Abstract
A number of APOBEC family DNA cytosine deaminases can induce mutations in tumor cells. APOBEC3H haplotype I is one of the deaminases that has been proposed to cause mutations in lung cancer. Here, we confirmed that APOBEC3H haplotype I can cause uracil-induced DNA damage in lung cancer cells that results in γH2AX foci. Interestingly, the database of cancer biomarkers in DNA repair genes (DNArCdb) identified a single-nucleotide polymorphism (rs139298) of APOBEC3H haplotype I that is involved in lung cancer. While we thought this may increase the activity of APOBEC3H haplotype I, instead we found through computational modeling and cell-based experiments that this single-nucleotide polymorphism causes the destabilization of APOBEC3H Haplotype I. Computational analysis suggests that the resulting K121E change affects the structure of APOBEC3H leading to active site disruption and destabilization of the RNA-mediated dimer interface. A K117E mutation in a K121E background stabilized the APOBEC3H haplotype I, thus enabling biochemical study. Subsequent analysis showed that K121E affected catalytic activity, single-stranded DNA binding and oligomerization on single-stranded DNA. The destabilization of a DNA mutator associated with lung cancer supports the model that too much APOBEC3-induced mutation could result in immune recognition or death of tumor cells.
INTRODUCTION
The 11-member Apolipoprotein B editing enzyme (APOBEC) family in humans is comprised of RNA and single-stranded (ss) DNA cytosine deaminases with diverse biological functions (1–3). Within this family is a subgroup of seven APOBEC3 family members that primarily deaminate cytosine in ssDNA, which forms promutagenic uracil. This C-to-U conversion is considered a DNA lesion and can result in a C-to-T mutation if uracil is used as a template during DNA replication or error-prone uracil repair can nucleate other mutations, such as C-to-G transversions (4). These enzymes are known for their ability to restrict the replication of various viruses, such as HIV-1, Epstein–Barr virus and hepatitis B virus, through the mutagenic fates of uracil in DNA (5–7). The host genomic DNA is usually safe from APOBEC3 cytosine deamination due to various protections such as cytoplasmic localization, low expression levels or cytoplasmic RNA binding that both sequesters and inhibits enzyme activity (8–12). However, when these checks are not in place, the APOBEC3 enzymes that can enter the nucleus, such as APOBEC3B (A3B), APOBEC3A (A3A) and APOBEC3H haplotype I (A3H Hap I) (8,13–18), can deaminate genomic DNA. This activity has linked these APOBEC3 enzymes to various cancers, such as lung, breast, bladder, cervical and others. The activity related to cancer is on transient genomic ssDNA during replication or transcription and is considered ‘off-target’ activity (19–23). Notably, the C-to-U conversion only occurs in a specific sequence context, e.g., 5′ RTCA (A3B, ‘R’ is A or G), 5′ YTCA (A3A, ‘Y’ is C or T) or 5′ TCT (A3H) (13,17,18). This 5′ TC sequence context has enabled the APOBEC3 mutation signature to be identified in at least 16 cancers and can be differentiated from chemical DNA damage, which may occur with lung cancer, for example (4,24). The main hypothesis is that APOBEC3 enzymes create uracils randomly throughout the genome and that the majority of these are repaired by base excision repair (BER), but some may not be repaired, resulting in transition mutations, and some may be repaired in an error-prone manner, resulting in transversion mutations (25–27).
Since APOBEC-induced mutations are stochastic, the effects may be variable and can range from cell death, enhanced immune recognition or tumor evolution (4,13,17,26–29). These fates depend on the cell cycle and the location of the mutations. The interplay between APOBEC3 enzymes in this process is not known. Although there have been multiple reports of A3B, A3A or A3H Hap I individually being involved in a cancer, it is not known if more than one APOBEC3 is expressed in a cancer cell at the same time and what would be the effects (13,16–18,30). What has recently been identified is that APOBEC3 mutations occur episodically in cancers (31). This is thought to be because constant expression and mutagenesis would result in cell death or recognition by the immune system, rather than creating a ‘just right’ level of mutagenesis for cancer evolution. It has also been reported that for lung cancer, A3H Hap I predominantly causes early mutations and A3B predominantly causes late-stage mutations (13).
Interestingly, there are seven haplotypes for A3H, defined by polymorphisms or deletions at positions 15, 18, 105, 121 and 178, but only A3H Hap I is primarily localized to the nucleus (32–34). The other A3H haplotypes are primarily in the cytoplasm (34). A3H has several determinants for stability, such as the propensity to become ubiquitinated and ability to bind cellular RNA (12,35). A3H forms a dimer that has no protein–protein contacts and is mediated by binding cellular RNA (8,12,36,37). A3H haplotypes that stably bind RNA are located in the cytoplasm, and mutants that do not stably bind RNA are nuclear, suggesting that dimerization mediated by RNA suppresses potential off-target deaminations by some A3H haplotypes by maintaining cytoplasmic localization, since only those APOBEC3 members that have access to the nucleus can contribute to cancer mutagenesis (8). In addition, some A3H haplotypes are rapidly ubiquitinated and degraded in cells (III, IV, VI), some are ubiquitinated and degraded on a slower timescale, enabling activity in cells (I), and others are not ubiquitinated and have a long half-life in cells (II, V, VII) (32–34,38). The short half-life of A3H Hap I is due to a G105, since mutation of this to an R105 forms an enzyme with a longer half-life in cells and is A3H Hap VII, which has been used as an A3H Hap I proxy in vitro (39). How the amino acid at position 105 affects ubiquitination and cellular stability is not completely understood. Despite A3H Hap I being hypo-active in comparison to A3H Hap II, V or VII (38), it is the only A3H that has been implicated in cancer mutagenesis (13). Starrett et al. have shown that A3H Hap I is an enzymatic contributor to ‘APOBEC signature’ mutations in lung cancer and likely contributor to breast cancer (13). This was a surprising result since A3H Hap I has a half-life in mammalian cells of ∼30 min (32). The short A3H Hap I half-life also precludes the purification of significant quantities of A3H Hap I from eukaryotic cells for use in biochemical assays, although A3H tagged with maltose-binding protein fusion can be purified from Escherichia coli and demonstrates activity (38). When mammalian cell lysates are used and A3H Hap I is overexpressed at equivalent protein levels to more stable haplotypes, the activity is comparable.
One single-nucleotide polymorphism (SNP), rs139298, resulting in A3H Hap I K121E has been recently reported to be associated with lung cancer (40). Lung cancer results in over 1.6 million deaths per year and nearly 2 million new cases annually, with pulmonary adenocarcinoma comprising ∼40% of the total cases of non-smoking lung cancer (41–44). Surprisingly, we found that this K121E mutation further destabilized A3H Hap I, giving the counter-intuitive result that the absence of A3H Hap I is associated with lung cancer. In this study, we examine the A3H Hap I K121E variant and its stabilization by a novel A3H Hap I stabilizing mutation, K117E, using computational, biochemical and cell-based techniques. Our results, considered in conjunction with the genetic data, strongly support the hypothesis that although APOBEC3-induced mutations can contribute to cancer evolution, constant exposure to APOBEC3 mutagenesis may be detrimental to cancer cells.
MATERIALS AND METHODS
Computational methods
Molecular dynamics (MD) simulations were performed with the ff14SB (45), OL15 (46), YIL (47) and TIP3P (48) force fields using the pmemd.cuda (49,50) and OpenMM (51) programs. All systems were constructed from pdbid 5W45, corresponding to A3H Hap I based on the UniProt consensus sequence (52,53). A search of 12 possible binding orientations of the DNA substrate on one single monomer was performed to determine the most probable orientation for the DNA substrate (see the Supplementary Data for coordinates of dimer in holo form) (54).
The most probable orientation was subsequently used to construct the biologically relevant dimer systems using the rhesus macaque APOBEC3H dimer structure (pdb id 5W3V) as a template for UCSF Chimera and taking into account the polarity of the substrate (55–57). The monomer crystal of human A3H was overlaid on the dimer crystal of the macaque A3H and aligned. The protonation states for the dimer systems were determined using H++, neutralized with K+ (Supplementary Table S1) and solvated in a box of TIP3P water of ∼143 Å on each side to allow free movement without self-interaction across the periodic boundary (58–60). Particle mesh Ewald was employed for long-range electrostatics, with a cutoff distance of 10 Å and an error tolerance of 10−4 kJ mol−1 Å−2 (50).
All simulations were performed in the NPT ensemble at 1.0 bar and 300 K using a Monte Carlo barostat, and a Langevin thermostat with a 1 fs time step (51,61,62). Simulation frames were saved at 10 ps intervals. To maintain active site geometry in the simulations, distance restraints of 20 kcal mol−1 Å−2 at 2.0 Å were applied between the Zn2+ atoms and their respective coordinating histidines, with additional 10 kcal mol−1 Å−2 restraints at 5.0 Å between the Zn2+ and water-coordinating glutamate to prevent incursion into the active site. Each system was minimized for 2000 steps and equilibrated for 20 ns before beginning production. Additional simulations using distance constraints of 2.0 Å between the Zn2+ and coordinating atoms were also run and found to give quantitatively similar results to the restrained simulations (data not shown).
Five systems were constructed including A3H Hap I wild type (WT), A3H Hap I K121E (cancer variant), A3H Hap I K117E/K121E, A3H Hap I K117E and A3H Hap I G105R (A3H Hap VII). Every system was run for 250 ns in triplicate (750 ns total simulation time per variant). Energy decomposition analysis was performed using an in-house FORTRAN90 program (63–65). Root mean square fluctuation (RMSF), root mean square deviation (RMSD), correlation, hydrogen bond (HBond) analysis, clustering, normal mode analysis and distance calculations were performed using cpptraj in the AmberTools suite (58,66).
Immunoblotting
Transfection of 1 × 105 293T cells per well of a six-well plate was carried out using GeneJuice (Novagen) transfection reagent as per the manufacturer’s protocol. Cells were maintained in Dulbecco’s modified Eagle medium with 10% fetal bovine serum (FBS). Plasmids transfected were empty pcDNA3.1 (mock) and the following C-terminally tagged A3H-3× HA constructs in pcDNA3.1: A3H Hap I, A3H Hap I K117E, A3H Hap I K121E and A3H Hap I K117E/K121E. After 48 h, cells were lysed using 2× Laemmli buffer and 30 μg total protein was used. A3H was detected using an anti-HA mouse antibody (Sigma) and loading control for cell lysate detected α-tubulin using an anti-α-tubulin rabbit antibody (Invitrogen). Secondary detection was performed using Licor IRDye antibodies produced in goat (IRDye 680-labeled anti-rabbit and IRDye 800-labeled anti-mouse).
Purification of A3H from Sf9 insect cells
The pFAST-bac1 vectors were used to produce recombinant baculovirus according to the protocol for the Bac-to-Bac system (Life Technologies). Expression and purification of GST-A3H Hap II and GST-A3H Hap VII were previously described (39,67). Recombinant GST-A3H Hap I WT and mutant baculovirus (K117E, K117E/K121E) were used to infect Sf9 cells at a multiplicity of infection of 1 and cells were harvested after 72 h. Cells lysates were treated with 100 μg ml−1 of RNase A (Qiagen). Lysates were cleared by centrifugation and then incubated with Glutathione Sepharose 4B resin (GE Healthcare) at 4°C overnight and subjected to a series of salt washes (0.25–1 M NaCl) as described previously (68). On-column cleavage from the GST tag with Thrombin (GE Healthcare) was performed at 21°C for 18 h in thrombin digestion buffer [20 mM HEPES, pH 7.5, 150 mM NaCl, 10% glycerol and 2 mM dithiothreitol (DTT)]. Proteins were assessed to be 90% pure by SDS-PAGE (Supplementary Figure S1).
Deamination assays
The 118-nt ssDNA substrate had the following sequence: 5′ GAA TAT AGT TTT TAG CTC AAA GTA AGT GAA GAT AAT [Fluorescein dT] TAG AGA GTT GTA ATG TGA TAT ATG TGT ATG AAA GAT ATA AGA CTC AAA GTG AAA AGT TGT TAA TGT GTG TAG ATA TGT TAA. The ssDNA substrate (100 nM), which contained two A3H deamination motifs (5′ CTC, with the underlined C being deaminated), was incubated with 200 nM of A3H Hap I mutants (K117E, K117E/K121E) or 50 nM of A3H Hap II, Hap VII and Hap I mutants for 60 min at 37°C in buffer that contained (50 mM Tris, pH 7.5, 40 mM KCl, 10 mM MgCl2 and 1 mM DTT). The A3H was inactivated and the ssDNA extracted with a phenol:chloroform extraction and subsequent chloroform washes. Then, A3H-catalyzed deaminations were detected by treating the ssDNA with Uracil DNA Glycosylase (UNG; New England Biolabs) and heating under alkaline conditions to fragment the ssDNA at abasic sites before resolving the Fluorescein dT-labeled ssDNA on a 10% (v/v) denaturing polyacrylamide gel. Due to the internal fluorescein label, deaminations at the 5′ CTC, 3′ CTC or both 5′ and 3′ CTC (5′ and 3′ C) were detected on the gel as ssDNA fragments of specific lengths. Gel images were obtained using a Chemidoc-MP imaging system (Bio-Rad) and integrated gel band intensities were analyzed using ImageQuant (GE Healthcare). For determination of specific activity, the pmoles of DNA deaminated per microgram of enzyme per minute was calculated for the 2.5 min time for all A3H variants. Values for three independent trials were averaged and the standard deviation calculated. For determination of enzyme processivity, reactions were carried out under single-hit conditions, i.e. <15% substrate usage, to ensure that deaminations on each ssDNA were catalyzed by a single enzyme (69). Under these conditions, a processivity factor can be determined by comparing the total amount of deaminations occurring at two sites on same ssDNA to a calculated theoretical value of deaminations at these two sites if the deamination events were uncorrelated (not processive) (70).
Detection of RNA bound to A3H
To examine the RNA present in purified A3H, 3.5 μg A3H was or was not treated with RNase A (Roche Applied Science) for 15 min at room temperature. At the end of the reactions, samples were mixed with an equal volume of formamide containing 5 mM EDTA and resolved on a 20% (v/v) denaturing polyacrylamide gel. The resolved nucleic acids were stained with SYBR Gold (Invitrogen) and gel images were obtained using a Chemidoc-MP imaging system (Bio-Rad).
Size exclusion chromatography
The oligomerization states of A3H haplotypes and mutants were determined by loading 120–300 μg of the purified enzymes onto a Superdex 200 Increase 10/300 (GE Healthcare). The running buffer contained 20 mM Tris, pH 8.0, 300 mM NaCl, 10% (v/v) glycerol and 1 mM DTT. The Bio-Rad gel filtration standard set was used to generate a calibration curve from which the apparent molecular masses and oligomerization states of the enzymes were determined.
Steady-state fluorescence polarization
The apparent dissociation constant (Kd) values of A3H Hap II, A3H Hap VII and A3H Hap I mutants K117E and K117E/K121E for fluorescein-labeled ssDNA (118 nt) were determined using steady-state fluorescence polarization to measure rotational anisotropy. Reactions (50 μl) were conducted in deamination buffer (50 mM Tris, pH 7.5, 40 mM KCl, 10 mM MgCl2 and 2 mM DTT) and contained 50 nM fluorescein-labeled ssDNA and increasing amounts of A3H. A QuantaMaster QM-4 spectrofluorometer (Photon Technology International) with a dual emission channel was used to collect data and calculate anisotropy. Measurements were performed at 21°C. Samples were excited with vertically polarized light at 495 nm (6 nm bandpass) and vertical and horizontal emissions were measured at 520 nm (6 nm bandpass). The Kd was obtained by fitting to a rectangular hyperbola or sigmoidal curve equation using SigmaPlot 11.2 software.
Cell culture and generation of stable cell lines
Lentivirus was generated by co-transfecting psPAX2, pVSV-G and the pLVX lentiviral vector containing A3H Hap I-Flag with GeneJuice (EMD Millipore) in HEK-293T cells. Medium was changed after 16 h and viral particles were harvested 48 h post-transfection. NCI-H1563 was obtained from ATCC and cultured in RPMI-1460 supplemented with 10% FBS, 10 mM HEPES and 1 mM sodium pyruvate. The NCI-H1563 cells were transduced with the resulting lentivirus by incubation for 16 h in a medium containing 8 μg ml−1 polybrene. Transduced cells were selected with 1 μg ml−1 puromycin for a week and maintained with 0.25 μg ml−1 puromycin.
Immunofluorescence microscopy
NCI-H1563 cells were treated with 2 μg ml−1 doxycycline (dox) for 24 h to induce A3H Hap I-Flag and fixed with 100% cold methanol for 10 min. Cells were permeabilized with 100% cold acetone for 1 min and 0.5% Triton X-100 for 10 min. Anti-Flag and anti-γH2AX antibodies (Invitrogen) in 3% bovine serum albumin in 4× saline–sodium citrate buffer were incubated 1 h. Primary antibodies were detected with Alexafluor-594- and Alexafluor-488-conjugated secondary antibodies (Invitrogen). Nuclei were stained with DAPI and cells were imaged using the Zeiss LSM700 system. Data were compiled from 373 cells from three trials.
qPCR
The RNA preparation, cDNA synthesis and qPCR were carried out according to Refsland et al. (71).
RESULTS
Expression of A3H Hap I in a lung cancer cell line induces γH2AX foci
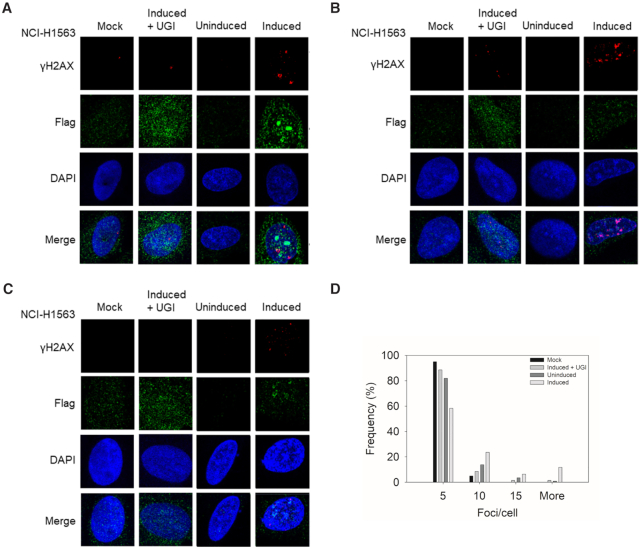
A3H Hap I has been implicated genetically in lung cancer, although no direct evidence of DNA damage has been shown in lung cells (13). To better understand the A3H Hap I SNP (rs139298), we first characterized the level of DNA damage that could be induced by A3H Hap I WT. Since current models indicate that APOBEC3 enzymes do not induce cancers, but contribute to cancer mutagenesis in already transformed cells, we used NCI-H1563, a lung adenocarcinoma cell line derived from a male non-smoker (13). We first determined whether NCI-H1563 cells expressed any endogenous A3s that have been implicated in inducing DNA damage. We found that NCI-H1563 did not express A3A, but did express A3B and very low levels of A3H mRNA, in comparison to the TBP mRNA control (Supplementary Figure S2). The NCI-H1563 cells expressed 5-fold more A3B mRNA than TBP mRNA. The endogenous A3H was not genotyped since the expression was low. Instead, this provided an opportunity to transduce these cells with an A3H Hap I-Flag expression construct to make dox-inducible stable cell lines to study the effect of A3H Hap I expression (Supplementary Figure S3). A key marker of high A3 activity is replication fork stalling from base excision of uracil, leaving an abasic site, or formation of double-stranded (ds) DNA breaks that are induced by base excision of closely located uracils (72). To avoid any possible cell death, we only induced cells with dox for 24 h, and then cells were fixed and antibodies used to detect γH2AX foci as a marker of stalled replication forks and dsDNA breaks (73,74). The uninduced condition had low amounts of γH2AX foci/cell suggesting that the A3B mRNA detected did not correlate with the protein level or the cell conditions inhibited high A3B activity (Figure 1A–D and Supplementary Figure S2) (30). As a result, the cells were used to study the difference between uninduced and induced conditions that enabled expression of A3H Hap I-Flag (Supplementary Figure S3). The uninduced A3H Hap I-Flag condition had ∼82% of cells having one to five γH2AX foci/cell (Figure 1A–D). However, when A3H Hap I-Flag expression was induced with dox, the number of γH2AX foci/cell increased with 12% of cells having over 15 γH2AX foci/cell (Figure 1A–D, More, range was between 19 and 61 per cell). The other major populations were 24% and 58% of cells with 6–10 and 1–5 γH2AX foci/cell, respectively (Figure 1D). The dox used to induce A3H Hap I-Flag expression was also tested in NCI-H1563 that were not transduced and showed that dox treatment by itself did not cause any γH2AX foci to form above background (Figure 1A–D, Mock). DNA repair is responsible for inducing the stalled replication and dsDNA breaks after the formation of abasic sites from the BER enzyme, UNG, which removes the APOBEC-induced uracil. To confirm the connection of uracil formation to DNA damage, we conducted the experiment in the presence of a bacteriophage protein that is an inhibitor of UNG (UGI). In the presence of A3H Hap I-Flag and UGI, the γH2AX foci/cell were similar to the uninduced condition, which demonstrates that A3H Hap I-Flag deamination activity forms uracils in DNA that are processed by DNA repair to induce abasic sites and/or DNA breaks that lead to γH2AX accumulation. Interestingly, A3H Hap I-Flag did not induce any γH2AX foci above the background in MRC-5 cells that are normal, fetal-derived cells (Supplementary Figure S4). Although the reason for this is not known, we speculate that different DNA repair mechanisms in these cells may be responsible. These data demonstrate that A3H Hap I is able to induce formation of uracils that become DNA damage-inducing events after the action of UNG and, as a result, we sought to further characterize the effects of the A3H SNP resulting in the A3H K121E variant.
Figure 1.
DNA damage induced by A3H Hap I. (A–C) The γH2AX foci detected by immunofluorescence microscopy in NCI-H1563 cells that were or were not transduced to express a dox-inducible A3H Hap I-Flag protein. Different cell treatment conditions were dox in non-transduced cells (Mock), dox treatment to induce A3H Hap I, but with transfection of a plasmid expressing UGI (Induced + UGI), transduced, but uninduced cells (Uninduced), and transduced and induced cells (Induced). The NCI-H1563 cells were analyzed 24 h after the treatment. Three representative images are shown. (D) Results were quantified and plotted as a histogram. Foci/cell are represented as 1–5 (Bin 5), 6–10 (Bin 10), 11–15 (Bin 15) and ≥15 (More) γH2AX foci/cell.
A3H Hap I K121E results in the formation of a new HBond network
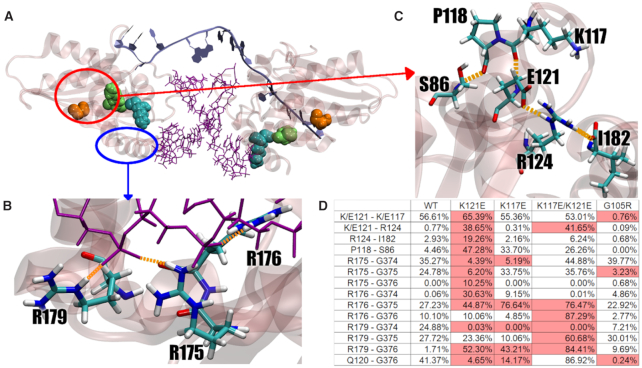
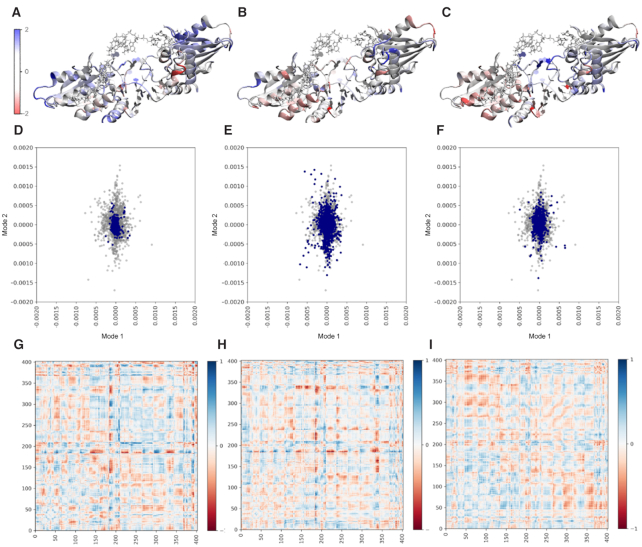
To investigate the effect of the SNP resulting in the A3H Hap I K121E mutation on the protein structure and dynamics compared with A3H Hap I WT, we used classical MD. Pairwise HBond analysis of the K121E indicates the formation of a new HBond network across part of the protein surface compared with the WT. This HBond network induces strain on the active site suggesting a change in stability or activity (Figure 2). HBond interactions between protein residues that differed by >30% of the simulation time were identified (Supplementary Table S2). The HBond network in the K121E system connects two existing networks (R124 to I182 and K117 to P118 to S87) from the WT into a single larger network in the K121E variant. This extended overstabilized network effectively ‘freezes’ the helix where the mutation occurs, disrupting both the active site and RNA-binding residues on the C-terminal helix. Changes in RMSF are also observed in the K121E system, with residues involved in the HBond network exhibiting reduced fluctuation while the rest of the protein exhibits increased fluctuation (Figure 2 and Supplementary Figure S5). Difference correlation matrices reveal that K121E has large regions of changing correlated movement, especially with respect to the region around the mutation point (Supplementary Figure S6). Principal component analysis (PCA) shows that the first two modes of motion in the K121E system are constrained and tightly correlated in comparison to the WT (Figure 3 and Supplementary Figure S7).
Figure 2.
Features of A3H Hap I WT and mutant enzymes. (A) A3H Hap I (transparent pink) with RNA interface (purple) and DNA substrate (ice blue). G105 is shown in orange, K117 in cyan and K121 in green, with corresponding residues on opposite monomer shown in transparency. (B) RNA-binding residues on terminal helix. (C) Formed HBond network as a result of K121E mutation. (D) Table of largest contributors in each HBond interaction, shown as percentage of total simulation time.
Figure 3.
Difference RMSF for A3H Hap I WT and mutants. The analysis with respect to A3H Hap I WT overlaid on protein structures for (A) K121E, (B) K121E/K117E and (C) K117E. Residues in blue exhibit increasing RMSF compared to A3H Hap I WT; residues in red exhibit decreasing RMSF compared to A3H Hap I WT. PCA for mutant systems (blue) over WT (gray) for (D) K121E, (E) K121E/K117E and (F) K117E. Difference correlation matrices against WT reference by residue for (G) K121E, (H) K121E/K117E and (I) K117E. Correlation differences range from complete anticorrelation in red (−1) to complete correlation in blue (+1).
The table in Figure 2D shows an increase in HBonds in several regions, including an increase in HBond prevalence between E121 and K117/R124. Based on this, we hypothesized that a second mutation that would eliminate the new interactions with E121 could rescue the cancer mutant. Thus, based on the increased HBond interactions from E121 to R124 and K117, these are the two likely candidates as mutation sites. In the case of R124, the Hbond to E121 is formed via the side chains. In addition, R124 also interacts with the C-terminus, I182. Thus, mutating R124 could result in negative effects. In the case of K117, the Hbond to E121 is formed between the backbone atoms. Therefore, it was hypothesized that a mutant that would provide a repulsive electrostatic interaction between the side chains at sites 117 and 121 could destabilize these backbone Hbond interactions to levels similar to those observed in the WT, and restore the Hbond network.
The simulation data (Supplementary Figures S7 and S8) suggest that a K117E mutation could rescue the K121E variant by restoring the two HBond networks. The K121E/K117E variant removes the HBond between the side chains of 121 and 117, and partially restores the remaining interactions in the network toward their WT levels (Figure 2). The K121E/K117E system shows less of a change in fluctuation, with residues in the network exhibiting near WT fluctuation, similarly to what is observed for the stabilized A3H Hap I G105R (A3H Hap VII) (Figure 3). The PCA shows that the first two modes are similar to those of the WT, as does the comparatively minor change in correlated movement.
K121E destabilizes A3H Hap I
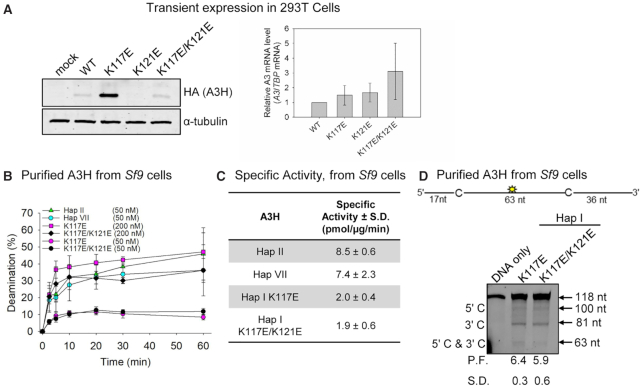
To test the computational results, we produced the A3H Hap I variants in 293T cells for cell-based analysis. To determine the steady-state expression levels of the A3H Hap I WT, K121E cancer variant, putative rescue mutant K117E/K121E and rescue mutant control K117E, we transfected 3× HA-tagged A3H expression constructs into 293T cells. Immunoblotting showed that steady-state expression levels for the cancer variant were not detectable by immunoblotting, suggesting that the K121E mutation destabilized A3H Hap I WT (Figure 4A), in agreement with the computational predictions (Figure 2). In support of this, A3H mRNA levels measured by qPCR showed that A3H Hap I WT, K117E, K121E and K117E/K121E mRNA levels were not significantly different, demonstrating that the destabilization was at the level of protein (Figure 4A). We were also unable to detect any deamination activity for A3H Hap I K121E in cell lysates (data not shown). We found that the K117E mutation stabilized A3H Hap I in the presence of the otherwise destabilizing G105 amino acid (32) and, as predicted, can stabilize the cancer variant (K117E/K121E; Figure 4A). Since the destabilizing SNP, K121E, was associated with lung cancer (40), these data suggested that A3H Hap I somatic mutations may be detrimental to cancer progression perhaps due to extensive mutagenesis leading to cell dysfunction or immune recognition. While beyond the scope of this study, the A3H Hap I K121E instability is consistent with A3H Hap I mutations being identified only early in cancer, the episodic nature of APOBEC3-induced mutations in cancer and the ability of A3H Hap I to cause DNA damage (Figure 1) (13,31,40).
Figure 4.
Expression and deamination activity of A3H and mutants. (A) Transient expression of HA-tagged A3H Hap I expression constructs in 293T cells was detected at the protein level by immunoblotting (left) and mRNA level by qPCR (right). The A3H Hap I WT, K117E, K121E and K117E/K121E had different steady-state protein expression levels, but the mRNA levels in cells were not significantly different, indicating that differences in stability were at the protein level. For immunoblotting, the α-tubulin served as the loading control. For qPCR, TBP mRNA served as the control and results are displayed relative to A3H Hap I WT. (B) Time course of deamination of A3H Hap II, A3H Hap VII and A3H Hap I mutants. Deamination was tested on a 118-nt ssDNA (100 nM) and gels analyzed for each plot are shown in Supplementary Figure S9. (C) Specific activity of A3H Hap II, A3H Hap VII and A3H Hap I mutants. (D) Processivity of A3H Hap I K117E and K117E/K121E. The processivity factor (P.F.) measures the likelihood of a processive deamination over a nonprocessive deamination and is described further in the ‘Materials and Methods’ section. Both K117E and K117E/K121E are ∼6-fold more likely to undergo processive deamination than a nonprocessive deamination. The standard deviation for three independent experiments is shown as error bars (B), in the table (C) or below the gel (D).
A G105R mutation results in a more active and stable A3H than a K117E mutation
To understand the reason for the instability of the A3H K121E variant, we conducted a biochemical analysis of A3H using an Sf9/baculovirus expression system. Although a K117E mutation can stabilize A3H Hap I, it is not a naturally occurring variant. The naturally occurring stabilization of A3H Hap I is a G105R mutation, which is known as A3H Hap VII (32). The reason for the instability induced by G105 is not entirely understood. Computational models of the G105R system exhibit dynamic motion and HBond patterns similar to the WT, indicating that this mutation does not negatively affect the structural stability of the protein (see Supplementary Figures S6–S8 for G105R computational data). The G105R may simply increase the propensity of the protein to become polyubiquitinated and degraded (35), in contrast to K121E that destabilizes the structural integrity of A3H Hap I (Figures 2 and 3). To test whether the longer steady-state half-life of A3H K117E and K117E/K121E in cells correlates with increased catalytic activity similar to A3H Hap VII, we conducted an in vitro deamination assay using protein purified from Sf9 insect cells to obtain a quantitative measurement. A3H deaminase activity was observed by adding a 118-nt ssDNA with two A3H deamination motifs (5′ CTC) and an internal fluorescein label to purified A3H Hap II, A3H Hap VII, A3H Hap I K117E and A3H Hap I K117E/K121E. The two 5′ CTC motifs account for the preference of some A3H variants to preferentially deaminate the cytosine motif nearest the 3′ end of the ssDNA (67). We found that despite recovery of protein stability the A3H Hap I K117E and A3H Hap I K117E/K121E were ∼4-fold less active than A3H Hap VII (relative to Hap I, G105R) or A3H Hap II (relative to Hap I, G105R, E178D) (Figure 4B and C, and Supplementary Figure S9). These data suggest that the A3H Hap I HBond network is important for catalytic activity and compensatory mutations can correct the instability, but not fully reinstate catalytic activity.
A second component important for APOBEC3 activity is processivity, which describes the search process the enzyme undergoes to deaminate multiple cytosines in a single enzyme–substrate encounter. The low activity can be due to a less efficient search on the ssDNA, which precludes finding any cytosines for deamination or can be due to changes to the active site that influence the chemistry of the deamination (1). The computational data predict that the active site structure would be affected, but cannot determine effects on activity or processivity, which are mitigated by ssDNA interactions with helix 6 on A3H (67). The processivity assay is conducted under single-hit conditions, so only if an enzyme can undergo multiple deamination reactions in a single enzyme–substrate encounter, a double deamination band (5′ C and 3′ C) should be observed (see the ‘Materials and Methods’ section). The processivity factor represents the likelihood of a processive deamination occurring relative to a nonprocessive deamination, which for the A3H Hap I mutants is ∼6-fold. Consistent with the determinants of processivity being outside of the active site, the mutants were processive, similar to what has been reported for A3H Hap VII and A3H Hap II (Figure 4D) (67).
A3H cancer variant destabilizes dimer interface
From monkeys to greater apes, the A3 family has lost activity. Although A3B activity can be lost through a gene deletion (75), other A3s have lost activity because of decreased catalytic activity due to mutation of residues near the active site, e.g. A3D, or loss of dimerization, e.g. A3C (76,77). Based on recent crystal structures, the association of A3H with RNA is concomitant with enzyme stability and dimerization (8,12,36,37,78). This is a unique feature of A3H compared to other A3s, in that it uses a dsRNA molecule to form a dimer interface without any protein–protein contacts and that this imparts stability to the enzyme. As a result, we hypothesized that the reason for the A3H cancer variant instability (Figure 4A) may be due to the inability to bind RNA and dimerize. RNA-mediated dimerization can be detected by treating a purified protein preparation with RNase A and observing whether an ∼12-nt RNA is protected from digestion. We confirmed that recombinant A3H Hap I mutants K117E and K117E/K121E purified from Sf9 cells with RNA and RNase A treatment resulted in an ∼12-nt RNA being protected from degradation by the protein (Figure 5A). The A3H Hap VII and A3H Hap II also bound cellular RNA and protected an ∼12-nt RNA in the presence of RNase A, although the A3H Hap II bound less RNA than the other A3H variants. The reason for this is not known; although several studies have confirmed that A3H Hap II binds RNA in order to dimerize, the amount of bound RNA was not compared in parallel to other A3H haplotypes (8,12,36,37,78). However, consistent with RNA binding, all four A3H variants formed primarily a dimer (44 kDa eluting at ∼17 ml, Figure 5B–D). Similar to A3G and as previously shown for A3H (67,68), the size exclusion chromatography (SEC) profile peaks were broad, indicating polydispersity. There were smaller peaks on either side of the dimer peak, indicating smaller populations of monomers and tetramers. A3H Hap II and A3H Hap I K117E/K121E also had larger MW fractions than the other A3H variants (Figure 5D, 14–16 ml).
Figure 5.
A3H Hap I mutant dimerization, oligomerization and ssDNA binding. (A) A3H dimerization is mediated by RNA. Purified A3H was or was not treated with RNase A and then denatured in formamide buffer and samples were resolved by urea denaturing PAGE. The gel was stained with SYBR Gold to detect nucleic acids. The A3H Hap I K117E, A3H Hap I K117E/K121E, A3H Hap VII and A3H Hap II protect an ∼12–15-nt RNA. Representative image shown from three independent experiments. (B) Standard curve and (C,D) SEC profile for A3H Hap I K117E, A3H Hap I K117E/K121E, A3H Hap VII and A3H Hap II demonstrating elution profiles that are composed of a dimer peak (44 kDa, 17 ml elution volume) and monomer peak (22 kDa, 19 ml elution volume). The 44 and 17 kDa elution volumes are shown on the (C) graph and (D) gels. (E–H) The apparent Kd of A3H enzymes from a 118-nt ssDNA was analyzed by steady-state rotational anisotropy. (E) The A3H Hap I K117E (3005 ± 1768 nM) and (F) A3H Hap I K117E/K121E (1239 ± 88 nM) bind the ssDNA with different affinities than (G) A3H Hap VII (522 ± 4 nM) and (H) A3H Hap II (492 ± 28 nM). The apparent Kd was calculated by determining the best fit by least-squares analysis, which was a hyperbolic fit for (E, F) and a sigmoidal fit for (G, H). The Hill coefficient for A3H Hap VII was 1.3 and for A3H Hap II was 1.8. (E–H) Error bars represent the standard deviation from three independent experiments.
Since we cannot purify the K121E variant, we used computational analysis to predict whether the change at amino acid 121 would destabilize dimerization. RMSF analysis for WT, K121E and K117E/K121E (Figure 3) showed that residues in the K121E system exhibited increased fluctuation compared with WT, except those residues involved in the new HBond network and many that interact with the RNA interface. These RNA-binding residues (R175, R176, R179) exhibit changing HBond interactions with the RNA interface. In the simulations, these changes cause the RNA to change positions, affecting additional protein–RNA interaction at loop 1 residues. Correlation matrices were calculated for all systems, with the K121E variant system showing increased correlation between the monomer subunits and decreased correlation within each monomer when compared to the WT (Supplementary Figure S6), suggesting that the dimer interface is destabilized by the newly formed HBond network. This prediction is supported by the SEC data that showed that the A3H Hap I K117E/K121E mutant had a distinct monomer peak, whereas A3H Hap I K117E had more polydisperse peak and was similar to A3H Hap I stabilized by G105R (A3H Hap VII) (Figure 5C and D). This was distinct from A3H Hap II that had the most predominant dimer peak (Figure 5C and D). This is in agreement with RMSF analysis where the K117E system shows qualitatively similar zones of correlation and anticorrelation at the dimer interface with respect to the WT. The K117E/K121E mutant correlation matrices reveal that this system more closely resembles the K121E than either the K117E or WT dimer systems, with qualitatively similar correlation matrices. These data are also in agreement with poor expression of A3H Hap I K121E in cells (Figure 4A). Further, consistent with the biochemical data, the K117E/K121E variant system exhibits fewer changes in fluctuation, and there is not a general trend of increased fluctuation as seen in the K121E variant.
A3H variants have weakened interactions with the substrate
We observed that both the A3H Hap I K117E and K117E/K121E had ∼6-fold (K117E) and ∼2.5-fold (K117E/K121E) higher apparent dissociation constants from ssDNA than A3H Hap VII or A3H Hap II (Figure 5E–H). Further, both the A3H Hap I K117E and K117E/K121E binding curves best fit to a rectangular hyperbola by least-squares analysis, in contrast to A3H Hap VII and A3H Hap II that best fit to a sigmoid. The sigmoidal binding relationship for A3H Hap II has been shown to be due to further oligomerization of A3H dimers on ssDNA (78). This does not appear to occur with A3H Hap I K117E or K117E/K121E (Figure 5E and F), although they can form dimers in solution (Figure 5B–D). The hindered oligomerization on ssDNA may be due to a faster off-rate from the substrate, exemplified in steady state by a higher apparent Kd (Figure 5E–H). Altogether, the data indicate that the A3H Hap I K121E variant results in an inability to use RNA for dimerization, emphasizing the importance of cellular RNA to the structural stability of A3H. Further, the HBond network is important for interactions with ssDNA and oligomerization on ssDNA, providing a reasoning for the decreased catalytic activity in A3H Hap I K117E and K117E/K121E.
DISCUSSION
A3H Hap I was identified to induce genomic mutations specifically in lung cancer (13). Interestingly, the activity of A3H Hap I was found because researchers wanted to investigate how genomic mutations with a cytidine deaminase signature were still occurring in people with an A3B−/− deletion (79–82), which occurs in the world population at 22.5%, although it is primarily found only in Oceanic populations (75). Although A3A is also involved in cancer mutagenesis, it appears to be strongly associated with breast cancer (30,83). The association of A3H Hap I expression and a mutation signature was strong in early clonal variants of lung cancers. Further supporting a role of A3H Hap I in lung cancer was a computational study that identified the association of SNP rs139298 in A3H Hap I with lung cancer (40). In this work, we confirm a role of A3H Hap I in inducing DNA damage in lung cells (Figure 1). However, we also demonstrate in this work that the A3H Hap I cancer-associated SNP makes the resulting A3H Hap I K121E completely unstable (Figures 3 and 4A), suggesting that loss of this enzyme specifically promoted cancer.
While the effects arising from the combination of environmental, e.g. smoking, and A3 mutations remain unknown, the instability of the cancer-associated A3H Hap I K121E variant naturally leads to the hypothesis that if A3 enzymes cause too many mutations it would be detrimental to cancer evolution. Recent reports support the seemingly contradictory idea that A3 enzymes, specifically APOBEC3B, can both contribute to cancer and contribute to success of cancer immune therapy due to A3-mutation created neoepitopes (84) or efficiency of platinum-based drugs due to contributions to DNA damage (85). This is usually also dependent on the type of cancer. In this report, we came to an equivalent conclusion since we determined that the previously identified rs139298 SNP that creates a K121E mutation in A3H Hap I is associated with lung cancer and resulted in a loss of enzyme stability in cells (Figure 4A). While in many A3/cancer studies it is difficult to determine the fate of A3 mutations, the genetic data (40) combined with computational and biochemical study of the K121E mutation demonstrate that A3H Hap I has the potential for these detrimental effects on cell growth and proliferation in lung cancer.
The inactivation of A3 enzymes through SNPs is not unique to A3H, but A3H does have the highest number of inactivating SNPs in the A3 family. In particular, A3H has been lost multiple times in primate evolution, presumably due to nuclear localization and acquisition of genomic mutations (32). While it is thought that A3H is kept active at a certain level in the population due to the antiviral effects of the enzyme, there are strong population stratifications in A3H active/hypo-active/non-active forms that correlate with the historical levels of HIV infection, a pathogen that active A3Hs can restrict efficiently (86,87). The antiviral properties of these enzymes are likely why they are maintained despite the negative effect of their off-target activity. The destabilizing SNPs for A3H other than the one studied here are not thought to destabilize protein structure, but to promote A3H ubiquitination and proteosomal degradation (35). Thus, the SNP studied in this work for A3H Hap I is unique since it appears to destabilize the enzyme by disrupting an HBond network. While this disruption decreased catalytic activity, it did not cause protein unfolding. Rather, it decreased the interaction strength with a dsRNA molecule that A3H acquires from the cell and uses to dimerize (Figure 3 and Supplementary Figure S6) (8,12,36,37,78). This dimerization using an RNA intermediate is essential to A3H thermodynamic stability (12,78). While the nature of dimerization for different A3s is unique, e.g. only A3H uses an RNA intermediate, an SNP that regulates activity through dimerization has also been found for A3C (77,88). This type of functional inactivation enables rapid evolutionary toggling of activity for times when enzyme activity is needed again (89).
The characterization of the A3H Hap I K121E variant and its stabilizing mutant, A3H Hap I K117E/K121E, demonstrates the importance of the A3H HBond network for catalytic activity and ability to bind RNA. Moreover, since the identification of distinct mutational signatures and high A3 expression in cancers (4,16,17), a number of research groups have contributed to our understanding of how A3 enzymes access ssDNA, which A3 enzymes are involved and, specifically for A3B, the clinical effects of the induced mutations (18,21,23,90–93). The data presented here support the idea that A3H Hap I contributes to mutations in lung cancer. The combined genetic (40) and biochemical data presented here support the conclusion that the loss of A3H Hap I activity through the K121E variant may benefit the cancer and be detrimental to the host.
Supplementary Material
ACKNOWLEDGEMENTS
Computing resources from CASCaM and the Department of Chemistry are gratefully acknowledged. We thank Milaid Granadillo Rodríguez for generating the A3H Hap I lentivirus.
Contributor Information
Mark A Hix, Department of Chemistry, University of North Texas, Denton, TX 76201, USA.
Lai Wong, Department of Biochemistry, Microbiology, and Immunology, University of Saskatchewan, Saskatoon, SK S7N 5E5, Canada.
Ben Flath, Department of Biochemistry, Microbiology, and Immunology, University of Saskatchewan, Saskatoon, SK S7N 5E5, Canada.
Linda Chelico, Department of Biochemistry, Microbiology, and Immunology, University of Saskatchewan, Saskatoon, SK S7N 5E5, Canada.
G Andrés Cisneros, Department of Chemistry, University of North Texas, Denton, TX 76201, USA; Center for Advanced Scientific Computing and Modeling (CASCaM), University of North Texas, Denton, TX 76201, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Cancer Online.
FUNDING
National Institutes of Health [R01GM108583 to G.A.C.]; National Science Foundation [CHE-1531468, in part]; Canadian Institutes of Health Research [PJT159560 to L.C.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Adolph M.B., Love R.P., Chelico L. Biochemical basis of APOBEC3 deoxycytidine deaminase activity on diverse DNA substrates. ACS Infect. Dis. 2018; 4:224–238. [DOI] [PubMed] [Google Scholar]
- 2. Smith H.C. RNA binding to APOBEC deaminases; not simply a substrate for C to U editing. RNA Biol. 2017; 14:1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Venkatesan S., Rosenthal R., Kanu N., McGranahan N., Bartek J., Quezada S.A., Hare J., Harris R.S., Swanton C. Perspective: APOBEC mutagenesis in drug resistance and immune escape in HIV and cancer evolution. Ann. Oncol. 2018; 29:563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Borresen-Dale A.L.et al. Signatures of mutational processes in human cancer. Nature. 2013; 500:415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harris R.S., Hultquist J.F., Evans D.T. The restriction factors of human immunodeficiency virus. J. Biol. Chem. 2012; 287:40875–40883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheng A.Z., Yockteng-Melgar J., Jarvis M.C., Malik-Soni N., Borozan I., Carpenter M.A., McCann J.L., Ebrahimi D., Shaban N.M., Marcon E.et al. Epstein–Barr virus BORF2 inhibits cellular APOBEC3B to preserve viral genome integrity. Nat. Microbiol. 2019; 4:78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Suspene R., Guetard D., Henry M., Sommer P., Wain-Hobson S., Vartanian J.P. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl Acad. Sci. U.S.A. 2005; 102:8321–8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shaban N.M., Shi K., Lauer K.V., Carpenter M.A., Richards C.M., Salamango D., Wang J., Lopresti M.W., Banerjee S., Levin-Klein R.et al. The antiviral and cancer genomic DNA deaminase APOBEC3H is regulated by an RNA-mediated dimerization mechanism. Mol. Cell. 2018; 69:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salamango D.J., Becker J.T., McCann J.L., Cheng A.Z., Demir O., Amaro R.E., Brown W.L., Shaban N.M., Harris R.S. APOBEC3H subcellular localization determinants define zipcode for targeting HIV-1 for restriction. Mol. Cell. Biol. 2018; 38:e00356-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bennett R.P., Presnyak V., Wedekind J.E., Smith H.C. Nuclear exclusion of the HIV-1 host defense factor APOBEC3G requires a novel cytoplasmic retention signal and is not dependent on RNA binding. J. Biol. Chem. 2008; 283:7320–7327. [DOI] [PubMed] [Google Scholar]
- 11. Soros V.B., Yonemoto W., Greene W.C. Newly synthesized APOBEC3G is incorporated into HIV virions, inhibited by HIV RNA, and subsequently activated by RNase H. PLoS Pathog. 2007; 3:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matsuoka T., Nagae T., Ode H., Awazu H., Kurosawa T., Hamano A., Matsuoka K., Hachiya A., Imahashi M., Yokomaku Y.et al. Structural basis of chimpanzee APOBEC3H dimerization stabilized by double-stranded RNA. Nucleic Acids Res. 2018; 46:10368–10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Starrett G.J., Luengas E.M., McCann J.L., Ebrahimi D., Temiz N.A., Love R.P., Feng Y., Adolph M.B., Chelico L., Law E.K.et al. The DNA cytosine deaminase APOBEC3H haplotype I likely contributes to breast and lung cancer mutagenesis. Nat. Commun. 2016; 7:12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu M., Wang Y., Wang C., Shen W., Liu J., Geng L., Cheng Y., Dai J., Jin G., Ma H.et al. The eQTL-missense polymorphisms of APOBEC3H are associated with lung cancer risk in a Han Chinese population. Sci. Rep. 2015; 5:14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Salter J.D., Bennett R.P., Smith H.C. The APOBEC protein family: united by structure, divergent in function. Trends Biochem. Sci. 2016; 41:578–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Burns M.B., Temiz N.A., Harris R.S. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 2013; 45:977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burns M.B., Lackey L., Carpenter M.A., Rathore A., Land A.M., Leonard B., Refsland E.W., Kotandeniya D., Tretyakova N., Nikas J.B.et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013; 494:366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chan K., Roberts S.A., Klimczak L.J., Sterling J.F., Saini N., Malc E.P., Kim J., Kwiatkowski D.J., Fargo D.C., Mieczkowski P.A.et al. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat. Genet. 2015; 47:1067–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoopes J.I., Cortez L.M., Mertz T.M., Malc E.P., Mieczkowski P.A., Roberts S.A. APOBEC3A and APOBEC3B preferentially deaminate the lagging strand template during DNA replication. Cell Rep. 2016; 14:1273–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kazanov M.D., Roberts S.A., Polak P., Stamatoyannopoulos J., Klimczak L.J., Gordenin D.A., Sunyaev S.R. APOBEC-induced cancer mutations are uniquely enriched in early-replicating, gene-dense, and active chromatin regions. Cell Rep. 2015; 13:1103–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kanu N., Cerone M.A., Goh G., Zalmas L.P., Bartkova J., Dietzen M., McGranahan N., Rogers R., Law E.K., Gromova I.et al. DNA replication stress mediates APOBEC3 family mutagenesis in breast cancer. Genome Biol. 2016; 17:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haradhvala N.J., Polak P., Stojanov P., Covington K.R., Shinbrot E., Hess J.M., Rheinbay E., Kim J., Maruvka Y.E., Braunstein L.Z.et al. Mutational strand asymmetries in cancer genomes reveal mechanisms of DNA damage and repair. Cell. 2016; 164:538–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seplyarskiy V.B., Soldatov R.A., Popadin K.Y., Antonarakis S.E., Bazykin G.A., Nikolaev S.I. APOBEC-induced mutations in human cancers are strongly enriched on the lagging DNA strand during replication. Genome Res. 2016; 26:174–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jarvis M.C., Ebrahimi D., Temiz N.A., Harris R.S. Mutation signatures including APOBEC in cancer cell lines. JNCI Cancer Spectr. 2018; 2:pky002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adolph M.B., Ara A., Chelico L. APOBEC3 host restriction factors of HIV-1 can change the template switching frequency of reverse transcriptase. J. Mol. Biol. 2019; 431:1339–1352. [DOI] [PubMed] [Google Scholar]
- 26. Roberts S.A., Gordenin D.A. Clustered and genome-wide transient mutagenesis in human cancers: hypermutation without permanent mutators or loss of fitness. BioEssays. 2014; 36:382–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Burns M.B., Leonard B., Harris R.S. APOBEC3B: pathological consequences of an innate immune DNA mutator. Biomed. J. 2015; 38:102–110. [DOI] [PubMed] [Google Scholar]
- 28. Roberts S.A., Lawrence M.S., Klimczak L.J., Grimm S.A., Fargo D., Stojanov P., Kiezun A., Kryukov G.V., Carter S.L., Saksena G.et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013; 45:970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stephens P.J., Tarpey P.S., Davies H., Van Loo P., Greenman C., Wedge D.C., Nik-Zainal S., Martin S., Varela I., Bignell G.R.et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012; 486:400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cortez L.M., Brown A.L., Dennis M.A., Collins C.D., Brown A.J., Mitchell D., Mertz T.M., Roberts S.A. APOBEC3A is a prominent cytidine deaminase in breast cancer. PLoS Genet. 2019; 15:e1008545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Petljak M., Alexandrov L.B., Brammeld J.S., Price S., Wedge D.C., Grossmann S., Dawson K.J., Ju Y.S., Iorio F., Tubio J.M.C.et al. Characterizing mutational signatures in human cancer cell lines reveals episodic APOBEC mutagenesis. Cell. 2019; 176:1282–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. OhAinle M., Kerns J.A., Li M.M., Malik H.S., Emerman M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe. 2008; 4:249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang X., Abudu A., Son S., Dang Y., Venta P.J., Zheng Y.H. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J. Virol. 2011; 85:3142–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li M.M., Emerman M. Polymorphism in human APOBEC3H affects a phenotype dominant for subcellular localization and antiviral activity. J. Virol. 2011; 85:8197–8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chesarino N.M., Emerman M. Polymorphisms in human APOBEC3H differentially regulate ubiquitination and antiviral activity. Viruses. 2020; 12:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bohn J.A., Thummar K., York A., Raymond A., Brown W.C., Bieniasz P.D., Hatziioannou T., Smith J.L. APOBEC3H structure reveals an unusual mechanism of interaction with duplex RNA. Nat. Commun. 2017; 8:1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ito F., Yang H., Xiao X., Li S.X., Wolfe A., Zirkle B., Arutiunian V., Chen X.S. Understanding the structure, multimerization, subcellular localization and mC selectivity of a genomic mutator and anti-HIV factor APOBEC3H. Sci. Rep. 2018; 8:3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gu J., Chen Q., Xiao X., Ito F., Wolfe A., Chen X.S. Biochemical characterization of APOBEC3H variants: implications for their HIV-1 restriction activity and mC modification. J. Mol. Biol. 2016; 428:4626–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adolph M.B., Love R.P., Feng Y., Chelico L. Enzyme cycling contributes to efficient induction of genome mutagenesis by the cytidine deaminase APOBEC3B. Nucleic Acids Res. 2017; 45:11925–11940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Silvestrov P., Maier S.J., Fang M., Cisneros G.A. DNArCdb: a database of cancer biomarkers in DNA repair genes that includes variants related to multiple cancer phenotypes. DNA Repair. 2018; 70:10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stewart B.W., Wild C.P. World Cancer Report 2014. 2014; Geneva: World Health Organization. [Google Scholar]
- 42. MacKinnon A.C., Jens K., Tariq S. The molecular and cellular biology of lung cancer: identifying novel therapeutic strategies. Br. Med. Bull. 2010; 95:47–61. [DOI] [PubMed] [Google Scholar]
- 43. Ferlay J., Ervik M., Lam F., Colombet M., Mery L., Piñeros M., Znaor A., Soerjomataram I., Bray F. Global Cancer Observatory: Cancer Today. 2018; Lyon: International Agency for Research on Cancer. [Google Scholar]
- 44. Kerr K.M. Pulmonary adenocarcinomas: classification and reporting. Histopathology. 2009; 54:12–27. [DOI] [PubMed] [Google Scholar]
- 45. Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015; 11:3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zgarbov Refinement of the sugar–phosphate backbone torsion beta for AMBER force fields improves the description of Z- and B-DNA. J. Chem. Theory Comput. 2015; 11:5723–5736. [DOI] [PubMed] [Google Scholar]
- 47. Yildirim I., Stern H.A., Kennedy S.D., Tubbs J.D., Turner D.H. Reparameterization of RNA χ torsion parameters for the AMBER force field and comparison to NMR spectra for cytidine and uridine. J. Chem. Theory Comput. 2010; 6:1520–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983; 79:926–935. [Google Scholar]
- 49. Götz A.W., Williamson M.J., Xu D., Poole D., Grand S.L., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 2012; 8:1542–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Salomon-Ferrer R., Götz A.W., Poole D., Grand S.L., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 2013; 9:3878–3888. [DOI] [PubMed] [Google Scholar]
- 51. Eastman P., Swails J., Chodera J.D., McGibbon R.T., Zhao Y., Beauchamp K.A., Wang L.P., Simmonett A.C., Harrigan M.P., Stern C.D.et al. OpenMM 7: rapid development of high performance algorithms for molecular dynamics. PLoS Comput. Biol. 2017; 13:e1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ito F., Yang H., Xiao X., Li S.X., Wolfe A., Zirkle B., Arutiunian V., Chen X.S. Understanding the structure, multimerization, subcellular localization and mC selectivity of a genomic mutator and anti-HIV factor APOBEC3H. Sci. Rep. 2018; 8:3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bateman A., Jesus M.M., Claire O.D., Michele M., Emanuele A., Ricardo A., Benoit B., Mark B., Carlos B., Ramona B.et al. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hix M.A., Cisneros G.A. Computational investigation of APOBEC3H substrate orientation and selectivity. J. Phys. Chem. 2020; 124:3903–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bohn J.A., Thummar K., York A., Raymond A., Brown W.C., Bieniasz P.D., Hatziioannou T., Smith J.L. APOBEC3H structure reveals an unusual mechanism of interaction with duplex RNA. Nat. Commun. 2017; 8:1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 57. Feng Y., Love R.P., Ara A., Baig T.T., Adolph M.B., Chelico L. Natural polymorphisms and oligomerization of human APOBEC3H contribute to single-stranded DNA scanning ability. J. Biol. Chem. 2015; 290:27188–27203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Case D.A., Ben-Shalom I.Y., Brozell S.R., Cerutti D.S., Cheatham I.T.E., Cruzeiro V.W.D., Darden T.A., Duke R.E., Ghoreishi D., Gilson M.K.et al. AMBER 2018. 2018; San Francisco: University of California. [Google Scholar]
- 59. Schafmeister C.E.A.F., Ross W.S., Romanovski V. LEAP. 1995; San Francisco: University of California. [Google Scholar]
- 60. Anandakrishnan R., Aguilar B., Onufriev A.V. H++ 3.0: automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 2012; 40:W537–W541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995; 103:8577. [Google Scholar]
- 62. Izaguirre J.A., Sweet C.R., Pande V.S. Multiscale dynamics of macromolecules using normal mode Langevin. Pac. Symp. Biocomput. 2010; 240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Graham S.E., Syeda F., Cisneros G.A. Computational prediction of residues involved in fidelity checking for DNA synthesis in DNA polymerase I. Biochemistry. 2012; 51:2569–2578. [DOI] [PubMed] [Google Scholar]
- 64. Elias A.A., Cisneros G.A. Karabencheva-Christova T. Computational study of putative residues involved in DNA synthesis fidelity checking in Thermus aquaticus DNA polymerase I. Biomolecular Modelling and Simulations. 2014; 96:NY: Academic Press; 39–75. [DOI] [PubMed] [Google Scholar]
- 65. Dewage S.W., Cisneros G.A. Computational analysis of ammonia transfer along two intramolecular tunnels in Staphylococcus aureus glutamine-dependent amidotransferase (GatCAB). J. Phys. Chem. B. 2015; 119:3669–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Roe D.R., Cheatham T.E. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013; 9:3084–3095. [DOI] [PubMed] [Google Scholar]
- 67. Feng Y., Love R.P., Ara A., Baig T.T., Adolph M.B., Chelico L. Natural polymorphisms and oligomerization of human APOBEC3H contribute to single-stranded DNA scanning ability. J. Biol. Chem. 2015; 290:27188–27203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chelico L., Prochnow C., Erie D.A., Chen X.S., Goodman M.F. Structural model for deoxycytidine deamination mechanisms of the HIV-1 inactivation enzyme APOBEC3G. J. Biol. Chem. 2010; 285:16195–16205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Creighton S., Bloom L.B., Goodman M.F. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 1995; 262:232–256. [DOI] [PubMed] [Google Scholar]
- 70. Chelico L., Pham P., Calabrese P., Goodman M.F. APOBEC3G DNA deaminase acts processively 3′ → 5′ on single-stranded DNA. Nat. Struct. Mol. Biol. 2006; 13:392–399. [DOI] [PubMed] [Google Scholar]
- 71. Refsland E.W., Stenglein M.D., Shindo K., Albin J.S., Brown W.L., Harris R.S. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 2010; 38:4274–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Siriwardena S.U., Chen K., Bhagwat A.S. Functions and malfunctions of mammalian DNA-cytosine deaminases. Chem. Rev. 2016; 116:12688–12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998; 273:5858–5868. [DOI] [PubMed] [Google Scholar]
- 74. Ward I.M., Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 2001; 276:47759–47762. [DOI] [PubMed] [Google Scholar]
- 75. Kidd J.M., Newman T.L., Tuzun E., Kaul R., Eichler E.E. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007; 3:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Duggal N.K., Malik H.S., Emerman M. The breadth of antiviral activity of Apobec3DE in chimpanzees has been driven by positive selection. J. Virol. 2011; 85:11361–11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wittkopp C.J., Adolph M.B., Wu L.I., Chelico L., Emerman M. A single nucleotide polymorphism in human APOBEC3C enhances restriction of lentiviruses. PLoS Pathog. 2016; 12:e1005865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Feng Y., Wong L., Morse M., Rouzina I., Williams M.C., Chelico L. RNA-mediated dimerization of the human deoxycytidine deaminase APOBEC3H influences enzyme activity and interaction with nucleic acids. J. Mol. Biol. 2018; 430:4891–4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cescon D.W., Haibe-Kains B., Mak T.W. APOBEC3B expression in breast cancer reflects cellular proliferation, while a deletion polymorphism is associated with immune activation. Proc. Natl Acad. Sci. U.S.A. 2015; 112:2841–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Klonowska K., Kluzniak W., Rusak B., Jakubowska A., Ratajska M., Krawczynska N., Vasilevska D., Czubak K., Wojciechowska M., Cybulski C.et al. The 30 kb deletion in the APOBEC3 cluster decreases APOBEC3A and APOBEC3B expression and creates a transcriptionally active hybrid gene but does not associate with breast cancer in the European population. Oncotarget. 2017; 8:76357–76374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Liu J., Sieuwerts A.M., Look M.P., van der Vlugt-Daane M., Meijer-van Gelder M.E., Foekens J.A., Hollestelle A., Martens J.W. The 29.5 kb APOBEC3B deletion polymorphism is not associated with clinical outcome of breast cancer. PLoS One. 2016; 11:e0161731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wen W.X., Soo J.S., Kwan P.Y., Hong E., Khang T.F., Mariapun S., Lee C.S., Hasan S.N., Rajadurai P., Yip C.H.et al. Germline APOBEC3B deletion is associated with breast cancer risk in an Asian multi-ethnic cohort and with immune cell presentation. Breast Cancer Res. 2016; 18:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Buisson R., Langenbucher A., Bowen D., Kwan E.E., Benes C.H., Zou L., Lawrence M.S. Passenger hotspot mutations in cancer driven by APOBEC3A and mesoscale genomic features. Science. 2019; 364:eaaw2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Barroso-Sousa R., Jain E., Cohen O., Kim D., Buendia-Buendia J., Winer E., Lin N., Tolaney S.M., Wagle N. Prevalence and mutational determinants of high tumor mutation burden in breast cancer. Ann. Oncol. 2020; 31:387–394. [DOI] [PubMed] [Google Scholar]
- 85. Harris R.S., Serebrenik A.A., Argyris P., Jarvis M.C., Brown W.L., Bazzaro M., Vogel R.I., Erickson B.K., Lee S.H., Goergen K.M.et al. The DNA cytosine deaminase APOBEC3B is a molecular determinant of platinum responsiveness in clear cell ovarian cancer. Clin. Cancer Res. 2020; 26:3397–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ooms M., Brayton B., Letko M., Maio S.M., Pilcher C.D., Hecht F.M., Barbour J.D., Simon V. HIV-1 Vif adaptation to human APOBEC3H haplotypes. Cell Host Microbe. 2013; 14:411–421. [DOI] [PubMed] [Google Scholar]
- 87. Refsland E.W., Hultquist J.F., Luengas E.M., Ikeda T., Shaban N.M., Law E.K., Brown W.L., Reilly C., Emerman M., Harris R.S. Natural polymorphisms in human APOBEC3H and HIV-1 Vif combine in primary T lymphocytes to affect viral G-to-A mutation levels and infectivity. PLoS Genet. 2014; 10:e1004761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Adolph M.B., Ara A., Feng Y., Wittkopp C.J., Emerman M., Fraser J.S., Chelico L. Cytidine deaminase efficiency of the lentiviral viral restriction factor APOBEC3C correlates with dimerization. Nucleic Acids Res. 2017; 45:3378–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Duggal N.K., Emerman M. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat. Rev. Immunol. 2012; 12:687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hoopes J.I., Cortez L.M., Mertz T.M., Malc E.P., Mieczkowski P.A., Roberts S.A. APOBEC3A and APOBEC3B preferentially deaminate the lagging strand template during DNA replication. Cell Rep. 2016; 14:1273–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Law E.K., Sieuwerts A.M., LaPara K., Leonard B., Starrett G.J., Molan A.M., Temiz N.A., Vogel R.I., Meijer-van Gelder M.E., Sweep F.C.et al. The DNA cytosine deaminase APOBEC3B promotes tamoxifen resistance in ER-positive breast cancer. Sci. Adv. 2016; 2:e1601737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Cescon D.W., Haibe-Kains B. DNA replication stress: a source of APOBEC3B expression in breast cancer. Genome Biol. 2016; 17:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Green A.M., Landry S., Budagyan K., Avgousti D.C., Shalhout S., Bhagwat A.S., Weitzman M.D. APOBEC3A damages the cellular genome during DNA replication. Cell Cycle. 2016; 15:998–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.