Abstract
Creativity represents one of the most important and partially heritable human characteristics, yet little is known about its genetic basis. Epidemiological studies reveal associations between creativity and psychiatric disorders as well as multiple personality and behavioral traits. To test whether creativity and these disorders or traits share genetic basis, we performed genome-wide association studies (GWAS) followed by polygenic risk score (PRS) analyses. Two cohorts of Han Chinese subjects (4,834 individuals in total) aged 18–45 were recruited for creativity measurement using typical performance test. After exclusion of the outliers with significantly deviated creativity scores and low-quality genotyping results, 4,664 participants were proceeded for GWAS. We conducted PRS analyses using both the classical pruning and thresholding (P+T) method and the LDpred method. The extent of polygenic risk was estimated through linear regression adjusting for the top 3 genotyping principal components. R2 was used as a measurement of the explained variance. PRS analyses demonstrated significantly positive genetic overlap, respectively, between creativity with schizophrenia ((P+T) method: R2(max) ~ .196%, P = .00245; LDpred method: R2(max) ~ .226%, P = .00114), depression ((P+T) method: R2(max) ~ .178%, P = .00389; LDpred method: R2(max) ~ .093%, P = .03675), general risk tolerance ((P+T) method: R2(max) ~ .177%, P = .00399; LDpred method: R2(max) ~ .305%, P = .00016), and risky behaviors ((P+T) method: R2(max) ~ .187%, P = .00307; LDpred method: R2(max) ~ .155%, P = .00715). Our results suggest that human creativity is probably a polygenic trait affected by numerous variations with tiny effects. Genetic variations that predispose to psychiatric disorders and risky behaviors may underlie part of the genetic basis of creativity, confirming the epidemiological associations between creativity and these traits.
Keywords: creativity, GWAS, polygenic risk score, psychiatric disorders, risky behaviors
Introduction
Creativity, generally recognized as a significant driving force for the mankind, has been widely studied by researchers from multiple fields yet remains difficult to accurately define or explain for scientific purposes.1 One of the most commonly accepted definitions for creativity is “the capacity to produce new or original ideas, insights, or inventions” 2 or elaborated as “a process of becoming sensitive to problems, deficiencies, gaps in knowledge, missing elements, disharmonies, and so on; identifying the difficulty; searching for solutions, making guesses, or formulating hypotheses about the deficiencies: testing and retesting these hypotheses and possibly modifying and retesting them; and finally communicating the results.” 3 Creativity is also regarded as an array of multidimensional intrinsic and stable characteristics4 affected by genetic factors, personality traits, cognitive abilities, environment, and certain social context.5
Although creative individuals are usually described as seekers who take novel or insightful approaches to solve problems,6,7 many of such people may be also well known for the accompanying unstable emotional and psychological characteristics. Indeed, the association between creativity and mental illnesses is one of the oldest notions in the psychiatric field,8,9 which has been epitomized by Aristotle who remarked that “no great genius has ever existed without a strain of madness.” 10 This hypothesis is initially generated from early studies assessing the prevalence of mental illnesses (eg, depression and bipolar disorder) among eminent creative individuals and investigations of the creative abilities in psychiatric patients and their relatives,11–14 and has also gained support from recent epidemiological analyses with large sample size.15,16 Moreover, MacCabe et al found that university students in artistic majors are at increased risk of developing schizophrenia, bipolar disorder, and unipolar depression later in life.16 In addition, creative persons are often described as risk takers, and accumulating epidemiological evidence suggests that creativity is associated with greater tendencies for risk-taking behaviors.17,18
Tremendous studies have attempted to explain the concomitant presence of creativity and certain psychiatric or psychological traits (such as psychiatric disorders, risk taking, and affected cognition) and thereby to explore factors affecting creativity.18–20 Earlier studies proposed that the shared divergent thinking patterns or cognitive styles might explain their associations.21,22 In a recent landmark study by Power et al,20 the authors demonstrated significant genetic overlap between the diagnosis of schizophrenia and bipolar disorder and career choices of creative occupations (ie, actors, dancers, musicians, visual artists, and writers) in Icelandic subjects (1,024 artists and 85,268 nonartists) and European replication cohorts (273 artists and 27,072 nonartists) using polygenic risk score (PRS) analysis. They showed that higher creativity was partially explained by common variations that putatively participate in the genetic risk of psychiatric disorders. Therefore, uncovering the genetic basis of creativity could prompt our understanding of its biological foundation and provide insights into psychiatric disorders and even other traits potentially linked with creativity. However, despite the prominent success so far in disentangle the puzzle of creativity in the mankind, amended strategies are still considered necessary for future studies.23 For example, artistic or scientific professions have been used as the proxy phenotype of creativity in earlier studies given the difficulty in its measurement.12,15,20 Although it is commonly agreed that these occupations do require high level of creativity and ensure generation of creative products, career choices are not always made independent of any mood or psychological factors. For instance, a recent study argued that “art or music may be used as a therapeutic treatment approach, or an artistic lifestyle may be more compatible with individuals of psychiatric disorders.” 16
Here, we apply a standardized self-assessment test24 to obtain the quantified creativity scores from 2 independent cohorts of general Han Chinese subjects. The creativity scores of participants from both cohorts follow a normal distribution. We have conducted genome-wide association studies (GWAS) for 4,664 participants who passed quality control (QC) using the linear regression analysis and have performed PRS analyses using the classical pruning and thresholding (P+T) approach and LDpred method to explore the shared genetic attributes between creativity and psychiatric disorders, multiple psychological and personality traits, as well as subcortical structural features. Through thorough genetic analyses, this study provides hints for the genetic foundations of creativity and the correlated phenotypes in humans.
Materials and Methods
Sample Recruitment
Creativity is considered an array of multidimensional intrinsic and stable characteristics with moderate heritability (~22%).25 To evaluate an individual’s creativity based on their typical habits and behaviors,24 Luo et al (coauthor of the present study) developed a typical performance test (TPT), a questionnaire constructed from 10 dimensions (seizing keystone, synthesis, association, syraesthesia, resolution-incongruity, originality, insight, summarily explaining, evaluation, and pointing on future) summarized according to the 13 measurement indices from the Torrance Tests of Creative Thinking (TTCT).26,27 The full score of TPT is 276, which is the score sum of 46 specific items (the full TPT questionnaire is included in supplementary material). The authors demonstrated satisfying internal consistency of TPT (α = .93)24 and an appropriate criterion validity, and the creativity scores measured with TPT are also significantly associated (r ~ .5) with those measured with the Creativity Assessment Packet developed by Frank Williams.28
In the present study, creative thinking abilities of subjects from 2 independent cohorts of general Han Chinese subjects were measured using the self-assessment TPT (cohort 1: 2,815 subjects; cohort 2: 2,019 subjects)24; all participants were aged 18–45 and have either finished or enrolled in undergraduate education. Individuals without major psychiatric conditions (ie, psychosis or depression) in their self-reported health record during the sample collection were included in the present study. Among these testees, 13 subjects (cohort 1: 9 subjects; cohort 2: 4 subjects) reported scores significantly deviated from others probably due to erroneous understanding of the rating scale or gross errors in task execution rather than an authentic reflection of their characteristics (eg, TPT score < 70), and were therefore excluded from further analyses. The internal consistency among 10 dimensions of TPT was tested by calculating the standardized Cronbach’s α using “alpha” function in R psych package. All protocols and methods used in this study were approved by the institutional review board of the Kunming Institute of Zoology, Chinese Academy of Sciences. All participants have provided informed consents before any study-related procedures were conducted.
Genotyping, Quality Control, and Imputation
Genomic DNA of all participants were extracted from saliva according to the manufacturer’s protocol. Subjects in cohort 1 (n = 2,806) were genotyped using the Illumina Infinium Global Screening Array (GSA) Chip (Beijing Guoke Biotechnology Co., LTD; www.bioguoke.com). Six lakh seventy thousand nine hundred and five single-nucleotide polymorphisms (SNPs) were genotyped and mapped to the known physical locations in the hg19 reference genome. QC was performed according to the standard suggested by Anderson et al.29 Individuals with discordance between their estimated sex according to the homozygosity rate across all X-chromosome SNPs and recorded sex (self-reported) were removed (n = 39). Subjects having more than 5% genotyping failure rate or more than 6 SD heterozygosity rates were also excluded (n = 7). The identity by descent (IBD) of each pair of individuals were calculated using PLINK v1.9,30 and a total of 19 individuals were removed after IBD analysis (IBD ≥ 0.1875). Principal component analysis (PCA) was conducted using the EIGENSTRAT software31 to examine ancestral divergence and population stratification of the participants. Outliers whose principal components (PCs) were distal from Asia cluster (n = 6) or most other participants (>6 SD from the mean on any one of the top 10 PCs, n = 58) were removed. A total of 99 nonoverlapped individuals were removed from further analyses after these QC procedures. After the above individual level QC, SNPs with call rate<95% (n = 7,094), minor allele frequency (MAF) < 1% (n = 282,140), and Hardy–Weinberg equilibrium (HWE) P < 1 × 10–5 (n = 7,933) were excluded. Finally, 373,738 SNPs in 2,707 individuals of cohort 1 were proceeded for genotype imputation.
Subjects in cohort 2 (n = 2,015) were genotyped using the Illumina Genome-Wide Asian Screening Array (ASA-CHIA) Chip (Beijing Guoke Biotechnology co., LTD; www.bioguoke.com) on 711,727 SNPs. We conducted the QC using the same criteria as cohort 1. Briefly, we removed ineligible individuals based on the validity of their sex (n = 16), their heterozygosity rates or missing rate (n = 3), IBD (n = 12), ancestral divergence (n = 2), and whether they were outliers (n = 32). These processes yielded 58 nonoverlapped individuals to be removed from further analyses. SNPs with call rate < 95% (n = 28,149), MAF < 1% (n = 175,714), and HWE P < 1 × 10−5 (n = 5,351) were excluded. Finally, a set of 502,513 SNPs in 1,957 individuals of cohort 2 were kept for genotype imputation.
The imputation of both cohorts 1 and 2 was conducted using the following criteria: the post-QC genotypes were phased using SHAPEIT for each chromosome.32,33 The haplotypes of pre-phasing 1000 Genomes Project Phase 3 were used as the reference data,34 and imputation was performed for every 5-Mb interval of each chromosome using IMPUTE2.35 The SNPs with INFO > 0.8, MAF > 1%, call rate > 95%, and HWE P > 1 × 10−5 were considered suitable for further analyses. In sum, 5,481,607 autosomal SNPs in 2,707 subjects from cohort 1 and 5,375,141 autosomal SNPs in 1,957 subjects from cohort 2 passed QC.
Statistical Analysis and Meta-analysis
Linear regression analyses were conducted for each cohort separately, adjusting for the top 3 genotyping PCs, and a meta-analysis was then carried out to combine the 2 independent cohorts. We estimated the between-sample heterogeneity using the Cochran’s (Q) χ 2-test based on beta and SE and then calculated the pooled beta and combined P-values using the appropriate effect model. In brief, if there was no significant heterogeneity between samples (Q-statistic > 0.1), we utilized a fixed-effect model to combine the samples; otherwise, we would use a random-effect model. All the statistical analyses were performed using PLINK v1.9.30
Polygenic Risk Scoring Analysis
We estimated the predictive power of polygenic scores for creativity in our combined samples (cohorts 1 and 2) based on the GWAS summary statistics of multiple disorders/traits. Each training data set was collected, respectively, from GWAS studies of bipolar disorder (20,352 cases and 31,358 controls),36 schizophrenia (40,675 cases and 64,643 controls),37 depression (170,756 cases and 329,443 controls),38 general risk tolerance (466,571 individuals),39 risky behaviors (315,894 individuals),39 intelligence (269,867 individuals),40 subcortical structures (eg, putamen [37,571 individuals], nucleus accumbens [32,562 individuals]),41–43 and other traits (shown in supplementary table S3 and S4). As described in the previous study, risk tolerance was defined as an individual’s tendency, preparedness, or willingness to take risks in general (eg, “would you describe yourself as someone who takes risks?”); risky behavior was calculated from the first PC of 4 risky behaviors including “automobile speeding propensity,” “drinks per week,” “ever smoker,” and “number of sexual partners.” 39 GWAS summary statistics of the above complex disorders/traits are available in public data sets (https://www.med.unc.edu/pgc/results-and-downloads; http://enigma.ini.usc.edu/research/download-enigma-gwas-results/; https://www.thessgac.org/data; http://walters.psycm.cf.ac.uk/; https://ctg.cncr.nl/software/summary_statistics).
We first constructed polygenic scores using the classical method (pruning and thresholding, P + T)44,45: a series of independent SNPs (r2 < .1 within 500 kb) was extracted based on LD information using PLINK “--clump” function; whether a pruned SNP was included to calculate risk score in further analyses was determined depending on whether its P-value was lower than the specific threshold (P(thresholds) = 1.00 × 10−6, 1.00 × 10−4, 5.00 × 10−4, 0.001, 0.01, 0.05, 0.1, 0.2). We also re-estimated effect sizes of SNPs based on their LD using LDpred method (v1.0.8) by measuring the posterior mean effect size of each marker based on its raw effect size and LD information, using default parameters (the fraction P of nonzero effects = 1, 0.3, 0.1, 0.03, 0.01, 0.003, 0.001).46 The infinitesimal model was also used to measure posterior mean for effect sizes when assuming distant markers were unlinked. Notably, LDpred method included all available SNPs (after mapping to HapMap 3) rather than considering only independent SNPs in the (P+T) method. The risk score of each individual in the target samples was generated according to the raw (or re-estimated by LDpred) effect size (ie, ln(OR) or Beta) of pruned (or each) SNP in the training data sets using the PLINK “--score” function.30 The extent of polygenic risk was estimated using linear regression through comparing the full model (creativity score ~ polygenic score + PC1 + PC2 + PC3) and a reduced model (creativity score ~ PC1 + PC2 + PC3). Finally, R2 was used as a measurement of the explained variance.47
Results
Sample Characteristics and Quality Control
We carefully measured the creative thinking abilities of individuals aged 18–45 using TPT in 2 independent Han Chinese cohorts from general population. After excluding obvious outliers assuming that scores of the participants followed a normal distribution (eg, the creativity scores < 70 were removed), 2,806 subjects in cohort 1 and 2,015 participants in cohort 2 were proceeded for further analyses. The internal consistency rating across the 10 dimensions of TPT creativity scores was α = .91 in cohort 1 and α = .90 in cohort 2, indicating a strong internal reliability. To further assess the validity of our TPT scores, we used an additional cohort (including a total of 951 individuals after QC, age ranged 18–28) in which the individuals underwent both TPT test (to measure their creative thinking abilities) and Creative Achievement Questionnaire (CAQ,48 to measure their creative achievements), and we found that their TPT scores showed a significantly positive correlation with their CAQ scores (Pearson r = .35, P < 2.2 × 10−16, supplementary figure S1).
We then conducted QC analyses using the genome-wide SNP data through evaluating sex discordance, missing rate, relatedness, heterozygosity rate, and ancestral divergence from Han Chinese. After this QC analysis, the remaining sample size was 2,707 for cohort 1 and 1,957 for cohort 2, with an average age of 26.96 (cohort 1) and 27.90 (cohort 2), respectively. The average TPT creativity score of individuals in cohort 1 was 189.4 (SD = 28.19, minimum = 88, maximum = 273), and that of cohort 2 was 190.6 (SD = 28.55, minimum = 78, maximum = 272) (supplementary table S1). Supplementary figure S2 depicts the distribution of creativity scores of individuals who passed QC analyses. After imputation to the 1000 Genomes Project data and systematic QC analyses, we obtained genotype data of 5,481,607 autosomal SNPs in cohort 1, and 5,375,141 autosomal SNPs in cohort 2.
GWAS Analyses of Creativity Scores
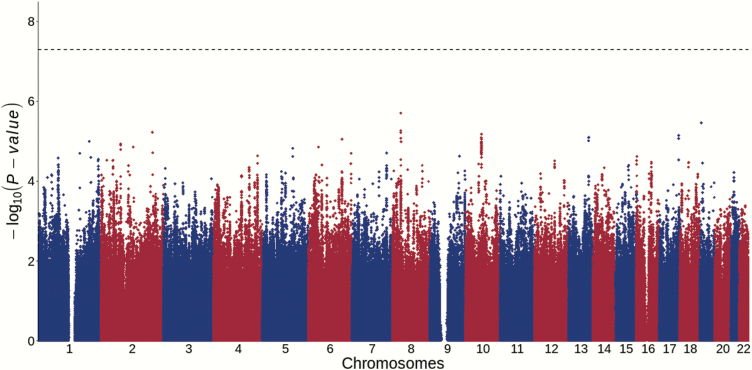
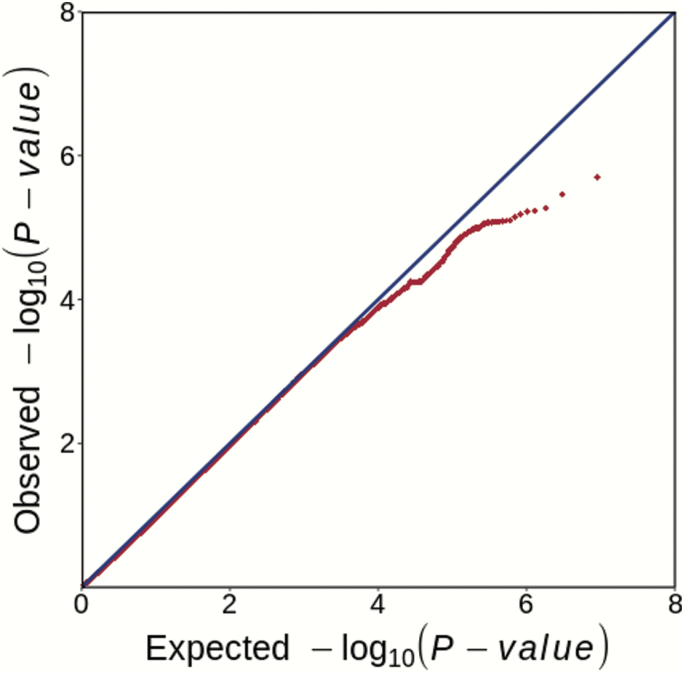
PCA using EIGENSTRAT comparing the genetic architectures between our GWAS samples and the Han Chinese samples from 1000 Genomes Project34 confirmed that the participants were of Han Chinese ancestry (supplementary figure S3). PCA was also used to assess the population substructures of cohort 1 and 2, and negligible population stratification was confirmed in both cohorts (supplementary figure S4). The genomic inflation λ was 1.005 for cohort 1 and 1.000 for cohort 2, further proving that no obvious population stratification exists in our samples and suggesting the polygenic inheritance of creativity. GWAS association analysis of each cohort was performed using linear regression covaring for top 3 genotyping PCs. Quantile–quantile (Q–Q) and Manhattan plots of each individual cohort are shown in supplementary figures S5 and S6. The GWAS statistics derived from each cohort were then subjected to the inverse-variance-weighted meta-analysis based on an appropriate effect model. Manhattan and Q–Q plots for the GWAS meta-analysis of 4,664 individuals are presented in figures 1 and 2, respectively. However, we did not observe any locus showing genome-wide significant association with creativity scores in each individual cohort (supplementary figures S5 and S6) or in the meta-analysis (figures 1 and 2). SNPs showing meta-analysis P-value lower than 5.00 × 10−5 are shown in supplementary table S2, and the nearest genes to each genomic variation were also indicated.
Fig. 1.
Manhattan plots for the GWAS meta-analysis of creativity among all individuals (n = 4,664). The dashed line indicates the threshold for genome-wide significance (P = 5.0 × 10−8).
Fig. 2.
Quantile–quantile (Q–Q) plot for the GWAS meta-analysis of creativity among all individuals (n = 4,664). The Q–Q plot displays the relationship between the experimentally observed P-values (vertical axis) to the expected P-values of a null distribution (horizontal axis).
Polygenic Prediction Analyses of Creativity by GWAS of Psychiatric Disorders, Risk Tolerance, and Risky Behaviors
We conducted polygenic score analyses to predict creativity in our samples (4,664 subjects from both cohorts 1 and 2 were included for PRS analyses to increase the statistical power) based on the published GWAS summary statistics of complex diseases/traits using both (P+T) and LDpred methods. We observed that the risk-profile SNPs of schizophrenia GWAS significantly and positively predicted creativity in our samples ((P+T): R2(max) ~ .196%, P = .00245; LDpred: R2(max) ~ .226%, P = .00114; table 1). Risk-profile SNPs of depression GWAS also significantly and positively predicted creativity in these subjects ((P+T): R2(max) ~ .178%, P = .00389; LDpred: R2(max) ~ .093%, P = .03675; table 1). These results further confirm previous findings by Power et al20 regarding the shared genetic basis between creativity and psychiatric disorders. However, the PRS of bipolar disorder did not predict creativity in our samples using either methods (supplementary tables S3 and S4). Nevertheless, the nonsignificant result regarding bipolar disorder and creativity does not deny their potential correlation because the PRS analysis is normally sensitive to the statistical power of the training data set (ie, bipolar disorder GWAS), and the sample size of the bipolar disorder GWAS is smaller compared with that of schizophrenia or depression. No significant correlation with creativity was seen in the PRS analyses using GWAS of other brain disorders (eg, Alzheimer’s disease, attention deficit hyperactivity disorder, Parkinson’s disease, obsessive–compulsive disorder) (supplementary tables S3 and S4).
Table 1.
Polygenic Risk Score (PRS) Analyses of Creativity
| (P+T) method | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Schizophrenia | Depression | General risk tolerance | Risky behaviors | |||||||||||||
| P threshold | Beta | SE | R 2 | P-value | Beta | SE | R 2 | P-value | Beta | SE | R 2 | P-value | Beta | SE | R 2 | P-value |
| 0.000001 | 0.020 | 0.015 | .00041 | .16851 | 0.027 | 0.015 | .00074 | .06231 | 0.035 | 0.015 | .00126 | .01545 | 0.043 | 0.015 | .00187 | .00307 |
| 0.0001 | 0.032 | 0.015 | .00102 | .02910 | 0.019 | 0.015 | .00037 | .18658 | 0.013 | 0.015 | .00018 | .36575 | 0.03 | 0.015 | .00090 | .04041 |
| 0.0005 | 0.04 | 0.015 | .00161 | .00605 | 0.021 | 0.015 | .00042 | .15895 | 0.015 | 0.015 | .00021 | .32297 | 0.028 | 0.015 | .00076 | .05937 |
| 0.001 | 0.044 | 0.015 | .00196 | .00245 | 0.016 | 0.015 | .00025 | .27875 | 0.029 | 0.015 | .00084 | .04698 | 0.029 | 0.015 | .00082 | .05103 |
| 0.01 | 0.035 | 0.015 | .00122 | .01714 | 0.042 | 0.015 | .00178 | .00389 | 0.042 | 0.015 | .00177 | .00399 | 0.002 | 0.015 | .00000 | .91235 |
| 0.05 | 0.027 | 0.015 | .00072 | .06604 | 0.037 | 0.015 | .00134 | .01225 | 0.035 | 0.015 | .00121 | .01745 | -0.002 | 0.015 | .00000 | .91142 |
| 0.1 | 0.021 | 0.015 | .00042 | .16140 | 0.042 | 0.015 | .00174 | .00430 | 0.033 | 0.015 | .00110 | .02336 | -0.001 | 0.015 | .00000 | .92002 |
| 0.2 | 0.024 | 0.015 | .00055 | .10802 | 0.032 | 0.015 | .00102 | .02892 | 0.041 | 0.015 | .00168 | .00503 | 0.000 | 0.015 | .00000 | .97302 |
| LDpred method | ||||||||||||||||
| Schizophrenia | Depression | General risk tolerance | Risky behaviors | |||||||||||||
| Prior | Beta | SE | R 2 | P-value | Beta | SE | R 2 | P-value | Beta | SE | R 2 | P-value | Beta | SE | R 2 | P-value |
| 0.001 | 0.002 | 0.015 | .00000 | .91748 | −0.007 | 0.015 | .00005 | .61850 | 0.046 | 0.015 | .00215 | .00151 | 0.006 | 0.015 | .00004 | .68110 |
| 0.003 | 0.012 | 0.015 | .00015 | .39926 | 0.003 | 0.015 | .00001 | .84053 | 0.038 | 0.015 | .00143 | .00985 | 0.008 | 0.015 | .00007 | .57422 |
| 0.01 | 0.013 | 0.015 | .00016 | .39174 | 0.014 | 0.015 | .00020 | .33373 | 0.035 | 0.015 | .00121 | .01760 | 0.016 | 0.015 | .00026 | .27429 |
| 0.03 | 0.031 | 0.015 | .00097 | .03317 | 0.021 | 0.015 | .00045 | .14908 | 0.033 | 0.015 | .00111 | .02307 | 0.018 | 0.015 | .00032 | .22352 |
| 0.1 | 0.035 | 0.015 | .00123 | .01636 | 0.029 | 0.015 | .00081 | .05126 | 0.034 | 0.015 | .00119 | .01859 | 0.024 | 0.015 | .00057 | .10393 |
| 0.3 | 0.036 | 0.015 | .00127 | .01477 | 0.031 | 0.015 | .00093 | .03675 | 0.045 | 0.015 | .00198 | .00237 | 0.039 | 0.015 | .00155 | .00715 |
| 1.0 | 0.042 | 0.015 | .00171 | .00471 | 0.028 | 0.015 | .00081 | .05155 | 0.055 | 0.015 | .00297 | .00019 | 0.023 | 0.015 | .00052 | .11888 |
| Infinitesimal | 0.048 | 0.015 | .00226 | .00114 | 0.027 | 0.015 | .00074 | .06252 | 0.055 | 0.015 | .00305 | .00016 | 0.018 | 0.015 | .00032 | .21975 |
The estimation of the proportion of variance was from R2 (computed by comparison of a full model including top 3 PCs and polygenic risk scores to a reduced model including PCs only). Bold represents the maximum R2 under distinct parameters using each method.
There has also been robust evidence supporting a positive epidemiological correlation between creativity and risky behaviors.18 To investigate whether this is (partially) due to their shared genetic roots, we conducted PRS analysis of creativity using GWAS data of general risk tolerance and risky behaviors. We found that the risk-profile SNPs of risk tolerance and risky behaviors both significantly and positively predicted creativity (for risk tolerance, (P+T): R2(max) ~ .177%, P = .00399; LDpred: R2(max) ~ .305%, P = .00016; for risky behaviors, (P+T): R2(max) ~ .187%, P = .00307; LDpred: R2(max) ~ .155%, P = .00715; table 1). These results support the shared genetic basis between creativity and risky behaviors. Because risky behaviors were also found associated with the risk of psychiatric illnesses, our data suggest that creativity and tendencies for risky behaviors may reflect 2 aspects of the multidimensional personality traits that are relevant to psychiatric disorders. However, no significant genetic overlap between creativity and other personality traits (eg, neuroticism, extraversion, and openness) was seen in the present study (supplementary tables S3 and S4).
We also examined whether there was shared genetic basis between creativity and subcortical structures as well as cognition-related phenotypes. Although the (P+T) PRS analysis revealed significantly positive correlation of creativity with common variations associated with putamen volumes and those associated with nucleus accumbens volumes (putamen: R2(max) ~ .237%, P = .00088; nucleus accumbens: R2(max) ~ .171%, P = .00464; supplementary table S3), these polygenic associations were not confirmed in the LDpred analysis (supplementary table S4). By contrast, the LDpred PRS analysis revealed significantly negative polygenic association between intelligence and creativity (R2(max) ~.120%, P = .01802; supplementary table S4), which was not significant in the (P+T) PRS analysis (supplementary table S3). Therefore, whether there are polygenic associations between creativity, subcortical structures, and intelligence remains inconclusive.
Discussion
Although widely recognized as an important trait playing key roles during human evolution, creativity remains relatively opaque regarding its mechanisms, especially the genetic basis.49 As discussed, there are numerous idioms in every culture in the world about the unstable psychological and emotional states in some talented high-achieving people, investigating the intrinsic and extrinsic factors affecting creativity, especially those simultaneously involved in mood and psychological characteristics, is of great interest.
The recent development of high-throughput genotyping technology offers the opportunity to investigate genetic basis of creativity and its genetic correlations with other traits at the genome-wide level. In fact, a previous study suggested that PRS of bipolar disorder and schizophrenia were significantly associated with creative occupations (ie, artistic professions), indicating shared genetic origins between creativity and psychiatric disorders.20 Enlightened by this study, we hypothesize that there is probably shared genetic basis between creativity and other psychological traits involved in psychiatric disorders. We herein investigated correlations between the genotypes of individuals with quantified creativity scores and the published GWAS summary statistics of multiple traits (ie, psychiatric disorders, personality behaviors, cognitive functions, and subcortical volumes) using the PRS method. Notably, we have applied a different approach of quantifying creativity and have further confirmed the shared genetic basis between psychiatric disorders and creativity, although a slight inconsistency (PRS of bipolar disorder did not predict creativity in our samples) exists between our results and the Power et al’s study.20 We have also identified genetic overlap between depression and creativity, which is consistent with a previous study showing that university students of artistic subjects are at an increased risk of developing unipolar depression later in life.16 These results are intriguing, and further investigations of the neural mechanisms underlying the genetic overlap between psychiatric disorders and creativity are warranted. Although limited evidence is available for such mechanisms, altered brain functional connectivity, which has been found to be involved in creativity,23 and to be altered in schizophrenia and depression,50–52 might be a candidate for future studies.
In addition, we for the first time find significant genetic overlap between creativity and risky behaviors through PRS analysis. Although biological and physiological explanations for this overlap is lacking, the current results still lend strong support for the direct impact of certain genetic factors on both creativity and risky behaviors. This is probably because some common SNPs, while contribute to increased creativity in humans, might also underlie processes resulting in a higher tendency of risky activities. Overall, a possible genetic link among creativity, risky behaviors, and psychiatric abnormalities is indicated in the present study.
We attempted to delve into the correlation among creativity, risky behaviors, and psychiatric abnormalities by exploring the association between creativity and some brain imaging and cognitive characteristics. As mentioned, 2 major methods for PRS analyses, the (P + T) and LDpred methods, were applied. These 2 methods differ at the calculation of effect sizes for SNPs. Specifically, the LDpred method re-estimates effect sizes of SNPs accounting for LD using the Bayesian approach and thereby results in potentially different raw effect sizes for SNPs. Nevertheless, our analyses revealed that both the diagnosis of schizophrenia and depression, and risky behaviors were significantly correlated with creativity regardless of the method used. Therefore, we sought to define additional brain imaging and cognitive phenotypes associated with creativity through both methods, which are considered more reliable and likely to be the true factors affecting creativity. Unfortunately, although significant associations between creativity and the volumes of putamen and nucleus accumbens were observed through the (P+T) PRS analysis, the LDpred analysis did not support these correlations. Meanwhile, intelligence predicted creativity only in the LDpred PRS analysis. As a result, the correlation between creativity and these phenotypes remain less convincing, and further investigations are needed. However, the negative associations between creativity and intelligence in the LDpred PRS analysis might still have some implications. Although creativity and intelligence have long been considered related to some extent, the fundamental nature of their interplay is still under debate.53 Specifically, Jung et al argued that intelligence and creativity occupied 2 extremes of a dichotomy,54 and they considered intelligence to be a “dedicated reasoning capacity” for problems that possess rule-based, cause–effect relationships, whereas they recognized creativity as an ability of “improvisational reasoning” and generating novel and unsighted solutions to problems. It is also raised that creativity is more than likely to be evolutionarily novel compared with intelligence, as creativity might appear between 60,000 and 30,000 years ago in the Middle/Upper Paleolithic, probably corroborating the emergence of art, science, politics, religion, and syntactical language in humans.55
Although the present study presents intriguing discoveries of the genetic foundation for creativity, certain limitations should be acknowledged. First, despite using 2 independent cohorts, the overall sample size of the present study is relatively small compared with other large-scale association studies. Owing to this limited sample size, the GWAS meta-analysis defined multiple variations showing nominal significance in both samples but no genome-wide significant loci. Nevertheless, this result might, on the other hand, support the polygenic nature of creativity, as previous GWAS of other complex traits, such as childhood intelligence,56 also identified variations showing nominal significance in multiple independent cohorts (eg, rs236330 in FNBP1L) rather than SNPs with genome-wide significance. Nevertheless, further analyses using much larger samples might allow identification of individual variations that are genome-wide significantly associated with creativity. Second, although we are confident that the recent technological development in genetic analytical tools will promote investigations of creativity as a complex and heritable trait, creativity remains a complicated readout to be accurately quantified for scientific purposes. Actually, multiple approaches have been proposed to measure the different domains of creativity in the past few decades, each has its merits and limitations. Applying different methods to systemically investigate creativity is therefore important to clarify its mechanisms from various perspectives. It should also be noted that participants of the present study finished the TPT creativity questionnaire online, and their mental health statuses (ie, having no diagnosis or family history of psychosis or depression) were self-reported rather than screened by psychiatrists according to the criteria of DSM-5 or ICD-10. This is a potential limitation and future analyses in subjects clinically ensured to be “mentally healthy” might provide more accurate results. In addition, the training and target data sets in our PRS analyses are from distinct populations due to the limited availability of large GWAS data of psychiatric disorders and related traits in Han Chinese. However, previous GWAS studies have also shown that genetic effects of psychiatric disorders (eg, schizophrenia) are highly consistent between different populations. For example, Lam et al showed that common genetic variations conferring risk of schizophrenia exerted highly similar effects in populations of East Asian and European ancestries (genetic correlation = 0.98 ± 0.03),57 and Li et al found that approximately 95% of the genome-wide significant index alleles (or their proxies) from the European schizophrenia PGC2 GWAS were over-represented in Chinese schizophrenia cases (~50% of the alleles achieved nominal significance).58 Similarly, a previous PRS study found significant effects of the polygenic risk derived from European schizophrenia GWAS on cortical gyrification in the inferior parietal lobules in 2 independent Han Chinese samples.59 In some nonpsychiatric disorders, Zhou et al performed the transethnic PRS analyses between height/body mass index/bone mineral density (training data sets, based on GWAS summary statistics from European and Asian, respectively) and lumbar disc degeneration/herniation (target data sets in Asian),60 and observed strong positive genetic correlations across traits, which is consistent with the biological underpinnings of lumbar disc degeneration. Taken together, transpopulation polygenic score analyses based on large-scale GWAS data sets might still provide useful insights.
Supplementary Material
Funding
This work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB13000000 to M.L.). X.X. was also supported by the Chinese Academy of Sciences Western Light Program and Youth Innovation Promotion Association, CAS. M.L. was also supported by CAS Pioneer Hundred Talents Program and the 1000 Young Talents Program.
Acknowledgment
The authors declare no competing financial interests.
Author Contributions
X.X. and M.L. designed the study and interpreted the results. H.L. conducted the primary statistical analysis. C.Z., X.C., L.W., and H.C. contributed to design and analysis of genomic. GeseDNA Research Team carried out subject recruitment. F.L. and Y.M. contributed to design the creativity test. H.L., Y.M., X.X., and M.L. drafted the manuscript, and all authors contributed to the final version of the article.
References
- 1. Kaufman JC, Sternberg RJ.. The Cambridge Handbook of Creativity. Cambridge: Cambridge University Press; 2010. [Google Scholar]
- 2. Vernon PE. The nature-nurture problem in creativity. In: Glover JA, Ronning RR, Reynolds C, eds. Handbook of Creativity: Perspectives on Individual Differences. New York: Plenum; 1989:93–110. [Google Scholar]
- 3. Sternberg RJ. The Nature of Creativity: Contemporary Psychological Perspectives. New York, NY: Cambridge University Press; 1988. [Google Scholar]
- 4. Harris JA. Measured intelligence, achievement, openness to experience, and creativity. Pers Indiv Differ. 2004;36:913–929. [Google Scholar]
- 5. Kandler C, Riemann R, Angleitner A, Spinath FM, Borkenau P, Penke L. The nature of creativity: the roles of genetic factors, personality traits, cognitive abilities, and environmental sources. J Pers Soc Psychol. 2016;111(2):230–249. [DOI] [PubMed] [Google Scholar]
- 6. Heilman KM, Nadeau SE, Beversdorf DO. Creative innovation: possible brain mechanisms. Neurocase. 2003;9(5):369–379. [DOI] [PubMed] [Google Scholar]
- 7. Ellamil M, Dobson C, Beeman M, Christoff K. Evaluative and generative modes of thought during the creative process. Neuroimage. 2012;59(2):1783–1794. [DOI] [PubMed] [Google Scholar]
- 8. Greenwood TA. Positive traits in the bipolar spectrum: the space between madness and genius. Mol Neuropsychiatry. 2017;2(4):198–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Silvia PJ, Kaufman JC. Creativity and mental illness. In: J. C. Kaufman JC, Sternberg RJ, eds. Cambridge Handbook of Creativity. New York: Cambridge University Press; 2010:381–394. [Google Scholar]
- 10. Clark JR, de la Motto AL. The paradox of genius and madness: seneca and his influence. Cuadernos Filología Clásica: Estudios latinos. 1992:189–200. [Google Scholar]
- 11. Post F. Creativity and psychopathology. A study of 291 world-famous men. Br J Psychiatry. 1994;165(1):22–34. [DOI] [PubMed] [Google Scholar]
- 12. Andreasen NC. Creativity and mental illness: prevalence rates in writers and their first-degree relatives. Am J Psychiatry. 1987;144(10):1288–1292. [DOI] [PubMed] [Google Scholar]
- 13. Taylor CL. Creativity and mood disorder: a systematic review and meta-analysis. Perspect Psychol Sci. 2017;12(6):1040–1076. [DOI] [PubMed] [Google Scholar]
- 14. Simeonova DI, Chang KD, Strong C, Ketter TA. Creativity in familial bipolar disorder. J Psychiatr Res. 2005;39(6):623–631. [DOI] [PubMed] [Google Scholar]
- 15. Kyaga S, Lichtenstein P, Boman M, Hultman C, Långström N, Landén M. Creativity and mental disorder: family study of 300,000 people with severe mental disorder. Br J Psychiatry. 2011;199(5):373–379. [DOI] [PubMed] [Google Scholar]
- 16. MacCabe JH, Sariaslan A, Almqvist C, Lichtenstein P, Larsson H, Kyaga S. Artistic creativity and risk for schizophrenia, bipolar disorder and unipolar depression: a Swedish population-based case-control study and sib-pair analysis. Br J Psychiatry. 2018;212(6):370–376. [DOI] [PubMed] [Google Scholar]
- 17. Pankove E, Kogan N. Creative ability and risk-taking in elementary school children. J Pers. 1968;36(3):420–439. [DOI] [PubMed] [Google Scholar]
- 18. Tyagi V, Hanoch Y, Hall SD, Runco M, Denham SL. The risky side of creativity: domain specific risk taking in creative individuals. Front Psychol. 2017;8:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Khalil R, Godde B, Karim AA. The link between creativity, cognition, and creative drives and underlying neural mechanisms. Front Neural Circuits. 2019;13:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Power RA, Steinberg S, Bjornsdottir G, et al. . Polygenic risk scores for schizophrenia and bipolar disorder predict creativity. Nat Neurosci. 2015;18(7):953–955. [DOI] [PubMed] [Google Scholar]
- 21. Folley BS, Park S. Verbal creativity and schizotypal personality in relation to prefrontal hemispheric laterality: a behavioral and near-infrared optical imaging study. Schizophr Res. 2005;80(2–3):271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Owen GS, Cutting J, David AS. Are people with schizophrenia more logical than healthy volunteers? Br J Psychiatry. 2007;191:453–454. [DOI] [PubMed] [Google Scholar]
- 23. Liu Z, Zhang J, Xie X, et al. . Neural and genetic determinants of creativity. Neuroimage. 2018;174:164–176. [DOI] [PubMed] [Google Scholar]
- 24. Luo F, Meng QM. Construction of self-assessment test on creative thinking abilities of high school students. Psychol Dev Educ. 2005;21:94–98. [Google Scholar]
- 25. Bouchard TJ, Lykken DT, Tellegen A, Blacker DM, Waller NG. Creativity, heritability, familiarity: which word does not belong? Psychol Inq. 1993;4:235–237. [Google Scholar]
- 26. Torrance EP. Predictive validity of the Torrance Tests of Creative Thinking. J Creat Behav. 1972;6:236–262. [Google Scholar]
- 27. Torrance EP. Empirical validation of criterion-referenced indicators of creative ability through a longitudinal study. Creative Child Adult Quart. 1981;6:136–140. [Google Scholar]
- 28. Williams FE. Creativity Assessment Packet: CAP. Buffalo, NY: DOK Publishers; 1980. [Google Scholar]
- 29. Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5(9):1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Purcell S, Neale B, Todd-Brown K, et al. . PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. [DOI] [PubMed] [Google Scholar]
- 32. Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9(2):179–181. [DOI] [PubMed] [Google Scholar]
- 33. Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10(1):5–6. [DOI] [PubMed] [Google Scholar]
- 34. Genomes Project Consortium, Auton A, Brooks LD, et al. . A global reference for human genetic variation. Nature. 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stahl EA, Breen G, Forstner AJ, et al. ; eQTLGen Consortium; BIOS Consortium; Bipolar Disorder Working Group of the Psychiatric Genomics Consortium Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51(5):793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pardiñas AF, Holmans P, Pocklington AJ, et al. ; GERAD1 Consortium; CRESTAR Consortium Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50(3):381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Howard DM, Adams MJ, Clarke TK, et al. ; 23andMe Research Team; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22(3):343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Karlsson Linnér R, Biroli P, Kong E, et al. ; 23andMe Research Team; eQTLgen Consortium; International Cannabis Consortium; Social Science Genetic Association Consortium Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet. 2019;51(2):245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Savage JE, Jansen PR, Stringer S, et al. . Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet. 2018;50(7):912–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hibar DP, Adams HHH, Jahanshad N, et al. . Novel genetic loci associated with hippocampal volume. Nat Commun. 2017;8:13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Satizabal CL, Adams HHH, Hibar DP, et al. . Genetic architecture of subcortical brain structures in 38,851 individuals. Nat Genet. 2019;51(11):1624–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Adams HH, Hibar DP, Chouraki V, et al. . Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat Neurosci. 2016;19(12):1569–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. International Schizophrenia Consortium, Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9(3):e1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vilhjálmsson BJ, Yang J, Finucane HK, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium, Discovery, Biology, and Risk of Inherited Variants in Breast Cancer (DRIVE) Study Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am J Hum Genet. 2015;97(4):576–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee SH, Goddard ME, Wray NR, Visscher PM. A better coefficient of determination for genetic profile analysis. Genet Epidemiol. 2012;36(3):214–224. [DOI] [PubMed] [Google Scholar]
- 48. Carson SH, Peterson JB, Higgins DM. Reliability, validity, and factor structure of the creative achievement questionnaire. Creativity Res J. 2005;17:37–50. [Google Scholar]
- 49. Piffer D. The genetics of creativity: the underdog of behavior genetics? In: Jung RE, Vartanian O, eds. The Cambridge Handbook of the Neuroscience of Creativity. Cambridge: Cambridge University Press; 2018:437–450. [Google Scholar]
- 50. Li T, Wang Q, Zhang J, et al. . Brain-wide analysis of functional connectivity in first-episode and chronic stages of schizophrenia. Schizophr Bull. 2017;43(2):436–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang Q, Zhang J, Liu Z, et al. . “Brain connectivity deviates by sex and hemisphere in the first episode of schizophrenia” – a route to the genetic basis of language and psychosis? Schizophr Bull. 2019;45(2):484–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rolls ET, Cheng W, Gong W, et al. . Functional connectivity of the anterior cingulate cortex in depression and in health. Cereb Cortex. 2019;29(8):3617–3630. [DOI] [PubMed] [Google Scholar]
- 53. Kaufman JC, Plucker JA. Intelligence and creativity. In: Sternberg RJ, Kaufman SB, eds. The Cambridge Handbook of Intelligence. Cambridge: Cambridge University Press; 2011:771–783. [Google Scholar]
- 54. Jung RE. Evolution, creativity, intelligence, and madness: “Here Be Dragons”. Front Psychol. 2014;5:784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kaufman JC, Sternberg RJ. Evolutionary approaches to creativity. In: The Cambridge Handbook of Creativity. 1st ed Cambridge and New York: Cambridge University Press; 2010. [Google Scholar]
- 56. Benyamin B, Pourcain B, Davis OS, et al. ; Wellcome Trust Case Control Consortium 2 (WTCCC2) Childhood intelligence is heritable, highly polygenic and associated with FNBP1L. Mol Psychiatry. 2014;19(2):253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lam M, Chen CY, Li Z, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Indonesia Schizophrenia Consortium; Genetic REsearch on schizophreniA neTwork-China and the Netherlands (GREAT-CN) Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat Genet. 2019;51(12):1670–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li Z, Chen J, Yu H, et al. . Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat Genet. 2017;49(11):1576–1583. [DOI] [PubMed] [Google Scholar]
- 59. Liu B, Zhang X, Cui Y, et al. . Polygenic risk for schizophrenia influences cortical gyrification in 2 independent general populations. Schizophr Bull. 2017;43(3):673–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhou X, Cheung CL, Karasugi T, et al. . Trans-ethnic polygenic analysis supports genetic overlaps of lumbar disc degeneration with height, body mass index, and bone mineral density. Front Genet. 2018;9:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.