Abstract
Background
An outbreak of a febrile respiratory illness due to the newly discovered Coronavirus, SARS-CoV-2, was initially detected in mid-December 2019 in the city of Wuhan, Hubei province (China). The virus then spread to most countries in the world. As an RNA virus, SARS-CoV-2 may acquire mutations that may be fixed. The aim of this study was to evaluate the selective pressure acting on SARS-CoV-2 protein coding genes.
Methods
Mutations and glycosylation site prediction were analyzed in SARS-CoV-2 genomes (from 464 to 477 sequences).
Results
Selective pressure on surface glycoprotein (S) revealed one positively selected site (AA 943), located outside the receptor binding domain (RBD). Mutation analysis identified five residues on the surface glycoprotein, with variations (AA positions 367, 458, 477, 483, 491) located inside the RDB. Positive selective pressure was identified in nsp2, nsp3, nsp4, nsp6, nsp12, helicase, ORF3a, ORF8, and N sub-sets. A total of 22 predicted N-glycosylation positions were found in the SARS-CoV-2 surface glycoprotein; one of them, 343N, was located within the RBD. One predicted N-glycosylation site was found in the M protein and 4 potential O-glycosylation sites in specific protein 3a sequences.
Conclusion
Overall, the data showed positive pressure and mutations acting on specific protein coding genes. These findings may provide useful information on: i) markers for vaccine design, ii) new therapeutic approach, iii) information to implement mutagenesis experiments to inhibit SARS-CoV-2 cell entry. The negative selection identified in SARS-CoV-2 protein coding genes may help the identification of highly conserved regions useful to implement new future diagnostic protocols.
Keywords: Bioinformatics, Microbiology, Public health, Virology, Infectious disease, Evolutionary biology, SARS-CoV-2, Selective pressure, Mutation, Glycosylation, Receptor binding domain
Bioinformatics; Microbiology; Public health; Virology; Infectious disease; Evolutionary biology; SARS-CoV-2; Selective pressure; Mutation; Glycosylation; Receptor binding domain
1. Introduction
Human coronaviruses (CoV) are enveloped positive-stranded RNA viruses belonging to the order Nidovirales, mostly responsible for upper respiratory and digestive tract infections (Fehr and Perlman, 2005).
An outbreak of a febrile respiratory illness due to the newly discovered Coronavirus (officially named by the World Health Organization as SARS-CoV-2) occurred in mid-December 2019, in the city of Wuhan, Hubei province (China). The virus spread to most countries in all the continents, causing a pandemic event (Wu et al., 2020; WHO a; WHO b).
Previous studies have examined the SARS-CoV-2 mutations, even though the studies were based on small sample size (Benvenuto et al., 2020; Phan, 2020; Tang et al., 2020; Pachetti et al., 2020).
At the molecular level, amino-acid changes that result in reduced fitness are generally removed by negative selection, whereas changes that increase virus fitness are maintained by positive selection. Differently, when amino-acid changes do not decrease or increase fitness, the changes are considered neutral. Thus, it is important to understand which sites evolve under selective pressure, especially in case of a new pathogen, because the presence of negative or positive selection implies that the sites are functionally important.
Hereby, we report data regarding the selective pressure on SARS-CoV-2 protein coding genes and their glycosylation site prediction on a large number of SARS-CoV-2 genomes (ranging from 464 to 477) downloaded from Gen Bank (NCBI, https://www.ncbi.nlm.nih.gov/pubmed) and from the GISAID platform (GISAID, https://www.gisaid.org/). We described the main results of a molecular evolutionary analysis aimed to: i) identify the selective pressure on the SARS-CoV-2 protein coding genes; ii) identify the mutations in SARS-CoV-2 surface glycoprotein (also known with the synonym: spike glycoprotein) sequences; iii) compare the specific positions belonging to the surface glycoprotein, among SARS-CoV-2, SARS-CoV and Bat SARS like sequences, previously reported to be critical for cross-species, human-to-human transmission in SARS-CoV (Li et al., 2005a, Li et al., 2005b); iv) evaluate and predict potential glycosylation sites, as already considered in the case of SARS-CoV (Chakraborti et al., 2005; Zhou et al., 2010).
2. Materials and methods
2.1. Phylogenetic analysis
A total of 500 SARS-CoV-2 sequences (complete and partial sequences) were downloaded from Gen Bank (NCBI, https://www.ncbi.nlm.nih.gov/pubmed) and GISAID database (GISAID, https://www.gisaid.org/) to constitute the starting dataset (Table S1) represented geographically and temporally and suitable in number to computational calculation time. To the purpose of selective pressure and mutation analysis the following protein –coding genes sequence sub-sets were defined, after excluding short sequences or those showing extensive presence of ambiguity codes: nsp1 (n = 468), nsp2 (n = 464), nsp3 (n = 464), nsp4 (n = 468), 3C-like proteinase (n = 468), nsp6 (n = 465), nsp7 (n = 469), nsp8 (n = 469), nsp9 (n = 467), nsp10 (n = 469), nsp11 (n = 460), nsp12 (n = 470), helicase (n = 469), 3′-to-5′-exonuclease (n = 468), endoRNAse (n = 466), 2′-O-ribose methyltransferase (n = 465), S (surface glycoprotein) (n = 460), ORF3a (n = 465), E (n = 468), M (n = 470), ORF6 (n = 469), ORF7a (n = 465), ORF8 (n = 467), N (n = 477) and ORF10 (n = 467). All the nucleotide sequence alignments were performed by using the multiple sequence alignment program MAFFT v.7 (Katoh and Standley, 2013) with the Galaxy platform (Galaxy, https://usegalaxy.org/; Afgan et al., 2018) and manually edited by Bioedit program (Hall, 1999).
2.2. Selective pressure analysis
The selective pressure analysis was performed on the above reported SARS-CoV-2 protein coding sequence sub-sets through the Datamonkey Adaptive Evolution Server (Delport et al., 2010; Pond and Frost, 2005a; Weaver et al., 2018), in order to characterize the SARS-CoV-2 variations, the evolutionary dynamics and to identify and localize statistically supported positive and negative selective pressure sites. If sites are statistically significant for a positive value of non synonymous to synonymous substitution ω > 1, positive diversifying selection is inferred, while purifying selection is inferred for ω < 1 (Zhang et al., 2005). On the contrary, neutrality is inferred for ω = 1 (Zhang et al., 2005). Three models were applied and the results were merged: i) FEL (Fixed Effects Likelihood): uses a maximum-likelihood (ML) approach to infer non- synoymous (dN) and synonymous (dS) substitution rates on a per-site basis for a given coding alignment and corresponding phylogeny. This method assumes that the selection pressure for each site is constant along the entire phylogeny; ii) FUBAR (Fast, Unconstrained Bayesian AppRoximation): uses a Bayesian approach to infer non-synoymous (dN) and synonymous (dS) substitution rates on a per-site basis for a given coding alignment and corresponding phylogeny. This method assumes that the selection pressure for each site is constant along with the entire phylogeny; iii) SLAC (Single-Likelihood Ancestor Counting) uses a combination of maximum-likelihood (ML) and counting approaches to infer non-synonymous (dN) and synonymous (dS) substitution rates on a per-site basis for a given coding alignment and corresponding phylogeny. This method assumes that the selection pressure for each site is constant along with the entire phylogeny (Pond and Frost, 2005a).
The positively selected sites with the corresponding amino acid variations identified in the Italian sequences of our sub-sets were highlighted.
The surface glycoprotein sub-set (gene S) was also analyzed for the identification of mutations.
A p-value < 0.1 for SLAC and FEL, and a posterior probability >0.90 for FUBAR have been used as statistical support for the amino acids sites found under selection, as previously reported (Lo Presti et al., 2016; Hu et al., 2016; Ebranati et al., 2015; Pond and Frost, 2005b) and these sites were considered candidates for selection. Only the statistically supported selective pressure sites were reported.
The positions of the selective pressure sites and mutations in the different SARS-CoV-2 sub-sets were referred respect to the protein products obtained from the SARS-CoV-2 Reference Sequence isolate Wuhan-Hu-1, Accession Number: NC_045512.2 and specifically respect to the protein _ id: YP_009725297.1 (nsp1), YP_009725298.1 (nsp2), YP_009725299.1 (nsp3), YP_009725300.1 (nsp4), YP_009725301.1 (3C-like proteinase), YP_009725302.1 (nsp6), YP_009725303.1 (nsp7), YP_009725304.1 (nsp8), YP_009725305.1 (nsp9), YP_009725306.1 (nsp10), YP_009725312.1 (nsp11), YP_009725307.1 (nsp12), YP_009725308.1 (helicase), YP_009725309.1 (3′-to-5′-exonuclease), YP_009725310.1 (endoRNAse), YP_009725311.1 (2′-O-ribose methyltransferase), YP_009724390.1 (surface glycoprotein), YP_009724391.1 (ORF3a), YP_009724392.1 (envelope), YP_009724393.1 (membrane glycoprotein), YP_009724394.1 (ORF6), YP_009724395.1 (ORF7a), YP_009724396.1 (ORF8), YP_009724397.2 (nucleocapsid phosphoprotein) and YP_009725255.1 (ORF10).
2.3. Glycosylation pattern
The glycosylation pattern of the SARS-CoV-2 surface glycoprotein, M and E protein sequences were analyzed through the N-GlycoSite l (Zhang et al., 2004; N-Glycosite, https://www.hiv.lanl.gov/content/sequence/GLYCOSITE/glycosite.html) to characterize and predict potential N-linked glycosylation sites. Furthermore, we aimed to perform the prediction of the potential O-glycosylation sites in the SARS-CoV-2 protein 3a, surface glycoprotein, E and M protein sub-sets by using NetOGlyc v. 4.0.0.13 software (Steentoft et al., 2013).
3. Results
3.1. Selective pressure analysis
Overall, the selective pressure analysis varied considerably across the genes.
The analysis conducted on nsp1, 3C-like proteinase, nsp10, 3′-to-5′ exonuclease, endoRNAse, 2′-O-ribose methyltransferase, E, M, ORF6, ORF7a, and ORF 10 sub-sets indicated only negatively selected sites (positions and amino acids reported in Table 1). In contrast, nsp7, nsp8, nsp9 and nsp11 showed neither positive nor negative sites. Table 1 showed five supported positively and three negatively selected sites in nsp 2, in contrast to nsp3, where a major number of negatively sites (n = 15) and fewer (n = 3) positively sites, were found.
Table 1.
Selective pressure analysis on SARS-CoV-2 protein coding gene sub-sets.
| sub- set | Positively selected sites (ω for sites >1)∗ | Negatively selected sites (ω for sites <1)∗ |
|---|---|---|
| nsp1 | ∖ | 65 (E); 83 (H) |
| nsp2 | 198 (V; I), 248 (S; G), 347 (K; C), 348 (S; V), 559 (I; V) | 287 (F); 488 (A); 565 (E) |
| nsp3 | 1454 (N; Y; D); 1507 (A; E), 1527 (A; V; E) | 106(F); 152 (Q), 353 (T), 380 (Q), 432 (T), 561 (L), 995 (Q), 1047 (D), |
| 1138 (K), 1303 (T), 1455 (S), 1456 (T), 1502 (A), 1544 (S), 1719 (P) | ||
| nsp4 | 33 (M; I) | 15 (L), 71 (F), 212 (V), 235 (V) |
| 3C-like proteinase | ∖ | 239 (Y) |
| nsp6 | 37 (L; F) | 222 (T), 289 (V) |
| nsp7 | ∖ | ∖ |
| nsp8 | ∖ | ∖ |
| nsp9 | ∖ | ∖ |
| nsp10 | ∖ | 128 (C) |
| nsp11 | ∖ | ∖ |
| nsp12 | 25 (G; Y); 323 (P; L); 644 (T; M) | 24 (T), 28 (T), 85 (T), 105 (R), 142 (L), 455 (Y), 591 (T), 643 (T), 896 (T) |
| helicase | 504 (P; L); 598 (A; S; V) | 337 (R); 521 (V); 547 (T), 553 (A) |
| 3′-to-5′ exonuclease | ∖ | 7 (L); 490 (E) |
| endoRNAse | ∖ | 73 (N), 127 (V), 216 (L) |
| 2′-O-ribose | ∖ | 4 (A); 36 (L), 138 (N), 163 (L) |
| methyltransferase | ||
| S | ||
| surface glycoprotein | 943 (S; P) | 348 (A); 669 (G); 681 (P); 795 (K); 853 (Q); 890 (A); 921 (K); 982 (S), 1044 (G), 1100 (T), 1166 (L) |
| ORF3a | 99 (A; S; V) | ∖ |
| E | ∖ | 63 (K) |
| M | ∖ | 69 (A) |
| ORF6 | ∖ | 61 (D) |
| ORF7a | ∖ | 69 (D); 70 (G); 92 (E) |
| ORF8 | 62 (V; L) | ∖ |
| N | 13 (P; L; S); 103 (D; Y) | 173 (A); 274 (F) |
| ORF10 | ∖ | 15 (S); 19 (C) |
Only the sites with a p-value < 0.1 (FEL, SLAC) and with a posterior probability >0.90 (FUBAR) were considered as candidates for selection and statistically supported.
Selective pressure analysis conducted on nsp4 sub-set revealed one positive and four negative selective sites (Table 1).
Nsp 6 revealed one positive 37 (L; F) and two negative sites 222 (T), 289 (V).
Selective pressure analysis conducted on nsp12 found three positively selected sites 25 (G; Y); 323 (P; L); 644 (T; M) and eight negative.
Selective pressure analysis on the helicase sub-set indicated two positive sites 504 (P; L); 598 (A; S; V) and four negative sites 337 (R); 521 (V); 547 (T), 553 (A) and in the SARS-CoV-2 S (surface glycoprotein) protein coding gene sub-set revealed one positive 943 (S; P) and 11 negatively selected sites. SARS-CoV-2 bind to ACE2 through the RBD (receptor binding domain for virus entry into the cells) of the spike protein in order to initiate membrane fusion and enter human cell. The positively selected site here identified (AA 943) appeared located outside the RBD of the spike glycoprotein (Chen et al., 2020).
Only one positively selected site 99 (A; S; V) has been identified in ORF3a. In ORF8 the AA position 62 (V; L) has been found subjected to positive selection (Table 1). Finally, the nucleocapsid phosphoprotein sub-set revealed two positive: 13 (P; L; S); 103 (D; Y) and two negative selective sites 173 (A); 274 (F) (Table 1).
3.2. Landscape of the positively selected sites/mutations on genomes collected in Italy
The positively selected sites, identified in this study, were represented in the Italian sequences included in our sub-sets (14 sequences from Italy for N sub-set; 13 sequences for nsp2, nsp3, nsp4, nsp6, nsp12, helicase, S, ORF3a, ORF8sub-sets) in order to monitor the variations.
All the Italian sequences showed the 198V; 248S; 347K; 348S; 559I aminoacid sites in the nsp2 gene and 1454N, 1507A, 1527A in the nsp3 gene with the exception of EPI_ISL_417446 genome showing the 1507E, and EPI_ISL_417446 showing 1527E variation.
In nsp4 all the Italian strains showed amino acid 33M.
In nsp6 all the Italian isolates showed 37L except for the genomes EPI_ISL_410546 and EPI_ISL_412974 showing the 37F variation.
All the genomes presented the 25G, 644T, 323L in the nsp12 gene except for three genomes (EPI_ISL_410546, EPI_ISL_410545 and EPI_ISL_412974) presenting 323P. In the helicase protein coding gene all the Italian isolates presented 504P and 598A. At amino acidic position 943 of surface glycoprotein all the Italian genomes showed the amino acid S. The residues 99A and 62V were found in all the Italian genomes for orf3a and ORF8, respectively. Regarding N sub-set all the genomes showed 13P (except sequence Id. EPI_ISL_408068 showing a gap) and 103D.
3.3. Identification of mutations in the surface glycoprotein (S) sub-set
The detailed results of mutation analysis performed on the surface glycoprotein (S) sub-set alignment are reported in Table 2 and Table S2.
Table 2.
Results of the mutation analysis performed on the surface glycoprotein (S) sub-set.
| Amino acid position | Reference Accession Number and residue identified | Accession Id and mutation identified |
|---|---|---|
| 27 | A | EPI_ISL_419885: V |
| 29 | T | EPI_ISL_418869: I |
| 32 | F | EPI_ISL_402132: I |
| 49 | H | EPI_ISL_403936: Y |
| EPI_ISL_403937: Y | ||
| EPI_ISL_406531: Y | ||
| EPI_ISL_408010: Y | ||
| 71 | S | EPI_ISL_417142: F |
| 146 | EPI_ISL_417977: Y | |
| 167 | T | EPI_ISL_408978: F |
| 184 | G | EPI_ISL_422298: D |
| 197 | I | EPI_ISL_418216: V |
| EPI_ISL_418265: V | ||
| 202 | K | EPI_ISL_413023: N |
| 215 | S | EPI_ISL_418409: H |
| 247 | S | EPI_ISL_406844: R |
| 255 | S | EPI_ISL_420877: F |
| 258 | W | EPI_ISL_417976: L |
| 367 | V | EPI_ISL_406596: F |
| EPI_ISL_406597: F | ||
| 458 | K | EPI_ISL_415159: R |
| 477 | S | EPI_ISL_419662: G |
| 483 | V | EPI_ISL_417139: A |
| EPI_ISL_413652: A | ||
| EPI_ISL_417076: A | ||
| 491 | P | EPI_ISL_419737: R |
| 519 | H | EPI_ISL_415159: P |
| 522 | A | EPI_ISL_421654: V |
| 574 | D | EPI_ISL_418421: Y |
| 614 | D | 614 G∗ |
| 615 | V | EPI_ISL_412983: L |
| 630 | T | EPI_ISL_417446: S |
| 631 | P | EPI_ISL_419704: S |
| 655 | H | EPI_ISL_413486: Y |
| 675 | Q | EPI_ISL_419709: R |
| 809 | P | EPI_ISL_417408: S |
| 879 | A | EPI_ISL_418401: S |
| 936 | D | EPI_ISL_418432: Y |
| 939 | S | EPI_ISL_420814: F |
| 941 | T | EPI_ISL_415159: A |
| 943 | S | EPI_ISL_415159: P |
| EPI_ISL_420335: P | ||
| 954 | Q | EPI_ISL_417978: K |
| 1132 | I | EPI_ISL_414628: V |
| 1143 | P | EPI_ISL_407896: L |
| 1229 | M | EPI_ISL_417575: I |
| 1247 | C | EPI_ISL_416655: F |
| 1254 | C | EPI_ISL_413594: F |
| 1263 | P | EPI_ISL_415133: L |
The list of sequences harbouring the mutation 614G has been reported in Table S2.
Overall, 41 AA residues (41/1273) representing the 3.2 % of the entire surface glycoprotein length has been found undergoing variation, indicating the presence of different variants.
The amino acidic position 614 (mutation D – G) has been found most frequently mutated in the sequences of our subset (Table S2).
Five residues (367, 458, 477, 483 and 491) which belonged to the RDB of the surface glycoprotein are subjected to variations in the sequences reported in Table 2. These amino acidic positions are subject to variations, they were not located within the residues interacting with ACE2 in the SARS-CoV RBD and conserved in SARS-CoV2 as highlighted in red in Chen et al. (2020). The AA position 49, 483 and 943 were also found most frequently mutated in our sub-set (Table 2).
The surface glycoprotein protein must likely be cleaved at both S1/S2 sand S2′ cleavage sites for virus entry, as previously described (Coutard et al., 2020). We investigated the surface glycoprotein sub-set alignment in the AA regions of the protein cleavage sites (SARS-CoV-2 S1/S2 site 1, site 2 and S2′) that appeared conserved in all the sequences of our sub-set.
3.4. Comparison of the S protein alignment among SARS-CoV-2, SARS-CoV and Bat SARS - like virus
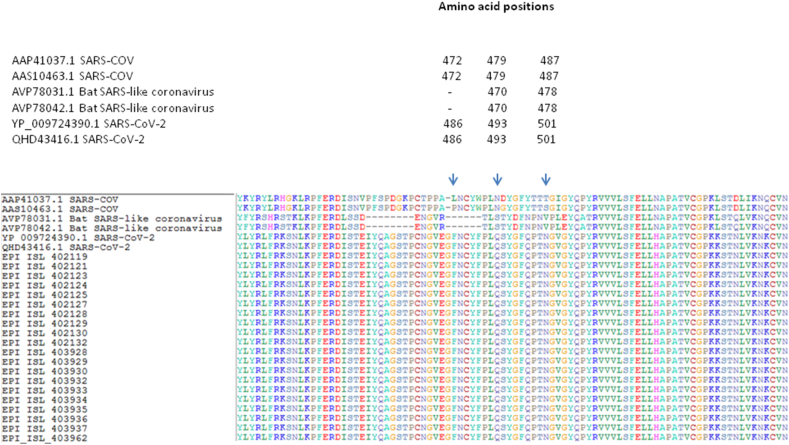
We analyzed the protein alignment of surface glycoprotein sub-set of SARS-CoV-2, compared to two sequences from SARS-COV (AAP41037.1 and AAS10463.1) and two Bat SARS-like coronavirus spike protein (AVP78031.1 and AVP78042.1), focusing the attention on the relevant positions 472 (amino acid L or P in SARS COV), 479 (amino acid N in SARS CoV) and 487 (amino acid T or S) of SARS CoV (Figure 1). These amino acid positions were previously reported (Li et al., 2005a, Li et al., 2005b) to be critical for cross-species and human-to-human transmission in SARS-COV. In the comparison of the paired positions in our alignment (Figure 1), differences in the amino acids harbored by SARS-CoV-2 surface glycoprotein sequences were identified that is: 486F, 493Q and 501 N (referred to SARS-CoV-2 Accession YP_009724390.1) aligned respectively to the amino acidic positions 472, 479 and 487 of SARS – CoV. In Bat SARS-like coronavirus spike protein sequences, in the paired positions of the previous alignment, we found: a gap (paired with the position 472 os SARS-CoV and with the position 486F of SARS_CoV-2), 470S (referred to Accession Number: AVP78031.1 and paired with the position 479N of SARS-COV and 493Q of SARS-CoV-2), and 478V (referred to Accession Number: AVP78031.1, paired with 487 T/S of SARS-COV and to 501N of SARS-CoV-2 (Figure 1).
Figure 1.
The representative alignment for the comparison of the surface glycoprotein between SARS-CoV-2, SARS-CoV and Bat SARS - like virus (including the first 20 SARS-CoV-2 sequences, in addition to SARS-CoV-2 references), focusing the attention on the relevant positions 472 (amino acid L or P in SARS COV), 479 (amino acid N in SARS CoV) and 487 (amino acid T or S) of SARS CoV.
3.5. Glycosylation pattern
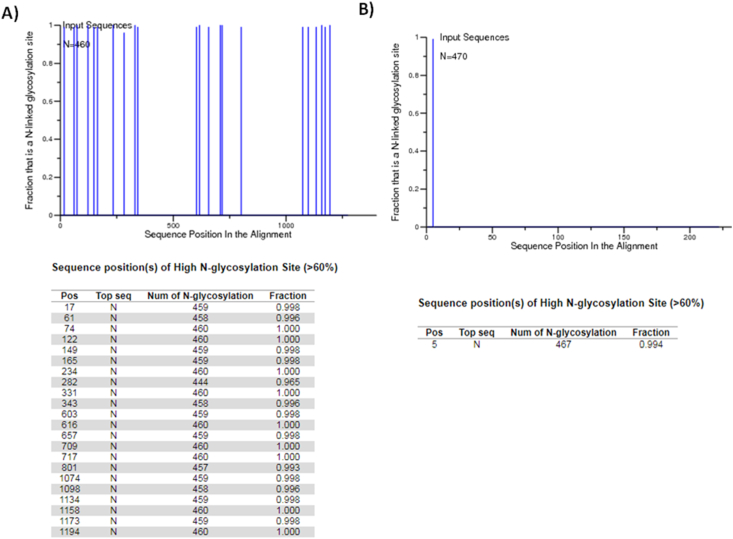
A total of 22 predicted N-glycosylation positions were found in SARS-CoV-2 surface glycoprotein sub-set by using N-GlycoSite. The positions, number and fraction of the predicted N-glycosylation sites in the alignment of SARS-CoV-2 surface glycoprotein sub-set were reported (Figure 2A). A total of 10087 N-glycosylation sites in 460 sequences have been found (considering that some sequences have deletions). In particular, we found that the sequence Id: EPI_ISL_408978 (derived from a throat swab collected from a 65 years old, female patient from Hubei/Wuhan) did not have a predicted N-glycosylation site on position 165. We also noted that the sequence Id: EPI_ISL_417439 (derived from an oro-pharyngeal swab from a 38 years old male patient, from Democratic Republic of the Congo/Kinshasa) did not show a predicted N-glycosylation site on position 1074.
Figure 2.
A. The predicted N-glycosylation sites in SARS-CoV-2 surface glycoprotein sub-set, obtained by using N-GlycoSite tool. The positions, number and fraction of the predicted N-glycosylation sites were reported. B. The predicted N-glycosylation sites in SARS-CoV-2 M protein sub-set, obtained by using N-GlycoSite tool. The position, number and fraction of the predicted N-glycosylation sites were reported.
In particular, three SARS-CoV-2 N-Glyc predicted sites 234N, 343N and 603N, corresponded to the SARS CoV N-glycosylation sites 227N, 330N and 589N (by exploring the paired alignment positions) (Zhou et al., 2010). Of these sites, one (343N) was located within the SARS-CoV-2 RDB.
Regarding the M protein sub-set, one predicted N-glycosylation position was found for SARS-CoV-2 M sub-set by using N-GlycoSite tool (Figure 2B). A total of 467 N-glycosylation sites in 470 sequences have been found (three sequences Id: EPI_ISL_406959, 406960 and 416464 were shorter). The position, graphic, number and fraction of the predicted N-glycosylation sites for M sub-set were reported (Figure 2B).
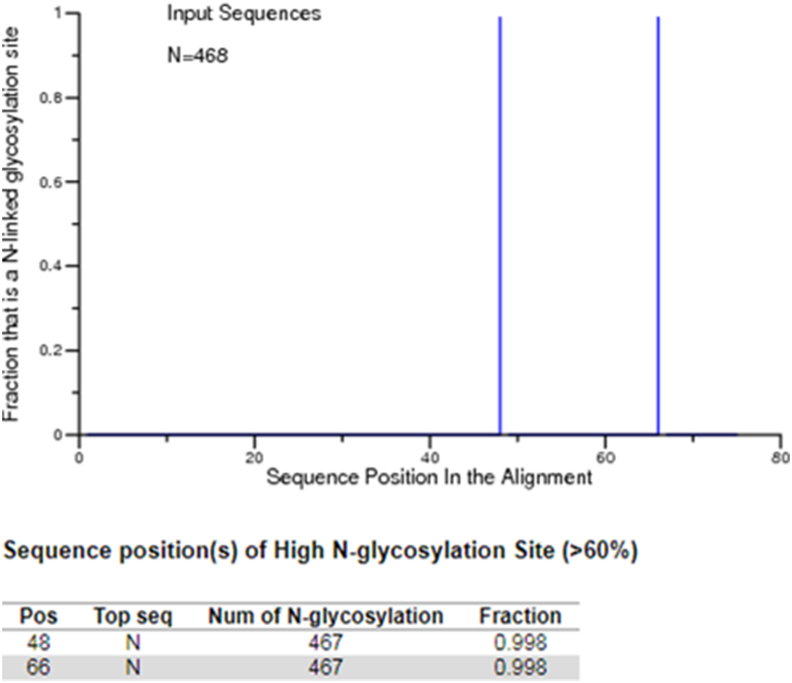
The analysis of the N-Glycosylation pattern on E protein sub-set revealed two potential predicted N-Glycosylation sites (AA. 48 and 66, Figure 3). A total of 934 N-glycosylation sites in 468 sequences have been found. The sequence EPI_ISL_418200 (derived from a 57 years old male patient from USA/New York/Manhattan), did not show a predicted N-glycosylation site at amino acidic position 48. Meanwhile, the N-GlycoSite analysis performed on the protein 3a sub-set showed no N-glycosylation sites predicted for this protein.
Figure 3.
The predicted N-glycosylation sites in SARS-CoV-2 E protein sub-set obtained by using N-GlycoSite tool. The positions, number and fraction of the predicted N-glycosylation sites were reported.
In contrast, the results obtained through Net O-Glyc 4.0 on protein 3a sub-set indicated the following four potential O-glycosylation sites, with confidence scores higher than 0.5: amino acid position 32 in the sequence Id number: EPI_ISL_416464 (USA); amino acid position 253 in the sequences Id number: EPI_ISL_419690 and EPI_ISL_419683(Spain/Valencia); finally, amino acid position 171 in sequence Id number: EPI_ISL_408978 (Wuhan, China). The predicted O –glycosylation sites for SARS-CoV-2 surface glycoprotein sub-set indicated sites 673 (serine), 678 (threonine) and 686 (serine) the most frequently predicted as glycosylated in our sub-set (~89–90% of the sequences). Other sites (19T, 22T, 29T, 250T, 349S) were found predicted O –glycosylated at lower frequency (between 2% and 6 % of the sequences) (data not shown). The M and E protein sub-set were not predicted to be O –glycosylated by Net O-Glyc.
4. Discussion
This work provides a large-scale genomics analysis towards understanding the selective pressure, mutation and glycosylation patterns of SARS-CoV-2.
Selective pressure analysis on the SARS-CoV-2 nsp2 and nsp3 sub-set revealed positive selection in five sites in nsp2 and three in nsp3. Nsp2 may have a role in modulating host cell survival, likely by altering host cell environment (Cromwell et al., 2009; SWISS-MODEL Repository, https://swissmodel.expasy.org/repository/species/2697049; UNIPROT, https://www.uniprot.org/uniprot/?query=taxonomy:2697049). Nsp3 is the papain-like protease that plays an important role in viral genome replication and in antagonize the host's innate immunity (Dong et al., 2020). In this study, a large set of genomes confirmed some amino acid changes, as previously described, i.e. amino acid change V198I in nsp2 (Pachetti et al., 2020), but described also the occurrence of hotspot mutations, driven by positive selection, in additional nsp2 and nsp3 sites (nsp2: 248; 347; 599; nsp3: 1454; 1507; 1527), suggesting a highly dynamic pattern and their possible role in viral genome replication.
Here, the analysis conducted on nsp1, 3C-like proteinase, nsp10, 3′-to-5′ exonuclease, endoRNAse, 2′-O-ribose methyltransferase, E, M, ORF6, ORF7a, and ORF 10 sub-sets indicated only negatively selected sites, suggesting a scenario of purifying selection. By contrary, nsp7, nsp8, nsp9 and nsp11 showed neither positive nor negative sites indicating that evolution and divergence can be constant across all the evolutionary lineages and that these genes can be considered neutral. These data can help identifying highly conserved regions, useful for implementing new diagnostic protocols.
In this study, one positive selected site (33 M; I) in nsp4 protein was identified. This protein acts in the assembly of virally-induced cytoplasmic double-membrane vesicles, essential for viral replication. This finding may imply a genetic “hot-spot” in SARS-CoV-2 viral replication and need to be further evaluated.
Here, we confirmed the positive selective site at amino-acid position 37 (L; F) in nsp6 previously reported on a smaller dataset by some authors (Benvenuto et al., 2020; Pachetti et al., 2020), but also observed in a recent study performed on a large dataset (Mercatelli and Giorgi, 2020). This protein plays a role in the initial induction of autophagosomes from host reticulum endoplasmic and later limits the expansion of these phagosomes, that are no longer able to deliver viral components to lysosomes (SWISS-MODEL Repository, https://swissmodel.expasy.org/repository/species/2697049).
Interestingly, two additional positive selective sites (25 G- Y; 644 T - M) in the RNA-dependent RNA polymerase (nsp12) were identified, in addition to confirming the residue at position 323 (P; L), previously reported (Pachetti et al., 2020). RNA-dependent RNA polymerase is an optimal target of choice for treatment because of its crucial role in RNA synthesis, lack of homolog host and high sequence and structural conservation. In particular, Remdesivir has recently been advanced to phase 3 clinical trials for SARS-CoV-2 (Shannon et al., 2020) due to its mechanism to interact with the active replication site and to the viral genome, thus inhibiting the replication. The identification of positively selected sites in the RNA-dependent RNA polymerase could be useful for therapeutic approaches.
We were able to update the evolutionary changes on the helicase by reporting an additional “hot-spot” (598 A-S-V) as well as confirming the residue in the previously reported residue at position 504 (P; L) (Pachetti et al., 2020).
The SARS-CoV-2 surface glycoprotein (S) is subjected to both positive and negative selection. Other authors, in agreement with our study, have identified some mutations within the surface glycoprotein (Phan, 2020; Tang et al., 2020; Pachetti et al., 2020; Mercatelli and Giorgi, 2020), confirming that this portion is subject to more frequent variation on position 614 D-G. This mutation is consistent with several hypotheses regarding a fitness advantage, a greater susceptibility to re-infection (with the new G614 change of the virus), a greater infectivity due to its spread, and a probable greater transmissibility with a potential impact on the severity of the disease, as previously reported (Korber et al., 2020). Surface glycoprotein plays a crucial role in binding of virus to the host receptor and subsequent membranes fusion for virus entry (Chen et al., 2020). The positive selection identified here is in agreement with the studies conducted on SARS-CoV (Chinese, 2004; Song et al., 2005; Zhang et al., 2006; Tang et al., 2009). We highlighted a positive selected site at position 943 (S; P), located outside the SARS-CoV-2 RBD for ACE2 (Wan et al., 2020; Li et al., 2003; Wrapp et al., 2020), suggesting this probably does not affect the RBD structure and the binding capacity of the virus to the host cell receptor, but has been linked to a suggestive model of recombination (Korber et al., 2020). Five additional mutations in the surface glycoprotein were reported in this study, which appeared within the RBD, indicating that changes in this portion may occur and should be carefully monitored, given the potential impact on viral binding capacity and infectivity. Among these mutations, the V367 site deserves attention because it is located on the same face as the epitope of CR3022, a neutralizing antibody isolated from a SARS-CoV convalescent patient though no direct contacts between V367 and CR3022 were observed, and for a potential interaction with ACE2 (Korber et al., 2020; Yuan et al., 2020).
The 62 (V-L) mutation in ORF8 (Tang et al., 2009) was confirmed as positive selected site, furthermore we were able to highlight an additional positive selected residue 99 A-S-V in orf3a. As for the N sub-set, we found two new sites (13 P-L-S and 103 D-Y) subjected to positive selection. This gene has been used in SARS–CoV-2 diagnostic tests. For this reason, it is important to monitor the selective pressure to highlight new variations useful to update, eventually, the diagnostic protocols.
A recent study (Mercatelli and Giorgi, 2020) analyzed a large SARS-CoV-2 dataset focusing the attention at single-nucleotide polymorphisms (SNPs). These authors highlighted a massive prevalence of SNPs over short insertion/deletion events (indels) worldwide and in every country. Moreover they reported that the aa-changing SNPs are the most prevalent mutational events in SARS-CoV-2 genomes, supporting our study and confirming the importance to monitor selective pressure and mutations.
Compared to our data, even if the two studies were based on different methodological approaches, we were able to confirm six mutation events as subjected to positive or negative selection, among the mutations that occur most frequently according to Mercatelli and Giorgi (2020). In addition, we also found the D614G mutation as the most frequent in our surface glycoprotein dataset.
The comparative analysis of the S protein alignment between SARS-CoV-2, SARS-CoV and Bat SARS - like coronavirus, was analyzed in three critical positions, previously described by Li et al., 2005a, Li et al., 2005b, to be crucial for cross-species and human-to-human transmission in SARS-CoV, the authors highlighted differences in the amino acids present at these sites.
All three positions were located within the SARS-CoV-2 RBD, the critical determinant of virus-receptor interaction and, therefore, of the viral host range and tropism (Li et al., 2005a, Li et al., 2005b). A previous study conducted on SARS (Chakraborti et al., 2005) identified some RBD amino acid residues that influence the binding with ACE2 expressing and testing their binding to ACE2. A similar procedure could also be hypothesized for SARS-CoV-2, performing the expression of mutants and trying to identify the residues that could significantly reduce the RBD-ACE2 interaction.
SARS-CoV-2 uses a densely glycosylated surface protein to gain entry into host cells. This study identified 22 N-glycosylation predicted positions for SARS-CoV-2 surface glycoprotein alignment which must be confirmed by mass spectrometric or biochemical analyses, as already done for SARS-CoV (Chakraborti et al., 2005; Ying et al., 2004; Krokhin et al., 2003). Interestingly, one of the predicted N-glycosylation position in SARS-CoV-2 surface glycoprotein is located inside the RBD. Seventeen of the twenty-two predicted N-glycosylation sites had previously been reported from a study conducted on a small sample (Kumar et al., 2020) and some of them were also reported as unique of SARS-CoV2 compared to SARS – CoV (Vankadari and Wilce, 2020). In this study, we therefore identified some additional predicted N-glycosylation sites of the SARS-CoV-2 spike glycoprotein, suggesting that the virus may use different glycosylation to interact with its receptors and may underlie the differences in host immunity.
A literature article (Zhao et al., 2020) defined the glycomics-informed, site-specific micro heterogeneity of 22 N-linked sites (confirming our predicted sites) using a combination of mass spectrometry approaches coupled with evolutionary and variant sequence analyses. These authors (Zhao et al., 2020) have suggested essential roles for glycosylation in mediating receptor binding, antigenic shielding, and potentially the evolution/divergence of these glycoproteins. The 22 predicted N-glycosylation positions here investigated in the spike glycoprotein, were also in line with those reported in a previous study (Shajahan et al., 2020) which identified by high resolution mass spectrometry the composition of glycans at 17 out of the 22 SARS-CoV-2 predicted sites of the spike glycoprotein reporting the remaining five sites as unoccupied. Other authors (Watanabe et al., 2020) have focused attention on the 22 N-linked gly-can sites, confirming our prediction results, but they have used a site-specific mass spectrometric approach revealing the glycan compositions on a recombinant SARS-CoV-2 S immunogen.
Four SARS-CoV-2 N-glycosylation predicted sites (234N, 343N, 370N and 603N) here identified, corresponded to the following aligned positions of the SARS-CoV N- glycosylation sites (227N, 330N, 357N and 589N) (Zhou et al., 2010). Mannose-binding lectin (MBL) is an important serum protein in the host's defenses. Zhou et al. (2010) reported the specificity of the site for glycosylation at position N330 (SARS-CoV) in the ability of MBL to inhibit SARS-CoV entry and infection in susceptible cell lines and it could be assumed a similar model for SARS-CoV-2 (Zhou et al., 2010).
Our study may indicate that site – directed mutagenesis and in vitro studies must be applied in order to clarify whether individual SARS-CoV-2 glycosylation sites are directly involved in DC-SIGN(R)–mediated binding and entry (Zhou et al., 2010) and if the glycan at 343N or others reported in this study, were critical in the ability of MBL to inhibit SARS-CoV-2 entry.
The predicted N-glycosylation sites here identified in SARS-CoV-2 M and E sub-set need to be confirmed by experiments and their role better clarified in further studies. The N-glycosylation profile and the absence of O-glycosylation on M protein refer to the SARS-CoV data (Nal et al., 2005). In contrast, the SARS-CoV E protein is not glycosylated.
The expected O-glycosylation sites must be confirmed through specific experiments, together with their roles. Many different functions have been assigned to the side chains of oligosaccharide. Carbohydrates have been shown to be important for the folding, structure, stability, and intracellular sorting of proteins and to play a role in evoking the immune responses. Our data are in agreement with O-glycosylation profile of SARS-CoV 3a protein (Nal et al., 2005), but in contrast with SARS-CoV surface glycoprotein that seems not to be O-glycosylated. A previous study (Shajahan et al., 2020) confirmed our O-glycosylation results on SARS-CoV-2 surface glycoprotein for sites 673, 678 and 686. In contrast, we did not identify O-glycosylation on surface glycoprotein at sites Thr 323 and Ser 325 but we found the predicted O-glycosylation, at lower percentage, in different positions. The O-glycosylation on the SARS-CoV-2 surface glycoprotein is also predicted in several recent reports (Andersen et al., 2020). Although it is unclear what the functions of these predicted O-linked glycans, it has been suggested to create a ‘mucin-like domain’ capable of protecting SARS-CoV-2 spike protein epitopes or key residues (Bagdonaite and Wandall, 2018). Since some viruses may use mucin-like domains as glycan shields for immunoevasion, further studies and experiments could better clarify the specific role of SARS-CoV-2 spike protein O-glycosylation and if predicted sites can be experimentally confirmed.
Limits and possible bias of the study should be mentioned. First, the analysis here presented depends on the genomes available in the database at the time of the last access. Second, the circulation period of the virus can affect the evaluation of the evolution of the virus.
The goal of this study was to identify the evolutionary differences between a large set of SARS-CoV-2 available genomes and to predict their possible implications. The data, which show positive selective pressure and mutations that act on specific gene encoding protein (i.e. surface glycoprotein), could provide markers for vaccine design and/or for therapeutic agents (i.e. nsp12). The negative selection identified in some SARS-CoV-2 protein encoding genes could help to implement new diagnostic protocols. Finally, the identification of specific SARS-CoV-2 glycosylation sites could help to understand the interaction of the virus with its receptor and implement future mutagenesis experiments that are fundamental for strategies aimed at inhibiting the entry of SARS-CoV-2 in the cells.
Declarations
Author contribution statement
Alessandra Lo Presti: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Giovanni Rezza, Paola Stefanelli: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We gratefully thank the Collaborators of ISS COVID-19 Study Group: Angela Di Martino, Marzia Facchini, Fabiola Mancini, Simona Puzelli, Giuseppina Di Mario, Laura Calzoletti, Stefano Fontana, Stefano Fiore, Antonella Marchi, Eleonora Benedetti, Concetta Fabiani, Giulietta Venturi, Claudia Fortuna, Giulia Marsili, Antonello Amendola, Fabrizio Barbanti, Maria Scaturro, Alessandra Ciervo, Maria Rita Castrucci, Daniela Fortini, Marco Di Luca, Luciano Toma, Francesco Severini, Patrizio Pezzotti, Antonino Bella, Flavia Riccardo, Alessia Caratelli, Dept. of Infectious Diseases, Istituto Superiore di Sanità, Rome, Italy.
We gratefully acknowledge the Authors, the Originating and Submitting Laboratories for their sequence and metadata shared through GISAID, on which this research is based. All submitters of data may be contacted directly via www.gisaid.org. We gratefully acknowledge the Authors, the Originating and Submitting Laboratories for their sequence and metadata shared through NCBI database. The Acknowledgment Table is reported as Table S1.
We gratefully acknowledge Dr. Maria Rita Gismondo - Clinical Microbiology, Virology and Bioemergency, L. Sacco University Hospital, Milan, Italy.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Afgan E., Baker D., Batut B., van den Beek M., Bouvier D., Čech M. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46(W1):W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020 doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdonaite I., Wandall H.H. Global aspects of viral glycosylation. Glycobiology. 2018;28:443–467. doi: 10.1093/glycob/cwy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuto D., Angeletti S., Giovanetti M., Bianchi M., Pascarella S., Cauda R., Ciccozzi M., Cassone A. Evolutionary analysis of SARS-CoV-2: how mutation of Non-Structural Protein 6 (NSP6) could affect viral autophagy. J. Infect. 2020;10 doi: 10.1016/j.jinf.2020.03.058. pii: S0163-4453(20)30186-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborti S., Prabakaran P., Xiao X., Dimitrov D.S. The SARS coronavirus S glycoprotein receptor binding domain: fine mapping and functional characterization. Virol. J. 2005;2:73. doi: 10.1186/1743-422X-2-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Guo Y., Pan Y., Zhao Z.J. Structure analysis of the receptor binding of 2019-nCoV. Biochem. Biophys. Res. Commun. 2020;17 doi: 10.1016/j.bbrc.2020.02.071. pii: S0006-291X(20)30339-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinese S.M.E.C. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromwell T., Cornillez-Ty, Liao L., Yates J.R., III, Kuhn P., Buchmeier Michael J. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J. Virol. 2009;83(19):10314–10318. doi: 10.1128/JVI.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delport W., Poon A.F., Frost S.D., Kosakovsky Pond S.L. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics. 2010;26(19):2455–2457. doi: 10.1093/bioinformatics/btq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S., Sun J., Mao Z., Wang L., Lu Y., Li J. A guideline for homology modeling of the proteins from newly discovered betacoronavirus, 2019 novel coronavirus(2019-nCoV) J. Med. Virol. 2020:1–7. doi: 10.1002/jmv.25768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebranati E., Pariani E., Piralla A., Gozualo-Marguello M., Veo C., Bubba L., Amendola A., Ciccozzi M., Galli M., Zanetti A.R., Baldanti F., Zehender G. 2015. Reconstruction of the Evolutionary Dynamics of A(H3N2) Influenza Viruses Circulating in Italy from 2004 to 2012. Published: September 2, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galaxy platform https://usegalaxy.org/ URL:
- GISAID Platform Global Initiative on Sharing All Influenza Data. https://www.gisaid.org/Last URL: [DOI] [PMC free article] [PubMed]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hu D., Lv L., Gu J., Chen T., Xiao Y., Liu S. Genetic diversity and positive selection Analysis of classical swine Fever Virus Envelope protein gene E2 in East China under C-Strain vaccination. Front. Microbiol. 2016;7:85. doi: 10.3389/fmicb.2016.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Foley B., Giorgi E.E., Bhattacharya T., Parker M.D., Partridge D.G., Evans C.M., de Silva T.I., on behalf of the Sheffield COVID-19 Genomics Group, LaBranche CC, and Montefiori DC Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv. 2020 [Google Scholar]
- Krokhin O., Li Y., Andonov A., Feldmann H., Flick R., Jones S. Mass spectrometric characterization of proteins from the SARS virus: a preliminary report. Mol. Cell. Proteomics. 2003;2(5):346–356. doi: 10.1074/mcp.M300048-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Maurya V., Prasad A., Bhatt M.L.B., Saxena S.K. Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV) Virus Dis. 2020 doi: 10.1007/s13337-020-00571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24(8):1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Presti A., Cella E., Giovanetti M., Lai A., Angeletti S., Zehender G., Ciccozzi M. Origin and evolution of nipah virus. J. Med. Virol. 2016;88(3):380–388. doi: 10.1002/jmv.24345. Epub 2015 Aug 14. [DOI] [PubMed] [Google Scholar]
- Mercatelli D., Giorgi F.L. Geographic and genomic distribution of SARS-CoV-2 mutations. Front. Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nal B., Chan C., Kien F., Siu L., Tse J., Chu K., Kam J., Staropoli I., Crescenzo-Chaigne B., Escriou N., van der Werf S., Yuen K.Y., Altmeyer R. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 2005;86:1423–1434. doi: 10.1099/vir.0.80671-0. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnology Information U.S. National Library of Medicine. https://www.ncbi.nlm.nih.gov/pubmed NCBI. (URL:
- N-Glycosite https://www.hiv.lanl.gov/content/sequence/GLYCOSITE/glycosite.html URL:
- Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C., Zella D., Ippodrino R. Emerging SARS-CoV- 2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18(1):179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evol. 2020;81:104260. doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond S.L., Frost S.D. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21(10):2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- Pond S.L.K., Frost S.D.W. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 2005;22(5):1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Shannon A., Le N.T., Selisko B., Eydoux C., Alvarez K., Guillemot J.C., Decroly E., Peersen O., Ferron F., Canard B. Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antivir. Res. 2020;178:104793. doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajahan A., Supekar N.T., Gleinich A.S., Azadi P. 2020. Deducing the N- and O- Glycosylation Profile of the Spike Protein of Novel Coronavirus SARS-CoV-2. this version posted April 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. U.S.A. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steentoft C., Vakhrushev S.Y., Joshi H.J., Kong Y., Vester-Christensen M.B., Schjoldager K.T. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32(10):1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SWISS-MODEL repository. https://swissmodel.expasy.org/repository/species/2697049 Available at:
- Tang X., Wu C., Li X., Song Y., Yao X., Wu X., Duan Y., Zhang H., Wang Y., Qian Z., Cui J., Lu J. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020:nwaa036. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Li G., Vasilakis N., Zhang Y., Shi Z., Zhong Y., Wang L.F., Zhang S. Differential stepwise evolution of SARS coronavirus functional proteins in different host species. BMC Evol. Biol. 2009;5(9):52. doi: 10.1186/1471-2148-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNIPROT https://www.uniprot.org/uniprot/?query=taxonomy:2697049 Available at:
- Vankadari N., Wilce J.A. Emerging WuHan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg. Microb. Infect. 2020;9(1):601–604. doi: 10.1080/22221751.2020.1739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J. Virol. 2020;29 doi: 10.1128/JVI.00127-20. pii: JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020 doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S., Shank S.D., Spielman S.J., Li M., Muse S.V., Kosakovsky Pond S.L. Datamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 2018;35(3):773–777. doi: 10.1093/molbev/msx335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO) 2020. Statement on the Second Meeting of the International Health Regulations (2005) Emergency Committee Regarding the Outbreak of Novel Coronavirus (2019-nCoV) (Press release) Archived from the original on 31 January 2020. [Google Scholar]
- World Health Organization (WHO) 2020. WHO Director-General's Opening Remarks at the media Briefing on COVID-19 - 11 March 2020. (Press release). Archived from the original on 11 March 2020. [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. Epub 2020 Feb 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W., Hao Y., Zhang Y., Peng W., Qin E., Cai Y. Proteomic analysis on structural proteins of severe acute respiratory syndrome coronavirus. Proteomics. 2004;4:492–504. doi: 10.1002/pmic.200300676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020 doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.Y., Wei J.F., He S.H. Adaptive evolution of the spike gene of SARS coronavirus: changes in positively selected sites in different epidemic groups. BMC Microbiol. 2006;6:88–97. doi: 10.1186/1471-2180-6-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Nielsen R., Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- Zhang M., Gaschen B., Blay W., Foley B., Haigwood N. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology. 2004;14:1229–1246. doi: 10.1093/glycob/cwh106. [DOI] [PubMed] [Google Scholar]
- Zhao P., Praissman J.L., Grant O.C., Cai Y., Xiao T., Rosenbalm K.E., Aoki K., Kellman B.P., Bridger R., Barouch D.H., Brindley M.A., Lewis N.E., Tiemeyer M., Chen B., Woods R.J., Wells L. Virus-receptor interactions of glycosylated 1 SARS-CoV-2 spike and human ACE2 receptor. bioRxiv. 2020 doi: 10.1016/j.chom.2020.08.004. preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Lu K., Pfefferle S., Bertram S., Glowacka I., Drosten C. A single asparagine-linked glycosylation site of the severe acute respiratory syndrome coronavirus spike glycoprotein facilitates inhibition by mannose-binding lectin through multiple mechanisms. J. Virol. 2010;84(17):8753–8764. doi: 10.1128/JVI.00554-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.