Abstract
The AT1 receptor is a G-protein-coupled receptor (GPCR); its activation from the basal state (R) requires an interaction between Asn111 in transmembrane helix III (TM-III) of the receptor and the Tyr4 residue of angiotensin II (Ang II). Asn111 to Gly111 mutation (N111G) results in constitutive activation of the AT1 receptor (Noda et al. (1996) Biochemistry, 35, 16435–16442). We show here that replacement of the AT1 receptors TM-III with a topologically identical 16-residue segment (Cys101-Val116) from the AT2 receptor induces constitutive activity, although Asn111 is preserved in the resulting chimera, CR18. Effects of CR18 and N111G mutations are neither additive nor synergistic. The conformation(s) induced in either mutant mimics the partially activated state (R′), and transition to the fully activated R* conformation in both no longer requires the Tyr4 of Ang II. Both the R state of the receptor and the Tyr4 Ang II dependence of receptor activation can be reinstated by introduction of a larger sized Phe side chain at the 111 position in CR18, suggesting that the CR18 mutation generated an effect similar to the reduction of side chain size in the N111G mutation. Consistently in the native AT1 receptor, R′ conformation is generated by replacement with residues smaller but not larger than the Asn111. However, size substitution of several other TM-III residues in both receptors did not affect transitions between R, R′, and R* states. Thus, the property responsible for Asn111 function as a conformational switch is neither polarity nor hydrogen bonding potential but the side chain size. We conclude that the fundamental mechanism responsible for constitutive activation of the AT1 receptor is to increase the entropy of the key agonist-switch binding residue, Asn111. As a result, the normally agonist-dependent R → R′ transition occurs spontaneously. This mechanism may be applicable to many other GPCRs.
Members of the G-protein-coupled receptor (GPCR1) superfamily each contain a seven-transmembrane helical structural motif and couple to a homologous family of G proteins. Because of these similarities, the molecular mechanism of signal transduction in GPCRs is thought to be fundamentally similar (1, 2). In its initial state, the GPCR polypeptide chain appears to be in an inactive conformation (R), and binding of agonists provides the impetus for the release from the constrained conformation. Recent studies indicate that constitutively active mutants of a large number of GPCRs can be created by natural mutations and through engineered mutagenesis (3–6). These mutations induce a conformation (R′) that partially mimics the agonist-dependent activated conformation (R*) of GPCRs. This fact suggests that the R state and agonist dependence of transition to the R* state of the GPCR is a general paradigm. Therefore, defining the structural basis of constraining interactions in relation to agonist-mediated activation is clearly important for understanding the molecular mechanism of receptor activation.
The peptide-mediated GPCR activation process is poorly understood. AT1 and AT2 receptors are members of the GPCR superfamily that mediate the actions of octapeptide hormone angiotensin II (Ang II) (7–10). The rat AT1 receptor is a single polypeptide of 359 residues predicted to consist of seven hydrophobic transmembrane α-helical segments connected by three loop regions on both the cytoplasmic and extracellular sides of the embedding membrane. Ang II-dependent activation of the AT1 receptor leads to intracellular inositol phosphate (IP) production through the activation of a G protein that is pertussis toxin insensitive (7–10). High-affinity binding of Ang II to tissue receptors was shown to involve several electrostatic and hydrophobic interactions (11–13). Site-directed mutagenesis of recombinantly expressed AT1 receptor (12–20) combined with group-specific modification of Ang II has identified several independent contacts between Ang II and the AT1 receptor (11–18). Two salt bridges, one between the Ang II side chain Arg2 and the AT1 residue Asp281, the other between Ang II α-COOH and the AT1 residue Lys199, are important for docking the hormone to the receptor (12, 13, 16). These salt-bridge interactions do not play a role in AT1 receptor activation. The Ang II residues Tyr4 and Phe8 are considered agonist switches because they are essential for agonism. Previously, we have shown that two interaction–one between Phe8 of Ang II and His256 in AT1 receptor transmembrane helix VI, the other between Ang II Tyr4 side chain and Asn111 in transmembrane helix III–are necessary to activate the receptor (14, 15). Asn111 is the key switch residue; the obliteration of its side chain not only causes the generation of a constitutively active form of the receptor (R′) but also obviates the need for Tyr4 and Phe8 in agonist-dependent induction of the active state (R*), implying that the Asn111 side chain may have a complex regulatory role in the AT1 receptor.
We now show that mutations of residues not directly involved in Ang II interaction lead to basal activation. When a segment of TM-III of the AT1 receptor is exchanged with an identical segment from the AT2 receptor without affecting the Asn111 residue (see Figure 1), the resulting chimera is constitutively active. There is no synergism between the basal activation induced by this chimera and the N111G mutation. However, when combined with Asn111 → Phe mutation, the chimera shows restoration of native receptor properties. We demonstrate that the active-state isomerization in this system is modulated by changes in tertiary interactions of a single switch, i.e., the position 111 side chain.
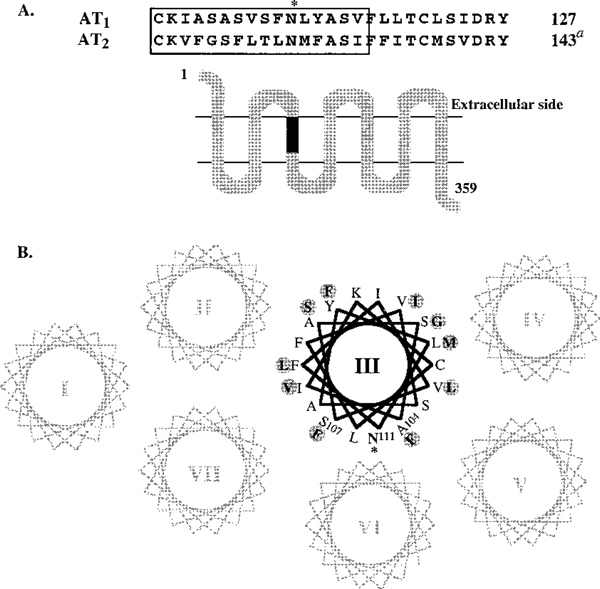
FIGURE 1:
(A) Secondary structure model of a 359-residue CR18 chimera of rat AT1 angiotensin II receptor. The transmembrane segments are putative α-helices forming the ligand pocket. The residues 101–116 of transmembrane helix III of rat AT1 receptor replaced with corresponding region of rat AT2 receptor (117–132, AT2 receptor residue numberingα) are shown within the box. The conservation of Asn111 in both sequences is indicated by an asterisk. (B) Helical wheel representation of transmembrane helical bundle of the AT1 receptor based on the Baldwin model of rhodopsin (32). The shaded circles show AT2-receptor-derived residues in CR18 chimera that influence the context around the conserved Asn111, highlighted by an asterisk.
MATERIALS AND METHODS
The monoclonal antibody 1D4 was supplied by the Cell Culture Center, Endotronic Inc., (Minneapolis, MN). Oligonucleotides were obtained from the oligonucleotide synthesis core facility of the Research Institute, The Cleveland Clinic Foundation. [Sar1,Ile8]Ang II, Ang II, and Ang II analogues were either synthesized by the peptide synthesis core facility of The Cleveland Clinic Foundation or obtained from Bachem. Losartan was a gift from DuPont-Merck, Wilmington, DE.
Experimental procedures for mutagenesis, expression, transfection, and preparation of transfected cell membrane were as described earlier (13, 14, 16). The membrane pellet in 50 mM HEPES, pH 7.2, 12.5 mM MgCl2, 1.5 mM EGTA, and 10% glycerol were used in ligand binding studies. The postnuclear supernatant of COS1 cells solubilized in 50 mM Tris-HCl, pH 6.8, 1% 3-[(3-chloramidopropyl)dimethylammonio]propanesulfonate (CHAPS), 50 μg/mL of PMSF, and 5 mM ethylenediaminetetraacetic acid (EDTA) pH 8.0 and centrifuged at 40 000 rpm was used for SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting analysis. The AT1 receptor polypeptides were visualized using the mouse monoclonal antibody 1D4 and the secondary antibody 125I-anti-mouse IgG (Amersham, Arlington Heights, IL), as described earlier (13–16). The ligand-binding experiments were carried out under equilibrium conditions. Membranes expressing wild-type receptors were incubated with 0.03–3 nM 125I-[Sar1,Ile8]Ang II (Sp. Act. 2200 Ci/mmol) in 50 mM sodium phosphate pH 7.2, 100 mM NaCl, 10 mM MgCl2, 1 mM EGTA, 0.2% bovine serum albumin, and 10 μg of bacitracin at 22 °C for 1 h. Nonspecific binding to the membranes was determined from 125I-[Sar1,Ile8]Ang II binding in the presence of 10−5 M 127I-[Sar1,Ile8]Ang II. The binding reaction was stopped by filtering under vacuum (Brandel Type M-24R) on FP-200 GF/C filters (Whatman Inc. Fairfield, NJ). Filter bound 125I-[Sar1-Ile8]Ang II was quantitated in a γ-counter (Packard). For competition binding studies, membranes expressing the wild-type receptor or the mutants were incubated at room temperature for 1 h with 300 pM 125I-[Sar1,Ile8]Ang II and various concentrations of the agonist Ang II or the peptide antagonist [Sar1,Ile8]Ang II. Equilibrium binding kinetics were determined using the computer program Ligand. The Kd values represent the mean ± standard error of the mean (SEM) of three or more independent determinations. For functional analysis of receptors, transfected COS1 cells labeled for 24 h with [3H]-myo-inositol, treated with or without [Sar1]Ang II for 45 min, were used to derive concentration–response curves. The total IP production is expressed as a percentage compared to maximum stimulation of the transfected wild-type AT1 receptor. Student T test was used for statistical analysis in Figure 2.
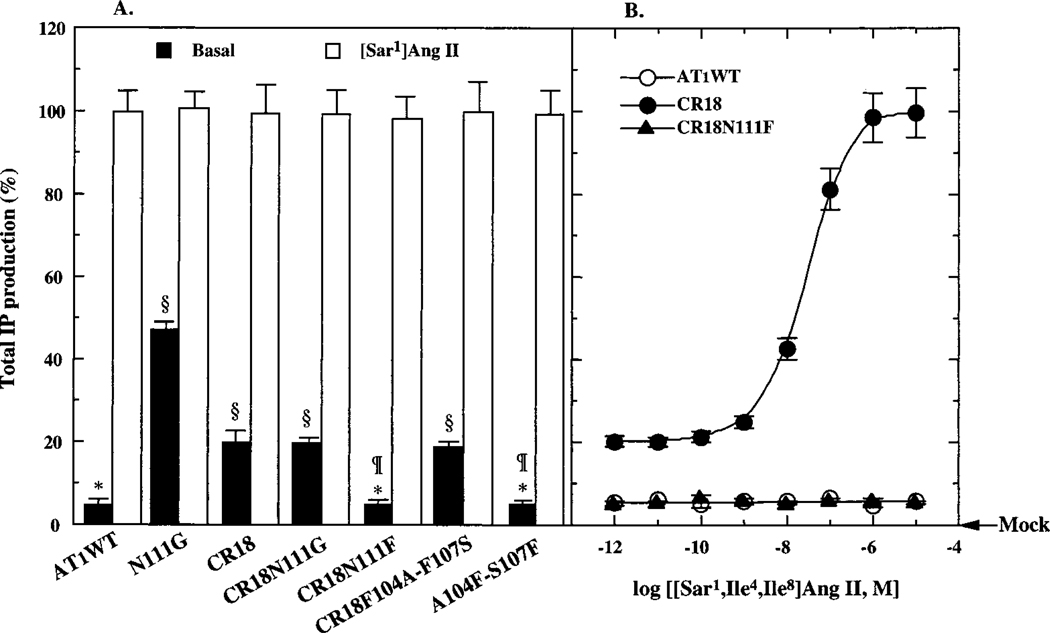
FIGURE 2:
(A) Basal and [Sar1]Ang II-stimulated maximal activation of wild-type and mutant AT1 receptors. The COS1 transfection conditions were optimized to achieve expression levels ranging in 3–5 pmol/mg of membrane protein ((~1.3–1.7) × 105 surface receptors/cell). Basal stimulation of IP accumulation was measured in transfected and mock-transfected COS1 cells. The [Sar1]Ang II-stimulated IP accumulation for all mutants shown was 100 ± 8% of that of the wild-type AT1 receptor (~50 000 ± 1200 cpm/107 cells). Results are the mean ± the standard error of three or more independent determinations. Asterisks indicate significant change (p < 0.01) compared with mock-transected cells; ¶ indicate values are not significantly (p > 0.05) different from wild-type basal; and § indicate values significantly different (p < 0.001) from the wild-type basal. (B) Activation of wild-type and mutant receptors by various concentrations of the inactive analogue, [Sar1,Ile4,Ile8]Ang II. The ligand-affinity properties of the mutant receptors are given in Table 1.
RESULTS
Experimental System.
Expression of the wild-type and mutant AT1 receptor in transiently transfected COS1 cells (at >25% transfection efficiency) was employed for structure–function analysis as described earlier (13–16). Expression in each case was measured by immunoblotting with C-terminal epitope directed monoclonal antibody 1D4 followed by saturation binding analysis using 125I-[Sar1,Ile8]Ang II. Folding was analyzed by competition binding to Ang II analogues and AT1 receptor-selective non-peptide antagonist losartan (Tables 1–3). Statistical analysis of 125I-[Sar1,Ile8]Ang II binding kinetics was best fit to a one-site model which indicated that homogeneous populations of receptors are produced in COS1 cells. Affinity of the wild-type AT1 receptor to the radioligand 125I-[Sar1,Ile8]Ang II was 0.37 ± 0.02 and 10 ± 2 nM for the nonpeptide antagonist, losartan. The Kd values estimated from competition binding for the agonist [Sar1]Ang II and the native hormone Ang II were 0.33 ± 0.02 and 1.48 ± 0.05 nM, respectively. The estimated Kd represent the intrinsic affinity of the receptor because the values did not significantly vary upon the addition of analogues of GTP to membrane preparations during the competition binding analysis. The protein expression of all the mutants described in this report was ± 20% of the level of the wild-type receptor expression by Western blot. The transient expression did not cause significant variation in cell surface receptor numbers (≈(1.3–1.7) × 105 sites per cell, in this study). Therefore we believe, the maximal IP response measured in each case truly reflects the functional activation.
Table 1:
Ligand Affinity of CR18 and Related Mutants
| receptor | expression Bmax(pmol/mg) | Kda (nM) | |||
|---|---|---|---|---|---|
| [Sar1,Ile8]Ang II | [Sar1,Ile4]Ang II | [Sar1,Ile4,Ile8]Ang II | losartan | ||
| wild-type | 5.4 ± 0.1 | 0.23 ± 0.02 | 275 ± 42 | 301 ± 33b | 10.0 ± 1.5 |
| CR18 | 4.1 ± 0.3 | 3.36 ± 0.84 | 15.9 ± 3.8 | 70.8 ± 5.7 | >1,000,000 |
| CR18N111G | 4.3 ± 0.4 | 1.04 ± 0.26 | 4.40 ± 1.2 | 24.0 ± 4.9 | >1,000,000 |
| CR18N111F | 5.3 ± 0.2 | 10.0 ± 2.36 | 3300 ± 235 | 3900 ± 215 | 590 ± 67 |
| CR18F104A-F107S | 5.1 ± 0.1 | 0.48 ± 0.23 | 3.95 ± 1.4 | 31.6 ± 5.2 | >1,000,000 |
| A104F-S107F | 5.2 ± 0.2 | 2.91 ± 1.42 | 322 ± 65 | 2850 ± 207 | 27.4 ± 8.9 |
The Kd values represent the mean ± SEM obtained from three to five independent transfection experiments performed in duplicate.
Note that the Kd of [Sar1,Ile4,Ile8]Ang II reported here is the corrected value, which is different from that reported in ref 15.
Table 3:
Ligand Affinity Properties of N111 Substitution Mutantsa
| receptor | expression Bmax (pmol/mg) | Kda (nM) | ||||
|---|---|---|---|---|---|---|
| [Sar1]Ang II | [Sar1,Ile4]Ang II | [Sar1,Ile8]Ang II | [Sar1,Ile4,Ile8]Ang II | losartan | ||
| wild-type | 5.4 ± 0.10 | 0.33 ± 0.02 | 234 ± 4.3 | 0.37 ± 0.07 | 301 ± 33 | 10 ± 1.5 |
| N111G | 3.6 ± 0.07 | 0.36 ± 0.02 | 0.3 ± 0.03 | 0.43 ± 0.07 | 5.9 ± 0.7 | 230 ± 30 |
| N111S | 4.9 ± 0.05 | 0.19 ± 0.01 | 2.1 ± 0.09 | 0.52 ± 0.05 | 4.7 ± 0.1 | 540 ± 40 |
| N111A | 6.8 ± 0.14 | 0.34 ± 0.05 | 0.4 ± 0.05 | 0.51 ± 0.03 | 3.8 ± 0.6 | 340 ± 20 |
| N111C | 3.4 ± 0.05 | 0.33 ± 0.01 | 1.3 ± 0.02 | 0.37 ± 0.02 | 11 ± 2.0 | 430 ± 30 |
| N111I | 3.1 ± 0.13 | 0.13 ± 0.02 | 41 ± 2.0 | 0.44 ± 0.13 | NDb | 3150 ± 480 |
| N111Q | 3.5 ± 0.03 | 0.24 ± 0.01 | 78 ± 0.8 | 0.58 ± 0.02 | NDb | 30 ± 0.7 |
| N111H | 5.1 ± 0.01 | 0.24 ± 0.01 | 52 ± 0.9 | 0.98 ± 0.13 | NDb | 3130 ± 180 |
| N111K | 5.8 ± 0.16 | 0.77 ± 0.01 | 367 ± 25 | 1.29 ± 0.29 | NDb | 1900 ± 390 |
| N111F | 3.8 ± 0.77 | 1.67 ± 0.21 | 43.2 ± 0.2 | 4.57 ± 0.77 | NDb | 920 ± 120 |
| N111Y | 4.1 ± 0.70 | 0.45 ± 0.04 | 33.9 ± 0.1 | 1.09 ± 0.17 | NDb | 1840 ± 180 |
The Kd and Bmax values represent the mean ± SEM obtained from three independent transfection experiments performed in duplicate. The Bmax values represent the receptor present in total membrane preparations.
ND, not determined because, in these mutants, the Kd of the particular ligand was >10−7 nM (n = 1).
The ability of the AT1 receptor to activate IP production in COS1 cells is shown in Figures 2 and 3. The basal IP production in transfected COS1 cells without [Sar1]Ang II treatment is 5 ± 0.5% when compared to the maximal IP response elicited by [Sar1]Ang II concentrations >10−7 M (taken as 100%) and is significantly higher than that measured in the mock transfected cells. Alterations in the basal activity of different mutant receptors expressed at comparable levels could be accurately measured, since the maximal IP response elicited in this system is very high (≈ 50 000 cpm/107 cells). The analogue [Sar1,Ile4,Ile8]Ang II is completely inactive in eliciting IP response from the wild-type AT1 receptor even at concentrations 300-fold more than the Kd value (301 ± 30 nM).
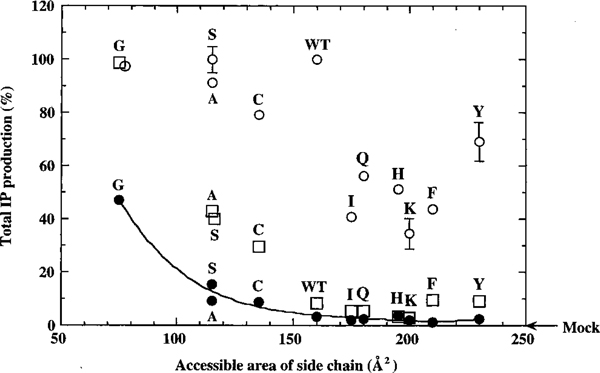
FIGURE 3:
Correlation between constitutive activity (basal IP production shown in filled circles) and accessible surface area (Å2) of the side chain substituted at position 111 in transmembrane helix III. The accessible surface area of amino acid residues in the receptor are as defined and derived from Creighton (33). This analysis identifies the accessible surface area of Asn111 as the constraining factor in the AT1 angiotensin II receptor with high confidence (r = 0.992) with a minor influence from the hydrogen bonding property in addition. The open circles represent [Sar1]Ang II-stimulated and open squares represent [Sar1,Ile4,Ile8]Ang II-stimulated IP production by the corresponding mutants. The values given are the mean ± the standard error of three or more independent determinations. The basal activity of each mutant was measured as accurately as possible within the limitations of technique. In each instance, to eliminate the possibility that any increase in activity may be because of random increase of receptor density, we expressed each construct at 0.5–1.0 and 3.0–5.0 pmol/mg of protein in parallel experiments and accurately determined the basal activity. This indicated that the significantly increased basal activity (p < 0.05) in each mutant is directly related to constitutive activation. The ligand-affinity properties of these mutant receptors are given in Table 2. Under these conditions, basal IP production in Ile111, Gln111, His111, Lys111, Phe111, and Tyr111 transfected COS1 cells ranged between 1.2 and 3.7% (in these comparisons, zero was the level of IP production in mock-transfected COS1 cells) compared with that of the wild-type receptor. The relationship between Basal IP production and surface area of the residue at the position best fit polynomial function (Y = m0 + m1x + m2x2 + m3x3 + … + m9x9) to fourth order, using the least-squares error method. Y and x parameters are as indicated in the figure; m represents polynomial coefficients: m0 = 36 254, m1 = −687.94, m2 = 5.5607, m3 = −0.020246, and m4 = 2.7817 × 10−5 in our analysis. It is important to emphasize here that the model is consistent with Asn111 harboring interactions with several residues, and the number of residues displaced in the transitions governing different mutations is unknown. Only the net displacements involved in transitions is anticipated to be deduced from the data. The basal activity read-out data (IP levels) in this paper were best fit using least-squares analysis. This variable did vary significantly among mutants where constitutive activation was noticeably higher than the matched wild-type control. Much of the variation could be accounted for by differences in the expression levels resulting from variable transfection efficiencies. The expression levels were determined by Bmax calculations and western blot analysis; both of these methods indirectly predict the plasma membrane receptor density. Therefore, it is possible that these are real variations in the basal activity for different mutants on the order of <10–12% of the mean value shown in each instance.
Effect of the Size of the Residue 111 on the Constitutive Activity of an AT1/AT2 Chimeric Receptor CR18.
The CR18 chimeric receptor (see Figure 1 for details) is constitutively active. The basal IP production in the CR18 chimera transfected cells was ≈10-fold higher than that (p < 0.001) in the mock transfected cells. This is nearly a 6-fold increase over the basal activity exhibited by the wild-type AT1 receptor (p < 0.001). The maximal Ang II-dependent IP production stimulated by the wild-type AT1 receptor and CR18 chimera (98 ± 8%) in transfected COS1 cells were nearly identical (Figure 2). The CR18 chimera was activated (to 90 ± 11%) by the Ang II analogue [Sar1,Ile4,Ile8]Ang II, but the wild-type AT1 receptor was not (Figure 2B). The level of expression of CR18 and the wild-type receptors are not significantly different (Table 1), eliminating the possibility that the increased basal activity of CR18 is due to a coincidental increase of cell-surface receptor density.
The ligand affinity profile of the CR18 chimera was comparable to that of constitutively activated N111G mutant AT1 receptor (15). The affinity was increased significantly (p < 0.001) for the peptide agonist [Sar1]Ang II (3–10-fold), for the partial agonist, [Sar1,Ile4]Ang II (17-fold) and for the inactive Ang II-analogue [Sar1,Ile4,Ile8]Ang II (33-fold). This increased affinity does not arise from an effect due to enhanced G protein coupling, since competition binding analysis in the presence of 100 μM GTP-γ-S which uncouples receptor/G protein interactions, did not alter the binding affinity (data not shown). The binding affinity of the nonpeptide antagonist losartan (>190 000-fold, p < 0.001) was substantially reduced (Table 1).
Substitution of the conserved Asn111 residue in CR18 with Gly111 did not synergistically increase the degree of constitutive activity of the CR18 chimeric receptor. The measured basal activity of CR18N111G is not equivalent to CR18 + N111G basal activities but is identical to that of CR18, which indicates that the effect of CR18 mutation is dominant over the effect of N111G mutation on the AT1 receptor. The CR18N111G mutation caused significant differences in the ligand affinity profiles compared to CR18 (Figure 2, Table 1).
In contrast, not only did substitution of the Asn111 residue with Phe111 in CR18 chimera restore the signaling property but also the ligand binding profiles observed with CR18 chimera were suppressed back to the levels comparable to those in the wild-type AT1 receptor (shown in Figure 2 and Table 1). The basal and maximal Ang II-stimulated IP production was comparable (p < 0.001) to that observed with the wild-type AT1 receptor. The EC50 of the CR18N111F mutant for [Sar1]Ang II-stimulated IP production was 89 ± 9 nM compared to 28.3 ± 0.3 nM for the CR18 receptor. The binding affinity of the CR18N111F mutant for [Sar1]Ang II was 3.1 nM and for Ang II was 14 nM. The CR18N111F mutant was not activated by 10–100 μM [Sar1, Ile4,Ile8]Ang II (Figure 2B). The affinity for [Sar1,Ile4,Ile8]Ang II decreased ≈-fold compared with the affinity of the CR18 receptor. An ≈5-fold lower affinity of CR18N111F mutant toward [Sar1]Ang II than that of the wild-type AT1 receptor (also for Ang II, not shown) is consistent with an unfavorable interaction between Tyr4 of the ligand and the Phe111 side chain of the receptor as described earlier (15). The affinity of CR18N111F mutant for losartan is >1000-fold increased when compared to the affinity of CR18 chimera.
Properties of two additional mutants, the F104A-F107S mutant of CR18 and the A104F-S107F mutant of AT1 receptor, suggest that control of the basal activity is a specific function of the position 111 side chain size (Figure 2, Table 1). In the CR18F104A-F107S mutant, two Phe side chains, located on the same helical surface of the transmembrane helix III (TM-III) as Asn111, were mutated to AT1 receptor residues at the same position (Figure 1B). This mutant receptor bound the Ang II analogues with 2- to 10-fold higher affinity and losartan with 5-fold lower affinity compared to CR18 chimera. The high basal and Ang II-analogue-dependent activation characteristics of CR18 were retained (Figure 2A, Table 1), indicating that Phe104 and Phe107 are not responsible for constitutive activation of CR18. Furthermore, in the reciprocal mutant, the A104F-S107F mutant of the AT1 receptor, the affinity of various ligands was reduced and the mutant did not exhibit significant changes in basal activity and Ang II-stimulated activation. Thus, alteration of several different residues within the TM-III helix could alter the ligand-binding profiles, but individual residue changes per se did not affect the basal inactive state of the AT1 receptor. These observations are consistent with previous studies of the AT1 receptor which demonstrated that mutagenesis of several residues (except Asn111) of transmembrane helix III do not affect receptor activation (15, 17).
Effect of Residue Size at Positions 292 and 295 in TM-VII of the AT1 Receptor on Constitutive Activation.
In molecular models of the AT1 receptor, Asn111 appears to interact with either Asn295 (20) or Tyr292 (18–20). Both of these residues are located in TM-VII. To examine whether the size of either Tyr292 or Asn295 is responsible for normal interaction between TM-VII and TM-III and disruption of this interaction is responsible for generating constitutive activation, we prepared the mutants shown in Table 2. The Y292Q, Y292A, and Y292G mutants did not show significantly different properties from the wild-type AT1 receptor in basal activation, interaction with agonists and nonpeptide antagonists. Furthermore, these mutants were able to fully activate IP production to within 74–83% of that of the wild-type AT1 receptor. The N295G, N295S, N295A, N295C, N295I, and N295F mutants did not show any significant increase in basal activity, and most mutants were like the wild-type in being stimulated by [Sar1]Ang II. Since Balmforth et al. (19) reported that N295S mutation produced constitutive activation in HEK cells, we examined agonist affinity profiles for this mutant. As shown in Table 2, the mutants showed a slight decrease of affinity for [Sar1]Ang II. In addition, the N295S mutant (reported as constitutively active (19)) affinity for the [Sar1,Ile4,Ile8]Ang II was 2410 ± 100 nM. Further, the agonist-dependent activation properties of this mutant were not significantly altered.
Table 2:
Ligand Binding and Activation Properties of Tyr292 and Asn295 Mutants
| receptor | Kda (nM) | IP (%) | ||
|---|---|---|---|---|
| [Sar1] Ang II | Losartan | Basal | [Sar1] Ang II stimulated | |
| wild-type | 0.33 ± 0.10 | 10 ± 1.5 | 5.0 ± 0.5 | 100 |
| Y292G | 1.75 ± 0.20 | 149 ± 21 | 6.6 ± 0.9 | 74 ± 4 |
| Y292A | 0.63 ± 0.07 | 32 ± 0.4 | 4.1 ± 1.0 | 83 ± 4 |
| Y292Q | 0.59 ± 0.07 | 9 ± 1.0 | 3.3 ± 0.3 | 75 ± 1 |
| N295G | 1.92 ± 0.22 | 489 ± 32 | 5.8 ± 2.2 | 105 ± 5 |
| N295A | 2.62 ± 0.39 | 602 ± 28 | 5.4 ± 0.5 | 100 ± 5 |
| N295C | 1.08 ± 0.19 | 45 ± 5.9 | 5.7 ± 1.3 | 99 ± 3 |
| N295S | 0.53 ± 0.10 | 341 ± 32 | 6.0 ± 2.3 | 94 ± 0.7 |
| N295I | 0.73 ± 0.10 | 26 ± 6 | 5.0 ± 2.7 | 86 ± 2 |
| N295F | 0.47 ± 0.11 | 1657 ± 291 | 5.3 ± 0.7 | 50 ± 6 |
The Kd values represent the mean ± SEM obtained from three to five independent transfection experiments performed in duplicate.
Influence of Residue Size at Position 111 on Basal and Ang II-Dependent Activation of the AT1 Receptor.
The basal activity of the AT1 receptor is highly influenced by the particular amino acid substituted at the Asn111 position (15). To further explore this effect, we examined a total of 10 different amino acids at position 111 in the AT1 receptor (Figure 3, Table 3). Gly-, Ala-, Ser-, and Cys-substituted mutant receptors were constitutively active: The IP stimulation in the absence of agonist by these mutants was significantly (p > 0.001) increased over the wild-type receptor but varied considerably between different mutants. All of these mutant receptors expressed well (<2-fold variability), bound [Sar1]Ang II, [Sar1,Ile8]Ang II nearly with the same affinity as the wild-type. Affinity of these mutants was increased >100-fold toward the partial agonist analogue [Sar1,Ile4]Ang II and >200-fold toward the inactive analogue [Sar1,Ile4,Ile8]Ang II. The [Sar1]Ang II-dependent maximum IP formation was not changed in these mutants (reduced by <20% in Cys111). Thus, correlation between the maximum agonistic effect of [Sar1]Ang II and side chain hydrophobicity, hydrogen bonding potential, or charge were not evident.
In contrast, the Ile-, Gln-, His-, Lys-, Phe-, and Tyr-substituted mutant receptors were not constitutively active. The [Sar1]Ang II-dependent maximum IP formation in these mutants was reduced by 35–70%. The basal IP production without agonist stimulation in cells transfected with these mutant receptors was not significantly different (p > 0.05) from that observed in wild-type receptor transfected cells. The expression level of these mutants varied (Table 3), but the surface receptor expression was not significantly different. The affinity of these mutants toward the agonist [Sar1]Ang II was within 4-fold (N111F), and for the partial agonist [Sar1,Ile8]Ang II was within 13-fold (N111F). However, in distinct contrast to the constitutively activated mutant AT1 receptors, Ile-, Gln-, His-, Lys-, Phe-, and Tyr-substituted mutant receptors did not show a dramatic increase of affinity toward the partial agonist analogue [Sar1,Ile4]Ang II and the inactive analogue [Sar1,Ile4,Ile8]Ang II. The ≈3- to 5-fold increased affinity of [Sar1,Ile4]Ang II toward some of the mutants is consistent with an interaction between position 4 of the ligand and the position 111 residue of the receptor (15). The Kd for the nonpeptide antagonist losartan was decreased (3- to 315-fold) in all of the position 111 mutants without significant correlation with any of the substituted side chain properties. This latter finding is consistent with the earlier observations (15, 17–19).
Figure 3 demonstrates that the basal constraint (inverse of magnitude of basal activity) correlates well (R2 = 0.992) with the surface area (and volume) of the amino acid side chain with a bimodal relationship. Correlation with hydrophobicity and hydrogen bonding ability is poor (not shown). Thus, residues larger than Asn at position 111 favor the basal inactive state, and smaller residues favor the intermediate active state. The degree to which this state is favored and the reduction of residue 111 surface area best fit to a power function, indicating a direct relationship between the two parameters (see Figure 3 legend for details).
DISCUSSION
Like other members of the GPCR family, the AT1 receptor is stabilized in an R state and the agonist-dependent transition to R* state is believed to proceed through a relaxed intermediate activated R′ state (15). The R′ state is characterized by increased basal activity, a preferential increase of affinity for partial agonist analogues of Ang II combined with the ability to be fully activated by the inactive peptide analogue [Sar1,Ile4,Ile8]Ang II. Additionally, the R′ state of the AT1 receptor decreases the affinity for nonpeptide antagonists but can be nearly completely inhibited by the antagonists at high concentration. The predicted secondary structure of the AT1 and AT2 receptors is similar. Since critical residues such as Asn111, Lys199, and Asp281 of the AT1 receptor that are involved in Ang II binding are conserved in the AT2 receptor, it is anticipated that AT1/AT2 chimera fold properly and bind Ang II with high affinity (7–10). Rather unexpectedly a number of AT1/AT2 receptor chimera stimulated IP production in transiently transfected COS1 cells without agonist stimulation (unpublished observations of Y. H. Feng and S. Karnik). The smallest region derived from the AT2 receptor that is common to all of the constitutively active AT1/AT2 receptor chimera (CR18) corresponded to 16 residues within TM-III shown in Figure 1. The CR18 chimera is in the R′ state; its properties are the result of a rearrangement in the Ang II binding pocket similar to a phenomenon seen with constitutively active mutations of the AT1 receptor that is also reflected in the pharmacological specificity of losartan (15, 18, 19).
The most straightforward explanation is that as a consequence of the disrupted interhelical interactions mimicry occurs conforming to the R′ state. This is not unique to the AT1 receptor since constitutively activated mutants have been discovered in several different GPCRs (3–6). But previous attempts to engineer constitutively activated mutants have failed with the sole exception of Asn111 mutations. Therefore, the ability of a 16 residue segment within TM-III of CR18 to induce the R′ state intrigued us. In this segment 11 of 16 residues (i.e., reflecting the overall homology of <32% between AT1 and AT2 receptors) are different. The resulting changes include substantial differences in size due to Ala104 → Phe, Ser105 → Gly, and Ser107 → Phe alterations, while the remaining eight residue changes are conservative. The phenotype of two reciprocal mutants, the CR18F104A-F107A and A104F-S107F AT1 receptor (see Table 1), indicate that these changes are not responsible for the constitutive activity of CR18. Consistent with these observations, previous studies of the AT1 receptor have indicated that mutagenesis of several residues (except Asn111) of transmembrane helix III do not affect receptor activation (15, 17).
The critical agonist switch binding residue Asn111 located within this segment is retained in this chimera, implying that preservation of an Asn side chain per se is not sufficient to guarantee the R state in CR18. This is surprising because previous mutagenesis and modeling studies suggested that tertiary interactions of Asn111 through hydrogen bonding constrains the AT1 receptor to basal inactive state (18–20). Alternatively, studies of Noda et al. (15) from this laboratory indicated that the size of the Asn111 side chain is an important constraining factor in the AT1 receptor. This hypothesis predicts that the size of the Asn111 side chain is insufficient to constrain the CR18 chimeric receptor to the R state; hence, an increased size of position 111 residue in CR18 should restore native wild-type-like signaling properties. To test whether an increase of the residue 111 side chain size could restore the properties of the R state, Asn111 of the CR18 chimera was substituted with a Phe111. This substitution causes an ≈50 Å2 increase in size, and increases side chain hydrophobicity and van der Waals radius. All three parameters of the R′ state were suppressed to nearly wild-type level in this mutant; basal inactivity and high affinity for losartan were restored. Furthermore, the mutant was insensitive to activation by [Sar1,Ile4,Ile8]Ang II. On the other hand, the introduction of the N111G mutation had no influence on the phenotype of CR18. Evaluation of database of protein structure–function has revealed that functions in proteins are the culmination of a set of cooperative interactions. Additivity of mutational effect suggests independence of individual amino acids in the function. Deviation from additivity suggests coupled interactive effects which can either reduce (negative interaction) or enhance (synergistic) the measured function (21). The lack of synergistic (or additive) effect on the basal activity suggests a coupled interactive influence between CR18 and N111G mutations. Alternatively, the CR18 mutation may have two independent effects, one promoting R → R′ transition and a second effect that interferes with stability of R′ and R* states or interaction with G protein.
We conclude that the conformation of the CR18N111F mutant chimera conforms to the inactive R state of the wild-type AT1 receptor. It is not the alteration of the TM-III helix sequence; rather it is the influence of sequence alteration on the “switching properties” of Asn111 that leads to constitutive activation of the CR18 receptor, which is restored by CR18N111F mutation. The amino acid composition influencing conformation of an α-helix as well as single amino acids in an α-helix perturbing interhelical interactions is wellestablished (22). In this regard, the effects on binding and signaling observed with different mutants described above, as well as in earlier studies, indicate that the Asn111 residue likely influences TM-III helical conformation and vice versa.
The conclusion above suggests that neither polarity nor hydrogen bonding potential but the size of Asn111 side chain may be critical for generating basal inactive conformation in the native AT1 receptor as well. This conclusion is significantly different from the suggested interactions for Asn111 in the wild-type AT1 receptor; i.e., Asn111 interacted with either Asn295 (19) or Tyr292 (18, 20) in TM-VII in the R state. In molecular models of the AT1 receptor, an interaction appears possible between Asn111 (TM-III) and Tyr292 (TM-VII) in the R state and between Asp74 (TM-II) and Tyr292 (TM-VII) in the R* state. The receptor activation would cause a disruption of Asn111–Tyr292 interaction, allowing an Asp74–Tyr292 interaction to be formed. The model prompts the existence in the wild-type receptor of another residue that forms an intramolecular bond with Asn111. Groblewski et al. (18) proposed that constitutive activation results when the Asn111–Tyr292 bond is disrupted favoring conformational flexibility. Balmforth et al. (19) reported constitutive activation by mutating Asn111 (TM-III) and Asn295 (TM-VII), suggesting that these two Asn residues have a complementary role in the active-state isomerization. Hence, the Asn111–Asn292 bond may be involved in the R* state. These AT1 receptor activation models are similar to the mechanism described in the rhodopsin family (1, 4) and in biogenic amine receptors (2, 23) where disruption of a salt bridge between TM-III and TM-VII is found to induce constitutive activation. These experimental observations lead us to suggest that the interaction between TM-III and TM-VII (either through the Asn111–Tyr292 or the Asn111–Asn295 interaction) may involve a size complementary of the proposed interactions (18–20). Pharmacological characterization of Tyr292 and Asn295 mutations (Table 2) demonstrated that these residues in TM-VII of the AT1 receptor do not interact with Asn111 in a side-chain-size-dependent fashion. The observation however does not exclude the possibility that several amino acids located on multiple TM helices are involved in stabilizing Asn111 in the inactive state.
Size of the position 111 side chain has two distinct effects on AT1 receptor activation (Figure 3, Table 3). Side chains larger than Asn, confer basal inactivity and resistance to activation by [Sar1,Ile4,Ile8]Ang II and prevent full activation by Ang II. We speculate that in the wild-type AT1 receptor Ang–Tyr4 must be capable of displacing the Asn111 side chain just to the point that is required for the subsequent R′ → R* transition by nonaromatic Ang II interactions. This calibration is defective in these mutants. In contrast, the constitutively active Gly-, Ala-, Ser-, and Cys-substituted AT1 receptor mutants could be activated (by 25–100% of [Sar1]Ang II induced activation) by [Sar1,Ile4,Ile8]Ang II, implying that different degrees of mimicry to the R′ state could occur. Figure 3 demonstrates that the degree to which the R′ state is favored and the reduction of the residue 111 surface area are directly related (see Figure 3 legend for details). The model to explain the best fit data in Figure 3 is consistent with multiple interactions, which is also supported by experiments described in the previous section, which indicates that Asn111 may not interact with a unique partner in TM-VII. Ang II binding normally releases Asn111, but in mutants stabilized at the R′ state the release becomes possible without agonist stimulation (presumably due to lowering of the energy barrier for the R → R′ transition). In such a model, the basal activity is the outcome of net changes, which is expected to be unique in each mutant. Changes in the number and nature of the contacts governing the basal activation in different mutations cannot be precisely measured. Thus, mutations are not abnormalities but very likely mimic transient states of the native mechanism of activation. The difference in the basal activation from the smallest (Gly111) to the largest (Tyr111) residue examined was 40% of the maximal achievable activation using [Sar1]Ang II.
The mechanism by which the size of the amino acid exerts a constraining effect is not clear. But it is unlikely to be due to a hydrogen bonding or hydrophobic packing effect since the correlation is poor. It is noteworthy that an alternative mechanism is possible based on Matthews and co-worker’s (24) observation in lysozyme mutants that 20 cal/Å2 of stabilization energy is lost by cavity created by mutation of residues. The possible mechanism for the AT1 receptor activation, therefore, could be that van der Waals contact between Asn111 and several residues is lost during normal activation and the activating mutants emulated this transition. Then what is the contribution of the CONH2 group in Asn111 to the inactive and active states? It is difficult to determine its role unambiguously in the stabilization of R state, but a casual analysis is presented: There are seven residues capable of hydrogen bonding interaction (Ser, Cys, Asn, Gln, His, Lys, and Tyr) in our analysis. From crude pairwise comparison the hydrogen bonding property appears to be not important. For example, if hydrogen bonding is the stabilizing conformational constraint, then we would expect lower constitutive activity associated with the hydrogen bonding residue when other side chain parameters are nearly matched. But from the regression line in Figure 3, the measured basal activity of the Gln111 mutant is similar to that of the Ile111 mutant. Comparison between Ser111 and Ala111 suggests that the hydrogen bonding potential is not constraining the basal activity. Most remarkably, Gln a structural analogue of Asn in hydrogen bonding ability, but, with a one methylene unit longer side chain, the comparison indicates that an increase of size is detrimental and cannot override the hydrogen bonding potential that is preserved. This relatively unimportant role of the CONH2 group in constraining the receptor should be contrasted with its importance in interaction with the “agonist-switch” Tyr4 group of Ang II suggested earlier (15). Thus, the Asn side chain at position 111 is not uniquely important for either the maintenance of the basal or the generation of the fully active state of the receptor. However, an important reason Asn111 is conserved in all Ang II receptors (25) may be that the Asn side chain allows three effects: maximal signal amplification between receptor “off” and “on” states, minimal noise when the receptor is in the “off” state, and enhanced specificity provided by Tyr4-dependent switching between inactive and active states.
The framework provided by studies of various GPCRs suggest that the transmembrane, the cytoplasmic, and the extracellular domains each have specific tertiary structures, which are stabilized cooperatively, would generate the binding pocket for the native ligand and the cognate G protein (1–6, 15, 23–26). Therefore it is not surprising that mutations in all three domains can disrupt the constraint leading to activation. In addition, GPCRs are activatable through the binding of agonists or antibody in the extracellular and transmembrane domains or enhanced G-protein binding in the cytoplasmic domain (1–6, 27–30). On the basis of these types of observations, it has been suggested that the activation of GPCRs do not necessarily involve unlocking of key residues by agonists but depends on unconstraining the intramolecular interactions that normally dictate inactive conformation. Since only partial activation is achieved by antibody binding or activating mutations or G proteins, the spontaneous conversion from R to R* states by these mechanisms in vivo must be insufficient to cause function and related pathophysiology. Receptor activation by native hormones and full agonists perhaps not only disrupts the R state but also initiates and promotes sharp transition to the R* state through coordinated movement of helices and loops, to provide newly formed binding sites for G proteins. Inevitably, the chemistry responsible for triggering the function involves disruption of intramolecular bonds within receptor and formation of transient bonds with the agonist. For example, in the light transducing rhodopsin, activation depends on deprotonation of the Schiff’s base followed by disruption of a salt bridge with Glu113 (1, 4). In adrenergic receptors the capture of the acidic counterion (Asp113 in the β adrenergic receptor) by the basic amine group of agonists is crucial (2, 23).
Unlike these electrostatically regulated systems, in the AT1 receptor, the Asn111 is considered as an “agonist-switch-binding” residue because it functionally interacts with the Tyr4 agonist group of Ang II, for the purpose of discrimination from those residues that are important for agonist binding but not critical for receptor activation (13–16). Our results here show that the activation of the AT1 receptor from mutagenesis of residues not directly involved in agonist interaction is because of the displacement of the agonist-switch-binding residue that normally controls the switching between R, R′, and R* states. This indicates that receptor tertiary structure and the putative agonist-switch-binding residues mutually regulate each other through a conformational coupling mechanism (30). Ang II-mediated activation of the AT1 receptor must involve destabilization of Asn111 directly by the hormone (15). The finding that the Asn111 → Phe111 mutation in CR18 can recapitulate the trilogy of effects observed with the Asn side chain in the wild-type receptor, that is, maximal signal amplification between receptor “off” and “on” states, minimal noise when the receptor is in the “off” state, and enhanced specificity provided by Tyr4-dependent switching underscores the importance of the side chain surface area, rather than the hydrogen-bonding potential, as the key structural feature of this switch residue. The discovery that the side chain surface area is the critical determinant of a switch residue responsible for receptor activation is unique, but likely to be a general paradigm in several GPCRs, which is worth exploring. We believe our study is important, generally to GPCR mechanism, because it challenges the view that the agonist pocket “does not matter” in the exploration of receptor activation mechanisms (31). Furthermore, it suggests that drug development efforts should target agonist-switch-binding residues far more than currently believed.
ACKNOWLEDGMENT
We thank Yasser Saad, Xiao-pu Liu, Dennis Wilk, and Dianne DiBattisti for excellent technical assistance, Robert M. Graham and Ramaswamy Ramachandran for critical reading of the manuscript, and Robin Lewis and Christine Kassuba for assistance in manuscript preparation.
Research supported in part by the National Institutes of Health Grants EY09704 and HL57470.
1. Abbreviations:
- GPCR
G-protein-coupled receptor
- Ang II or AII
the octapeptide hormone angiotensin II, NH2–D–R–V–Y–I–H–P–F–COOH
- IP
inositol phosphate
- GPCRs
G-protein-coupled receptors
- TM-III
transmembrane helix III
- PAGE
SDS-polyacrylamide gel electrophoresis
- AT1
Ang II type 1
REFERENCES
- 1.Khorana HG (1992) J. Biol. Chem. 267, 1–4. [PubMed] [Google Scholar]
- 2.Strader CD, Fong TM, Tota MR, Underwood D, and Dixon RAF (1994) Annu. ReV. Biochem. 63, 101–132. [DOI] [PubMed] [Google Scholar]
- 3.Kjelsberg MA, Cotecchia S, Ostrowski J, Caron MG, and Lefkowitz RJ (1992) J. Biol. Chem. 268, 4625–4636. [Google Scholar]
- 4.Robinson PR, Cohen GB, Zhukovsky EA, and Oprian DD (1992) Neuron 9, 719–725. [DOI] [PubMed] [Google Scholar]
- 5.Kenakin T. (1995) Trends Pharmacol. Sci. 16, 256–258. [DOI] [PubMed] [Google Scholar]
- 6.Shenker A. (1995) Baillieres Clin. Endocrinol. Metab. 9, 427–451. [DOI] [PubMed] [Google Scholar]
- 7.Murphy TJ, Alexander RW, Griendling KK, Runge MS, and Bernstein KE (1991) Nature 351, 233–236. [DOI] [PubMed] [Google Scholar]
- 8.Sasaki K, Yamano Y, Bardhan S, Iwai N, Murray JJ, Hosegawa M, Matsuda Y, and Inagami T. (1991) Nature 351, 230–232. [DOI] [PubMed] [Google Scholar]
- 9.Mukoyama M, Nakajima M, Horiuchi M, Sasamura H, Pratt RE, and Dzau VJ (1993) J. Biol. Chem. 268, 24539–24542. [PubMed] [Google Scholar]
- 10.Kambyashi Y, Bardhan S, Takahashi K, Tsuzuki S, Hamakubo T, and Inagami T. (1993) J. Biol. Chem. 268, 24543–24546. [PubMed] [Google Scholar]
- 11.Bumpus FM, and Khosla MC (1977) in Hypertension: Physiology and Treatment (Genest J, Koiw E, and Kuchel O, Eds.) pp 183–201, McGraw-Hill, New York. [Google Scholar]
- 12.Yamano Y, Ohyama K, Chaki S, Guo DF, and Inagami T. (1992) Biochem. Biophys. Res. Commun. 187, 1426–1431. [DOI] [PubMed] [Google Scholar]
- 13.Noda K, Saad Y, Kinoshita A, Boyle TP, Grahams RM, Husain A, and Karnik SS (1995). J. Biol. Chem. 270, 2284–2289. [DOI] [PubMed] [Google Scholar]
- 14.Noda K, Saad Y, and Karnik SS (1995) J. Biol. Chem. 270, 28511–28514. [DOI] [PubMed] [Google Scholar]
- 15.Noda K, Feng Y-H, Liu XP, Saad Y, Husain A, and Karnik SS (1996) Biochemistry 35, 16435–16442. [DOI] [PubMed] [Google Scholar]
- 16.Feng Y-H, Noda K, Saad Y, Liu X, Husain A, and Karnik SS (1995) J. Biol. Chem. 270, 12846–12850. [DOI] [PubMed] [Google Scholar]
- 17.Monnot C, Bihoreau C, Conchon S, Curnow KM, Corvol P, and Clauser E. (1996) J. Biol. Chem. 271, 1507–1513. [DOI] [PubMed] [Google Scholar]
- 18.Groblewski T, Maigret B, Languier R, Lombard C, Bonnafores JC, and Marie J. (1997) J. Biol. Chem. 292, 1822–1826. [DOI] [PubMed] [Google Scholar]
- 19.Balmforth AJ, Lee LJ, Warburton P, Donnelly D, and Ball SG (1997) J. Biol. Chem. 272, 4245–4251. [DOI] [PubMed] [Google Scholar]
- 20.Joseph M-P, Maigret B, Bonnafous J-C, Marie J, and Scheraga HH (1995) J. Protein Chem. 14, 381–3908. [DOI] [PubMed] [Google Scholar]
- 21.Wells JA (1990) Biochemistry 29, 8509–8516. [DOI] [PubMed] [Google Scholar]
- 22.O’Neil KT, and DeGrado WF (1990) Science 250, 646–651. [DOI] [PubMed] [Google Scholar]
- 23.Porter JE, Hwa J, and Perez DM (1996) J. Biol. Chem. 271, 28318–28323. [DOI] [PubMed] [Google Scholar]
- 24.Eriksson AE, Baase WA, Zhang X-J, Heinz DW, Blaber M, Baldwin EP, and Matthews BW (1992) Science 255, 178–183. [DOI] [PubMed] [Google Scholar]
- 25.Karnik SS, Husain A, and Graham RM (1996) Clin. Exp. Pharmacol. Physiol. Suppl. 3, S58–S66. [PubMed] [Google Scholar]
- 26.Gether U, Ballesteros JA, Seifert R, Sander-Bush E, Weinstein H, and Kobilka BK (1997) J. Biol. Chem. 272, 2587–2590. [DOI] [PubMed] [Google Scholar]
- 27.Leiber D, Harbon S, Guillet JG, Andre C, and Strasberg AD (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 4331–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magnusson Y, Wallukat G, Waagstein F, Hjalmarson A, and Hoebeke J. (1994) Circulation 89, 2760–2767. [DOI] [PubMed] [Google Scholar]
- 29.Nanevicz T, Wang L, Chen M, Ishii M, and Coughlin SR (1996) J. Biol. Chem. 271, 702–706. [DOI] [PubMed] [Google Scholar]
- 30.Gavish B. (1986) in Fluctuating Enzyme (Welch G, Ed.) pp 263, Wiley-Interscience, New York. [Google Scholar]
- 31.Schwartz TW, and Rosenkilde MM (1996) Trends Pharmacol. Sci. 17, 213–216. [DOI] [PubMed] [Google Scholar]
- 32.Baldwin JM (1993) EMBO J. 12, 1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Creighton TE (1984) in Proteins: Structural and Molecular Principles pp 2–60, Freeman and Co., New York. [Google Scholar]