Abstract
Bacterial pathogens that cause invasive disease in the vertebrate host must adapt to host efforts to cripple their viability. Major host insults are reactive oxygen and reactive nitrogen species as well as cellular stress induced by antibiotics. Hydrogen sulfide (H2S) is emerging as an important player in cytoprotection against these stressors, which may well be attributed to downstream more oxidized sulfur species termed reactive sulfur species (RSS). In this review, we summarize recent work that suggests that H2S/RSS impacts bacterial survival in infected cells and animals. We discuss the mechanisms of biogenesis and clearance of RSS in the context of a bacterial H2S/RSS homeostasis model and the bacterial transcriptional regulatory proteins that act as “sensors” of cellular RSS that maintain H2S/RSS homeostasis. In addition, we cover fluorescence imaging– and MS–based approaches used to detect and quantify RSS in bacterial cells. Last, we discuss proteome persulfidation (S-sulfuration) as a potential mediator of H2S/RSS signaling in bacteria in the context of the writer-reader-eraser paradigm, and progress toward ascribing regulatory significance to this widespread post-translational modification.
Keywords: hydrogen sulfide, reactive sulfur species, persulfidation, thiol chemistry, cysteine, host-pathogen interface, antibiotics, antibiotic resistance, post-translational modification (PTM), proteomics, microbial pathogenesis, microbiology, sulfhydryl, persulfide, S-sulfuration
Infectious disease is a global and significant threat to human health. There is an increasingly urgent need to develop new antimicrobial strategies to combat these increasingly drug-resistant and life-threatening pathogens (1, 2). One important approach to do this is to understand bacterial adaptation to the myriad of host immune responses that have evolved to clear bacterial infections. For example, transition metal homeostasis (metallostasis) (3) effectively controls the metalation status of the proteome (4, 5). Upon infection, the host actively disrupts metallostasis by restricting access (6–8) or intoxicating cells with metals (9, 10) to limit bacterial growth. Pathogens, in turn, adapt by employing specialized transcriptional regulators, metallosensors, that sense metals and regulate the expression of genes encoding proteins that collectively maintain bioavailable metal in a range compatible with physiological needs (Fig. 1A, top) (11–14). In an analogous fashion, bacteria encode specialized transcriptional regulators that sense oxidized or “reactive” sulfur species (RSS), derived from hydrogen sulfide (H2S) (15–20). As cellular concentrations of RSS rise, RSS sensors turn on the expression of genes that encode enzymes that reduce cellular loads of H2S/RSS to avoid H2S toxicity and overpersulfidation of the metabolome and proteome (Fig. 1A, bottom). These RSS sensors, like metallosensors, control H2S/RSS homeostasis, allowing bacterial cells access to these molecules at low concentrations to meet physiological needs. In the infected host, H2S and RSS are derived from host cell metabolism, from commensal bacteria in polymicrobial communities, or from the pathogen itself. Recent studies that build on prior work (21) suggest that bacterial H2S biogenesis may well be a clinically important adaptive response during infections (22–26).
Figure 1.

Set-point homeostasis models and speciation in bacteria. A, transition metal homeostasis (top) is orchestrated by a panel of metal-specific sensors that prevent metal starvation or toxicity by regulating the expression of proteins involved in the uptake, efflux, storage, or allocation of metals in cells (3, 4, 11–14). Transcriptional response curves are shown for a pair of sensors that detect a specific metal (e.g. ZnII). These dual sensors collaboratively control metal bioavailability in a concentration range that is compatible with cellular physiology (gray box). H2S/RSS homeostasis (bottom) is achieved by a single RSS sensor that transcriptionally regulates the expression of enzymes involved in the biogenesis, clearance, transport, and assimilation of H2S/RSS (16–20). The transcriptional response of an RSS sensor detects a concentration range (gray box) that prevents cellular toxicity, while maintaining access to H2S/RSS that is physiologically beneficial at lower concentrations. B, metal speciation (top) of first row, late d-block transition metals is defined by the metallome, a descriptor of all oxidation states and coordination complexes in the cell, ranging from exchange-labile small-molecule metal complexes to protein cofactors (shown in cartoon form). Reactive sulfur speciation (bottom) is defined by all inorganic and organic small molecules that harbor sulfur atoms in oxidation states more positive than –2 (see key) (27) and are collectively termed reactive sulfur species (RSS).
A second feature that is common to metallostasis and H2S/RSS homeostasis, beyond the sensors themselves, is the concept of speciation (Fig. 1B). In metallostasis, speciation defines the metallome, or all coordination complexes, both small molecule and protein, and oxidation states of all transition metals in the cell (Fig. 1B, top). Metallosensors surveil the cytoplasm for some specific feature of the metallome (e.g. zinc in exchange-labile complexes) and alter gene expression upon metal binding. In H2S/RSS homeostasis, speciation is defined by the components of the RSS pool, which encompasses organic and inorganic molecules containing sulfur in oxidation states higher than H2S, many of which contain sulfur-bonded or “sulfane” sulfur (Fig. 1B, bottom) (27). Analogous to a metallosensor, known RSS sensors specifically surveil the cytoplasm for a particular feature of the RSS pool, in this case sulfane sulfur (15–20).
In this review, we summarize the biogenesis and clearance of H2S/RSS and the potential role these molecules play in bacterial infections. In addition, we discuss the molecular mechanisms of RSS sensors that maintain H2S/RSS homeostasis in bacteria. Elucidation of how H2S/RSS are leveraged in bacteria at the host-pathogen interface relies on the development of molecular tools to identify, detect, and quantify H2S and RSS as well as small-molecule probes to generate these species in vitro or in vivo. Last, we discuss recent efforts to detect and understand the regulatory significance of protein persulfidation (S-sulfuration) in bacteria.
Hydrogen sulfide and reactive sulfur species in bacteria
H2S is an electron-rich molecule historically well-known to drive photosynthesis (28) and energy metabolism in sulfide-oxidizing and sulfate- or sulfite-reducing microorganisms (29, 30). In 2011, Nudler and co-workers (21) reported that endogenously synthesized H2S or application of exogenous sulfide salts protected multiple bacterial pathogens against a broad array of mechanistically distinct antibiotics when grown in culture. This initial report, despite few insights into a possible mechanism, suggested that H2S might have beneficial properties in human disease–causing microorganisms and has thus inspired considerable research over the last 10 years. These bacteria endogenously synthesize H2S utilizing bacterial homologs of the mammalian reverse transsulfuration pathway via “side” reactions catalyzed by cystathionine β-synthase (CBS) and cystathionine γ-lyase (CSE) (31–34) or from cysteine catabolism to 3-mercaptopyruvate (3-MP) via cysteine aminotransferase (CAT) (35) (Fig. 2A). 3-MP is then converted to pyruvate and H2S by 3-MP sulfurtransferase (3MST) (36–38) via the intermediacy of a protein persulfide, E-SSH (Fig. 2A). Bacteria generally encode either 3MST or CBS/CSE, and it was recently demonstrated that L-cysteine desulfhydrases and cysteine desulfurases also contribute to H2S biogenesis in Escherichia coli (39, 40). In addition, two groups recently reported the discovery of a glycyl radical enzyme from Bilophila wadsworthia that catalyzes C–S bond cleavage in the catabolism of tissue-abundant taurine and the analogous alcohol isethionate (2-hydroxyethanesulfonate) (41, 42). This reaction produces sulfite (), which is reduced to H2S by a dissimilatory sulfite reductase, thus defining a novel pathway for H2S production by gut microbiota.
Figure 2.
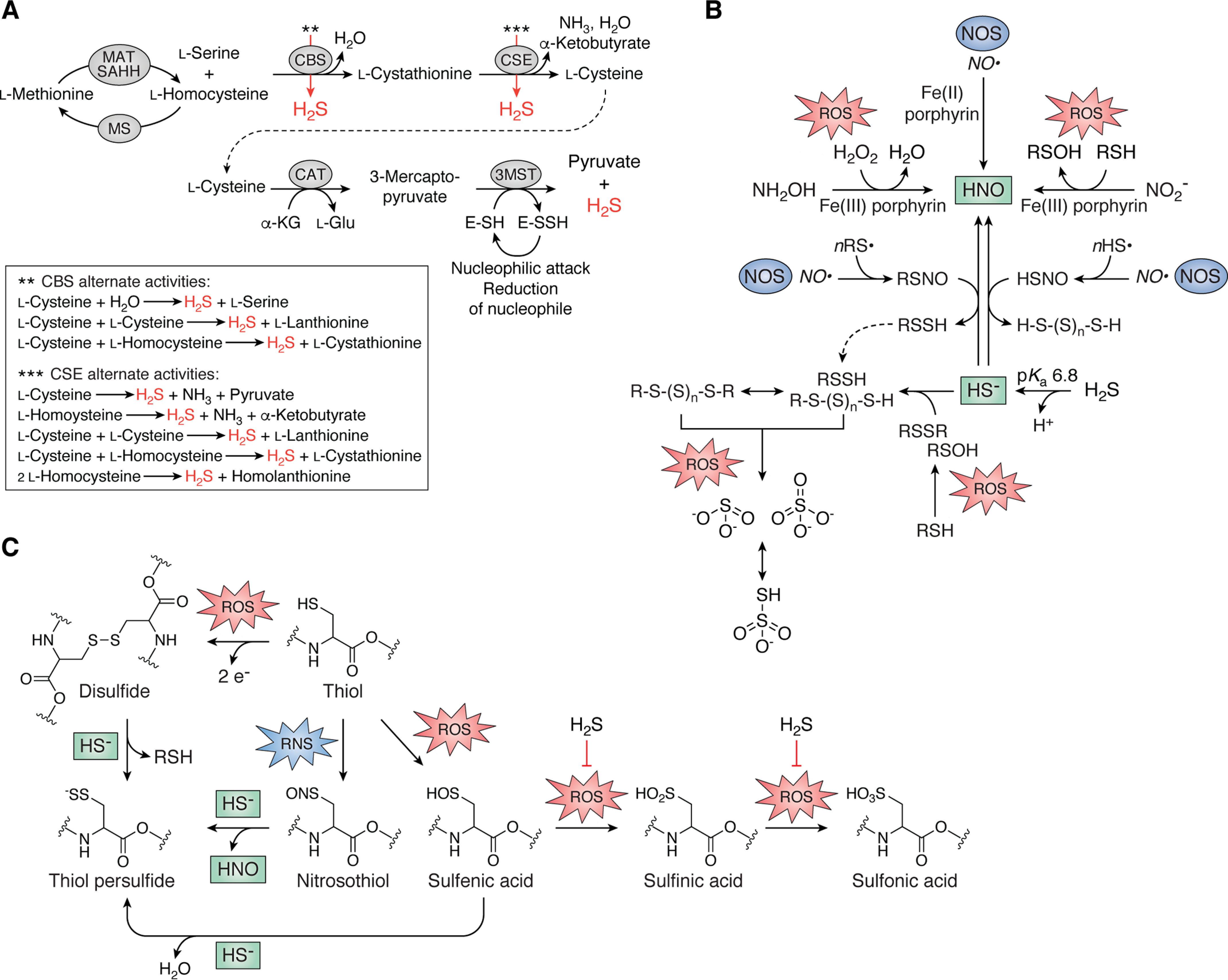
Endogenous production of H2S and cross-talk between H2S/RSS and ROS/RNS in bacteria. A, the reverse transsulfuration pathway synthesizes L-cysteine from L-homocysteine via the intermediacy of L-cystathionine, which is catabolized by CAT, whose product is utilized by 3MST to generate pyruvate and H2S (31–38). L-Homocysteine is also the immediate precursor to L-methionine. Both CBS and CSE catalyze a number of additional reactions with alternative substrates (red arrows) that generate endogenous H2S (43), illustrated in the dashed box. CBS, cystathionine-β-synthase, CSE, cystathionine-γ-lyase; MAT, methionine adenosyltransferase; MS, methionine synthase; SAHH, SAH hydrolase. B, small-molecule cross-talk between H2S/RSS and RNS and ROS. C, direct reaction of HS– with protein nitrosothiols or disulfides and sulfenic acids induced by RNS and ROS, respectively, all result in the formation of the thiol persulfide with release of HNO, protein thiol (RSH), and water, respectively. Reaction of HS– with sulfenic acids is believed to protect these protein thiols from irreversible overoxidation, indicated by the block arrow (right).
With a sulfur oxidation state of “–2”, H2S and organic thiols (e.g. cysteine or GSH (RSH)) are in their most reduced forms and can only function as cellular reductants (27, 43). RSS harbor higher sulfur oxidation states, ranging from “–1” to “+6” (Fig. 1B, bottom). The organic thiol persulfide (hydropersulfide, RSSH) is of particular interest because of its “Janus” character and can function as either a nucleophile when deprotonated (RSS−) or an electrophile when protonated (RSSH). Due to a considerably lower pKa than the corresponding thiol, the anionic form predominates at physiological pH (43–45). Persulfides also have enhanced nucleophilicity compared with their corresponding thiolate because of the α-effect (46), which increases the reactivity of the terminal sulfur atom because of unpaired electrons in the adjacent atom.
Persulfides readily react with oxidants such as hydrogen peroxide (H2O2) and peroxynitrite (44, 47) and are superior one-electron reductants to thiols and H2S as reviewed elsewhere (48–50). Their Janus character, enhanced nucleophilicity, and superior reducing capabilities make RSSH, along with organic polysulfides and their inorganic counterparts, potent antioxidants (Fig. 1B, bottom) (43, 51, 52). These properties may well be responsible for many of the beneficial traits attributed to H2S, including protection against oxidative stress and antibiotics in the infected host (21, 26, 47).
Recent work by several groups reveals significant physiological overlap or cross-talk between H2S/RSS and reactive oxygen (ROS) and reactive nitrogen (RNS) species. Oxidation of RSS results in the production of inorganic sulfur-containing molecules sulfite, thiosulfate, and sulfate (Fig. 2B) (51). ROS can also drive the formation of low-molecular weight (LMW) thiol disulfides (RSSR) and sulfenic acids (RSOH), a major physiological marker of H2O2 reactivity, which reacts with HS– to form organic RSS (Fig. 2B). H2S and nitric oxide (NO·) intersect via nitroxyl (HNO), and incubation of bacterial cells with a nitroxyl donor, Angeli's salt, results in an increase in cellular levels of RSS in Staphylococcus aureus (53) possibly via thionitrous acid (HSNO) or nitrosopersulfide (SSNO–) formation (Fig. 2B) (54–56). In addition, polysulfides can by synthesized from incubation of RSH with sodium nitrite () to form organic nitrosothiols (RSNO), which readily react with HS− at acidic pH to form a mixture of RS–Sn–SR, consistent with proposed H2S/NO· cross-talk (57). Last, protein persulfidation (S-sulfuration) is now a widely recognized post-translational modification (PTM) believed to function in H2S signaling alongside, and possibly interconverting with, other thiol modifications, including S-thiolation (RSSR′), S-nitrosation (RSNO), or oxidation to sulfenic, sulfinic (RSO2H), and sulfonic acids (RSO3H) (Fig. 2C) (43, 58–60). It is important to note that the chemistry presented here between H2S/RSS and ROS/RNS can potentially occur on both small-molecule and protein thiols (Fig. 2, B and C). Furthermore, the onslaught of host-generated ROS and RNS at sites of infection suggests this chemical cross-talk may be biologically relevant in the infected host (61–64).
Physiological conditions for the production, regulation, and signaling of H2S/RSS in bacteria
H2S and the gut microbiome
The gut microbiome is a complex, nutrient-rich environment and host to well over 100 bacterial species (65, 66). The microorganisms that inhabit this niche are a significant endogenous source of sulfur-containing compounds and H2S, the latter estimated to range from ∼0.2 to 2.4 mm (Fig. 3A) (67). Methionine catabolism to cysteine via the reverse transsulfuration pathway (Fig. 2A) and the catabolism of organic sulfonates, notably taurine, are known to be catalyzed by gut microbiota (41, 42, 68–72). This niche is also home to sulfate-reducing bacteria that are responsible for significant production of H2S (73, 74), in addition to the reduction of tetrathionate and thiosulfate to H2S that also occurs in the gut (75). H2S produced by the gut microbiota is oxidized by gastrointestinal epithelial cells to thiosulfate and tetrathionate (76, 77); these molecules in turn are utilized by the microbiota as electron acceptors, resulting in a symbiotic relationship derived from interconversion of sulfur species (Fig. 3A) (72). Perturbation of this symbiotic relationship results in the accumulation of H2S now linked to several gut-derived diseases (78–80). Interestingly, gut inflammation caused by the pathogen Salmonella enterica serovar Typhimurium results in increased production of tetrathionate from the oxidation of thiosulfate by gut inflammation–derived ROS, which this bacterium uses as an alternate electron acceptor, thus providing a growth advantage in this niche (81).
Figure 3.
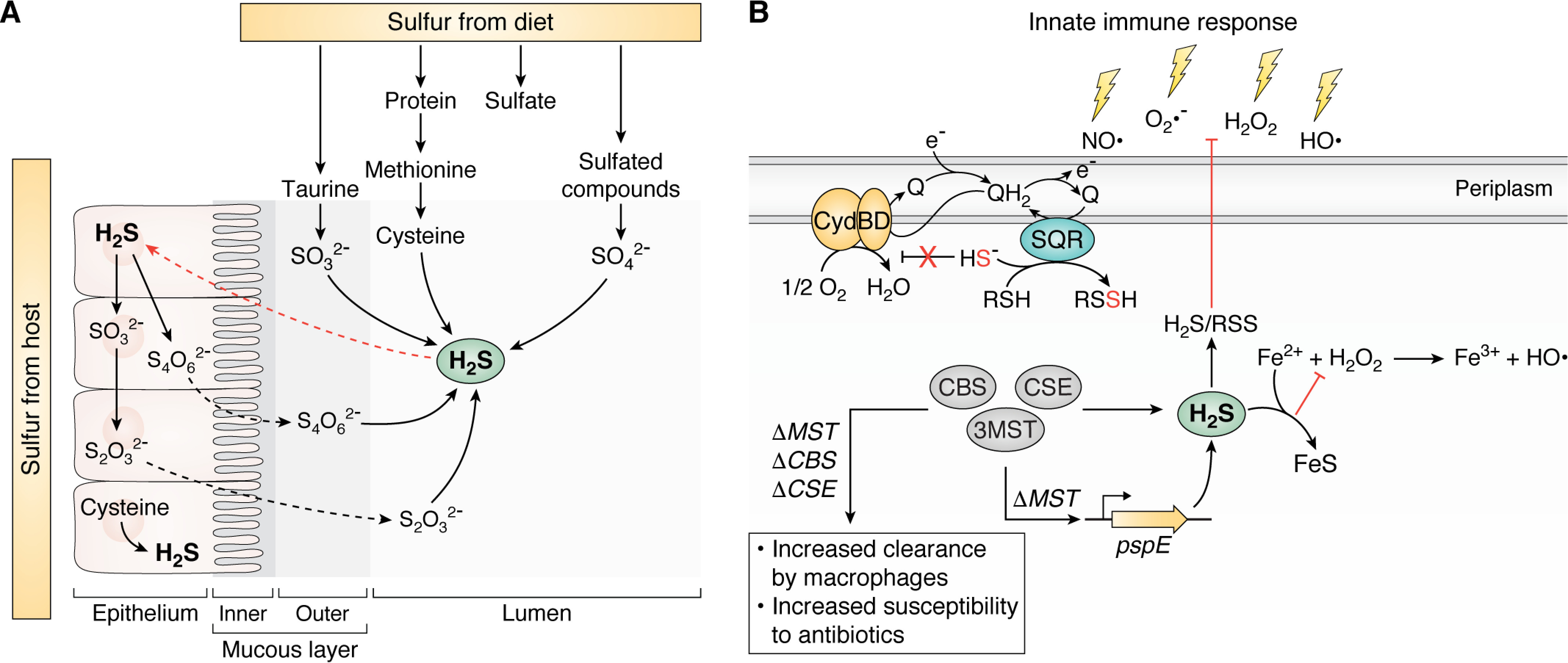
H2S and RSS at the host-pathogen interface. A, common sulfur sources in the gastrointestinal tract derived from host epithelial cells and dietary sulfur metabolized by the gut microbiota. B, endogenous production of H2S via CBS, CSE, and/or 3MST (see Fig. 2A) or other enzymatic processes and more oxidized RSS are cytoprotective against myriad stressors of innate immune response or by antibiotics. See section on physiological conditions for the production, regulation, and signaling of H2S/RSS in bacteria for details. PspE, single-domain sulfurtransferase in E. coli; Q/QH2, quinone; SQR, sulfide:quinone oxidoreductase.
H2S at the host-pathogen interface
There are two major physiological conditions within the infected host where H2S and downstream RSS may enhance bacterial survival in what is otherwise a hostile microenvironment. These are (i) resistance against myriad oxidative stressors and antibiotic challenge and (ii) H2S/RSS-dependent regulation of biofilm dynamics. The host immune system produces diverse ROS, including (82–84), hydrogen peroxide (H2O2) (85), and hydroxy radical (OH·) (86) to combat bacterial infections (87). Antibiotics are also thought to induce generalized oxidative stress (88–91), although this has been widely debated in recent years (92, 93). The first documentation that H2S impacted antibiotic resistance in E. coli was published 47 years ago (94). Renewed interest came in 2011 when it was demonstrated that bacterially derived H2S conferred resistance to a broad range of antibiotics in several bacterial pathogens (21). Bacterially derived NO· has also been found to provide protection against antibiotics by the same group (95) perhaps because of H2S/NO· cross-talk, which has only been investigated in recent years (Fig. 2B) (53–56). It was not until 2014 that RSS were shown to function as antioxidants in mammalian cells (47), which may partly explain H2S-enhanced bacterial resistance to antibiotics (21). RSS have been detected in a number of bacterial pathogens (17, 20, 53, 96) and are the subject of ongoing work to better understand the role of RSS in the bacterial response to the host immune system.
Recent work has investigated the mechanism by which H2S/RSS might confer antibiotic resistance and protection against ROS and sulfide toxicity (Fig. 3B). In E. coli, increased H2S results in a respiratory flux switch from that of the primary cytochrome bo oxidase to the alternate cytochrome bd oxidase, a copper-free enzyme that is far less susceptible to inhibition by H2S (26, 97). Although cytochrome bd oxidase does not pump protons, it still enables aerobic metabolism and robust growth. This respiratory switch in response to H2S may well occur in several other bacterial pathogens, including Acinetobacter baumannii (20) and Mycobacterium tuberculosis; in the latter case, low levels of H2S enhance the respiration, energy production, and survival of M. tuberculosis in infected mice (25). Further studies are required to establish the generality of this adaptive response across a wider range of organisms. Others have posited that H2S-mediated cytoprotection occurs via sequestration of the prooxidant free Fe(II) (23, 98); however, this remains incompletely understood.
A number of recent reports have described a potential role of H2S at the host-pathogen interface beyond protection against antibiotic and oxidative stress (22, 24, 25, 99). In infected macrophages and in mice, Helicobacter pylori was found to induce the expression of the host transsulfuration pathway enzyme CSE (Fig. 2A), resulting in increased cystathionine production that enhances H. pylori survival in these models (99). Any connection of cystathionine to host or bacterially derived H2S was not elucidated in this work. S. aureus and E. coli strains lacking enzymes involved in H2S biogenesis are more readily cleared in infected macrophages and are less resistant to leukocyte-mediated killing in a burn-infection model (22). In addition, E. coli strains lacking the H2S-generating enzyme 3MST, when challenged with antibiotics, give rise to a suppressor mutation that recovers H2S biogenesis via up-regulation of the single-domain sulfurtransferase PspE (24). Together, these studies suggest that H2S biogenesis reduces the efficacy of antibiotics and that up-regulation of H2S may be a clinically important adaptive response during infections. These studies support the proposal that H2S functions as an infection-relevant antioxidant or pro-antioxidant, in the latter case, as a precursor to oxidized RSS (47).
Whereas H2S/RSS-dependent regulation of biofilm dynamics remains largely unknown, recent studies suggest a potential connection. Biofilms are often polymicrobial communities that assemble on both abiotic (e.g. catheters and implants) and biotic (e.g. cells and cell debris) surfaces while conferring increased resistance to antibiotics (100–102). Cells near the base of biofilm structures are often nutrient-poor, and some reside at an oxic/anoxic boundary. Low-O2 (hypoxic) conditions can also result from increased O2 consumption by host immune cells to produce superoxide anion () (103, 104). In these low-O2 regions, bacteria respire via reduction of nitrate (), producing NO· on pathway to nitrous oxide (N2O) and dinitrogen (N2) (105). These nitrogen-containing species have been reported to lead to biofilm dissemination of S. aureus (106) and P. aeruginosa (107, 108), consistent with an impact on biofilm dynamics.
Redox homeostasis is also implicated in proper biofilm formation in P. aeruginosa (109), whereas cysteine and GSH-deficient uropathogenic E. coli exhibit dysregulated biofilm formation that is restored upon the addition of exogenous thiols (110). Although the connection between biofilm regulation and H2S/RSS homeostasis is largely speculation at this point, H2S has been detected in cystic fibrosis sputum, a complex biofilm (111), and H2S has been found to promote formation of biofilms by intestinal microbiota while reducing the proliferation of planktonic bacterial cells (112). We recently characterized the biofilm growth–associated repressor, BigR, in A. baumannii as an RSS sensor (20, 113), as previously characterized in plant pathogens (114–116). That work also identified two transcriptional regulators in A. baumannii known or projected to be involved in biofilm regulation that were characterized by significantly increased protein persulfidation mediated by exogenous sulfide (20). Whereas these studies suggest that H2S/RSS homeostasis impacts biofilm dynamics, more studies are needed to better understand this connection at a mechanistic level.
Biogenesis and clearance of organic RSS in bacteria
The endogenous production of H2S in bacteria suggests that more oxidized forms of sulfur may be present in cells and formed via enzymatic and possibly nonenzymatic mechanisms (Fig. 4, A–C). The extent to which these pathways, particularly nonenzymatic routes, contribute to RSS pools in bacteria is not known and may well differ among organisms. Emerging evidence in mammalian systems demonstrates the role of ferric (FeIII)-heme in the oxidation of H2S, which reduces the FeIII to FeII and forms the one-electron oxidized radical HS· upon dissociation (Fig. 4A) (117–121). Recent work reveals that this mechanism is used to reactivate enzymes requiring a catalytically active ferrous heme from the inactive ferric state, formed during turnover (121). Additionally, HS· can be formed by the reaction of H2S with superoxide radical anion or with cysteine-coordinated Zn(II) sites in proteins (122). FeIII-heme has also been shown to result in formation of thiosulfate and hydropolysulfide species in mammalian systems (Fig. 4A) (117, 118). Formation of organic thiyl radical, RS·, and related organic polysulfide species may occur via similar chemistry, supported by recent work using an LMW thiol for the reactivation of a catalytically active ferrous heme (Fig. 4B) (121). The extent to which heme-based biogenesis of RSS occurs in bacteria is not yet known.
Figure 4.

Biogenesis and clearance of organic and inorganic RSS in bacteria. A and B, biogenesis of inorganic (A) and organic (B) RSS via enzymatic or nonenzymatic processes (43, 51). In A, the biogenesis of thiosulfate shown is a schematic rendering only. C, transsulfuration reactions that may impact sulfur speciation and assimilation catalyzed by sulfurtransferases (STR) (126), peroxiredoxins (Prx) (136), or other thiol-containing enzymes or via nonenzymatic interconversion among organic LMW thiol/persulfides or inorganic RSS. D, enzymatic clearance of RSS known to occur in bacteria (17, 76, 138, 139). Red sulfurs indicate inorganic sulfur additions. Characterized enzymes are represented by blue ovals, whereas incompletely understood enzymatic reactions are shown as gray ovals and dashed arrows. CoAPR, CoA persulfide reductase; CstB, S. aureus cst operon-encoded persulfide dioxygenase-rhodanese fusion; 3MST, 3-mercaptopyruvate sulfurtransferase; PDO, persulfide dioxygenase; PRF, persulfide dioxygenase-rhodanese fusion; Q/QH2, quinone; SQR, sulfide:quinone oxidoreductase.
An important enzymatic route to the biogenesis of RSS in bacteria is the sulfide:quinone oxidoreductase (SQR) (76, 123–125). SQR catalyzes the two-electron oxidation of H2S to sulfane sulfur fixed as organic and inorganic per- and polysulfides (Fig. 4B), concomitant with reduction of the quinone pool (126). This enzyme may well provide a source of electrons for the alternative cytochrome bd oxidase in organisms that encode this alternate oxidase, analogous to that observed for SQR with complex III/IV when the concentration of H2S is low (Fig. 3B) (76). In organisms (e.g. Enteroccocus faecalis) that do not appear to encode an SQR but where RSS have been detected and quantified (17), the mechanism of RSS biogenesis is not known, and may well suggest a role for nonenzymatic or as yet uncharacterized enzymatic processes in these organisms. In addition to SQR, recent work in A. baumannii reveals that 3MST may also contribute to pools of LMW persulfides, although there are clearly other contributors (20).
Sulfurtransferases
Major structural classes of sulfurtransferases (STRs) adopt either a rhodanese or TusA (tRNA 2-thiouridine–synthesizing protein A)-like fold (126) and harbor an active-site cysteine that is known or projected to function in interdomain or intermolecular persulfide transfer, termed transsulfuration (Fig. 4C) (127). Although rhodanese domains were originally believed to function in cyanide (CN−) detoxification by forming thiocyanide (SCN−) (128), it is well-established that Fe-S cluster biogenesis, molybdenum cofactor biosynthesis, 2-thiouridine synthesis, and thiamine pyrophosphate biosynthesis are known or proposed to use STRs as persulfide transfer catalysts (129–133). Such “targeted” transsulfuration reactions require specific, albeit likely transient, interactions between donor and acceptor and an exposed active site, as described for TSTD1 and thioredoxin in colon epithelial cells (134).
“Orphan” STRs, which we define as not yet connected to any biosynthetic pathway, particularly those regulated by RSS sensors in bacteria, may well play roles in sulfide detoxification or assimilation (17, 135), but their biological functions remain enigmatic. This remains a significant challenge in the field. RSS sensor–regulated STRs are often kinetically characterized in vitro as sulfurtransferases from a thiosulfate donor to a CN− acceptor; however, their physiological donors and acceptors, whether they be small molecules or proteins, have generally not been identified, and any role in targeted transsulfuration has not been established (Fig. 4C). Recently, a single cysteine peroxiredoxin (a major H2O2-detoxifying enzyme) characterized by a long-lived sulfenylated intermediate was shown to rapidly react with H2S to form a protein persulfide, which participated in persulfide transfer to a thiol acceptor (136). This suggests that peroxiredoxins may function in transsulfuration, but this requires further investigation (Fig. 4C). Similarly, the extent to which small-molecule RSS species, particularly those containing sulfane sulfur, participate in transsulfuration reactions with each other, LMW thiols, or even protein thiols is largely unknown (Fig. 4C).
RSS clearance enzymes
In addition to the biogenesis of RSS, a number of bacterial enzymes have been characterized that function in their clearance (Fig. 4D). A well-known player in the clearance of organic persulfides is persulfide dioxygenase (PDO), which harbors a mononuclear, nonheme FeII center (76, 137–140). In bacteria, PDOs have been characterized as single or multidomain enzymes, and the presence of additional domains appears to impact the distribution of products. All PDOs, regardless of their domain organization, use molecular oxygen to oxidize the terminal sulfur of an RSSH substrate to sulfite, which, for a single-domain PDO, is the final product. Some PDOs have an appended STR domain, and these have been designated PDO-rhodanese fusion proteins (PRFs) (Fig. 4D). In the case of the PRF characterized in Burkholderia phytofirmans, the C-terminal rhodanese domain generates the GSH persulfide substrate that the PDO domain then oxidizes to sulfite (138). In contrast, the multidomain PRF CstB from S. aureus oxidizes two equivalents of persulfide substrate to thiosulfate as the final product; the C-terminal rhodanese domain also possesses transsulfuration and thiosulfate transferase activity (139). In contrast to the oxidative chemistry of PDOs, E. faecalis encodes a CoA disulfide reductase-rhodanese homology domain fusion protein (CoADR-RHD) that specifically reduces CoA persulfide to form the reduced thiol and H2S and is thus a CoA persulfide reductase (CoAPR) (Fig. 4D) (17, 141–143).
Regulatory sensing of RSS in bacteria
The discovery of endogenous H2S production and pathways for the biogenesis and clearance of RSS in bacteria requires a mechanism to establish cellular H2S/RSS homeostasis. This is mediated by RSS sensors (Fig. 1A, bottom). We and others have discovered and characterized structurally diverse transcriptional regulators that react with RSS to drive transcriptional derepression or activation of genes encoding common sulfide detoxification or oxidation enzymes described above. These RSS sensors are widespread and have been identified in both Gram-positive and Gram-negative organisms. They include CstR from S. aureus (15, 16) and E. faecalis (17), SqrR from Rhodobacter capsulatus (18, 113), the SqrR homolog BigR from Xylella fastidiosa (114–116) and A. baumannii (20), and FisR from Cupriavidus pinatubonensis (19) and A. baumannii (20).
CstR
The CsoR-like sulfurtransferase repressor, CstR, is a member of the CsoR (copper-sensitive operon repressor) family of transcriptional repressors (144) and was first discovered in S. aureus (15, 16). S. aureus CstR regulates the cst operon encoding a multidomain STR (CstA), a PRF (CstB), and a type II SQR, rather analogous to the well-studied mitochondrial sulfide detoxification system (76, 123, 135, 139). The enzymes encoded by the S. aureus cst operon oxidize sulfide to thiosulfate via persulfide intermediates and have been extensively reviewed elsewhere (126). Recently, we characterized CstR from a second human pathogen E. faecalis that regulates a cst-like operon encoding two orphan STRs and a CoAPR (Fig. 5A, left) (17). CstR is homotetrameric in solution and is anticipated to harbor four peripherally arranged dithiol “sensing” sites found between protomers in a dimer-of-dimers D2-symmetric architecture, like CsoR (Fig. 5A, middle) (145). CstR represses transcription in the reduced state, whereas reaction with sulfane sulfur–containing RSS, but not H2S itself, results in a mixture of di-, tri-, and tetrasulfide interprotomer cross-links that negatively regulates DNA operator-promoter binding, allowing for transcription initiation (Fig. 5A, right) (16). In both S. aureus and E. faecalis, the operons are also inducible by the nitroxyl (HNO) donor Angeli's salt, but not by an NO· donor (17, 53). In S. aureus this may be the result of increased cellular per- and polysulfides after the addition of nitroxyl to cells, supporting the notion of H2S/NO· cross-talk in this organism (Fig. 2B). Although CstR appears selective for H2S and RSS in cell-based transcription reactions (16), the structure/reactivity factors that enforce this apparent specificity are not yet known.
Figure 5.

Regulated operons, modes of regulation, structure, and RSS reactivity products of bacterial RSS sensors. A, CstR in its reduced form transcriptionally represses the cst and cst-like operons in S. aureus and E. faecalis, respectively (left) (16). A structural model of CstR using Protein Data Bank entry 5FMN as a template reveals an all α-helical protein with four peripheral dithiol-sensing sites in the tetrameric structure (middle). The reaction products of CstR with RSS reveal a mixture of di-, tri-, and tetrasulfide interprotomer linkages (right). B, SqrR (operon not shown), like its homologs BigR and PigS, function as transcriptional repressors in their reduced forms and regulate the expression of typical sulfide oxidation and detoxification enzymes (left) (18, 20, 116, 147). The homodimeric structure of SqrR reveals a pair of dithiol-sensing sites (middle, Protein Data Bank entry 6O8L), which readily form intraprotomer sulfur bridges of four (SqrR) or five (BigR) sulfur atoms upon reaction with RSS (right) (113). C, FisR is a σ54-dependent transcriptional activator and activates the expression of a sulfide detoxification operon that is similar to that encoded by the cst operon (left). FisR is organized into three domains (regulatory, ATPase, and DNA-binding domain), but to date, there are no structures of a functionally characterized RSS-sensing FisR (middle). C. pinatubonensis FisR reacts with RSS in vitro to form a mixture of di- and tetrasulfide linkages that weakly stimulate the ATPase activity, resulting in transcriptional activation from σ54-RNA polymerase (RNAP)-transcribed promoters (right) (19). The regulatory mechanism operative in A. baumannii FisR has not yet been determined (20).
SqrR and homologs
The sulfide:quinone reductase repressor, SqrR, was originally characterized in the photosynthetic bacterium R. capsulatus and is responsible for the regulation of 45% of all sulfide-responsive genes in this organism, including an SQR (18). SqrR is a member of the arsenic repressor (ArsR) superfamily (146) in striking structural contrast to CstR. SqrR adopts the ArsR family α1-α2-α3-α4-β1-β2-α5 “winged-helical” dimeric fold, where one Cys in the α2 helix and one Cys in the α5 helix from the same subunit create a pair of dithiol RSS-sensing sites on the dimer (Fig. 5B, middle) (113). Other RSS-responsive ArsR family repressors include the biofilm growth-associated repressor BigR, characterized in X. fastidiosa (114–116) and A. baumannii (20), and PigS characterized in Serratia spp. (147). A. baumannii BigR regulates a secondary RSS detoxification system that includes a second PDO and two transmembrane proteins proposed to be involved in the transport of sulfur-containing molecules (20, 147). Although PigS has not been functionally characterized as an RSS sensor, it regulates several enzymes known to function in H2S/RSS clearance, including a single-domain PDO and a CoAPR encoded by coaP (Fig. 5B, left) (147). It is interesting to note that PigS and its regulon are part of the larger PigP regulon involved in the biosynthesis of the antibiotic prodigiosin (Pig), thus implying a regulatory connection between antibiotic biosynthesis and H2S/RSS homeostasis. The ArsR-family RSS sensors that have been functionally characterized behave analogously to CstR, functioning as repressors in the reduced state and dissociating from the DNA upon reaction with sulfane sulfur–containing RSS, to readily form nearly exclusively tetrasulfide (SqrR) and pentasulfide (BigR) bridges, respectively (Fig. 5B, right) (113). In contrast to CstR, these (poly)sulfur bridges are formed within a subunit, and although other linkages are made, they are far less abundant compared with the mixture of products found in CstR (16).
Recent work from our laboratory utilized SqrR as a model dithiol transcriptional regulator to investigate the structural and reactivity features that govern its oxidant selectivity and specificity (113). Indeed, SqrR is specific for sulfane sulfur and only forms a disulfide when treated with potent diazirene electrophile TMAD (diamide), but not with more common cellular oxidants including GSH disulfide or H2O2. Whereas this low reactivity toward cellular oxidants can be partially explained by the relatively high apparent pKa of the dithiol pair, the high selectivity toward RSS is enforced by structural features of SqrR in various oxidation states. These structures reveal a high energetic barrier to form the disulfide because of large rearrangements that must occur in order to form the disulfide; in addition, the disulfide is not on pathway to form the major tetrasulfide product. In contrast, formation of the tetrasulfide does not require large structural rearrangements; on the contrary, this linkage results in the collapse of the dithiol pocket that completely shields the tetrasulfide linkage from solvent. This study demonstrates that SqrR-like dithiol-based repressors achieve high RSS specificity from the conformational landscape of the protein ensemble, which favors installation of a PTM that minimizes local structural frustration (113). It will be interesting to determine whether these principles apply to other structural classes of RSS sensors or if there are additional determinants that dictate their specificity.
FisR
A third structural class of RSS-sensing transcriptional regulators first characterized in C. pinatubonensis (19), and more recently in A. baumannii (20), is FisR (Fis family transcriptional regulator). In both bacteria, FisR transcriptionally activates the expression of a PDO and SQR and a putative sulfite/sulfonate effluxer, TauE (148), in only A. baumannii (Fig. 5C, left). In contrast to CstR and SqrR-like RSS sensors, FisR is a canonical σ54-dependent transcriptional activator that harbors an N-terminal regulatory domain, a central AAA+ ATPase domain, and a C-terminal DNA-binding domain (Fig. 5C, middle) (149). In C. pinatubonensis FisR, reaction with inorganic RSS appears to result in the formation of di- and tetrasulfide cross-links between two cysteine residues in the regulatory domain, which in turn stimulates the ATPase activity of the central AAA+ domain, which likely activates hexameric assembly and promoter melting by σ54-RNA polymerase holoenzyme (Fig. 5C, right) (19). In A. baumannii FisR, these cysteines are not present, and as a result, H2S/RSS is likely sensed using an alternate mechanism (20), which includes heme-based (117–121) and mononuclear, nonheme Fe-based RSS-sensing regulatory models (150–152).
Chemical tools for generation, detection, and quantification of RSS
To understand the role of H2S and RSS in signaling at the host-pathogen interface, tools must be available that allow for the generation, detection, and quantification of these species in vivo. The type and number of molecular probes used for the generation of H2S and RSS have substantially increased over the past several years, and they are now being used in bacteria to provide critical insights into H2S signaling in these organisms (22, 25, 26). Fluorescence-based probes provide rapid and sensitive detection of H2S or sulfane sulfur with several options now commercially available. In addition, recent efforts to quantify H2S and RSS in complex cellular mixtures have provided new insights into this process. As many of these molecular tools have been extensively reviewed elsewhere (153–159), here we provide only a summary of the available approaches, while pointing out specific challenges to their use.
H2S and RSS donors
H2S donors largely fall into three main classes. These are hydrolysis-based, thiol-dependent, and carbonyl sulfide (COS)-based donors (Fig. 6A). Hydrolysis-based donors function over a wide range of pH, with GYY4137 as a widely used and commercially available donor employed by several groups to study the mechanism of H2S cytoprotection in bacteria (22, 25, 160). Concerns over the relatively slow release rates have led to second-generation GYY4137 derivatives, including JK-2, that more efficiently release H2S at physiological pH (161). The use of hydrolysis-based donors requires careful consideration of the pH dependence and kinetics of H2S release, and many donors are not commercially available. Thiol-dependent H2S donors are attractive tools because of their use of typically cell-abundant cellular reducing thiols, including GSH and cysteine (Fig. 6A). However, some of these probes are quite slow, require high concentrations of thiol, or are activated by a specific thiol (154). As the type and concentrations of thiols in bacteria have only recently been investigated and only in a small sampling of bacteria (17, 20, 53, 96), this potentially limits the broad applicability of such probes in bacteria. In addition, the use of cellular thiols to activate compounds may disrupt the cellular thiol/disulfide redox balance, leading to physiological impacts not specifically due to H2S release. Furthermore, some thiol-activated donors proceed through a highly reactive persulfide intermediate, making it difficult to attribute a physiological impact to the persulfide intermediate or to H2S itself. Last, COS-based donors have been designed with various release mechanisms and rely on an endogenous carbonic anhydrase to produce H2S in high catalytic yields from probe-dependent release of COS (Fig. 6A) (162). The use of these compounds, like other enzyme-activated H2S donors (26), obviously relies on broad cell permeability and constitutive expression of a carbonic anhydrase (163), neither of which has been systematically investigated in bacteria.
Figure 6.
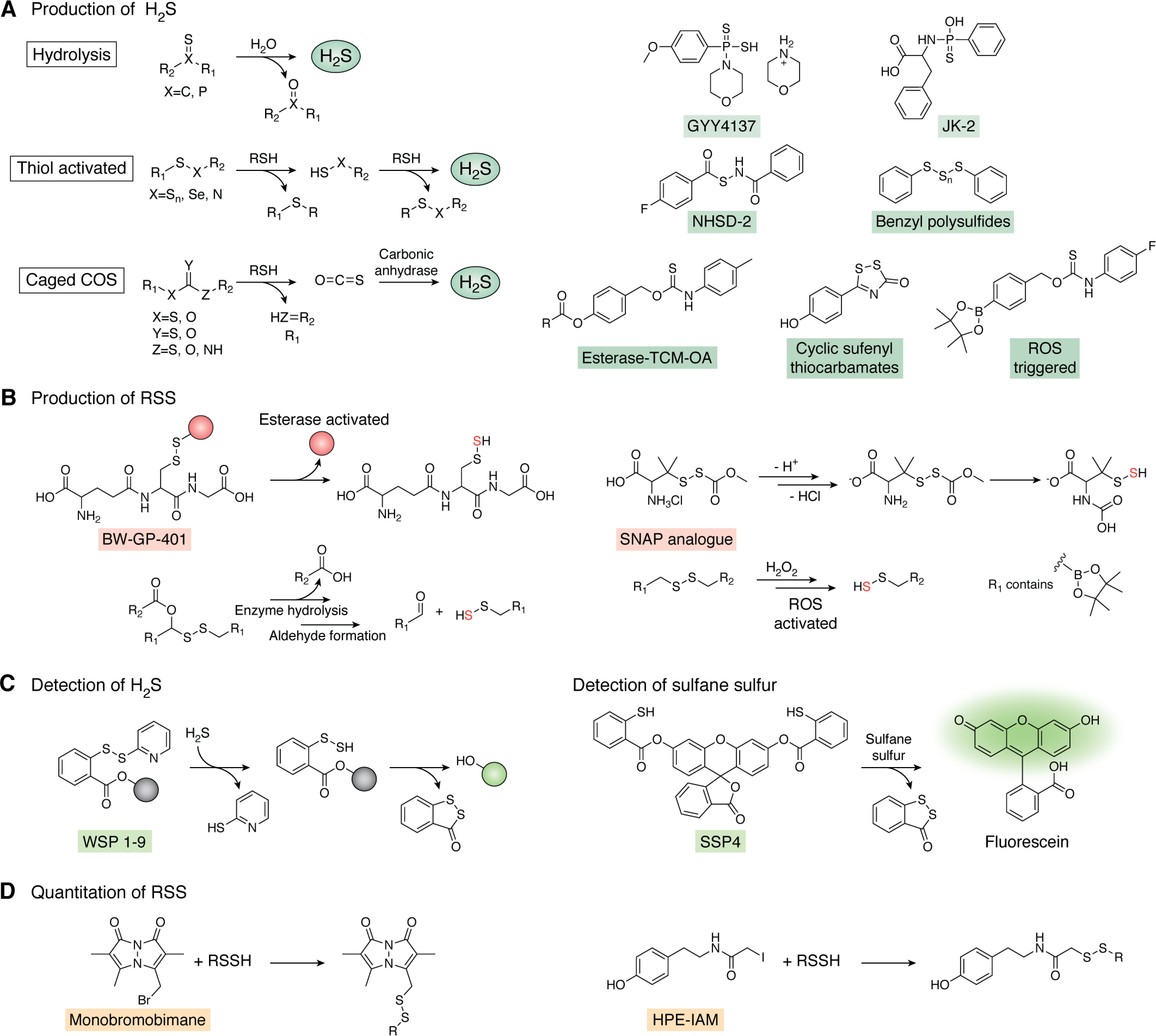
Molecular tools for the production, detection, and quantification of H2S and RSS. A, H2S donors are generally categorized as hydrolysis-based, thiol-activated, or caged COS donors. A subset of representative probes in each category are shown (right). B, representative group of persulfide donors activated by an esterase, ROS, or spontaneous rearrangement at physiological pH. C, WSP and SSP, series of fluorescent probes used for detection of H2S and sulfane sulfur, respectively. D, quantitation of RSS (e.g. RSSH) via electrophilic trapping by monobromobimane or HPE-IAM. See section on chemical tools for generation, detection, and quantification of RSS for additional details.
Whereas H2S donors have been extensively developed over the last 10 years, RSS donors typically found in the form of persulfide donors are comparatively less so (153, 158). For in vitro chemical reactivity studies, we and others have relied on in situ thiol persulfide generation from the reaction of excess H2S with a disulfide, resulting in a mixture containing the thiol persulfide that is not easily separated from the remaining reactants of H2S, disulfide, and thiol (45). Although in situ thiol persulfide generation may also result in formation of the more stable trisulfide species (164), we observed no reaction of SqrR (Fig. 5B) with cysteine trisulfide (113). Furthermore, whereas this work also revealed somewhat faster kinetics of SqrR with the in situ generated persulfide versus a persulfide donor analog of S-nitrosoacetyl-penicillamine (SNAP), the SNAP analog persulfide donor has the added benefit that it is soluble in aqueous solution and spontaneously generates the persulfide species after S to N carbonyl transfer (Fig. 6B) (165). In addition, Wang and co-workers (166, 167) recently developed a series of esterase-sensitive persulfide donors, including GSH persulfide, whose physiological effects have thus far only been studied in mammalian cells. These cell-permeable persulfide donors generate persulfide species under physiological conditions with minimal disruption to cellular redox status (i.e. they are not thiol-activated, an important feature).
ROS-activated H2S and persulfide donors have also garnered recent attention (Fig. 6B) (154, 158) because these types of probes are particularly useful for studying ROS/H2S cross-talk, which is likely relevant at sites of infection. These donors are specific for H2O2 activation and react readily to generate H2S or a persulfide species. Recently, two groups have developed H2O2-specific persulfide-generating probes that exhibit greater cytoprotective effects in cells challenged with oxidative stress than H2S only–generating probes, such as GYY4137 (168, 169). This finding is consistent with the idea that RSS rather than H2S per se are important effector molecules of H2S signaling and cytoprotection and merits further study in bacterial cells.
Detection of H2S/RSS
Although historically many groups have utilized methylene blue (170) or lead acetate (21) paper strips to measure H2S in growing bacterial cells, the latter is not quantitative, and both methods are unable to detect RSS. More recently, several groups have developed sensitive fluorescent probes for detection of H2S and RSS, and this has been comprehensively reviewed elsewhere (155, 157, 159). Of these tools, the WSP and SSP series of fluorescent molecules that detect H2S and sulfane sulfur, respectively, are most commonly used in both mammalian and bacterial cells (Fig. 6C) (24, 77). When used by themselves, these probes cannot be used to perform absolute quantitation of H2S/RSS; however, one group recently coupled SSP4 cell labeling with MS to quantitate sulfane sulfur, thus establishing the possibility of making these optical methods quantitative (171). This approach, however, would only allow for quantitation of total sulfane sulfur without molecule-specific identification. Regardless of their inability to provide absolute quantitation, these probes provide a rapid and simple readout for production of H2S/RSS appropriate for comparing bacterial strains and growth conditions. In addition, the SSP series provide a rapid means for quantifying sulfane sulfur from enzymatic activity in vitro, a strategy recently used to measure RSS biogenesis and clearance activities of catalase and superoxide dismutase, respectively (172, 173).
Quantification of H2S/RSS in cell lysates
Isotope-dilution LC–MS–based methods to detect and quantify H2S and RSS have considerable advantages over optical imaging modalities, including the ability to perform molecular identification and absolute quantitation; however, these methods are not easily applied to living bacterial cells and therefore rely on post-cell growth sample work-up and chemical derivatization of RSS. We and others have traditionally employed electrophilic trapping by monobromobimane (MBB) with quantitation originally via fluorescence detection and more recently via the addition of isotopically labeled internal standards necessary for LC–MS quantitation (Fig. 6D) (17, 20, 47, 53, 96). It was recently shown that quantitation of H2S by MBB is sensitive to small variations in pH, alkylation time, and temperature, thus emphasizing the importance of controlling reaction conditions when comparing samples (174). Bogdandi and co-workers (175) also showed that the electrophilicity of the alkylating agent also impacts quantitation of RSS to varying degrees. N-Ethylmaleimide, the most electrophilic agent tested, resulted in cleavage of per- and polysulfide chains, whereas β-(4-hydroxyphenyl)ethyl iodoacetamide (HPE-IAM) (Fig. 6D) showed little to no cleavage under the same experimental conditions. MBB has an intermediate impact resulting in slightly lower yields of per- and polysulfide species relative to HPE-IAM but exhibited significantly better quantitation than N-ethylmaleimide. As a result, many groups now use HPE-IAM for quantification of RSS (175–177).
A more general consideration of electrophile-based trapping approaches for RSS quantitation is that these methods provide only a snapshot of cellular RSS speciation, particularly in light of our incomplete understanding of sulfane sulfur “scrambling” that will conspire against any method of quantitative analysis. Better understanding of this will provide complementary information to quantitative techniques and valuable insights on the lifetime and speciation of RSS in cells.
H2S signaling via protein S-sulfuration
PTMs of cysteine thiols are known to impact signaling and play regulatory roles in proteins (43, 178–180). Persulfidation of cysteine residues can occur via reduction of nitrosated or sulfenylated protein thiols, while also protecting these latter thiols from overoxidation (59, 60). Persulfidation may also function in H2S signaling as a regulatory modification by introducing altered chemistry or “blocking” of active-site thiols, thiols in a regulatory or allosteric domain, or those found in transcriptional regulators, as shown for the virulence regulator MgrA in S. aureus (96). However, the relevance and regulatory nature of this PTM remain largely unexplored in bacteria, leaving large gaps in our understanding of protein persulfidation as a regulatory modification in H2S/RSS signaling. For example, persulfidation of the active-site thiol in the cell-abundant glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH) has been reported by several groups; however, there are conflicting reports to whether this modification activates (58) or inhibits (96, 181) the activity of this enzyme. Furthermore, the same active site thiol in GAPDH is the target of many other thiol modifications, including S-nitrosation (182), S-sulfenylation, S-sulfinylation (183), and S-thiolation (184). To better understand the role of protein persulfidation in H2S/RSS signaling, it becomes important to understand how this modification is installed and removed from proteins and to identify regulatory targets of this PTM.
It is instructive to discuss thiol-based PTMs, including persulfidation, S-sulfenylation, or S-nitrosation, in the “writer, reader, eraser” paradigm, which represents the formation, signal transduction, and removal of PTMs (Fig. 7A). The writer refers to the process(es) that install the PTM to the protein target, such as S-nitrosoglutathione for protein S-nitrosation (185). The reader includes PTM-dependent interacting partners, whether it be a protein or small molecule, thus transducing the signal in response to the PTM (e.g. S-nitrosation of procaspase-3 promotes its interaction with acid sphingomyelinase and prevents apoptosis) (186). Last, the eraser refers to the proteins or small molecules responsible for removing the PTM after signal transduction has occurred. In S-nitrosation, several denitrosylases have been characterized, including the thioredoxin system (187); other enzymes reduce S-nitrosoglutathione or S-nitroso-CoA directly to sulfenamides, thus lowering the steady-state pools of these small-molecule NO donors (188–190). For bacterial protein persulfidation, the writers and erasers are perhaps slightly better understood, whereas we have little to no knowledge of the readers. We note also that writers and erasers are typically restricted to enzymes (e.g. kinase-phosphatase and/or acetylase-deacetylase pairs), but here we explicitly consider nonenzymatic pathways as well.
Figure 7.
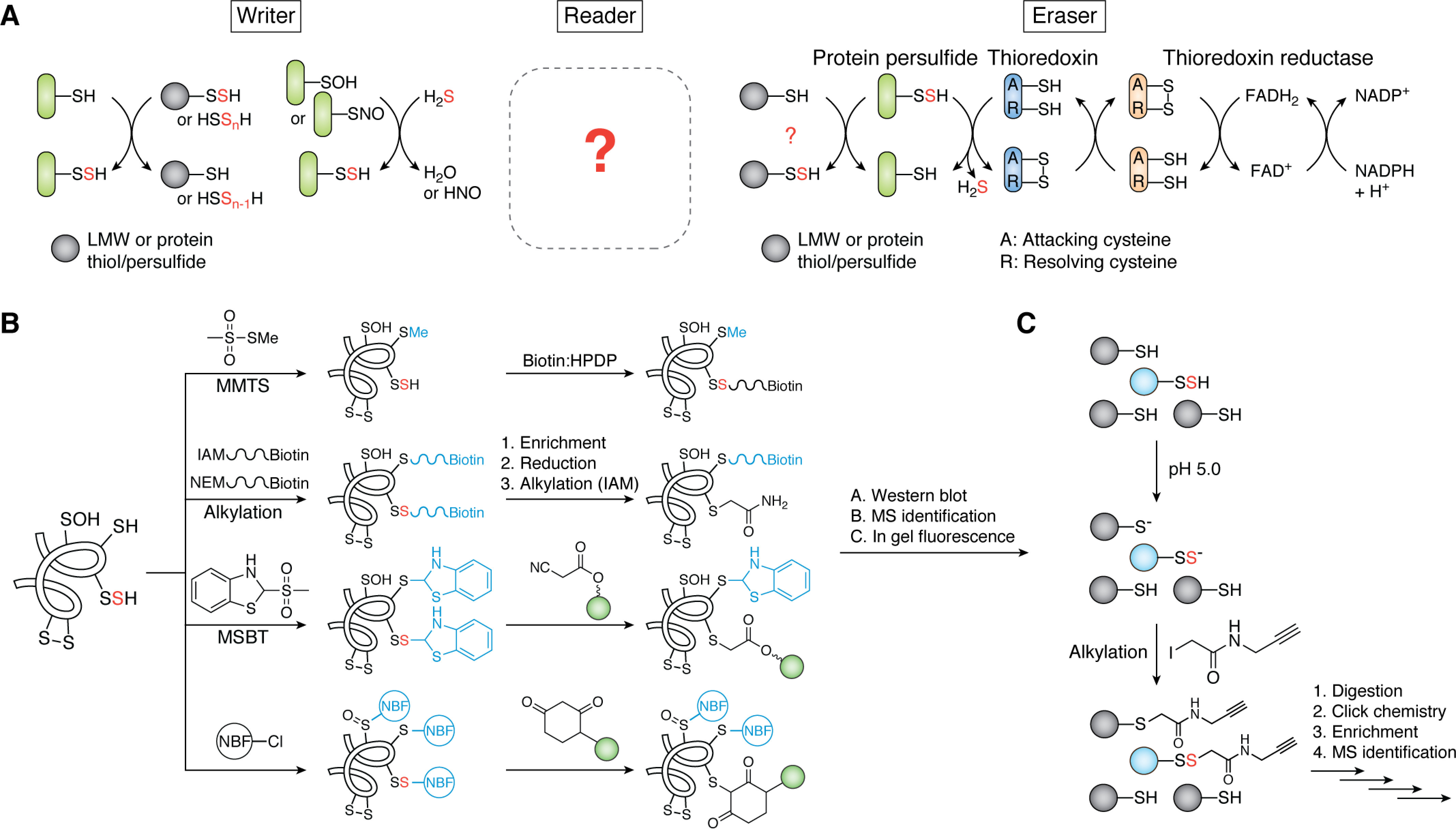
H2S signaling via protein persulfidation. A, reversible protein persulfidation of a target protein (green) in the context of the formation (writer), transduction (reader), and removal (eraser) paradigm of sulfane sulfur flow. Candidate or known writers and erasers are shown; there are no concrete examples of readers in bacteria (indicated by the question mark). B, overview of methodologies developed to detect global proteome persulfidation via alkylation/reduction or “tag-switch” approaches (43, 198). Protein persulfides are indicated with a red S. Sulfur adducts generated in the first step are shown in blue, whereas green spheres indicate functional groups not shown here (e.g. a fluorescent moiety). C, schematic of the low-pH QTRP method that allows for the direct detection of protein persulfides (red S, blue sphere) (206). See section on H2S signaling via protein S-sulfiration for additional details.
The writers of protein persulfidation may include nonenzyme-catalyzed transsulfuration with RSS or direct reaction of H2S with more oxidized cysteines or, enzymatically, via transsulfuration by STRs or other enzymes (Fig. 7A, left). Although the reactivity of STRs strongly suggests a role in protein transsulfuration, little is known regarding the rates of these reactions, specificity, and physiological impact of many STRs. These small-molecule and protein writers can also potentially function in reverse, as the erasers, by removing the sulfane sulfur in a process that is not fully understood. In S. aureus, the cst operon–encoded STR CstA was shown to facilely remove the persulfide moiety on the cysteine desulfurase SufS in support of the idea that STRs can also catalyze the removal of protein persulfides (135). In addition, the thioredoxin (Trx)/thioredoxin reductase cascade may be largely responsible for the removal of protein persulfides for which there is now evidence (Fig. 7A, right) (191, 192). The characterization of two thioredoxins in S. aureus that have significant activities on protein persulfides relative to disulfides versus the canonical TrxA certainly suggests a role for Trx in this process, as well as the possibility that bacteria encode specific thioredoxin-like proteins for this purpose (96). Furthermore, a thioredoxin-based proteomic profiling strategy was used to identify potential cellular targets for these persulfide-reducing thioredoxins in S. aureus (193). Analogous strategies might be applied to STRs in an effort to identify protein targets that could function as donors or acceptors in transsulfuration reactions.
Detection of global proteome persulfidation
To evaluate proteome persulfidation, a number of methods have been developed over the past several years to identify candidate targets of this PTM. The first method proposed for detection of protein persulfides relies on S-methylmethanethiosulfonate to selectively methylate protein thiols over persulfides, followed by persulfide capture by biotin-HPDP (N-[6-(biotinamido)-hexyl]-3′-(2-pyridyldithio)propionamide) and enrichment by streptavidin beads (Fig. 7B) (58). However, the selectivity of S-methylmethanthiosulfonate toward thiols over persulfides has been challenged and shown not to be sufficient for general application (194). Several groups then developed approaches that rely on derivatizing both protein thiols and persulfides with a biotinylated alkylating agent (96, 192, 195, 196). After enrichment with streptavidin, the mixed disulfide that characterizes only persulfide-containing peptides or proteins is selectively reduced, alkylated, and identified by LC–MS/MS. Enrichment of whole proteins versus peptides is far more prone to artifacts (192, 197) and should be avoided. Enriching peptides rather than proteins will also tend to minimize the presence of false positives that derive from endogenous disulfide bonds in the lysate, although this may be expected to be low for cytoplasmic lysates, given the high reducing capacity of the cytoplasm (96, 195, 196).
Several “tag-switch” approaches have also been developed for identifying protein persulfides (191, 197–200). These methods do not rely on the reduction of mixed disulfides from alkylated persulfides but instead exploit the unique reactivity of the mixed disulfide toward a specific nucleophile. The first tag-switch method reported utilized an electrophilic blocking reagent, methylsulfonylbenzo-thiazole, to react with both protein thiols and persulfides (199). These residues when blocked have very different reactivities toward nucleophiles (i.e. thiol adducts are unreactive, whereas persulfide adducts as mixed disulfides retain significant reactivity). The persulfide adduct is then reacted with a cyanoacetate-based reagent (e.g. CN-biotin) to “switch” the tag on these residues (Fig. 7B) (47, 198, 199). Several cyanoacetate-based reagents have been developed, including fluorescent adducts, and each relies on its high specificity toward the mixed disulfide of protein persulfides over protein disulfides (191). Most recently, Zivanovic et al. (200) reported a new tag-switch workflow where 4-chloro-7-nitrobenzofuran (201, 202) is used to label thiols, persulfides, sulfenic acids, and free amines, followed by selective switching of the persulfide-derived mixed disulfide by dimedone, a reagent commonly used to detect sulfenylated cysteines (Fig. 7B) (203, 204). The advantage of this workflow is that the availability of many derivatized dimedones makes it possible to modify the workflow to fit the application, be it fluorescence imaging, streptavidin enrichment, or click chemistry–based enrichment approaches and downstream MS (205).
Although these workflows are powerful and are expected to be broadly applied, none achieve direct detection of protein persulfides. A recent study by Fu et al. (206) approached this challenge by leveraging the inherent pKa difference between persulfides and thiols in their development of low-pH quantitative thiol reactivity profiling (QTRP) (Fig. 7C). QTRP incorporates an initial alkylation step in low-pH conditions, where most thiols are protonated and unreactive, thus allowing selective alkylation of persulfides (206). Use of isotopically labeled reagents permits estimation of the fraction persulfide versus thiol for all Cys-containing peptides. An important shortcoming of this method is that pKa estimations for thiols and persulfides (44) are not generally known on a proteome scale and may well vary widely. Thus far, QTRP has only been applied in mammalian cells; it would be of interest to determine how many proteins can be detected with high fractional persulfidation levels in less complex bacterial proteomes, which may aid in the identification of regulatory persulfides, characterized by high relative quantitation of persulfide versus thiol.
Finally, regardless of the workflow used to detect bacterial proteome persulfidation, these studies seem to suggest that the proteome may function as a “sink” for sulfane sulfur, given that upward of ∼15% of the proteins in a bacterial proteome are persulfidated (at some fraction) in a way that is not greatly impacted by the addition of exogenous sulfide to growing cells (20, 96). Taken at face value, this suggests that most of these proteome persulfidation events are unlikely to be regulatory, consistent with a lack of strongly compelling persulfidation consensus sequence motif, implying that these cysteines may be persulfidated at random (20, 206, 207). The challenge we now face is not in the detection of sites of persulfidation in a proteome but to distinguish those regulatory sites from those that are persulfidated collaterally on solvent-accessible, nonconserved, and highly reactive cysteines. This is a key aspect of the “writer, reader, eraser” paradigm of H2S/RSS signaling that is thus far largely unexplored. We would expect regulatory persulfidation events to be found on conserved cysteines in enzymes that catalyze committed steps or define metabolic hubs or branch points in a metabolic pathway(s) (195), in transcriptional regulators (111), as allosteric modulators, or in enzyme active sites (e.g. GAPDH) (58, 96, 181). Indeed, a subset of these regulatory cysteines may be subject to other PTMs, which would increase the biological complexity of the regulatory response.
Future outlook
Whereas studies of the chemical biology of H2S and RSS have greatly expanded in recent years, our molecular understanding of the underlying mechanisms that drive regulatory signaling and cytoprotection of bacterial cells against antibiotic and host assault remains poorly defined. The high reactivity and potentially facile interconversion of RSS represents a particularly challenging analytical problem. Recent advances in methodologies used to quantify RSS will allow us to further elucidate their roles as potent antioxidants (47), modulators of antibiotic efficacy (21), and the “currency” of H2S signaling, the latter analogous to LMW organic nitrosothiols in S-nitrosation–based signaling pathways (185, 188–190).
Recent studies have begun to establish the physiological importance of H2S/RSS biogenesis in bacteria at the host-pathogen interface and the central role played by distinct classes of transcriptional regulators that act as molecular sensors that survey cellular H2S/RSS required, in some cases, to retain full pathogen virulence (17, 96). We do not yet fully understand the mechanisms that make these sensors specific for RSS relative to other oxidants (16), although these data are beginning to appear (113). Finally, although global S-sulfuration profiling has successfully mapped hundreds of sites of protein persulfidation in many different contexts and systems, the next major challenge is to identify and characterize regulatory sites of protein persulfidation in bacteria, like that recently found in human aquaporin-8, a gated H2O2 transmembrane channel (208). These thiols represent physiologically relevant targets of reversible persulfidation that impact enzyme activity, a metabolic pathway(s) or gene expression program (195), beyond simple detoxification and homeostasis of H2S/RSS. We suspect that as yet uncharacterized “orphan” STRs that are regulated by RSS sensors (17, 135) and perhaps cell-abundant peroxiredoxins (136) and thioredoxins may be excellent candidates as writers and/or erasers in H2S/RSS signaling (Fig. 7A), where specific protein-protein interactions may well “direct traffic” in the dynamic flow of regulatory sulfur in cells. Addressing these challenges will better strengthen the foundation of H2S/RSS signaling as a bona fide signaling process in bacterial cells in the infected host, thus representing new physiology prime for the development of novel antimicrobial interventions.
Note added in proof—In a recent paper, Ng et al. (2020) Front. Microbiol. 11, 1875 showed that two multidrug-resistant strains, including one clinical isolate, of Acinetobacter baumannii, do not biosynthesize appreciable hydrogen sulfide and that exogenous NaHS appears to potentiate the killing efficacy of a number of diverse antibiotics, in contrast to nearly all other findings discussed here. Interestingly, neither A. baumannii strain used by Ng et al. encodes 3-mercaptopyruvate sulfurtransferase (3MST) or RSS sensors FisR or BigR (see Fig. 5) or FisR- and BigR-regulated genes as described in A. baumannii ATCC 17978 (20).
Funding and additional information—This work was supported by National Institutes of Health Grant R35 GM118157 (to D. P. G.). B. J. C. W. was supported by a fellowship from the Graduate Training Program Quantitative and Chemical Biology (QCB) at Indiana University (Grants T32 GM131994 and GM109825) and the Kratz Fellowship awarded by the Department of Chemistry, Indiana University. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- RSS
- reactive sulfur species
- CBS
- cystathionine β-synthase
- CSE
- cystathionine γ-lyase
- 3-MP
- 3-mercaptopyruvate
- CAT
- cysteine aminotransferase
- 3MST
- 3-MP sulfurtransferase
- ROS
- reactive oxygen species
- RNS
- reactive nitrogen species
- LMW
- low-molecular weight
- PTM
- post-translational modification
- SQR
- sulfide:quinone oxidoreductase
- STR
- sulfurtransferase
- PDO
- persulfide dioxygenase
- PRF
- PDO-rhodanese fusion protein
- SNAP
- S-nitrosoacetyl-penicillamine
- MBB
- monobromobimane
- HPE-IAM
- β-(4-hydroxyphenyl)ethyl iodoacetamide
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- QTRP
- quantitative thiol reactivity profiling.
References
- 1. Lusti-Narasimhan M., Pessoa-Silva C. L., and Temmerman M. (2013) Moving forward in tackling antimicrobial resistance: WHO actions. Sex. Transm. Infect. 89, iv57–iv59 10.1136/sextrans-2012-050910 [DOI] [PubMed] [Google Scholar]
- 2. Chang H. H., Cohen T., Grad Y. H., Hanage W. P., O'Brien T. F., and Lipsitch M. (2015) Origin and proliferation of multiple-drug resistance in bacterial pathogens. Microbiol. Mol. Biol. Rev. 79, 101–116 10.1128/MMBR.00039-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ma Z., Jacobsen F. E., and Giedroc D. P. (2009) Coordination chemistry of bacterial metal transport and sensing. Chem. Rev. 109, 4644–4681 10.1021/cr900077w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Waldron K. J., Rutherford J. C., Ford D., and Robinson N. J. (2009) Metalloproteins and metal sensing. Nature 460, 823–830 10.1038/nature08300 [DOI] [PubMed] [Google Scholar]
- 5. Wang Y., Weisenhorn E., MacDiarmid C. W., Andreini C., Bucci M., Taggart J., Banci L., Russell J., Coon J. J., and Eide D. J. (2018) The cellular economy of the Saccharomyces cerevisiae zinc proteome. Metallomics 10, 1755–1776 10.1039/c8mt00269j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zygiel E. M., and Nolan E. M. (2018) Transition metal sequestration by the host-defense protein calprotectin. Annu. Rev. Biochem. 87, 621–643 10.1146/annurev-biochem-062917-012312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Damo S. M., Kehl-Fie T. E., Sugitani N., Holt M. E., Rathi S., Murphy W. J., Zhang Y., Betz C., Hench L., Fritz G., Skaar E. P., and Chazin W. J. (2013) Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc. Natl. Acad. Sci. U. S. A. 110, 3841–3846 10.1073/pnas.1220341110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cunden L. S., and Nolan E. M. (2018) Bioinorganic explorations of Zn(II) sequestration by human S100 host-defense proteins. Biochemistry 57, 1673–1680 10.1021/acs.biochem.7b01305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sheldon J. R., and Skaar E. P. (2019) Metals as phagocyte antimicrobial effectors. Curr. Opin. Immunol. 60, 1–9 10.1016/j.coi.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu Z., Wang P., Wang H., Yu Z. H., Au-Yeung H. Y., Hirayama T., Sun H., and Yan A. (2019) Zinc excess increases cellular demand for iron and decreases tolerance to copper in Escherichia coli. J. Biol. Chem. 294, 16978–16991 10.1074/jbc.RA119.010023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Capdevila D. A., Edmonds K. A., and Giedroc D. P. (2017) Metallochaperones and metalloregulation in bacteria. Essays Biochem. 61, 177–200 10.1042/EBC20160076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Capdevila D. A., Wang J., and Giedroc D. P. (2016) Bacterial strategies to maintain zinc metallostasis at the host-pathogen interface. J. Biol. Chem. 291, 20858–20868 10.1074/jbc.R116.742023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jordan M. R., Wang J., Capdevila D. A., and Giedroc D. P. (2020) Multi-metal nutrient restriction and crosstalk in metallostasis systems in microbial pathogens. Curr. Opin. Microbiol. 55, 17–25 10.1016/j.mib.2020.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Osman D., Martini M. A., Foster A. W., Chen J., Scott A. J. P., Morton R. J., Steed J. W., Lurie-Luke E., Huggins T. G., Lawrence A. D., Warren M. J., Chivers P. T., and Robinson N. J. (2019) Bacterial sensors define intracellular free energies for correct enzyme metalation. Nat Chem Biol. 15, 241–249 10.1038/s41589-018-0211-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grossoehme N., Kehl-Fie T. E., Ma Z., Adams K. W., Cowart D. M., Scott R. A., Skaar E. P., and Giedroc D. P. (2011) Control of copper resistance and inorganic sulfur metabolism by paralogous regulators in Staphylococcus aureus. J. Biol. Chem. 286, 13522–13531 10.1074/jbc.M111.220012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luebke J. L., Shen J., Bruce K. E., Kehl-Fie T. E., Peng H., Skaar E. P., and Giedroc D. P. (2014) The CsoR-like sulfurtransferase repressor (CstR) is a persulfide sensor in Staphylococcus aureus. Mol. Microbiol. 94, 1343–1360 10.1111/mmi.12835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shen J., Walsh B. J. C., Flores-Mireles A. L., Peng H., Zhang Y., Zhang Y., Trinidad J. C., Hultgren S. J., and Giedroc D. P. (2018) Hydrogen sulfide sensing through reactive sulfur species (RSS) and nitroxyl (HNO) in Enterococcus faecalis. ACS Chem. Biol. 13, 1610–1620 10.1021/acschembio.8b00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shimizu T., Shen J., Fang M., Zhang Y., Hori K., Trinidad J. C., Bauer C. E., Giedroc D. P., and Masuda S. (2017) Sulfide-responsive transcriptional repressor SqrR functions as a master regulator of sulfide-dependent photosynthesis. Proc. Natl. Acad. Sci. U. S. A. 114, 2355–2360 10.1073/pnas.1614133114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li H., Li J., Lu C., Xia Y., Xin Y., Liu H., Xun L., and Liu H. (2017) FisR activates σ54 -dependent transcription of sulfide-oxidizing genes in Cupriavidus pinatubonensis JMP134. Mol. Microbiol. 105, 373–384 10.1111/mmi.13725 [DOI] [PubMed] [Google Scholar]
- 20. Walsh B. J. C., Wang J., Edmonds K. A., Palmer L. D., Zhang Y., Trinidad J. C., Skaar E. P., and Giedroc D. P. (2020) The response of Acinetobacter baumannii to hydrogen sulfide reveals two independent persulfide sensing systems and a connection to biofilm regulation. mBio 11, e01254–20 10.1128/mBio.01254-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shatalin K., Shatalina E., Mironov A., and Nudler E. (2011) H2S: a universal defense against antibiotics in bacteria. Science 334, 986–990 10.1126/science.1209855 [DOI] [PubMed] [Google Scholar]
- 22. Toliver-Kinsky T., Cui W., Törö G., Lee S.-J., Shatalin K., Nudler E., and Szabo C. (2019) H2S, a bacterial defense mechanism against the host immune response. Infect. Immun. 87, e00272–18 10.1128/iai.00272-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mironov A., Seregina T., Nagornykh M., Luhachack L. G., Korolkova N., Lopes L. E., Kotova V., Zavilgelsky G., Shakulov R., Shatalin K., and Nudler E. (2017) Mechanism of H2S-mediated protection against oxidative stress in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 114, 6022–6027 10.1073/pnas.1703576114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luhachack L., Rasouly A., Shamovsky I., and Nudler E. (2019) Transcription factor YcjW controls the emergency H2S production in E. coli. Nat. Commun. 10, 2868 10.1038/s41467-019-10785-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Saini V., Chinta K. C., Reddy V. P., Glasgow J. N., Stein A., Lamprecht D. A., Rahman M. A., Mackenzie J. S., Truebody B. E., Adamson J. H., Kunota T. T. R., Bailey S. M., Moellering D. R., Lancaster J. R. Jr., and Steyn A. J. C. (2020) Hydrogen sulfide stimulates Mycobacterium tuberculosis respiration, growth and pathogenesis. Nat. Commun. 11, 557 10.1038/s41467-019-14132-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shukla P., Khodade V. S., SharathChandra M., Chauhan P., Mishra S., Siddaramappa S., Pradeep B. E., Singh A., and Chakrapani H. (2017) “On demand” redox buffering by H2S contributes to antibiotic resistance revealed by a bacteria-specific H2S donor. Chem. Sci. 8, 4967–4972 10.1039/c7sc00873b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mishanina T. V., Libiad M., and Banerjee R. (2015) Biogenesis of reactive sulfur species for signaling by hydrogen sulfide oxidation pathways. Nat. Chem. Biol. 11, 457–464 10.1038/nchembio.1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bronstein M., Schütz M., Hauska G., Padan E., and Shahak Y. (2000) Cyanobacterial sulfide-quinone reductase: cloning and heterologous expression. J. Bacteriol. 182, 3336–3344 10.1128/jb.182.12.3336-3344.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Friedrich C. G., Bardischewsky F., Rother D., Quentmeier A., and Fischer J. (2005) Prokaryotic sulfur oxidation. Curr. Opin. Microbiol. 8, 253–259 10.1016/j.mib.2005.04.005 [DOI] [PubMed] [Google Scholar]
- 30. Vigneron A., Cruaud P., Alsop E., de Rezende J. R., Head I. M., and Tsesmetzis N. (2018) Beyond the tip of the iceberg; a new view of the diversity of sulfite- and sulfate-reducing microorganisms. J. ISME 12, 2096–2099 10.1038/s41396-018-0155-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen X., Jhee K. H., and Kruger W. D. (2004) Production of the neuromodulator H2S by cystathionine β-synthase via the condensation of cysteine and homocysteine. J. Biol. Chem. 279, 52082–52086 10.1074/jbc.C400481200 [DOI] [PubMed] [Google Scholar]
- 32. Chiku T., Padovani D., Zhu W., Singh S., Vitvitsky V., and Banerjee R. (2009) H2S biogenesis by human cystathionine γ-lyase leads to the novel sulfur metabolites lanthionine and homolanthionine and is responsive to the grade of hyperhomocysteinemia. J. Biol. Chem. 284, 11601–11612 10.1074/jbc.M808026200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Singh S., and Banerjee R. (2011) PLP-dependent H2S biogenesis. Biochim. Biophys. Acta 1814, 1518–1527 10.1016/j.bbapap.2011.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Singh S., Padovani D., Leslie R. A., Chiku T., and Banerjee R. (2009) Relative contributions of cystathionine β-synthase and γ-cystathionase to H2S biogenesis via alternative trans-sulfuration reactions. J. Biol. Chem. 284, 22457–22466 10.1074/jbc.M109.010868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miyamoto R., Otsuguro K., Yamaguchi S., and Ito S. (2014) Contribution of cysteine aminotransferase and mercaptopyruvate sulfurtransferase to hydrogen sulfide production in peripheral neurons. J. Neurochem. 130, 29–40 10.1111/jnc.12698 [DOI] [PubMed] [Google Scholar]
- 36. Kimura Y., Koike S., Shibuya N., Lefer D., Ogasawara Y., and Kimura H. (2017) 3-Mercaptopyruvate sulfurtransferase produces potential redox regulators cysteine- and glutathione-persulfide (Cys-SSH and GSSH) together with signaling molecules H2S2, H2S3 and H2S. Sci. Rep. 7, 10459 10.1038/s41598-017-11004-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kimura Y., Toyofuku Y., Koike S., Shibuya N., Nagahara N., Lefer D., Ogasawara Y., and Kimura H. (2015) Identification of H2S3 and H2S produced by 3-mercaptopyruvate sulfurtransferase in the brain. Sci. Rep. 5, 14774 10.1038/srep14774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yadav P. K., Yamada K., Chiku T., Koutmos M., and Banerjee R. (2013) Structure and kinetic analysis of H2S production by human mercaptopyruvate sulfurtransferase. J. Biol. Chem. 288, 20002–20013 10.1074/jbc.M113.466177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li K., Xin Y., Xuan G., Zhao R., Liu H., Xia Y., and Xun L. (2019) Escherichia coli uses separate enzymes to produce H2S and reactive sulfane sulfur from l-cysteine. Front. Microbiol. 10, 298 10.3389/fmicb.2019.00298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang J., Guo X., Li H., Qi H., Qian J., Yan S., Shi J., and Niu W. (2019) Hydrogen sulfide from cysteine desulfurase, not 3-mercaptopyruvate sulfurtransferase, contributes to sustaining cell growth and bioenergetics in E. coli under anaerobic conditions. Front. Microbiol. 10, 2357 10.3389/fmicb.2019.02357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peck S. C., Denger K., Burrichter A., Irwin S. M., Balskus E. P., and Schleheck D. (2019) A glycyl radical enzyme enables hydrogen sulfide production by the human intestinal bacterium Bilophila wadsworthia. Proc. Natl. Acad. Sci. U. S. A. 116, 3171–3176 10.1073/pnas.1815661116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xing M., Wei Y., Zhou Y., Zhang J., Lin L., Hu Y., Hua G., A N. N. U., Liu D., Wang F., Guo C., Tong Y., Li M., Liu Y., Ang E. L., et al. (2019) Radical-mediated C-S bond cleavage in C2 sulfonate degradation by anaerobic bacteria. Nat. Commun. 10, 1609 10.1038/s41467-019-09618-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Filipovic M. R., Zivanovic J., Alvarez B., and Banerjee R. (2018) Chemical biology of H2S signaling through persulfidation. Chem. Rev. 118, 1253–1337 10.1021/acs.chemrev.7b00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cuevasanta E., Lange M., Bonanata J., Coitiño E. L., Ferrer-Sueta G., Filipovic M. R., and Alvarez B. (2015) Reaction of hydrogen sulfide with disulfide and sulfenic acid to form the strongly nucleophilic persulfide. J. Biol. Chem. 290, 26866–26880 10.1074/jbc.M115.672816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Francoleon N. E., Carrington S. J., and Fukuto J. M. (2011) The reaction of H2S with oxidized thiols: generation of persulfides and implications to H2S biology. Arch. Biochem. Biophys. 516, 146–153 10.1016/j.abb.2011.09.015 [DOI] [PubMed] [Google Scholar]
- 46. Edwards J. O., and Pearson R. G. (1962) The factors determining nucleophilic reactivities. J. Am. Chem. Soc. 84, 16–24 10.1021/ja00860a005 [DOI] [Google Scholar]
- 47. Ida T., Sawa T., Ihara H., Tsuchiya Y., Watanabe Y., Kumagai Y., Suematsu M., Motohashi H., Fujii S., Matsunaga T., Yamamoto M., Ono K., Devarie-Baez N. O., Xian M., Fukuto J. M., et al. (2014) Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. U. S. A. 111, 7606–7611 10.1073/pnas.1321232111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Benson S. W. (1978) Thermochemistry and kinetics of sulfur-containing molecules and radicals. Chem. Rev. 78, 23–35 10.1021/cr60311a003 [DOI] [Google Scholar]
- 49. Everett S. A., and Wardman P. (1995) Perthiols as antioxidants: radical-scavenging and prooxidative mechanisms. Methods Enzymol. 251, 55–69 10.1016/0076-6879(95)51110-5 [DOI] [PubMed] [Google Scholar]
- 50. Koppenol W. H., and Bounds P. L. (2017) Signaling by sulfur-containing molecules: quantitative aspects. Arch. Biochem. Biophys. 617, 3–8 10.1016/j.abb.2016.09.012 [DOI] [PubMed] [Google Scholar]
- 51. Benchoam D., Cuevasanta E., Möller M. N., and Alvarez B. (2019) Hydrogen sulfide and persulfides oxidation by biologically relevant oxidizing species. Antioxidants (Basel) 8, 48 10.3390/antiox8020048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ezerina D., Takano Y., Hanaoka K., Urano Y., and Dick T. P. (2018) N-Acetyl cysteine functions as a fast-acting antioxidant by triggering intracellular H2S and sulfane sulfur production. Cell. Chem. Biol. 25, 1–13 10.1016/j.chembiol.2018.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Peng H., Shen J., Edmonds K. A., Luebke J. L., Hickey A. K., Palmer L. D., Chang F. J., Bruce K. A., Kehl-Fie T. E., Skaar E. P., and Giedroc D. P. (2017) Sulfide homeostasis and nitroxyl intersect via formation of reactive sulfur species in Staphylococcus aureus. mSphere 2, e00082–17 10.1128/mSphere.00082-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bailey T. S., Henthorn H. A., and Pluth M. D. (2016) The intersection of NO and H2S: persulfides generate NO from nitrite through polysulfide formation. Inorg. Chem. 55, 12618–12625 10.1021/acs.inorgchem.6b01660 [DOI] [PubMed] [Google Scholar]
- 55. Cortese-Krott M. M., Kuhnle G. G. C., Dyson A., Fernandez B. O., Grman M., DuMond J. F., Barrow M. P., McLeod G., Nakagawa H., Ondrias K., Nagy P., King S. B., Saavedra J. E., Keefer L. K., Singer M., et al. (2015) Key bioactive reaction products of the NO/H2S interaction are S/N-hybrid species, polysulfides, and nitroxyl. Proc. Natl. Acad. Sci. U. S. A. 112, E4651–E4660 10.1073/pnas.1509277112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ivanovic-Burmazovic I., and Filipovic M. R. (2019) Saying NO to H2S: a Story of HNO, HSNO, and SSNO. Inorg. Chem. 58, 4039–4051 10.1021/acs.inorgchem.8b02592 [DOI] [PubMed] [Google Scholar]
- 57. Zhang T., Ono K., Tsutsuki H., Ihara H., Islam W., Akaike T., and Sawa T. (2019) Enhanced cellular polysulfides negatively regulate TLR4 signaling and mitigate lethal endotoxin shock. Cell. Chem. Biol. 26, 686–698e684 10.1016/j.chembiol.2019.02.003 [DOI] [PubMed] [Google Scholar]
- 58. Mustafa A. K., Gadalla M. M., Sen N., Kim S., Mu W., Gazi S. K., Barrow R. K., Yang G., Wang R., and Snyder S. H. (2009) H2S signals through protein S-sulfhydration. Sci. Signal. 2, ra72 10.1126/scisignal.2000464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Paul B. D., and Snyder S. H. (2012) H2S signalling through protein sulfhydration and beyond. Nat. Rev. Mol. Cell Biol. 13, 499–507 10.1038/nrm3391 [DOI] [PubMed] [Google Scholar]
- 60. Paul B. D., and Snyder S. H. (2015) Protein sulfhydration. Methods Enzymol. 555, 79–90 10.1016/bs.mie.2014.11.021 [DOI] [PubMed] [Google Scholar]
- 61. Luebke J. L., and Giedroc D. P. (2015) Cysteine sulfur chemistry in transcriptional regulators at the host-bacterial pathogen interface. Biochemistry 54, 3235–3249 10.1021/acs.biochem.5b00085 [DOI] [PubMed] [Google Scholar]
- 62. Tharmalingam S., Alhasawi A., Appanna V. P., Lemire J., and Appanna V. D. (2017) Reactive nitrogen species (RNS)-resistant microbes: adaptation and medical implications. Biol. Chem. 398, 1193–1208 10.1515/hsz-2017-0152 [DOI] [PubMed] [Google Scholar]
- 63. Winterbourn C. C., Kettle A. J., and Hampton M. B. (2016) Reactive oxygen species and neutrophil function. Annu. Rev. Biochem. 85, 765–792 10.1146/annurev-biochem-060815-014442 [DOI] [PubMed] [Google Scholar]
- 64. Yang Y., Bazhin A. V., Werner J., and Karakhanova S. (2013) Reactive oxygen species in the immune system. Int. Rev. Immunol. 32, 249–270 10.3109/08830185.2012.755176 [DOI] [PubMed] [Google Scholar]
- 65. Ley R. E., Peterson D. A., and Gordon J. I. (2006) Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848 10.1016/j.cell.2006.02.017 [DOI] [PubMed] [Google Scholar]
- 66. Qin J., Li R., Raes J., Arumugam M., Burgdorf K. S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., Mende D. R., Li J., Xu J., Li S., Li D., et al. . MetaHIT Consortium, (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Macfarlane G. T., Gibson G. R., and Cummings J. H. (1992) Comparison of fermentation reactions in different regions of the human colon. J. Appl. Bacteriol. 72, 57–64 10.1111/j.1365-2672.1992.tb05187.x [DOI] [PubMed] [Google Scholar]
- 68. Riedijk M. A., Stoll B., Chacko S., Schierbeek H., Sunehag A. L., van Goudoever J. B., and Burrin D. G. (2007) Methionine transmethylation and transsulfuration in the piglet gastrointestinal tract. Proc. Natl. Acad. Sci. U. S. A. 104, 3408–3413 10.1073/pnas.0607965104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shoveller A. K., Brunton J. A., House J. D., Pencharz P. B., and Ball R. O. (2003) Dietary cysteine reduces the methionine requirement by an equal proportion in both parenterally and enterally fed piglets. J. Nutr. 133, 4215–4224 10.1093/jn/133.12.4215 [DOI] [PubMed] [Google Scholar]
- 70. Stegink L. D., and Besten L. D. (1972) Synthesis of cysteine from methionine in normal adult subjects: effect of route of alimentation. Science 178, 514–516 10.1126/science.178.4060.514 [DOI] [PubMed] [Google Scholar]
- 71. Zlotkin S. H., Bryan M. H., and Anderson G. H. (1981) Cysteine supplementation to cysteine-free intravenous feeding regimens in newborn infants. Am. J. Clin. Nutr. 34, 914–923 10.1093/ajcn/34.5.914 [DOI] [PubMed] [Google Scholar]
- 72. Barton L. L., Ritz N. L., Fauque G. D., and Lin H. C. (2017) Sulfur cycling and the intestinal microbiome. Dig. Dis. Sci. 62, 2241–2257 10.1007/s10620-017-4689-5 [DOI] [PubMed] [Google Scholar]
- 73. Gibson G. R., Macfarlane G. T., and Cummings J. H. (1988) Occurrence of sulphate-reducing bacteria in human faeces and the relationship of dissimilatory sulphate reduction to methanogenesis in the large gut. J. Appl. Bacteriol. 65, 103–111 10.1111/j.1365-2672.1988.tb01498.x [DOI] [PubMed] [Google Scholar]
- 74. Macfarlane G. T., Cummings J. H., and Macfarlane S. (2007) Sulphate-reducing bacteria and the human large intestine. in Sulphate-reducing Bacteria: Environmental and Engineered Systems (Barton L. L., and Hamilton W. A., eds) pp. 503–522, Cambridge University Press, Cambridge, UK [Google Scholar]
- 75. Barrett E. L., and Clark M. A. (1987) Tetrathionate reduction and production of hydrogen sulfide from thiosulfate. Microbiol. Rev. 51, 192–205 10.1128/MMBR.51.2.192-205.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Libiad M., Yadav P. K., Vitvitsky V., Martinov M., and Banerjee R. (2014) Organization of the human mitochondrial hydrogen sulfide oxidation pathway. J. Biol. Chem. 289, 30901–30910 10.1074/jbc.M114.602664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Libiad M., Vitvitsky V., Bostelaar T., Bak D. W., Lee H. J., Sakamoto N., Fearon E., Lyssiotis C. A., Weerapana E., and Banerjee R. (2019) Hydrogen sulfide perturbs mitochondrial bioenergetics and triggers metabolic reprogramming in colon cells. J. Biol. Chem. 294, 12077–12090 10.1074/jbc.RA119.009442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pitcher M. C., and Cummings J. H. (1996) Hydrogen sulphide: a bacterial toxin in ulcerative colitis? Gut 39, 1–4 10.1136/gut.39.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Attene-Ramos M. S., Wagner E. D., Plewa M. J., and Gaskins H. R. (2006) Evidence that hydrogen sulfide is a genotoxic agent. Mol. Cancer Res. 4, 9–14 10.1158/1541-7786.MCR-05-0126 [DOI] [PubMed] [Google Scholar]
- 80. Xu G. Y., Winston J. H., Shenoy M., Zhou S., Chen J. D., and Pasricha P. J. (2009) The endogenous hydrogen sulfide producing enzyme cystathionine-β synthase contributes to visceral hypersensitivity in a rat model of irritable bowel syndrome. Mol. Pain 5, 44 10.1186/1744-8069-5-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Winter S. E., Thiennimitr P., Winter M. G., Butler B. P., Huseby D. L., Crawford R. W., Russell J. M., Bevins C. L., Adams L. G., Tsolis R. M., Roth J. R., and Bäumler A. J. (2010) Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429 10.1038/nature09415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Babior B. M., Kipnes R. S., and Curnutte J. T. (1973) Biological defense mechanisms: the production by leukocytes of superoxide, a potential bactericidal agent. J. Clin. Invest. 52, 741–744 10.1172/JCI107236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Goldstein I. M., Roos D., Kaplan H. B., and Weissmann G. (1975) Complement and immunoglobulins stimulate superoxide production by human leukocytes independently of phagocytosis. J. Clin. Invest. 56, 1155–1163 10.1172/JCI108191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Johnston R. B. Jr., Keele B. B. Jr., Misra H. P., Lehmeyer J. E., Webb L. S., Baehner R. L., and RaJagopalan K. V. (1975) The role of superoxide anion generation in phagocytic bactericidal activity: studies with normal and chronic granulomatous disease leukocytes. J. Clin. Invest. 55, 1357–1372 10.1172/JCI108055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Root R. K., Metcalf J., Oshino N., and Chance B. (1975) H2O2 release from human granulocytes during phagocytosis. I. Documentation, quantitation, and some regulating factors. J. Clin. Invest. 55, 945–955 10.1172/JCI108024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tauber A. I., and Babior B. M. (1977) Evidence for hydroxyl radical production by human neutrophils. J. Clin. Invest. 60, 374–379 10.1172/JCI108786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Mundi H., Björkstén B., Svanborg C., Ohman L., and Dahlgren C. (1991) Extracellular release of reactive oxygen species from human neutrophils upon interaction with Escherichia coli strains causing renal scarring. Infect. Immun. 59, 4168–4172 10.1128/IAI.59.11.4168-4172.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Albesa I., Becerra M. C., Battán P. C., and Páez P. L. (2004) Oxidative stress involved in the antibacterial action of different antibiotics. Biochem. Biophys. Res. Commun. 317, 605–609 10.1016/j.bbrc.2004.03.085 [DOI] [PubMed] [Google Scholar]
- 89. Kohanski M. A., Dwyer D. J., Hayete B., Lawrence C. A., and Collins J. J. (2007) A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130, 797–810 10.1016/j.cell.2007.06.049 [DOI] [PubMed] [Google Scholar]
- 90. Dwyer D. J., Kohanski M. A., Hayete B., and Collins J. J. (2007) Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Systems Biol. 3, 91 10.1038/msb4100135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wang X., and Zhao X. (2009) Contribution of oxidative damage to antimicrobial lethality. Antimicrob. Agents Chemother. 53, 1395–1402 10.1128/AAC.01087-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Keren I., Wu Y., Inocencio J., Mulcahy L. R., and Lewis K. (2013) Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339, 1213–1216 10.1126/science.1232688 [DOI] [PubMed] [Google Scholar]
- 93. Liu Y., and Imlay J. A. (2013) Cell death from antibiotics without the involvement of reactive oxygen species. Science 339, 1210–1213 10.1126/science.1232751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Darland G., and Davis B. R. (1974) Biochemical and serological characterization of hydrogen sulfide-positive variants of Escherichia coli. Appl. Microbiol. 27, 54–58 10.1128/AEM.27.1.54-58.1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Gusarov I., Shatalin K., Starodubtseva M., and Nudler E. (2009) Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science 325, 1380–1384 10.1126/science.1175439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Peng H., Zhang Y., Palmer L. D., Kehl-Fie T. E., Skaar E. P., Trinidad J. C., and Giedroc D. P. (2017) Hydrogen sulfide and reactive sulfur species impact proteome S-sulfhydration and global virulence regulation in Staphylococcus aureus. ACS Infect. Dis. 3, 744–755 10.1021/acsinfecdis.7b00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Korshunov S., Imlay K. R., and Imlay J. A. (2016) The cytochrome bd oxidase of Escherichia coli prevents respiratory inhibition by endogenous and exogenous hydrogen sulfide. Mol. Microbiol. 101, 62–77 10.1111/mmi.13372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ooi X. J., and Tan K. S. (2016) Reduced glutathione mediates resistance to H2S toxicity in oral Streptococci. Appl. Environ. Microbiol. 82, 2078–2085 10.1128/AEM.03946-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Gobert A. P., Latour Y. L., Asim M., Finley J. L., Verriere T. G., Barry D. P., Milne G. L., Luis P. B., Schneider C., Rivera E. S., Lindsey-Rose K., Schey K. L., Delgado A. G., Sierra J. C., Piazuelo M. B., et al. (2019) Bacterial pathogens hijack the innate immune response by activation of the reverse transsulfuration pathway. mBio 10, e02174–19 10.1128/mBio.02174-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Davies D. (2003) Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2, 114–122 10.1038/nrd1008 [DOI] [PubMed] [Google Scholar]
- 101. Prax M., and Bertram R. (2014) Metabolic aspects of bacterial persisters. Front. Cell. Infect. Microbiol. 4, 148–148 10.3389/fcimb.2014.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Sonderholm M., Bjarnsholt T., Alhede M., Kolpen M., Jensen P. O., Kuhl M., and Kragh K. N. (2017) The consequences of being in an infectious biofilm: microenvironmental conditions governing antibiotic tolerance. Int. J. Mol. Sci. 18, 2688 10.3390/ijms18122688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Babior B. M., Curnutte J. T., and McMurrich B. J. (1976) The particulate superoxide-forming system from human neutrophils: properties of the system and further evidence supporting its participation in the respiratory burst. J. Clin. Invest. 58, 989–996 10.1172/JCI108553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kolpen M., Hansen C. R., Bjarnsholt T., Moser C., Christensen L. D., van Gennip M., Ciofu O., Mandsberg L., Kharazmi A., Döring G., Givskov M., Høiby N., and Jensen P. Ø. (2010) Polymorphonuclear leucocytes consume oxygen in sputum from chronic Pseudomonas aeruginosa pneumonia in cystic fibrosis. Thorax 65, 57–62 10.1136/thx.2009.114512 [DOI] [PubMed] [Google Scholar]
- 105. Line L., Alhede M., Kolpen M., Kühl M., Ciofu O., Bjarnsholt T., Moser C., Toyofuku M., Nomura N., Høiby N., and Jensen P. Ã. (2014) Physiological levels of nitrate support anoxic growth by denitrification of Pseudomonas aeruginosa at growth rates reported in cystic fibrosis lungs and sputum. Front. Microbiol. 5, 554 10.3389/fmicb.2014.00554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Schlag S., Nerz C., Birkenstock T. A., Altenberend F., and Götz F. (2007) Inhibition of staphylococcal biofilm formation by nitrite. J. Bacteriol. 189, 7911–7919 10.1128/JB.00598-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Barraud N., Hassett D. J., Hwang S.-H., Rice S. A., Kjelleberg S., and Webb J. S. (2006) Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J. Bacteriol. 188, 7344–7353 10.1128/JB.00779-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Barraud N., Schleheck D., Klebensberger J., Webb J. S., Hassett D. J., Rice S. A., and Kjelleberg S. (2009) Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J. Bacteriol. 191, 7333–7342 10.1128/JB.00975-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Klare W., Das T., Ibugo A., Buckle E., Manefield M., and Manos J. (2016) Glutathione-disrupted biofilms of clinical Pseudomonas aeruginosa strains exhibit an enhanced antibiotic effect and a novel biofilm transcriptome. Antimicrob. Agents Chemother. 60, 4539–4551 10.1128/AAC.02919-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Hufnagel D. A., Price J. E., Stephenson R. E., Kelley J., Benoit M. F., and Chapman M. R. (2018) Thiol starvation induces redox-mediated dysregulation of Escherichia coli biofilm components. J. Bacteriol. 200, e00389–17 10.1128/jb.00389-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Cowley E. S., Kopf S. H., LaRiviere A., Ziebis W., and Newman D. K. (2015) Pediatric cystic fibrosis sputum can be chemically dynamic, anoxic, and extremely reduced due to hydrogen sulfide formation. mBio 6, e00767–15 10.1128/mBio.00767-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Motta J.-P., Flannigan K. L., Agbor T. A., Beatty J. K., Blackler R. W., Workentine M. L., Da Silva G. J., Wang R., Buret A. G., and Wallace J. L. (2015) Hydrogen sulfide protects from colitis and restores intestinal microbiota biofilm and mucus production. Inflamm. Bowel Dis. 21, 1006–1017 10.1097/MIB.0000000000000345 [DOI] [PubMed] [Google Scholar]
- 113. Capdevila D. A., Walsh B. J. C., Zhang Y., Dietrich C., Gonzalez-Gutierrez G., and Giedroc D. P. (2020) Structural determinants of persulfide-sensing specificity in a dithiol-based transcriptional regulator. bioRxiv 10.1101/2020.03.22.001966 10.1101/2020.03.22.001966 [DOI] [PMC free article] [PubMed]
- 114. Barbosa R. L., and Benedetti C. E. (2007) BigR, a transcriptional repressor from plant-associated bacteria, regulates an operon implicated in biofilm growth. J. Bacteriol. 189, 6185–6194 10.1128/JB.00331-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Guimarães B. G., Barbosa R. L., Soprano A. S., Campos B. M., de Souza T. A., Tonoli C. C. C., Leme A. F. P., Murakami M. T., and Benedetti C. E. (2011) Plant pathogenic bacteria utilize biofilm growth-associated repressor (BigR), a novel winged-helix redox switch, to control hydrogen sulfide detoxification under hypoxia. J. Biol. Chem. 286, 26148–26157 10.1074/jbc.M111.234039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. de Lira N. P. V., Pauletti B. A., Marques A. C., Perez C. A., Caserta R., de Souza A. A., Vercesi A. E., Paes Leme A. F., and Benedetti C. E. (2018) BigR is a sulfide sensor that regulates a sulfur transferase/dioxygenase required for aerobic respiration of plant bacteria under sulfide stress. Sci. Rep. 8, 3508 10.1038/s41598-018-21974-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Vitvitsky V., Yadav P. K., Kurthen A., and Banerjee R. (2015) Sulfide oxidation by a noncanonical pathway in red blood cells generates thiosulfate and polysulfides. J. Biol. Chem. 290, 8310–8320 10.1074/jbc.M115.639831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Bostelaar T., Vitvitsky V., Kumutima J., Lewis B. E., Yadav P. K., Brunold T. C., Filipovic M., Lehnert N., Stemmler T. L., and Banerjee R. (2016) Hydrogen sulfide oxidation by myoglobin. J. Am. Chem. Soc. 138, 8476–8488 10.1021/jacs.6b03456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Ruetz M., Kumutima J., Lewis B. E., Filipovic M. R., Lehnert N., Stemmler T. L., and Banerjee R. (2017) A distal ligand mutes the interaction of hydrogen sulfide with human neuroglobin. J. Biol. Chem. 292, 6512–6528 10.1074/jbc.M116.770370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Vitvitsky V., Miljkovic J. L., Bostelaar T., Adhikari B., Yadav P. K., Steiger A. K., Torregrossa R., Pluth M. D., Whiteman M., Banerjee R., and Filipovic M. R. (2018) Cytochrome c reduction by H2S potentiates sulfide signaling. ACS Chem. Biol. 13, 2300–2307 10.1021/acschembio.8b00463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Nelp M. T., Zheng V., Davis K. M., Stiefel K. J. E., and Groves J. T. (2019) Potent activation of indoleamine 2,3-dioxygenase by polysulfides. J. Am. Chem. Soc. 141, 15288–15300 10.1021/jacs.9b07338 [DOI] [PubMed] [Google Scholar]
- 122. Lange M., Ok K., Shimberg G. D., Bursac B., Markó L., Ivanović-Burmazović I., Michel S. L. J., and Filipovic M. R. (2019) Direct zinc finger protein persulfidation by H2S is facilitated by Zn(2). Angew. Chem. Int. Ed. Engl. 58, 7997–8001 10.1002/anie.201900823 [DOI] [PubMed] [Google Scholar]
- 123. Shen J., Peng H., Zhang Y., Trinidad J. C., and Giedroc D. P. (2016) Staphylococcus aureus sqr encodes a type II sulfide:quinone oxidoreductase and impacts reactive sulfur speciation in cells. Biochemistry 55, 6524–6534 10.1021/acs.biochem.6b00714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Duzs Á., Tóth A., Németh B., Balogh T., Kós P. B., and Rákhely G. (2018) A novel enzyme of type VI sulfide:quinone oxidoreductases in purple sulfur photosynthetic bacteria. Appl. Microbiol. Biotechnol. 102, 5133–5147 10.1007/s00253-018-8973-x [DOI] [PubMed] [Google Scholar]
- 125. Landry A. P., Moon S., Kim H., Yadav P. K., Guha A., Cho U.-S., and Banerjee R. (2019) A catalytic trisulfide in human sulfide quinone oxidoreductase catalyzes coenzyme a persulfide synthesis and inhibits butyrate oxidation. Cell Chem Biol. 26, 1515–1525 10.1016/j.chembiol.2019.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Walsh B. J. C., Brito J. A., and Giedroc D. P. (2020) Hydrogen sulfide signaling and enzymology. in Comprehensive Natural Products III (Liu H.-W., and Begley T. P., eds), Vol 4, pp 430–473, Elsevier, Oxford [Google Scholar]
- 127. Mueller E. G. (2006) Trafficking in persulfides: delivering sulfur in biosynthetic pathways. Nat. Chem. Biol. 2, 185–194 10.1038/nchembio779 [DOI] [PubMed] [Google Scholar]
- 128. Cipollone R., Ascenzi P., and Visca P. (2007) Common themes and variations in the rhodanese superfamily. IUBMB Life 59, 51–59 10.1080/15216540701206859 [DOI] [PubMed] [Google Scholar]
- 129. Dahl J. U., Radon C., Bühning M., Nimtz M., Leichert L. I., Denis Y., Jourlin-Castelli C., Iobbi-Nivol C., Méjean V., and Leimkühler S. (2013) The sulfur carrier protein TusA has a pleiotropic role in Escherichia coli that also affects molybdenum cofactor biosynthesis. J. Biol. Chem. 288, 5426–5442 10.1074/jbc.M112.431569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Dahl J. U., Urban A., Bolte A., Sriyabhaya P., Donahue J. L., Nimtz M., Larson T. J., and Leimkühler S. (2011) The identification of a novel protein involved in molybdenum cofactor biosynthesis in Escherichia coli. J. Biol. Chem. 286, 35801–35812 10.1074/jbc.M111.282368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Palmer L. D., Leung M. H., and Downs D. M. (2014) The cysteine desulfhydrase CdsH is conditionally required for sulfur mobilization to the thiamine thiazole in Salmonella enterica. J. Bacteriol. 196, 3964–3970 10.1128/JB.02159-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Shi R., Proteau A., Villarroya M., Moukadiri I., Zhang L., Trempe J. F., Matte A., Armengod M. E., and Cygler M. (2010) Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions. PLoS Biol. 8, e1000354 10.1371/journal.pbio.1000354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Shigi N. (2018) Recent advances in our understanding of the biosynthesis of sulfur modifications in tRNAs. Front. Microbiol. 9, 2679 10.3389/fmicb.2018.02679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Libiad M., Motl N., Akey D. L., Sakamoto N., Fearon E. R., Smith J. L., and Banerjee R. (2018) Thiosulfate sulfurtransferase-like domain-containing 1 protein interacts with thioredoxin. J. Biol. Chem. 293, 2675–2686 10.1074/jbc.RA117.000826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Higgins K. A., Peng H., Luebke J. L., Chang F. M., and Giedroc D. P. (2015) Conformational analysis and chemical reactivity of the multidomain sulfurtransferase, Staphylococcus aureus CstA. Biochemistry 54, 2385–2398 10.1021/acs.biochem.5b00056 [DOI] [PubMed] [Google Scholar]
- 136. Cuevasanta E., Reyes A. M., Zeida A., Mastrogiovanni M., De Armas M. I., Radi R., Alvarez B., and Trujillo M. (2019) Kinetics of formation and reactivity of the persulfide in the one-cysteine peroxiredoxin from Mycobacterium tuberculosis. J. Biol. Chem. 294, 13593–13605 10.1074/jbc.RA119.008883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. McCoy J. G., Bingman C. A., Bitto E., Holdorf M. M., Makaroff C. A., and Phillips G. N. Jr. (2006) Structure of an ETHE1-like protein from Arabidopsis thaliana. Acta Crystallogr. D Biol. Crystallogr. 62, 964–970. 10.1107/S0907444906020592 [DOI] [PubMed] [Google Scholar]
- 138. Motl N., Skiba M. A., Kabil O., Smith J. L., and Banerjee R. (2017) Structural and biochemical analyses indicate that a bacterial persulfide dioxygenase-rhodanese fusion protein functions in sulfur assimilation. J. Biol. Chem. 292, 14026–14038 10.1074/jbc.M117.790170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Shen J., Keithly M. E., Armstrong R. N., Higgins K. A., Edmonds K. A., and Giedroc D. P. (2015) Staphylococcus aureus CstB is a novel multidomain persulfide dioxygenase-sulfurtransferase involved in hydrogen sulfide detoxification. Biochemistry 54, 4542–4554 10.1021/acs.biochem.5b00584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Sattler S. A., Wang X., Lewis K. M., DeHan P. J., Park C. M., Xin Y., Liu H., Xian M., Xun L., and Kang C. (2015) Characterizations of two bacterial persulfide dioxygenases of the metallo-β-lactamase superfamily. J. Biol. Chem. 290, 18914–18923 10.1074/jbc.M115.652537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Lee K. H., Humbarger S., Bahnvadia R., Sazinsky M. H., and Crane E. J. 3rd (2014) Characterization of the mechanism of the NADH-dependent polysulfide reductase (Npsr) from Shewanella loihica PV-4: formation of a productive NADH-enzyme complex and its role in the general mechanism of NADH and FAD-dependent enzymes. Biochim. Biophys. Acta 1844, 1708–1717 10.1016/j.bbapap.2014.06.013 [DOI] [PubMed] [Google Scholar]
- 142. Warner M. D., Lukose V., Lee K. H., Lopez K., M H. S., and Crane E. J. 3rd (2011) Characterization of an NADH-dependent persulfide reductase from Shewanella loihica PV-4: implications for the mechanism of sulfur respiration via FAD-dependent enzymes. Biochemistry 50, 194–206 10.1021/bi101232y [DOI] [PubMed] [Google Scholar]
- 143. Wallen J. R., Mallett T. C., Boles W., Parsonage D., Furdui C. M., Karplus P. A., and Claiborne A. (2009) Crystal structure and catalytic properties of Bacillus anthracis CoADR-RHD: implications for flavin-linked sulfur trafficking. Biochemistry 48, 9650–9667 10.1021/bi900887k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Liu T., Ramesh A., Ma Z., Ward S. K., Zhang L., George G. N., Talaat A. M., Sacchettini J. C., and Giedroc D. P. (2007) CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat. Chem. Biol. 3, 60–68 10.1038/nchembio844 [DOI] [PubMed] [Google Scholar]
- 145. Chang F. M., Coyne H. J., Cubillas C., Vinuesa P., Fang X., Ma Z., Ma D., Helmann J. D., García-de los Santos A., Wang Y. X., Dann C. E. 3rd, and Giedroc D. P. (2014) Cu(I)-mediated allosteric switching in a copper-sensing operon repressor (CsoR). J. Biol. Chem. 289, 19204–19217 10.1074/jbc.M114.556704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Busenlehner L. S., Pennella M. A., and Giedroc D. P. (2003) The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27, 131–143 10.1016/S0168-6445(03)00054-8 [DOI] [PubMed] [Google Scholar]
- 147. Gristwood T., McNeil M. B., Clulow J. S., Salmond G. P., and Fineran P. C. (2011) PigS and PigP regulate prodigiosin biosynthesis in Serratia via differential control of divergent operons, which include predicted transporters of sulfur-containing molecules. J. Bacteriol. 193, 1076–1085 10.1128/JB.00352-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Weinitschke S., Denger K., Cook A. M., and Smits T. H. M. (2007) The DUF81 protein TauE in Cupriavidus necator H16, a sulfite exporter in the metabolism of C2 sulfonates. Microbiology 153, 3055–3060 10.1099/mic.0.2007/009845-0 [DOI] [PubMed] [Google Scholar]
- 149. Studholme D. J., and Dixon R. (2003) Domain architectures of σ54-dependent transcriptional activators. J. Bacteriol. 185, 1757–1767 10.1128/jb.185.6.1757-1767.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. D'Autreaux B., Tucker N. P., Dixon R., and Spiro S. (2005) A non-haem iron centre in the transcription factor NorR senses nitric oxide. Nature 437, 769–772 10.1038/nature03953 [DOI] [PubMed] [Google Scholar]
- 151. Tucker N. P., D'Autreaux B., Yousafzai F. K., Fairhurst S. A., Spiro S., and Dixon R. (2008) Analysis of the nitric oxide-sensing non-heme iron center in the NorR regulatory protein. J. Biol. Chem. 283, 908–918 10.1074/jbc.M705850200 [DOI] [PubMed] [Google Scholar]
- 152. Yang B., Nie X., Xiao Y., Gu Y., Jiang W., and Yang C. (2020) Ferrous-iron-activated transcriptional factor AdhR regulates redox homeostasis in Clostridium beijerinckii. Appl. Environ. Microbiol. 86, e02782–19 10.1128/AEM.02782-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Bora P., Chauhan P., Pardeshi K. A., and Chakrapani H. (2018) Small molecule generators of biologically reactive sulfur species. RSC Adv. 8, 27359–27374 10.1039/C8RA03658F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Levinn C. M., Cerda M. M., and Pluth M. D. (2020) Activatable small-molecule hydrogen sulfide donors. Antioxid. Redox Signal. 32, 96–109 10.1089/ars.2019.7841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Lin V. S., Chen W., Xian M., and Chang C. J. (2015) Chemical probes for molecular imaging and detection of hydrogen sulfide and reactive sulfur species in biological systems. Chem. Soc. Rev. 44, 4596–4618 10.1039/c4cs00298a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Powell C. R., Dillon K. M., and Matson J. B. (2018) A review of hydrogen sulfide (H2S) donors: chemistry and potential therapeutic applications. Biochem. Pharmacol. 149, 110–123 10.1016/j.bcp.2017.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Takano Y., Echizen H., and Hanaoka K. (2017) Fluorescent probes and selective inhibitors for biological studies of hydrogen sulfide- and polysulfide-mediated signaling. Antioxid. Redox. Signal. 27, 669–683 10.1089/ars.2017.7070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Xu S., Hamsath A., Neill D. L., Wang Y., Yang C. T., and Xian M. (2019) Strategies for the design of donors and precursors of reactive sulfur species. Chemistry 25, 4005–4016 10.1002/chem.201804895 [DOI] [PubMed] [Google Scholar]
- 159. Jiao X., Li Y., Niu J., Xie X., Wang X., and Tang B. (2018) Small-molecule fluorescent probes for imaging and detection of reactive oxygen, nitrogen, and sulfur species in biological systems. Anal. Chem. 90, 533–555 10.1021/acs.analchem.7b04234 [DOI] [PubMed] [Google Scholar]
- 160. Li L., Whiteman M., Guan Y. Y., Neo K. L., Cheng Y., Lee S. W., Zhao Y., Baskar R., Tan C. H., and Moore P. K. (2008) Characterization of a novel, water-soluble hydrogen sulfide-releasing molecule (GYY4137): new insights into the biology of hydrogen sulfide. Circulation 117, 2351–2360 10.1161/CIRCULATIONAHA.107.753467 [DOI] [PubMed] [Google Scholar]
- 161. Kang J., Li Z., Organ C. L., Park C.-M., Yang C-T., Pacheco A., Wang D., Lefer D. J., and Xian M. (2016) pH-controlled hydrogen sulfide release for myocardial ischemia-reperfusion injury. J. Am. Chem. Soc. 138, 6336–6339 10.1021/jacs.6b01373 [DOI] [PubMed] [Google Scholar]
- 162. Levinn C. M., Cerda M. M., and Pluth M. D. (2019) Development and application of carbonyl sulfide-based donors for H2S delivery. Acc. Chem. Res. 52, 2723–2731 10.1021/acs.accounts.9b00315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Supuran C. T., and Capasso C. (2017) An overview of the bacterial carbonic anhydrases. Metabolites 7, 56 10.3390/metabo7040056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Bianco C. L., Akaike T., Ida T., Nagy P., Bogdandi V., Toscano J. P., Kumagai Y., Henderson C. F., Goddu R. N., Lin J., and Fukuto J. M. (2019) The reaction of hydrogen sulfide with disulfides: formation of a stable trisulfide and implications for biological systems. Br. J. Pharmacol. 176, 671–683 10.1111/bph.14372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Artaud I., and Galardon E. (2014) A persulfide analogue of the nitrosothiol SNAP: formation, characterization and reactivity. Chembiochem 15, 2361–2364 10.1002/cbic.201402312 [DOI] [PubMed] [Google Scholar]
- 166. Yuan Z., Zheng Y., Yu B., Wang S., Yang X., and Wang B. (2018) Esterase-sensitive glutathione persulfide donor. Org. Lett. 20, 6364–6367 10.1021/acs.orglett.8b02611 [DOI] [PubMed] [Google Scholar]
- 167. Zheng Y., Yu B., Li Z., Yuan Z., Organ C. L., Trivedi R. K., Wang S., Lefer D. J., and Wang B. (2017) An esterase-sensitive prodrug approach for controllable delivery of persulfide species. Angew. Chem. Int. Ed. Engl. 56, 11749–11753 10.1002/anie.201704117 [DOI] [PubMed] [Google Scholar]
- 168. Bora P., Chauhan P., Manna S., and Chakrapani H. (2018) A vinyl-boronate ester-based persulfide donor controllable by hydrogen peroxide, a reactive oxygen species (ROS). Org. Lett. 20, 7916–7920 10.1021/acs.orglett.8b03471 [DOI] [PubMed] [Google Scholar]
- 169. Powell C. R., Dillon K. M., Wang Y., Carrazzone R. J., and Matson J. B. (2018) A persulfide donor responsive to reactive oxygen species: insights into reactivity and therapeutic potential. Angew. Chem. Int. Ed. Engl. 57, 6324–6328 10.1002/anie.201803087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Moest R. R. (1975) Hydrogen sulfide determination by the methylene blue method. Anal. Chem. 47, 1204–1205 10.1021/ac60357a008 [DOI] [Google Scholar]
- 171. Bibli S.-I., Luck B., Zukunft S., Wittig J., Chen W., Xian M., Papapetropoulos A., Hu J., and Fleming I. (2018) A selective and sensitive method for quantification of endogenous polysulfide production in biological samples. Redox Biol. 18, 295–304 10.1016/j.redox.2018.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Olson K. R., Gao Y., Arif F., Arora K., Patel S., DeLeon E. R., Sutton T. R., Feelisch M., Cortese-Krott M. M., and Straub K. D. (2018) Metabolism of hydrogen sulfide (H2S) and production of reactive sulfur species (RSS) by superoxide dismutase. Redox Biol. 15, 74–85 10.1016/j.redox.2017.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Olson K. R., Gao Y., DeLeon E. R., Arif M., Arif F., Arora N., and Straub K. D. (2017) Catalase as a sulfide-sulfur oxido-reductase: an ancient (and modern?) regulator of reactive sulfur species (RSS). Redox Biol. 12, 325–339 10.1016/j.redox.2017.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Ditrói T., Nagy A., Martinelli D., Rosta A., Kožich V., and Nagy P. (2019) Comprehensive analysis of how experimental parameters affect H2S measurements by the monobromobimane method. Free Radic. Biol. Med. 136, 146–158 10.1016/j.freeradbiomed.2019.04.006 [DOI] [PubMed] [Google Scholar]
- 175. Bogdándi V., Ida T., Sutton T. R., Bianco C., Ditrói T., Koster G., Henthorn H. A., Minnion M., Toscano J. P., van der Vliet A., Pluth M. D., Feelisch M., Fukuto J. M., Akaike T., and Nagy P. (2019) Speciation of reactive sulfur species and their reactions with alkylating agents: do we have any clue about what is present inside the cell? Br. J. Pharmacol. 176, 646–670 10.1111/bph.14394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Hamid H. A., Tanaka A., Ida T., Nishimura A., Matsunaga T., Fujii S., Morita M., Sawa T., Fukuto J. M., Nagy P., Tsutsumi R., Motohashi H., Ihara H., and Akaike T. (2019) Polysulfide stabilization by tyrosine and hydroxyphenyl-containing derivatives that is important for a reactive sulfur metabolomics analysis. Redox Biol. 21, 101096 10.1016/j.redox.2019.101096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Lin J., Akiyama M., Bica I., Long F. T., Henderson C. F., Goddu R. N., Suarez V., Baker B., Ida T., Shinkai Y., Nagy P., Akaike T., Fukuto J. M., and Kumagai Y. (2019) The uptake and release of polysulfur cysteine species by cells: physiological and toxicological implications. Chem. Res. Toxicol. 32, 447–455 10.1021/acs.chemrestox.8b00340 [DOI] [PubMed] [Google Scholar]
- 178. Lo Conte M., and Carroll K. S. (2013) The redox biochemistry of protein sulfenylation and sulfinylation. J. Biol. Chem. 288, 26480–26488 10.1074/jbc.R113.467738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Loi V. V., Rossius M., and Antelmann H. (2015) Redox regulation by reversible protein S-thiolation in bacteria. Front. Microbiol. 6, 187 10.3389/fmicb.2015.00187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Fernando V., Zheng X., Walia Y., Sharma V., Letson J., and Furuta S. (2019) S-Nitrosylation: an emerging paradigm of redox signaling. Antioxidants (Basel) 8, 404 10.3390/antiox8090404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Jarosz A. P., Wei W., Gauld J. W., Auld J., Özcan F., Aslan M., and Mutus B. (2015) Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is inactivated by S-sulfuration in vitro. Free Radic. Biol. Med. 89, 512–521 10.1016/j.freeradbiomed.2015.09.007 [DOI] [PubMed] [Google Scholar]
- 182. Padgett C. M., and Whorton A. R. (1995) S-Nitrosoglutathione reversibly inhibits GAPDH by S-nitrosylation. Am. J. Physiol. Cell Physiol. 269, C739–C749 10.1152/ajpcell.1995.269.3.C739 [DOI] [PubMed] [Google Scholar]
- 183. Woo H. A., Jeong W., Chang T.-S., Park K. J., Park S. J., Yang J. S., and Rhee S. G. (2005) Reduction of cysteine sulfinic acid by sulfiredoxin is specific to 2-Cys peroxiredoxins. J. Biol. Chem. 280, 3125–3128 10.1074/jbc.C400496200 [DOI] [PubMed] [Google Scholar]
- 184. Ravichandran V., Seres T., Moriguchi T., Thomas J. A., and Johnston R. B. (1994) S-Thiolation of glyceraldehyde-3-phosphate dehydrogenase induced by the phagocytosis-associated respiratory burst in blood monocytes. J. Biol. Chem. 269, 25010–25015 [PubMed] [Google Scholar]
- 185. Broniowska K. A., Diers A. R., and Hogg N. (2013) S-Nitrosoglutathione. Biochim. Biophys. Acta 1830, 3173–3181 10.1016/j.bbagen.2013.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Matsumoto A., Comatas K. E., Liu L., and Stamler J. S. (2003) Screening for nitric oxide-dependent protein-protein interactions. Science 301, 657–661 10.1126/science.1079319 [DOI] [PubMed] [Google Scholar]
- 187. Benhar M., Forrester M. T., and Stamler J. S. (2009) Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat. Rev. Mol. Cell Biol. 10, 721–732 10.1038/nrm2764 [DOI] [PubMed] [Google Scholar]
- 188. Anand P., Hausladen A., Wang Y.-J., Zhang G.-F., Stomberski C., Brunengraber H., Hess D. T., and Stamler J. S. (2014) Identification of S-nitroso-CoA reductases that regulate protein S-nitrosylation. Proc. Natl. Acad. Sci. U. S. A. 111, 18572–18577 10.1073/pnas.1417816112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Zhou H.-L., Zhang R., Anand P., Stomberski C. T., Qian Z., Hausladen A., Wang L., Rhee E. P., Parikh S. M., Karumanchi S. A., and Stamler J. S. (2019) Metabolic reprogramming by the S-nitroso-CoA reductase system protects against kidney injury. Nature 565, 96–100 10.1038/s41586-018-0749-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Liu L., Hausladen A., Zeng M., Que L., Heitman J., and Stamler J. S. (2001) A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410, 490–494 10.1038/35068596 [DOI] [PubMed] [Google Scholar]
- 191. Wedmann R., Onderka C., Wei S., Szijártó I. A., Miljkovic J. L., Mitrovic A., Lange M., Savitsky S., Yadav P. K., Torregrossa R., Harrer E. G., Harrer T., Ishii I., Gollasch M., Wood M. E., et al. (2016) Improved tag-switch method reveals that thioredoxin acts as depersulfidase and controls the intracellular levels of protein persulfidation. Chem. Sci. 7, 3414–3426 10.1039/c5sc04818d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Dóka E., Pader I., Biró A., Johansson K., Cheng Q., Ballagó K., Prigge J. R., Pastor-Flores D., Dick T. P., Schmidt E. E., Arnér E. S., and Nagy P. (2016) A novel persulfide detection method reveals protein persulfide- and polysulfide-reducing functions of thioredoxin and glutathione systems. Sci. Adv. 2, e1500968 10.1126/sciadv.1500968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Peng H., Zhang Y., Trinidad J. C., and Giedroc D. P. (2018) Thioredoxin profiling of multiple thioredoxin-like proteins in Staphylococcus aureus. Front. Microbiol. 9, 2385 10.3389/fmicb.2018.02385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Pan J., and Carroll K. S. (2013) Persulfide reactivity in the detection of protein S-sulfhydration. ACS Chem. Biol. 8, 1110–1116 10.1021/cb4001052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Gao X. H., Krokowski D., Guan B. J., Bederman I., Majumder M., Parisien M., Diatchenko L., Kabil O., Willard B., Banerjee R., Wang B., Bebek G., Evans C. R., Fox P. L., Gerson S. L., et al. (2015) Quantitative H2S-mediated protein sulfhydration reveals metabolic reprogramming during the integrated stress response. Elife 4, e10067 10.7554/eLife.10067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Longen S., Richter F., Köhler Y., Wittig I., Beck K.-F., and Pfeilschifter J. (2016) Quantitative persulfide site identification (qPerS-SID) reveals protein targets of H2S releasing donors in mammalian cells. Sci. Rep. 6, 29808 10.1038/srep29808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Kouroussis E., Adhikari B., Zivanovic J., and Filipovic M. R. (2019) Measurement of protein persulfidation: improved tag-switch method. in Vascular Effects of Hydrogen Sulfide: Methods and Protocols (Bełtowski J., ed) pp. 37–50, Springer, New York: [DOI] [PubMed] [Google Scholar]
- 198. Park C.-M., Macinkovic I., Filipovic M. R., and Xian M. (2015) Use of the “tag-switch” method for the detection of protein S-sulfhydration. Methods Enzymol. 555, 39–56 10.1016/bs.mie.2014.11.033 [DOI] [PubMed] [Google Scholar]
- 199. Zhang D., Macinkovic I., Devarie-Baez N. O., Pan J., Park C.-M., Carroll K. S., Filipovic M. R., and Xian M. (2014) Detection of protein S-sulfhydration by a tag-switch technique. Angew. Chem. Int. Ed. Engl. 53, 575–581 10.1002/anie.201305876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Zivanovic J., Kouroussis E., Kohl J. B., Adhikari B., Bursac B., Schott-Roux S., Petrovic D., Miljkovic J. L., Thomas-Lopez D., Jung Y., Miler M., Mitchell S., Milosevic V., Gomes J. E., Benhar M., et al. (2019) Selective persulfide detection reveals evolutionarily conserved antiaging effects of S-sulfhydration. Cell Metab. 30, 1152–1170.e13 10.1016/j.cmet.2019.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Bernal-Perez L. F., Prokai L., and Ryu Y. (2012) Selective N-terminal fluorescent labeling of proteins using 4-chloro-7-nitrobenzofurazan: a method to distinguish protein N-terminal acetylation. Anal. Biochem. 428, 13–15 10.1016/j.ab.2012.05.026 [DOI] [PubMed] [Google Scholar]
- 202. Ellis H. R., and Poole L. B. (1997) Novel application of 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole to identify cysteine sulfenic acid in the AhpC component of alkyl hydroperoxide reductase. Biochemistry 36, 15013–15018 10.1021/bi972191x [DOI] [PubMed] [Google Scholar]
- 203. Paulsen C. E., and Carroll K. S. (2013) Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem. Rev. 113, 4633–4679 10.1021/cr300163e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Furdui C. M., and Poole L. B. (2014) Chemical approaches to detect and analyze protein sulfenic acids. Mass Spectrom. Rev. 33, 126–146 10.1002/mas.21384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Pan J., and Carroll K. S. (2014) Chemical biology approaches to study protein cysteine sulfenylation. Biopolymers 101, 165–172 10.1002/bip.22255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Fu L., Liu K., He J., Tian C., Yu X., and Yang J. (2019) Direct proteomic mapping of cysteine persulfidation. Antioxid. Redox Signal. 10.1089/ars.2019.7777 10.1089/ars.2019.7777 [DOI] [PubMed] [Google Scholar]
- 207. Yang J., Gupta V., Carroll K. S., and Liebler D. C. (2014) Site-specific mapping and quantification of protein S-sulphenylation in cells. Nat. Commun. 5, 4776 10.1038/ncomms5776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Bestetti S., Medraño-Fernandez I., Galli M., Ghitti M., Bienert G. P., Musco G., Orsi A., Rubartelli A., and Sitia R. (2018) A persulfidation-based mechanism controls aquaporin-8 conductance. Sci. Adv. 4, eaar5770 10.1126/sciadv.aar5770 [DOI] [PMC free article] [PubMed] [Google Scholar]


