Abstract
Matrix metalloprotease (MMP) activation contributes to degradation of the extracellular matrix (ECM), resulting in a multitude of pathologies. Low-density lipoprotein receptor–related protein 1 (LRP1) is a multifaceted endocytic and signaling receptor responsible for internalization and lysosomal degradation of diverse proteases, protease inhibitors and lipoproteins along with numerous other proteins. In the current study, we identified MMP-1 as a novel LRP1 ligand. Binding studies employing surface plasmon resonance revealed that both proMMP-1 and active MMP-1 bind to purified LRP1 with equilibrium dissociation constants (KD) of 19 nM and 25 nM, respectively. We observed that human aortic smooth muscle cells readily internalize and degrade 125I-labeled proMMP-1 in an LRP1-mediated process. Our binding data also revealed that all tissue inhibitors of metalloproteases (TIMPs) bind to LRP1 with KD values ranging from 23 nM to 33 nM. Interestingly, the MMP-1/TIMP-1 complex bound to LRP1 with 30-fold higher affinity (KD = 0.6 nM) than either component alone, revealing that LRP1 prefers the protease:inhibitor complex as a ligand. Of note, modification of lysine residues on either proMMP-1 or TIMP-1 ablated the ability of the MMP-1/TIMP-1 complex to bind to LRP1. LRP1’s preferential binding to enzyme:inhibitor complexes was further supported by higher binding affinity for proMMP-9/TIMP-1 complexes compared with either of these two components alone. LRP1 has four clusters of ligand-binding repeats, and MMP-1, TIMP-1 and MMP-1/TIMP-1 complexes bound to cluster III most avidly. Our results reveal an important role for LRP1 in controlling ECM homeostasis by regulating MMP-1 and MMP-9 levels.
INTRODUCTION
As an essential contributor to tissue homeostasis, the extracellular matrix (ECM) constantly undergoes remodeling by ECM-modifying enzymes and proteases. These events are complex and are tightly regulated processes that are initiated by environmental cues. Dysregulation of ECM remodeling due to an imbalance between matrix production, secretion, alteration, and degradation is a crucial part of pathogenesis in various diseases. The ECM plays a key role in Alzheimer’s disease and other neurodegenerative diseases 1–3, fibrotic diseases, and tumor development and metastasis (reviewed in 4,5). Tissue fibrosis is the abnormal response to injury or aging and is typically characterized by hyperproliferation and excessive ECM synthesis and secretion. Remodeling of the ECM usually changes the properties of the matrix and, in the case of aortic aneurysms, pathological remodeling of the aortic ECM contributes to disease progression 6–9. ECM remodeling is driven by proteolytic degradation mediated by a variety of proteases that include members of the matrix metalloprotease (MMP) family, which affect ECM-cell interactions to regulate cell proliferation and differentiation 10. We 11 and others 12–17 have reported increased abundance of MMP-2, MMP-9, and MT1-MMP in patients with aortic aneurysms and in mouse models of aneurysms, which exhibit a significant disruption of prominent members of the ECM of elastic arteries, specifically collagens and elastic fibers.
MMPs are synthesized as proenzymes that require activation. MMP activity is regulated by a family of proteins called tissue inhibitors of metalloproteases (TIMPs), which form tight non-covalent complexes with target MMPs. The levels of several MMPs, including MMP-2 18,19, MMP-9 20,21, MMP-13 22,23, ADAMTS-5 24 and ADAMTS-4 25 are regulated by the endocytic receptor, low-density lipoprotein receptor-related protein 1 (LRP1), which mediates their internalization and delivery to lysosomal compartments where they are degraded. In addition, LRP1 has been reported to directly bind several TIMP family members, including TIMP-1 26,27 and TIMP-3 28.
LRP1 is a multifunctional receptor that is involved in receptor-mediated endocytosis and various cellular signaling pathways. LRP1 was first recognized as a member of the LDL receptor (LDLR) family (reviewed in 29). The receptor localizes to lipid rafts and clathrin-coated pits where it undergoes constitutive endocytosis and recycling 30,31. Originally, LRP1 was identified as the hepatic receptor responsible for the catabolism of alpha-2-macroglobulin (α2M)-protease complexes 32,33 and was subsequently shown to be responsible for the hepatic removal of complexes of serine proteases and their complementary serpins 34. LRP1 is now known to bind and mediate the internalization of numerous ligands and to function in signaling pathways 35–37.
Published data thus far reveal that complexes of proteases and their target inhibitors bind much tighter to LRP1 than either component alone. For example, LRP1 directly interacts with plasminogen activator inhibitor-1 (PAI-1), a serpin that regulates the activity of two plasminogen activators, urokinase-type plasminogen activator (uPA) and tissue-type plasminogen activator. The binding affinities of PAI-1 and uPA alone to LRP1 are much weaker than that of the uPA:PAI-1 complex to LRP1, which exhibits an approximate 100-fold increase in affinity for LRP1 38–41. Further, native forms of α2M are not recognized by LRP1, whereas the α2M-protease complex binds to LRP1 with nanomolar (nM) affinity 32.
To gain insight into the mechanisms by which LRP1 regulates levels of MMPs, we initiated studies to investigate the binding of MMPs and their target inhibitors with LRP1. Since genetic deletion of LRP1 in vascular smooth muscle cells leads to aneurysm formation 11,42, we focused our studies on MMP-9 and MMP-1, both of which have been implicated in aneurysm formation 43,44. Our studies identify MMP-1 as a novel LRP1 ligand and reveal that both MMP-1 and proMMP-9 complexed with TIMP-1 bind much tighter to LRP1 than either protease alone. These results reveal that the physiological MMP ligands for LRP1 are likely the MMPs in complex with their TIMP inhibitor.
MATERIALS/EXPERIMENTAL DETAILS
Proteins
Human His-tagged proMMP-1 protein (Cat#: 10532-H08H, Uniprot entry P03956) and recombinant human MMP-3 catalytic domain (MMP-3cd) protein (Cat#: 10467-HNAE, Uniprot entry P08254) were purchased from Sino Biological. Anti-MMP-3 antibody (Cat#: MAB3306) and purified human proMMP-9 (Cat#: CC079, Uniprot entry P14780) were purchased from Chemicon. Rabbit anti-LRP1 IgG R2629 was prepared as previously described 45. Receptor associated protein (RAP) (Uniprot entry P30533) was expressed in Escherichia coli and purified as described 46. LRP1 (Uniprot entry Q07954) was purified from human placenta as previously described 32. TIMP-1 (Uniprot entry P01033), TIMP-2 (Uniprot entry P16035), TIMP-3 (Uniprot entry P35625), TIMP-4 (Uniprot entry Q99727) were prepared as described previously 47. Recombinant human LRP1 cluster II, III, and IV Fc chimera proteins were obtained from Molecular Innovations.
Cell culture
MMP-9−/− mouse embryonic fibroblasts (MMP-9 KO MEFs) and TIMP-1−/− MEFs (TIMP-1 KO MEFs) were kindly gifted by Zena Werb (University of California, San Francisco) and used to examine proMMP-9/TIMP-1 uptake in vitro. Mouse embryonic fibroblasts (MEFs), MMP-9 KO MEFs, and TIMP-1 KO MEFs were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Corning) containing Nutridoma (Sigma). Human aortic smooth muscle cells (hAoSMCs) isolated from a healthy male donor aged 16-years-old were purchased from Lonza (Cat#: CC-2571; Walkersville, Maryland) and cultured in Clonetics™ SmGM-2™ growth medium (Lonza) containing 10% fetal bovine serum (FBS; Atlanta Biologicals). Cultured cells were maintained at 37°C, 5% CO2 in a humidified atmosphere.
MMP activation and complex formation with TIMP-1
ProMMP-1 was dialyzed prior to any manipulations into 0.01 M HEPES, 0.15 M NaCl, 1 mM CaCl2, pH 7.4 with four exchanges of 250 mL each in D-Tube Dialyzer Mini spin columns with molecular weight cut-off of 6-8 kDa (Millipore, Cat# 71504-3). For proMMP-1 activation, MMP-3 was activated by incubation with 1.5 mM p-aminophenylmercuric acetate (APMA, Sigma, Cat#: A-8514) at 37°C for 2 hours which yielded active catalytic domain MMP-3 (MMP-3cd). Subsequently, proMMP-1 was activated by incubation with a 0.1 molar ratio of active MMP-3cd and 1 mM APMA at 37°C for 2 hours. MMP-3cd was removed by immunoprecipitation with anti-MMP-3 antibody (Chemicon, Cat#: MAB3306) and protein G beads (New England BioLabs, Cat#: S1430S) at a concentration of 0.2 ug antibody/mg beads. APMA was removed by size exclusion spin columns (Spin-X centrifuge tube, 0.45 µM pore, Costar, Cat#: CLS8163). To form MMP-1/TIMP-1 complexes, active MMP-1 was incubated with an equimolar amount of TIMP-1 at 37°C for 1 hour.
Purified TIMP-free MMP-9 (75 μg) was activated by incubation at 37°C for various times with either 2 mM APMA or 100 mM MMP-3cd. To form proMMP-9/TIMP-1 complexes, proMMP-9 was incubated with TIMP-1 at a 2.5:1 ratio by weight.
Protein Analysis
Receptor blotting was performed as previously described 20 with changes denoted below. Briefly, proteins were analyzed by SDS-PAGE using 4-12% Tris-glycine gradient gels (Novex) under non-reducing conditions. SDS-PAGE gels were then either stained with Coomassie or Colloidal Coomassie protein stain overnight and destained for 2 hours at room temperature or transferred to nitrocellulose membranes. Membranes were blocked in 3% nonfat milk and incubated with the monoclonal anti-LRP1 antibody 8G1 and goat anti-mouse IgG conjugated to HRP and visualized by chemiluminescence.
Fluorescence substrate assay
To measure activity of MMP-1, a quenched fluorogenic substrate (DNP-Pro-Cha-Abu-Cys(Me)-His-Ala-Lys(N-Me-Abz)-NH₂, [Cha = β-cyclohexylalanyl; Abu = L-α-aminobutyryl; Abz = 2-aminobenzoyl], Calbiochem, Cat#: 444219) was used according to manufacturer protocol. 200 μM fluorogenic substrate was added to 20 nM of indicated forms of MMP-1, and fluorescence measurements were recorded every 20 seconds for 2 hours using an excitation wavelength of 365 nm and emission wavelength of 450 nm.
Surface plasmon resonance
Surface plasmon resonance (SPR) experiments were performed as described in Migliorini et al. 41. Briefly, purified LRP1, LRP1 cluster II, LRP1 cluster III, or LRP1 cluster IV were immobilized on a CM5 sensor chip (GE Healthcare Life Sciences) surface at 9619 response units, 5501 response units, 4407 response units, 4166 response units, respectively, using a working solution of 20 μg/ml of each protein. All SPR experiments were performed in triplicate except those for proMMP-9 and the proMMP-9/TIMP-1 complex, which were performed in duplicate. All SPR experiments were performed on a Biacore 3000 instrument (GE Healthcare Life Sciences). All ligands were assessed by dilution into HBS-P or HBS-EP buffer (GE Healthcare Life Sciences) at a flow rate of 20 μl/min at 25°C. Regeneration of surfaces was achieved by 30 second injections of 100 mM phosphoric acid at a 100 μl/min flow rate.
Surface plasmon resonance data analysis
Kinetic data were analyzed by a bivalent model using BIAevaluation software as described previously 41. Equilibrium binding data was determined by fitting the association rates to a pseudo-first order process to obtain Req. Req was then plotted against total ligand concentration and fit to a binding isotherm using non-linear regression analysis in GraphPad 8.0 software as described 41.
Alkylation of proMMP-1 and TIMP-1
Alkylation of proMMP-1 and TIMP-1 were performed as previously described in Migliorini, et al., 41. Briefly, primary amines of lysine residue side chains on proMMP-1 or TIMP-1 were chemically modified by 50-fold excess of Sulfo-NHS-acetate (Thermo-Fisher Scientific) for 3 hours at 4°C. Alkylated proMMP-1 or TIMP-1 was then desalted.
Cell-mediated internalization assays
Either proMMP-1, proMMP-9, or TIMP-1 were labeled with the iodine-125 isotope (125I; Perkin Elmer NEZ-033) using Pierce™ Iodination Reagent (Thermo Scientific). Iodinated ligands (125I-proMMP-1, 125I-proMMP-9, 125I-TIMP-1) were desalted using a PD-10 column (GE Healthcare). For experiments examining uptake of complexes, either 125I-proMMP-9 or 125I-TIMP-1 were incubated with an equimolar amount of TIMP-1 or proMMP-9, respectively, for one hour on ice. For experiments with 125I-proMMP-1, hAoSMCs were used in passage 3-6. Cells were trypsinized and resuspended in Clonetics™ SmGM-2™ growth medium containing 10% FBS for hAoSMCs or DMEM containing Nutridoma for MEFs, and seeded at 7.2 × 104 cells/well for hAoSMCs or 1 × 105 cells for MEFs, in a 12-well tissue culture plate. Cultures were maintained overnight for 16-18 hours at 37°C, 5% CO2 in a humidified atmosphere. The following day, cultures at 60-70% confluency were washed with DPBS and incubated in serum-free DMEM, 20 mM HEPES, 1 mM CaCl2, 1.5% bovine serum albumin (BSA; Sigma-Aldrich) (assay media) for one hour at 37°C. After one hour, assay media was aspirated from each well and cells were incubated with 500 µL of assay media containing 125I-proMMP-1 (25 nM), 125I-proMMP-9 (5 nM), TIMP-1/125I-proMMP-9 (5 nM), 125I-TIMP-1 (5 nM), or proMMP-9/125I-TIMP-1 (5 nM) for 0, 1, 3, 6, 12, or 24 hours at 37°C. At each time point, 400 µL of assay media from each well was added to a microcentrifuge tube containing 100 µL of 50% trichloroacetic acid (TCA; Sigma-Aldrich) to determine degradation of the iodinated ligand. Cells were washed twice with DPBS and the wash was carefully aspirated. Cells were then detached from the plate with 0.05% trypsin, 0.53 mM EDTA (Corning) containing 50 µg/mL proteinase K (Thermo Scientific), collected from each well and transferred to a microcentrifuge tube, and centrifuged at 1,200 rpm for 5 minutes at room temperature. The supernatant was then removed from the cell pellet. Internalization and degradation of iodinated ligand was determined by radioactivity in the cell pellet and TCA-precipitated assay media, respectively. The total number of cells was counted in three separate wells and the average cell number was used for normalization.
Statistical Analysis
All graphs were plotted as the mean ± standard error mean (SEM). Data was analyzed by one-way ANOVA with a post-hoc Tukey’s test for data groups of more than two samples. Significance was recognized as a p-value of less than 0.05.
RESULTS
MMP-1 is a novel ligand for LRP1.
MMP-1 is a collagenase with emerging importance since abnormally high levels are associated with ECM breakdown and pathogenesis. Although multiple MMP family members have been identified as ligands for LRP1, the interaction between MMP-1 and LRP1 has not been investigated. Previous studies have demonstrated that the hemopexin domain of MMP-9 is recognized by LRP1 21, and since MMP-1 also contains a hemopexin-like domain, we initiated studies to determine if LRP1 recognizes various forms of MMP-1. The purity of proMMP-1 was assessed by SDS-PAGE. The results (Fig 1A, inset) reveal that proMMP-1 migrates as a doublet which is expected since proMMP-1 is secreted as a glycosylated as well as unglycosylated protein 48. In addition, small amounts of a 25 kDa fragment were detected which is generated by cleavage of the hinge region of proMMP1 49. proMMP-1 was activated by incubation with active MMP-3 and APMA, and then complexed with TIMP-1 to form the MMP-1/TIMP-1 complex. Activity assays using a quenched fluorescent MMP substrate 50 confirmed activation of MMP-1 and subsequent inhibition by TIMP-1 (Fig 1A). Further, the data reveal that proMMP-1 was unable to cleave this substrate.
Figure 1. All forms of MMP-1 bind to LRP1 by known LRP1-mediated mechanisms.
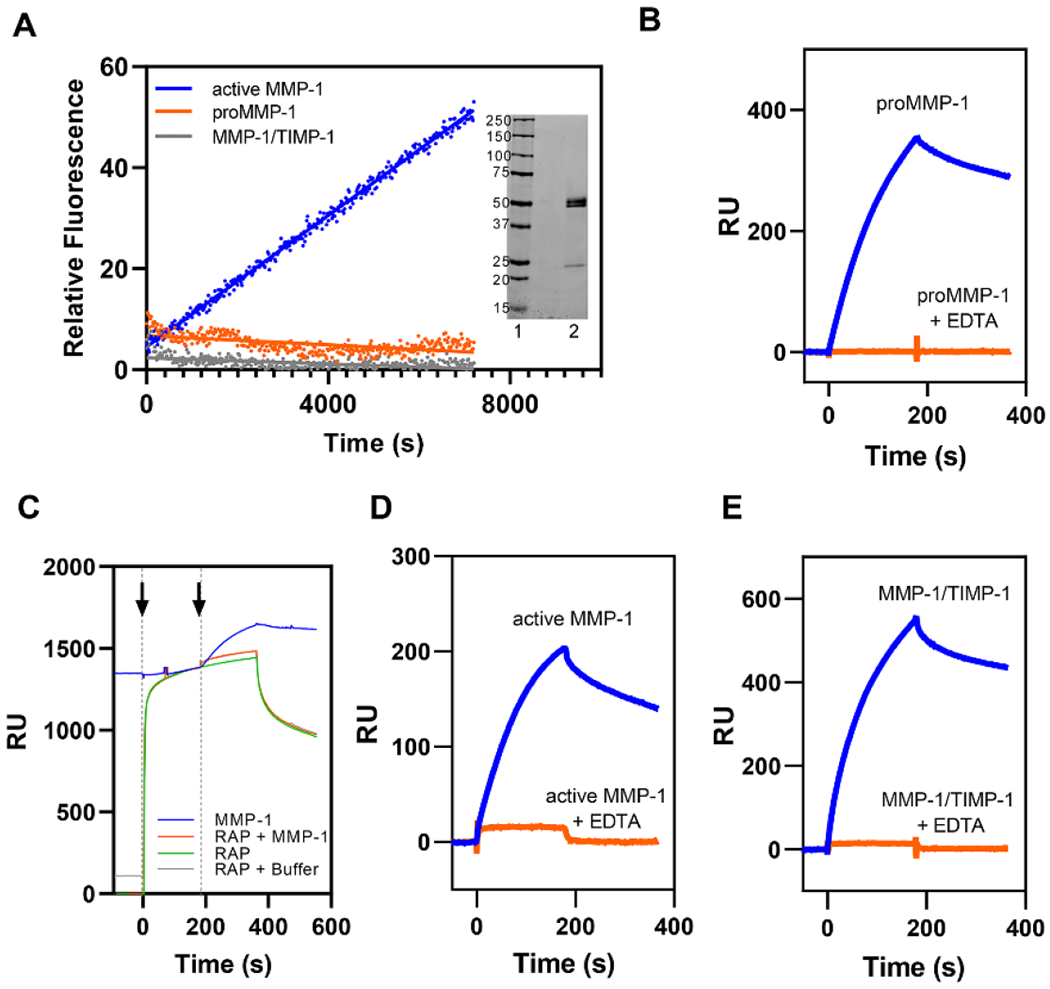
A. MMP-1/TIMP-1 complex formation abolished MMP-1 protease activity as assessed by fluorescent substrate assay. Each form of MMP-1 (20 nM) was incubated with 200 µM of quenched fluorescent substrate. Fluorescence was measured every 20 seconds for 2 hours. All measurements were controlled for by background subtraction and then the relative fluorescence intensity was averaged across all replicates (n = 3). proMMP-1 (orange), active MMP-1 (blue), and MMP-1/TIMP-1 complex (gray). Inset. SDS PAGE analysis of proMMP-1 under non-reducing conditions. Lane 1, Standards, Lane 2, proMMP-1. B. Purified LRP1 was immobilized on a CM5 sensor chip and 75 nM proMMP-1 was injected in the absence (blue lines) or presence (orange lines) of 3 mM EDTA. C. RAP (1 μM, green) was injected (first arrow) on an LRP1-coated CM5 sensor chip (green) followed by either a co-injection (second arrow) of 75 nM proMMP-1 and 1 μM RAP (orange), a co-injection of buffer and 1 μM RAP (gray), or another injection of 1 μM RAP (green). These traces were compared to an injection of 75 nM proMMP-1 (blue) in the absence of RAP. D-E. Purified LRP1 was immobilized on a CM5 sensor chip and active MMP-1 (D) or MMP-1/TIMP-1 complex (E) were injected in the absence (blue lines) or presence (orange lines) of 3 mM EDTA. The data shown is a representative experiment from three independent experiments that were performed.
To assess if MMP-1 is capable of binding LRP1, we employed surface plasmon resonance (SPR) experiments and injected 75 nM of proMMP-1 over an SPR surface to which purified LRP1 was coupled. The results confirmed binding of proMMP-1 to LRP1 (Fig 1B, blue). We next performed an SPR competition assay in the presence of receptor associated protein (RAP). RAP is an endogenous chaperone protein found in the endoplasmic reticulum that is responsible for facilitating protein folding of LRP1. RAP has a binding affinity for LRP1 in the nanomolar range and it competitively inhibits binding of all known LRP1 ligands identified to date 46,51. In this experiment, RAP was first injected over the LRP1-coated surface and allowed to bind (Fig 1C, RAP, green). Then, 75 nM of proMMP-1 complex along with RAP (Fig 1C, RAP + MMP-1, orange) was co-injected and the results compared to injection of 75 nM proMMP-1 alone (Fig 1C, MMP-1, blue). Blocking available LRP1 binding sites with RAP pre-incubation completely abolishes MMP-1 binding, suggesting that the MMP-1 interaction is LRP1-specific. Together, the data demonstrate that MMP-1 is a novel ligand for LRP1 interacting with this receptor in a process inhibited by RAP.
All forms of MMP-1 bind to LRP1.
We next conducted experiments to determine if various forms of MMP-1 also bind to LRP1. The results of these experiments reveal that, in addition to proMMP-1 (Fig 1B), both active MMP-1 (Fig 1D) and the MMP-1/TIMP-1 complex (Fig 1E) also bind to LRP1. Importantly, binding of all three forms of MMP-1 to LRP1 was completely inhibited when EDTA was added (Fig 1B, D, E) to chelate the essential calcium ions that are required to stabilize the ligand binding repeats of LRP1 52,53. This result provided further support that MMP-1 has specific binding to LRP1. Together, the data reveal that LRP1 recognizes proMMP-1, active MMP-1, and the MMP-1/TIMP-1 complex.
All TIMP family members bind directly to LRP1.
We next examined the binding of TIMP-1, TIMP-2, TIMP-3, and TIMP-4 to LRP1 by measuring equilibrium binding to LRP1 using SPR. The results confirmed that all TIMPs bind to LRP1 (Fig 2) with binding affinities ranging from 23 nM to 33 nM for various TIMPs (Table 1). The KD values we measured for TIMP-1 binding to LRP1 is similar to the reported binding affinity for TIMP-1 to cluster II of LRP1 26,27 while the KD values we measured for TIMP-3 binding to LRP1 is similar to earlier reports 54.
Figure 2. All TIMP family members bind to LRP1 with similar binding affinities.

SPR experiments examined binding of TIMP-1 (A), TIMP-2 (B), TIMP-3 (C), or TIMP-4 (D) to full length LRP1. Concentrations used were: 0.58, 1.17, 2.34, 4.68, 9.38, 18.75, 37.5, 75, and 150 nM for TIMP-1; 4.68, 9.38, 18.75, 37.5, 75, and 150 nM for TIMP-2; 0.29, 0.58, 1.17, 2.34, 4.68, 9.38, 18.75, 37.5, 75, and 150 nM for TIMP-3 and TIMP-4. At each concentration, the binding association curves were fit to a pseudo-first order process to determine Req. Req values were then plotted versus concentration, and the data fit to a binding isotherm using GraphPad Prism 8.0 software. To normalize the data from different experiments, Req/Rmax was plotted versus ligand concentrations, and the data plotted shows mean ± SEM (n=3).
Table 1.
Equilibrium binding constants for the interaction of TIMP-1, TIMP-2, TIMP-3, and TIMP-4 with LRP1. Equilibrium binding constants were calculated from equilibrium SPR measurements, in which Req was determined by fitting the association data to a pseudo-first-order process. Req was then plotted versus concentration and analyzed by non-linear regression analysis to determine the KD using GraphPad 8.0 software.
| Ligand | KD (nM) |
|---|---|
| TIMP-1 | 23 ± 5 |
| TIMP-2 | 30 ± 14 |
| TIMP-3 | 33 ± 12 |
| TIMP-4 | 24 ± 10 |
Lysine residues on MMP-1 and TIMP-1 are essential for binding to LRP1.
Since lysine residues have been reported to be critical for the binding of ligands to LRP1 41,55–57 we initiated experiments to chemically modify these residues on proMMP-1 and TIMP-1 with Sulfo-NHS-Acetate. Initially, we conducted experiments to assure that modified MMP-1 and TIMP-1 retained functional activity. The results (Fig 3A) reveal that following activation of alkylated MMP-1, the enzyme was still active, and formed a complex with TIMP-1 as revealed by loss of enzymatic activity. Further, alkylated TIMP-1 still retained its ability to inhibit the enzymatic activity of active MMP-1.
Figure 3. Alkylation of either MMP-1 or TIMP-1 prevents binding of MMP-1/TIMP-1 complexes to LRP1.
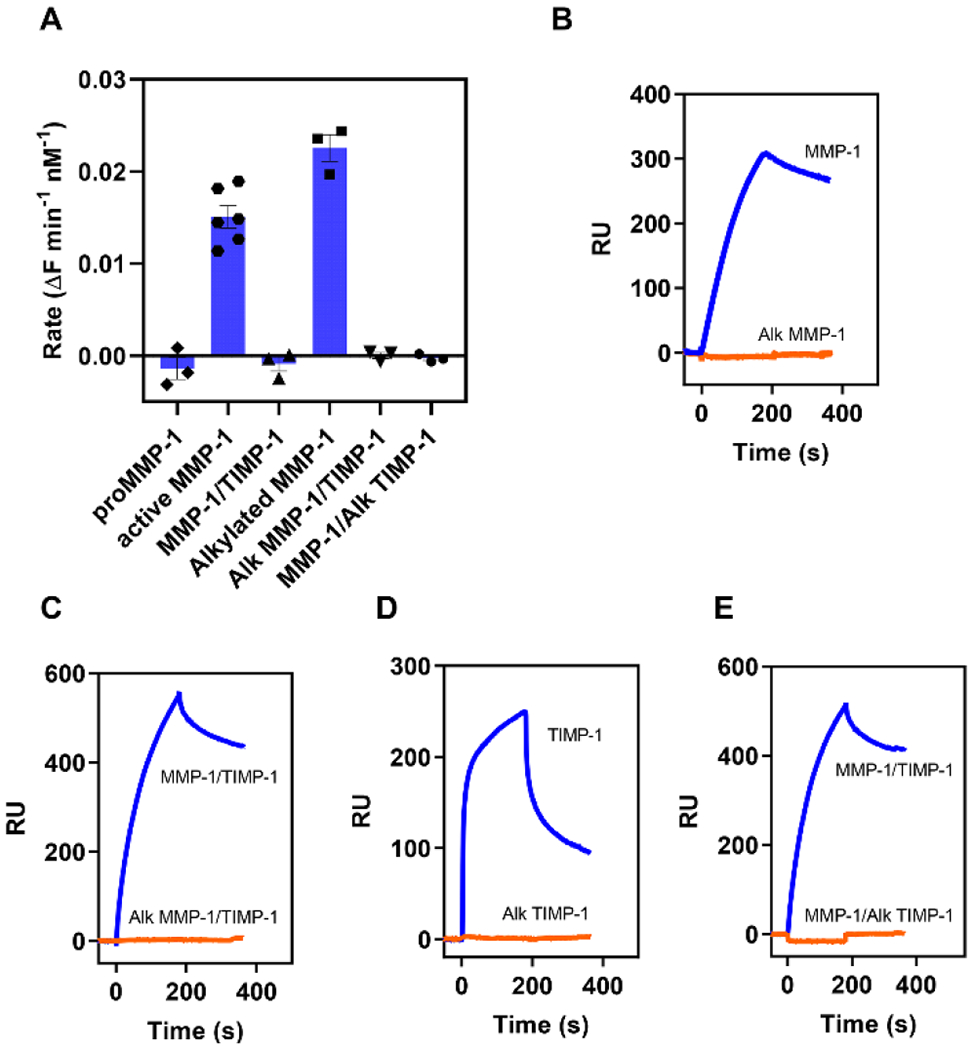
Lysine residues on proMMP-1 or TIMP-1 were alkylated (Alk MMP-1 or Alk TIMP-1) by incubation with a 50-fold molar excess of Sulfo-NHS-acetate at 4°C for 2 hours. The alkylated proteins were then dialyzed into HBS + 1 mM CaCl2. Alk or unmodified proMMP-1 was then activated and complexed with either unmodified TIMP-1 or Alk TIMP-1. The experiment was repeated 3 times. (A) Activity for each MMP-1 species or complex was determined by fluorescent substrate assay using either 20 nM of unmodified or 40 nM of alkylated species or complex. For each replicate, background hydrolysis of the substrate was subtracted and the rate determined from the slope of the linear regression of the data. (B-E) LRP1 was immobilized on a CM5 chip and 75 nM of unmodified (blue) or 100 nM alkylated (orange) MMP-1 or TIMP-1 species or complex were injected over the chip. The data shown is a representative experiment from three independent experiments that were performed.
We next assessed the ability of modified proMMP-1 to bind LRP1. The results revealed that modification of lysine residues on MMP-1 prevented the binding of this molecule to LRP1 (Fig 3B). Interestingly, the complex formed by incubation of alkylated MMP-1 and unalkylated TIMP-1 (Alk MMP-1/TIMP-1) also failed to bind to LRP1 (Fig 3C). Our studies also revealed that alkylated TIMP-1 failed to bind LRP1 (Fig 3D) and that the complex of MMP-1/alkylated TIMP-1 also failed to bind LRP1 (Fig 3E). These results demonstrate that lysine residues on MMP-1 and TIMP-1 contribute to their binding to LRP1. In addition, determinants on both MMP-1 and TIMP-1 appear important for interaction of the MMP-1/TIMP-1 complex with LRP1.
MMP-1/TIMP-1 complexes are the preferred over MMP-1 and active MMP-1 for LRP1 binding.
The data presented in Figure 3 suggests the involvement of lysine residues on MMP-1 in the interaction with LRP1. This is consistent with the canonical model for the binding of ligands to LDLR family members in which two (or more) ε-amino groups of specific lysine residues located on the ligand form salt bridges with carboxylates of aspartate residues within the LDLR class a (LDLa) repeats 58. These observations raise the possibility that binding of MMP-1 to LRP1 would adhere to a bivalent kinetic model in which high-affinity binding results from avidity effects mediated by the interaction of two separate regions on MMP-1, each containing charged residues, with two LDLa repeats on LRP1 (Fig 4A). Similar models have been proposed for the binding of FVIII 56 and PAI-1 41 to LRP1. To test this model, we performed kinetic measurements examining the binding of proMMP-1 (Fig 4B), active MMP-1 (Fig 4C) and the MMP-1/TIMP-1 complex (Fig 4D) to LRP1 using SPR experiments. The results revealed that the experimental data are well described by a bivalent binding model.
Figure 4. Binding of all forms of MMP-1 to LRP1 is well described by a bivalent binding model.
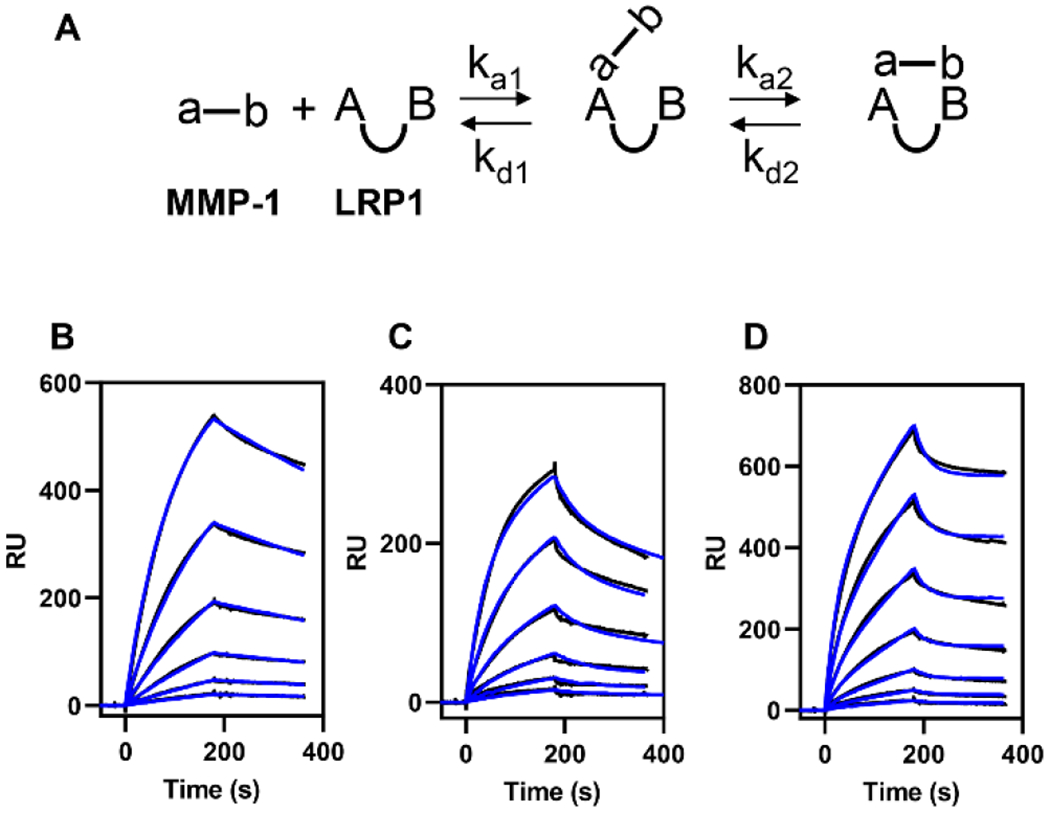
(A) Schematic of the bivalent binding model used to fit the data. In this model, the MMP-1 ligand contains two regions (a-b) that interact with two LDLa ligand binding repeats on LRP1 (AB). The first region on MMP-1 (a) docks into an LDLa repeat (A) to form the initial complex. Then, the second region on MMP-1 (b) docks into the remaining LDLa repeat (B) to form the bivalent complex. (B) Increasing concentrations (4.7, 9.4, 18.7, 37.5, 75, and 150 nM) of proMMP-1 were injected over the LRP1-coated surface. (C) Increasing concentrations (4.7, 9.4, 18.7, 37.5, 75, and 150 nM) of active MMP-1 were injected over the LRP1-coated surface. (D) Increasing concentrations (2.3, 4.7, 9.4, 18.7, 37.5, 75, and 150 nM) of MMP-1/TIMP-1 complex were injected over the LRP1-coated surface. Fits of the experimental data (black lines) to a bivalent binding model are shown as blue lines. The data shown is a representative experiment from three independent experiments that were performed.
The kinetic data derived from the best fit of the experimental data to the bivalent model (Table 2) reveal that all forms of MMP-1 bind with high affinity under 30 nM. Interestingly, MMP-1/TIMP-1 complexes bind to LRP1 with approximately 30-fold increased affinity (KD = 0.6 ± 0.2 nM) over that of proMMP-1 (KD = 19 ± 1 nM) or active MMP-1 (KD = 25 ± 2 nM). Inspection of the kinetic constants reveal that the initial association (ka1) is similar for proMMP-1, active MMP-1, and the MMP-1/TIMP-1 complex. The higher affinity of MMP-1/TIMP-1 complex for LRP1 primarily results from approximately 100-fold slower dissociation rate (kd2) from complex II (Fig 4A, Table 2).
aTable 2.
Kinetic and equilibrium constants for the binding of proMMP-1, proMMP-9, MMP-1/TIMP-1 complexes, and proMMP-9/TIMP-1 complexes to LRP1.
| Ligand | ka1 (M−1s−1) | kd1 (s−1) | ka2 (s−1) | kd2 (s−1) | bKD (nM) |
|---|---|---|---|---|---|
| ProMMP-1 | 9.9 ± 2.0 X 104 | 1.4 ± 0.5 X 10−1 | 2.0 ± 0.2 X 10−1 | 3.2 ± 0.7 X 10−3 | 19 ± 1 |
| Active MMP-1 | 1.0 ± 0.1 X 105 | 1.7 ± 0.4 X 10−2 | 8.4 ± 0.9 X 10−3 | 1.5 ± 0.2 X 10−3 | 25 ± 4 |
| MMP-1/TIMP-1 complex | 9.3 ± 0.5 X 104 | 2.6 ± 0.1 X 10−2 | 1.2 ± 0.1 X 10−2 | 2.7 ± 1.5 X 10−5 | 0.6 ± 0.4 |
| ProMMP-9 | 2.1 ± 0.3 X 105 | 1.1 ± 0.2 X 10−1 | 1.1 ± 0.1 X 10−2 | 2.6 ± 0.1 X 10−3 | 95 ± 2 |
| ProMMP-9/TIMP-1 complex | 2.0 ± 0.3 X 105 | 4.2 ± 0.9 X 10−2 | 6.8 ± 1.0 X 10−3 | 8.6 ± 1.1 X 10−4 | 23 ± 1 |
Kinetic constants were obtained by fitting the data to a bivalent model (Fig 4A). Three independent experiments were performed, and the values shown are the average ± SEM.
The equilibrium binding constant KA was calculated using the following equation: KA = (ka1/kd1)*(1 + (ka2/kd2)) and KD was calculated as: KD= 1/KA
LRP1 expressed in human aortic smooth muscle cells mediates the endocytosis of proMMP-1.
Significant increases in activity of various MMPs have been implicated in the pathogenesis of aortic aneurysms and thus it is essential to define mechanisms regulating MMP activity that maintain vascular homeostasis. To determine if LRP1 mediates the endocytosis of MMP-1, we examined the cellular uptake of 125I-labeled proMMP-1 (125I-proMMP-1) by human aortic smooth muscle cells (hAoSMCs). To confirm the contribution of LRP1 to this process we also measured the uptake in the presence of excess RAP and an anti-LRP1 IgG. The results reveal that treatment with either RAP or anti-LRP1 IgG significantly inhibit the cellular internalization (Fig 5A) and degradation (Fig 5B) of 125I-proMMP-1. These results confirm that LRP1 functions as an endocytic receptor for MMP-1 at the cellular level. Interestingly, incubation of cells with anti-LRP1 IgG to block LRP1 function results in a slight increase in proMMP-1 expression on the cell surface (Fig 5C). These results are similar to what we have observed for the LRP1-mediated VLDL uptake induced by lipoprotein lipase which is sequestered on the cell surface by association with cell surface proteoglycans 59.
Figure 5. proMMP-1 is internalized and degraded in an LRP1-dependent manner.
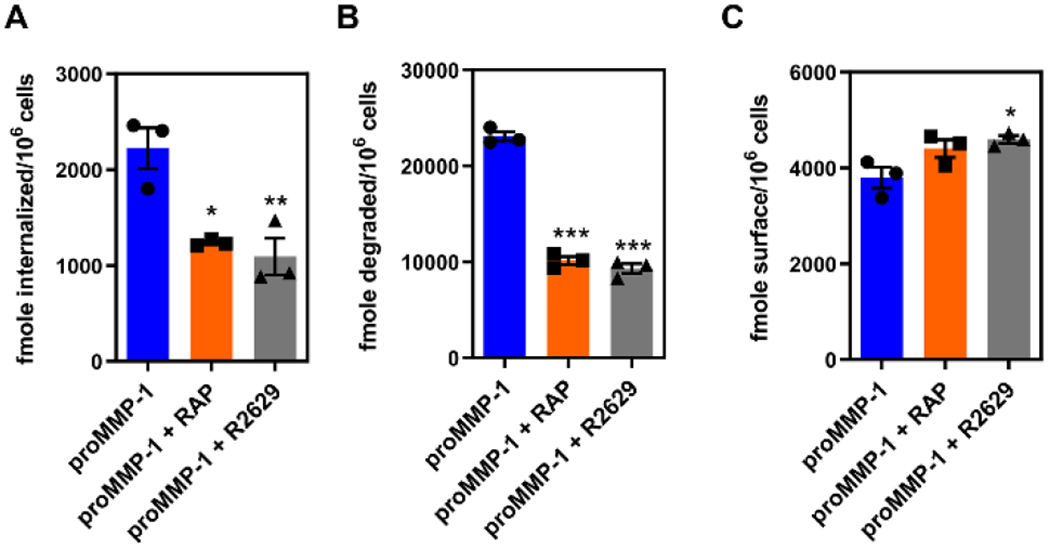
hAoSMCs were plated at 7.2 × 10⁴ cells per well in a 12-well plate and were incubated with 125I-labeled proMMP-1 (25 nM) for 24 hours at 37°C in the absence or presence of RAP (2.5 μM) or the LRP1-specific polyclonal antibody R2629 (300 mg/mL). Following incubation, the amount of proMMP-1 internalized (A) degraded (B) or located on the cell surface (C) was measured. All data are plotted as mean ± SEM (n=3). Statistical significance was determined by one-way ANOVA with post-hoc Tukey’s test (*p<0.05, **p<0.01, ***p<0.0001).
MMP-1 and TIMP-1 preferentially bind to cluster III in LRP1.
Virtually all LRP1 ligands bind to clusters of LDLa repeats, termed clusters I, II, III and IV. Of these four clusters, most ligands prefer either cluster II or IV 60. We next examined the ability of MMP-1 and TIMP-1 to bind to clusters II, III, and IV immobilized on the SPR chip. The data reveal that proMMP-1 preferentially binds to cluster III of LRP1, while its binding to cluster II and cluster IV is 5-fold and 10-fold weaker, respectively (Fig 6A, Table 3). Likewise, TIMP-1 prefers cluster III, while its binding to cluster II and cluster IV is 3-fold and 10-fold weaker, respectively (Fig 6B, Table 3). The apparent preference of TIMP-1 and MMP-1 in binding to cluster III of LRP1 is interesting in light of the findings that TIMP-3 preferentially binds to cluster II of LRP1 54, while proMMP-13 binds equally well to cluster II and III of LRP1 and only weakly to cluster IV 23.
Figure 6. Both proMMP-1 and TIMP-1 bind with greatest affinity to Cluster III binding region of LRP1.
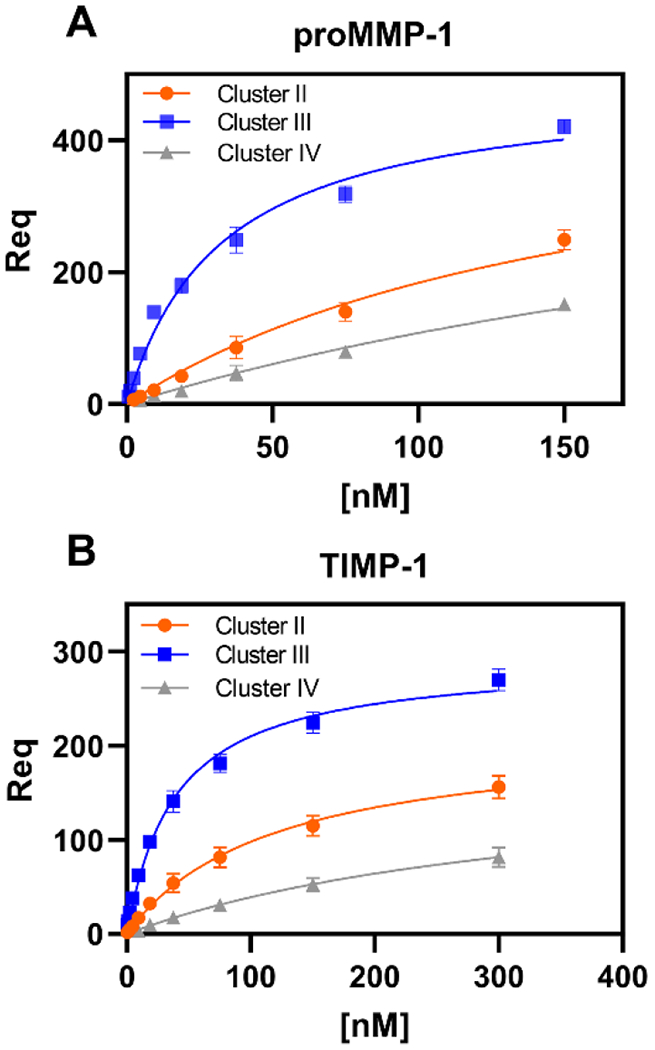
Recombinant LRP1 fragments of either Cluster II, III, or IV were immobilized to three individual flow cells of a CM5 sensor chip using an amine-reactive coupling process. SPR experiments tested binding of proMMP-1 (A) or TIMP-1 (B) to Cluster II (orange), III (blue), and IV (gray) with increasing concentrations of ligand (proMMP-1: 0.29, 0.58, 1.17, 2.34, 4.68, 9.38, 18.75, 37.5, 75, and 150 nM; TIMP-1: 0.58, 1.17, 2.34, 4.68, 9.38, 18.75, 37.5, 75, 150, and 300 nM). At each concentration, the binding association curves were fit to a pseudo-first order process. Req was estimated as the maximum number of response units at equilibrium for each concentration. Shown are the plots of Req versus ligand concentration. Data is plotted as mean ± SEM (n=3). The data were normalized to the amount of cluster coated on the CM5 sensor chip. GraphPad Prism 8.0 software was used to fit the data to the specific binding non-linear regression model to determine the KD of each ligand for each cluster. All calculated KD values are reported in Table 3.
aTable 3.
Equilibrium binding constants for the interaction of proMMP-1 and TIMP-1 with LRP1 clusters.
| Ligand | LRP1 cluster | KD (nM) |
|---|---|---|
| ProMMP-1 | Cluster II | 169 ± 21 |
| ProMMP-1 | Cluster III | 34 ± 5 |
| ProMMP-1 | Cluster IV | 358 ± 38 |
| TIMP-1 | Cluster II | 129 ± 28 |
| TIMP-1 | Cluster III | 39 ± 2 |
| TIMP-1 | Cluster IV | 391 ± 67 |
Equilibrium binding constants were calculated from equilibrium SPR measurements, in which Req was determined by the fitting the association data to a pseudo-first-order process. Req was then plotted versus concentration and analyzed by non-linear regression analysis to determine the KD using GraphPad 8.0 software.
We also examined the binding of MMP-1/TIMP-1 complexes to LRP1 ligand binding cluster and the results are summarized in Table 4. The data confirm a preference of the MMP-1/TIMP-1 complex for Cluster III, and reveal that MMP-1/TIMP-1 complexes do not bind as tightly to the clusters as to the intact LRP1 molecule. These data suggest that MMP-1/TIMP-1 complexes may also interact with regions of LRP1 that are outside of the specific cluster used in the experiment. Similar results have been noted for the interaction of low molecular weight uPA:PAI-1 complexes with LRP1 clusters 41.
aTable 4.
Kinetic and equilibrium constants for the binding of MMP-1/TIMP-1 complexes to LRP1 ligand binding clusters.
| LRP1 cluster | ka1 (M−1s−1) | kd1 (s−1) | ka2 (s−1) | kd2 (s−1) | bKD (nM) |
|---|---|---|---|---|---|
| Custer II | 1.2 ± 0.1 X 105 | 7.7 ± 0.1 X 10−2 | 2.0 ± 0.1 X 10−2 | 8.5 ± 0.9 X 10−4 | 27 ± 2 |
| Cluster III | 2.0 ± 0.1 X 105 | 7.9 ± 0.2 X 10−2 | 1.9 ± 0.1 X 10−2 | 6.2 ± 0.4 X 10−4 | 12 ± 1 |
| Cluster IV | 9.9 ± 1.3 X 104 | 6.4 ± 0.3 X 10−2 | 2.0 ± 0.1 X 10−2 | 1.0 ± 0.1 X 10−3 | 32 ± 5 |
Kinetic constants were obtained by fitting the data to a bivalent model (Fig 4A). Three independent experiments were performed, and the values shown are the average ± SEM.
The equilibrium binding constant KA was calculated using the following equation: KA = (ka1/kd1)*(1 + (ka2/kd2)) and KD was calculated as: KD= 1/KA
LRP1 also shows a preference for the proMMP-9/TIMP-1 complex.
Given the enhanced binding affinity for LRP1 that we observed for MMP-1 in complex with TIMP-1 when compared to either component alone, we decided to investigate other MMP family members that are known ligands for LRP1, in particular, MMP-9. In the experiments shown in Figure 7, proMMP-9 was activated either with MMP-3 or autocatalytically by treatment with a mercurial compound, APMA. We then employed a receptor blotting procedure which was successful in our initial characterization of the binding of MMP-9 to LRP1 20. The domain organization of MMP-9 is shown in Figure 7A. To visualize the cleavage of MMP-9 over time, the protease was treated with either MMP-3 catalytic domain (MMP-3cd) (Fig 7B) or APMA (Fig 7C) for the varying times and subjected to SDS-PAGE and Coomassie blue staining. Since activation of proMMP-9 by APMA results in a C-terminal cleavage event that does not occur by activation with MMP-3, we could further localize the binding site of LRP1 to MMP-9. When the cleavage products were analyzed by receptor blotting, we showed that all the amino terminally truncated species were able to bind to LRP1. The only cleavage product of MMP-9 that LRP1 does not bind to is the approximately 70 kDa MMP-9 species from which the C-terminus is removed (Fig 7, *). This is consistent with data revealing that the LRP1 binding site in MMP-9 is located within the C-terminal hemopexin domain 21.
Figure 7. Binding of LRP1 to MMP-9 requires an intact C-terminal hemopexin-like domain.

(A) Domain structure of MMP-9 with schematic representation of intact 92 kDa proMMP-9 and predicted fragment sizes by activation with MMP-3cd and APMA indicated. Purified TIMP-free MMP-9 (75 μg) was treated with either the MMP-3cd at 100 μM (B) or 2 mM APMA (C) for the indicated times. At each time point, 15 μg was removed for analysis by Coomassie protein staining or receptor blotting as indicated. Receptor binding was detected using monoclonal anti-LRP1 IgG 8G1 and goat anti-mouse IgG conjugated to HRP and visualized using chemiluminescence. MMP-9 is a 92 kDa zymogen. In the presence of either MMP-3cd or APMA, N-terminal cleavage of MMP-9 occurs yielding 86 and 82 kDa enzyme species (bracket). In the presence of APMA, the 82 kDa species undergoes C-terminal processing yielding a 68 kDa enzyme species (*).
To assess the effect of TIMP-1 on the binding of proMMP-9 to LRP1, we performed SPR experiments. Quantification of the interaction with proMMP-9/TIMP-1 complexes with LRP1 is especially important because proMMP-9 is often associated directly with its inhibitor, TIMP-1 61,62. We found that the binding data for both proMMP-9 and proMMP-9/TIMP-1 complexes to LRP1 fit well to a bivalent binding model as observed for MMP-1 (Fig 8). The kinetic parameters determined by fitting to a bivalent model are summarized in Table 2. These data revealed that LRP1 bound to MMP-9/TIMP-1 complexes (KD = 23 ± 1 nM) with a 4-fold greater affinity than for proMMP-9 alone (KD = 95 ± 1 nM) but with a similar binding affinity as that of TIMP-1 (Table 1, KD = 24 ± 3 nM). Thus, in the case of proMMP-9, complex formation with TIMP-1 increases the binding affinity of proMMP-9 to LRP1 but not more than that of TIMP-1, suggesting the binding epitope may be localized to TIMP-1 in the proMMP-9/TIMP-1 complex.
Figure 8. proMMP-9/TIMP-1 complexes bind with greater affinity to LRP1 than proMMP-9 alone.

Full length LRP1 was immobilized on a CM5 sensor chip using an amine-reactive coupling process. (A) proMMP-9 was injected over the chip in increasing concentrations of ligand (11.1, 33.3, 100, and 300 nM). (B) proMMP-9/TIMP-1 complexes were injected over the chip in increasing concentrations of ligand (3.7, 11.1, 33.3, 100, and 300 nM). Fits of the experimental data (black lines) to a bivalent binding model are shown as blue lines. The data shown is a representative experiment from two independent experiments that were performed.
LRP1-mediated catabolism of both proMMP-9 and TIMP-1 are enhanced by complex formation.
Since TIMP-1 increases the affinity of proMMP-9 for LRP1, experiments were performed to investigate LRP1-mediated internalization of free proMMP-9 and free TIMP-1, or either of these proteins in a complex. For these experiments, mouse embryonic fibroblasts (MEFs) were used in internalization experiments along with MEFs from TIMP-1 and MMP-9 knockout (KO) mice to compare the time course of internalization and degradation for the individual proteins versus the proMMP-9/TIMP-1 complex. The results confirm that both free proMMP-9 and free TIMP-1 were internalized and degraded by MEFs (Fig 9A). However, when 125I-labeled proMMP-9 in complex with TIMP-1 was examined, a significant increase in both internalization and degradation were observed, compared to either proMMP-9 or TIMP-1 alone (Fig 9A). To ensure that proMMP-9 internalization was not due to endogenously produced TIMP-1, the experiments were repeated in TIMP-1 KO fibroblasts (Fig 9B) which confirm that complexes of proMMP-9 with TIMP-1 are more efficiently internalized.
Figure 9. ProMMP-9/TIMP-1 complexes are internalized more effectively than proMMP-9 or TIMP-1 alone.

Cells were plated at 1 × 105 cells/well in a 12-well plate. A, B. 5 nM of 125I-labeled proMMP-9 (blue circles) or 125I-labeled proMMP-9/TIMP-1 (orange triangles) were incubated with mouse embryonic fibroblasts (MEFs) (A) or TIMP-1 KO MEFs (B) in the presence or absence of 1 μM RAP for indicated times and the amount internalized (left panel) or degraded (right panel) determined (n=2). C, D. 5 nM of 125I-labeled TIMP-1 (blue circles) or proMMP-9/125I-labeled TIMP-1 complex (orange triangles) were incubated with MEFs (C) or MMP-9 KO MEFs (D) in the presence or absence of 1 μM RAP for indicated times and the amount internalized (left panel) or degraded (right panel) determined (n=3). The data shown represent the RAP inhibitable fraction of internalized or degraded protein
These experiments were also performed in MEFs (Fig 9C) and MMP-9 KO fibroblasts (Fig 9D) using 125I-labeled TIMP-1 which confirm that complexes of proMMP-9 with TIMP-1 are more effectively internalized by LRP1 independent of endogenously produced MMP-9. These data support our SPR data revealing that high affinity binding of proMMP-9 to LRP1 requires complex formation with TIMP-1 in a system with biological relevance.
DISCUSSION
In the current investigation, we identified MMP-1 as a novel ligand for LRP1 that is internalized and degraded by cells in a process inhibited by anti-LRP1 IgG and RAP. The activity of MMPs are regulated by their TIMP inhibitors, and while proMMP-1, active MMP-1 and TIMP-1 bind to LRP1 with KD values of 19 nM, 25 nM, and 23 nM, respectively, our data reveal that the MMP-1/TIMP-1 complex binds with approximately 30-fold higher affinity to LRP1 (KD = 0.6 nM) than either component alone. This indicates that the physiological form of MMP-1 recognized by LRP1 for clearance is most likely the MMP-1/TIMP-1 complex. We extended these studies to investigate proMMP-9 and the proMMP-9/TIMP-1 complex and also noted an increased affinity of the complex for LRP1. The importance of this finding to regulation of MMPs in the extracellular environment was further demonstrated by cell uptake experiments where proMMP-9/TIMP-1 complexes are internalized more efficiently by LRP1 in vitro than proMMP-9 or TIMP-1 alone. Together, the data suggest that regulation of MMP-1 and proMMP-9 in the extracellular environment likely requires complex formation with inhibitors as the preferred ligands for LRP1. The identification of MMP-1 as a novel LRP1 ligand and the concept that TIMPs regulate LRP1-mediated clearance is in excellent agreement with the recent studies by Carreca et al. 63 whose data confirms that LRP1 binds MMP-1 with a 7-fold increased affinity after forming a complex with TIMP-3 complex (KD = 5 nM).
The data accumulated thus far identify that LRP1 is a critical regulator of protease activity with the preferred (i.e. high affinity) ligands represented as protease:inhibitor complexes. LRP1 was originally identified as the hepatic receptor for complexes of proteases with α2M 32,33,64. Further, studies confirmed that LRP1 functions as a hepatic receptor for protease:serpin complexes 34,65. Data from the current study and from the work of Carreca et al. 63 and Emonard et al. 19 confirm that MMPs are no exception.
A question that arises from these studies is how complex formation of MMP-1 with TIMP-1 enhances binding of the complex to LRP1. A canonical model of ligand binding to LDL receptor family members has emerged from the structural studies examining the third domain of RAP, which recognized that K256 and K270 on RAP are essential for the binding of this molecule to LRP1 66. A crystal structure of the third domain of RAP in complex with two LDLa repeats from the LDL receptor determined that the ε-amino groups of K256 and K270 on RAP form salt-bridges with carboxylates of aspartate residues that form acidic pockets within the LDLa repeats on the receptor 58. These studies suggest that contacts of multiple lysine residues contribute to avidity effects that represents a general binding model for ligand association to this class of receptors.
The current studies reveal that lysine residues on MMP-1 and TIMP-1 are critical for the interaction of these proteins with LRP1. The involvement of critical lysines on TIMP-3 has been previously demonstrated by mutagenesis experiments 67. We propose that the enhanced affinity of the complex for LRP1 results from additional contact sites in TIMP-1 for the following reasons: i) TIMP-1 is recognized by LRP1, ii) both TIMP-1 and MMP-1 are preferentially recognized by cluster III of LRP1, and iii) modification of lysine residues on either TIMP-1 or MMP-1 prevents the complex from binding to LRP1. Identification of specific lysine residues on MMP-1 and TIMP-1 that are involved in interacting with LRP1 represent important future work. Our binding studies to the LRP1 clusters reveal that while the MMP-1/TIMP-1 complex still has a preference for Cluster III, the complex does bind to Cluster II, III, and Cluster IV with higher affinity than either component alone. In addition, the data reveal that the MMP-1/TIMP-1 complex binds weaker to Cluster III (KD=12 nM) than to the full-length molecule (KD=0.6 nM) suggesting that other regions of LRP1 might also be involved in the interaction with the complex. Further, defining the exact molecular mechanism resulting in higher affinity of MMP-1/TIMP-1 complex for LRP1 than either ligand alone will require additional structural studies.
Our work identifies a potential critical regulation pathway for MMP-1 activity, an MMP that degrades collagen and is commonly upregulated in states of disease and injury. MMP-1 is only detectable at very low levels in plasma and in tissue in healthy individuals, but is upregulated in inflammation or disease (for review see 68). While MMP-1 has a crucial role in regulating tissue homeostasis in transient conditions, unregulated or excessive MMP-1 activity can cause extensive tissue damage that can promote disease conditions and pathologies highly associated with abnormal ECM breakdown. MMP-1 is known to digest native fibrillar collagens type Ⅰ, II, and Ⅲ and abnormally high levels of heart-specific MMP-1 expression result in collagen loss and diminished contractility that leads to cardiomyopathy 69. Upregulation of MMP-2 and MMP-9 has been identified as a key event occurring during aneurysm growth 43,70. More recently, MMP-1 has been implicated as an associating factor for the often fatal outcomes of aneurysms including dissections and ruptures 50,70. MMP-1 has been shown to be highly expressed in a variety of cancers 71,72, and it is well known that ECM breakdown can promote tumor progression. Tumors with high expression of protease inhibitors correlate with good prognosis, whereas those with high MMPs correlate with poor prognosis and increased risk of recurrence 73.
The novel role for LRP1 in regulating MMP-1 activity underlines the important role of LRP1 in regulating ECM remodeling by regulating the activities of protease and protease:inhibitor complexes. Increased understanding of how LRP1 regulates ECM remodeling and affects disease progression will contribute to the development of new therapeutics.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R35 HL135743 (DKS), F30 HL145952 (ALA), and T32 HL007698 (ALA), and American Heart Association Grant AHA 15SDG24470170 (SCM). SCM contributed to this article as an employee of the University of Maryland, Baltimore. The content is solely the responsibility of the authors and do not necessarily represent the views of the National Institutes of Health or the United States Government.
Footnotes
REFERENCES
- (1).Heck N, Garwood J, Larmet Y, and Faissner A (2004) Differential upregulation of extracellular matrix molecules associated with the appearance of granule cell dispersion and mossy fiber sprouting during epileptogenesis in a murine model of temporal lobe epilepsy. Neuroscience 129, 309–324. [DOI] [PubMed] [Google Scholar]
- (2).Morawski M, Brückner G, Jäger C, Seeger G, Matthews RT, and Arendt T (2012) Involvement of Perineuronal and Perisynaptic Extracellular Matrix in Alzheimer’s Disease Neuropathology. Brain Pathol. 22, 547–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Dzyubenko E, Gottschling C, and Faissner A (2016) Neuron-Glia Interactions in Neural Plasticity : Contributions of Neural Extracellular Matrix and Perineuronal Nets. Neural Plast. 2016, 5214961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Cox TR, and Erler JT (2011) Remodeling and homeostasis of the extracellular matrix : implications for fibrotic diseases and cancer 178, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Lu P, Takai K, Weaver VM, and Werb Z (2011) Extracellular Matrix Degradation and Remodeling in Development and Disease. Cold Spring Harb. Perspect. Biol. 3, a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Didangelos A, Yin X, Mandal K, Saje A, Smith A, Xu Q, Jahangiri M, and Mayr M (2011) Extracellular matrix composition and remodeling in human abdominal aortic aneurysms: a proteomics approach. Mol. Cell. Proteomics 10, M111.008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Fava M, Barallobre-Barreiro J, Mayr U, Lu R, Didangelos A, Baig F, Lynch M, Catibog N, Joshi A, Barwari T, Yin X, Jahangiri M, and Mayr M (2018) Role of ADAMTS-5 in aortic dilatation and extracellular matrix remodeling. Arterioscler. Thromb. Vasc. Biol 38, 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Fedak PWM, De Sa MPL, Verma S, Nili N, Kazemian P, Butany J, Strauss BH, Weisel RD, David TE, Yacoub MH, Sundt TM, Sellke FW, and Pizarro C (2003) Vascular matrix remodeling in patients with bicuspid aortic valve malformations: Implications for aortic dilatation. J. Thorac. Cardiovasc. Surg 126, 797–805. [DOI] [PubMed] [Google Scholar]
- (9).Mimler T, Nebert C, Eichmair E, Winter B, Aschacher T, Stelzmueller ME, Andreas M, Ehrlich M, Laufer G, and Messner B (2019) Extracellular matrix in ascending aortic aneurysms and dissections – What we learn from decellularization and scanning electron microscopy. PLoS One 14, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Bonnans C, Chou J, and Werb Z (2014) Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol 15, 786–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Muratoglu SC, Belgrave S, Hampton B, Migliorini M, Coksaygan T, Chen L, Mikhailenko I, and Strickland DK (2013) LRP1 protects the vasculature by regulating levels of connective tissue growth factor and HtrA1. Arterioscler. Thromb. Vasc. Biol 33, 2137–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Sakalihasan N, Delvenne P, Nusgens BV, Limet R, and Lapière CM (1996) Activated forms of MMP2 and MMP9 in abdominal aortic aneurysms. J. Vasc. Surg 24, 127–133. [DOI] [PubMed] [Google Scholar]
- (13).Xiong W, Knispel RA, Dietz HC, Ramirez F, and Baxter BT (2014) Doxycycline delays aneurysm rupture in a mouse model of Marfan syndrome. J. Vasc. Surg 47, 166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Xiong W, Knispel R, MacTaggart J, Greiner TC, Weiss SJ, and Baxter BT (2009) Membrane-type 1 Matrix Metalloproteinase Regulates Macrophage-dependent Elastolytic Activity and Aneurysm Formation in Vivo. J. Biol. Chem 284, 1765–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Jones JA, Ruddy JM, Bouges S, Zavadzkas JA, Brinsa TA, Stroud RE, Mukherjee R, Spinale FG, and Ikonomidis JS (2010) Alterations in membrane type-1 matrix metalloproteinase abundance after the induction of thoracic aortic aneurysm in a murine model. Am. J. Physiol. - Hear. Circ. Physiol 299, 114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Nataatmadja M, West M, West J, Summers K, Walker P, Nagata M, and Watanabe T (2003) Abnormal Extracellular Matrix Protein Transport Associated With Increased Apoptosis of Vascular Smooth Muscle Cells in Marfan Syndrome and Bicuspid Aortic Valve Thoracic Aortic Aneurysm. Circulation 108, II-329-II–334. [DOI] [PubMed] [Google Scholar]
- (17).Chung AWY, Au Yeung K, Sandor GGS, Judge DP, Dietz HC, and van Breemen C (2007) Loss of elastic fiber integrity and reduction of vascular smooth muscle contraction resulting from the upregulated activities of matrix metalloproteinase-2 and −9 in the thoracic aortic aneurysm in Marfan syndrome. Circ. Res 101, 512–522. [DOI] [PubMed] [Google Scholar]
- (18).Yang Z, Strickland DK, and Bornstein P (2001) Extracellular Matrix Metalloproteinase 2 Levels are Regulated by the Low Density Lipoprotein-related Scavenger Receptor and Thrombospondin 2. J. Biol. Chem 276, 8403–8408. [DOI] [PubMed] [Google Scholar]
- (19).Emonard H, Bellon G, Troeberg L, Berton A, Robinet A, Henriet P, Marbaix E, Kirkegaard K, Patthy L, Eeckhout Y, Nagase H, Hornebeck W, and Courtoy PJ (2004) Low density lipoprotein receptor-related protein mediates endocytic clearance of pro-MMP-2-TIMP-2 complex through a thrombospondin-independent mechanism. J. Biol. Chem 279, 54944–54951. [DOI] [PubMed] [Google Scholar]
- (20).Hahn-Dantona E, Ruiz JF, Bornstein P, and Strickland DK (2001) The Low Density Lipoprotein Receptor-related Protein Modulates Levels of Matrix Metalloproteinase 9 (MMP-9) by Mediating Its Cellular Catabolism. J. Biol. Chem 276, 15498–15503. [DOI] [PubMed] [Google Scholar]
- (21).Van Den Steen PE, Van Aelst I, Hvidberg V, Piccard H, Fiten P, Jacobsen C, Moestrup SK, Fry S, Royle L, Wormald MR, Wallis R, Rudd PM, Dwek RA, and Opdenakker G (2006) The hemopexin and O-glycosylated domains tune gelatinase B/MMP-9 bioavailability via inhibition and binding to cargo receptors. J. Biol. Chem 281, 18626–18637. [DOI] [PubMed] [Google Scholar]
- (22).Barmina OY, Walling HW, Fiacco GJ, Freije JMP, López-Otín C, Jeffrey JJ, and Partridge NC (1999) Collagenase-3 binds to a specific receptor and requires the low density lipoprotein receptor-related protein for internalization. J. Biol. Chem 274, 30087–30093. [DOI] [PubMed] [Google Scholar]
- (23).Yamamoto K, Okano H, Miyagawa W, Visse R, Shitomi Y, Santamaria S, Dudhia J, Troeberg L, Strickland DK, Hirohata S, and Nagase H (2016) MMP-13 is constitutively produced in human chondrocytes and co-endocytosed with ADAMTS-5 and TIMP-3 by the endocytic receptor LRP1. Matrix Biol. 56, 57–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Yamamoto K, Troeberg L, Scilabra SD, Pelosi M, Murphy CL, Strickland DK, and Nagase H (2013) LRP-1-mediated endocytosis regulates extracellular activity of ADAMTS-5 in articular cartilage. FASEB J. 27, 511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Yamamoto K, Owen K, Parker AE, Scilabra SD, Dudhia J, Strickland DK, Troeberg L, and Nagase H (2014) Low density lipoprotein receptor-related protein 1 (LRP1)-mediated endocytic clearance of a disintegrin and metalloproteinase with thrombospondin motifs-4 (ADAMTS-4): Functional differences of non-catalytic domains of ADAMTS-4 and ADAMTS-5 in LRP1 binding. J. Biol. Chem 289, 6462–6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Thevenard J, Verzeaux L, Devy J, Etique N, Jeanne A, Schneider C, Hachet C, Ferracci G, David M, Martiny L, Charpentier E, Khrestchatisky M, Rivera S, Dedieu S, and Emonard H (2014) Low-density lipoprotein receptor-related protein-1 mediates endocytic clearance of tissue inhibitor of metalloproteinases-1 and promotes its cytokine-like activities. PLoS One 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Verzeaux L, Belloy N, Thevenard-Devy J, Devy J, Ferracci G, Martiny L, Dedieu S, Dauchez M, Emonard H, Etique N, and Devarenne-Charpentier E (2017) Intrinsic dynamics study identifies two amino acids of TIMP-1 critical for its LRP-1-mediated endocytosis in neurons. Sci. Rep 7, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Scilabra SD, Yamamoto K, Pigoni M, Sakamoto K, Müller SA, Papadopoulou A, Lichtenthaler SF, Troeberg L, Nagase H, and Kadomatsu K (2017) Dissecting the interaction between tissue inhibitor of metalloproteinases-3 (TIMP-3) and low density lipoprotein receptor-related protein-1 (LRP-1): Development of a “TRAP” to increase levels of TIMP-3 in the tissue. Matrix Biol. 59, 69–79. [DOI] [PubMed] [Google Scholar]
- (29).Strickland DK, Gonias SL, and Argraves WS (2002) Diverse roles for the LDL receptor family. Trends Endocrinol. Metab 13, 66–74. [DOI] [PubMed] [Google Scholar]
- (30).Weaver AM, Hussaini IM, Mazar A, Henkin J, and Gonias SL (1997) Embryonic fibroblasts that are genetically deficient in low density lipoprotein receptor-related protein demonstrate increased activity of the urokinase receptor system and accelerated migration on vitronectin. J. Biol. Chem 272, 14372–14379. [DOI] [PubMed] [Google Scholar]
- (31).Boucher P, Liu P, Gotthardt M, Hiesberger T, Anderson RGW, and Herz J (2002) Platelet-derived growth factor mediates tyrosine phosphorylation of the cytoplasmic domain of the low density lipoprotein receptor-related protein in caveolae. J. Biol. Chem 277, 15507–15513. [DOI] [PubMed] [Google Scholar]
- (32).Ashcom JD, Tiller SE, Dickerson K, Cravens JL, Argraves WS, and Strickland DK (1990) The human a2-macroglobulin receptor: Identification of a 420-kD cell surface glycoprotein specific for the activated conformation of a2-macroglobulin. J. Cell Biol 110, 1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Strickland DK, Ashcom JD, Williams S, Burgess WH, Migliorini M, and Scott Argraves W (1990) Sequence identity between the α2-macroglobulin receptor and low density lipoprotein receptor-related protein suggests that this molecule is a multifunctional receptor. J. Biol. Chem 265, 17401–17404. [PubMed] [Google Scholar]
- (34).Kounnas MZ, Church FC, Argraves WS, and Strickland DK (1996) Cellular internalization and degradation of antithrombin III-thrombin, heparin cofactor II-thrombin, and α1-antitrypsin-trypsin complexes is mediated by the low density lipoprotein receptor-related protein. J. Biol. Chem 271, 6523–6529. [DOI] [PubMed] [Google Scholar]
- (35).Herz J, and Strickland DK (2001) Multiligand receptors LRP : a multifunctional scavenger and signaling receptor. J. Clin. Invest 108, 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Lillis AP, Mikhailenko I, and Strickland DK (2005) Beyond endocytosis: LRP function in cell migration, proliferation and vascular permeability. J. Thromb. Haemost 3, 1884–1893. [DOI] [PubMed] [Google Scholar]
- (37).Gonias SL, and Campana WM (2014) LDL receptor-related protein-1: A regulator of inflammation in atherosclerosis, cancer, and injury to the nervous system. Am. J. Pathol 184, 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Herz J, Clouthier DE, and Hammer RE (1992) LDL receptor-related protein internalizes and degrades uPA-PAI-1 complexes and is essential for embryo implantation. Cell 71, 411–421. [DOI] [PubMed] [Google Scholar]
- (39).Nykjaer A, Conese M, Christensen EI, Olson D, Cremona O, Gliemann J, and Blasi F (1997) Recycling of the urokinase receptor upon internalization of the uPA:serpin complexes. EMBO J 16, 2610–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Stefansson S, Muhammad S, Cheng X, Battey FD, Strickland DK, and Lawrence DA (1998) Plasminogen Activator Inhibitor-1 Contains a Cryptic High Affinity Binding Site for the Low Density Lipoprotein Receptor-related Protein * 273, 6358–6366. [DOI] [PubMed] [Google Scholar]
- (41).Migliorini M, Li S, Zhou A, Emal CD, Lawrence DA, and Strickland DK (2020) High-affinity binding of plasminogen-activator inhibitor 1 complexes to LDL receptor-related protein 1 requires lysines. J. Biol. Chem 295, 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Au DT, Ying Z, Hernández-ochoa EO, Fondrie WE, Hampton B, Migliorini M, Galisteo R, Schneider MF, Daugherty A, Rateri DL, Strickland DK, and Muratoglu SC (2018) LRP1 (Low-Density Lipoprotein Receptor-Related Protein 1) Regulates Smooth Muscle Contractility by Modulating Ca 2 + Signaling and Expression of Cytoskeleton-Related Proteins. Arter. Thromb Vasc Biol 38, 2651–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Wilson WRW, Anderton M, Choke EC, Dawson J, Loftus IM, and Thompson MM (2008) Elevated plasma MMP1 and MMP9 are associated with abdominal aortic aneurysm rupture. Eur. J. Vasc. Endovasc. Surg 35, 580–584. [DOI] [PubMed] [Google Scholar]
- (44).Ameku T, Taura D, Sone M, Numata T, Nakamura M, Shiota F, Toyoda T, Matsui S, Araoka T, Yasuno T, Mae S-I, Kobayashi H, Kondo N, Kitaoka F, Amano N, Arai S, Ichisaka T, Matsuura N, Inoue S, Yamamoto T, Takahashi K, Asaka I, Yamada Y, Ubara Y, Muso E, Fukatsu A, Watanabe A, Sato Y, Nakahata T, Mori Y, Koizumi A, Nakao K, Yamanaka S, and Osafune K (2016) Identification of MMP1 as a novel risk factor for intracranial aneurysms in ADPKD using iPSC models. Sci. Rep 6, 30013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Newton CS, Loukinova E, Mikhailenko I, Ranganathan S, Gao Y, Haudenschild C, and Strickland DK (2005) Platelet-derived growth factor receptor-β (PDGFR-β) activation promotes its association with the low density lipoprotein receptor-related protein (LRP): Evidence for co-receptor function. J. Biol. Chem 280, 27872–27878. [DOI] [PubMed] [Google Scholar]
- (46).Williams SE, Ashcom JD, Argraves WS, and Strickland DK (1992) A novel mechanism for controlling the activity of α2-macroglobulin receptor/low density lipoprotein receptor-related protein. Multiple regulatory sites for 39-kDa receptor-associated protein. J. Biol. Chem 267, 9035–9040. [PubMed] [Google Scholar]
- (47).Chowdhury A, Brinson R, Wei B, and Stetler-Stevenson WG (2017) Tissue Inhibitor of Metalloprotease-2 (TIMP-2): Bioprocess Development, Physicochemical, Biochemical, and Biological Characterization of Highly Expressed Recombinant Protein. Biochemistry 56, 6423–6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Wilhelm CM, Eisen AZ, Teter M, Clark SD, Kronberger A, and Goldberg G (1986) Human fibroblast collagenase: Glycosylation and tissue-specific levels of enzyme synthesis 3756–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Clark IM, and Cawston TE (1989) Fragments of human fibroblast collagenase: purification and characterisation. Biochem J 263, 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).McGeehan GM, Bickett DM, Green M, Kassel D, Wiseman JS, and Berman J (1994) Characterization of the peptide substrate specificities of interstitial collagenase and 92-kDa gelatinase. Implications for substrate optimization. J. Biol. Chem 269, 32814–32820. [PubMed] [Google Scholar]
- (51).Herz J, Goldstein JL, Strickland DK, Ho YK, and Brown MS (1991) 39-kDa Protein Modulates Binding of Ligands to Low Density Lipoprotein Receptor-related Protein/a2-Macroglobulin Receptor*. J. Biol. Chem 266, 21232–21236. [PubMed] [Google Scholar]
- (52).Herz J, Hamann U, Rogne S, Myklebost O, Gausepohl H, and Stanley KK (1988) Surface location and high affinity for calcium of a 500-kd liver membrane protein closely related to the LDL-receptor suggest a physiological role as lipoprotein receptor. EMBO J. 7, 4119–4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Fass D, Blacklow S, Kim PS, and Berger JM (1997) Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module. Nature 388, 691–693. [DOI] [PubMed] [Google Scholar]
- (54).Scilabra SD, Troebergs L, Yamamoto K, Emonard H, Thgøersen I, Enghild JJ, Strickland DK, and Nagases H (2013) Differential regulation of extracellular tissue inhibitor of metalloproteinases-3 levels by cell membrane-bound and shed low density lipoprotein receptor-related protein 1. J. Biol. Chem 288, 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Prasad JM, Young PA, and Strickland DK (2016) High affinity binding of the receptor-associated protein D1D2 domains with the low density lipoprotein receptor related protein (LRP1) Involves bivalent complex formation: Critical roles of lysines 60 and 191. J. Biol. Chem 291, 18430–18439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Young PA, Migliorini M, and Strickland DK (2016) Evidence that factor VIII forms a bivalent complex with the Low Density Lipoprotein (LDL) Receptor-related Protein 1 (LRP1): Identification of cluster IV on LRP1 as the major binding site. J. Biol. Chem 291, 26035–26044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Arandjelovic S, Hall BD, and Gonias SL (2005) Mutation of lysine 1370 in full-length human α2- macroglobulin blocks binding to the low density lipoprotein receptor-related protein-1. Arch. Biochem. Biophys 438, 29–35. [DOI] [PubMed] [Google Scholar]
- (58).Fisher C, Beglova N, and Blacklow SC (2006) Structure of an LDLR-RAP Complex Reveals a General Mode for Ligand Recognition by Lipoprotein Receptors. Mol. Cell 22, 277–283. [DOI] [PubMed] [Google Scholar]
- (59).Chappell DA, Inoue I, Fry GL, Pladet MW, Bowen SL, Iverius P-H, Lalouel J-M, and Strickland DK (1994) Cellular catabolism of normal very low density lipoproteins via the low density lipoprotein receptor-related protein/α2-macroglobulin receptor is induced by the c-terminal domain of lipoprotein lipase. J. Biol. Chem 269, 18001–18006. [PubMed] [Google Scholar]
- (60).Neels JG, Van Den Berg BMM, Lookene A, Olivecrona G, Pannekoekt H, and Van Zonneveld AJ (1999) The second and fourth cluster of class A cysteine-rich repeats of the low density lipoprotein receptor-related protein share ligand-binding properties. J. Biol. Chem 274, 31305–31311. [DOI] [PubMed] [Google Scholar]
- (61).Welgus HG, Campbell EJ, Cury JD, Eisen AZ, Senior RM, Wilhelm SM, and Goldberg GI (1990) Neutral metalloproteinases produced by human mononuclear phagocytes. Enzyme profile, regulation, and expression during cellular development. J. Clin. Invest 86, 1496–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Roderfeld M, Graf J, Giese B, Salguero-Palacios R, Tschuschner A, Müller-Newen G, and Roeb E (2007) Latent MMP-9 is bound to TIMP-1 before secretion. Biol Chem 388, 1227–1234. [DOI] [PubMed] [Google Scholar]
- (63).Carreca AP, Pravatà V, Murphy G, Nagase H, Troeberg L, and Scilabra SD (2019) TIMP-3 facilitates binding of target metalloproteinases to the endocytic receptor LRP-1 and promotes scavenging of MMP-1. bioRxiv 1, 2019.12.23.886762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Moestrup SK, and Gliemann J (1989) Purification of the rat hepatic α2-macroglobulin receptor as an approximately 440-kDa single chain protein. J. Biol. Chem 264, 15574–15577. [PubMed] [Google Scholar]
- (65).Moestrup SK, Holtet TL, Etzerodt M, Thøgersen HC, Nykjær A, Andreasen PA, Rasmussen HH, Sottrup-Jensen L, and Gliemann J (1993) Α2-Macroglobulin-Proteinase Complexes, Plasminogen Activatorinhibitor Type-1-Plasminogen Activator Complexes, and Receptor-Associated Protein Bind To a Region of the Α2-Macroglobulin Receptor Containing a Cluster of Eight Complement-Type Repeats. J. Biol. Chem 268, 13691–13696. [PubMed] [Google Scholar]
- (66).Migliorini MM, Behre EH, Brew S, Ingham KC, and Strickland DK (2003) Allosteric modulation of ligand binding to low density lipoprotein receptor-related protein by the receptor-associated protein requires critical lysine residues within its carboxyl-terminal domain. J. Biol. Chem 278, 17986–17992. [DOI] [PubMed] [Google Scholar]
- (67).Doherty CM, Visse R, Dinakarpandian D, Strickland DK, Nagase H, and Troeberg L (2016) Engineered tissue inhibitor of metalloproteinases-3 variants resistant to endocytosis have prolonged chondroprotective activity. J. Biol. Chem 291, 22160–22172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Mittal R, Patel AP, Debs LH, Nguyen D, Patel K, Grati M, Mittal J, Yan D, Chapagain P, and Liu XZ (2016) Intricate Functions of Matrix Metalloproteinases in Physiological and Pathological Conditions. J. Cell. Physiol 231, 2599–2621. [DOI] [PubMed] [Google Scholar]
- (69).Kim HE, Dalal SS, Young E, Legato MJ, Weisfeldt ML, and D’Armiento J (2000) Disruption of the myocardial extracellular matrix leads to cardiac dysfunction. J. Clin. Invest 106, 857–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Vianello E, Dozio E, Rigolini R, Marrocco-Trischitta MM, Tacchini L, Trimarchi S, and Corsi Romanelli MM (2016) Acute phase of aortic dissection: A pilot study on CD40L, MPO, and MMP-1, −2, 9 and TIMP-1 circulating levels in elderly patients. Immun. Ageing 13, 7–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Rao J, and Pulukuri S (2008) Matrix metalloproteinase-1 promotes prostate tumor growth and metastasis. Int. J. Oncol 32, 757–765. [PMC free article] [PubMed] [Google Scholar]
- (72).Poola I, DeWitty RL, Marshalleck JJ, Bhatnagar R, Abraham J, and Leffall LSD (2005) Identification of MMP-1 as a putative breast cancer predictive marker by global gene expression analysis. Nat. Med 11, 481–483. [DOI] [PubMed] [Google Scholar]
- (73).Bergamaschi A, Tagliabue E, Sorlie T, Naume B, Triulzi T, Orlandi R, Russness H, Nesland J, Tammi R, Auvinen P, Kosma V-M, Menard S, and Børresen-Dale A-L (2008) Extracellular matrix signature identifies breast cancer subgroups with different clinical outcome. J. Pathol 214, 357–367. [DOI] [PubMed] [Google Scholar]


