Abstract
Background
The coronavirus disease (COVID-19) outbreak in China was first reported to the World Health Organization (WHO) on December 31, 2019, and the first cases were officially identified around December 8, 2019. Although the origin of COVID-19 has not been confirmed, approximately half of the early cases were linked to a seafood market in Wuhan. However, the first two documented patients did not visit the seafood market. News reports, social media, and informal sources may provide information about outbreaks prior to formal notification.
Objective
The aim of this study was to identify early signals of pneumonia or severe acute respiratory illness (SARI) in China prior to official recognition of the COVID-19 outbreak in December 2019 using open-source data.
Methods
To capture early reports, we searched an open source epidemic observatory, EpiWatch, for SARI or pneumonia-related illnesses in China from October 1, 2019. The searches were conducted using Google and the Chinese search engine Baidu.
Results
There was an increase in reports following the official notification of COVID-19 to the WHO on December 31, 2019, and a report that appeared on December 26, 2019 was retracted. A report of severe pneumonia on November 22, 2019, in Xiangyang was identified, and a potential index patient was retrospectively identified on November 17.
Conclusions
The lack of reports of SARI outbreaks prior to December 31, 2019, with a retracted report on December 26, suggests media censorship, given that formal reports indicate that cases began appearing on December 8. However, the findings also support a relatively recent origin of COVID-19 in November 2019. The case reported on November 22 was transferred to Wuhan approximately one incubation period before the first identified cases on December 8; this case should be further investigated, as only half of the early cases were exposed to the seafood market in Wuhan. Another case of COVID-19 has since been retrospectively identified in Hubei on November 17, 2019, suggesting that the infection was present prior to December.
Keywords: COVID-19, infectious disease, surveillance, epidemiology, biosecurity
Introduction
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a new betacoronavirus that was first reported in Wuhan, China, in December 2019; this virus has caused the worst pandemic of the past 100 years [1-3]. On December 31, 2019, Chinese authorities notified the World Health Organization (WHO) of an outbreak of pneumonia in Wuhan [2]. The WHO declared the coronavirus disease (COVID-19) outbreak to be a public health emergency of international concern on January 30, 2020, and it was declared a pandemic on March 12 [2,4]. It is commonly believed that the outbreak began in early December 2019.
Coronaviruses are a large family of viruses that are found in many different species of animals, including camels, cattle, cats, and bats. Zoonotic coronaviruses that have emerged in humans are Middle Eastern respiratory syndrome coronavirus (MERS-CoV), sudden acute respiratory syndrome coronavirus (SARS-CoV), and now SARS-CoV-2. This is the third time in two decades that a zoonotic coronavirus has emerged from animals to infect humans [4]. Of the betacoronaviruses, SARS-CoV-2 is more closely related to SARS-CoV than to MERS-CoV [5].
The origin of SARS-CoV-2, its intermediary animal host, and the mechanism of its species jump to humans are not known [6,7]. Initially, it was believed that the COVID-19 pandemic originated at the Huanan Seafood Wholesale Market located in Wuhan, China, where farm animals, bats, and snakes were also sold [8]; this is still believed by many people. Approximately half of the initial cases were exposed to the seafood market; however, the first two identified cases did not visit the seafood market [9]. Viral RNA was found in environmental samples from the wet market, such as surfaces [10]. Phylogenetic analysis revealed that the viral RNA found in the environmental samples was very closely related to viruses sampled from the earliest Wuhan patients, suggesting that the market played a role in the early spread of the virus [10]. The source of positive environmental samples from the market is unknown, and animal samples from the market are not available. Therefore, it has not been possible to identify an animal source at the market [10].
On March 2020, however, the timeline of the pandemic was questioned when it was determined that the first person infected with the new disease may have been a Hubei resident who was infected on November 17, 2019 [11]. However, official information states that the first patient presented on December 8, 2019, and that the first exposure may have been around December 1 in the Huanan Seafood Wholesale Market [2]. Local health authorities initially failed to report the coronavirus epidemic, resulting in a delay in reporting it to the WHO until December 31, 2019.
Emerging infectious diseases are becoming increasingly common [12,13]. The world is increasingly interconnected; therefore, it is essential to identify epidemics early [14]. A disease with true epidemic potential can grow exponentially within weeks or months; thus, each day of delay is a lost opportunity for prevention [15]. Rapid prediction, detection, and surveillance of outbreaks are critical in fighting emerging infectious diseases with epidemic potential [12]. The media may have reported a surge of unknown or undiagnosed cases of severe pneumonia, severe acute respiratory illness (SARI), or other related diseases prior to the official reporting of confirmed COVID-19 confirmed cases in China. Epidemic intelligence from open-source, informal data can provide early warnings of public health emergencies [12-14,16,17].
EpiWatch is a curated epidemic observatory that searches media reports, press releases, official reports, and social media for early detection of outbreaks of infectious diseases; it can be tailored for different languages [18]. EpiWatch provides early outbreak alerts and can be used to detect and monitor early reports of potential COVID-19 outbreaks through publicly available sources in settings with poor disease surveillance or censorship of information [19]. Early reports of unknown pneumonia in Hubei Province in China that appeared prior to official reports can be identified using open-source data, providing insight into whether COVID-19 was present in China before December 2019.
Aim
The aim of our study was to use open-source data to identify early signals of pneumonia and SARI in China prior to official recognition of the COVID-19 outbreak in December 2019.
Methods
EpiWatch is an open-source epidemic observatory that was developed at the University of New South Wales as a management web application enhanced by machine learning; it has been used to collect outbreak data since 2016. The principle of EpiWatch is that cases of infectious diseases or outbreaks may be reported in the news or discussed on social media before official notification by health authorities. EpiWatch mines open-source data to detect early signals, which can be customized for common clinical infectious disease syndromes. Many countries have weak or delayed surveillance systems and poor reporting. In other countries, censorship may prevent notification of serious epidemics. Open-source data can be used to help identify epidemic signals in such circumstances.
The system includes three major features. First, reports are gathered from international organizations and news outlets by an intelligent and modular system. An administrator can easily add new sources without requiring further development of the application. The data collected include news reports and social media posts as well as grey literature, such as government reports. If the format in which data is delivered changes for a given source, an administrator can promptly modify the system to adapt to this change. This includes adding or changing the languages used for searching. The system is set up to support a variety of intelligent data gathering elements, such as natural language processing algorithms, regular expression matching, and supervised machine learning algorithms, to process reports and attempt to identify important data points such as outcomes, locations, and diseases mentioned within the gathered data.
Second, EpiWatch reports are reviewed by a team of epidemiologists, ensuring a good level of quality control as well as increased accuracy and relevance. The EpiWatch management system is a web application that enables the internal team to log on and review reports and key data points identified by the automated data gathering system. The team can check the data collected by the automated system and correct any mistakes that are present. A machine learning system learns from this human input and corrections and uses that feedback to improve its ability to group reports and identify key information over time.
Finally, the EpiWatch management web application consists of two software programs. One is a web application that is built on the Vue framework, and the other is a server-side application built on the NodeJS framework. Both applications are written in JavaScript. The third software program is the data-gathering program, which is also a NodeJS application written in JavaScript. This program is scheduled to run on a regular basis to re-scan sources at intervals chosen by the system administrator. Searches can be tailored for specific languages or regions as well as for specific infectious disease syndromes. The data are stored in a PostgreSQL database. Most of the data is textual in nature and is easily compressed; therefore, the storage requirements are currently very modest (<100 MB). The EpiWatch observatory is managed and funded by the Australian National Health and Medical Research Council (NHMRC) Centre of Research Excellence, Integrated Systems for Epidemic Response (ISER) and is managed by staff at the Biosecurity Program, The Kirby Institute, University of New South Wales Sydney.
To capture early reports of SARI or pneumonia-related illnesses in China, searches were performed in the Chinese language using keywords reflecting severe acute respiratory syndrome or pneumonia as well as Wuhan and China as geolocations. We performed searches from October 1, 2019, to February 14, 2020. Any relevant news reports with the keywords pneumonia, SARI and related terms, and coronavirus were extracted. The information before December 31, 2019 (the date on which the WHO was notified of the COVID-19 outbreak) was reviewed for potential early signals of COVID-19. Google and the Chinese search engine Baidu were used [20,21]. Reports in Chinese were retrieved and reviewed by EK, XC, and MZ and translated to English.
Results
Between October 2019 and February 2020, a total of 218 reports were found and included in the study. There were no duplicates. We identified two potentially relevant news reports prior to December 31, 2019. Figure 1 shows the number of pneumonia and/or SARI reports from October 1, 2019, to February 14, 2020. It shows an increase in reports after the official notification to the WHO on December 31, 2019. A report appeared on December 26, 2019, with the heading “One sample is suspected as novel coronavirus”; this report appears to have been retracted, as the link to the news item has become invalid [22]. We found 11 reports of cases of pneumonia between October 1 and December 31, 2019, including a case identified retrospectively in March 2020, which is believed to be an index case. The number of reports in the same period one year prior was determined for comparison; there were 12 reports in 2018. Of the 11 reports in 2019, 3 (27%) were cases of pneumonia of unknown cause, and 7 (64%) had known causes; 2 reports (18%) were related to pulmonary nodules, 3 (27%) were caused by lung cancer, cerebral infarction, or asthma, and 2 cases (18%) were caused by bacterial infection. The information of interest was the single report of unknown serious pneumonia in November 2019 in Hubei, the province in China where the COVID-19 pandemic arose.
Figure 1.
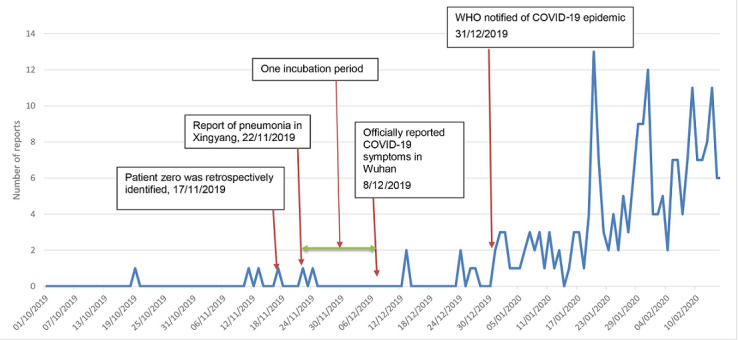
Reports of pneumonia, severe acute respiratory illness, or coronavirus from October 1, 2019, to February 14, 2020. COVID-19: coronavirus disease; WHO: World Health Organization.
On November 22, 2019, a local newspaper, the Wuhan Evening News, reported that a patient with severe pneumonia of unknown cause was taken to Wuhan as an emergency transfer by helicopter from Xiangyang in Hubei Province, 325 kilometers from Wuhan [23]. Figure 2 shows the location of Xiangyang in relation to Wuhan. After November 22, there were no reports of pneumonia in the local media, although it was later confirmed that by December 30, 2019, there were 27 cases of pneumonia of unknown cause in Wuhan.
Figure 2.
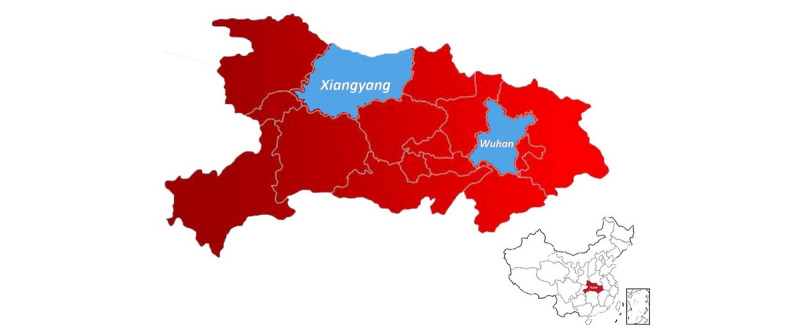
Location of Xiangyang relative to Wuhan within Hubei Province.
Discussion
The origin of the COVID-19 epidemic is unknown. Only half the initial patients were exposed to the Huanan Seafood Wholesale Market [9], and the first two cases in Wuhan did not visit the market. No pneumonia or SARI signals in Wuhan were identified prior to December 31, which supports the relatively recent emergence of COVID-19. However, open-source intelligence identified a case of severe pneumonia in Xiangyang, Hubei Province, 325 km from Wuhan, who was transferred to Wuhan for treatment on November 21, 2019. This case may be part of an early outbreak cluster. In early March, it was reported that the first case of COVID-19, a different case identified retrospectively, may have been observed on November 17 [24-26]. Approximately one COVID-19 incubation period (2 weeks) [9] after November 17 to November 21, the first formally reported cases in Wuhan became symptomatic (around December 1-8). If no definitive diagnosis was made, further diagnostic investigation of the case from Xiangyang and epidemiological investigation is warranted to determine if this case did have COVID-19. There may be a connection between the Xiangyang patient and an unidentified early cluster of COVID-19.
From December 31, 2019, through January 3, 2020, a total of 44 case patients with pneumonia of unknown etiology were detected by syndromic surveillance by the China Center for Disease Control and Prevention. Exposure to the Huanan Seafood Wholesale Market was initially suspected to be the origin of the virus, and the market was closed on January 1, 2020. At least 35 environmental samples from the seafood section of the Huanan Seafood Market in Wuhan tested positive for the virus [27,28]. However, the first two cases did not report visiting the seafood market, and there is no epidemiological link between the first patient and later cases [5,28]. This, together with the identification of at least two severe pneumonia cases in November (the one identified in this study and the case on November 17, 2019), suggests that the epidemic originated earlier than December 2019.
The absence of news reports in December is curious given that the outbreak appears to have been recognized in early December. It is possible that media reporting was censored; this is supported by what appears to be a retracted news item on December 26. The findings also support the relatively recent origin of COVID-19 in November 2019. The case reported on November 22 was transferred to Wuhan approximately one incubation period before the first cases were reported on December 8. This case should be further investigated, as only half of the early cases were exposed to the Huanan Seafood Market. The Chinese government has been questioned about its failure to identify and report the epidemic early, which resulted in worldwide spread of the disease and led to a pandemic [29]. Surveillance of waste water may also shed light on the origin. A sample of stored waste water in Spain tested positive for SARS-CoV-2 in March 2019, raising questions about whether the infection was present much earlier than December that year [30].
Epidemic diseases grow exponentially and rapidly [31], as seen in China, Europe, and the United States [32]. Early detection and epidemic control can reduce epidemic growth and prevent further spread. Open-source intelligence is a potential tool to aid early detection, especially where formal surveillance data are lacking. Although these data are not validated, once a signal is detected, it can and should be formally investigated, tested, and validated. The use of open-source epidemic intelligence can supplement conventional surveillance to provide early detection of serious emerging epidemics, especially where official disease surveillance reporting is lacking.
Acknowledgments
EpiWatch is funded by the Australian NHMRC Centre for Research Excellence, Integrated Systems for Epidemic Response (ISER) (grant number APP1107393).
Abbreviations
- COVID-19
coronavirus disease
- ISER
Integrated Systems for Epidemic Response
- MERS-CoV
Middle Eastern respiratory syndrome coronavirus
- NHMRC
National Health and Medical Research Council
- SARI
severe acute respiratory illness
- SARS-CoV
severe acute respiratory syndrome coronavirus
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- WHO
World Health Organization
Footnotes
Conflicts of Interest: EK works as a research associate at ISER. She receives a salary from ISER (grant number APP1107393).
References
- 1.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 Feb;395(10223):497–506. doi: 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronavirus disease (COVID-19) pandemic. World Health Organization. [2020-07-02]. https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- 3.Zhao S, Lin Q, Ran J, Musa SS, Yang G, Wang W, Lou Y, Gao D, Yang L, He D, Wang MH. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: A data-driven analysis in the early phase of the outbreak. Int J Infect Dis. 2020 Mar;92:214–217. doi: 10.1016/j.ijid.2020.01.050. https://linkinghub.elsevier.com/retrieve/pii/S1201-9712(20)30053-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long JB, Ehrenfeld JM. The Role of Augmented Intelligence (AI) in Detecting and Preventing the Spread of Novel Coronavirus. J Med Syst. 2020 Feb 04;44(3):59. doi: 10.1007/s10916-020-1536-6. http://europepmc.org/abstract/MED/32020374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ralph R, Lew J, Zeng T, Francis M, Xue B, Roux M, Toloue Ostadgavahi A, Rubino S, Dawe NJ, Al-Ahdal MN, Kelvin DJ, Richardson CD, Kindrachuk J, Falzarano D, Kelvin AA. 2019-nCoV (Wuhan virus), a novel Coronavirus: human-to-human transmission, travel-related cases, and vaccine readiness. J Infect Dev Ctries. 2020 Jan 31;14(1):3–17. doi: 10.3855/jidc.12425. http://www.jidc.org/index.php/journal/article/view/32088679. [DOI] [PubMed] [Google Scholar]
- 6.Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020 Jul;24:91–98. doi: 10.1016/j.jare.2020.03.005. https://linkinghub.elsevier.com/retrieve/pii/S2090-1232(20)30054-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuen K, Ye ZW, Fung S, Chan C, Jin D. SARS-CoV-2 and COVID-19: The most important research questions. Cell Biosci. 2020;10:40. doi: 10.1186/s13578-020-00404-4. https://cellandbioscience.biomedcentral.com/articles/10.1186/s13578-020-00404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biscayart C, Angeleri P, Lloveras S, Chaves TDSS, Schlagenhauf P, Rodríguez-Morales AJ. The next big threat to global health? 2019 novel coronavirus (2019-nCoV): What advice can we give to travellers? - Interim recommendations January 2020, from the Latin-American society for Travel Medicine (SLAMVI) Travel Med Infect Dis. 2020 Jan;33:101567. doi: 10.1016/j.tmaid.2020.101567. http://europepmc.org/abstract/MED/32006657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020 Mar 26;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. http://europepmc.org/abstract/MED/31995857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Holmes EC. A Genomic Perspective on the Origin and Emergence of SARS-CoV-2. Cell. 2020 Apr 16;181(2):223–227. doi: 10.1016/j.cell.2020.03.035. https://linkinghub.elsevier.com/retrieve/pii/S0092-8674(20)30328-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma J. Coronavirus: China’s first confirmed Covid-19 case traced back to November 17. South China Morning Post. 2020. Mar 13, [2020-03-15]. https://www.scmp.com/news/china/society/article/3074991/coronavirus-chinas-first-confirmed-covid-19-case-traced-back.
- 12.Ellwanger JH, Kaminski VDL, Chies JA. Emerging infectious disease prevention: Where should we invest our resources and efforts? J Infect Public Health. 2019 May;12(3):313–316. doi: 10.1016/j.jiph.2019.03.010. https://linkinghub.elsevier.com/retrieve/pii/S1876-0341(19)30119-4. [DOI] [PubMed] [Google Scholar]
- 13.Rees E, Ng V, Gachon P, Mawudeku A, McKenney D, Pedlar J, Yemshanov D, Parmely J, Knox J. Risk assessment strategies for early detection and prediction of infectious disease outbreaks associated with climate change. Can Commun Dis Rep. 2019 May 02;45(5):119–126. doi: 10.14745/ccdr.v45i05a02. doi: 10.14745/ccdr.v45i05a02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan K. Warning system for infectious diseases and method therefor. WIPO patent WO2013120199A1. Google Patents. 2013. [2020-07-02]. https://patents.google.com/patent/WO2013120199A1/en.
- 15.MacIntyre R. The risk of selective investment in downstream pandemic planning. Global Biosecurity. 2019 Aug 13;1(2):85. doi: 10.31646/gbio.36. [DOI] [Google Scholar]
- 16.Joshi A, Karimi S, Sparks R, Paris C, Macintyre CR. Survey of Text-based Epidemic Intelligence. ACM Comput Surv. 2020 Jan 21;52(6):1–19. doi: 10.1145/3361141. [DOI] [Google Scholar]
- 17.Yan S, Chughtai A, Macintyre C. Utility and potential of rapid epidemic intelligence from internet-based sources. Int J Infect Dis. 2017 Oct;63:77–87. doi: 10.1016/j.ijid.2017.07.020. https://linkinghub.elsevier.com/retrieve/pii/S1201-9712(17)30199-6. [DOI] [PubMed] [Google Scholar]
- 18.Hii A, Chughtai AA, Housen T, Saketa S, Kunasekaran MP, Sulaiman F, Yanti NS, MacIntyre CR. Epidemic intelligence needs of stakeholders in the Asia-Pacific region. Western Pac Surveill Response J. 2018 Dec 18;9(4):28–36. doi: 10.5365/wpsar.2018.9.2.009. http://europepmc.org/abstract/MED/30766745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puca C, Trent M. Using the Surveillance Tool EpiWATCH to Rapidly Detect Global Mumps Outbreaks. Global Biosecurity. 2020 Apr 30;1(3) doi: 10.31646/gbio.54. [DOI] [Google Scholar]
- 20.Baidu. [2019-11-01]. https://www.baidu.com/
- 21.Google. [2019-11-01]. https://www.google.com/
- 22.Wuhan, China, one sample is suspected as novel coronavirus 2019. Baidu. 2019. Dec 26, [2019-12-26]. https://tinyurl.com/ya7ar3v8.
- 23.Wuhan Evening News Xiangyang severe pneumonia patient moves to wuhan emergency by helicopter. Article in Chinese. Sohu. 2019. Nov 22, [2019-11-22]. https://www.sohu.com/a/355347300_120424456.
- 24.Hale T. The First COVID-19 Case Was Earlier Than Initially Thought, Suggests New Report. IFLScience. 2020. Mar 17, [2020-03-17]. https://www.iflscience.com/health-and-medicine/the-first-covid19-case-was-earlier-than-intially-thought-suggests-new-report/
- 25.Luchini S, Teschl M, Pintus P, Baunez C, Moatti JP. Urgently Needed for Policy Guidance: An Operational Tool for Monitoring the COVID-19 Pandemic. SSRN Journal. 2020 Mar 20; doi: 10.2139/ssrn.3563688. [DOI] [Google Scholar]
- 26.Mimouni F, Lakshminrusimha S, Pearlman SA, Raju T, Gallagher PG, Mendlovic J. Perinatal aspects on the covid-19 pandemic: a practical resource for perinatal-neonatal specialists. J Perinatol. 2020 May 10;40(5):820–826. doi: 10.1038/s41372-020-0665-6. http://europepmc.org/abstract/MED/32277162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacIntyre CR. Wuhan novel coronavirus 2019nCoV – update January 27th 2020. Global Biosecurity. 2020 Jan 28;1(3) doi: 10.31646/gbio.51. [DOI] [Google Scholar]
- 28.She J, Jiang J, Ye L, Hu L, Bai C, Song Y. 2019 novel coronavirus of pneumonia in Wuhan, China: emerging attack and management strategies. Clin Transl Med. 2020 Feb 20;9(1):19. doi: 10.1186/s40169-020-00271-z. doi: 10.1186/s40169-020-00271-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan L. Coronavirus Crisis Shows China’s Governance Failure. New York Times. 2020. Feb 04, [2020-07-02]. https://www.nytimes.com/2020/02/04/business/china-coronavirus-government.html.
- 30.Chavarria-Miró G, Anfruns-Estrada E, Guix S. Sentinel surveillance of SARS-CoV-2 in wastewater anticipates the occurrence of COVID-19 cases. medRxiv. 2020 Jun 13; https://www.medrxiv.org/content/10.1101/2020.06.13.20129627v1.article-info. [Google Scholar]
- 31.MacIntyre R. The risk of selective investment in downstream pandemic planning. Global Biosecurity. 2019 Aug 13;1(2):85. doi: 10.31646/gbio.36. [DOI] [Google Scholar]
- 32.Coronavirus disease (COVID-19) Situation Report – 107. World Health Organization. 2020. May 06, [2020-05-06]. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200506covid-19-sitrep-107.pdf?sfvrsn=159c3dc_2.


