Abstract
BACKGROUND
Long noncoding RNAs (lncRNAs) and mRNAs are widely involved in various physiological and pathological processes. The use of glucagon-like peptide-1 receptor agonists (GLP-1RAs) is a novel therapeutic strategy that could promote insulin secretion and decrease the rate of β-cell apoptosis in type 2 diabetes mellitus (T2DM) patients. However, the specific lncRNAs and mRNAs and their functions in these processes have not been fully identified and elucidated.
AIM
To identify the lncRNAs and mRNAs that are involved in the protective effect of GLP-1RA in β cells, and their roles.
METHODS
Rat gene microarray was used to screen differentially expressed (DE) lncRNAs and mRNAs in β cells treated with geniposide, a GLP-1RA. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed to assess the underlying functions of DE mRNAs. Hub mRNAs were filtered using the STRING database and the Cytoscape plugin, CytoHubba. In order to reveal the regulatory relationship between lncRNAs and hub mRNAs, their co-expression network was constructed based on the Pearson coefficient of DE lncRNAs and mRNAs, and competing endogenous RNA (ceRNA) mechanism was explored through miRanda and TargetScan databases.
RESULTS
We identified 308 DE lncRNAs and 128 DE mRNAs with a fold change filter of ≥ 1.5 and P value < 0.05. GO and KEGG pathway enrichment analyses indicated that the most enriched terms were G-protein coupled receptor signaling pathway, inflammatory response, calcium signaling pathway, positive regulation of cell proliferation, and ERK1 and ERK2 cascade. Pomc, Htr2a, and Agtr1a were screened as hub mRNAs using the STRING database and the Cytoscape plugin, CytoHubba. This result was further verified using SwissTargetPrediction tool. Through the co-expression network and competing endogenous (ceRNA) mechanism, we identified seven lncRNAs (NONRATT027738, NONRATT027888, NONRATT030038, etc.) co-expressed with the three hub mRNAs (Pomc, Htr2a, and Agtr1a) based on the Pearson coefficient of the expression levels. These lncRNAs regulated hub mRNA functions by competing with six miRNAs (rno-miR-5132-3p, rno-miR-344g, rno-miR-3075, etc.) via the ceRNA mechanism. Further analysis indicated that lncRNA NONRATT027738 interacts with all the three hub mRNAs, suggesting that it is at a core position within the ceRNA network.
CONCLUSION
We have identified key lncRNAs and mRNAs, and highlighted here how they interact through the ceRNA mechanism to mediate the protective effect of GLP-1RA in β cells.
Keywords: Type 2 diabetes, β cell, Long noncoding RNA, Competing endogenous RNA, Co-expression analysis, Glucagon-like peptide-1 receptor agonist
Core Tip: This study investigated the long noncoding RNA (lncRNA) regulatory network involved in the protective effects of geniposide, a glucagon-like peptide-1 receptor agonist (GLP-1RA), in pancreatic β cells using a microarray. We identified key lncRNAs and mRNAs, and highlighted how they interact through the competing endogenous RNA mechanism to mediate GLP-1RA-mediated protection in β cells. Our study has contributed to a deeper understanding of the molecular mechanism of β cell protection by GLP-1RA at the transcriptional level.
INTRODUCTION
The impaired function and diminished mass of β cells in type 2 diabetes mellitus (T2DM) patients lead to insufficient insulin secretion and hyperglycemia[1]. Additionally, malfunction, de-differentiation, and apoptosis of β cells are the key characteristics of T2DM[2]. Impaired insulin secretion is a key contributor to chronic hyperglycemia in T2DM patients[2,3]. Hence, a strategy to block β cell apoptosis and restore β cell function is urgently needed. As a class of promising anti-diabetic drugs, glucagon-like peptide-1 receptor agonists (GLP-1RAs) have been shown to potentiate insulin secretion in a glucose-dependent manner, which can decrease blood glucose levels without the risk of hypoglycemia[4-6]. Moreover, studies have demonstrated that GLP-1RAs prevent β cells from premature apoptosis and promote their function[7,8]. Recent studies reported that geniposide, a monomer extracted from gardenia, is a novel GLP-1RA with multiple protective effects in human diseases, such as Alzheimer’s disease[9], obesity-related cardiac injury[10], and myocardial ischemia[11]. We and other researchers found that geniposide potentiates insulin secretion, promotes proliferation, and decreases the rate of β cell apoptosis by stimulating the GLP-1 receptor[12-14].
Long noncoding RNAs (lncRNA) are some of the recently studied regulatory molecules[15]. These RNAs are transcripts that are longer than 200 nucleotides and do not code for proteins[16]. Mechanically, lncRNAs exert their regulatory effects through communication with other molecules. Growing evidence demonstrates that lncRNAs regulate mRNA expression by competing with microRNAs (miRNAs)[17,18]. The competition among lncRNAs, miRNAs, and mRNAs was termed the competing endogenous RNA mechanism or “ceRNA mechanism”, which is widely involved in multiple biological processes, including insulin signal transduction that may affect diabetes development[19]. Currently, the mechanisms of the protective effect of GLP-1RA in β cells have been widely investigated, but their potential relationship with mRNAs and lncRNAs is yet to be explored.
We previously demonstrated that geniposide protects pancreatic β cells via GLP-1R[13]. In this study, we examined the expression profiles of lncRNAs and mRNAs in INS-1 cells treated with or without geniposide via microarray. We further explored, via biological information analysis, the interaction between lncRNAs and mRNAs, and whether the ceRNA network was involved in their regulatory relationship. We aimed to identify the roles of lncRNAs and mRNAs in mediating the protective effect of GLP-1RA in β cells.
MATERIALS AND METHODS
Cell culture and treatment
Rat pancreatic INS-1 cells were purchased from the National Infrastructure of Cell Line Resource (Identification number: 3111C0001CCC000378). The cells grew irregularly, polygonally, and adherently. Mycoplasma detection was negative. INS-1 cells were grown in high-glucose Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 µg/mL streptomycin, 100 µg/mL penicillin, and 50 µmol/L β-mercaptoethanol at 37 °C in an atmosphere containing 5% CO2. INS-1 cells were seeded in 12-well plates to approximately 80% confluence and treated with or without 10 μmol/L geniposide for 24 h. Three technical replicates were performed on each independent sample.
RNA extraction, purification, and quality control
Total RNA was extracted and purified using the RNeasy Mini Kit (Cat#74106, QIAGEN, GmBH, Germany) following the manufacturer’s instructions and checked for an RIN number to inspect RNA integration with an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, United States).
The initial sample of the chip experiment was total RNA that was subjected to quality inspection using a NanoDrop ND-2000 spectrophotometer and Agilent Bioanalyzer 2100 (Agilent Technologies). Then, quality-qualified RNA was subjected to subsequent chip experiments.
Microarray and data analysis
Rat microarray: The Rat microarray Agilent-074571 RAT_LNCRNA_20150413 was developed by Shanghai Bohao Company. The probe information was queried from the GEO database with platform number GPL27603. This microarray was used to profile the lncRNAs and mRNAs. The probe design was based on the latest version of the genome covering core lncRNA and mRNA databases, such as GENCODE V21, Ensembl, UCSC, NONCODE, LNCipedia, and lncRNAdb.
RNA labeling and array hybridization: Total RNA was amplified and labeled with the Low Input Quick Amp Labeling Kit, One-Color (Cat. # 5190-2305, Agilent Technologies), following the manufacturer’s instructions. Labeled cRNAs were purified with the RNeasy Mini Kit (Cat. # 74106, QIAGEN).
Each slide was hybridized using 600 ng Cy3-labeled cRNA and a Gene Expression Hybridization Kit (Cat. # 5188-5242, Agilent Technologies) in a hybridization oven (Cat. # G2545A, Agilent Technologies), according to the manufacturer’s instructions. After 17 h of hybridization, slides were washed in staining dishes (Cat. # 121, Thermo Shandon, Waltham, MA, USA) with the Gene Expression Wash Buffer Kit (Cat. # 5188-5327, Agilent Technologies) according to the manufacturer’s instructions.
Data acquisition and analysis: Slides were scanned with an Agilent Microarray Scanner (Cat. # G2565CA, Agilent Technologies) with default settings: Dye channel, green; scan resolution = 3 μm; PMT 100%; 20 bit. Data were extracted with Feature Extraction software 10.7 (Agilent Technologies). Raw data were normalized using Robust Multichip Average (RMA) algorithm and limma packages in R. Significantly differentially expressed (DE) lncRNAs and mRNAs between the two groups were selected if the fold changes of the threshold values were ≥ 1.5 and P value < 0.05.
Gene Ontology analysis
Gene Ontology (GO) analysis can be divided into three parts: Molecular function, biological process, and cellular component, which respectively describe the molecular functions of potential gene products, the biological processes involved, and the cellular environment in which they are located. Enrichment analysis was performed via David 6.8 (https://david.ncifcrf.gov/) database[20,21]. David 6.8 for annotation, visualization, and integrated discovery provides a comprehensive set of functional annotation tools to understand the biological meaning behind the long list of genes. The GO terms obtained in the drawing are arranged in descending order according to the –log10 (P value) of enrichment, and we take the first 10 if there are more than 10 results.
Pathway analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DE genes can enrich the significant pathways and help to find the biological regulatory pathways for significant differences in experimental conditions. The David 6.8 (https://david.ncifcrf.gov/) database also can be used for the enrichment of pathway. The KEGG pathway terms obtained in the drawing are arranged in descending order according to –log10 (P value) of enrichment, and we take the first 10 if there are more than 10 results.
Screening hub mRNAs
Based on the GO and KEGG analyses, a protein-protein interaction (PPI) network was constructed through the STRING (v11.0, https://string-db.org/) database for all DE mRNAs that were enriched in the GO and KEGG terms. STRING is a database of known and predicted protein-protein interactions, including direct (physical) and indirect (functional) associations, which stem from computational prediction, knowledge transfer between organisms, and interactions aggregated from other (primary) databases. Further improvements in version 11.0 include a completely redesigned prediction pipeline for inferring protein-protein associations from co-expression data, an API interface for the R computing environment, and improved statistical analysis for enrichment tests in user-provided networks. The co-expression scores in STRING v11.0 are computed using a revised and improved pipeline, making use of all microarray gene expression experiments deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus[22]. Since our data came from microarray, the protein-protein interactions could be reflected through the STRING database based on the co-expression relationship, and a confidence score ≥ 0.4 was set as the cut-off criterion.
Next, mRNAs in the PPI network were ranked with the Cytoscape (Cytoscape_v3.7.2) plugin CytoHubba, which provides 11 topological analysis methods, including Degree, Edge Percolated Component, Maximum Neighborhood Component, Density of Maximum Neighborhood Component, Maximal Clique Centrality, and six centralities (Bottleneck, EcCentricity, Closeness, Radiality, Betweenness, and Stress), based on the shortest paths. CytoHubba provides a user-friendly interface for exploring important nodes in biological networks[23]. The hub mRNAs were selected from the top three via integrated scores of the 11 algorithms.
Prediction of GLP-1 and geniposide targets
SwissTargetPrediction (http://www.swisstargetprediction.ch/) is an online analysis software for small molecule target prediction, which can predict the target of small molecule compounds based on the principle of molecular similarity. We used this tool to predict the GLP-1 and geniposide targets by converting GLP-1 and geniposide into the standard SMILES format (Canonical SMILES) via the PubChem database (https://pubchem.ncbi.nlm.nih.gov/), importing the SMILES format file into the SwissTargetPrediction online analysis platform, setting the property to “Rattus norvegicus”, and predicting the targets.
Construction of co-expression network
The Pearson coefficient was calculated based on the normalized chip expression matrix of DE lncRNAs and mRNAs. Molecules were considered with a strong correlation with the filters set at P < 0.05 and |R| > 0.9. These molecules including lncRNAs and hub mRNAs were constructed into a co-expression network.
Construction of ceRNA network
The ceRNA network was constructed based on the relationships among lncRNAs, miRNAs, and mRNAs. It is established that post-transcriptional regulation of mRNAs could be bound by single-stranded miRNAs, and lncRNAs can directly interact by invoking the miRNA sponge to regulate mRNA expression and activity[24]. The specific steps are as follows:
Prediction of hub mRNA-miRNA pairs: The MiRanda (http://www.microrna.org/) and TargetScan (http://www.targetscan.org/vert_71/) databases provide two algorithms for finding genomic targets for miRNAs. The input file included rat miRNA sequences and 3' untranslated region (UTR) sequences of hub mRNAs. The energy and score threshold filters set for MiRanda were -20 kcal/mol and 50, respectively, and the TargetScan binding type filters were set as 8-mer and 7-mer. The intersections of the results from both databases offered the final prediction of hub mRNA-miRNA pairs.
Prediction of lncRNA-miRNA pairs: Rat lncRNA sequences were downloaded from the NONCODE (http://www.noncode.org/index.php) database. The input file included 3' UTR sequences of lncRNAs from the co-expression network and miRNA sequences from last step, which could interact with hub mRNAs. MiRanda and TargetScan were used as described before to screen lncRNA-miRNA pairs. The intersection of the results from both databases offered the final lncRNA-miRNA pairs.
ceRNA network construction: Based on common miRNAs, the ceRNA network was constructed among lncRNAs, miRNAs, and hub mRNAs, indicating that these lncRNAs could co-express with and regulate hub mRNAs through miRNAs. Cytoscape v3.7.1 was used to construct and visualize the ceRNA network.
Statistical analysis
IBM SPSS 25.0 software was used to analyze all statistical data. The random variance model t-test was employed to identify DE mRNAs and lncRNAs between the control and geniposide-treated groups. Fisher’s exact test was applied for the GO and pathway analyses. P values < 0.05 were considered statistically significant.
RESULTS
Microarray data profile
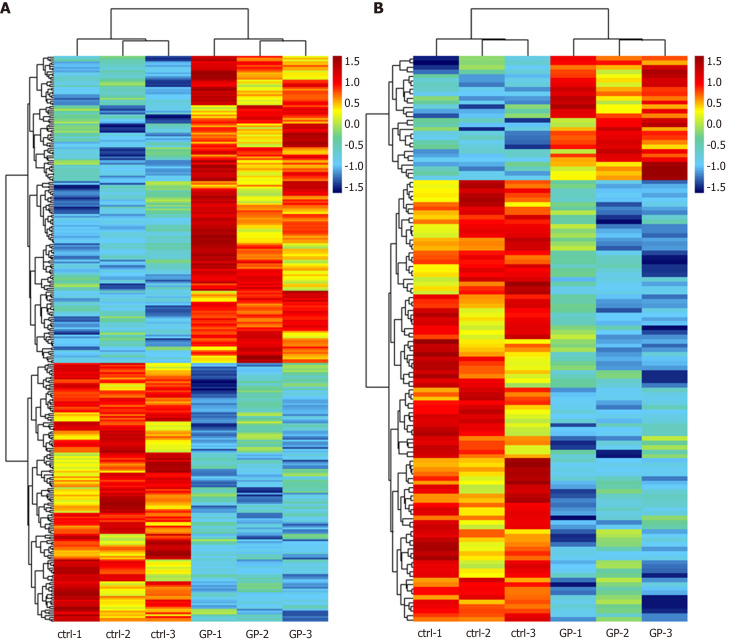
According to the microarray expression profiling data, a total of 167 upregulated and 141 downregulated lncRNAs were identified in the geniposide-treated group compared with those in the control group with a set filter fold change ≥ 1.5 and P value < 0.05. Meanwhile, 28 upregulated and 100 downregulated mRNAs were identified with the same filter settings (Figure 1A and B). This result showed that geniposide treatment induced differential expression of lncRNAs and mRNAs in β cells.
Figure 1.
Heatmap of differentially expressed long noncoding RNAs (A) and mRNAs (B). A: A total of 167 upregulated and 141 downregulated long noncoding RNAs were identified in the geniposide-treated group (GP1 to GP3) compared to the control group (ctrl-1 to ctrl-3) with fold change filter set at ≥ 1.5 and P value < 0.05. B: A total of 28 upregulated and 100 downregulated mRNAs were identified with the same filter settings.
GO and KEGG pathway analyses
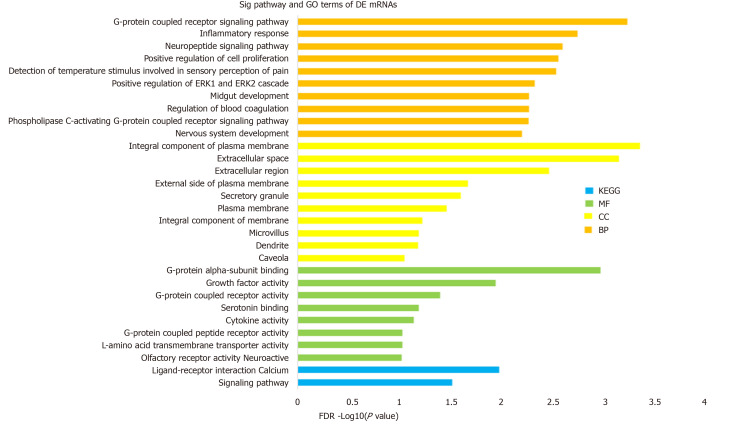
To investigate the biological function and potential mechanism of DE mRNAs, GO and KEGG pathway enrichment analyses were performed via the DAVID database (Figure 2). Biological process analysis was mainly enriched in terms of inflammatory response and positive regulation of the ERK1 and ERK2 cascade. Molecular function analysis was primarily enriched in terms of G-protein coupled receptor (GPCR) activity, and the KEGG pathway was mainly enriched in terms of the calcium signaling pathway. Detailed information is presented in Table 1. These findings showed that DE mRNAs participated in biological functions that were closely related to insulin secretion and β cell viability (Figure 2). Next, we investigated the core mRNAs that perform these functions.
Figure 2.
Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment of differentially expressed mRNAs. Gene Ontology (GO) analysis can be divided into three parts: Molecular function, biological process, and cellular component, which respectively describe the molecular functions of potential gene products, the biological processes involved, and the cellular environments in which they are located. Enrichment analysis was performed via the DAVID 6.8 database. The enriched terms of GO and Kyoto Encyclopedia of Genes and Genomes pathway analysis are arranged in descending order according to -log10 (P value). BP: Biological process; MF: Molecular function; CC: Cellular component; KEGG: Kyoto Encyclopedia of Genes and Genomes.
Table 1.
Significantly enriched Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway terms of differentially expressed long noncoding RNAs
| Category | Term | –log10 (P value) |
| GOTERM_BP | G-protein coupled receptor signaling pathway | 3.23 |
| GOTERM_BP | Inflammatory response | 2.73 |
| GOTERM_BP | Neuropeptide signaling pathway | 2.59 |
| GOTERM_BP | Positive regulation of cell proliferation | 2.55 |
| GOTERM_BP | Detection of temperature stimulus involved | 2.53 |
| GOTERM_BP | Positive regulation of ERK1 and ERK2 cascade | 2.32 |
| GOTERM_BP | Regulation of blood coagulation | 2.26 |
| GOTERM_BP | Midgut development | 2.26 |
| GOTERM_BP | Phospholipase G-protein coupled receptor signaling pathway | 2.26 |
| GOTERM_BP | Nervous system development | 2.19 |
| GOTERM_CC | Integral component of plasma membrane | 3.35 |
| GOTERM_CC | Extracellular space | 3.14 |
| GOTERM_CC | Extracellular region | 2.46 |
| GOTERM_CC | External side of plasma membrane | 1.67 |
| GOTERM_CC | Secretory granule | 1.60 |
| GOTERM_CC | Plasma membrane | 1.46 |
| GOTERM_CC | Integral component of membrane | 1.22 |
| GOTERM_CC | Microvillus | 1.19 |
| GOTERM_CC | Dendrite | 1.17 |
| GOTERM_CC | Caveola | 1.04 |
| GOTERM_MF | G-protein alpha-subunit binding | 2.96 |
| GOTERM_MF | Growth factor activity | 1.93 |
| GOTERM_MF | G-protein coupled receptor activity | 1.39 |
| GOTERM_MF | Serotonin binding | 1.18 |
| GOTERM_MF | Cytokine activity | 1.13 |
| GOTERM_MF | L-amino acid transmembrane transporter activity | 1.02 |
| GOTERM_MF | G-protein coupled peptide receptor activity | 1.02 |
| GOTERM_MF | Olfactory receptor activity | 1.01 |
| KEGG PATHWAY | Neuroactive ligand-receptor interaction | 1.97 |
| KEGG PATHWAY | Calcium signaling pathway | 1.51 |
Construction of PPI network and screening hub mRNAs
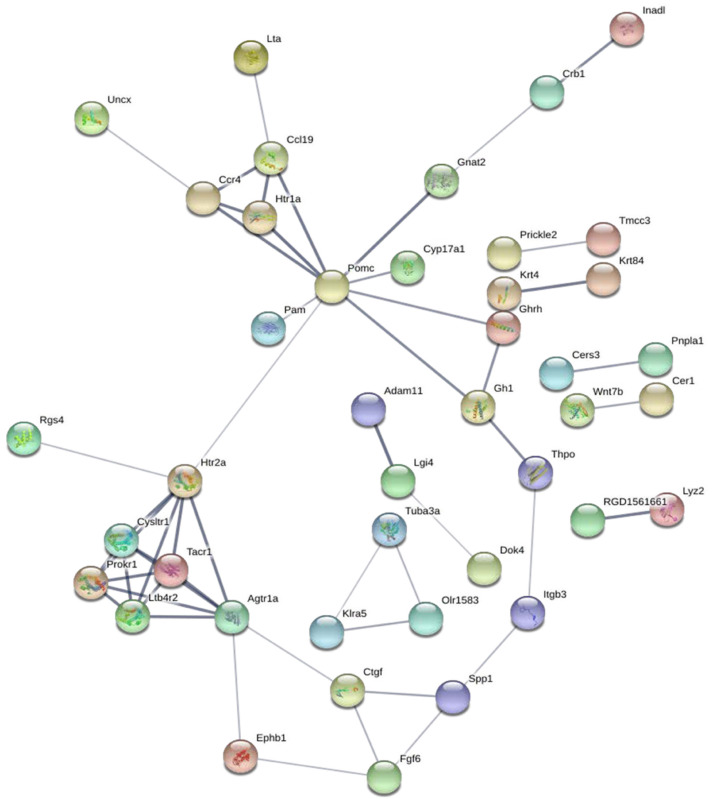
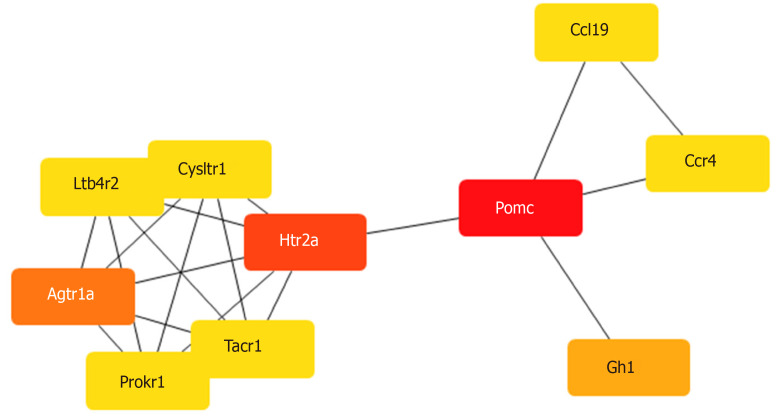
To construct a DE mRNA interaction network, we studied the PPI relationship via the STRING database. This network was comprised of DE mRNAs enriched in the GO and KEGG terms, including 120 nodes and 52 edges (Figure 3). To discover the core mRNAs in this network, CytoHubba was used to screen the hub mRNAs. Based on the CytoHubba scores, the top three hub mRNAs, Pomc, Htr2a, and Agtr1a, were identified (Figure 4), indicating that they had more interactions with other mRNAs and participated in various functions and pathways than other hub mRNAs.
Figure 3.
Protein-protein interaction network of differentially expressed mRNAs. The protein-protein interaction network was constructed through the STRING database for all differentially expressed mRNAs that were enriched in the Gene Ontology and Kyoto Encyclopedia of Genes and Genomes terms. This network includes 120 nodes and 52 edges.
Figure 4.
Hub mRNAs. The Cytoscape plugin, CytoHubba, was used to identify hub mRNAs of the complex network. The mRNAs were colored according to their scores in CytoHubba. The darker the color, the higher the score. By combining the scores of the 11 algorithms of CytoHubba, the mRNAs Pomc, Htr2a, and Agtr1a got the top three scores and were thus considered to be hub mRNAs.
Verification of hub mRNA prediction
To confirm the accuracy of the hub mRNA prediction, we queried the GLP-1 and geniposide targets using SwissTargetPrediction tool. One hundred GLP-1 and 80 geniposide targets were predicted with SwissTargetPrediction. Among them, 22 common targets were shared by both GLP-1 and geniposide (Table 2). The prediction results showed that Agtr1 and Agtr1b were among the most common targets. Agtr1 and Agtr1b, as well as our hub mRNA Agtr1a, all belong to the Agtr family. Additionally, Htr2a was also present in the GLP-1 targets. This was consistent with our mining of Htr2a and Agtr1a as target mRNAs that mediate the functions of geniposide, confirming the accuracy of our analysis. On the other hand, Pomc was not found as a GLP-1 or geniposide target per SwissTargetPrediction, but was predicted as the hub mRNA with the highest score by CytoHubba, suggesting that Pomc was a potential GLP-1 and geniposide target.
Table 2.
Common molecular targets of glucagon-like peptide-1 and geniposide from SwissTargetPrediction
| Common name | Uniprot ID | Target class |
| Ace | P47820 | Ligand-gated ion channel |
| Adrb3 | P26255 | Electrochemical transporter |
| Agtr1 | P25095 | Family A G protein-coupled receptor |
| Agtr1b | P29089 | Family A G protein-coupled receptor |
| App | P08592 | Secreted protein |
| Ca2 | P27139 | Transferase |
| Chrna7 | Q05941 | Hydrolase |
| Ctsk | O35186 | Protease |
| Ednra | P26684 | Enzyme |
| Ednrb | P21451 | Family A G protein-coupled receptor |
| Hdac1 | Q4QQW4 | Nuclear receptor |
| Hmgcr | P51639 | Enzyme |
| Mme | P07861 | Enzyme |
| Oprm1 | P33535 | Family A G protein-coupled receptor |
| Pparg | O88275 | Nuclear receptor |
| Prkcg | P63319 | Structural protein |
| Ptgs2 | P35355 | Family A G protein-coupled receptor |
| Pygl | P09811 | Enzyme |
| Ren1 | P08424 | Family A G protein-coupled receptor |
| Slc6a3 | P23977 | Electrochemical transporter |
| Tert | Q673L6 | Enzyme |
| Trpv1 | O35433 | Voltage-gated ion channel |
Co-expression network among hub mRNAs and DE lncRNAs
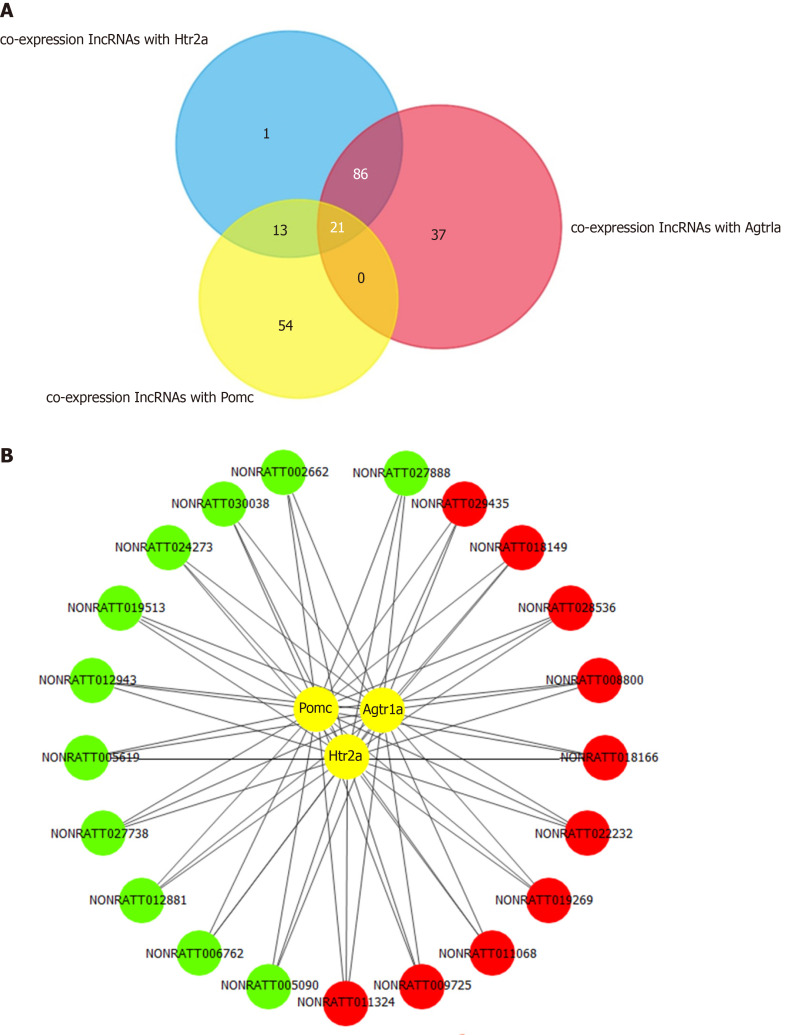
To explore the key lncRNAs that are closely related to the hub mRNAs, a co-expression network was constructed based on the Pearson coefficient of DE lncRNAs and mRNA expression levels. The number of lncRNAs identified to be co-expressed with Pomc, Htr2a, and Agtr1a was 21 (Figure 5A and 5B). The fold change and the Pearson coefficient of each lncRNA are shown in Table 3.
Figure 5.
Co-expression network of long noncoding RNAs and hub mRNAs. A: Venn diagram of co-expressed long noncoding RNAs (lncRNAs) with Pomc, Htr2a, and Agtr1a. A co-expression network was constructed based on the Pearson coefficient with the filter condition as P value < 0.05 and |R| > 0.90. Twenty-one common lncRNAs were obtained by intersecting the co-expressed lncRNAs of Pomc, Htr2a, and Agtr1a. B: The 21 common lncRNAs co-expressed with Pomc, Htr2a, and Agtr1a, including 11 downregulated and 10 upregulated lncRNAs. Red circles represent upregulated lncRNAs, whereas the green circles represent downregulated lncRNAs.
Table 3.
Long noncoding RNAs co-expressed with Pomc, Htr2a, and Agtr1a
| Accession No. | Fold change |
Pearson coefficient |
||
| Pomc_R | Htr2a_R | Agtr1a_R | ||
| NONRATT002662 | 0.1959 | 0.9090 | 0.9755 | 0.9817 |
| NONRATT024273 | 0.2602 | 0.9154 | 0.9771 | 0.9545 |
| NONRATT012943 | 0.3411 | 0.9204 | 0.9472 | 0.9517 |
| NONRATT027738 | 0.3471 | 0.9406 | 0.9727 | 0.9301 |
| NONRATT005090 | 0.3603 | 0.9784 | 0.9478 | 0.9135 |
| NONRATT012881 | 0.4662 | 0.9546 | 0.9744 | 0.9543 |
| NONRATT005619 | 0.5677 | 0.9246 | 0.9325 | 0.9156 |
| NONRATT019513 | 0.5766 | 0.9175 | 0.9455 | 0.9540 |
| NONRATT027888 | 0.6136 | 0.9008 | 0.9814 | 0.9888 |
| NONRATT006762 | 0.6532 | 0.9681 | 0.9586 | 0.9307 |
| NONRATT030038 | 0.6647 | 0.9150 | 0.9693 | 0.9579 |
| NONRATT018166 | 1.5013 | -0.9275 | -0.9451 | -0.9448 |
| NONRATT009725 | 1.5275 | -0.9611 | -0.9396 | -0.9083 |
| NONRATT029435 | 1.5916 | -0.9026 | -0.9437 | -0.9207 |
| NONRATT011324 | 1.6616 | -0.9657 | -0.9531 | -0.9340 |
| NONRATT028536 | 1.7515 | -0.9181 | -0.9419 | -0.9257 |
| NONRATT022232 | 1.8792 | -0.9313 | -0.9539 | -0.9562 |
| NONRATT008800 | 1.9416 | -0.9273 | -0.9485 | -0.9313 |
| NONRATT019269 | 2.1925 | -0.9377 | -0.9152 | -0.9126 |
| NONRATT011068 | 2.4783 | -0.9561 | -0.9828 | -0.9551 |
| NONRATT018149 | 3.9636 | -0.9114 | -0.9435 | -0.9337 |
Pearson coefficient < 0 means a negative correlation, and Pearson coefficient > 0 means a positive correlation.
The expression of the three hub mRNAs (Pomc, Htr2a, and Agtr1a) was downregulated in geniposide-treated INS-1 cells compared to that in untreated cells. Among the 21 co-expressed lncRNAs, 11 (NONRATT002662, NONRATT024273, NONRATT012943, NONRATT027738, NONRATT005090, NONRATT012881, NONRATT005619, NONRATT019513, NONRATT027888, NONRATT006762, and NONRATT030038) were downregulated and positively correlated with hub mRNAs. The correlation between the expression levels of the 11 lncRNAs and hub mRNAs prompted us to explore whether they are functionally related.
Construction of ceRNA network
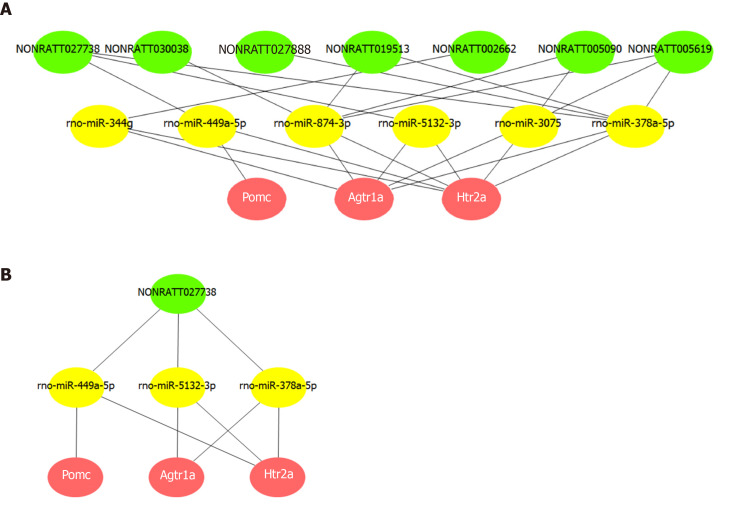
A ceRNA network was constructed based on the common miRNAs that could bind to the 11 lncRNAs, as well as the three hub mRNAs. Based on the intersection of the prediction results from MiRanda and TargetScan, we obtained 77, 16, and 23 miRNAs that could bind to the 3' UTR of Htr2a, Pomc, and Agtr1a, respectively. Among these miRNAs, we found that rno-miR-449a-5p could interact with both Htr2a and Pomc, while rno-miR-5132-3p, rno-miR-344g, rno-miR-3075, rno-miR-378a-5p, and rno-miR-874-3p could interact with both Htr2a and Agtr1a.
Then, we screened lncRNAs that could be bound by these six miRNAs from the 11 co-expressed lncRNAs and found seven such lncRNAs. Based on this analysis, a ceRNA network composed of three hub mRNAs, six miRNAs, and seven lncRNAs was constructed (Figure 6A). This network suggested that lncRNAs (NONRATT002662, NONRATT005090, NONRATT005619, NONRATT019513, NONRATT027738, NONRATT027888, and NONRATT030038) may competitively bind to miRNAs (rno-miR-449a-5p, rno-miR-5132-3p, rno-miR-344g, rno-miR-3075, rno-miR-378a-5p, and rno-miR-874-3p), and thereby affect the expression and function of hub mRNAs (Pomc, Htr2a, and Agtr1a). Further analysis of the network indicated that NONRATT027738 can regulate all the three hub mRNAs (Pomc, Htr2a, and Agtr1a) through miRNAs (rno-miR-449a-5p, rno-miR-5132-3p, and rno-miR-378a-5p) (Figure 6B).
Figure 6.
Competing endogenous RNA network of key long noncoding RNAs and Pomc, Htr2a, and Agtr1a. A: The competing endogenous RNA network was constructed via common miRNAs that could bind to these 11 long noncoding RNAs (lncRNAs), as well as the three hub mRNAs. Based on the intersection of the predictions from miRanda and TargetScan, seven lncRNAs were found to competitively bind six miRNAs, thereby affecting the expression and function of the hub mRNAs. B: Further network analysis indicated that NONRATT027738 can regulate all the three hub mRNAs (Pomc, Htr2a, and Agtr1a) through miRNAs (rno-miR-449a-5p, rno-miR-5132-3p, and rno-miR-378a-5p).
DISCUSSION
GLP-1RA increases β cell sensitivity to glucose and protects β cells from apoptosis[25]. The insulinotropic effects of GLP-1RA are glucose-dependent, posing a low risk for hypoglycemia[26]. To further understand the functions of GLP-1RA in β cells, it is essential to identify the molecular mechanisms involved. Increasing evidence indicates that lncRNAs play key roles in many biological processes, including insulin secretion and cell proliferation by the ceRNA mechanism[27]. In this study, we analyzed the microarray data of INS-1 cells treated with geniposide, which is a GLP-1RA confirmed by plenty of studies[10,13,14]. Our input data were analyzed using bioinformatic tools, including analyses of GO/KEGG pathway, PPI network, co-expression network, and the ceRNA network. With the bioinformatic tools, we identified three hub mRNAs, seven lncRNAs, and six miRNAs that mediated the protective effects of GLP-1RA in β cells through the ceRNA mechanism.
Pomc, Htr2a, and Agtr1a were also identified as hub mRNAs in the ceRNA network of pancreatic islet-like cell clusters. Pomc expression was downregulated in T3pi cells, which could increase β cell proportions and insulin synthesis[28]. Dominguez et al[29] compared the mRNA expression in pancreatic islets from type 2 diabetic and non-diabetic patients. The mRNA expression levels were higher in T2DM patients; however, their findings confirmed that the lower expression of Pomc was protective to β cells. It was reported that the expression of 5-hydroxytryptamine (5-HT) participates in the regulation of insulin secretion, and overexpression of Htr2a is associated with islet dysfunction in T2DM[30]. Testosterone was shown to prevent pancreatic β cell apoptosis by suppressing Agtr1a expression[31]. These studies further confirm our finding that the downregulation of Pomc, Htr2a, and Agtr1a is protective to β cells.
Based on the GO and KEGG analyses, Pomc, Htr2a, and Agtr1a were enriched in the G-protein-coupled receptor signaling pathway, serotonin receptor signaling pathway, inflammatory response, positive regulation of cell proliferation, ERK1 and ERK2 cascade, and cytosolic calcium ion concentration. More studies have shown that the ERK signaling pathway[32], calcium ion concentration[33], inflammatory response[34], and cell proliferation are essential for augmenting insulin secretion and β cell mass protection from premature apoptosis.
Additionally, through the SwissTargetPrediction database, we verified that Htr2a and Agtr1a are GLP-1 and geniposide targets. This result strongly confirmed the accuracy of our screening for hub mRNAs. Specifically, Pomc scored highest among the hub mRNAs based on our CytoHubba analysis, suggesting that Pomc is a potential GLP-1RA target.
Our analysis showed that six miRNAs were involved in mediating GLP-1RA function within the ceRNA network. In support of our results, four of the six miRNAs (miR-449a, miR-378a, miR-344, and miR-874) have already been implicated in regulating insulin signaling, improving metabolic dysregulation, and activating insulin synthesis[35-38]. The other two miRNAs (miR-5132-3p and miR-3075) have been suggested to play a regulatory role in the proliferation and migration of osteoblasts and Schwann cells[39,40]. Hence, we propose that miR-5132-3p and miR-3075 may act as new effector molecules in β-cell regulation.
LncRNAs can act as miRNA sponges to regulate mRNA expression and activity via the ceRNA mechanism[24]. Recently, studies have revealed that lncRNAs are involved in the process of insulin secretion and β cell apoptosis through the ceRNA mechanism[41,42]. In this study, we identified seven lncRNAs (NONRATT002662, NONRATT005090, NONRATT005619, NONRATT019513, NONRATT027738, NONRATT027888, and NONRATT030038) that competitively bind to six miRNAs (rno-miR-449a-5p, rno-miR-5132-3p, rno-miR-344g, rno-miR-3075, rno-miR-378a-5p, and rno-miR-874-3p), thereby influencing the expression and function of hub mRNAs (Pomc, Htr2a, and Agtr1a). Deciphering this lncRNA-miRNA-mRNA network deepens our understanding of the ceRNA mechanism in the protective effect of GLP-1RA in β cells.
Considering that therapeutic RNAi technology has now been tested in humans[43,44], we believe that our report provides novel RNAs as potential therapeutic targets in the GLP-1RA-mediated protection of β cells. However, further studies are required to better understand and confirm the specific function of these RNAs in β cells.
In summary, this study revealed the expression profiles of lncRNAs and mRNAs in geniposide-treated INS-1 cells. Further exploration via biological information analysis demonstrated that the ceRNA mechanism is involved in the regulatory relationship between lncRNAs and mRNAs in β cells. These findings provide significant insight in understanding the mechanisms of GLP-1RA function at the transcription level.
ARTICLE HIGHLIGHTS
Research background
As a class of promising anti-diabetic drugs, glucagon-like peptide-1 receptor agonists (GLP-1RAs) have been shown to prevent β cells from apoptosis and potentiate insulin secretion in a glucose-dependent manner, which can decrease blood glucose levels without the risk of hypoglycemia. Long noncoding RNAs (lncRNA) are transcripts that are longer than 200 nucleotides and do not code for proteins. Growing evidence demonstrates that lncRNAs regulate mRNA expression by competing with miRNAs, which was termed as “ceRNA mechanism”. Studies have demonstrated that ceRNA mechanism is widely involved in multiple biological processes, including insulin signal transduction that may affect diabetes development. Currently, the mechanisms of the protective effect of GLP-1RA on β cells have been widely investigated; however, the specific lncRNAs and mRNAs and their functions in these processes have not been fully identified and elucidated.
Research motivation
Is there any specific lncRNAs that participate in the protective effect of GLP-1RAs in β cells? What is the mechanism of lncRNAs involved in this process? Answering these questions will provide significant insight in understanding the mechanisms of GLP-1RA function at the transcription level.
Research objectives
We and other researchers found that geniposide potentiates insulin secretion, promotes proliferation, and decreases the rate of β cell apoptosis by stimulating the GLP-1 receptor. In this study, we further identified the lncRNAs and mRNAs that were involved in the protective effect of geniposide on β cells, and their roles. This will be helpful for in-depth exploration of the mechanism of GLP-1RAs function in β cells.
Research methods
Rat gene microarray was used to screen differentially expressed (DE) lncRNAs and mRNAs in β cells treated with geniposide, a GLP-1RA. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed to assess the underlying functions of DE mRNAs. Hub mRNAs were filtered using the STRING database and the Cytoscape plugin, CytoHubba. In order to reveal the regulatory relationship between lncRNAs and hub mRNAs, their co-expression network was constructed based on the Pearson coefficient of DE lncRNAs and mRNAs, and competing endogenous (ceRNA) mechanism was explored through miRanda and TargetScan databases.
Research results
We identified 308 DE lncRNAs and 128 DE mRNAs with a fold change filter of ≥ 1.5 and P value < 0.05. GO and KEGG pathway enrichment analyses indicated that the most enriched terms were G-protein coupled receptor signaling pathway, inflammatory response, calcium signaling pathway, positive regulation of cell proliferation, and ERK1 and ERK2 cascade. Pomc, Htr2a, and Agtr1a were screened as hub mRNAs using the STRING database and the Cytoscape plugin, CytoHubba. This result was further verified using SwissTargetPrediction tool. Through the co-expression network and competing endogenous (ceRNA) mechanism, we identified seven lncRNAs (NONRATT027738, NONRATT027888, NONRATT030038, etc.) co-expressed with the three hub mRNAs (Pomc, Htr2a, and Agtr1a) based on the Pearson coefficient of the expression levels. These lncRNAs regulated hub mRNA functions by competing with six miRNAs (rno-miR-5132-3p, rno-miR-344g, rno-miR-3075, etc.) via the ceRNA mechanism. Further analysis indicated that lncRNA NONRATT027738 interacts with all the three hub mRNAs, suggesting that it is at a core position within the ceRNA network.
Research conclusions
We have identified key lncRNAs and mRNAs, and highlighted here how they interact through the ceRNA mechanism to mediate the protective effect of GLP-1RA in β cells.
Research perspectives
The “ceRNA mechanism”, which is widely involved in multiple biological processes, mediates the protective effect of GLP-1RAs in β cells. The value of bioinformatics allows scientists to create comprehensive databases of biological and health information that can be used to test theories and generate solutions to medical problems that affect us all.
ACKNOWLEDGEMENTS
The authors would like to acknowledge Tao Bai for skillful technical assistance.
Footnotes
Institutional review board statement: This study did not involve human studies and/or animal experiments.
Institutional animal care and use committee statement: This study did not involve human studies and/or animal experiments.
Conflict-of-interest statement: The authors declare that they have no competing interests and have nothing to disclose.
Manuscript source: Unsolicited manuscript
Peer-review started: April 17, 2020
First decision: April 22, 2020
Article in press: August 4, 2020
Specialty type: Endocrinology and metabolism
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gupta MK S-Editor: Liu JH L-Editor: Wang TQ E-Editor: Ma YJ
Contributor Information
Li-Juan Cui, Department of Pharmacology, Basic Medical College, Shanxi Medical University, Taiyuan 030001, Shanxi Province, China.
Tao Bai, Department of Endocrinology, The First Clinical Hospital of Shanxi Medical University, Taiyuan 030001, Shanxi Province, China.
Lin-Ping Zhi, Department of Pharmacology, Basic Medical College, Shanxi Medical University, Taiyuan 030001, Shanxi Province, China.
Zhi-Hong Liu, Department of Respiratory Medicine, The First Clinical Hospital of Shanxi Medical University, Taiyuan 030001, Shanxi Province, China.
Tao Liu, Department of General Surgery, Shanxi Bethune Hospital, Taiyuan 030006, Shanxi Province, China.
Huan Xue, Department of Pharmacology, Basic Medical College, Shanxi Medical University, Taiyuan 030001, Shanxi Province, China.
Huan-Huan Yang, Department of Pharmacology, Basic Medical College, Shanxi Medical University, Taiyuan 030001, Shanxi Province, China.
Xiao-Hua Yang, Department of Pharmacology, Basic Medical College, Shanxi Medical University, Taiyuan 030001, Shanxi Province, China.
Min Zhang, College of Pharmacy, Shanxi Medical University, Taiyuan 030001, Shanxi Province, China.
Ya-Ru Niu, Second Clinical Medical College, Shanxi Medical University, Taiyuan 030001, Shanxi Province, China.
Yun-Feng Liu, Department of Endocrinology, The First Clinical Hospital of Shanxi Medical University, Taiyuan 030001, Shanxi Province, China.
Yi Zhang, Department of Pharmacology, Basic Medical College, Shanxi Medical University, Taiyuan 030001, Shanxi Province, China. yizhang313@163.com.
Data sharing statement
The datasets (Series GSE138744) generated and analyzed during the current study are available in the (GEO) repository, (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138744). To review the data, please enter token: opslyaomvhmlzkb into the box. Our data in GSE138744 remains in private status now. You can review it through the pathway above, and we will make the data publicly available prior to the publication of this article.
References
- 1.Xiong X, Wang X, Li B, Chowdhury S, Lu Y, Srikant CB, Ning G, Liu JL. Pancreatic islet-specific overexpression of Reg3β protein induced the expression of pro-islet genes and protected the mice against streptozotocin-induced diabetes mellitus. Am J Physiol Endocrinol Metab. 2011;300:E669–E680. doi: 10.1152/ajpendo.00600.2010. [DOI] [PubMed] [Google Scholar]
- 2.Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52:102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 3.Remedi MS, Emfinger C. Pancreatic β-cell identity in diabetes. Diabetes Obes Metab. 2016;18 Suppl 1:110–116. doi: 10.1111/dom.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drucker DJ, Habener JF, Holst JJ. Discovery, characterization, and clinical development of the glucagon-like peptides. J Clin Invest. 2017;127:4217–4227. doi: 10.1172/JCI97233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drucker DJ, Nauck MA. The incretin system: glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. Lancet. 2006;368:1696–1705. doi: 10.1016/S0140-6736(06)69705-5. [DOI] [PubMed] [Google Scholar]
- 6.Drucker DJ, Yusta B. Physiology and pharmacology of the enteroendocrine hormone glucagon-like peptide-2. Annu Rev Physiol. 2014;76:561–583. doi: 10.1146/annurev-physiol-021113-170317. [DOI] [PubMed] [Google Scholar]
- 7.Lamont BJ, Li Y, Kwan E, Brown TJ, Gaisano H, Drucker DJ. Pancreatic GLP-1 receptor activation is sufficient for incretin control of glucose metabolism in mice. J Clin Invest. 2012;122:388–402. doi: 10.1172/JCI42497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith EP, An Z, Wagner C, Lewis AG, Cohen EB, Li B, Mahbod P, Sandoval D, Perez-Tilve D, Tamarina N, Philipson LH, Stoffers DA, Seeley RJ, D'Alessio DA. The role of β cell glucagon-like peptide-1 signaling in glucose regulation and response to diabetes drugs. Cell Metab. 2014;19:1050–1057. doi: 10.1016/j.cmet.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z, Wang X, Zhang D, Liu Y, Li L. Geniposide-mediated protection against amyloid deposition and behavioral impairment correlates with downregulation of mTOR signaling and enhanced autophagy in a mouse model of Alzheimer's disease. Aging (Albany NY) 2019;11:536–548. doi: 10.18632/aging.101759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma ZG, Kong CY, Song P, Zhang X, Yuan YP, Tang QZ. Geniposide Protects against Obesity-Related Cardiac Injury through AMPKα- and Sirt1-Dependent Mechanisms. Oxid Med Cell Longev. 2018;2018:6053727. doi: 10.1155/2018/6053727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang YQ, Chang GL, Wang Y, Zhang DY, Cao L, Liu J. Geniposide Prevents Hypoxia/Reoxygenation-Induced Apoptosis in H9c2 Cells: Improvement of Mitochondrial Dysfunction and Activation of GLP-1R and the PI3K/AKT Signaling Pathway. Cell Physiol Biochem. 2016;39:407–421. doi: 10.1159/000445634. [DOI] [PubMed] [Google Scholar]
- 12.Liu C, Hao Y, Yin F, Zhang Y, Liu J. Geniposide protects pancreatic β cells from high glucose-mediated injury by activation of AMP-activated protein kinase. Cell Biol Int. 2017;41:544–554. doi: 10.1002/cbin.10758. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Ding Y, Zhong X, Guo Q, Wang H, Gao J, Bai T, Ren L, Guo Y, Jiao X, Liu Y. Geniposide acutely stimulates insulin secretion in pancreatic β-cells by regulating GLP-1 receptor/cAMP signaling and ion channels. Mol Cell Endocrinol. 2016;430:89–96. doi: 10.1016/j.mce.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 14.Guo LX, Liu JH, Zheng XX, Yin ZY, Kosaraju J, Tam KY. Geniposide improves insulin production and reduces apoptosis in high glucose-induced glucotoxic insulinoma cells. Eur J Pharm Sci. 2017;110:70–76. doi: 10.1016/j.ejps.2017.03.038. [DOI] [PubMed] [Google Scholar]
- 15.St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singer RA, Sussel L. Islet Long Noncoding RNAs: A Playbook for Discovery and Characterization. Diabetes. 2018;67:1461–1470. doi: 10.2337/dbi18-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8 Suppl 4:S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo LL, Song CH, Wang P, Dai LP, Zhang JY, Wang KJ. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol. 2015;21:11680–11687. doi: 10.3748/wjg.v21.i41.11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lovshin JA, Drucker DJ. Incretin-based therapies for type 2 diabetes mellitus. Nat Rev Endocrinol. 2009;5:262–269. doi: 10.1038/nrendo.2009.48. [DOI] [PubMed] [Google Scholar]
- 26.Lovshin JA. Glucagon-like Peptide-1 Receptor Agonists: A Class Update for Treating Type 2 Diabetes. Can J Diabetes. 2017;41:524–535. doi: 10.1016/j.jcjd.2017.08.242. [DOI] [PubMed] [Google Scholar]
- 27.Fu Z, Gilbert ER, Liu D. Regulation of insulin synthesis and secretion and pancreatic Beta-cell dysfunction in diabetes. Curr Diabetes Rev. 2013;9:25–53. [PMC free article] [PubMed] [Google Scholar]
- 28.Chen BZ, Yu SL, Singh S, Kao LP, Tsai ZY, Yang PC, Chen BH, Shoei-Lung Li S. Identification of microRNAs expressed highly in pancreatic islet-like cell clusters differentiated from human embryonic stem cells. Cell Biol Int. 2011;35:29–37. doi: 10.1042/CBI20090081. [DOI] [PubMed] [Google Scholar]
- 29.Dominguez V, Raimondi C, Somanath S, Bugliani M, Loder MK, Edling CE, Divecha N, da Silva-Xavier G, Marselli L, Persaud SJ, Turner MD, Rutter GA, Marchetti P, Falasca M, Maffucci T. Class II phosphoinositide 3-kinase regulates exocytosis of insulin granules in pancreatic beta cells. J Biol Chem. 2011;286:4216–4225. doi: 10.1074/jbc.M110.200295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennet H, Balhuizen A, Medina A, Dekker Nitert M, Ottosson Laakso E, Essén S, Spégel P, Storm P, Krus U, Wierup N, Fex M. Altered serotonin (5-HT) 1D and 2A receptor expression may contribute to defective insulin and glucagon secretion in human type 2 diabetes. Peptides. 2015;71:113–120. doi: 10.1016/j.peptides.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 31.Kooptiwut S, Hanchang W, Semprasert N, Junking M, Limjindaporn T, Yenchitsomanus PT. Testosterone reduces AGTR1 expression to prevent β-cell and islet apoptosis from glucotoxicity. J Endocrinol. 2015;224:215–224. doi: 10.1530/JOE-14-0397. [DOI] [PubMed] [Google Scholar]
- 32.Lee YS, Jun HS. Anti-diabetic actions of glucagon-like peptide-1 on pancreatic beta-cells. Metabolism. 2014;63:9–19. doi: 10.1016/j.metabol.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 33.Gao J, Bai T, Ren L, Ding Y, Zhong X, Wang H, Guo Y, Li J, Liu Y, Zhang Y. The PLC/PKC/Ras/MEK/Kv channel pathway is involved in uncarboxylated osteocalcin-regulated insulin secretion in rats. Peptides. 2016;86:72–79. doi: 10.1016/j.peptides.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 34.Kahn SE, Cooper ME, Del Prato S. Pathophysiology and treatment of type 2 diabetes: perspectives on the past, present, and future. Lancet. 2014;383:1068–1083. doi: 10.1016/S0140-6736(13)62154-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poddar S, Kesharwani D, Datta M. miR-449a regulates insulin signalling by targeting the Notch ligand, Jag1 in skeletal muscle cells. Cell Commun Signal. 2019;17:84. doi: 10.1186/s12964-019-0394-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Machado IF, Teodoro JS, Palmeira CM, Rolo AP. miR-378a: a new emerging microRNA in metabolism. Cell Mol Life Sci. 2020;77:1947–1958. doi: 10.1007/s00018-019-03375-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng LQ, Wei SB, Sun YM, Qin WY, Cheng J, Mitchelson K, Xie L. Systematic profiling of mRNA and miRNA expression in the pancreatic islets of spontaneously diabetic Goto-Kakizaki rats. Mol Med Rep. 2015;11:67–74. doi: 10.3892/mmr.2014.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Q, Xiao X, Li M, Li W, Yu M, Zhang H, Ping F, Wang Z, Zheng J, Xiang H. miR-375 and miR-30d in the effect of chromium-containing Chinese medicine moderating glucose metabolism. J Diabetes Res. 2014;2014:862473. doi: 10.1155/2014/862473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan Z, Guo Y, Wang Y, Li Y, Wang J. MicroRNA profiles of BMSCs induced into osteoblasts with osteoinductive medium. Exp Ther Med. 2018;15:2589–2596. doi: 10.3892/etm.2018.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang P, He J, Wang S, Wang X, Liu Q, Peng W, Qian T. miR-3075 Inhibited the Migration of Schwann Cells by Targeting Cntn2. Neurochem Res. 2018;43:1879–1886. doi: 10.1007/s11064-018-2605-9. [DOI] [PubMed] [Google Scholar]
- 41.Kong X, Liu CX, Wang GD, Yang H, Yao XM, Hua Q, Li XY, Zhang HM, Ma MZ, Su Q, Lv K. LncRNA LEGLTBC Functions as a ceRNA to Antagonize the Effects of miR-34a on the Downregulation of SIRT1 in Glucolipotoxicity-Induced INS-1 Beta Cell Oxidative Stress and Apoptosis. Oxid Med Cell Longev. 2019;2019:4010764. doi: 10.1155/2019/4010764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin F, Wang N, Zhu Y, You L, Wang L, De W, Tang W. Downregulation of Long Noncoding RNA Gas5 Affects Cell Cycle and Insulin Secretion in Mouse Pancreatic β Cells. Cell Physiol Biochem. 2017;43:2062–2073. doi: 10.1159/000484191. [DOI] [PubMed] [Google Scholar]
- 43.Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, Judge AD, Lam K, McClintock K, Nechev LV, Palmer LR, Racie T, Röhl I, Seiffert S, Shanmugam S, Sood V, Soutschek J, Toudjarska I, Wheat AJ, Yaworski E, Zedalis W, Koteliansky V, Manoharan M, Vornlocher HP, MacLachlan I. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 44.Tabernero J, Shapiro GI, LoRusso PM, Cervantes A, Schwartz GK, Weiss GJ, Paz-Ares L, Cho DC, Infante JR, Alsina M, Gounder MM, Falzone R, Harrop J, White AC, Toudjarska I, Bumcrot D, Meyers RE, Hinkle G, Svrzikapa N, Hutabarat RM, Clausen VA, Cehelsky J, Nochur SV, Gamba-Vitalo C, Vaishnaw AK, Sah DW, Gollob JA, Burris HA., 3rd First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov. 2013;3:406–417. doi: 10.1158/2159-8290.CD-12-0429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets (Series GSE138744) generated and analyzed during the current study are available in the (GEO) repository, (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138744). To review the data, please enter token: opslyaomvhmlzkb into the box. Our data in GSE138744 remains in private status now. You can review it through the pathway above, and we will make the data publicly available prior to the publication of this article.