the presence of substantial affinity maturation in l-CDR1 of VRC01-class bnAbs.
Deletion events, which are often considered rare, were also observed in HuGL18 BGC cells, in HC or LC. Seven HuGL18 BGC clones, isolated from two separate mice, showed deletions of 2 to 3 aa in H-CDR3 on d36 postimmunization (Fig. 5G), whereas an H-CDR3 insertion has been implicated in the maturation pathway of the VRC01-class bnAb VRC08 (19). Strikingly, 13 clones, isolated from two independent mice, contained single aa deletions in l-CDR1 (Fig. 5H). Interestingly, 12 of these deletions were of S30 and one deletion was found in Y33, which aligns with the deletion found in the mature VRC01 bnAb and a minimally mutated VRC01 bnAb construct (10). Interestingly, all of the clones that contained deletions had an accompanying V29G mutation. V29G is present in the mature VRC01 and in the minimally mutated VRC01 bnAb construct, MinVRC01 (10, 34). Both of these observations are of considerable interest, as l-CDR1 deletions or mutations are critical for most VRC01-class Abs to develop breadth, to circumvent clashing with the Env N276 glycan. Deletions or mutations in l-CDR1 have been considered to be a major hurdle for VRC01-class bnAb vaccine development (5, 19). Here, we show that l-CDR1 deletions and mutations can occur in a HuGL B cell in as few as 16 d following eOD-GT8 60mer immunization.
Lastly, we sought to assess if HuGL18 BGC cells underwent affinity maturation (Fig. 5I and Dataset S1). To answer this question, we chose 15 representative paired HC and LC sequences from multiple mice at day 36 and expressed the antibodies for affinity measurements. Three antibodies had undetectable affinity for eOD-GT8, perhaps representing random deleterious mutations in GCs; two of these nonbinders were clonally related and contained an introduced unpaired cysteine, Y59C, which may have caused folding problems. Of the 12 mAbs assessed with detectable affinity, 9 showed appreciable affinity maturation toward eOD-GT8, reaching 0.87 nM KD affinity. The average geometric mean affinity of the 12 binding antibodies was 12 nM KD (Fig. 5I). This represented an average affinity improvement of 10-fold for antibodies with measurable affinity for eOD-GT8.
In summary, authentic VRC01-class naive B cells primed at physiologically rare precursor frequencies were able to undergo substantial SHM in both the HC and LC and accrue specific VRC01-class mutations, including deletions in l-CDR1, in response to a single priming immunization. Thus, germline-targeting immunization strategies can both prime and affinity mature rare cells possessing epitope-specific naive human B cell BCRs, with a single immunization.
A Medium Affinity, Vk1-5+ VRC01-Class BCR Knockin Mouse Model.
Immune responses by Vk1-5+ VRC01-class naive B cells are of specific interest because a Vk1-5+ VRC01-class bnAb was recently identified, and that bnAb (PCIN63) has very appealing characteristics of low SHM% and the absence of any indels (34). Using the adoptive transfer approach, we assessed the immune response of HuGL17 B cells (Vk1-5+) (Fig. 6). At a precursor frequency of 1 in 1,000, a strong GC response of the donor cells was elicited by eOD-GT8 60mer but not the eOD-GT8-knockout (KO) 60mer (Fig. 6A). Responding donor BGC cells underwent a robust IgG1 class switch comparable to that of the host response (Fig. 6B). We then assessed the response of HuGL17 cells seeded at a precursor frequency of 1 in 106 (Fig. 6 C–I). The total (host + donor) GC response peaked at ∼d8 (Fig. 6C). The HuGL17 cells proved able to participate in the GC response (Fig. 6D), with HuGL17 BGC cells increasing to d16 and then remaining stable to d36 (Fig. 6E). The HuGL17 VRC01-class B cell response plateaued at ∼0.1% of the total BGC and 0.005% of total B cells, representing a ∼50-fold expansion from input on average (Fig. 6F and SI Appendix, Fig. S5E). The variable nature of the outgrowth of HuGL17 B cells was observed across experiments. At this low precursor frequency, we were unable to consistently enumerate HuGL17 memory B cells, but HuGL17 memory B cells were consistently observed in experiments starting from a precursor frequency of 1 in 105 (SI Appendix, Fig. S5 A–D). These data indicate that, when primed by eOD-GT8 60mer, Vk1-5+ VRC01-class naive B cells were able to participate and persist in the highly competitive GC response under physiological precursor frequency conditions.
Fig. 6.
Characterization of HuGL17, a medium-affinity VK1-5+ VRC01-class BCR model. Analysis of total and HuGL17 splenic BGC cells. (A and B) Analysis of HuGL17 responsiveness to eOD-GT8 in vivo at a HuGL17 B cell precursor frequency of 10−3 (“1K” = 1,000). (A) Total (Left) and donor-derived fraction (Right, boxes) of BGC cells after immunization with eOD-GT8 or eOD-GT8-KO. (B) Representative flow plots of antigen-specific and IgG1 class-switched HuGL17 and endogenous (CD45.1+) BGC cells (TCRβ–CD19+CD38–GL7+). (C–L) Longitudinal analysis of mice immunized with eOD-GT8 60mer when HuGL17 precursor frequencies were 1 in 1 million B cells. (C) Longitudinal analysis of total frequency of BGC cells. (D) Representative flow plots enumerating HuGL17 BGC cells. (E) Longitudinal analysis of GC occupation by HuGL17 B cells. Each data point indicates the value measured in one recipient spleen. (F) HuGL17 BGC cell percent among all B cells. (G) Analysis of the number of amino acid replacements per sequence in the HC and LC, respectively, at d16 (n = 197, 172) and d36 (n = 178, 226). (H and I) Analysis of HC VRC01-class mutations on d16 and d36. Analysis was conducted as in Fig. 5D. (J and K) Distribution of replacement mutations as a function of amino acid position. (L) Quantitation of HC replacement mutations d36 (n = 178). (M) SPR measured KD affinities for mAbs recovered from paired d36 HuGL17 GC B cell sequences, with dotted blue line indicating affinity of the HuGL17 precursor. Asterisks (*) mark VRC01-class mutations. n = 3. n = 2 to 7 mice per experiment.
Antibody gene sequencing analysis of HuGL17 BGC cells at d16 and d36 revealed extensive SHM, particularly in the HC (Fig. 6 G–L and SI Appendix, Fig. S6A). Many HC mutations within VH1-2 were identical to mutations seen in VRC01-class bnAbs (Fig. 6 H, I, and L and SI Appendix, Figs. S6C and S4D). Statistical analysis of these VRC01-class mutations found no significant difference above observed patterns for antigen agnostic amino acid mutation accumulation in human VH1-2 memory B cells (Materials and Methods), comparable to the findings for HuGL18, above. However, multiple HuGL17 clones achieved perfect or near perfect VRC01-class mutational directionality (Fig. 6I). VRC01-class mutational analysis with an expanded set of recently discovered VRC01-class bnAbs showed nearly identical results (SI Appendix, Fig. S4D). The l-CDR3 was barely mutated (Fig. 6K and SI Appendix, Fig. S6B), preserving the critical residues found in VRC01-class bnAbs. In contrast, the HuGL17 H-CDR3 was heavily mutated (Fig. 6J and SI Appendix, Fig. S6A), especially at R107, an arginine lying at the center of the H-CDR3 loop that may be subject to negative selection. In the Vk1-5+ bnAb PCIN63, four glycine replacements are present in the l-CDR1 instead of the small l-CDR1 deletion observed in the bnAb VRC01. Although eOD-GT8 is not designed to specifically select for VK1-5 l-CDR1 features, six HuGL17 BGC cells were found to have introduced glycine replacement mutations in l-CDR1, which is predicted to facilitate evolution to neutralization breadth. One HuGL17 BGC clone contained a double glycine replacement in l-CDR1 positions identical to that of the bnAb PCIN63.
We next sought to assess the affinity of HuGL17 BGC cells. To do this, we expressed heavy and light pairs from day 36 sorted GC B cells (Dataset S1). We found that all antibodies that were expressed had detectable affinity for eOD-GT8 and, on average, showed a 500-fold improvement of affinity. Nine out of 10 HuGL17 antibodies improved to low nanomolar range affinity (Fig. 6M).
We conclude that, at these physiological precursor frequencies, HuGL17 B cells participate in the GC response and undergo affinity maturation, indicating that the Vk1-5+ VRC01-class naive B cells identified in humans are likely targets, and interesting targets, for priming by eOD-GT8 60mer in humans.
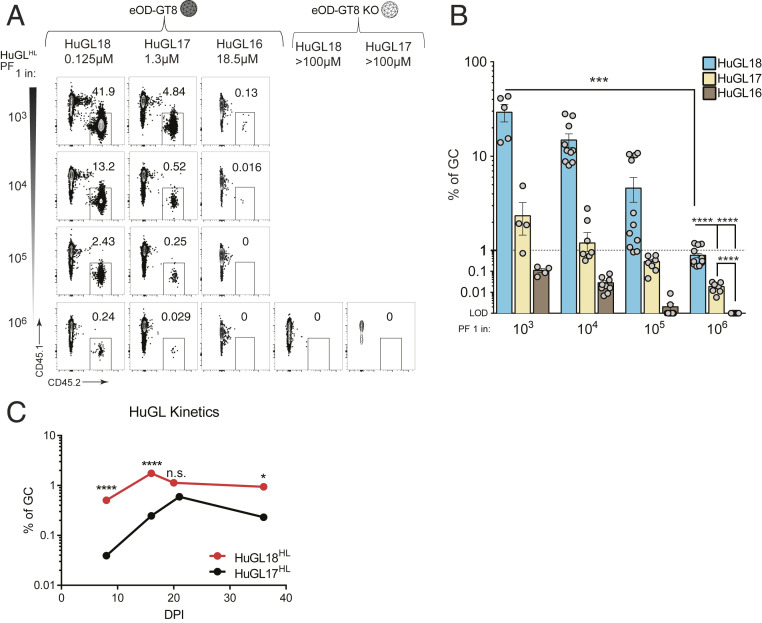
Precursor Frequency and Affinity Are Interdependent in Determining Competitive Success for B Cells Possessing Authentic Human Naive VCR01-Class BCRs.
There is evidence that both B cell precursor frequency and BCR affinity influence B cell competition following immunization (24, 27, 45, 52, 53). To quantitatively assess this prediction with B cells expressing authentic naive human BCR sequences, we transferred 10-fold serial dilutions of HuGL B cells into congenic recipients and immunized with eOD-GT8 60mer. These HuGL B cells not only utilize three LC V genes found in VRC01-class bnAbs (VK3-20, VK1-5, and VK1-33), but HuGL18, HuGL17, and HuGL16 also span a physiological range of affinities (0.125 μM → 18.5 μM) found in humans. At day 8 postimmunization, a clear hierarchy in GC occupancy was observed based both on the starting precursor frequencies of the naive B cells and the BCR affinity (Fig. 7 A and B). Combining the available data, the competitive fitness of high (HuGL18) and medium (HuGL17) affinity cells over time can be compared. Using experiments with HuGL18 or HuGL17 seeded in recipient mice at the 1 in 106 physiological precursor frequency (Figs. 3E and 6E), the magnitude of the VRC01-class BGC cell responses correlated with starting BCR affinity (Fig. 7C). While HuGL18 B cells were present in GCs in higher numbers than HuGL17 as early as d8 (P < 0.001, Fig. 7C), on average the HuGL17 B cells did shrink the gap with the HuGL18 BGC response (d16, 10-fold difference; d21, 2-fold difference; d36, 5-fold difference) (Fig. 7C). HuGL18 cells exhibited rapid population of GCs by d8, calculated to be a ∼100-fold outgrowth of cells from the d0 HuGL18 precursor frequency. HuGL17 B cells showed greater outgrowth from d8 to d16 within GCs (Fig. 7C). This highlights the affinity of precursors as well as Vκ gene usage as possibly affecting the numbers of initial GCs seeded and/or the kinetics within GCs. Taken together, these data highlight the importance of precursor frequency and antigen affinity in vaccine design. This preclinical model predicts, with certain caveats, that the ongoing clinical trial of the germline-targeting HIV vaccine antigen eOD-GT8 60mer will prime desired human VRC01-class B cells and recruit those cells to GCs in a manner dependent both on B cell precursor frequency and antigen affinity.
Fig. 7.
Precursor frequency and affinity are interdependent in determining competitive success for B cells possessing authentic human naive VCR01-class BCRs. (A) Frequency of HuGL B cells in GCs d8 postimmunization following immunization with eOD-GT8 60mer or eOD-GT8KO 60mer. BGC cells gated as SSL/B220+/CD4−/CD38−/GL7+/CD45.1−/CD45.2+. For comparison purposes between mouse strains, HuGL18 plots (Fig. 3) are reshown here. (B) Quantitation of HuGL B cells in GCs as shown in A. All available data are shown. Data were pooled from multiple independent experiments. Dashed line at 1% is provided as visual aide only for the reader to denote responses greater or less than 1%. (C) Statistical comparisons of HuGL GC B cells over time. Kinetics data shown in C are the average of all data shown in Figs. 3E and 6E. Mice were immunized with eOD-GT8 containing HuGL17 or HuGL18 B cells at a precursor frequency of 1 in 106 B cells.*P < 0.05 ***P < 0.001, ****P < 0.0001. n.s., not significant; n = 3, n = 3 to 4 mice per experiment for A and B. n = 4 to 5 for HuGL17 and HuGL18 each, n = 3 to 7 mice per time point for C.
Discussion
Here, BCRs isolated from authentic antigen-specific naive human B cells were tested in mouse models for their ability to respond to antigen, somatically hypermutate, affinity mature, and compete in germinal centers. VRC01-class naive B cells from all three BCR H/L knockin mouse models were able to be primed in vivo and participate in GCs. High-affinity and medium-affinity VRC01-class B cells (HuGL18 and HuGL17) were able to successfully compete in GCs and affinity mature—including acquisition of bnAb-type somatic mutations—even when the HuGL B cells started from rare physiological precursor frequencies. Taken together, these data validate a key tenet of the germline-targeting approach to vaccine design. These data suggest that eOD-GT8 60mer, and germline-targeting immunogens with similar properties, are likely to be successful in human clinical trials.
Germline-targeting vaccine design is based on the concept of designing vaccine antigens to bind precursors of highly potent neutralizing antibodies isolated from infected humans. Cycles of antigen design and immunization strategy iteration are almost certainly required for germline-targeting immunogen designs to be applicable for human vaccine needs. One strategy is to design germline-targeting antigens based on iGL sequences from bnAbs and then use the germline-targeting immunogen to screen the human B cell repertoire for the capacity of the putative germline-targeting immunogen to identify (bind) bnAb precursors in normal healthy donors, selecting top candidate immunogens to move forward into clinical trials (6, 7). A significant gap in this strategy has been the absence of an in vivo model to demonstrate the in vivo specificity and responsiveness of the human antigen-specific naive B cell BCR sequences prior to a full clinical trial. One solution to this problem, employed here, is to express antigen-specific human naive B cell BCRs in mice and thus provide a direct preclinical platform for evaluating relevant antigen design and immunization strategy iterations. The approach of identifying antigen-specific human naive B cells and developing HuGL mouse models of preclinical vaccine testing is also applicable more broadly to other, nongermline-targeting, reverse vaccinology antigen design strategies.
We found precursor frequency and antigen affinity to be critical in dictating B cell outcomes following vaccination. We observed that these factors were interdependent in determining recruitment to GCs, competition within GCs, and exit to the memory B cell pool. These HuGL results were consistent with our previous study (27). While HuGL B cell competitive success in GCs past day 8 was more stochastic between individual mice than seen for VRC01gHL iGL B cells, BCR affinity was a determining factor in competitive success of HuGL B cells over time. The interanimal variance of HuGL GC B cell responses may be a strength of this model by being reflective of variance that may be seen in humans. This stochasticity reinforces the importance of interclonal competition and immunodominance as hurdles in vaccine development to complex antigens (54–56). As the precursor frequency of virtually all bnAb B cells is predicted to be low (10), the present study indicates that a preimmune BCR KD of >10 μM would be insufficient for efficient recruitment into the nanoparticle response, and that vaccine immunogens should be designed with considerably tighter binding to BCRs of authentic naive human B cell targets (0.1 to 1 μM or better).
HuGL17 B cells underwent a more substantial affinity gain (∼500-fold) within 36 d than HuGL18 B cells did (∼10-fold). Experiment selection may have skewed the apparent affinity gain of HuGL17 B cells because mice with poor HuGL17 responses had too few cells for sorting, and as such were not sampled. A separate possibility is that some B cell precursors may have a more challenging mutational trajectory than others. The antibodies selected for HuGL17 contained more mutations on average than HuGL18 mAbs. This observation may represent an intrinsic advantage of VK1-5 antibodies (34) over VK3-20 antibodies in ease of affinity maturation pathway in a competitive microenvironment starting from rare precursors. Alternatively, these outcomes could reflect an affinity ceiling in the response of HuGL cells to eOD-GT8. Both HuGL18 and HuGL17 BGC cells ended up with median affinities in the nanomolar range. Taken together, these differences highlight the importance of assessing authentic human precursors for their competitive fitness in stringent preclinical models to inform vaccine immunogen design efforts.
A major goal of mouse immunology is to model human immune responses; however, mice and humans have substantially different immunoglobulin gene repertoires, limiting the ability to test reverse vaccinology design concepts outside of humans. While multiple mouse lines have been designed to express diverse human immunoglobulin genes, a limitation of those models is that they fail to properly represent the precursor frequencies found in humans. Precursor frequency can be a major factor in B cell responses, and immunoglobulin-locus transgenic animals generally have greatly underrepresented (23) or overrepresented B cells of interest (12–14, 22, 25, 26, 46, 48, 57). In contrast, transfer models such as those used here can titrate the cell numbers to match the physiological precursor frequencies found in humans (27). In addition to precursor frequency issues, BCR knockin models can depend on iGL sequences of bnAbs (13–15, 25, 26, 58). Such models have key caveats for interpreting how a candidate antigen may perform in humans. First, the H-CDR3 and l-CDR3 generally remain unchanged from the somatically hypermutated bnAb (because the original CDR3s cannot be accurately predicted due to stochastically determined sequence variations generated in each naive B cell during development). This is generally expected to result in significant unintended advantage to those iGL BCR B cells compared to authentic naive B cell BCRs. Second, bnAb iGL sequences are not known to be reliable proxies for true naive B cell BCR sequences represented in most humans. Lastly, the bnAb iGL sequence in the mouse model is frequently an exact match to a bnAb iGL sequence used in the immunogen design. Here, we provide a more rigorous test. eOD-GT8 was designed based on a broad panel of bnAb iGLs (13). eOD-GT8 was then tested for its ability to “fish out” epitope-specific human naive B cells (28, 43). To our knowledge, eOD-GT8 is only one of two engineered vaccine antigens to successfully pass the test of binding epitope-specific human naive B cells directly from multiple human blood samples (28, 29, 43). [The predecessor of eOD-GT8, eOD-GT6, was designed based on a narrower panel of bnAb iGLs and did not successfully pass the test of binding VRC01-class precursor naive B cells in human blood samples (28), highlighting the magnitude of the protein design challenge for binding diverse authentic human B cell BCRs.] By isolating dozens of epitope-specific naive human B cells with eOD-GT8 (43), we were able to categorize and stratify the BCRs.
The use of CRISPR technology greatly accelerates the speed of BCR knockin mouse generation at the HC locus (59, 60). In this work, we used CRISPR-facilitated targeting in zygotes to introduce functional knockins into the κ-locus, developing models for three distinct LCs along with their corresponding HC models.
Here, we selected three representative VRC01-class naive B cell BCRs for construction of BCR knockin mice, in an effort to make a multipronged set of mouse models for predicting potential outcomes of germline-targeting vaccine immunization in humans. Based on the experiments reported herein, these models indicate that a human clinical trial with eOD-GT8 60mer should be successful, based on the parameter of activating and expanding VRC01-class naive B cells. Both high- and intermediate-affinity VRC01-class naive B cell BCRs successfully expanded upon eOD-GT8 immunization in HuGL transfer models, when HuGL B cells were present at a precursor frequency of 1 in 1 million. The available data indicate that high- and intermediate-affinity BCRs (<3 µM) are present in the human repertoire at a frequency of ∼1 in 0.9 million. Low-affinity VRC01-class BCR naive B cells are severalfold more abundant, but, at least for HuGL16, that does not suffice to overcome the competitive disadvantage of the weaker affinity cells.
Interesting VRC01-class B cell SHMs were observed in this study using HuGL B cells. VRC01-class mutations were observed in HCs and LCs of HuGL18 and HuGL17 B cells after only a single immunization with eOD-GT8 60mer. The H-CDR3s in both HuGL18 and HuGL17 B cells underwent substantial SHM. While most VRC01-class bnAbs have limited engagement of HIV Env via H-CDR3, recent analysis suggests that development of the VRC01 bnAb and the related VRC08 bnAb involved extensive mutation of H-CDR3 that affected the development of breadth. Deletions are generally considered very rare events, and deletions in l-CDR1 are thought of as a major hurdle to overcome in the development of VRC01-class bnAbs. Intriguingly, we found that deletions in l-CDR1 of HuGL18 B cells could be found in BGC cells in as little as 16 d after eOD-GT8 60mer immunization. Taken together, these data highlight the importance of studying B cell responses by B cells expressing human BCRs from authentic naive B cells.
It remains unclear how to induce a full VRC01-class bnAb maturation path by immunization, and it remains unclear what fraction of VRC01-class naive B cells could successfully navigate such a path under optimal conditions. VRC01-class B cells refers to all B cells with the core HC and LC sequence characteristics of VRC01-class bnAbs: VH1-2*02 (or *03 or *04) paired with a LC possessing a 5-aa CDR3. A subset of naive B cells with those characteristics binds to eOD-GT8, generally reflecting clearer relatedness to VRC01-class bnAbs (i.e., l-CDR3 with a QQYxx motif and a preference for a short l-CDR1) (10, 43). Development of a VRC01-class B cell into a bnAb over time is not a given, as even in HIV+ donors with identified VRC01-class B cell responses, lineages or branches of lineages can fail to develop breadth (19, 32, 61). Whether some VRC01-class naive B cells are more likely to develop breadth remains an important open question for future investigation. For example, data shown here indicate that both VK3-20+ and VK1-5+ VRC01-class B cells have the ability to rapidly acquire mutations in l-CDR1 that are frequently considered important for accommodating Env N276. In the case of VK3-20+, l-CDR1 deletions were detected among BGC cells. In the case of VK1-5+, two glycine mutations were found in l-CDR1. Previously it was thought that such mutations may be extremely rare. Lastly, the recently identified VK1-5+ VRC01-class bnAb PCIN63 lineage acquired breadth with as little as 19% amino acid mutations in the HC (34). It was encouraging to observe up to 12.7% amino acid mutations in both HuGL18 and HuGL17 BGC cells after only a single immunization with eOD-GT8 60mer (Figs. 5 and 6).
The HuGL models have limitations. The best assessment of those limitations will come with the completion of the full eOD-GT8 60mer clinical trial (the trial is currently ongoing). Human adaptive immunity cell biology is not identical to that of mice, and the B cell repertoires of the species are different, resulting in differences in the competitive environment that cannot be calculated and may or may not be sufficiently different to impact postimmunization outcomes. Overall, human versus mouse differences could theoretically cause a range of differences in the outcomes between mouse and human immunization with a germline-targeting antigen. Given the theoretical nature of those differences, we feel that the most productive current use of HuGL mouse models is to make specific predictions, allowing for clear assessment of whether specific predictions were useful, accurate, or inaccurate when the full clinical trial results become public. At least six different mouse models of VRC01-class B cell responses to eOD-GT have been tested, including the HuGL model reported here (12, 22, 23, 27, 46). It will be of great value to compare each of the mouse models to the human clinical trial results. Experimental advantages of other models include the use of truly polyclonal antigen-specific repertoires (22, 23), or the presence of high precursor frequencies or affinities making positive results in the model more likely (12–14, 46), or the use of the prototypic VRC01 iGL (12, 13, 27, 46).
The HuGL approach reported here is the only mouse model utilizing exact naive BCR sequences known to exist in the naive B cell repertoires of HIV-negative humans with truly authentic H-CDR3s. It is also the only model to match the known precursor frequency and affinity range in humans. The HuGL strategy has the added advantage of straightforward cell transfers to modulate the precursor frequencies, unlike models using genetically modified mice in a mixed background (22, 23). It is conceivable these HuGL models may underestimate how well VRC01-class B cells can respond in humans due to challenges of introducing human BCR sequences into mice, as we observed that a fraction of transgenic B cells in all three mice coexpressed endogenous mouse LCs and had somewhat reduced overall BCR levels. Lastly, the experiments demonstrate the power of new CRISPR-based approaches to make HuGL-type mouse models, because of the speed of new mouse generation.
Overall, these data indicate that VRC01-class B cells with VK3-20 or VK1-5 are likely to respond to eOD-GT8 immunizations of humans, and the data imply that VRC01-class B cells of any recognized subtype (VK3-20, VK1-5, VK1-33, VK3-15, and VK4-1) are likely to respond in humans in a manner that is interdependent of precursor frequency and affinity. Furthermore, the HuGL mouse model data predict that the expansion of high and intermediate VCR01-class B cells should be accompanied by substantial SHM and possibly rare events such as deletions, even after a single immunization. These data complete a key loop of germline-targeting preclinical development strategy, and this strategy is generalizable to other germline-targeting and reverse vaccinology strategies for HIV or other pathogens.
Materials and Methods
Extended materials and methods have been added at the end of the SI Appendix immediately after SI Appendix, Table S1. Methods used for the generation of mice, flow cytometry, immunogen production, immunizations, ELISAs, histology, and single-cell sequencing are all largely based on previously published studies (13, 47, 48), and extensive details are provided in SI Appendix, Materials and Methods. All animal studies were completed under approved Institutional Animal Care and Use Committee protocols at La Jolla Institute for Immunology or The Scripps Research Institute.
Supplementary Material
Acknowledgments
This work was supported in part by the NIH National Institute of Allergy and Infectious Diseases under awards Al100663 (Scripps Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery) and UM1 AI144462 (Scripps Consortium for HIV/AIDS Vaccine Development) (to S.C., W.R.S.); NIH Grant K99 AI145762 (R.K.A.); by the Ragon Institute of Massachusetts General Hospital, Massachusetts Instititue of Technology, and Harvard University (W.R.S.); and by the International AIDS Vaccine Initiative (IAVI) Neutralizing Antibody Consortium (NAC) and Center (W.R.S.); and through the Collaboration for AIDS Vaccine Discovery funding for the IAVI NAC Center (W.R.S.).
Footnotes
Competing interest statement: W.R.S. is an inventor on a patent application submitted by IAVI and The Scripps Research Institute that covers the eOD-GT8 immunogen. W.R.S. is involved as a principal investigator of a human clinical trial involving eOD-GT8 60mer (G001). As part of that involvement, W.R.S. is privy to certain clinical trial data, and he was forbidden from sharing that data with the authors of this study during the course of this HuGL study, as is standard for blinded clinical trials. Any conclusions in this manuscript regarding relationships between HuGL mouse models and humans were made by the other authors, without input from W.R.S. related to any clinical trial activities.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004489117/-/DCSupplemental.
Data Availability.
BCR sequences are provided in supplementary information (Dataset S1).
References
- 1.Bloom D. E., Fan V. Y., Sevilla J. P., The broad socioeconomic benefits of vaccination. Sci. Transl. Med. 10, eaaj2345 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Plotkin S. A., Orenstein W. A., Offit P. A., Plotkin’s Vaccines (Elsevier, Philadelphia, PA, ed. 7, 2018), p. 1691, pp. xxi. [Google Scholar]
- 3.Havenar-Daughton C., Lee J. H., Crotty S., Tfh cells and HIV bnAbs, an immunodominance model of the HIV neutralizing antibody generation problem. Immunol. Rev. 275, 49–61 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Piot P.et al., Immunization: Vital progress, unfinished agenda. Nature 575, 119–129 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Kwong P. D., Mascola J. R., HIV-1 vaccines based on antibody identification, B cell ontogeny, and epitope structure. Immunity 48, 855–871 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Burton D. R., What are the most powerful immunogen design vaccine strategies? Reverse vaccinology 2.0 shows great promise. Cold Spring Harb. Perspect. Biol. 9, a030262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rappuoli R., Bottomley M. J., D’Oro U., Finco O., De Gregorio E., Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design. J. Exp. Med. 213, 469–481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jardine J.et al., Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGuire A. T.et al., Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J. Exp. Med. 210, 655–663 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jardine J. G.et al., Minimally mutated HIV-1 broadly neutralizing antibodies to guide reductionist vaccine design. PLoS Pathog. 12, e1005815 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landais E., Moore P. L., Development of broadly neutralizing antibodies in HIV-1 infected elite neutralizers. Retrovirology 15, 61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dosenovic P.et al., Immunization for HIV-1 broadly neutralizing antibodies in human Ig knockin mice. Cell 161, 1505–1515 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jardine J. G.et al., HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 349, 156–161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Medina-Ramírez M.et al., Design and crystal structure of a native-like HIV-1 envelope trimer that engages multiple broadly neutralizing antibody precursors in vivo. J. Exp. Med. 214, 2573–2590 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steichen J. M.et al., HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity 45, 483–496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Escolano A.et al., Immunization expands B cells specific to HIV-1 V3 glycan in mice and macaques. Nature 570, 468–473 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LaBranche C. C.et al., Neutralization-guided design of HIV-1 envelope trimers with high affinity for the unmutated common ancestor of CH235 lineage CD4bs broadly neutralizing antibodies. PLoS Pathog. 15, e1008026 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parks K. R.et al., Overcoming steric restrictions of VRC01 HIV-1 neutralizing antibodies through immunization. Cell Rep. 29, 3060–3072.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonsignori M.et al., Inference of the HIV-1 VRC01 antibody lineage unmutated common ancestor reveals alternative pathways to overcome a key glycan barrier. Immunity 49, 1162–1174.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Julien J. P.et al., Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 9, e1003342 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mouquet H.et al., Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc. Natl. Acad. Sci. U.S.A. 109, E3268–E3277 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian M.et al., Induction of HIV neutralizing antibody lineages in mice with diverse precursor repertoires. Cell 166, 1471–1484.e18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sok D.et al., Priming HIV-1 broadly neutralizing antibody precursors in human Ig loci transgenic mice. Science 353, 1557–1560 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dosenovic P.et al., Anti-HIV-1 B cell responses are dependent on B cell precursor frequency and antigen-binding affinity. Proc. Natl. Acad. Sci. U.S.A. 115, 4743–4748 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Escolano A.et al., Sequential immunization elicits broadly neutralizing anti-HIV-1 antibodies in Ig knockin mice. Cell 166, 1445–1458.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGuire A. T.et al., Specifically modified Env immunogens activate B-cell precursors of broadly neutralizing HIV-1 antibodies in transgenic mice. Nat. Commun. 7, 10618 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abbott R. K.et al., Precursor frequency and affinity determine B cell competitive fitness in germinal centers, tested with germline-targeting HIV vaccine immunogens. Immunity 48, 133–146.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jardine J. G.et al., HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science 351, 1458–1463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steichen J. M.et al., A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses. Science 366, eaax4380 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anonymous, A Phase I Trial to Evaluate the Safety and Immunogenicity of eOD-GT8 60mer Vaccine, Adjuvanted. ClinicalTrials.gov, NCT03547245 G001 (2018).
- 31.Zhou T.et al., Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811–817 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou T.et al.; NISC Comparative Sequencing Program , Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 39, 245–258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang J.et al., Identification of a CD4-binding-site antibody to HIV that evolved near-Pan neutralization breadth. Immunity 45, 1108–1121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Umotoy J.et al., Rapid and focused maturation of a VRC01-class HIV broadly neutralizing antibody lineage involves both binding and accommodation of the N276-glycan. Immunity 51, 141–154.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sajadi M. M.et al., Identification of near-pan-neutralizing antibodies against HIV-1 by deconvolution of plasma humoral responses. Cell 173, 1783–1795.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu X.et al., Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Victora G. D., Nussenzweig M. C., Germinal centers. Annu. Rev. Immunol. 30, 429–457 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Cyster J. G., Allen C. D. C., B cell responses: Cell interaction dynamics and decisions. Cell 177, 524–540 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crotty S., T follicular helper cell biology: A decade of discovery and diseases. Immunity 50, 1132–1148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corcoran L. M., Tarlinton D. M., Regulation of germinal center responses, memory B cells and plasma cell formation-an update. Curr. Opin. Immunol. 39, 59–67 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Bannard O., Cyster J. G., Germinal centers: Programmed for affinity maturation and antibody diversification. Curr. Opin. Immunol. 45, 21–30 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Victora G. D., Mouquet H., What are the primary limitations in B-cell affinity maturation, and how much affinity maturation can we drive with vaccination? Lessons from the antibody response to HIV-1. Cold Spring Harb. Perspect. Biol. 10, a029389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Havenar-Daughton C.et al., The human naive B cell repertoire contains distinct subclasses for a germline-targeting HIV-1 vaccine immunogen. Sci. Transl. Med. 10, eaat0381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duan H.et al., Glycan masking focuses immune responses to the HIV-1 CD4-binding site and enhances elicitation of VRC01-class precursor antibodies. Immunity 49, 301–311.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Havenar-Daughton C., Abbott R. K., Schief W. R., Crotty S., When designing vaccines, consider the starting material: The human B cell repertoire. Curr. Opin. Immunol. 53, 209–216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Briney B.et al., Tailored immunogens direct affinity maturation toward HIV neutralizing antibodies. Cell 166, 1459–1470.e11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doyle-Cooper C.et al., Immune tolerance negatively regulates B cells in knock-in mice expressing broadly neutralizing HIV antibody 4E10. J. Immunol. 191, 3186–3191 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ota T.et al., B cells from knock-in mice expressing broadly neutralizing HIV antibody b12 carry an innocuous B cell receptor responsive to HIV vaccine candidates. J. Immunol. 191, 3179–3185 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saunders K. O.et al., Targeted selection of HIV-specific antibody mutations by engineering B cell maturation. Science 366, eaay7199 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.D’Souza L., Gupta S. L., Bal V., Rath S., George A., CD73 expression identifies a subset of IgM+ antigen-experienced cells with memory attributes that is T cell and CD40 signalling dependent. Immunology 152, 602–612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weisel F., Shlomchik M., Memory B., Memory B cells of mice and humans. Annu. Rev. Immunol. 35, 255–284 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Angeletti D., Yewdell J. W., Understanding and manipulating viral immunity: Antibody immunodominance enters center stage. Trends Immunol. 39, 549–561 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Sangesland M.et al., Germline-encoded affinity for cognate antigen enables vaccine amplification of a human broadly neutralizing response against influenza virus. Immunity 51, 735–749.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuraoka M.et al., Complex antigens drive permissive clonal selection in germinal centers. Immunity 44, 542–552 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Angeletti D.et al., Defining B cell immunodominance to viruses. Nat. Immunol. 18, 456–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Victora G. D., Wilson P. C., Germinal center selection and the antibody response to influenza. Cell 163, 545–548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Verkoczy L.et al., Rescue of HIV-1 broad neutralizing antibody-expressing B cells in 2F5 VH x VL knockin mice reveals multiple tolerance controls. J. Immunol. 187, 3785–3797 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verkoczy L.et al., Induction of HIV-1 broad neutralizing antibodies in 2F5 knock-in mice: Selection against membrane proximal external region-associated autoreactivity limits T-dependent responses. J. Immunol. 191, 2538–2550 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin Y. C.et al., One-step CRISPR/Cas9 method for the rapid generation of human antibody heavy chain knock-in mice. EMBO J. 37, e99243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jacobsen J. T.et al., One-step generation of monoclonal B cell receptor mice capable of isotype switching and somatic hypermutation. J. Exp. Med. 215, 2686–2695 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kong L.et al., Key gp120 glycans pose roadblocks to the rapid development of VRC01-class antibodies in an HIV-1-infected chinese donor. Immunity 44, 939–950 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
BCR sequences are provided in supplementary information (Dataset S1).