Abstract
Purpose
Vitamin D exerts its inhibitory influence on colon cancer growth by inhibiting Wnt signaling and angiogenesis. We hypothesized that SNPs in genes involved in vitamin D transport, metabolism, and signaling are associated with outcome in metastatic colorectal cancer (mCRC) patients treated with first-line FOLFIRI and bevacizumab.
Experimental Design
522 mCRC patients enrolled in the FIRE-3 (discovery cohort) and TRIBE (validation set) trials treated with FOLFIRI/bevacizumab were included in this study. 278 patients receiving FOLFIRI and cetuximab (FIRE-3) served as a control cohort. Six SNPs in 6 genes (GC, CYP24A1, CYP27B1, VDR, DKK1, CST5) were analyzed.
Results
In the discovery cohort, AA carriers of the GC rs4588 SNP encoding for the vitamin D–binding protein, and treated with FOLFIRI/bevacizumab had a shorter overall survival (OS) than those harboring any C allele (15.9 vs. 25.1 months) in both univariable (P = 0.001) and multivariable analyses (P = 0.047). This association was confirmed in the validation cohort in multivariable analysis (OS 18.1 vs. 26.2 months, HR, 1.83; P = 0.037). Interestingly, AA carriers in the control set exhibited a longer OS (48.0 vs. 25.2 months, HR, 0.50; P = 0.021). This association was further confirmed in a second validation cohort comprising refractory mCRC patients treated with cetuximab ± irinotecan (PFS 8.7 vs. 3.7 months) in univariable (P = 0.033) and multivariable analyses (P = 0.046).
Conclusions
GC rs4588 SNP might serve as a predictive marker in mCRC patients treated with FOLFIRI/bevacizumab or FOLFIRI/cetuximab. Whereas AA carriers derive a survival benefit with FOLFIRI/cetuximab, treatment with FOLFIRI/bevacizumab is associated with a worse outcome.
Introduction
Colorectal cancer is the fourth leading cause of cancer-related death worldwide. With the addition of biologicals to 5-fluorouracil (5-FU)–based treatment regimens, the outcome of metastatic colorectal cancer (mCRC) patients has markedly improved during the last decade (1). However, to further optimize treatment, new effective drugs are highly warranted.
Accumulating evidence points to a critical role of vitamin D not only in maintenance of calcium and phosphate homeostasis, but also in several other biological processes such as cell proliferation, differentiation, and the immune response (2). Interestingly, several studies indicate that vitamin D deficiency might facilitate the development of diabetes mellitus type 2, cardiovascular diseases, autoimmune disorders and cancer (3). A meta-analysis comprising nine studies showed that higher vitamin D intake and blood 25-hydroxyvitamin D (25-OHD) levels are associated with lower colorectal cancer risk (4). Another study demonstrated that increased plasma 25-OHD levels were associated with prolonged overall survival (OS) in colorectal cancer patients with a trend toward a decreased cancer-specific mortality (5). Previous studies have also shown a tight link between higher vitamin D levels and longer cancer-specific survival in patients with prostate, breast, lung cancer, and lymphoma (6–8).
However, the exact underlying mechanism by which vitamin D exerts its antitumor effect remains largely elusive. Alvarez-Diaz and colleagues proposed that the antitumor effect of vitamin D in colon cancer cells is mediated by cystatin D, a cysteine proteinase inhibitor encoded by the CST5 gene, which inhibits proliferation, migration and antagonizes Wnt signaling (9). Another group reported that in colorectal cancer, vitamin D exerts its antineoplastic activity by increasing the expression of DKK1, a potent inhibitor of the Wnt pathway (10).
Cholecalciferol derived from sunlight exposure or dietary intake binds to vitamin D binding protein (DBP; 80%) or albumin (20%) and is metabolized in the liver to 25-hydroxyvitamin D, which is further converted in the kidney by 25-hydroxyvitamin D1α-hydroxylase (CYP27B1) to its biologically active form, 1,25-dihydroxyvitamin D. The biological functions of 1,25-dihydroxyvitamin D are mediated by the vitamin D receptor (VDR), which translocates as vitamin D-bound VDR to the nucleus and regulates gene expression (2).
The degradation of 25-hydroxyvitamin D and 1,25-dihydroxyvitamin D is initiated by the enzyme, 25-hydroxyvitamin D 24-hydroxylase (CYP24A1; ref. 2).
The literature regarding variations within genes encoding for critical proteins involved in vitamin D transport, metabolism, and signaling as well as their associations with outcome in colorectal cancer patients is scarce. A few studies have demonstrated a correlation between SNPs within the VDR gene and increased colon cancer risk (11), yet no association with outcome has been observed (12). Interestingly, preclinical studies suggest that vitamin D possesses antiangiogenic properties (2, 13), by decreasing both VEGF-dependent and -independent angiogenesis (14). With bevacizumab treatment, VEGF-independent escape mechanisms might occur to circumvent the angiogenesis blockade (15). Given the association between the AA genotype of the GC rs4588 SNP and lower circulating 25-OHD in several studies (16–21), we hypothesized that AA carriers treated with FOLFIRI and bevacizumab may have a worse outcome compared with C allele carriers, as the inhibitory effect on VEGF- independent angiogenesis might be smaller among AA carriers. Conversely, we hypothesized that AA carriers treated with FOLFIRI and cetuximab might have a better outcome, as low 25-OHD levels have been associated with a more activated adaptive immune response, and cetuximab exerts a more pronounced anticancer effect when adaptive immunity is activated (22–24). Furthermore, given the inhibitory influence of 25-OHD on the adaptive immune response (22), we hypothesized that in AA carriers with expectedly decreased 25-OHD levels, the inhibitory effect on the adaptive immune response might be less compared with C allele carriers, who have constitutively higher 25-OHD levels. In addition to our main SNP of interest, GC rs4588, we investigated five SNPs within genes, involved in 25-OHD metabolism, transport or signaling which may have an impact on circulating 25-OHD levels and therefore, outcome.
Patients and Methods
Study design and patient population
This study was comprised of 893 mCRC patients. 800 mCRC patients were enrolled in the randomized phase III FIRE-3 and TRIBE trials, and received either first-line FOLFIRI/bevacizumab (discovery and validation cohorts) or FOLFIRI and cetuximab (FIRE-3, control cohort). 93 patients with refractory mCRC enrolled in a prospective cohort (Department of Medical Oncology, Pisa, Italy) and a phase II trial (IMCL-0144), treated with either irinotecan plus cetuximab (Pisa, Italy, 2008–2011) or cetuximab monotherapy (University of Southern California, Norris Comprehensive Cancer Center, Los Angeles, CA, 2002–2003), served as a second validation cohort (25, 26). Because we expected the opposite effect on outcome between AA carriers of the GC rs4588 SNP treated with FOLFIRI/bevacizumab (worse outcome) versus FOLFIRI/cetuximab (better outcome), we arbitrarily chose the FIRE-3 FOLFIRI/bevacizumab group as the discovery and the FIRE-3 FOLFIRI/cetuximab group as the control cohort. In the FIRE-3 FOLFIRI/bevacizumab cohort, bevacizumab was given at a dose of 5 mg/kg biweekly. In the FIRE-3 FOLFIRI/cetuximab group, the first cetuximab infusion was administered at a dose of 400 mg/m2, thereafter 250 mg/m2 once a week. The FOLFIRI backbone therapy consisted of 180 mg/m2 irinotecan, 400 mg/m2 leucovorin, 400 mg/m2 fluorouracil (5-FU) bolus infusion and 2,400 mg/m2 infusion over 46 hours. Each cycle was repeated every 2 weeks until disease progression or intolerable toxic effects occurred (27). Validation cohort 1 included mCRC patients in the randomized phase III TRIBE trial treated with the same regimen (FOLFIRI/bevacizumab) as described above, with the exception of leucovorin, which was given at a dose of 200 mg/m2. After 12 cycles, patients in TRIBE underwent a maintenance therapy with 5-FU/bevacizumab until disease progression (28). Validation cohort 2 was comprised of treatment refractory mCRC patients who received 500 mg/m2 cetuximab and 180 mg/m2 irinotecan on day 1 every 2 weeks, or cetuximab monotherapy (first dose 400 mg/m2 followed by 250 mg/m2 weekly; refs. 25, 26). This study was approved by the local review boards for each participating site. All patients provided informed consent for the analysis of molecular correlates, which were conducted at the USC / Norris Comprehensive Cancer Center in Los Angeles, CA. Our study was performed according to the reporting recommendations for tumor marker prognostic studies (REMARK; ref. 29).
Candidate polymorphisms
Potentially functional variations within genes involved in vitamin D transport, metabolism and signaling were chosen according to the following criteria: minor allele frequency >10% in Caucasians; potential to alter the function of a gene according to public databases (National Institute of Environmental Health Science SNP Function Prediction, snpinfo.niehs.nih.gov, Queen’s University FSNP, compbio.cs.queensu.ca, NCBI-Pubmed and dbSNP, www.ncbi.nlm.nih.gov as well as Genecards, genecards.org).
Genotyping
Genomic DNA was extracted from formalin-fixed paraffin-embedded (FFPE) tissue in the discovery and control cohorts, and from blood in both validation cohorts, using the QIAmp DNA Easy Kit (Qiagen). In the FIRE-3 study (discovery and control cohorts), only FFPE tissue was available for analyses, whereas in TRIBE (validation cohort 1) and validation cohort 2 (USA and Italy cetuximab cohort), only blood was available. The assays to extract DNA from both blood and FFPE tissue were performed with the same extraction kit (QIAamp DNA Mini Kit, Qiagen), using the identical standardized workflow according to the manufacturer’s protocol. Briefly, for DNA extraction from FFPE tissue, we transferred the tissue from the glass slide into a tube, added 180 μL of tissue lysis buffer, followed by 40 μL proteinase and incubated the tubes at 56ºC overnight in a heated water bath to dissolve the paraffin from the tissue. For DNA extraction from blood, we pipetted 200 μL blood into a tube and added 40 μL proteinase. All subsequent steps were identical for both DNA extraction from FFPE tissue and from blood according to the manufacturer’s protocol (QIAamp DNA Mini Kit, sample and assay technologies). Because of lack of blood in the discovery and control cohorts (FIRE-3) and FFPE tissue in the validation cohorts (TRIBE and USA/Italy cohorts), we did not perform the analyses in both tissue and blood on the same patient. Six potentially functional SNPs in 6 genes (GC, CYP24A1, CYP27B1, VDR, DKK1, and CST5) were analyzed by PCR-based direct sequencing. Forward and reverse primers (Supplementary Table S1) were used for PCR amplification. PCR fragments were then sequenced on an ABI 3100A Capillary Genetic Analyzer (Applied Biosystem) to identify the SNP. The investigator (M.D. Berger) reading the sequence was blinded to the clinical outcome data.
Statistical analysis
The aim of this study was to identify associations between SNPs within genes involved in vitamin D transport, metabolism, and signaling and clinical outcome in mCRC patients enrolled in two phase III randomized trials, FIRE-3, and TRIBE. The discovery cohort consisted of patients receiving first-line FOLFIRI/bevacizumab within the FIRE-3 trial, whereas patients treated with the same regimen in TRIBE served as a validation set. The control cohort consisted of patients receiving first-line FOLFIRI/cetuximab in FIRE-3, and the second validation cohort was comprised of patients treated with cetuximab ± irinotecan.
The primary endpoint was progression-free survival (PFS), and secondary endpoints were OS and overall tumor response rate (ORR). PFS was defined as the time from randomization until disease progression, death or until last follow-up in patients who were alive and remained free of disease progression. OS was defined as the time from randomization until death. Patients still alive were censored at the last date of follow-up. ORR represented the percentage of patients who achieved either a complete (CR) or a partial (PR) response according to the Response Evaluation Criteria in Solid Tumors (RECIST). Allelic distribution of genetic variants was tested for deviation from Hardy–Weinberg equilibrium (HWE) using the χ2 test. Differences between baseline characteristics among the cohorts were compared by using the χ2 test. To evaluate the effects of different SNPs on PFS and OS, the log-rank test was used in the univariable analysis and Wald test in the multivariable Cox proportional regression model. The associations between SNPs and tumor response were evaluated using the Fisher’s exact test. Moreover, we used the Fisher’s exact test to assess, whether there is an association of potentially significant and validated SNPs and genomic subtypes such as BRAF and (K)RAS mutational status and other clinical variables such as location, age and sex. SNPs significantly associated with outcome in the discovery cohort (FIRE-3) were tested in the validation set (TRIBE) and a control cohort (FIRE-3). With 295 patients (251 PFS events) in the FOLFIRI/bevacizumab arm of FIRE-3 (discovery cohort) successfully sequenced, we would have 80% power to detect a minimum hazard ratio (HR) of 1.43 to 1.59 for PFS for a SNP with a minor allele frequency from 0.1 to 0.5, using a two-sided 0.05 level log-rank test. A HR of 1.44 to 1.60 would be detected in the FOLFIRI/cetuximab arm of FIRE-3 (control set, N = 278, 241 PFS events), 1.54 to 1.73 in the FOLFIRI/bevacizumab arm of TRIBE (validation set 1, N = 227, 173 events), and 1.88 to 2.32 in the second validation cohort treated with cetuximab ± irinotecan (N = 93, 83 events) for the same SNP with similar allele frequencies using the same test and power. The adjusting factors for multivariate analyses were sex, age, ECOG performance status, primary tumor site, liver limited metastases, primary tumor resection, previous adjuvant chemotherapy, RAS and BRAF status in the discovery, control, and combined cohorts and sex, age, ECOG performance status, primary tumor site, primary tumor resection, prior adjuvant chemotherapy and BRAF status in the validation cohort. Because of the limited data available for validation cohort 2 (studies were conducted in 2002–2003 (Los Angeles) and 2008–2011 (Italy), when information regarding BRAF/RAS mutational status and sidedness was not routinely collected in clinical trials) and the lack of specimen available to conduct additional molecular analyses, we could only include age and sex as adjusting factors for multivariate analysis. All P values were from two-sided Wald tests at a 0.05 significance level. All tests were conducted by using the SAS statistical package version 9.4 (SAS Institute).
Results
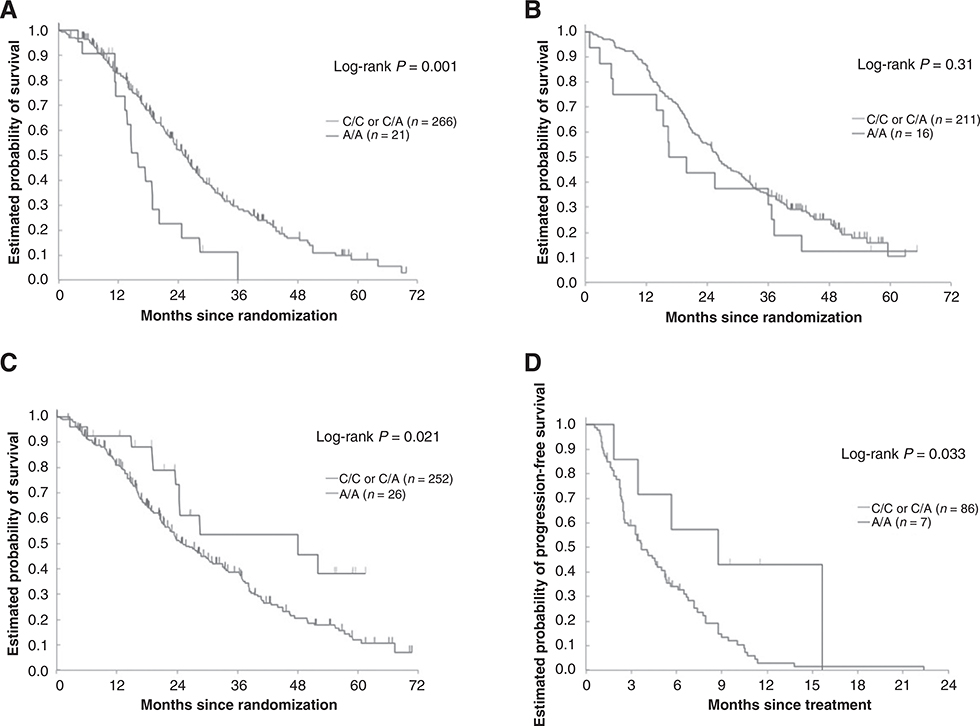
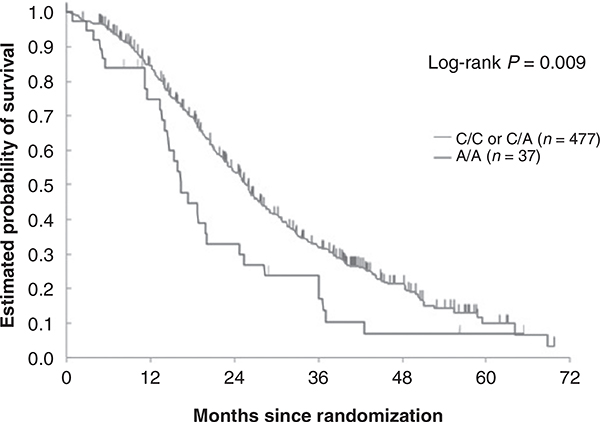
Patient characteristics of the four study cohorts (one discovery, one control, and two validation sets) are outlined in Table 1. Briefly, 893 patients with mCRC receiving either FOLFIRI/bevacizumab (discovery cohort FIRE-3, N = 295 and validation cohort 1 TRIBE, N = 227), FOLFIRI/cetuximab (control cohort FIRE-3, N = 278) or cetuximab ± irinotecan (validation set 2, N = 93) were included in this study. Genotyping was successful in >94% of the cases for each polymorphism. Causes of failure included limited quality or quantity of extracted DNA. All tested SNPs were within HWE. A genotyping quality control showed 99% concordance. The frequency of genotypes of the GC rs4588 SNP in our study corresponded well with those observed in the general population according to the Ensembl database (https://www.ensembl.org/). In the discovery cohort (FIRE-3 FOLFIRI/bevacizumab), the GC rs4588 SNP was significantly associated with OS. Patients carrying the AA genotype had a shorter OS (15.9 vs. 25.1 months) compared to those harboring any C allele in both univariable [HR, 2.19; 95% confidence interval (CI), 1.32–3.64; P = 0.001] and multivariable analyses (HR, 1.72; 95% CI, 1.01–2.92; P = 0.047; Table 2, Fig. 1A). In the validation cohort (TRIBE FOLFIRI/bevacizumab), no statistically significant difference in OS was observed between AA carriers and patients harboring any C allele of the GC rs4588 SNP in univariable analysis, as shown in the Kaplan–Meier curve in Fig. 1B and outlined in Table 2 (18.1 vs. 26.2 months, HR, 1.32; 95% CI, 0.76–2.29; P = 0.31). However, after adjusting for sex, age, ECOG performance status, primary tumor site, primary tumor resection, BRAF status, and previous adjuvant chemotherapy, the association between the GC rs4588 SNP and OS was statistically significant in multivariate analysis (HR, 1.83; 95% CI, 1.04–3.23; P = 0.037; Table 2). Because of the small number of patients harboring the AA genotype, we combined both FOLFIRI/bevacizumab cohorts. As expected, the association between the GC rs4588 SNP and OS was more evident. AA genotype carriers still exhibited a shorter OS in comparison with those having any C allele (16.4 vs. 25.8 months) in univariable (HR, 1.63; 95% CI, 1.12–2.35; P = 0.009) and multivariable analyses (HR, 1.59; 95% CI, 1.08–2.33; P = 0.019; Table 2, Fig. 2). Conversely, mCRC patients with the AA genotype receiving FOLFIRI/cetuximab (control cohort, FIRE-3) had a longer OS (48.0 vs. 25.2 months) in comparison with those with any C allele in univariable analysis (HR, 0.50; 95%CI, 0.27–0.91; P = 0.021; Table 2, Fig. 1C). This favorable outcome observed among AA carriers treated with first-line FOLFIRI and cetuximab was also confirmed in an independent second validation cohort comprising refractory mCRC patients who were treated with cetuximab ± irinotecan (PFS 8.7 vs. 3.7 months), in both univariable (HR, 0.40; 95% CI, 0.16–0.99; P = 0.033) and multivariable analyses (HR, 0.39; 95% CI, 0.16–0.98; P = 0.046; Table 2, Fig. 1D). However, in mCRC patients treated with FOLFIRI and bevacizumab, no difference in PFS could be observed between AA carriers and those harboring any C allele in the discovery, validation and combined cohorts (10.5 vs. 10.1 months, HR, 1.14; 95% CI, 0.71–1.82; P = 0.59; 7.5 vs. 9.7 months, HR, 1.27; 95% CI, 0.75–2.16; P = 0.37; and 9.3 vs. 10.1 months, HR, 1.19; 95% CI, 0.84–1.70; P = 0.32, respectively). Likewise, the PFS of mCRC patients receiving FOLFIRI and cetuximab was similar in AA genotype versus any C allele carriers in the control cohort (10.4 vs. 9.5 months, HR, 0.96; 95% CI, 0.63–1.47; P = 0.86; Table 2). Furthermore, the VDR rs731236 SNP was associated with OS in mCRC patients of the discovery cohort treated with FOLFIRI/bevacizumab (Supplementary Table S2). Here, GG genotype carriers had a significantly better OS (28.8 vs. 23.7 months) in multivariable analysis (HR, 0.62; 95% CI, 0.39–0.99; P = 0.046). However, this association was not observed in validation cohort 1 (Supplementary Table S3). No associations between the GC rs4588 SNP and BRAF and (K)RAS status or other clinical variables such as location, age and sex were observed in the discovery, validation or control cohorts (Supplementary Table S4). There was an insufficient number of patients with right-sided, RAS mutant/wild-type or BRAF mutant cancers harboring the AA genotype of the GC rs4588 SNP in the discovery and validation cohorts. Therefore, we restricted our analysis to the overall population and performed an exploratory subgroup analysis in patients with left-sided cancers, in whom sufficient numbers of AA carriers were present. Here, patients in the discovery group harboring the AA genotype (N = 13) had a shorter OS than C allele carriers (N = 193), but only in univariate analysis (17.4 vs. 27.4 months, HR, 2.40; 95% CI, 1.28–4.50; P = 0.004). PFS was similar in AA genotype and C allele carriers (10.8 vs. 10.7 months, HR, 1.41; 95% CI, 0.76–2.61; P = 0.26). Neither PFS nor OS differed between AA and C allele carriers in the validation cohort (data not shown).
Table 1.
Baseline characteristics
| Treatment | N | FIRE-3 FOLFIRI+Bev n = 295 |
TRIBE FOLFIRI+Bev n = 227 |
FIRE-3 FOLFIRI+Cet n = 278 |
Italy + US Cet ± irinotecan n = 93 |
Pa |
|---|---|---|---|---|---|---|
| Sex | 0.44 | |||||
| Male | 578 | 193 (65%) | 138 (61%) | 188 (68%) | 59 (63%) | |
| Female | 315 | 102 (35%) | 89 (39%) | 90 (32%) | 34 (37%) | |
| Age | <0.001 | |||||
| Median (range) | 65 (31–76) | 60 (29–75) | 65 (38–79) | 63 (35–82) | ||
| ≤65 | 516 | 155 (53%) | 162 (71%) | 146 (53%) | 53 (57%) | |
| >65 | 377 | 140 (47%) | 65 (29%) | 132 (47%) | 40 (43%) | |
| ECOG performance status | <0.001 | |||||
| 0 | 491 | 162 (55%) | 188 (83%) | 141 (51%) | ||
| 1–2 | 308 | 133 (45%) | 38 (17%) | 137 (49%) | ||
| Unspecified | 1 | – | 1 (1%) | – | ||
| Tumor site | 0.073 | |||||
| Right colon | 182 | 75 (25%) | 56 (25%) | 51 (18%) | ||
| Left colon or rectum | 588 | 213 (72%) | 156 (69%) | 219 (79%) | ||
| Unspecified | 30 | 7 (3%) | 15 (7%) | 8 (3%) | ||
| Liver limited metastases | 0.88 | |||||
| Yes | 259 | 95 (32%) | 71 (31%) | 93 (33%) | ||
| No | 541 | 200 (68%) | 156 (69%) | 185 (67%) | ||
| Primary tumor resected | <0.001 | |||||
| Yes | 631 | 255 (86%) | 143 (63%) | 233 (84%) | ||
| No | 167 | 40 (14%) | 84 (37%) | 43 (15%) | ||
| Unspecified | 2 | – | – | 2 (1%) | ||
| Adjuvant therapy | 0.009 | |||||
| Yes | 145 | 54 (18%) | 28 (12%) | 63 (23%) | ||
| No | 653 | 241 (82%) | 199 (88%) | 213 (77%) | ||
| Unspecified | 2 | – | – | 2 (1%) | ||
| KRAS status | <0.001 | |||||
| Wildtype | 570 | 248 (84%) | 96 (42%) | 226 (81%) | ||
| Mutant | 192 | 47 (16%) | 93 (41%) | 52 (19%) | ||
| Unspecified | 38 | – | 38 (17%) | – | ||
| RAS status | <0.001 | |||||
| Wild-type | 432 | 199 (67%) | 55 (24%) | 178 (64%) | ||
| Mutant | 288 | 83 (28%) | 116 (51%) | 89 (32%) | ||
| Unspecified | 80 | 13 (4%) | 56 (25%) | 11 (4%) | ||
| BRAF status | 0.61 | |||||
| Wild-type | 690 | 261 (88%) | 177 (78%) | 252 (91%) | ||
| Mutant | 56 | 25 (8%) | 12 (5%) | 19 (7%) | ||
| Unspecified | 54 | 9 (3%) | 38 (17%) | 7 (2%) |
Abbreviations: Bev, bevacizumab; Cet, cetuximab.
The P value was based on the χ2 test. The unspecified group was not included in the analysis.
Table 2.
Association between GC rs4588 and clinical outcomes in mCRC patients
| Tumor response |
Progression-free survival |
OS |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | PR+CR | SDb PD | Pa | Median, months (95% CI) | HR (95% CI)a(i) | Pa | HR (95% CI)a(ii) | Pa | Median, months (95% CI) | HR (95% Cl)a(i) | Pa | HR (95% CI)a(ii) | P valuea | |
| Discovery cohort | 0.42 | 0.48 | 0.65 | 0.004 | 0.13 | |||||||||
| C/C | 149 | 89 (65%) | 48 (35%) | 10.3 (9.2–12.2) | 1 (Reference) | 1 (Reference) | 26.4 (23.0–29.1) | 1 (Reference) | 1 (Reference) | |||||
| C/A | 117 | 61 (57%) | 46 (43%) | 9.8 (8.9–11.9) | 1.16 (0.89–1.50) | 1.13 (0.87–1.48) | 23.6 (20.6–26.5) | 1.15 (0.86–1.55) | 1.05 (0.77–1.43) | |||||
| A/A | 21 | 12 (57%) | 9 (43%) | 10.5 (6.9–12.0) | 1.21 (0.75–1.96) | 1.10 (0.66–1.83) | 15.9 (11.5–18.9) | 2.32 (1.37–3.93) | 1.76 (1.01–3.05) | |||||
| 0.70 | 0.59 | 0.91 | 0.001 | 0.047 | ||||||||||
| Any C | 266 | 150 (61%) | 94 (39%) | 10.1 (9.2–11.5) | 1 (Reference) | 1 (Reference) | 25.1 (22.7–27.5) | 1 (Reference) | 1 (Reference) | |||||
| A/A | 21 | 12 (57%) | 9 (43%) | 10.5 (6.9–12.0) | 1.14 (0.71–1.82) | 1.03 (0.63–1.69) | 15.9 (11.5–18.9) | 2.19 (1.32–3.64) | 1.72 (1.01–2.92) | |||||
| Validation cohort 1 | 0.20 | 0.62 | 0.16 | 0.46 | 0.052 | |||||||||
| C/C | 124 | 73 (61%) | 47 (39%) | 10.3 (8.8–11.1) | 1 (Reference) | 1 (Reference) | 26.2 (22.4–32.9) | 1 (Reference) | 1 (Reference) | |||||
| C/A | 87 | 50 (58%) | 36 (42%) | 9.6 (8.8–11.1) | 0.94 (0.68–1.29) | 0.90 (0.65–1.25) | 26.1 (20.5–30.8) | 1.12 (0.82–1.54) | 1.24 (0.89–1.71) | |||||
| A/A | 16 | 5 (36%) | 9 (64%) | 7.5 (3.6–14.1) | 1.24 (0.71–2.14) | 1.66 (0.90–3.05) | 18.1 (5.5–36.7) | 1.39 (0.79–2.43) | 2.01 (1.12–3.62) | |||||
| 0.078 | 0.37 | 0.070 | 0.31 | 0.037 | ||||||||||
| Any C | 211 | 123 (60%) | 83 (40%) | 9.7 (9.3–10.8) | 1 (Reference) | 1 (Reference) | 26.2 (22.7–30.8) | 1 (Reference) | 1 (Reference) | |||||
| A/A | 16 | 5 (36%) | 9 (64%) | 7.5 (3.6–14.1) | 1.27 (0.75–2.16) | 1.73 (0.96–3.14) | 18.1 (5.5–36.7) | 1.32 (0.76–2.29) | 1.83 (1.04–3.23) | |||||
| Combined cohort | 0.19 | 0.52 | 0.54 | 0.015 | 0.044 | |||||||||
| C/C | 273 | 162 (63%) | 95 (37%) | 10.3 (9.4–11.1) | 1 (Reference) | 1 (Reference) | 26.3 (23.8–30.2) | 1 (Reference) | 1 (Reference) | |||||
| C/A | 204 | 111 (58%) | 82 (42%) | 9.7 (9.2–10.9) | 1.06 (0.87–1.30) | 1.04 (0.84–1.28) | 24.8 (21.3–27.3) | 1.14 (0.92–1.42) | 1.10 (0.88–1.37) | |||||
| A/A | 37 | 17 (49%) | 18 (51%) | 9.3 (6.9–11.2) | 1.23 (0.85–1.76) | 1.24 (0.85–1.81) | 16.4 (14.1–20.1) | 1.72 (1.18–2.52) | 1.66 (1.12–2.47) | |||||
| 0.16 | 0.32 | 0.29 | 0.009 | 0.019 | ||||||||||
| Any C | 477 | 273 (61%) | 177 (39%) | 10.1 (9.5–10.8) | 1 (Reference) | 1 (Reference) | 25.8 (23.6–27.4) | 1 (Reference) | 1 (Reference) | |||||
| A/A | 37 | 17 (49%) | 18 (51%) | 9.3 (6.9–11.2) | 1.19 (0.84–1.70) | 1.22 (0.85–1.76) | 16.4 (14.1–20.1) | 1.63 (1.12–2.35) | 1.59 (1.08–2.33) | |||||
| Control cohort | 0.65 | 0.72 | 0.64 | 0.061 | 0.090 | |||||||||
| C/C | 128 | 74 (66%) | 38 (34%) | 9.7 (8.0–10.8) | 1 (Reference) | 1 (Reference) | 22.6 (19.3–28.7) | 1 (Reference) | 1 (Reference) | |||||
| C/A | 124 | 66 (63%) | 38 (37%) | 9.3 (7.3–10.6) | 1.11 (0.85–1.45) | 1.06 (0.80–1.40) | 28.8 (23.0–37.1) | 0.92 (0.68–1.26) | 0.79 (0.57–1.09) | |||||
| A/A | 26 | 19 (73%) | 7 (27%) | 10.4 (7.9–13.3) | 1.02 (0.65–1.58) | 1.26 (0.79–2.01) | 48.0 (23.8–61.6) | 0.48 (0.25–0.90) | 0.52 (0.27–1.00) | |||||
| 0.40 | 0.86 | 0.38 | 0.021 | 0.10 | ||||||||||
| Any C | 252 | 140 (65%) | 76 (35%) | 9.5 (8.2–10.3) | 1 (Reference) | 1 (Reference) | 25.2 (21.7–30.4) | 1 (Reference) | 1 (Reference) | |||||
| A/A | 26 | 19 (73%) | 7 (27%) | 10.4 (7.9–13.3) | 0.96 (0.63–1.47) | 1.22 (0.78–1.90) | 48.0 (23.8–61.6) | 0.50 (0.27–0.91) | 0.60 (0.32–1.11) | |||||
| Validation cohort 2 | 0.51 | 0.082 | 0.089 | 0.49 | 0.45 | |||||||||
| C/C | 48 | 10 (22%) | 36 (78%) | 3.4 (2.4–5.3) | 1 (Reference) | 1 (Reference) | 9.1 (7.2–13.3) | 1 (Reference) | 1 (Reference) | |||||
| C/A | 38 | 8 (22%) | 29 (78%) | 4.1 (2.5–5.6) | 0.86 (0.55–1.36) | 0.80 (0.50–1.29) | 12.1 (5.5–15.7) | 1.01 (0.60–1.72) | 0.96 (0.56–1.63) | |||||
| A/A | 7 | 3 (43%) | 4 (57%) | 8.7 (3.4–15.6b) | 0.37 (0.15–0.94) | 0.36 (0.14–0.92) | 18.4b (4.7–18.4b) | 0.51 (0.15–1.66) | 0.46 (0.14–1.53) | |||||
| 0.35 | 0.033 | 0.046 | 0.23 | 0.21 | ||||||||||
| Any C | 86 | 18 (22%) | 65 (78%) | 3.7 (2.6–5.2) | 1 (Reference) | 1 (Reference) | 10.5 (7.4–13.3) | 1 (Reference) | 1 (Reference) | |||||
| A/A | 7 | 3 (43%) | 4 (57%) | 8.7 (3.4–15.6b) | 0.40 (0.16–0.99) | 0.39 (0.16–0.98) | 18.4b (4.7–18.4b) | 0.50 (0.16–1.61) | 0.47 (0.15–1.52) | |||||
Abbreviations: CR, complete response; PD, progressive disease; PR, partial response; SD, stable disease.
The P value was based on χ2 test or Fisher exact test for tumor response, whenever appropriate, log-rank test for PFS and OS in the univariable analysis (i), and Wald test in the multivariable Cox proportional hazards regression model (ii) adjusting for sex, age, ECOG performance status, primary tumor site, liver limited metastases, primary tumor resection, adjuvant chemotherapy, RAS status, and BRAF status in the discovery, control, and combined cohorts; adjusting for sex, age, ECOG performance status, primary tumor site, primary tumor resection, BRAF status, and stratified by adjuvant chemotherapy in the validation cohort 1; adjusting for sex and age in the validation cohort 2. P values <0.050 are shown in bold.
Estimate was not reached yet.
Figure 1.
A, Discovery cohort: GC rs4588 and OS (FIRE-3 FOLFIRI/bevacizumab arm). B, Validation cohort 1: GC rs4588 and OS (TRIBE FOLFIRI/bevacizumab arm). C, Control cohort: GC rs4588 and OS (FIRE-3 FOLFIRI/cetuximab arm). D, Validation cohort 2: GC rs4588 and PFS in refractory mCRC patients treated with cetuximab ± irinotecan.
Figure 2.
Combined cohort: GC rs4588 and OS in FIRE-3 and TRIBE FOLFIRI/bevacizumab arms.
Discussion
Previous studies have demonstrated that 25-OHD inhibits proliferation, induces differentiation and stimulates apoptosis in colon cancer cells (30). Furthermore, 25-OHD decreases tumor progression by inhibiting angiogenesis through downregulation of angiogenic factors such as VEGF, IL8, and prostaglandin E2 (PGE2; ref. 14). Although vitamin D3 exerts its inhibitory influence on RAS signaling through downregulation of EGFR, it simultaneously inhibits proliferation of endothelial cells in vitro and decreases angiogenesis in vivo (2, 13, 31). A previous study demonstrated that the addition of 25-OHD resulted in upregulation of both the antiangiogenic factor, thrombospondin 1, and the proangiogenic factor, VEGF, in the SW480 human colon cancer cell line, leading to a change in the angiogenetic profile (32). Iseki and colleagues (33) observed a significant decrease in vessel counts and VEGF staining in colon tumors after prolonged administration of 25-OHD. Similarly, another study demonstrated that 25-OHD decreases mRNA and protein expression of HIF1alpha and VEGF in colon cancer cells and inhibits cell proliferation under hypoxia (34). A previous study also observed a critical role of 25-OHD in suppressing angiogenesis through downregulation of IL8 in prostate cancer (35). Currently, several clinical trials are evaluating the impact of vitamin D supplementation in patients receiving both adjuvant chemotherapy for stage III colon cancer as well as palliative therapy for mCRC (NCT02603757 and NCT01516216). Just recently, Ng and colleagues (36) presented preliminary results from the SUNSHINE trial (NCT01516216) at the 2017 ASCO Annual Meeting, demonstrating that mCRC patients treated with first-line FOLFOX/bevacizumab plus high-dose vitamin D achieved a longer PFS than those receiving FOLFOX/bevacizumab plus low-dose vitamin D.
According to our findings, genotyping of the GC gene might help identify which subset of patients benefits from adding bevacizumab to first-line chemotherapy.
Powe and colleagues (37) demonstrated a positive correlation between 25-OHD and DBP levels. Beside its role as a vitamin D transporter, DBP serves as a precursor for the macrophage-activating factor which exerts its antineoplastic effects by inhibiting angiogenesis (38).
The GC gene, located on chromosome 4, encodes for DBP and consists of 13 exons. The missense SNP rs4588, located on exon 11, results in an amino acid change of threonine to lysine at position 436 that affects the binding affinity of DBP.
Previous studies have shown that rs4588 AA carriers have the lowest affinity of DBP for vitamin D and lowest plasma 25-hydroxyvitamin D levels (16, 17). This functional association was further confirmed by Janssens and colleagues (18) who demonstrated that COPD patients harboring the AA genotype had significantly lower baseline 25-OHD levels than those carrying a C allele. In addition, the same trend was observed in a healthy control group, albeit without statistical significance. Similarly, lower 25-OHD levels among AA carriers of the rs4588 SNP was shown in a large cohort of 741 premenopausal Caucasian women (19). In a meta-analysis comprising nine studies, the ancestral C allele was found to be significantly associated with higher 25-OHD concentrations (20). Chinese children carrying an AA genotype exhibited significantly lower 25-OHD levels than C allele carriers. This association persisted after adjusting for age, sex, BMI, and vitamin D intake (21). Notably, these functional correlations between the GC rs4588 SNP and 25-OHD levels were consistent across all studies (20). On the basis of these mechanistic data and our validated clinical associations, we hypothesize that mCRC patients carrying the AA genotype of the GC rs4588 SNP might have constitutively lower 25-OHD levels, which translates into an inferior OS with FOLFIRI/bevacizumab, but an improved OS with FOLFIRI/cetuximab.
Importantly, we should note that previous findings of associations between the GC rs4588 AA genotype and lower concentrations of circulating vitamin D levels are derived from populations of healthy individuals and patients with diseases other than cancer (16–21). Therefore, we cannot state with absolute certainty, whether these findings can be completely translated to our study population of mCRC patients.
To our knowledge, this is the first study demonstrating that the rs4588 SNP within the GC gene, encoding for the DBP, might serve as a predictive biomarker in mCRC patients treated with both FOLFIRI/bevacizumab and FOLFIRI/cetuximab. Carriers of the AA genotype had a shorter OS compared with those harboring any C allele when receiving FOLFIRI/bevacizumab in both the discovery (FIRE-3) and validation (TRIBE) cohorts. On combining these cohorts, this association became even more significant. Conversely, AA carriers treated with FOLFIRI/cetuximab (FIRE-3 control cohort) had a better OS compared with those patients harboring any C allele. This favorable outcome was also observed in a second validation set comprising refractory mCRC patients treated with either cetuximab monotherapy or cetuximab and irinotecan. That the second validation cohort consisted of patients who received cetuximab ± irinotecan as later line therapy (vs. the other cohorts who underwent first-line therapy) might explain the significant association between GC rs4588 SNP and PFS but not OS. Nevertheless, the favorable effect on outcome among AA carriers treated with cetuximab ± irinotecan or FOLFIRI/cetuximab was consistent, regardless of the number of prior treatment lines.
The inverse associations with outcome among AA carriers treated with FOLFIRI/bevacizumab and FOLFIRI/cetuximab or cetuximab ± irinotecan suggest opposing effects of the GC gene with either bevacizumab or cetuximab treatment. With blockade of the VEGF angiogenic pathway by bevacizumab, VEGF-independent angiogenic escape mechanisms will be activated, mainly through the IL8 and Wnt signaling pathways (14, 15). Moreover, mounting evidence has shown that 25-OHD exerts its anticancer effect not only by decreasing VEGF-dependent and -independent angiogenesis, but also through downregulating Wnt signaling, which in turn further impairs tumor angiogenesis (14, 15). Given the association between the AA genotype of the GC rs4588 SNP with lower circulating 25-OHD (16–21), we hypothesized that in AA carriers the inhibitory effect on VEGF-independent angiogenesis, mainly driven by Wnt signaling but also IL8 and other angiogenic factors (14, 15), may be smaller compared with those with a C allele (higher circulating 25-OHD levels). This may serve as an explanation as to why mCRC patients harboring the AA genotype treated with FOLFIRI and bevacizumab have a worse outcome compared with C allele carriers. Given the inhibitory effect of 25-OHD on the adaptive immune system (22), we speculate that in AA carriers with constitutively lower 25-OHD levels, the inhibitory influence on the adaptive immune response might be less than in C allele carriers, who exhibit higher 25-OHD levels. Thus, in AA carriers, the equilibrium might slightly shift toward an activated adaptive immune state, where proinflammatory Th1 and Th17 cells are more predominant than immunosuppressive Tregs and Th2 cells (22, 23). Moreover, accumulating data indicate that the EGFR inhibitor, cetuximab, exhibits an improved anticancer effect when the adaptive immune system is activated (24). This phenomenon has not been described with bevacizumab. This distinction may explain why mCRC patients carrying the AA genotype treated with FOLFIRI and cetuximab have a better outcome compared to those harboring any C allele. Our results support our hypothesis of opposing effects on outcome between AA carriers treated with FOLFIRI/bevacizumab (worse outcome) or FOLFIRI/cetuximab (better outcome). Importantly, our study demonstrates that the selection of appropriate first-line treatment among patients with the same genotype of the GC rs4588 SNP might have a meaningful impact on survival. Although AA carriers with mCRC benefit most from adding cetuximab to FOLFIRI backbone chemotherapy in the first-line setting, bevacizumab may even cause harm. As we report about a new potential prognostic and predictive biomarker, it is important to keep in mind that in recent years a number of both prognostic and predictive markers have been identified in mCRC. For example, patients harboring a BRAF mutation have a shorter PFS and OS than those with BRAF wild-type tumors (39). Even though there are conflicting results from two meta-analyses investigating the clinical benefit of anti-EGFR treatment in mCRC patients with RAS wild-type and BRAF mutant tumors, the vast majority of clinicians do not treat patients with BRAF mutant tumors with cetuximab or panitumumab (40–43). Furthermore, it is well established that mCRC patients with RAS-mutant tumors do not derive benefit from anti-EGFR therapy (44). In addition, mounting evidence has shown that mCRC patients with right-sided primary tumors have a shorter OS compared with those with left-sided primary tumors and that patients with right-sided cancers might not derive the same benefit from cetuximab as those with left-sided primary tumors (45, 46). Interestingly, as shown in Supplementary Table S4, GC rs4588 genotypes were not associated with BRAF or RAS mutational status, nor primary tumor location, suggesting the GC rs4588 SNP to be an independent predictive and prognostic biomarker. Limitations of our study are that we neither assessed circulating 25-OHD levels, nor evaluated the inhibitory effect of vitamin D on angiogenesis. Moreover, we did not determine the ability of vitamin D to downregulate EGFR expression directly on the human tissue in this study. The lack of adequate specimen, including fresh-frozen tissue and the paucity of blood in validation cohorts 1 and 2, precluded us from conducting these assays. In addition, although in the discovery and control cohorts (FIRE-3) only FFPE tissue was available, we tested blood in the validation cohorts (TRIBE and USA/Italy). Despite the different origin of tissue (FFPE samples versus blood) used in our study, there is increasing evidence from several comparison studies, that DNA from FFPE tissue can serve as a valid proxy for germline DNA in pharmacogenetic association studies, not only in colorectal cancer (47, 48) but also in several other malignancies (49, 50). Furthermore, validation cohort 2 was comprised of total only 93 patients—86 with a C allele and 7 with the AA genotype. The number of AA genotype carriers (N=7) in validation cohort 2 is relatively small due to the low minor allele frequency (MAF = 0.28) of the GC rs4588 SNP. Therefore, the results obtained in the second validation cohort must be interpreted within the limitations of these small numbers. However, the findings in the second validation cohort resonate with the ones showed in the FIRE-3 cetuximab cohort, as we observed the same statistically significant difference in outcome in these two independent cohorts, whose patients were uniformly treated with cetuximab ± irinotecan-based chemotherapy. Strengths of our study include the total sample size of 893 mCRC patients and the inclusion of a discovery, control and two validation cohorts from two phase III randomized trials, one phase II study and one cohort with prospectively enrolled patients. Second, the results we obtained in both the discovery and control cohorts were confirmed in two independent validation sets. Furthermore, the rationale to test the impact of the rs4588 SNP within the GC gene, encoding for the DBP, on outcome is based on previously published mechanistic data. Sample size limitations and the low frequency of GC rs4588 AA genotype carriers (7%–9.4%) precluded us from conducting subgroup analyses in RAS wild-type/mutant or BRAF-mutant patients, and in those with right-sided primary tumors, as it would not allow us to draw any firm conclusions. Associations between GC SNP rs4588 and outcome with either bevacizumab or cetuximab should be further evaluated before clinical use, and should take into account known prognostic factors, particularly RAS/BRAF mutation status and sidedness. Retrospective, pooled biomarker studies using larger datasets, preferentially from randomized trials, are needed to define which subgroups of patients will derive the most benefit from either bevacizumab- or cetuximab-based chemotherapy, according to GC rs4588 SNP (AA versus any C allele). For example, there is increasing evidence from recent studies that mCRC patients with right-sided tumors do not derive benefit from cetuximab (45, 46). Thus, our findings might provide an impetus to further explore whether AA carriers of the GC rs4588 SNP with right-sided, RAS wild-type mCRC would benefit from treatment with cetuximab. As our study design did not allow for evaluation of circulating 25-OHD, it would be interesting to conduct a study exploring whether vitamin D supplementation might result in increased vitamin D levels among AA carriers of the GC rs4588 SNP, as the AA genotype has been shown to be associated with constitutively decreased 25-OHD levels (16–21). Looking forward, to use the GC rs4588 SNP in the context of already established prognostic and predictive biomarkers, we will need future algorithms combining this SNP with information about RAS/BRAF mutational status and sidedness, which will help us to further optimize personalized treatment for mCRC patients. In conclusion, the GC rs4588 SNP might serve as both a prognostic and predictive biomarker in mCRC patients. Our results suggest that genotyping of the rs4588 polymorphism within the GC gene may help identify patients who will derive the most benefit from adding either bevacizumab or cetuximab to irinotecan-based chemotherapy.
Supplementary Material
Translational Relevance.
Epidemiologic studies have shown that higher blood 25-hydroxyvitamin D (25-OHD) levels are associated with lower colorectal cancer risk. In addition, preclinical studies have demonstrated that 25-OHD inhibits colon cancer growth by stimulating apoptosis and decreasing cancer cell proliferation and tumor angiogenesis. A variation of the vitamin D–binding protein gene, GC rs4588, might serve as a predictive marker in mCRC patients treated with FOLFIRI and bevacizumab or FOLFIRI and cetuximab. Whereas AA genotype carriers derive a survival benefit from FOLFIRI/cetuximab, treatment with FOLFIRI/bevacizumab is associated with worse outcome. Genotyping of the rs4588 polymorphism within the GC gene may help identify patients who will benefit most from adding either bevacizumab or cetuximab to irinotecan-based chemotherapy.
Acknowledgments
M.D. Berger received a grant from the Werner and Hedy Berger-Janser Foundation for cancer research and the Swiss Cancer League (BIL KLS-3334-02-2014). This work was partly supported by the National Cancer Institute (grant number P30CA014089), the Gloria Borges Wunderglo Project, Daniel Butler Research Fund, Dhont Family Foundation and Call to Cure. Y. Miyamoto received a grant from the Japan Society for the Promotion of Science (S2606).
Disclosure of Potential Conflicts of Interest
S. Stintzing reports receiving speakers bureau honoraria from AMGEN, Bayer, Lilly, Merck KgaA, Nordic Pharma, Roche, Sanofi, and Takeda, and is a consultant/advisory board member for AMGEN, Bayer, Lilly, Merck KgaA, Nordic Pharma, Roche, and Sanofi. V. Heinemann reports receiving commercial research grants from AMGEN, Boehringer Ingelheim, Celgene, MERCK KgaA, Pfizer, Roche, and Shire, speakers bureau honoraria from Amgen, Baxalta, Merck KgaA, Roche AG, Sanofi, Servier, and SIRTEX, and is a consultant/advisory board member for Amgen, Baxalta, Boehringer Ingelheim, Lilly, Merck KgaA, Merrimack, Roche AG, Sanofi, Servier, SIRTEX, and Taiho. A. Falcone is a consultant/advisory board member for Amgen, Bayer, Bristol, Lilly, Merck, Roche, and Servier. H. Lenz is a consultant/advisory board member for Merck Serono and Roche. No potential conflicts of interest were disclosed by the other authors.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
References
- 1.Berger MD, Lenz HJ. The safety of monoclonal antibodies for treatment of colorectal cancer. Expert Opin Drug Saf 2016;15:799–808. [DOI] [PubMed] [Google Scholar]
- 2.Christakos S, Dhawan P, Verstuyf A, Verlinden L, Carmeliet G. Vitamin D: metabolism, molecular mechanism of action, and pleiotropic effects. Physiol Rev 2016;96:365–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holick MF. Vitamin D deficiency. N Engl J Med 2007;357:266–81. [DOI] [PubMed] [Google Scholar]
- 4.Ma Y, Zhang P, Wang F, Yang J, Liu Z, Qin H. Association between vitamin D and risk of colorectal cancer: a systematic review of prospective studies. J Clin Oncol 2011;29:3775–82. [DOI] [PubMed] [Google Scholar]
- 5.Ng K, Meyerhardt JA, Wu K, Feskanich D, Hollis BW, Giovannucci EL, et al. Circulating 25-hydroxyvitamin d levels and survival in patients with colorectal cancer. J Clin Oncol 2008;26:2984–91. [DOI] [PubMed] [Google Scholar]
- 6.Tretli S, Hernes E, Berg JP, Hestvik UE, Robsahm TE. Association between serum 25(OH)D and death from prostate cancer. Br J Cancer 2009; 100:450–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim Y, Je Y. VitaminDintake, blood 25(OH)D levels, and breast cancer risk or mortality: a meta-analysis. Br J Cancer 2014;110:2772–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tretli S, Schwartz GG, Torjesen PA, Robsahm TE. Serum levels of 25-hydroxyvitamin D and survival in Norwegian patients with cancer of breast, colon, lung, and lymphoma: a population-based study. Cancer Causes Control 2012;23:363–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alvarez-Díaz S, Valle N, García JM, Peña C, Freije JM, Quesada V, et al. Cystatin D is a candidate tumor suppressor gene induced by vitamin D in human colon cancer cells. J Clin Invest 2009;119:2343–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aguilera O, Peña C, García JM, Larriba MJ, Ordóñez-Morán P, Navarro D, et al. The Wnt antagonist DICKKOPF-1 gene is induced by 1alpha,25-dihydroxyvitamin D3 associated to the differentiation of human colon cancer cells. Carcinogenesis 2007;28:1877–84. [DOI] [PubMed] [Google Scholar]
- 11.Bai YH, Lu H, Hong D, Lin CC, Yu Z, Chen BC. Vitamin D receptor gene polymorphisms and colorectal cancer risk: a systematic meta-analysis. World J Gastroenterol 2012;18:1672–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perna L, Hoffmeister M, Schöttker B, Arndt V, Haug U, Holleczek B, et al. Vitamin D receptor polymorphism and colorectal cancer-specific and all-cause mortality. Cancer Epidemiol 2013;37:905–7. [DOI] [PubMed] [Google Scholar]
- 13.Mantell DJ, Owens PE, Bundred NJ, Mawer EB, Canfield AE. 1 alpha,25-dihydroxyvitamin D(3) inhibits angiogenesis in vitro and in vivo. Circ Res 2000;87:214–20. [DOI] [PubMed] [Google Scholar]
- 14.Feldman D, Krishnan AV, Swami S, Giovannucci E, Feldman BJ. The role of vitamin D in reducing cancer risk and progression. Nat Rev Cancer 2014;14:342–57. [DOI] [PubMed] [Google Scholar]
- 15.Gacche RN. Compensatory angiogenesis and tumor refractoriness. Oncogenesis 2015;4:e153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nissen J, Rasmussen LB, Ravn-Haren G, Andersen EW, Hansen B, Andersen R, et al. Common variants in CYP2R1 and GC genes predict vitamin D concentrations in healthy Danish children and adults. PLoS One 2014;9: e89907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnaud J, Constans J. Affinity differences for vitamin D metabolites associated with the genetic isoforms of the human serum carrier protein (DBP). Hum Genet 1993;92:183–8. [DOI] [PubMed] [Google Scholar]
- 18.Janssens W, Bouillon R, Claes B, Carremans C, Lehouck A, Buysschaert I, et al. Vitamin D deficiency is highly prevalent in COPD and correlates with variants in the vitamin D-binding gene. Thorax 2010; 65:215–20. [DOI] [PubMed] [Google Scholar]
- 19.Sinotte M, Diorio C, Bérubé S, Pollak M, Brisson J. Genetic polymorphisms of the vitamin D binding protein and plasma concentrations of 25-hydroxyvitamin D in premenopausal women. Am J Clin Nutr 2009; 89:634–40. [DOI] [PubMed] [Google Scholar]
- 20.McGrath JJ, Saha S, Burne TH, Eyles DW. A systematic review of the association between common single nucleotide polymorphisms and 25-hydroxyvitamin D concentrations. J Steroid Biochem Mol Biol 2010; 121:471–7. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Wang X, Liu Y, Qu H, Qu S, Wang W, et al. The GC, CYP2R1 and DHCR7 genes are associated with vitamin D levels in northeastern Han Chinese children. Swiss Med Wkly 2012;142:w13636. [DOI] [PubMed] [Google Scholar]
- 22.Bikle DD. Vitamin D metabolism, mechanism of action, and clinical applications. Chem Biol 2014;21:319–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hewison M Vitamin D and immune function: an overview. Proc Nutr Soc 2012;71:50–61. [DOI] [PubMed] [Google Scholar]
- 24.Yang X, Zhang X, Mortenson ED, Radkevich-Brown O, Wang Y, Fu YX. Cetuximab-mediated tumor regression depends on innate and adaptive immune responses. Mol Ther 2013;21:91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loupakis F, Antoniotti C, Cremolini C, Zhang W, Yang D, Wakatsuki T, et al. Prospective study of EGFR intron 1 (CA)n repeats variants as predictors of benefit from cetuximab and irinotecan in chemo-refractory metastatic colorectal cancer (mCRC) patients. Pharmacogenomics J 2014;14:322–7. [DOI] [PubMed] [Google Scholar]
- 26.Zhang W, Gordon M, Schultheis AM, Yang DY, Nagashima F, Azuma M, et al. FCGR2A and FCGR3A polymorphisms associated with clinical outcome of epidermal growth factor receptor expressing metastatic colorectal cancer patients treated with single-agent cetuximab. J Clin Oncol 2007;25:3712–8. [DOI] [PubMed] [Google Scholar]
- 27.Stintzing S, Modest DP, Rossius L, Lerch MM, von Weikersthal LF, Decker T, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab for metastatic colorectal cancer (FIRE-3): a post-hoc analysis of tumour dynamics in the final RAS wild-type subgroup of this randomised open-label phase 3 trial. Lancet Oncol 2016;17:1426–1434. [DOI] [PubMed] [Google Scholar]
- 28.Loupakis F, Cremolini C, Masi G, Lonardi S, Zagonel V, Salvatore L, et al. Initial therapy with FOLFOXIRI and bevacizumab for metastatic colorectal cancer. N Engl J Med 2014;371:1609–18. [DOI] [PubMed] [Google Scholar]
- 29.Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. BMC Med 2012;10:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Padi SK, Zhang Q, Rustum YM, Morrison C, Guo B. MicroRNA-627 mediates the epigenetic mechanisms of vitamin D to suppress proliferation of human colorectal cancer cells and growth of xenograft tumors in mice. Gastroenterology 2013;145:437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deeb KK, Trump DL, Johnson CS. Vitamin D signalling pathways in cancer: potential for anticancer therapeutics. Nat Rev Cancer 2007;7:684–700. [DOI] [PubMed] [Google Scholar]
- 32.Fernandez-Garcia NI, Palmer HG, Garcia M, Gonzalez-Martin A, del Rio M, Barettino D, et al. 1alpha,25-Dihydroxyvitamin D3 regulates the expression of Id1 and Id2 genes and the angiogenic phenotype of human colon carcinoma cells. Oncogene 2005;24:6533–44. [DOI] [PubMed] [Google Scholar]
- 33.Iseki K, Tatsuta M, Uehara H, Iishi H, Yano H, Sakai N, et al. Inhibition of angiogenesis as a mechanism for inhibition by 1alpha-hydroxyvitamin D3 and 1,25-dihydroxyvitamin D3 of colon carcinogenesis induced by azoxymethane in Wistar rats. Int J Cancer 1999;81:730–3. [DOI] [PubMed] [Google Scholar]
- 34.Ben-Shoshan M, Amir S, Dang DT, Dang LH, Weisman Y, Mabjeesh NJ. 1alpha,25-dihydroxyvitamin D3 (Calcitriol) inhibits hypoxia-inducible factor-1/vascular endothelial growth factor pathway in human cancer cells. Mol Cancer Ther 2007;6:1433–9. [DOI] [PubMed] [Google Scholar]
- 35.Bao BY, Yao J, Lee YF. 1alpha, 25-dihydroxyvitamin D3 suppresses interleukin-8-mediated prostate cancer cell angiogenesis. Carcinogenesis 2006; 27:1883–93. [DOI] [PubMed] [Google Scholar]
- 36.Ng K, Nimeiri HS, McCleary NJ, Abrams TA, Yurgelun MB, Cleary JM, et al. SUNSHINE: Randomized double-blind phase II trial of vitamin D supplementation in patients with previously untreated metastatic colorectal cancer. J Clin Oncol 35:15s, 2017(suppl; abstr 3506). [Google Scholar]
- 37.Powe CE, Ricciardi C, Berg AH, Erdenesanaa D, Collerone G, Ankers E, et al. Vitamin D-binding protein modifies the vitamin D-bone mineral density relationship. J Bone Miner Res 2011;26:1609–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kisker O, Onizuka S, Becker CM, Fannon M, Flynn E, D’Amato R, et al. Vitamin D binding protein-macrophage activating factor (DBP-maf) inhibits angiogenesis and tumor growth in mice. Neoplasia 2003;5: 32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Modest DP, Ricard I, Heinemann V, Hegewisch-Becker S, Schmiegel W, Porschen R, et al. Outcome according to KRAS-, NRAS- and BRAF-mutation as well as KRAS mutation variants: pooled analysis of five randomized trials in metastatic colorectal cancer by the AIO colorectal cancer study group. Ann Oncol 2016;27:1746–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rowland A, Dias MM, Wiese MD, Kichenadasse G, McKinnon RA, Karapetis CS, et al. Meta-analysis of BRAF mutation as a predictive biomarker of benefit from anti-EGFR monoclonal antibody therapy for RAS wild-type metastatic colorectal cancer. Br J Cancer 2015;112:1888–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pietrantonio F, Petrelli F, Coinu A, Di Bartolomeo M, Borgonovo K, Maggi C, et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: a meta-analysis. Eur J Cancer 2015;51:587–94. [DOI] [PubMed] [Google Scholar]
- 42.van Brummelen EMJ, de Boer A, Beijnen JH, Schellens JHM. BRAF mutations as predictive biomarker for response to anti-EGFR monoclonal antibodies. Oncologist 2017;22:864–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cremolini C, Antoniotti C, Moretto R, Masi G, Falcone A. First-line therapy for mCRC - the influence of primary tumour location on the therapeutic algorithm. Nat Rev Clin Oncol 2017;14:113. [DOI] [PubMed] [Google Scholar]
- 44.Allegra CJ, Rumble RB, Hamilton SR, Mangu PB, Roach N, Hantel A, et al. Extended RAS gene mutation testing in metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy: American Society of Clinical Oncology Provisional Clinical Opinion Update 2015. J Clin Oncol 2016;34:179–85. [DOI] [PubMed] [Google Scholar]
- 45.Venook AP, Niedzwicki D, Innocenti F, Fruth B, Green C, O’Neil BH, et al. Impact of primary (1º) tumor location on overall survival (OS) and progression-free survival (PFS) in patients (pts) with metastatic colorectal cancer (mCRC): analysis of CALGB/SWOG 80405 (Alliance). J Clin Oncol 34:15s, 2016(suppl; abstr 3504). [Google Scholar]
- 46.Tejpar S, Stintzing S, Ciardiello F, Tabernero J, Van CutsemE, Beier F, et al. Prognostic and predictive relevance of primary tumor location in patients with RAS wild-type metastatic colorectal cancer: retrospective analyses of the CRYSTAL and FIRE-3 trials. JAMA Oncol. 2016. October 10 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Huis-Tanja L, Kweekel D, Gelderblom H, Koopman M, Punt K, Guchelaar HJ, et al. Concordance of genotype for polymorphisms in DNA isolated from peripheral blood and colorectal cancer tumor samples. Pharmacogenomics 2013;14:2005–12. [DOI] [PubMed] [Google Scholar]
- 48.Marsh S, Mallon MA, Goodfellow P, McLeod HL. Concordance of pharmacogenetic markers in germline and colorectal tumor DNA. Pharmacogenomics 2005;6:873–7. [DOI] [PubMed] [Google Scholar]
- 49.Goetz MP, Rae JM, Suman VJ, Safgren SL, Ames MM, Visscher DW, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol 2005;23:9312–8. [DOI] [PubMed] [Google Scholar]
- 50.McWhinney SR, McLeod HL. Using germline genotype in cancer pharmacogenetic studies. Pharmacogenomics 2009;10:489–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.