Abstract
ICK (also known as CILK1) is a mitogen-activated protein kinase–like kinase localized at the ciliary tip. Its deficiency is known to result in the elongation of cilia and causes ciliopathies in humans. However, little is known about how ICK is transported to the ciliary tip. We here show that the C-terminal noncatalytic region of ICK interacts with the intraflagellar transport (IFT)–B complex of the IFT machinery and participates in its transport to the ciliary tip. Furthermore, total internal reflection fluorescence microscopy demonstrated that ICK undergoes bidirectional movement within cilia, similarly to IFT particles. Analysis of ICK knockout cells demonstrated that ICK deficiency severely impairs the retrograde trafficking of IFT particles and ciliary G protein–coupled receptors. In addition, we found that in ICK knockout cells, ciliary proteins are accumulated at the bulged ciliary tip, which appeared to be torn off and released into the environment as an extracellular vesicle. The exogenous expression of various ICK constructs in ICK knockout cells indicated that the IFT-dependent transport of ICK, as well as its kinase activity and phosphorylation at the canonical TDY motif, is essential for ICK function. Thus, we unequivocally show that ICK transported to the ciliary tip is required for retrograde ciliary protein trafficking and consequently for normal ciliary function.
Keywords: cilia, extracellular vesicle, ICK, IFT-B complex, intraflagellar transport, intracellular trafficking, protein kinase, extracellular vesicles, CRISPR/Cas
Primary cilia are microtubule-based projections found on a variety of eukaryotic cells and serve as mechanosensors for extracellular stimuli, such as fluid flow, and as chemosensors for signaling molecules, such as the Hedgehog (Hh) morphogen (1). To fulfill these functions, the ciliary membrane has specific receptors and ion channels. In addition, to maintain the integrity of the axonemal microtubules, αβ-tubulin dimers and associated molecules are continuously transported toward the ciliary tip (2). The protein and lipid compositions of the membrane and interior of cilia are distinguished from those of the plasma membrane and cytoplasm, respectively, by the presence of the transition zone (TZ) as a permeability/diffusion barrier (3). The functions and assembly of cilia are therefore dependent not only on the bidirectional trafficking of proteins along the axonemal microtubules but also on their restrictive import and export across the TZ. Reflecting their crucial roles, defects in protein trafficking within cilia result in various hereditary disorders, which are generally termed as the ciliopathies (4, 5). These include Bardet–Biedl syndrome (BBS), Meckel syndrome, Joubert syndrome, and short-rib thoracic dysplasia (SRTD).
The intraflagellar transport (IFT) machinery, often referred to as IFT particles or IFT trains, mediates ciliary protein trafficking along the axoneme in both anterograde and retrograde directions, as well as their import and export across the TZ (6, 7). The IFT machinery is composed of two large protein complexes, IFT-B and IFT-A. The IFT-B complex composed of 16 subunits mediates anterograde protein trafficking from the base to the tip driven by the kinesin-2 motor, and the IFT-A complex composed of six subunits together with the TULP3 adaptor protein mediates retrograde trafficking from the tip to the base driven by the dynein-2 motor complex (8, 9). In addition to the IFT complexes, the BBSome complex composed of eight BBS proteins, along with the ARL6/BBS3 GTPase, mediates the exit of ciliary membrane proteins by connecting them with the IFT machinery (10–13).
Compared with the mechanisms of the IFT-B–mediated anterograde trafficking and the IFT-A–mediated retrograde trafficking, little is known about the turnaround event at the ciliary tip, where the IFT machinery is remodeled and the anterograde kinesin-2 motor is replaced with the retrograde dynein-2 motor (6, 14–16). A recent cryoelectron tomographic study of Chlamydomonas IFT trains indicated that the anterograde and retrograde trains adopt completely distinct architectures (17). The study, together with a PhotoGate imaging study of Chlamydomonas IFT trains, suggested that dynein-2 is transported as an inactive cargo of the anterograde train (16, 17); this is supported by a more recent cryoelectron microscopic study of the structure of the human dynein-2 complex (18).
A candidate key regulator of the turnaround event is intestinal cell kinase (ICK; recently renamed CILK1 for ciliogenesis-associated kinase 1), which belongs to the mitogen-activated protein kinase superfamily (19). ICK is ubiquitously expressed (20, 21) and is often found at the distal tip of cilia (22). An ICK paralog, MAK, is expressed specifically in the retina and testis and is also localized mainly at the distal portion of cilia (23). ICK is activated via phosphorylation at the canonical TDY motif by another kinase, cell cycle–related kinase (CCRK; also known as CDK20) (24, 25). Mutation/knockdown/knockout (KO) of ICK and its homologs in a variety of organisms results in a long cilia/flagella phenotype (26–31). Furthermore, genetic mutations in the ICK gene in humans cause ciliopathies, such as endocrine-cerebro-osteodysplasia syndrome and SRTD (32–34). A recent study also indicated that mutations in the ICK gene are associated with juvenile myoclonic epilepsy (35).
Although ICK appears to function at the ciliary tip (22, 36), where ICK has been proposed to phosphorylate KIF3A (22, 37), which is a motor subunit of heterotrimeric kinesin-II, it is unknown as to how ICK is transported from the ciliary base toward the tip. Utilizing the visible immunoprecipitation (VIP) assay, we here show that ICK interacts with the IFT-B complex via its C-terminal noncatalytic region; this is in agreement with a recent study showing that an ICK construct lacking the noncatalytic region could not localize to cilia (37). In addition, by analyzing the phenotypes of ICK-KO cells and those exogenously expressing WT ICK and its mutants, we show that the IFT-dependent anterograde transport of ICK, as well as its kinase activity, is required for normal ciliogenesis and retrograde ciliary protein trafficking, probably by regulating the turnaround event at the tip.
Results
Interaction of ICK with the IFT-B complex
ICK has a kinase domain in its N-terminal (NT) region and a C-terminal (CT) noncatalytic region (Fig. 1A). We divided the ICK protein into the NT (residues 1–287) and CT (residues 285–632) regions. When the ICK constructs fused to mCherry (mChe) were expressed in human telomerase reverse transcriptase–immortalized retinal pigment epithelial 1 (hTERT-RPE1) cells, which are then serum-starved to induce ciliogenesis, mChe-ICK(CT) and mChe-ICK(WT), but not mChe-ICK(NT), were often found localized at the ciliary tip (Fig. 1B, panels a–c), indicating that the noncatalytic region is responsible for the localization of ICK to the ciliary tip. In support of our observations, while this study was in progress, Oh et al. (37) reported that an ICK construct (residues 1–291) lacking the noncatalytic region did not show ciliary localization.
Figure 1.
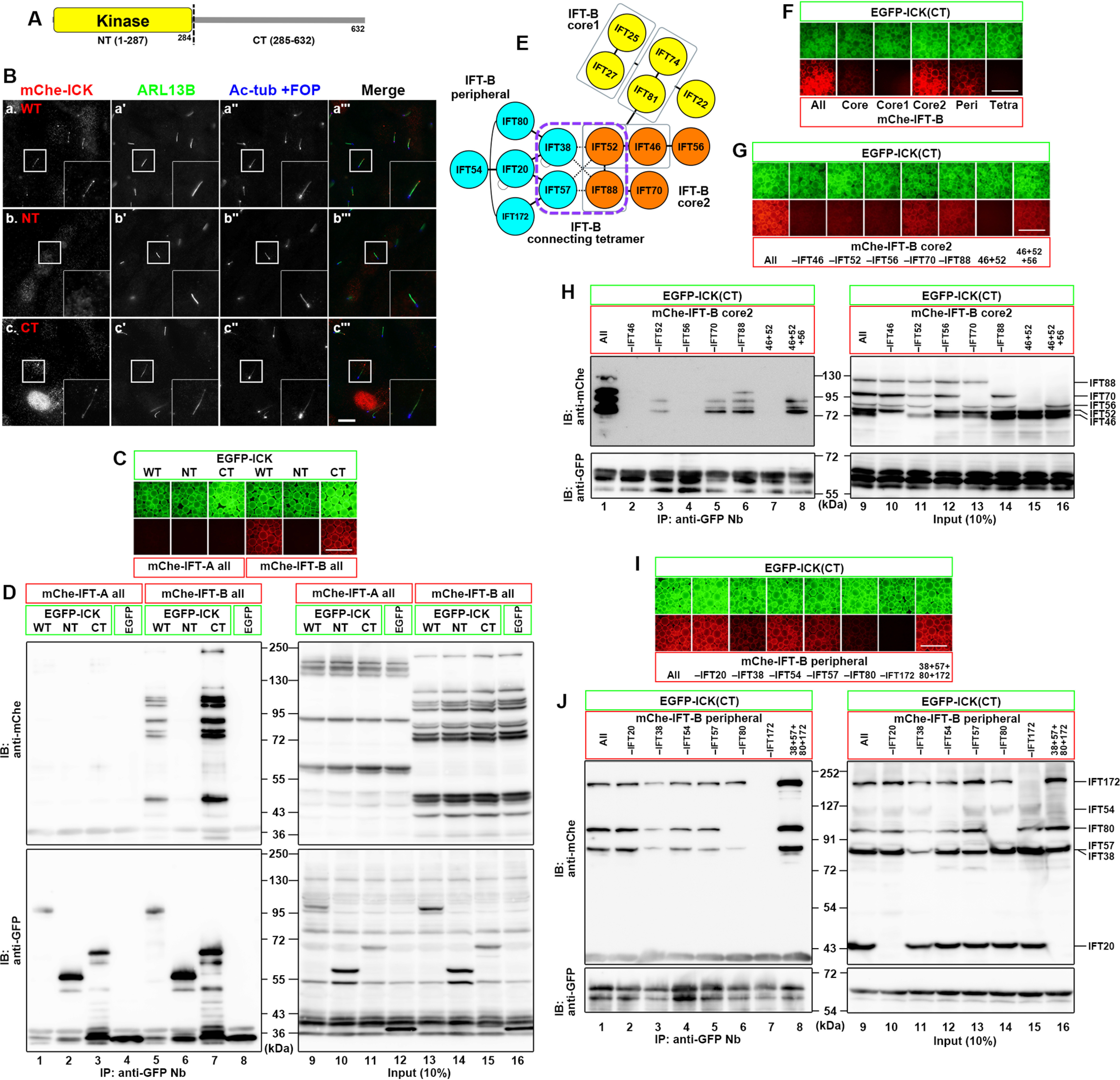
Interaction of ICK with the IFT-B complex. A, schematic representation of the domain organization of ICK. B, ciliary localization of the ICK constructs in hTERT-RPE1 cells. Cells expressing mChe-ICK(WT) (panel a), mChe-ICK(NT) (panel b), or mChe-ICK(CT) (panel c) were serum-starved for 24 h to induce ciliogenesis and immunostained for ARL13B (panels á–c´), and acetylated α-tubulin (Ac-tub) + FOP (panels á´–c´´). Insets are 2.5-fold enlarged images of the boxed regions. C and D, interaction of the C-terminal noncatalytic region of ICK with the IFT-B complex. Lysates of HEK293T cells coexpressing EGFP-fused ICK(WT), ICK(NT), or ICK(CT) and all the IFT-A or IFT-B subunits fused to mChe were processed for the “one-to-many” VIP assay using GST-fused anti-GFP Nbs (C) followed by immunoblotting analysis using an anti-mChe or anti-GFP antibody (D). E, schematic representation of the overall architecture of the IFT-B complex. F, interaction of ICK with the core 2 and peripheral subcomplexes of the IFT-B complex. Lysates of HEK293T cells coexpressing EGFP-ICK(CT) and mChe-fused IFT-B subunits, as indicated, were processed for the VIP assay using GST-fused anti-GFP Nbs. G–J, subtraction assays to determine the core 2 subunits (G and H) or the peripheral subunits (I and J) of the IFT-B complex required for the interaction with ICK. Lysates of cells coexpressing EGFP-ICK(CT) and mChe-fused IFT-B subunits, as indicated, were processed for the VIP assay using GST-fused anti-GFP Nbs (G and I) followed by immunoblotting analysis using an anti-mChe or anti-GFP antibody (H and J). Scale bars, 10 μm in B and 500 μm in C, F, G, and I. Peri, peripheral; Tetra, tetramer; IB, immunoblot; IP, immunoprecipitation.
We recently showed that KIF17, which is homodimeric kinesin-2, and is localized at the ciliary tip in mammalian cells, is transported to the tip via its binding to the IFT-B complex (38). Furthermore, an interactome study suggested that ICK interacts, either directly or indirectly, with some IFT-B subunits (39). In addition, as described below, ICK undergoes bidirectional movement within cilia, similarly to IFT particles. We therefore analyzed whether ICK is also transported to the ciliary tip via binding to the IFT machinery. To this end, we utilized the VIP assay, which we recently developed as a convenient and versatile screening strategy that allows visual detection of not only binary protein–protein interactions but also one-to-many and many-to-many protein interactions (40–42); however, as described in previous studies (40–42), caution is required regarding the results of the VIP assay, because “not detected” does not necessarily mean “no interaction” and because the interactions detected do not necessarily represent interactions that could occur under physiological circumstances.
When lysates of HEK293T cells coexpressing EGFP-ICK and all the 6 IFT-A subunits or all the 16 IFT-B subunits fused to mChe were processed for immunoprecipitation with GSH S-transferase (GST)–tagged anti-GFP nanobodies (Nbs) prebound to GSH-Sepharose beads, EGFP-ICK coprecipitated mChe-IFT-B (Fig. 1C, fourth column) but not mChe-IFT-A (first column). When the ICK protein was divided into the NT and CT regions, only the ICK(CT) construct fused to EGFP coimmunoprecipitated mChe-IFT-B (sixth column). The VIP results were confirmed by immunoblotting analysis; many bands for the mChe-fused IFT-B subunits were detected when coexpressed with EGFP-fused ICK(WT) and ICK(CT) (Fig. 1D, lanes 5 and 7). These results of the VIP assay and immunoblotting are compatible with the above observation that ICK(CT) and ICK(WT) are transported to the ciliary tip (Fig. 1B).
When the IFT-B subunits were grouped into those of the core 1 (IFT-B1-1), core 2 (IFT-B1-2), and peripheral (IFT-B2) subcomplexes, and those of the connecting tetramer (42, 43) (Fig. 1E), the subunits of the core 2 and peripheral subcomplexes fused to mChe were coprecipitated with EGFP-ICK(CT) (Fig. 1F, fourth and fifth columns). We then made use of the subtraction VIP assay to determine which of the core 2 subunits contribute to the ICK(CT) interaction. When one of the core 2 subunits was omitted from the coimmunoprecipitation assay, the signals were considerably decreased in the absence of IFT46, IFT52, or IFT56 (Fig. 1, G, second through fourth columns, and H, lanes 2–4). However, IFT46 + IFT52 + IFT56 was not sufficient, because the signals in the presence of all the core 2 subunits were much more intense than those in the presence of only IFT46+IFT52+IFT56 (compare lane 1 with lane 8). We also performed the subtraction VIP assay for subunits of the peripheral subcomplex. The results showed that among the peripheral subunits, IFT172 makes a major contribution to the ICK(CT) interaction (Fig. 1, I, seventh column, and J, lane 7), and IFT38 and IFT80 also contribute to the interaction (third and sixth columns and lanes 3 and 6).
Transport of ICK to the ciliary tip correlates with its binding to the IFT-B complex
We then narrowed down the area within the ICK CT region that is responsible for its interaction with IFT-B core 2 and the peripheral subcomplexes. Truncation of ICK(CT) to residue 561 from the C terminus (residue 632) did not apparently diminish its interaction with the IFT-B core 2 subunits (Fig. 2, A, second column, and B, lane 2). Truncation to residue 537 slightly diminished the interaction (Fig. 2, compare third column in A and lane 3 in B with first column in A and lane 1 in B, respectively). When the ICK(CT) construct was truncated to residue 501, virtually no interaction with the core 2 subunits was detected (Fig. 2, A, fourth column, and B, lane 4). In the case of the IFT-B peripheral subunits, truncation of ICK(CT) to residue 501 from the C terminus did not significantly affect its interaction with the peripheral subunits (Fig. 2, C, first through third columns, and D, lanes 1–3), and truncation to residue 473 slightly diminished the interaction (fourth column and lane 4). However, truncation to residue 361 abolished the interaction of ICK(CT) with the IFT-B peripheral subunits (fifth column and lane 5). Thus, distinct parts of the broad noncatalytic region participate in the interaction of ICK with the core 2 and peripheral subunits of the IFT-B complex, although it is unknown whether or not these regions overlap with each other. Because the noncatalytic region of ICK was suggested to have characteristics of intrinsically disordered proteins (19, 37), the broad noncatalytic region might have the flexibility to interact with multiple subunits of the IFT-B core 2 and peripheral subcomplexes.
Figure 2.
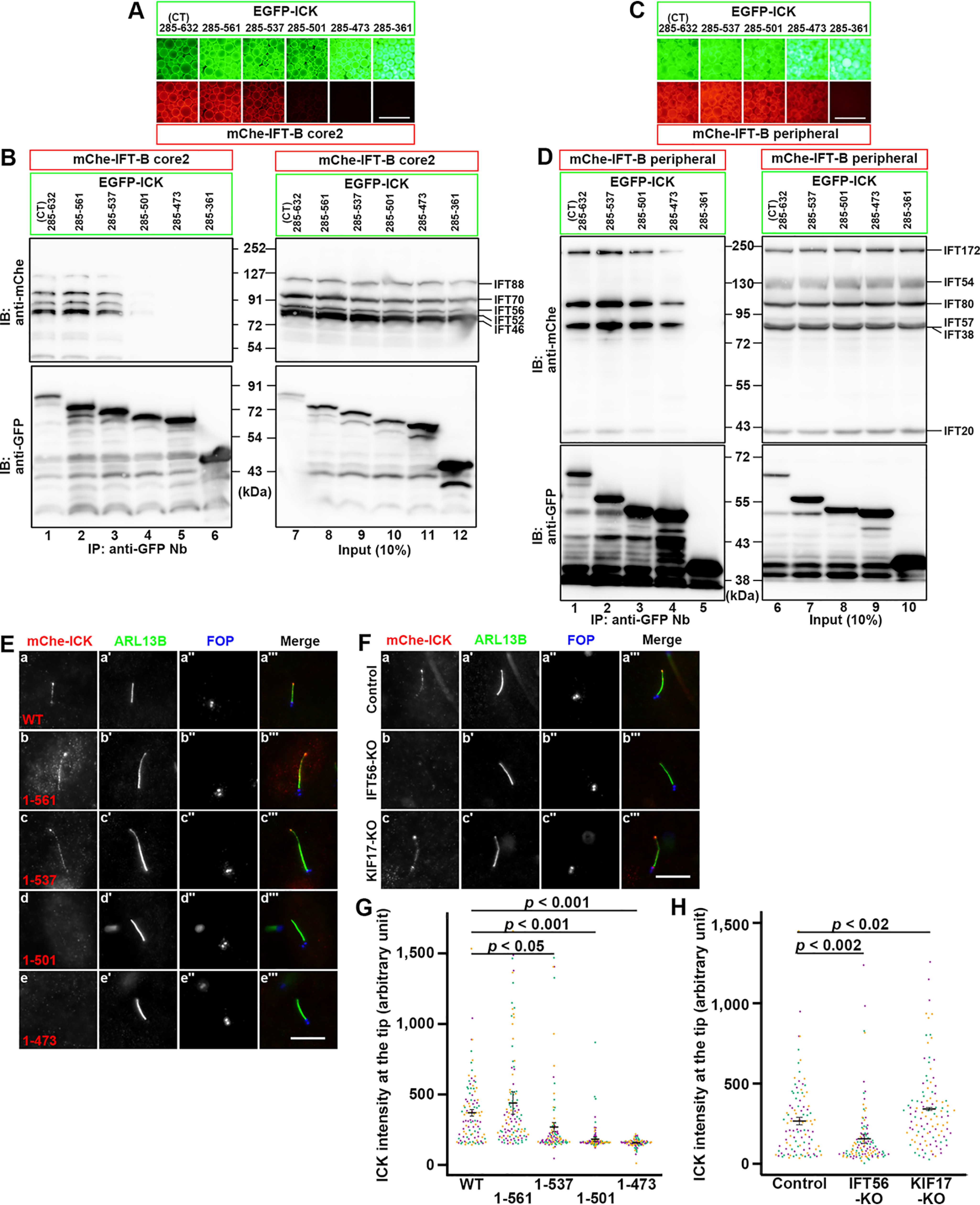
Correlation of the ciliary tip localization of ICK with its IFT-B binding. A–D, participation of distinct parts of the ICK noncatalytic region in its interaction with IFT-B core 2 and peripheral subcomplexes. Lysates of HEK293T cells coexpressing EGFP-fused ICK constructs, as indicated, and mChe-fused IFT-B core 2 (A and B) or peripheral (C and D) subunits processed for the VIP assay using GST-fused anti-GFP Nbs (A and C) followed by immunoblotting using an anti-mChe or anti-GFP antibody (B and D). E, ciliary localization of ICK deletion constructs. hTERT-RPE1 cells expressing the mChe-fused ICK construct, as indicated (panels a–e), were serum-starved for 24 h to induce ciliogenesis, and immunostained for ARL13B (panels á–é) and FOP (panels á´–é´). F, IFT56 deficiency compromises the localization of ICK to the ciliary tip. Control hTERT-RPE1 cells (panel a), IFT56-KO cells (panel b), or KIF17-KO cells (panel c) expressing mChe-ICK were immunostained as described for E. G, relative intensities of mChe-fused ICK constructs at the ciliary tip shown in E were estimated and expressed as scatter plots. H, relative intensities of mChe-ICK constructs at the ciliary tip in control, IFT56-KO, and KIF17-KO cells shown in F were estimated and expressed as scatter plots. In the scatter plots, different colored dots represent three independent experiments, horizontal lines are means, and error bars are S.D. Statistical significances were calculated using one-way ANOVA followed by the Dunnett multiple comparison test. Scale bars, 500 μm in A and C and 5 μm in E and F. IB, immunoblot; IP, immunoprecipitation.
We then analyzed whether the ICK truncation constructs can localize to the ciliary tip. When expressed in hTERT-RPE1 cells, mChe-ICK(1–561) and mChe-ICK(WT) were found at the distal tip (Fig. 2E, panels a and b). mChe-ICK(1–537) was also able to localize to the tip (Fig. 2E, panel c), but with much lower efficiency (Fig. 2G). In striking contrast, virtually no ciliary signal was detected for mChe-ICK(1–501) and mChe-ICK(1–473) (Fig. 2E, panels d and e; also see Fig. 2G). Taken together with the interaction data (Fig. 2, A–D), the localization of ICK to the ciliary tip appears to correlate with its binding to the IFT-B complex, at least to the core 2 subcomplex.
To confirm the IFT-B–dependent transport of ICK to the tip, we used IFT56-KO cells. Although the absence of an IFT-B subunit in many cases results in a cilia-lacking phenotype (9), we previously showed that ciliogenesis is normal in IFT56-KO cells (38). However, KO of IFT56 abolishes the ciliary tip localization of KIF17, which binds to the IFT-B complex via the IFT46–IFT56 dimer (38). Because ICK interacts with the IFT-B core 2 subcomplex, which contains IFT56 (Fig. 1E), we expected that the KO of IFT56 also affects the ciliary tip localization of ICK. As shown in Fig. 2F, the ciliary tip localization of mChe-ICK was greatly diminished, although not completely abolished, in IFT56-KO cells, whereas its localization was not significantly affected in KIF17-KO cells, which were used as a negative control (Fig. 2F, panels b and c; also see Fig. 2H); it is noteworthy that, in contrast to the established role of the Caenorhabditis elegans KIF17 ortholog OSM-3 in anterograde trafficking in the distal singlet segment of neuronal cilia (44), the role of mammalian KIF17 remains unclear, because cells knocked out of a subunit of heterotrimeric kinesin-II (45), but not KIF17-KO cells (38), show abnormal phenotype with respect to ciliogenesis and anterograde ciliary protein trafficking (46). Thus, these data support that ICK is transported to the ciliary tip in a manner dependent on its binding to the IFT-B complex.
ICK deficiency results in impaired retrograde ciliary protein trafficking and the release of extracellular vesicles from the bulged ciliary tip
We established ICK-KO hTERT-RPE1 cell lines and characterized their phenotypes. Two independent KO cell lines (#ICK-3-14 and #ICK-4-6), which were established using distinct target sequences (Fig. S1), were selected for the following analyses. When immunostained for the ARL13B small GTPase, a marker for the ciliary membrane, the cilia of both KO cell lines were demonstrated to be longer than those of control RPE1 cells (Fig. 3A, compare panels b and c with panel a; also see Fig. 3C), which was consistent with previous studies of ICK homologs in other organisms (26–31, 47–49). The long cilia often appeared to be bending or winding, suggesting that these cilia might grow on the ventral cell surface (50). It is noteworthy that the variation in ciliary length was larger in the ICK-KO cell lines than in control cells (Fig. 3C) (see below). In these ICK-KO cells, IFT88 (an IFT-B subunit) and IFT140 (an IFT-A subunit) were considerably accumulated at the bulged ciliary tip, whereas they were mainly found at the base of cilia in control RPE1 cells (Fig. 3, A and B, compare panels b′ and c′ with panel a′; also see D and E), which is compatible with a previous study of Ick-KO MEFs (22). It is thus likely that, in the absence of ICK, the IFT machinery remains at the distal tip after its anterograde trafficking.
Figure 3.
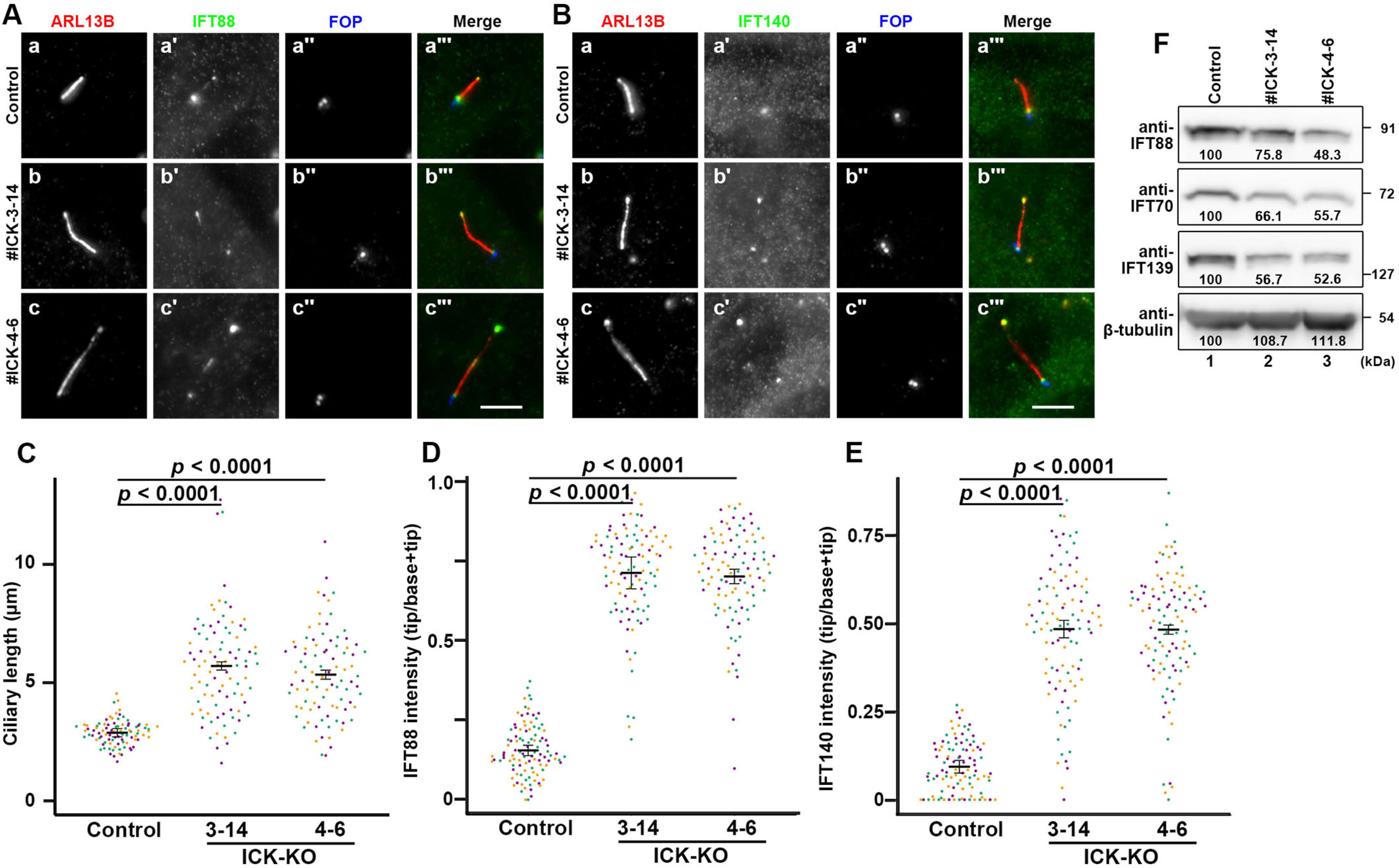
Accumulation of IFT proteins at the ciliary tip in ICK-KO cells. A and B, control RPE1 cells (panels a) and the ICK-KO cell lines #ICK-3-14 (panels b) and #ICK-4-6 (panels c) were serum-starved for 24 h to induce ciliogenesis and triple immunostained for ARL13B (panels a–c), either IFT88 (A) or IFT140 (B, panels a′–c′), and FOP (panels a″–c″). Scale bars, 5 μm. C, ciliary lengths of control cells and ICK-KO cells shown in A were measured and expressed as scatter plots. D and E, the relative staining intensities of IFT88 and IFT140 at the ciliary tip and base, shown in A and B, respectively, were estimated, and the intensity ratios of tip/(tip + base) were expressed as scatter plots. In all the scatter plots, different colored dots represent three independent experiments, horizontal lines are means, and error bars are S.D. Statistical significances were calculated using one-way ANOVA followed by the Dunnett multiple comparison test. F, comparison of the levels of the IFT proteins in control RPE1 cells and in ICK-KO cells. Lysates were prepared from serum-starved control RPE1 cells (lane 1), and the ICK-KO cell lines #ICK-3-14 (lane 2) and #ICK-4-6 (lane 3). The lysates containing 30 µg of protein were separated by SDS-PAGE and subjected to immunoblotting analysis using antibodies against IFT88, IFT70, IFT139, and β-tubulin as indicated. The band intensities were measured, and the relative intensities are represented as the intensity in control RPE1 cells as 100%.
We also compared the levels of the IFT-B and IFT-A proteins in control RPE1 cells and in the ICK-KO cells. As shown in Fig. 3F, the levels of IFT88 and IFT70 (IFT-B subunits) and IFT139 (an IFT-A subunit) in the ICK-KO cells were substantially lower than those in control RPE1 cells. Although we do not know the exact reason for the reduction in the IFT protein levels in ICK-KO cells, the reduction might reflect the instability of the IFT machinery in the absence of ICK or the elimination of the excessively accumulated IFT proteins at the bulged ciliary tip as extracellular vesicles (ECVs) (see below).
We then analyzed the localization of GPR161 and Smoothened (SMO), both of which are ciliary G protein–coupled receptors involved in Hh signaling (51, 52). Under basal conditions in control RPE1 cells, GPR161 is localized within cilia (Fig. 4A, panel a) and negatively regulates Hh signaling, whereas SMO is not found within cilia (Fig. 4B, panel a). When control cells are stimulated with the small molecule activator SAG (Smoothened Agonist), SMO enters cilia (Fig. 4B, panel d), whereas GPR161 exits cilia (Fig. 4A, panel d), resulting in the cancelation of the negative regulation. In marked contrast to the control cells, GPR161 remained within cilia even upon SAG stimulation in both ICK-KO cell lines (Fig. 4A, panels e and f; also see Fig. 4C). On the other hand, a substantial amount of SMO was found within cilia, often at the bulged tip, under both nonstimulated and SAG-stimulated conditions in the ICK-KO cell lines (Fig. 4B, panels b and c; also see Fig. 4D). These observations indicate that the retrograde trafficking of G protein–coupled receptors and/or their exit from cilia is impaired in the absence of ICK.
Figure 4.
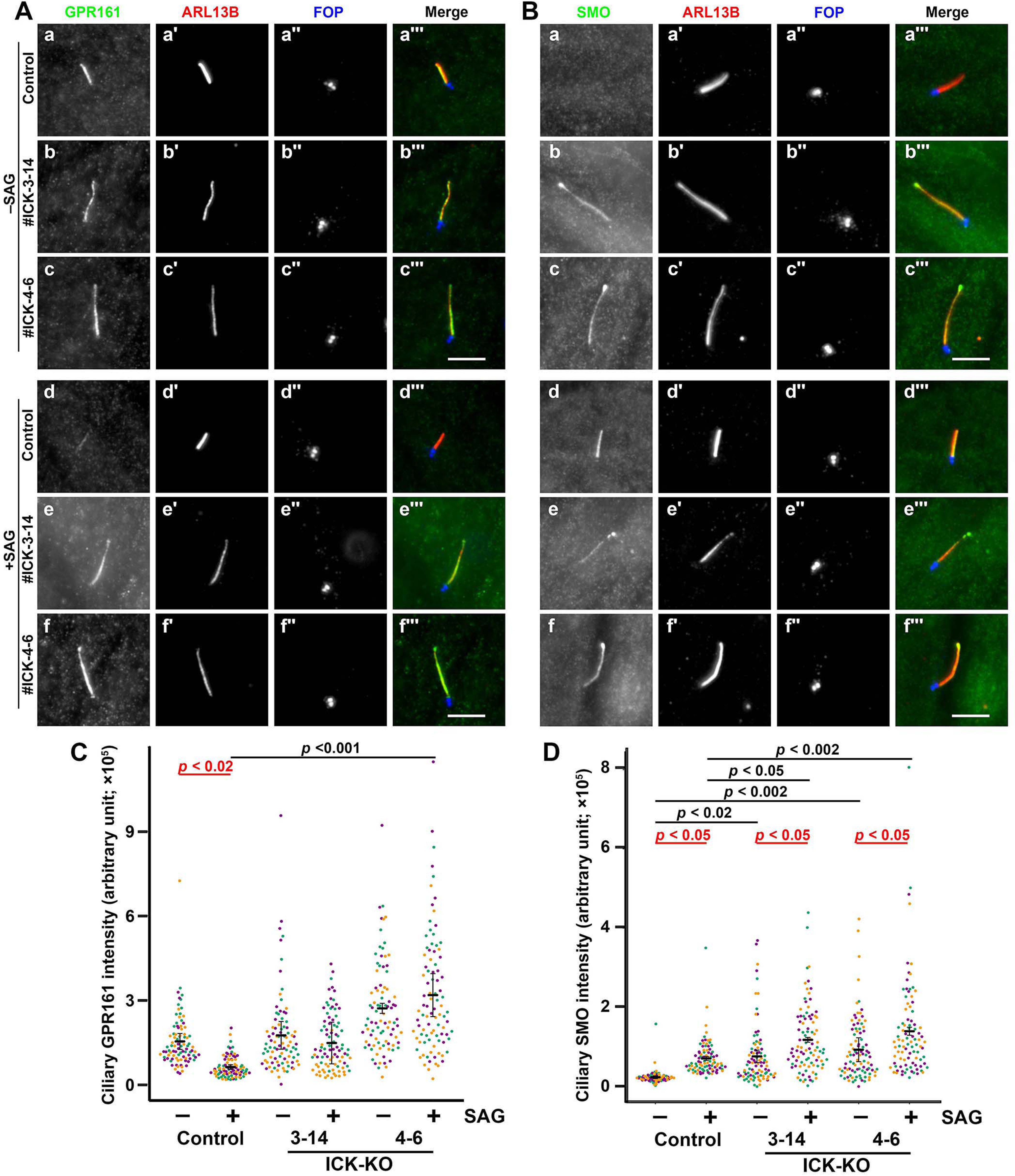
Accumulation of GPR161 and SMO within cilia in ICK-KO cells. A and B, control RPE1 cells (panels a and d) and the ICK-KO cell lines #ICK-3-14 (panels b and e) and #ICK-4-6 (panels c and f) were serum-starved for 24 h to induce ciliogenesis and further incubated for 24 h without (−SAG; panels a–c) or with (+SAG; panels d–f) 200 nm SAG. The cells were triple immunostained for either GPR161 (A) or SMO (B) (panels a–f), ARL13B (panels a′–f′), and FOP (panels a″–f″). Scale bars, 5 μm. C and D, the relative ciliary staining intensities of GPR161 and SMO shown in A and B, respectively, were estimated and expressed as scatter plots. In the scatter plots, different colored dots represent three independent experiments, horizontal lines are means, and error bars are S.D. Statistical significances among multiple cell lines were calculated using one-way ANOVA followed by the Tukey multiple comparison test, and those between two groups (−SAG and +SAG) were calculated using the Bonferroni-adjusted Student's t test.
During the immunostaining analysis of ICK-KO cells, we noticed unusual immunostaining patterns for ciliary proteins. When control RPE1 cells were triply immunostained for IFT88, ARL13B, and FOP (also known as FGFR1OP and recently renamed as CEP43; a marker for the basal body and mother and daughter centrioles), IFT88 was mainly found around the ciliary base, marked by FOP staining (Fig. 5A; also see Fig. 3A, panel á). When ICK-KO cells were stained for these markers, IFT88 was mainly found at the end (tip) of cilia opposite to the end stained for FOP (Fig. 5B, panels a–d). However, we also noticed the presence of punctate structures, which are positive for both IFT88 and ARL13B but negative for the basal body marker, FOP (Fig. 5B, panels e–h). We speculated that these punctate structures represent vesicles released from the ciliary tip, where IFT88 is excessively accumulated. Because cilia can emerge not only on the dorsal surface but also on the ventral surface in RPE1 cells (50), it is possible that the released vesicles were trapped in the small space between the ventral cell surface and the coverslip.
Figure 5.
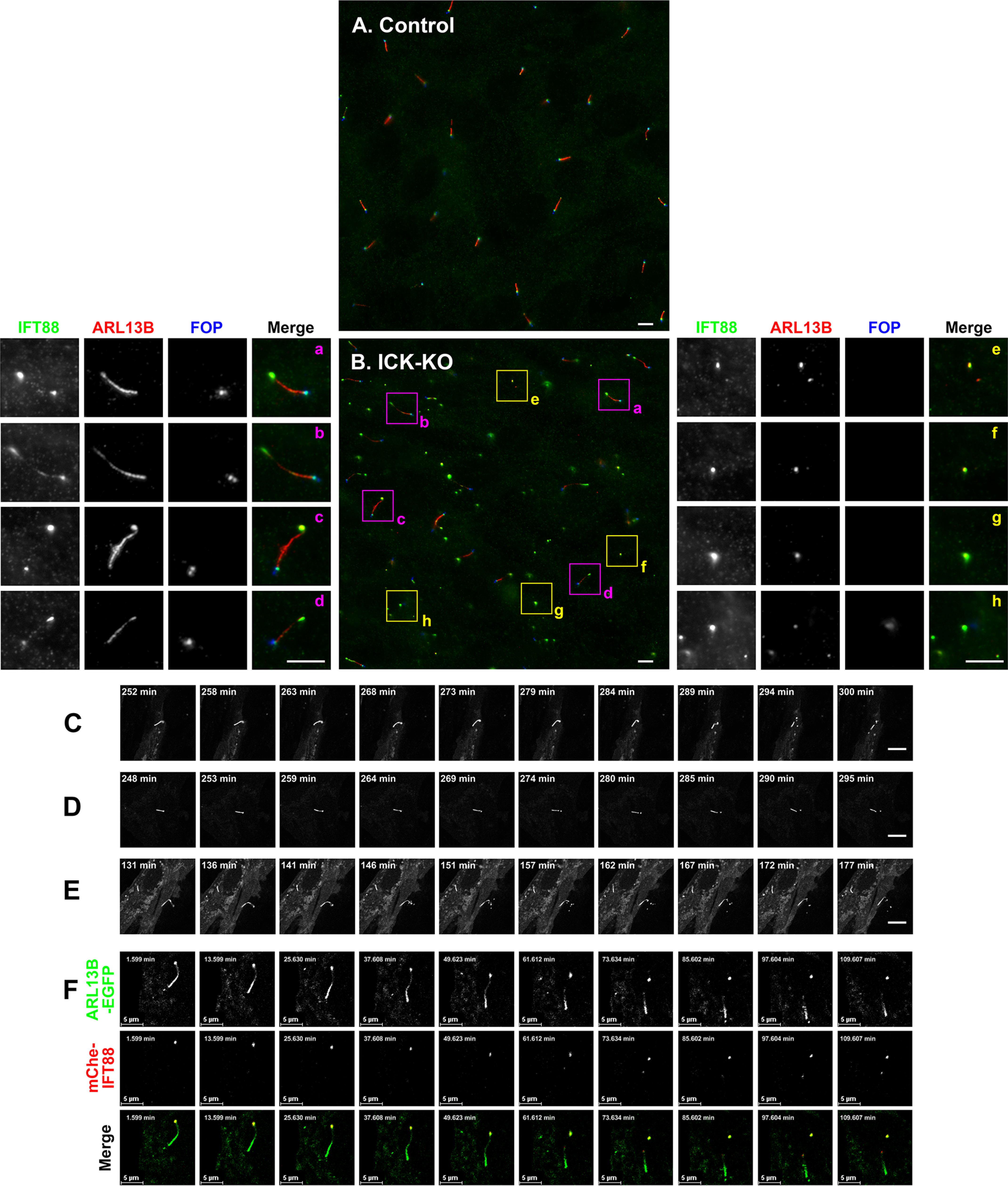
Formation of extracellular vesicles at the bulged ciliary tip of ICK-KO cells. A and B, control RPE1 cells (A) and the ICK-KO cell line #ICK-4-6 (B) were treated and immunostained as described in the legend for Fig. 3A. Three-fold enlarged images of the boxed regions are shown in panels a–h. C–E, release of ECVs from the bulged ciliary tips of the ICK-KO cell line (#ICK-4-6) stably expressing ARL13B(ΔGD)-EGFP. Time-lapse sequences of Videos S1–S3, respectively, are shown. F, release of ECVs from the bulged ciliary tips of the #ICK-4-6 cell line coexpressing ARL13B(ΔGD)-EGFP and mChe-IFT88. A time-lapse sequence of Video S4 is shown. Scale bars, 5 μm.
To address this speculation, we established ICK-KO cells expressing ARL13B(ΔGD)-EGFP and observed the cilia of these cells by time-lapse recording; for the labeling of cilia, instead of ARL13B(WT), we used ARL13B(ΔGD), which lacks the GTPase domain but retains the ability to localize on the ciliary membrane, because exogenous expression of ARL13B(WT) causes the elongation of cilia (53, 54). As shown in Videos S1–S3 and in Fig. 5 (C–E), ARL13B-positive vesicles appeared to be torn off and released from the bulged ciliary tip in ICK-KO cells; it is noteworthy that vesicles were independently released from two distinct cilia in Video S3. When ICK-KO cells coexpressing ARL13B(ΔGD)-EGFP and mChe-IFT88 were examined, released vesicles were shown to contain both ARL13B and IFT88 (Video S4 and Fig. 5F). Thus, it is highly likely that ciliary proteins that excessively accumulate at the tip are eliminated as ECVs to liberate cilia from their aberrant overaccumulation. Our observations are compatible with a previous proteomic profiling study of ECV contents released under growth-stimulated conditions; the ECVs contained ICK and most components of the IFT machinery, as well as membrane proteins, including ARL13B (55).
ICK undergoes IFT particle–like movement, and its anterograde trafficking is required for retrograde ciliary protein trafficking
The long cilia phenotype and accumulation of IFT88 at the bulged ciliary tip in ICK-KO cells were rescued by the exogenous expression of mChe-ICK(WT) (Fig. 6A, compare panel c with panel b; also see Fig. 6, B and C). When ICK-KO cells expressing mChe-ICK(WT) were observed by total internal reflection fluorescence (TIRF) microscopy, the bidirectional movement of mChe-ICK–positive particles was observed (Video S5). This observation supports our conclusion that ICK is delivered to the ciliary tip as a cargo of IFT particles and is consistent with a previous study (36). When the C-terminally truncated constructs were expressed in ICK-KO cells, the elongation of cilia and accumulation of IFT88 at the ciliary tip was substantially rescued by mChe-fused ICK(1–561), ICK(1–537), and ICK(1–501) (Fig. 6A, panels d–f; also see Fig. 6, B and C). mChe-ICK(1–473) rescued the long cilia phenotype and IFT88 accumulation much less efficiently than ICK(WT) (Fig. 6A, panel g; also see Fig. 6, B and C).
Figure 6.
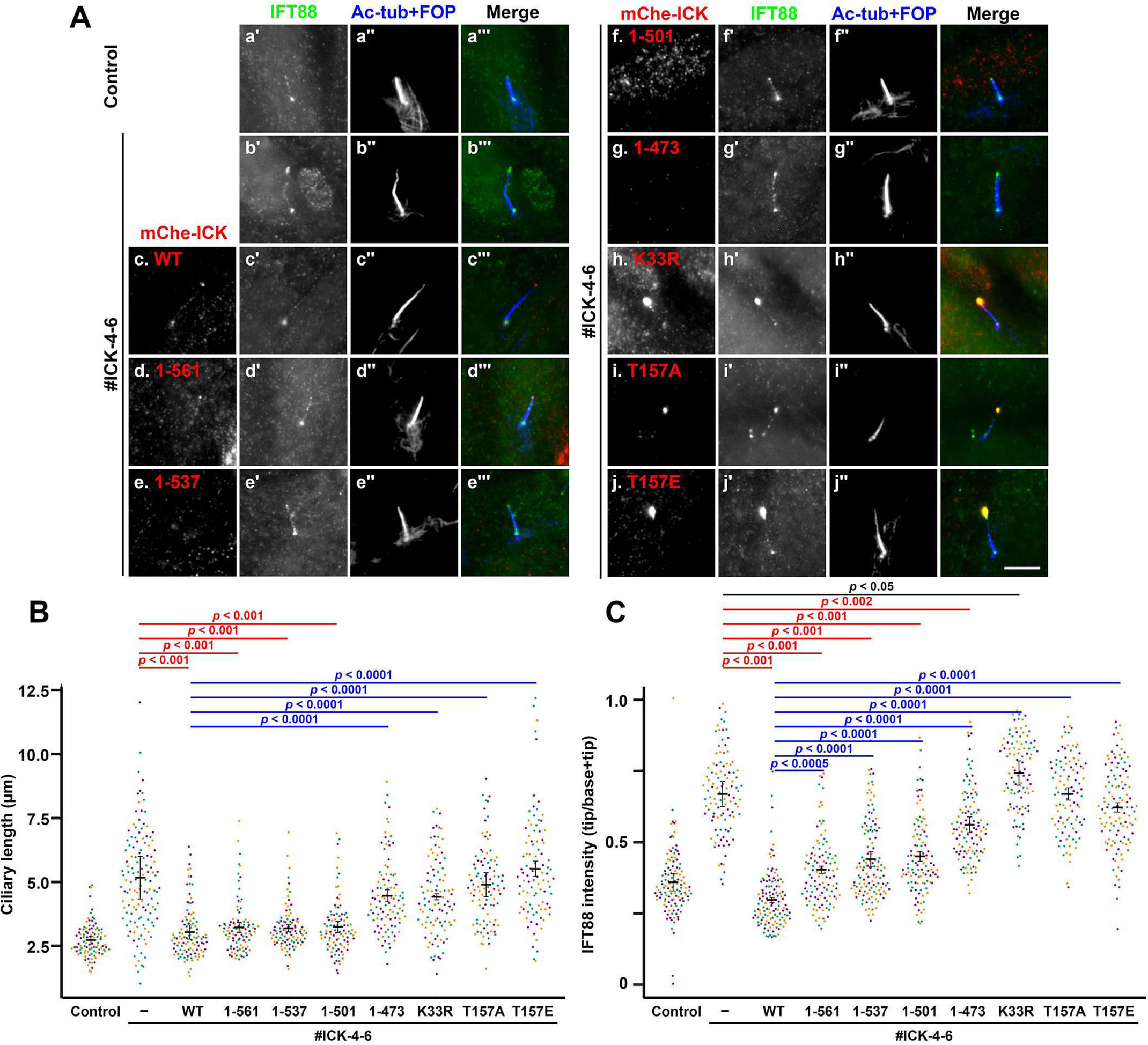
Ciliary tip localization, kinase activity, and phosphorylation at the TDY motif of ICK are required for its functions. A, ICK-KO cells (#ICK-4-6) stably expressing mChe-fused ICK(WT) (panel c), ICK(1–567) (panel d), ICK(1–537) (panel e), ICK(1–506) (panel f), ICK(1–473) (panel g), ICK(K33R) (panel h), ICK(T157A) (panel i), or ICK(T157E) (panel j) were immunostained for IFT88 (panels a′–i′) and acetylated α-tubulin (Ac-tub) + FOP (panels a″–i″). Original RPE1 cells (panel a) and the ICK-KO cell line #ICK-4-6 (panel b) were used as positive and negative controls, respectively. Scale bars, 5 μm. B and C, ciliary lengths (B) and relative staining intensities of IFT88 and IFT140 at the ciliary tip and base (C) in ICK-KO cells stably expressing the ICK construct, as shown in A, was analyzed as described in the legend for Fig. 3 (C and D), respectively.
The expression of mChe-fused ICK(K33R), a kinase-dead mutant (22, 25, 29), did not rescue the elongation of cilia or the accumulation of IFT88 at the tip (Fig. 6A, panel h). It is worth noting that the ICK(K33R) mutant itself was accumulated at the bulged tip (Fig. 6A, panel h), and there was substantially greater accumulation of IFT88 in these cells than in the ICK-KO cells without exogenous ICK expression (Fig. 6C, compare the eighth plot with the second plot). These results confirm that the kinase activity of ICK is crucial for its function (22, 29).
We then analyzed ICK mutants at the TDY motif, because ICK was reported to undergo activation by phosphorylation at this motif by CCRK/CDK20 (24,25). The exogenous expression of mChe-fused ICK(T157A) and ICK(T157E), which are a phosphorylation-defective mutant and a phosphomimetic mutant, respectively, was unable to rescue cilia elongation and IFT88 accumulation at the ciliary tip in ICK-KO cells, although these mutants were able to localize at the ciliary tip (Fig. 6A, panels i and j; also see Fig. 6, B and C), suggesting that the phosphorylation cycle at the TDY motif is important for the function of ICK. These observations are compatible with previous studies in mouse and other organisms, demonstrating that mutations in the genes of not only ICK orthologs but also CCRK orthologs result in cilia elongation (30, 47–49, 56). These observations altogether demonstrate that the kinase activity of ICK, delivered to the ciliary tip via binding to the IFT machinery, is essential for its function, which is probably regulated by its phosphorylation within the TDY motif by CCRK/CDK20.
Discussion
The localization of ICK at the ciliary tip had suggested that this protein kinase might be transported to the tip as a cargo of the IFT machinery. In this study, we demonstrated the direct binding of ICK to the IFT-B complex via its C-terminal noncatalytic region (Fig. 1). Because the noncatalytic region of ICK has characteristics of intrinsically disordered proteins (19, 37), it may be able to flexibly interact with multiple IFT-B subunits. We also found that ICK constructs compromised with respect to their IFT-B–binding ability were unable to localize to the ciliary tip (Fig. 2) and that ICK undergoes intraciliary movements, similarly to IFT particles (Video S5). We thus conclude that ICK is delivered to the ciliary tip via its direct binding to the IFT-B complex.
Although many proteins are believed to be transported within cilia (57), only a limited number of cargo proteins have been demonstrated to directly bind to the IFT machinery, as follows (46): (i) the best characterized cargo transported on IFT trains is the αβ-tubulin heterodimer, which binds to IFT74–IFT81 of the IFT-B complex (2, 58, 59); (ii) in Chlamydomonas, ODA16, a cargo-specific adaptor for outer dynein arm proteins, was shown to be transported via interacting with IFT46 (60–62); and (iii) we recently showed that KIF17 is transported to the ciliary tip via binding to IFT46–IFT56 (38). Thus, to our knowledge, ICK is the fourth protein demonstrated to be transported via its direct binding to the IFT machinery. In addition, a recent cryoelectron tomographic study of anterograde IFT particles in Chlamydomonas flagella demonstrated that the dynein-2 complex is positioned away from the axonemal microtubules and suggested that it is transported as an inactive cargo of anterograde IFT trains (17).
ICK-KO cells showed not only the elongation of cilia but also the aberrant accumulation of IFT-A and IFT-B complexes at the bulged ciliary tip (Fig. 3). The considerable accumulation of IFT components at the tip is reminiscent of the phenotypes observed in KO cells of the subunits of dynein-2 and IFT-A complexes, as reported by us and others (63–66). In view of the fact that the dynein-2 complex in conjunction with IFT-A mediates retrograde trafficking of the IFT machinery, it is likely that, in the absence of ICK, although the IFT machinery can undergo anterograde trafficking from the base to the tip, it is unable to move retrogradely from the ciliary tip. In this context, it is also worth noting that mutations not only in the genes of the dynein-2 and IFT-A subunits (67, 68) but also in the ICK gene (32) result in the same skeletal ciliopathy, i.e. SRTD.
A kinase-dead ICK mutant was unable to rescue the abnormal phenotype of ICK-KO cells, although it was able to reach the ciliary tip, indicating that phosphorylation of some substrates at the tip by ICK is crucial for retrograde trafficking of the IFT machinery. Candidate ICK substrates are motor proteins (kinesin and dynein) and IFT subunits that are crucial for assembly of the IFT machinery itself or involved in binding of the IFT machinery to motor proteins. The most likely protein under the regulation of ICK is heterotrimeric kinesin-II, as the C. elegans ICK ortholog DYF-5 was proposed to participate in the undocking of kinesin-II from IFT particles (28). In addition, a previous study showed that KIF3A, a motor subunit of heterotrimeric kinesin-II, is phosphorylated by ICK in vitro (22, 37, 69). Furthermore, in Chlamydomonas, phosphorylation of FLA8/KIF3B by a calcium-dependent protein kinase was proposed to regulate the binding of kinesin-II to the IFT-B complex and thereby control the entry of the IFT machinery and its turnaround at the ciliary tip (70). In view of the aberrant accumulation of IFT components at the bulged ciliary tip in ICK-KO cells, it is likely that ICK regulates the turnaround event of the IFT machinery by phosphorylating kinesin-II subunits or other components of the IFT machinery.
During our experiments, we noticed that in ICK-KO cells, not only is the mean ciliary length longer, but the variation in ciliary length is also larger than in control cells (Fig. 3C), and we found the presence of vesicles containing ciliary components around the ICK-KO cells (Fig. 5B). We finally demonstrated that ECVs are released by the bulged ciliary tip by being pinched off (Videos S1–S3; and Fig. 5, C–E). Thus, it is likely that in the absence of ICK, cilia are elongated to a limited length, and then the accumulated proteins are eliminated by excision of the bulged tip. Previous studies showed the release of ECVs by excision of the distal ciliary tip under certain conditions (55, 71–74). Our observations are consistent with a previous proteomic analysis showing the presence of not only components of the IFT machinery, including IFT-A, IFT-B, kinesin-II, and membrane proteins, including ARL13B, but also ICK in ECVs (55). As proposed in a previous study (55), the excision of cilia may serve as an outlet for the IFT machinery, in particular, the IFT-B complex, which directly participates in anterograde trafficking of the αβ-tubulin heterodimer, which is a building block required for axoneme elongation. It is thus possible that the release of ECVs containing IFT components is a mechanism that prevents the excessive elongation of cilia.
Experimental procedures
Plasmids, antibodies, and reagents
Expression vectors for human ICK (22) and its point mutants and deletion mutants were constructed in this study as listed in Table S1. Many of expression vectors for other proteins were used in our previous studies. Antibodies used in this study are listed in Table S2. GST-tagged anti-GFP Nbs prebound to GSH–Sepharose 4B beads were prepared as described previously (40, 41). SAG was purchased from Enzo Life Sciences.
VIP assay and immunoblotting analysis
The VIP assay and subsequent immunoblotting analysis were performed as described previously (41, 75); experimental details of the VIP assay were recently described (40). HEK293T cells grown on a 6-well plate were cotransfected with expression vectors for EGFP-fused and mChe-fused proteins using Polyethylenimine Max (Polysciences) and cultured for 24 h in high glucose Dulbecco's modified Eagle's medium (Nacalai Tesque) supplemented with 5% fetal bovine serum (FBS). The transfected cells were then lysed in 250 μl of HMDEKN cell-lysis buffer (10 mm HEPES, pH 7.4, 5 mm MgSO4, 1 mm DTT, 0.5 mm EDTA, 25 mm KCl, and 0.05% Nonidet P-40) containing protease inhibitor mixture (Nacalai Tesque) for 15 min on ice. The lysates were then centrifuged at 16,100 × g for 15 min, and the supernatant (200 μl) was transferred to a 0.2-ml eight-tube strip, to which GST-tagged anti-GFP Nbs prebound to GSH–Sepharose 4B beads (∼5-μl bed volume of the beads; GE Healthcare) was added and incubated for 1 h at 4 °C with constant rotation of the tube strip. After brief centrifugation at 2,000 × g for 10 s, the beads precipitated were washed three times with lysis buffer (180 μl), transferred to a 96-well plate or a 96-well glass-bottomed plate (AGC Techno Glass), and observed under a BZ-8000 all-in-one type microscope (Keyence) with a 20×, 0.75 NA objective lens under fixed conditions (sensitivity ISO 400, exposure 1/10 s for green fluorescence; and sensitivity ISO 800, exposure 1/5 s for red fluorescence).
The beads bearing fluorescent proteins were then subjected to immunoblotting analysis. Proteins on the beads were separated by SDS-PAGE and electroblotted onto an Immobilon-P membrane (Merck Millipore). The membrane was blocked in 3% skimmed milk and incubated sequentially with primary antibody and peroxidase-conjugated secondary antibody. The protein bands were detected using a Chemi-Lumi One L kit (Nacalai Tesque).
Establishment of ICK-KO cell lines
The strategy for the disruption of genes in hTERT-RPE1 cells (American Type Culture Collection, CRL-4000) by the CRISPR/Cas9 system using homology-independent DNA repair was previously described in detail (Ref. 76; also see Refs. 12, 77, and 78). In brief, single-guide RNA (sgRNA) sequences targeting the human ICK gene (Table S3) were designed using CRISPOR (79). Double-stranded oligonucleotides for the target sequences were inserted into the all-in-one sgRNA expression vector peSpCAS9(1.1)-2×sgRNA (Addgene 80768). hTERT-RPE1 cells grown on a 12-well plate to ∼3.0 × 105 cells were transfected with the sgRNA vector (1 µg) and the donor knock-in vector, pDonor-tBFP-NLS-Neo(universal) (0.25 µg; Addgene 80767), using X-tremeGENE9 Reagent (Roche Applied Science). The cells with nuclear tBFP signals were isolated after culturing in the presence of G418 (600 µg/ml). Genomic DNA extracted from the isolated cells was analyzed by PCR using KOD FX Neo DNA polymerase (Toyobo) and three sets of primers (Table S3) to distinguish the following three states of integration of the donor knock-in vector: forward integration (Fig. S1, A and C, lane b), reverse integration (lane c), and no integration with a small indel (lane a) (63). The disruption of both ICK alleles was confirmed by direct sequencing of the PCR products (Fig. S1, B and D). IFT56-KO and KIF17-KO cell lines were established as described previously (38).
Preparation of cells stably expressing mChe-fused ICK constructs
Lentiviral vectors for the expression of ICK constructs were prepared by a previously described method (80) with minor modifications; to keep the expression level of the mChe-ICK protein low, a short upstream ORF was inserted immediately before the mChe ORF (81). pRRLsinPPT–upstream ORF–mChe–ICK or its mutant construct was transfected into HEK293T cells along with packaging plasmids (pRSV-REV, pMD2.g, and pMDLg/pRRE; kind gifts from Peter McPherson, McGill University) (82). Culture media were replaced 8 h after transfection, and those containing viral particles were collected at 24, 36, and 48 h after transfection. The collected media were passed through a 0.45-µm filter and centrifuged at 32,000 × g at 4 °C for 4 h. The precipitates containing lentiviral particles were resuspended in Opti-MEM (Invitrogen). The lentiviral suspension was added to the culture medium of hTERT-RPE1 cells or ICK-KO cell lines established as described above. After a 24-h incubation, the cells were used for subsequent analyses.
Immunofluorescence analysis, live cell imaging, and TIRF microscopy
hTERT-RPE1 cells or the ICK-KO cell lines were cultured in Dulbecco's modified Eagle's medium/F-12 (Nacalai Tesque) supplemented with 10% FBS and 0.348% sodium bicarbonate. To induce ciliogenesis, the cells were grown on coverslips to 100% confluence and starved for 24 h in Opti-MEM containing 0.2% BSA.
Unless otherwise stated, immunofluorescence analysis was performed as described previously (53, 63). The cells on coverslips were fixed with 3% paraformaldehyde for 5 min at 37 °C, permeabilized with 100% methanol for 5 min at −20°C (for experiments shown in Figs. 3–5), or fixed and permeabilized with 100% methanol for 5 min at −20°C (for experiments shown in Figs. 1, 2, and 6), and washed three times with PBS. The fixed/permeabilized cells were blocked with 10% FBS, incubated sequentially with primary and secondary antibodies diluted in Can Get Signal immunostain solution A (Toyobo) (for the detection of SMO) or in 5% FBS (for the detection of the other proteins), and observed using an AxioObserver microscope (Carl Zeiss).
Live cell imaging was performed as described previously (80). Cells expressing EGFP-fused ARL13B(ΔGD) or those coexpressing EGFP-fused ARL13B(ΔGD) and mChe-fused IFT88 were serum-starved for 24 h on a glass-bottomed culture dish (Greiner Bio-One) and observed under an A1R-MP confocal laser-scanning microscope (Nikon). Time-lapse images were acquired sequentially every 5 min and analyzed using the NIS-Elements imaging software (Nikon).
TIRF microscopy was performed as described previously (64, 83). Cells expressing an mChe-fused ICK construct were serum-starved for 24 h on a glass-bottomed culture dish, placed on a microscope stage prewarmed to 37 °C, and observed using a TIRFM ECLIPSE Ti microscope (Nikon) at a video rate using the NIS-Elements imaging software (Nikon).
Data availability
All data are contained within the article.
Supplementary Material
Acknowledgments
We thank Peter McPherson for providing plasmids for lentiviral production; Shohei Nozaki and Taro Chaya for technical advice; and Helena Akiko Popiel for critical reading of the manuscript.
This article contains supporting information.
Author contributions—K. Nakamura, Y. O., T. F., Y. K., and K. Nakayama conceptualization; K. Nakamura data curation; K. Nakamura, T. N., and M. T. formal analysis; K. Nakamura investigation; K. Nakamura, Y. O., and T. F. writing-review and editing; Y. O., Y. K., and K. Nakayama funding acquisition; Y. K. and K. Nakayama supervision; Y. K. and K. Nakayama writing-original draft.
Funding and additional information—This work was supported in part by Japan Society for the Promotion of Science (JSPS) Grants 15H04370 and 19H00980 (to K. Nakayama), 18H02403 (to Y. K.), and 19H03420 (to Y. O.); a JSPS International Joint Research Program grant-LEAD with UK Research and Innovation from JSPS (to K. Nakayama); a Uehara Memorial Foundation grant (to Y. K.); and Kyoto University Internal Grant ISHIZUE (to K. Nakayama).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- BBS
- Bardet–Biedl syndrome
- CCRK
- cell cycle–related kinase
- CILK1
- ciliogenesis-associated kinase 1
- ECV
- extracellular vesicle
- GST
- glutathione S-transferase
- Hh
- Hedgehog
- hTERT-RPE1
- human telomerase reverse transcriptase-immortalized retinal pigment epithelial 1
- ICK
- intestinal cell kinase
- IFT
- intraflagellar transport
- mChe
- mCherry
- Nb
- nanobody
- SMO
- Smoothened
- SRTD
- short-rib thoracic dysplasia
- TIRF
- total internal reflection fluorescence
- TZ
- transition zone
- KO
- knockout
- VIP
- visible immunoprecipitation
- NT
- N-terminal
- CT
- C-terminal
- EGFP
- enhanced green fluorescent protein
- FBS
- fetal bovine serum
- sgRNA
- single-guide RNA
- ANOVA
- analysis of variance.
References
- 1. Bangs F., and Anderson K. V. (2017) Primary cilia and mammalian hedgehog signaling. Cold Spring Harb. Perspect. Biol. 9, a028175 10.1101/cshperspect.a028175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bhogaraju S., Weber K., Engel B. D., Lechtreck K.-F., and Lorentzen E. (2014) Getting tubulin to the tip of the cilium: one IFT train, many different tubulin cargo binding sites? Bioessays 36, 463–467 10.1002/bies.201400007 [DOI] [PubMed] [Google Scholar]
- 3. Garcia-Gonzalo F. R., and Reiter J. F. (2017) Open sesame: how transition fibers and the transition zone control ciliary composition. Cold Spring Harb. Perspect. Biol. 9, a028134 10.1101/cshperspect.a028134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reiter J. F., and Leroux M. R. (2017) Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 18, 533–547 10.1038/nrm.2017.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Braun D. A., and Hildebrandt F. (2017) Ciliopathies. Cold Spring Harb. Perspect. Biol. 9, a028191 10.1101/cshperspect.a028191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ishikawa H., and Marshall W. F. (2011) Ciliogenesis: building the cell's antenna. Nat. Rev. Mol. Cell Biol. 12, 222–234 10.1038/nrm3085 [DOI] [PubMed] [Google Scholar]
- 7. Sung C.-H., and Leroux M. R. (2013) The roles of evolutionarily conserved functional modules in cilia-related trafficking. Nat. Cell Biol. 15, 1387–1397 10.1038/ncb2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Taschner M., and Lorentzen E. (2016) The intraflagellar transport machinery. Cold Spring Harb. Perspect. Biol. 8, a028092 10.1101/cshperspect.a028092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nakayama K., and Katoh Y. (2018) Ciliary protein trafficking mediated by IFT and BBSome complexes with the aid of kinesin-2 and dynein-2 motors. J. Biochem. 163, 155–164 10.1093/jb/mvx087 [DOI] [PubMed] [Google Scholar]
- 10. Liu P., and Lechtreck K. F. (2018) The Bardet–Biedl syndrome protein complex is an adapter expanding the cargo of range of intraflagellar transport trains for ciliary export. Proc. Natl. Acad. Sci. U.S.A. 115, E934–E943 10.1073/pnas.1713226115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ye F., Nager A. R., and Nachury M. V. (2018) BBSome trains remove activated GPCRs from cilia by enabling passage through the transition zone. J. Cell Biol. 217, 1847–1868 10.1083/jcb.201709041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nozaki S., Castro Araya R. F., Katoh Y., and Nakayama K. (2019) Requirement of IFT-B–BBSome complex interaction in export of GPR161 from cilia. Biol. Open 8, bio043786 10.1242/bio.043786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nozaki S., Katoh Y., Kobayashi T., and Nakayama K. (2018) BBS1 is involved in retrograde trafficking of ciliary GPCRs in the context of the BBSome complex. PLoS One 13, e0195005 10.1371/journal.pone.0195005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mijalkovic J., van Krugten J., Oswald F., Acar S., and Peterman E. J. G. (2018) Single-molecule turnarounds of intraflagellar transport at the C. elegans ciliary tip. Cell Rep. 25, 1701–1707 10.1016/j.celrep.2018.10.050 [DOI] [PubMed] [Google Scholar]
- 15. Bertiaux E., Mallet A., Fort C., Blisnick T., Bonnefoy S., Jung J., Lemos M., Marco S., Vaughan S., Trépout S., Tinevez J.-Y., and Bastin P. (2018) Bidirectional intraflagellar transport is restricted to two sets of microtubule doublets in the trypanosome flagellum. J. Cell Biol. 217, 4284–4297 10.1083/jcb.201805030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chien A. S., Shih S. M., Bower R., Tritschler D., Porter M. E., and Yildiz A. (2017) Dynamics of the IFT machinery at the ciliary tip. eLife 6, e28606 10.7554/eLife.28606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jordan M. A., Diener D. R., Stepanek L., and Pigino G. (2018) The cryo-EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional anterogarde movement in cilia. Nat. Cell Biol. 20, 1250–1255 10.1038/s41556-018-0213-1 [DOI] [PubMed] [Google Scholar]
- 18. Toropova K., Zalyte R., Mukhopadhyay A. G., Mladenov M., Carter A. P., and Roberts A. J. (2019) Structure of the dynein-2 complex and its assembly with intraflagellar transport trains. Nat. Struct. Mol. Biol. 26, 823–829 10.1038/s41594-019-0286-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fu Z., Gailey C. D., Wang E. J., and Brautigan D. L. (2019) Ciliogenesis associated kinase 1: targets and functions in various organ systems. FEBS Lett. 593, 2990–3002 10.1002/1873-3468.13600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Togawa K., Yan Y.-X., Inomoto T., Slaugenhaupt S., and Rustgi A. K. (2000) Intestinal cell kinase (ICK) localizes to the crypt region and requires a dual phosphorylation site found in MAP kinases. J. Cell. Physiol. 183, 129–139 [DOI] [PubMed] [Google Scholar]
- 21. Chen T., Wu D., Moskaluk C. A., and Fu Z. (2013) Distinct expression patterns of ICK/MAK/MOK protein kinases in the intestine implicate functional diversity. PLoS One 8, e79359 10.1371/journal.pone.0079359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chaya T., Omori Y., Kuwahara R., and Furukawa T. (2014) ICK is essential for cell type-specific ciliogenesis and the regulation of ciliary transport. EMBO J. 33, 1227–1242 10.1002/embj.201488175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Omori Y., Chaya T., Katoh K., Kajimura N., Sato S., Muraoka K., Ueno S., Koyasu T., Kondo M., and Furukawa T. (2010) Negative regulation of ciliary length by ciliary male germ cell-associated kinase (Mak) is required for retinal photoreceptor survival. Proc. Natl. Acad. Sci. U.S.A. 107, 22671–22676 10.1073/pnas.1009437108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fu Z., Larson K. A., Chitta R. K., Parker S. A., Turk B. E., Lawrence M. W., Kaldis P., Galaktionov K., Cohn S. M., Shabanowitz J., Hunt D. F., and Sturgill T. W. (2006) Identification of yin-yang regulators and a phosphorylation consensus for male germ cell-associated kinase (MAK)-related kinase. Mol. Cell Biol. 26, 8639–8654 10.1128/MCB.00816-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fu Z., Schroeder M. J., Shabanowitz J., Kaldis P., Togawa K., Rustgi A. K., Hunt D. F., and Sturgill T. W. (2005) Activation of a nuclear Cdc2-related kinase within a mitogen-activated protein kinase-like TDY motif by autophosphorylation and cyclin-dependent protein kinase-activating kinase. Mol. Cell Biol. 25, 6047–6064 10.1128/MCB.25.14.6047-6064.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bengs F., Scholz A., Kuhn D., and Wiese M. (2005) LmxMPK9, a mitogen-activated protein kinase homologue affects flagellar length in Leishmania mexicana. Mol. Microbiol. 55, 1606–1615 10.1111/j.1365-2958.2005.04498.x [DOI] [PubMed] [Google Scholar]
- 27. Berman S. A., Wilson N. F., Haas N. A., and Lefebvre P. A. (2003) A novel MAP kinase regulates flagellar length in Chlamydomonas. Curr. Biol. 13, 1145–1149 10.1016/S0960-9822(03)00415-9 [DOI] [PubMed] [Google Scholar]
- 28. Burghoorn J., Dekkers M. P. J., Rademakers S., de Jong T., Willemsen R., and Jansen G. (2007) Mutation of the MAP kinase DYF-5 affects docking and undocking of kinesin-2 motors and reduces their speed in the cilia of Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 104, 7157–7162 10.1073/pnas.0606974104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moon H., Song J., Shin J.-O., Lee H., Kim H.-K., Eggenschwiller J. T., Bok J., and Ko H. W. (2014) Intestinal cell kinase, a protein associated with endocrine-cerebro-osteodysplasia syndrome, is a key regulator of cilia length and Hedgehog signaling. Proc. Natl. Acad. Sci. U.S.A. 111, 8541–8546 10.1073/pnas.1323161111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jiang Y.-Y., Maier W., Baumeister R., Minevich G., Joachimiak E., Wloga D., Ruan Z., Kannan N., Bocarro S., Bahraini A., Vasudevan K. K., Lechtreck K., Orias E., and Gaertig J. (2019) LF4/MOK and a CDK-related kinase regulate the number and length of cilia in Tetrahymena. PLoS Genet. 15, e1008099 10.1371/journal.pgen.1008099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Okamoto S., Chaya T., Omori Y., Kuwahara R., Kubo S., Sakaguchi H., and Furukawa T. (2017) ICK ciliary kinase is essential for planar cell polarity formation in inner ear hair cells and hearing function. J. Neurosci. 37, 2073–2085 10.1523/JNEUROSCI.3067-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paige Taylor S., Kunova Bosakova M., Varecha M., Balek L., Barta T., Trantirek L., Jelinkova I., Duran I., Vesela I., Forlenza K. N., Martin J. H., Hampl A., University of Washington Center for Mendelian Genomics, Bamshad M., Nickerson D., et al. (2016) An inactivating mutation in intestinal cell kinase, ICK, impairs hedgehog signalling and causes short rib-polydactyly syndrome. Hum. Mol. Genet. 25, 3998–4011 10.1093/hmg/ddw240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oud M., Bonnard C., Mans D., Altunoglu U., Tohari S., Ng A., Eskin A., Lee H., Rupar C., de Wagenaar N., Wu K., Lahiry P., Pazour G., Nelson S., Hegele R., et al. (2016) A novel ICK mutation causes ciliary disruption and lethal endocrine-cerebro-osteodysplasia syndrome. Cilia 5, 8 10.1186/s13630-016-0029-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lahiry P., Wang B., Robinson J. F., Turowec J. P., Litchfield D. W., Lanktree M. B., Gloor G. B., Puffenberger E. G., Strauss K. A., Martens M. B., Ramsay D. A., Rupar C. A., Siu V., and Hegele R. (2009) A multiplex human syndrome implicates a key role for intestinal cell kinase in development of central nervous, skeletal, and endocrine systems. Am. J. Hum. Genet. 84, 134–147 10.1016/j.ajhg.2008.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bailey J. N., de Nijs L., Bai D., Suzuki T., Miyamoto H., Tanaka M., Patterson C., Lin Y. C., Medina M. T., Alonso M. E., Serratosa J. M., Durón R. M., Nguyen V. H., Wight J. E., Martínez-Juárez I. E., et al. (2018) Variant intestinal-cell kinase in juvenile myoclonic epilepsy. New Engl. J. Med. 378, 1018–1028 10.1056/NEJMoa1700175 [DOI] [PubMed] [Google Scholar]
- 36. Broekhuis J. R., Verhey K. J., and Jansen G. (2014) Regulation of cilium length and intraflagellar transport by the RCK-kinase ICK and MOK in renal epithelial cells. PLoS One 9, e108470 10.1371/journal.pone.0108470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oh Y. S., Wang E. J., Gailey C. D., Brautigan D. L., Allen B. L., and Fu Z. (2019) Ciliopathy-associated protein kinase ICK requires its non-catalytic carboxyl-terminal domain for regulation of ciliogenesis. Cells 8, 677 10.3390/cells8070677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Funabashi T., Katoh Y., Michisaka S., Terada M., Sugawa M., and Nakayama K. (2017) Ciliary entry of KIF17 is dependent on its binding to the IFT-B complex via IFT46-IFT56 as well as on its nuclear localization signal. Mol. Biol. Cell 28, 624–633 10.1091/mbc.E16-09-0648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boldt K., van Reeuwijk J., Lu Q., Koutroumpas K., Nguyen T. M., Texier Y., van Beersum S. E. C., Horn N., Willer J. R., Mans D., Dougherty G., Lamers I. J., Coene K. L., Arts H. H., Betts M. J., et al. (2016) An organelle-specific protein landscape identifies novel diseases and molecular mechanisms. Nat. Commun. 7, 11491 10.1038/ncomms11491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Katoh Y., Nakamura K., and Nakayama K. (2018) Visible immunoprecipitation (VIP) assay: a simple and versatile method for visual detection of protein-protein interactions. Bio-protocol 8, e2687 10.21769/BioProtoc.2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Katoh Y., Nozaki S., Hartanto D., Miyano R., and Nakayama K. (2015) Architectures of multisubunit complexes revealed by a visible immunoprecipitation assay using fluorescent fusion proteins. J. Cell Sci. 128, 2351–2362 10.1242/jcs.168740 [DOI] [PubMed] [Google Scholar]
- 42. Katoh Y., Terada M., Nishijima Y., Takei R., Nozaki S., Hamada H., and Nakayama K. (2016) Overall architecture of the intraflagellar transport (IFT)-B complex containing Cluap1/IFT38 as an essential component of the IFT-B peripheral subcomplex. J. Biol. Chem. 291, 10962–10975 10.1074/jbc.M116.713883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Taschner M., Weber K., Mourão A., Vetter M., Awasthi M., Stiegler M., Bhogaraju S., and Lorentzen E. (2016) Intraflagellar transport proteins 172, 80, 57, 54, 38, and 20 form a stable tubulin-binding IFT-B2 complex. EMBO J. 35, 773–790 10.15252/embj.201593164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Prevo B., Scholey J. M., and Peterman E. J. G. (2017) Intraflagellar transport: mechanisms of motor action, cooperation, and cargo delivery. FEBS J. 284, 2905–2931 10.1111/febs.14068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Funabashi T., Katoh Y., Okazaki M., Sugawa M., and Nakayama K. (2018) Interaction of heterotrimeric kinesin-II with IFT-B–connecting tetramer is crucial for ciliogenesis. J. Cell Biol. 217, 2867–2876 10.1083/jcb.201801039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakayama K., and Katoh Y. (2020) Architecture of the IFT ciliary trafficking machinery and interplay between its components. Crit. Rev. Biochem. Mol. Biol. 55, 179–196 10.1080/10409238.2020.1768206 [DOI] [PubMed] [Google Scholar]
- 47. Yi P., Xie C., and Ou G. (2018) The kinases male germ cell-associated kinase and cell cycle–related kinase regulate kinesin-2 motility in Caenorhabditis elegans neuronal cilia. Traffic 19, 522–535 10.1111/tra.12572 [DOI] [PubMed] [Google Scholar]
- 48. Asleson C. M., and Lefebvre P. A. (1998) Genetic analysis of flagellar length control in Chlamydomonas reinhardtii: a new long-flagella locus and extragenic suppressor mutations. Genetics 148, 693–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang Y., Ren Y., and Pan J. (2019) Regulation of flagellar assembly and length in Chlamydomonas by LF4, a MAPK-related kinase. FASEB J. 33, 6431–6441 10.1096/fj.201802375RR [DOI] [PubMed] [Google Scholar]
- 50. Kukic I., Rivera-Molina F., and Toomre D. (2016) The IN/OUT assay: a new tool to study ciliogenesis. Cilia 5, 23 10.1186/s13630-016-0044-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Briscoe J., and Thérond P. P. (2013) The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14, 416–429 10.1038/nrm3598 [DOI] [PubMed] [Google Scholar]
- 52. Mukhopadhyay S., and Rohatgi R. (2014) G-protein–coupled receptors, Hedgehog signaling and primary cilia. Semin. Cell Dev. Biol. 33, 63–72 10.1016/j.semcdb.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nozaki S., Katoh Y., Terada M., Michisaka S., Funabashi T., Takahashi S., Kontani K., and Nakayama K. (2017) Regulation of ciliary retrograde protein trafficking by the Joubert syndrome proteins ARL13B and INPP5E. J. Cell Sci. 130, 563–576 10.1242/jcs.197004 [DOI] [PubMed] [Google Scholar]
- 54. Larkins C. E., Aviles G. D. G., East M. P., Kahn R. A., and Caspary T. (2011) Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol. Biol. Cell 22, 4694–4703 10.1091/mbc.E10-12-0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Phua S. C., Chiba S., Suzuki M., Su E., Roberson E. C., Pusapati G. V., Schurmans S., Setou M., Rohatgi R., Reiter J. F., Ikegami K., and Inoue T. (2017) Dynamic remodeling of membrane composition drives cell cycle through primary cilia excision. Cell 168, 264–279 10.1016/j.cell.2016.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Snouffer A., Brown D., Lee H., Walsh J., Lupu F., Norman R., Lechtreck K., Ko H. W., and Eggenschwiler J. (2017) Cell cycle–related kinase (CCRK) regulates ciliogenesis and Hedgehog signaling in mice. PLoS Genet. 13, e1006912 10.1371/journal.pgen.1006912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lechtreck K. F. (2015) IFT-cargo interactions and protein transport in cilia. Trends Biochem. Sci. 40, 765–778 10.1016/j.tibs.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bhogaraju S., Cajanek L., Fort C., Blisnick T., Weber K., Taschner M., Mizuno N., Lamla S., Bastin P., Nigg E. A., and Lorentzen E. (2013) Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science 341, 1009–1012 10.1126/science.1240985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kubo T., Brown J. M., Bellve K., Craige B., Craft J. M., Fogarty K., Lechtreck K.-F., and Witman G. B. (2016) The IFT81 and IFT74 N-termini together form the main module for intraflagellar transport of tubulin. J. Cell Sci. 129, 2106–2119 10.1242/jcs.187120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ahmed N. T., Gao C., Lucker B. F., Cole D. G., and Mitchell D. R. (2008) ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery. J. Cell Biol. 183, 313–322 10.1083/jcb.200802025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hou Y., Qin H., Follit J. A., Pazour G. J., Rosenbaum J. L., and Witman G. B. (2007) Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J. Cell Biol. 176, 653–665 10.1083/jcb.200608041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Taschner M., Mourão A., Awasthi M., Basquin J., and Lorentzen E. (2017) Structural basis of outer dynein arm intraflagellar transport by the transport adaptor protein ODA16 and the intraflagellar transport protein IFT46. J. Biol. Chem. 292, 7462–7473 10.1074/jbc.M117.780155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hirano T., Katoh Y., and Nakayama K. (2017) Intraflagellar transport–A complex mediates ciliary entry and retrograde trafficking of ciliary G protein–coupled receptors. Mol. Biol. Cell 28, 429–439 10.1091/mbc.E16-11-0813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hamada Y., Tsurumi Y., Nozaki S., Katoh Y., and Nakayama K. (2018) Interaction of WDR60 intermediate chain with TCTEX1D2 light chain of the dynein-2 complex is crucial for ciliary protein trafficking. Mol. Biol. Cell 29, 1628–1639 10.1091/mbc.E18-03-0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Takahara M., Katoh Y., Nakamura K., Hirano T., Sugawa M., Tsurumi Y., and Nakayama K. (2018) Ciliopathy-associated mutations of IFT122 impair ciliary protein trafficking but not ciliogenesis. Hum. Mol. Genet. 27, 516–528 10.1093/hmg/ddx421 [DOI] [PubMed] [Google Scholar]
- 66. Wang W., Allard B. A., Pottorf T. S., Wang H. H., Vivian J. L., and Tran P. V. (2020) Genetic interaction of mammalian IFT-A paralogs regulates cilia disassembly, ciliary entry of membrane protein, Hedgehog signaling, and embryogenesis. FASEB J. 34, 6369–6381 10.1096/fj.201902611r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schmidts M. (2014) Clinical genetics and pathobiology of ciliary chondrodysplasias. J. Pediatr. Genet. 3, 49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. McInerney-Leo A. M., Harris J. E., Leo P. J., Marshall M. S., Gardiner B., Kinning E., Leong H. Y., McKenzie F., Ong W. P., Vodopiutz J., Wicking C., Brown M. A., Zankl A., and Duncan E. L. (2015) Whole exome sequencing is an efficient, sensitive and specific method for determining the genetic cause of short-rib thoracic dystrophies. Clin. Genet. 88, 550–557 10.1111/cge.12550 [DOI] [PubMed] [Google Scholar]
- 69. Wang E. J., Gailey C. D., Brautigan D. L., and Fu Z. (2020) Functional alterations in ciliogenesis-associated kinase 1 (CILK1) that result from mutations linked to juvenile myoclonic epilepsy. Cells 9, 694 10.3390/cells9030694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Liang Y., Pang Y., Wu Q., Hu Z., Han X., Xu Y., Deng H., and Pan J. (2014) FLA8/KIF3B phosphorylation regulates kinesin-II interaction with IFT-B to control IFT entry and turnaround. Dev. Cell 30, 585–597 10.1016/j.devcel.2014.07.019 [DOI] [PubMed] [Google Scholar]
- 71. Wood C. R., Huang K., Diener D. R., and Rosenbaum J. L. (2013) The cilium secretes bioactive ectosomes. Curr. Biol. 23, 906–911 10.1016/j.cub.2013.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hoang-Minh L. B., Dutra-Clarke M., Breunig J. J., and Sarkisian M. R. (2018) Glioma cell proliferation is enhanced in the presence of tumor-derived cilia vesicles. Cilia 7, 6 10.1186/s13630-018-0060-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nager A. R., Goldstein J. S., Herranz-Pérez V., Portran D., Ye F., Garcia-Verdugo J. M., and Nachury M. V. (2017) An actin network dispatches ciliary GPCRs into extracellular vesicles to modulate signaling. Cell 168, 252–263 10.1016/j.cell.2016.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang J., Silva M., Haas L. A., Morsci N. S., Nguyen K. C. Q., Hall D. H., and Barr M. M. (2014) C elegans ciliated sensory neurons release extracellular vesicles that function in animal communication. Curr. Biol. 24, 519–525 10.1016/j.cub.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nishijima Y., Hagiya Y., Kubo T., Takei R., Katoh Y., and Nakayama K. (2017) RABL2 interacts with the intraflagellar transport B complex and CEP19 and participates in ciliary assembly. Mol. Biol. Cell 28, 1652–1666 10.1091/mbc.E17-01-0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Katoh Y., Michisaka S., Nozaki S., Funabashi T., Hirano T., Takei R., and Nakayama K. (2017) Practical method for targeted disruption of cilia-related genes by using CRISPR/Cas9-mediated homology-independent knock-in system. Mol. Biol. Cell 28, 898–906 10.1091/mbc.E17-01-0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tsurumi Y., Hamada Y., Katoh Y., and Nakayama K. (2019) Interactions of the dynein-2 intermediate chain WDR34 with the light chains are required for ciliary retrograde protein trafficking. Mol. Biol. Cell 30, 658–670 10.1091/mbc.E18-10-0678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Takahara M., Kunii M., Nakamura K., Harada A., Hirano T., Katoh Y., and Nakayama K. (2019) C11ORF74 interacts with the IFT-A complex and participates in ciliary BBSome localization. J. Biochem. 165, 257–267 10.1093/jb/mvy100 [DOI] [PubMed] [Google Scholar]
- 79. Haeussler M., Schönig K., Eckert H., Eschstruth A., Mianné J., Renaud J. B., Schneider-Maunoury S., Shkumatava A., Teboul L., Kent J., Joly J. S., and Concordet J. P. (2016) Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 10.1186/s13059-016-1012-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Takahashi S., Kubo K., Waguri S., Yabashi A., Shin H.-W., Katoh Y., and Nakayama K. (2012) Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. J. Cell Sci. 125, 4049–4057 10.1242/jcs.102913 [DOI] [PubMed] [Google Scholar]
- 81. Ferreira J. P., Overton K. W., and Wang C. L. (2013) Tuning gene expression with synthetic upstream open reading frames. Proc. Natl. Acad. Sci. U.S.A. 110, 11284–11289 10.1073/pnas.1305590110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Thomas S., Ritter B., Verbich D., Sanson C., Bourbonnière L., McKinney R. A., and McPherson P. S. (2009) Intersectin regulates dendritic spine development and somatodendritic endocytosis but not synaptic vesicle recycling in hippocampal neurons. J. Biol. Chem. 284, 12410–12419 10.1074/jbc.M809746200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kubo K., Kobayashi M., Nozaki S., Yagi C., Hatsuzawa K., Katoh Y., Shin H.-W., Takahashi S., and Nakayama K. (2015) SNAP23/25 and VAMP2 mediate exocytic event of transferrin receptor-containing recycling vesicles. Biol. Open 4, 910–920 10.1242/bio.012146 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.


