Abstract
A recent genome-wide association study identified seven single-nucleotide polymorphisms (SNPs) in region 16q24, near the Forkhead box-F1 (FOXF1) gene, which confer susceptibility to esophageal adenocarcinoma. We examined whether these SNPs are associated with clinical outcomes in gastric cancer (GC) patients in Japan and the United States. A total of 362 patients were included in this study: 151 Japanese GC patients treated with first-line S1 plus CDDP (training cohort) and 211 GC patients from Los Angeles County (LAC; validation cohort). Genomic DNA was isolated from whole blood or tumor tissue and analyzed by PCR-based direct DNA sequencing. Cox proportional hazard regression analyses were used to assess relationships between FOXF1 SNPs and progression-free survival (PFS) and overall survival (OS). FOXF1 rs3950627 was significantly associated with survival in both the training and validation cohorts. Japanese patients with the C/C genotype had a longer PFS (median 8.2 vs 5.3 months, hazard ratio (HR) 1.44, P = 0.037) and OS (median 16.4 vs 12.2 months, HR 1.44, P = 0.043) compared to patients with any A allele. Similarly, LAC patients with the C/C genotype had improved OS (3.9 vs 2.3 years, HR 1.5, P = 0.022). Subgroup analyses showed these associations were specific to male patients and primary tumor subsite. Our findings suggest that FOXF1 rs3950627 might be a promising prognostic marker in GC patients.
INTRODUCTION
Forkhead box-F1 (FOXF1) belongs to a family of transcription factors that regulate embryonic development, and is implicated in carcinogenesis. FOXF1 is expressed in organs derived from the primitive gut,1,2 and gene knockout mice models have demonstrated an essential role for FOXF1 in the formation of the esophagus, liver, gallbladder and lung.3 The link between FOXF1 and carcinogenesis is not fully understood, and both oncogenic and tumor suppressor roles have been proposed in different tumor types. For instance, in breast cancer murine xenografts,4 FOXF1 expression has been associated with an invasive, mesenchymal phenotype and enhanced tumor growth. Conversely, in colorectal cancer cell lines, FOXF1 inhibits cell invasion and migration in a p53-dependent manner.5
The mechanisms underlying FOXF1-mediated gastric tumorigenesis are not well characterized, though FOXF1 promotes cancer cell motility and invasiveness, partly by regulating E-cadherin expression.5 Furthermore, hedgehog (Hh) signaling, which is integral to gastric gland differentiation,6,7 has been shown to induce FOXF1 expression during gut development (including stomach and intestinal organogenesis), vasculogenesis1,3,8,9 as well as in cancer-associated fibroblasts.10 Notably, Hh pathway activation and resulting tumor cell proliferation are thought to be regulated by ERα,11 which is more highly expressed in diffuse-type or undifferentiated GC variants.12
A recent genome-wide association study identified single-nucleotide polymorphisms (SNPs) in region 16q24, near the FOXF1 gene, to confer susceptibility to esophageal adenocarcinoma.13 The relationship between these SNPs and gastric cancer remains unknown, and understanding such associations may shed insight on molecular determinants of treatment efficacy and patient outcomes. We hypothesized that FOXF1 SNPs may predict clinical outcomes in GC patients and that these associations may vary by gender, ethnicity, primary tumor subsite and histology. Accordingly, we tested the prognostic value of seven previously reported FOXF1 SNPs in two independent cohorts of GC patients from Japan and the United States (US).
MATERIALS AND METHODS
Patients and treatment protocols
A total of 362 patients were included in this study. The training cohort consisted of 151 patients with histologically confirmed metastatic and recurrent gastric adenocarcinoma, treated with first-line S1 plus CDDP at the Cancer Institute Hospital in Japan. Concurrent chemotherapy consisted of infusion of cisplatin (60 mg m−2) on days 1 and 8 plus oral S1 (80 mg m− 2 day− 1) on days 1–21.14 Treatment was administered to patients unless there was evidence of disease progression, intolerable toxicities or early withdrawal. Response was measured by contrast-enhanced computed tomography (CT) scans every 8 weeks according to Response Evaluation Criteria in Solid Tumors (RECIST) 1.0.
The validation cohort consisted of 211 patients with gastric adenocarcinoma treated at Los Angeles County-University of Southern California (LAC-USC) Medical Center. These patients were recruited from a population-based case–control study that included White, African-American, Latino and Asian-American patients diagnosed with gastric cardia adenocarcinoma between 1992 and 1997 or with distal gastric cancer between 1992 and 1994.15
This study was approved by the Institutional Review Boards of each institute, and all patients signed an informed consent for the analysis of molecular correlates.
Genotyping
Genomic DNA was extracted from formalin-fixed paraffin-embedded (FFPE) tissues of 151 Japanese GC patients in the training cohort and from peripheral whole blood of 211 GC patients in the LAC validation cohort, using the QIAmp Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol (www.qiagen.com). PCR-based direct DNA sequence analysis using ABI 3100A Capillary Genetic Analyzer and Sequencing Scanner v1.0 (Applied Biosystems, Waltham, MA, USA) was performed for genotyping the SNPs. Extracted DNA was amplified using the following primer set: forward: 5′-CCTAAGAATTCGCCTCCACA-3′ and reverse: 5′-CCAAATCTCCGTGAATGTGA-3′ for rs1490865; forward: 5′-CCCAAAGGGCTGAGATTACA-3′ and reverse: 5′-AATTGGGCAAATAAAGGATGG-3′ for rs3111601; forward: 5′-GTCAGAGCCACAGTGCTACA-3′ and reverse: 5′-GAGGGTGGTAGAGAGTGGC-3′ for rs9936833; forward: 5′-AGTCGTCATGTTTGCCACCT-3′ and reverse: 5′-ATGACATCAGGGCACAGCTT-3′ for rs1728400; forward: 5′-TTCTGGTGCTGGCAATCCTT-3′ and reverse: 5′-AGGATGACTGGCACTCTCAT-3′ for rs3950627; forward: 5′-AAGGGAGGTGCATAGGTACG-3′ and reverse: 5′-GATTGTTAGGGCAGGCAAAA-3′ for rs2178146; forward: 5′-CAAATGGGCTCAAAGAGGTT-3′ and reverse: 5′-GGTCCTGGACTCTCGAATG-3′ for rs13332095.
Statistical analysis
The primary endpoint of this retrospective study was overall survival (OS), and the secondary endpoints were tumor response and progression-free survival (PFS). OS was defined between the date of initiation of therapy and death from any cause in Japanese cohort, whereas OS was defined between the date of diagnosis and death from any cause in LAC cohort. If death was not observed, OS was censored on the date of last contact. PFS was defined as the interval from first day of first-line chemotherapy to first day of documented disease progression or death in Japanese cohort. If progression was not observed, PFS was censored on the day of the last CT scan. Tumor responses were grouped into responders, including complete or partial response, and non-responders, including stable or progressive disease. In the LAC cohort, information regarding OS but not PFS or tumor responses was available.
The allelic distribution of the polymorphisms for each race/ethnic group was tested for deviation from Hardy–Weinberg equilibrium using the exact test with one degree of freedom. Linkage disequilibrium (LD) among candidate SNPs for each race/ethnic group was assessed using D′ and r2 values, and the haplotype frequencies were inferred using Haploview version 4.2 (www.broad.mit.edu/mpg/haploview).
The baseline characteristics between Japanese and LAC cohorts were compared by χ2 test for categorical variables and the Wilcoxon rank sum test for continuous variable. The associations between the SNPs with OS and PFS were examined by Kaplan–Meier estimation and log-rank test, using codominant, dominant or recessive genetic models, when appropriate. The multivariable Cox proportional hazards regression model with stratification factors was used, to evaluate the association between analyzed SNPs, considering the imbalance in the distributions of baseline characteristics in each cohort. The baseline characteristics that remained statistically significantly associated with endpoints in the model selection procedures (P < 0.1) were included in multivariable analyses of the SNPs and clinical outcome. The relationships between SNPs and tumor responses were assessed using the Fisher’s exact test. With 151 patients in the training cohort (OS events = 124), there was 80% power to detect a hazard ratio (HR) of 1.66–2.42 for a SNP associated with OS in the dominant model whether the frequency of the minor allele is from 0.05 to 0.3 using a two-sided log-rank test at a significance level of 0.05. Using the same test among the validation cohort (N = 211, OS events = 151), the power would exceed 87% for the same range of HRs (1.66–2.42) with the same allele frequencies.
SAS 9.4 (SAS Institute, Cary, NC, USA) was used to conduct all analyses. Case-wide deletion was applied when patients with missing values for all candidate SNPs in each cohort in univariable and multivariable analyses. All tests were two-sided with a significance level of 0.05.
RESULTS
Patient and tumor characteristics
The baseline demographic and clinical characteristics of patients in the Japanese and LAC are summarized in Table 1. Associations between baseline characteristics and clinical outcomes are presented for each cohort (Supplementary Tables 1A and B). The median follow-up period was 3.4 years in the Japanese cohort and 8.6 years in the LAC cohort. The median OS was 2.6 years for the LAC cohort. The median PFS and OS were 7.1 months and 14.1 months in the Japanese cohort.
Table 1.
Baseline clinical characteristics of patients in the Japanese and LAC cohorts
|
Japanese cohort (n = 151) |
LAC cohort (n = 211) |
P-valuea | |||
|---|---|---|---|---|---|
| n | % | n | % | ||
| Gender | 0.59 | ||||
| Male | 107 | 71 | 144 | 68 | |
| Female | 44 | 29 | 67 | 32 | |
| Age (year) | |||||
| Median (range) | 61 (16–78) | 62 (26–74) | 0.92 | ||
| < 65 | 91 | 61 | 123 | 58 | |
| ⩾ 65 | 58 | 39 | 88 | 42 | 0.60 |
| Stage | |||||
| I–III | 20 | 13 | 158 | 75 | < 0.001 |
| IV | 131 | 87 | 39 | 18 | |
| Unknownb | 14 | 7 | |||
| Lymph nodes | |||||
| No | 57 | 38 | 66 | 31 | 0.81 |
| Yes | 94 | 62 | 103 | 49 | |
| Unknownb | 42 | 20 | |||
| Tumor site | |||||
| Cardia | 47 | 31 | 93 | 44 | 0.030 |
| Distal | 97 | 64 | 118 | 56 | |
| Unknownb | 7 | 5 | |||
| Tumor differentiation | |||||
| Differentiated | 45 | 30 | 51 | 24 | 0.47 |
| Undifferentiated | 106 | 70 | 143 | 68 | |
| Unknownb | 17 | 8 | |||
| Ethnicity | |||||
| Asian | 151 | 100 | 108 | 51 | NA |
| Caucasian | 18 | 9 | |||
| Hispanic | 51 | 24 | |||
| African-American | 31 | 15 | |||
Abbreviation: NA, not applicable.
On the basis of χ2 test, or the Wilcoxon rank sum test whenever appropriate.
Not included in the test.
Associations between genetic variants and clinical outcomes in the Japanese training cohort
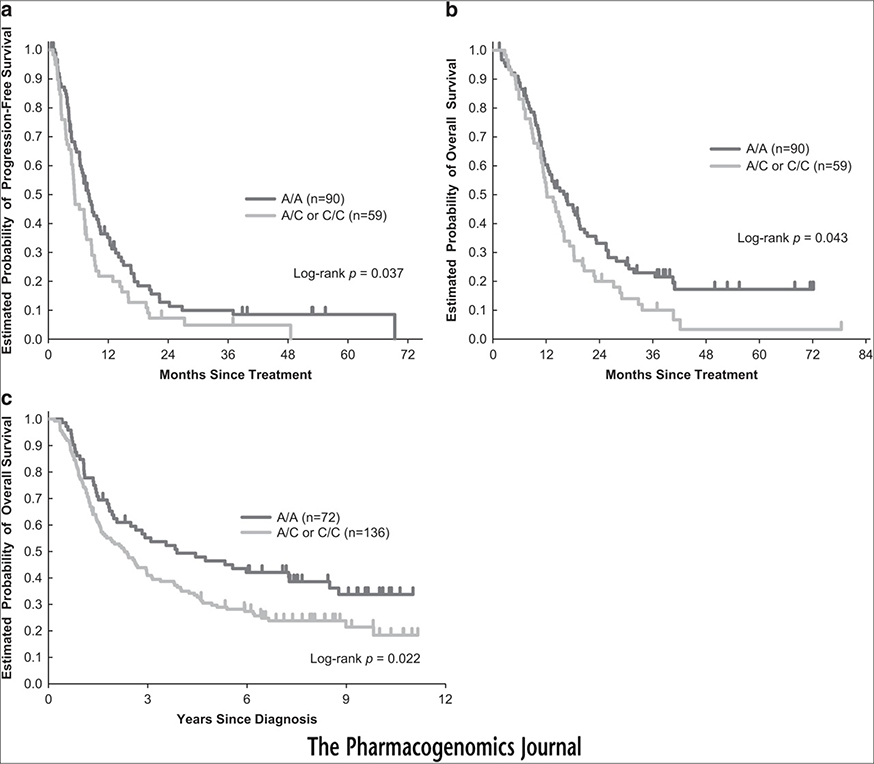
Associations between the selected FOXF1 SNPs and outcomes are shown in Table 2. Among the seven examined SNPs, only FOXF1 rs3950627 significantly influenced survival. Patients with the C/C genotype had significantly longer median PFS and OS compared to those with any A allele (Figures 1a and b). FOXF1 rs3950627 remained statistically significantly associated with PFS in multivariable analysis when adjusted for Eastern Cooperative Oncology Group (ECOG) performance status, liver metastases, lymph node metastases and number of metastasis sites. There was also a trend towards improved OS in patients carrying the C/C genotype compared to those with an A allele in multivariable analysis. No minor allele of FOXF1 rs1490865 was observed in Japanese patients, thus, it was excluded from further analysis. No LD was found between the analyzed polymorphisms for each ethnic/race group.
Table 2.
Associations between FOXF1 SNPs and clinical outcomes in the Japanese training cohort
|
Tumor response |
PFS |
OS |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | CR+PR | SD+PD | P-valuea | Median (95% CI), months | HR (95% CI)† | P-valuea | HR (95% CI)‡ | P-valuea | Median (95% CI), months | HR (95% CI)† | P-valuea | HR (95% CI)‡ | P-valuea | |
| rs3950627 | 0.45 | 0.073 | 0.097 | 0.10 | 0.14 | |||||||||
| C/C | 90 | 32 (37%) | 55 (63%) | 8.2 (6.3, 10.1) | 1 (Reference) | 1 (Reference) | 16.4 (11.7, 19.5) | 1 (Reference) | 1 (Reference) | |||||
| C/A | 48 | 13 (28%) | 34 (72%) | 5.1 (3.7, 7.3) | 1.53 (1.05, 2.23) | 1.52 (1.04, 2.21) | 12.1 (9.2, 16) | 1.50 (1.02, 2.19) | 1.47 (1, 2.16) | |||||
| A/A | 11 | 2 (20%) | 8 (80%) | 7.4 (2.7, 19.9) | 1.15 (0.59, 2.22) | 1.21 (0.62, 2.36) | 15 (9, 40.7) | 1.22 (0.63, 2.38) | 1.29 (0.66, 2.53) | |||||
| 0.21 | 0.037 | 0.040 | 0.043 | 0.053 | ||||||||||
| C/C | 90 | 32 (37%) | 55 (63%) | 8.2 (6.3, 10.1) | 1 (Reference) | 1 (Reference) | 16.4 (11.7, 19.5) | 1 (Reference) | 1 (Reference) | |||||
| Any A | 59 | 15 (26%) | 42 (74%) | 5.3 (4.6, 7.4) | 1.44 (1.02, 2.05) | 1.45 (1.02, 2.06) | 12.2 (10.8, 15.7) | 1.44 (1.01, 2.06) | 1.43 (1, 2.05) | |||||
| rs2178146 | 0.69 | 0.67 | 0.68 | 0.98 | 0.92 | |||||||||
| T/T | 96 | 29 (32%) | 62 (68%) | 7.2 (5.3, 8.8) | 1 (Reference) | 1 (Reference) | 14.1 (11.5, 18) | 1 (Reference) | 1 (Reference) | |||||
| T/Cb | 46 | 18 (39%) | 28 (61%) | 7.1 (4.9, 8.6) | 1.08 (0.76, 1.55) | 1.08 (0.75, 1.57) | 15 (11, 18.3) | 1 (0.69, 1.44) | 1.02 (0.70, 1.48) | |||||
| C/Cb | 8 | 2 (25%) | 6 (75%) | |||||||||||
| rs13332095 | 0.085 | 0.90 | 0.67 | 0.47 | 0.34 | |||||||||
| G/G | 127 | 37 (30%) | 86 (70%) | 7.1 (5.3, 8.2) | 1 (Reference) | 1 (Reference) | 13.8 (11.7, 16.1) | 1 (Reference) | 1 (Reference) | |||||
| G/Ab | 9 | 5 (56%) | 4 (44%) | 6.9 (2.1, 16) | 0.96 (0.47, 1.96) | 0.85 (0.41, 1.78) | 14.8 (4.2, 68+) | 0.76 (0.35, 1.63) | 0.68 (0.31, 1.50) | |||||
| A/Ab | 1 | 1 (100%) | 0 | |||||||||||
| rs1728400 | 0.58 | 0.19 | 0.20 | 0.18 | 0.13 | |||||||||
| A/A | 92 | 33 (37%) | 56 (63%) | 8 (6.2, 10) | 1 (Reference) | 1 (Reference) | 16 (11.7, 19.4) | 1 (Reference) | 1 (Reference) | |||||
| A/C | 47 | 13 (28%) | 33 (72%) | 5.2 (3.7, 7.7) | 1.38 (0.95, 2) | 1.40 (0.96, 2.04) | 13.6 (10.7, 16.1) | 1.41 (0.96, 2.07) | 1.48 (1.01, 2.18) | |||||
| C/C | 12 | 3 (27%) | 8 (73%) | 7.2 (2.6, 9.2) | 1.38 (0.73, 2.60) | 1.28 (0.68, 2.44) | 13.6 (9, 32.8) | 1.32 (0.70, 2.51) | 1.05 (0.54, 2.05) | |||||
| 0.29 | 0.068 | 0.077 | 0.066 | 0.085 | ||||||||||
| A/A | 92 | 33 (37%) | 56 (63%) | 8 (6.2, 10) | 1 (Reference) | 1 (Reference) | 16 (11.7, 19.4) | 1 (Reference) | 1 (Reference) | |||||
| Any C | 59 | 16 (28%) | 41 (72%) | 5.3 (4.6, 7.4) | 1.38 (0.97, 1.95) | 1.37 (0.97, 1.95) | 13.6 (11, 16) | 1.39 (0.97, 1.99) | 1.37 (0.96, 1.97) | |||||
| rs9936833 | 0.88 | 0.15 | 0.17 | 0.37 | 0.53 | |||||||||
| T/T | 123 | 40 (33%) | 80 (67%) | 7.5 (5.4, 9.1) | 1 (Reference) | 1 (Reference) | 14.8 (11.5, 18.2) | 1 (Reference) | 1 (Reference) | |||||
| T/Cb | 27 | 9 (36%) | 16 (64%) | 5.4 (3.7, 7.3) | 1.37 (0.89, 2.11) | 1.36 (0.88, 2.10) | 13.1 (11.2, 16.1) | 1.22 (0.79, 1.89) | 1.15 (0.74, 1.79) | |||||
| C/Cb | 1 | 0 | 1 (100%) | |||||||||||
| rs3111601 | 0.41 | 0.68 | 0.75 | 0.29 | 0.20 | |||||||||
| T/T | 104 | 29 (29%) | 72 (71%) | 7.2 (5.3, 9.2) | 1 (Reference) | 1 (Reference) | 13.9 (11.5, 17.3) | 1 (Reference) | 1 (Reference) | |||||
| T/Cb | 29 | 12 (41%) | 17 (59%) | 6.6 (5, 11.9) | 0.92 (0.60, 1.41) | 0.93 (0.60, 1.44) | 17.1 (11, 25.5) | 0.79 (0.51, 1.23) | 0.75 (0.48, 1.17) | |||||
| C/Cb | 5 | 1 (25%) | 3 (75%) | |||||||||||
Abbreviations: CI, confidence interval; HR, hazard ratio; OS, overall survival; PFS, progression-free survival.
The P-values were based on Fisher’s exact test for response, the log-rank test for PFS and OS in the univariate analyses (†) and the Wald test for PFS and OS in the multivariate Cox regression model adjusted for ECOG performance, liver metastases, lymph node metastases, and number of metastasis sites (‡).
Combined for estimates of HR.
Figure 1.
Kaplan–Meier cumulative survival curves by FOXF1 rs3950627 genotype for PFS (a) and overall survival (OS) (b) in the Japanese cohort, and OS (c) in the LAC cohort.
Associations between FOXF1 rs3950627 and clinical outcomes in the LAC validation cohort
The association between FOXF1 rs3950627 polymorphisms and outcomes in the LAC cohort is shown in Table 3. In univariable analyses, the C/C genotype of rs3950627 was associated with significantly longer OS. Patients with the C/C genotype had a median OS of 3.9 years, compared with a median OS of 2.3 years for those with any A allele (Figure 1c). This relationship remained statistically significant in multivariable analysis which adjusted for primary tumor site, stage, lymph node metastasis, differentiation and stratified by race.
Table 3.
Associations between FOXF1 SNPs and clinical outcomes in the LAC validation cohort
|
OS |
||||||
|---|---|---|---|---|---|---|
| N | Median, years (95% CI) | HR (95% CI)† | P-valuea | HR (95% CI)‡ | P-valuea | |
| rs3950627 | 0.068 | 0.043 | ||||
| C/C | 72 | 3.9 (2.1, 7.3) | 1 (Reference) | 1 (Reference) | ||
| C/A | 90 | 2.4 (1.6, 3.4) | 1.46 (1, 2.14) | 1.71 (1.12, 2.60) | ||
| A/A | 46 | 2.2 (1.3, 3.8) | 1.56 (1.01, 2.42) | 1.38 (0.83, 2.30) | ||
| 0.022 | 0.022 | |||||
| C/C | 72 | 3.9 (2.1, 7.3) | 1 (Reference) | 1 (Reference) | ||
| Any A | 136 | 2.3 (1.6, 3) | 1.50 (1.06, 2.13) | 1.60 (1.07, 2.40) | ||
Abbreviations: CI, confidence interval; HR, hazard ratio; OS, overall survival.
The P-values were based on the log-rank test for OS in the univariate analyses (†) and the Wald test in the multivariate Cox proportional hazards regression model adjusted for primary tumor site, stage, lymph node metastases and differentiation, and stratified by race (‡).
Associations between FOXF1 rs3950627 and clinical outcomes stratified by gender, tumor location and histology
Within the Japanese cohort, men with the FOXF1 rs3950627 C/C genotype had a significantly longer median PFS and OS than those with any A allele (Table 4). In multivariable analysis, FOXF1 rs3950627 remained significantly associated with PFS and OS in Japanese men (Table 4). Similarly, in the LAC cohort, men with any A allele genotype had a significantly improved OS than those with any A allele (Table 5). This association maintained significance in multivariable analysis. In analyses by gastric subsite (cardia vs distal GC), Japanese patients with distal GC and the FOXF1 rs3950627 C/C genotype had median OS of 8.6 months, compared with 6.2 months among those with any A allele. In multivariable analysis, FOXF1 rs3950627 remained significantly associated with OS for distal GC patients (Table 4). Within the LAC cohort, the C/C genotype was not significantly associated with OS for distal GC patients but was associated with significantly longer OS among patients with gastric cardia cancers (Table 5). Patients with the C/C genotype had a median OS of 4.4 years, compared with those with any A allele who had median PFS of 1.5 years. This association remained significant for OS in multivariable analysis (Table 5).
Table 4.
Associations between FOXF1 rs3950627 and clinical outcomes in the Japanese cohort by gender, tumor location and histology
|
PFS |
OS |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Median (95% CI), months | HR (95% CI)† | P-valuea | HR (95% CI)‡ | P-valuea | Median (95% CI), months | HR (95% CI)† | P-valuea | HR (95% CI)‡ | P-valuea | |
| Gender | |||||||||||
| Male | 0.012 | 0.014 | 0.009 | 0.024 | |||||||
| C/C | 67 | 8.2 (6.4, 10.4) | 1 (Reference) | 1 (Reference) | 16.6 (11.7, 23.2) | 1 (Reference) | 1 (Reference) | ||||
| C/Ab | 32 | 5.1 (3.5, 7.7) | 1.69 (1.10, 2.61) | 1.77 (1.12, 2.78) | 12.1 (8.9, 15) | 1.75 (1.13, 2.71) | 1.68 (1.07, 2.63) | ||||
| A/Ab | 6 | ||||||||||
| Female | 0.93 | 0.88 | 0.78 | 0.92 | |||||||
| C/C | 23 | 7.5 (4.3, 17.2) | 1 (Reference) | 1 (Reference) | 13.9 (10.9, 20.7) | 1 (Reference) | 1 (Reference) | ||||
| C/Ab | 16 | 7.2 (3.7, 10.1) | 1.03 (0.54, 1.97) | 1.05 (0.52, 2.14) | 15.3 (10.7, 28.6) | 0.91 (0.48, 1.75) | 0.96 (0.48, 1.93) | ||||
| A/Ab | 5 | ||||||||||
| Tumor location | |||||||||||
| Cardia | 0.61 | 0.70 | 0.25 | 0.79 | |||||||
| C/C | 29 | 8.2 (4.6, 10.4) | 1 (Reference) | 1 (Reference) | 16.4 (11.7, 27.8) | 1 (Reference) | 1 (Reference) | ||||
| C/Ab | 14 | 5 (2.5, 9.3) | 1.18(0.61, 2.27) | 0.87(0.43, 1.76) | 12 (5.9, 28.6) | 1.47 (0.75, 2.85) | 1.11 (0.53, 2.32) | ||||
| A/Ab | 3 | ||||||||||
| Distal | 0.017 | 0.005 | 0.087 | 0.064 | |||||||
| C/C | 55 | 8.6 (6.4, 12.7) | 1 (Reference) | 1 (Reference) | 16.4 (10.9, 19.7) | 1 (Reference) | 1 (Reference) | ||||
| C/Ab | 33 | 6.2 (3.7, 8.6) | 1.67 (1.07, 2.60) | 1.92 (1.22, 3.02) | 14.2 (11, 17.3) | 1.45 (0.93, 2.26) | 1.53 (0.98, 2.39) | ||||
| A/Ab | 8 | ||||||||||
| Histology | |||||||||||
| Differentiated | 0.66 | 0.58 | 0.66 | 0.54 | |||||||
| C/C | 31 | 7.1 (4.3, 12.3) | 1 (Reference) | 1 (Reference) | 18.2 (9.6, 27.8) | 1 (Reference) | 1 (Reference) | ||||
| C/Ab | 9 | 7.5 (2.5, 10.1) | 1.17 (0.57, 2.38) | 1.23 (0.59, 2.55) | 14.6 (5.3, 33.6) | 1.17 (0.57, 2.41) | 1.26 (0.60, 2.66) | ||||
| A/Ab | 3 | ||||||||||
| Undifferentiated | 0.038 | 0.12 | 0.052 | 0.070 | |||||||
| C/C | 59 | 8.2 (6.3, 11.9) | 1 (Reference) | 1 (Reference) | 15.1 (11.7, 19.4) | 1 (Reference) | 1 (Reference) | ||||
| C/Ab | 39 | 5.2 (3.7, 7.3) | 1.53 (1.01, 2.32) | 1.40 (0.92, 2.13) | 11.8 (10.1, 15.7) | 1.50 (0.99, 2.28) | 1.49 (0.97, 2.29) | ||||
| A/Ab | 8 | ||||||||||
Abbreviations: CI, confidence interval; HR, hazard ratio; OS, overall survival; PFS, progression-free survival.
The P-values were based on Fisher’s exact test for response, the log-rank test for PFS and OS in the univariate analyses (†) and the Wald test for PFS and OS in the multivariate Cox regression model adjusted for ECOG performance, liver metastases, lymph node metastases, and number of metastases (‡).
Combined for estimates of HR.
Table 5.
Associations between FOXF1 rs3950627 and clinical outcomes in the LAC cohort by gender, location and histology
|
OS |
||||||
|---|---|---|---|---|---|---|
| N | Median, years (95% CI) | HR (95% CI)† | P-valuea | HR (95% CI)‡ | P-valuea | |
| Gender | ||||||
| Male | 0.041 | 0.046 | ||||
| C/C | 53 | 3.6 (1.8, 8.8) | 1 (Reference) | 1 (Reference) | ||
| A/C | 65 | 1.9 (1.2, 3.4) | 1.50 (0.96, 2.33) | 1.89 (1.13, 3.15) | ||
| A/A | 25 | 1.4 (1.2, 3) | 1.92 (1.12, 3.31) | 1.78 (0.91, 3.47) | ||
| 0.022 | 0.014 | |||||
| C/C | 53 | 3.6 (1.8, 8.8) | 1 (Reference) | 1 (Reference) | ||
| Any A | 90 | 1.6 (1.3, 2.7) | 1.61 (1.06, 2.43) | 1.86 (1.14, 3.06) | ||
| Female | 0.65 | 0.376 | ||||
| C/C | 19 | 6 (1.5, 11+) | 1 (Reference) | 1 (Reference) | ||
| A/C | 25 | 2.9 (1.6, 6.4) | 1.39 (0.66, 2.93) | 1.54 (0.61, 3.88) | ||
| A/A | 21 | 3.1 (1.6, 9.8) | 1.31 (0.61, 2.81) | 0.87 (0.35, 2.20) | ||
| 0.37 | 0.75 | |||||
| C/C | 19 | 6 (1.5, 11+) | 1 (Reference) | 1 (Reference) | ||
| Any A | 46 | 2.9 (2, 6.2) | 1.35 (0.70, 2.64) | 1.14 (0.50, 2.61) | ||
| Tumor location | ||||||
| Cardia | 0.11 | 0.11 | ||||
| C/C | 27 | 4.4 (1.5, 10.4+) | 1 (Reference) | 1 (Reference) | ||
| A/C | 42 | 1.5 (1, 3.4) | 1.78 (0.98, 3.24) | 1.96 (1.02, 3.76) | ||
| A/A | 23 | 1.5 (0.9, 2.6) | 1.84 (0.94, 3.60) | 1.87 (0.89, 3.95) | ||
| 0.035 | 0.036 | |||||
| C/C | 27 | 4.4 (1.5, 10.4+) | 1 (Reference) | 1 (Reference) | ||
| Any A | 65 | 1.5 (1.2, 2.6) | 1.80 (1.03, 3.16) | 1.93 (1.05, 3.56) | ||
| Distal | 0.58 | 0.38 | ||||
| C/C | 45 | 3.9 (1.8, 8.5) | 1 (Reference) | 1 (Reference) | ||
| A/C | 48 | 2.8 (1.8, 5.1) | 1.25 (0.76, 2.05) | 1.44 (0.81, 2.55) | ||
| A/A | 23 | 3 (1.3, 4.7) | 1.31 (0.72, 2.35) | 1.06 (0.51, 2.18) | ||
| 0.30 | 0.32 | |||||
| C/C | 45 | 3.9 (1.8, 8.5) | 1 (Reference) | 1 (Reference) | ||
| Any A | 71 | 2.9 (1.9, 4.3) | 1.27 (0.80, 2) | 1.32 (0.76, 2.28) | ||
| Histology | ||||||
| Differentiated | 0.20 | 0.59 | ||||
| C/C | 18 | 7.3 (1.8, 11+) | 1 (Reference) | 1 (Reference) | ||
| A/Ca | 24 | 3.8 (1.8, 6.2) | 1.65 (0.76, 3.59) | 1.34 (0.46, 3.86) | ||
| A/Aa | 9 | |||||
| Undifferentiated | 0.059 | 0.19 | ||||
| C/C | 45 | 3.8 (1.8, 8.5) | 1 (Reference) | 1 (Reference) | ||
| A/C | 60 | 2.1 (1.1, 2.9) | 1.59 (1, 2.52) | 1.60 (0.95, 2.69) | ||
| A/A | 35 | 1.5 (1.3, 3) | 1.74 (1.05, 2.90) | 1.56 (0.86, 2.85) | ||
| 0.020 | 0.067 | |||||
| C/C | 45 | 3.8 (1.8, 8.5) | 1 (Reference) | 1 (Reference) | ||
| Any A | 95 | 1.9 (1.3, 2.6) | 1.64 (1.07, 2.52) | 1.59 (0.97, 2.60) | ||
Abbreviations: CI, confidence interval; HR, hazard ratio; OS, overall survival.
The P-values were based on the log-rank test for OS in the univariate analyses (†) and the Wald test in the multivariate Cox proportional hazards regression model adjusted for primary tumor site, stage, lymph node metastases and differentiation, and stratified by race (‡).
The association between the FOXF1 rs3950627 C/C genotype and outcomes also appeared to be stronger among Japanese (Table 4) and LAC patients with undifferentiated GC (Table 5). Within the Japanese cohort, patients with undifferentiated GC and the C/C genotype had significantly longer median PFS with a trend towards improved OS compared with those with any A allele (Table 4). In multivariable analysis, there was also a trend towards improved OS in Japanese patients with undifferentiated tumors and the FOXF1 rs3950627 C/C genotype. Among the LAC cohort, patients with undifferentiated tumors and the C/C genotype had a significantly longer OS than those with any A allele; there was a similar but non-significant trend in multivariable analysis (Table 5).
DISCUSSION
The role of FOXF1 in gut development and carcinogenesis has been established, and a potential link between SNPs near the FOXF1 gene and a predilection for esophageal adenocarcinoma has been recently reported.13 Among FOXF1 SNPs, rs3950627 has the most significant impact on susceptibility to esophageal adenocarcinoma.13 However, the clinical relevance of FOXF1 SNPs in gastric cancer has not been explored. Using two independent cohorts from Japan and the US, we investigated associations between FOXF1 SNPs and outcomes in GC patients. To our knowledge, our study is the first to demonstrate and validate FOXF1 rs3950627 as a biomarker for survival in Japanese and US GC patients. Furthermore, we found these prognostic associations to be influenced by gender and primary tumor subsite.
Although the FOXF1 protein has putative oncogenic4 and tumor suppressor16,17 roles, the function of FOXF1 rs3950627 remains unclear. As rs3950627 is a tag SNP, it may exert its effects through linked polymorphisms at other loci. In vitro functional assays are necessary to better define associations between rs3950627, gastric carcinogenesis and patient outcomes.
Consistent with prior studies, our gender-stratified analysis showed FOXF1 rs3950627 to specifically predict survival in men but not women, independent of ethnicity and after accounting for known prognostic factors in multivariable analysis. In a case–control study, Dura et al.18 reported another FOXF1 SNP (rs9936833) to be associated with esophageal adenocarcinoma, particularly in males. Gender-based differences in gastric cancer outcomes are well established, as men have a two-fold greater incidence and increased mortality compared to women,19,20 and estrogen has been shown to confer a potential protective effect against GC.21 Our data support a possible role for FOXF1 in mediating these effects, as it lies downstream of both ERα and sonic Hh pathway activation.11
Insights into the genetic heterogeneity of GC have revealed subsite specific etiologic and histologic molecular profiles.22 Motivated by these findings, we assessed whether the prognostic impact of FOXF1 polymorphisms varied by primary tumor site and found unique relationships in the Japanese and LAC cohorts. In the Japanese cohort, the rs3950627 C/C genotype was associated with longer OS in distal GC patients, whereas in the US cohort, the C/C genotype was associated with improved OS among patients with cardiac GC. The significance of these findings is not entirely clear and need to be confirmed in subsequent studies, but may reflect distinct GC etiologies between different ethnicities.
Certain limitations of our study should be acknowledged. Given the retrospective design and relatively small sample size of our study, our findings need to be prospectively validated in larger populations. Information regarding specific treatment and PFS was not available for the LAC cohort, and this limits the comparisons that can be made between the training and validation cohorts. Furthermore, the two cohorts differed in that the validation cohort included patients with localized and metastatic disease, whereas the training set only included patients with metastatic disease. In a separate analysis, we investigated differences between patients with localized and metastatic disease within the validation cohort. In patients with localized disease, the C/C genotype of rs3950627 was associated with significantly longer OS. In metastatic disease, there was also a trend towards improved OS with rs3950627 C/C genotype, which did not reach statistical significance, likely due to small sample size (data not shown). Although we adjusted for stage in our primary multivariable analysis within the validation cohort, whether these polymorphisms are more clinically relevant in patients with more advanced disease remains to be determined.
In conclusion, our study is the first to show that FOXF1 rs3950627 may be a promising prognostic marker in two independent cohorts of GC patients. Future investigations are needed to determine the functional significance of FOXF1 rs3950627 and define the clinical utility of FOXF1 polymorphisms in the management of GC patients.
Supplementary Material
ACKNOWLEDGMENTS
Satoshi Matsusaka is a recipient of Takashi Tsuruo Memorial Fund. Stefan Stremitzer is a recipient of an Erwin Schrödinger fellowship of the Austrian Science Fund (J3501-B13). Martin D. Berger received a grant from the Swiss Cancer League (BIL KLS-3334-02-2014). The project described was supported in part by Award Number P30CA014089 from the National Cancer Institute.
DISCLAIMER
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. This project was also supported in part by the San Pedro Guild Foundation.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website (http://www.nature.com/tpj)
REFERENCES
- 1.Ormestad M, Astorga J, Landgren H, Wang T, Johansson BR, Miura N et al. Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development 2006; 133: 833–843. [DOI] [PubMed] [Google Scholar]
- 2.De Wever O, Demetter P, Mareel M, Bracke M. Stromal myofibroblasts are drivers of invasive cancer growth. Int J Cancer 2008; 123: 2229–2238. [DOI] [PubMed] [Google Scholar]
- 3.Mahlapuu M, Enerback S, Carlsson P. Haploinsufficiency of the forkhead gene Foxf1, a target for sonic hedgehog signaling, causes lung and foregut malformations. Development 2001; 128: 2397–2406. [DOI] [PubMed] [Google Scholar]
- 4.Lo PK, Lee JS, Sukumar S. The p53-p21WAF1 checkpoint pathway plays a protective role in preventing DNA rereplication induced by abrogation of FOXF1 function. Cell Signal 2012; 24: 316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tamura M, Sasaki Y, Koyama R, Takeda K, Idogawa M, Tokino T. Forkhead transcription factor FOXF1 is a novel target gene of the p53 family and regulates cancer cell migration and invasiveness. Oncogene 2014; 33: 4837–4846. [DOI] [PubMed] [Google Scholar]
- 6.van den Brink GR, Hardwick JC, Tytgat GN, Brink MA, Ten Kate FJ, Van Deventer SJ et al. Sonic hedgehog regulates gastric gland morphogenesis in man and mouse. Gastroenterology 2001; 121: 317–328. [DOI] [PubMed] [Google Scholar]
- 7.van den Brink GR, Hardwick JC, Nielsen C, Xu C, ten Kate FJ, Glickman J et al. Sonic hedgehog expression correlates with fundic gland differentiation in the adult gastrointestinal tract. Gut 2002; 51: 628–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Astorga J, Carlsson P. Hedgehog induction of murine vasculogenesis is mediated by Foxf1 and Bmp4. Development 2007; 134: 3753–3761. [DOI] [PubMed] [Google Scholar]
- 9.Madison BB, McKenna LB, Dolson D, Epstein DJ, Kaestner KH. FoxF1 and FoxL1 link hedgehog signaling and the control of epithelial proliferation in the developing stomach and intestine. J Biol Chem 2009; 284: 5936–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saito RA, Micke P, Paulsson J, Augsten M, Pena C, Jonsson P et al. Forkhead box F1 regulates tumor-promoting properties of cancer-associated fibroblasts in lung cancer. Cancer Res 2010; 70: 2644–2654. [DOI] [PubMed] [Google Scholar]
- 11.Kameda C, Nakamura M, Tanaka H, Yamasaki A, Kubo M, Tanaka M et al. Oestrogen receptor-alpha contributes to the regulation of the hedgehog signalling pathway in ERalpha-positive gastric cancer. Br J Cancer 2010; 102: 738–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao XH, Gu SZ, Liu SX, Pan BR. Expression of estrogen receptor and estrogen receptor messenger RNA in gastric carcinoma tissues. World J Gastroenterol 2003; 9: 665–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levine DM, Ek WE, Zhang R, Liu X, Onstad L, Sather C et al. A genome-wide association study identifies new susceptibility loci for esophageal adenocarcinoma and Barrett’s esophagus. Nat Genet 2013; 45: 1487–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koizumi W, Narahara H, Hara T, Takagane A, Akiya T, Takagi M et al. S-1 plus cisplatin versus S-1 alone for first-line treatment of advanced gastric cancer (SPIRITS trial): a phase III trial. Lancet Oncol 2008; 9: 215–221. [DOI] [PubMed] [Google Scholar]
- 15.Wu AH, Wan P, Bernstein L. A multiethnic population-based study of smoking, alcohol and body size and risk of adenocarcinomas of the stomach and esophagus (United States). Cancer Causes Control 2001; 12: 721–732. [DOI] [PubMed] [Google Scholar]
- 16.Watson JE, Doggett NA, Albertson DG, Andaya A, Chinnaiyan A, van Dekken H et al. Integration of high-resolution array comparative genomic hybridization analysis of chromosome 16q with expression array data refines common regions of loss at 16q23-qter and identifies underlying candidate tumor suppressor genes in prostate cancer. Oncogene 2004; 23: 3487–3494. [DOI] [PubMed] [Google Scholar]
- 17.Lo PK, Lee JS, Liang X, Han L, Mori T, Fackler MJ et al. Epigenetic inactivation of the potential tumor suppressor gene FOXF1 in breast cancer. Cancer Res 2010; 70: 6047–6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dura P, van Veen EM, Salomon J, te Morsche RH, Roelofs HM, Kristinsson JO et al. Barrett associated MHC and FOXF1 variants also increase esophageal carcinoma risk. Int J Cancer 2013; 133: 1751–1755. [DOI] [PubMed] [Google Scholar]
- 19.Yang D, Hendifar A, Lenz C, Togawa K, Lenz F, Lurje G et al. Survival of metastatic gastric cancer: Significance of age, sex and race/ethnicity. J Gastrointest Oncol 2011; 2: 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandanos E, Lagergren J. Oestrogen and the enigmatic male predominance of gastric cancer. Eur J Cancer 2008; 44: 2397–2403. [DOI] [PubMed] [Google Scholar]
- 21.Sipponen P, Correa P. Delayed rise in incidence of gastric cancer in females results in unique sex ratio (M/F) pattern: etiologic hypothesis. Gastric Cancer 2002; 5: 213–219. [DOI] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014; 513: 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.