Haemophilus influenzae is a Gram-negative bacterium that can be classified into typeable (types a through f) and nontypeable (NTHi) groups. This opportunistic pathogen asymptomatically colonizes the mucosal epithelium of the upper respiratory tract, from where it spreads to other neighboring regions, potentially leading to disease. Infection with NTHi can cause otitis media, sinusitis, conjunctivitis, exacerbations of chronic obstructive pulmonary disease, and pneumonia, but it is increasingly causing invasive disease, including bacteremia and meningitis.
KEYWORDS: Haemophilus influenzae, IgG, IgM, complement evasion, sepsis, meningitis, bacteremia
ABSTRACT
Haemophilus influenzae is a Gram-negative bacterium that can be classified into typeable (types a through f) and nontypeable (NTHi) groups. This opportunistic pathogen asymptomatically colonizes the mucosal epithelium of the upper respiratory tract, from where it spreads to other neighboring regions, potentially leading to disease. Infection with NTHi can cause otitis media, sinusitis, conjunctivitis, exacerbations of chronic obstructive pulmonary disease, and pneumonia, but it is increasingly causing invasive disease, including bacteremia and meningitis. Invasive NTHi strains are more resistant to complement-mediated killing. However, the mechanisms of complement resistance have never been studied in large numbers of invasive NTHi strains. In this study, we determined the relationship between binding of IgG or IgM and the bacterial survival in normal human serum for 267 invasive H. influenzae strains from Spain, Portugal, and the Netherlands, of which the majority (200 [75%]) were NTHi. NTHi bacteria opsonized with high levels of IgM had the lowest survival in human serum. IgM binding to the bacterial surface, but not IgG binding, was shown to be associated with complement-mediated killing of NTHi strains. We conclude that evasion of IgM binding by NTHi strains increases survival in blood, thereby potentially contributing to their ability to cause severe invasive diseases.
INTRODUCTION
Before the introduction of Haemophilus influenzae type b (Hib) polysaccharide conjugate vaccination, Hib was responsible for a large proportion of invasive H. influenzae disease cases, especially in young children (1). While the number of Hib infections drastically declined after the introduction of the Hib polysaccharide conjugate vaccine in the 1990s, the number of non-type b H. influenzae strains, especially nontypeable H. influenzae (NTHi), has been noticeably increasing over the past 2 decades (2, 3).
NTHi is a microorganism with several unique bacteriological features. Unlike capsulated H. influenzae, NTHi strains are genetically and phenotypically heterogeneous. In the host organism, NTHi is exposed to the antimicrobial action of the complement system, which is an important defense mechanism against bacterial infections. Direct complement-mediated killing takes place when the membrane attack complex (MAC) is formed in the bacterial membrane as a final result of the multistep process of complement activation.
Previously, we showed that invasive NTHi strains are more resistant to complement-mediated killing (4). It has been reported that the binding of IgM antibodies to the bacterial surface is an influential factor concerning the complement-mediated killing of NTHi (5, 6). This finding is in agreement with a study demonstrating that individuals with the hyper-IgM syndrome are less prone to be carriers of NTHi (7). Conversely, patients with common variable immune deficiency (CVID) having low serum IgM levels (<0.05 g/liter) are at increased risk for H. influenzae colonization (8).
The genetic and phenotypic heterogeneity of NTHi strains requires research using large numbers of strains to allow a better understanding of the role of IgG and IgM antibody binding as the first step in the complement-mediated killing of NTHi. In this study, we investigated the association between binding of IgG or IgM to the bacterial surface of invasive H. influenzae strains and bacterial survival in human serum.
RESULTS
Characteristics of invasive H. influenzae strains collected from the Netherlands, Spain, and Portugal.
In total, 267 invasive H. influenzae strains were used in this study, 161 from The Netherlands, 51 from Spain, and 55 from Portugal (Table 1). Most were collected from blood (239/267 [89.5%]), 17 strains were collected from cerebrospinal fluid (6.4%), 8 strains were collected from pleural fluid (3.0%), 2 strains were collected from joint fluid (0.7%), and a single strain was collected from undefined tissue (0.4%). Most of the collected invasive H. influenzae strains (200/267 [74.9%]) were NTHi, followed by 40 of type b (15.0%), 20 of type f (7.5%), 3 of type a (1.1%), 3 of type e (1.1%), and 1 of type d (0.4%).
TABLE 1.
Characteristics of analyzed invasive H. influenzae strains
| Parameter | Value for: |
|||
|---|---|---|---|---|
| The Netherlands | Spain | Portugal | Combined | |
| No. of strainsa | 161 | 51 | 55 | 267 |
| Serotype a | 2 (1.2) | 1 (1.8) | 3 (1.1) | |
| Serotype b | 30 (18.6) | 4 (7.8) | 6 (10.9) | 40 (15.0) |
| Serotype d | 1 (0.6) | 1 (0.4) | ||
| Serotype e | 3 (1.9) | 3 (1.1) | ||
| Serotype f | 8 (5.0) | 9 (17.6) | 3 (5.5) | 20 (7.5) |
| NTHi | 117 (72.7) | 38 (74.5) | 45 (81.8) | 200 (74.9) |
| Collection sitea | ||||
| Blood | 146 (90.7) | 45 (88.2) | 48 (87.3) | 239 (89.5) |
| CSFb | 13 (8.1) | 2 (3.9) | 2 (3,6) | 17 (6.4) |
| Pleural fluid | 4 (7.8) | 4 (7.3) | 8 (3.0) | |
| Joint fluid | 1 (0.6) | 1 (1.8) | ||
| Undefined tissue | 1 (0.6) | 1 (0.4) | ||
| Agec | 64 (38–75) | 66 (56–76) | 56 (24–76) | 63 (40–76) |
| Sexa | ||||
| Male | 80 (49.7) | 23 (45.1) | 20 (36.4) | 123 (46.1) |
| Female | 79 (49.1) | 28 (54.9) | 35 (63.6) | 142 (53.2) |
| Unknown | 2 (1.2) | 2 (0.7) | ||
Presented as number (percent).
CSF, cerebrospinal fluid.
Presented as median age in years (25% to 75% quantiles).
The H. influenzae-infected patients in Spain were older (median, 66 years [25% to 75% quantiles, 56 to 76 years]) than the patients from Portugal (median, 56 years [25% to 75% quantiles, 24 to 76 years]) and the Netherlands (median, 64 years [25% to 75% quantiles, 38 to 75 years]) (Table 1). This difference in age is largely due to the fact that patients from the Hospital Universitario Bellvitge (Spain) were mainly adults, resulting in a low number of patients of <18 years in Spain (3/51; 6%) compared to Portugal (14/55 [25%]) or the Netherlands (28/161 [17%]).
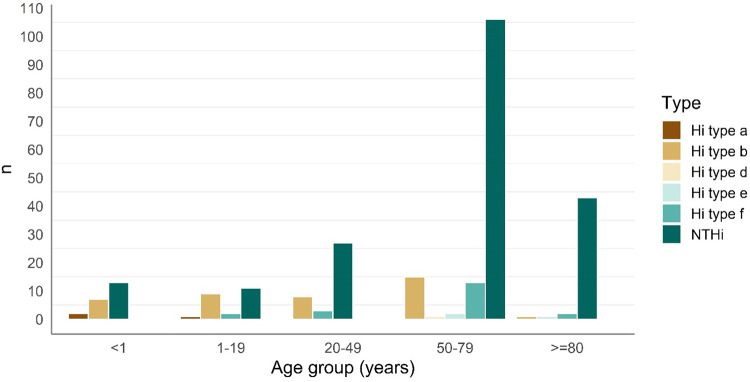
The distributions of H. influenzae types were different according to patient age. Type a or b strains were mainly isolated from young patients. Among patients <1 year of age, of all isolated strains, 9 were H. influenzae type a or b (9/22 [40.9%]). In the age group ≥80 years, only 1 strain of type b (1/47 [2.1%]) was isolated. Infections with NTHi were most prevalent in patients ≥80 years of age (43/47 [91.5%]), whereas they were responsible for 54.5% (12/22) of infections in the patients <1 year of age (Fig. 1). H. influenzae type f was found to cause infections in all age categories except in the youngest age group (Fig. 1). Haemophilus influenzae type a was found only in children <2 years of age, whereas the only H. influenzae type d and three H. influenzae type e strains were found in adults >55 years of age.
FIG 1.
Numbers of H. influenzae type a, b, d, e, and f and NTHi strains found among patients in different age groups.
Binding of serum IgG and serum IgM and bacterial survival in normal human serum of 260 invasive H. influenzae strains.
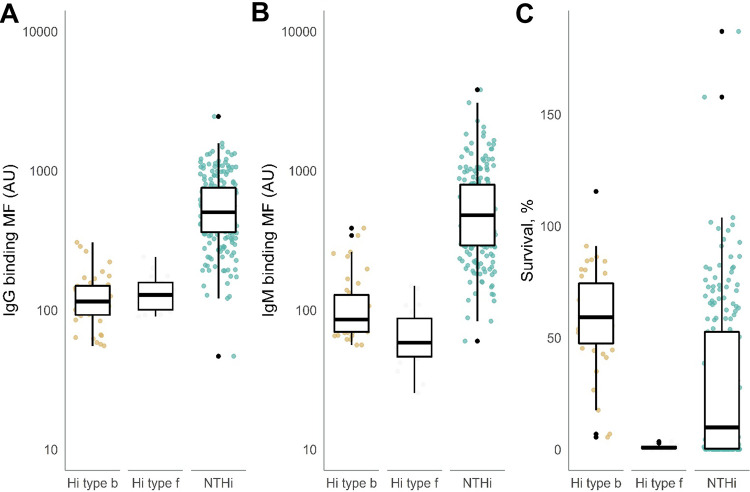
We performed analysis for H. influenzae type b (n = 40), H. influenzae type f (n = 20), and NTHi strains (n = 200), excluding type a (n = 3), type d (n = 1), and type e (n = 3) due to low numbers. Binding of IgG to the surface of NTHi strains was higher than for H. influenzae type b and f strains (Fig. 2A). Within the NTHi strains, the median binding of IgG to the surface measured as mean fluorescence intensity (MFI) was 504 (25% to 75% quantiles, 363 to 754) arbitrary units (AU), whereas median binding of IgG to the surface of capsulated H. influenzae strains was lower (115 AU; 25% to 75% quantiles, 92 to 149 AU) (Fig. 2A). The median binding of IgM to the bacterial surface of NTHi strains was 480 (25% to 75% quantiles, 291 to 794) AU. Median IgM binding of H. influenzae type b (85.5 AU; 25% to 75% quantiles, 70 to 129 AU) and f (58 AU; 25% to 75% quantiles, 46 to 87 AU) strains was lower than that of NTHi (Fig. 2B). The strains collected in the different countries had similar patterns of IgG or IgM binding to H. influenzae types b and f and to NTHi (data not shown).
FIG 2.
IgG and IgM antibody binding and bacterial survival of 260 H. influenzae strains. H. influenzae strains were incubated with 10% HI-NHS, and binding of IgG (A) and IgM (B) was analyzed by flow cytometry (n = 3). The IgG and IgM antibody binding is presented on log10 scale. (C) H. influenzae strains were incubated with 10% (typeable Hi strains) or 5% (NTHi) NHS or HI-NHS for 60 min, and survival was determined by dividing the number of CFU after exposure to NHS with the CFU after exposure to HI-NHS (n = 3). We used analysis of variance (ANOVA) to compare the groups.
We determined bacterial survival in normal human serum (NHS) for all H. influenzae strains. In the serum bactericidal assay for NTHi strains, we used 5% NHS since these strains are, in general, more susceptible to complement-mediated killing. For capsulated H. influenzae, we used 10% NHS. Nearly all H. influenzae type b isolates showed a high survival rate (median, 59.2% [25% to 75% quantiles, 47.5% to 74.3%]), with two strains showing survival rates as low as 5% and 7% (Fig. 2C). The median survival rate of H. influenzae type f (0.9% [25% to 75% quantiles, 0.4% to 1.3%]) was significantly lower than that of H. influenzae type b strains (Fig. 2C). The median survival rate of NTHi strains in 5% NHS was 9.9% (25% to 75% quantiles, 0.3% to 52.6%), with several strains showing survival of >100%. Although only 5% NHS was used for NTHi strains, survival was significantly lower than that of H. influenzae type b strains, for which 10% NHS was used (Fig. 2C). H. influenzae type b and type f and NTHi collected in the different countries showed similar patterns of survival in NHS (data not shown).
Association of serum IgG and serum IgM binding with bacterial survival in normal human serum of 200 NTHi strains.
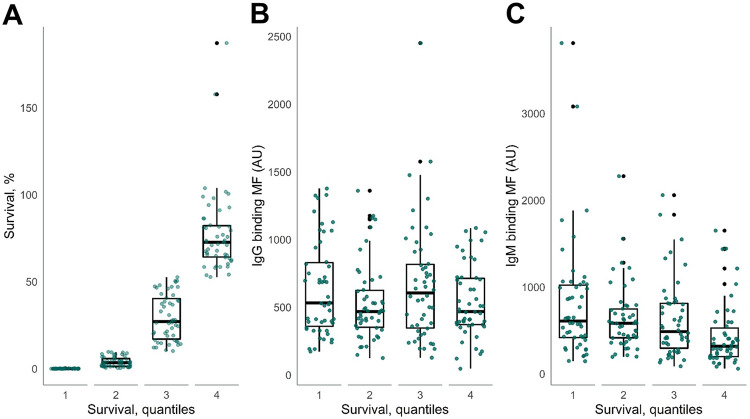
We divided the NTHi strains into four equal groups based on the survival quantiles in 5% NHS (Fig. 3A) and evaluated the binding of IgG and IgM to the bacterial surface. Binding of IgG to the surface of NTHi strains was not associated with bacterial survival (Fig. 3B). Binding of IgM to the surface of NTHi strains was inversely associated with bacterial survival. NTHi quantile 4 had the highest survival (72.7%) and had the lowest median IgM binding (320 AU [25% to 75% quantiles, 199 to 528 AU]) (Fig. 3C).
FIG 3.
IgG and IgM antibody binding to 200 NTHi strains divided into quantiles based on bacterial survival. (A) NTHi strains were divided into quantiles based on bacterial survival in 5% NHS. (B and C) Binding of IgG (B) and IgM (C) to the surface of NTHi strains divided into quantiles. We used ANOVA to compare the groups.
Binding of IgM to the bacterial surface is associated with increased bacterial killing of NTHi.
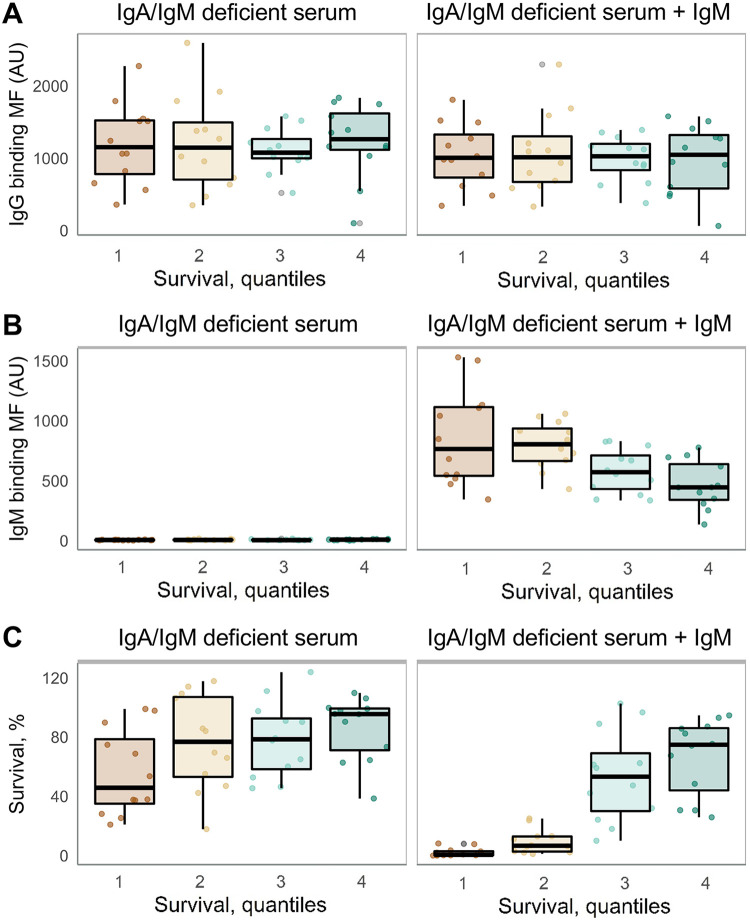
Previously, we used IgA/IgM-deficient serum to determine the contribution of IgM in complement-mediated killing of NTHi strains (4, 9, 10). In order to determine the contribution of IgM in complement-mediated killing of invasive NTHi strains, we determined binding of IgG and IgM to the bacterial surface and bacterial killing of 48 NTHi strains divided into four equal groups based on the quantiles for survival in 5% NHS (Fig. 3A).
Binding of IgG to the surface of NTHi strains was not associated with bacterial survival using IgA/IgM-deficient serum with or without supplementation of IgM (Fig. 4A). Binding of IgM was absent with IgA/IgM-deficient serum, but supplementation of IgA/IgM-deficient serum with IgM resulted in binding of IgM to the surface of NTHi strains, which was inversely associated with bacterial survival (Fig. 4B). Bacterial survival rates in IgA/IgM-deficient serum for quantiles 1 through 4 were 45.8% (25% to 75% quantiles, 20.9 to 86.0), 76.8% (25% to 75% quantiles, 17.8 to 108), 78.4% (25% to 75% quantiles, 45.5 to 95.8), and 95.5% (25% to 75% quantiles, 38.5 to 99.5), respectively (Fig. 4C). Supplementation of IgA/IgM-deficient serum with IgM decreased survival for quantiles 1 through 4 to 0.56% (25% to 75% quantiles, 0.18 to 3.31), 6.6% (25% to 75% quantiles, 2.36 to 13.0), 53.2% (25% to 75% quantiles, 26.6 to 82.3), and 74.8% (25% to 75% quantiles, 35.1 to 86.7), respectively (Fig. 4C). These results show that binding of IgM to the surface of NTHi has a major contribution in complement-mediated killing and that NTHi strains with low binding of IgM present the highest survival rates in human serum.
FIG 4.
IgG antibody binding, IgM antibody binding, and survival of 48 NTHi strains using IgA/IgM-deficient serum supplemented or not with IgM. NTHi strains were divided into quantiles based on bacterial survival in 5% NHS. Shown are binding of IgG (A) and IgM (B) to the bacterial surface and survival (C) of NTHi strains divided into quantiles. We used ANOVA to compare the groups.
DISCUSSION
We examined the association between binding of IgG or IgM to the surface of H. influenzae strains and survival in normal human serum. Survival of H. influenzae in the blood of patients is largely dependent on two mechanisms: (i) bacterial resistance to complement-mediated killing and (ii) the immune competence of the host. In this study, we focused on bacterial resistance to complement-mediated killing. We have used a single batch of pooled NHS containing opsonizing IgG and IgM antibodies recognizing NTHi protein and lipooligosaccharide (LOS) epitopes, as tested previously (11). We could not take into account the patients’ immune status in the development of invasive NTHi infection due to the unavailability of serum samples from affected individuals.
We showed that binding of IgM to the surface of NTHi strains has a small impact on bacterial survival in normal human serum, whereas we found no association between IgG binding and bacterial survival (Fig. 3). The results of this study are consistent with previously published studies revealing the impact of IgM opsonization on the immune response against NTHi, including opsonophagocytosis and complement-mediated killing (4–6). Previously, Nakamura et al. showed that NTHi strains from the lower respiratory tract were significantly more resistant to complement-mediated killing than NTHi strains from the upper respiratory tract, and this was associated with reduced binding of serum IgM to the bacterial surface (6). We showed that NTHi strains collected from the middle ear fluid showed increased resistance to complement-mediated killing, which coincided with a lower binding of serum IgM to the bacterial surface (5). We also observed that NTHi strains collected from blood were more resistant to complement-mediated killing and bound less IgM than strains collected from the upper respiratory tracts of age-matched healthy adults (4).
Bacterial mechanisms that result in evasion of IgM binding to the bacterial surface are diverse. For instance, incorporation of sialic acids decreases the binding of IgM to the bacterial surface (9), possibly due to covering galactose residues extending on heptose III of the LOS structure (12). Decreased incorporation of phosphorylcholine into the LOS of NTHi decreased the binding of IgM as well (10), which was consistent with having a lower phosphorylcholine expression and IgM binding to the surface of NTHi strains collected from the blood of a patient with bacteremia (4). Outer membrane protein P5 (OmpP5) has been shown to decrease the binding of IgM to the bacterial surface (13). Finally, Nakamura and colleagues showed that retrograde trafficking of phospholipids from the outer to the inner leaflet of the cell envelope decreased the binding of IgM to the bacterial surface (6).
All of the above-mentioned mechanisms were identified by in vitro experiments, but the importance of anti-NTHi IgM antibodies in the immune defense against NTHi infections is also supported by clinical observations in patients. Individuals with hyper-IgM syndrome had a significantly lower risk of NTHi carriage, confirming that IgM can play a protective role independently of other antibody isotypes (7). Conversely, low serum IgM (<0.05 g/liter) was a risk factor for respiratory tract colonization by NTHi in patients with CVID (8).
We found no association between IgG binding and bacterial survival, indicating that binding of IgG to the surface of NTHi, at least in our in vitro experimental setting using 5% serum, is not sufficient to initiate complement-mediated killing. The lack of complement activation by IgG might be partly explained by the fact that IgG recognizing sugar epitopes such as LOS, but also type B polysaccharide, is mainly IgG2 (14, 15), which is poor in complement activation and killing of H. influenzae (16). Therefore, binding of IgM, which is more efficient in complement activation (17), has a larger contribution in overall complement-mediated killing of NTHi.
The limited role for IgG in killing of NTHi is in contrast to serum bactericidal activity (SBA) assays for Neisseria meningitidis (18, 19) or Hib (20) used to determine vaccine responses. However, in these SBA assays, the individual contribution of either IgG or IgM is not determined because serum containing both IgG and IgM is used. Kim et al. showed that postvaccination anti-Hib IgG levels correlated well with Hib SBA titers (21). Baggett et al. showed that SBA Hib prevaccination titers were positively correlated with log IgM levels, indicating that IgM can contribute in SBA activity, at least in unvaccinated individuals (22).
Although this study found that binding of IgM to the bacterial surface may be a determinant for the complement-mediated killing of NTHi strains, the association was weak. The distributions of IgM binding were largely overlapping for NTHi strains in all survival quantile groups. Other mechanisms are known to play a role in bacterial survival in human serum. For instance, binding of IgG was shown to be decreased by incorporation of phosphorylcholine (23), and binding of human complement regulatory proteins, including factor H and C4-binding protein, can affect complement-mediated killing (13, 24, 25). Besides the known studied mechanisms, other, yet-unknown mechanisms can contribute to complement evasion, as illustrated by a large number of genes identified in genome-wide mutant screening studies (5, 6).
An interesting observation is that Hib and Hif bind equal levels of IgG and IgM, yet killing of Hif strains is much more pronounced than that of Hib strains (Fig. 2). This can, at least partly, be explained by the fact that the Hib capsule prevents SBA activity better than Hif in capsule switch mutants (26), but other immune evasion mechanisms different between Hib and Hif are likely to contribute as well. In addition, differences in IgG subclass might also contribute to differences in complement-mediated killing; for instance, binding of IgG2 to Hib polysaccharide capsule limits complement-mediated killing (14).
Since a compromised immune status is expected to contribute to the increased risk of bacterial infections, it would be beneficial to understand whether there are particular immune dysfunctions that make the host susceptible to acquiring invasive NTHi. Considering this, it is interesting to investigate whether or not underlying medical conditions influence the immune response against NTHi and lead to invasive infection, especially among the elderly, who are mostly affected according to our data and other literature (27, 28). Moreover, it is known that serum IgG levels against pathogens are reduced in the elderly (29). However, it is not yet clearly established whether the level of antibody binding to the bacterial surface alone might be a sufficient reason for the development of invasive disease caused by NTHi or whether the decline in immune function requires a combination with increased bacterial virulence to cause invasive infections. The contribution of bacterial virulence is supported by the fact that healthy adults without known underlying medical conditions can also present with invasive NTHi infections (30, 31). A large genetic variability among NTHi strains, in combination with phase-variable expression of genes, and the ability to utilize multiple mechanisms of escaping immune response makes NTHi an interesting and challenging subject for research.
Although most invasive nontypeable Haemophilus strains are found to be H. influenzae, invasive Haemophilus haemolyticus has also been found (32, 33). Haemophilus strains from Spain were typed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), which showed concordance with the absence of the lgtC gene in 65 out of 68 H. haemolyticus strains (32). However, strains from Portugal were not always determined by MALDI-TOF MS, and for Haemophilus strains from the Netherlands, a hemolysis test was performed to differentiate between Haemophilus species. Therefore, it is possible that our collection contains H. haemolyticus strains. However, since H. haemolyticus strains are rarely seen in blood (32–34), we expect that most strains are actually NTHi.
In summary, we have shown that invasive NTHi bacteria opsonized with low levels of IgM showed the highest survival in normal human serum. Evasion of IgM binding may be an important mechanism for invasive NTHi strains to prevent the complement-mediated killing.
MATERIALS AND METHODS
Ethics statement.
This was a retrospective observational study using bacterial strains obtained as part of the Reference Laboratory for Bacterial Meningitis routine work. Written informed consent is not required by the Dutch legislation for this type of research. All the data were anonymized and protected according to Dutch national guidelines.
Strain collections and culture conditions.
One hundred sixty-one invasive H. influenzae strains from the Netherlands were collected in 2014 by the Reference Laboratory for Bacterial Meningitis. Haemophilus strains were typed by slide agglutination. When negative, factor V and X growth requirement, delta-aminolevulinic acid differentiation disk test, and hemolysis test were performed to differentiate between Haemophilus species. Fifty-one invasive H. influenzae strains from Spain were obtained from the collections in the Hospital Universitario Bellvitge (Barcelona) and Hospital Universitario Donostia (San Sebastian) from 2013 to 2015. Capsulated and noncapsulated strains were determined by slide agglutination using omniserum and capsulated H. influenzae strains were typed by PCR (35). Discrimination between H. influenzae and H. haemolyticus was determined by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) using a MALDI Biotyper (Bruker Daltonics) (32). Fifty-five invasive H. influenzae strains from Portugal were obtained from the collection of the Portuguese National Institute of Health from August 2013 to June 2015. Haemophilus strains were typed by PCR (35). Whenever necessary, H. influenzae identification was confirmed by MALDI-TOF MS (32). Available clinical data included source and date of the sample collected and age and sex of the patients.
We transferred H. influenzae strains from the primary stock to brain heart infusion (BHI; BD Biosciences) supplemented with 10 μg/ml of hemin (Sigma) and 2 μg/ml of NAD (Merck) (sBHI) agar plates, where they were grown at 37°C with 5% CO2 overnight. Next, we grew bacteria in sBHI liquid medium at 37˚C with shaking at 250 rpm to an optical density of 0.9 to 1.1 measured at 620 nm (Tecan; Infinite F50). We stored 1-ml aliquots of bacteria in sBHI with 20% glycerol at −80°C.
Serum sources.
We conducted experiments with a single batch of pooled normal human serum (NHS) (Immucor; lot no. 2131U; PHS-N100). Serum from an agammaglobulinemia patient on IgG replacement therapy was used as IgA/IgM-deficient serum (36). Purified IgM from human serum (Sigma-Aldrich; I8260) was washed with phosphate-buffered saline (PBS) on an Amicon Ultra-0.5 centrifugal filter unit column (Millipore) to remove the preservative sodium azide and suspended in PBS at a concentration of 1 mg/ml. For IgM supplementation experiments, 10% IgA/IgM-deficient serum was mixed with 0.1 mg/ml of IgM.
Serum bactericidal assay.
For capsulated strains, we used a final concentration of 10% NHS, and for NTHi strains, we used a final concentration of 5% NHS. We washed H. influenzae from −80°C storage with PBS and diluted in Hank’s balanced salt solution (HBSS+Ca2+/Mg2+) with 0.1% gelatin (HBSS3+) such that the bacterial concentration would be ∼2 × 105 CFU/ml. We mixed 50 μl of diluted H. influenzae with either 50 μl of NHS, IgA/IgM-deficient serum, IgA/IgM-deficient serum plus IgM, heat-inactivated (30 min at 56°C) NHS (HI-NHS), HI-IgA/IgM-deficient serum, or HI-IgA/IgM-deficient serum plus IgM and incubated the mixture for 60 min at 37°C with 5% CO2. After incubation, series of 10-fold dilutions were made and 3 droplets with a total volume of 60 μl of each dilution of H. influenzae exposed to either NHS or HI-NHS was plated on sBHI agar plates and incubated overnight at 37°C with 5% CO2; the CFU counts were determined the following morning. We computed the survival percentage of H. influenzae strains by dividing the number of CFU after exposure to NHS by the number of CFU after exposure to HI-NHS. We repeated serum bactericidal assays three times for each strain and calculated geometric means of percent survival in NHS.
Flow cytometry.
We transferred 50 μl of H. influenzae from −80°C storage to a v-bottom 96-well plate, pelleted the organisms by centrifugation at 3,200 × g, washed them once with PBS, suspended them in 100 μl of 10% HI-NHS, HI-IgA/IgM-deficient serum, or HI-IgA/IgM-deficient serum plus IgM diluted in HBSS3+ as a source of IgG and IgM antibodies, and incubated the strains for 30 min at 37˚C with 5% CO2. We pelleted the bacteria by centrifugation at 3,200 × g. To determine binding of IgG and IgM to the bacterial surface, we suspended the bacterial strains in 100 μl of PBS with 2% bovine serum albumin (BSA) containing either 1:100-diluted fluorescein isothiocyanate (FITC)-labeled anti-human IgG (Sigma; F0132), 1:100-diluted FITC-labeled anti-human IgM (Sigma; F5384), or 1:500-diluted phycoerythrin (PE)-labeled anti-human IgG (Jackson ImmunoResearch) with 1:500-diluted Alexa Fluor 647-labeled anti-human IgM (Jackson ImmunoResearch) at 4°C for 30 min. We pelleted the bacteria by centrifugation at 3,200 × g and fixed the bacteria with 100 μl of PBS containing 2% paraformaldehyde for 20 min at room temperature (RT). Next, we pelleted the bacteria by centrifugation at 3,200 × g and suspended them in 1:2,500-diluted nucleic acid stain SYTO40 (Thermo Fisher Scientific Inc.) in PBS as bacterial DNA staining and incubated them for 5 min. We diluted stained samples 20-fold in PBS and performed flow cytometry with a FACS LSRII (BD Biosciences) using the following settings: forward scatter (FSC) = 200, side scatter (SSC) = 200, and FSC threshold = 200. We repeated flow cytometry three times for each of the strains and computed geometric means of IgG and IgM binding for each bacterial strain.
Statistical analyses.
Statistical analyses were performed with R (37).
ACKNOWLEDGMENTS
Author contributions are as follows: conceptualization, M.I.D.J. and J.D.L.; investigation, E.D., L.D.S., G.J.A.V., and J.D.L.; formal analysis, E.D. and J.D.L.; resources, A.V.D.E., J.M.M., P.B.-L., C.A., and S.M.; writing of original draft, J.D.L.; writing review and editing, all authors.
We declare that we have no competing financial interest.
REFERENCES
- 1.Adams WG, Deaver KA, Cochi SL, Plikaytis BD, Zell ER, Broome CV, Wenger JD. 1993. Decline of childhood Haemophilus influenzae type b (Hib) disease in the Hib vaccine era. JAMA 269:221–226. doi: 10.1001/jama.1993.03500020055031. [DOI] [PubMed] [Google Scholar]
- 2.Langereis JD, de Jonge MI. 2015. Invasive disease caused by nontypeable Haemophilus influenzae. Emerg Infect Dis 21:1711–1718. doi: 10.3201/eid2110.150004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whittaker R, Economopoulou A, Dias JG, Bancroft E, Ramliden M, Celentano LP, European Centre for Disease Prevention and Control Country Experts for Invasive Haemophilus influenzae Disease. 2017. Epidemiology of invasive Haemophilus influenzae disease, Europe, 2007–2014. Emerg Infect Dis 23:396–404. doi: 10.3201/eid2303.161552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langereis JD, Cremers AJH, Vissers M, van Beek J, Meis JF, de Jonge MI. 2018. Nontypeable Haemophilus influenzae invasive blood isolates are mainly phosphorylcholine negative and show decreased complement-mediated killing that is associated with lower binding of IgM and CRP in comparison to colonizing isolates from the oropharynx. Infect Immun 87:e00604-18. doi: 10.1128/IAI.00604-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langereis JD, Stol K, Schweda EK, Twelkmeyer B, Bootsma HJ, de Vries SP, Burghout P, Diavatopoulos DA, Hermans PW. 2012. Modified lipooligosaccharide structure protects nontypeable Haemophilus influenzae from IgM-mediated complement killing in experimental otitis media. mBio 3:e00079-12. doi: 10.1128/mBio.00079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura S, Shchepetov M, Dalia AB, Clark SE, Murphy TF, Sethi S, Gilsdorf JR, Smith AL, Weiser JN. 2011. Molecular basis of increased serum resistance among pulmonary isolates of non-typeable Haemophilus influenzae. PLoS Pathog 7:e1001247. doi: 10.1371/journal.ppat.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Micol R, Kayal S, Mahlaoui N, Beauté J, Brosselin P, Dudoit Y, Obenga G, Barlogis V, Aladjidi N, Kebaili K, Thomas C, Dulieu F, Monpoux F, Nové-Josserand R, Pellier I, Lambotte O, Salmon A, Masseau A, Galanaud P, Oksenhendler E, Tabone M-D, Teira P, Coignard-Biehler H, Lanternier F, Join-Lambert O, Mouillot G, Theodorou I, Lecron J-C, Alyanakian M-A, Picard C, Blanche S, Hermine O, Suarez F, Debré M, Lecuit M, Lortholary O, Durandy A, Fischer A. 2012. Protective effect of IgM against colonization of the respiratory tract by nontypeable Haemophilus influenzae in patients with hypogammaglobulinemia. J Allergy Clin Immunol 129:770–777. doi: 10.1016/j.jaci.2011.09.047. [DOI] [PubMed] [Google Scholar]
- 8.Pulvirenti F, Camilli R, Giufre M, Milito C, Pimentel de Araujo F, Mancini F, Cardines R, Ciervo A, Pantosti A, Cerquetti M, Quinti I. 2018. Risk factors for Haemophilus influenzae and pneumococcal respiratory tract colonization in CVID. J Allergy Clin Immunol 142:1999–2002.e1993. doi: 10.1016/j.jaci.2018.08.014. [DOI] [PubMed] [Google Scholar]
- 9.Oerlemans MMP, Moons SJ, Heming JJA, Boltje TJ, de Jonge MI, Langereis JD. 2019. Uptake of sialic acid by nontypeable Haemophilus influenzae increases complement resistance through decreasing IgM-dependent complement activation. Infect Immun 87:e00077-19. doi: 10.1128/IAI.00077-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langereis JD, van der Pasch ES, de Jonge MI. 2019. Serum IgM and C-reactive protein binding to phosphorylcholine of non-typeable Haemophilus influenzae increases complement-mediated killing. Infect Immun 87:e00299-19. doi: 10.1128/IAI.00299-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langereis JD, van Dongen TM, Stol K, Venekamp RP, Schilder AG, Hermans PW. 2013. Resistance to complement-mediated killing and IgM binding to non-typeable Haemophilus influenzae is not altered when ascending from the nasopharynx to the middle ears in children with otitis media. Med Microbiol Immunol 202:407–415. doi: 10.1007/s00430-013-0302-5. [DOI] [PubMed] [Google Scholar]
- 12.Jackson MD, Wong SM, Akerley BJ. 2019. Underlying glycans determine the ability of sialylated lipooligosaccharide to protect nontypeable Haemophilus influenzae from serum IgM and complement. Infect Immun 87:e00456-19. doi: 10.1128/IAI.00456-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosadini CV, Ram S, Akerley BJ. 2014. Outer membrane protein P5 is required for resistance of nontypeable Haemophilus influenzae to both the classical and alternative complement pathways. Infect Immun 82:640–649. doi: 10.1128/IAI.01224-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston RB Jr, Anderson P, Rosen FS, Smith DH. 1973. Characterization of human antibody to polyribophosphate, the capsular antigen of Hemophilus influenzae, type B. Clin Immunol Immunopathol 1:234–240. doi: 10.1016/0090-1229(73)90024-X. [DOI] [PubMed] [Google Scholar]
- 15.Goldenberg HB, McCool TL, Weiser JN. 2004. Cross-reactivity of human immunoglobulin G2 recognizing phosphorylcholine and evidence for protection against major bacterial pathogens of the human respiratory tract. J Infect Dis 190:1254–1263. doi: 10.1086/424517. [DOI] [PubMed] [Google Scholar]
- 16.Amir J, Scott MG, Nahm MH, Granoff DM. 1990. Bactericidal and opsonic activity of IgG1 and IgG2 anticapsular antibodies to Haemophilus influenzae type b. J Infect Dis 162:163–171. doi: 10.1093/infdis/162.1.163. [DOI] [PubMed] [Google Scholar]
- 17.Raff HV, Bradley C, Brady W, Donaldson K, Lipsich L, Maloney G, Shuford W, Walls M, Ward P, Wolff E. 1991. Comparison of functional activities between IgG1 and IgM class-switched human monoclonal antibodies reactive with group B streptococci or Escherichia coli K1. J Infect Dis 163:346–354. doi: 10.1093/infdis/163.2.346. [DOI] [PubMed] [Google Scholar]
- 18.Borrow R, Balmer P, Miller E. 2005. Meningococcal surrogates of protection—serum bactericidal antibody activity. Vaccine 23:2222–2227. doi: 10.1016/j.vaccine.2005.01.051. [DOI] [PubMed] [Google Scholar]
- 19.Goldschneider I, Gotschlich EC, Artenstein MS. 1969. Human immunity to the meningococcus. I. The role of humoral antibodies. J Exp Med 129:1307–1326. doi: 10.1084/jem.129.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlesinger Y, Granoff DM, Murphy TV, Osterholm MT, McHugh JE, Anderson R, Esbenshade J, Blatter M, Reisinger KS, Keyserling H, Guito K, Meschievitz CK, the Vaccine Study Group. 1992. Avidity and bactericidal activity of antibody elicited by different Haemophilus influenzae type b conjugate vaccines. JAMA 267:1489–1494. doi: 10.1001/jama.267.11.1489. [DOI] [PubMed] [Google Scholar]
- 21.Kim HW, Kim KH, Kim J, Nahm MH. 2016. A high throughput serum bactericidal assay for antibodies to Haemophilus influenzae type b. BMC Infect Dis 16:473. doi: 10.1186/s12879-016-1808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baggett HC, Hennessy TW, Bulkow L, Romero-Steiner S, Hurlburt D, Holder P, Parkinson AJ, Singleton RJ, Levine O, Carlone GM, Butler JC. 2006. Immunologic response to Haemophilus influenzae type b (Hib) conjugate vaccine and risk factors for carriage among Hib carriers and noncarriers in southwestern Alaska. Clin Vaccine Immunol 13:620–626. doi: 10.1128/CVI.00077-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark SE, Snow J, Li J, Zola TA, Weiser JN. 2012. Phosphorylcholine allows for evasion of bactericidal antibody by Haemophilus influenzae. PLoS Pathog 8:e1002521. doi: 10.1371/journal.ppat.1002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hallstrom T, Jarva H, Riesbeck K, Blom AM. 2007. Interaction with C4b-binding protein contributes to nontypeable Haemophilus influenzae serum resistance. J Immunol 178:6359–6366. doi: 10.4049/jimmunol.178.10.6359. [DOI] [PubMed] [Google Scholar]
- 25.Langereis JD, de Jonge MI, Weiser JN. 2014. Binding of human factor H to outer membrane protein P5 of non-typeable Haemophilus influenzae contributes to complement resistance. Mol Microbiol 94:89–106. doi: 10.1111/mmi.12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swift AJ, Moxon ER, Zwahlen A, Winkelstein JA. 1991. Complement-mediated serum activities against genetically defined capsular transformants of Haemophilus influenzae. Microb Pathog 10:261–269. doi: 10.1016/0882-4010(91)90010-8. [DOI] [PubMed] [Google Scholar]
- 27.Blain A, MacNeil J, Wang X, Bennett N, Farley MM, Harrison LH, Lexau C, Miller L, Nichols M, Petit S, Reingold A, Schaffner W, Thomas A, Clark T, Cohn A, Briere E. 2014. Invasive Haemophilus influenzae disease in adults >/=65 years, United States, 2011. Open Forum Infect Dis 1:ofu044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ladhani S, Slack MP, Heath PT, von Gottberg A, Chandra M, Ramsay ME, European Union Invasive Bacterial Infection Surveillance participants. 2010. Invasive Haemophilus influenzae disease, Europe, 1996–2006. Emerg Infect Dis 16:455–463. doi: 10.3201/eid1603.090290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hawdon N, Biman B, McCready W, Brigden M, Malik S, Vergidis D, Kisselgoff O, Ulanova M. 2012. Antibody against Haemophilus influenzae protein D in patients with chronic conditions causing secondary immunodeficiency. Vaccine 30:1235–1238. doi: 10.1016/j.vaccine.2011.12.113. [DOI] [PubMed] [Google Scholar]
- 30.RIVM/AMC. 2014. Bacterial meningitis in the Netherlands. Reference Laboratory for Bacterial Meningitis in the Netherlands, Amsterdam, the Netherlands. [Google Scholar]
- 31.Brown VM, Madden S, Kelly L, Jamieson FB, Tsang RS, Ulanova M. 2009. Invasive Haemophilus influenzae disease caused by non-type b strains in Northwestern Ontario, Canada, 2002–2008. Clin Infect Dis 49:1240–1243. doi: 10.1086/605671. [DOI] [PubMed] [Google Scholar]
- 32.Marti S, Puig C, de la Campa AG, Tirado-Velez JM, Tubau F, Domenech A, Calatayud L, Garcia-Somoza D, Ayats J, Liñares J, Ardanuy C. 2016. Identification of Haemophilus haemolyticus in clinical samples and characterization of their mechanisms of antimicrobial resistance. J Antimicrob Chemother 71:80–84. doi: 10.1093/jac/dkv307. [DOI] [PubMed] [Google Scholar]
- 33.Anderson R, Wang X, Briere EC, Katz LS, Cohn AC, Clark TA, Messonnier NE, Mayer LW. 2012. Haemophilus haemolyticus isolates causing clinical disease. J Clin Microbiol 50:2462–2465. doi: 10.1128/JCM.06575-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy TF, Brauer AL, Sethi S, Kilian M, Cai X, Lesse AJ. 2007. Haemophilus haemolyticus: a human respiratory tract commensal to be distinguished from Haemophilus influenzae. J Infect Dis 195:81–89. doi: 10.1086/509824. [DOI] [PubMed] [Google Scholar]
- 35.Falla TJ, Crook DW, Brophy LN, Maskell D, Kroll JS, Moxon ER. 1994. PCR for capsular typing of Haemophilus influenzae. J Clin Microbiol 32:2382–2386. doi: 10.1128/JCM.32.10.2382-2386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langereis JD, Henriet SS, Kuipers S, Weemaes CMR, van der Burg M, de Jonge MI, van der Flier M. 2018. IgM augments complement bactericidal activity with serum from a patient with a novel CD79a mutation. J Clin Immunol 38:185–192. doi: 10.1007/s10875-017-0474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]