Abstract
BACKGROUND
It is unclear whether immune escape-associated mutations in the major hydrophilic region of hepatitis B virus surface antigen (HBsAg) are associated with nucleoside/nucleotide analog resistance.
AIM
To evaluate the association between immune escape-associated mutations and nucleoside/nucleotide analog resistance mutations.
METHODS
In total, 19440 patients with chronic hepatitis B virus infection, who underwent resistance testing at the Fifth Medical Center of Chinese PLA General Hospital between July 2007 and December 2017, were enrolled. As determined by sequence analysis, 6982 patients harbored a virus with resistance mutations and 12458 harbored a virus lacking resistance mutations. Phenotypic analyses were performed to evaluate HBsAg production, replication capacity, and drug-induced viral inhibition of patient-derived drug-resistant mutants with or without the coexistence of sA159V.
RESULTS
The rate of immune escape-associated mutation was significantly higher in 9 of the 39 analyzed mutation sites in patients with resistance mutations than in patients without resistance mutations. In particular, these mutations were sQ101H/K/R, sS114A/L/T, sT118A/K/M/R/S/V, sP120A/L/Q/S/T, sT/I126A/N/P/S, sM133I/L/T, sC137W/Y, sG145A/R, and sA159G/V. Among these, sA159V was detected in 1.95% (136/6982) of patients with resistance mutations and 1.08% (134/12,458) of patients lacking resistance mutations (P < 0.05). The coexistence of sA159V with lamivudine (LAM) and entecavir (ETV)-resistance mutations in the same viral genome was identified during follow-up in some patients with drug resistance. HBsAg production was significantly lower and the replication capacity was significantly higher, without a significant difference in LAM/ETV susceptibility, in sA159V-containing LAM/ETV-resistant mutants than in their sA159V-lacking counterparts.
CONCLUSION
In summary, we observed a close link between the increase in certain immune escape-associated mutations and the development of resistance mutations. sA159V might increase the fitness of LAM/ETV-resistant mutants under environmental pressure in some cases.
Keywords: Hepatitis B virus, Immune escape-associated mutation, Drug-resistance mutation, Nucleoside/nucleotide analogs, Hepatitis B surface antigen, Major hydrophilic region
Core Tip: A large number of patients were surveyed for immune escape-associated mutations, and the link between immune escape-associated and resistant mutations was identified. Contribution of sA159V to resistance was found in multiple followed up patients, in particular sA159V reduced hepatitis B surface antigen but raised the replication capacity of lamivudine/entecavir-resistant mutants.
INTRODUCTION
It is estimated that 292 million people worldwide are chronically infected with hepatitis B virus (HBV), including 86 million residing in China[1]. The treatment of chronic HBV infection is aimed towards the long-term suppression of viral replication to prevent disease progression[2]. Currently, nucleoside/nucleotide analogs (NAs) including lamivudine (LAM), adefovir dipivoxil (ADV), entecavir (ETV), telbivudine (LdT), tenofovir disoproxil fumarate (TDF), and tenofovir alafenamide (TAF) are approved for the treatment of HBV infection. However, a concern is the drug resistance caused by mutations in the reverse transcriptase (RT) region of the HBV genome. Drug-resistance mutations tend to arise in patients treated with LAM, LdT, and ADV as well as in LAM-refractory patients subsequently treated with ETV[3,4]. Classical primary resistance mutations include rtM204I/V (LAM-r) for LAM (rtM204I also confers resistance to LdT), rtA181V/rtN236T for ADV as well as LAM-r along with at least one substitution at rt184 (A/C/F/G/I/L/M/S), rt202 (C/G/I), and rtM250 (I/L/V) for ETV[3,5,6]. In addition, rtS106C+rtH126Y+rtD134E+rtL269I quadruple mutations have recently been reported to confer TDF resistance[7]. TAF, also known as TDF II, has a higher intrahepatic drug concentration and lower plasma drug concentration than TDF as well as a lower probability of kidney and bone abnormalities during therapy[8].
The rapid selection of drug-resistant HBV mutants may depend on viral fitness, which could be influenced by the host immune response in addition to drug pressure[9,10,11]. A few drug-resistance mutations, such as rtS78T and rtA181T, introduce a stop codon in the overlapping S region and affect the immune response, thereby influencing the clinical presentation of NA-treated patients[12-15]. Hepatitis B surface antigen (HBsAg) is diagnostic marker of HBV infection and an important index for predicting the effects of antiviral treatment[16,17]. HBV immune escape-associated mutations, located mainly in the major hydrophilic region (MHR, amino acids 99−169), have the potential to weaken the immune response. Currently, it is unclear whether these mutations influence drug resistance. Only a few studies on a limited number of patients reference this issue, showing that the frequency of some immune escape-associated mutations is higher in LAM-treated patients than in NA-naive patients, suggesting that selection of drug-resistance mutations is associated with immune escape-associated mutation enrichment[18,19].
We recently identified several novel immune escape-associated mutations in patients with occult HBV infection; a summary of previously documented immune escape-associated mutations is provided in Table 1[20,21]. Notably, for many previously documented mutations, phenotypic information is lacking. In this study, we evaluated a large number of patients to determine whether immune escape-associated mutations are associated with drug-resistance mutations, with a focus on the sA159V mutation.
Table 1.
Summary of immune escape-associated mutations in the major hydrophilic region of hepatitis B surface antigen
| Region in major hydrophilic region (aa 99–169) | Mutation pattern |
| Upstream “a” determinant (aa 99-123) | sY100S, sQ101H/K/R, sM103I/T, sL109I/P/R, sP111L/Q/S, sG112K/R, sS114A/L/T, sT115A/N, sT116N, sS117G/N/R, sT118A/K/M/R/S/V, sG119E/R/T, sP120A/L/Q/S/T, sC121R/S, sK122R, sT123A/I/N/S/V |
| Within “a” determinant (aa 124-147) | sC124R/Y, sT125A/M, sI/T126A/N/P/S, sP127H/L/S/T, sA128T/V, sQ129N/H/P/R, sG130A/K/N/R/S, sT131A/I/K/N, sS132F/P, sM133I/L/T, sF/Y134H/L/R/S/V, sS136F/P/Y, sC137W/Y, sC139R/S/Y, sT140I, sK141E/R, sP142L, sD144A/E, sG145A/R, sC147R/Y |
| Downstream “a” determinant (aa 148-169) | sS154P, sA159G/V, sV168A |
aa: Amino acid.
The rapidity of selection of drug-resistant HBV mutants may depend on viral fitness, which could be influenced by the host immune response in addition to drug pressure[9,10,11]. A few drug-resistance mutations, such as rtS78T and rtA181T, introduce a stop codon in the overlapping S region and affect the immune response, thereby influencing the clinical presentation of NA-treated patients[12-15]. HBsAg is diagnostic marker of HBV infection and an important index for predicting the effects of antiviral treatment[16,17]. HBV immune escape-associated mutations, located mainly in the MHR (amino acids 99−169), have the potential to weaken the immune response. Currently, it is unclear whether these mutations influence drug resistance. Only a few studies on a limited number of patients reference this issue, showing that the frequency of some immune escape-associated mutations is higher in LAM-treated patients than in NA-naïve patients, suggesting that selection of drug-resistance mutations is associated with immune escape-associated mutation enrichment[18,19].
We recently identified several novel immune escape-associated mutations in patients with occult HBV infection; a summary of previously documented immune escape-associated mutations is provided in Table 1[20,21]. Notably, for many previously documented mutations, phenotypic information is lacking. In this study, we evaluated a large number of patients to determine whether immune escape-associated mutations are associated with drug-resistance mutations, with a focus on the sA159V mutation.
MATERIALS AND METHODS
Patient samples
From July 2007 to December 2017, 19440 patients with chronic HBV infection who underwent resistance testing (by direct sequencing) at the Fifth Medical Center of Chinese PLA General Hospital (originally named Beijing 302 Hospital) were enrolled in the study, and their serum samples were collected. All patients were previously treated with NAs. Illness categories included chronic hepatitis B, HBV-related liver cirrhosis, and hepatocellular carcinoma. The diagnostic criteria were based on the guidelines for the prevention and treatment of chronic hepatitis B in China (2005)[22], and the updated guidelines were used according to the time of patient enrollment. Patients who were co-infected with other hepatitis viruses or human immuno-deficiency virus were excluded from the study. All patients were from the Database of Beijing 302 Hospital and provided informed consent for the use of their samples for research before enrollment. The study was approved by the Ethics Committee of Beijing 302 Hospital.
Detection of serological markers and HBV deoxyribonucleic acid
Biochemical and serological markers as well as HBV deoxyribonucleic acid (DNA) levels in the serum samples were routinely detected at the Central Clinical Laboratory of the Fifth Medical Center of the Chinese PLA General Hospital. Roche Elecsys reagents (Basel, Switzerland) were used to measure the serum HBsAg levels, and the threshold for negativity was < 0.05 IU/mL in the quantitative assay or a cut-off index of < 1.00 in the chemiluminescent immunoassay assay.
Sequence analysis of HBV reverse transcriptase/S genes and phylogenetic tree analysis
Sequence and phylogenetic analyses were performed as previously described[23,24]. In brief, a 1225-bp fragment [nucleotides (nt) 54-1278] spanning the full-length RT region (nt 130-1161) and S region (nt 155-835) of the viral genome was analyzed. Drug-resistance and immune escape-associated mutations were analyzed by direct sequencing using an in-house nested PCR method with a lower detection limit of 10 IU/mL. Clonal sequencing of the samples of interest was performed (20 clones per sample). Phylogenetic trees were constructed using MEGA 7 software.
Construction of 1.1-mer HBV reverse genome vectors and site-directed mutagenesis
Replication-competent vectors containing various mutant or wild-type (WT) RT/S genes were constructed for a phenotypic analysis based on the pTriEx-mod-1.1 vector, which was used for antigenicity analyses as previously described[21,24]. Eight recombinant vectors harboring RT/S genes from eight viral strains of a representative patient (patient A) were constructed. The eight strains were: WT, sA159V (M1), rtM204I (M2), sA159V+rtM204I (M3), rtL180M+rtM204V (M4), sA159V+rtL180M+rtM204V (M5), rtL180M+rtT184L+rtM204V (M6), and sA159V+rtL180M+rtT184L+rtM204V (M7). M6 was modified from M7 by the elimina-tion of the sA159V mutation using the QuikChange Lightning Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, United States). The primer (sense) was 5′-CCTGGGCTTTCGCAAAATTCCTATG-3′.
Phenotypic analysis of HBsAg, replication capacity, and drug-induced viral inhibition
Experiments were performed as described previously, with minor modifications[21,24]. Briefly, recombinant vectors were individually transfected into HepG2 cells. At 3 d after cultivation, the supernatant was harvested for the measurement of HBsAg by two assays, i.e. a chemiluminescence immunoassay (Roche) and an enzyme-linked immunosorbent assay (ELISA; Wantai Bio Pharm., Beijing, China).
To assess drug-induced viral inhibition, transfected HepG2 cells were cultured in the presence or absence of NAs for 4 d. Cells were lysed, and viral core particles were immunoprecipitated using anti-HBc/protein A+G. HBV replicative intermediates in the core particles were released and quantified by real-time PCR. The relative replication capacity of a mutant vs WT strain was determined in the absence of NAs. Approximately 90% effective concentrations of the four NAs were used, as previously determined[25,26]. These were 0.05 μmol/L for LAM, 0.05 μmol/L for ETV, 3.0 μmol/L for ADV, and 5.0 μmol/L for TDF. Viral inhibition was determined as the relative value of the NA-treated samples vs the NA-untreated samples. Experiments were performed at least three times independently.
Statistical analysis
Data are presented as the means (standard deviation) or medians (interquartile range). Differences between groups were examined by the Student’s t-test (two-tailed) or chi-squared tests. Multivariate regression was used to determine independent risk factors. Statistical analyses were performed using SPSS 23.0 for Windows (SPSS Inc., Chicago, IL, United States). A value of P < 0.05 was considered statistically significant.
RESULTS
Clinical profile of immune escape-associated mutations in patients with and without resistance mutations
Drug-resistance mutations were detected in 35.92% (6,982/19,440) of all patients included in the study. Moreover, patients harboring resistance mutations had higher HBV DNA levels than patients lacking resistance mutations. The rate of immune escape-associated mutation was significantly higher at 9 of 39 analyzed mutation sites in patients with resistance mutations than in patients without resistance mutations. These mutations were sQ101H/K/R, sS114A/L/T, sT118A/K/M/R/S/V, sP120A/L/Q/S/T, sT/I126A/N/P/S, sM133I/L/T, sC137W/Y, sG145A/R, and sA159G/V. The percentage of patients with MHR mutations was significantly higher in the resistance mutation-positive group than in the resistance mutation-negative group (23.32% vs 18.51%, P < 0.05) (Table 2). In particular, mutations were detected in 67/63/168 of 298 sQ101H/K/R-positive patients, 39/2/76 of 117 sS114A/L/T-positive patients, 5/5/15/3/1/2 of 31 sT118A/K/M/R/S/V-positive patients, 3/0/0/23/81 of 107 sP120A/L/Q/S/T-positive patients, 94/58/0/214 of 366 sT/I126A/N/P/S-positive patients, 14/46/136 of 196 sM133I/L/T-positive patients, 1/8 of 9 sC137W/Y-positive patients, 77/46 of 123 sG145A/R-positive patients, and 87/136 of 223 sA159G/V-positive patients.
Table 2.
Analysis of immune escape-associated mutations in the major hydrophilic region in patients with and without resistance mutations
| Clinical features | Resistance mutation (+), n = 6982 | Resistance mutation (-), n = 12458 | P value | The MHR Mutation occurrence [(+) vs (-)] |
| Age in year | 44.01 ± 11.73 | 39.01 ± 13.08 | < 0.05 | |
| Gender, male | 5709 (81.77%) | 9982 (80.13%) | < 0.05 | |
| HBV genotype, C%/B% | 86.79/12.52 | 83.00/16.05 | < 0.05 | |
| HBV DNA as log10 IU/mL | 4.62 (3.17, 6.47) | 4.24 (2.83, 6.16) | < 0.05 | |
| sY100S | 20 (0.29%) | 29 (0.23%) | NS | |
| sQ101H/K/R | 298 (4.27%) | 229 (1.84%) | < 0.05 | ↑ |
| sM103I/T | 8 (0.11%) | 14 (0.11%) | NS | |
| sL109I/P/R | 12 (0.17%) | 24 (0.19%) | NS | |
| sP111L/Q/S | 10 (0.14%) | 21 (0.17%) | NS | |
| sG112K/R | 4 (0.06%) | 9 (0.07%) | NS | |
| sS114A/L/T | 117 (1.68%) | 137 (1.10%) | < 0.05 | ↑ |
| sT115A/N | 2 (0.03%) | 8 (0.06%) | NS | |
| sT116N | 6 (0.09%) | 32 (0.26%) | < 0.05 | ↑ |
| sS117G/N/R | 5 (0.07%) | 19 (0.15%) | NS | |
| sT118A/K/M/R/S/V | 31 (0.44%) | 32 (0.26%) | < 0.05 | ↑ |
| sG119E/R/T | 6 (0.09%) | 10 (0.08%) | NS | |
| sP120A/L/Q/S/T | 107 (1.53%) | 35 (0.28%) | < 0.05 | ↑ |
| sC121R/S | 1 (0.01%) | 1 (0.01%) | NS | |
| sK122R | 154 (2.21%) | 315 (2.53%) | NS | |
| sT123A/I/N/S/V | 56 (0.80%) | 91 (0.73%) | NS | |
| sC124R/Y | 4 (0.06%) | 9 (0.07%) | NS | |
| sT125A/M | 8 (0.11%) | 21 (0.17%) | NS | |
| sT/I126A/N/P/S | 366 (5.24%) | 534 (4.29%) | < 0.05 | ↑ |
| sP127H/L/S/T | 91 (1.30%) | 165 (1.32%) | NS | |
| sA128T/V | 7 (0.10%) | 15 (0.12%) | NS | |
| sQ129N/H/P/R | 90 (1.29%) | 129 (1.04%) | NS | |
| sG130A/K/N/R/S | 65 (0.93%) | 85 (0.68%) | NS | |
| sT131A/I/K/N | 133 (1.90%) | 219 (1.76%) | NS | |
| sS132F/P | 2 (0.03%) | 9 (0.07%) | NS | |
| sM133I/L/T | 196 (2.81%) | 277 (2.22%) | < 0.05 | ↑ |
| sF/Y134H/L/R/S/V | 65 (0.93%) | 51 (0.41%) | NS | |
| sS136F/P/Y | 6 (0.09%) | 4 (0.03%) | NS | |
| sC137W/Y | 9 (0.13%) | 5 (0.04%) | < 0.05 | ↑ |
| sC139R/S/Y | 3 (0.04%) | 9 (0.07%) | NS | |
| sT140I | 39 (0.56%) | 61 (0.49%) | NS | |
| sK141E/R | 3 (0.04%) | 4 (0.03%) | NS | |
| sP142L | 11 (0.16%) | 9 (0.07%) | NS | |
| sD144A/E | 17 (0.24%) | 26 (0.21%) | NS | |
| sG145A/R | 123 (1.76%) | 163 (1.31%) | < 0.05 | ↑ |
| sC147R/Y | 0 | 0 | NS | |
| sS154P | 1 (0.01%) | 2 (0.02%) | NS | |
| sA159G/V | 223 (3.19%) | 319 (2.56%) | < 0.05 | ↑ |
| sV168A | 143 (2.05%) | 234 (1.88%) | NS | |
| Average number/patient | 0.35 | 0.27 | < 0.05 | ↑ |
| Patient percentage with the MHR mutation(s) | 23.32% (1628/ 6982) | 18.51% (2306/12458) | < 0.05 | ↑ |
(+): Positive; (-): Negative; DNA: Deoxyribonucleic acid; HBV: Hepatitis B virus; MHR: Major hydrophilic region; NS: Not significant.
Clinical incidence and features of the sA159V mutation
Restricted by the scale of the study, sA159V was selected as a representative immune escape-associated mutation for further analyses. The detection rate of sA159V was significantly higher in patients with resistance mutations than in patients lacking resistance mutations [1.95% (136/6982) vs 1.08% (134/12458), P < 0.05]. In contrast, the detection rate of sA159G did not differ significantly between the two patient groups (1.25% (87/6982) vs 1.48% (185/12458), P > 0.05). The clinical features of the sA159V-positive and sA159V-negative patients are summarized in Table 3. A multivariate analysis showed that age and the coexistence of ADV-r/LAM-r mutations were independently associated with the sA159V mutation. The sA159V-positive patients had higher rates of coexisting drug-resistance mutations than the sA159V-negative patients.
Table 3.
Analysis of clinical features of sA159V-positive and sA159V-negative patients
| Clinical features | sA159V-positive, n = 270 | sA159V-negative, n = 19170 | Univariate, P value | Univariate, P | Multivariate, P value | Multivariate , P |
| Age in year | 46.67 ± 12.19 | 40.72 ± 12.83 | 0.00 | < 0.05 | 0.00 | < 0.05 |
| Gender, male | 214 (79.26%) | 15477 (80.74%) | 0.54 | > 0.05 | 0.97 | > 0.05 |
| Genotype, C%/B% | 90.00/10.00 | 84.31/14.82 | 0.03 | < 0.05 | 0.40 | > 0.05 |
| HBV DNA as log10 IU/mL | 4.54 (3.18, 6.55) | 4.37(2.95, 6.27) | 0.09 | > 0.05 | 0.40 | > 0.05 |
| ALT in U/L | 42 (25, 87) | 42 (26, 82) | 0.67 | > 0.05 | 0.32 | > 0.05 |
| AST in U/L | 41 (28, 83) | 38 (26,71) | 0.33 | > 0.05 | 0.61 | > 0.05 |
| TBIL in μmol/L | 16.10 (10.95, 25.45) | 14.30 (10.40, 22.10) | 0.13 | > 0.05 | 0.58 | > 0.05 |
| CHE in U/L | 5940 (3114, 8091) | 6764 (4276, 8469) | 0.11 | > 0.05 | 0.87 | > 0.05 |
| HBsAg, COI | 4987.83 ± 3128.43 | 5064.50 ± 2931.79 | 0.05 | > 0.05 | 0.15 | > 0.05 |
| Coexistent with ADV-r mutation | 41 (15.19%) | 1663 (8.68%) | 0.00 | < 0.05 | 0.00 | < 0.05 |
| Coexistent with LAM-r mutation | 97 (35.93%) | 5316 (27.73%) | 0.00 | < 0.05 | 0.01 | < 0.05 |
| Coexistent with ETV-r mutation | 20 (7.41%) | 868 (4.53%) | 0.04 | < 0.05 | 0.40 | > 0.05 |
ADV-r: Adefovir resistance; ALT: Alanine aminotransferase; AST: Aspartate aminotransferase; CHE: Cholinesterase; COI: Cut-off index; DNA: Deoxyribonucleic acid; ETV-r: Entecavir resistance; HBsAg: Hepatitis B surface antigen; HBV: Hepatitis B virus; LAM-r: Lamivudine resistance; TBIL: Total bilirubin.
Longitudinal analysis of the clinical course of patients with HBV mutations during nucleotide analog therapy
Five representative sA159V-positive patients with available serial serum samples were subjected to clonal analysis of HBV RT/S genes. The five patients were infected with genotype C HBV and diagnosed with chronic hepatitis B or HBV-related liver cirrhosis.
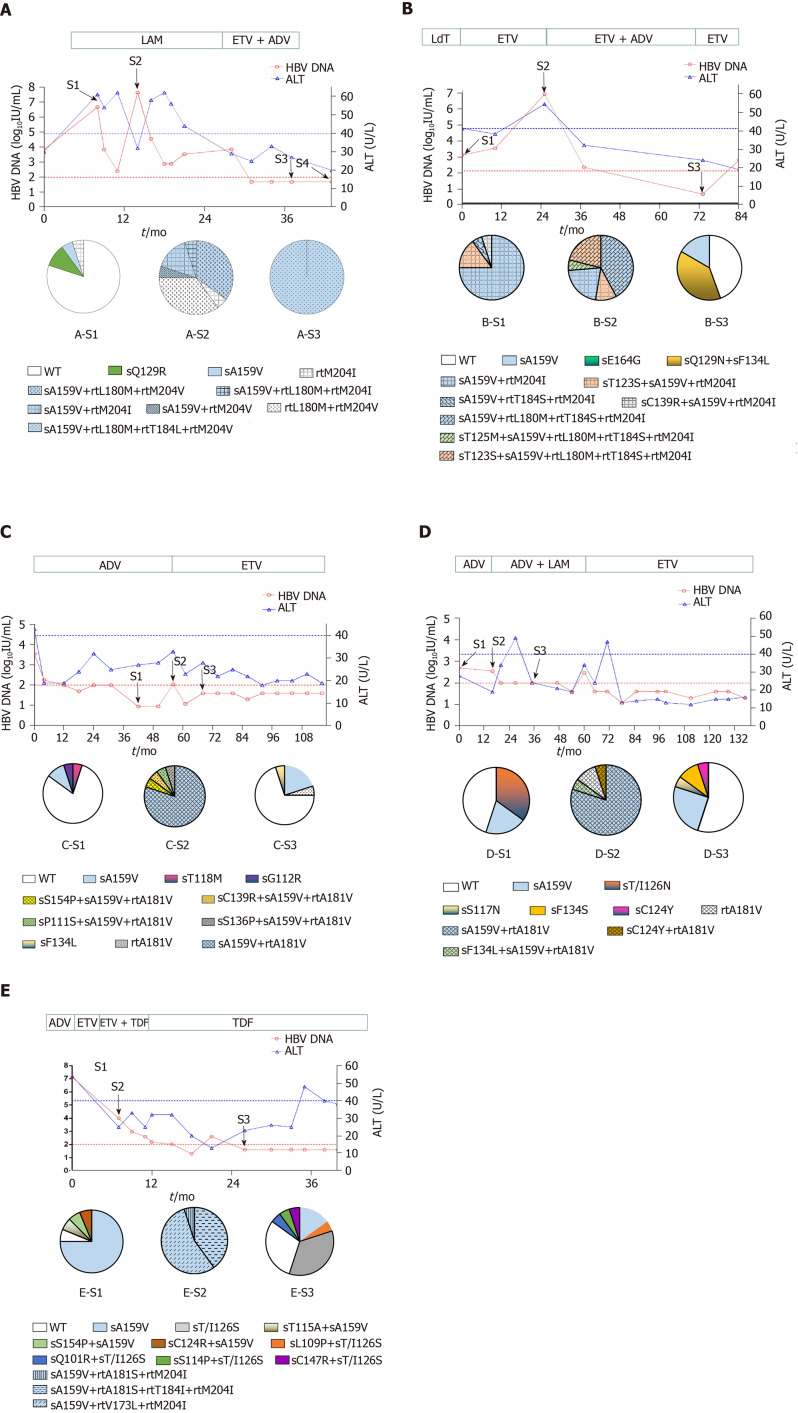
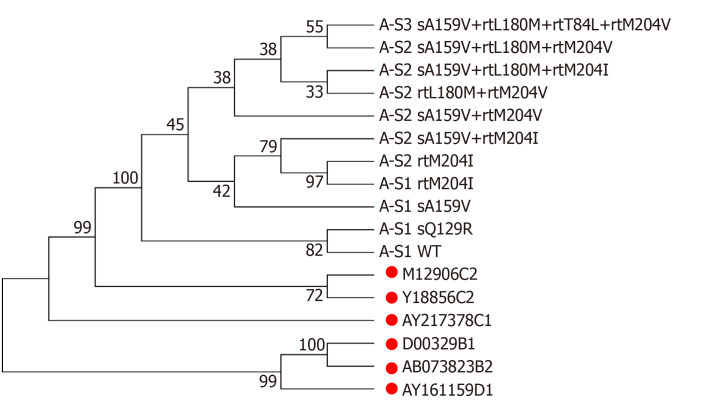
Patient A, a 52-year-old male, was first admitted in October 2007 with chronic hepatitis B. The patient received LAM (from May 2008 to October 2009) and ETV+ADV (from October 2009 to June 2011). In tested clones of sample A-S1, WT, sA159V, sQ129R, and rtM204I detection rates were 80%, 10%, 5%, and 5%, respectively. In sample A-S2, six mutants were detected, i.e. sA159V+rtL180M+rtM204V (35%), rtL180M+rtM204V (35%), sA159V+rtM204I (15%), sA159V+rtL180M+rtM204I (5%), sA159V+rtM204V (5%), and rtM204I (5%). In sample A-S3, sA159V+rtL180M+rtM204V+rtT184L was the most abundant (Figure 1A). Clonal sequencing of sample A-S4 failed due to an extremely low viral load. The 10 viral clonal sequences from the patient were deposited in GenBank (MN642606-MN642615) and used to construct a phylogenetic tree (Figure 2).
Figure 1.
Evolution of drug-resistant hepatitis B virus strains and clinical responses during antiviral therapy in five representative patients. Dynamic changes in serum hepatitis B virus deoxyribonucleic acid (HBV DNA) and alanine aminotransferase (ALT) levels are shown along with antiviral therapies. The duration (months) of antiviral therapy is represented by bars above the graph, and the serum samples from the patient for cloning are indicated by arrows. Two dashed lines show the lower detection limit of HBV DNA (100 IU/mL) and normal ALT levels (40 U/L). Proportions of wild-type and mutant HBV reverse transcriptase from each sample are depicted by a series of pie charts. A: Evolution of drug-resistant hepatitis B virus strains and clinical responses during antiviral therapy in patient A; B:Evolution of drug-resistant hepatitis B virus strains and clinical responses during antiviral therapy in patient B; C:Evolution of drug-resistant hepatitis B virus strains and clinical responses during antiviral therapy in patient C; D:Evolution of drug-resistant hepatitis B virus strains and clinical responses during antiviral therapy in patient D; E:Evolution of drug-resistant hepatitis B virus strains and clinical responses during antiviral therapy in patient E.
Figure 2.
Phylogenetic tree analysis of hepatitis B virus reverse transcriptase sequences from a patient with sA159V + resistance mutations. Reference sequences are marked with solid red circles.
Four patients received various NAs: LAM, ADV, or ETV, as monotherapy or in combination. Patient B had seven sA159V-containing mutants in samples B-S1 and B-S2 during ETV therapy. Resistant mutants were subsequently suppressed by ETV+ADV, whereas the sA159V mutation was observed in sample B-S3 (Figure 1B). Patient C and Patient D initially received ADV and failed to exhibit virological breakthrough. In both patients, the sA159V mutant existed before virological breakthrough (samples C-S1 and D-S1), and sA159V-containing ADV-resistant mutants were dominant in samples C-S2 and D-S2 at virological breakthrough (Figure 1C and D). Samples from Patient E included three sA159V-containing ETV/LAM-resistant mutants (sample E-S2) when the virological response was inadequate upon ETV+TDF therapy (Figure 1E).
Quantitative HBsAg levels
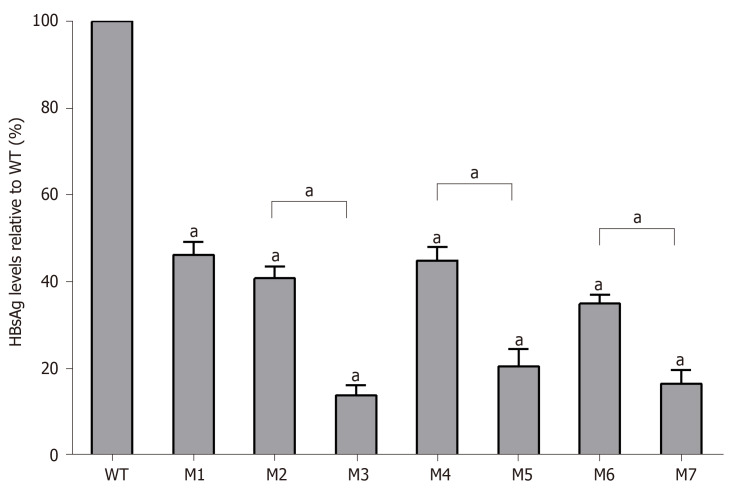
As determined by the Roche quantitative assay, the HBsAg levels in the supernatant of the seven mutants decreased significantly to 46.2% (M1), 40.8% (M2), 14.0% (M3), 44.9% (M4), 20.6% (M5), 35.1% (M6), and 16.6% (M7) of the WT level. Three sA159V-containing resistant mutants had significantly lower HBsAg levels than their sA159V-lacking counterparts (M2 vs M3, M4 vs M5, M6 vs M7, all P < 0.05) (Figure 3). Consistent results were obtained by ELISA (data not shown).
Figure 3.
Quantitative analysis of hepatitis B surface antigen levels in individual viral vector-transfected human hepatocellular carcinoma cells. Relative values (%) of mutant hepatitis B surface antigen levels vs wild-type levels are shown. Data are expressed as means ± standard deviations. M1, sA159V; M2, rtM204I; M3, sA159V+rtM204I; M4, rtL180M+rtM204V; M5, sA159V+rtL180M+rtM204V; M6, rtL180M+rtT184L+rtM204V; M7,sA159V+rtL180M+rtT184L+rtM204V. aP < 0.05 (mutant vs wild-type or other indicated mutant).
Assessment of viral replication capacity
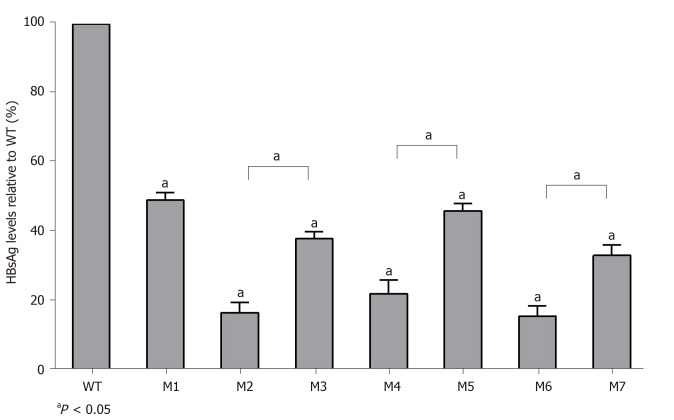
The replication capacities of the seven mutants (M1–M7) decreased significantly to 49.0% (M1), 16.4% (M2), 37.8% (M3), 22.0% (M4), 46.0% (M5), 15.4% (M6), and 33.0% (M7) of the WT level. Three sA159V-containing resistant mutants had significantly higher replication capacities than their sA159V-lacking counterparts (M2 vs M3, M4 vs M5, M6 vs M7, all P < 0.05) (Figure 4).
Figure 4.
Quantitative analysis of hepatitis B virus deoxyribonucleic acid production levels in individual viral vector-transfected human hepatocellular carcinoma cells. Relative values (%) of mutant hepatitis B virus deoxyribonucleic acid levels vs wild-type levels are shown. Data are expressed as the means ± standard deviations. M1, sA159V; M2, rtM204I; M3, sA159V+rtM204I; M4, rtL180M+rtM204V; M5, sA159V+rtL180M+rtM204V; M6,sA159V+rtL180M+rtT184L+rtM204V; M7, rtL180M+rtT184L+rtM204V. aP < 0.05 (mutant vs wild-type or other indicated mutants).
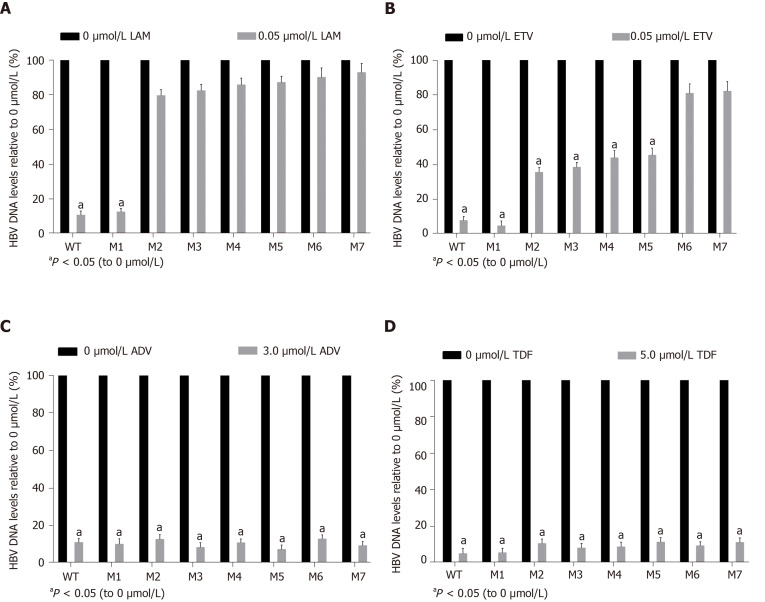
Assessment of drug-induced viral inhibition
A viral inhibition test was performed using the WT and seven mutant strains. Inhibition was evaluated by the levels of HBV replicative intermediates in the samples treated with NAs relative to those in the untreated samples. LAM strongly inhibited WT and M1, with rates of inhibition of 90.5% and 87.5%, respectively. In contrast, LAM-resistant mutants (M2–M5) and ETV-resistant mutants (M6-M7) were highly resistant to LAM, regardless of the presence or absence of the sA159V mutation in the viral genome (Figure 5A). ETV also strongly inhibited WT and M1 with rates of inhibition of 92.5% and 95.6%, respectively. M2–M5 were partially resistant to ETV, with 54.9%-64.7% inhibition. M6–M7 were highly resistant to ETV (Figure 5B). All the tested viral strains were highly sensitive to ADV and TDF, with rates of inhibition of 87.29%-95.04% (Figure 5C and D).
Figure 5.
Assessment of drug-induced viral inhibition. Human hepatocellular carcinomas cells were transiently transfected with wild-type or individual viral vectors and cultured with or without one of the following four drugs: (A) 0.05 μmol/L lamivudine, (B) 0.05 μmol/L entecavir, (C) 3.0 μmol/L adefovir, and (D) 5.0 μmol/L tenofovir disoproxil fumarate. Viral inhibition was evaluated as the relative hepatitis B virus deoxyribonucleic acid level of samples with the drug to that without the drug. M1, sA159V; M2, rtM204I; M3, sA159V+rtM204I; M4, rtL180M+rtM204V; M5, sA159V+tL180M+rtM204V; M6, rtL180M+rtT184L+rtM204V; M7, sA159V+rtL180M+rtT184L+rtM204V. aP < 0.05.
DISCUSSION
The clinical implications of immune escape-associated mutations arise from their relationship with occult HBV infections, HBV reactivation, and HB vaccination failure[27-30]. There is a paucity of data from population-based clinical investigations about the link between immune escape-associated mutations and drug-resistance mutations. Our analysis of the largest number of resistance mutation-positive patients to date clearly showed that the frequency of immune escape-associated mutations is significantly higher in resistance mutation-positive patients than in resistance mutation-negative patients.
We selected the sA159V mutation for detailed analyses because: (1) Its frequency was significantly higher in resistance mutation-positive patients than in resistance mutation-negative patients; (2) It was frequently detected together with LAM-, ADV-, and ETV-resistance mutations; and (3) Its virological features have not been documented by phenotypic analyses.
In a longitudinal analysis of five patients, the coexistence of sA159V with LAM/ADV/ETV-resistance mutations was frequently detected in the viral pool along with virological breakthrough or an inadequate virological response upon NA therapy. In addition, the sA159V mutation alone was detected before the emergence of the resistance mutation and was recovered after the sA159V-containing resistant mutants were effectively suppressed by rescue therapy. A phylogenetic analysis of Patient A-derived viral strains showed that sA159V+rtM204I (rtL180M) and sA159V+rtM204V (rtL180M) mutants are likely derived from the sA159V mutant as an adaptation to LAM pressure. The sA159V+rtL180M+rtT184L+rtM204V mutant is probably derived from sA159V+rtL180M+rtM204V as an adaptation to ETV pressure.
sA159V mutants had lower HBsAg levels than the WT strain. Viral strains harboring both sA159V and LAM- or ETV-resistance mutations exhibited significantly lower HBsAg levels than their counterpart strains lacking sA159V mutations. In contrast, the HBV DNA levels were partially restored in the LAM- or ETV-resistance viral strains with sA159V compared to the levels in the WT strain, suggesting that the sA159V mutation has a compensatory effect on replication in resistant viral strains. Notably, the sA159V mutation had no effect on LAM and ETV sensitivity.
In view of these results, it is possible that the sA159V mutation increases the fitness of resistant mutants by alleviating anti-HB immune stress and enhancing viral replication competency rather than by directly increasing drug resistance. This is supported by a previous study demonstrating that two classical immune escape-associated mutations, sG145R and sP120T, significantly reduce HBsAg production and increase the replication capacity of LAM-resistant HBV mutants[31]. Of note, our study was based on patient-derived viral strains rather than artificially generated strains, thus providing convincing evidence.
Nevertheless, in vitro experimental data may not always fully reflect in vivo processes. In our study, the sA159V mutant had lower HBsAg production than the WT strain. However, sA159V-positive patients had similar serum HBsAg levels to those of sA159V-negative patients. This bias could be explained by the frequent coexistence of the sA159V mutant with the WT virus in these patients. Although a large number of patients were investigated, the study was restricted by the inability to collect serial samples from many patients.
In summary, we provide evidence supporting the influence of HBV immune escape-associated mutations on drug resistance based on a large-scale clinical investigation. We also found that the sA159V mutation might increase the fitness of LAM/ETV-resistant mutants by decreasing the HBsAg levels and increasing the viral replication capacity. These results provide new insights into the association of HBV immune escape with HBV drug resistance.
ARTICLE HIGHLIGHTS
Research background
A large number of patients were surveyed for both immune escape-associated and drug-resistance mutations.
Research motivation
A link between immune escape-associated and resistance mutations was identified.
Research objectives
The association between immune escape-associated mutations and nucleotide analog resistance mutations was evaluated.
Research methods
Upon follow-up, hepatitis B virus (HBV) sA159V was found to have contributed to resistance in several patients.
Research results
HBV sA159V reduced the hepatitis B surface antigen production but increased the replication capacity of lamivudine (LAM)/entecavir (ETV)-resistant mutants.
Research conclusions
sA159V might increase the fitness of LAM/ETV-resistant mutants under environmental pressure in some cases.
Research perspectives
Immune escape-associated and drug-resistance mutations.
ACKNOWLEDGEMENTS
All authors have read and approve the final manuscript, and the authors would like to thank all the individuals who participated in this study. We are grateful to Dai JZ and Yao ZT for their excellent technical assistance.
Footnotes
Institutional review board statement: The study was reviewed and approved by the Ethics Committee Approval Document of 302 Hospital, Institutional Review Board Approval No. 2012020D; The study was reviewed and approved by the Ethics Committee Approval Document of 302 Hospital, Institutional Review Board Approval No. 2013052D.
Informed consent statement: All study participants, or their legal guardian, provided informed written consent prior to study enrollment.
Conflict-of-interest statement: We do not have any patents, whether planned, pending or issued, broadly relevant to the work.
Manuscript source: Unsolicited manuscript
Peer-review started: April 23, 2020
First decision: May 1, 2020
Article in press: August 12, 2020
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Hossain MG, Yang R S-Editor: Wang DM L-Editor: Filipodia P-Editor: Li JH
Contributor Information
Bi-Xia Huang, Guangdong Provincial Key Laboratory of Medical Molecular Diagnostics/Institute of Biochemistry & Molecular Biology, Guangdong Medical University, Dongguan 523808, Guangdong Province, China.
Yan Liu, Institute of Infectious Diseases, the Fifth Medical Center of Chinese PLA General Hospital, Beijing 100039, China.
Zhen-Ping Fan, Institute of Infectious Diseases, the Fifth Medical Center of Chinese PLA General Hospital, Beijing 100039, China.
Lan-Lan Si, Institute of Infectious Diseases, the Fifth Medical Center of Chinese PLA General Hospital, Beijing 100039, China.
Rong-Juan Chen, Institute of Infectious Diseases, the Fifth Medical Center of Chinese PLA General Hospital, Beijing 100039, China.
Jun Wang, Institute of Infectious Diseases, the Fifth Medical Center of Chinese PLA General Hospital, Beijing 100039, China.
Dan Luo, Department of Infectious Diseases, the First Affiliated Hospital of Xi’an Jiaotong University, Xi’an 710061, Shaanxi Province, China.
Fu-Sheng Wang, Institute of Infectious Diseases, the Fifth Medical Center of Chinese PLA General Hospital, Beijing 100039, China.
Dong-Ping Xu, Institute of Infectious Diseases, the Fifth Medical Center of Chinese PLA General Hospital, Beijing 100039, China. xudongping302@sina.com.
Xin-Guang Liu, Guangdong Provincial Key Laboratory of Medical Molecular Diagnostics/Institute of Biochemistry & Molecular Biology, Guangdong Medical University, Dongguan 523808, Guangdong Province, China.
Data sharing statement
No additional data are available.
References
- 1.Polaris Observatory Collaborators. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol. 2018;3:383–403. doi: 10.1016/S2468-1253(18)30056-6. [DOI] [PubMed] [Google Scholar]
- 2.Tang LSY, Covert E, Wilson E, Kottilil S. Chronic Hepatitis B Infection: A Review. JAMA. 2018;319:1802–1813. doi: 10.1001/jama.2018.3795. [DOI] [PubMed] [Google Scholar]
- 3.Lok AS, Zoulim F, Locarnini S, Bartholomeusz A, Ghany MG, Pawlotsky JM, Liaw YF, Mizokami M, Kuiken C Hepatitis B Virus Drug Resistance Working Group. Antiviral drug-resistant HBV: standardization of nomenclature and assays and recommendations for management. Hepatology. 2007;46:254–265. doi: 10.1002/hep.21698. [DOI] [PubMed] [Google Scholar]
- 4.Tenney DJ, Rose RE, Baldick CJ, Pokornowski KA, Eggers BJ, Fang J, Wichroski MJ, Xu D, Yang J, Wilber RB, Colonno RJ. Long-term monitoring shows hepatitis B virus resistance to entecavir in nucleoside-naïve patients is rare through 5 years of therapy. Hepatology. 2009;49:1503–1514. doi: 10.1002/hep.22841. [DOI] [PubMed] [Google Scholar]
- 5.Zoulim F, Locarnini S. Optimal management of chronic hepatitis B patients with treatment failure and antiviral drug resistance. Liver Int. 2013;33 Suppl 1:116–124. doi: 10.1111/liv.12069. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Y, Wu J, Sun L, Liu G, Li B, Zheng Y, Li X, Tao J. Prevalence of mutations in HBV DNA polymerase gene associated with nucleos(t)ide resistance in treatment-naive patients with Chronic Hepatitis B in Central China. Braz J Infect Dis. 2016;20:173–178. doi: 10.1016/j.bjid.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park ES, Lee AR, Kim DH, Lee JH, Yoo JJ, Ahn SH, Sim H, Park S, Kang HS, Won J, Ha YN, Shin GC, Kwon SY, Park YK, Choi BS, Lee YB, Jeong N, An Y, Ju YS, Yu SJ, Chae HB, Yu KS, Kim YJ, Yoon JH, Zoulim F, Kim KH. Identification of a quadruple mutation that confers tenofovir resistance in chronic hepatitis B patients. J Hepatol. 2019;70:1093–1102. doi: 10.1016/j.jhep.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 8.De Clercq E. Tenofovir alafenamide (TAF) as the successor of tenofovir disoproxil fumarate (TDF) Biochem Pharmacol. 2016;119:1–7. doi: 10.1016/j.bcp.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 9.Carrouée-Durantel S, Durantel D, Werle-Lapostolle B, Pichoud C, Naesens L, Neyts J, Trépo C, Zoulim F. Suboptimal response to adefovir dipivoxil therapy for chronic hepatitis B in nucleoside-naive patients is not due to pre-existing drug-resistant mutants. Antivir Ther. 2008;13:381–388. [PubMed] [Google Scholar]
- 10.Rajoriya N, Combet C, Zoulim F, Janssen HLA. How viral genetic variants and genotypes influence disease and treatment outcome of chronic hepatitis B. Time for an individualised approach? J Hepatol. 2017;67:1281–1297. doi: 10.1016/j.jhep.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 11.Xue Y, Wang MJ, Yang ZT, Yu DM, Han Y, Huang D, Zhang DH, Zhang XX. Clinical features and viral quasispecies characteristics associated with infection by the hepatitis B virus G145R immune escape mutant. Emerg Microbes Infect. 2017;6:e15. doi: 10.1038/emi.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shirvani-Dastgerdi E, Winer BY, Celià-Terrassa T, Kang Y, Tabernero D, Yagmur E, Rodríguez-Frías F, Gregori J, Luedde T, Trautwein C, Ploss A, Tacke F. Selection of the highly replicative and partially multidrug resistant rtS78T HBV polymerase mutation during TDF-ETV combination therapy. J Hepatol. 2017;67:246–254. doi: 10.1016/j.jhep.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colagrossi L, Hermans LE, Salpini R, Di Carlo D, Pas SD, Alvarez M, Ben-Ari Z, Boland G, Bruzzone B, Coppola N, Seguin-Devaux C, Dyda T, Garcia F, Kaiser R, Köse S, Krarup H, Lazarevic I, Lunar MM, Maylin S, Micheli V, Mor O, Paraschiv S, Paraskevis D, Poljak M, Puchhammer-Stöckl E, Simon F, Stanojevic M, Stene-Johansen K, Tihic N, Trimoulet P, Verheyen J, Vince A, Lepej SZ, Weis N, Yalcinkaya T, Boucher CAB, Wensing AMJ, Perno CF, Svicher V HEPVIR working group of the European Society for translational antiviral research (ESAR) Immune-escape mutations and stop-codons in HBsAg develop in a large proportion of patients with chronic HBV infection exposed to anti-HBV drugs in Europe. BMC Infect Dis. 2018;18:251. doi: 10.1186/s12879-018-3161-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao L, Li X, Cheng Y, Chen R, Shao J, Zhou Y, Li Q, Liao H, Zhao Y, Liu L, Su H, Liu Y, Liu Y, Xu D. Hepatitis B virus rtA181T/sW172non-stop mutation may increase resistance fold to adefovir- and entecavir-resistant mutants compared to rtA181T/sW172* mutation. Antiviral Res. 2018;154:26–34. doi: 10.1016/j.antiviral.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Luo D, Liu Y, Chen R, Niu M, Liu L, Li X, Li Q, Huang B, Wang J, Xu D, Lin S. Investigation of hepatitis B virus (HBV) rtS78T/sC69* mutation in a large cohort of chronic HBV-infected patients with nucleoside/nucleotide analogue treatment. Antiviral Res. 2019;170:104579. doi: 10.1016/j.antiviral.2019.104579. [DOI] [PubMed] [Google Scholar]
- 16.Ganji A, Esmaeilzadeh A, Ghafarzadegan K, Helalat H, Rafatpanah H, Mokhtarifar A. Correlation between HBsAg quantitative assay results and HBV DNA levels in chronic HBV. Hepat Mon. 2011;11:342–345. [PMC free article] [PubMed] [Google Scholar]
- 17.Chen BF. Hepatitis B virus pre-S/S variants in liver diseases. World J Gastroenterol. 2018;24:1507–1520. doi: 10.3748/wjg.v24.i14.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding H, Liu B, Zhao C, Yang J, Yan C, Yan L, Zhuang H, Li T. Amino acid similarities and divergences in the small surface proteins of genotype C hepatitis B viruses between nucleos(t)ide analogue-naïve and lamivudine-treated patients with chronic hepatitis B. Antiviral Res. 2014;102:29–34. doi: 10.1016/j.antiviral.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 19.Shan M, Shen Z, Sun H, Zheng J, Zhang M. The enrichment of HBV immune-escape mutations during nucleoside/nucleotide analogue therapy. Antivir Ther. 2017;22:717–720. doi: 10.3851/IMP3156. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Liu Y, Zhao J, Xu Z, Chen R, Si L, Lu S, Li X, Wang S, Zhang K, Li J, Han J, Xu D. Characterization of Novel Hepatitis B Virus PreS/S-Gene Mutations in a Patient with Occult Hepatitis B Virus Infection. PLoS One. 2016;11:e0155654. doi: 10.1371/journal.pone.0155654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang K, Liu Y, Chen R, Li Q, Xu Z, Si L, Cheng Y, Yang Y, Chen J, Xu D, Lin S. Antigenicity reduction contributes mostly to poor detectability of HBsAg by hepatitis B virus (HBV) S-gene mutants isolated from individuals with occult HBV infection. J Med Virol. 2018;90:263–270. doi: 10.1002/jmv.24936. [DOI] [PubMed] [Google Scholar]
- 22.Chinese Society of Hepatology, Chinese Medical Association. Chinese Society of Infectious Diseases, Chinese Medical Association. Guideline on prevention and treatment of chronic hepatitis B in China (2005) Chin Med J (Engl) 2007;120:2159–2173. [PubMed] [Google Scholar]
- 23.Xu Z, Liu Y, Xu T, Chen L, Si L, Wang Y, Ren X, Zhong Y, Zhao J, Xu D. Acute hepatitis B infection associated with drug-resistant hepatitis B virus. J Clin Virol. 2010;48:270–274. doi: 10.1016/j.jcv.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Zhou Y, Li X, Niu M, Chen R, Shao J, Si L, Luo D, Lin Y, Li L, Zhang K, Xiao X, Xu Z, Liu M, Lu M, Zoulim F, Xu D. Hepatitis B virus mutation pattern rtL180M+A181C+M204V may contribute to entecavir resistance in clinical practice. Emerg Microbes Infect. 2019;8:354–365. doi: 10.1080/22221751.2019.1584018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L, Liu Y, Chen R, Li X, Luo D, Zhao Y, Li Q, Huang B, Wang FS, Liu X, Xu D. Prevalence of the entecavir-resistance-inducing mutation rtA186T in a large cohort of Chinese hepatitis B virus patients. Antiviral Res. 2019;164:131–138. doi: 10.1016/j.antiviral.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Wu C, Chen R, Li X, Xu Z, Li Q, Li L, Wang FS, Yang D, Lu M, Xu D. Molecular cloning and phenotypic analysis of drug-resistance mutants with relevant S-region variants of HBV for a patient during 189-month anti-HBV treatment. Antivir Ther. 2019;24:237–246. doi: 10.3851/IMP3305. [DOI] [PubMed] [Google Scholar]
- 27.Pollicino T, Cacciola I, Saffioti F, Raimondo G. Hepatitis B virus PreS/S gene variants: pathobiology and clinical implications. J Hepatol. 2014;61:408–417. doi: 10.1016/j.jhep.2014.04.041. [DOI] [PubMed] [Google Scholar]
- 28.Tang Z, Li X, Wu S, Liu Y, Qiao Y, Xu D, Li J. Risk of hepatitis B reactivation in HBsAg-negative/HBcAb-positive patients with undetectable serum HBV DNA after treatment with rituximab for lymphoma: a meta-analysis. Hepatol Int. 2017;11:429–433. doi: 10.1007/s12072-017-9817-y. [DOI] [PubMed] [Google Scholar]
- 29.Hsu HY, Chang MH, Ni YH, Chiang CL, Wu JF, Chen HL. Universal infant immunization and occult hepatitis B virus infection in children and adolescents: a population-based study. Hepatology. 2015;61:1183–1191. doi: 10.1002/hep.27650. [DOI] [PubMed] [Google Scholar]
- 30.Wu CC, Chen YS, Cao L, Chen XW, Lu MJ. Hepatitis B virus infection: Defective surface antigen expression and pathogenesis. World J Gastroenterol. 2018;24:3488–3499. doi: 10.3748/wjg.v24.i31.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amini-Bavil-Olyaee S, Vucur M, Luedde T, Trautwein C, Tacke F. Differential impact of immune escape mutations G145R and P120T on the replication of lamivudine-resistant hepatitis B virus e antigen-positive and -negative strains. J Virol. 2010;84:1026–1033. doi: 10.1128/JVI.01796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No additional data are available.