Abstract
Currently, coronavirus disease 2019 (COVID-19), has posed an imminent threat to global public health. Although some current therapeutic agents have showed potential prevention or treatment, a growing number of associated adverse events have occurred on patients with COVID-19 in the course of medical treatment. Therefore, a comprehensive assessment of the safety profile of therapeutic agents against COVID-19 is urgently needed. In this study, we proposed a network-based framework to identify the potential side effects of current COVID-19 drugs in clinical trials. We established the associations between 116 COVID-19 drugs and 30 kinds of human tissues based on network proximity and gene-set enrichment analysis (GSEA) approaches. Additionally, we focused on four types of drug-induced toxicities targeting four tissues, including hepatotoxicity, renal toxicity, lung toxicity, and neurotoxicity, and validated our network-based predictions by preclinical and clinical evidence available. Finally, we further performed pharmacovigilance analysis to validate several drug-tissue toxicities via data mining adverse event reporting data, and we identified several new drug-induced side effects without labeling in Food and Drug Administration (FDA) drug instructions. Overall, this study provides forceful approaches to assess potential side effects on COVID-19 drugs, which will be helpful for their safe use in clinical practice and promoting the discovery of antiviral therapeutics against SARS-CoV-2.
Keywords: Side effect, COVID-19 drug, Network proximity, CMap, OpenVigil, Pharmacovigilance
Highlights
-
•
Network-based framework identifies the potential side effects of current COVID-19 drugs.
-
•
Network proximity and GSEA approaches predict the associations between COVID-19 drugs and human tissues.
-
•
Pharmacovigilance analysis validates several drug-tissue toxicities via data mining adverse event reporting data.
-
•
Several tissue toxicities of drugs that hadn't been labeled by FDA drug instructions were uncovered.
1. Introduction
The current Coronavirus disease 2019 (COVID-19) has presented a serious threat to global public health (Del Rio and Malani, 2020). As of July 18th, 2020, the confirmed cases have reached up to 14 million and caused more than 600,000 deaths (Yang, 2020). However, many aspects of transmission, infection, and treatment of COVID-19 remain unclear (Wiersinga et al., 2020). Nowadays, a large number of clinical trials are being conducted on repurposed or experimental drugs to identify prophylactic and therapeutic treatments against COVID-19. Although some current therapeutic agents have showed potential in ameliorating symptoms and reducing mortality in patients (Ledford, 2020), a growing number of associated adverse events have occurred on patients with COVID-19 in the course of medical treatment. For example, the malaria medication chloroquine, has been brought into multiple clinical trials (e.g. ClinicalTrials.gov number, NCT04353336) to determine its effect on inhibiting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Unfortunately, a latest research has demonstrated that hydroxychloroquine or chloroquine used alone or with a macrolide, do not show a benefit on in-hospital outcomes for COVID-19 and on the contrary, were relevant to decrease in-hospital survival as well as increase frequency of ventricular arrhythmias (Mehra et al., 2020). Therefore, a comprehensive evaluation of the safety profile of potential therapeutic agents against COVID-19 is of paramount importance (Javorac et al., 2020).
Generally, conventional experimentally approaches including in vivo and in vitro assays for toxicity determination are time-consuming and costly (Merz et al., 2014). Thus, computational strategies to assess side effects of therapeutic drugs are propitious to improve efficiency and accelerate the development of drug discovery (Cai et al., 2018, 2019; Wu et al., 2019). Recent technological and computational advances in genomics and systems biology have offered possibilities for identifying side effects or therapeutic agents by integrating disease proteins and drug-target interactions in human protein–protein interactome (Cheng et al., 2018, 2019; Fang et al., 2019). Since drug targets do not work in isolation from the complex system of proteins that comprise the molecular environment of the cell, each drug-target interaction must be examined in an appropriate integrative context (Menche et al., 2015). Several network-based methodologies, such as network proximity, offer powerful tools for efficient identification of potentially adverse effects or new indications for existing drugs (Fang et al., 2020). In order to prioritize these drug-side effect pairs identified by network-based approaches, rigorous validation is mandatory (Cheng et al., 2018). Since approved drugs are already in clinical practice, their potential side effect can be validated via pharmacovigilance analysis that utilizes adverse drug event data available (Böhm et al., 2012).
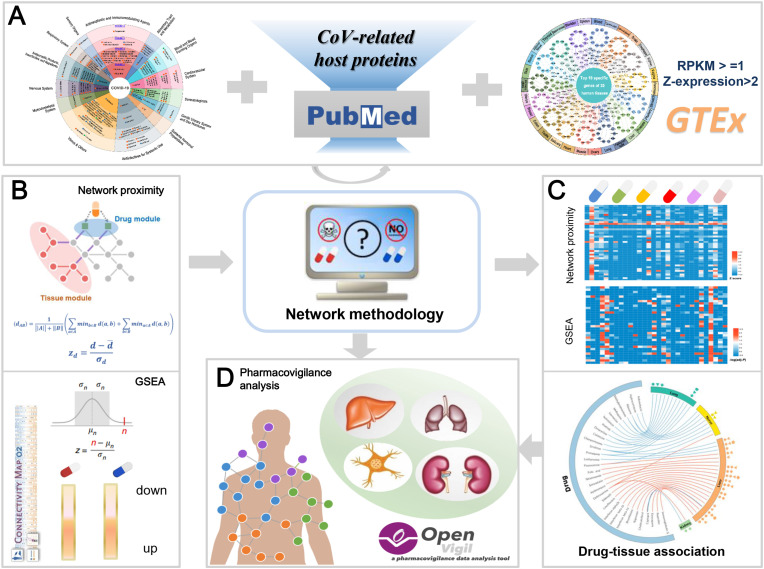
The Genotype-Tissue Expression (GTEx) is a functional genomics database that provides genotype-tissue expression to study the relationship between genetic variation and gene expression in human tissues (GTEx project, 2013). In this study, we developed a network-based framework to identify side effects of the ongoing drugs in COVID-19 pipeline (Fig. 1). We posited that systematic examination of the relationship between drug targets (or drug regulatory genes) and specific expression genes in each tissue from GTEx, will serve as a foundation for generating predictive models to identify potential side effects in tissue. Specifically, we first collected the ongoing drugs in COVID-19 through searching for clinical trials and built their drug-target network from six data sources available as well as drug-gene signature from the Connectivity Map (CMap). To determine the potential adverse events related to COVID-19 drugs, we established the associations between COVID-19 drugs and 30 kinds of human tissues based on network proximity and gene-set enrichment analysis (GSEA) approaches. Moreover, we concentrated on side effects of four tissues including liver, kidney, lung and nerve, and validated several drug-tissue associations utilizing large-scale adverse drug event data and more than half of them supported our network-based prediction. Our overall framework is summarized in Fig. 1.
Fig. 1.
A schematic diagram illustrating network methodology and adverse drug event data-based validation for in silico prediction of side effects of ongoing drugs in COVID-19 pipeline. A Integration of ongoing drugs in COVID-19 pipeline, coronavirus (CoV)-related host proteins and specific high-expressed genes in tissues; B Network proximity and gene-set enrichment analysis (GSEA) approaches; CIn silico prediction of side effects of drugs on 30 tissues; D Pharmacovigilance analysis across four specific tissues (liver, lung, nerve and kidney)-induced side effects.
2. Material and method
2.1. Collection of ongoing drugs in COVID-19 pipeline
The ongoing drugs in COVID-19 pipeline were integrated from current clinical trials. We first searched for clinical trials (accessed in 21st April 2020) related to COVID-19 medication by using terms ‘COVID OR COVID-19 OR coronavirus OR SARS-COV-2’ in ClinicalTrials.gov database (https://clinicaltrials.gov/). Only trials labeled with “Drug” in “Interventions” column were preserved. In total, we collected 116 drugs from 308 COVID-19 clinical trials after removing duplicates (Table S1). Each drug was subsequently matched by the unified DrugBank ID and classified by the Anatomical Therapeutic Chemical Classification (ATC) code.
2.2. Integration of coronavirus (CoV) host proteins
The host proteins of coronavirus (CoV) were collected from four recent public references (Gordon et al., 2020; Maryam Al-Motawa, 2020a; Shen et al., 2020; Zhou et al., 2020). Zhou et al. has integrated 119 high-quality experimental validated host proteins for drug repurposing for human CoV (Zhou et al., 2020). Additionally, a latest research has identified 332 high-confidence SARS-CoV-2-human protein-protein interactions (PPIs) which were connected to multiple biological processes (Gordon et al., 2020). Moreover, 204 and 50 host proteins of SARS-CoV-2 have been separately acquired from two recent proteomics publication (Maryam Al-Motawa, 2020a; Shen et al., 2020). In total, 705 CoV-related host proteins were consequently integrated (Table S2).
2.3. Construction of drug-target network
We assembled high-quality physical drug target interactions (DTIs) for ongoing COVID-19 drugs from six authoritative databases, including DrugBank database (v4.3) (Wishart et al., 2018), the Therapeutic Target Database (TTD, v4.3.02) (Yan et al., 2018), the PharmGKB database (Barbarino et al., 2018), ChEMBL (v20) (Gaulton et al., 2017), BindingDB (Gilson et al., 2016), and IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018). A physical DTI was defined by using reported binding affinity data: inhibition constant/potency (Ki), dissociation constant (Kd), median effective concentration (EC50), or median inhibitory concentration (IC50) ≤ 10 μM. Only physical DTIs that meet the three criteria were preserved: (i) proteins targets owning unique UniProt accession number; (ii) proteins targets labeled as “reviewed” in the UniProt database; and (iii) proteins targets marked as Homo sapiens species. Eventually, 1222 DTIs covering 81 ongoing COVID-19 drugs were obtained (Table S3).
2.4. Tissue-specific protein network
The RNA-seq data (RPKM value) of 30 tissues were extracted from the GTEx V6 release (https://gtexportal.org/home/). We asserted that a gene is expressed in a specific tissue if its RPKM value is larger or equal to 1 in over 80% of samples, and vice versa. To quantify the expression significance of a tissue-expressed gene i in a tissue t, the average expression E(i) and the standard deviation of a gene's expression across all considered tissues were calculated. The significance of gene expression in tissue t is defined as below:
The gene was considered as high expressed in a specific tissue if its Z-expression was higher than 2 in this tissue.
2.5. Network-based approaches
2.5.1. Network proximity
Given A, the module formed by specific high expressed proteins in a given tissue (tissue module), and B, the module formed by target proteins of a given drug (drug module), the closest distance represents the average shortest path length of all the nodes in tissue module (A) to drug module (B) in the human protein–protein interactome, which can be described as:
where indicates the shortest path length between tissue protein and drug target .
To assess the significance of the network distance between each drug module and tissue module, we constructed a reference distance distribution corresponded to the expected distance of each two randomly selected groups of proteins from human protein–protein interactome which have the same size and degree distribution as the original protein modules. This process was repeated 1000 times. According to the mean and standard deviation () of the reference distribution, the Z-score () was computed by converting an observed (non-Euclidean) distance to a normalized distance:
2.5.2. Gene-set enrichment analysis (GSEA)
The gene set enrichment analysis (GSEA) was performed as an additional prioritization method. Drug-gene signatures were collected from the Connectivity Map (CMap2.0), a pharmacogenomics database inducing over 7000 gene expression profiles from cultured human cell lines treated with 1309 bioactive molecules at different concentrations, covering 6100 individual instances (Lamb et al., 2006). CMap offers a measure to evaluate the extent of differential expression for a given probe set. The amplitude () is defined as follows:
where refers to the scaled and threshold average difference value for the drug treatment group, and represents the threshold average difference value for the control group. Therefore, and indicate increased expression (upregulation) and decreased expression (downregulation) upon treatment respectively, while means no differential expression. In this study, drug gene signatures with absolute value of a greater than two-fold induction (|| > 0.67) were defined as drug-gene pairs. Finally, we obtained the drug regulatory gene network for 38 COVID-19 ongoing drugs, covering 91,164 drug-gene pairs. (Table S4).
We assumed that a drug would have a correlation with a specific tissue if its regulatory genes tend to be the specific high expressed proteins in this tissue. For each drug-tissue pair, we calculated the number of proteins in each tissue-specific protein network, drug regulatory gene network, as well as their overlap. The statistical significance (P-values) of the gene-set enrichment was calculated by Fisher's exact test and corrected by Benjamini–Hochberg method (Benjamini and Hochberg, 1995). We set corrected P-value (q) < 0.05 as a threshold to define significantly predicted drug–tissue pairs.
2.6. Pharmacovigilance analysis
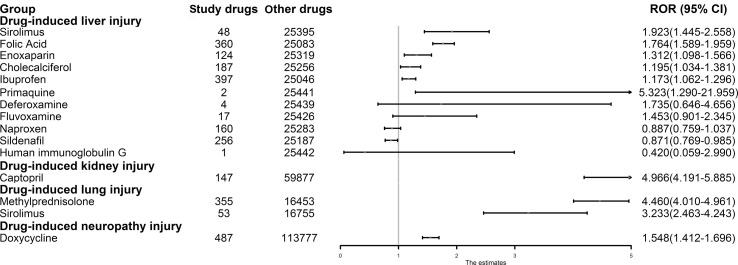
We retrieved adult adverse drug event data from the US Food and Drug Administration's (FDA) Adverse Event Reporting System (AERS) compiled from the first quarter of 2010 to the last quarter of 2019 using OpenVigil 2.1 (Böhm et al., 2012) (http://openvigil.sourceforge.net), which is a platform for data mining and analysis of adverse drug event data. In the AERS, adverse events can be specified at different levels of the medical dictionary for regulatory activities (MedDRA) terminology. We employed 9 preferred terms (PTs) for collecting relevant cases associated with “drug-induced liver injury”, 7 PTs for cases associated with “drug-induced kidney injury”, 4 PTs for cases associated with “drug-induced lung injury”, and 4 PTs for cases associated with “drug-induced neuropathy”. Detailed PTs and their number for each drug-induced organ injury were listed in Table 1 . Moreover, the major advantage of the OpenVigil 2.1 is that the platform could perform Yates' chi-square test and calculate disproportionality (like reporting odds ratio (ROR), proportional reporting ratios (PRRs), and relative reporting ratio (RRR)) from adverse drug event data to determine the relationship between the drug and adverse event.Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7 and figs1.
Table 1.
Preferred terms for collecting relevant cases associated with adverse drug events.
| Drug-induced tissue toxicity | Preferred terms | MEDDRA | Number | Drugs for validation |
|---|---|---|---|---|
| Drug-induced liver injury (Number = 25,443) | Drug-induced liver injury | 10072268 | 6388 | sirolimus, folic acid, enoxaparin, cholecalciferol, ibuprofen, primaquine, deferoxamine, fluvoxamine, naproxen, sildenafil, human immunoglobulin G |
| Hepatotoxicity | 10019851 | 5942 | ||
| Acute hepatic failure | 10000804 | 4312 | ||
| Hepatic failure | 10019663 | 8409 | ||
| Liver disorder | 10024670 | 12,389 | ||
| Hepatic lesion | 10061998 | 1181 | ||
| Liver function test increased | 10077692 | 4074 | ||
| Transaminases increased | 10054889 | 5654 | ||
| Hepatic enzyme increased | 10060795 | 15,087 | ||
| Drug-induced kidney injury (Number = 60,024) | Renal failure | 10038435 | 41,907 | captopril |
| Renal failure acute | 10038436 | 15,616 | ||
| Acute kidney injury | 10069339 | 37,440 | ||
| Blood creatinine increased | 10005483 | 15,069 | ||
| Urinary nitrogen increased | 10046547 | 8 | ||
| Renal injury | 10061481 | 16,385 | ||
| Renal tubular injury | 10078933 | 124 | ||
| Drug-induced lung injury (Number = 16,808) | Acute lung injury | 10069351 | 487 | methylprednisolone, sirolimus |
| Acute respiratory failure | 10001053 | 3930 | ||
| Respiratory failure | 10038695 | 19,653 | ||
| Pulmonary function test decreased | 10061922 | 726 | ||
| Headache | 10019211 | 170,461 | doxycycline | |
| Drug-induced neuropathy | Acute polyneuropathy | 10066699 | 69 | |
| (Number = 114,264) | Neuropathy peripheral | 10029331 | 22,716 | |
| Nervous system disorder | 10029202 | 14,631 |
Note: MEDDRA: medical dictionary for regulatory activities terminology.
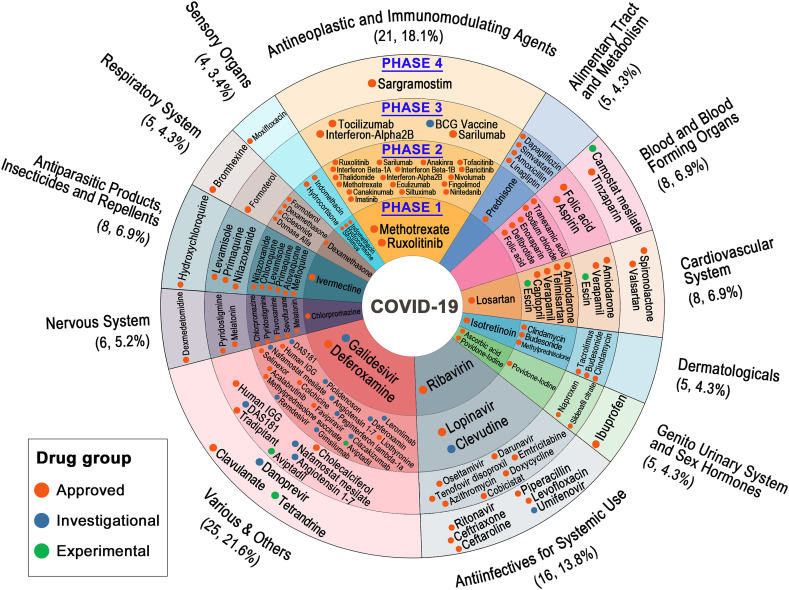
Fig. 2.
Overview of ongoing drugs in COVID-19 clinical trials (as of April 21, 2020). Drugs are classified by the Anatomical Therapeutic Chemical Classification (ATC) code and clinical trial phase. The ATC label, number of drugs and percentage of each drug classification are presented on the outer circle. The four different levels from internal to external represent the corresponding drugs in phase 1, phase 2, phase 3 and phase 4 of COVID-19 clinical trials, respectively. The high-resolution version is provided in the Supporting Information.
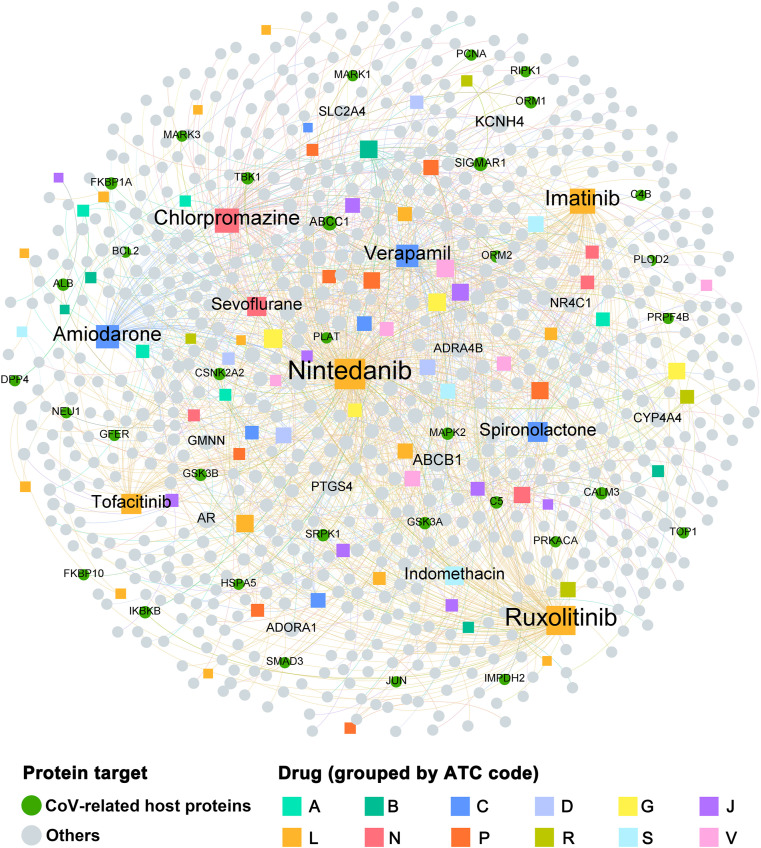
Fig. 3.
Bipartite drug-target network covering 81 drugs with 719 human target proteins. The dot stands for the protein target while the square node represents the drug. The sizes of node and label are proportional to degree. Note: A: alimentary tract and metabolism; B: blood and blood forming organs; C: cardiovascular system; D: dermatologicals; G: genito urinary system and sex hormones; J: antiinfectives for systemic use; L: antineoplastic and immunomodulating agents; N: nervous system; P: antiparasitic products: insecticides and repellents; R: respiratory system; S: sensory organs; V: various.
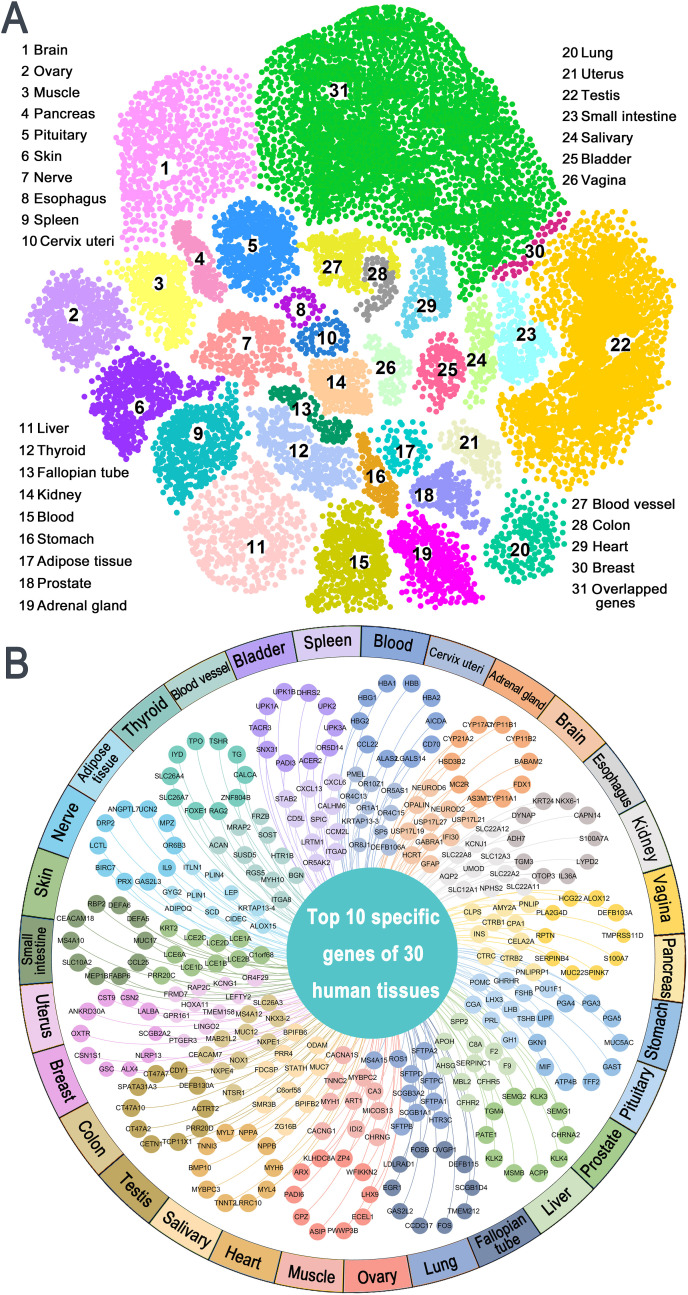
Fig. 4.
Tissue-specific protein network analysis. A High-expressed genes with Z-expression higher than 2 across 30 human tissues. Overlapped genes refer to the genes highly expressed in multiple tissues simultaneously; B Top 10 specific high-expressed genes in each tissue.
Fig. 5.
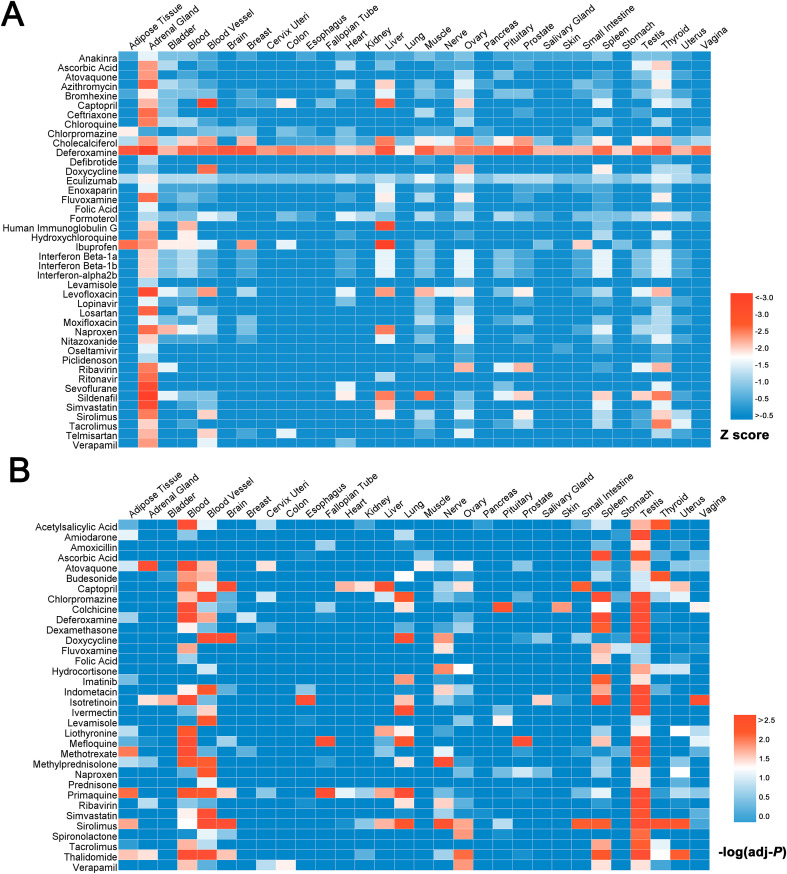
Heatmap presenting the associations between COVID-19 ongoing drugs and 30 human tissues predicted by network proximity approach (A) and GSEA approach (B). Hypothesis for the two methods: A Network proximity, a drug would have a potential specific tissue toxicity if there is a significant network distance between its drug module and tissue module in human protein–protein interactome; B GSEA, a drug would have a correlation with a specific tissue if its regulatory proteins tend to be the specific high expressed proteins in this tissue. The color key indicates the significance (-log(adj-P) value or Z-score) of drug-tissue toxicity associations, red: highest and blue: lowest. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 6.
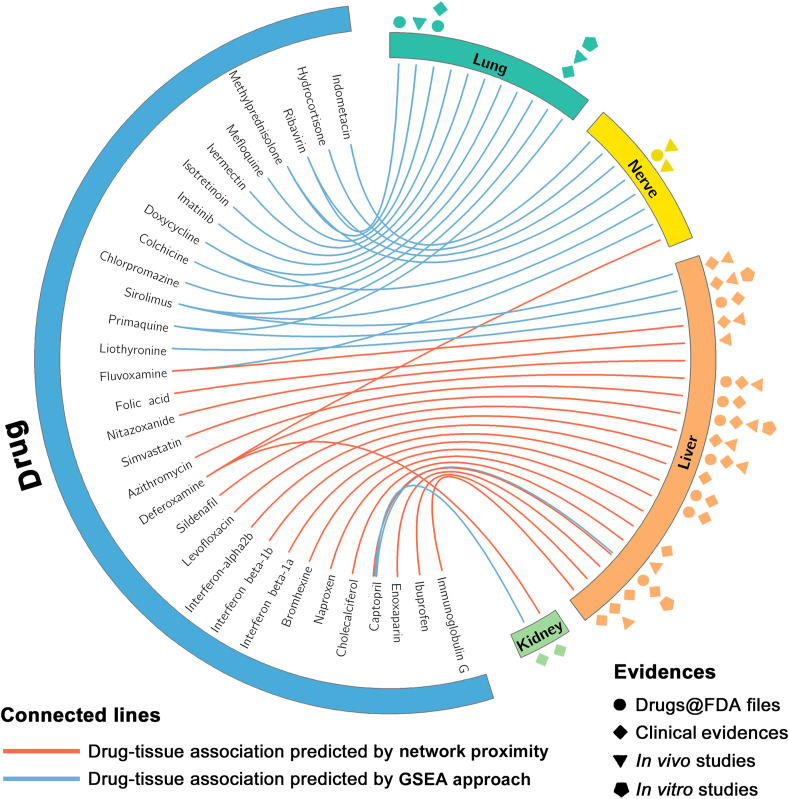
Circos plot exhibiting drug-tissue associations focusing on four specific tissues (liver, lung, nerve and kidney) predicted by network proximity and GSEA approaches.
Fig. 7.
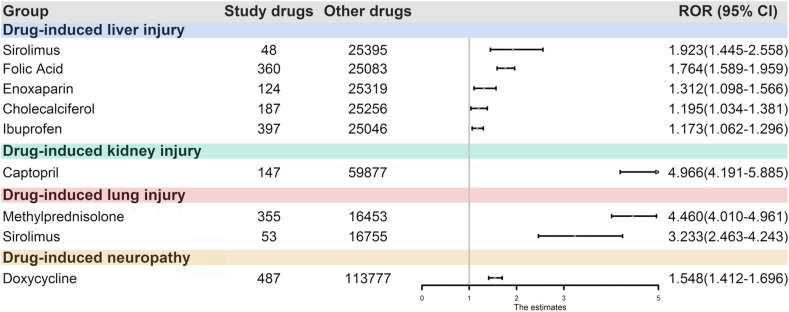
Drug-Forest plot illustrating reporting odds ratios (RORs) and 95% confidence intervals (CIs) for drugs and adverse events of interest.
In this study, number of adverse events of drugs >4, Chi square values > 4, and the lower limit of the 95% confidence interval (CI) of the ROR>1 indicate that the adverse event and the drug is related and can be considered as “likely an adverse reaction”. The RORs and 95% CIs for drugs and adverse events of interest were shown using the forest plots.
2.7. Statistical analysis and network visualization
The statistical analysis in this study was performed by Python (v3.2, http://www.python.org/) and R platforms (v3.01, http://www.r-project.org/). Networks were generated by Cytoscape (v3.2.1, http://www.cytoscape.org/) and Gephi (v0.9.2, https://gephi.org/). In the graphical network, drugs or genes were presented by nodes and interactions were encoded by edges. The degree of each node was calculated to evaluate its topological property, as this characterizes the most important nodes in a network.
3. Results
3.1. Overview of ongoing drugs in COVID-19 pipeline
As of April 21, 2020, there were 116 agents in 308 clinical trials of anti- SARS-CoV-2 therapies. Here, a comprehensive overview of available information on potential drugs for treatment of COVID-19 was presented (Fig. 2). Most of ongoing drugs in COVID-19 clinical trials are approved drugs since existing drug could shorten the time and reduce the cost in comparison with developing novel compounds from scratch (Kupferschmidt and Cohen, 2020). In addition, there are still 16 investigational agents (e.g. remdesivir) and 4 experimental agents (e.g. tetrandrine) enrolled into the clinical trials for their potential therapeutical effects on COVID-19. As shown in Fig. 2, there are 21 antineoplastic and immunomodulating agents in the clinical trials for COVID-19 treatment, occupying the highest proportion (18.1%) among all the ongoing drug classes except “various & others”. These agents are mainly designed to act on the coronavirus directly (such as inhibiting crucial viral enzyme and blocking viral entry to human cells) or modulate the human immune system (such as boosting the innate response and inhibiting the inflammatory processes) (Wiersinga et al., 2020). As recent study demonstrated that chemotherapy could produce selective toxicity to SARS-CoV-2 via improving the levels of endogenous reactive metabolites (Maryam Al-Motawa, 2020b), scientists have turned their attention to antineoplastic agents. Besides, coronavirus was able to decrease immune capacity since it primarily exerts cytotoxic action on the hematopoietic and immune systems with reducing the number of neutrophils (Remuzzi and Remuzzi, 2020). Thus, immunomodulating agents were also brought into the clinical trials in view of its positive regulation on immunity of the organism. Furthermore, Fig. 2 shows that 16 drugs intend to be classified as antiinfectives for systemic use. Since SARS-CoV-2 is a single-stranded RNA-enveloped virus that can be infected from person to person, drugs for antiinfectives for systemic use such as azithromycin (Parra-Lara et al., 2020) are also considered to own the potential of suppressing the virus.
Among the 116 ongoing agents in pipeline, 76% of them (88 agents) are being tested in phase 2 and phase 3 trials with uncertain effects rather than confirmed therapeutic effects on COVID-19. Although a few agents in COVID-19 trials (Fig. 2) seem to show promising results based on current researches and clinical trials, they may play a limited role in COVID-19 treatment or even remain controversial (Ko et al., 2020). Meanwhile, it also needs to be alerted for the potential toxicity of these drugs. For example, case reports study in France has demonstrated that four patients of COVID-19 developed adverse events including elevations of alanine transaminase (ALT) levels and renal failures after treatment with remdesivir (Dubert et al., 2020).
3.2. Drug-target network analysis
We constructed a comprehensive drug-target network of ongoing drugs in COVID-19 pipeline through integrating curated physical DTIs and CoV host proteins. As depicted in Fig. 3, the drug-target network contains 1222 DTIs connecting 81 drugs with 719 human target proteins. Among these human target proteins, 34 of them were CoV host proteins. The CoV host protein with the highest degree is ABCC1(K=5), which are targeted by 5 drugs, including indomethacin, methotrexate, imatinib, verapamil and colchicine. Besides, the average target degree (D) of ongoing drugs is 8.88. Among the 81 drugs, the top 10 with largest target degree are: nintedanib (D = 215), ruxolitinib (D = 137), imatinib (D = 74), chlorpromazine (D = 62), amiodarone (D = 54), verapamil (D = 46), tofacitinib (D = 32), spironolactone (D = 31), sevoflurane (D = 27), and indomethacin (D = 25). Recent evidences have demonstrated their potential correlation with COVID-19 treatment. For instance, Goker et al. has revealed that ruxolitinib shows the promising efficacy on COVID-19 by inhibiting JAK/STAT pathway (Goker Bagca and Biray Avci, 2020). Imatinib might play its potentially beneficial immunomodulatory role in patients with COVID-19 by multiple mechanisms, such as reducing the transcription factor NF-κB signaling pathway and modulating tumor necrosis factor-α (TNF-α) and IL-6 production (Bernal-Bello et al., 2020). However, according to the network analysis, the COVID-19 ongoing drugs could act on multiple targets, including both CoV and non-CoV host proteins. As most of the non-CoV host proteins targeted by COVID-19 drug candidates are likely to be highly expressed in different tissues of human, they may bring the potential to cause adverse side effects.
3.3. Tissue-specific protein network analysis
In this study, the expression level of genes across 30 human tissues is quantified by Z-expression, including colon, esophagus, fallopian tube, heart, kidney, liver, lung, muscle, nerve, ovary, adipose tissue, pancreas, pituitary, prostate, salivary, skin, small intestine, spleen, stomach, testis, thyroid, adrenal gland, uterus, vagina, bladder, blood vessel, blood, brain, breast, and cervix uteri. Genes in a tissue-protein network with Z-expression greater than 2 are considered as high expression. As shown in Fig. 4A, testis tissue possesses the largest number of high expressed genes (n = 3521), followed by brain (n = 733) and liver (n = 465).
We further prioritized the high-expressed genes in each tissue by Z-expression and highlighted the top 10 specific genes of the 30 human tissues (Fig. 4B). Interestingly, we find that among these genes, there are no overlap between any two tissues, implying they tend to have a high multiplicity and tissue specificity. These high-expressed genes seem to possess high association with the occurrence of drug-induced tissue toxicity. For example, the top 10 specific genes of kidney are UMOD, SLC22A8, SLC22A12, SLC12A1, SLC22A11, AQP2, SLC12A3, NPHS2, KCNJ1, and SLC22A2. As previous studies reported, uromodulin (UMOD) is a kidney-specific protein (Tachibana et al., 2019) expressed by epithelial cells lining the thick ascending limb of the loop of Henle, the accumulation of mutated UMOD in endoplasmic reticulum can lead to renal injury (Johnson et al., 2017; Trudu et al., 2017). In addtion, recent literature also demonstrated that mutation of aquaporin 2 (AQP2) is able to cause nephrogenic diabetes insipidus since AQP2 is responsible for the water reabsorption by kidney collecting duct cells (Saglar Ozer et al., 2020).
3.4. Uncovering significant associations between COVID-19 ongoing drugs and human tissues
To identify the potential tissue toxicities of COVID-19 ongoing drugs, we applied two complementary approaches to compute the significance of the association between each tissue and drug, including: (i) network proximity approach which quantifies the network distance between each drug module and tissue module in human protein–protein interactome; and (ii) GSEA approach that calculates the statistical significance of the gene-set enrichment between each tissue-specific protein network and drug regulatory gene network. We herein assumed that a drug may induce potential toxicity on specific tissue if they have significant associations (P network proximity or q GESA < 0.05). As illustrated in Fig. 5A, there are in total 42 drugs significantly associated with multiple tissues via network proximity approach (P network proximity < 0.05), covering 140 significant drug-tissue pairs. It suggests that those drugs may cause corresponding predicted tissue toxicities. For example, deferoxamine, which targets kidney organ and exhibits great tissue specificity (Z-score = −2.01), has been demonstrated the potential risk of inducing acute renal failure (Prasannan et al., 2003). Moreover, deferoxamine also has significant association with liver tissue (Z-score = −1.97). In vitro studies have suggested that deferoxamine was toxic to hepatoma cells in the absence of iron (Christensen et al., 2001). Meanwhile, GSEA approach has predicted that 34 potential candidates (q GESA < 0.05, Fig. 5B) have significant associations with different human tissues, including 132 significant drug-tissue pairs. Taking sirolimus (also known as rapamycin) as an example, GSEA gave a q GESA value of 0.003 with lung tissue, indicating its potential lung toxicity. An in vivo study has revealed that sirolimus could augment lipopolysaccharide-induced lung injury and apoptosis in C57BL/6J mice through involving signal transducer and activator of transcription 1 (STAT1) and the induction of STAT1-dependent apoptosis genes (Fielhaber et al., 2012). Notably, we found that 9 drug-tissue pairs were simultaneously identified by network proximity and GSEA approaches, alerting their potential tissue toxicities, including captopril-liver, sirolimus-blood vessel, atovaquone-adrenal gland, deferoxamine-blood, doxycycline-blood vessel, deferoxamine-testis, deferoxamine-spleen, deferoxamine-blood vessel, and captopril-ovary. The detailed information of the network-based prediction results are provided in Table S5.
To present specifically how drug candidate manifests itself and targets tissue toxicity within the drug-target networks and gene expression profiles, we further focus on adverse events on four tissues including lung, nerve, liver and kidney. As exhibited in Fig. 6, there are 42 drug-tissue pairs covering 32 agents predicted by network proximity or GSEA approaches. Among them, 64.3% (27/42) of the predicted tissue toxicities were validated by drug labels retrieved from Drug@FDA database, clinical evidences including clinical studies and case reports, as well as preclinical evidence available (Table S6). Moreover, 8 out of the 32 drugs are predicted to yield potential toxicities in at least 2 tissues. They are sirolimus, captopril, deferoxamine, fluvoxamine, ribavirin, primaquine, methylprednisolone and doxycycline. Despite drug-tissue associations which had been validated by Drug@FDA files, clinical studies, in vivo or in vitro studies, it is worth noting that those drug-tissue associations without validations (15/42) deserve further exploration.
Taken together, these results mentioned above initially support the effectiveness of our approaches including network proximity and GSEA for identification of drug-tissue associations that are potentially relevant for side effects.
3.5. Validating the predicted drug-tissue associations via pharmacovigilance analysis
Given the significance of clinical data, we further assessed whether these drug-tissue associations mentioned above were of statistical difference regarding the side effects of drug in specific tissue via pharmacovigilance analysis. Among 42 predicted significant drug-tissue pairs above, we selected drug-tissue pairs for further validation using subject matter expertise based on a combination of factors: (i) not being labeled by FDA drug instructions; (ii) strength of the network-based prediction; and (iv) availability of supportive experimental evidence. Applying these criteria resulted in identifying 15 potential drug-tissue toxicities across four tissues for further validation (Table 1).
Specifically, we first retrieved a total of 3,004,759 adverse events from the AERS. We identified 25,443, 60,024, 16,808, and 114,264 cases who developed drug-induced liver injury, drug-induced kidney injury, drug-induced lung injury, and drug-induced neuropathy while receiving any drugs, respectively. The statistical significant drug-induced toxicity across four tissues together with their RORs and 95% CIs were shown in Fig. 7 and the detailed information on all drug-tissue associations can be found in Figure S1. According to the criteria, drug-induced liver injury was significantly related to ibuprofen, enoxaparin, cholecalciferol, folic acid, and sirolimus. Indeed, recent literatures have reported that these drugs do cause liver injury, such as sirolimus (Neff et al., 2004; Umemura et al., 2014). Surprisingly, we found some drug-tissue associations which had not been labeled by FDA drug instructions were of statistical difference, such as the potential hepatotoxicity of enoxaparin (Hahn et al., 2015; Pivarnik et al., 2016). According to the pharmacovigilance analysis of adverse drug event data, enoxaparin seems to possess significant association with hepatotoxicity (ROR = 1.312, 95% CI:1.098–1.566). Moreover, we also found that captopril (ROR = 4.966, 95% CI: 4.191–5.885) was potentially related to kidney injury. Besides, methylprednisolone and sirolimus were significantly related to drug-induced lung injury (ROR = 4.46, 95% CI: 4.01–4.961; ROR = 3.233, 95% CI: 2.463–4.243), while doxycycline may lead to neuropathy (ROR = 1.548, 95% CI: 1.412–1.696). Overall, 60% (9/15) of our network-based predictions are confirmed by pharmacovigilance analysis, indicating the accuracy of our network-based approaches and implicating the potential tissue toxicities of these COVID-19 ongoing drugs.
4. Discussion and conclusion
The current 2019-nCoV outbreak is still moving fast, and a rebound of victims of COVID-19 in some areas become a more serious concern (Zhu et al., 2020). Although numerous drugs are under therapeutic evaluation in clinical trials toward SARS-CoV-2, several medications such as hydroxychloroquine and azithromycin have been reported to exacerbate cardiotoxicity and liver injury in COVID-19 patients (Boeckmans et al., 2020). Additionally, the pharmacological and toxicity properties of some investigational or experimental drugs in clinical trials have not been fully explored, meaning that there is greater safety risk within these drugs. Therefore, it is necessary to comprehensively assess side effects of ongoing COVID-19 drugs, which could benefit for the discovery and clinical application of pharmacotherapy for COVID-19.
In this study, we utilized network-based approaches for rapid identification of potential side effects for ongoing COVID-19 drugs. Specifically, after integration of drug-target network, drug-gene signatures, tissue-specific gene expression profiles from GTEx, and human protein-protein interactome, we established the associations between 116 COVID-19 drugs and 30 kinds of human tissues based on network proximity and GSEA approaches. Our network proximity and GSEA approaches achieved a high accuracy to identify the associations between COVID-19 drugs and human tissues. For example, ibuprofen was deemed to own potential toxicity on liver based on our network-based prediction (Fig. 6). Emerging evidence shows that ibuprofen may elicit hepatotoxicity via altering several major pathways including energy metabolism, protein degradation, fatty acid metabolism as well as antioxidant system in mice (Tiwari et al., 2020). Additionally, an in vivo study (Wen et al., 2018) also revealed that ibuprofen treatment on rats increased the levels of alanine transaminase (ALT) and aspartate aminotransferase (AST), causing more serious liver injury than did aspirin. Moreover, we found several drugs could induce potential toxicities in multiple tissues. Taking captopril as an example, network-based prediction indicates that captopril have correlations with both liver and kidney tissues, which are consistent with previous clinical reports (LiverTox, 2012; Tan et al., 2011).
In addition, we further validated the predicted drug-tissue association via data mining and analysis of adverse drug event data. Evidences from Drug@FDA files, clinical studies, in vivo and in vitro studies have primarily validated the drug-tissue associations predicted by our in silico approaches. For instance, fluvoxamine is predicted to highly associate with liver tissue. It is reported that fluvoxamine therapy is associated with both transient asymptomatic elevations in serum aminotransferase levels and rare instances of clinically apparent acute liver injury (LiverTox, 2018). Besides, network-based prediction indicates that captopril have correlations with both liver and kidney tissues. Clinical case reports of hepatic injury (LiverTox, 2012) and acute renal failure (Tan et al., 2011) induced by captopril to some extent verified our predictions. Interestingly, we found that captopril-liver pair was simultaneously identified by network proximity and GSEA methods, further indicating its high probability of liver injury. In fact, Drug@FDA files on captopril have clearly labled its potential hepatoxicity. Moreover, we found several potential tissue toxicities of drugs which hadn't been labeled by FDA drug instructions. For example, several clinical case reports have indicated the potential hepatotoxicity of enoxaparin, though there is no clear annotation of liver injury in FDA drug instructions yet. It is reported that patients with deep vein thrombosis (DVT) or dural venous thrombosis can develop drug-induced liver injury during treatment with enoxaparin (Hahn et al., 2015; Pivarnik et al., 2016). Pharmacovigilance analysis conducted on AERS data further support these findings. Overall, our findings offered a comprehensive assessment of drug safety profiles for current COVID-19 medications, which should be further validated in animal studies and large scale patient data.
However, several limitations should be acknowledged in present study. First, both network proximity and GSEA approaches were applied to predict tissue toxicity of drugs by integrating physical drug-target interactions and transcriptome data on CMap. Given lacking of large scale published available experimental data, whether combining the two methods (network proximity and GSEA) synergistically enhance the prediction performance remain further evaluation. Moreover, this study mainly focuses on the prediction and validation of drug-tissue associations, the specific mechanism of actions (MOAs) related with tissue toxicity need to be explored in future. In addition, adverse drug events obtained through the adverse reaction reporting system should be treated with caution, since there are inherent limitations, such as selective reporting, under-reporting, and lack of information about total drug consumption. The results of disproportionality analyses using AERS data should not be considered as the quantification of the true risk, but rather as the assessment of signal detection. Finally, as current COVID-19 medications are being renewed very quickly, we can not cover all the ongoing drugs in COVID-19 pipeline.
In summary, this study provides a powerful network-based methodology for efficient identification of drug-induced potential side effects in COVID-19 drug pipeline, followed by pharmacovigilance analysis that utilized large scale clinical adverse drug event data. If broadly applied, our computational framework can be used to assess potential tissue toxicity for all drugs, such as cancer or cardiovascular drugs. In this manner, we can build bridges between basic medical research and clinical outcomes, which could shorten the time to prioritize side effects associated with drugs and alert the potential adverse effects of drugs during pharmacotherapy course of COVID-19.
CRediT authorship contribution statement
Qihui Wu: Methodology, Validation, Writing - original draft. Xiude Fan: Investigation, Visualization. Honghai Hong: Writing - review & editing. Yong Gu: Software. Zhihong Liu: Formal analysis. Shuhuan Fang: Resources. Qi Wang: Data curation. Chuipu Cai: Conceptualization, Writing - review & editing. Jiansong Fang: Conceptualization, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 81903912 and 81902693), the Youth Scientific Research Training Project of GZUCM (Grant No. 2019QNPY05), Provincial Key R&D Program of Hainan (Grant No. ZDYF2019196), China Postdoctoral Science Foundation funded project (Grant No. 2019M662878), Guangdong province science and technology plan international cooperation project (No.2020A0505100052), and the Natural Science Foundation of Guangdong Province (No. 2018A030310298).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fct.2020.111767.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
References
- Böhm R., Höcker J., Cascorbi I., Herdegen T. OpenVigil--free eyeballs on AERS pharmacovigilance data. Nat. Biotechnol. 2012;30:137–138. doi: 10.1038/nbt.2113. [DOI] [PubMed] [Google Scholar]
- Barbarino J.M., Whirl-Carrillo M., Altman R.B., Klein T.E. PharmGKB: a worldwide resource for pharmacogenomic information. Wiley Interdiscip Rev Syst Biol Med. 2018;10:e1417. doi: 10.1002/wsbm.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series. 1995;57:289–300. [Google Scholar]
- Bernal-Bello D., Jaenes-Barrios B., Morales-Ortega A., Ruiz-Giardin J.M., García-Bermúdez V., Frutos-Pérez B., Farfán-Sedano A.I., de Ancos-Aracil C., Bermejo F., García-Gil M., Zapatero-Gaviria A., San Martín-López J.V. Imatinib might constitute a treatment option for lung involvement in COVID-19. Autoimmun. Rev. 2020;19 doi: 10.1016/j.autrev.2020.102565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckmans J., Rodrigues R.M., Demuyser T., Piérard D., Vanhaecke T., Rogiers V. COVID-19 and drug-induced liver injury: a problem of plenty or a petty point? Arch. Toxicol. 2020;94:1367–1369. doi: 10.1007/s00204-020-02734-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C., Fang J., Guo P., Wang Q., Hong H., Moslehi J., Cheng F. In silico pharmacoepidemiologic evaluation of drug-induced cardiovascular complications using combined classifiers. J. Chem. Inf. Model. 2018;58:943–956. doi: 10.1021/acs.jcim.7b00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C., Guo P., Zhou Y., Zhou J., Wang Q., Zhang F., Fang J., Cheng F. Deep learning-based prediction of drug-induced cardiotoxicity. J. Chem. Inf. Model. 2019;59:1073–1084. doi: 10.1021/acs.jcim.8b00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Desai R.J., Handy D.E., Wang R., Schneeweiss S., Barabási A., Loscalzo J. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat. Commun. 2018;9:2691. doi: 10.1038/s41467-018-05116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Lu W., Liu C., Fang J., Hou Y., Handy D.E., Wang R., Zhao Y., Yang Y., Huang J., Hill D.E., Vidal M., Eng C., Loscalzo J. A genome-wide positioning systems network algorithm for in silico drug repurposing. Nat. Commun. 2019;10:3476. doi: 10.1038/s41467-019-10744-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen D.W., Kisling R., Thompson J., Kirby M.A. Deferoxamine toxicity in hepatoma and primary rat cortical brain cultures. Hum. Exp. Toxicol. 2001;20:365–372. doi: 10.1191/096032701680350532. [DOI] [PubMed] [Google Scholar]
- Del Rio C., Malani P.N. 2019 novel coronavirus-important information for clinicians. J. Am. Med. Assoc. 2020;323:1039–1040. doi: 10.1001/jama.2020.1490. [DOI] [PubMed] [Google Scholar]
- Dubert M., Visseaux B., Isernia V., Bouadma L., Deconinck L., Patrier J., Wicky P.-H., Le Pluart D., Kramer L., Rioux C., Le Hingrat Q., Houhou-Fidouh N., Yazdanpanah Y., Ghosn J., Lescure F.-X. Case reports study of the first five patients COVID-19 treated with remdesivir in France. Int. J. Infect. Dis. 2020;98:290–293. doi: 10.1016/j.ijid.2020.06.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J., Cai C., Chai Y., Zhou J., Huang Y., Gao L., Wang Q., Cheng F. Quantitative and systems pharmacology 4. Network-based analysis of drug pleiotropy on coronary artery disease. Eur. J. Med. Chem. 2019;161:192–204. doi: 10.1016/j.ejmech.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J., Pieper A.A., Nussinov R., Lee G., Bekris L., Leverenz J.B., Cummings J., Cheng F. Harnessing endophenotypes and network medicine for Alzheimer’s drug repurposing. Med. Res. Rev. 2020 doi: 10.1002/med.21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielhaber J.A., Carroll S.F., Dydensborg A.B., Shourian M., Triantafillopoulos A., Harel S., Hussain S.N., Bouchard M., Qureshi S.T., Kristof A.S. Inhibition of mammalian target of rapamycin augments lipopolysaccharide-induced lung injury and apoptosis. J. Immunol. 2012;188:4535–4542. doi: 10.4049/jimmunol.1003655. [DOI] [PubMed] [Google Scholar]
- Gaulton A., Hersey A., Nowotka M., Bento A.P., Chambers J., Mendez D., Mutowo P., Atkinson F., Bellis L.J., Cibriánuhalte E. The ChEMBL database in 2017. Nucleic Acids Res. 2017;45:D945–D954. doi: 10.1093/nar/gkw1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson M.K., Liu T., Baitaluk M., Nicola G., Hwang L., Chong J. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016;44:D1045–D1053. doi: 10.1093/nar/gkv1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goker Bagca B., Biray Avci C. The potential of JAK/STAT pathway inhibition by ruxolitinib in the treatment of COVID-19. Cytokine Growth Factor Rev. 2020;54:51–62. doi: 10.1016/j.cytogfr.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.-P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.-Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn K.J., Morales S.J., Lewis J.H. Enoxaparin-induced liver injury: case report and review of the literature and FDA adverse event reporting system (FAERS) Drug Saf Case Rep. 2015;2:17. doi: 10.1007/s40800-015-0018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding S.D., Sharman J.L., Faccenda E., Southan C., Pawson A.J., Ireland S., Gray A.J.G., Bruce L., Alexander S.P.H., Anderton S., Bryant C., Davenport A.P., Doerig C., Fabbro D., Levi-Schaffer F., Spedding M., Davies J.A. The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Res. 2018;46:D1091–D1106. doi: 10.1093/nar/gkx1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javorac D., Grahovac L., Manić L., Stojilković N., Anđelković M., Bulat Z., Đukić-Ćosić D., Curcic M., Djordjevic A.B. An overview of safety assessment of the medicines currently used in the treatment of COVID-19 disease. Food Chem. Toxicol. 2020 doi: 10.1016/j.fct.2020.111639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B.G., Dang L.T., Marsh G., Roach A.M., Levine Z.G., Monti A., Reyon D., Feigenbaum L., Duffield J.S. Uromodulin p.Cys147Trp mutation drives kidney disease by activating ER stress and apoptosis. J. Clin. Invest. 2017;127:3954–3969. doi: 10.1172/JCI93817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko W.-C., Rolain J.-M., Lee N.-Y., Chen P.-L., Huang C.-T., Lee P.-I., Hsueh P.-R. Arguments in favour of remdesivir for treating SARS-CoV-2 infections. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupferschmidt K., Cohen J. Race to find COVID-19 treatments accelerates. Science. 2020;367:1412–1413. doi: 10.1126/science.367.6485.1412. [DOI] [PubMed] [Google Scholar]
- Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J., Lerner J., Brunet J.-P., Subramanian A., Ross K.N., Reich M., Hieronymus H., Wei G., Armstrong S.A., Haggarty S.J., Clemons P.A., Wei R., Carr S.A., Lander E.S., Golub T.R. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Ledford H. Coronavirus breakthrough: dexamethasone is first drug shown to save lives. Nature. 2020;582:469. doi: 10.1038/d41586-020-01824-5. [DOI] [PubMed] [Google Scholar]
- LiverTox . 2012. Captopril. LiverTox: Clinical and Research Information on Drug-Induced Liver Injury. [PubMed] [Google Scholar]
- LiverTox . LiverTox: Clinical and Research Information on Drug-Induced Liver Injury. 2018. Fluvoxamine. [Google Scholar]
- Maryam Al-Motawa H.A., Wijten Patrick, Alberto de la Fuente, Xue Mingzhan, Rabbani Naila, Thornalley Paul J. 2020. Virus-host Interactome and Proteomic Survey of PBMCs from COVID-19 Patients Reveal Potential Virulence Factors Influencing SARS-CoV-2 Pathogenesis. Med (N Y) [Google Scholar]
- Maryam Al-Motawa H.A., Wijten Patrick, Alberto de la Fuente, Xue Mingzhan, Rabbani Naila, Thornalley Paul J. 2020. Vulnerabilities of the SARS-CoV-2 Virus to Proteotoxicity – Opportunity for Repurposed Chemotherapy of COVID-19 Infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra M.R., Desai S.S., Ruschitzka F., Patel A.N. RETRACTED: hydroxychloroquine or chloroquine with or without a macrolide for treatment of COVID-19: a multinational registry analysis. Lancet. 2020;S0140-6736:31180–31186. doi: 10.1016/S0140-6736(20)31180-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Menche J., Sharma A., Kitsak M., Ghiassian S.D., Vidal M., Loscalzo J., Barabási A.-L. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347 doi: 10.1126/science.1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz M., Lee K.R., Kullak-Ublick G.A., Brueckner A., Watkins P.B. Methodology to assess clinical liver safety data. Drug Saf. 2014;37:33–45. doi: 10.1007/s40264-014-0184-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff G.W., Ruiz P., Madariaga J.R., Nishida S., Montalbano M., Meyer D., Levi D.M., Tzakis A.G., O'Brien C.B. Sirolimus-associated hepatotoxicity in liver transplantation. Ann. Pharmacother. 2004;38:1593–1596. doi: 10.1345/aph.1E165. [DOI] [PubMed] [Google Scholar]
- Parra-Lara L.G., Martínez-Arboleda J.J., Rosso F. Azithromycin and SARS-CoV-2 infection: where we are now and where we are going. J Glob Antimicrob Resist. 2020;22:680–684. doi: 10.1016/j.jgar.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pivarnik K.A., Schiffman F., Sullivan J., Finn A. Enoxaparin-induced hepatotoxicity: an under-recognised complication of enoxaparin therapy. BMJ Case Rep. 2016;2016 doi: 10.1136/bcr-2016-216592. bcr2016216592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasannan L., Flynn J.T., Levine J.E. Acute renal failure following deferoxamine overdose. Pediatr. Nephrol. 2003;18:283–285. doi: 10.1007/s00467-002-1051-7. [DOI] [PubMed] [Google Scholar]
- project G. The genotype-tissue expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remuzzi A., Remuzzi G. COVID-19 and Italy: what next? Lancet. 2020;395:1225–1228. doi: 10.1016/S0140-6736(20)30627-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saglar Ozer E., Moeller H.B., Karaduman T., Fenton R.A., Mergen H. Molecular characterization of an aquaporin-2 mutation causing a severe form of nephrogenic diabetes insipidus. Cell. Mol. Life Sci. 2020;77:953–962. doi: 10.1007/s00018-019-03219-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., Quan S., Zhang F., Sun R., Qian L., Ge W., Liu W., Liang S., Chen H., Zhang Y., Li J., Xu J., He Z., Chen B., Wang J., Yan H., Zheng Y., Wang D., Zhu J., Kong Z., Kang Z., Liang X., Ding X., Ruan G., Xiang N., Cai X., Gao H., Li L., Li S., Xiao Q., Lu T., Zhu Y., Liu H., Chen H., Guo T. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182 doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana S., Iyoda M., Suzuki T., Kanazawa N., Iseri K., Wada Y., Matsumoto K., Shibata T. Serum uromodulin is associated with the severity of clinicopathological findings in ANCA-associated glomerulonephritis. PloS One. 2019;14 doi: 10.1371/journal.pone.0224690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L.-H., Du L.-Z., Carr M.R., Kuzin J.K., Moffett B.S., Chang A.C. Captopril induced reversible acute renal failure in a premature neonate with double outlet right ventricle and congestive heart failure. World J Pediatr. 2011;7:89–91. doi: 10.1007/s12519-011-0252-1. [DOI] [PubMed] [Google Scholar]
- Tiwari S., Mishra M., Salemi M.R., Phinney B.S., Newens J.L., Gomes A.V. Gender-specific changes in energy metabolism and protein degradation as major pathways affected in livers of mice treated with ibuprofen. Sci. Rep. 2020;10:3386. doi: 10.1038/s41598-020-60053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudu M., Schaeffer C., Riba M., Ikehata M., Brambilla P., Messa P., Martinelli-Boneschi F., Rastaldi M.P., Rampoldi L. Early involvement of cellular stress and inflammatory signals in the pathogenesis of tubulointerstitial kidney disease due to UMOD mutations. Sci. Rep. 2017;7:7383. doi: 10.1038/s41598-017-07804-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura A., Park E.J., Taniguchi K., Lee J.H., Shalapour S., Valasek M.A., Aghajan M., Nakagawa H., Seki E., Hall M.N., Karin M. Liver damage, inflammation, and enhanced tumorigenesis after persistent mTORC1 inhibition. Cell Metabol. 2014;20:133–144. doi: 10.1016/j.cmet.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C., Zhuang Z., Song H., Tong S., Wang X., Lin Y., Zhan H., Chen Z., Hu L. Metabolism of liver CYP450 and ultrastructural changes after long-term administration of aspirin and ibuprofen. Biomed. Pharmacother. 2018;108:208–215. doi: 10.1016/j.biopha.2018.08.162. [DOI] [PubMed] [Google Scholar]
- Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. J. Am. Med. Assoc. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., Assempour N., Iynkkaran I., Liu Y., Maciejewski A., Gale N., Wilson A., Chin L., Cummings R., Le D., Pon A., Knox C., Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Cai C., Guo P., Chen M., Wu X., Zhou J., Luo Y., Zou Y., Liu A.L., Wang Q., Kuang Z., Fang J. Identification and mechanism exploration of hepatotoxic ingredients in traditional Chinese medicine. Front. Pharmacol. 2019;10:458. doi: 10.3389/fphar.2019.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Ye L., Yin S., Lu X., Liu X., Lu S., Cui J., Fan L., Kaplowitz N., Hu H. Glycycoumarin protects mice against acetaminophen-induced liver injury predominantly via activating sustained autophagy. Br. J. Pharmacol. 2018;175:3747–3757. doi: 10.1111/bph.14444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K.S., He Sixuan, Li Enyu, Sun Peter, Zuo Lin, Hu Jiayue, Mo Yiwen, Zhang Weiwei, Chen Pingying, Zhang Haonan, Chen Jingxue, Guo Yu. 2020. CovidNet: to Bring the Data Transparency in Era of COVID-19. [Google Scholar]
- Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.