Abstract
The ongoing SARS-CoV-2 pandemic is a global public health emergency posing a high burden on nations’ health care systems and economies. Despite the great effort put in the development of vaccines and specific treatments, no prophylaxis or effective therapeutics are currently available. Nitric oxide (NO) is a broad-spectrum antimicrobial and a potent vasodilator that has proved to be effective in reducing SARS-CoV replication and hypoxia in patients with severe acute respiratory syndrome. Given the potential of NO as treatment for SARS-CoV-2 infection, we have evaluated the in vitro antiviral effect of NO on SARS-CoV-2 replication. The NO-donor S-nitroso-N-acetylpenicillamine (SNAP) had a dose dependent inhibitory effect on SARS-CoV-2 replication, while the non S-nitrosated NAP was not active, as expected. Although the viral replication was not completely abolished (at 200 μM and 400 μM), SNAP delayed or completely prevented the development of viral cytopathic effect in treated cells, and the observed protective effect correlated with the level of inhibition of the viral replication. The capacity of the NO released from SNAP to covalently bind and inhibit SARS-CoV-2 3CL recombinant protease in vitro was also tested. The observed reduction in SARS-CoV-2 protease activity was consistent with S-nitrosation of the enzyme active site cysteine.
Keywords: SARS-CoV-2, COVID-19, Nitric oxide, 3CL protease, FRET
Graphical abstract
1. Introduction
In late 2019 a viral pneumonia, the coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) rapidly spread around the world and became classified as the second pandemic of the 21st century by the World Health Organization on March 11, 2020 [1]. SARS-CoV-2 belongs to the betacoronavirus genus and is, together with the severe acute respiratory syndrome coronavirus (SARS-CoV) and the middle-east respiratory syndrome (MERS), the third coronavirus (CoV) emerging from animals to humans within less than two decades [1]. The nucleotide sequence similarity between SARS-CoV-2 and SARS-CoV is about 79% and between SARS-CoV-2 and MERS-CoV about 50% [2].
Given the severity and the rapid spread of SARS-CoV-2, including a high proportion of asymptomatic carriers [3], it is of clear importance to find effective therapeutics against COVID-19. Despite extensive efforts to treat the disease and given the limitation in usage and effectiveness of remdesivir [4], the development of therapeutic interventions is hindered by the lack of effective antiviral drugs against SARS-CoV-2.
However, nitric oxide (NO), known to have a broad antimicrobial effect against bacteria, fungi, helminths, protozoa and viruses, is a promising compound [5]. Antiviral effects have been described against both DNA and RNA viruses including Herpesvirus [6], Coxsackie virus [7], and Hantavirus [8]. Moreover, NO has been demonstrated as an effective antiviral against SARS-CoV in vitro [9,10] and in vivo by inhalation in very low concentrations in a small clinical trial [11]. Furthermore, inhaled NO improved arterial oxygenation in hypoxemic patients by redistributing blood flow in the lung to better ventilated regions, and counteracted blood clotting, both effects being of importance for COVID-19 patients [12]. The role of NO-inhalation in the prevention and treatment of COVID-19 has been proposed [13], but no clinical data has yet been reported.
In the present study the antiviral effect of NO on SARS-CoV-2 infected cells was tested in vitro. The potential mechanism of action of NO as cysteine protease inhibitor, previously reported for Coxsackievirus 3C cysteine protease [7], was furthermore examined on SARS-CoV-2 3CL cysteine protease.
2. Material and methods
2.1. Virus yield reduction assay
The inhibitory effect of nitric oxide (NO) on the replication of SARS-CoV-2 was assessed performing a yield reduction assay combined with RT-qPCR for the quantification of viral RNA. Confluent Vero E6 cells, seeded in 12-wells plates, were inoculated with 0.005 multiplicity of infection (MOI) of SARS-CoV-2 isolated in our laboratory [3] and at the same time treated with either 200 μM or 400 μM of the nitric oxide (NO) donor S-nitroso-N-acetylpenicillamine (SNAP) (Sigma-Aldrich, product number N3398) or its non S-nitrosated version NAP (Sigma-Aldrich, product number 01423). The chosen SNAP and NAP concentrations were in the same range as used in previous SARS-CoV in vitro tests [9,10].
The experiment was carried out in triplicates keeping the final volume in each well at 1 ml. Viral replication kinetics and development of cytopathic effect (CPE) were followed up to 72 h post infection (hpi). During the first 36 h, supernatants were collected, and the cells were re-treated with SNAP or NAP every 4 h. At each time-point 500 μl of supernatants were collected and subsequently 500 μl new cell media containing 200 μM or 400 μM of SNAP or NAP, respectively, were added to the wells, which effectively adds half of the final tested concentration and accounts for SNAP's short half-life of approximately 4.6 h [14]. At 48 and 72 hpi 500 μl supernatant was collected and subsequently cell media without any added compounds was added. Infected but non-treated cells, treated but non-infected cells, as well as cell controls were handled as explained above by collecting 500 μl of supernatant and adding 500 μl of cell media with or without compounds. Viral RNA was extracted from the collected supernatants and the copy numbers of the viral genome was quantified by RT-qPCR. All experiments involving infection of cells with SARS-CoV-2 virus were performed in a BSL3 lab at the Zoonosis Science Center, IMBIM, Uppsala University.
2.2. RT-qPCR assay
RNA was extracted using the Direct-zol™-96 RNA kit according to the manufacturer's protocol (Zymo Research, USA). The Envelope (E) gene was quantified by the RT-qPCR assay as described previously [3,15]. The primer pair and probe were based on sequences published by Corman et al. [16]. Forward primer ACAGGTACGTTA ATAGTTAATAGCGT; reverse primer TGTGTGCGTACTGCTGCAATAT; and the probe 5′-FAM-ACACTAGCCATCC TTACTGCGCTTCG-TAMRA-3′ were used. The RT-qPCR was run using the Thermofisher/Invitrogen SuperScript III OneStep RT-PCR System with Platinum® Taq DNA Polymerase kit (Cat. No. 12574026) according to manufacturer's instructions with an adjusted magnesium sulfate concentration. The reaction mixture contained 12.5 μl of reaction buffer, 0.5 μl of enzyme solution, 1.25 μl of Probe primers solution, 2.4 μl 25 nM magnesium sulfate, 3.35 μL of nuclease-free water, and 5 μl of RNA template. The RT-PCR was performed under the following conditions: incubation at 55 °C for 30 min and 95 °C for 3 min followed by 45 cycles of denaturation at 95 °C for 15 s, extension at 57 °C for 30 s and collecting the fluorescence signal at 68 °C for 30 s.
The corresponding viral genome copy number to each Ct value was calculated based on a curve equation generated with known standards: DNA gene fragments corresponding to the amplified region of the viral genome were designed as templates and diluted to known concentrations of 106 to 100 copies per μl. A standard curve plotted to the known concentrations was then created by performing qPCR on serial dilutions of the templates.
2.3. Expression and purification of SARS-CoV-2 3CL protease
The recombinant SARS-CoV-2 3CL protease was produced adopting a published construct used for the expression of SARS-CoV 3CL protease [17], containing nucleotide sequences corresponding to residues S1-Q306 (Chinese isolate, NCBI accession number YP_009725301). A detailed protocol is described in the supporting information.
2.4. In vitro enzymatic assay
Recombinant SARS-CoV-2 3CL protease was incubated with either SNAP or NAP at different concentrations for 10 min at room temperature. The FRET substrate DABCYL-Lys-Thr-Ser-Ala-Val-Leu-Gln-Ser-Gly-Phe-Arg-Lys-Met-Glu-EDANS (Bachem Holding AG, Switzerland) was added to start the enzymatic reaction. The fluorescence emission was monitored every 60 s for 35 min at 37 °C. The relative fluorescence units (RFU) per second were plotted and the initial velocities were calculated, normalized, and used to calculate the half maximal inhibitory concentration (IC50). A detailed protocol is provided in the supplementary material. The data analysis was conducted in GraphPad Prism (v.6.0).
2.5. Statistical analysis
Sars-CoV-2 replication kinetic curves were analyzed by calculating the mean of all measurements for treated and untreated groups replicates while SARS-CoV-2 3CL protease enzymatic assay time response curves were analyzed by calculating the area under the curve for each replicate. The calculated means of all measurements and areas under the curves were compared for statistical significance by One-Way ANOVA followed by Tukey's test for multiple comparison. Mean inhibition values (%) of treated and control groups from inhibition kinetic curve and dose response column graph were analyzed by One-Way ANOVA followed by Tukey's test for multiple comparison. Differences in mean areas under the curves and inhibition values were considered statistically significant when p < 0.05. Statistical analysis was performed using GraphPad Prism (v.6.0).
3. Results and discussion
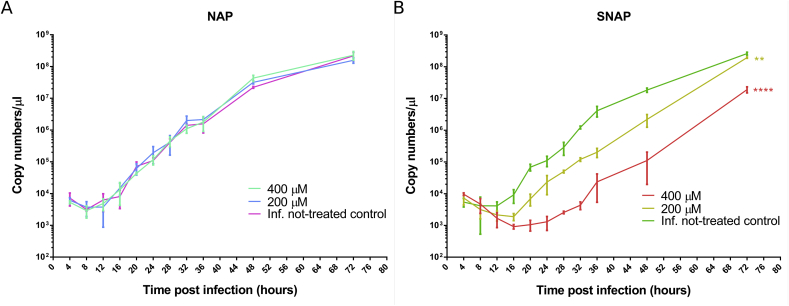
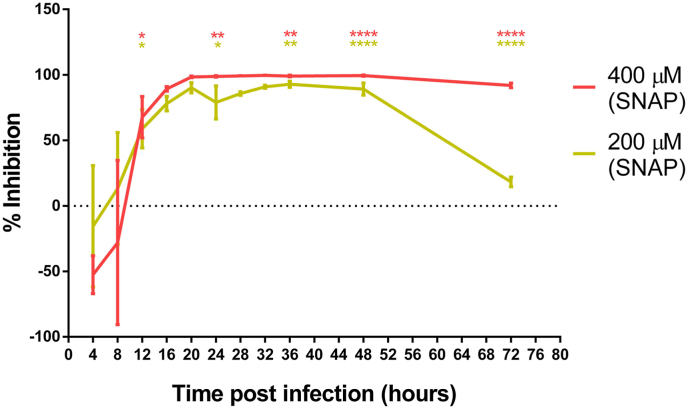
The inhibitory effect of NO on the replication of SARS-CoV-2 was evaluated by performing a yield reduction assay by RT-qPCR for the quantification of the viral RNA (Supplementary Table 1). Viral replication kinetics and development of CPE were followed up to 72 hpi. SNAP (NO donor), but not NAP (NO-lacking version of SNAP) showed a dose dependent reduction of SARS-CoV-2 viral RNA copy numbers (Fig. 1), which proves that NO has an antiviral effect against SARS-CoV-2 and probably in the similar manner as previously described for SARS-CoV and other viruses [[6], [7], [8], [9]]. The viral replication life cycle was almost blocked by 400 μM NO up to 24 hpi. After termination of the SNAP treatment at 36 hpi, the level of viral genome copy numbers significantly increased (Fig. 1). At 36 hpi (last time point for re-treatment), treatment by 400 μM and 200 μM of SNAP resulted in 99.42% (±0.44 SD) and 95.07% (±1.58 SD) SARS-CoV-2 replication inhibition respectively (Fig. 2) as compared to the infected not-treated controls. At 72 hpi (36 h after the last re-treatment), the inhibition of viral replication for the 200 μM SNAP treatment decreased to 25.10% (±3.37 SD), while for the 400 μM SNAP treatment, the inhibition was still above 90% (92.64 ± 1.59 SD). Given that the cell treatment slowed and not stopped viral replication, the rebound of viral replication after the interruption of treatment could be explained with the gradual decrease in SNAP (due to the release of NO) and NO concentrations.
Fig. 1.
SARS-CoV-2 replication kinetics in Vero-E6 cells in the presence and absence of NAP (A) and SNAP (B) as expressed by viral RNA copy numbers/μl (mean values from triplicates with standard deviation are indicated at each time point). In the presence of NAP no significant difference in viral replication between treated cells and not-treated controls was observed. A dose dependent inhibitory effect of SNAP on SARS-CoV-2 replication was observed. Data was analyzed by One-Way ANOVA of the calculated mean of the copy numbers for each replicate. Statistical significance is reported as **p < 0.001, ****p < 0.0001 (as compared with the controls).
Fig. 2.
SNAP inhibitory effect plotted as percentage of the viral replication reduction over time. At a concentration of 400 μM of SNAP inhibited the viral replication after the treatment was terminated (36 hpi). For the two tested concentrations, mean values from triplicates with standard deviation are indicated at each time point. Average viral replication inhibition values (%) at 12, 24, 36, 48 and 72 hpi were compared by One-Way ANOVA. Statistical significance is reported as *p < 0.05, **p < 0.001, ****p < 0.0001 (as compared with the control).
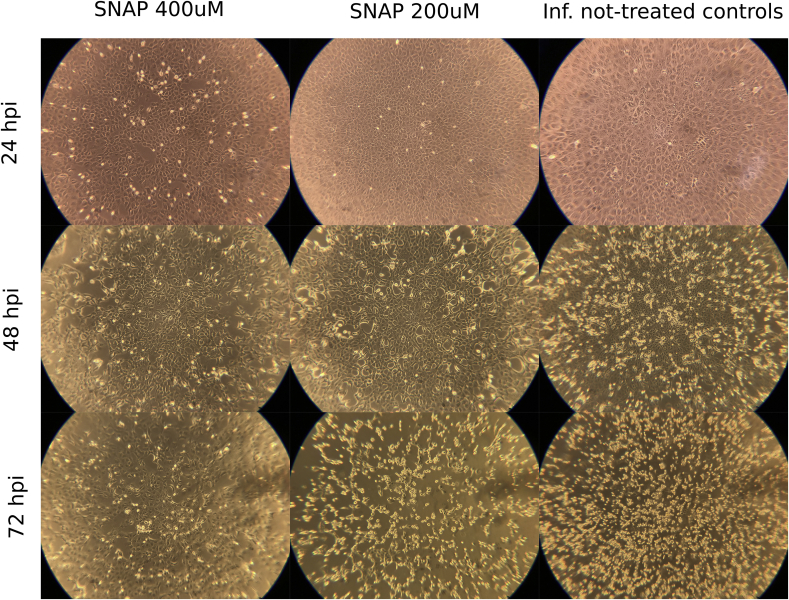
Similarly to the reduction in viral replication induced by SNAP, we observed an inverse dose dependent correlation between the concentration of SNAP and the development of viral CPE (Fig. 3). In untreated, but virus-infected, cell controls CPE started to appear at 24 hpi, while cells treated with 200 μM of SNAP developed CPE at 36 hpi (Supplementary Figure 1). Cells treated with 400 μM of SNAP did not develop any visible viral CPE up to 72 hpi. However, SNAP exhibited a cytotoxic effect, in the treated non-infected control (supplementary Figure 2). At 72 hpi, a clear protective effect of SNAP could still be observed in the wells treated by 400 μM SNAP (Fig. 3), although the amount of viral RNA increased in the 400 μM SNAP-treated group after 36 hpi. Several antiviral mechanisms of NO have been reported. Peroxynitrite, formed by the interaction between free NO in solution and superoxide anion radical (O2-) can increase viral RNA mutation rate and reduce viral particles infectivity [8,18]. Dinitrogen trioxide (N2O3), formed during the interaction of free NO and nitrogen dioxide (NO2), can donate a nitrosonium ion (NO+) and mediate the nitrosation of viral proteins and cellular host factors essential for the virus life cycle [19,20]. These mechanisms may explain the observed capacity of SNAP to prevent CPE at high concentrations without completely abolishing viral replication.
Fig. 3.
Comparison of the CPE development between cells treated with SNAP and untreated controls at 24 h intervals. The shown wells are representative for all replicates, the total magnification used to observe the cells was 100×.
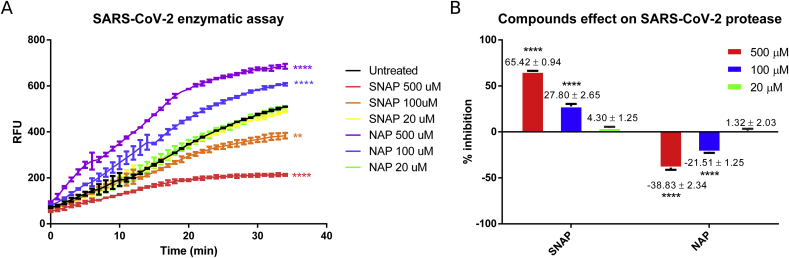
SARS-CoV-2 3CL cysteine protease could be a possible target for S-nitrosation, resulting in inhibition of the protease activity and consequent reduction in viral replication, as reported for Coxsackievirus 3C cysteine protease [7]. In order to evaluate if SARS-CoV-2 3CL protease can be directly inhibited by S-nitrosation, a FRET-based enzymatic assay was performed. For the assay, recombinant SARS-CoV-2 3CL protease was incubated for 10 min with different concentration of SNAP and NAP at room temperature prior to the addition of the substrate DABCYL-Lys-Thr-Ser-Ala-Val-Leu-Gln-Ser-Gly-Phe-Arg-Lys-Met-Glu-EDANS. The cleavage of the substrate and the relative increase in emitted fluorescence was monitored over time. SNAP displayed a dose dependent inhibition of the 3CL protease (IC50 = 440.95 μM ± 36.15 SE), while NAP acted as a reducing agent, increasing the protease activity as expected (Fig. 4). The reduction of the maximum fluorescence detected together with the profile of the curves observed in the enzymatic reactions where SNAP was added, was consistent with the covalent inhibition of the protease. The enzymatic assay results suggest that SARS-CoV-2 protease could be covalently inhibited through SNAP transnitration (direct transfer on a NO + ion) to the protease active site cysteine. In fact, the more enzyme is S-nitrosated and therefore removed from the pool of active protease, the less substrate will be cleaved. However, the proposed S-nitrosation mechanism of action remains to be proven. Moreover, whether inhaled NO exerts antiviral effects by a similar mechanism, and at what concentration, remains also to be shown, an issue recently touched by Ignarro [13].
Fig. 4.
Effect of SNAP and NAP on the activity of recombinant SARS-CoV-2 protease. The protease activity (A) is shown as the amount of relative fluorescence units (RFU) per minute. SNAP and NAP effect was also quantified (B). SNAP clearly inhibited SARS-CoV-2 protease in a dose dependent manner. In contrast, NAP acts as a reducing agent increasing the protease activity. Observed effects of compounds on SARS-CoV-2 protease activity was evaluated by One-Way ANOVA of the calculated mean area under the curves, the difference in average inhibitory activity (column graph) of the compounds was evaluated by One-Way ANOVA. Statistical significance is reported as ****p < 0.0001, **p < 0.001 (as compared with the controls).
4. Conclusions
In this study, we demonstrated that NO can inhibit the replication of SARS-CoV-2 in Vero E6 and we identified the SARS-CoV-2 main protease as a target for NO. There is a great need for effective antivirals against SARS-CoV-2 to be used in the on-going COVID-19 pandemic. Based on this study and previous studies on SARS-CoV in vitro [8,9], and in a small clinical trial [10], we conclude that NO may be applied for clinical use in the treatment of COVID-19 and other human coronavirus infections.
Declaration of competing interest
The authors declare no competing financial or non-financial interests.
Acknowledgments
We thank Petra Lukacik, Frank von Delft and Martin Walsh at University of Oxford, and the Protein Science Facility (PSF) at the Karolinska Institute, SciLifeLab for help with the production of the SARS-CoV-2 virus protease. We also thank Johan Lindbäck, Uppsala Clinical Research Center, for his valuable help with the statistical analysis. This work was supported by grants from The Swedish Research Council (VR: 2017-05807 and 2018-02569) and Knut and Alice Wallenberg Foundation and Science for Life Laboratory Uppsala (project “Nevermore Covid”).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101734.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Guarner J. Three emerging coronaviruses in two DecadesThe story of SARS, MERS, and now COVID-19. Am. J. Clin. Pathol. 2020;153:420–421. doi: 10.1093/ajcp/aqaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nissen K., Hagbom M., Krambrich J., Akaberi D., Sharma S., Ling J., Hoffman T., Bondeson K., Svensson L., Lundkvist Å., Salaneck E. Presymptomatic viral shedding and infective ability of Severe Acute Respiratory Syndrome coronavirus 2. 2020. [DOI]
- 4.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., de Castilla D.L., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. Remdesivir for the treatment of covid-19 — preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nathan C., Xie Q.W. Nitric oxide synthases: roles, tolls, and controls. Cell. 1994;78:915–918. doi: 10.1016/0092-8674(94)90266-6. [DOI] [PubMed] [Google Scholar]
- 6.Croen K.D. Evidence for antiviral effect of nitric oxide. Inhibition of herpes simplex virus type 1 replication. J. Clin. Invest. 1993;91:2446–2452. doi: 10.1172/JCI116479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saura M., Zaragoza C., McMillan A., Quick R.A., Hohenadl C., Lowenstein J.M., Lowenstein C.J. An antiviral mechanism of nitric oxide: inhibition of a viral protease. Immunity. 1999;10:21–28. doi: 10.1016/S1074-7613(00)80003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klingström J., Åkerström S., Hardestam J., Stoltz M., Simon M., Falk K.I., Mirazimi A., Rottenberg M., Lundkvist Å. Nitric oxide and peroxynitrite have different antiviral effects against hantavirus replication and free mature virions. Eur. J. Immunol. 2006;36:2649–2657. doi: 10.1002/eji.200535587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keyaerts E., Vijgen L., Chen L., Maes P., Hedenstierna G., Van Ranst M. Inhibition of SARS-coronavirus infection in vitro by S-nitroso-N-acetylpenicillamine, a nitric oxide donor compound. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2004;8:223–226. doi: 10.1016/j.ijid.2004.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Åkerström S., Mousavi-Jazi M., Klingström J., Leijon M., Lundkvist Å., Mirazimi A. Nitric oxide inhibits the replication cycle of severe acute respiratory syndrome coronavirus. J. Virol. 2005;79:1966–1969. doi: 10.1128/JVI.79.3.1966-1969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L., Liu P., Gao H., Sun B., Chao D., Wang F., Zhu Y., Hedenstierna G., Wang C.G. Inhalation of nitric oxide in the treatment of severe acute respiratory syndrome: a rescue trial in beijing. Clin. Infect. Dis. 2004;39:1531–1535. doi: 10.1086/425357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu B., Ichinose F., Bloch D.B., Zapol W.M. Inhaled nitric oxide. Br. J. Pharmacol. 2019;176:246–255. doi: 10.1111/bph.14512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ignarro L.J. Inhaled NO and covid-19. Br. J. Pharmacol. 2020 doi: 10.1111/bph.15085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ignarro L.J., Lippton H., Edwards J.C., Baricos W.H., Hyman A.L., Kadowitz P.J., Gruetter C.A. Mechanism of vascular smooth muscle relaxation by organic nitrates, nitrites, nitroprusside and nitric oxide: evidence for the involvement of S-nitrosothiols as active intermediates. J. Pharmacol. Exp. Therapeut. 1981;218:739–749. [PubMed] [Google Scholar]
- 15.Nissen K., Krambrich J., Akaberi D., Hoffman T., Ling J., Lundkvist Å., Salaneck E. Long-distance airborne dispersal of SARS-CoV-2 in COVID-19 wards. 2020. [DOI] [PMC free article] [PubMed]
- 16.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue X., Yang H., Shen W., Zhao Q., Li J., Yang K., Chen C., Jin Y., Bartlam M., Rao Z. Production of authentic SARS-CoV mpro with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction. J. Mol. Biol. 2007;366:965–975. doi: 10.1016/j.jmb.2006.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akaike T., Fujii S., Kato A., Yoshitake J., Miyamoto Y., Sawa T., Okamoto S., Suga M., Asakawa M., Nagai Y., Maeda H. Viral mutation accelerated by nitric oxide production during infection in vivo. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2000;14:1447–1454. doi: 10.1096/fj.14.10.1447. [DOI] [PubMed] [Google Scholar]
- 19.Bi Z., Reiss C.S. Inhibition of vesicular stomatitis virus infection by nitric oxide. J. Virol. 1995;69:2208–2213. doi: 10.1128/jvi.69.4.2208-2213.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colasanti M., Persichini T., Venturini G., Ascenzi P. S-nitrosylation of viral proteins: molecular bases for antiviral effect of nitric oxide. IUBMB Life. 1999;48:25–31. doi: 10.1080/713803459. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.