Abstract
In patients with colorectal liver metastases (CLM), liver resection offers the possibility of cure and long-term survival. The liver is a highly immunogenic organ harboring ~80% of the body’s tissue macrophages. Emerging data demonstrate a critical role of the immune response for cancer treatment. We investigated variations within genes involved in immune response checkpoints and their association with outcomes in patients with CLM who underwent neoadjuvant chemotherapy including bevacizumab and liver resection. Single-nucleotide polymorphisms (SNPs) in nine genes (CCL2, CCR2, LAG3, NT5E, PDCD1, CD274, IDO1, CTLA4 and CD24) were analyzed in genomic DNA from 149 patients with resected bevacizumab-pretreated CLM by direct Sanger DNA sequencing, and correlated with response, recurrence-free survival (RFS), overall survival (OS), probability of cure and recurrence patterns. IDO1 (indoleamine 2, 3-dioxygenase) rs3739319 G>A and CD24 rs8734 G>A showed a significant difference in 3-year OS rates. In addition, IDO1 rs3739319 G>A was significantly associated with extrahepatic recurrence. Recursive partitioning analyses revealed that IDO1 rs3739319 G>A was the dominant SNP predicting RFS and OS. Our data suggest that variants within genes involved in immune response checkpoints are associated with outcomes in patients with resected CLM and might lead to improved treatment strategies modulating anti-tumor immune response by targeting novel immune checkpoints.
INTRODUCTION
Approximately 50% of the patients with colorectal cancer present with metastases either at the time of diagnosis or at a later time.1,2 If metastases are limited to the liver, patients can be cured or survival can be prolonged in a multidisciplinary treatment approach.3 In patients with resectable colorectal liver metastases (CLM), perioperative chemotherapy has demonstrated to prolong progression-free survival, and in patients with unresectable CLM, chemotherapy can reduce tumor burden to allow secondary liver resection.4,5 The addition of the monoclonal antibody bevacizumab to chemotherapy, which inhibits angiogenesis by targeting the vascular endothelial growth factor (VEGF), has been approved in the metastatic setting and is being used successfully in patients with liver-limited disease.6,7
The aim of this multidisciplinary treatment approach is cure, which can be achieved in ~ 25% of the patients.4 However, predictive and prognostic biomarkers are needed to select patients benefiting from treatment. The liver is a highly immunogenic organ harboring ~80% of the body’s tissue macrophages. In addition, T cells and other immune cells infiltrate the microenvironment of CLM, representing the tumor’s immunogenic nature.8 Thereby, investigating the immune system is a promising approach to identify biomarkers in patients with CLM. Previously, it has been demonstrated that these cells facilitate the anti-tumoral immune response and thereby influence clinical outcome in the treatment of CRC.9 Emerging data show that VEGF and other factors involved in angiogenesis are having a significant role in modulation of the immune system, suggesting a potential role of immune response on the efficacy of anti-VEGF targeted therapy.10
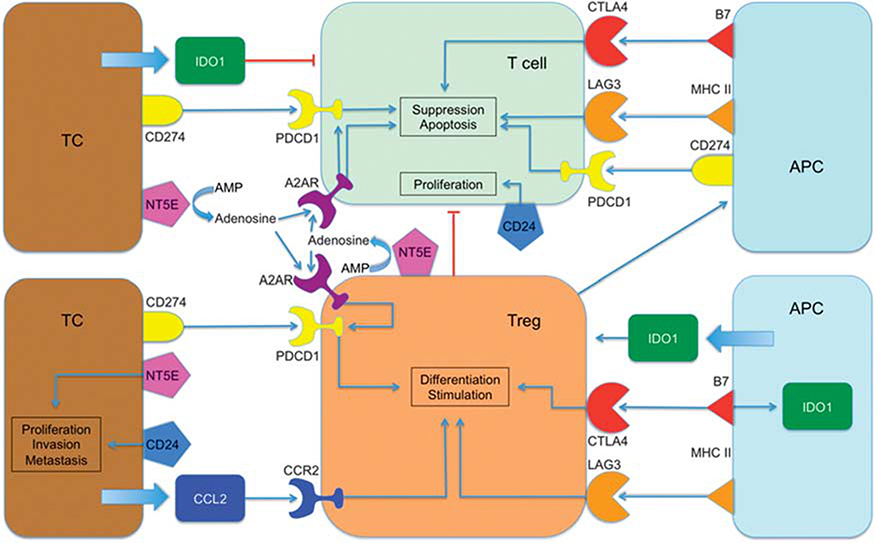
The regulation of the immune response upon activation by antigens is facilitated via immune checkpoints. They represent an entity of cellular immune regulators and are considered attractive targets for drug development.11 Therefore, these immune checkpoints may be predictive or prognostic biomarkers in the treatment of metastatic CRC. However, the influence of these checkpoints on clinical outcome in patients is still unclear and specifically, in patients with CLM undergoing perioperative bevacizumab-based chemotherapy has not yet been studied. Our goal was to test the clinical significance of single-nucleotide polymorphisms (SNPs) in genes coding for immune checkpoints on response and clinical outcome to bevacizumab-based perioperative chemotherapy. We investigated 10 SNPs in a panel of nine genes involved in immune response checkpoints (Figure 1). This comprised genes that are involved in inhibition of cytotoxic T cells and activation of inhibitory T-regulatory cells (Tregs). These mechanisms of action are facilitated via adenosine induced through adenosine monophosphate dephosphorylation by NT5E, via cell–cell interaction by CD274, PDCD1, LAG3 and CTLA4, via tryptophan degeneration by IDO1 (indoleamine 2, 3-dioxygenase), via the CCL2/CCR2 axis and via homeostatic proliferation by CD24.
Figure 1.
Network of action of the investigated genes. A2AR, adenosine A2A receptor; AMP, adenosine monophosphate; APC, antigen-presenting cell; B7, B-lymphocyte-activation antigen B7; CCL2, chemokine (C-C motif) ligand 2; CCR2, C-C chemokine receptor 2; CTLA4, cytotoxic T-lymphocyte antigen 4; MHC II, major histocompatibility complex II; PDCD1, programmed cell death 1; TC, tumor cell; Treg, T-regulatory cell.
MATERIALS AND METHODS
In this study, 149 patients from a single institution (Medical University Vienna) who underwent 3 months of neoadjuvant and 3 months of adjuvant fluoropyrimidine-based combination chemotherapy (oxaliplatin, n = 124; irinotecan, n =18; both, n = 7) including bevacizumab were enrolled, of whom 87 (58.4%) were male. The last dose of bevacizumab was administered 5 weeks before liver resection. Clinical data were obtained from a prospectively maintained database. The patients’ median age was 62 years (range 30–80). For all patients, the median recurrence-free survival (RFS) was 11.7 months (95% confidence interval 10.3, 15.1). The 3-year overall survival (OS) rate was 71% (s.e. ± 4%). The median follow-up in this study was 3.9 years (range 0.02–7.7). Patient demographic data are given in Table 1. The study was approved by the local institutional review board.
Table 1.
Patient and tumor characteristics
| n | % | |
|---|---|---|
| Sex | ||
| Male | 87 | 58.4 |
| Female | 62 | 41.6 |
| Age | ||
| Median (range) | 62 years (30–80) | |
| < 65 years | 90 | 60.4 |
| ⩾ 65 years | 59 | 39.6 |
| Size of metastasesa | ||
| 1–50 mm | 127 | 85.8 |
| > 50 mm | 21 | 14.2 |
| Number of metastasesa | ||
| 1–2 | 88 | 59.5 |
| > 2 | 60 | 40.5 |
| Distribution of metastases | ||
| Unilobar | 76 | 51.0 |
| Bilobar | 73 | 49.0 |
| Timing of metastases | ||
| Metachronous | 55 | 36.9 |
| Synchronous | 94 | 63.1 |
| Primary tumor sitea | ||
| Right colon | 39 | 26.4 |
| Left colon | 60 | 40.5 |
| Rectum | 49 | 33.1 |
One patient had missing information.
Radiological response after neoadjuvant chemotherapy was assessed by two radiologists according to Response Evaluation Criteria In Solid Tumours (RECIST 1.1) using computed tomography.12 Only patients with radiological response or disease stabilization underwent liver resection and were included in this study.
The assessment of histological response was performed according to Rubbia-Brandt et al.13 Major histological response comprised tumor regression grade 1 and 2, partial histological response tumor regression grade 3 and no histological response tumor regression grade 4 and 5.13
After completion of treatment, clinical examinations, computed tomography scans of thorax, liver and abdomen and blood tests including carcinoembryonic antigen were routinely performed every 3 months during the first 3 years, and then every 6 months.
SNP selection and genotyping
Candidate SNPs with a minor allele frequency of ⩾ 10% in Caucasians according to the Ensembl database were chosen for analyses in this study, when having functional relevance according to Queen’s University F-SNP and National Institute of Environmental Health Sciences SNP Function Prediction, representing a set of other SNPs (tag) or have been previously described to be clinically relevant according to the literature if no functional data were available (Table 2).14–22
Table 2.
Genes and SNPs of immune checkpoints
| rs number | MAFa | Base change | Location | Function | Primer sequence |
|---|---|---|---|---|---|
| CCL2 | |||||
| rs4586 | 39% | A > G | Exon 2 | Protein coding, splicing regulation and post translation | F: 5′-TCAATGCCCCAGTCACCT-3′ R: 5′-TCACATCACAGCTTCTTTGGG-3′ |
| LAG3 | |||||
| rs3782735 | 42% | A > G | Intronic | Tag | F: 5′-GGACTCCCCTGCTCTCAATT-3′ R: TCACTCCTCCCAAATCCAGG-3′ |
| NT5E | |||||
| rs6922 | 33% | G > T | 3′-UTR | Transcriptional regulation | F: 5′-TTGTTGCAGCAAAATAATAGCC-3′ R: 5′-CCCTATTTTACTGGCCAAGTGT-3′ |
| PDCD1 | |||||
| rs10204525 | 12% | C > T | 3′-UTR | Tag | F: 5′-GGTGTTGGGAGGGCAGAA-3′ R: 5′-GTGTGGATGTGAGGAGTGGA-3′ |
| CCR2 | |||||
| rs3092964 | 22% | T > C | Near 5′-UTR | Transcriptional regulation | F: 5′-CCTGTATCTCCGCCTTCACT-3′ R: 5′-AATGCCTACAACTCACCAGC-3′ |
| IDO1 | |||||
| rs9657182 | 45% | A > G | Upstream | NA | F: 5′-GGTCATAAAAGGAGAAAATGACC-3′ R: 5′-TTGCAAAGAGCTTTCAGAAAAA-3′ |
| rs3739319 | 40% | G > A | Intronic | Transcriptional regulation | F: 5′-GACCTTGACCTCAGTGAATGC-3′ R: 5′-ATGTCCTGGAGGAACTGAGC-3′ |
| CD274 | |||||
| rs2297137 | 27% | G > A | Intronic | Transcriptional regulation | F: 5′-ATTCTCCTGCCCCTCACC-3′ R: 5′-AAGGGGTGAGAATTGGATCA-3′ |
| CTLA4 | |||||
| rs231777 | 15% | C > T | Intronic | Transcriptional regulation | F: 5′-TGACACCAGGTTTGTTACACG-3′ R: 5′-TCTTCTCCAGTAAAAATTAAATCATCA-3′ |
| CD24 | |||||
| rs8734 | 32% | G > A | Exon 1 | Protein coding, splicing regulation and post translation | F: 5′-TGGAGGTAGCTGTGATGTGG-3′ R: 5′-ACCACGAAGAGACTGGCTGT-3′ |
Abbreviations: A, adenine; C, cytosine; G, guanine; MAF, minor allele frequency; NA, not available; rs, reference SNP number; T, thymine; 3′-UTR, 3′-untranslated region; 5′-UTR, 5′-untranslated region.
According to the Ensembl database (phase 1 of the 1000 Genomes Project) for Europeans.
Genomic DNA from formalin-fixed paraffin-embedded CLM was extracted using the QIAamp DNAeasy Kit (Qiagen, Hilden, Germany). Primers were used for PCR as given in Table 2. DNA from CLM was analyzed by direct Sanger sequencing and genotype was determined using the ABI Sequencing Scanner v1.0 (Applied Biosystems, Foster City, CA, USA). Patients’ clinical data were unknown to investigators involved in SNP analyses.
Statistical analyses
SNPs involved in immune response checkpoints were tested for associations with radiological and histological response, RFS and OS. The primary end point of the study was RFS. RFS was defined as the time from the day of liver resection to the first day of documented disease recurrence or death. If recurrence or death was not observed, RFS was censored on the day of the last computed tomography scan. The secondary end points were radiological and histological response and OS. OS was defined as the time from liver resection to the date of death or censored on the date last known to be alive. All possible modes of inheritance of SNPs were considered. The distribution of the SNP alleles was tested for Hardy–Weinberg equilibrium using a 1° freedom χ2-test. Kaplan–Meier estimation and log-rank tests were used in univariable analysis and Cox proportional hazards regression models were used in multivariable analysis to the test for associations of SNPs with RFS and OS. The multivariable analysis was adjusted for age (< 65 vs 65+ years), number of metastases (1–2 vs >2), distribution of metastases (unilobar vs bilobar) and timing of metastases (metachronous vs synchronous) because these characteristics were associated with RFS or OS at a 0.10 significance level. χ2-tests were used to investigate associations between SNPs and reponse in the univariable analysis. In the multivariable analysis, logistic regression model were used controlling for potential predictive variables. SNP profiles for radiological and histological response, RFS and OS were investigated using recursive partitioning.
Cure rate by individual SNPs was estimated by mixture cure model.23 The patterns of recurrence were estimated using cumulative incidences of intrahepatic only or extrahepatic recurrence using competing risks model.
SAS/STAT 12.3 (SAS Institute, Cary, NC, USA), a SAS macro (%pspmcm), along with rpart and cmprsk functions in R package (Version 3.1.2, R Foundation for Statistical Computing, Vienna, Austria) were used to perform all the analyses. Case-wise deletion was applied when patients with missing polymorphisms were excluded in the analyses. All tests were two-sided at a significance level of 0.05. P-values were not adjusted for multiple hypothesis testing. For internal validation, leave-one-out cross validation were used as previously described.24,25
RESULTS
Hardy–Weinberg equilibrium
All tested SNPs were in Hardy–Weinberg equilibrium except CD24 rs8734 G >A.
Gene variants and OS
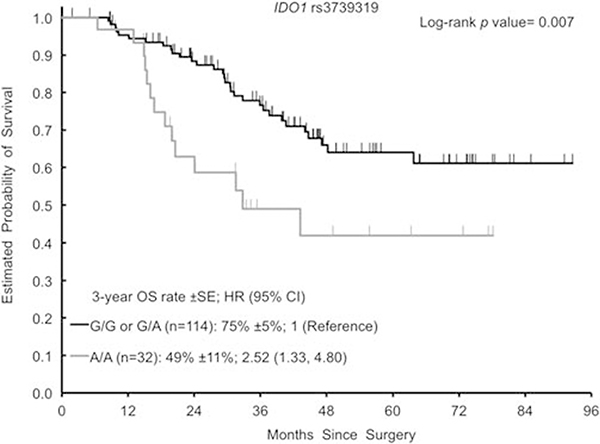
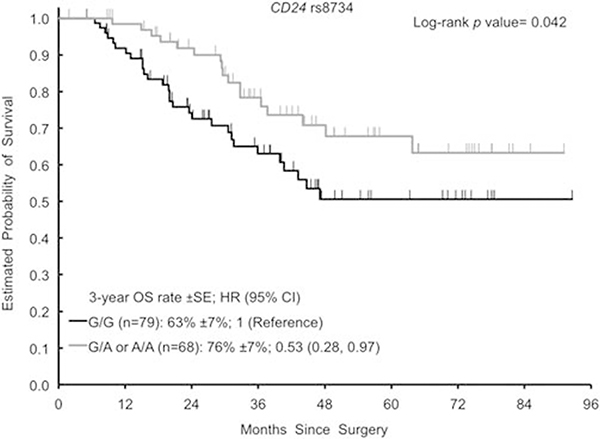
The homozygous variant genotype (A/A) of IDO1 rs3739319 G >A was associated with shorter OS in this study (Figure 2). The 3-year OS rate was 49% (±11%) for the patients harboring an A/A genotype compared with 75% (±5%) for patients with a G/G or G/A genotypes. The hazard ratio was 2.52 (1.33, 4.80; P = 0.007) in univariable analysis and 2.04 (1.03, 4.03; P = 0.040) in multivariable analysis. The variant genotypes (G/A or A/A) of CD24 rs8734 G > A were associated with longer OS (Figure 3). The 3-year OS rate was 76% (±7%) for G/A or A/A compared with 63% (±7%) for G/G. The hazard ratio was 0.53 (0.28, 0.97; P = 0.042) in univariable analysis. In multivariable analysis, CD24 rs8734 G>A did not remain significant for OS (hazard ratio 0.55 (0.30, 1.01; P = 0.054). Data on OS are given in Table 3.
Figure 2.
Kaplan–Meier curves for IDO1 rs3739319 G>A for OS. CI, confidence interval; HR, hazard ratio; OS, overall survival.
Figure 3.
Kaplan–Meier curves for CD24 rs8734 G>A for OS. CI, confidence interval; HR, hazard ratio; OS, overall survival.
Table 3.
Gene variants and recurrence-free survival and overall survival
| Recurrence-free survival | Overall survival | ||||||
|---|---|---|---|---|---|---|---|
| n | Median (95%CI) | Univariable HR (95% CI) | Multivariable HR (95% CI) | 3-year rate | Univariable HR (95% CI) | Multivariable HR (95% CI) | |
| CCL2 rs4586 | |||||||
| A/A | 55 | 11.7 (8.3, 16.0) | 1 (reference) | 1 (reference) | 0.63 ± 0.08 | 1 (reference) | 1 (reference) |
| A/G | 68 | 13.5 (10.1, 22.5) | 0.87 (0.57, 1.35) | 1.05 (0.67, 1.64) | 0.72 ± 0.07 | 0.84 (0.45, 1.59) | 1.08 (0.56, 2.07) |
| G/G | 22 | 10.1 (4.6, 14.3) | 1.31 (0.73, 2.34) | 1.47 (0.80, 2.69) | 0.83 ± 0.14 | 0.61 (0.21, 1.79) | 1.05 (0.38, 2.89) |
| P-valuea | 0.37 | 0.44 | 0.83 | 0.97 | |||
| LAG3 rs3782735 | |||||||
| A/A | 58 | 11.2 (8.4, 18.5) | 1 (reference) | 1 (reference) | 0.66 ± 0.08 | 1 (reference) | 1 (reference) |
| A/G | 70 | 13.0 (9.5, 16.5) | 0.92 (0.60, 1.40) | 0.89 (0.58, 1.37) | 0.77 ± 0.07 | 0.82 (0.42, 1.59) | 0.96 (0.50, 1.85) |
| G/G | 20 | 11.0 (4.1, 81.2) | 0.99 (0.53, 1.84) | 0.97 (0.51, 1.85) | 0.59 ± 0.14 | 1.61 (0.72, 3.58) | 1.68 (0.74, 3.83) |
| P-valuea | 0.92 | 0.87 | 0.35 | 0.38 | |||
| NT5E rs6922 | |||||||
| G/G | 61 | 13.0 (9.2, 18.6) | 1 (reference) | 1 (reference) | 0.72 ± 0.08 | 1 (reference) | 1 (reference) |
| G/T | 72 | 10.3 (8.4, 16.3) | 1.11 (0.73, 1.68) | 0.89 (0.58, 1.37) | 0.66 ± 0.07 | 1.28 (0.68, 2.40) | 1.09 (0.58, 2.07) |
| T/T | 15 | 14.0 (6.5, 16.6) | 1.02 (0.51, 2.05) | 0.87 (0.42, 1.79) | 0.75 ± 0.17 | 0.74 (0.22, 2.52) | 0.64 (0.18, 2.24) |
| P-valuea | 0.89 | 0.85 | 0.54 | 0.70 | |||
| PDCD1 rs10204525 | |||||||
| C/C | 119 | 11.1 (9.5, 14.3) | 1 (reference) | 1 (reference) | 0.69 ± 0.05 | 1 (reference) | 1 (reference) |
| C/T | 25 | ||||||
| T/T | 3 | ||||||
| C/T or T/T | 28 | 16.0 (9.2, 77.3) | 0.68 (0.39, 1.17) | 0.69 (0.40, 1.21) | 0.74 ± 0.13 | 0.56 (0.22, 1.41) | 0.61 (0.24, 1.58) |
| P-valuea | 0.16 | 0.20 | 0.17 | 0.31 | |||
| CCR2 rs3092964 | |||||||
| T/T | 90 | 10.8 (8.4, 14.4) | 1 (reference) | 1 (reference) | 0.68 ± 0.06 | 1 (reference) | 1 (reference) |
| T/C | 47 | 14.3 (10.4, 22.1) | 0.85 (0.55, 1.29) | 0.87 (0.56, 1.36) | 0.78 ± 0.08 | 0.91 (0.47, 1.77) | 0.82 (0.42, 1.61) |
| C/C | 10 | 7.8 (4.0, 46.3) | 1.17 (0.53, 2.55) | 1.09 (0.49, 2.41) | 0.40 ± 0.15 | 2.17 (0.83, 5.66) | 1.79 (0.66, 4.85) |
| P-valuea | 0.64 | 0.78 | 0.24 | 0.38 | |||
| IDO1 rs9657182 | |||||||
| A/A | 48 | 11.7 (7.6, 27.9) | 1 (reference) | 1 (reference) | 0.66 ± 0.09 | 1 (reference) | 1 (reference) |
| A/G | 75 | 11.0 (8.7, 15.1) | 1.19 (0.76, 1.88) | 1.00 (0.63, 1.59) | 0.70 ± 0.07 | 0.86 (0.44, 1.68) | 0.77 (0.40, 1.50) |
| G/G | 26 | 16.5 (6.5, 29.1) | 0.96 (0.53, 1.76) | 0.80 (0.41, 1.54) | 0.75 ± 0.13 | 0.76 (0.31, 1.90) | 0.80 (0.32, 2.01) |
| P-valuea | 0.64 | 0.74 | 0.75 | 0.74 | |||
| IDO1 rs3739319 | |||||||
| G/G or G/A | 114 | 13.0 (10.5,17.8) | 1 (reference) | 1 (reference) | 0.75 ± 0.05 | 1 (reference) | 1 (reference) |
| G/G | 43 | ||||||
| G/A | 71 | ||||||
| A/A | 32 | 8.4 (4.9,14.0) | 1.48 (0.93,2.34) | 1.13 (0.68,1.88) | 0.49 ± 0.11 | 2.52 (1.33,4.80) | 2.04 (1.03,4.03) |
| P-valuea | 0.093 | 0.64 | 0.007 | 0.040 | |||
| CD274 rs2297137 | |||||||
| G/G | 88 | 12.3 (10.1, 16.3) | 1 (reference) | 1 (reference) | 0.71 ± 0.06 | 1 (reference) | 1 (reference) |
| G/A | 49 | ||||||
| A/A | 8 | ||||||
| G/A or A/A | 57 | 11.2 (8.4, 19.5) | 0.87 (0.57, 1.31) | 0.84 (0.54, 1.31) | 0.66 ± 0.08 | 1.08 (0.58, 2.00) | 1.11 (0.59, 2.10) |
| P-valuea | 0.50 | 0.44 | 0.76 | 0.74 | |||
| CTLA4 rs231777 | |||||||
| C/C | 111 | 12.8 (10.1, 15.1) | 1 (reference) | 1 (reference) | 0.67 ± 0.06 | 1 (reference) | 1 (reference) |
| C/T | 33 | ||||||
| T/T | 4 | ||||||
| C/T or T/T | 37 | 11.7 (6.7, 18.6) | 1.17 (0.75, 1.81) | 1.11 (0.70, 1.75) | 0.80 ± 0.10 | 0.85 (0.42, 1.73) | 0.65 (0.31, 1.36) |
| P-valuea | 0.48 | 0.65 | 0.55 | 0.25 | |||
| CD24 rs8734 | |||||||
| G/G | 79 | 10.8 (7.7, 14.3) | 1 (reference) | 1 (reference) | 0.63 ± 0.07 | 1 (reference) | 1 (reference) |
| G/A | 64 | ||||||
| A/A | 4 | ||||||
| G/A or A/A | 68 | 11.7 (10.1, 22.6) | 0.79 (0.53, 1.17) | 0.74 (0.49, 1.13) | 0.76 ± 0.07 | 0.53 (0.28, 0.97) | 0.55 (0.30, 1.01) |
| P-valuea | 0.24 | 0.17 | 0.042 | 0.054 | |||
Abbreviations: A, adenine; C, cytosine; CI, confidence interval; G, guanine; HR, hazard ratio; T, thymine.
Based on log-rank test in the univariable analysis and Wald test in the multivariable analysis within Cox regression model. Multivariable Cox regression model was adjusted for age (< 65 vs ⩾ 65 years), number of metastases (1–2 vs > 2), size of metastases (1–50 mm vs > 50 mm), and time since diagnosis of CRC (< 6 months, 6–12 months, 1–2 years and > 2 years).
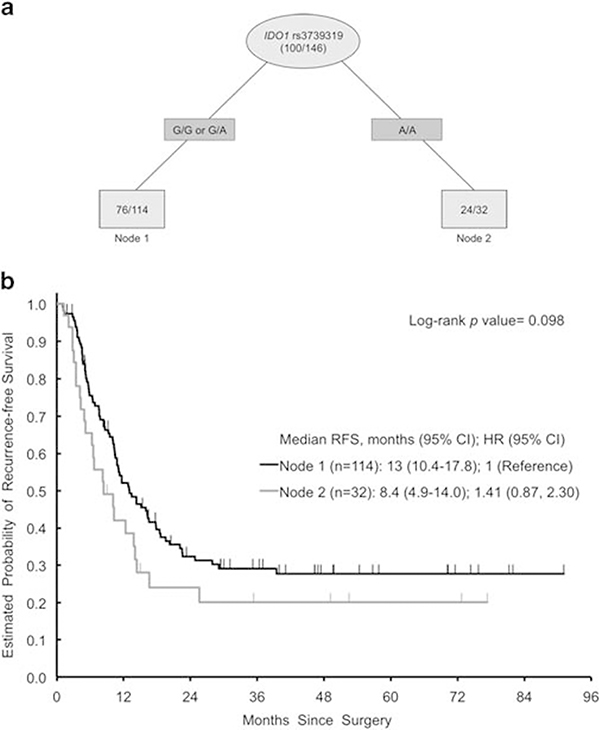
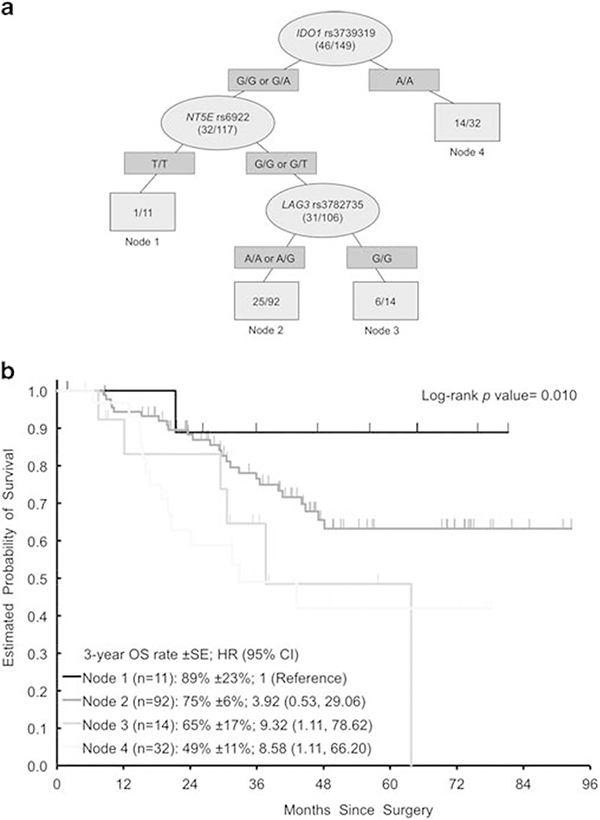
Recursive partitioning for RFS and OS
IDO1 rs3739319 G>A was the most important SNP to predict RFS and OS upon recursive partitioning analysis (Figures 4 and 5). Other SNPs predicting OS in subgroups were NT5E rs6922 G>T and LAG3 (lymphocyte-activation gene 3) rs3782735 A>G. For RFS, node 1 showed a trend toward longer RFS (log rank, P = 0.098). Recursive partitioning analyses did not create decision trees for radiological and histological response.
Figure 4.
(a) Recursive Partitioning for RFS. Light gray ovals represent intermediate subgroups; light gray squares represent terminal nodes. Dark gray rectangles indicate predictive polymorphism. Fractions within nodes indicate patients who relapsed per total patients with that node. Node 1 represents intermediate-risk patients. Node 2 represents high-risk patients. (b) Kaplan–Meier curves for RFS nodes. HR, hazard ratio; RFS, recurrence-free survival.
Figure 5.
(a) Recursive Partitioning for OS. Light gray ovals represent intermediate subgroups; light gray squares represent terminal nodes. Dark gray rectangles indicate predictive polymorphism. Fractions within nodes indicate patients who died per total patients with that node. Node 1 represents low-risk patients; Node 2 represents intermediate-risk patients. Node 3, 4 represent high-risk patients. (b) Kaplan–Meier curves for OS nodes. HR, hazard ratio; OS, overall survival.
Gene variants and pattern of recurrence
IDO1 rs3739319 G>A was associated with extrahepatic recurrence. The 2-year incidence of extrahepatic recurrence was 72% (s.e. ± 9%) for A/A and 56% (±5%) for G/G or G/A (P =0.024).
Gene variants and radiological and histological response, RFS and probability of cure
None of the SNPs investigated in this study was significantly associated with radiological or histological response, probability of cure or RFS in univariable or multivariable analysis (Tables 3 and 4).
Table 4.
Gene variants and radiological and histological response
| Radiological response |
Histological response |
|||||
|---|---|---|---|---|---|---|
| n | PR | SD | MjHR | PHR | NHR | |
| CCL2 rs4586 | ||||||
| A/A | 55 | 42 (76%) | 13 (24%) | 20 (37%) | 14 (26%) | 20 (37%) |
| A/G | 68 | 56 (85%) | 10 (15%) | 25 (38%) | 21 (32%) | 20 (30%) |
| G/G | 22 | 20 (95%) | 1 (5%) | 9 (41%) | 4 (18%) | 9 (41%) |
| P-valuea | 0.14 | 0.87 | ||||
| LAG3 rs3782735 | ||||||
| A/A | 58 | 44 (77%) | 13 (23%) | 19 (34%) | 19 (34%) | 18 (32%) |
| A/G | 70 | 60 (87%) | 9 (13%) | 26 (38%) | 17 (25%) | 26 (38%) |
| G/G | 20 | 16 (84%) | 3 (16%) | 10 (50%) | 4 (20%) | 6 (30%) |
| P-valuea | 0.35 | 0.64 | ||||
| NT5E rs6922 | ||||||
| G/G | 61 | 51 (85%) | 9 (15%) | 21 (34%) | 18 (30%) | 22 (36%) |
| G/T | 72 | 57 (81%) | 13 (19%) | 27 (39%) | 18 (26%) | 24 (35%) |
| T/T | 15 | 13 (87%) | 2 (13%) | 7 (47%) | 4 (27%) | 4 (27%) |
| P-valuea | 0.85 | 0.67 | ||||
| PDCD1 rs10204525 | ||||||
| C/C | 119 | 97 (83%) | 20 (17%) | 41 (35%) | 34 (29%) | 41 (35%) |
| C/T | 25 | |||||
| T/T | 3 | |||||
| C/T or T/T | 28 | 22 (81%) | 5 (19%) | 14 (50%) | 6 (21%) | 8 (29%) |
| P-valuea | 0.79 | 0.23 | ||||
| CCR2 rs3092964 | ||||||
| T/T | 90 | 76 (85%) | 13 (15%) | 32 (36%) | 29 (33%) | 28 (31%) |
| T/C | 47 | 36 (80%) | 9 (20%) | 20 (43%) | 8 (17%) | 18 (39%) |
| C/C | 10 | 7 (70%) | 3 (30%) | 3 (33%) | 2 (22%) | 4 (44%) |
| P-valuea | 0.36 | 0.87 | ||||
| IDO1 rs9657182 | ||||||
| A/A | 48 | 37 (77%) | 11 (23%) | 14 (29%) | 15 (31%) | 19 (40%) |
| A/G | 75 | 62 (83%) | 13 (17%) | 32 (44%) | 18 (25%) | 23 (32%) |
| G/G | 26 | 22 (96%) | 1 (4%) | 10 (40%) | 7 (28%) | 8 (32%) |
| P-valuea | 0.14 | 0.35 | ||||
| IDO1 rs3739319 | ||||||
| G/G or G/A | 114 | 92 (83%) | 19 (17%) | 44 (39%) | 30 (27%) | 39 (35%) |
| G/G | 43 | |||||
| G/A | 71 | |||||
| A/A | 32 | 27 (84%) | 5 (16%) | 12 (40%) | 9 (30%) | 9 (30%) |
| P-valuea | 1.00 | 0.75 | ||||
| CD274 rs2297137 | ||||||
| G/G | 88 | 74 (85%) | 13 (15%) | 28 (33%) | 28 (33%) | 30 (35%) |
| G/A | 49 | |||||
| A/A | 8 | |||||
| G/A or A/A | 57 | 44 (80%) | 11 (20%) | 26 (46%) | 11 (20%) | 19 (34%) |
| P-valuea | 0.49 | 0.31 | ||||
| CTLA4 rs231777 | ||||||
| C/C | 111 | 92 (85%) | 16 (15%) | 42 (39%) | 27 (25%) | 40 (37%) |
| C/T | 33 | |||||
| T/T | 4 | |||||
| C/T or T/T | 37 | 29 (78%) | 8 (22%) | 14 (39%) | 12 (33%) | 10 (28%) |
| P-valuea | 0.44 | 0.57 | ||||
| CD24 rs8734 | ||||||
| G/G | 79 | 62 (79%) | 16 (21%) | 26 (34%) | 27 (35%) | 24 (31%) |
| G/A | 64 | |||||
| A/A | 4 | |||||
| G/A or A/A | 68 | 57 (86%) | 9 (14%) | 28 (42%) | 13 (19%) | 26 (39%) |
| P-valuea | 0.38 | 0.98 | ||||
Abbreviations: A, adenine; C, cytosine; G, guanine; MjHR, major histological response; NHR, no histological response; PHR, partial histological response; PR, partial response; SD, stable disease; T, thymine.
Based on the Cochran–Mantel–Haenszel test for histological response and Fisher’s exact test for radiological response.
Internal validation
Upon leave-one-out cross validation, IDO1 rs3739319 G>A was significantly associated with OS in both univariable and multivariable analysis for the recessive model (132 of 149 leave-one-out data sets). For the dominant model of CD24 rs8734 G>A, 101 of 149 leave-one-out data sets remained significant in univariable analysis and 43 of 149 in multivariable analysis (42 of 149 in both univariable and multivariable analysis).
DISCUSSION
We investigated a unique cohort of patients with bevacizumab-pretreated CLM who underwent liver resection with curative intent. This study confirms that the immune response influences clinical outcome and that SNPs within genes of immune checkpoints may serve as biomarkers in the treatment of colorectal cancer. The most important SNP to predict clinical outcome was IDO1 rs3739319 G>A. IDO1 rs3739319 G>A identified patients who benefited most from this multidisciplinary treatment approach with respect to OS.
IDO1 rs3739319 G> A is an intronic SNP and is associated with transcriptional regulation, suggesting functional relevance.15 IDO1 is an enzyme that converts the essential amino acid tryptophan into kynurenine. Tryptophan metabolites are involved in the regulation of the immune response. Toxic kynurenine activates Tregs and inhibits cytotoxic T cells.26,27 In turn, Tregs can also suppress T-cell activation. Moreover, tryptophan deficiency has been shown to inhibit the Mechanistic Target of Rapamycin pathway and activate the General Control Nonderepressible 2 pathway, leading to upregulation of stress response pathways, which suppresses T cells.28–30 Under physiological conditions, these processes provide immune tolerance at the fetal–maternal interface.31 In cancer, IDO1 is expressed by cancer cells and antigen-presenting cells and is associated with poor prognosis.32 Due to the mechanisms of action of IDO1, the host’s immune response is attenuated, which facilitates tumor growth. VEGF has been shown to increase IDO1 expression in dendritic cells, therefore anti-VEGF targeted treatment may potentially influence IDO1 and immune response.33 The importance of IDO1 rs3739319 G>A is further confirmed by the fact that it predicted the probability of extrahepatic recurrence. This finding that the A/A genotype was significantly associated with a higher 2-year extrahepatic recurrence rate and a lower 3-year OS rate is in line with a previous study demonstrating that extrahepatic recurrence after liver resection of CLM is a negative prognostic marker.34 However, the investigation of the clinical value of extrahepatic recurrence prediction warrants further studies. IDO1 is a promising drug target and its inhibition has demonstrated anti-proliferative efficacy in murine cancer models in vitro and in vivo.35–37 Clinical studies are currently conducted to investigate the IDO1 inhibitors, INCB024360 and indoximod, in combination with immunotherapy or chemotherapy in patients with melanoma, breast cancer and pancreatic cancer (NCT01604889, NCT01792050 and NCT02077881). In liver-limited metastatic colorectal cancer, the introduction of activators of a T-cell-driven immune response, such as IDO1 inhibitors, to the treatment concept appears especially promising due to the liver’s highly immunogenic nature.
Besides IDO1 rs3739319 G>A, CD24 rs8734 G>A was associated with OS in univariable analysis. CD24 is a cell surface glycosyl-phosphatidylinositol-anchored protein expressed on T cells and on cancer cells. In T cells, CD24 mediates homeostatic proliferation, a mechanism to replace activated and discarded lymphocytes.38 In addition, CD24 rs8734 G>A has previously been associated with inflammatory bowel disease susceptibility.17 In cancer cells, CD24 expression has been associated with cancer stem cells and more aggressive cancer behavior.39,40 Interestingly, hypoxia inducible factor-1 α is up-regulating CD24 expression and in turn CD24 salvages cell proliferation in hypoxia inducible factor-1α-depleted cancer cells, suggesting a protective effect of CD24 against hypoxia.41 The connection of hypoxia inducible factor-1α to CD24 is not surprising, as angiogenesis has been closely linked to the immune system. In colorectal cancer, selective blockade of the VEGF-A/VEGFR axis resulted in an attenuated proliferation of T-cell-inhibiting Tregs in vivo.10 Furthermore, anti-VEGF treatment was associated with a polarization of M2-immunosuppressive toward M1-antitumoral macrophages in a breast cancer model.42 However, it is more likely that the effect of CD24 observed in this study is related to immune response and VEGF-modulation by bevacizumab treatment rather than to a cancer stem cell-related effect. This hypothesis is supported by the fact that CD24 rs8734 G>A did not affect survival in a previous study in the adjuvant setting.43 However, the interpretation of the results for CD24 rs8734 G>A has to be made with caution, as this SNP was not in Hardy–Weinberg equilibrium. Moreover, the statistical significance for OS was lost in multivariable analysis.
In addition to univariable and multivariable analyses, we performed recursive partitioning analyses that identified SNPs that were associated with RFS and OS in population subgroups in an unbiased, hierarchical manner. IDO1 rs3739319 G>A was the strongest predictor of RFS and OS, whereas NT5E rs6922 G>T and LAG3 rs3782735 A>G predicted OS in subgroups.
NT5E is an ecto-nucleotidase expressed on cancer cells and Tregs that converts extracellular adenosine monophosphate to adenosine. Adenosine binds to its receptor A2AR on T cells leading to their suppression and also to the A2AR receptor on Tregs leading to their stimulation.44 By suppression of T cells and stimulation of Tregs, NT5E inhibits immune response. In previous studies, NT5E expression has been shown be affected by hypoxia inducible factor-1 a and, VEGF expression by CD74 and adenosine demonstrating its relevance for angio- genesis.45,46 These results suggest that NT5E rs6922 G>T may be associated with outcome in patients treated with anti-VEGF targeted treatment.
In patients harboring any G allele in NT5E rs6922 G>T, LAG3 rs3782735 A>G separated patient with intermediate risk for death. LAG3, which is expressed on T cells and Tregs and binds to class II major histocompatibility complex, has been associated with inhibited immune response. LAG3 leads to T-cell suppression and Treg stimulation.47 Anti-LAG3 targeted therapy has demonstrated anti-tumor activity in a murine model by interrupting its immunosuppressive effect.48 So far, LAG3 has not been associated with VEGF and angiogenesis, therefore it is unknown if the differences in OS found in one of the three analysis subgroup can be considered to be treatment related.
The findings from recursive partitioning analyses demonstrate that some genes may not be relevant for all patients but only for some patients in distinct subgroups. This statistical approach allows stratifying patients according to their clinical outcome in a comprehensive manner and may help to develop genetic risk profiles for future studies. However, the identified risk groups require further validation in prospective trials before they can be introduced into clinical practice. The fact that a recursive partitioning tree was created for IDO1 rs3739319 G>A for RFS while the results from the log-rank test for this SNP were not statistically significant can be explained with the different statistical methods used. Nevertheless, as the two methods yielded different results, the interpretation of the association should be performed with caution. Moreover, no trees were created for radiological and histological response. This can also be explained by the statistical method that defines the terminal nodes (responders vs non-responders) depending on the patient majority. The method creates a tree only when, in one genotype group, the majority of patients are responders and non-responders in the other. However, in all genotype groups for all investigated SNPs, the majority were responders.
Interestingly, the SNPs investigated in this study were related to OS and not to response or RFS. However, it is unclear whether these SNPs are prognostic or predictive, as the differences seen between the groups may be a treatment effect of bevacizumab and subsequent immune modulation. The hypothesis that these findings are due to a treatment effect is supported by the results from the pivotal study that investigated the anti-CTLA4 antibody ipilimumab and led to its approval in melanoma. Patients receiving ipilimumab had prolonged OS, but not an improved RFS compared with those receiving glycoprotein 100.49
A limitation of this study is that the results could not be validated in an independent patient population as the presented cohort underwent a unique treatment regimen. However, the results for IDO1 rs3739319 G>A could be validated in internal cross validation. Nevertheless, when interpreting the results, it has to be considered that no correction for multiple testing has been performed.
In conclusion, this study shows for the first time that variations within genes involved in immune response checkpoints predict clinical outcome in this uniformly treated patient cohort. As currently most of the patients who undergo liver resection will develop recurrent disease, biomarkers that predict cure are needed. The results of this study may help to identify those patients who can be cured by this aggressive treatment concept and, moreover, may help to identify those who do not benefit and should therefore be referred to other treatment strategies. Furthermore, the close link of angiogenesis and immune response may make these SNPs potentially relevant biomarkers in the treatment of metastatic colorectal cancer with anti-VEGF targeted treatment. Apart from the identification of SNPs as biomarkers, these results may be relevant to identify targets for novel treatment strategies to enhance immune response.
ACKNOWLEDGMENTS
SS is a recipient of an Erwin Schrödinger fellowship of the Austrian Science Fund (J 3501-B13). AS is a recipient of a Rio Hortega Research Grant from the Insituto de Salud Carlos III (CM11/00102). SS is supported by a postdoctoral fellowship from the German Cancer Aid (Mildred-Scheel Foundation). TG receives financial support by Roche, Merck-Serono, Sanofi-Aventis, Bayer and Amgen (all Vienna, Austria). H-JL receives financial support by the P30CA014089 NIH grant, and the Daniel Butler Research Fund.
Footnotes
CONFLICT OF INTEREST
TG is a member of the advisory boards of Roche, Merck-Serono and Sanofi-Aventis (all Vienna, Austria). H-JL is a member of the advisory board of Genentech (San Francisco, CA). The remaining authors declare no conflict of interest.
REFERENCES
- 1.Howlader N, Noone AM, Krapcho M, Garshell J, Neyman N, Altekruse SF et al. (based on November 2012 SEER data submission, posted to the SEER web site, April 2013). SEER Cancer Statistics Review, 1975–2010. National Cancer Institute: Bethesda, MD, USA. [Google Scholar]
- 2.Andre T, Boni C, Mounedji-Boudiaf L, Navarro M, Tabernero J, Hickish T et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med 2004; 350: 2343–2351. [DOI] [PubMed] [Google Scholar]
- 3.Kopetz S, Chang GJ, Overman MJ, Eng C, Sargent DJ, Larson DW et al. Improved survival in metastatic colorectal cancer is associated with adoption of hepatic resection and improved chemotherapy. J Clin Oncol 2009; 27: 3677–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nordlinger B, Sorbye H, Glimelius B, Poston GJ, Schlag PM, Rougier P et al. Perioperative chemotherapy with FOLFOX4 and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC Intergroup trial 40983): a randomised controlled trial. Lancet 2008; 371: 1007–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adam R, Delvart V, Pascal G, Valeanu A, Castaing D, Azoulay D et al. Rescue surgery for unresectable colorectal liver metastases downstaged by chemotherapy: a model to predict long-term survival. Ann Surg 2004; 240: 644–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med 2004; 350: 2335–2342. [DOI] [PubMed] [Google Scholar]
- 7.Gruenberger B, Tamandl D, Schueller J, Scheithauer W, Zielinski C, Herbst F et al. Bevacizumab, capecitabine, and oxaliplatin as neoadjuvant therapy for patients with potentially curable metastatic colorectal cancer. J Clin Oncol 2008; 26: 1830–1835. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA.. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- 9.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006; 313: 1960–1964. [DOI] [PubMed] [Google Scholar]
- 10.Terme M, Pernot S, Marcheteau E, Sandoval F, Benhamouda N, Colussi O et al. VEGFA-VEGFR pathway blockade inhibits tumor-induced regulatory T-cell proliferation in colorectal cancer. Cancer Res 2013; 73: 539–549. [DOI] [PubMed] [Google Scholar]
- 11.Butt AQ, Mills KH.. Immunosuppressive networks and checkpoints controlling antitumor immunity and their blockade in the development of cancer immunotherapeutics and vaccines. Oncogene 2013; 33: 4623–4631. [DOI] [PubMed] [Google Scholar]
- 12.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009; 45: 228–247. [DOI] [PubMed] [Google Scholar]
- 13.Rubbia-Brandt L, Giostra E, Brezault C, Roth AD, Andres A, Audard V et al. Importance of histological tumor response assessment in predicting the outcome in patients with colorectal liver metastases treated with neo-adjuvant chemotherapy followed by liver surgery. Ann Oncol 2007; 18: 299–304. [DOI] [PubMed] [Google Scholar]
- 14.Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S et al. Ensembl 2014. Nucleic Acids Res 2014; 42: D749–D755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee PH, Shatkay H F-SNP: computationally predicted functional SNPs for disease association studies. Nucleic Acids Res 2008; 36: D820–D824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Z, Taylor JA.. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res 2009; 37: W600–W605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lisiansky V, Kraus S, Naumov I, Kazanov D, Nabiochtchikov I, Toledano O et al. Role of CD24 polymorphisms in the susceptibility to inflammatory bowel disease. Int J Biol Markers 2014; 29: e62–e68. [DOI] [PubMed] [Google Scholar]
- 18.Smith AK, Simon JS, Gustafson EL, Noviello S, Cubells JF, Epstein MP et al. Association of a polymorphism in the indoleamine- 2,3-dioxygenase gene and interferon-alpha-induced depression in patients with chronic hepatitis C. Mol Psychiatry 2012; 17: 781–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu H, Zheng L, Tang W, Yin P, Cheng F, Wang L Programmed death-1 (PD-1) polymorphisms in Chinese patients with esophageal cancer. Clin Biochem 2014; 47: 612–617. [DOI] [PubMed] [Google Scholar]
- 20.Anand A, Sharma NK, Gupta A, Prabhakar S, Sharma SK, Singh R et al. Single nucleotide polymorphisms in MCP-1 and its receptor are associated with the risk of age related macular degeneration. PLoS One 2012; 7: e49905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pizarro C, Garcia-Diaz DF, Codner E, Salas-Perez F, Carrasco E, Perez-Bravo F. PD-L1 gene polymorphisms and low serum level of PD-L1 protein are associated to T1D in Chilean population. Diabetes Metab Res Rev 2014; 30: 761–766. [DOI] [PubMed] [Google Scholar]
- 22.Aiba Y, Nakamura M, Joshita S, Inamine T, Komori A, Yoshizawa K et al. Genetic polymorphisms in CTLA4 and SLC4A2 are differentially associated with the pathogenesis of primary biliary cirrhosis in Japanese patients. J Gastroenterol 2011; 46: 1203–1212. [DOI] [PubMed] [Google Scholar]
- 23.Corbiere F, Joly P A SAS macro for parametric and semiparametric mixture cure models. Comput Methods Programs Biomed 2007; 85: 173–180. [DOI] [PubMed] [Google Scholar]
- 24.Molinaro AM, Simon R, Pfeiffer RM. Prediction error estimation: a comparison of resampling methods. Bioinformatics 2005; 21: 3301–3307. [DOI] [PubMed] [Google Scholar]
- 25.Hu-Lieskovan S, Vallbohmer D, Zhang W, Yang D, Pohl A, Labonte MJ et al. EGF61 polymorphism predicts complete pathologic response to cetuximab-based chemoradiation independent of KRAS status in locally advanced rectal cancer patients. Clin Cancer Res 2011; 17: 5161–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mezrich JD, Fechner JH, Zhang X, Johnson BP, Burlingham WJ, Bradfield CA. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J Immunol 2010; 185: 3190–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fallarino F, Grohmann U, Vacca C, Bianchi R, Orabona C, Spreca A et al. T cell apoptosis by tryptophan catabolism. Cell Death Differ 2002; 9: 1069–1077. [DOI] [PubMed] [Google Scholar]
- 28.Rohde J, Heitman J, Cardenas ME.. The TOR kinases link nutrient sensing to cell growth. J Biol Chem 2001; 276: 9583–9586. [DOI] [PubMed] [Google Scholar]
- 29.Gao X, Zhang Y, Arrazola P, Hino O, Kobayashi T, Yeung RS et al. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat Cell Biol 2002; 4: 699–704. [DOI] [PubMed] [Google Scholar]
- 30.Munn DH, Sharma MD, Baban B, Harding HP, Zhang Y, Ron D et al. GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity 2005; 22: 633–642. [DOI] [PubMed] [Google Scholar]
- 31.Munn DH, Zhou M, Attwood JT, Bondarev I, Conway SJ, Marshall B et al. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science 1998; 281: 1191–1193. [DOI] [PubMed] [Google Scholar]
- 32.Munn DH. Blocking IDO activity to enhance anti-tumor immunity. Front Biosci 2012; 4: 734–745. [DOI] [PubMed] [Google Scholar]
- 33.Marti LC, Pavon L, Severino P, Sibov T, Guilhen D, Moreira-Filho CA.. Vascular endothelial growth factor-A enhances indoleamine 2,3-dioxygenase expression by dendritic cells and subsequently impacts lymphocyte proliferation. Memorias do Instituto Oswaldo Cruz 2013; 109: 70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayashi M, Inoue Y, Komeda K, Shimizu T, Asakuma M, Hirokawa F et al. Clinicopathological analysis of recurrence patterns and prognostic factors for survival after hepatectomy for colorectal liver metastasis. BMC Surg 2010; 10: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muller AJ, DuHadaway JB, Donover PS, Sutanto-Ward E, Prendergast GC. Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene Bin1, potentiates cancer chemotherapy. Nat Med 2005; 11: 312–319. [DOI] [PubMed] [Google Scholar]
- 36.Hou DY, Muller AJ, Sharma MD, DuHadaway J, Banerjee T, Johnson M et al. Inhibition of indoleamine 2,3-dioxygenase in dendritic cells by stereoisomers of 1-methyl-tryptophan correlates with antitumor responses. Cancer Res 2007; 67: 792–801. [DOI] [PubMed] [Google Scholar]
- 37.Koblish HK, Hansbury MJ, Bowman KJ, Yang G, Neilan CL, Haley PJ et al. Hydroxyamidine inhibitors of indoleamine-2,3-dioxygenase potently suppress systemic tryptophan catabolism and the growth of IDO-expressing tumors. Mol Cancer Ther 2010; 9: 489–498. [DOI] [PubMed] [Google Scholar]
- 38.Li O, Zheng P, Liu Y. CD24 expression on T cells is required for optimal T cell proliferation in lymphopenic host. J Exp Med 2004; 200: 1083–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee TK, Castilho A, Cheung VC, Tang KH, Ma S, Ng IO. CD24(+) liver tumor-initiating cells drive self-renewal and tumor initiation through STAT3-mediated NANOG regulation. Cell Stem Cell 2011; 9: 50–63. [DOI] [PubMed] [Google Scholar]
- 40.Kristiansen G, Pilarsky C, Pervan J, Sturzebecher B, Stephan C, Jung K et al. CD24 expression is a significant predictor of PSA relapse and poor prognosis in low grade or organ confined prostate cancer. Prostate 2004; 58: 183–192. [DOI] [PubMed] [Google Scholar]
- 41.Thomas S, Harding MA, Smith SC, Overdevest JB, Nitz MD, Frierson HF et al. CD24 is an effector of HIF-1-driven primary tumor growth and metastasis. Cancer Res 2012; 72: 5600–5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang Y, Yuan J, Righi E, Kamoun WS, Ancukiewicz M, Nezivar J et al. Vascular normalizing doses of antiangiogenic treatment reprogram the immune-suppressive tumor microenvironment and enhance immunotherapy. Proc Natl Acad Sci USA 2012; 109: 17561–17566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerger A, Zhang W, Yang D, Bohanes P, Ning Y, Winder T et al. Common cancer stem cell gene variants predict colon cancer recurrence. Clin Cancer Res 2011; 17: 6934–6943. [DOI] [PubMed] [Google Scholar]
- 44.Stagg J The double-edge sword effect of anti-CD73 cancer therapy. Oncoimmunology 2012; 1: 217–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Synnestvedt K, Furuta GT, Comerford KM, Louis N, Karhausen J, Eltzschig HK et al. Ecto-5’-nucleotidase (CD73) regulation by hypoxia-inducible factor-1 mediates permeability changes in intestinal epithelia. J Clin Invest 2002; 110: 993–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allard B, Turcotte M, Spring K, Pommey S, Royal I, Stagg J. Anti-CD73 therapy impairs tumor angiogenesis. Int J Cancer 2014; 134: 1466–1473. [DOI] [PubMed] [Google Scholar]
- 47.Goldberg MV, Drake CG. LAG-3 in cancer immunotherapy. Curr Top Microbiol Immunol 2011; 344: 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woo SR, Turnis ME, Goldberg MV, Bankoti J, Selby M, Nirschl CJ et al. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res 2012; 72: 917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010; 363: 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]