Abstract
α1-Antitrypsin (AAT) encoded by the SERPINA1 gene is an acute-phase protein synthesized in the liver and secreted into the circulation. Its primary role is to protect lung tissue by inhibiting neutrophil elastase. The Z allele of SERPINA1 encodes a mutant AAT, named ATZ, that changes the protein structure and leads to its misfolding and polymerization, which cause endoplasmic reticulum (ER) stress and liver disease through a gain-of-function toxic mechanism. Hepatic retention of ATZ results in deficiency of one of the most important circulating proteinase inhibitors and predisposes to early-onset emphysema through a loss-of-function mechanism. The pathogenetic mechanisms underlying the liver disease are not completely understood. C/EBP-homologous protein (CHOP), a transcription factor induced by ER stress, was found among the most up-regulated genes in livers of PiZ mice that express ATZ and in human livers of patients homozygous for the Z allele. Compared with controls, juvenile PiZ/Chop−/− mice showed reduced hepatic ATZ and a transcriptional response indicative of decreased ER stress by RNA-Seq analysis. Livers of PiZ/Chop−/− mice also showed reduced SERPINA1 mRNA levels. By chromatin immunoprecipitations and luciferase reporter–based transfection assays, CHOP was found to up-regulate SERPINA1 cooperating with c-JUN, which was previously shown to up-regulate SERPINA1, thus aggravating hepatic accumulation of ATZ. Increased CHOP levels were detected in diseased livers of children homozygous for the Z allele. In summary, CHOP and c-JUN up-regulate SERPINA1 transcription and play an important role in hepatic disease by increasing the burden of proteotoxic ATZ, particularly in the pediatric population.
Keywords: alpha1-antitrypsin deficiency, CHOP, SERPINA1, c-JUN, liver, liver injury, c-Jun transcription factor, serpin, transcription regulation, alpha1-antitrypsin
α1-Antitrypsin (AAT) encoded by the SERPINA1 gene is an acute-phase protein and one of the major circulating proteinase inhibitors mainly synthesized and secreted by hepatocytes. AAT deficiency is a common genetic cause of lung and liver diseases. The vast majority of patients with AAT deficiency (∼95%) are homozygous for the Z mutation in the SERPINA1 gene (PiZZ), which results in a single glutamic acid–to–lysine substitution at amino acid position 342 (p.Glu342Lys) (1). This mutation changes the protein structure, leading to misfolding, polymerization, and accumulation of mutant AAT (ATZ) in the endoplasmic reticulum (ER) (1). In PiZZ individuals, intrahepatic retention of ATZ results in low circulating levels of AAT and inadequate anti-protease protection in the lower respiratory tract that can lead to progressive lung emphysema (1). PiZZ individuals are also at risk of developing liver disease due to intracellular retention and accumulation of aberrantly folded ATZ leading to hepatitis, cirrhosis, liver failure, and hepatocellular carcinoma (2, 3). There is great variability in the severity of liver disease among PiZZ homozygotes, and a survey of a unique cohort of homozygote individuals identified by an unbiased newborn screening carried out in Sweden showed that about 8% of homozygotes develop clinically significant liver disease in the first 4 decades of life (3). A greater proportion of this population will likely develop liver injury as they reach older ages. Nevertheless, a number of PiZZ individuals do not manifest clinical symptoms of liver disease throughout their lifetime (4). These data suggest that genetic and/or environmental modifiers play a major role in susceptibility to liver disease. Understanding the mechanisms involved in the pathogenesis of the liver disorder is important to identify disease modifiers and to develop preventive and therapeutic strategies.
Combining results in mouse and human livers expressing ATZ, we identified CHOP as an important transcriptional regulator of SERPINA1 expression aggravating the liver disease. For this study, we used transgenic PiZ mice, that have been a valuable model for studying the liver disease induced by ATZ because in the livers of these mice, ATZ accumulates within the ER of hepatocytes as periodic acid–Schiff–diastase–resistant (PAS-D) positive globules, in a nearly identical manner to livers of human patients (5). Because they have been genetically engineered to express the human SERPINA1 gene harboring the Z mutation that includes the human promoter region (6), they are also useful to investigate SERPINA1 transcriptional regulation. Importantly, mouse data were validated in human liver tissues from PiZZ patients of various ages with hepatic disease.
Results
CHOP is activated in mouse livers expressing ATZ
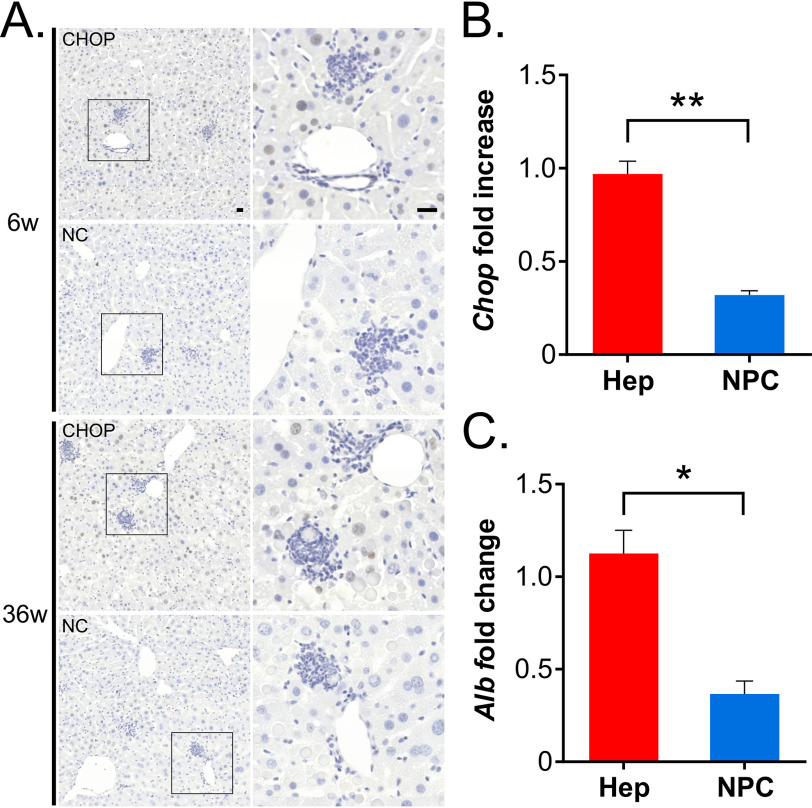
We previously found up-regulation of genes related to response to ER stress in PiZ livers (7). Among differentially expressed genes, the transcription factor Chop was among the most up-regulated. Under nonstress conditions, CHOP is typically not expressed at detectable levels, and its subcellular location is mainly in cytoplasm, whereas stress conditions induce its nuclear translocation (8). Compared with age-matched WT controls, Chop showed a significant 12-fold up-regulation in 6-week-old PiZ mouse livers by real-time PCR (Fig. 1A). Greater Chop up-regulation was detected in 6-week-old compared with 36-week-old PiZ mouse livers (Fig. 1A). Increased expression of CHOP was confirmed at the protein level in 6- and 36-week-old PiZ mouse livers (Fig. 1B and Fig. S1A). Compared with WT controls, PiZ mouse livers showed increased CHOP levels by Western blot analysis also on nuclear extracts, although at lower levels compared with mice injected with tunicamycin, an inducer of ER stress and CHOP expression (9) (Fig. 1C and Fig. S1B). Increased nuclear CHOP in PiZ livers was confirmed by immunohistochemistry (Fig. 1D), and gene set enrichment analysis (GSEA) on microarray expression data (7) showed enrichment of up-regulated CHOP target genes compared with WT controls (enrichment score = 0.58) (Fig. S2, supporting data and Fig. 1E). Moreover, by immunohistochemistry, CHOP-positive nuclei were found in hepatocytes, but they were not detected in infiltrates of inflammatory cells (Fig. 2A). In addition, greater Chop expression was detected in the parenchymal cell fraction compared with nonparenchymal fraction obtained by liver perfusion of PiZ mice (Fig. 2, B and C).
Figure 1.
CHOP is activated in PiZ mouse livers. A, Chop expression in livers of WT and PiZ mice of 6 and 36 weeks of age. B, Western blotting for CHOP in livers of WT and PiZ mice of 6 and 36 weeks of age (each lane corresponds to an independent mouse). A liver of a WT mouse injected intraperitoneally with 4 µg/g tunicamycin was used as a positive control. C, Western blotting for CHOP on nuclear extracts of 6-week-old WT and PiZ mouse livers. Lamin A was used as a nuclear marker and GAPDH as a cytosolic marker. D, representative immunohistochemistry with anti-CHOP antibody in 6- and 36-week-old WT and PiZ mouse livers (×40 magnification; scale bar, 100 μm; n = 3/group). E, heatmap from GSEA using a set of genes up-regulated by CHOP and differentially expressed genes between 6-week-old PiZ and WT mouse livers. The set of CHOP target genes was defined by combining ChIP-Seq and RNA-Seq data reported previously (44) from tunicamycin-treated WT mouse embryonic fibroblasts. Averages ± S. E. (error bars) are shown. Two-way ANOVA and Tukey's post-hoc tests were used: **, p < 0.001. 6w, 6 weeks old; 36w, 36 weeks old; N, nuclear extracts; C, cytosolic extracts; TUNI, tunicamycin.
Figure 2.
CHOP expression detected in nuclei of hepatocytes but not of inflammatory infiltrating cells. A, representative images from immunohistochemistry with anti-CHOP antibody and negative control (NC) in livers of PiZ mice of 6 and 36 weeks of age. Images on the right correspond to the insets of the left panels. Scale bar, 20 μm; n = 3/group). 6w, 6 weeks old; 36w, 36 weeks old. B, Chop expression is greater in the parenchymal fraction enriched for hepatocytes (Hep) compared with the nonparenchymal cell (NPC) fraction. C Albumin, Albumin (Alb) gene expression was evaluated to confirm enrichment for hepatocytes. Averages ± S. E. (error bars) are shown; t test: *, p < 0.05; **, p < 0.01.
To further investigate CHOP expression and hepatic ATZ accumulation, we injected 4-week-old PiZ mice with a recombinant serotype 8 adeno-associated viral (AAV) vector that incorporates an artificial microRNA targeting the human SERPINA1 gene (AAV8-CB-mir914) and results in ATZ hepatic knockdown (10). As controls, mice were injected with the same dose of an AAV8-CB-GFP expressing the GFP. By 4 weeks postinjection, mice injected with AAV8-CB-mir914 showed a reduction of hepatic ATZ by PAS-D staining (Fig. S3A), an ∼90% reduction of serum ATZ (Fig. S3B), and ∼50% reduction of Chop expression compared with mice injected with the control vector expressing GFP (Fig. S3, C and D). Taken together, these data support up-regulation and activation of CHOP in livers of mice expressing ATZ.
Reduced hepatic ATZ accumulation in Chop-deleted juvenile PiZ mice
To evaluate the role of CHOP in hepatic ATZ accumulation, we crossed PiZ mice with Chop−/− mice (9). Chop−/− mice do not exhibit a substantial phenotype unless they are subjected to stress signals (9). Compared with WT, PiZ mice have reduced weight (11), but PiZ/Chop−/− mice showed greater weight compared with PiZ mice (Fig. S4). Serum ATZ in PiZ/Chop−/− mice was lower compared with PiZ mice at 6 weeks of age, but no differences were detected in 36-week-old PiZ/Chop−/− mice (Fig. 3A). Compared with PiZ, livers of 6-week-old PiZ/Chop−/− mice showed marked reduction of hepatic ATZ by PAS-D staining and immunofluorescence with anti-polymer antibody (Fig. 3B). However, neither PAS-D staining nor ATZ polymer immunofluorescence detected any difference in ATZ accumulation in livers of 36-week-old PiZ/Chop−/− mice compared with age-matched PiZ controls (Fig. 3B and Fig. S5). PiZ/Chop−/− mouse livers did not show increased apoptosis like PiZ controls, consistent with previous reports (12) (Fig. S6). Targeted qPCR analysis showed down-regulation of SERPINA1 expression in PiZ/Chop−/− compared with age- and gender-matched 6-week-old PiZ mice, and consistent with liver ATZ content, no significant differences in SERPINA1 expression were detected in older mice (Fig. 3C). Although CHOP has been involved in liver fibrosis (13), compared with age-matched PiZ, liver fibrosis in 69-week-old PiZ/Chop−/− mice appeared to be unaffected by Sirius Red staining and expression of Col1a1, Timp1, and α-Sma (Fig. 3D and Fig. S7). Consistent with PAS-D staining and ATZ polymer immunofluorescence, compared with age-matched PiZ controls, PiZ/Chop−/− mice showed reduced ATZ protein by immunoblotting performed on soluble and insoluble fractions at 6 weeks of age but not at 36 weeks (Fig. 3E and Fig. S8).
Figure 3.
Genetic ablation of Chop reduces hepatic ATZ accumulation in juvenile PiZ mice. A, ELISA for serum ATZ in PiZ and PiZ/Chop−/− mice of 6 and 36 weeks of age (n ≥ 4/group). Serum was collected from the same mice at different ages. Averages ± S. D. (error bars) are shown. Two-way ANOVA and Tukey's post hoc tests were used: *, p < 0.05. B, representative PAS-D staining (×20 magnification; scale bar, 100 μm; n = 19 for 6-week-old PiZ/Chop−/− mice and n = 10 for 36-week-old PiZ/Chop−/− mice) and immunofluorescence (IF) for polymeric ATZ (×63 magnification; scale bar, 50 μm; n = 3/group) of livers of PiZ and PiZ/Chop−/− mice of 6 and 36 weeks of age. C, SERPINA1 expression in PiZ and PiZ/Chop−/− mice at 6 and 36 weeks of age (n = 3/group). β2-Microglobulin was used for normalization. Averages ± S. E. (error bars) are shown. Two-way ANOVA and Tukey's post hoc test were used: *, p < 0.05. D, representative Sirius Red staining on livers of 69-week-old PiZ and PiZ/Chop−/− mice (×10 magnification; scale bar, 200 μm; n = 10 PiZ and n = 5 PiZ/Chop−/− mice). E, Western blotting on soluble total ATZ and insoluble ATZ polymer of 6- and 36-week-old PiZ and PiZ/Chop−/− mouse livers. GAPDH and H3 were used for normalization. ns, not statistically significant; 6w, 6 weeks old; 36W, 36 weeks old; 69W, 69 weeks old.
Unbiased RNA-Seq on liver RNA revealed that PiZ/Chop−/− exhibited a hepatic gene expression profile different from age- and gender-matched PiZ mice both at 6 and 36 weeks of age (Fig. S9). To specifically investigate the effect of CHOP on gene expression profiles of PiZ mice, we performed a comparison of all data sets, as shown by the VENN diagrams in which we evaluated differentially expressed genes obtained by three different comparisons: PiZ versus WT, PiZ/Chop−/− versus WT, and PiZ/Chop−/− versus PiZ at 6 and 36 weeks of age. This analysis allowed us to isolate only genes differentially expressed in PiZ versus WT and normalized to WT levels in PiZ/Chop−/− mice. Whereas at 36 weeks of age no differences in gene expression were detected, at 6 weeks of age, the analysis revealed 912 genes differentially expressed (468 up-regulated genes and 444 down-regulated genes, almost all in opposite correlation) in the intersection between PiZ versus WT and PiZ/Chop−/− versus PiZ (shown in red in Fig. 4A). Gene ontology functional analysis was performed by restricting the output to the biological processes in which differentially expressed genes (up- and down-regulated, separately) were functionally involved. This analysis showed several biological processes, mainly including ER stress, cell proliferation, and immune response for up-regulated genes and metabolic pathways for down-regulated genes among the most significant (enrichment score >1.5) (Figs. S10 and S11). Next, we dissected clusters relevant for the pathogenesis of liver disease induced by ATZ, including cell cycle, immune response, ER stress, cell death, and response to oxidative stress, that were up-regulated in PiZ mouse livers and normalized to WT levels in PiZ/Chop−/− mice (Fig. 4B and supporting data). In contrast, biological clusters of processes related to metabolic functions were down-regulated in PiZ mouse livers and normalized to WT levels in PiZ/Chop−/− mice (Fig. 4C and supporting data). Collectively, these data showed that deletion of Chop reduces expression of pathways involved ER in stress and immune response while restoring several metabolic functions induced by ATZ accumulation. CHOP is also involved in induction of autophagy genes, such as Atg5 and Atg7 upon nutrient deprivation (14). However, RNA-Seq studies did not reveal changes in expression of autophagy genes.
Figure 4.

A, Venn diagram comparing the gene data sets of PiZ versus WT, PiZ/Chop−/− versus WT, and PiZ/Chop−/− versus PiZ at 6 weeks of age. B and C, heatmaps showing the most relevant up- (B) and down-regulated (C) pathways in 6-week-old PiZ mouse livers and normalized to age-matched WT levels in PiZ/Chop−/− mice. DEG, differentially expressed genes; DW, down-regulated; UP, up-regulated.
CHOP cooperates with c-JUN to up-regulate human SERPINA1
Human SERPINA1 gene contains regulatory elements at both the 5′-UTR and 3′-UTR (15). As described previously, the 5′-UTR contains binding sites for AP-1 (shown in green in Fig. 5A) (15). CHOP can make stable heterodimers with the AP-1 complex (e.g. c-JUN and c-FOS), increasing AP-1–mediated gene expression without binding DNA (16). To investigate CHOP-mediated regulation of human SERPINA1 expression, we co-transfected HeLa cells with a plasmid expressing CHOP and the pAAT-Luc-AAT-3′UTR plasmid, which contains the SERPINA1 regulatory elements included in PiZ transgenic mice (7) (dashed line in Fig. 5A) upstream from the firefly luciferase coding region and human SERPINA1 3′-UTR. As positive control, cells were co-transfected with a plasmid expressing c-JUN that transactivates luciferase expression, as shown previously (7). Compared with cells co-transfected with the negative control, cells co-transfected with CHOP showed a mild increase in luciferase levels, suggesting that CHOP up-regulates SERPINA1 expression (Fig. 5B). In contrast, transfections of plasmids expressing c-FOS did not significantly increase luciferase expression (Fig. 5B). Co-transfection of CHOP and c-JUN resulted in greater increase of luciferase levels compared with cells transfected with plasmids expressing each of the two transcription factors alone (Fig. 5B). Mutagenesis of site 2 but not site 1 of the AP-1–binding sites (shown in green in Fig. 5A) reduced luciferase expression after co-transfection of CHOP and c-JUN (Fig. 5C). Taken together, these results show that site 2 of the AP-1–binding sites is required for transactivation by CHOP and c-JUN. Previous studies showed that CHOP forms a complex with c-JUN and enhances AP-1–mediated gene expression without direct binding to the DNA (16). Consistent with these studies, protein co-immunoprecipitation in HeLa cells co-transfected with plasmids expressing FLAG-tagged CHOP and MYC-tagged c-JUN showed that CHOP binds c-JUN (Fig. S12). However, we also found two putative binding sites for CHOP in the SERPINA1 5′-UTR (shown in red in Fig. 5A) (17) that were confirmed by real-time PCR on ChIP of PiZ mouse livers using an anti-CHOP antibody. CHOP-binding sites (shown in red in Fig. 5A) were indeed significantly enriched in PiZ mouse livers, whereas no enrichment was detected for AP-1–binding sites (shown in green in Fig. 5A) (Fig. 5D). As positive control of the ChIP with anti-CHOP antibody, p62 promoter region was found to be enriched in PiZ livers, whereas no signal was detected for the Rpl30 promoter used as negative control. Both CHOP-binding sites and the p62 promoter region were not enriched by ChIP with anti-CHOP antibody in PiZ/Chop−/− mouse livers (Fig. S13). Consistent with previous findings (7), enrichment of AP-1–binding site sequences (shown in green in Fig. 5A) was detected in the chromatin immunoprecipitates with anti-phospho-c-JUN antibody, whereas no enrichment was detected for the Gapdh promoter region used as a negative control (Fig. S14). Mutagenesis of CHOP-binding site 2 and, to a significantly lower extent, of site 1 (shown in red in Fig. 5A) reduced luciferase expression after co-transfection of CHOP and c-JUN (Fig. 5E). Mutagenesis of CHOP-binding site 2 strongly reduced transactivation mediated by CHOP alone, whereas mutagenized AP-1 site 2 abrogated transactivation by CHOP (Fig. 5F). Altogether, these results show that CHOP binds to the SERPINA1 regulatory region and cooperates with c-JUN in regulating SERPINA1 expression.
Figure 5.

CHOP and c-JUN up-regulate human SERPINA1 expression. A, schematic representation of AP-1–binding sites (1 and 2; shown in green) and CHOP-binding sites (1 and 2; shown in red) in the 5′-UTR of the human SERPINA1 gene (drawn to scale) retained in PiZ mouse genome. Nucleotide sequence is shown for the binding sites confirmed by luciferase assays. Nucleotide numbering is calculated as distance from the hepatic TSS. Arrows indicate the position of the primers for the qPCR on the liver chromatin immunoprecipitates shown in D and in Figs. S13 and S14. The dashed line indicates the promoter region cloned upstream of the luciferase reporter gene in the pAAT-Luc-AAT.3′UTR plasmid. Underlined nucleotides have been mutagenized. B, luciferase expression in HeLa cells co-transfected with the pAAT-Luc-AAT.3′UTR construct and the plasmids expressing human CHOP, c-JUN, c-FOS, or their combinations. An empty CMV-3XFLAG plasmid was used as control. Averages ± S. E. (error bars) are shown. One-way ANOVA and Tukey's post hoc test were used: a, p < 0.001 versus CMV-3XFLAG; b, p < 0.0001 versus c-JUN; c, p < 0.0001 versus CHOP; ns, not statistically significant versus CMV-3XFLAG. C, luciferase expression in HeLa cells co-transfected with the plasmids expressing human CHOP and c-JUN and the pAAT-Luc-AAT.3′UTR construct with mutagenized sites 1 and 2 of AP-1 sites. An empty CMV-3XFLAG plasmid was used as control. One-way ANOVA and Tukey's post hoc test were used: a, p < 0.001 versus CMV-3XFLAG; b, p < 0.0001 versus c-JUN + CHOP and versus AP-1 site 1wt-2M c-JUN + CHOP. D, ChIP on PiZ mouse livers using anti-CHOP antibody followed by qPCR on CHOP putative binding sites and AP-1 sites. p62 and Gapdh promoter regions were amplified as positive and negative control for CHOP enrichment, respectively. t test was used: *, p < 0.01. E, luciferase expression in HeLa cells co-transfected with the plasmids expressing human CHOP and c-JUN and the pAAT-Luc-AAT.3′UTR construct with mutagenized CHOP-binding sites 1 and 2. An empty CMV-3XFLAG plasmid was used as control. One-way ANOVA and Tukey's post hoc test were used: a, p < 0.001 versus CMV-3XFLAG; b, p < 0.0001 versus c-JUN + CHOP; c, p < 0.0001 versus AP-1 site 1wt-2M c-JUN + CHOP; d, p < 0.05 versus CMV-3XFLAG. F, luciferase expression in HeLa cells co-transfected with the plasmids expressing human CHOP, and the pAAT-Luc-AAT.3'UTR construct with mutagenized AP-1 site 2 and CHOP-binding site 2. An empty CMV-3XFLAG plasmid was used as control. One-way ANOVA and Tukey's post hoc test were used: a, p < 0.001 versus CMV-3XFLAG; b, p < 0.0001 versus CHOP; c, p < 0.0001 versus AP-1 site 1wt-2M CHOP. IP, immunoprecipitates; ns, not statistically significant.
Age-dependent up-regulation of CHOP in human PiZZ livers
To interrogate the clinical relevance of our findings, we investigated CHOP levels and activation in livers of PiZZ patients. In liver samples from PiZZ patients of less than 16 years of age with extensive PAS-D staining and end-stage liver disease requiring liver transplantation (7) (Table S1), increased CHOP nuclear signals were detected compared with controls (Fig. 6 and Fig. S15).
Figure 6.
Increased expression of CHOP in human livers expressing ATZ. Representative immunohistochemistry (IHC) with anti-CHOP antibody (top panels) and corresponding PAS-D staining (bottom panels) in the liver of a PiZZ patient with end-stage liver disease who underwent liver transplantation (PiZZ #3) and an age-matched PiMM control (PiMM #4) who also underwent liver transplantation as control (7) (×20 magnification; scale bar, 100 μm; insets, ×40 magnifications). Immunohistochemistry with anti-CHOP antibody and PAS-D staining for additional cases are shown in Fig. S15.
Based on the mouse data, showing a role of CHOP in ATZ accumulation in younger mice but not in older animals, we investigated CHOP expression in PiZZ children with liver disease compared with PiZZ adults with liver disease. CHOP up-regulation was indeed found to be greater in children compared with adults (Fig. 7 and Table S2). Taken together, these results supported a role of CHOP in the pathogenesis of ATZ-related liver disease in children.
Figure 7.
Age-dependent up-regulation of CHOP in human livers expressing ATZ. ddPCR for CHOP expression in human liver biopsies from healthy controls (adults: n = 6, age 50.7 ± 13.3 years; children: n = 12, age 6.8 ± 3.6 years), diseased controls (adults: n = 6, age 50.5 ± 13.0 years; children: n = 10, age 5.9 ± 4.3 years), and PiZZ patients (adults: n = 6, age 50.8 ± 13.0 years; children: n = 14, age 6.5 ± 3.7 years). Averages ± S. E. (error bars) are shown. Two-way ANOVA and Tukey's post hoc test were used: *, p < 0.05; **, p < 0.01.
Discussion
AAT is an acute-phase reactant, and its circulating levels may rise several fold in response to several inflammatory stimuli and stress conditions (15). However, in subjects carrying the Z allele, increased synthesis of ATZ augments the burden of mutant protein, which correlates with the degree of liver injury and fibrosis (18). To address the factors involved in the pathogenesis of the liver disease, most studies have been focused on ATZ protein degradation by the ubiquitin-proteasome system or autophagy (19), whereas the contribution of transcriptional regulation of SERPINA1 expression has been neglected for the most part. In this study, we found that CHOP is an important regulator of SERPINA1 expression; it is up-regulated and activated in livers of PiZ mice and PiZZ patients, and juvenile Chop-deleted mice show marked reduction of liver ATZ accumulation. Moreover, we found an important role of CHOP in regulating c-JUN–mediated up-regulation of SERPINA1. However, we also found that the lack of CHOP does not affect the progression of the hepatic disease because livers of older mice show ATZ accumulation and liver fibrosis similar to PiZ/Chop+/+ mice. Finally, we detected higher levels of CHOP in children with ATZ-related liver disease compared with adults.
Whereas the precise factors responsible for its activation in livers expressing ATZ remain to be identified, CHOP was found to play an important role in aggravating ATZ accumulation in juvenile PiZ mice through transcriptional up-regulation of SERPINA1 expression. CHOP is an important player in the unfolded protein response and ER-stress response. Moreover, it is activated by several stimuli, such as amino acid and glucose deprivation, DNA damage, cellular growth arrest, and hypoxia (8, 20). Previous studies showed that cells expressing ATZ are more sensitive to activate ER stress after a second hit (21). We also found previously that toxic accumulation of ATZ leads to dysfunctional liver zonation and metabolic alterations (22). Considering this evidence, we speculate that ATZ accumulation may induce such liver alterations, including perturbed metabolism, which in turn leads to CHOP activation.
We previously found that JNK-mediated activation of c-JUN results in up-regulation of SERPINA1 in human and mouse livers expressing ATZ (7). Here, we showed that CHOP enhances c-JUN–mediated transcriptional up-regulation of SERPINA1. These findings are consistent with previous studies proposing that CHOP enhances transcriptional activation of AP-1 by tethering to the AP-1 complex without direct binding of DNA (16). Previous data also showed that CHOP and c-JUN also cooperatively mediate the induction of PUMA (p53–up-regulated modulator of apoptosis) during hepatocyte lipoapoptosis (23). More broadly, these data raise the question of whether CHOP can potentiate c-JUN–mediated transcription in other cell types under different physiologic and disease conditions. Interestingly, CHOP is transcriptionally positively regulated by the AP-1 complex (24), suggesting a feedback-positive loop, in which c-JUN activation up-regulates CHOP, whereas CHOP activation in turn further enhances SERPINA1 expression. Such a feed-forward mechanism could increase ATZ overload thus worsening the liver disease. Moreover, JNK-mediated activation of c-JUN might also result in long-term effects on progression of the liver disease that deserve further investigation.
Previous studies showed that CHOP activates GADD34, which dephosphorylates eIF2α and increases protein load (25). Moreover, CHOP was also found to induce oxidative stress, which may affect protein folding (26–28). Therefore, in addition to the transcriptional effect, we cannot rule out a beneficial effect of CHOP deletion on improved protein folding by reduction of protein load and oxidative stress. By immunohistochemistry and analysis of parenchymal and nonparenchymal liver cell fractions, Chop expression was detected largely in hepatocytes. However, additional effects of CHOP in other cell types besides hepatocytes cannot be ruled out.
In contrast to older mice, juvenile PiZ mice deleted for Chop showed marked reduction of ATZ accumulation, suggesting a different role of CHOP at different disease stages. Interestingly, levels of blood ATZ in PiZZ patients appear to decline over time (29), a finding that parallels the mouse data shown in this and previous studies (21). The mechanisms underlying the age-dependent reduction of CHOP in livers expressing ATZ are unknown, but the reduced proteotoxic load due to recently recognized decreased of HNF-4α is a possible explanation (21).
Clinical data suggest that the human disease has different features in infants compared with adults. A survey of large transplantation databases found that AAT deficiency most commonly causes liver disease requiring liver transplantation in adults, but it also revealed that almost all transplants in children were performed under the age of 5 years (30). These differences suggest that pathogenetic mechanisms vary depending on the age and more potent modifiers play a role in the liver disease in children (30). Based on our findings, we propose that CHOP and factors regulating its expression might be among the modifiers responsible for the more severe disease occurring in the pediatric population. Interestingly, CHOP is up-regulated by nonsteroidal anti-inflammatory drugs (31), and indomethacin given to PiZ mice was previously shown to increase hepatic injury and SERPINA1 expression, resulting in greater accumulation of ATZ (32). Moreover, exposure of hepatocytes to acetaminophen activates JNK and CHOP, and disruption of Jnk2 or Chop reduces acetaminophen-induced liver injury (33). Therefore, environmental factors, including drugs, could aggravate ATZ-induced liver injury and may be dangerous particularly in children with AAT deficiency.
In conclusion, our study suggests that CHOP is an important player in aggravating hepatic accumulation of toxic ATZ in mice and in patients, especially at younger ages. Drugs inhibiting CHOP activity might have potential for therapy of liver disease in PiZZ children.
Experimental procedures
Mouse studies
PiZ transgenic mice (6) were maintained on a C57BL/6 background. Chop−/− mice were purchased from Jackson Laboratory (stock no. 005530) and bred with PiZ mice. Genotyping for the Chop allele was performed by PCR on genomic DNA from mouse tails. The mutant allele showed a band of 320 bp, whereas the WT allele was 544 bp. Primers used for genotyping were ATGCCCTTACCTATCGTG (common oligonucleotide), AACGCCAGGGTTTTCCCAGTCA (knockout reverse), and GCAGGGTCAAGAGTAGTG (WT reverse). As controls, C57BL/6 (Charles River Laboratories) were treated with DMSO (Sigma) or 4 μg/g of tunicamycin (Sigma). Only male mice were used for all experiments. Blood samples were collected by retro-orbital bleedings. The rAAV8pCB-mir914 and rAAV8pCB-GFP were generated, purified, and titered by the University of Massachusetts Gene Therapy Vector Core as described previously (10). Vectors were diluted in saline solution and intravenously injected in 4-week-old PiZ mice at the dose of 1 × 1013 genome copies/kg.
Liver protein extracts and sections, and serum samples were analyzed by PAS-D staining, immunofluorescence for polymeric ATZ, and ELISA for total human AAT, respectively, as described previously (7, 34). Stained liver sections were examined under a Zeiss LSM700 for fluorescence and Leica DM5000 microscope for bright field. The mean area and number of the ATZ globules by liver PAS-D staining were quantified using the ImageJ plug-in “Analyze particles” on the segmented images. Five different fields of view selected randomly for n = 3 PiZ and PiZ/Chop−/− mice were used for the quantification. Nuclear and cytoplasmic protein extracts were prepared using the NXTRACT kit (Sigma–Aldrich) according to the manufacturer's instructions.
For real-time PCR, total RNA was extracted using RNeasy kit (Qiagen), and 1 μg of RNA was reverse-transcribed using a high-capacity cDNA reverse transcription kit according to the manufacturer's protocol (Applied Biosystems). PCR reactions were performed using SYBR Green Master Mix (Roche Applied Science). PCR conditions were as follows: preheating, 5 min at 95 °C; cycling, 40 cycles of 15 s at 95 °C, 15 s at 60 °C, and 25 s at 72 °C. Results were expressed in terms of cycle threshold. Cycle threshold values were averaged for each duplicate. β2-Microglobulin was used as an endogenous control. Data were analyzed using LightCycler 480 software, version 1.5 (Roche Applied Science). Primer sequences are listed in Table S3.
For Western blotting, liver samples homogenized in RIPA buffer with protease and phosphatase inhibitor mixture (Roche Applied Science) were incubated for 20 min at 4 °C and centrifuged at 13,200 rpm for 10 min. 10–20 μg of lysates were electrophoresed on a 12% SDS-PAGE. After transfer to nitrocellulose membrane, blots were blocked in TBS-Tween 20 with 5% nonfat milk for 1 h at room temperature, and the primary antibody was applied overnight at 4 °C. Anti-rabbit IgG or anti-mouse IgG conjugated with horseradish peroxidase (GE Healthcare; 1:3,000) and ECL (Pierce) were used for detection. Primary antibodies are listed in Table S4.
For immunohistochemistry, 5-μm-thick sections were rehydrated and permeabilized in PBS, 0.5% Triton (Sigma) for 20 min. Antigen unmasking was performed in 0.01 m citrate buffer in a microwave oven. Next, sections underwent blocking of endogenous peroxidase activity in methanol plus 1.5% H2O2 (Sigma) for 30 min and were incubated with blocking solution (3% BSA (Sigma), 5% donkey serum (Millipore), 1.5% horse serum (Vector Laboratories), 20 mm MgCl2, 0.3% Triton (Sigma) in PBS) for 1 h. Sections were incubated with primary antibody (Table S4) overnight at 4 °C and with universal biotinylated horse anti-mouse/rabbit IgG secondary antibody (Vector Laboratories) for 1 h. Biotin/avidin-horseradish peroxidase signal amplification was achieved using the ABC Elite Kit (Vector Laboratories) according to the manufacturer's instructions. 3,30-Diaminobenzidine (Vector Laboratories) was used as peroxidase substrate. Mayer's hematoxylin (Bio-Optica) was used for counterstaining. Sections were dehydrated and mounted in Vectashield (Vector Laboratories). Image capture was performed using a Leica DM5000 microscope. Three mice per group and three sections per mouse were analyzed.
Sirius Red staining was performed on 5-μm liver sections rehydrated and stained for 1 h in 0.1% Sirius Red (Sigma) in saturated aqueous solution of picric acid. After two changes of acidified water (0.5% acetic acid in water), the sections were dehydrated, cleared in xylene, and mounted in a resinous medium. Image capture was performed using the Leica DM5000 microscope. The METAVIR scoring system (35) was used to determine the severity of hepatic fibrosis and necro-inflammatory stage. All samples were evaluated by the same pathologist (S. C.) who was blinded to the laboratory data.
Immunofluorescence staining of apoptotic cells was performed by terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick-end labeling using the In Situ Cell Death Detection Kit (Roche Molecular Biochemicals) according to the manufacturer's instructions. Mice injected with 0.5 μg/g of body weight of CD95-activating antibody (BD Biosciences) for 6 h were used as a positive control. The image capture was performed using Leica DM5000 microscope.
Isolation of parenchymal and nonparenchymal cells
Parenchymal and nonparenchymal cells were isolated from livers of 6-week-old PiZ mice by a modified protocol based on Pronase/collagenase digestion (36). In brief, mouse livers were perfused through the inferior vena cava with EGTA solution followed by enzymatic digestion with Pronase (Sigma–Aldrich) and then collagenase type D (Roche Applied Science). Next, livers were harvested, and liver cells were disassociated by digestion with Pronase/collagenase solution and filtered through a nylon filter (Corning) to remove undigested tissues and debris. The resulting cell suspension was centrifuged at 70 × g for 3 min at 4 °C. Supernatant containing nonparenchymal cells was collected. The cell pellet was washed three times with Williams' medium E (Gibco) and centrifuged at 70 × g for 3 min at 4 °C to obtain hepatocytes. The nonparenchymal liver cell fraction was obtained after the centrifugation at 600 × g for 10 min at 4 °C. The cell pellet was washed with Hanks' balanced salt solution without Ca2+ and Mg2+ one time (Gibco) and centrifuged at 70 × g for 3 min at 4 °C to minimize hepatocyte contamination. The final cell suspension was centrifuged at 600 × g for 10 min at 4 °C to obtain the nonparenchymal cell fraction. Real-time PCR for the albumin gene was performed to evaluate enrichment for the parenchymal fraction, and gene expression was normalized to 18S rRNA.
ATZ immunoblot
Preparation of soluble and insoluble fractions from livers was performed according to previous studies (37), and immunoblotting was performed using antibodies against total AAT and ATZ polymer (Table S4).
QuantSeq 3′ mRNA sequencing
Preparation of libraries was performed with a total of 100 ng of RNA from each sample using the QuantSeq 3′mRNA-Seq Library prep kit (Lexogen, Vienna, Austria) according to the manufacturer's instructions. Total RNA was quantified using the Qubit 2.0 fluorimetric Assay (Thermo Fisher Scientific). Libraries were prepared from 100 ng of total RNA using the QuantSeq 3′ mRNA-Seq Library Prep Kit FWD for Illumina (Lexogen GmbH). Quality of the libraries was assessed by screen tape high-sensitivity DNA D1000 (Agilent Technologies). Libraries were sequenced on a NovaSeq 6000 sequencing system using an S1, 100-cycle flow cell (Illumina Inc.). Amplified fragmented cDNA of 300 bp in size were sequenced in single-end mode with a read length of 100 bp. Illumina NovaSeq base call files were converted in a fastq file through bcl2fastq (https://support.illumina.com/downloads/bcl2fastq2-conversion-user-guide-v2.html) (version 2.20.0.422). For analysis, sequence reads were trimmed using bbduk software (https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-user-guide/usage-guide/) (bbmap suite 37.31) to remove adapter sequences, poly(A) tails, and low-quality end bases (regions with average quality below 6). Alignment was performed with STAR 2.6.0a3 (38) on the mm10 reference assembly obtained from the cellRanger website (https://support.10xgenomics.com/single-cell-gene-expression/software/release-notes/build#mm10_3.0.0; Ensembl assembly release 93). Expression levels of genes were determined with htseq-count (39) using the Gencode/Ensembl gene model. We filtered out all genes having <1 cpm in less than n_min samples and Perc MM reads >20% simultaneously. Differential expression analysis was performed using edgeR (40), a statistical package based on generalized linear models, suitable for multifactorial experiments. Gene ontology and functional annotation clustering analyses were performed using DAVID Bioinformatic Resources (41, 42) on up- and down-regulated genes separately. Data were deposited in GEO with accession number GSE141593.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed by GSEA software (www.broadinstitute.org/gsea) on expression data from QuantSeq analysis without filtering (43). An expression microarray of livers from PiZ mice and WT controls was obtained from previous studies (GSE93115), and the CHOP target gene set was defined by combining ChIP-Seq and RNA-Seq data reported previously from tunicamycin-treated WT mouse embryonic fibroblasts (44).
Protein immunoprecipitation
The pCMV-c-JUN-MYC plasmid was generated by cloning the human c-JUN coding sequence in the pCMV-MYC plasmid (Thermo Fisher Scientific). Primers used for generation of the construct are shown in Table S5. HeLa cells were co-transfected with plasmids expressing CHOP-FLAG and c-JUN-MYC. Untreated cells or cells co-transfected with the plasmid expressing CHOP-FLAG were used as controls. 48 h after transfection, cells were washed with PBS, and then protein cross-linking was performed using a solution of 1.5 mm dithiobis(succinimidyl propionate) (DSP) (Thermo Fisher Scientific) in PBS added to the dishes. DSP is a bifunctional cross-linking agent containing amine-reactive N-hydroxysuccinimide esters that react with primary amines to form stable amide bonds and is more efficient than formaldehyde as a protein-protein cross-linker (45). Dishes were incubated with DSP for 2 h at 4 °C, and the reaction was stopped with the addition for 15 min at room temperature of 1 m Tris, pH 7.8, to a final concentration of 20 mm. Dishes were washed with PBS and then lysed as described above. 1 mg of protein lysates was used for the immunoprecipitation with mouse anti-FLAG antibody or mouse IgG overnight at 4 °C. The day after, 20 μl of Dynabeads Protein G (Thermo Fisher Scientific) were added and incubated for 2 h at 4 °C. The flow-through was collected using the magnet, and the complex Dynabeads-Ab-Ag was washed five times using RIPA buffer with 0.1% Triton without SDS and once with RIPA buffer without Triton and SDS. Finally, the elution was performed by boiling tubes at 100 °C for 5 min in Laemmli buffer. Samples were analyzed by SDS-PAGE as described above. Primary antibodies used for immunoprecipitations are listed in Table S4.
Luciferase assays
The pAAT-Luc-AAT.3'UTR plasmid was generated by cloning SERPINA1 3′-UTR downstream of the firefly luciferase coding region in the previously generated pAAT-Luc vector. The cDNAs of human CHOP and c-FOS were amplified by RT-PCR from Huh7 cells and cloned into p3XFLAG-CMV-14 vector. The plasmid expressing human c-JUN was generated previously (7). AP-1 sites in the pAAT-Luc-AAT.3′UTR plasmid were mutagenized as described previously (7). Primers used for generation of the constructs are shown in Table S5. HeLa cells were cultured in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum and 5% penicillin/streptomycin. Cells were co-transfected with the plasmid containing the WT or mutagenized AP-1 consensus sequences and with a plasmid expressing the human CHOP and c-JUN. Each well was co-transfected with the pRL-TK plasmid (Promega) expressing the Renilla luciferase as control. Lipo D293 (SigmaGen Laboratories) was used for the DNA transfection according to the manufacturer's instructions. Cells were harvested 48 h after transfection and assayed for luciferase activity using the Dual-Luciferase reporter (DLR™) assay system (Promega). Data were expressed relative to Renilla luciferase activity to normalize for transfection efficiency. Transfections were repeated at least three times.
Chromatin immunoprecipitations
ChIP was performed as described previously (7). Briefly, PiZ mouse livers were mechanically homogenized in PBS in the presence of protein inhibitors and then cross-linked by formaldehyde to a final concentration of 1% for 10 min at room temperature. The cross-linking reaction was stopped by the addition of glycine at a final concentration of 0.125 mm for 5 min at room temperature. After three washes in PBS with protein inhibitors, liver homogenates were lysed in cell lysis buffer (5 mm PIPES (pH 8.0), 0.5% Igepal, 85 mm KCl) for 15 min. Nuclei were lysed in lysis buffer (50 mm Tris-HCl (pH 8.0), 10 mm EDTA, 0.8% SDS) for 30 min. Chromatin was sonicated to yield DNA fragments of ∼200–1,000 bp. DNA was co-immunoprecipitated using the ChIP-grade anti-phospho-c-JUN (Ser-73) and anti-CHOP antibodies overnight at 4 °C. No antibody was used in the negative control of the immunoprecipitation. Purified immunoprecipitated DNA samples and inputs were amplified by quantitative PCR with primers specific for the AP-1–binding sites of the 5′-UTR of the SERPINA1 gene. Antibodies for ChIP and primer sequences are listed in Tables S4 and S6, respectively.
Human liver samples
Liver samples for immunohistochemistry were collected anonymously from subjects with end-stage liver disease homozygous for the Z or M alleles of SERPINA1. Diseased liver controls were obtained from patients with diagnoses of biliary atresia or cirrhosis. Frozen liver powdered samples were homogenized in TRIzol (Invitrogen), and total RNA was extracted according to the manufacturer's recommendations. 500 ng of RNA were retrotranscribed using a high-capacity RNA to cDNA kit (Applied Biosystems), and 2 μl were used as an input for droplet digital PCR (ddPCR). For gene expression, the DDIT3 (CHOP) gene (unique assay ID: dHasEG5002009; Bio-Rad) was used as a target and the POLR2H gene (unique assay ID: dHasEG5005571; Bio-Rad) as a reference. ddPCR was run according to the PrimePCR ddPCR Gene expression assay (EvaGreen®) protocol (Bio-Rad).
Statistical analyses
Data are expressed as averages ± S.D or S.E. as indicated in the figure legends. Two-tailed Student's t test and analysis of variance (ANOVA) plus Tukey's post hoc analysis were used as statistical tests for mean comparisons. A time series was used for body weight changes.
Study approvals
Mouse procedures were performed in accordance with the regulations of the Italian Ministry of Health and conformed to the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health (NIH publication 86-23, revised 1985). Human liver samples were obtained from the St. Louis University School of Medicine (St. Louis, MO, USA) and University of Massachusetts (Boston, MA, USA). The ethics committees of St. Louis University School of Medicine and the University of Massachusetts approved the studies. Written informed consent was received from participants prior to inclusion in the study. The studies abide by the Declaration of Helsinki principles.
Data availability
Data were deposited in the Gene Expression Omnibus (GEO) with accession number GSE141593. All the other data described in this study are contained within the article and accompanying supporting information.
Supplementary Material
This article contains supporting information.
Author contributions—S. A., P. P., and N. B.-P. conceptualization; S. A., R. F., G. G., E. N., J. T., P. P., and N. B.-P. data curation; S. A., R. F., G. G., R. D. C., A. C., S. C., P. P., and N. B.-P. formal analysis; S. A. and N. B.-P. writing-original draft; C. M. and N. B.-P. investigation; P. P. and N. B.-P. supervision; P. P. and N. B.-P. funding acquisition; N. B.-P. writing-review and editing.
Funding and additional information—This work was supported by grants from Fondazione Telethon Italy (to N. B.-P.), the Alpha-1 Foundation (research grant (to N. B.-P.) and Gordon L. Snider Award 2016 (to P. P.), and the “Federico II” University of Naples (STAR Program) (to P. P.). S. A. is supported by an Alpha-1-Antitrypsin Laurell's Training Award (ALTA).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- AAT
- α1-antitrypsin
- AAV
- adeno-associated viral
- DSP
- dithiobis(succinimidyl propionate)
- ER
- endoplasmic reticulum
- GSEA
- gene set enrichment analysis
- PAS-D
- periodic acid–Schiff–diastase–resistant
- qPCR
- quantitative PCR
- RIPA
- radioimmune precipitation assay
- ddPCR
- droplet digital PCR
- ANOVA
- analysis of variance
- JNK
- c-JUN N-terminal kinase.
References
- 1. Carrell R. W., and Lomas D. A. (2002) α1-Antitrypsin deficiency—a model for conformational diseases. N. Engl. J. Med. 346, 45–53 10.1056/NEJMra010772 [DOI] [PubMed] [Google Scholar]
- 2. Sveger T. (1988) The natural history of liver disease in α1-antitrypsin deficient children. Acta Paediatr. Scand. 77, 847–851 10.1111/j.1651-2227.1988.tb10767.x [DOI] [PubMed] [Google Scholar]
- 3. Eriksson S., Carlson J., and Velez R. (1986) Risk of cirrhosis and primary liver cancer in alpha1-antitrypsin deficiency. N. Engl. J. Med. 314, 736–739 10.1056/NEJM198603203141202 [DOI] [PubMed] [Google Scholar]
- 4. Sveger T. (1976) Liver disease in α1-antitrypsin deficiency detected by screening of 200,000 infants. N. Engl. J. Med. 294, 1316–1321 10.1056/NEJM197606102942404 [DOI] [PubMed] [Google Scholar]
- 5. Perlmutter D. H. (2002) Liver injury in α1-antitrypsin deficiency: an aggregated protein induces mitochondrial injury. J. Clin. Invest. 110, 1579–1583 10.1172/JCI16787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carlson J. A., Rogers B. B., Sifers R. N., Finegold M. J., Clift S. M., DeMayo F. J., Bullock D. W., and Woo S. L. (1989) Accumulation of PiZ α1-antitrypsin causes liver damage in transgenic mice. J. Clin. Invest. 83, 1183–1190 10.1172/JCI113999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pastore N., Attanasio S., Granese B., Castello R., Teckman J., Wilson A. A., Ballabio A., and Brunetti-Pierri N. (2017) Activation of the c-Jun N-terminal kinase pathway aggravates proteotoxicity of hepatic mutant Z α1-antitrypsin. Hepatology 65, 1865–1874 10.1002/hep.29035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang Y., Liu L., Naik I., Braunstein Z., Zhong J., and Ren B. (2017) Transcription factor C/EBP homologous protein in health and diseases. Front. Immunol. 8, 1612 10.3389/fimmu.2017.01612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R. T., Remotti H., Stevens J. L., and Ron D. (1998) CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 12, 982–995 10.1101/gad.12.7.982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mueller C., Tang Q., Gruntman A., Blomenkamp K., Teckman J., Song L., Zamore P. D., and Flotte T. R. (2012) Sustained miRNA-mediated knockdown of mutant AAT with simultaneous augmentation of wild-type AAT has minimal effect on global liver miRNA profiles. Mol. Ther. J. Am. Soc. Gene Ther. 20, 590–600 10.1038/mt.2011.292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hubner R. H., Leopold P. L., Kiuru M., De B. P., Krause A., and Crystal R. G. (2009) Dysfunctional glycogen storage in a mouse model of α1-antitrypsin deficiency. Am. J. Respir. Cell Mol. Biol. 40, 239–247 10.1165/rcmb.2008-0029OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lindblad D., Blomenkamp K., and Teckman J. (2007) α1-Antitrypsin mutant Z protein content in individual hepatocytes correlates with cell death in a mouse model. Hepatology 46, 1228–1235 10.1002/hep.21822 [DOI] [PubMed] [Google Scholar]
- 13. DeZwaan-McCabe D., Riordan J. D., Arensdorf A. M., Icardi M. S., Dupuy A. J., and Rutkowski D. T. (2013) The stress-regulated transcription factor CHOP promotes hepatic inflammatory gene expression, fibrosis, and oncogenesis. PLoS Genet. 9, e1003937 10.1371/journal.pgen.1003937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. B'chir W., Maurin A.-C., Carraro V., Averous J., Jousse C., Muranishi Y., Parry L., Stepien G., Fafournoux P., and Bruhat A. (2013) The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 41, 7683–7699 10.1093/nar/gkt563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kalsheker N., Morley S., and Morgan K. (2002) Gene regulation of the serine proteinase inhibitors α1-antitrypsin and α1-antichymotrypsin. Biochem. Soc. Trans. 30, 93–98 10.1042/bst0300093 [DOI] [PubMed] [Google Scholar]
- 16. Ubeda M., Vallejo M., and Habener J. F. (1999) CHOP enhancement of gene transcription by interactions with Jun/Fos AP-1 complex proteins. Mol. Cell Biol. 19, 7589–7599 10.1128/mcb.19.11.7589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ubeda M., Wang X. Z., Zinszner H., Wu I., Habener J. F., and Ron D. (1996) Stress-induced binding of the transcriptional factor CHOP to a novel DNA control element. Mol. Cell Biol. 16, 1479–1489 10.1128/mcb.16.4.1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Clark V. C., Marek G., Liu C., Collinsworth A., Shuster J., Kurtz T., et al. (2018) Clinical and histologic features of adults with α1-antitrypsin deficiency in a non-cirrhotic cohort. J Hepatol. 69, 1357–1364 10.1016/j.jhep.2018.08.005 [DOI] [PubMed] [Google Scholar]
- 19. Perlmutter D. H. (2006) Pathogenesis of chronic liver injury and hepatocellular carcinoma in α1-antitrypsin deficiency. Pediatr. Res. 60, 233–238 10.1203/01.pdr.0000228350.61496.90 [DOI] [PubMed] [Google Scholar]
- 20. Fafournoux P., Bruhat A., and Jousse C. (2000) Amino acid regulation of gene expression. Biochem. J. 351, 1–12 10.1042/0264-6021:3510001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ordóñez A., Snapp E. L., Tan L., Miranda E., Marciniak S. J., and Lomas D. A. (2013) Endoplasmic reticulum polymers impair luminal protein mobility and sensitize to cellular stress in alpha1-antitrypsin deficiency. Hepatology 57, 2049–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Piccolo P., Annunziata P., Soria L. R., Attanasio S., Barbato A., Castello R., Carissimo A., Quagliata L., Terracciano L. M., and Brunetti-Pierri N. (2017) Down-regulation of hepatocyte nuclear factor-4α and defective zonation in livers expressing mutant Z α1-antitrypsin. Hepatology 66, 124–135 10.1002/hep.29160 [DOI] [PubMed] [Google Scholar]
- 23. Cazanave S. C., Elmi N. A., Akazawa Y., Bronk S. F., Mott J. L., and Gores G. J. (2010) CHOP and AP-1 cooperatively mediate PUMA expression during lipoapoptosis. Am. J. Physiol. Gastrointest. Liver Physiol. 299, G236–G243 10.1152/ajpgi.00091.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guyton K. Z., Xu Q., and Holbrook N. J. (1996) Induction of the mammalian stress response gene GADD153 by oxidative stress: role of AP-1 element. Biochem. J. 314, 547–554 10.1042/bj3140547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pakos-Zebrucka K., Koryga I., Mnich K., Ljujic M., Samali A., and Gorman A. M. (2016) The integrated stress response. EMBO Rep. 17, 1374–1395 10.15252/embr.201642195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Song B., Scheuner D., Ron D., Pennathur S., and Kaufman R. J. (2008) Chop deletion reduces oxidative stress, improves beta cell function, and promotes cell survival in multiple mouse models of diabetes. J. Clin. Invest. 118, 3378–3389 10.1172/JCI34587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Malhotra J. D., and Kaufman R. J. (2007) Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a double-edged sword? Antioxid. Redox Signal. 9, 2277–2293 10.1089/ars.2007.1782 [DOI] [PubMed] [Google Scholar]
- 28. Marciniak S. J., Yun C. Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H. P., and Ron D. (2004) CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18, 3066–3077 10.1101/gad.1250704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Donato L. J., Jenkins S. M., Smith C., Katzmann J. A., and Snyder M. R. (2012) Reference and interpretive ranges for α1-antitrypsin quantitation by phenotype in adult and pediatric populations. Am. J. Clin. Pathol. 138, 398–405 10.1309/AJCPMEEJK32ACYFP [DOI] [PubMed] [Google Scholar]
- 30. Chu A. S., Chopra K. B., and Perlmutter D. H. (2016) Is severe progressive liver disease caused by α1-antitrypsin deficiency more common in children or adults? Liver Transpl. 22, 886–894 10.1002/lt.24434 [DOI] [PubMed] [Google Scholar]
- 31. Tsutsumi S., Gotoh T., Tomisato W., Mima S., Hoshino T., Hwang H.-J., Takenaka H., Tsuchiya T., Mori M., and Mizushima T. (2004) Endoplasmic reticulum stress response is involved in nonsteroidal anti-inflammatory drug-induced apoptosis. Cell Death Differ. 11, 1009–1016 10.1038/sj.cdd.4401436 [DOI] [PubMed] [Google Scholar]
- 32. Rudnick D. A., Shikapwashya O., Blomenkamp K., and Teckman J. H. (2006) Indomethacin increases liver damage in a murine model of liver injury from α1-antitrypsin deficiency. Hepatology 44, 976–982 10.1002/hep.21326 [DOI] [PubMed] [Google Scholar]
- 33. Gunawan B. K., Liu Z. X., Han D., Hanawa N., Gaarde W. A., and Kaplowitz N. (2006) c-Jun N-terminal kinase plays a major role in murine acetaminophen hepatotoxicity. Gastroenterology 131, 165–178 10.1053/j.gastro.2006.03.045 [DOI] [PubMed] [Google Scholar]
- 34. Pastore N., Blomenkamp K., Annunziata F., Piccolo P., Mithbaokar P., Maria Sepe R., Vetrini F., Palmer D., Ng P., Polishchuk E., Iacobacci S., Polishchuk R., Teckman J., Ballabio A., Brunetti-Pierri N., et al. (2013) Gene transfer of master autophagy regulator TFEB results in clearance of toxic protein and correction of hepatic disease in α1-anti-trypsin deficiency. EMBO Mol. Med. 5, 397–412. 10.1002/emmm.201202046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goodman Z. D. (2007) Grading and staging systems for inflammation and fibrosis in chronic liver diseases. J. Hepatol. 47, 598–607 10.1016/j.jhep.2007.07.006 [DOI] [PubMed] [Google Scholar]
- 36. Mederacke I., Dapito D. H., Affo S., Uchinami H., and Schwabe R. F. (2015) High-yield and high-purity isolation of hepatic stellate cells from normal and fibrotic mouse livers. Nat. Protoc. 10, 305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schmidt B. Z., and Perlmutter D. H. (2005) Grp78, Grp94, and Grp170 interact with alpha1-antitrypsin mutants that are retained in the endoplasmic reticulum. Am. J. Physiol. Gastrointest. Liver Physiol. 289, G444–G455 10.1152/ajpgi.00237.2004 [DOI] [PubMed] [Google Scholar]
- 38. Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T. R. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Anders S., Pyl P. T., and Huber W. (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robinson M. D., McCarthy D. J., and Smyth G. K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 43. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., and Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Han J., Back S. H., Hur J., Lin Y.-H., Gildersleeve R., Shan J., Yuan C. L., Krokowski D., Wang S., Hatzoglou M., Kilberg M. S., Sartor M. A., and Kaufman R. J. (2013) ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15, 481–490 10.1038/ncb2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang L., Rayner S., Katoku-Kikyo N., Romanova L., and Kikyo N. (2007) Successful co-immunoprecipitation of Oct4 and Nanog using cross-linking. Biochem. Biophys. Res. Commun. 361, 611–614 10.1016/j.bbrc.2007.07.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data were deposited in the Gene Expression Omnibus (GEO) with accession number GSE141593. All the other data described in this study are contained within the article and accompanying supporting information.