Abstract
Objective
Papillary thyroid cancer (PTC) is one of the most prevalent endocrine malignancies and the fifth most common cancer in women. Circular RNAs (circRNAs) have been shown to play vital functions in cancers, but few studies have focused on the functions and mechanism of dysregulated circRNAs in PTC.
Methods
Quantitative reverse transcription PCR was used to measure circ-NCOR2 levels in PTC tissues and cell lines. The functions of circ-NCOR2 in PTC were examined by analysis using the cell counting kit-8, clone forming, flow cytometry, and Transwell experiments. Bioinformatic analysis and dual luciferase reporter gene testing were used to identify the mechanisms of circ-NCOR2.
Results
Circ-NCOR2 overexpression was observed in PTC tissues and cells. Silenced or overexpressed expression of circ-NCOR2 strikingly attenuated or facilitated, respectively, the growth, migration, and invasion of PTC cells. Mechanistically, miR-615a-5p was identified as the target of circ-NCOR2. Moreover, circ-NCOR2 enhanced the expression of metastasis-associated protein 2 (MTA2) by sponging miR-615a-5p, thereby facilitating PTC cell progression.
Conclusions
Taken together, our findings reveal a novel circ-NCOR2/miR-615a-5p/MTA2 axis in PTC, which could become a potential therapeutic target for this disease.
Keywords: Papillary thyroid cancer, circular RNA, circ-NCOR2, miR-516a-5p, metastasis-associated protein 2, sponging
Introduction
Papillary thyroid cancer (PTC) is a metastatic and invasive tumor of the endocrine system1 that has shown an increase in worldwide incidence in recent years.2 Most patients only show typical symptoms at an advanced stage of disease, and despite improvements in early diagnosis the postoperative recurrence remains high and the 5-year survival rate is still unsatisfactory.3,4 The molecular mechanisms underlying the pathogenesis of PTC are largely unknown, so it is important to understand them to help contribute to the discovery of new biomarkers and improve prognosis.
Circular RNAs (circRNAs) are covalently closed loop structures with limited protein-coding potential5 that cannot be degraded by RNA exonucleases, thus maintaining stable expression.5 They are extensively expressed in cells and their dysregulation may cause human diseases, including cancer.6 CircRNAs interact with micro (mi)RNAs to inhibit their expression and function, thus exerting regulatory effects in multiple diseases including esophageal cancer,7 oral cancer,8 cardiovascular disease,9 and Alzheimer’s disease.10 Such interactions are known as the competitive endogenous RNA (ceRNA) theory.11 Additionally, emerging evidence has indicated that circRNAs interact directly with some proteins to exert their biological effects in tumors. For example, circFOXP1 interacts with polypyrimidine tract-binding protein 1 that binds to the 3′ untranslated region (UTR) and coding region of enzyme pyruvate kinase to protect pyruvate kinase L/R (PKLR) mRNA from decay.12 Hence, determining the mechanisms underlying the progression of PTC and searching for new drugs from the perspective of circRNAs is imperative.
Hsa_circ_0000461, located on chr12:124911167-124934413, was previously screened as a circRNA upregulated in PTC tissues compared with healthy controls by circRNA microarray.13 However, its detailed functions and mechanisms are unknown. The spliced sequence length of hsa_circ_0000461 is 566 bp, and its genomic structure shows that it is looped by exons 9 to 13 of the NCOR2 gene (Figure 1a). Thus, hsa_circ_0000461 can also be termed circ-NCOR2. Herein, we confirm that circ-NCOR2 is upregulated in both PTC tissue specimens and cell lines. We also show that the malignant behaviors of circ-NCOR2 are partially attributed to its modulation of the miR-516a-5p/metastasis-associated protein 2 (MTA2) axis.
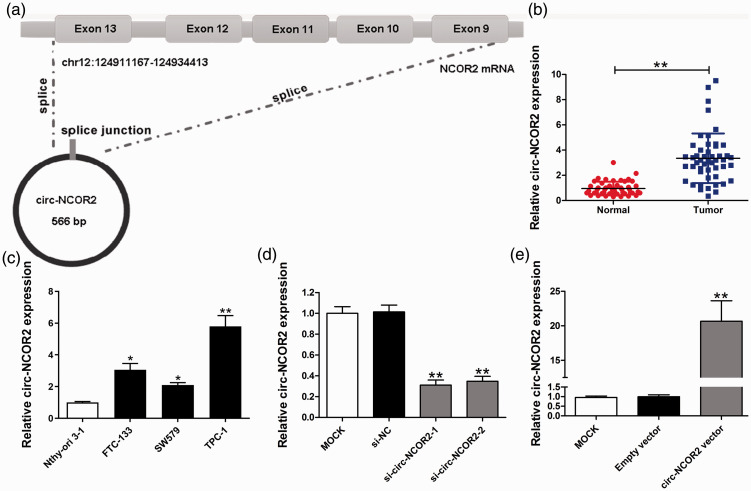
Figure 1.
Relative expression of circ-NCOR2 in PTC tissues and cell lines. (a) Schematic of the genomic location and splicing pattern of circ-NCOR2. (b) Relative expression of circ-NCOR2 in PTC tissue samples and paired non-cancerous tissue samples measured by qRT-PCR. (c) Relative expression of circ-NCOR2 in PTC cell lines and a normal cell line measured by qRT-PCR. (d) Circ-NCOR2 expression was detected after circ-NCOR2 silencing in TPC-1 cells by qRT-PCR. (e) Circ-NCOR2 expression was detected after circ-NCOR2 overexpression in SW579 cells by qRT-PCR. *p < 0.05, **p < 0.01.
Materials and methods
Patients and specimens
PTC specimens and matched non-cancerous samples (n = 49) were harvested from PTC patients diagnosed by two experienced pathologists during surgery at our hospital between May 2018 to April 2019. None of the patients had undergone radiotherapy or chemotherapy before surgery. This work was authorized by the Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University. Informed consent was obtained from each patient.
Cell culture
Human PTC cells (FTC-133, SW579,K1, and TPC-1) and a human thyroid epithelial cell line (Nthy-ori 3-1) were obtained from the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI-1640 medium (GIBCO® Cell Culture, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; HyClone Laboratories Inc., Logan, UT, USA) in a 5% CO2 atmosphere with humidified air at 37°C.
Cell transfection
Small interfering (si)RNA specifically targeting the back-spliced junction of circ-NCOR2 (si-circ-NCOR2-1/-2), and miR-516a-5p/NC mimics/inhibitor were obtained from GenePharma (Shanghai, China). The circ-NCOR2/MTA2 overexpression construct and corresponding negative controls were acquired from GeneChem (Shanghai, China). Cells were transfected using Lipofectamine 3000 (Invitrogen Corp., Carlsbad, CA, USA) according to the manufacturer’s instructions. The targeted sequences of siRNA-circ-NCOR2 are as follows: si-circ-NCOR2-1, 5′-GGGAGAACCGCTGTACAACCA-3′ and si-circ-NCOR2-2, 5′-TTCCGGGAGAACCGCTGTACA-3′.
RNA isolation and quantitative (q) reverse transcription (RT)-PCR
Total RNA was isolated by TRIzol (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions, then reverse-transcribed into cDNA after preparing the reaction mixture on ice and then maintaining at 50°C for 1 hour. (Roche Molecular Biochemicals, Mannheim, Germany). qRT-PCR was carried out on a 7500 Fast Real-Time PCR system using SYBR Green Master Mix (Roche Molecular Biochemicals). U6 and GAPDH were used as internal controls and all reactions were carried out in triplicate. Relative RNA expression was analyzed using the ΔΔCt method. PCR primers for circ-NCOR2 and GAPDH are listed below. circ-NCOR2: 5′-AACATGAACGGGCTTATGGC-3′ (forward) and 5′-TCTCATGATACTGCCGGGTG-3′ (reverse); GAPDH: 5′-GGGAGCCAAAAGGGTCAT-3′ (forward) and 5′-GAGTCCTTCCACGATACCAA-3′ (reverse).
Cell growth determination
Cell viability was evaluated by the cell counting kit-8 (CCK-8) and colony formation assay. For CCK-8, 1500 cells were placed in 96-well plates and then 10 μl of CCK-8 solution (Dojin do Molecular Technologies Inc., Beijing, China) was added to each well according to the manufacturer’s instructions. Absorbance at 450 nm was estimated using a spectrophotometer at the indicated time point.
For the clone formation assay, cells transfected for 48 hours were harvested and counted with a cell counter. The same number of cells was then seeded in 2.5-cm plates which were gently mixed to evenly distribute the cells. After incubating for about 12 days, the cells were fixed with paraformaldehyde and stained with crystal violet before counting with the naked eye.
Cell apoptosis assay
An apoptosis analysis kit (BD Biosciences, San Jose, CA, USA) was used to evaluate the apoptotic rate of transfected PTC cells. Transfected cells were washed twice with phosphate-buffered saline, then incubated with Annexin V-FITC and propidium iodide for 15 minutes in the dark and evaluated using a flow cytometer (FACScan, BD Biosciences).
Transwell assay
For the Transwell migration assay (Corning Inc., Corning, NY, USA), RPMI-1640 medium with 10% FBS was added to the lower chamber, while the upper chamber with a polycarbonate membrane was filled with transfected cells in serum-free medium. After incubation for 2 hours, cells were fixed with paraformaldehyde and stained with crystal violet before being imaged and counted. The protocol for analyzing cell invasion was similar except that the upper chamber was coated with Matrigel (BD Biosciences).
Target prediction and dual luciferase reporter test
The interaction between circ-NCOR2 and miR-516a-5p was predicted by the Circular RNA Interactome database (https://circinteractome.nia.nih.gov/) and downstream targets of miR-516a-5p were analyzed by TargetScan (http://www.targetscan.org/vert_72/). Luciferase reporter assays were used to confirm putative bindings. Fragment sequences containing potential binding sites were synthesized by PCR and inserted into a pmirGLO vector (Promega, Madison, WI, USA). Cells were cotransfected with the pmirGLO vector and miR-516a-5p mimics or mimics-NC using Lipofectomine 3000 reagent and incubated for 36 hours. The luciferase intensity was then measured using a Dual Luciferase Assay Kit (Promega).
Immunoblotting assay
PTC cells were lysed with radioimmunoprecipitation assay buffer containing protease inhibitor. Equal amounts of protein were separated via sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes. The membranes were blocked with nonfat milk and incubated with primary antibodies against MTA2 (1:800 dilution; Abcam, Cambridge, MA, USA) or GAPDH (1:10000 dilution; Abcam). After incubation with a secondary antibody (1:2000 dilution; Zhongshan Goldbridge, Beijing, China), they were treated with enhanced chemiluminescence solution (Beyotime Biotech Inc., Jiangsu, China) to detect the protein signals.
Determination of subcellular distribution
We used the PARIS Kit (Gibco BRL Life Technologies Inc., Gaithersburg, MD, USA) to separate RNAs in cytoplasmic and nuclear fractions, then qRT-PCR was carried out with U6 and GAPDH as nuclear and cytoplasmic controls, respectively.
Statistical analysis
The Cancer Genome Atlas (TCGA) database was used to analyze the expression of MTA2 in cancerous and healthy tissues. Multiple groups were compared by one-way analysis of variance, while two groups were compared by t-test using GraphPad Prism 5.01 software (GraphPad Software Inc., La Jolla, CA, USA). Pearson’s correlation coefficient was used to analyze the correlation between circRNA, miRNA, and mRNA expression. P values <0.05 were deemed statistically significant.
Results
Expression levels of circ-NCOR2 in PTC tissues and cells
qRT-PCR showed that circ-NCOR2 expression was significantly elevated in PTC specimens compared with normal tissue samples (p < 0.01; Figure 1b), while significantly higher circ-NCOR2 expression was detected in PTC cells relative to Nthy-ori 3-1 cells (Figure 1c). In particular, TPC-1 cells showed the highest expression of PTC cells, with SW579 cells the lowest, but both were significantly higher than controls (p < 0.05 and p < 0.01, respectively).
To investigate the functions of circ-NCOR2 in PTC progression, two siRNAs targeted to the back-spliced site of circ-NCOR2 were synthesized to inhibit circ-NCOR2 expression. The TPC-1 cell line was used for knockdown studies because of its high endogenous circ-NCOR2 expression. Following the transfection of both si-circ-NCOR2-1 and si-circ-NCOR2-2, circ-NCOR2 expression decreased significantly in TPC-1 cells suggesting that knockdown was successful (p < 0.01; Figure 1d). We also observed a significant elevation of circ-NCOR2 after transfecting circ-NCOR2 into SW579 cells (p < 0.01; Figure 1e).
Circ-NCOR2 contributes to PTC cell progression in vitro
CCK-8 was used to evaluate the influence of circ-NCOR2 on cell viability. Circ-NCOR2-siRNA-mediated circ-NCOR2 downregulation reduced cell viability, indicating that its knockdown suppressed the proliferation of TPC-1 cells. By contrast, increased expression of circ-NCOR2 boosted the growth of SW579 cells (p < 0.01; Figure 2a). Additionally, the clone formation assay showed that circ-NCOR2 silencing reduced the number of colonies in the two selected cell types. Circ-NCOR2 vector-transfected cells showed more colony growth relative to the control group (p < 0.01; Figure 2b).
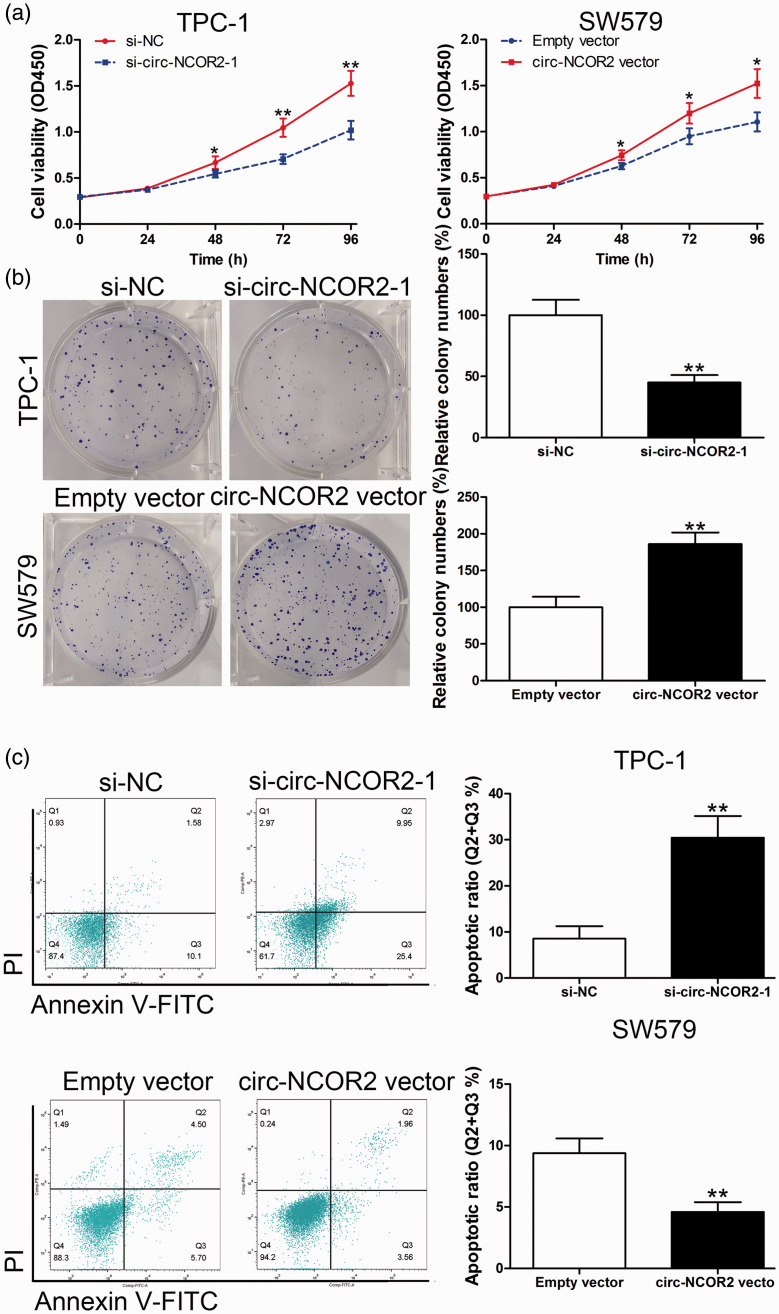
Figure 2.
Circ-NCOR2 promotes PTC cell proliferation. (a) CCK-8 assays were used to detect the viability of TPC-1 and SW579 cells after circ-NCOR2 knockdown/overexpression. (b) Colony formation assays were used to detect the clone forming ability of TPC-1 and SW579 cells after circ-NCOR2 knockdown/overexpression. (c) Flow cytometric analysis was used to detect apoptosis of TPC-1 and SW579 cells after circ-NCOR2 knockdown/overexpression. *p < 0.05, **p < 0.01.
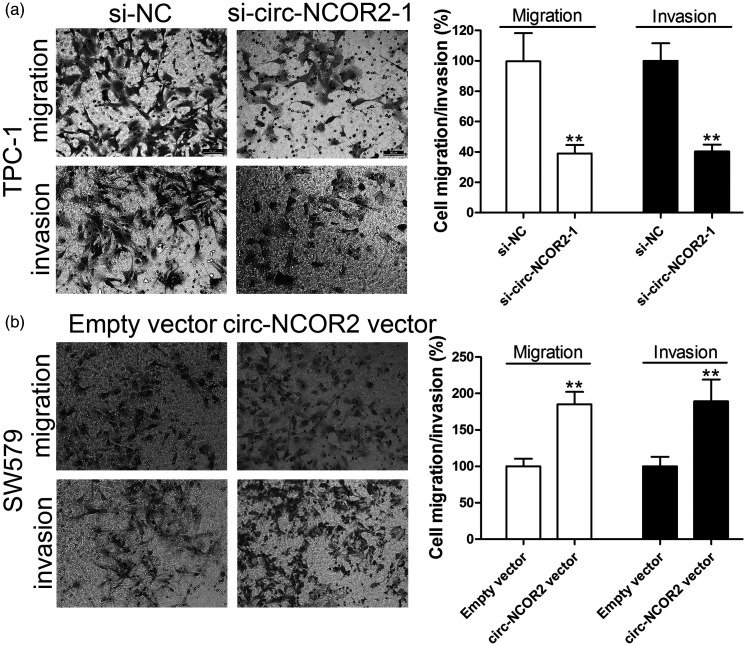
We then investigated whether the promotion of cell growth by circ-NCOR2 was dependent on its regulation of cell apoptosis. Flow cytometry showed that cells with decreased or increased circ-NCOR2 had significantly elevated or inhibited rates of cell apoptosis, respectively (p < 0.01; Figure 2c). Transwell assays showed that inhibiting circ-NCOR2 expression significantly reduced the migratory and invasive potential relative to controls (p < 0.01; Figure 3a), while the ectopic expression of circ-NCOR2 significantly enhanced the migratory and invasive potential of SW579 cells (p < 0.01; Figure 3b).
Figure 3.
Circ-NCOR2 promotes PTC cell migration and invasion. (a) Transwell assays were used to detect cell migration and invasion capacities of TPC-1 cells after circ-NCOR2 knockdown. (b) Transwell assays were used to detect cell migration and invasion capacities of SW579 cells after circ-NCOR2 overexpression. **p < 0.01.
Circ-NCOR2 sponges miR-516a-5p to upregulate MTA2 in PTC
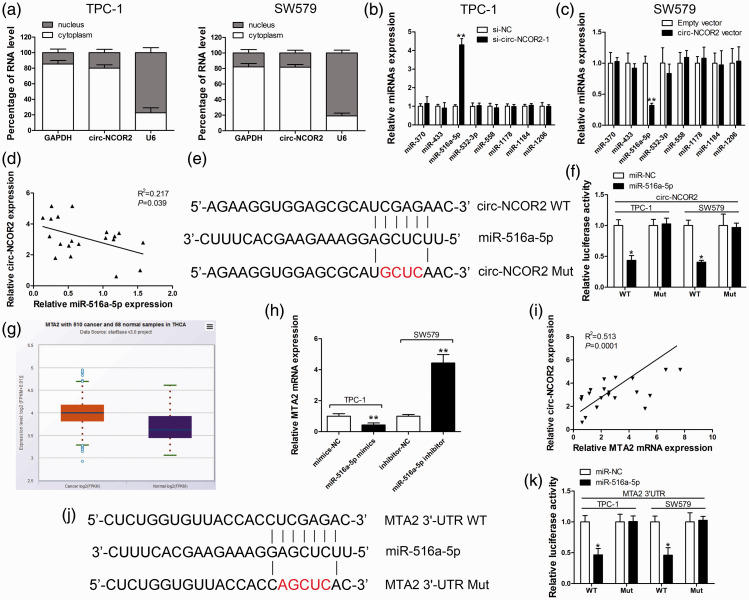
To investigate the potential mechanisms of circ-NCOR2 in PTC, we first detected its localization in TPC-1 and SW579 cells. The subcellular distribution assay revealed that circ-NCOR2 was primarily localized in the cytoplasm of PTC cells (Figure 4a), suggesting that it might be involved in posttranscriptional regulation. To further understand the mechanism of circ-NCOR2 in PTC, bioinformatic analysis was used to explore potential target miRNAs of circ-NCOR2. This predicted that miR-370, miR-433, miR-516a-5p, miR-532-3p, miR-558, miR-1178, miR-1184, and miR-1206 may be sponged by circ-NCOR2. qRT-PCR was then used to measure changes in the expression of putative miRNA targets after circ-NCOR2 silencing. miR-516a-5p was the only significantly upregulated miRNA in circ-NCOR2-silenced cells (p < 0.01; Figure 4b). As expected, circ-NCOR2 overexpression significantly induced the downregulation of miR-516a-5p in SW579 cells (p < 0.01; Figure 4c). Pearson’s correlation coefficient analysis demonstrated a negative correlation between circ-NCOR2 and miR-516a-5p expression in PTC tissue samples (Figure 4d).
Figure 4.
Circ-NCOR2 sponges miR-516a-5p to elevate MTA2 expression. (a) qRT-PCR detection of the percentage of circ-NCOR2, GAPDH, and U6 in the cytoplasm and nuclear fractions of TPC-1 and SW579 cells. GAPDH and U6 were used as cytoplasmic and nuclear localization markers, respectively. (b) Relative miRNA expression was detected in TPC-1 cells after circ-NCOR2 knockdown. (c) Relative miRNA expression was detected in SW579 cells after circ-NCOR2 overexpression. (d) Correlation analysis of circ-NCOR2 and miR-516a-5p in PTC patient tissues. (e) Diagram of the binding site for circ-NCOR2 and miR-516a-5p. (f) Luciferase reporter assay was conducted to evaluate the interaction between miR-516a-5p and circ-NCOR2. (g) Relative MTA2 expression was detected in PTC tissues and normal samples by the TCGA database. (h) MTA2 mRNA expression was detected by qRT-PCR after miR-516a-5p knockdown/overexpression in TPC-1 and SW579 cells. (i) Correlation analysis of circ-NCOR2 and MTA2 mRNA in PTC patient tissues. (j) Diagram of the binding sites for MTA2 3′-UTR and miR-516a-5p. (k) Luciferase reporter assay was conducted to evaluate the interaction between MTA2 3′-UTR and miR-516a-5p. *p < 0.05, **p < 0.01.
To validate the binding ability of circ-NCOR2 and miR-516a-5p, a luciferase reporter gene containing a wild-type or mutated circ-NCOR2 sequence was constructed and the luciferase reporter test used to confirm the predicted binding site (Figure 4e). Dual luciferase reporter analysis showed that the transfection of miR-516a-5p mimics significantly reduced the luciferase intensity (p < 0.05) of the wt-circ-NCOR2 reporter relative to the mimics-NC group, while the luciferase signal of the mut-circ-NCOR2 reporter was unchanged (Figure 4f). TargetScan predicted MTA2 as a putative target of circ-NCOR2, and data from the TCGA database indicated that MTA2 was markedly elevated in PTC samples relative to non-cancerous samples (Figure 4g). qRT-PCR found that miR-516a-5p mimics significantly attenuated the expression of MTA2 in TPC-1 cells, while an miR-516a-5p inhibitor had the opposite effect (p < 0.01; Figure 4h). Pearson’s correlation coefficient analysis identified a positive correlation between circ-NCOR2 and MTA2 mRNA expression in 20 pairs of tissues from PTC patients (Figure 4i). To validate this binding ability, we constructed wt-MTA2 3′UTR (Luc WT) and mut-MTA2 3′UTR reporter vectors (Figure 4j). Dual luciferase reporter analysis demonstrated that miR-516a-5p overexpression significantly suppressed the luciferase signal of the reporter containing the wt-MTA2 3′UTR sequence compared with the reporter with the mutated miR-516a-5p binding site (p < 0.05; Figure 4k).
Circ-NCOR2 promotes PTC cell growth and aggressiveness via the miR-516a-5p/MTA2 axis
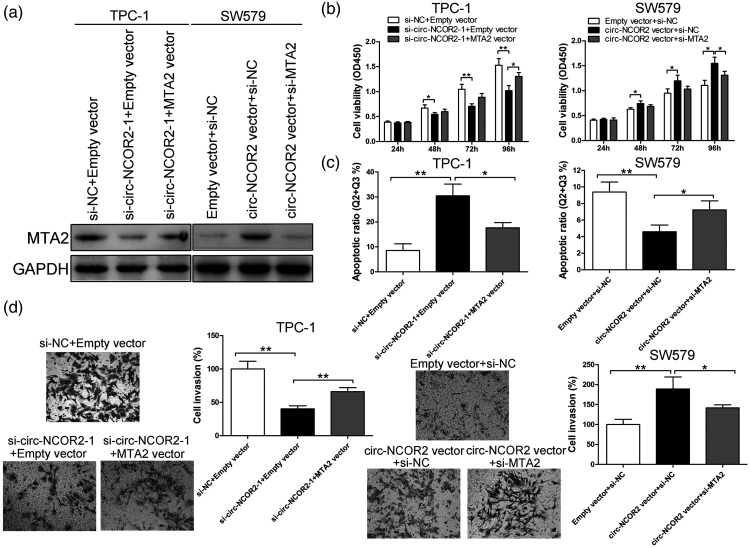
To further verify the existence of a circ-NCOR2/miR-516a-5p/MTA2 axis, we performed a rescue assay by cotransfecting si-circ-NCOR2-1 and the MTA2 vector into TPC-1 cells. Western blot analysis revealed that circ-NCOR2 downregulation decreased MTA2 protein expression, but that the MTA2 vector reversed this effect (Figure 5a). si-MTA2 partially reversed the upregulation of MTA2 induced by the circ-NCOR2 vector (Figure 5a). CCK-8 and Transwell assays showed that the MTA2 vector significantly reversed the inhibition of si-circ-NCOR2-1 on PTC cell proliferation and invasion (p < 0.01; Figure 5b and d). Cell growth and invasive capacity were significantly inhibited after the cotransfection of circ-NCOR2 and si-MTA2 compared with circ-NCOR2 and si-NC (p < 0.01; Figure 5b and d). Flow cytometry showed that circ-NCOR2 silencing significantly facilitated apoptosis in TPC-1 cells, and that this could be partially reversed by MTA2 upregulation (p < 0.01; Figure 5c). Moreover, circ-NCOR2 overexpression significantly attenuated cell apoptosis, while cotransfection with si-MTA2 partially rescued this (p < 0.01; Figure 5c). Taken together, the rescue experiments suggested that the circ-NCOR2/miR-516a-5p/MTA2 axis plays a key role in PTC cell development and progression, indicating a ceRNA mechanism for circ-NCOR2 targeting the miR-516a-5p/MTA2 axis.
Figure 5.
The oncogenic role of circ-NCOR2 is dependent on its regulation of MTA2. (a) MTA2 expression was detected by western blot in TPC-1 and SW579 cells after transfection. (b) CCK-8 assay was used to evaluate viability after transfection in TPC-1 and SW579 cells. (c) Flow cytometric assay was used to evaluate apoptosis after transfection in TPC-1 and SW579 cells. (d) Transwell assays were used to evaluate the invasive potential after transfection in TPC-1 and SW579 cells. *p < 0.05, **p < 0.01.
Discussion
PTC has always been a public health concern, and its underlying mechanisms remain unclear. Although surgery is the treatment of choice,3 therapeutic effects are not satisfactory and patient prognosis is unfavorable.4
Several genes have been found to associate with cancer cell development and progression and the role of circRNA in gene expression regulation is attracting increasing attention worldwide. CircRNA dysregulation is involved in the progression of multiple cancers, including PTC.6–8,13 For example, Yao et al.14 reported that hsa_circ_0058124 accelerates PTC carcinogenesis and progression by affecting the NOTCH3/GATAD2A pathway. In this work, we characterized circ-NCOR2, which originated from the back-splicing of NCOR2 that was markedly overexpressed in PTC samples and cells. Subsequent gain- and loss-of-function assays revealed that circ-NCOR2 contributed to the growth, invasion, and migration abilities of PTC cells.
Multiple evidence suggests that circRNAs possess dynamic and distinct functions in tumorigenesis and development via the ceRNA theory.15 For instance, circ-DENND2A directly sponges miR-625-5p to accelerate glioma cell aggressiveness,16 while circ-HIPK3 contributes to colorectal cancer growth and metastasis by sponging miR-7.17 Similarly, we found that circ-NCOR2 functions as an miRNA sponge to absorb miR-516a-5p.
Our qRT-PCR and luciferase reporter findings showed that miR-516a-5p is a target of circ-NCOR2. miR-516a-5p was found to be markedly downregulated in several tumors. Yao et al.18 reported that circ_0001955 contributed to hepatocellular carcinoma progression through absorbing miR-516a-5p to release TRAF6 and MAPK11, while another study identified HIST3H2A as the direct target of miR-516a-5p in non-small cell lung cancer.19 Similarly, we verified that miR-516a-5p was decreased in PTC, and propose that circ-NCOR2 exerts a tumorigenesis-promoting role mainly by binding and inhibiting the functions of miR-516a-5p.
We also found that miR-516a-5p binds directly to the MTA2 3′ UTR to repress MTA2 expression. MTA2 is a member of the metastasis tumor-associated family of transcriptional regulators and is a core scaffold for the Mi-2/NuRD complex.20,21 MTA2 acts as a central hub for cytoskeletal organization and transcription and provides a link between nuclear and cytoskeletal organization. It was also found to be significantly upregulated and to function as an oncogene in a wide range of malignancies such as renal cancer,22 hepatocellular carcinoma,23 and pancreatic ductal adenocarcinoma.24 Moreover, MTA2 expression is regulated at the posttransciptional level in gastric cancer,25 and the long noncoding RNA HOTAIR promotes the invasion and metastasis of oral squamous cell carcinoma through targeting MTA2.26
Here, the elevation of circ-NCOR2 significantly increased MTA2 expression to promote the proliferation and invasion of PTC cells. Importantly, the oncogenic role was abrogated by MTA2 knockdown, implying that the regulatory network of the circ-NCOR2/miR-516a-5p/MTA2 axis plays a key role in PTC progression.
Despite the limitation that this study did not conduct in vivo work or investigate the clinical value of circ-NCOR2, it nevertheless demonstrates that circ-NCOR2 reduced miR-516a-5p-mediated repression of MTA2 to facilitate the progression of PTC. These results reveal the functional implications of circ-NCOR2 and provide robust evidence of a crucial regulatory role for this circRNA in PTC progression. Future work may identify other differentially expressed circRNAs in PTC which will promote the discovery of molecular markers or therapeutic targets to aid diagnosis and treatment.
Authors’ contributions
Youmin Guo designed the study and provided the final approval of the version to be published. Sha Luan, Peng Fu, Xinyu Wang, and Yue Gao performed the experiments. Sha Luan and Peng Fu analyzed the data. Peng Fu and Ke Shi drafted the manuscript.
Availability of data and materials
The datasets used in the project are available from the corresponding author on request.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
This study was supported by the Health and Family Planning Commission Research Project of Heilongjiang Province (Grant No. 2017-141).
ORCID iD
Youmin Guo https://orcid.org/0000-0002-4953-3348
References
- 1.Xing M, Alzahrani AS, Carson KA, et al. Association between BRAF V600E mutation and mortality in patients with papillary thyroid cancer. J Am Med Assoc 2013; 309: 1493–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Canc 2010; 127: 2893–2917. [DOI] [PubMed] [Google Scholar]
- 3.Frohlich E, Wahl R. The current role of targeted therapies to induce radioiodine uptake in thyroid cancer. Cancer Treat Rev 2014; 40: 665–674. [DOI] [PubMed] [Google Scholar]
- 4.Lundgren CI, Hall P, Dickman PW, et al. Clinically significant prognostic factors for differentiated thyroid carcinoma: a population-based, nested case-control study. Cancer 2006; 106: 524–531. [DOI] [PubMed] [Google Scholar]
- 5.Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer 2017; 16: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Y, Yao Y, Zhong X, et al. Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells. Biochem Biophys Res Commun 2018; 496: 455–461. [DOI] [PubMed] [Google Scholar]
- 7.Su H, Lin F, Deng X, et al. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med 2016; 14: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Zhang S, Wu J, et al. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene 2017; 36: 4551–4561. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Tan WL, Lim T, Anene-Nzelu CG, et al. A landscape of circular RNA expression in the human heart. Cardiovasc Res 2017; 113: 298–309. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y, Alexandrov PN, Jaber V, et al. Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer’s disease (AD) is linked to deficits in a natural circular miRNA-7 sponge (circRNA; ciRS-7). Genes (Basel) 2016; 7: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li LJ, Zhao W, Tao SS, et al. Competitive endogenous RNA network: potential implication for systemic lupus erythematosus. Expert Opin Ther Targets 2017; 21: 63–648. [DOI] [PubMed] [Google Scholar]
- 12.Wang S, Zhang Y, Cai Q, et al. Circular RNA FOXP1 promotes tumor progression and Warburg effect in gallbladder cancer by regulating PKLR expression. Mol Cancer 2019; 18: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan Y, Xu T, Liu Y, et al. Upregulated circular RNA circ_0025033 promotes papillary thyroid cancer cell proliferation and invasion via sponging miR-1231 and miR-1304. Biochem Biophys Res Commun 2019; 510: 334–338. [DOI] [PubMed] [Google Scholar]
- 14.Yao Y, Chen X, Yang H, et al. Hsa_circ_0058124 promotes papillary thyroid cancer tumorigenesis and invasiveness through the NOTCH3/GATAD2A axis. J Exp Clin Cancer Res 2019; 38: 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kristensen LS, Hansen TB, Veno MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 2018; 37: 555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su H, Zou D, Sun Y, et al. Hypoxia-associated circDENND2A promotes glioma aggressiveness by sponging miR-625-5p. Cell Mol Biol Lett 2019; 24: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis 2018; 9: 417. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Yao Z, Xu R, Yuan L, et al. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell Death Dis 2019; 10: 945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye XY, Xu L, Lu S, et al. MiR-516a-5p inhibits the proliferation of non-small cell lung cancer by targeting HIST3H2A. Int J Immunopathol Pharmacol 2019; 33: 2058738419841481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowen NJ, Fujita N, Kajita M, et al. Mi-2/NuRD: multiple complexes for many purposes. Biochimica et Biophysica Acta 2004; 1677: 52–57. [DOI] [PubMed] [Google Scholar]
- 21.Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer 2011; 11: 588–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen YS, Hung TW, Su SC, et al. MTA2 as a potential biomarker and its involvement in metastatic progression of human renal cancer by miR-133b targeting MMP-9. Cancers (Basel) 2019; 11: 1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guan C, Chang Z, Gu X, et al. MTA2 promotes HCC progression through repressing FRMD6, a key upstream component of hippo signaling pathway. Biochem Biophys Res Commun 2019; 515 112–118. [DOI] [PubMed] [Google Scholar]
- 24.Si W, Liu X, Wei R, et al. MTA2-mediated inhibition of PTEN leads to pancreatic ductal adenocarcinoma carcinogenicity. Cell Death Dis 2019; 10: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An JX, Ma MH, Zhang CD, et al. miR-1236-3p inhibits invasion and metastasis in gastric cancer by targeting MTA2. Cancer Cell Int 2018; 18: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao D, Zhang Z, Liu X, et al. LncRNA HOTAIR promotes the invasion and metastasis of oral squamous cell carcinoma through metastasis-associated gene 2. Mol Carcinog 2020; 59: 353–364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used in the project are available from the corresponding author on request.