Abstract
Immunosuppression at tumor microenvironment (TME) is one of the major obstacles to be overcome for an effective therapeutic intervention against solid tumors. Tumor-associated macrophages (TAMs) comprise a sub-population that plays multiple pro-tumoral roles in tumor development including general immunosuppression, which can be identified in terms of high expression of mannose receptor (MR or CD206). Immunosuppressive TAMs, like other macrophage sub-populations, display functional plasticity that allows them to be re-programmed to inflammatory macrophages. In order to mitigate immunosuppression at the TME, several efforts are ongoing to effectively re-educate pro-tumoral TAMs. Extracellular vesicles (EVs), released by both normal and tumor cells types, are emerging as key mediators of the cell to cell communication and have been shown to have a role in the modulation of immune responses in the TME. Recent studies demonstrated the enrichment of high mannose glycans on the surface of small EVs (sEVs), a subtype of EVs of endosomal origin of 30–150 nm in diameter. This characteristic renders sEVs an ideal tool for the delivery of therapeutic molecules into MR/CD206-expressing TAMs. In this review, we report the most recent literature data highlighting the critical role of TAMs in tumor development, as well as the experimental evidences that has emerged from the biochemical characterization of sEV membranes. In addition, we propose an original way to target immunosuppressive TAMs at the TME by endogenously engineered sEVs for a new therapeutic approach against solid tumors.
Keywords: tumor-associated macrophages, tumor microenvironment, macrophage polarization, mannose receptor, exosomes, extracellular vesicles, HIV-1 Nef
1. Introduction
Both immunosuppression and genetic escape are formidable weapons through which tumors can elude host immune surveillance. Solid tumors develop in a quite complex context, referred to as tumor microenvironment (TME) [1,2], which is composed of both cellular and non-cellular elements, usually resulting in an immunosuppressive behavior. Counteracting such a general effect would favor both spontaneous and therapeutic anti-tumor immunity, hence critically contributing to control tumor cell growth. Therefore, subverting TME immunosuppression represents a major goal for anticancer immunotherapies.
Both normal and tumor cells constitutively release membrane-bilayered vesicles, commonly referred to as extracellular vesicles (EVs) [3,4]. They differ in the mechanisms of biogenesis and secretion, giving rise to the generation of a heterogeneous population of vesicles with different sizes and contents [5,6], which include small EVs (sEVs) or exosomes and microvesicles or ectosomes. Exosomes are vesicles of 30–150 nm diameter generated by inward budding of endosomal membranes to form intraluminal vesicles that accumulate in intracellular organelles called multivesicular bodies (MVBs). MVBs ultimately fuse with the plasma membrane, thereby releasing intraluminal vesicles into the extracellular environment (Figure 1). On the contrary, ectosomes are 100–500 nm vesicles shed by direct budding from the plasma membrane [7,8]. Different types of EVs often show overlapping features that make difficult to obtain relatively pure preparations when purified from cell-conditioned media or biological fluids. In this review, we will use the term sEV to refer to EV types co-isolated by typical purification methods and exosomes to distinguish EV whose subcellular biogenesis strictly derives from multivesicular bodies/endosomes [8].
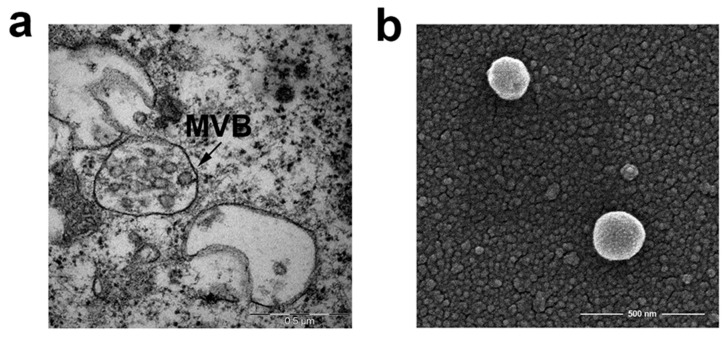
Figure 1.
Electron microscopy of multivesicular bodies (MVB) and small extracellular vesicles (sEVs) (a) TEM micrograph of multivesicular bodies with intraluminal vesicles in Mel501, a melanoma cell line (b) SEM (Scanning Electron Microscope). Micrograph of sEVs purified from conditioned medium of Mel501 cells by differential centrifugations. Courtesy of Francesca Iosi and the Microscopy Area of the ISS Core Facilities.
SEVs carry a complex cargo of nucleic acids, proteins, and lipids that largely reflects the characteristics and the functional state of the cells they originate from, and that will be delivered to neighboring or distant cells [9,10]. As a result, the functions of those recipient cells will be modulated by sEVs in a manner that is strictly dependent on the nature of producer cells, making sEVs central players in intercellular communication and reprogramming of target cells [11]. Ectosomes generation is a much less known process that requires the accumulation of their cargo at the cytosolic surface of specific plasma membrane microdomains [7,12].
SEVs-mediated transfer of molecular and genetic material from one cell to another, either locally or at long distance, is a key contributor to the mechanisms of intercellular communication involved in various physiological and pathological conditions [13,14,15]. Moreover, for these reasons, sEVs are now considered powerful tools for clinical applications, including advanced diagnostics, therapeutics, and regenerative medicine [16,17,18,19].
The molecular composition of sEVs is determined by the cell type of origin as well as by the intracellular pathway followed en route to their release into the extracellular space [8,20,21]. This heterogeneity confers to sEVs distinct properties, such as tropism to certain organs, and uptake by specific cell types. In the case of tumor-derived sEVs, these events often lead to the impairment of immune responses at TME [22], also favoring pre-metastatic niche formation and metastasis [23,24].
In tumor cells, sEV biogenesis and ultimately sEV composition is a complex and regulated process, which involves many different molecules associated with the sEV biogenesis pathway [4,25]. Whatever the cell type of origin, sEVs can be characterized, although not exclusively, by the presence of different types of cell surface proteins, such as tetraspanins, (i.e., CD9, CD81, CD63), ESCRT (endosomal sorting complex required for transport) proteins (Alix and TSG101), integrins, RNA, DNA, lipids such as ceramide and the atypical phospholipid, lysobisphosphatidic acid (LBPA) [26], and oligosaccharides [27].
TME exerts a key influence on tumor cells, and the resulting sEVs, responsible for proteins and genetic material transfer from primary tumor cells, play a crucial role in metastatic colonization and in the formation of the pre-metastatic niche, driving recipient cells to acquire a pro-tumorigenic phenotype [23,28]. The selective conditions present in TME, such as the generation of a hypoxic [29] and acidic environment [30], strongly influence sEV secretion by tumor cells, thus contributing to the malignant tumor phenotype. Furthermore, sEV membrane composition reflects TME changes and conceivably influence and control the different mechanisms of entry or interaction of sEVs with target cells supporting tumor growth [31,32].
Interestingly, it has been described that major players of immunosuppression at the TME, i.e., immunosuppressive tumor-associated macrophages (TAMs), express on their surface high levels of mannose receptor (MR, CD206) [33]. The MR is an endocytic receptor with a high affinity for high mannose oligosaccharides, glycans highly enriched on the surface of sEVs [34]. In this review, literature data regarding both TAM functions and the molecular structure of sEVs are reviewed. In addition, we propose an original way to exploit typical molecular signatures of both TAMs and sEVs to counteract the immunosuppression at the TME.
2. The Tumor Microenvironment
In solid tumors, cancer cells are embedded within a milieu that favors their proliferation and comprises both cellular and non-cellular components. Fibroblasts, endothelial cells, and essentially all types of immune cells are part of the TME [35,36]. Among non-cellular components, tumor-derived sEVs play a key role in immune suppression. TME composition can vary among different tumors, and between primary and metastatic neo-formations in the same patient, and is tightly associated with the clinical outcome of cancer patients.
TMEs can be categorized based on different criteria. In terms of abundance of tumor-infiltrating cytotoxic CD8+ T lymphocytes (CTLs), TMEs can be distinguished in either hot/inflamed, with the highest content of CTLs, or cold/desert, with a virtual absence of infiltrated CTLs [37]. TME core infiltrated by CTLs represents a favorable condition for an effective anticancer immune response, both spontaneous and induced by immunotherapeutic interventions.
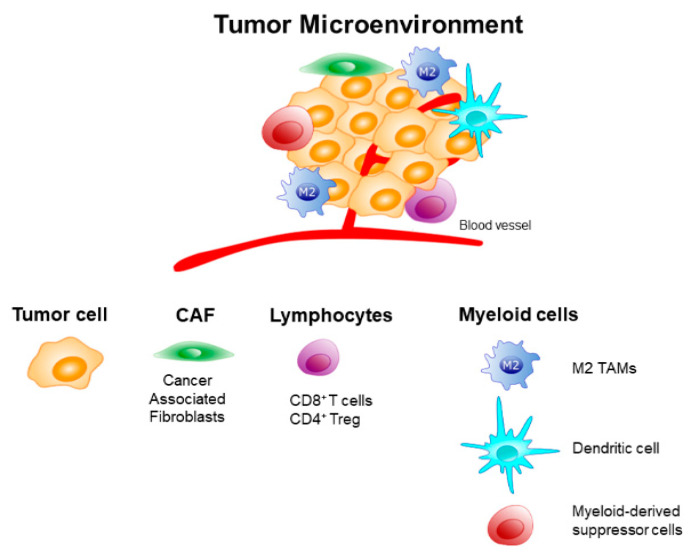
TME is populated by different kinds of immune cells having immune suppressive actions. Among these are myeloid-derived suppressor cells, neutrophils, CD4+ Treg lymphocytes, and immunosuppressive M2-like TAMs [38]. These latter cells can represent up to 50% of the tumor mass, and play a key role in the immune evasion at TME by secreting proteases, angiogenic factors, and pro-tumoral products. The functional plasticity of TAMs modifies their phenotype and activity in response to a great number of microenvironmental stimuli, although the mechanisms that determine the different polarization states are still to be elucidated [39]. These different functional states often coexist and can significantly vary between different tumors [40,41]. TAMs can also dispose at the tumor margin, where they can interact with CTLs, thus inhibiting their infiltration towards tumor cells [42]. For all these reasons, immunosuppressive TAMs have been identified as a major cell target for novel designs of cancer immunotherapies focused on improving the overall anti-tumor immune response. A schematic representation of cells populating TEM is illustrated in Figure 2.
Figure 2.
Schematic representation of cells populating the tumor microenvironment (TME).
3. The TAM-Mediated Immunosuppression at the TME
A large number of macrophages infiltrate solid tumors, thereby influencing several aspects of tumor development [43]. The most relevant effects include suppression of anticancer immunity, angiogenesis promotion, and support for metastasis. Macrophages are recruited at TME, in response to the secretion by tumor cells and other TME cell types, of a number of chemoattractant soluble factors, including vascular-endothelial growth factor A (VEGF-A) [44], chemokine ligand 2 (CCL2) [45], and colony-stimulating factor 1 (CSF-1) [46].
TME-populating macrophages can be schematically distinguished in M1- and M2-like macrophages. M1-like macrophages show both pro-inflammatory and immune-stimulatory properties, thus exerting an anti-tumor function. On the other hand, M2-like macrophages favor tumor angiogenesis and immunosuppression. Such a distinction, although useful from both therapeutic and diagnostic points of view, is now outdated, due to the identification of a large number of intermediate subclasses, i.e., up to 19 [47]. They have been identified through most recent transcriptomic techniques, e.g., single-cell mass cytometry and single-cell RNA sequencing [48,49,50], and in vivo represent a continuum of functional phenotypes with intermediates showing overlapping features.
TAMs can be characterized by the expression of different surface markers [47,51], distinct metabolic changes [52,53], and a broad transcriptional repertoire with the involvement of key transcription factors, which can be activated by the environmental signals received. In particular, members of the signal transducer and activator of transcription (STAT), peroxisome proliferator-activated receptors (PPARs), interferon regulatory factor (IRF), and nuclear transcription factor-κB (NF-κB) families are essential for macrophage polarization toward the M1 profile [41,54,55].
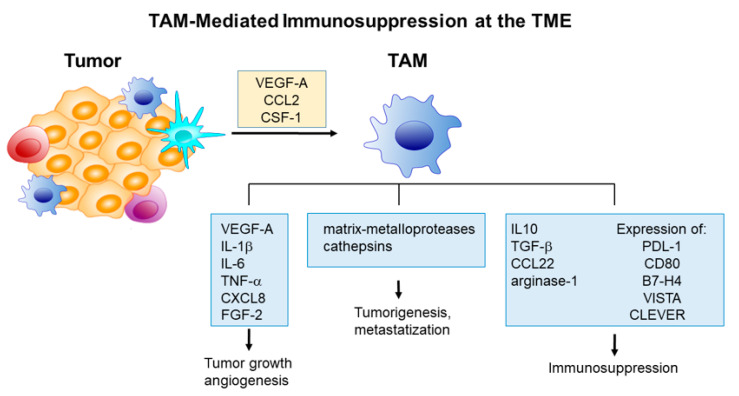
M2-like TAMs contribute to tumor angiogenesis by secreting soluble factors inducing endothelial cell proliferation, including VEGF-A, interleukin (IL)-1β, IL-6, tumor necrosis factor (TNF)α, CXCL8, and fibroblast growth factor (FGF)-2 [56]. In particular, the secretion of VEGF-A by perivascular TAMs can increase vascular permeability and access of tumor cells to peripheral blood circulation [57]. On the other hand, the production of proteases, e.g., matrix-metalloproteases, induces degradation of extracellular matrix and the consequent liberation of embedded soluble factors released by both cancer and stromal cells having pro-tumoral effects and favoring metastasization.
TAM-mediated immunosuppression at TME is essentially mediated by three concurrent mechanisms: (i) Release of soluble immunosuppressive factors, e.g., IL-10, CCL22, and transforming growth factor (TGF)-α as well as factors recruiting regulatory T cells (Treg) [58]; (ii) expression of ligands for lymphocyte suppressor factors PD-1 and CTLA-4, i.e., PDL-1 and CD80, as well as other checkpoint inhibitors with similar functions, including B7-H4, V-domain Ig suppressor of T cell activation (VISTA) [59], and vascular endothelial receptor (CLEVER) [60], and (iii) starving the TME of l-arginine, i.e., an essential factor for T-cell activity, through the release of arginase-1 [61]. Figure 3 illustrates the principal mechanisms of TAM mediated immunosuppression at the TME.
Figure 3.
Principal mechanisms of tumor-associated macrophage (TAM) mediated immunosuppression at the tumor microenvironment (TME).
The multiple immunosuppressive signals at play within the TME greatly reduce the efficacy of current immunotherapies. Therefore, new strategies to effectively reprogram the various immunosuppressive cell types at the TME are urgently needed.
4. Re-Programming of TAMs
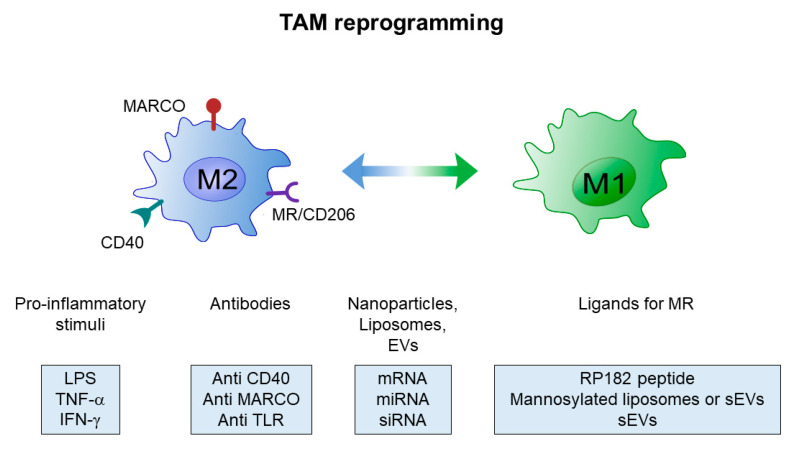
Immunosuppressive TAMs represent a privileged therapeutic target for the treatment of solid tumors, especially in the case immune checkpoint blockers (ICBs) are used. Given the enormous therapeutic value of TAMs re-education towards M1-like macrophages to promote tumor regression, much attention has focused on effective strategies aimed at targeting TAMs, including the blockade of the M2 phenotype, enhanced activation of M1 macrophages and reprogramming of TAMs toward M1-like phenotype [58,62,63,64,65]. Many of these different approaches against immunosuppressive TAMs have been summarized in Table 1 and strategies directed at TAM reprogramming illustrated in Figure 4.
Table 1.
Selected strategies to target tumor-associated macrophages (TAMs).
| Mechanism of Action | Active Agent | Vehicle Carrier | Target | References |
|---|---|---|---|---|
| Depletion of M2 TAMs | Shiga toxins | Shigella Flexneri attenuated strain | TAMs | [66] |
| Immunotoxins | TAMs Receptors | [67,68] | ||
| Bisphosphonates (e.g., clodrolip, zoledronic acid) | Liposomes | TAMs, Kupffer cells | [69,70] | |
| Trabectedin | TAMs | [71] | ||
| Tyrosine Kinase Inhibitors (e.g., Dasatinib, Bosutinib) | endothelial and myeloid cells in TEM, TAMs | [72,73] | ||
| Inhibition of circulating monocyte recruitment into tumor | CCR2 inhibitors; anti-CCR2/CCL2 blocking antibodies | TAMs CCR2 | [45,74,75,76] | |
| Antagonists of CXCL12/CXCR4 axis | TAMs CXCR4 | [77,78] | ||
| anti-CSF-1R antibody | TAMs CSF-1R | [79,80] | ||
| neutralizing CD11b antibody | CD11b on Myeloid Cells | [81,82] | ||
| Blockade of M2 Phenotype | Tyrosine kinase inhibitors or drugs blocking STAT3 | TAMs STAT3 | [83,84] | |
| drugs blocking STAT6 | TAMs STAT6 | [85] | ||
| Enhanced Activation of M1 Macrophages | Th1 cytokines like IFN-γ | TAMs STAT1 stimulation | [86,87] | |
| metformin | TAMs AMPKα1 stimulation | [88] | ||
| toll-like receptor agonists, CpG-ODNs; PI3Kγ deletion | TAMs NF-κB stimulation | [89,90,91] | ||
| Reprogramming TAMs Toward M1-Like Phenotype | mRNAs; miRNA | Targeted Nanocarriers | TAMs | [92,93] |
| siRNA | Different types of Nanoparticles | TAMs | [94,95,96] | |
| anti-CD40 antibody | TAMs CD40 | [97,98,99] | ||
| anti-MARCO antibody | TAMs MARCO | [100] | ||
| gefitinib/vorinostat | Trastuzumab-modified Mannosylated Liposomes | TAMs MR | [101] | |
| Drug free | Mannosylated Liposomes | TAMs MR | [102] | |
| RP-182 Peptide | TAMs MR | [103] |
AMPKα1, AMP-activated protein kinase; CCL2, C–C chemokine ligand 2; CCR2, C–C chemokine receptor type 2; CSF-1, Colony-Stimulating Factor 1; CSF-1R, colony-stimulating factor 1 receptor; CXCL12, C–X–C motif chemokine 12; CXCR4, C-X-C chemokine receptor type 4; CpG-ODN, unmethylated cytosineguanine (CpG) oligodeoxynucleotides; IFN-γ, interferon gamma; MARCO, macrophage receptor with collagenous structure; MR, mannose receptor/CD206; NF-κB, nuclear factor kappa B; PI3Kγ, phosphoinositide 3-kinase; STAT, signal transducer and activator of transcription.
Figure 4.
Scheme of current strategies for tumor-associated macrophage (TAM) reprogramming.
Promising results have been obtained with direct activation of M1-like macrophages by Th1 cytokines like IFN-γ [87], and by targeting toll-like receptors (TLR) and/or CD40 with agonists and monoclonal antibodies [97,98,99]. However, the onset of systemic inflammation limited the therapeutic efficacy of these approaches in vivo, and additional investigations are ongoing to circumvent this hurdle. In any case, considering the functional plasticity of macrophages, re-educating M2- versus an M1-like macrophage phenotype currently appears the most attractive therapeutic option.
A major hindrance to effective targeting of immunosuppressive TAMs is represented by the scarcity of specific protein markers expressed on M2 macrophages. Some potential targets, whose expression also correlates with poor prognosis, have been investigated. Among them, the selective targeting of MARCO (macrophage receptor with collagenous structure) with monoclonal antibodies has been recently used to promote a switch to an M1-activated phenotype [100].
Increasing evidence suggests that a valuable alternative is represented by targeting the MR, which is highly expressed on M2, but not M1 macrophages [92,103,104,105].
5. The Mannose Receptor in M1 Polarization
MR is expressed on TAMs where is a prototypical marker of M2-type activation. It is also expressed on the surface of immature dendritic cells (DCs), liver sinusoidal endothelial cells, and other tissue macrophages. Earlier studies have demonstrated that MR expression is strongly down-regulated by IFN-γ [106], and upregulated by interleukin-4 (IL-4) [107]. The MR is a 175 kDa Type I integral membrane protein that belongs to the family of C-type lectin receptors and binds glycoconjugates terminated in mannose, fucose, or N-acetil-β-d-glucosamine (GlcNAc) in a calcium-dependent manner [108,109,110]. The receptor contains three distinct extracellular domains, i.e., an N-terminal cysteine-rich domain (CR) that binds sulfated carbohydrates, a fibronectin type II domain (FNII) that binds collagen, and eight tandem C-type lectin carbohydrate-recognition domains (CRDs) [111,112]. CRDs have only weak affinity affinities for single sugars, and several CRDs need to be clustered to achieve high-affinity binding to oligosaccharides. This clustering allows for the internalization of mannosylated proteins and other exogenous molecules, including allergens and microbial products.
MR is a highly effective clathrin-dependent endocytic receptor that constantly recycles between the plasma membrane and the early endosomal compartment [111]. Most part of MRs is intracellular, while only ∼15% of the cellular pool can be found on the cell surface. Like other members of the C-type lectin receptor family, the MR undergoes conformational changes upon ligand binding or as pH decreases in intracellular compartments [111,113]. Once acidification takes place in the endosomal compartment, the MR dissociates from its ligands, and the empty receptor recycles back to the plasma membrane.
Several approaches have been adopted to target the MR and selectively deliver therapeutic nanoparticles. Among these, drug-free mannosylated liposomes have been shown to induce effective anti-tumor activity by enhancing the expression ratio of CD86/MR [102]. In another study, mannosylated nanoparticles suitable for intracellular delivery of drug carriers have been shown to selectively target with high specificity MR expressing macrophages [104].
MR conformational changes that occur upon ligand binding have been recently exploited to target M2 macrophages and induce reprogramming toward M1 phenotypes. For instance, precision targeting with short peptides showed some potential for the intracellular delivery of therapeutically relevant molecules [114]. In addition, a very recent report showed that direct binding of MR with a synthetic peptide (RP-182), i.e., an analogue of naturally occurring antimicrobial peptides, activates phagocytosis and autophagy in M2-like macrophages, reverting these cells into an anti-tumor M1-like phenotype with increased M1 cytokine production and phagocytosis of cancer cells [103].
On the other hand, also cell-secreted sEVs can be considered attractive candidates to specifically target M2-like macrophages via the MR, since they expose high mannose and other classes of N-linked oligosaccharides on their surface [115,116].
6. Extracellular Vesicles for Anti-Tumor Therapy
Pioneering studies have shown that sEVs secreted by DCs pulsed with cancer peptides successfully eradicate established tumors in mice [117]. Furthermore, tumor-derived EVs are a source of neoantigens that, once internalized by DCs, could cross-prime CD8+ T cells and lead to tumor rejection [118]. Since these early studies, the field of sEVs-based cancer therapeutics has attracted many efforts, and sEVs have emerged as promising tools for targeted drug delivery. Despite a growing interest in these nanovesicles as natural carriers, there are still many open questions that need further investigation. For example, specific recognition by target cells is of fundamental importance for an effective delivery of bioactive molecules. EVs uptake may occur via receptor-mediated endocytosis or phagocytosis, or direct fusion with the plasma membrane. Some studies have pointed at integrins [24] and scavenging receptors [119] as mediators of EVs targeting, but current knowledge on this matter is rapidly evolving and has been recently comprehensively reviewed [5,7,120,121,122].
The different modes of sEV uptake may result in distinct localization and functional effects of the sEVs components, but it is still unknown whether a specific route of entry is to be preferred for a successful transfer of EVs cargo. Thus, understanding through which mechanisms sEVs deliver their content into target cells is a central point that needs to be further elucidated. To the best of current knowledge, while either non-selective uptake or direct fusion with the plasma membrane of target cells seem to be the preferential mechanisms of bulk sEV incorporation (Figure 5) [123], alternative and more specific routes of uptake may depend on the characteristics of surface components of both sEV and target cells.
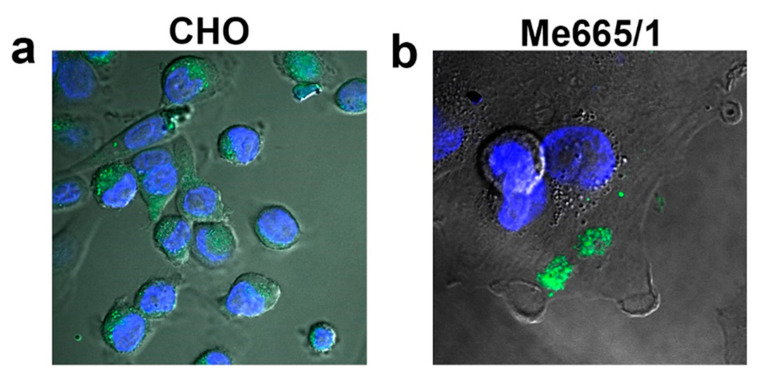
Figure 5.
Fluorescent sEVs uptake by different cell lines. Confocal fluorescence microscopy images of green fluorescent sEVs derived from melanoma Me665/1 cells transferred on (a) CHO cells and (b) Me665/1 cells in nonspecific conditions [66]. Blue-fluorescent nuclei are stained with DAPI.
Tumor-derived EVs can be efficiently taken up by DCs for antigen processing and cross-presentation to tumor-specific CTLs [124]. Immature DCs (iDCs) internalize EVs more efficiently than mature DCs, whereas mature DCs retain more EVs on the cell surface [125]. The surface of iDCs harbors sugar-binding C-type lectin receptors (CLRs) [126,127], which is a characteristic shared with M2-like macrophages.
Glycomic studies conducted to date demonstrate that surface glycoprofiles of sEVs contain high amounts of mannose and other classes of N-linked oligosaccharides [27,34,128,129] making them suitable ligands for the MR. Recently, it has also been shown that mannose-modified serum sEVs display elevated uptake by murine DCs [116]. This evidence, together with a number of studies showing that mannosylation of both liposomes [102] and synthetic nanocarriers [130,131] enhance cellular uptake by M2 macrophages, inducing stimulation and polarization of macrophages toward the M1 phenotype, point to sEVs as powerful ligands of M2 macrophages. Furthermore, the affinity of EVs membrane components (i.e., proteins, lipids, and glycans) for certain tissues greatly affects biodistribution of EVs in vivo, thus, encouraging studies aimed at altering the surface of EVs to improve targeting to selected organs. Many different strategies have been adopted, but there is now accumulating evidence that carbohydrates on the vesicle surface participate in the recognition and uptake of EVs by phagocytes. Interestingly, manipulation of surface glycans on EVs either by removal of sialic acid [132] or by treatment with N-glycosidases [133,134] alters the uptake capacity of different cells. In the first case, the change also affects the in vivo biodistribution of sEVs showing accumulation of desialylated sEVs in the lungs. Altogether, these results point to the importance of N-glycosylation in cellular uptake, but since different cell types respond differently to glycosylation changes appears evident that the cell to be targeted, with its endowment of specific protein receptors, represent the cornerstone of receptor ligand recognition.
7. Molecular Basis of TAM Re-Programming by Engineered sEVs
The subversion of TAM-mediated immunosuppression at TME through macrophage transcriptional reprogramming represents a quite attractive option for anticancer combined immunotherapy. Like normal cells, tumor cells also constitutively release sEVs, which interact primarily with TME cell constituents. SEVs, as naturally occurring vesicles, have a low intrinsic immunogenic profile, are able to avoid, at least in part, the degradative pathway and possess the ability to overcome the blood-brain barrier. Consequently, they have emerged as an important means to deliver therapeutic agents. In this context, engineering tumor-derived sEVs with molecules inducing an inflammatory-like macrophage phenotype would be instrumental in alleviating the immunosuppression at TME. Over the past few years, several engineering strategies have been devised to manipulate tumor-derived sEVs in order to induce cellular and innate immunity. SEV engineering can be carried out either at the level of producer cells or directly on purified sEVs. The different approaches used mostly depend on cargo properties, such as hydrophilicity, hydrophobicity, and molecular weight, as each method has a different loading capacity. For example, chemicophysical methods like electroporation are widely used for loading relatively large molecules. such as siRNA or miRNA into sEVs [19]. Particularly for cancer research, the observation that exosomal miRNAs effectively engage target mRNA and suppress gene expression in recipient cells has been heavily favored. Due to the availability of various cellular engineering methods, different types of RNAs that are released via sEVs have also been exploited and recently reviewed [11].
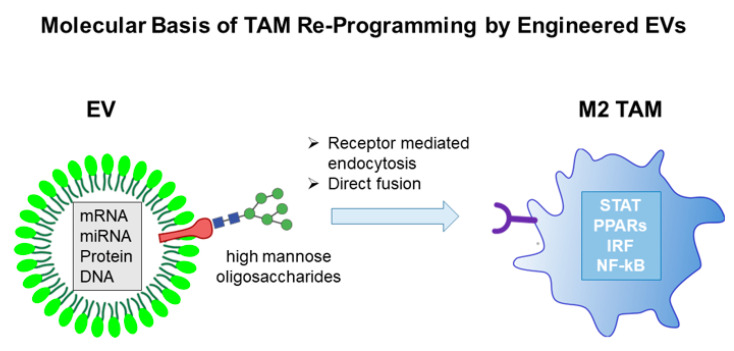
During the past decade, many efforts have been devoted to transfect sEV-producing cells with plasmids encoding protein sequences that, once uploaded in the nanovesicles, are able to alter the phenotype of target cells [135,136,137,138]. However, a method to target M2 macrophages with sEVs capable of inducing their reprogramming to M1 anti-tumor phenotype is not yet available. One major hurdle in EVs research for an effective therapeutic application is represented by the lack of optimized isolation, characterization and quantification procedures. Various purification techniques, such as differential ultracentrifugation, density gradients, precipitation, filtration, size exclusion chromatography, and immunoisolation are currently used to obtain less heterogeneous sEVs preparation; however, quantification often relies on the total protein content of EVs. Fluorescent labeling techniques are more accurate and have the main advantage that track EVs in vivo [139,140]. The different approaches for TAMs reprogramming by engineered EVs are schematically illustrated in Figure 6.
Figure 6.
TAMs reprogramming by engineered EVs.
8. HIV-1 Nef Protein as Effector of TAM Reprogramming
A plausible candidate for reprogramming M2 macrophages is the Human immunodeficiency virus (HIV)-1 Negative Regulatory Factor (Nef) protein [141], a 27 kilodalton (kDa) scaffold protein, which lacks enzymatic activities. After synthesis at free ribosomes, Nef reaches both intracellular and plasma membranes with which it tightly interacts through its N-terminal myristoylation. Nef acts as a scaffold/adaptor element in triggering activation of signal transducing molecules like p21 PAK-2, NF-κB, STATs, ERK1/2, Vav, and Src family kinases. In most cases, signal activation occurs upon Nef association with lipid raft microdomains at cell membranes [142,143,144]. The fact that also sEV membranes are enriched in lipid raft microdomains explains why Nef can be found in EVs [145,146,147,148,149].
Cumulate literature data demonstrate that the presence of Nef inside macrophages induces a strong pro-inflammatory response. In particular, Nef switches on the transcription of many inflammatory genes, as well as the release of inflammatory factors like CCL3, CCL4, IL-1β, IL-6, TNF-α [150,151], and interferon gamma (IFN)-γ [152]. This potent pro-inflammatory response is mediated by the activation of several signal transduction molecules, including STAT-1, 2, and 3, NF-κB, JNK, ERK1/2, and MAPK [153,154]. The inflammatory effects of Nef on macrophages depends on four glutamate-acidic cluster domain located at 62–65 amino acid position [155].
Data from many independent investigation groups strongly support the idea that Nef associates with EVs at low levels [156,157]. Conversely, we identified a Nef mutant incorporating in sEVs/EVs at quite high levels [158]. This Nef mutant (referred to as Nefmut) is defective for the most part of Nef functions, including down-regulation of cell membrane receptors, Nef-associated kinase (NAK) activation, an increase of HIV expression [159]. Nevertheless, it maintains an unaltered acidic cluster domain that correlates with the induction of cell activation in antigen-presenting cells when it is delivered by nanovesicles [160].
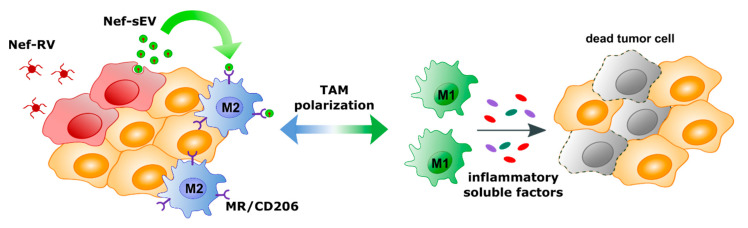
Considering this evidence, one may hypothesize that the delivery of Nefmut-engineered EVs inside M2-TAMs would be instrumental in re-educating macrophages at TME from an M2-like to an M1-like phenotype. In the case of solid tumors, this design could be applied through a quite simple strategy, i.e., tumor cell engineering for Nefmut expression through retroviral vector-mediated transduction. In this way, only actively replicating cells at the TME, hence preferably tumor cells, are expected to be transduced. Nef-engineered tumor-derived sEVs may diffuse within TME, thereby preferentially entering MR positive TAMs in view of the mannose expressed on the sEV surface. Once internalized by macrophages, Nefmut might switch on intracellular signals—ultimately leading to the release of pro-inflammatory factors. These factors might act in an autocrine/paracrine loop to induce the reprogramming of macrophage transcriptional profile toward the M1-like phenotype (Figure 7).
Figure 7.
Scheme of the proposed mechanism for TAM reprogramming. Tumor cells are transfected with retroviral vectors expressing Nefmut (Nef-RV). Nef-engineered sEVs (Nef-sEV) are then released into the TME infiltrated with M2 like macrophages expressing the mannose receptor (MR/CD206). MR mediated uptake of Nef-Sev might induce polarization of M2 into M1 like macrophages—ultimately leading to the release of pro-inflammatory factors.
Eventually, this mechanism, which essentially hijacks the sEV-mediated intercellular communication at the TME, is expected to alleviate the immune suppression at the TME, thereby favoring the action of anticancer adaptive immune responses.
Hopefully, once supported by experimental confirmation, this design would have a therapeutic utility in the battle against solid tumors.
9. Conclusions
Immunosuppression at TME protects cancer cells from both spontaneous and artificially generated host immune responses. Hence, subverting immunosuppression should be considered a priority for any anticancer immunotherapeutic strategy. Even if M2-like TAMs are major players, other cell types contribute to the general immunosuppression at TME, including CD4+ Tregs lymphocytes, myeloid-derived suppressor cells, and neutrophils [42]. Similar to macrophages, neutrophils can polarize in pro- and anti-tumor phenotypes, depending on the stimuli they receive at TME. Interestingly, it has been reported that IFN-γ can polarize neutrophils towards an anti-tumor phenotype [161]. Considering the quite high levels of IFN-γ transcripts induced by Nef in macrophages, the delivery of Nefmut-engineered, tumor-derived sEVs to M2-like TAMs is expected to have paracrine anti-tumor effects also on neutrophils that populate the TME.
The strategy we propose is certainly only one of many potential new anti-tumor therapeutic approaches that the manipulation of sEVs/EVs can offer. The increase in knowledge of the sEV/EVs biology, mainly regarding the mechanisms of cell entry, will favor the implementation of new and more efficient therapeutic approaches against tumors and infectious diseases.
Funding
This work was supported by the grant PGR00810 from Ministero degli Affari Esteri e della Cooperazione Internazionale, Italy.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- 1.Fridman W.H., Pages F., Sautes-Fridman C., Galon J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 2.Quail D.F., Joyce J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Niel G., D’Angelo G., Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018;19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 4.Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raposo G., Stahl P.D. Extracellular vesicles: A new communication paradigm? Nat. Rev. Mol. Cell Biol. 2019;20:509–510. doi: 10.1038/s41580-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 6.Colombo M., Raposo G., Théry C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cell Dev. Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 7.Meldolesi J. Exosomes and Ectosomes in Intercellular Communication. Curr. Biol. CB. 2018;28:R435–R444. doi: 10.1016/j.cub.2018.01.059. [DOI] [PubMed] [Google Scholar]
- 8.Thery C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeppesen D.K., Fenix A.M., Franklin J.L., Higginbotham J.N., Zhang Q., Zimmerman L.J., Liebler D.C., Ping J., Liu Q., Evans R., et al. Reassessment of Exosome Composition. Cell. 2019;177:428–445. doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kowal J., Arras G., Colombo M., Jouve M., Morath J.P., Primdal-Bengtson B., Dingli F., Loew D., Tkach M., Théry C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Brien K., Breyne K., Ughetto S., Laurent L.C., Breakefield X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020 doi: 10.1038/s41580-020-0251-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cocucci E., Meldolesi J. Ectosomes and exosomes: Shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25:364–372. doi: 10.1016/j.tcb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Mathivanan S., Ji H., Simpson R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Maia J., Caja S., Strano Moraes M.C., Couto N., Costa-Silva B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front. Cell Dev. Biol. 2018;6:18. doi: 10.3389/fcell.2018.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andaloussi S.E.L., Mager I., Breakefield X.O., Wood M.J. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 16.Tkach M., Thery C. Communication by Extracellular Vesicles: Where We are and Where We Need to Go. Cell. 2016;164:1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 17.Xu R., Greening D.W., Zhu H.J., Takahashi N., Simpson R.J. Extracellular vesicle isolation and characterization: Toward clinical application. J. Clin. Investig. 2016;126:1152–1162. doi: 10.1172/JCI81129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ratajczak M.Z., Kucia M., Jadczyk T., Greco N.J., Wojakowski W., Tendera M., Ratajczak J. Pivotal role of paracrine effects in stem cell therapies in regenerative medicine: Can we translate stem cell-secreted paracrine factors and microvesicles into better therapeutic strategies? Leukemia. 2012;26:1166–1173. doi: 10.1038/leu.2011.389. [DOI] [PubMed] [Google Scholar]
- 19.Luan X., Sansanaphongpricha K., Myers I., Chen H., Yuan H., Sun D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol. Sin. 2017;38:754–763. doi: 10.1038/aps.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nogues L., Benito-Martin A., Hergueta-Redondo M., Peinado H. The influence of tumour-derived extracellular vesicles on local and distal metastatic dissemination. Mol. Asp. Med. 2017 doi: 10.1016/j.mam.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maas S.L.N., Breakefield X.O., Weaver A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017;27:172–188. doi: 10.1016/j.tcb.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clayton A., Mitchell J.P., Court J., Mason M.D., Tabi Z. Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer Res. 2007;67:7458–7466. doi: 10.1158/0008-5472.CAN-06-3456. [DOI] [PubMed] [Google Scholar]
- 23.Peinado H., Aleckovic M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., Hergueta-Redondo M., Williams C., Garcia-Santos G., Ghajar C., et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoshino A., Costa-Silva B., Shen T.L., Rodrigues G., Hashimoto A., Tesic Mark M., Molina H., Kohsaka S., Di Giannatale A., Ceder S., et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015 doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hessvik N.P., Llorente A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2017 doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruenberg J. Life in the lumen: The multivesicular endosome. Traffic. 2020;21:76–93. doi: 10.1111/tra.12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams C., Royo F., Aizpurua-Olaizola O., Pazos R., Boons G.J., Reichardt N.C., Falcon-Perez J.M. Glycosylation of extracellular vesicles: Current knowledge, tools and clinical perspectives. J. Extracell Vesicles. 2018;7:1442985. doi: 10.1080/20013078.2018.1442985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costa-Silva B., Aiello N.M., Ocean A.J., Singh S., Zhang H., Thakur B.K., Becker A., Hoshino A., Mark M.T., Molina H., et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laoui D., Van Overmeire E., Di Conza G., Aldeni C., Keirsse J., Morias Y., Movahedi K., Houbracken I., Schouppe E., Elkrim Y., et al. Tumor hypoxia does not drive differentiation of tumor-associated macrophages but rather fine-tunes the M2-like macrophage population. Cancer Res. 2014;74:24–30. doi: 10.1158/0008-5472.CAN-13-1196. [DOI] [PubMed] [Google Scholar]
- 30.Colegio O.R., Chu N.Q., Szabo A.L., Chu T., Rhebergen A.M., Jairam V., Cyrus N., Brokowski C.E., Eisenbarth S.C., Phillips G.M., et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513:559–563. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whiteside T.L. Tumor-Derived Exosomes and Their Role in Cancer Progression. Adv. Clin. Chem. 2016;74:103–141. doi: 10.1016/bs.acc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han L., Lam E.W., Sun Y. Extracellular vesicles in the tumor microenvironment: Old stories, but new tales. Mol. Cancer. 2019;18:59. doi: 10.1186/s12943-019-0980-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murray P.J., Allen J.E., Biswas S.K., Fisher E.A., Gilroy D.W., Goerdt S., Gordon S., Hamilton J.A., Ivashkiv L.B., Lawrence T., et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerlach J.Q., Griffin M.D. Getting to know the extracellular vesicle glycome. Mol. Biosyst. 2016;12:1071–1081. doi: 10.1039/C5MB00835B. [DOI] [PubMed] [Google Scholar]
- 35.Nistico P., Ciliberto G. Biological mechanisms linked to inflammation in cancer: Discovery of tumor microenvironment-related biomarkers and their clinical application in solid tumors. Int. J. Biol. Markers. 2020;35:8–11. doi: 10.1177/1724600820906155. [DOI] [PubMed] [Google Scholar]
- 36.Hinshaw D.C., Shevde L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019;79:4557–4566. doi: 10.1158/0008-5472.CAN-18-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Binnewies M., Roberts E.W., Kersten K., Chan V., Fearon D.F., Merad M., Coussens L.M., Gabrilovich D.I., Ostrand-Rosenberg S., Hedrick C.C., et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beatty G.L., Winograd R., Evans R.A., Long K.B., Luque S.L., Lee J.W., Clendenin C., Gladney W.L., Knoblock D.M., Guirnalda P.D., et al. Exclusion of T Cells From Pancreatic Carcinomas in Mice Is Regulated by Ly6Clow F4/80+ Extratumoral Macrophages. Gastroenterology. 2015;149:201–210. doi: 10.1053/j.gastro.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mosser D.M., Edwards J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang M., McKay D., Pollard J.W., Lewis C.E. Diverse Functions of Macrophages in Different Tumor Microenvironments. Cancer Res. 2018;78:5492–5503. doi: 10.1158/0008-5472.CAN-18-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez F.O., Gordon S. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galdiero M.R., Marone G., Mantovani A. Cancer Inflammation and Cytokines. Cold Spring Harb. Perspect. Biol. 2018;10 doi: 10.1101/cshperspect.a028662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanahan D., Coussens L.M. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 44.Pollard J.W. Tumour-educated macrophages promote tumour progression and metastasis. Nat. Rev. Cancer. 2004;4:71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 45.Qian B.Z., Li J., Zhang H., Kitamura T., Zhang J., Campion L.R., Kaiser E.A., Snyder L.A., Pollard J.W. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin E.Y., Nguyen A.V., Russell R.G., Pollard J.W. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J. Exp. Med. 2001;193:727–740. doi: 10.1084/jem.193.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aras S., Zaidi M.R. TAMeless traitors: Macrophages in cancer progression and metastasis. Br. J. Cancer. 2017;117:1583–1591. doi: 10.1038/bjc.2017.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cuccarese M.F., Dubach J.M., Pfirschke C., Engblom C., Garris C., Miller M.A., Pittet M.J., Weissleder R. Heterogeneity of macrophage infiltration and therapeutic response in lung carcinoma revealed by 3D organ imaging. Nat. Commun. 2017;8:14293. doi: 10.1038/ncomms14293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Y.K., Wang M., Sun Y., Di Costanzo N., Mitchell C., Achuthan A., Hamilton J.A., Busuttil R.A., Boussioutas A. Macrophage spatial heterogeneity in gastric cancer defined by multiplex immunohistochemistry. Nat. Commun. 2019;10:3928. doi: 10.1038/s41467-019-11788-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Azizi E., Carr A.J., Plitas G., Cornish A.E., Konopacki C., Prabhakaran S., Nainys J., Wu K., Kiseliovas V., Setty M., et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell. 2018;174:1293–1308. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gordon S., Pluddemann A., Mukhopadhyay S. Plasma membrane receptors of tissue macrophages: Functions and role in pathology. J. Pathol. 2020;250:656–666. doi: 10.1002/path.5404. [DOI] [PubMed] [Google Scholar]
- 52.Langston P.K., Shibata M., Horng T. Metabolism Supports Macrophage Activation. Front. Immunol. 2017;8:61. doi: 10.3389/fimmu.2017.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Neill L.A., Pearce E.J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 2016;213:15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xue J., Schmidt S.V., Sander J., Draffehn A., Krebs W., Quester I., De Nardo D., Gohel T.D., Emde M., Schmidleithner L., et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lawrence T., Natoli G. Transcriptional regulation of macrophage polarization: Enabling diversity with identity. Nat. Rev. Immunol. 2011;11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 56.Chanmee T., Ontong P., Konno K., Itano N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers. 2014;6:1670–1690. doi: 10.3390/cancers6031670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Palma M., Biziato D., Petrova T.V. Microenvironmental regulation of tumour angiogenesis. Nat. Rev. Cancer. 2017;17:457–474. doi: 10.1038/nrc.2017.51. [DOI] [PubMed] [Google Scholar]
- 58.Beltraminelli T., De Palma M. Biology and therapeutic targeting of tumour-associated macrophages. J. Pathol. 2020;250:573–592. doi: 10.1002/path.5403. [DOI] [PubMed] [Google Scholar]
- 59.Blando J., Sharma A., Higa M.G., Zhao H., Vence L., Yadav S.S., Kim J., Sepulveda A.M., Sharp M., Maitra A., et al. Comparison of immune infiltrates in melanoma and pancreatic cancer highlights VISTA as a potential target in pancreatic cancer. Proc. Natl. Acad. Sci. USA. 2019;116:1692–1697. doi: 10.1073/pnas.1811067116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mantovani A., Bonecchi R. One Clever Macrophage Checkpoint. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019;25:3202–3204. doi: 10.1158/1078-0432.CCR-19-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodriguez P.C., Quiceno D.G., Zabaleta J., Ortiz B., Zea A.H., Piazuelo M.B., Delgado A., Correa P., Brayer J., Sotomayor E.M., et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004;64:5839–5849. doi: 10.1158/0008-5472.CAN-04-0465. [DOI] [PubMed] [Google Scholar]
- 62.Cassetta L., Pollard J.W. Targeting macrophages: Therapeutic approaches in cancer. Nat. Rev. Drug Discov. 2018;17:887–904. doi: 10.1038/nrd.2018.169. [DOI] [PubMed] [Google Scholar]
- 63.Mantovani A., Marchesi F., Malesci A., Laghi L., Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017;14:399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Genard G., Lucas S., Michiels C. Reprogramming of Tumor-Associated Macrophages with Anticancer Therapies: Radiotherapy versus Chemo- and Immunotherapies. Front. Immunol. 2017;8:828. doi: 10.3389/fimmu.2017.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anfray C., Ummarino A., Andon F.T., Allavena P. Current Strategies to Target Tumor-Associated-Macrophages to Improve Anti-Tumor Immune Responses. Cells. 2019;9:46. doi: 10.3390/cells9010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Galmbacher K., Heisig M., Hotz C., Wischhusen J., Galmiche A., Bergmann B., Gentschev I., Goebel W., Rapp U.R., Fensterle J. Shigella mediated depletion of macrophages in a murine breast cancer model is associated with tumor regression. PLoS ONE. 2010;5:e9572. doi: 10.1371/journal.pone.0009572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bak S.P., Walters J.J., Takeya M., Conejo-Garcia J.R., Berwin B.L. Scavenger Receptor-A–Targeted Leukocyte Depletion Inhibits Peritoneal Ovarian Tumor Progression. Cancer Res. 2007;67:4783. doi: 10.1158/0008-5472.CAN-06-4410. [DOI] [PubMed] [Google Scholar]
- 68.Nagai T., Tanaka M., Tsuneyoshi Y., Xu B., Michie S.A., Hasui K., Hirano H., Arita K., Matsuyama T. Targeting tumor-associated macrophages in an experimental glioma model with a recombinant immunotoxin to folate receptor beta. Cancer Immunol. Immunother. 2009;58:1577–1586. doi: 10.1007/s00262-009-0667-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lehenkari P.P., Kellinsalmi M., Näpänkangas J.P., Ylitalo K.V., Mönkkönen J., Rogers M.J., Azhayev A., Väänänen H.K., Hassinen I.E. Further insight into mechanism of action of clodronate: Inhibition of mitochondrial ADP/ATP translocase by a nonhydrolyzable, adenine-containing metabolite. Mol. Pharmacol. 2002;61:1255–1262. doi: 10.1124/mol.61.5.1255. [DOI] [PubMed] [Google Scholar]
- 70.Zhang W., Zhu X.D., Sun H.C., Xiong Y.Q., Zhuang P.Y., Xu H.X., Kong L.Q., Wang L., Wu W.Z., Tang Z.Y. Depletion of tumor-associated macrophages enhances the effect of sorafenib in metastatic liver cancer models by antimetastatic and antiangiogenic effects. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010;16:3420–3430. doi: 10.1158/1078-0432.CCR-09-2904. [DOI] [PubMed] [Google Scholar]
- 71.Allavena P., Signorelli M., Chieppa M., Erba E., Bianchi G., Marchesi F., Olimpio C.O., Bonardi C., Garbi A., Lissoni A., et al. Anti-inflammatory properties of the novel antitumor agent yondelis (trabectedin): Inhibition of macrophage differentiation and cytokine production. Cancer Res. 2005;65:2964–2971. doi: 10.1158/0008-5472.CAN-04-4037. [DOI] [PubMed] [Google Scholar]
- 72.Liang W., Kujawski M., Wu J., Lu J., Herrmann A., Loera S., Yen Y., Lee F., Yu H., Wen W., et al. Antitumor activity of targeting SRC kinases in endothelial and myeloid cell compartments of the tumor microenvironment. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010;16:924–935. doi: 10.1158/1078-0432.CCR-09-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ozanne J., Prescott A.R., Clark K. The clinically approved drugs dasatinib and bosutinib induce anti-inflammatory macrophages by inhibiting the salt-inducible kinases. Biochem. J. 2015;465:271–279. doi: 10.1042/BJ20141165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brana I., Calles A., LoRusso P.M., Yee L.K., Puchalski T.A., Seetharam S., Zhong B., de Boer C.J., Tabernero J., Calvo E. Carlumab, an anti-C-C chemokine ligand 2 monoclonal antibody, in combination with four chemotherapy regimens for the treatment of patients with solid tumors: An open-label, multicenter phase 1b study. Target. Oncol. 2015;10:111–123. doi: 10.1007/s11523-014-0320-2. [DOI] [PubMed] [Google Scholar]
- 75.Kitamura T., Qian B.-Z., Soong D., Cassetta L., Noy R., Sugano G., Kato Y., Li J., Pollard J.W. CCL2-induced chemokine cascade promotes breast cancer metastasis by enhancing retention of metastasis-associated macrophages. J. Exp. Med. 2015;212:1043–1059. doi: 10.1084/jem.20141836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bonapace L., Coissieux M.M., Wyckoff J., Mertz K.D., Varga Z., Junt T., Bentires-Alj M. Cessation of CCL2 inhibition accelerates breast cancer metastasis by promoting angiogenesis. Nature. 2014;515:130–133. doi: 10.1038/nature13862. [DOI] [PubMed] [Google Scholar]
- 77.Sánchez-Martín L., Estecha A., Samaniego R., Sánchez-Ramón S., Vega M.Á., Sánchez-Mateos P. The chemokine CXCL12 regulates monocyte-macrophage differentiation and RUNX3 expression. Blood. 2011;117:88–97. doi: 10.1182/blood-2009-12-258186. [DOI] [PubMed] [Google Scholar]
- 78.Hughes R., Qian B.Z., Rowan C., Muthana M., Keklikoglou I., Olson O.C., Tazzyman S., Danson S., Addison C., Clemons M., et al. Perivascular M2 Macrophages Stimulate Tumor Relapse after Chemotherapy. Cancer Res. 2015;75:3479–3491. doi: 10.1158/0008-5472.CAN-14-3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ao J.Y., Zhu X.D., Chai Z.T., Cai H., Zhang Y.Y., Zhang K.Z., Kong L.Q., Zhang N., Ye B.G., Ma D.N., et al. Colony-Stimulating Factor 1 Receptor Blockade Inhibits Tumor Growth by Altering the Polarization of Tumor-Associated Macrophages in Hepatocellular Carcinoma. Mol. Cancer Ther. 2017;16:1544–1554. doi: 10.1158/1535-7163.MCT-16-0866. [DOI] [PubMed] [Google Scholar]
- 80.Zhu Y., Knolhoff B.L., Meyer M.A., Nywening T.M., West B.L., Luo J., Wang-Gillam A., Goedegebuure S.P., Linehan D.C., De Nardo D.G. CSF1/CSF1R Blockade Reprograms Tumor-Infiltrating Macrophages and Improves Response to T-cell Checkpoint Immunotherapy in Pancreatic Cancer Models. Cancer Res. 2014;74:5057. doi: 10.1158/0008-5472.CAN-13-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Q.-Q., Hu X.-W., Liu Y.-L., Ye Z.-J., Gui Y.-H., Zhou D.-L., Qi C.-L., He X.-D., Wang H., Wang L.-J. CD11b deficiency suppresses intestinal tumor growth by reducing myeloid cell recruitment. Sci. Rep. 2015;5:15948. doi: 10.1038/srep15948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ahn G.O., Tseng D., Liao C.H., Dorie M.J., Czechowicz A., Brown J.M. Inhibition of Mac-1 (CD11b/CD18) enhances tumor response to radiation by reducing myeloid cell recruitment. Proc. Natl. Acad. Sci. USA. 2010;107:8363–8368. doi: 10.1073/pnas.0911378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun L., Chen B., Jiang R., Li J., Wang B. Resveratrol inhibits lung cancer growth by suppressing M2-like polarization of tumor associated macrophages. Cell. Immunol. 2017;311:86–93. doi: 10.1016/j.cellimm.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 84.Edwards J.P., Emens L.A. The multikinase inhibitor sorafenib reverses the suppression of IL-12 and enhancement of IL-10 by PGE₂ in murine macrophages. Int. Immunopharmacol. 2010;10:1220–1228. doi: 10.1016/j.intimp.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dong R., Gong Y., Meng W., Yuan M., Zhu H., Ying M., He Q., Cao J., Yang B. The involvement of M2 macrophage polarization inhibition in fenretinide-mediated chemopreventive effects on colon cancer. Cancer Lett. 2017;388:43–53. doi: 10.1016/j.canlet.2016.11.029. [DOI] [PubMed] [Google Scholar]
- 86.Dunn G.P., Koebel C.M., Schreiber R.D. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 87.Parker B.S., Rautela J., Hertzog P.J. Antitumour actions of interferons: Implications for cancer therapy. Nat. Rev. Cancer. 2016;16:131–144. doi: 10.1038/nrc.2016.14. [DOI] [PubMed] [Google Scholar]
- 88.Ding L., Liang G., Yao Z., Zhang J., Liu R., Chen H., Zhou Y., Wu H., Yang B., He Q. Metformin prevents cancer metastasis by inhibiting M2-like polarization of tumor associated macrophages. Oncotarget. 2015;6:36441–36455. doi: 10.18632/oncotarget.5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang L., Xu H., Peng G. TLR-mediated metabolic reprogramming in the tumor microenvironment: Potential novel strategies for cancer immunotherapy. Cell. Mol. Immunol. 2018;15:428–437. doi: 10.1038/cmi.2018.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen N., Wei M., Sun Y., Li F., Pei H., Li X., Su S., He Y., Wang L., Shi J., et al. Self-assembly of poly-adenine-tailed CpG oligonucleotide-gold nanoparticle nanoconjugates with immunostimulatory activity. Small. 2014;10:368–375. doi: 10.1002/smll.201300903. [DOI] [PubMed] [Google Scholar]
- 91.Kaneda M.M., Messer K.S., Ralainirina N., Li H., Leem C.J., Gorjestani S., Woo G., Nguyen A.V., Figueiredo C.C., Foubert P., et al. PI3Kγ is a molecular switch that controls immune suppression. Nature. 2016;539:437–442. doi: 10.1038/nature19834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang F., Parayath N.N., Ene C.I., Stephan S.B., Koehne A.L., Coon M.E., Holland E.C., Stephan M.T. Genetic programming of macrophages to perform anti-tumor functions using targeted mRNA nanocarriers. Nat. Commun. 2019;10:3974. doi: 10.1038/s41467-019-11911-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cai X., Yin Y., Li N., Zhu D., Zhang J., Zhang C.-Y., Zen K. Re-polarization of tumor-associated macrophages to pro-inflammatory M1 macrophages by microRNA-155. J. Mol. Cell Biol. 2012;4:341–343. doi: 10.1093/jmcb/mjs044. [DOI] [PubMed] [Google Scholar]
- 94.Zhang M., Gao Y., Caja K., Zhao B., Kim J.A. Non-viral nanoparticle delivers small interfering RNA to macrophages in vitro and in vivo. PLoS ONE. 2015;10:e0118472. doi: 10.1371/journal.pone.0118472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jia N., Wu H., Duan J., Wei C., Wang K., Zhang Y., Mao X. Polyethyleneimine-coated Iron Oxide Nanoparticles as a Vehicle for the Delivery of Small Interfering RNA to Macrophages In Vitro and In Vivo. JoVE. 2019:e58660. doi: 10.3791/58660. [DOI] [PubMed] [Google Scholar]
- 96.Liang S., Zheng J., Wu W., Li Q., Saw P.E., Chen J., Xu X., Yao H., Yao Y. A Robust Nanoparticle Platform for RNA Interference in Macrophages to Suppress Tumor Cell Migration. Front. Pharm. 2018;9:1465. doi: 10.3389/fphar.2018.01465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zippelius A., Schreiner J., Herzig P., Muller P. Induced PD-L1 expression mediates acquired resistance to agonistic anti-CD40 treatment. Cancer Immunol. Res. 2015;3:236–244. doi: 10.1158/2326-6066.CIR-14-0226. [DOI] [PubMed] [Google Scholar]
- 98.Rodell C.B., Arlauckas S.P., Cuccarese M.F., Garris C.S., Li R., Ahmed M.S., Kohler R.H., Pittet M.J., Weissleder R. TLR7/8-agonist-loaded nanoparticles promote the polarization of tumour-associated macrophages to enhance cancer immunotherapy. Nat. Biomed. Eng. 2018;2:578–588. doi: 10.1038/s41551-018-0236-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vogel D.Y., Glim J.E., Stavenuiter A.W., Breur M., Heijnen P., Amor S., Dijkstra C.D., Beelen R.H. Human macrophage polarization in vitro: Maturation and activation methods compared. Immunobiology. 2014;219:695–703. doi: 10.1016/j.imbio.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 100.Georgoudaki A.M., Prokopec K.E., Boura V.F., Hellqvist E., Sohn S., Ostling J., Dahan R., Harris R.A., Rantalainen M., Klevebring D., et al. Reprogramming Tumor-Associated Macrophages by Antibody Targeting Inhibits Cancer Progression and Metastasis. Cell Rep. 2016;15:2000–2011. doi: 10.1016/j.celrep.2016.04.084. [DOI] [PubMed] [Google Scholar]
- 101.Peng H., Chen B., Huang W., Tang Y., Jiang Y., Zhang W., Huang Y. Reprogramming Tumor-Associated Macrophages To Reverse EGFRT790M Resistance by Dual-Targeting Codelivery of Gefitinib/Vorinostat. Nano Lett. 2017;17:7684–7690. doi: 10.1021/acs.nanolett.7b03756. [DOI] [PubMed] [Google Scholar]
- 102.Ye J., Yang Y., Dong W., Gao Y., Meng Y., Wang H., Li L., Jin J., Ji M., Xia X., et al. Drug-free mannosylated liposomes inhibit tumor growth by promoting the polarization of tumor-associated macrophages. Int. J. Nanomed. 2019;14:3203–3220. doi: 10.2147/IJN.S207589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jaynes J.M., Sable R., Ronzetti M., Bautista W., Knotts Z., Abisoye-Ogunniyan A., Li D., Calvo R., Dashnyam M., Singh A., et al. Mannose receptor (CD206) activation in tumor-associated macrophages enhances adaptive and innate antitumor immune responses. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.aax6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen P., Zhang X., Venosa A., Lee I.H., Myers D., Holloway J.A., Prud’homme R.K., Gao D., Szekely Z., Laskin J.D., et al. A Novel Bivalent Mannosylated Targeting Ligand Displayed on Nanoparticles Selectively Targets Anti-Inflammatory M2 Macrophages. Pharmaceutics. 2020;12:243. doi: 10.3390/pharmaceutics12030243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Irache J.M., Salman H.H., Gamazo C., Espuelas S. Mannose-targeted systems for the delivery of therapeutics. Expert Opin. Drug Deliv. 2008;5:703–724. doi: 10.1517/17425247.5.6.703. [DOI] [PubMed] [Google Scholar]
- 106.Harris N., Super M., Rits M., Chang G., Ezekowitz R.A. Characterization of the murine macrophage mannose receptor: Demonstration that the downregulation of receptor expression mediated by interferon-gamma occurs at the level of transcription. Blood. 1992;80:2363–2373. doi: 10.1182/blood.V80.9.2363.bloodjournal8092363. [DOI] [PubMed] [Google Scholar]
- 107.Stein M., Keshav S., Harris N., Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: A marker of alternative immunologic macrophage activation. J. Exp. Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Taylor M.E., Drickamer K. Mammalian sugar-binding receptors: Known functions and unexplored roles. FEBS J. 2019;286:1800–1814. doi: 10.1111/febs.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pontow S.E., Kery V., Stahl P.D. Mannose receptor. Int. Rev. Cytol. 1992;137b:221–244. doi: 10.1016/s0074-7696(08)62606-6. [DOI] [PubMed] [Google Scholar]
- 110.Blum J.S., Stahl P.D., Diaz R., Fiani M.L. Purification and characterization of the D-mannose receptor from J774 mouse macrophage cells. Carbohydr. Res. 1991;213:145–153. doi: 10.1016/S0008-6215(00)90605-0. [DOI] [PubMed] [Google Scholar]
- 111.Martinez-Pomares L. The mannose receptor. J. Leukoc. Biol. 2012;92:1177–1186. doi: 10.1189/jlb.0512231. [DOI] [PubMed] [Google Scholar]
- 112.Stahl P.D., Ezekowitz R.A. The mannose receptor is a pattern recognition receptor involved in host defense. Curr. Opin. Immunol. 1998;10:50–55. doi: 10.1016/S0952-7915(98)80031-9. [DOI] [PubMed] [Google Scholar]
- 113.Hu Z., Shi X., Yu B., Li N., Huang Y., He Y. Structural Insights into the pH-Dependent Conformational Change and Collagen Recognition of the Human Mannose Receptor. Structure. 2018;26:60–71. doi: 10.1016/j.str.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 114.Scodeller P., Simon-Gracia L., Kopanchuk S., Tobi A., Kilk K., Saalik P., Kurm K., Squadrito M.L., Kotamraju V.R., Rinken A., et al. Precision Targeting of Tumor Macrophages with a CD206 Binding Peptide. Sci. Rep. 2017;7:14655. doi: 10.1038/s41598-017-14709-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Costa J. Glycoconjugates from extracellular vesicles: Structures, functions and emerging potential as cancer biomarkers. Biochim. Biophys. Acta. 2017;1868:157–166. doi: 10.1016/j.bbcan.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 116.Choi E.S., Song J., Kang Y.Y., Mok H. Mannose-Modified Serum Exosomes for the Elevated Uptake to Murine Dendritic Cells and Lymphatic Accumulation. Macromol. Biosci. 2019;19:e1900042. doi: 10.1002/mabi.201900042. [DOI] [PubMed] [Google Scholar]
- 117.Zitvogel L., Regnault A., Lozier A., Wolfers J., Flament C., Tenza D., Ricciardi-Castagnoli P., Raposo G., Amigorena S. Eradication of established murine tumors using a novel cell-free vaccine: Dendritic cell derived exosomes. Nat. Med. 1998;4:594–600. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]
- 118.Wolfers J., Lozier A., Raposo G., Regnault A., Théry C., Masurier C., Flament C., Pouzieux S., Faure F., Tursz T., et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat. Med. 2001;7:297–303. doi: 10.1038/85438. [DOI] [PubMed] [Google Scholar]
- 119.Plebanek M.P., Mutharasan R.K., Volpert O., Matov A., Gatlin J.C., Thaxton C.S. Nanoparticle Targeting and Cholesterol Flux Through Scavenger Receptor Type B-1 Inhibits Cellular Exosome Uptake. Sci. Rep. 2015;5:15724. doi: 10.1038/srep15724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mulcahy L.A., Pink R.C., Carter D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell Vesicles. 2014;3 doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mathieu M., Martin-Jaular L., Lavieu G., Thery C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019;21:9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- 122.Gonda A., Kabagwira J., Senthil G.N., Wall N.R. Internalization of Exosomes through Receptor-Mediated Endocytosis. Mol. Cancer Res. 2019;17:337–347. doi: 10.1158/1541-7786.MCR-18-0891. [DOI] [PubMed] [Google Scholar]
- 123.Zanetti C., Gallina A., Fabbri A., Parisi S., Palermo A., Fecchi K., Boussadia Z., Carollo M., Falchi M., Pasquini L., et al. Cell Propagation of Cholera Toxin CTA ADP-Ribosylating Factor by Exosome Mediated Transfer. Int. J. Mol. Sci. 2018;19:1521. doi: 10.3390/ijms19051521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Andre F., Schartz N.E.C., Movassagh M., Flament C., Pautier P., Morice P., Pomel C., Lhomme C., Escudier B., Le Chevalier T., et al. Malignant effusions and immunogenic tumour-derived exosomes. Lancet. 2002;360:295–305. doi: 10.1016/S0140-6736(02)09552-1. [DOI] [PubMed] [Google Scholar]
- 125.Montecalvo A., Shufesky W.J., Beer Stolz D., Sullivan M.G., Wang Z., Divito S.J., Papworth G.D., Watkins S.C., Robbins P.D., Larregina A.T., et al. Exosomes As a Short-Range Mechanism to Spread Alloantigen between Dendritic Cells during T Cell Allorecognition. J. Immunol. 2008;180:3081. doi: 10.4049/jimmunol.180.5.3081. [DOI] [PubMed] [Google Scholar]
- 126.Lee R.T., Hsu T.L., Huang S.K., Hsieh S.L., Wong C.H., Lee Y.C. Survey of immune-related, mannose/fucose-binding C-type lectin receptors reveals widely divergent sugar-binding specificities. Glycobiology. 2011;21:512–520. doi: 10.1093/glycob/cwq193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Robinson M.J., Sancho D., Slack E.C., LeibundGut-Landmann S., Reis e Sousa C. Myeloid C-type lectins in innate immunity. Nat. Immunol. 2006;7:1258–1265. doi: 10.1038/ni1417. [DOI] [PubMed] [Google Scholar]
- 128.Krishnamoorthy L., Bess J.W., Jr., Preston A.B., Nagashima K., Mahal L.K. HIV-1 and microvesicles from T cells share a common glycome, arguing for a common origin. Nat. Chem. Biol. 2009;5:244–250. doi: 10.1038/nchembio.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shimoda A., Sawada S.I., Sasaki Y., Akiyoshi K. Exosome surface glycans reflect osteogenic differentiation of mesenchymal stem cells: Profiling by an evanescent field fluorescence-assisted lectin array system. Sci. Rep. 2019;9:11497. doi: 10.1038/s41598-019-47760-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen P., Zhang X., Jia L., Prud’homme R.K., Szekely Z., Sinko P.J. Optimal structural design of mannosylated nanocarriers for macrophage targeting. J. Control. Release Off. J. Control. Release Soc. 2014;194:341–349. doi: 10.1016/j.jconrel.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ahowesso C., Black P.N., Saini N., Montefusco D., Chekal J., Malosh C., Lindsley C.W., Stauffer S.R., Di Russo C.C. Chemical inhibition of fatty acid absorption and cellular uptake limits lipotoxic cell death. Biochem. Pharmacol. 2015;98:167–181. doi: 10.1016/j.bcp.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Royo F., Cossío U., de Angulo A.R., Llop J., Falcon-Perez J.M. Modification of the glycosylation of extracellular vesicles alters their biodistribution in mice. Nanoscale. 2019;11:1531–1537. doi: 10.1039/C8NR03900C. [DOI] [PubMed] [Google Scholar]
- 133.Williams C., Pazos R., Royo F., Gonzalez E., Roura-Ferrer M., Martinez A., Gamiz J., Reichardt N.C., Falcon-Perez J.M. Assessing the role of surface glycans of extracellular vesicles on cellular uptake. Sci. Rep. 2019;9:11920. doi: 10.1038/s41598-019-48499-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lucchetti D., Colella F., Perelli L., Ricciardi-Tenore C., Calapa F., Fiori M.E., Carbone F., De Maria R., Sgambato A. CD147 Promotes Cell Small Extracellular Vesicles Release during Colon Cancer Stem Cells Differentiation and Triggers Cellular Changes in Recipient Cells. Cancers. 2020;12:260. doi: 10.3390/cancers12020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yuan Z., Kolluri K.K., Gowers K.H., Janes S.M. TRAIL delivery by MSC-derived extracellular vesicles is an effective anticancer therapy. J. Extracell Vesicles. 2017;6:1265291. doi: 10.1080/20013078.2017.1265291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sterzenbach U., Putz U., Low L.-H., Silke J., Tan S.-S., Howitt J. Engineered Exosomes as Vehicles for Biologically Active Proteins. Mol. Ther. J. Am. Soc. Gene Ther. 2017;25:1269–1278. doi: 10.1016/j.ymthe.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cooks T., Pateras I.S., Jenkins L.M., Patel K.M., Robles A.I., Morris J., Forshew T., Appella E., Gorgoulis V.G., Harris C.C. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat. Commun. 2018;9:771. doi: 10.1038/s41467-018-03224-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang Z., Dombroski J.A., King M.R. Engineering of Exosomes to Target Cancer Metastasis. Cell Mol. Bioeng. 2020;13:1–16. doi: 10.1007/s12195-019-00607-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Maas S.L.N., de Vrij J., van der Vlist E.J., Geragousian B., van Bloois L., Mastrobattista E., Schiffelers R.M., Wauben M.H.M., Broekman M.L.D., Nolte-’t Hoen E.N.M. Possibilities and limitations of current technologies for quantification of biological extracellular vesicles and synthetic mimics. J. Control. Release Off. J. Control. Release Soc. 2015;200:87–96. doi: 10.1016/j.jconrel.2014.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Coscia C., Parolini I., Sanchez M., Biffoni M., Boussadia Z., Zanetti C., Fiani M.L., Sargiacomo M. Generation, quantification, and tracing of metabolically labeled fluorescent exosomes. In: Federico M., editor. Lentiviral Vectors and Exosomes as Gene and Protein Delivery Tools. Springer; New York, NY, USA: 2016. pp. 217–235. [DOI] [PubMed] [Google Scholar]
- 141.Pereira E.A., daSilva L.L. HIV-1 Nef: Taking Control of Protein Trafficking. Traffic. 2016;17:976–996. doi: 10.1111/tra.12412. [DOI] [PubMed] [Google Scholar]
- 142.Mukhamedova N., Hoang A., Dragoljevic D., Dubrovsky L., Pushkarsky T., Low H., Ditiatkovski M., Fu Y., Ohkawa R., Meikle P.J., et al. Exosomes containing HIV protein Nef reorganize lipid rafts potentiating inflammatory response in bystander cells. PLoS Pathog. 2019;15:e1007907. doi: 10.1371/journal.ppat.1007907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zheng Y.H., Plemenitas A., Linnemann T., Fackler O.T., Peterlin B.M. Nef increases infectivity of HIV via lipid rafts. Curr. Biol. CB. 2001;11:875–879. doi: 10.1016/S0960-9822(01)00237-8. [DOI] [PubMed] [Google Scholar]
- 144.Zheng Y.H., Plemenitas A., Fielding C.J., Peterlin B.M. Nef increases the synthesis of and transports cholesterol to lipid rafts and HIV-1 progeny virions. Proc. Natl. Acad. Sci. USA. 2003;100:8460–8465. doi: 10.1073/pnas.1437453100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sami Saribas A., Cicalese S., Ahooyi T.M., Khalili K., Amini S., Sariyer I.K. HIV-1 Nef is released in extracellular vesicles derived from astrocytes: Evidence for Nef-mediated neurotoxicity. Cell Death Dis. 2017;8:e2542. doi: 10.1038/cddis.2016.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pužar Dominkuš P., Ferdin J., Plemenitaš A., Peterlin B.M., Lenassi M. Nef is secreted in exosomes from Nef.GFP-expressing and HIV-1-infected human astrocytes. J. Neurovirology. 2017;23:713–724. doi: 10.1007/s13365-017-0552-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.McNamara R.P., Costantini L.M., Myers T.A., Schouest B., Maness N.J., Griffith J.D., Damania B.A., MacLean A.G., Dittmer D.P. Nef Secretion into Extracellular Vesicles or Exosomes Is Conserved across Human and Simian Immunodeficiency Viruses. mBio. 2018;9 doi: 10.1128/mBio.02344-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lee J.H., Wittki S., Brau T., Dreyer F.S., Kratzel K., Dindorf J., Johnston I.C., Gross S., Kremmer E., Zeidler R., et al. HIV Nef, paxillin, and Pak1/2 regulate activation and secretion of TACE/ADAM10 proteases. Mol. Cell. 2013;49:668–679. doi: 10.1016/j.molcel.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 149.Arenaccio C., Chiozzini C., Columba-Cabezas S., Manfredi F., Affabris E., Baur A., Federico M. Exosomes from human immunodeficiency virus type 1 (HIV-1)-infected cells license quiescent CD4+ T lymphocytes to replicate HIV-1 through a Nef- and ADAM17-dependent mechanism. J. Virol. 2014;88:11529–11539. doi: 10.1128/JVI.01712-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Swingler S., Brichacek B., Jacque J.M., Ulich C., Zhou J., Stevenson M. HIV-1 Nef intersects the macrophage CD40L signalling pathway to promote resting-cell infection. Nature. 2003;424:213–219. doi: 10.1038/nature01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Olivetta E., Percario Z., Fiorucci G., Mattia G., Schiavoni I., Dennis C., Jager J., Harris M., Romeo G., Affabris E., et al. HIV-1 Nef induces the release of inflammatory factors from human monocyte/macrophages: Involvement of Nef endocytotic signals and NF-kappa B activation. J. Immunol. 2003;170:1716–1727. doi: 10.4049/jimmunol.170.4.1716. [DOI] [PubMed] [Google Scholar]
- 152.Mangino G., Percario Z.A., Fiorucci G., Vaccari G., Manrique S., Romeo G., Federico M., Geyer M., Affabris E. In vitro treatment of human monocytes/macrophages with myristoylated recombinant Nef of human immunodeficiency virus type 1 leads to the activation of mitogen-activated protein kinases, IkappaB kinases, and interferon regulatory factor 3 and to the release of beta interferon. J. Virol. 2007;81:2777–2791. doi: 10.1128/JVI.01640-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Federico M., Percario Z., Olivetta E., Fiorucci G., Muratori C., Micheli A., Romeo G., Affabris E. HIV-1 Nef activates STAT1 in human monocytes/macrophages through the release of soluble factors. Blood. 2001;98:2752–2761. doi: 10.1182/blood.V98.9.2752. [DOI] [PubMed] [Google Scholar]
- 154.Percario Z., Olivetta E., Fiorucci G., Mangino G., Peretti S., Romeo G., Affabris E., Federico M. Human immunodeficiency virus type 1 (HIV-1) Nef activates STAT3 in primary human monocyte/macrophages through the release of soluble factors: Involvement of Nef domains interacting with the cell endocytotic machinery. J. Leukoc. Biol. 2003;74:821–832. doi: 10.1189/jlb.0403161. [DOI] [PubMed] [Google Scholar]
- 155.Mangino G., Percario Z.A., Fiorucci G., Vaccari G., Acconcia F., Chiarabelli C., Leone S., Noto A., Horenkamp F.A., Manrique S., et al. HIV-1 Nef induces proinflammatory state in macrophages through its acidic cluster domain: Involvement of TNF alpha receptor associated factor 2. PLoS ONE. 2011;6:e22982. doi: 10.1371/journal.pone.0022982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Luo X., Fan Y., Park I.W., He J.J. Exosomes are unlikely involved in intercellular nef transfer. PLoS ONE. 2015;10:e0124436. doi: 10.1371/journal.pone.0124436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Olivetta E., Arenaccio C., Manfredi F., Anticoli S., Federico M. The Contribution of Extracellular Nef to HIV-Induced Pathogenesis. Curr. Drug Targets. 2016;17:46–53. doi: 10.2174/1389450116666151001110126. [DOI] [PubMed] [Google Scholar]
- 158.Lattanzi L., Federico M. A strategy of antigen incorporation into exosomes: Comparing cross-presentation levels of antigens delivered by engineered exosomes and by lentiviral virus-like particles. Vaccine. 2012;30:7229–7237. doi: 10.1016/j.vaccine.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 159.D’Aloja P., Santarcangelo A.C., Arold S., Baur A., Federico M. Genetic and functional analysis of the human immunodeficiency virus (HIV) type 1-inhibiting F12-HIVnef allele. J. Gen. Virol. 2001;82:2735–2745. doi: 10.1099/0022-1317-82-11-2735. [DOI] [PubMed] [Google Scholar]
- 160.Sistigu A., Bracci L., Valentini M., Proietti E., Bona R., Negri D.R., Ciccaglione A.R., Tritarelli E., Nisini R., Equestre M., et al. Strong CD8+ T cell antigenicity and immunogenicity of large foreign proteins incorporated in HIV-1 VLPs able to induce a Nef-dependent activation/maturation of dendritic cells. Vaccine. 2011;29:3465–3475. doi: 10.1016/j.vaccine.2011.02.059. [DOI] [PubMed] [Google Scholar]
- 161.Andzinski L., Kasnitz N., Stahnke S., Wu C.F., Gereke M., von Kockritz-Blickwede M., Schilling B., Brandau S., Weiss S., Jablonska J. Type I IFNs induce anti-tumor polarization of tumor associated neutrophils in mice and human. Int. J. Cancer. 2016;138:1982–1993. doi: 10.1002/ijc.29945. [DOI] [PubMed] [Google Scholar]