Abstract
The novel coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a major pandemic outbreak recently. Various diagnostic technologies have been under active development. The novel coronavirus disease (COVID-19) may induce pulmonary failures, and chest X-ray imaging becomes one of the major confirmed diagnostic technologies. The very limited number of publicly available samples has rendered the training of the deep neural networks unstable and inaccurate. This study proposed a two-step transfer learning pipeline and a deep residual network framework COVID19XrayNet for the COVID-19 detection problem based on chest X-ray images. COVID19XrayNet firstly tunes the transferred model on a large dataset of chest X-ray images, which is further tuned using a small dataset of annotated chest X-ray images. The final model achieved 0.9108 accuracy. The experimental data also suggested that the model may be improved with more training samples being released.
Graphic abstract
COVID19XrayNet, a two-step transfer learning framework designed for biomedical images.
Keywords: Two-step transfer learning, COVID19XrayNet, ResNet34, Feature smoothing layer (FSL), Feature extraction layer (FEL)
Introduction
The recent outbreak of the novel coronavirus disease (COVID-19) started in Wuhan at the end of the last year, and now COVID-19 has been spreading across the world [1]. The disease was caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which was a single-stranded RNA virus. SARS-CoV-2 may have been transmitted from animals such as bats to humans, and the respiratory droplet nuclei were believed to facilitate the inter-human transmissions [1, 2]. Various clinical symptoms were observed in COVID-19 patients, including mild cough and acute respiratory failure, etc. COVID-19 patients with mild symptoms were estimated to have a low mortality rate, but the exact number is difficult to summarize because they were usually not tested [3]. However, those with respiratory failures had to take mechanical ventilation treatment in hospital and their mortality rate could reach as high as 81% [4]. Because COVID-19 is a very recent outbreak epidemic event, both diagnosis technologies and treatment options are still under active development [1, 5].
Various diagnostic technologies of COVID-19 are under active development and clinical evaluation. There are roughly three groups of diagnostic technologies, i.e., nasopharyngeal and oropharyngeal swab (NOS) test, antibody-based blood (ABB) test and imaging test. The samples collected from the NOS test are screened for the existence of the genomic regions of SARS-CoV-2 using molecular biology technologies such as reverse-transcription polymerase chain reaction (RT-PCR) or real-time RT-PCR (rRT-PCR) [6]. Antibody-based test detects the IgM antibodies responding to SARS-CoV-2 in the blood within 15 min and demonstrates a very high detection accuracy even in the patient’s blood just a few days after the initial infection [7]. Chest computerized tomography (CT) serves as another informative diagnostic technology of COVID-19 patients even when the NOS tests are negative [8]. Various feature extraction algorithms were proposed to generate quantitative features from these medical images, e.g., local binary patterns (LBP) [9], histogram of gradients (HOG) [10], and rotation-invariant TriZ [11]. Medical image-based diagnosis models may be established by the classification algorithms using the image features generated from the above-mentioned feature extraction algorithms [12].
Pneumonia detection based on deep learning and X-ray images of lungs is a challenging problem due to the limited number of publicly available annotated images. At present, this study has only collected 189 COVID19-related X-ray images and 36 control images. This is insufficient for training a stable and well-performing neural network. At the same time, only a single chest X-ray series was obtained for each patient, meaning it was impossible to know the health and disease status of the same sample, which is unfair for model training.
This study investigated the problem of discriminating COVID-19 patients from the healthy and other pneumonia samples using a limited number of publicly available chest X-ray images of COVID-19 patients. A two-step transfer learning model (COVID19XrayNet) was proposed to provide a candidate solution for training an accurate neural network model using the existing small dataset of COVID-19 X-ray images. Firstly, a pre-trained deep residual network (DRN) model ResNet34 was fine-tuned on a large dataset of pneumonia chest X-ray images. Then the fine-tuned model was transferred to detect the COVID-19 chest X-ray images. The final model exhibited a satisfying detection performance of COVID-19 patients against both healthy and other pneumonia chest X-ray images. Our experimental data also suggested that the proposed model may be further refined with more COVID-19 chest X-ray images.
Materials and Methods
Datasets
This study utilized two datasets for training the COVID-19 detection model. The first dataset released 5860 chest X-ray images from the routine clinical examinations of patients aged between 1 and 5 years [13]. This dataset has been pre-split into the training and testing datasets, and there are two classes of samples, i.e., pneumonia patients and normal control persons, as shown in Table 1. The dataset was denoted as dsPneumonia, and publicly available at https://data.mendeley.com/datasets/rscbjbr9sj/2. All the images are released in the JPEG image format with varied image sizes. The detailed information may be found on its web site.
Table 1.
Chest X-ray images of pneumonia and normal samples in the dataset dsPneumonia
| Dataset | Normal | Pneumonia | Total |
|---|---|---|---|
| Training | 1350 | 3884 | 5234 |
| Testing | 235 | 391 | 626 |
| Total | 1585 | 4275 | 5860 |
The dataset was pre-split into the training and testing datasets by the original authors
The second publicly available dataset was recently released as the imaging data of various virus-infected pneumonia patients [14]. This dataset pays special attention to the SARS-CoV-2-infected patients and consists of both chest X-ray and CT images. Only the chest X-ray images were used in this study. The images annotated as “no finding” or being infected by multiple viruses were excluded from further analysis. The dataset does not have normal samples. So the 235 normal images from the Testing dataset of the dataset dsPneumonia [13] were added to this dataset, and a three-class dataset was generated as dsCOVID19, as shown in Table 2. A stratified strategy was conducted to randomly split the dataset dsCOVID19 into the Training dataset (70%), Validating dataset (10%) and Testing dataset (20%). This dataset was publicly available at https://github.com/ieee8023/covid-chestxray-dataset.
Table 2.
Summary of the three classes of samples in the dataset dsCOVID19
| dsCOVID19 | COVID19 | Other pneumonia | Total pneumonia | Normal |
|---|---|---|---|---|
| Training | 131 | 43 | 174 | 164 |
| Validating | 19 | 6 | 25 | 23 |
| Testing | 39 | 14 | 53 | 48 |
| Total | 189 | 63 | 252 | 235 |
Only the chest X-ray images of persons carrying one disease or no disease were kept for analysis
Some other chest X-ray image datasets may also be utilized to replace the first dataset. The Radiological Society of North America (RSNA) released the RSNA Pneumonia Detection Challenge in 2018, and their dataset may serve the same purpose as our first dataset [15]. The large dataset of chest X-rays CheXpert provides another comprehensive source for pre-training the models [16], and 2.44% of the CheXpert images are from the pneumonia patients. The third dataset MIMIC-CXR covers 297 class labels for its chest X-ray images, and 6.9% of its images are from pneumonia patients [17].This study serves as the proof-of-principle experiment for the two-stage transfer learning strategy. So these existing datasets are not evaluated in this study.
Convolution Neural Network (CNN)
Convolutional neural network (CNN) is a feedforward neural network framework inspired by the connected visual nerve system [18]. CNN is designed to abstract the visual components of images and to map the images to lower dimensions while retaining the essential image features. A typical CNN architecture has three types of layers, i.e., convolutional, pooling and fully connected layers. The convolutional layer utilizes the convolutional kernel to extract local features from the training images, such as extracting features in the human vision system [19]. The pooling layer may efficiently reduce the parameter dimensions by sub-sampling the previous layer, so that the overfitting may be avoided [20]. The fully connected layer serves as the output layer for the final prediction results, as similar in the traditional neural networks [21]. Our proposed algorithm framework is based on the CNN architecture.
Deep Residual Network (ResNet)
The depth of a deep neural network (DNN) is a crucial factor for the model performance [22]. A DNN with more layers may extract more complicated image features. So theoretically, better model performance may be achieved with deeper DNNs. However, the degradation problem renders the DNN saturated if the number of DNN layers increases. The deep residual network (ResNet) tries to solve this problem by bypassing the input information directly to the output layer, so that the output layer has access to the un-altered input data. ResNet consists of multiple residual blocks, each of which may be defined as H(x) = F(x) + x, where x is the input data, F(x) is the mapping function of the identity residuals and H(x) is the mapped solution function. ResNet34 is a pre-trained deep residual network (DRN) model for image recognition, and this study borrowed some residual blocks of ResNet34 to our proposed deep learning framework [23].
Transfer Learning and Model Tuning
Transfer learning is a supervised machine learning strategy to exert the pre-trained model using a small-scale dataset [24]. A large number of samples are required to effectively train a DNN model, e.g., ResNet34 was pre-trained using 1.28 million images [23]. We hypothesized that the growth patterns of pulmonary internal structures and the real-world objects followed the same physical rules. The pre-trained model ResNet34 may be used to detect COVID-19 patients based on the chest X-ray images through fine-tuning on a small dataset of COVID-19 images. The essence of the transfer learning is to map the pre-trained model f(x) through an additional mapping function g(x), so that a fine-tuned transfer learning model may be defined as g(f(x)). Because of the hierarchical representation nature of convolutional neural networks, the shallow layers usually learn basic features, such as texture and edges. The pre-training process f(x) enables the parameters learned by over a million images to be shared with downstream image-based learning tasks. This strategy speeds up the convergence of the downstream tasks g(x). Many recently published transfer learning studies suggested the validity of this strategy. This study transferred the pre-trained ResNet34 model to the COVID-19 detection problem based on the chest X-ray images.
Modified Neural Network Architecture, COVID19XrayNet
The collection and annotation procedure of medical images has the characteristics of labour intensiveness and difficult sample recruitment, etc. So, it is almost impossible to train a DNN model of biomedical images from scratch. The pre-trained ResNet34 was transferred to be fine-tuned for our investigated problem.
We proposed two types of novel layers to the pre-trained model ResNet34, as shown in Fig. 1. The feature smoothing layer (FSL) used the 1 × 1 convolution to keep the shape of the tensors, and smoothed the pre-trained tensors to learn features from the new training images. FSL was inserted after the pre-trained residual blocks. The feature extraction layer (FEL) doubled the number of channels using the first three operations and flattened the feature map into one dimension. FEL extracted the operation-based features and described the previous layer with abstracted information. So, we hypothesized that FEL will help improve the final fully connected layer with these abstracted features.
Fig. 1.
The framework COVID19XrayNet in this study. This study proposed two newly designed layers, i.e., feature smoothing layer (FSL) and feature extraction layer (FEL). The framework of the pre-trained ResNet34 model was revised to solve the COVID-19 detection problem in this study. The dashed-line box is the framework of ResNet34. The bottom part illustrates the framework proposed in this study
The bottom part of Fig. 1 illustrates the proposed network framework COVID19XrayNet in this study. The input medical image is 224 × 224 in pixels and three in channels, the same as ResNet34. This study used the input layer and the next three residual blocks of ResNet34. We inserted one FSL before and after the second residual block, respectively. An FEL was inserted after the third residual block. The final fully connected 512 × k layer facilitates the model training using the limited number of COVID-19 images, where k is the number of classes in the investigated problem. COVID19XrayNet(k) may be used for the biomedical image-based classification problem, where k is the number of classes.
The Experimental Pipeline of This Study
This study proposed a two-step transfer learning pipeline to firstly transfer and tune the ResNet34 model using the chest X-ray images dataset dsPneumonia without COVID-19 cases, and then further transfer the tuned model on the dataset dsCOVID19, as illustrated in Fig. 2.
Fig. 2.
Illustration of the experimental pipeline in this study. The first step trained and evaluated the framework COVID19XrayNet(2) using the dataset dsPneumonia, while the second step trained and evaluated the framework COVID19XrayNet(3) using the small dataset dsCOVID19
In the first step of our pipeline, the pre-trained model ResNet34 was transferred to the dataset dsPneumonia and the proposed framework COVID19XrayNet(2) was utilized to tune the parameters of the internal layers. This binary classification model was initialized using the pre-trained model ResNet34, and was further trained using the 5234 training to extract the pneumonia-specific features in the chest X-ray images. The model was trained for ten epochs. The batch size was set to 32. The optimizer AamW was used with the initial learning rate 0.0001. All the other parameters were set to the default values.
The second step used the model trained in the first step of this pipeline, and tuned the model using the framework COVID19XrayNet(3). The dataset dsCOVID19 consisted of three sample classes, i.e., COVID-19, other pneumonia, and normal. So the output layer was revised as the dimensions of 512 × 3. The model was trained on the training dataset and tuned on the validating dataset. The final test result was calculated on the testing dataset. Compared with the first step, this model has fewer training samples. So the batch size was reduced to 16, and the model was trained for 20 epochs with a smaller learning rate 0.00005. All the other parameters were set to the default values.
Although the upstream task is a two-class classification without the label COVID-19 and the downstream task is the three-class classification, both of the tasks abstract image features from the chest X-ray images. The proposed model COVID19XrayNet learns the inherent patterns from the X-ray images for similar purposes. So we hypothesize that the model tuned in the upstream task will be further refined in the downstream task.
Implementations
All the experiments were carried out in a Standard NC6 (6 CPUs, 56 GB Memory, and 1 K80 GPU card) Azure Virtual Machine. The code was implemented by PyTorch 1.4 and Scikit-learn 0.22, in the Python programming languages.
Experimental Results and Discussions
Data Augmentation
Deep neural networks rely on a large number of high-quality training samples to achieve accurate detection results. But compared with the publicly available training dataset in the area of computer vision, most of the biomedical image datasets release fewer than 10,000 images, including the two datasets used in this study [13, 14].
This study employed multiple image augmentation techniques to generate simulated images of the training dataset of dsPneumonia, so that the deep neural network COVID19XrayNet could be trained with a sufficient number of chest X-ray images. The image augmentation techniques used in this study were randomly resized crop, random rotation, random horizontal flip, and random vertical flip. These augmentation techniques ensure that the simulated images are actually the variants of the original images and carry similar image patterns as the original ones [25]. The crop ratio [0.8, 1.0] was set for the operation of the randomly resized crop, so that the lesion sites in the chest X-ray images were not excluded. For the same reason, the operation of random rotation used a random rotation angle [− 20, 20] in degrees. All data augmentation functions were provided by the Python package PyTorch and were integrated into the model training pipeline. These data augmentation functions were automatically utilized before generating each training batch at the pipeline.
Evaluating the Model of Pneumonia Versus Normal
The network framework COVID19XrayNet(2) was initialized from the pre-trained ResNet34 and further tuned using the augmented training dataset, as described in the above sections. The cross entropy loss measures the performance of a classification model with the probabilistic outputs [26]. The COVID19XrayNet(2) was evaluated for its cross entropy loss function and the running averages over the window size 5.
Figure 3 demonstrates that the model’s initial loss was very high and the first cross entropy loss was as high as 0.6736. The running average loss was 0.7308 over the window size 5. A radical decrease in both metrics was observed after a few batches of training. For example, the cross entropy loss and running average on the 100th batch dropped to 0.4702 and 0.4314. The training process stopped at the batch 1635, and the cross entropy loss was decreased to 0.0717, compared with the initial loss value 0.6736.
Fig. 3.
The loss values of the training models over different batches. The horizontal axis shows different batches, while the vertical axis shows the loss values for the two metrics, cross entropy loss and the running average
A binary classification model was trained and evaluated in this step, and its performance metrics are illustrated in Fig. 4. The pneumonia and normal images were regarded as positive and negative samples, respectively. The metrics specificity (Sp) and sensitivity (Sn) were percentages of correctly predicted negative and positive samples, respectively. The metric accuracy (Acc) was the percentage of all the correctly predicted samples. The area under the curve (receiver operating characteristic) was a parameter-independent metric to describe a binary classification model. The detailed definitions may be found in [27, 28].
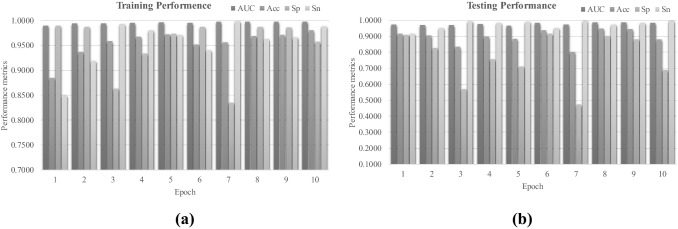
Fig. 4.
Performance metrics AUC, Acc, Sp and Sn of the network COVID19XrayNet(2) on the dataset dsPneumonia. The horizontal axis shows one of the ten epochs and the vertical axis shows the performance metric values. The performance metrics were calculated on the a training and b testing datasets of dsPneumonia
The COVID19XrayNet(2) was satisfyingly tuned to achieve the accurate detection of pneumonia samples in the dataset dsPneumonia, as shown in Fig. 4. The parameter-independent metric AUC reached as high as 0.9978 on the training dataset, as shown in Fig. 4a. The trained model achieved very good AUC (> 0.9660). The model trained in epoch 8 achieved a balance of Sp = 0.9017 and Sn = 0.9744, as shown in Fig. 4b.
So we hypothesized that the COVID19XrayNet(2) achieved the best testing performance at epoch 8. The model achieved 0.9977 in AUC on the training dataset, and 0.9857 in AUC on the testing dataset. The metrics Sp = 0.9017 and Sn = 0.9744 suggested that the model performed accurately and stably on both training and testing datasets. So the COVID19XrayNet(2) model at the epoch 8 was transferred to the second step for detecting COVID-19 patients in the following sections.
Evaluating the COVID-19 Detection Model
The best model transferred from the above section was further tuned using the dataset dsCOVID19, as shown in Fig. 5. The investigated problem had three classes of samples, i.e., COVID-19, other pneumonia, and normal ones. The metric Cohen’s kappa was introduced to evaluate the tuned models. The Cohen’s kappa (Kappa) was defined as K = (p0–pe)/(1–pe), where p0 is the empirical probability of agreeing on the label assignments of the samples, while pe is the expected agreement probability for a random prediction [29]. Kappa is considered as a more robust metric than the percentile agreement of predictions. Kappa ranges between − 1 and 1. A larger Kappa value suggested a better model performance. The prediction accuracy (Acc) was still defined as the overall percentage of correctly predicted samples over the three classes.
Fig. 5.
The COVID-19 detection model was evaluated on the training, validating and testing datasets. a Performances on the training and validating datasets. The horizontal axis shows one of the 20 epochs, while the vertical axis shows the performance metrics. The metrics Acc and Kappa on the validating dataset are plotted in solid lines, while these two metrics on the training dataset are in dotted lines. b Heatmap of the confusion matrix on the testing dataset. The rows indicate the real class labels, while the columns the predicted class labels
Firstly, the framework COVID19XrayNet(3) seemed to be overfitted after the epochs 18 of the training process, as shown in Fig. 5a. Although the training performance had a slight increase in both Acc and Kappa, the validating performance metrics kept decreasing on the epochs 19 and 20. So the model trained on the epoch 18 was retrieved for the evaluation on the testing dataset.
Our final model achieved 0.9108 in Acc and 0.8574 in Kappa on the testing dataset of the COVID detection problem, as shown in Fig. 5b. In particular, our model achieved 0.9231 in Acc for COVID-19 patients, while 0.9583 in Acc for normal persons. Non-COVID-19 pneumonia patients received less accurate predictions, which may be due to the very limited number of images in the dataset dsCOVID19.
Two-Step Transfer Learning Model was Necessary
This study proposed one more step of tuning the transferred model on a similar dataset for the COVID-19 images. Our hypothesis was that the pre-trained model ResNet34 was not optimized for the chest X-ray images, and the parameter tuning of ResNet34 on the same data type may help the model perform reasonably on the target dataset. The model training used both original images and the augmented images, while the model detection performance was calculated using only the original images.
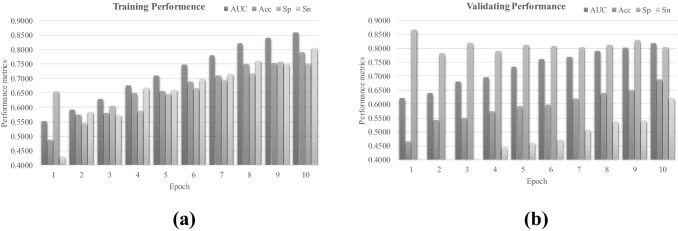
Figure 6 illustrates the experimental data of directly transferring the pre-trained model ResNet34 to the binary classification problem between pneumonia and normal samples. The model performance kept increasing with more epochs of training, as in Fig. 6a. But the model achieved both Acc and Sn smaller than 0.7000. This was not acceptable, since pneumonia patients need to be accurately detected. The AUC and Sp were 0.8176 and 0.8340, respectively. This may be because the class labels of the dataset dsPneumonia do not exist in the ImageNet dataset, and the pre-trained model ResNet34 cannot extract the effective features of these X-ray image.
Fig. 6.
Performance metrics of AUC, Acc, Sp and Sn of the network COVID19XrayNet(3) transferred from ResNet34 on the dataset dsPneumonia. The horizontal axis indicates one of the ten epochs and the vertical axis the performance metric values. The performance metrics were calculated on the a training and b validating datasets of dsCOVID19
COVID19XrayNet Outperformed ResNet34
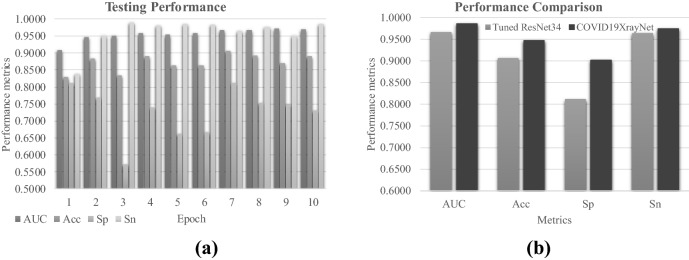
We evaluated whether the proposed new deep residual network COVID19XrayNet outperformed the existing ResNet34, as shown in Fig. 7. Firstly, the existing deep neural network framework ResNet34 performed very well on after the parameter tuning, as shown in Fig. 7a. The best model performance metric AUC reached 0.9718 at the epoch 9, but its overall accuracy was only Acc = 0.8690. Figure 7b illustrates that the proposed framework COVID19XrayNet outperformed ResNet34 in all the four performance metrics. The COVID19XrayNet also achieved a balanced pair of Sp and Sn. So the proposed COVID19XrayNet may serve as a good complementary framework for the medical image-based prediction problems.
Fig. 7.
Performance comparison between COVID19XrayNet and ResNet34. a Performance metrics of the fine-tuned ResNet34 for the pneumonia detection problem. b The four performance metrics AUC, Acc, Sp and Sn between the tuned ResNet34 model and the proposed COVID19XrayNet
We also evaluated how the existing framework ResNet34 performed on the two-step transfer learning pipeline, as shown in Fig. 8. ResNet34 performed slightly better on detecting the normal samples than the proposed model COVID19XrayNet. But ResNet34 achieved more than 0.1000 worse in detecting COVID-19 patients than COVID19XrayNet, and 0.2500 worse in detecting the other pneumonia patients than COVID19XrayNet.
Fig. 8.

Comparison of ResNet34 and COVID19XrayNet in the two-step transfer learning pipeline. The horizontal axis shows one of the three classes. The vertical axis shows the three-class prediction accuracy
The experimental data suggested that both the two-step transfer learning pipeline and the new framework COVID19XrayNet were necessary for the COVID-19 detection problem.
COVID19XrayNet may be Improved Using more Training Samples
The COVID19XrayNet was evaluated for its performances using different numbers of training samples, as shown in Fig. 9. The dataset dsCOVID19 was randomly split into the training (70%), validating (10%) and testing (20%) samples. The overall accuracy (Acc) and the Cohen’s kappa (Kappa) were calculated for the COVID19XrayNet-based two-step transfer learning pipeline using 20%, 40%, 60%, 80% and 100% of the training samples. Figure 9 illustrates that COVID19XrayNet achieved 0.8081 in Acc even using only 20% of the training samples. Both Acc and Kappa were increased with more training samples being utilized.
Fig. 9.

Performances of the COVID19XrayNet models trained using different numbers of training samples. The horizontal axis shows the percentage of training samples. The vertical axis shows the values of the performance metrics Acc and Kappa
So the experimental data suggested that our proposed COVID-19 detection model may be improved if more training data were available.
Future Directions
This study proposed to use a two-stage transfer learning strategy to solve the problem of medical imaging with a small amount of data. We modified the model structure of ResNet34 to obtain the neural network framework COVID19XrayNet, which enables it to migrate to multiple downstream tasks. But some issues can be discussed in detail to make it a general framework. For instance, what model is most suitable for migration? What kind of general algorithm is used to solve the structural modification from upstream model to downstream model? These problems are the directions that we need to continue to study in the future.
Conclusion
This study proposed a two-step transfer learning pipeline based on the deep neural network framework COVID19XrayNet for the COVID-19 detection problem. COVID19XrayNet is not a completely new neural network. Instead, COVID19XrayNet integrated two novel layers into the popular ResNet32, i.e., feature smoothing layer (FSL) and feature extraction layer (FEL). Our experimental data demonstrate that COVID19XrayNet outperforms the original version of ResNet32. The overall accuracy of 0.9192 was achieved. Our experimental data supported that the accurate prediction performance of the proposed model was achieved through the collaborations of all the three factors, i.e., the novel framework COVID19XrayNet, the two-step transfer learning pipeline and the sufficient number of training samples. The proposed model was anticipated to be improved if more chest X-ray images of COVID-19 patients were released.
This proof-of-principle study demonstrated that a deep learning model with satisfying prediction performance may be developed on a small dataset by the two-step transfer learning strategy. The experimental data showed the necessity of the two-step transfer learning strategy to improve the X-ray image-based deep learning model using only 189 annotated COVID19 X-ray images. It is anticipated that the model will be further improved with more available annotated images.
Acknowledgements
Insightful comments from the anonymous reviewers are greatly appreciated.
Funding
This work was supported by the Jilin Provincial Key Laboratory of Big Data Intelligent Computing (20180622002JC), the Education Department of Jilin Province (JJKH20180145KJ), and the startup grant of the Jilin University. This work was also partially supported by the Bioknow MedAI Institute (BMCPP-2018-001), the High Performance Computing Center of Jilin University, and by the Fundamental Research Funds for the Central Universities, JLU.
Compliance with Ethical Standards
Conflict of Interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
References
- 1.Zhai P, Ding Y, Wu X, Long J, Zhong Y, Li Y. The epidemiology, diagnosis and treatment of COVID-19. Int J Antimicrob Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Onder G, Rezza G, Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA. 2020;323(18):1775–1776. doi: 10.1001/jama.2020.4683. [DOI] [PubMed] [Google Scholar]
- 4.Weiss P, Murdoch DR. Clinical course and mortality risk of severe COVID-19. Lancet. 2020;395(10229):1014–1015. doi: 10.1016/S0140-6736(20)30633-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Velavan TP, Meyer CG. The COVID-19 epidemic. Trop Med Int Health. 2020;25(3):278–280. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, Ng DYM, Wan CKC, Yang P, Wang Q, Peiris M, Poon LLM. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020;66(4):549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, Sun R, Wang Y, Hu B, Chen W, Zhang Y, Wang J, Huang B, Lin Y, Yang J, Cai W, Wang X, Cheng J, Chen Z, Sun K, Pan W, Zhan Z, Chen L, Ye F. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020 doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020 doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang Y, Wang Z (2008) Improving LBP features for gender classification. In: 2008 International Conference on Wavelet Analysis and Pattern Recognition. IEEE, pp 373–377. 10.1109/ICWAPR.2008.4635807
- 10.Satpathy A, Jiang X, Eng H-L. Human detection by quadratic classification on subspace of extended histogram of gradients. IEEE Trans Image Process. 2013;23(1):287–297. doi: 10.1109/TIP.2013.2264677. [DOI] [PubMed] [Google Scholar]
- 11.Zhao R, Zhang R, Tang T, Feng X, Li J, Liu Y, Zhu R, Wang G, Li K, Zhou W, Yang Y, Wang Y, Ba Y, Zhang J, Liu Y, Zhou F. TriZ-a rotation-tolerant image feature and its application in endoscope-based disease diagnosis. Comput Biol Med. 2018;99:182–190. doi: 10.1016/j.compbiomed.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Zhang R, Zhao R, Zhao X, Wu D, Zheng W, Feng X, Zhou F. pyHIVE, a health-related image visualization and engineering system using Python. BMC Bioinf. 2018;19(1):452. doi: 10.1186/s12859-018-2477-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kermany DS, Goldbaum M, Cai W, Valentim CCS, Liang H, Baxter SL, McKeown A, Yang G, Wu X, Yan F, Dong J, Prasadha MK, Pei J, Ting MYL, Zhu J, Li C, Hewett S, Dong J, Ziyar I, Shi A, Zhang R, Zheng L, Hou R, Shi W, Fu X, Duan Y, Huu VAN, Wen C, Zhang ED, Zhang CL, Li O, Wang X, Singer MA, Sun X, Xu J, Tafreshi A, Lewis MA, Xia H, Zhang K. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Cohen JP, Morrison P, Dao L (2020) COVID-19 image data collection. arXiv preprint arXiv:2003.11597
- 15.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM (2017) Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp 2097–2106. 10.1109/CVPR.2017.369
- 16.Irvin J, Rajpurkar P, Ko M, Yu Y, Ciurea-Ilcus S, Chute C, Marklund H, Haghgoo B, Ball R, Shpanskaya K (2019) Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison. In: Proceedings of the AAAI Conference on Artificial Intelligence. pp 590–597. 10.1609/aaai.v33i01.3301590
- 17.Johnson AE, Pollard TJ, Berkowitz SJ, Greenbaum NR, Lungren MP, Deng C-y, Mark RG, Horng S. MIMIC-CXR, a de-identified publicly available database of chest radiographs with free-text reports. Sci Data. 2019 doi: 10.1038/s41597-019-0322-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence S, Giles CL, Tsoi AC, Back AD. Face recognition: a convolutional neural-network approach. IEEE Trans Neural Netw. 1997;8(1):98–113. doi: 10.1109/72.554195. [DOI] [PubMed] [Google Scholar]
- 19.LeCun Y, Kavukcuoglu K, Farabet C (2010) Convolutional networks and applications in vision. In: Proceedings of 2010 IEEE international symposium on circuits and systems. IEEE, pp 253–256. 10.1109/ISCAS.2010.5537907
- 20.Frazao X, Alexandre LA (2014) Dropall: Generalization of two convolutional neural network regularization methods. In: International Conference Image Analysis and Recognition. Springer, pp 282–289. 10.1007/978-3-319-11758-4_31
- 21.Liu K, Kang G, Zhang N, Hou B. Breast cancer classification based on fully-connected layer first convolutional neural networks. IEEE Access. 2018;6:23722–23732. doi: 10.1109/ACCESS.2018.2817593. [DOI] [Google Scholar]
- 22.Paszke A, Chaurasia A, Kim S, Culurciello E (2016) Enet: A deep neural network architecture for real-time semantic segmentation. arXiv preprint arXiv:1606.02147
- 23.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp 770–778. 10.1109/CVPR.2016.90
- 24.Ng H-W, Nguyen VD, Vonikakis V, Winkler S (2015) Deep learning for emotion recognition on small datasets using transfer learning. In: Proceedings of the 2015 ACM on international conference on multimodal interaction. pp 443–449. 10.1145/2818346.2830593
- 25.Hu B, Song R-J, Wei X-S, Yao Y, Hua X-S, Liu Y (2020) PyRetri: A PyTorch-based library for unsupervised image retrieval by Deep Convolutional Neural Networks. arXiv preprint arXiv:2005.02154
- 26.Yang H, Kim J-Y, Kim H, Adhikari SP. Guided soft attention network for classification of breast cancer histopathology images. IEEE Trans Med Imaging. 2019 doi: 10.1109/TMI.2019.2948026. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Chen C, Duan M, Liu S, Huang L, Zhou F. BioDog, biomarker detection for improving identification power of breast cancer histologic grade in methylomics. Epigenomics. 2019;11(15):1717–1732. doi: 10.2217/epi-2019-0230. [DOI] [PubMed] [Google Scholar]
- 28.Hao D, Peng J, Wang Y, Liu J, Zhou X, Zheng D. Evaluation of convolutional neural network for recognizing uterine contractions with electrohysterogram. Comput Biol Med. 2019;113:103394. doi: 10.1016/j.compbiomed.2019.103394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McHugh ML. Interrater reliability: the kappa statistic. Biochem Med (Zagreb) 2012;22(3):276–282. doi: 10.11613/BM.2012.031. [DOI] [PMC free article] [PubMed] [Google Scholar]