Abstract
Background:
The lack of molecular targets for triple negative breast cancer (TNBC) has limited treatment options and reduced survivorship. Identifying new molecular targets may help improve patient survival and decrease recurrence and metastasis. As DNA repair defects are prevalent in breast cancer, we evaluated the expression and repair capacities of DNA repair proteins in preclinical models.
Methods:
DNA repair capacity was analyzed in four TNBC cell lines, MDA-MB-157 (MDA-157), MDA-MB-231 (MDA-231), MDA-MB-468 (MDA-468), and HCC1806, using fluorescence multiplex host cell reactivation (FM-HCR) assays. Expression of DNA repair genes was analyzed with RNA-seq, and protein expression was evaluated with immunoblot. Responses to the combination of DNA damage response inhibitors and primary chemotherapy drugs doxorubicin or carboplatin were evaluated in the cell lines.
Results:
Defects in base excision and nucleotide excision repair were observed in preclinical TNBC models. Gene expression analysis showed a limited correlation between these defects. Loss in protein expression was a better indicator of these DNA repair defects. Over-expression of PARP1, XRCC1, RPA, DDB1, and ERCC1 was observed in TNBC preclinical models, and likely contributed to altered sensitivity to chemotherapy and DNA damage response (DDR) inhibitors. Improved cell killing was achieved when primary therapy was combined with DDR inhibitors for ATM, ATR, or CHK1.
Conclusion:
Base excision and nucleotide excision repair pathways may offer new molecular targets for TNBC. The functional status of DNA repair pathways should be considered when evaluating new therapies and may improve the targeting for primary and combination therapies with DDR inhibitors.
Keywords: chemotherapy, DNA damage, homologous recombination, nonhomologous end joining, nucleotide excision repair, small molecule inhibitor
Introduction
Treatments for breast cancer have advanced considerably over the past several decades with effective therapies targeting the estrogen receptor (ER), progesterone receptor (PR), and amplification of ERBB2 or HER2. However, triple-negative breast cancer (TNBC) is challenging to treat because of the lack of ER, PR, and amplified HER2, which limits the treatment options to surgical resection and cytotoxic radiotherapy or chemotherapy.1,2 Although some TNBCs respond to these therapies, 50% of patients have a recurrence within 3 years, and 37% of patients die within 5 years after surgery and chemotherapy.3–5 In addition, adverse events of radiotherapy and chemotherapy, such as cardiotoxicity and hepatotoxicity, can limit treatment duration and cause long-term sequelae in survivors.
Further characterization of TNBC tumors and cell lines is needed to improve therapeutic options and outcomes. It is necessary to identify TNBC-specific molecular targets for monotherapy or combination therapy and increase knowledge about the mechanisms of recurrence and therapeutic resistance. With the primary use of DNA damaging radiotherapy and chemotherapy in treating TNBC, the DNA damage response (DDR) and DNA repair pathways are important factors in determining cell fate. When these pathways fail, mutations may occur that promote therapeutic resistance. Although efforts to understand DNA repair defects in breast cancer frequently focus on mutational analysis, no robust associations between mutations and DNA repair defects are known except for mutations in breast cancer susceptibility gene (BRCA1/2) or tumor suppressor protein p53 (TP53).6–8
The major DNA repair pathways, including base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), nonhomologous end joining (NHEJ), and homologous recombination (HR), can have overlapping functions that preserve genomic fidelity, even when defects exist.9 As a result, it may be difficult to predict which drugs may be effective against a particular tumor. Gene expression signatures, coupled with mutational status, may provide additional information about DNA repair defects. However, these measures often fall short. What is needed is functional characterization of DNA repair capacity, which will more accurately identify DNA repair defects that could be targeted in TNBC therapy.
As preclinical cell line models are essential for the development and testing of new therapeutic agents, we examined the expression and repair capacities of DNA repair proteins in four TNBC cell lines. We used the information about repair capacities to evaluate the responses of the TNBC cell lines to DNA-damaging chemicals and assessed combination therapies with inhibitors to DDR proteins to increase cell killing.
Materials and methods
Cell culture
The TBNC cell lines MDA-MB-157 (MDA-157), MDA-MB-231 (MDA-231), MDA-MB-468 (MDA-468), and HCC1806 were purchased (HTB-24, HTB-26, HTB-132, and CRL-2335, American Type Culture Collection, Manassas, VA, USA) within the previous 24 months and passaged < 15 times for all experiments (Table 1). Cells were tested biweekly for mycoplasma contamination (MycoAlert, Lonza, Basel, Switzerland). MDA-157, MDA-231, MDA-468, and MCF10A cells were grown in Dulbecco Modified Eagle Medium (DMEM High Glucose with GlutaMAX, Life Technologies, Carlsbad, CA, USA) and supplemented with 1% sodium pyruvate (Life Technologies) and 10% fetal bovine serum (FBS) (Premium Select, Atlantic Biologicals, Miami, FL, USA). HCC1806 cells were grown in RPMI 1640 medium (Life Technologies) supplemented with 10% FBS. Cells were maintained in a humidified 37°C incubator with 5% carbon dioxide.
Table 1.
TNBC model cell lines used in this study.
| Cell line | Mutations | Subgroup |
|---|---|---|
| HCC1806 | TCF12-A482V | Basal A |
| MDA-MB-157 | FAT4-L4468P; MSH6-R644S | Basal B |
| MDA-MB-231 | BRAF-G4646V; CD79A-C106Y; KRAS-G13D; NF2-E231*; PBRM1-I228V; PDGFRA-Y172F; TP53-R280K | Basal B |
| MDA-MB-468 | CACNA1D-E953D; TP53-R273H, PTEN V85_splice | Basal A |
TNBC, triple-negative breast cancer.
Fluorescence multiplex host cell reactivation
Fluorescence multiplex host cell reactivation (FM-HCR) assays were performed as described previously.10,11 The cells were seeded 48 h before transfection into cell culture flasks (T25, ThermoFisher, Waltham, MA, USA) and collected for transfection at 85% confluence. Cells were electroporated (Neon Transfection System, ThermoFisher) with 2 pulses (20 ms each) at 1200 V. Transfected cells were seeded into 12-well culture plates and collected 24 h after transfection for flow cytometry. Transfection efficiency for each assay was controlled by the inclusion of an undamaged fluorescent reporter plasmid with the repair reporter constructs, as described previously.11 Fluorescent reporter expression was calculated as the percentage of the fluorescent reporter protein expressed versus the undamaged control plasmids from a second transfection.10,11 We normalized fluorescent reporter expression for each assay to the most repair-competent of the four cell lines to facilitate comparisons between cell lines.
Immunoblot
Immunoblot was performed as described previously.12 Cells were grown in 10-cm dishes and cultured to 70–80% confluence. Cells were rinsed with phosphate-buffered saline (PBS), scraped, stored overnight at −80°C, and lysed. Lysates were separated on a 4–15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel (Bio-Rad, Hercules, CA, USA) and transferred to a nitrocellulose membrane. The membrane was probed with antibodies diluted in 5% nonfat dry milk in Tris-buffered saline (VWR, Radnor, PA, USA) and 0.1% Tween 20 (Fisher Scientific, Waltham, MA, USA) and raised against reagents for immunoblot (Table 2). Antibodies were incubated at 4°C overnight on a rocker. The blots were washed and incubated with horseradish peroxidase (HRP)-labeled secondary antibodies (goat anti-rabbit-HRP or goat anti-mouse-HRP) (Cell Signaling Technology) and diluted 1:5000 for 1 h at room temperature on a rocker. HRP antibody target proteins were detected by incubating with an HRP substrate (WesternBright Sirius, Advansta, San Jose, CA, USA). All immunoblots were performed in two or more biological replicates.
Table 2.
Immunoblot reagents and dilutions used in this study.
| Dilution reagent | Source |
|---|---|
| 1:20,000 | |
| β-actin | Invitrogen, Carlsbad, CA, USA |
| 1:1000 | |
| APE1 | Abcam, Cambridge, UK |
| ATM | Cell Signaling, Danvers, MA, USA |
| CHK1 | Cell Signaling |
| CHK2 | Cell Signaling |
| DDB1 | Cell Signaling |
| DNA-PKcs | Cell Signaling |
| ERCC1 | Cell Signaling |
| FEN1 | Abcam |
| GAPDH | Santa Cruz Biotechnology, Dallas, TX, USA |
| Ku70 | Abcam |
| Ligase 4 | Abcam |
| MSH2 | Cell Signaling |
| p-ATM S1981 | Cell Signaling |
| RPA70 | Cell Signaling |
| XPA | Cell Signaling |
| XPD | Cell Signaling |
| XPF | Cell Signaling |
| 1:500 | |
| Ku80 | Cell Signaling |
| Ligase 1 | Novus Biologicals, Littleton, CO, USA |
| MLH1 | Santa Cruz Biotechnology |
| p-CHK1 S345 | Cell Signaling |
| p-CHK2 T68 | Cell Signaling |
| PMS2 | Santa Cruz Biotechnology |
| RAD51 | Santa Cruz Biotechnology |
| UNG | GeneTex, Irvine, CA, USA |
Cytotoxicity
Cytotoxicity was determined with cell growth inhibition assays for DNA-damaging drugs and combination treatment or a cell viability assay (CellTiter-Glo, Promega, Madison, WI, USA) for small molecule inhibitors. For cell growth inhibition assays, MDA-157, MDA-231, and MDA-468 cells (2 × 104 cells per well) and HCC1806 cells (1 × 104 cells per well) were seeded in 12-well dishes and incubated for 48 h. Cells were treated with carboplatin (Sigma-Aldrich, St. Louis, MO, USA) or doxorubicin (Selleck Chemicals, Houston, TX, USA) diluted to 10 mM in dimethyl sulfoxide (DMSO) and further diluted to the experimental concentrations in the growth medium. The cells were exposed to DMSO vehicle, carboplatin, or doxorubicin continuously for 5 days and counted with an automated cell counter (TC20, Bio-Rad).
For coexposure experiments, MDA-231 cells were exposed to DMSO vehicle control, doxorubicin, and 10 μM KU-55933 (ATM inhibitor, ATMi), 1 μM AZD6738 (ATR inhibitor, ATRi), 5 μM NU7026 (DNAPK inhibitor, DNAPKi), or 10 nM prexasertib (LY2606368, Selleck Chemicals). Cells were coexposed continuously for 5 days and counted. Results were normalized to values for cells exposed to DMSO vehicle or medium control and graphed to generate values of half-maximal inhibitory concentration (IC50) using software (Prism, GraphPad, San Diego, CA, USA).
For the small molecule inhibitor viability assays (CellTiter-Glo), MDA-231 and HCC1806 cells (2 × 103 cells per well) and MDA-157 and MDA-468 cells (5 × 103 cells per well) were seeded in white, clear bottom 96-well plates and cultured for 48 h. Cells were treated with ATMi, ATRi, NU7026, or prexasertib diluted to 10 mM in DMSO and further diluted to the experimental concentrations in the growth medium. The cells were exposed to DMSO vehicle, ATMi, ATRi, or prexasertib for 4 days. The assay reagent (CellTiter-Glo) was added to plates and incubated according to instructions from the manufacturer. Luminescence was read on a multimodal plate reader (M1000, Tecan, Männedorf, Switzerland). Results were normalized to values for cells exposed to DMSO vehicle control and graphed to generate IC50 values. All growth inhibition and viability assays were performed with technical triplicates over three biological replicates.
RNA-seq
Cell pellets containing 106 cells were sent to a genomics service (Genewiz, South Plainfield, NJ, USA) for library preparation, RNA sequencing, and analysis. Heatmaps were created for transcripts per million (TPM) from the analyzed samples. TPM values were calculated by normalizing to gene length for comparisons between samples.
Statistical analysis
Mean IC50 values ± standard error of the mean (SEM) were determined from replicate experiments. The fluorescent signal from the repair of damaged and undamaged substrates was quantitated in each triplicate and reported as mean ± SEM. Cell line values were evaluated with 1-way analysis of variance (ANOVA), and means were compared with Dunnett’s post hoc test. Statistical significance was defined by *p < 0.05, **p < 0.01, and ***p < 0.001.
Results
Characterization of DNA repair defects in TNBC cell lines
We previously used the flow cytometric host cell reactivation assay (FM-HCR) to characterize the BER repair capacity of a panel of commonly used TNBC cell line models, MDA-157, MDA-231, HCC1806, and MDA-468.13 From this analysis, we observed that MDA-468 cells were the most BER competent of the four cell lines.13 MDA-157 cells were the next most BER competent, with only a slight defect in the repair of 8-oxo-2′-deoxyguanosine (8-oxo-dG) opposite cytosine recognized by the oxidative glycosylases OGG1, NEIL1, or NEIL2. HCC1806 cells showed defects in BER catalyzed by AAG (also known as MPG) glycosylase, MUTYH glycosylase, and the oxidative glycosylases. MDA-231 cells were the least competent in BER with major defects in repair initiated by AAG glycosylase, MUTYH glycosylase, the oxidative glycosylases, and UNG glycosylase.
The previous BER analysis primarily evaluated lesion recognition by DNA glycosylases. Although glycosylase activity determines the rate at which BER is initiated, apurinic/apyrimidinic endonuclease 1 (APE1) also is important in BER initiation.14 To complete the analysis of BER defects in the TNBC panel, we examined the repair of tetrahydrofuran (THF), which is recognized by APE1 and processed by long patch BER, using FM-HCR. A THF-opposite-cytosine (THF:C) base pair was incorporated into the coding sequence of the fluorescent reporter gene, blocking transcription and expression of the reporter. When APE1 recognizes and nicks the DNA at the THF site, DNA polymerase β (POLβ) dRP-lyase activity may remove the abasic site, but reduced abasic sites also may be processed by long patch BER repair.15,16 After the DNA is repaired, transcription proceeds and fluorescent reporter expression is detected. The percentage recovery of the fluorescence signal from the damaged versus an undamaged plasmid provides the efficiency of repair within the cell.11
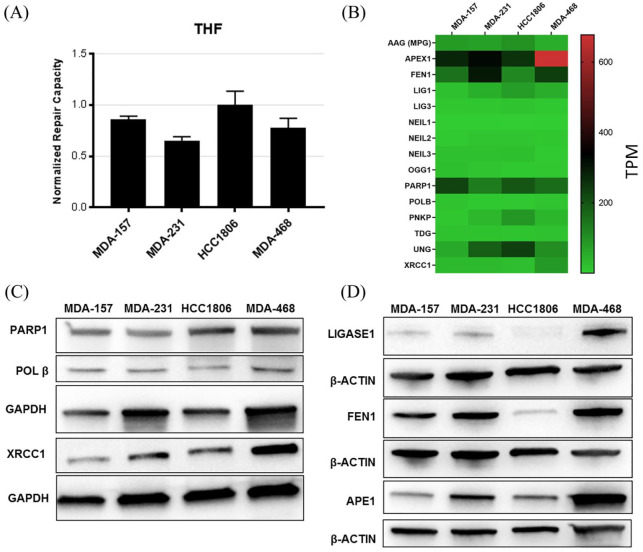
Evaluation of the efficiency of THF repair by APE1 between the TNBC cell lines showed efficient processing of THF lesions in all TNBC cell lines, unlike the glycosylase substrates (Figure 1).13 The highest level of expression for the THF reporter was observed in HCC1806 cells. MDA157 and HCC1806 also showed the highest levels of THF activity and lowest amounts of APE1 at the protein and mRNA levels in the cell lines (Figure 1).
Figure 1.
Key factors involved in BER. (A) Abasic site repair measured by FM-HCR in the TNBC cell lines. (B) Heatmap of mRNA expression (TPM) of key BER proteins in the TNBC cell lines. Row scaled TPM shown in Supplemental Figure S1. (C, D) Immunoblot confirming expression of BER proteins in the cell lines.19 Loading controls GAPDH or β-actin are shown for each blot.
BER, base excision repair; FM-HCR, fluorescence multiplex host cell reactivation; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; TNBC, triple negative breast cancer; TPM, transcripts per million.
Except for MDA-468, the TNBC cell lines showed defects in BER that correlated with dysregulation of BER gene and protein expression (Figure 1 and Supplemental Figure S1). MDA-468 showed high expression levels of BER factors, including APE1, FEN1, PARP1, POLβ, and XRCC1, consistent with the high BER capacity and resistance to alkylating DNA damage observed previously.13 Low oxidative DNA damage repair capacity was reported previously for several TNBC cell lines, including the four cell lines in this study, consistent with the low expression of OGG1 and NEIL family proteins in these cell lines.17
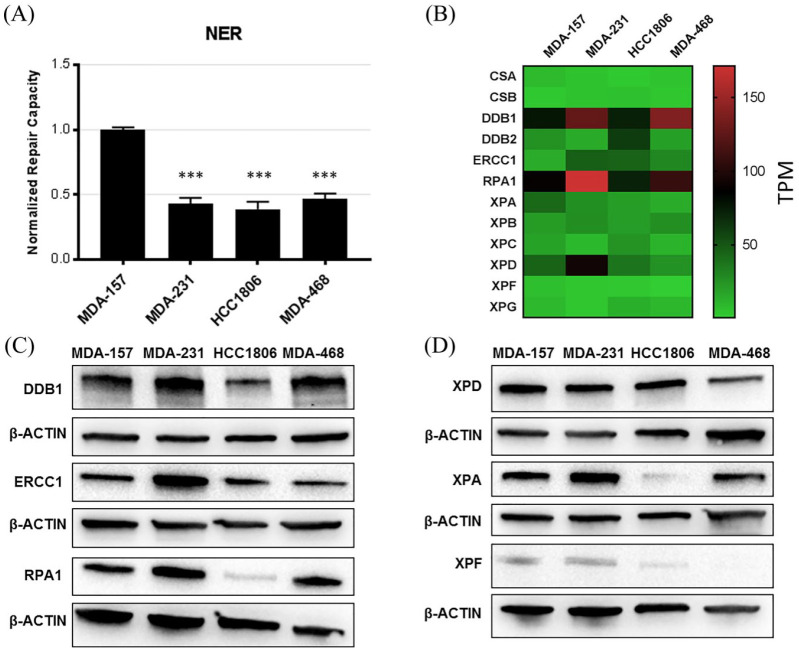
We next evaluated repair by the NER pathway. NER substrates were generated by exposing a fluorescent reporter plasmid to ultraviolet (UV) radiation to induce helix distorting bulky photoproducts. Repair by NER machinery removes transcription blocking lesions and restores fluorescent reporter expression, and NER proteins XPA-XPG are involved in damage recognition, unwinding, and excision.18 Evaluation of repair by the NER pathway showed that MDA-231, HCC1806, and MDA-468 had significantly lower NER capacity than MDA-157 cells (Figure 2). The mRNA expression of XPA-XPG and other accessory proteins did not show any specific expression signature that could explain this finding (Figure 2 and Supplemental Figure S2). However, protein expression was variable among the cell lines tested, which differed from mRNA expression observed in critical NER proteins (Figure 2C and D and Supplemental Figure S2). HCC1806 cells had markedly lower levels of ERCC1, RPA, XPA, and XPF than the other cell lines (Figure 2C and D and Supplemental Figure S2) that would affect multiple steps in NER lesion processing and repair.19 MDA-468 also showed deficits in XPF protein levels. MDA-231 cells did not show expression changes in these proteins but had a defect in NER (Figure 2A). MDA-231 cells had low gene expression of XPC, XPG, and DDB2, which was not observed in the other TNBC cells, and which may contribute to the NER defect (Figure 2B). Evaluation of these NER protein levels was not feasible because the antibodies were not sufficiently specific. Elevated expression of other NER factors DDB1, ERCC1, RPA, and XPA was observed in MDA-231 versus the other cell lines. MDA-157 and MDA-468 cells also showed elevated DDB1 expression.
Figure 2.
Key factors involved in NER. (A) NER substrate repair measured by FM-HCR in the TNBC cell lines. (B) Heatmap of mRNA expression (TPM) of key NER proteins in the TNBC cell lines. Row scaled TPM shown in Supplemental Figure S2. (C, D) Immunoblot confirming highly expressed BER proteins in the cell lines. ***p < 0.001 compared with MDA-157 cells. Loading control is shown for each blot.
FM-HCR, fluorescence multiplex host cell reactivation; NER, nucleotide excision repair; TNBC, triple negative breast cancer; TPM, transcripts per million.
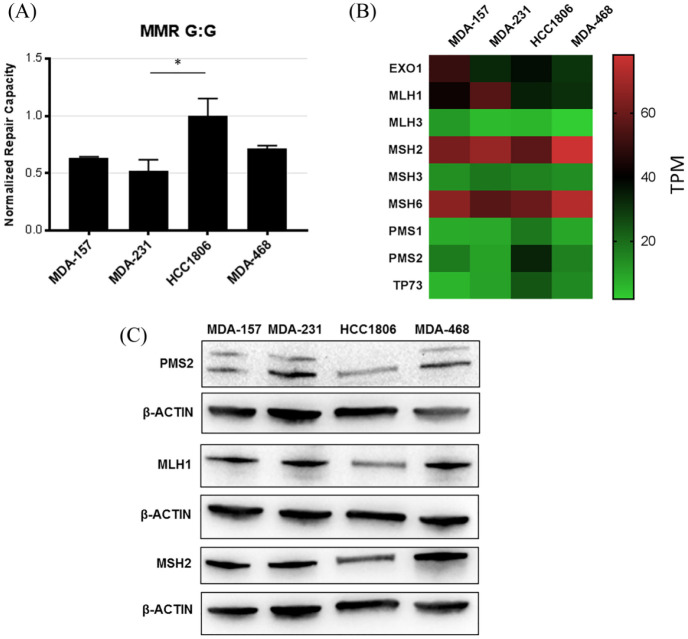
We evaluated MMR with a G:G mismatch-containing plasmid that expresses a nonfluorescent mutant protein until the wild type cytosine in the transcribed strand is restored by MMR.11 All TNBC cell lines showed MMR activity, highest in HCC1806 cells (Figure 3A). The mRNA levels of the MMR proteins were similar between cell lines (Figure 3B and Supplemental Figure S3), with higher levels of gene and protein expression levels for MLH1, MSH2, and MSH6 relative to the other MMR proteins (Figure 3B and C).
Figure 3.
Key factors involved in MMR. (A) MMR activity measured by FM-HCR in the TNBC cell lines. (B) Heatmap of mRNA expression (TPM) of key MMR proteins in the TNBC cell lines. Row scaled TPM shown in Supplemental Figure S3. (C) Immunoblot confirming highly expressed MMR proteins in the cell lines. Loading control is shown for each blot. *p < 0.05 compared with HCC1806 cells.
FM-HCR, fluorescence multiplex host cell reactivation; MMR, mismatch repair; TNBC, triple negative breast cancer; TPM, transcripts per million.
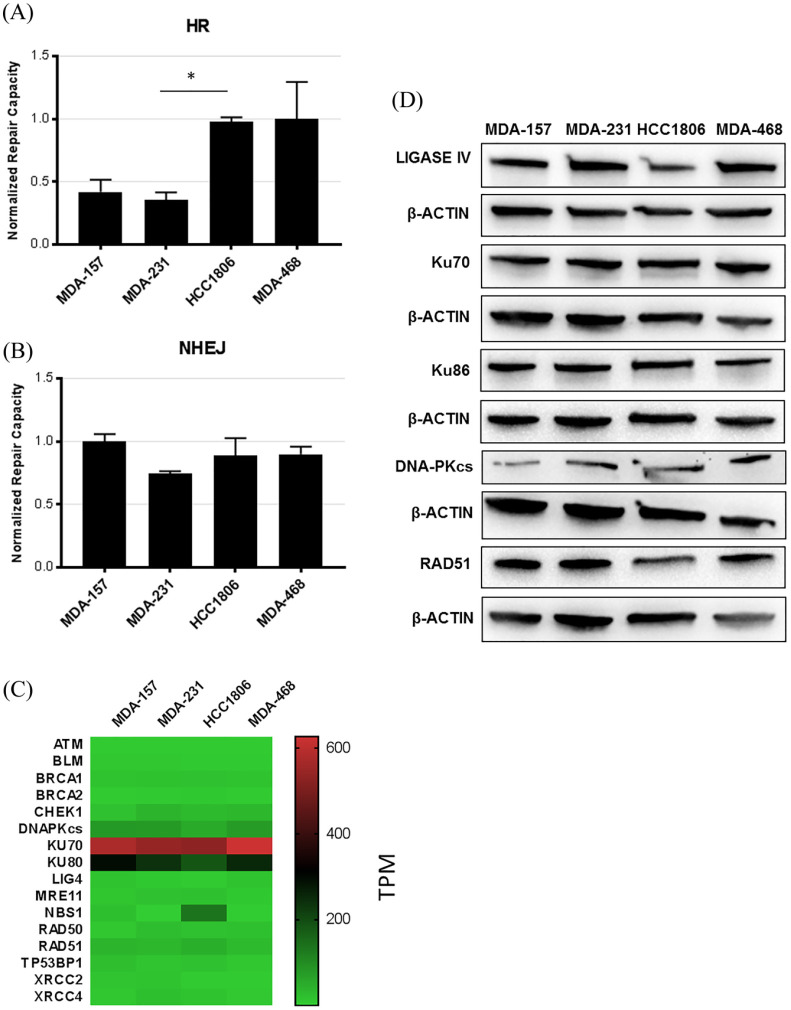
We measured the double-strand break repair (DSBR) capacity of the TNBC cell lines using HR and NHEJ substrates. HR was measured using a two-plasmid system that contains a truncated green fluoresence protein (GFP) reporter plasmid that requires a homologous donor sequence from the second donor plasmid; this direct ligation does not produce GFP. Successful HR events result in the restoration of the 5′ WT GFP sequence in the truncated plasmid to generate a functional GFP protein.20 To measure NHEJ, a BFP plasmid is cut within the promoter region using the ScaI restriction enzyme to linearize the plasmid. Accurate DSBR restores the promoter region and drives transcription of the reporter.11,21
FM-HCR showed that the HR capacity varied between TNBC cell lines. HCC1806 and MDA-468 cells had higher levels, and MDA-157 and MDA-231 had lower levels of HR (Figure 4 and Supplemental Figure S4). HR defects commonly result from mutations in BRCA1/2. The four TNBC cell lines are wild type for BRCA1/2.22–24 However, MDA-157 and MDA-231 cells have allelic loss in BRCA1 and low levels of protein expression.13,22 HCC1806 and MDA-468 cells also showed allelic loss of BRCA1 and low levels of BRCA1 mRNA (Figure 4) but high protein.13,22 The NHEJ repair capacity was similar, with no significant differences in repair between the four TNBC cell lines.
Figure 4.
Key factors involved in DSBR. (A) HR activity measured by FM-HCR in the TNBC cell lines. (B) NHEJ activity measured by FM-HCR in the TNBC cell lines. (C) Heatmap of mRNA expression (TPM) of key HR and NHEJ proteins in the TNBC cell lines. Row scale heat map is shown in Supplemental Figure S4. (D) Immunoblot confirming highly expressed HR and NHEJ proteins in the cell lines. Loading control is shown for each blot. *p < 0.05 compared with HCC1806 cells.
DSBR, double-strand break repair; FM-HCR, fluorescence multiplex host cell reactivation; HR, homologous recombination; MMR, mismatch repair; NHEJ, nonhomologous end joining; TNBC, triple negative breast cancer; TPM, transcripts per million.
In sum, MDA-157 was the most proficient of the four cell lines in DNA repair, with only subtle defects in BER and HR. MDA-231 cells were the least competent of the four cell lines, with lower repair capacity observed in most pathways. MDA-468 had the highest BER capacity, competent NHEJ and HR, and lower NER. HCC1806 cells had lower NER capacity and mixed defects in BER. HCC1806 cells were competent in excision of uracil, primarily performed by UNG, and repair of the abasic site analog THF. HCC1806 cells also were competent in both DSBR pathways.
Drug sensitivity in TNBC cell lines
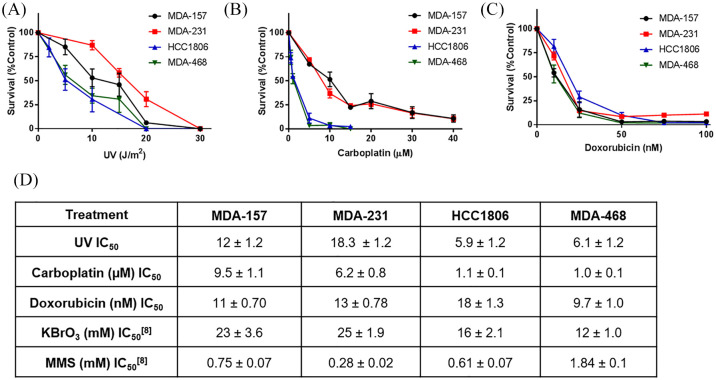
With the observed DNA repair defects in BER and NER, we examined the cytotoxicity of UV radiation and three DNA damaging chemotherapy drugs used to treat TNBC that induce DNA lesions processed by these pathways (Figure 5). Despite being deficient for NER, which is the pathway responsible for repairing UV-induced DNA damage, MDA-231 cells were the most resistant to UV radiation (254 nm). MDA-157 cells also had resistance, consistent with their robust NER capacity. HCC1806 and MDA-468 cells had similar sensitivity to UV damage, consistent with their low NER capacity (Figure 2). Similarly, MDA-157 and MDA-231 cells had less sensitivity to carboplatin than MDA-468 and HCC1806 cells (Figure 5). All four TNBC cell lines show similar sensitivity to doxorubicin, with only small variations in IC50.
Figure 5.
TNBC cell line sensitivity to DNA damaging agents measured by growth inhibition. (A) Cells exposed to UV-C radiation (wavelength, 254 nm). (B) Cells continuously exposed to carboplatin. (C) Cells continuously exposed doxorubicin. (D) IC50 values for DNA damaging agents. KBrO3 and MMS were previously reported.8
MMS, methyl methanesulfonate; TNBC, triple negative breast cancer.
Increasing cytotoxicity of DNA damaging agents with small molecular inhibitors to DDR proteins
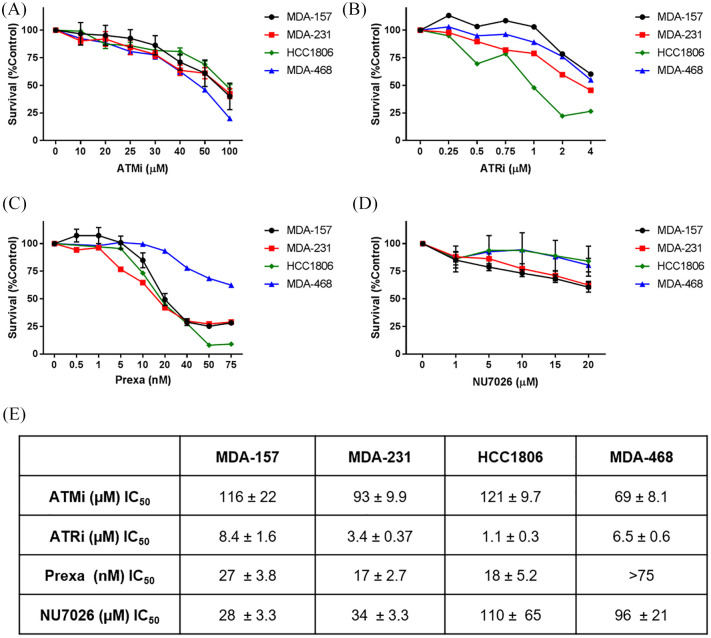
The spectrum of DNA repair defects in the TNBC cell lines and varied responses to DNA damaging agents suggested that regulatory proteins for DDR may be good molecular targets for TNBC. As the major DNA repair pathways in the TNBC cell lines are regulated by ATM, ATR, DNA-PK, and CHK1, we examined the sensitivity of the TNBC cell lines to inhibitors of these proteins (Figure 6).25 We verified there was phosphorylation activity for these DDR proteins to ensure that no defect in protein expression or activation was present (Supplemental Figure S5).
Figure 6.
TNBC cell line sensitivity to DDR small molecule inhibitors. (A) Cells continuously exposed to ATM inhibitor KU-55933 (ATMi). (B) Cells continuously exposed to ATR inhibitor AZD6738 (ATRi). (C) Cells continuously exposed to CHK1/2 inhibitor prexasertib (LY2606368). (D) Cells continuously exposed to DNA-PK inhibitor NU7062. (E) IC50 values for DDR inhibitors.
DDR, DNA damage response; TNBC, triple negative breast cancer.
Cytotoxicity was low when TNBC cells were exposed to ATMi. ATM predominantly senses double-strand breaks, and deficiency in ATM induces HR defects in cells.26 Both MDA-157 and MDA-231 cells showed defects in HR (Figure 4), but only MDA-468 cells showed sensitivity to ATMi (IC50, 69 ± 8 µM).
ATR recognizes single-strand breaks, and also is activated by ATM in the presence of double strand breaks.25 Inhibition with ATRi induced cytotoxicity at low micromolar concentrations in all four TNBC cell lines, but the sensitivity between the cell lines varied (Figure 6B and E). MDA-157 and MDA-468 cells were more resistant to ATR inhibition, consistent with repair competencies in BER and NER. The greatest sensitivity to ATR inhibition was observed in HCC1806 (IC50, 1.1 μM) and MDA-231 cells (IC50, 3.4 μM), and these cell lines contained more defects in BER and NER pathways (Figures 1 and 2).13
ATR and CHK1 stabilize and protect the replication fork under replication stress induced by oncogenes or DNA damage. We examined the use of CHK1 inhibitor prexasertib in the TNBC cell lines. CHK1 inhibition was effective at nanomolar concentrations for MDA-157, MDA-231, and HCC1806 cells. The greatest sensitivity to prexasertib was observed in HCC1806 (IC50, 17 nM) and MDA-231 cells (IC50, 18 nM), but MDA-468 cells were resistant.
MDA-157 and MDA-231 cells were sensitive to DNAPKi and had lower levels of DNA-PKcs than the other TNBC cell lines (Figure 4D and Supplemental Figure S4), consistent with the greater reliance of MDA-157 and MDA-231 cells on NHEJ because of their HR defects.
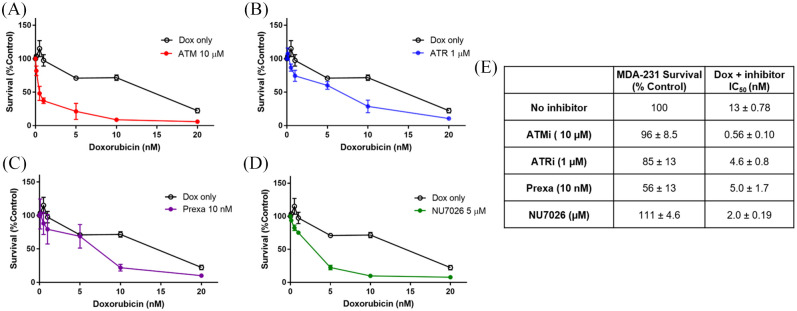
DDR inhibitors also can be used as potent sensitizers to DNA damaging agents, and their effectiveness is often markedly increased by the presence of DNA repair defects within the cell, as shown by the synthetic lethality of PARP in HR-defective cell lines.9,27–29 As MDA-231 cells showed the least efficient DNA repair capacity in several pathways and the most resistance in response to DNA damaging agents, we examined the combination of DDR inhibitors with doxorubicin in MDA-231 cells (Figure 7). Fixed doses of the DDR inhibitors were selected based on cytotoxicity, and the results of previous studies examining coexposures.30,31 All four inhibitors increased cell killing by doxorubicin when used in combination. Prexasertib, DNAPKi, and ATRi increased cell killing by doxorubicin. However, ATMi showed the highest increase in cell killing by doxorubicin (23-fold increase), consistent with the impaired repair of doxorubicin-induced double-strand breaks.
Figure 7.
Continuous coexposure of DDR inhibitors with doxorubicin in MDA-231 cells.
(A) 10 µM ATMi. (B) 1 µM ATRi. (C) 10 nM prexasertib. (D) 5 µM NU7026. (E) Survival of MDA-231 exposed to DDR inhibitors at their fixed doses, and IC50 values for coexposure.
DDR, DNA damage response.
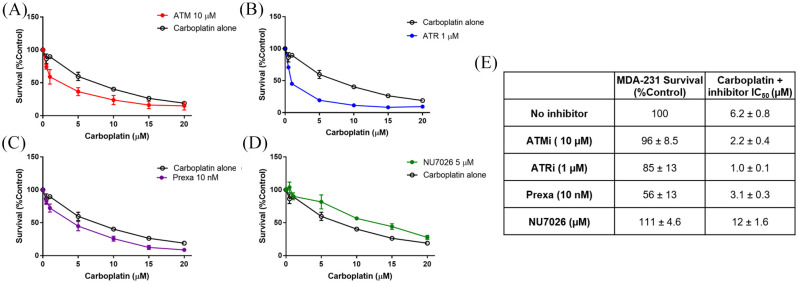
We tested the ability to increase cell killing by combining DDR inhibitors with carboplatin (Figure 8). All inhibitors except DNAPKi increased the sensitivity of MDA-231 cells to carboplatin, but the increase in cell killing was modest. ATRi showed the most marked change in sensitivity, with a six-fold increase in cell killing. As NHEJ is not important in the repair of platinum adducts, the lack of sensitization by DNAPKi was expected.
Figure 8.
Continuous coexposure of DDR inhibitors with carboplatin in MDA-231 cells. (A) 10 µM ATMi. (B) 1 µM ATRi. (C) 10 nM prexasertib. (D) 5 µM NU7026. (E) Survival of MDA-231 exposed to DDR inhibitors at their fixed doses, and IC50 values for coexposure.
DDR, DNA damage response.
Discussion
Gene expression profiles do not fully capture DNA repair defects in cancer cells. Discordance between gene and protein expression patterns results from post-translational modifications, epigenetic modifications, genetic mutations, and many other factors. The variety and complexity of these changes in tumor cells makes assessing DNA repair efficiency challenging without functional measures. The functional characterization of DNA repair pathways using the FM-HCR showed that preclinical models of TNBC have a spectrum of DNA repair defects that may offer new therapeutic targets for the treatment of TNBC.
In BER, defects in the glycosylase-driven repair of DNA lesions were observed previously and correspond to the low gene expression of the different DNA glycosylases, POL β and DNA ligases in the TNBC cells lines (Figure 1).13,17 These BER defects also increase the sensitivity of the cell lines to methyl methanesulfonate (MMS) and potassium bromate (KBrO3), which induce alkylation and oxidative DNA damage addressed by DNA glycosylases (Figure 5). Competent repair of the THF abasic site mimic was achieved by all of the cell lines and is supported by the high expression of APE1, PARP1, and XRCC1 across the cell line panel (Figure 1).13,17 In general, mRNA levels of BER proteins lacked correlation with protein levels, and the functional assessment of BER did not track with any single DNA repair protein or transcript.
Target BER deficiency in TNBC and other breast cancers has been promoted through the use of PARP inhibitors and combination therapies with PARP inhibitors.32,33 PARP inhibitors directly impact BER because they reduce PARP1 signaling and the coordination of BER proteins at single-strand breaks. The high gene and protein expression of PARP1 and XRCC1 in the cell line panel is consistent with TCGA datasets and findings from the I-SPY trials, where PARP inhibitor monotherapy has only shown modest efficacy.13,34 The functional analysis of BER substrates indicates that BER deficiency in TNBC is significantly linked to oxidative or alkylating glycosylases through low protein production.13,17 Therefore, combining PARP inhibitors with therapies that generate reactive oxygen species and strain the defective glycosylase activities would offer more targeted therapies for TNBC.
Similar to BER, discordance between expression and function was also observed in the NER analysis (Figure 2). HCC1806 and MDA-468 cells showed protein and gene expression loss of critical proteins in the recognition and excision of bulky base lesions ERCC1, XPA, and XPF (Figure 2C and D). Loss of XPA, which is an essential scaffold protein in NER, causes severe sensitivity to UV radiation and a high risk of carcinogenesis.19 The ERCC1-XPF complex is responsible for DNA incision. Expression levels of XPF have been used to functionally characterize NER in breast cancer cell lines and patient samples.35 NER deficiency was associated with epigenetic silencing of XPF (ERCC4) and decreased expression of this NER nuclease, which is consistent with the results shown for HCC1806 and MDA-468 cells (Figure 2C and D). However, the NER defect in MDA-231 cells did not show these expression changes (Figure 2). There is no clear driver of the NER defect in this cell line because the observed gene expression levels were low for XPC, XPG, and DDB2 and elevated for DDB1, ERCC1, RPA, and XPA.
The functional analysis indicates that NER, like BER, is a potential therapeutic target in TNBC that has not been thoroughly investigated. ERCC1 and XPF have been suggested as biomarkers for therapeutic responses to crosslinking agents like cisplatin or irinotecan, but specific inhibitors for NER proteins are still being developed.35–37 One recent report used RNAi and spironolactone to inhibit XPB to induce sensitivity to alkylating agents and overcome alkylating agent resistance in multiple myeloma.38 Triptolide also inhibits the ATPase activity of XPB and has been shown to sensitize breast cancer cell lines to doxorubicin and cisplatin.39–41 Spironolactone and triptolide have other mechanisms of action and off-target effects that may make them unsuitable for TNBC treatment long term.42–44 However, these studies demonstrate that exploiting NER defects in TNBC could offer new targets and combination therapies for TNBC.
Although defects in BER and NER were observed, MMR and NHEJ repair pathways were functional. MMR proteins were slightly overexpressed in all four cell lines and readily repaired the G:G mismatched substrate (Figure 3). This result is consistent with the low level of microsatellite instability observed in breast cancers, including TNBC.45,46 NHEJ function and mRNA and protein expression of NHEJ proteins also were consistent between the cell lines, with only MDA-231 cells showing a slightly lower capacity for the NHEJ substrate (Figure 4). We observed high levels of mRNA expression for the Ku heterodimer, but the protein level of Ku was not overexpressed. NHEJ can compensate, in part, for defects in BER and HR, therefore, when challenges to BER or HR occur, the use of the error-susceptible BER pathway by the TNBC cell lines to survive may drive mutagenesis and chromosomal aberrations.47,48
Defects in HR are commonly associated with TNBC. Approximately 80% of breast cancers with germline mutations in BRCA1/2 are TNBC.49 Germline and sporadic mutations in BRCA1/2 only account for ~15% of all TNBCs cases.50–52 However, HR defects or BRCAness occurs in ~50% of TNBC tumors.53–55 While the selected TNBC cell lines are wild-type for BRCA1, we still observed functional defects in HR in the TNBC cell lines. MDA-157 and MDA-231 cells had low HR, consistent with their allelic loss in BRCA1.22 HCC1806 and MDA-468 cells showed higher levels of HR and HR-related gene expression (Figure 4), consistent with the high protein expression of BRCA1 observed previously.13 As no other deficits in HR gene or protein expression were observed in the four cell lines, the BRCA1 protein level likely drives the observed HR defects in MDA-157 and MDA-231 cells.
Exploiting HR defects in breast cancer has been extensively investigated through the use of PARP inhibitors.56–59 While most effective when used in combination with BRCA1/2 mutations, PARP inhibitors have shown efficacy in the treatment of TNBC without BRCA1/2 mutations and in combination with carboplatin.58 Resistance to PARP inhibitors is a growing concern with increased efflux of the drug, mutations in PARP1, and restoration of HR proficiency or replication fork stability all observed in resistant tumors.60
With replication fork stability playing a critical role in HR defects and PARP inhibitor sensitivity, inhibitors of DNA damage response and cell cycle checkpoint proteins ATM, ATR, and CHK1 have naturally emerged as targeted therapies for HR and other DNA repair-deficient cell lines.58,60–62 These inhibitors may also restore sensitivity to PARP inhibitors by destabilizing the replication fork and causing premature entry into mitosis.60 The strong dysregulation of DNA repair proteins in the four cell lines supports the targeting of DNA repair defects in TNBC with these inhibitors. However, as shown in Figure 5, monotherapy alone was sufficient to overcome the compensatory repair mechanisms from dysregulated repair in these cell lines. The cells were insensitive to ATM inhibition, which compromises HR and DNAPK inhibition, which compromise NHEJ (Figure 5 and Supplemental Figure 5). ATR inhibition was slightly more successful than ATM and DNAPK inhibition, with some sensitivity observed. CHK1 inhibition by prexasertib had the best efficacy as a monotherapy in the TNBC cells except for MDA-468 cells, which showed resistance. Resistance to prexasertib was observed previously for HCT116 and PANC-1 cells, but the mechanisms underlying resistance are not understood.63
Cross-talk between repair pathways may impair targeted cell death by DNA damaging agents and decrease the efficacy of DDR inhibitors. We observed mRNA and protein expression level differences in proteins that have cross talk between DNA repair pathways. PARP1, XRCC1, and DDB1 were highly expressed in these cell lines and have multiple roles in different DNA repair pathways. PARP1 is involved in BER, single-strand break repair (SSBR), HR, and alternative NHEJ (a-NHEJ) or microhomology-mediated end joining (MMEJ).64 XRCC1 also is involved in BER, SSBR, NER, and a-NHEJ.65,66 UV-DBB (DDB1/DDB2) is linked to BER because it stimulates OGG1 and APE1 activities.67 MDA-157 and MDA-468 cells have the highest competency at BER, which may be stimulated by increased protein levels of DDB2.13 DDB1 also interacts with the E3-ubiquitin ligase Cul4A and functions in cell cycle regulation and replication.68,69
The overexpression of these proteins may explain the observed divergence in sensitivity to DNA damaging agents and the insensitivity to DDR inhibitors (Figure 5). Despite having a clear defect in NER, MDA-231 cells were more resistant to UV-C- and cisplatin-induced DNA damage than HCC1806 and MDA-468 cells (Figure 2). These lesions are repaired primarily by NER and interstrand crosslink (ICL) repair, but BER, NHEJ, and HR also may address these lesions.70,71 We did not evaluate the repair capacity for ICL, but mRNA expression analysis of Fanconi anemia proteins involved in ICL did not show an mRNA gene expression loss or an overexpression pattern that would explain the resistance of MDA-231 cells (Supplemental Figure S5). However, the high expression of ERCC1, RPA, and XRCC1 may markedly contribute to the resistance of MDA-231 cells, despite the observed deficiency in BER and NER (Figures 1 and 2). Overexpression of ERCC1 is associated with increased resistance to topoisomerase poisons and cisplatin.37,72 Overexpression of RPA also is observed in several cancers, and correlates with decreased cisplatin sensitivity and stimulation of NER.73 Overexpression of RPA may also stabilize the replication fork, promoting resistance to the DDR inhibitors. Additionally, the presence of the R280K p53 mutation in MDA-231 cells, which disrupts DNA binding and leads to alterations in p53-mediated transcriptional activation, favoring proliferation and reduced cell cycle arrest, would favor survival and contribute to the resistance of MDA-231 cells to DNA damaging agents and DDR inhibitors.74
Combining DDR inhibitors with DNA damaging therapy was more effective at inducing cell death and overcoming potential cross talk between DNA repair pathways. MDA-231 cells showed significantly increased doxorubicin sensitivity when combined with ATM inhibition (Figure 7). Combinations of carboplatin with ATM, ATR, and CHK1 inhibitors also showed increased cell killing in MDA-231. The present results suggest that combination therapies need to be targeted to DNA repair defects, account for overexpression of cell cycle regulators, and may depend on the mutational status of p53.
Although the functional characterization of DNA repair pathways in clinical tumors is technically challenging and unfeasible at present, the functional characterization of preclinical tumor models is achievable and provides insight into DDR proteins and DNA repair defects that can be considered as potential targets for therapeutic intervention. Molecular targets such as RPA, DDB1, or XRCC1 should be reevaluated as biomarkers for resistance and potentially targeted to increase the sensitivity to DNA damaging agents and DDR inhibitors.68,69,75–77 Furthermore, the characterization of DNA repair defects in preclinical models and patients may provide ways to improve the effectiveness of immune checkpoint blockade. The interactions between genomic instability and immune response have been reported previously but are poorly understood.78,79 The present study highlights the potential importance of DNA repair in preclinical models toward improving stratification and combination therapy selection for breast cancer patients.
Conclusion
In summary, we evaluated DNA repair defects that may influence therapeutic responses in four TNBC cell lines. Based on the response of the TNBC cell lines to DNA damaging chemicals and experiments with small molecule inhibitors to DDR proteins as monotherapies and combination therapies, we suggest that knowledge about DNA repair defects potentially can be used to overcome resistance to therapy. Further evaluation of DNA repair defects is justified toward using them as therapeutic targets in TNBC.
Supplemental Material
Supplemental material, Supplemental_Figure1 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure2 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure3 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure4 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure6 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure_10 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure_11 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure_12 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure_5 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure_7 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure_8 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure_9 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Acknowledgments
The authors thank John V. Marymont, Mary I. Townsley, and Elly Trepman for editorial support.
Footnotes
Conflict of interest statement: The authors declare that there is no conflict of interest.
Ethics Statement: No human or animal subjects were used in this work, so ethical approval was not required.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by an Innovation Award from the Breast Cancer Research Foundation of Alabama to NRG and P01CA092584 from the National Institutes of Health, National Cancer Institute to ZDN. Editing was provided by the Dean’s Office, University of South Alabama College of Medicine.
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Kevin J. Lee, College of Medicine, Mitchell Cancer Institute, University of South Alabama, Mobile, AL, USA
Elise Mann, College of Medicine, University of South Alabama, Mobile, AL, USA.
Griffin Wright, College of Medicine, Mitchell Cancer Institute, University of South Alabama, Mobile, AL, USA.
Cortt G. Piett, Harvard T.H. Chan School of Public Health, Harvard University, Boston, MA, USA
Zachary D. Nagel, Harvard T.H. Chan School of Public Health, Harvard University, Boston, MA, USA
Natalie R. Gassman, Mitchell Cancer Institute, University of South Alabama, 1660 Springhill Avenue, Mobile, AL 36607, USA; University of South Alabama College of Medicine, 307 N. University Blvd, Mobile, AL 36688, USA.
References
- 1. Pareja F, Geyer FC, Marchio C, et al. Triple-negative breast cancer: the importance of molecular and histologic subtyping, and recognition of low-grade variants. NPJ Breast Cancer 2016; 2: 16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med 2010; 363: 1938–1948. [DOI] [PubMed] [Google Scholar]
- 3. Liedtke C, Mazouni C, Hess KR, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol 2008; 26: 1275–1281. [DOI] [PubMed] [Google Scholar]
- 4. Guarneri V, Broglio K, Kau SW, et al. Prognostic value of pathologic complete response after primary chemotherapy in relation to hormone receptor status and other factors. J Clin Oncol 2006; 24: 1037–1044. [DOI] [PubMed] [Google Scholar]
- 5. Rimawi MF, Shetty PB, Weiss HL, et al. Epidermal growth factor receptor expression in breast cancer association with biologic phenotype and clinical outcomes. Cancer 2010; 116: 1234–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ellis MJ. Mutational analysis of breast cancer: guiding personalized treatments. Breast 2013; 22(Suppl. 2): S19–S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lim LY, Vidnovic N, Ellisen LW, et al. Mutant p53 mediates survival of breast cancer cells. Br J Cancer 2009; 101: 1606–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pirie A, Guo Q, Kraft P, et al. Common germline polymorphisms associated with breast cancer-specific survival. Breast Cancer Res 2015; 17: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nickoloff JA, Jones D, Lee SH, et al. Drugging the cancers addicted to DNA repair. J Natl Cancer Inst 2017; 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chaim IA, Nagel ZD, Jordan JJ, et al. In vivo measurements of interindividual differences in DNA glycosylases and APE1 activities. Proc Natl Acad Sci U S A 2017; 114: E10379–E10388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nagel ZD, Margulies CM, Chaim IA, et al. Multiplexed DNA repair assays for multiple lesions and multiple doses via transcription inhibition and transcriptional mutagenesis. Proc Natl Acad Sci U S A 2014; 111: E1823–E1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sonavane M, Sykora P, Andrews JF, et al. Camptothecin efficacy to poison top1 is altered by bisphenol A in mouse embryonic fibroblasts. Chem Res Toxicol 2018; 31: 510–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee KJ, Piett CG, Andrews JF, et al. Defective base excision repair in the response to DNA damaging agents in triple negative breast cancer. PLoS One 2019; 14: e0223725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat Struct Biol 2000; 7: 176–178. [DOI] [PubMed] [Google Scholar]
- 15. Wei W, Englander EW. DNA polymerase beta-catalyzed-PCNA independent long patch base excision repair synthesis: a mechanism for repair of oxidatively damaged DNA ends in post-mitotic brain. J Neurochem 2008; 107: 734–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J 1997; 16: 3341–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alli E, Sharma VB, Sunderesakumar P, et al. Defective repair of oxidative dna damage in triple-negative breast cancer confers sensitivity to inhibition of poly(ADP-ribose) polymerase. Cancer Res 2009; 69: 3589–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Spivak G. Nucleotide excision repair in humans. DNA Repair 2015; 36: 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van Steeg H, de Vries A, van Oostrom C, et al. DNA repair-deficient Xpa and Xpa/p53+/- knock-out mice: nature of the models. Toxicol Pathol 2001; 29(Suppl): 109–116. [DOI] [PubMed] [Google Scholar]
- 20. Beharry AA, Nagel ZD, Samson LD, et al. Fluorogenic real-time reporters of DNA repair by MGMT, a clinical predictor of antitumor drug response. PLoS One 2016; 11: e0152684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nagel ZD, Kitange GJ, Gupta SK, et al. DNA repair capacity in multiple pathways predicts chemoresistance in glioblastoma multiforme. Cancer Res 2017; 77: 198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chavez KJ, Garimella SV, Lipkowitz S. Triple negative breast cancer cell lines: one tool in the search for better treatment of triple negative breast cancer. Breast Dis 2010; 32: 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smith SE, Mellor P, Ward AK, et al. Molecular characterization of breast cancer cell lines through multiple omic approaches. Breast Cancer Res 2017; 19: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tate JG, Bamford S, Jubb HC, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res 2019; 47: D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marechal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol 2013; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paull TT. Mechanisms of ATM activation. Annu Rev Biochem 2015; 84: 711–738. [DOI] [PubMed] [Google Scholar]
- 27. Gavande NS, VanderVere-Carozza PS, Hinshaw HD, et al. DNA repair targeted therapy: The past or future of cancer treatment? Pharmacol Ther 2016; 160: 65–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brandsma I, Fleuren EDG, Williamson CT, et al. Directing the use of DDR kinase inhibitors in cancer treatment. Expert Opin Investig Drugs 2017; 26: 1341–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brown JS, O’Carrigan B, Jackson SP, et al. Targeting DNA repair in cancer: beyond PARP inhibitors. Cancer Discov 2017; 7: 20–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fok JHL, Ramos-Montoya A, Vazquez-Chantada M, et al. AZD7648 is a potent and selective DNA-PK inhibitor that enhances radiation, chemotherapy and olaparib activity. Nat Commun 2019; 10: 5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shaheen FS, Znojek P, Fisher A, et al. Targeting the DNA double strand break repair machinery in prostate cancer. PLoS One 2011; 6: e20311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sultana R, Abdel-Fatah T, Abbotts R, et al. Targeting XRCC1 deficiency in breast cancer for personalized therapy. Cancer Res 2013; 73: 1621–1634. [DOI] [PubMed] [Google Scholar]
- 33. De Summa S, Pinto R, Pilato B, et al. Expression of base excision repair key factors and miR17 in familial and sporadic breast cancer. Cell Death Dis 2014; 5: e1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wolf DM, Yau C, Sanil A, et al. DNA repair deficiency biomarkers and the 70-gene ultra-high risk signature as predictors of veliparib/carboplatin response in the I-SPY 2 breast cancer trial. NPJ Breast Cancer 2017; 3: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rajkumar-Calkins AS, Szalat R, Dreze M, et al. Functional profiling of nucleotide excision repair in breast cancer. DNA Repair 2019; 82: 102697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ribeiro E, Ganzinelli M, Andreis D, et al. Triple negative breast cancers have a reduced expression of DNA repair genes. PLoS One 2013; 8: e66243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boerner JL, Nechiporchik N, Mueller KL, et al. Protein expression of DNA damage repair proteins dictates response to topoisomerase and PARP inhibitors in triple-negative breast cancer. PLoS One 2015; 10: e0119614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Szalat R, Samur MK, Fulciniti M, et al. Nucleotide excision repair is a potential therapeutic target in multiple myeloma. Leukemia 2018; 32: 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Deng Y, Li F, He P, et al. Triptolide sensitizes breast cancer cells to Doxorubicin through the DNA damage response inhibition. Mol Carcinog 2018; 57: 807–814. [DOI] [PubMed] [Google Scholar]
- 40. Titov DV, Gilman B, He QL, et al. XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat Chem Biol 2011; 7: 182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xiong J, Su T, Qu Z, et al. Triptolide has anticancer and chemosensitization effects by down-regulating Akt activation through the MDM2/REST pathway in human breast cancer. Oncotarget 2016; 7: 23933–23946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gabbard RD, Hoopes RR, Kemp MG. Spironolactone and XPB: an old drug with a new molecular target. Biomolecules 2020; 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Biggar RJ, Andersen EW, Wohlfahrt J, et al. Spironolactone use and the risk of breast and gynecologic cancers. Cancer Epidemiol 2013; 37: 870–875. [DOI] [PubMed] [Google Scholar]
- 44. Ruan Q, Xu Y, Xu R, et al. The adverse effects of triptolide on the reproductive system of caenorhabditis elegans: oogenesis impairment and decreased oocyte quality. Int J Mol Sci 2017; 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cortes-Ciriano I, Lee S, Park WY, et al. A molecular portrait of microsatellite instability across multiple cancers. Nat Commun 2017; 8: 15180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marra A, Viale G, Curigliano G. Recent advances in triple negative breast cancer: the immunotherapy era. BMC Med 2019; 17: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sporikova Z, Koudelakova V, Trojanec R, et al. Genetic markers in triple-negative breast cancer. Clin Breast Cancer 2018; 18: e841–e850. [DOI] [PubMed] [Google Scholar]
- 48. Ben-David U, Ha G, Khadka P, et al. The landscape of chromosomal aberrations in breast cancer mouse models reveals driver-specific routes to tumorigenesis. Nat Commun 2016; 7: 12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lakhani SR, Van De Vijver MJ, Jacquemier J, et al. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol 2002; 20: 2310–2318. [DOI] [PubMed] [Google Scholar]
- 50. Couch FJ, Hart SN, Sharma P, et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J Clin Oncol 2015; 33: 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hartman AR, Kaldate RR, Sailer LM, et al. Prevalence of BRCA mutations in an unselected population of triple-negative breast cancer. Cancer 2012; 118: 2787–2795. [DOI] [PubMed] [Google Scholar]
- 52. Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 2001; 98: 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Akashi-Tanaka S, Watanabe C, Takamaru T, et al. BRCAness predicts resistance to taxane-containing regimens in triple negative breast cancer during neoadjuvant chemotherapy. Clin Breast Cancer 2015; 15: 80–85. [DOI] [PubMed] [Google Scholar]
- 54. Lips EH, Mulder L, Oonk A, et al. Triple-negative breast cancer: BRCAness and concordance of clinical features with BRCA1-mutation carriers. Br J Cancer 2013; 108: 2172–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nicolas E, Bertucci F, Sabatier R, et al. Targeting BRCA deficiency in breast cancer: what are the clinical evidences and the next perspectives? Cancers 2018; 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bianchini G, Balko JM, Mayer IA, et al. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol 2016; 13: 674–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Beniey M, Haque T, Hassan S. Translating the role of PARP inhibitors in triple-negative breast cancer. Oncoscience 2019; 6: 287–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee JS, Yost SE, Yuan Y. Neoadjuvant treatment for triple negative breast cancer: recent progresses and challenges. Cancers 2020; 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ohmoto A, Yachida S. Current status of poly(ADP-ribose) polymerase inhibitors and future directions. Onco Targets Ther 2017; 10: 5195–5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cleary JM, Aguirre AJ, Shapiro GI, et al. Biomarker-guided development of DNA repair inhibitors. Mol Cell 2020; 78: 1070–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ronco C, Martin AR, Demange L, et al. ATM, ATR, CHK1, CHK2 and WEE1 inhibitors in cancer and cancer stem cells. Medchemcomm 2017; 8: 295–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sanjiv K, Hagenkort A, Calderon-Montano JM, et al. Cancer-specific synthetic lethality between ATR and CHK1 kinase activities. Cell Rep 2016; 14: 298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. King C, Diaz HB, McNeely S, et al. LY2606368 causes replication catastrophe and antitumor effects through CHK1-dependent mechanisms. Mol Cancer Ther 2015; 14: 2004–2013. [DOI] [PubMed] [Google Scholar]
- 64. Ray Chaudhuri A, Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol 2017; 18: 610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. London RE. The structural basis of XRCC1-mediated DNA repair. DNA Repair 2015; 30: 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair 2003; 2: 955–969. [DOI] [PubMed] [Google Scholar]
- 67. Jang S, Kumar N, Beckwitt EC, et al. Damage sensor role of UV-DDB during base excision repair. Nat Struct Mol Biol 2019; 26: 695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li J, Wang QE, Zhu Q, et al. DNA damage binding protein component DDB1 participates in nucleotide excision repair through DDB2 DNAbinding and cullin 4A ubiquitin ligase activity. Cancer Res 2006; 66: 8590–8597. [DOI] [PubMed] [Google Scholar]
- 69. Iovine B, Iannella ML, Bevilacqua MA. Damage-specific DNA binding protein 1 (DDB1): a protein with a wide range of functions. Int J Biochem Cell Biol 2011; 43: 1664–1667. [DOI] [PubMed] [Google Scholar]
- 70. Torgovnick A, Schumacher B. DNA repair mechanisms in cancer development and therapy. Front Genet 2015; 6: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang LC, Gautier J. The Fanconi anemia pathway and ICL repair: implications for cancer therapy. Crit Rev Biochem Mol Biol 2010; 45: 424–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Britten RA, Liu D, Tessier A, et al. ERCC1 expression as a molecular marker of cisplatin resistance in human cervical tumor cells. Int J Cancer 2000; 89: 453–457. [PubMed] [Google Scholar]
- 73. Belanger F, Fortier E, Dube M, et al. Replication protein A availability during DNA replication stress is a major determinant of cisplatin resistance in ovarian cancer cells. Cancer Res 2018; 78: 5561–5573. [DOI] [PubMed] [Google Scholar]
- 74. Bae YH, Shin JM, Park HJ, et al. Gain-offunction mutant p53-R280K mediates survival of breast cancer cells. Genes Genom 2014; 36: 171–178. [Google Scholar]
- 75. Shuck SC, Turchi JJ. Targeted inhibition of replication protein A reveals cytotoxic activity, synergy with chemotherapeutic DNA-damaging agents, and insight into cellular function. Cancer Res 2010; 70: 3189–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Weterings E, Gallegos AC, Dominick LN, et al. A novel small molecule inhibitor of the DNA repair protein Ku70/80. DNA Repair 2016; 43: 98–106. [DOI] [PubMed] [Google Scholar]
- 77. Mohiuddin IS, Kang MH. DNA-PK as an emerging therapeutic target in cancer. Front Oncol 2019; 9: 635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mouw KW, Goldberg MS, Konstantinopoulos PA, et al. DNA damage and repair biomarkers of immunotherapy response. Cancer Discov 2017; 7: 675–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang Z, Zhao J, Wang G, et al. Comutations in DNA damage response pathways serve as potential biomarkers for immune checkpoint blockade. Cancer Res 2018; 78: 6486–6496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplemental_Figure1 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure2 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure3 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure4 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure6 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure_10 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure_11 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure_12 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure_5 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure_7 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure_8 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology
Supplemental material, Supplemental_Figure_9 for Exploiting DNA repair defects in triple negative breast cancer to improve cell killing by Kevin J. Lee, Elise Mann, Griffin Wright, Cortt G. Piett, Zachary D. Nagel and Natalie R. Gassman in Therapeutic Advances in Medical Oncology