Abstract
Objective
This study was to investigate the CT quantification of COVID-19 pneumonia and its impacts on the assessment of disease severity and the prediction of clinical outcomes in the management of COVID-19 patients.
Materials Methods
Ninety-nine COVID-19 patients who were confirmed by positive nucleic acid test (NAT) of RT-PCR and hospitalized from January 19, 2020 to February 19, 2020 were collected for this retrospective study. All patients underwent arterial blood gas test, routine blood test, chest CT examination, and physical examination on admission. In addition, follow-up clinical data including the disease severity, clinical treatment, and clinical outcomes were collected for each patient. Lung volume, lesion volume, nonlesion lung volume (NLLV) (lung volume – lesion volume), and fraction of nonlesion lung volume (%NLLV) (nonlesion lung volume / lung volume) were quantified in CT images by using two U-Net models trained for segmentation of lung and COVID-19 lesions in CT images. Furthermore, we calculated 20 histogram textures for lesions volume and NLLV, respectively. To investigate the validity of CT quantification in the management of COVID-19, we built random forest (RF) models for the purpose of classification and regression to assess the disease severity (Moderate, Severe, and Critical) and to predict the need and length of ICU stay, the duration of oxygen inhalation, hospitalization, sputum NAT-positive, and patient prognosis. The performance of RF classifiers was evaluated using the area under the receiver operating characteristic curves (AUC) and that of RF regressors using the root-mean-square error.
Results
Patients were classified into three groups of disease severity: moderate (n = 25), severe (n = 47) and critical (n = 27), according to the clinical staging. Of which, a total of 32 patients, 1 (1/25) moderate, 6 (6/47) severe, and 25 critical (25/27), respectively, were admitted to ICU. The median values of ICU stay were 0, 0, and 12 days, the duration of oxygen inhalation 10, 15, and 28 days, the hospitalization 12, 16, and 28 days, and the sputum NAT-positive 8, 9, and 13 days, in three severity groups, respectively. The clinical outcomes were complete recovery (n = 3), partial recovery with residual pulmonary damage (n = 80), prolonged recovery (n = 15), and death (n = 1). The %NLLV in three severity groups were 92.18 ± 9.89%, 82.94 ± 16.49%, and 66.19 ± 24.15% with p value <0.05 among each two groups. The AUCs of RF classifiers using hybrid models were 0.927 and 0.929 in classification of moderate vs (severe + critical), and severe vs critical, respectively, which were significantly higher than either radiomics models or clinical models (p < 0.05). The root-mean-square errors of RF regressors were 0.88 weeks for prediction of duration of hospitalization (mean: 2.60 ± 1.01 weeks), 0.92 weeks for duration of oxygen inhalation (mean: 2.44 ± 1.08 weeks), 0.90 weeks for duration of sputum NAT-positive (mean: 1.59 ± 0.98 weeks), and 0.69 weeks for stay of ICU (mean: 1.32 ± 0.67 weeks), respectively. The AUCs for prediction of ICU treatment and prognosis (partial recovery vs prolonged recovery) were 0.945 and 0.960, respectively.
Conclusion
CT quantification and machine-learning models show great potentials for assisting decision-making in the management of COVID-19 patients by assessing disease severity and predicting clinical outcomes.
Key Words: COVID-19, Novel coronavirus pneumonia, Computed tomography, Quantitative image analysis, Machine-learning
INTRODUCTION
The outbreak of viral pneumonia caused by the 2019 novel coronavirus originally identified in China (1,2), named COVID-19 (3) and officially labeled as a pandemic by World Health Organization (WHO) (4), is spreading rapidly over 200 countries and more than 5 million confirmed cases worldwide as of May 2020 (5). At present, nucleic acid test (NAT) of RT-PCR (reverse transcription polymerase chain reaction) remains the gold standard for diagnosis of COVID-19 infection (6,7). CT findings of viral pneumonia is listed as one of the three clinical evidences (the other two are respiratory symptoms and blood test) to identify suspected cases in the guideline of Chinese Health Commission (CHC) (8). Typical CT signs of COVID-19 infection include ground-glass opacities (GGO), GGO with lung consolidation, bronchial dilation, bilateral involvement, and peripheral distribution (9, 10, 11). These abnormal CT findings play an important role in assisting the diagnosis of COVID-19 patients. However, some studies observed that asymptomatic or mild symptomatic patients might have atypical or normal CT findings (12,13). Thereby, the primary role of CT imaging is limited to identify the suspected patients if a patient has clinical symptoms suggestive of COVID-19 infection. As opposed to China, chest radiography has been deemed as the modality of choice for assessing and monitoring COVID-19 by American College of Radiology and Society of Thoracic Radiology in the United States (14).
Chest CT plays a vital role in the diagnosis and management of various lung diseases (15). The advent of high-resolution CT images has led to its increasing use in the management of chronic and acute pulmonary diseases such as chronic obstructive pulmonary disease (16), hypersensitivity pneumonitis (15), and interstitial lung diseases (17). Chest CT scans can assist diagnosis and guide treatment decision for community-acquired pneumonia (18,19). Quantitative imaging provided reliable and objective biomarkers in the management of severe acute respiratory syndrome and middle-east respiratory syndrome (20,21). Recent studies reported that AI-powered CT diagnosis may outperform lab testing for screening of COVID-19 with sensitivity of 97% (22), and CT quantification as indictors may assess disease severity and predict prognosis in the management of COVID-19 (23,24).
According to the clinical staging and disease progression, COVID-19 patients are classified into four groups, mild, moderate, severe, and critical groups, in terms of CHC guideline (8). Mild patients tend to have mild clinical symptoms and scarcely identified pneumonia lesions on CT. Studies observed that disease progression is generally associated with the increasing of numbers and sizes of GGO lesions in CT (25). Thus, CT quantification as an advanced imaging technique is more effective for advanced progressive cases or those with complications. However, due to the short outbreak of COVID-19, little is known regarding CT imaging characteristics specific to the disease (26), and the validity of CT quantification in the management of COVID-19. The clinical roles of CT quantification have not been explored for COVID-19 yet, in particular, in assisting decision-making in the management of COVID-19. It is expected that quantitative imaging analysis combined with AI and deep-learning technology will play an important role in the management of COVID-19 patients such as assessment of disease severity and the prediction of prognosis (27).
The objectives of this study were (1) to investigate quantitative imaging analysis techniques by using deep-learning for quantification of COVID-19 pneumonia in CT images, (2) to correlate quantitative image biomarkers to the clinical manifestations of disease severity and clinical outcomes, and (3) to build up machine-learning (ML) models with quantitative imaging biomarkers for stratification of disease severity and prediction of clinical outcomes in the management of COVID-19 patients.
MATERIALS AND METHODS
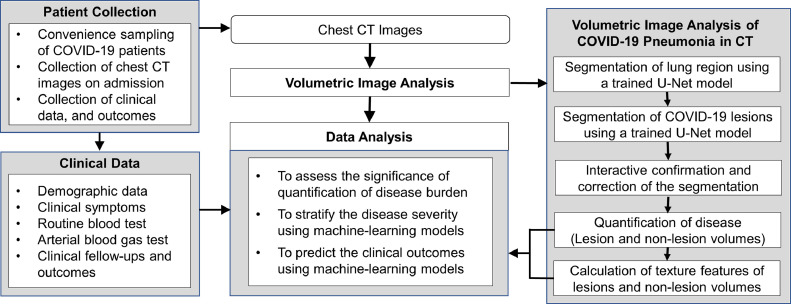
The Ethics Committees of both institutions have approved this retrospective study, in which informed consent was waived, but patient confidentiality was protected. The working procedure of the study is illustrated in Figure 1 .
Figure 1.
Flow chart of the study.
Patient Cohort
All patients who were diagnosed of COVID-19 infection by positive NAT of RT-PCR and hospitalized from January 19, 2020 to February 19, 2020 were collected in a convenience sample of patients seen at the First Affiliated Hospital at Zhejiang University School of Medicine. Inclusion criteria for this study were: (1) an adult patient over 18 years old; (2) confirmed by positive NAT of RT-PCR for COVID-19; (3) hospitalized for the treatment of COVID-19; (4) evaluated as moderate, severe, or critical disease severity; and (5) underwent chest CT examination within 1 day on admission.
The exclusion criteria were as follows: (1) patients who were evaluated as mild stage; (2) patients who were not hospitalized for treatment; (3) no baseline CT examination was performed within 1 day of admission; and (4) incomplete clinical record when transferred from other hospitals.
A total of 99 patients were selected from initial identification of 105 hospitalized patients after exclusion of patients who were classified into mild group (n = 2), those had no CT scanning or underwent CT examination within 1 day of admission (n = 3), and those with incomplete clinical data (n = 1).
Clinical Data
The patient's clinical data including demographic data, clinical symptoms, arterial blood gas test, routine blood test, treatment, and outcomes were retrieved from the hospital's medical record system. A total of 40 clinical parameters that are associated with COVID-19 patient management were collected, including
-
•
Demographic data (nine parameters): gender, age, days from illness to clinical visit, Wuhan contact history, and other medical history (hypertension, diabetes, history of surgery, coronary heart disease, hepatitis B);
-
•
Clinical symptoms (nine parameters): fever, chills, cough, sputum, dizziness, headache, fatigue, body ache, chest tightness, diarrhea;
-
•
Routine blood test (seven parameters): white blood cells, lymphocytes, eosinophils, neutrophils, lymphocyte counts, eosinophil count, C-reactive protein;
-
•
Arterial blood gas test (15 parameters): blood oxygen saturation, partial pressure of arterial blood oxygen (PO2), partial pressure of arterial carbon dioxide (PCO2), PH value, activated partial prothrombin time, prothrombin time, D-dimer, lactate dehydrogenase, phosphomuscular acid kinase, creatine kinase isoenzyme, Alanine Aminotransferease (ALT), aspartate aminotransferase (AST), blood creatinine, blood urea nitrogen, procalcitonin.
For clinical treatment and outcomes, these patients were followed and outcome data were collected including the ICU support and length of ICU stay, the duration of oxygen inhalation, the duration of sputum NAT-positive, the duration of hospital stay, and the final outcome in terms of complete recovery, partial recovery with residual pulmonary damage, prolonged recovery, and death. The duration of sputum NAT-positive refers to the period from the hospitalization to the clear of infection for patients who were tested negative for COVID-19 for two consecutive days in respiratory samples, which is an important clinical outcome indicating the clearing of the virus infection in the patients. In our study, viral RNA was extracted using the MagNA Pure 96 (Roche, Basel, Switzerland), and RT-PCR was performed using a commercial kit specific for COVID-19 virus detection (BioGerm, Shanghai, China).
CT Image Acquisition
All subjects underwent chest CT examinations on a multidetector CT scanner (GE Revolution EVO 64-slice CT scanner, GE Healthcare, Milwaukee, WI) within 1 day on admission with the following CT scanning parameters: supine position, 120 kVp tube voltage, automatic tube current modulation, 0.725 mm collimation, 1 mm and 5 mm reconstruction intervals. The scanning range was from thoracic inlet to upper abdominal.
Volumetric Image Analysis
Quantification of lung and pneumonia lesions was performed on a volumetric image analysis platform 3DQI (3D quantitative imaging: https://3dqi.mgh.harvard.edu), which was developed on a collection of open-source packages, including Qt (v4.8.4), VTK (v5.10.1), DCMTK (v3.6.5), R (v3.6.3), and OpenCV (v2.4.11). 3DQI platform consists of three major components: (1) 3DQI console for management of the study, (2) 3DQI imaging analysis for segmentation of organs and lesions, and (3) 3DQI radiomics with a rich set of tools for data visualization, statistical analysis, and ML classification based on extracted image features and clinical parameters.
The volumetric image analysis of CT quantification of COVID-19 consisted of five steps: (1) segmentation of lung region, (2) detection and segmentation of pneumonia regions, (3) interactive confirmation and correction of the segmentation results, (4) quantification of disease (lung volume, lesion volume, non-lesion lung volume (NLLV), and fraction of NLLV [%NLLV]), and (5) calculation of histogram textures of lesion and NLLVs. This pipeline is illustrated in the right part of Figure 1.
Segmentation of Lung and Lesions
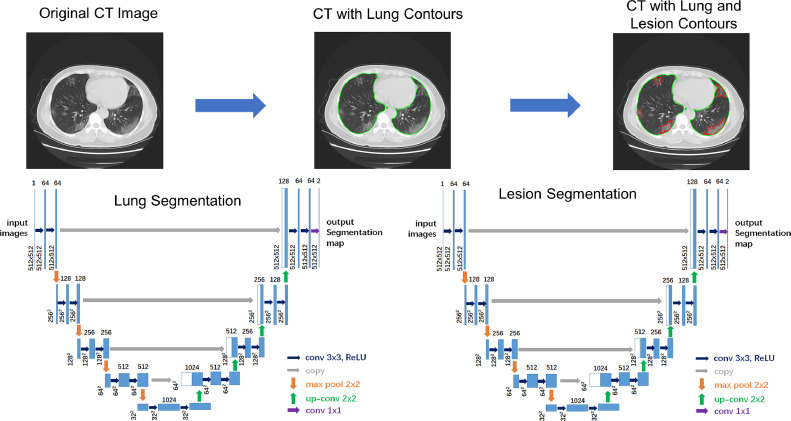
The segmentation of lung and lesions was carried out by using the U-Net model (28), which represents one of the most well-known convolutional neural network architectures for medical image segmentation. U-Net consists of convolutions path for feature extraction followed by up-convolutions path for resolution restoration and an identity skip connection. A schematic workflow of the U-Net architecture is shown in Figure 2 .
Figure 2.
U-Net architecture and resulting segmentation of lung and lesions in CT images. (Color version of figure is available online.)
In this study, we trained two end-to-end 2D cascading U-Net segmentation models of lung and lesions. As a preprocessing step of the segmentation, the 12 bits CT images were first mapped to 8 bit images by using a lung CT window-level setting (WW: 1000 HU, WL: −500 HU). All convolutions and up-convolutions were performed with two 3 × 3 convolutions layers with padding to keep the size unchanged followed by batch normalization, activation with the rectified linear unit and 2 × 2 max pooling. The bottom most layer mediates between the convolutions path and the up-convolution path. After that, the resultant mapping passes through another 3 × 3 convolutional neural network layer with the number of feature maps equal to the number of segments desired. Data augmentation was performed by adding noise and random elastic deformation including random rotation, random shift, random shear, and random zoom. The weighted cross-entropy was used as the loss function. We applied mini-batch Adam optimizer to train the model. The learning rate was set to 1e-4 and the batch size was set to 8. The training process stopped when the training loss did not improve in 10 epochs. The dice similarity coefficient (DSC) was used as our evaluation metrics.
The U-Net models were trained on 250 chest CT scans randomly selected from the follow-up CT examinations of the COVID-19 patients, of which lung regions and pneumonia regions were annotated by the consensus of a junior and a senior radiologists using the semi-automated contouring tools on 3DQI platform. We applied 10-fold cross-validation in the training procedure. The extracted lung regions were fed into the U-Net model for lesion segmentation which was trained by the same datasets with the annotation of COVID-19 pneumonia lesions.
Quantification of Disease
After the automated segmentation of the U-Net models, the resulting images were reviewed by the consensus of a senior radiologist and an image analyst, who were blinded to the patient clinical data including disease severity and clinical outcomes. The CT quantification of disease was determined by calculating the lung volume, the lesion volume, the NLLV (lung volume – lesion volume), and fraction of NLLV (%NLLV) (non-lesion lung volume / lung volume) by using the segmentation results.
After completion of segmentation and quantification, clinical data became visible to the image analyst, who calculated the imaging texture features of lung and lesion regions and correlated to the clinical data and prognosis for the further data analysis.
Calculation of Image Texture Features
For the analysis of additional quantitative image features of the diseased and nondiseased lung, we calculated the histogram features for both lesion volume and NLLV. The histogram was constructed with bin size of 10 HU in the range between −1000 HU and 200 HU, resulting 120 bins. The histogram was normalized in terms of the size of the lesion volume and NLLV (the number of voxels), respectively. A set of 20 histogram statistics features such as mean, standard deviation, skewness, kurtosis, energy, and entropy (29) was calculated for both lesions and NLLV. (The list of histogram textures refers to Appendix 1.)
Data Analysis
Data analysis was focused on two aspects: first, the role of CT quantification in the assessment of disease severity and prediction of clinical outcome, and second, the predictive power of ML models which combine clinical data, CT quantification, and image textures to predict the need and duration of ICU, duration of oxygen inhalation, duration of hospitalization, duration of sputum NAT-positive, and clinical prognosis.
To assess whether CT quantification is a significant parameter in the assessment of disease severity and prognosis, we calculated the significance of the quantitative imaging biomarkers of the disease (lesion volume, NLLV, and %NLLV) for three disease severity groups (moderate, severe, and critical), and four prognosis groups (complete recovery, partial recovery with residual pulmonary damage, prolonged recovery, and death). A chi-squared test was performed to evaluate whether a biomarker is significantly different with the null hypothesis (H0) that there is no difference for this biomarker among groups of different diseases severity and outcomes. This null hypothesis will be rejected if p value <0.05 whereby this biomarker is considered significantly different among different disease severity groups and different clinical outcome groups, which is an indicator that the biomarker is a significant parameter of disease severity and clinical outcomes.
To assess significant parameters in the prediction of clinical outcomes, we performed Boruta algorithm to select significant features related to disease severity and prognosis. In Boruta, each original feature vector (i = 1…n, where n is the number of features) is randomly permuted to create a “shadow” counterpart . The original and shadow features are combined together + that are used to train a Boruta built-in random forest (RF) model (30). For each one of the original and shadow features involved, a pseudo feature is crafted by randomly permuting . The RF models are then trained using and , respectively, and their accuracies and obtained. The loss of accuracy is used as an indicator of feature importance. Features are selected by Boruta only if their importance values are significantly greater than the maximum of shadow feature importance. In this study, those selected features were sorted in descending order of importance. To maintain consistency, only the top five features, if any, were used to train the ML models. A higher place of a feature among selected features indicates greater significance and predictive value of the feature for the ML model created.
To establish the ML model using clinical parameters, CT quantification and imaging texture features calculated from the lesions and NLLVs, we built RF models for the purpose of classification and regression using the top five selected features to classify and predict the clinical outcomes. To reduce the bias that may be caused by an unbalanced number of positive and negative samples, we applied the Synthetic Minority Oversampling Technique resampling method (31), which combines informed oversampling of the minority class (patients with small number of sampling data) with random undersampling of the majority class (patients with large number of sampling data) to balance the samples between different patient groups.
The performance of the RF classifiers and regressors trained in this study was evaluated using repeated cross-validation (100 repetitions, 10-fold partition for each repetition). The RF model performance was compared using the area under the receiver operating characteristic curves (AUC) values with 95% confidence interval and that of RF regressors using the root-mean-square error (RMSE).
Statistical Analysis
All statistical analyses were performed using our 3DQI radiomics tool, whose core components are based on the statistical programming language R (V3.6.3) and a large collection of open-source R libraries. A chi-square test or Fisher's exact test was used for the nominal variable. A Mann-Whitney U test was used for the unordered categorical variable. A Student's t test was used for the continuous variable. A p value less than 0.05 was considered statistically significant.
Data Availability
Anonymized data will be shared by request from any qualified investigators: (1) clinical data of 99 patients, (2) chest CT images of 99 patients, (3) segmentation results of lung and lesions of COVID-19 pneumonia of 99 patients, and (4) imaging features calculated from the segmented lesions and NLLVs.
RESULTS
The chest CT images of 99 patients were subjected to the process of volumetric image analysis of COVID-19 pneumonia. Subsequently, the significance of CT quantification and the performance of prediction models were evaluated.
Clinical Data
Ninety-nine patients (58 males; 41 females; mean age 54.5 ± 15.4 years; range 24-96 years) were finally enrolled in the study. All patients were confirmed by positive NAT of RT-PCR and hospitalized for treatment. As per disease severity, these patients were classified into three groups: moderate (n = 25), severe (n = 47), and critical (n = 27), respectively, according to the disease progress criteria (Version 6) of CHC (8). The ratios of males to females in three group were 0.92 (12:13), 1.47 (28:19), and 2.00 (18:9), respectively, while the ratio was 1.41 (58:41) in the study cohort. In addition, the ages of patients were increased significantly as the disease becomes more severe for instance the medium ages were 45 years (moderate), 53 years (severe), and 66 years (critical), respectively. The demographic and clinical data of the study are summarized in Table 1 .
Table 1.
Demographic, Symptom, Arterial Blood Gas Test, and Routine Blood Test on Admission
| Moderate | Severe | Critical | |
|---|---|---|---|
| Number of patients | 25 | 47 | 27 |
| Age (y) (median, range) | 45 (24-67) | 53 (29-96) | 66 (36-90) |
| Mean ± SD | 46.6 ± 12.2 | 52.4 ± 14.3 | 65.6 ± 14.3 |
| Gender (M:F) | 12:13 | 28:19 | 18:9 |
| Symptom (count, %) | |||
| Illness days (mean ± SD, median) | 7.8 ± 5.8 (7) | 7.3 ± 4.0 (7) | 8.5 ± 3.9 (7) |
| Wu Han contact | 4 (16%) | 16 (34%) | 6 (22%) |
| Hypertension | 5 (20%) | 11 (23%) | 18 (67%) |
| Diabetes | 0 (0%) | 2 (4%) | 4 (15%) |
| History of surgery | 1 (4%) | 0 (0%) | 3 (11%) |
| Coronary heart disease | 0 (0%) | 3 (6%) | 2 (7%) |
| Hepatitis B | 2 (8%) | 1 (2%) | 1 (4%) |
| Fever | 21 (84%) | 38 (81%) | 23 (85%) |
| Chill | 1 (4%) | 4 (9%) | 3 (11%) |
| Cough and sputum | 16 (64%) | 29 (62%) | 17 (63%) |
| Dizziness and headache | 1 (4%) | 0 (0%) | 2 (7%) |
| Fatigue | 1 (4%) | 9 (19%) | 5 (19%) |
| Body aches | 4 (16%) | 7 (15%) | 2 (7%) |
| Chest tightness | 3 (12%) | 8 (17%) | 5 (19%) |
| Diarrhea | 0 (0%) | 3 (6%) | 4 (15%) |
| Routine blood test (mean ± SD, median) | |||
| White blood cell count | 6.3 ± 3.9 (5.1) | 7.2 ± 4.3 (6.0) | 9.3 ± 5.6 (7.8) |
| Lymphocyte percentage | 0.197 ± 0.115 (0.199) | 0.161 ± 0.112 (0.138) | 0.104 ± 0.069 (0.089) |
| Eosinophil ratio | 0.025 ± 0.070 (0.001) | 0.012 ± 0.052 (0.000) | 0.000 ± 0.001 (0.000) |
| Neutrophil | 4.8 ± 3.9 (3.4) | 5.9 ± 4.1 (4.4) | 8.1 ± 5.5 (7.2) |
| Lymphocyte count | 1.0 ± 0.5 (1.0) | 0.9 ± 0.5 (0.8) | 0.7 ± 0.4 (0.6) |
| Eosinophil count | 0.027 ± 0.073 (0.000) | 0.010 ± 0.030 (0.000) | 0.080 ± 0.382 (0.000) |
| C-reactive protein | 25.6 ± 35.3 (13.3) | 36.9 ± 38.0 (25.1) | 48.0 ± 40.6 (50.0) |
| Arterial blood gas test (mean ± SD, median) | |||
| Blood oxygen saturation | 96.9 ± 4.4 (98.2) | 96.1 ± 7.4 (97.8) | 95.9 ± 2.7 (96.3) |
| PO2 | 117.9 ± 38.9 (106.5) | 107.6 ± 40.9 (91.6) | 91.4 ± 37.8 (84.4) |
| PCO2 | 35.9 ± 3.2 (35.0) | 36.5 ± 4.4 (36.8) | 35.4 ± 5.2 (35.2) |
| PH | 7.4 ± 0.0 (7.4) | 7.4 ± 0.0 (7.4) | 7.4 ± 0.0 (7.4) |
| Activated partial prothrombin time | 31.7 ± 3.5 (31.3) | 32.2 ± 4.0 (31.9) | 32.8 ± 6.1 (33.6) |
| Prothrombin time | 12.2 ± 1.5 (12.1) | 11.9 ± 1.2 (11.7) | 11.9 ± 0.9 (11.8) |
| D-dimer | 403.3 ± 430.4 (281.0) | 553.6 ± 550.9 (380.0) | 2530.3 ± 9070.3 (608.0) |
| Lactate dehydrogenase | 244.0 ± 71.4 (223.0) | 267.0 ± 81.0 (246.0) | 341.7 ± 134.4 (330.0) |
| Phosphocreatine kinase | 97.6 ± 86.8 (60.0) | 96.8 ± 70.6 (70.0) | 143.1 ± 132.6 (97.0) |
| Creatine kinase isoenzyme | 22.1 ± 9.9 (19.0) | 18.8 ± 4.6 (20.0) | 22.7 ± 11.4 (22.0) |
| ALT | 38.5 ± 37.7 (26.0) | 29.1 ± 40.7 (20.0) | 25.2 ± 18.7 (19.0) |
| AST | 29.8 ± 24.4 (22.0) | 26.3 ± 18.0 (20.0) | 31.9 ± 28.6 (25.0) |
| Serum creatinine | 74.3 ± 22.9 (72.0) | 76.6 ± 13.5 (75.0) | 119.7 ± 183.8 (79.0) |
| Blood urea nitrogen | 5.0 ± 2.6 (4.5) | 5.8 ± 2.7 (5.2) | 7.9 ± 6.7 (5.6) |
| Procalcitonin | 0.09 ± 0.18 (0.05) | 0.08 ± 0.12 (0.06) | 0.23 ± 0.41 (0.07) |
Of 14 clinical symptoms, fever, cough, and sputum were high frequent symptoms in all three groups, as well as the same illness days to hospital visit. Comparing to moderate and sever groups, critical group had significantly higher ratio of hypertension, diabetes, and diarrhea. Whereas moderate group had lower ratio of diabetes, heart disease, fatigue, chest tightness, and diarrhea. In routine blood test, while the disease progressed from moderate to critical, we observed that white blood cell count (p = 0.0325), Neutrophil (p = 0.0179), C-reactive protein (p = 0.0153) were significantly high in the critical group, whereas lymphocyte count (p = 0.0446) was significantly lower. In arterial blood gas test, we observed that blood oxygen saturation (p = 0.219), PO2 (p = 0.0141) were decreased, whereas D-dimer (p = 0.234), phosphocreatine kinase (p = 0.147), and blood urea nitrogen (p = 0.0413) were increased as disease became more severe.
Of 99 patients, 32 patients were admitted to the ICU, out of which 1 (1/25) was in moderate, 6 (6/47) in severe, and 25 (25/27) in critical group, respectively. Table 2 lists the distribution of the duration of hospitalization (days), oxygen inhalation (days), ICU stay (days), and sputum NAT-positive (days) in terms of disease severity and the clinical outcomes. The median lengths of the hospitalization duration were 12, 16.5, and 28 days, the oxygen inhalation 10, 15, and 28 days, the ICU stay 0, 0, 12 days, and the sputum NAT-positive 7.5, 9, 13 days, respectively. These outcomes were statistically significant different with p value <0.05 except for duration of ICU (moderate vs severe, p = 0.211), oxygen inhalation (moderate vs severe, p = 0.0814), hospitalization duration (moderate vs severe, p = 0.0747), sputum NAT-positive (moderate vs severe, p = 0.486; severe vs critical, p = 0.106).
Table 2.
Clinical Outcomes in Terms of Disease Severity and Prognosis
| Disease Severity | Moderate | Severe | Critical |
|---|---|---|---|
| Hospitalization (days) | 14.60 ± 7.57 (Median: 12) | 18.11 ± 8.20 (Median: 16) | 33.26 ± 17.38 (Median: 28) |
| ICU length (days) | 0.24 ± 1.20 (Median: 0) (1pt) | 1.34 ± 5.72 (Median: 0) (6pt) | 25.07 ± 23.64 (Median: 12) (25pt) |
| Oxygen inhalation (days) | 13.08 ± 8.10 (Median: 10) | 16.64 ± 8.03 (Median: 15) | 32.93 ± 17.62 (Median: 28) |
| Sputum NAT-positive (days) | 9.56 ± 7.56 (Median: 8) | 10.89 ± 7.88 (Median: 9) | 13.70 ± 6.60 (Median: 13) |
| Prognosis (category) | 1.92 ± 0.28 (Median: 2) | 2.00 ± 0.21 (Median: 2) | 2.59 ± 0.57 (Median: 3) |
| (Prognosis, Complete Recovery, Partial Recovery, Prolonged Recovery) | |||
| Hospitalization (days) | 14.7 ± 2.3 (Median: 16) | 16.7 ± 7.3 (Median: 15) | 46.6 ± 12.1 (Median: 50) |
| ICU length (days) | 0.0 ± 0.0 (Median: 0) | 1.0 ± 2.3 (Median: 0) | 42.3 ± 18.6 (Median:49) |
| Oxygen inhalation (days) | 13.7 ± 2.5 (Median: 14) | 15.4 ± 7.3 (Median: 15) | 46.4 ± 12.4 (Median: 50) |
| Sputum NAT-positive (days) | 8.7 ± 3.2 (Median: 10) | 10.4 ± 7.6 (Median: 8) | 15.8 ± 4.6 (Median: 17) |
As per disease severity, 99 patients were categorized into three groups: moderate (n = 25), severe (n = 47), and critical (n = 27), respectively. As per prognosis, patients were categorized into 1: complete recovery (n = 3), 2: partial recovery with residual pulmonary damage (n = 80), 3: prolonged recovery (n = 15), and 4: death (n = 1).
The clinical prognoses were complete recovery (n = 3), partial recovery with residual pulmonary damage (n = 80), prolonged recovery (n = 15), and death (n = 1), respectively. The differences between partial recovery and prolonged recovery groups were statistically significant including the ICU days (p < 0.0001), the oxygen inhalation days (p < 0.0001), the hospitalization days (p < 0.0001), and the days of sputum NAT-positive (p = 0.0008). The numbers of patients in the complete recovery (n = 3) and death (n = 1) were too small to calculate the statistical significance.
Volumetric Image Analysis of COVID-19 Pneumonia
We evaluated the performance of our two trained U-Net models for segmentation of lung and lesions by using 99 chest CT scans in this study. The mean DSCs were 0.981 for the lung segmentation, and 0.778 for the lesion segmentation. In the moderate, sever, and critical groups, the average DSCs were 0.990, 0.987, 0.961 for segmentation of lungs, and 0.746, 0.790, 0.826 for segmentation of lesions, respectively. Table 3 lists the performance of lung and lesion segmentation. In general, lung segmentation performed better in less severely diseased groups, whereas lesion segmentation performed better in more severely diseased group in term of DSC and Jaccard index. With the disease progression, a decrease in GGO and an increase in consolidation were observed. In terms of relative volume difference (RVD), these differences changed from over-segmentation (positive RVD) to under-segmentation (negative RVD). On average, the segmentation process of each CT case took 0.8 seconds.
Table 3.
Performance of U-Net Models for Segmentation of Lung and Lesions
| Lung Segmentation | DSC | Jaccard | RVD |
|---|---|---|---|
| Overall | 0.981 | 0.965 | −0.46% |
| Moderate | 0.990 | 0.980 | 0.63% |
| Severe | 0.987 | 0.975 | 0.16% |
| Critical | 0.961 | 0.935 | −2.56% |
| (Lesion Segmentation, DSC, Jaccard, RVD) | |||
| Overall | 0.778 | 0.663 | 2.1% |
| Moderate | 0.746 | 0.619 | 3.1% |
| Severe | 0.790 | 0.671 | 2.6% |
| Critical | 0.826 | 0.723 | −3.3% |
Note:
• Dice similarity coefficient (DSC): 2*TP / ( 2*TP + FP + FN ).
• Jaccard index: TP / ( TP + FP + FN ).
• Relative volume difference (RVD): (Vol(res) / Vol(ref) -1) *100%.
res: automated segmentation results exported by U-Net.
ref: reference segmentation results contoured by radiologists.
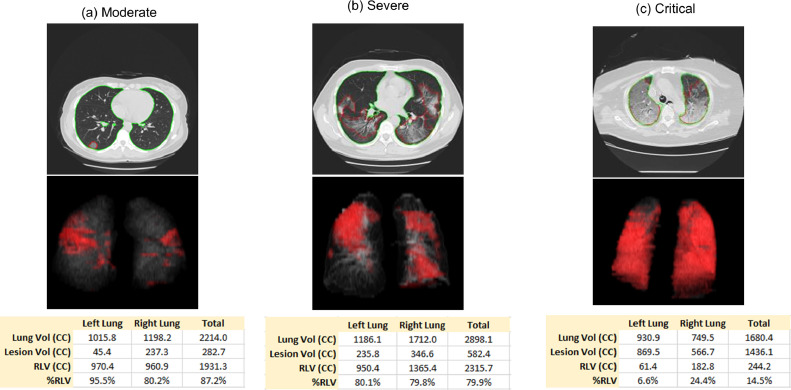
The lung volume, lesion volume, NLLV, and %NLLV were calculated based on the segmentation results. Figure 3 shows three example cases, one for each severity group. The quantification of disease in terms of disease severity and prognosis are listed in Table 4 . There were no statistically significant differences of lung volume with p values of 0.269 (moderate vs severe), 0.125 (moderate vs critical), and 0.437 (severe vs critical). The lesion volume increased significantly, whereas NLLV and %NLLV decreased significantly when disease became more severe with p value <0.05 among each two groups (with the only exception of NLLV between moderate vs severe where p = 0.0575). In addition, the lesion volume, NLLV, and %NLLV were statistically significant different between partial recovery and prolonged recovery groups with pvalue <0.05. The number of patients in the complete recovery (n = 3) and death (n = 1) were too small to calculate the statistical significance.
Figure 3.
Examples of resulting images of lung and lesions segmentation. (Color version of figure is available online.)
Table 4.
CT Quantification of Lung Volume, Lesion Volume, Nonlesion Lung Volume (NLLV) (Lung Volume – Lesion Volume) and Fraction of NLLV (%NLLV) (Nonlesion Lung Volume / Lung Volume), as well as the Mean CT Value of Lesions and NLLV in Terms of Disease Severity and Prognosis
| Disease Severity | Moderate | Severe | Critical |
|---|---|---|---|
| Lung volume (CC) | 3988.60 ± 1504.96 (Median: 3547.33) | 3613.43 ± 989.67 (Median: 3589.73) | 3412.80 ± 1098.01 (Median: 3451.62) |
| Lesion volume (CC) | 260.95 ± 279.30 (Median: 179.26) | 578.32 ± 569.79 (Median: 358.94) | 1022.59 ± 707.11 (Median: 880.90) |
| Nonlesion lung volume (NLLV) (CC) | 3727.64 ± 1556.42 (Median: 3386.40) | 3035.11 ± 1151.38 (Median: 2936.06) | 2390.21 ± 1387.89 (Median: 2285.69) |
| Fraction of nonlesion lung volume (%NLLV) (%) | 92.18 ± 9.89 (Median: 96.92) | 82.94 ± 16.49 (Median: 87.54) | 66.19 ± 24.15 (Median: 67.70) |
| Mean CT value of lesion (HU) | −534.1 ± 123.1 (Median: −560.9 ) | −484.4 ± 117.5 (Median: −487.7) | −462.6 ± 126.9 (Median: −495.1) |
| Mean CT value of NLLV (HU) | −785.6 ± 43.5 (Median: −787.5) | −768.2 ± 39.9 (Median: −775.2) | −750.6 ± 67.6 (Median: −760.1) |
| Prognosis | Complete Recovery | Partial Recovery | Prolonged Recovery |
| Lung volume (CC) | 5222.44 ± 2659.42 (Median: 5349.15) | 3658.69 ± 1090.57 (Median: 3578.03) | 3273.01 ± 1108.67 (Median: 3075.67) |
| Lesion volume (CC) | 140.30 ± 220.81 (Median: 14.38) | 527.24 ± 509.20 (Median: 346.45) | 1224.90 ± 843.52 (Median: 1013.60) |
| Nonlesion lung volume (NLLV) (CC) | 5082.13 ± 2476.64 (Median: 5337.89) | 3131.45 ± 1250.39 (Median: 3017.44) | 2048.11 ± 1413.66 (Median: 1868.99) |
| Fraction of nonlesion lung volume (%NLLV) (%) | 98.05 ± 2.70 (Median: 99.43) | 84.07 ± 15.66 (Median: 88.53) | 58.59 ± 27.50 (Median: 61.44) |
| Mean CT value of lesion (HU) | −620.5 ± 130.7 (Median: −660.3) | −489.8 ± 122.7 (Median: −494.8) | −476.4 ± 120.9 (Median: −503.7) |
| Mean CT value of NLLV (HU) | −789.1 ± 60.0 (Median: −817.9) | −773.3 ± 43.3 (Median: −775.3) | −730.7 ± 71.4 (Median: −742.8) |
Among three severity groups, the mean CT values of lesions between groups of moderate vs critical were significantly increased (p = 0.0444), whereas the increase of CT values between other two groups (moderate vs severe, p = 0.104; severe vs critical, p = 0.469) were not statistically significant. For the mean CT values of NLLV, we observed an increase for the more severely diseased group; however, the differences between any two groups were not statistically significant (moderate vs severe, p = 0.103; severe vs critical, p = 0.226) except (moderate vs critical; p = 0.0305). Between the partial recovery and prolonged recovery groups, the increase of mean CT values of NLLV were statistically significant (p = 0.0398), whereas those of lesions were not statistically significant (p = 0.697).
Classification of Disease Severity
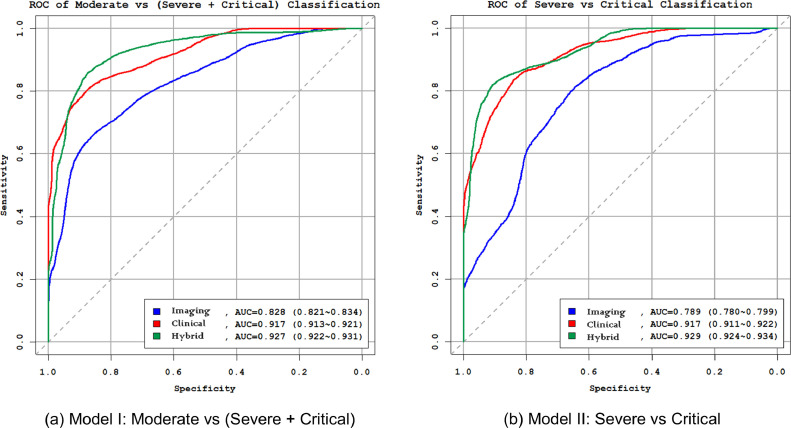
We applied the feature selection method to build the RF models for classification of disease severity. Table 5 lists the top five selected features, if any, and the performances of the radiomics models, clinical data models, and the hybrid models. Figure 4 shows the receiver operating characteristic curves of three models for classification of disease severity.
Table 5.
The Selected Features and the Performance for Classification of Moderate vs (Severe + Critical) (Model I), and Severe vs Critical (Model II)
| Model I: Moderate vs (Severe + Critical) | Model II: Severe vs Critical | |||
|---|---|---|---|---|
| Selected features (radiomics) |
|
|
||
| Performance (radiomics) | AUC | 0.828 (0.821-0.834) | AUC | 0.789 (0.780-0.799) |
| Specificity | 0.703 | Specificity | 0.662 | |
| Sensitivity | 0.797 | Sensitivity | 0.791 | |
| Accuracy | 0.750 (0.743-0.757) | Accuracy | 0.726 (0.717-0.735) | |
| Selected features (clinical) |
|
|
||
| Performance (clinical) | AUC | 0.917 (0.913-0.921) | AUC | 0.917 (0.911-0.922) |
| Specificity | 0.801 | Specificity | 0.854 | |
| Sensitivity | 0.877 | Sensitivity | 0.812 | |
| Accuracy | 0.839 (0.833-0.845) | Accuracy | 0.833 (0.826-0.841) | |
| Selected features (hybrid) |
|
|
||
| Performance (hybrid) | AUC | 0.927 (0.922-0.931) | AUC | 0.929 (0.924-0.934) |
| Specificity | 0.809 | Specificity | 0.872 | |
| Sensitivity | 0.901 | Sensitivity | 0.842 | |
| Accuracy | 0.855 (0.849-0.860) | Accuracy | 0.857 (0.850-0.864) | |
Note: a suffix with “residual” indicates the nonlesion lung.
Figure 4.
Receiver operating characteristic (ROC) curves of radiomics, clinical and multiomics models for classification of disease severity: (a) Model I: moderate vs (severe + critical); (b) Model II: severe vs critical. (Color version of figure is available online.)
First, to build the radiomics models for classification of moderate vs (severe + critical) (Model I radiomics), and severe vs critical (Model II radiomics), we applied our feature selection methods to histogram features calculated from the lesions and NLLVs including the quantification of disease (lesion volume, NLLV, and %NLLV). Five and three features were identified for two models, respectively. In both models, %NLLV was selected in the top five important features. The AUCs of two models were 0.828 and 0.789.
Second, we applied the same feature selection procedure to clinical data to build the clinical models for classification of moderate vs (severe + critical) (Model I clinical), and severe vs critical (Model II clinical). In both models, age was identified in both models. Top five features were identified for the two clinical models, of which the AUCs were 0.917 for both models.
Third, we established the hybrid models by mixing both the radiomics feature and clinical features and using the same feature selection procedure. Histogram uniformity in NLLV was the only image feature that was reserved in Model I, whereas both %NLLV and NLLV were selected in Model II. The AUCs were 0.927 and 0.929 for (Model I hybrid) and (Model II hybrid), respectively. This performance was significantly higher than either the radiomic models or clinical models with p < 0.01.
Prediction of Clinical Outcomes and Prognosis
We applied the RF classification and regression models for the prediction of various clinical outcomes: need and duration of ICU, duration of hospitalization, duration of oxygen inhalation, duration of sputum NAT-positive, and the prediction of prognosis. Table 6 lists the top five selected features and the prediction performance of four RF regression models and two RF classification models. We observed that %NLLV was among the top five features in four out of six RF models, which was ranked second after age. %NLLV was the only selected quantification biomarker related to the disease size. In addition, we observed that some histogram features such as variance, mean absolute deviation of NLLV, had significant contributes to the prediction of duration of ICU and recovery.
Table 6.
Comparison of Model Performance With and Without CT Features for Prediction of Clinical Outcomes
| Clinical Outcomes | Selected features | Performance |
||
|---|---|---|---|---|
| With CT Features | Without CT Features | |||
| Duration of hospitalization (≤4 weeks) | %NLLV * (12) | CV RMSE | 0.878 | 0.916 |
| Age (11) | ||||
| Creatine_kinase_isoenzyme (7.9) | ||||
| Hypertension (6.9) | ||||
| PO2 (6.9) | ||||
| Duration of oxygen inhalation (≤4 weeks) | Age (15) | CV RMSE | 0.920 | 0.940 |
| %NLLV * (11) | ||||
| Creatine_kinase_isoenzyme (8.1) | ||||
| PO2 (7.6) | ||||
| Procalcitonin (6.7) | ||||
| Duration of sputum nucleic acid test positive (≤4 weeks) | Age (14) | CV RMSE | 0.901 | NA |
| History_of_surgery (7.8) | ||||
| Creatine_kinase_isoenzyme (7.0) | ||||
| Need of ICU | Age (23) | |||
| Procalcitonin (17) | AUC | 0.945 (0.941-0.948) | 0.932 (0.927-0.936) | |
| Hypertension (16) | Specificity | 0.843 | 0.845 | |
| %NLLV * (12) | Sensitivity | 0.884 | 0.879 | |
| C_reactive_protein (11) | Accuracy | 0.864 (0.858-0.870) | 0.862 (0.856-0.868) | |
| Duration of ICU (≤2 weeks) | HIST_var_lesion * (5.9) | CV RMSE | 0.688 | 0.798 |
| Blood_oxygen_saturation (5.3) | ||||
| Prediction of prognosis (partial recovery vs prolonged recovery) | Age (19) | |||
| %NLLV * (15) | AUC | 0.960 (0.957-0.963) | 0.806 (0.800-0.813) | |
| HIST_mad_residual * (12) | Specificity | 0.892 | 0.792 | |
| HIST_kurt_residual * (9.4) | Sensitivity | 0.907 | 0.659 | |
| HIST_quant_range_residual * (8.5) | Accuracy | 0.899 (0.895-0.904) | 0.726 (0.719-0.733) | |
Features with suffix “residual” are those for the nonlesion lung. CT features are marked with asterisks. Numbers in the parenthesis are the importance of the features, obtained from Boruta's selection process. For regression problems, the importance was evaluated based on the mean increase in MSE. For classification problems, it was based on the mean decrease in accuracy.
For the duration of hospitalization, duration of oxygen inhalation and the duration of sputum NAT-positive, we applied the upper bound of 4 weeks to clamp the maximum values. The RF regression models achieved an RMSE between 0.69 and 0.92 weeks, that is, approximately ±5-7 days of length of stay. For the binary classification problem to determine if a patient shall be admitted to ICU, the RF classification models achieved an AUC of 0.945. For the 32 patients treated in ICU, the RF regression model to predict the length of ICU stay achieved an RMSE of 0.69 weeks, approximately ±5 days, using upper bound of 2 weeks. To clarify, the RMSEs reported were derived from the repeated 10-fold cross-validation to avoid underestimation that would otherwise occur if the performance is evaluated simply by applying the RF models to the full training set. Although the RMSEs of our RF models are not found to be considerably smaller than the standard deviation, they are, however, consistently better than the values obtained by other ML methods such as Least Absolute Shrinkage and Selection Operator regression (32).
Due to the limited number of patients in the prognosis of complete recovery (n = 3) and death (n = 1) group, we only performed the prediction of partial recovery and prolonged recovery. The RF model achieved a high AUC of 0.960 for prediction of patient prognosis.
DISCUSSION
In this study, we investigated the AI-assisted CT quantification of COVID-19 pneumonia and the ML models on stratification of disease severity and prediction of clinical outcomes in the management of COVID-19 patients. Deep-learning-based U-Net architecture provides a feasible and efficient technique to detect and segment pneumonia lesions in CT images. More importantly, ML models may provide an accurate assessment of disease severity and prediction of clinical outcomes for the decision-making in the management of COVID-19 patients.
Chest CT findings tend to be used as one of clinical manifestations in the confirmation of the diagnosis of COVID-19 infection (8). Many clinical studies have extensively investigated the CT imaging signs related to COVID-19 infection such as GGO, GGO with lung consolidation, interlobular septal thickening, and pulmonary fibrosis for patients at different stages and severity (9, 10, 11). Although chest CT has high sensitivity in identifying COVID-19 infection, it has low specificity to discriminate from other viral phenomena (22,33). Majority of the recently published studies focused on the detection and diagnosis of COVID-19 such as using U-Net for automated detection of GGO areas (34,35), differentiation of COVID-19 pneumonia from other viral pneumonia using radiomics or deep-learning methods (36,37). However, there have been few studies to investigate the validity of CT for assisting decision-making in the management of COVID-19: stratification of disease severity and prediction of clinical outcomes.
To investigate the disease progression, qualitative evaluation method was developed in terms of the percentage of lung involvement of the abnormal findings on a scale of 0-4 (38), and found CT score differed significantly between different stages that disease progression was associated with both increased numbers and sizes of GGO combined with consolidative opacities and septal thickening. The combination of radiological indices such as mass of infection and percentage of infection with other clinical data to predict disease severity (severe vs nonsevere) of COVID-19 achieved an performance of AUC of 0.89 (39).
CT quantitation and ML models demonstrated potentials for prediction of disease severity or clinical outcomes. Our study achieved a better performance to stratify the severity into moderate, sever, and critical groups with AUC more than 0.925. According to the CHC infection (8), severe groups show obvious lesion progression >50% within 24-48 hours in CT. Our models may precisely classify severe or critically ill patients using one CT scan on admission, thereby saving time on the second follow-up CT scan and patients may receive early treatment.
Although studies reported that 97% of COVID-19 patients showed positive in CT by using an AI-based detection model (22); however, some studies also found that the typical CT signs such as GGO might not observed in some mild or asymptomatic patients that were confirmed by positive NAT of RT-PCR (12). Thus, NAT is still the gold standard for clinical diagnosis of COVID-19. Considering the fact that CT has high sensitivity in identifying GGO lesions, we believe that CT is being and will be used in the management of severely ill patients in particular for advanced progressive cases or those with complications for the decision-making of ICU treatment, and the prediction of ICU stay. This will directly assist physicians in the management of COVID-19 patients. This is also the primary aim of our study to demonstrate the validity of CT quantification in the decision-making and prediction of clinical outcomes in the management of COVID-19 patients.
We investigated the ML models combining CT quantification and image textures in classifying disease severity with AUC >0.925, prediction of clinical outcomes such as the need of ICU treatment on admission with AUC of 0.945, and the prediction of prognosis of partial recovery vs prolonged recovery with AUC of 0.960. In addition, we demonstrated that neither radiomics models nor clinical models could achieve this high performance as hybrid models. This may indicate that some of the disease characteristics might not be captured by CT imaging alone. For instance, age has been selected by both classification models in Table 5, and five out of six models in Table 6. For better understanding the importance of the CT features in the predicting outcomes of the disease, we compared the performance between the models with the CT features and without CT features in Table 6. CT features were found to offer consistent performance improvement for ML models. For the three regression models, the RMSE (lower value indicates better performance) became higher if the models were trained without the CT features. In particular, the model to predict the duration of ICU was degraded in performance by as much as 16%. For the two binary classification problems, the AUC and accuracy (higher value indicates better performance) were both negatively affected by removal of CT features. Whereas the model to predict the need of ICU became slightly worse, performance of the model to predict prognosis dropped precipitously. We will explore comprehensive textures instead of only histogram features in future.
Considering the imbalanced number of patients in each group, which may bias our observations due to "within-patient clustering" artifact, we used the Synthetic Minority Oversampling Technique resampling method (31) to balance the radiomics sampling number in the statistical analysis. As a result, larger and less specific regions are learned, thus, paying attention to minority class samples without causing overfitting and bias in training ML models. Thus, our models are stable and not biased to the number of patients in positive and negative groups.
This study had several limitations. The first limitation was the relatively small number of cases for sufficiently training of our U-Net models for lesion segmentation. This caused some of the interaction efforts for modifying the results of the automated segmentation. We will collect more cases to improve the accuracy of the U-Net models for lesion segmentation, as well as using other deep-learning models such as ResNet to classify COVID-19 lesions from other viral pneumonia lesions and lung tissues. In addition, since the segmentation of lung had a very high accuracy, we will also work on the imaging biomarker calculated in the lung region instead of lesions for classification and prediction. Another limitation is that our study used single-center data. We plan to collect multicenter cases to train and validate our models for segmentation and prediction. Our models need multicenter data for further external validation.
Overall, the validity of CT in the management of COVID-19 patients has been held back by its controversial specificity in the diagnosis of COVID-19 pneumonia, whereas NAT remains the gold standard of the diagnosis. Our studies imposed that AI-assisted CT quantification and ML models may be an effective tool assisting the decision-making in the management of hospitalized patients such as prediction of ICU treatment, the duration of oxygen inhalation, and prognosis, which are critical questions in the management of patients, in particular for severely or critically ill patients. We observed that %NLLV and other imaging textures are significant imaging biomarkers in the management of the COVID-19, and ML models may achieve significance high performance for prediction of clinical outcomes.
Although the findings of this study warrant validation by larger multicenter studies, it may provide a new dimension for investigating the validity of CT focusing on clinical management of hospitalized severely ill patients, including the decision-making of ICU treatment, the duration of ICU, oxygen inhalation, and hospitalization, which are essential for clinical management.
Acknowledgments
The authors thank Mark Vangel, PhD, from the General Clinical Research Center at Massachusetts General Hospital for statistical consultant with this manuscript.
This study has partly funded by internal department funds of Massachusetts General Hospital and Zhejiang University School of Medicine the First Affiliated Hospital.
Appendix 1 The Abbreviation of Histogram Texture Features
| Abbreviation | Texture Feature Name |
|---|---|
| HIST_mpp | histogram_mean positive value |
| HIST_energy | histogram_energy |
| HIST_rms | histogram_root mean square |
| HIST_uniformity | histogram_uniformity |
| HIST_entropy | histogram_entropy |
| HIST_kurt | histogram_kurtosis |
| HIST_skew | histogram_skewness |
| HIST_mean | histogram_mean |
| HIST_median | histogram_median |
| HIST_min | histogram_minimum |
| HIST_max | histogram_maximum |
| HIST_range | histogram_range |
| HIST_var | histogram_variance |
| HIST_std | histogram_standard deviation |
| HIST_mad | histogram_mean absolute deviation |
| HIST_quant0.25 | histogram_quantile0.25 |
| HIST_quant0.75 | histogram_quantile0.75 |
| HIST_quant0.025 | histogram_quantile0.025 |
| HIST_quant0.975 | histogram_quantile0.975 |
| HIST_quant_range | histogram_quantile_range |
References
- 1.Huang C, Wang Y, Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munster VJ, Koopmans M, van Doremalen N. A novel coronavirus emerging in China—key questions for impact assessment. N Engl J Med. 2020;382:692–694. doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- 3.WHO Director General's Remarks at the Media Briefing on 2019-nCoV on 11 February 2020. 2020. Available at: https://www.who.int/dg/speeches/detail/who-director-general-s-remarks-at-the-media-briefing-on-2019-ncov-on-11-february-2020.)
- 4.Cucinotta D, Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.COVID-19 coronavirus pandemic. Available at: https://wwwworldometersinfo/coronavirus/2020.
- 6.World Health Organization, Clinical management of severe acute respiratory infection when novel coronavirus (2019-nCoV) infection is suspected: interim guidance. 2020. Available at: https://www.who.int/publications/i/item/clinical-management-of-covid-19.
- 7.Centers for Disease Control and Prevention, CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel. 2020. Available at: https://www.fda.gov/media/134922/download.
- 8.Office of State Administration of Traditional Chinese Medicine. Notice on the issuance of a program for the diagnosis and treatment of novel coronavirus (2019-nCoV) infected pneumonia (trial sixth edition) (2020-02-19). 2020. Available at: http://yzs.satcm.gov.cn/zhengcewenjian/2020-02-19/13221.html.)
- 9.Chung M, Bernheim A, Mei X. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295:202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Y, Gao ZH, Liu YL. Clinical and CT imaging features of 2019 novel coronavirus disease (COVID-19) J Infect. 2020;81:147–178. doi: 10.1016/j.jinf.2020.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi H, Han X, Jiang N. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20:425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling Z, Xu X, Gan Q. Asymptomatic SARS-CoV-2 infected patients with persistent negative CT findings. Eur J Radiol. 2020;126 doi: 10.1016/j.ejrad.2020.108956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Z, Song C, Xu C. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci. 2020;63:706–711. doi: 10.1007/s11427-020-1661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.ACR recommendations for the use of chest radiography and computed tomography (CT) for suspected COVID-19 infection. 2020. Available at:https://www.acr.org/Advocacy-and-Economics/ACR-Position-Statements/Recommendations-for-Chest-Radiography-and-CT-for-Suspected-COVID19-Infection.)
- 15.Dias OM, Baldi BG, Pennati F. Computed tomography in hypersensitivity pneumonitis: main findings, differential diagnosis and pitfalls. Expert Rev Respir Med. 2018;12:5–13. doi: 10.1080/17476348.2018.1395282. [DOI] [PubMed] [Google Scholar]
- 16.Galban CJ, Han MK, Boes JL. Computed tomography-based biomarker provides unique signature for diagnosis of COPD phenotypes and disease progression. Nat Med. 2012;18:1711–1715. doi: 10.1038/nm.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartholmai BJ, Raghunath S, Karwoski RA. Quantitative computed tomography imaging of interstitial lung diseases. J Thorac Imaging. 2013;28:298–307. doi: 10.1097/RTI.0b013e3182a21969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Claessens YE, Debray MP, Tubach F. Early chest computed tomography scan to assist diagnosis and guide treatment decision for suspected community-acquired pneumonia. Am J Respir Crit Care Med. 2015;192:974–982. doi: 10.1164/rccm.201501-0017OC. [DOI] [PubMed] [Google Scholar]
- 19.Garin N, Marti C, Scheffler M. Computed tomography scan contribution to the diagnosis of community-acquired pneumonia. Curr Opin Pulm Med. 2019;25:242–248. doi: 10.1097/MCP.0000000000000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong CK, Lai V, Wong YC. Comparison of initial high resolution computed tomography features in viral pneumonia between metapneumovirus infection and severe acute respiratory syndrome. Eur J Radiol. 2012;81:1083–1087. doi: 10.1016/j.ejrad.2011.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ooi GC, Daqing M. SARS: radiological features. Respirology. 2003;8(suppl):S15–S19. doi: 10.1046/j.1440-1843.2003.00519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ai T, Yang Z, Hou H. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296:e32–e40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng Z, Qin L, Cao Q. Quantitative computed tomography of the coronavirus disease 2019 (COVID-19) pneumonia. Radiol Infect Dis. 2020;7:55–61. doi: 10.1016/j.jrid.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee KS. Pneumonia associated with 2019 novel coronavirus: can computed tomographic findings help predict the prognosis of the disease? Korean J Radiol. 2020;21:257–258. doi: 10.3348/kjr.2020.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salehi S, Abedi A, Balakrishnan S. Coronavirus disease 2019 (COVID-19): a systematic review of imaging findings in 919 patients. AJR Am J Roentgenol. 2020;215:87–93. doi: 10.2214/AJR.20.23034. [DOI] [PubMed] [Google Scholar]
- 26.Zu ZY, Jiang MD, Xu PP. Coronavirus Disease 2019 (COVID-19): A Perspective from China. Radiology. 2020;296:e15–e25. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong D, Tang Z, Wang S. The role of imaging in the detection and management of COVID-19: a review. IEEE Rev Biomed Eng. 2020 doi: 10.1109/RBME.2020.2990959. [DOI] [PubMed] [Google Scholar]
- 28.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Navab N., Hornegger J., Wells W., Frangi A., editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI. Lecture Notes in Computer Science, vol 9351; Springer, Cham: 2015. [DOI] [Google Scholar]
- 29.Sonka M, Hlavac V, Boyle R. 3rd ed. Thompson Learning; Toronto: 2008. Image processing, analysis, and machine vision. [Google Scholar]
- 30.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 31.Chawla NV, Bowyer KW, Hall LO. SMOTE: Synthetic Minority Over-sampling Technique. J Artif Intell Res. 2002;16:321–357. [Google Scholar]
- 32.Tibshirani R. Regression shrinkage and selection via the LASSO. J R Stat Soc B. 1996;58:267–288. [Google Scholar]
- 33.Yang W, Sirajuddin A, Zhang X. The role of imaging in 2019 novel coronavirus pneumonia (COVID-19) Eur Radiol. 2020;30:4874–4882. doi: 10.1007/s00330-020-06827-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Wu L, Zhang J. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study in 27 patients. medRxiv. 2020 doi: 10.1101/2020.02.25.20021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng C, Deng X, Fu Q. Deep learning-based detection for COVID-19 from chest CT using weak label. IEEE Transactions on Medical Imaging, 2020;39:2615–2625. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- 36.Fang M, He B, Li L. CT radiomics can help screen the coronavirus disease 2019 (COVID-19): a preliminary study. Sci China Inf Sci. 2020;63 172103. [Google Scholar]
- 37.Li L, Qin L, Xu Z. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020;296:e65–e71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang X, Zhao M, Li S. Changes of CT findings in a 2019 novel coronavirus (2019-nCoV) pneumonia patient. QJM. 2020;113:271–272. doi: 10.1093/qjmed/hcaa038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi W, Peng X, Liu T. Deep learning-based quantitative computed tomography model in predicting the severity of COVID-19: a retrospective study in 196 patients. SSRN Electron J. 2020 doi: 10.2139/ssrn.3546089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Anonymized data will be shared by request from any qualified investigators: (1) clinical data of 99 patients, (2) chest CT images of 99 patients, (3) segmentation results of lung and lesions of COVID-19 pneumonia of 99 patients, and (4) imaging features calculated from the segmented lesions and NLLVs.