Biofilm formation by Streptococcus pyogenes (group A streptococcus [GAS]) in model systems mimicking the respiratory tract is poorly documented. Most studies have been conducted on abiotic surfaces, which poorly represent human tissues. We have previously shown that GAS forms mature and antibiotic-resistant biofilms on physiologically relevant epithelial cells. However, the roles of the substratum, extracellular matrix (ECM) components, and GAS virulence factors in biofilm formation and structure are unclear.
KEYWORDS: antibiotic resistance, aggregation, biofilm formation, biofilm structure, epithelial cells, extracellular matrix, Streptococcus pyogenes, virulence factors, keratinocytes, respiratory tract, adherence, biofilms, mucosal pathogens
ABSTRACT
Biofilm formation by Streptococcus pyogenes (group A streptococcus [GAS]) in model systems mimicking the respiratory tract is poorly documented. Most studies have been conducted on abiotic surfaces, which poorly represent human tissues. We have previously shown that GAS forms mature and antibiotic-resistant biofilms on physiologically relevant epithelial cells. However, the roles of the substratum, extracellular matrix (ECM) components, and GAS virulence factors in biofilm formation and structure are unclear. In this study, biofilm formation was measured on respiratory epithelial cells and keratinocytes by determining biomass and antibiotic resistance, and biofilm morphology was visualized using scanning electron microscopy. All GAS isolates tested formed biofilms that had similar, albeit not identical, biomass and antibiotic resistance for both cell types. Interestingly, functionally mature biofilms formed more rapidly on keratinocytes but were structurally denser and coated with more ECM on respiratory epithelial cells. The ECM was crucial for biofilm integrity, as protein- and DNA-degrading enzymes induced bacterial release from biofilms. Abiotic surfaces supported biofilm formation, but these biofilms were structurally less dense and organized. No major role for M protein, capsule, or streptolysin O was observed in biofilm formation on epithelial cells, although some morphological differences were detected. NAD-glycohydrolase was required for optimal biofilm formation, whereas streptolysin S and cysteine protease SpeB impaired this process. Finally, no correlation was found between cell adherence or autoaggregation and GAS biofilm formation. Combined, these results provide a better understanding of the role of biofilm formation in GAS pathogenesis and can potentially provide novel targets for future treatments against GAS infections.
INTRODUCTION
Streptococcus pyogenes (group A streptococcus [GAS]) is a strictly human pathogen causing around 18 million severe infections annually, of which 517,000 cases are fatal (1, 2). However, the majority of infections are less severe and include either skin infections, such as impetigo or pyoderma, which are responsible for about 100 million cases annually worldwide, or pharyngitis (strep throat), causing up to 600 million cases worldwide every year (1). Additionally, GAS is responsible for an increasing number of cases of otitis media and pneumonia.
A first step in the pathogenesis of GAS is adherence to and colonization of the respiratory tract or skin surfaces (3, 4). Asymptomatic colonization with GAS is common, with approximately 20% of children carrying GAS in the oropharynx for extended periods of time (5). Persistent colonization requires the expression of various bacterial factors, such as M protein and hyaluronic acid capsule, that are associated with adherence and persistence in the host niche (6–8). Additionally, colonization and subsequent GAS infection are highly associated with formation of microcolonies on mucosal surfaces that expand to form complex three-dimensional structures, or biofilms (5, 9).
A number of studies have investigated biofilm formation by GAS (10, 11). However, biofilm formation has consistently been studied on abiotic (plastic or glass) surfaces that do not represent or mimic the mucosal surfaces occupied by GAS during colonization and infection. Biofilms formed on abiotic surfaces produce an extracellular matrix (ECM) composed of primarily DNA and proteins (12, 13), and the ability of GAS strains to autoaggregate in culture has been found to correlate with their ability to form biofilms (10, 14). It has therefore been proposed that autoaggregation as well as attachment to structures on biological surfaces may play a role in GAS biofilm formation in vivo (9).
We previously demonstrated the ability of the oropharyngeal GAS strain 771 (M-type 3) to form biofilms on prefixed keratinocytes using conditions that resemble the physiological environment that GAS colonizes (15). In that study, gene expression of virulence factors, such as capsule, M protein, streptolysin S (SLS), and the cysteine protease SpeB, was lower in biofilm bacteria than in planktonic bacteria grown in broth, suggesting a potential role for these factors in the biofilm-forming process (15). Similar, but not identical, results were obtained in a separate study measuring the global gene expression in biofilms formed on abiotic surfaces (10). The direct contribution of GAS virulence factors (M protein, capsule, SpeB, and NADase) in biofilms formed on abiotic surfaces has also been studied, and M protein and SpeB have been implicated as important factors for GAS biofilm formation (10, 14). The direct roles of these and other factors, such as streptolysin O (SLO), during biofilm formation on biological surfaces, such as epithelial cells, have not yet been studied.
The objective of this study was to better understand the growth kinetic and morphology of GAS biofilms formed on surfaces representing the local niches where these organisms colonize and cause infection and to investigate the role of specific virulence factors in this process. To that end, we used epithelial cells of bronchial origin (respiratory epithelial cells) as well as from the oropharynx (keratinocytes) combined with GAS strains of various serotypes and mutants lacking virulence factors of potential importance for biofilm formation. Understanding the mechanisms involved in biofilm formation on relevant surfaces has the potential to help identify potential future therapeutic targets against GAS infection.
RESULTS
GAS biofilm formation and functionality on epithelial cells.
To date, GAS biofilm formation has been studied mostly on abiotic surfaces in vitro, surfaces which do not accurately represent the physiological environment of these bacteria (9). Here, we used a well-established biofilm model, originally developed for Streptococcus pneumoniae. Bacteria were allowed to form biofilms on prefixed respiratory epithelial cells (NCI-H292 bronchial epithelial cells) in the presence of a nutrient-limited chemically defined medium (CDM) at the nasopharyngeal temperature of 34°C (15–18). After 48 and 72 h of growth, the numbers of CFU were determined in the absence (to measure the total biomass) or presence (as a measure of the functionality and integrity of the biofilms) of gentamicin. S. pyogenes strain GAS-771 (M3) was used throughout the studies, as this strain was used in our initial study on biofilm formation and has been used by us and others to investigate bacterium-host interactions (6, 15, 19).
GAS-771 biofilms formed on cells showed increases in biomass over time (from 48 to 72 h) (Fig. 1A), which correlated with an increased resistance to gentamicin, as monitored by decreased killing (Fig. 1B), indicating development of mature and functional biofilms. Compared to GAS biofilms, S. pneumoniae strain D39 formed biofilms that displayed higher biomass and developed gentamicin resistance earlier (see Fig. S1 in the supplemental material). Structurally, as observed by scanning electron microscopy (SEM), both species (GAS and S. pneumoniae) formed complex biofilms composed of a network of bacterial chains on H292 epithelial cells, suggesting functional and structural integrity of the biofilms (Fig. 1C and Fig. S1). Combined, these results indicate the ability of GAS to form developed and functional biofilms over time on H292 cells.
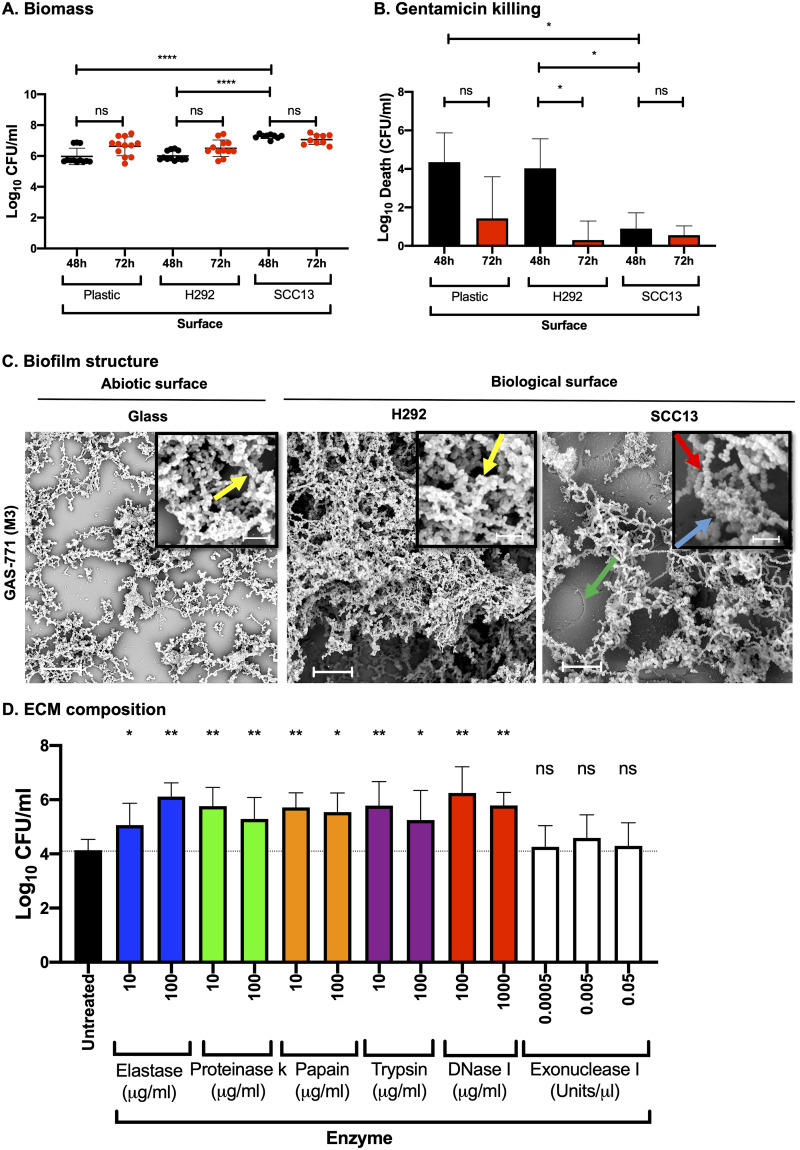
FIG 1.
Streptococcus pyogenes biofilm development, structure, and composition on abiotic versus biological surfaces. Biofilms were formed using S. pyogenes strain GAS-771 on abiotic (plastic) or biological (prefixed H292 cells or SCC13 keratinocytes) surfaces for various times at 34°C in CDM. (A) Biomass formation was evaluated by determining the log10 CFU per milliliter at 48 h (black dots) or 72 h (red dots). Data are from three separate experiments with three individual biofilms each (with standard deviations [SD]; n = 9), and data were compared using the Mann-Whitney U test. (B) Determination of gentamicin killing in GAS-771 biofilms was measured by calculating death in log10 CFU per milliliter (i.e., total biomass [CFU/ml] − biofilm biomass [CFU/ml] after treatment with 500 μg/ml gentamicin for 3 h) after 48 h (black bars) or 72 h (red bars). The results are means from three separate experiments of three individual biofilms each (with SD; n = 9), and data were compared using Student's t test. (C) The structure of GAS-771 biofilms formed on abiotic (glass) or biological (prefixed H292 epithelial cells or SCC13 keratinocytes) for 72 h was inspected using SEM (bar = 5 μm). Where visible, underlying cells are indicated by a green arrow. (Insets) Higher magnification (bar = 2 μm) of the biofilms, with arrows highlighting bacterial chains (red), ECM aggregates (blue), or bacterial chains coated with ECM (yellow). (D) To identify components present in the ECM, GAS-771 biofilms formed over 72 h at 34°C on prefixed epithelial H292 cells were treated with a set of proteases (elastase, proteinase K, papain, and trypsin) and DNases (DNase I and exonuclease I) at the indicated concentrations. After incubation, the log10 CFU/ml of bacteria released into the supernatant was determined. The results represent data from three separate experiments of two individual biofilms each (with SD; n = 6), and data were compared using the Mann-Whitney U test. *, P < 0.05; **, P < 0.01; ****, P < 0.0001; ns, nonsignificant difference.
GAS biofilm formation on biological versus abiotic surfaces.
To determine the potential importance of the growth surface for biofilm development, GAS-771 biofilms were formed on both abiotic (plastic) and biological (prefixed H292 respiratory epithelial cells or SCC13 keratinocytes) surfaces. Biomass and gentamicin resistance (as measured by decreased bacterial killing) were monitored after 48 or 72 h of growth. No significant differences were detected in the biomass of GAS-771 biofilms formed on plastic or prefixed H292 cells. Interestingly, biofilms formed on prefixed SCC13 keratinocytes had a significantly higher biomass than those formed on either plastic or H292 cells (Fig. 1A). Gentamicin resistance, a measure of integrity and maturity, of the biofilms formed on all three surfaces increased over time (seen by a reduced killing of bacteria at 72 h compared to 48 h [Fig. 1B]). However, biofilms grown on SCC13 cells formed mature, antibiotic-resistant biofilms earlier (at 48 h) than biofilms grown on either plastic or H292 cells, and biofilms grown on plastic showed lower resistance levels at 72 h, suggesting that GAS form more functional biofilms on biological surfaces than on abiotic surfaces.
Biofilm morphology and role and composition of extracellular matrix for structural integrity.
In previous studies, the structure of GAS biofilms formed on abiotic surfaces, as visualized by SEM, has been revealed as a network of bacterial chains either lacking or exhibiting ECM (13, 20, 21). Here, GAS-771 biofilms grown on either glass and those on prefixed H292 cells both contained complex structures of aggregating and interlacing bacterial chains of similar length (Fig. 1C, yellow arrows) and coated with ECM. However, more developed and abundant growth with a more three-dimensional and complex structure was observed on H292 cells, despite similar biomass levels on both surfaces prior to electron microscopy imaging (Fig. S2).
Biofilms formed on prefixed SCC13 keratinocytes (Fig. 1C, green arrow) appeared similarly denser and more structurally complex than biofilms formed on glass and also formed longer chains than biofilms formed on either H292 cells or glass. In addition, the ECM in biofilms formed on prefixed SCC13 keratinocytes was present in aggregate-like structures (Fig. 1C, blue arrow) surrounded by noncoated bacterial chains (Fig. 1C, red arrow). These results suggest that the growth surface directly affects the morphology and structural complexity of GAS-771 biofilms.
The ECM in GAS biofilms formed on abiotic surfaces has previously been shown to contain protein and DNA (12). Since ECM also formed an integral part of GAS-771 biofilms grown on prefixed H292 cells (Fig. 1C), we investigated the potential role of ECM in GAS biofilm integrity. GAS-771 biofilms grown for 72 h on prefixed H292 cells were treated for 2 h with a set of proteases (elastase, proteinase K, papain, or trypsin) or DNases (DNase I or exonuclease I), and the number of bacteria released into the supernatant (Fig. 1D) and the remaining biomass (results not shown) were determined. Passive dispersion of bacteria from biofilms occurs as the biofilm grows and develops. At any given time, approximately 0.1 to 1% of the total biomass (∼104 CFU/ml) (Fig. 1D) can be found released into the medium. A significantly increased release of bacteria into the supernatant (10 to 100 times the passive dispersion) was detected in biofilms treated with each of the proteases compared to untreated biofilms (Fig. 1D). Treatment with DNase I, an endonuclease that cleaves both single- and double-stranded DNA (22), also resulted in significant and comparable bacterial release, whereas no significant release was detected using exonuclease I, a nuclease known to cleave only single-stranded DNA in the 3′–5′ direction (23) (Fig. 1D). Together, these results indicate that the ECM of biofilms formed on epithelial cells contains proteins and DNA and that degrading these molecules affects biofilm integrity, which leads to the release of bacteria from the biofilm.
Association of biofilm formation with GAS serotype.
To investigate the general ability of GAS to form biofilms on biological surfaces, we first examined three strains expressing surface M proteins associated with respiratory tract infections (M1, M5, and M18). In general, and similar to the M3 (GAS-771) strain described above (Fig. 1), GAS biofilms formed on SCC13 cells produced higher biomass and developed significantly higher gentamicin resistance at an earlier time point than biofilms formed on H292 cells (Fig. 2A and B). Although all three strains formed biofilms on both cell types, biofilm growth kinetics and development of antibiotic resistance varied between strains.
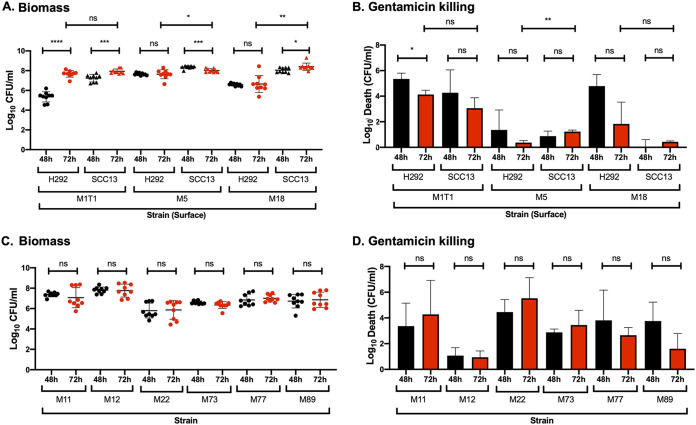
FIG 2.
Biofilm growth and antibiotic resistance of different GAS serotypes on prefixed H292 epithelial cells or SCC13 keratinocytes. (A and B) Biofilms were formed over 48 h (black) and 72 h (red) at 34°C for the isolates M1T1 (941079), M5 (Manfredo), and M18 (87-282) on prefixed respiratory H292 cells or SCC13 keratinocytes, and biomass (A) and gentamicin killing (B) were measured by determining log10 CFU per milliliter and log10 death (i.e., the total biomass [CFU/ml] − biofilm biomass [CFU/ml] after treatment with 500 μg/ml gentamicin for 3 h), respectively. (C and D) Biofilms of the erythromycin-resistant clinical isolates M11 (GAS-53), M12 (GAS-6), M22 (GAS-8), M73 (GAS-138), M77 (GAS-125), and M89 (GAS-128) were formed over 48 h (black) and 72 h (red) at 34°C on prefixed epithelial H292 cells and evaluated for biomass (C) and gentamicin killing (D). Data are from three separate experiments with three individual biofilms each. Biomass (A and C) is plotted as individual data points (with SD; n = 9), and groups were compared using the Mann-Whitney U test. Gentamicin killing data are means (with SD; n = 3), and groups were compared using Student's t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant difference.
Compared to GAS-771 (M3), biofilms formed by the M1T1 (941079) strain reached similar biomass on both cell types but failed to reach comparable gentamicin resistance levels even after 72 h of incubation (Fig. 2B). A similar phenotype was observed for another M1 strain, SF370 (Fig. 3B). The M18 (87-282) strain showed a phenotype similar to that of M3 (GAS-771), with higher levels of biomass formed on SCC13 cells than on H292 cells, which was associated with increased gentamicin resistance over time that was highest at 72 h on SCC13 cells (Fig. 2A and B). Visually, the M18 biofilms appeared denser and were macroscopically more visible than M3 biofilms (results not shown), suggesting a potential difference in the amount of extracellular matrix produced by each strain. Finally, the M5 (Manfredo) strain rapidly developed biofilms superior to those of M3 (GAS-771), with higher biomass and full gentamicin resistance being reached at 48 h on both SCC13 and H292 cells and sustained at 72 h (Fig. 2A and B). These biofilms were visibly dense and thick (even before 48 h), similar to those of the M18 strain (results not shown). When observed by SEM, M5 biofilms formed on prefixed SCC13 cells exhibited longer chains than those formed on either H292 cells or glass, similar to what was observed for the M3 (GAS-771) biofilms (Fig. S3). Furthermore, M5 biofilms formed on prefixed SCC13 cells, but not on prefixed H292 cells, formed shorter bacterial chains when in closer proximity to the cells, whereas longer chains were detected further away from the cell surface (Fig. S3). Additionally, and supporting the observations seen in M3 biofilms, M5 biofilms formed on biological surfaces appeared denser and more structurally complex with more abundant growth than those formed on glass, despite the similar biomass levels on all surfaces prior to electron microscopy imaging (Fig. S3).
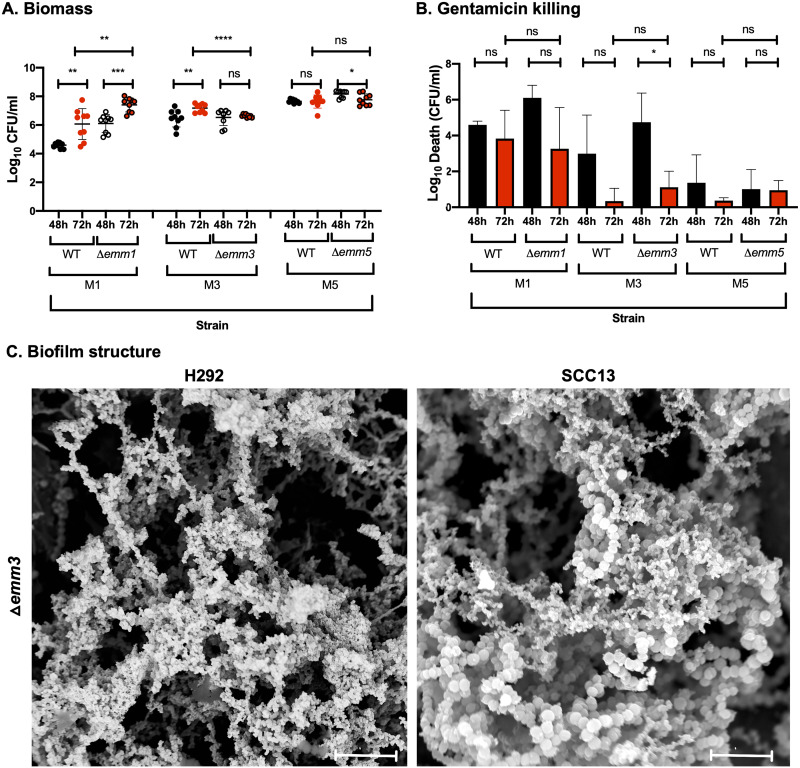
FIG 3.
Role of the M protein in GAS biofilm biomass, antibiotic resistance, and structure in strains lacking or expressing M protein. (A and B) Biofilms of M1 (SF370), M3 (GAS-771), and M5 (Manfredo) strains expressing wild-type M protein (WT) or lacking the M protein (Δemm) were formed over 48 (black) and 72 h (red) at 34°C on prefixed epithelial H292 cells and evaluated for biomass (A) or gentamicin killing (B) by measuring the log10 CFU per milliliter or the log10 death (i.e., total biomass [CFU/ml] − biofilm biomass [CFU/ml] after treatment with 500 μg/ml gentamicin for 3 h), respectively. Data are from three separate experiments with three individual biofilms each. Biomass is plotted as individual data points (with SD; n = 9), and groups were compared using the Mann-Whitney U test. Gentamicin killing data are means (with SD; n = 3), and groups were compared using Student's t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant difference. (C) The structure of biofilms formed by the Δemm3 mutant on different epithelial cells (respiratory H292 cells or SCC13 keratinocytes) for 72 h was viewed using SEM (bar = 5 μm). The cell substratum is not visible in the image due to the focus on capturing the biofilm structures.
Additionally, GAS biofilm formation was also investigated on H292 epithelial cells using a set of erythromycin-resistant clinical isolates (Table 1) (Fig. 2C and D). All isolates except M12 (GAS-6) and M22 (GAS-8) formed biofilms with biomass and gentamicin resistance levels that were comparable to those of the M3 strain (GAS-771) (Fig. 2D). M12 GAS developed highly resistant biofilms with high biomass at 48 h, similar to the M5 (Manfredo) strain. On the other hand, the nonencapsulated M22 isolate formed less developed and less resistant biofilms with significantly lower biomass than the M3 strain GAS-771 (Fig. 1A and B; Fig. 2C and D). Combined, these results indicate that all clinical GAS isolates tested have the ability to form biofilms in vitro, but with variable growth kinetics and development of gentamicin resistance.
TABLE 1.
Strains used in this study
| Strain or genotypea | Reference name | Phenotype (gene) | Source |
|---|---|---|---|
| D39 | D39 | Serotype 2 wild-type S. pneumoniae strain | 48 |
| M1T1 | 941079 | M type 1 wild-type strain with low capsule expression | 49 |
| M1 | SF370 | M type 1 wild-type strain | 50 |
| Δemm1 | SF370 △emm1 | M1 protein (emm1) deletion mutant in SF370 | 50 |
| M5 | Manfredo | M type 5 wild-type strain | 51 |
| Δemm5 | Manfredo △emm5 | M5 protein (emm5) deletion mutant in Manfredo | 51 |
| M6 | JRS4 | M type 6 wild-type strain from June Scott | 52 |
| M6ΔhasA | JRS46 | Capsule-deficient (hasA) mutant in JRS4 | June R. Scott |
| M11 | GAS-53 | Erythromycin-resistant clinical isolate expressing an ermB methylase | 53 |
| M12 | GAS-6 | Erythromycin-resistant clinical isolate expressing a mefA + msrD efflux pump | 53 |
| M18 | 87-282 | M type 18 wild-type strain with high capsule expression | 54 |
| M22 | GAS-8 | Erythromycin-resistant clinical isolate expressing a mefA + msrD efflux pump. This strain lacks the hasABC genes encoding hyaluronic acid capsule. | 53, 55 |
| M73 | GAS-138 | Erythromycin-resistant clinical isolate expressing a mefA + msrD efflux pump | 53 |
| M77 | GAS-125 | Erythromycin-resistant clinical isolate expressing an ermTR methylase | 53 |
| M89 | GAS-128 | Erythromycin-resistant clinical isolate expressing an ermB methylase | 53 |
| M3 strains | |||
| AM3 (H) | AM3 (H) | M type 3 strain with high capsule expression | 56 |
| SS-90 (H) | SS-90 | M type 3 strain with high capsule expression | 57 |
| SS-1271 (M) | SS-1271 | M type 3 strain with medium capsule expression | 58 |
| 94421 (M) | 94421 | M type 3 strain with medium capsule expression | 57 |
| 950802 (L) | 950802 | M type 3 strain with low capsule expression | 57 |
| 87-136 (L) | 87-136 | M type 3 strain with low capsule expression | 57 |
| M3 (M) | 950771 (GAS-771) | M type 3 wild-type strain, medium encapsulated, from child with necrotizing fasciitis and sepsis | 59 |
| Δemm3 | 355 | M type 3 (emm3) deletion mutant derived from 950771 | 59 |
| M3ΔhasA | 188 | Nonencapsulated isogen of 950771 with a Km insertion in hasA | 59 |
| Δslo | 500 | Streptolysin O (slo) deletion mutant derived from 950771 | 60 |
| Δslo ΔhasA | 781 | Nonencapsulated (deletion) and SLO-deficient (Km insertion) double mutant derived from 950771 | 19 |
| Δnga | C3 | NADase (nga) mutant of 950771 | 60 |
| Δslo Δnga | C42 | 950771 with deletion of SLO (slo) and NADase (nga) | 60 |
| ΔspeB | 241 | Cysteine protease (speB)-deficient mutant derived from 950771, Km insertion in speB | 59 |
| ΔsagA | 775 | 950771 with deletion of 60 bp in SLS (sagA) | 61 |
Capsule amount scale: 0 to 2 fg/CFU, low (L); 2.1 to 25 fg/CFU, medium (M); 26 to 90 fg/CFU, high (H).
Specific role of M protein in biofilm formation by different GAS serotypes.
As GAS strains expressing different M proteins formed biofilms with variable biomass and phenotype, the direct role of the M protein for biofilm development was evaluated. We used isogenic mutants lacking the emm genes in the M1 (SF-370), M3 (GAS-771), and M5 (Manfredo) strain backgrounds that formed less, intermediately, and highly mature biofilms over time, respectively. A significantly higher level of biomass was detected for biofilms formed by the SF-370 Δemm1 strain at both 48 and 72 h compared to M1-expressing wild-type (WT) biofilms (Fig. 3A). This correlated with higher maturity, while the M1-negative strain was also slightly more resistant to gentamicin at 72 h (Fig. 3B). For GAS-771, no differences in biomass was observed between the WT and Δemm3 mutant at 48 h regardless of cell type, although a significantly lower biomass was seen at 72 h, but only with H292 cells. A slightly lower gentamicin resistance was observed for the M3-negative strain at both time points on H292 cells but not on SCC13 cells, on which mature biofilms were formed more quickly (Fig. 3A and B; Fig. S4). Structurally, GAS-771 lacking the M3 protein formed biofilms with a less compact “honeycomb” structure than the wild-type strain on prefixed H292 cells, whereas no change in morphology was observed on SCC13 cells (Fig. 1C; Fig. 3C). The absence of the M5 protein had no impact on biofilm structure (results not shown), biomass, or gentamicin resistance (Fig. 3A and B). These results suggest different roles for the three M proteins in biofilm development on epithelial cells.
Positive correlation between capsule expression and GAS biofilm formation.
Although capsule expression is required for optimal colonization, GAS has been shown to downregulate capsule during biofilm formation (10, 15), suggesting that a thinner capsule may be beneficial for bacterium-bacterium and bacterium-host cell interactions. However, when the biofilm-forming ability of the M1 (941079), M3 (GAS-771), and M18 (87-282) strains (Fig. 1 and 2) was correlated with the amount of capsule expressed on their surface during growth in medium (Table 2), biofilm growth and phenotype were positively correlated with capsule expression (Fig. 2 and Fig. S5). To confirm this correlation, we examined the biofilm-forming ability of a set of M3 strains expressing various amounts of capsule after growth in THY medium (Table 2; Fig. 4A and B). Indeed, strains expressing medium or high capsule levels in medium (except for strain 94421) formed highly antibiotic-resistant biofilms with a high biomass level after 72 h, whereas strains expressing low capsule levels in medium formed biofilms that were more sensitive to gentamicin despite having similar biomass levels. Closer inspection of the data showed a positive and significant correlation between capsule expression and antibiotic resistance (as seen by reduced antibiotic sensitivity) for strains producing less than 8 fg/CFU (linear regression, P < 0.05) (Fig. S5). For strains expressing capsule levels above 8 fg/CFU, antibiotic resistance had reached high enough levels that no correlation could be observed. Overall, these results suggest a positive correlation between the ability to form functional biofilms and the amount of capsule expressed.
TABLE 2.
Quantification of hyaluronic acid capsule levels in GAS isolates
| Strain name | Capsule amt (fg/CFU)a |
|---|---|
| M1T1 (L) | 0.108 |
| M3 (771; M) | 8.44 |
| M6 (L) | 0.128 |
| M18 (H) | 81.096 |
| AM3 (H) | 35.294 |
| SS-90 (H) | 28.018 |
| SS-1271 (M) | 19.262 |
| 94421 (M) | 5.546 |
| 950802 (L) | 1.748 |
| 87-136 (L) | 0b |
Capsule amount scale: 0 to 2 fg/CFU, low (L); 2.1 to 25 fg/CFU, medium (M); 26 to 90 fg/CFU, high (H).
Value below detection limit.
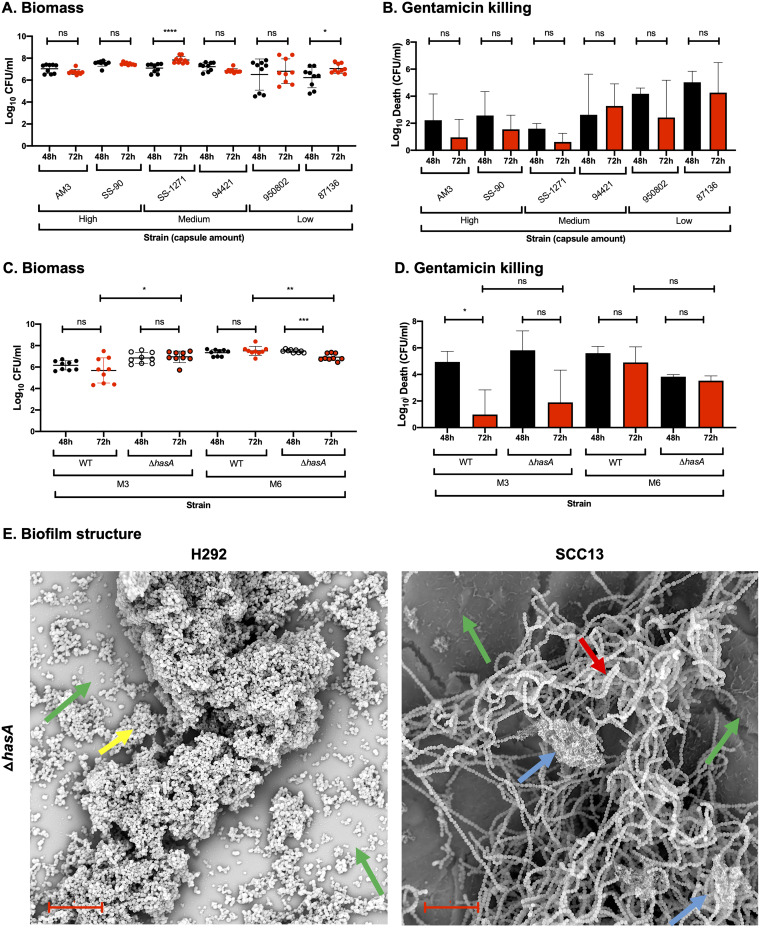
FIG 4.
Role of capsule in biofilm structure, biomass, and antibiotic resistance in strains lacking or expressing different capsule levels. (A and B) Biofilms of M3 strains expressing various capsule amounts, i.e., high (AM3 and SS-90), medium (SS-1271 and 94421), or low (950802 and 87-136), were formed over 48 (black) and 72 h (red) at 34°C on prefixed epithelial H292 cells and evaluated for biomass (A) or gentamicin killing (B) by measuring the log10 CFU per ml or the log10 death (i.e., the total biomass [CFU/ml] − biofilm biomass [CFU/ml] after treatment with 500 μg/ml gentamicin for 3 h), respectively. (C and D) Biofilms of M3 and M6 strains expressing wild-type capsule (WT) or lacking capsule (ΔhasA) were formed over 48 and 72 h at 34°C on prefixed epithelial H292 cells and evaluated for biomass (C) and gentamicin killing (D), as described above. Data are from three separate experiments with three individual biofilms each. Biomass (A and C) is plotted as individual data points (with SD; n = 9), and groups were compared using the Mann-Whitney U test. Gentamicin killing data (B and D) are means (with SD; n = 3), and groups were compared using Student's t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant difference. (E) Structure of biofilms formed by the M3ΔhasA mutant on different epithelial cells (respiratory H292 cells or SCC13 keratinocytes) for 72 h, viewed using SEM (bar = 10 μm). The underlying cell substratum is indicated by a green arrow, the red arrow highlights bacterial chains, and blue and yellow arrows indicate ECM aggregates alone and cells coated with ECM, respectively.
Capsule does not play a direct role in biofilm formation.
To directly test the role of capsule during biofilm formation, we investigated biofilm formation on prefixed H292 cells for WT GAS-771 (M3; medium capsule level) or GAS JRS4 (M6; low capsule level) and their corresponding capsule-negative mutants (Table 1, Table 2, and Fig. S5). The GAS M6 strain, expressing smaller capsule amounts in liquid medium, showed higher biomass levels but failed to develop gentamicin resistance to the same degree as the GAS M3 strain (Fig. 4C and D), consistent with the data above. Biofilm biomass levels of the encapsulated (GAS M6 WT) and nonencapsulated (GAS M6 ΔhasA) strains were similar. However, a higher level of gentamicin resistance (even at 48 h) developed in M6 ΔhasA than in the M6 WT.
No major differences in biomass or gentamicin resistance were detected between the encapsulated M3 WT and the nonencapsulated M3ΔhasA strain on either cell type (Fig. 4C and D; Fig. S4). However, morphologically, a clear difference in biofilm structure was observed on H292 cells, where the nonencapsulated M3ΔhasA strain formed grape-clustered formations that predominantly lacked visible ECM (Fig. 4E) rather than a bacterial chain lattice covered with ECM, as seen for the M3 WT strain (Fig. 1C). In contrast, no major morphological differences were observed when M3 WT and M3ΔhasA biofilms were formed on prefixed SCC13 cells (Fig. 1C; Fig. 4E). In conclusion, despite the positive correlation between capsule expression and biofilm formation observed above, capsule per se does not appear to be directly involved in the ability of GAS to form biofilms in vitro, except for a potential role in biofilm structure on H292 cells.
Role of other virulence factors in GAS biofilm formation.
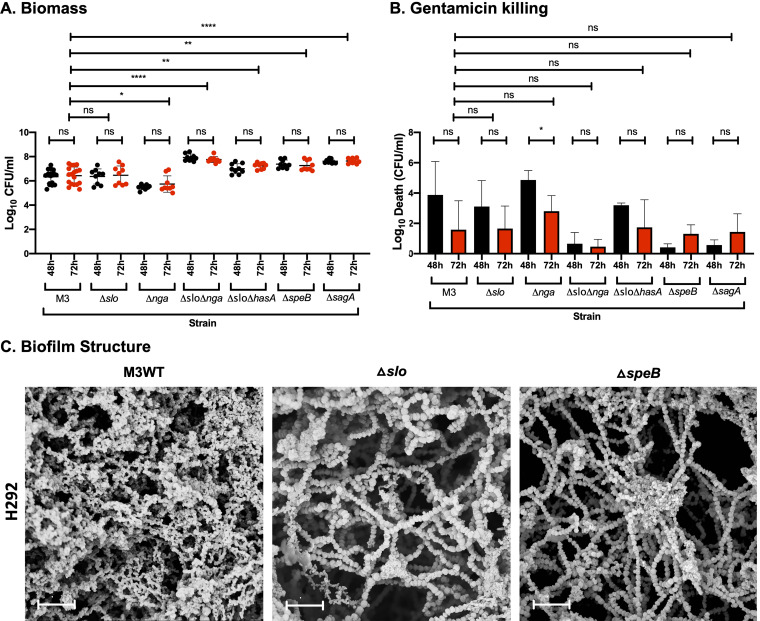
Prior experiments using GAS-771 (M3) showed that biofilm bacteria downregulated the gene expression of the virulence factors SLS (encoded by sagA), cysteine protease SpeB (speB), and capsule (hasA), whereas expression of SLO (slo) was unaffected (15). To determine whether these virulence factors (as well as NADase, encoded by the nga gene and downregulated in biofilms formed on abiotic surfaces [10]) play a role during biofilm formation, GAS-771 WT and isogenic mutants lacking these factors were assessed for biofilm formation (Fig. 5A and B) and visualized by SEM (Fig. 5C).
FIG 5.
Role of virulence factors in biofilm formation, structure, and antibiotic resistance on prefixed H292 cells. (A and B) Biofilms of GAS-771 expressing or lacking SLO (Δslo), NADase (Δnga), both SLO and NADase (Δslo Δnga), capsule and SLO (Δslo ΔhasA), SpeB (ΔspeB), or SLS (ΔsagA) were formed over 48 (black) and 72 h (red) and evaluated for biomass (A) or gentamicin killing (B) by measuring the log10 CFU per ml or the log10 death (i.e., total biomass [CFU/ml] − biofilm biomass [CFU/ml] after treatment with 500 μg/ml gentamicin for 3 h), respectively. Data are from three separate experiments with three individual biofilms each. Biomass is plotted as individual data points (with SD; n = 9, except for M3, where n = 18), and groups were compared using the Mann-Whitney U test. Gentamicin killing data are means (with SD; n = 3, except for M3, where n = 6), and groups were compared using Student's t test. *, P < 0.05; **, P < 0.01; ****, P < 0.0001; ns, nonsignificant difference. (C) Biofilms of the GAS-771 strain and its isogenic mutants (Δslo or ΔspeB) were formed on prefixed H292 cells and visualized by SEM (bar = 5 μm). The cell substratum is not visible in the image due to the focus on capturing the biofilm structures.
In agreement with the prior gene expression results, absence of SLO alone did not affect the development and functionality of GAS-771 biofilms (Fig. 5A and B). On the other hand, structural differences were detected where absence of SLO resulted in longer bacterial chains with less visible ECM than the WT biofilms (Fig. 5C). Additionally, absence of capsule alone (Fig. 4) did not affect the maturation and functionality of GAS-771 biofilms. In contrast to the synergistic effect of SLO and capsule that has previously been observed to inhibit cell internalization (19), GAS-771 lacking both capsule and SLO exhibited an increase in biofilm biomass but no difference in antibiotic resistance compared to the WT strain (Fig. 5A and B). On the other hand, NADase-negative GAS-771 formed biofilms with a significantly reduced biomass and a slightly reduced gentamicin resistance, suggesting a role for this enzyme in biofilm formation. Interestingly, the GAS-771 Δslo Δnga double mutant (lacking both SLO and NADase) formed biofilms that more closely resembled S. pneumoniae D39 biofilms, as biomass and gentamicin resistance levels peaked as early as 48 h after seeding (Fig. 5A and B) and reached biomass levels similar to those of S. pneumoniae D39 biofilms (Fig. S1). Thus, mutants lacking only NADase formed suboptimal biofilms, but those lacking both NADase and SLO formed more mature and functional biofilms (Fig. 5A and B).
Consistent with the downregulation of their corresponding genes in biofilms (15), strains lacking SpeB or SLS established gentamicin resistance faster (at 48 h) and developed more biomass than the corresponding GAS-771 WT strain (Fig. 5A and B). Morphologically, biofilms lacking SpeB showed characteristics similar to those of biofilms lacking SLO, with longer chains and less visible ECM (Fig. 5C). In conclusion, an SLO-dependent role of NADase was observed during biofilm formation on epithelial cells, and in agreement with previous gene expression results, both SpeB and SLS inhibited GAS biofilm formation.
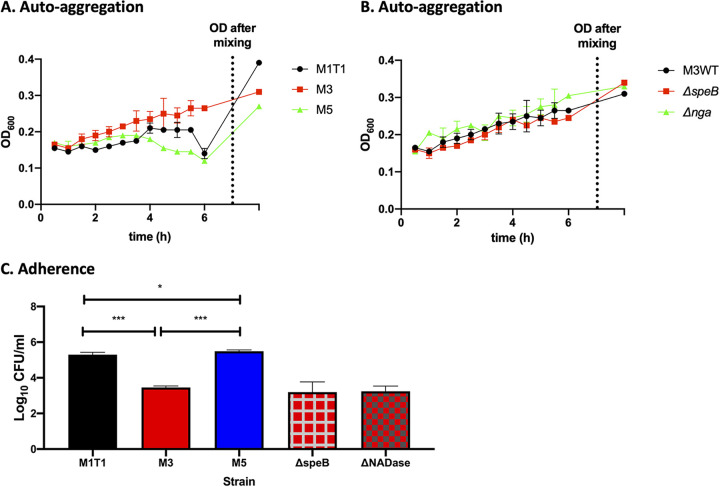
Biofilm formation was not correlated with initial adherence or autoaggregation of GAS during planktonic growth.
Autoaggregation of planktonic bacteria during growth in liquid medium was previously found to directly correlate with, and potentially act as a precursor for, biofilm formation on abiotic surfaces (9, 14). Autoaggregation may increase initial adherence to the mucosal surface and induce microcolony formation and subsequent biofilm colonization in vivo. However, our results here showed no clear correlation between cell adherence or autoaggregation and biofilm formation in vitro (Fig. 2; Fig. 6). Two of the three strains tested, M1 (941079) and M5 (Manfredo), readily autoaggregated during growth in liquid media (Fig. 6A) and adhered in higher numbers to live epithelial cells (Fig. 6C) but showed opposite biofilm-forming abilities (Fig. 2A and B). Despite a high autoaggregation and cell adherence, the M1 strain formed poor biofilms on both cell types, whereas the M5 strain formed highly developed biofilms. The third strain, M3 (GAS-771), showed significantly lower cell adherence and autoaggregation during growth than the M1 strain (Fig. 6A and C) but produced biofilms with higher biomass and antibiotic resistance (Fig. 1A and B).
FIG 6.
Correlation between autoaggregation and adhesion of broth-grown planktonic bacteria and biofilm formation in GAS. Aggregation (as determined by sedimentation rate during growth) was monitored by measuring the OD600 of actively growing planktonic bacteria in CDM over time for the M1, M3, and M5 serotypes (A) or the M3 WT (GAS-771) strain and its isogenic ΔspeB and Δnga mutants (B). The final OD600 data point represents the OD600 reading after inversion of each tube at the 6-h time point to produce a homogenous suspension, which provides a measure of the extent of aggregation. (C) To determine the cell adherence properties of these strains, actively growing planktonic bacteria resuspended in RPMI with 2% fetal bovine serum were used to infect live respiratory H292 cells for 1 h at 34°C. Adherence to cells was determined by assessing the log10 CFU per milliliter after plating cell lysates on blood agar plates and determining viable plate counts after growth at 37°C for 48 h. Data are from two separate experiments; n = 2 (A and B), n = 4 (C). One-way ANOVA using Tukey’s multiple-comparison test was used to compare the data sets in panel C, and only significant differences between the wild-type strains are presented. There were no significant changes between M3 and its mutant strains. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In support of these results, we also compared cell adherence and autoaggregation during growth of GAS-771 WT and mutant strains with different biofilm-forming abilities, as described above (Fig. 5A and B). Despite showing significantly decreased (NADase mutant) or increased (SpeB mutant) biofilm formation ability compared to the WT strain, no difference in cell adherence or autoaggregation kinetics during growth could be detected (Fig. 6B and C). These results therefore suggest that initial cell adherence and autoaggregation by GAS in liquid medium might not be an accurate predictor of structurally complex biofilm formation on biological surfaces.
DISCUSSION
Infections caused by S. pyogenes (GAS) depend on its colonization niche within the human body and are serotype dependent (4, 24–27). Colonization by upper respiratory tract pathogens, including GAS, has been linked to their ability to form biofilms, i.e., communities of bacteria that form complex structures encased in an extracellular, self-produced matrix conferring protection against antibacterial host factors as well as antibiotics (28–32). This insensitivity or resistance to antimicrobial agents in biofilms is a phenotypic characteristic of their structural complexity that is reversible and should not be confused with hereditary antibiotic resistance due to genetic mutations or acquisition of resistance cassettes.
Our previous results have indicated that biofilm formation plays an important role in colonization during GAS pathogenesis (15). Although GAS virulence and biofilm formation have been studied by various groups, these studies were carried out on abiotic surfaces rather than in the context of human cells, which more closely represent the environmental niche where the bacteria reside (9). Similar to what we have observed for pneumococci, the results in this study showed that GAS effectively formed structurally complex biofilms on prefixed respiratory epithelial cells. These biofilms were associated with a biofilm matrix containing double-stranded DNA and proteins, similar to what has previously been reported for GAS biofilms formed on abiotic surfaces (12). Furthermore, we were able to show that the presence of ECM in GAS biofilms was essential for biofilm integrity, as enzymatic treatment led to dispersal and release of GAS from the biofilms. This emphasizes the role of biofilm-produced matrix, not only for protection against host factors, but also as an essential part of the biofilm structure.
The specific role of the underlying surface in GAS biofilm formation has not previously been determined. Here, we found that GAS strains formed phenotypically and structurally different biofilms when grown on biological surfaces (respiratory epithelial H292 cells or SCC13 keratinocytes) compared with abiotic (glass or plastic) surfaces, despite producing biofilms of approximately similar biomass. Biofilms formed on abiotic surfaces developed lower levels of antibiotic resistance than biofilms formed on cells, which correlated well with the thinner biofilms of less complexity observed by SEM. Biofilms formed on cells developed more mature and complex biofilms, although the phenotypes were somewhat cell dependent. Structurally, biofilms formed on biological surfaces generally displayed increased three-dimensional morphology, suggesting the ability of the bacteria to attach and organize better and more quickly on an epithelial surface. Still, biofilms formed on prefixed respiratory H292 cells showed more abundant, dense, and homogenous growth than biofilms grown on prefixed SCC13 keratinocytes. In contrast, biofilms formed on SCC13 keratinocytes, displayed longer bacterial chains, and the biofilm ECM was deposited in clusters rather than the more homogenous pattern observed on H292 respiratory cells. Despite their less compact morphology with less dense ECM, GAS isolates grown on prefixed SCC13 keratinocytes matured earlier and developed resistance to antibiotic (gentamicin) faster than biofilms formed on prefixed H292 respiratory cells. This suggests that ECM alone does not explain the antibiotic resistance phenotype. Instead, bacterial phenotypes, such as persister cell formation and metabolic changes, are likely also involved in protecting the biofilm from antimicrobial agents, as has been suggested for other bacterial organisms (33, 34). How these morphological and phenotypic differences are associated with niche-related advantages for optimal colonization and persistence is an interesting question for further studies.
Biofilm formation was a general characteristic of GAS strains, although biofilm-forming ability, structural phenotypes, and antibiotic resistance levels varied. These differences in biomass and maturity when formed on epithelial surfaces may be associated both with the expression of various virulence determinants combined with their ability to colonize or cause infection in vivo, as has been shown previously for S. pneumoniae (35).
M protein and capsule are two of several virulence factors that have been implicated in GAS biofilm formation and colonization (6, 10, 14). M protein is known for its role as a cell adhesion factor and for its ability to bind to various serum factors, including complement proteins (36–38). Bacteria lacking the global regulator gene mga and thus showing significantly decreased M protein expression also showed a decreased ability to form biofilms on abiotic surfaces (14). However, studies using different serotypes lacking or overexpressing M protein have resulted in inconclusive evidence for a direct role of M protein in biofilm formation on plastic (39). In this study, the role of M protein was variable and depended on serotype. The absence of M5 protein did not affect any of the phenotypic biofilm parameters measured in this study, and the absence of M3 protein resulted in only minor structural and functional deficiencies. In contrast, strains lacking M1 protein formed biofilms with higher biomass that matured and developed antibiotic resistance somewhat faster than those of the isogenic wild-type strains, suggesting an inhibitory role for this M protein in biofilm formation. These results indicate that M protein plays no role or even an inhibitory role in biofilm formation, which is consistent with previous results that showed a downregulation of M protein expression in biofilms (15). The varied phenotypes of biofilms formed by these strains could be explained by the fact that M proteins bind to different host structures (40). The requirement for M protein during in vivo colonization may therefore be either associated with its role in initial attachment to mucosal surfaces during biofilm colonization or for ensuring persistence through the serotype-dependent ability to inhibit complement activation (6, 41).
The capsule did not appear to have any direct role in biofilm formation in this study, as mutants lacking capsule expression formed biofilms as functional as those of their encapsulated counterparts. However, strains with the ability to express medium and high capsule levels in liquid medium developed more antibiotic-resistant biofilms than those expressing smaller amounts of capsule, showing a direct positive correlation between the ability to express capsule and the ability to form functional biofilms. The discrepancy between the two sets of results therefore points toward a more important role of the genetic background of these strains and suggests that other factors, potentially those coregulated with the capsule locus through CovR/S (CsrR/S) or other regulatory mechanisms, contribute to variability in maturation of biofilms between and within serotypes. Such differences in genetic backgrounds between isolates with various capsule expression are of great interest for future studies and will be pursued through transcriptomic and proteomic analyses. The data also support prior results showing that biofilm bacteria downregulate capsule expression when forming biofilms both on epithelial cells and abiotic surfaces. These results support the requirement for some level of capsule expression for colonization in the host and highlight the importance of capsule downregulation, also in highly encapsulated strains, to allow rapid adherence and biofilm formation on mucosal surfaces as a way to persist in the colonizing niche, as was proposed earlier both for GAS and S. pneumoniae (10, 15, 35).
Downregulation of other virulence factors in biofilm bacteria, such as SLS and SpeB, has been documented in a GAS M3 isolate (15). However, the direct role of these factors (as well as the important virulence factors SLO and NADase) during biofilm formation on biological surfaces has not been addressed. Using a set of isogenic mutants, we found that, similar to capsule, the absence of SLO did not affect biofilm formation on biological surfaces. On the other hand, the absence of SpeB or SLS resulted in mature and resistant biofilms that formed faster than that of the wild-type strain. Thus, these results are consistent with the previously reported expression of these genes during biofilm formation on keratinocytes (15). Morphologically, biofilms lacking SLO or SpeB exhibited different biofilm structure compared to biofilms formed by their corresponding WT strain. On the other hand, the increased biofilm formation induced by the SLO and NADase double mutant suggested that expression of NADase negatively impacted biofilm formation. This was, however, the case only when SLO was also absent. For unknown reasons, the absence of NADase alone did not result in the same phenotype as the SLO-NADase double deletion but did result in impaired biofilm formation. As SLO and NADase act both in unison and separately, depending on the situation (42–44), one possibility could be that SLO and NADase, when present together or alone, display different functions and/or affect the function of other virulence factors that might play a role during GAS biofilm formation. Our results are of major interest for future studies to better understand the role of these virulence factors in biofilm formation and colonization. Besides the factors tested in this study, so-far-unidentified factors are likely also involved in both the biofilm maturity and functionality process. Therefore, proteomic analysis of biofilms at different stages in development as well as biofilms formed by mutants or WT with various biofilm formation abilities are ongoing to potentially identify novel factors of importance for biofilm formation in the mucosal niche.
Autoaggregation of planktonic bacteria grown in liquid medium is thought to play a role in biofilm formation of GAS in vitro (9, 14) and is also thought to be associated with microcolony formation in vivo, which precedes biofilm colonization (5, 9). In a prior study using GAS mutants lacking various virulence factors, a direct correlation was observed between the ability to aggregate during planktonic growth and the ability to form biofilms on abiotic surfaces (14). The same results were not observed in this study. The M1 strain (941079) that has a high autoaggregation capacity in growth medium adhered in high numbers to cells but developed biofilms of lower functionality than an M3 strain that displayed lower levels of autoaggregation and cell adherence. On the other hand, the M5 strain exhibited autoaggregation and cell adherence levels similar to those of the M1 strain but formed highly developed and mature biofilms. Moreover, adherence and autoaggregation kinetics did not differ between the SpeB mutant, which formed more developed biofilms, and the NADase mutant, which formed less developed biofilms than their corresponding wild-type strain. This indicates that the ability of bacteria to form microcolonies in various niches in vivo is neither related to autoaggregation in vitro nor related to the in vitro adherence ability of various strains when in contact with relevant physiological surfaces.
In conclusion, the results presented in this work are of clinical importance and will help in better understanding the colonization process and the role of biofilms during GAS pathogenesis, which in turn has the potential to aid in improving treatments of GAS infections, such as strep throat, otitis media, and pneumonia. Identifying virulence mechanisms involved in biofilm formation will help with identifying potential therapeutic targets involved in these infections.
MATERIALS AND METHODS
Reagents.
Reagents for preparation of chemically defined medium (CDM) were purchased from Sigma. The medium was prepared as previously described (16). Todd-Hewitt broth, yeast extract, paraformaldehyde (PFA), and hyaluronic acid (HA) from human umbilical cord were purchased from Sigma. RPMI medium, fetal bovine serum (FBS), and other cell culture reagents were purchased from HyClone, Thermo Fisher. Keratinocyte serum-free medium (K-SFM, 1×) together with the supplements (human recombinant epidermal growth factor [rEGF] and bovine pituitary extract [BPE]), anhydrous calcium chloride (CaCl2), streptavidin-horseradish peroxidase, and gentamicin were purchased from Thermo Fisher Scientific. DNase I and proteases (proteinase K, papain, and elastase) were purchased from Sigma. Exonuclease I was purchased from Thermo Fisher, and trypsin was purchased from HyClone, Thermo Fisher. Hyaluronic acid-binding protein (HABP) and biotinylated HABP were purchased from Seikagaku.
Epithelial cell culture.
A liquid nitrogen frozen stock (−80°C) of the bronchial mucoepidermoid carcinoma cell line NCI-H292 from ATCC (ATCC CRL-1848) was inoculated into small flasks containing sRPMI, i.e., RPMI supplemented with 10% FBS, 100 mM sodium pyruvate, and a mixture of penicillin (100 U/ml) and streptomycin (100 μg/ml), and cells were incubated at 37°C (with 5% CO2) and grown to confluence. Similarly, frozen stocks of SCC13 keratinocytes (squamous cell carcinoma keratinocytes) (45) were inoculated into small flasks containing sK-SFM, i.e., K-SFM supplemented with 0.2 ng/ml rEGF, 50 μg/ml BPE, and 0.3 mM CaCl2, and cells were incubated at 37°C (with 5% CO2) and grown to confluence. Medium was replaced every 72 h. For cell propagation, attached cells were dissociated by 1% trypsin treatment, detached cells were diluted in sRPMI or washed and diluted in sK-SFM, and 0.5 ml was seeded into 24-well plates and incubated for 5 days. For cell fixation, confluent cells were treated with 4% PFA, incubated at room temperature (RT) for 1 h, and washed 4 times with PBS prior to biofilm formation experiments.
Bacterial strains.
The bacterial strains used in this study are listed in Table 1. SF370 and Manfredo together with their corresponding M protein mutants (Δemm1 and Δemm5) were kindly provided by Gunnar Lindahl (Lund University, Lund, Sweden). Additionally, the clinical GAS isolates were kindly provided by Gunnar Kahlmeter (EUCAST, Växjö, Sweden). Virulence factor mutants in the M3 (GAS-771) and M6 (JRS4) backgrounds as well as M3 isolates expressing different capsule amounts were obtained from Michael R. Wessels (Harvard Medical School, USA).
Planktonic growth, autoaggregation, and sensitivity to gentamicin.
Planktonic sensitivity assay using the aminoglycoside gentamicin was performed to be able to determine the increase in gentamicin resistance of GAS upon biofilm formation relative to that in planktonic bacteria, which in turn indicates the development of functional biofilms. From a blood agar plate streaked with bacteria, an inoculum was taken and immersed in a glass tube containing CDM. The inoculated bacteria were incubated at 34°C until they reached an optical density at 600 nm (OD600) of 0.6 (∼108 CFU/ml) with OD measurement every 30 min during incubation. Autoaggregation of GAS was measured by the sedimentation amount and rate of bacteria during planktonic growth in broth cultures and compared with the OD measurement after the bacterial suspension was vortexed to homogeneity after sedimentation. To prepare the bacterial suspension for the gentamicin sensitivity or adherence assay, planktonic bacteria were centrifuged, and the pellet was resuspended in PBS or RPMI supplemented with 2% serum (RPMI-2%), respectively.
In a short time-kill assay, planktonic bacteria were added to a 96-well plate containing 2-fold serially diluted gentamicin in PBS (pH 7.2). The plate was then incubated at 34°C for 3 h (in the presence of 5% CO2). To determine the number of CFU after treatment, a 10-fold serial dilution of the treated bacteria was performed in PBS (pH 7.2), and dilutions were spread on blood agar plates to be further incubated for 48 h at 37°C before colonies were counted to determine the number of CFU per milliliter. Results of experiments performed in triplicate (n = 3) for each strain are presented in Fig. S6.
Biofilm formation.
Biofilm formation and biofilm assessment were performed as described earlier (17). A bacterial inoculum of ∼105 CFUs from frozen bacterial stocks (at −80°C) in Todd-Hewitt broth with 0.5% yeast extract (THY) was diluted in CDM and used to seed 24-well plates covered with PFA-prefixed H292 or SCC13 cells, having no cell substratum, or carrying a glass coverslip insert. The seeded bacteria were incubated at 34°C (in the presence of 5% CO2) over a period of 48 to 72 h to allow formation of biofilms. Medium was replaced every 12 h.
Biofilm phenotype assessment.
Biofilms incubated for 48 or 72 h were assessed for biomass and gentamicin sensitivity as indications of biofilm maturation and functionality, respectively. Biofilms were treated in PBS in the absence (for total biomass) or presence (for gentamicin sensitivity as an indication of antibiotic resistance) of 500 μg/ml gentamicin dissolved in PBS for 3 h at 34°C (in the presence of 5% CO2). To disrupt biofilm structures and dissociate bacteria, biofilms were sonicated 3 times using a water bath sonicator (1-s interval between sonications). To determine the number of CFU after treatment, a 10-fold serial dilution of the sonicated bacteria was performed in PBS (pH 7.2), and dilutions were spread on blood agar plates to be further incubated for 48 h at 37°C before colonies were counted to determine the number of CFU per milliliter.
Cell adherence assay.
To assess the correlation between autoaggregation and adhesion to epithelial cells, live H292 cells were infected with actively growing planktonic bacteria at a multiplicity of infection (MOI) of 10 for 1 h. After incubation, the cells were washed three times with PBS (pH 7.2), detached with 1% trypsin, resuspended in PBS, collected, and centrifuged at 9,000 rpm for 2 min. After centrifugation, the supernatant was discarded, the pellet was treated with 0.1% Triton X-100 to disrupt cells, and the lysates were diluted (10-fold) and spread on blood agar plates to determine the number CFU of cell-associated bacteria per milliliter, after incubation of the plates at 37°C for 48 h.
Enzyme treatment assay.
Assessment of ECM composition and role in biofilm integrity was performed using an enzyme treatment assay in which biofilms incubated for 72 h at 34°C were treated with cysteine protease (papain), a set of serine proteases (proteinase K, elastase, and trypsin), or DNases (DNase I and exonuclease I) and bacterial release into the supernatant was measured. Biofilms were washed with PBS once and treated with enzymes diluted in PBS (pH 7.2) for 2 h at 34°C. After incubation, supernatants were collected and serially diluted 10-fold in PBS (pH 7.2), and the dilutions were spread on blood agar plates to be further incubated for 48 h at 37°C before colonies were counted to determine the number of CFU per milliliter.
HABP solid-phase binding assay.
The HABP solid-phase binding assay was performed as described earlier (46). A 96-well plate was coated with 0.06 μg/ml of HABP dissolved in PBS and incubated overnight at 4°C. HA from human umbilical cord was diluted in 3-fold dilutions ranging from 0 to 3,000 ng/ml to generate a standard curve (bacterial strains were assayed using optimal dilutions by first testing each strain in 10-fold dilutions). The wells were washed three times with 0.1% Tween (in PBS) between steps in the assay. The coated plate was blocked with 1% bovine serum albumin (BSA) in PBS and left at RT for 1 h. HA was added to wells at different concentrations to produce a standard curve, and bacterial strain extracts were added to test wells and incubated for 1 h at RT. After incubation, 0.03 μg/ml of biotinylated HABP was added to each well of the plate and incubated for an additional 1 h at RT. Streptavidin-horseradish peroxidase (1 μg/ml) was added to the plate followed by 30 min incubation at RT. After incubation, Sure-Blue peroxidase substrate (KPL) was added and left until blue color was established. The reaction was then stopped with H2SO4 (4 N), yielding a yellow color where the color intensity was detected at an OD450 using a plate reader (Perkin Elmer HTS 7000 Plus Bioassay reader). Results were calculated based on the HA standard curve and are presented as femtograms of HA per CFU. Results are shown in Table 2.
Scanning electron microscopy.
Biofilm and planktonic samples were prepared for SEM as described previously (17). Planktonic bacteria were grown as described above. Biofilms were grown using the conditions described above but on round glass coverslips that were uncoated or coated with prefixed H292 or SCC13 cells in a 24-well plate. Ruthenium red (RR; 0.075%) and lysine-acetate (0.075 M) are thought to retain carbohydrate structures and improve preservation of biofilm structures and were therefore used in a fixation solution containing glutaraldehyde (2.5%) and sodium cacodylate buffer (0.1 M; pH 7.2) (47). Samples were treated with fixation solution overnight at RT, washed 3 times with wash solution containing RR (0.075%) and sodium cacodylate buffer (0.2 M, pH 7.2), and then dehydrated with increasing ethanol concentrations (50 to 99.89% [vol/vol]) prior to critical-point drying with liquid carbon dioxide, using ethanol (99.89%) as the intermediate solvent. Samples were then mounted onto aluminum holders, painted with silver around the sides for better conductivity, and sputter coated with 20-nm palladium-gold. Sample imaging was done using a DELPHI correlative light and electron microscope (Phenom-World; IQ Biotechnology Platform, Infection Medicine, Lund University).
M protein gene (emm) typing.
The M protein serotype in clinical GAS isolates (Table 1) was identified by PCR analysis of the emm gene using primers indicated in Table 3 and an annealing temperature of 55°C. PCR reagents were purchased from Sigma. The variable part of the M protein gene is closest to the 5′ primer; therefore, PCR products from the 5′ primers (F primers) were sequenced, and sequence results were matched with the CDC (Centers for Disease Control and Prevention) database for interpretation of results and M protein identification.
TABLE 3.
Primers used for M protein determination
Statistical analysis.
GraphPad Prism 8 software (GraphPad Software LLC) was used to graph data and perform statistical analyses. Biofilm biomass and ECM composition data were assessed for statistical significance using the Mann-Whitney U tests. Planktonic and biofilm gentamicin sensitivity data as well as biofilm biomass data of SEM figures were analyzed using Student’s t test. Adherence data were analyzed using one-way analysis of variance (ANOVA) with Tukey’s multiple-comparison test. P values of <0.05 were considered statistically significant. Data are presented as mean values, with error bars representing the standard deviation, i.e., the error of mean values for multiple biological data points, as described in the figure legends. Simple linear regression was used to analyze the significant correlation between expressed capsule amounts and the biofilm functionality.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ravi Bhongir (IQ Biotechnology Platform, Infection Medicine, Lund University) for dehydrating and mounting samples on SCC13 keratinocytes for SEM. We are grateful to the Clinical Microbiological Laboratory at Labmedicin Skåne (Region Skåne) for the M typing of the clinical GAS isolates. Quantification of capsular hyaluronic acid was conducted by Emelia Anne DeForce (University of Massachusetts, Boston, MA) during preparation of her master’s thesis. We are grateful to Gunnar Lindahl (Professor Emeritus, Lund University, Sweden), Gunnar Kahlmeter (Technical Data Coordinator and Webmaster, EUCAST, Sweden), Michael Wessels (Children’s Hospital Boston, Boston, MA), and June R. Scott (Emory University, Atlanta, GA) for providing GAS strains for this study. SCC13 keratinocytes used in this study were purchased and kindly provided by James Rheinwald (Harvard Medical School, Boston, MA, USA).
Financial support allowing us to conduct this project was provided by the Royal Physiographic Society in Lund (Hedda and John Forssmans Foundation; to F.A.) and the Swedish Research Council (VR; to A.P.H.).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Carapetis JR, Steer AC, Mulholland EK, Weber M. 2005. The global burden of group A streptococcal diseases. Lancet Infect Dis 5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 2.Tan LK, Eccersley LR, Sriskandan S. 2014. Current views of haemolytic streptococcal pathogenesis. Curr Opin Infect Dis 27:155–164. doi: 10.1097/QCO.0000000000000047. [DOI] [PubMed] [Google Scholar]
- 3.Brouwer S, Barnett TC, Rivera-Hernandez T, Rohde M, Walker MJ. 2016. Streptococcus pyogenes adhesion and colonization. FEBS Lett 590:3739–3757. doi: 10.1002/1873-3468.12254. [DOI] [PubMed] [Google Scholar]
- 4.Muotiala A, Seppälä H, Huovinen P, Vuopio-Varkila J. 1997. Molecular comparison of group A streptococci of T1M1 serotype from invasive and noninvasive infections in Finland. J Infect Dis 175:392–399. doi: 10.1093/infdis/175.2.392. [DOI] [PubMed] [Google Scholar]
- 5.Roberts AL, Connolly KL, Kirse DJ, Evans AK, Poehling KA, Peters TR, Reid SD. 2012. Detection of group A Streptococcus in tonsils from pediatric patients reveals high rate of asymptomatic streptococcal carriage. BMC Pediatr 12:3. doi: 10.1186/1471-2431-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashbaugh CD, Moser TJ, Shearer MH, White GL, Kennedy RC, Wessels MR. 2000. Bacterial determinants of persistent throat colonization and the associated immune response in a primate model of human group A streptococcal pharyngeal infection. Cell Microbiol 2:283–292. doi: 10.1046/j.1462-5822.2000.00050.x. [DOI] [PubMed] [Google Scholar]
- 7.Courtney HS, Hasty DL, Dale JB. 2002. Molecular mechanisms of adhesion, colonization, and invasion of group A streptococci. Ann Med 34:77–87. doi: 10.1080/07853890252953464. [DOI] [PubMed] [Google Scholar]
- 8.Nobbs AH, Lamont RJ, Jenkinson HF. 2009. Streptococcus adherence and colonization. Microbiol Mol Biol Rev 73:407–450. doi: 10.1128/MMBR.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiedler T, Köller T, Kreikemeyer B. 2015. Streptococcus pyogenes biofilms-formation, biology, and clinical relevance. Front Cell Infect Microbiol 5:15. doi: 10.3389/fcimb.2015.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho KH, Caparon MG. 2005. Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes. Mol Microbiol 57:1545–1556. doi: 10.1111/j.1365-2958.2005.04786.x. [DOI] [PubMed] [Google Scholar]
- 11.Baldassarri L, Creti R, Recchia S, Imperi M, Facinelli B, Giovanetti E, Pataracchia M, Alfarone G, Orefici G. 2006. Therapeutic failures of antibiotics used to treat macrolide-susceptible Streptococcus pyogenes infections may be due to biofilm formation. J Clin Microbiol 44:2721–2727. doi: 10.1128/JCM.00512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doern CD, Roberts AL, Hong W, Nelson J, Lukomski S, Swords WE, Reid SD. 2009. Biofilm formation by group A Streptococcus: a role for the streptococcal regulator of virulence (Srv) and streptococcal cysteine protease (SpeB). Microbiology 155:46–52. doi: 10.1099/mic.0.021048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matysik A, Kline KA. 2019. Streptococcus pyogenes capsule promotes microcolony-independent biofilm formation. J Bacteriol 201:e00052-19. doi: 10.1128/JB.00052-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo F, Lizano S, Banik S, Zhang H, Bessen DE. 2008. Role of Mga in group A streptococcal infection at the skin epithelium. Microb Pathog 45:217–224. doi: 10.1016/j.micpath.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marks LR, Mashburn-Warren L, Federle MJ, Hakansson AP. 2014. Streptococcus pyogenes biofilm growth in vitro and in vivo and its role in colonization, virulence, and genetic exchange. J Infect Dis 210:25–34. doi: 10.1093/infdis/jiu058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van de Rijn I, Kessler RE. 1980. Growth characteristics of group A streptococci in a new chemically defined medium. Infect Immun 27:444–448. doi: 10.1128/IAI.27.2.444-448.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chao Y, Bergenfelz C, Hakansson AP. 2019. Growing and characterizing biofilms formed by Streptococcus pneumoniae. Methods Mol Biol 1968:147–171. doi: 10.1007/978-1-4939-9199-0_13. [DOI] [PubMed] [Google Scholar]
- 18.Keck T, Leiacker R, Riechelmann H, Rettinger G. 2000. Temperature profile in the nasal cavity. Laryngoscope 110:651–654. doi: 10.1097/00005537-200004000-00021. [DOI] [PubMed] [Google Scholar]
- 19.Håkansson A, Bentley CC, Shakhnovic EA, Wessels MR. 2005. Cytolysin-dependent evasion of lysosomal killing. Proc Natl Acad Sci U S A 102:5192–5197. doi: 10.1073/pnas.0408721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts AL, Holder RC, Reid SD. 2010. Allelic replacement of the streptococcal cysteine protease SpeB in a Δsrv mutant background restores biofilm formation. BMC Res Notes 3:281. doi: 10.1186/1756-0500-3-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lembke C, Podbielski A, Hidalgo-Grass C, Jonas L, Hanski E, Kreikemeyer B. 2006. Characterization of biofilm formation by clinically relevant serotypes of group A streptococci. Appl Environ Microbiol 72:2864–2875. doi: 10.1128/AEM.72.4.2864-2875.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varela-Ramirez A, Abendroth J, Mejia AA, Phan IQ, Lorimer DD, Edwards TE, Aguilera RJ. 2017. Structure of acid deoxyribonuclease. Nucleic Acids Res 45:6217–6227. doi: 10.1093/nar/gkx222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehman IR, Nussbaum AL. 1964. The deoxyribonucleases of Escherichia coli. V. On the specificity of exonuclease I (phosphodiesterase). J Biol Chem 239:2628–2636. [PubMed] [Google Scholar]
- 24.Anthony BF, Kaplan EL, Wannamaker LW, Chapman SS. 1976. The dynamics of streptococcal infections in a defined population of children: serotypes are associated with skin and respiratory infections. Am J Epidemiol 104:652–666. doi: 10.1093/oxfordjournals.aje.a112344. [DOI] [PubMed] [Google Scholar]
- 25.Wannamaker LW. 1970. Differences between streptococcal infections of the throat and of the skin. I. N Engl J Med 282:23–31. doi: 10.1056/NEJM197001012820106. [DOI] [PubMed] [Google Scholar]
- 26.Cunningham MW. 2000. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev 13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wannamaker LW. 1970. Differences between streptococcal infections of the throat and of the skin (second of two parts). N Engl J Med 282:78–85. doi: 10.1056/NEJM197001082820206. [DOI] [PubMed] [Google Scholar]
- 28.Fux CA, Costerton JW, Stewart PS, Stoodley P. 2005. Survival strategies of infectious biofilms. Trends Microbiol 13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Stoodley P, Sauer K, Davies DG, Costerton JW. 2002. Biofilms as complex differentiated communities. Annu Rev Microbiol 56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 30.Wolcott RD, Ehrlich GD. 2008. Biofilms and chronic infections. JAMA 299:2682–2684. doi: 10.1001/jama.299.22.2682. [DOI] [PubMed] [Google Scholar]
- 31.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 32.Reid SD, Hong W, Dew KE, Winn DR, Pang B, Watt J, Glover DT, Hollingshead SK, Swords WE. 2009. Streptococcus pneumoniae forms surface-attached communities in the middle ear of experimentally infected chinchillas. J Infect Dis 199:786–794. doi: 10.1086/597042. [DOI] [PubMed] [Google Scholar]
- 33.Lewis K. 2008. Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol 322:107–131. doi: 10.1007/978-3-540-75418-3_6. [DOI] [PubMed] [Google Scholar]
- 34.Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 35.Marks LR, Parameswaran GI, Hakansson AP. 2012. Pneumococcal interactions with epithelial cells are crucial for optimal biofilm formation and colonization in vitro and in vivo. Infect Immun 80:2744–2760. doi: 10.1128/IAI.00488-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Areschoug T, Carlsson F, Stålhammar-Carlemalm M, Lindahl G. 2004. Host-pathogen interactions in Streptococcus pyogenes infections, with special reference to puerperal fever and a comment on vaccine development. Vaccine 22:S9–S14. doi: 10.1016/j.vaccine.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 37.Ermert D, Shaughnessy J, Joeris T, Kaplan J, Pang CJ, Kurt-Jones EA, Rice PA, Ram S, Blom AM. 2015. Virulence of group A streptococci is enhanced by human complement inhibitors. PLoS Pathog 11:e1005043. doi: 10.1371/journal.ppat.1005043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okada N, Liszewski MK, Atkinson JP, Caparon M. 1995. Membrane cofactor protein (CD46) is a keratinocyte receptor for the M protein of the group A streptococcus. Proc Natl Acad Sci U S A 92:2489–2493. doi: 10.1073/pnas.92.7.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Courtney HS, Ofek I, Penfound T, Nizet V, Pence MA, Kreikemeyer B, Podbielski A, Podbielbski A, Hasty DL, Dale JB. 2009. Relationship between expression of the family of M proteins and lipoteichoic acid to hydrophobicity and biofilm formation in Streptococcus pyogenes. PLoS One 4:e4166. doi: 10.1371/journal.pone.0004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gustafsson MC, Lannergård J, Nilsson OR, Kristensen BM, Olsen JE, Harris CL, Ufret-Vincenty RL, Stålhammar-Carlemalm M, Lindahl G. 2013. Factor H binds to the hypervariable region of many Streptococcus pyogenes M proteins but does not promote phagocytosis resistance or acute virulence. PLoS Pathog 9:e1003323. doi: 10.1371/journal.ppat.1003323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laabei M, Ermert D. 2019. Catch me if you can: Streptococcus pyogenes complement evasion strategies. J Innate Immun 11:3–12. doi: 10.1159/000492944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madden JC, Ruiz N, Caparon M. 2001. Cytolysin-mediated translocation (CMT): a functional equivalent of type III secretion in gram-positive bacteria. Cell 104:143–152. doi: 10.1016/s0092-8674(01)00198-2. [DOI] [PubMed] [Google Scholar]
- 43.Michos A, Gryllos I, Håkansson A, Srivastava A, Kokkotou E, Wessels MR. 2006. Enhancement of streptolysin O activity and intrinsic cytotoxic effects of the group A streptococcal toxin, NAD-glycohydrolase. J Biol Chem 281:8216–8223. doi: 10.1074/jbc.M511674200. [DOI] [PubMed] [Google Scholar]
- 44.Hancz D, Westerlund E, Bastiat-Sempe B, Sharma O, Valfridsson C, Meyer L, Love JF, O’Seaghdha M, Wessels MR, Persson JJ. 2017. Inhibition of inflammasome-dependent interleukin 1β production by streptococcal NAD+-glycohydrolase: evidence for extracellular activity. mBio 8:e00756-17. doi: 10.1128/mBio.00756-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rheinwald JG, Beckett MA. 1981. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultured from human squamous cell carcinomas. Cancer Res 41:1657–1663. [PubMed] [Google Scholar]
- 46.Gryllos I, Grifantini R, Colaprico A, Jiang S, Deforce E, Hakansson A, Telford JL, Grandi G, Wessels MR. 2007. Mg(2+) signalling defines the group A streptococcal CsrRS (CovRS) regulon. Mol Microbiol 65:671–683. doi: 10.1111/j.1365-2958.2007.05818.x. [DOI] [PubMed] [Google Scholar]
- 47.Hammerschmidt S, Wolff S, Hocke A, Rosseau S, Müller E, Rohde M. 2005. Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect Immun 73:4653–4667. doi: 10.1128/IAI.73.8.4653-4667.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Avery OT, Macleod CM, McCarty M. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med 79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chatellier S, Ihendyane N, Kansal RG, Khambaty F, Basma H, Norrby-Teglund A, Low DE, McGeer A, Kotb M. 2000. Genetic relatedness and superantigen expression in group A streptococcus serotype M1 isolates from patients with severe and nonsevere invasive diseases. Infect Immun 68:3523–3534. doi: 10.1128/iai.68.6.3523-3534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abbot EL, Smith WD, Siou GP, Chiriboga C, Smith RJ, Wilson JA, Hirst BH, Kehoe MA. 2007. Pili mediate specific adhesion of Streptococcus pyogenes to human tonsil and skin. Cell Microbiol 9:1822–1833. doi: 10.1111/j.1462-5822.2007.00918.x. [DOI] [PubMed] [Google Scholar]
- 51.Johnsson E, Berggård K, Kotarsky H, Hellwage J, Zipfel PF, Sjöbring U, Lindahl G. 1998. Role of the hypervariable region in streptococcal M proteins: binding of a human complement inhibitor. J Immunol 161:4894–4901. [PubMed] [Google Scholar]
- 52.Biswas I, Scott JR. 2003. Identification of rocA, a positive regulator of covR expression in the group A streptococcus. J Bacteriol 185:3081–3090. doi: 10.1128/jb.185.10.3081-3090.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alamiri F, Riesbeck K, Hakansson AP. 2019. HAMLET, a protein complex from human milk has bactericidal activity and enhances the activity of antibiotics against pathogenic. Antimicrob Agents Chemother 63:e01193-19. doi: 10.1128/AAC.01193-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wessels MR, Moses AE, Goldberg JB, DiCesare TJ. 1991. Hyaluronic acid capsule is a virulence factor for mucoid group A streptococci. Proc Natl Acad Sci U S A 88:8317–8321. doi: 10.1073/pnas.88.19.8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flores AR, Jewell BE, Fittipaldi N, Beres SB, Musser JM. 2012. Human disease isolates of serotype M4 and M22 group A streptococcus lack genes required for hyaluronic acid capsule biosynthesis. mBio 3:e00413-12. doi: 10.1128/mBio.00413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ashbaugh CD, Wessels MR. 2001. Absence of a cysteine protease effect on bacterial virulence in two murine models of human invasive group A streptococcal infection. Infect Immun 69:6683–6688. doi: 10.1128/IAI.69.11.6683-6686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schrager HM, Rheinwald JG, Wessels MR. 1996. Hyaluronic acid capsule and the role of streptococcal entry into keratinocytes in invasive skin infection. J Clin Invest 98:1954–1958. doi: 10.1172/JCI118998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pérez-Caballero D, Albertí S, Vivanco F, Sánchez-Corral P, Rodríguez de Córdoba S. 2000. Assessment of the interaction of human complement regulatory proteins with group A Streptococcus. Identification of a high-affinity group A Streptococcus binding site in FHL-1. Eur J Immunol 30:1243–1253. doi:. [DOI] [PubMed] [Google Scholar]
- 59.Ashbaugh CD, Warren HB, Carey VJ, Wessels MR. 1998. Molecular analysis of the role of the group A streptococcal cysteine protease, hyaluronic acid capsule, and M protein in a murine model of human invasive soft-tissue infection. J Clin Invest 102:550–560. doi: 10.1172/JCI3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bricker AL, Cywes C, Ashbaugh CD, Wessels MR. 2002. NAD+-glycohydrolase acts as an intracellular toxin to enhance the extracellular survival of group A streptococci. Mol Microbiol 44:257–269. doi: 10.1046/j.1365-2958.2002.02876.x. [DOI] [PubMed] [Google Scholar]
- 61.Sierig G, Cywes C, Wessels MR, Ashbaugh CD. 2003. Cytotoxic effects of streptolysin O and streptolysin S enhance the virulence of poorly encapsulated group A streptococci. Infect Immun 71:446–455. doi: 10.1128/iai.71.1.446-455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Centers for Disease Control and Prevention. 2018. M protein gene (emm) typing. https://www.cdc.gov/streplab/groupa-strep/emm-background.html.
- 63.Kahn F, Linder A, Petersson AC, Christensson B, Rasmussen M. 2010. Axillary abscess complicated by venous thrombosis: identification of Streptococcus pyogenes by 16S PCR. J Clin Microbiol 48:3435–3437. doi: 10.1128/JCM.00373-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jasir A, Tanna A, Efstratiou A, Schalén C. 2001. Unusual occurrence of M type 77, antibiotic-resistant group A streptococci in southern Sweden. J Clin Microbiol 39:586–590. doi: 10.1128/JCM.39.2.586-590.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.