Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) disease, more commonly COVID-19 has emerged as a world health pandemic. There are couples of treatment methods for COVID-19, however, well-established drugs and vaccines are urgently needed to treat the COVID-19. The new drug discovery is a tremendous challenge; repurposing of existing drugs could shorten the time and expense compared with de novo drug development. In this study, we aimed to decode molecular signatures and pathways of the host cells in response to SARS-CoV-2 and the rapid identification of repurposable drugs using bioinformatics and network biology strategies. We have analyzed available transcriptomic RNA-seq COVID-19 data to identify differentially expressed genes (DEGs). We detected 177 DEGs specific for COVID-19 where 122 were upregulated and 55 were downregulated compared to control (FDR<0.05 and logFC ≥ 1). The DEGs were significantly involved in the immune and inflammatory response. The pathway analysis revealed the DEGs were found in influenza A, measles, cytokine signaling in the immune system, interleukin-4, interleukin −13, interleukin −17 signaling, and TNF signaling pathways. Protein-protein interaction analysis showed 10 hub genes (BIRC3, ICAM1, IRAK2, MAP3K8, S100A8, SOCS3, STAT5A, TNF, TNFAIP3, TNIP1). The regulatory network analysis showed significant transcription factors (TFs) that target DEGs, namely FOXC1, GATA2, YY1, FOXL1, NFKB1. Finally, drug repositioning analysis was performed with these 10 hub genes and showed that in silico validated three drugs with molecular docking. The transcriptomics signatures, molecular pathways, and regulatory biomolecules shed light on candidate biomarkers and drug targets which have potential roles to manage COVID-19. ICAM1 and TNFAIP3 were the key hubs that have demonstrated good binding affinities with repurposed drug candidates. Dabrafenib, radicicol, and AT-7519 were the top-scored repurposed drugs that showed efficient docking results when they tested with hub genes. The identified drugs should be further evaluated in molecular level wet-lab experiments in prior to clinical studies in the treatment of COVID-19.
Keywords: COVID-19; SARS-CoV-2,transcriptomics; Drug repositioning,
Highlights
-
•
Molecular transcriptomes alterations of the lung epithelial cells in response to SARS-CoV-2 were detected.
-
•
Significant molecular pathways altered in COVID-19 revealed.
-
•
Hub proteins were detected as novel drug targets for COVID-19.
-
•
Molecular regulatory signatures were identified.
-
•
Repurposable drugs were identified for COVID-19.
1. Introduction
The novel coronavirus-2019 (SARS-CoV-2) has emerged as global public health threats with massive loss of lives worldwide (Cucinotta and Vanelli, 2020). The number of infections currently (07/17/2020) 13, 378, 853with 580,045 deaths caused by this virus (WHO COVID-19 Situation Reports, 07/17/2020)/). The infection of SARS-CoV-2 (COVID-19) first appeared in Wuhan, China, in December 2019. Now, SARS-CoV-2 has been declared as pandemic by WHO (Cucinotta and Vanelli, 2020; Sanders et al., 2020). The infections with this virus result in mild cold symptoms, fever, cough, and pneumonia in patients (N. Chen et al., 2020; Huang et al., 2020). The host immune system response to SARS-CoV-2 infection is still unclear, given increasing global challenges to COVID-19. The immune systems recognize the viral RNAs via cytoplasmic pattern recognition receptors. This recognition results in interferon activity (IFN) expression and anti-viral effectors modulation (Fehr et al., 2017; Nelemans and Kikkert, 2019; Newton et al., 2016). Coronaviruses, such as SARS-CoV and MERS-CoV (Middle East respiratory syndrome coronavirus), have evolved mechanisms to slow down and postpone the production of IFNs that normally result in excessive inflammatory host reactions that result in serious lung inflammation (Chen et al., 2020; Fehr et al., 2017; Kindler and Thiel, 2016). Recent reports suggest that excessive production of inflammatory cytokines in response to dysregulated host response correlated with the severity during SARS-CoV and MERS-CoV infection (de Wit et al., 2016; Fehr et al., 2017; Newton et al., 2016). Some studies demonstrated the increased levels of - inflammatory cytokines release-is crucial in COVID-19 disease progression ( Chen et al., 2020; Huang et al., 2020). However, the immunological mechanisms are still unclear.
Analyses of cellular transcriptomes in response to viral infections are very important to detect host immune response (Monaco et al., 2019; Wilson et al., 2017; Cavalli et al., 2020a). Blanco-Melo et al. recently examined SARS-Cov-2 infected lung cells' transcriptional responses using RNA sequencing (RNA-Seq) gene expression data of COVID-19 infected lung epithelial cells (Blanco-Melo et al., 2020). Their efforts were made to detect core transcriptional signatures. Fagone et al. also recently characterized differentially expressed genes and enrichment analysis of transcriptional responses to COVID-19 in lung epithelial cells (Fagone et al., 2020). Cavalli and colleagues have pointed out the transcriptional responses of COVID-19 may be associated with mTOR signaling (Cavalli et al., 2020a). Despite the important findings from these previous studies, the comprehensive evaluation of host transcriptional response to COVID-19, and the integration of biomolecular networks are required to uncover molecular signatures and potential candidate repurposable drugs for COVID-19. However, the functional characterization, regulatory molecular signatures, and potential candidate drug signature have yet to be determined. Therefore, we decided to integrate the transcriptional expression with pathway analysis, hub gene analysis, transcription factor analysis, gene-drug interaction analysis. Hence, we employed comprehensive systems biology and network bioinformatics analyses to decode core genes, pathways, and repurposable drugs for COVID-19.
In this study, we have performed an integrative analysis of transcriptomic RNA-seq data of COVID-19 to detect differentially expressed genes. Then, we performed gene ontology-based pathway enrichment analysis. Protein-protein interaction (PPI) network analysis was also conducted and revealed hub genes. Hub protein-drug interaction showed several candidate repurposable drugs for COVID-19 and their in silico validations were carried out via molecular docking analysis.
2. Materials and methods
2.1. Identification of differentially expressed genes lung epithelial cells infected with SARS-CoV-2
The transcriptomic data of lung epithelial cells, infected with SARS-CoV-2, and documented in the Gene Expression Omnibus (GEO) with GSE147507 accession number was recently analyzed by Blanc-Melo et al. (Blanco-Melo et al., 2020). Raw data have been downloaded from the GEO database (Accessed and downloaded on 5 April 2020). We have only taken into consideration the raw counts of primary cells in the lung and the expression of alveolar adenocarcinoma cells is omitted. These data contain 20 samples, but we only consider six samples, which contain three primary lung epithelial samples infected by SARS-CoV-2 and three samples of lung mock epithelial (i.e. uninfected).
The RNA-Seq dataset was analyzed in R using edgeR package. We utilized the filtering criteria to include genes that have non-zero counts per million (CPM) reads in at least n samples and the group size of n is small. Then, in this study, the read count data were normalized using the logarithm of counts per million (log CPM) of the trimmed mean of M values (TMM) (Robinson and Oshlack, 2010). To classify differentially expressed genes in COVID-19, a generalized linear model (GLM) was used to implement a likelihood ratio test using the edgeR package. The TMM normalization was applied to normalize read counts among different samples. To determine the statistical significance, FDR <0.05 and log2FC ≥ 1 were used, and genes that passed from these cut-offs were selected as DEGs and used for further consideration in the network construction.
2.2. Gene enrichment and pathway analysis
To clarify the possible biological process and pathways behind SARS-CoV-2, gene functional annotation analysis was carried out via g: profiler to determine the biological process and pathways involved in the DEGs., We have conducted pathway analyses using enrichment analysis protocol g: GOSt method in g: Profiler suggested by Raudvere and coworkers (Raudvere et al., 2019). g: GOSt method, embedded in g: Profiler, performs computational enrichment analysis and integrates user-input gene functional annotation based on their biological functions, thus defining over-represented (significantly enriched) biological process and pathways for DEGs. Notably, g: GOSt databases, including Gene Ontology and pathways (Reactome and KEGG) are regularly updated. An adjusted p-value <0.05 was considered as a cut of criterion to designate statistical significance in enrichment analysis.
2.3. Hub gene network analysis
Data from the DifferentialNet Database (Basha et al., 2018) were collected for tissue-specific protein-protein interactions (PPI) analysis. The differential protein-protein interactions in lung tissues were performed through NetworkAnalyst tool (Zhou et al., 2019). The hub genes derived from the PPI may represent essential biological signaling molecules. The network was built with a degree filter value > 15. The degree (greater than 30) was considered for the identification of highly interacting hub proteins from PPI analysis. The interactions among the hub proteins were further evaluated via GeneMania-online tool (Franz et al., 2018).
2.4. Identification of transcription factors that may regulate differentially expressed genes
DEGs were checked with JASPAR database which is a publicly available curated and non-redundant DNA binding transcription factors (TFs) repository. The DEGs-TFs interaction analyses retrieved from the JASPAR database via NetworkAnalyst have defined transcriptional regulatory TFs (Zhou et al., 2019). The significant hub TFs were selected based on degree cutoff ≥ 45.
2.5. Hub proteins specific drug repositioning
We used the Library of Integrated Network-based Cellular Signatures (LINCS) – L1000 data which covers gene expression data from ~50 human cell lines in response to ~20,000 compounds (Campillos et al., 2008). To identify small molecules that can potentially reverse gene expression of COVID-19, we queried top-scored 10 hub genes as differential expression vectors which are all up-regulated as input. We employed the L1000CDS2 (Duan et al., 2016) search engine, which provides 30,000 significant signatures with reverse and mimic modes that were gathered from the LINCS L1000 data, to determine small molecule signatures related with COVID-19 specific hub proteins. The cosine distances between input and all the gene expression signatures in L1000CDS2 database were calculated by the search engine. The top-ranked signatures will be those of small cosine distances which comes from mimic mode, or those with larger cosine distances which represent reverse mode. Since we aimed to identify small molecules that reverse the gene expression of COVID-19, we used larger cosine distances which represent the reverse mode. The resultant drugs were ranked according to their cosine distance scores and the top 10 were selected for further investigation. Drugs were checked through literature and publicly available datasets such as CTD (Davis et al., 2017), geneXpharma (Turanli et al., 2017), ChEBI (Degtyarenko et al., 2008) to identify their mechanism of action and associated conditions.
2.6. Molecular docking analysis of hub proteins and repurposed drug candidates
The structure of hub proteins (ICAM1, TNFAIP3, BIRC3, S100A8, MAP3K8, TNF) were retrieved from Protein Data Bank (PDB) (Berman et al., 2002) with the codes of 5MZA, 2VFJ, 3EB5, 1MR8, 5IU2, 1TNF respectively. The structure of candidate drugs which were Radicicol, Dabrafenib, AT-7519 and inhibitors of each hub proteins, A-205804 (ICAM1), Lenalidomide (TNFAIP3), LCL161 (BIRC3), JQ1 (S100A8), TPL2 Kinase (MAP3K8) and Thalidomide (TNF) were obtained from PubChem database (Kim et al., 2019). Molecular docking analyses were performed via Autodock Vina software (Trot et al., 2009). Interactions between candidate drugs and targets were calculated based on their binding affinities (kcal/mol). Inhibitors of each hub proteins were considered as positive control so that significance of docking was determined according to binding affinities of inhibitors for each hub proteins.
3. Results
3.1. Differential gene identification and biological process analysis
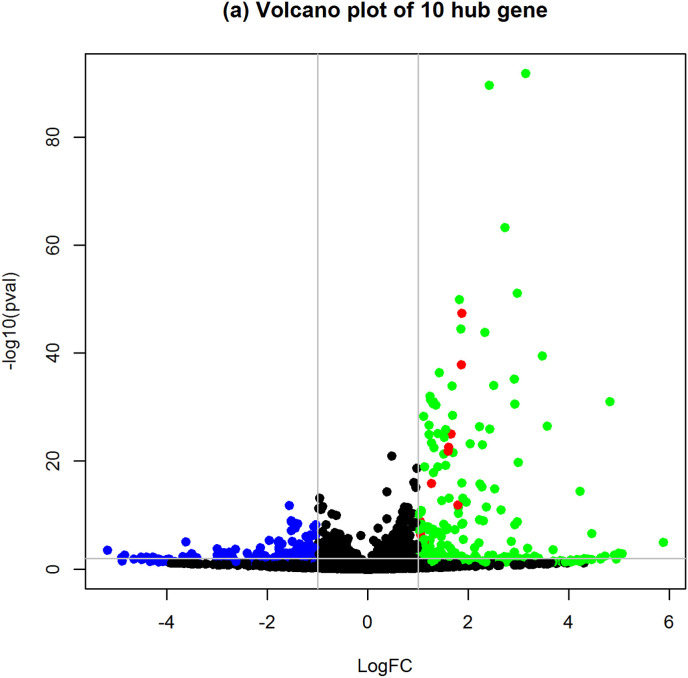
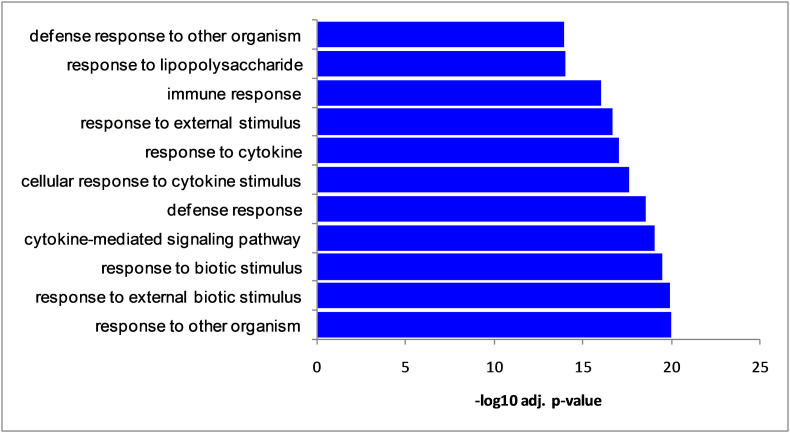
We have analyzed the transcriptomics of SARS-CoV-2 infected epithelial cells of the lung. The integrative analysis by edgeR package revealed 177 differentially expressed genes (DEGs) in epithelial cells of the lung infected with SARS-CoV-2. Among the 177 DEGs, 122 DEGs were upregulated and 55 DEGs were downregulated (Fig. 1 ). Table 1 shows the top 25 upregulated DEGs and Table 2 represents 25 top down-regulated DEGs. The total DEGs are provided as in Supplementary Table S1. Then, we conducted a biological process (BP) analysis of the identified DEGs. The significant BP terms were enriched in cytokine-mediated signaling pathway, response to other organisms, response to external biotic stimulus, and defense response (Fig. 2 ).
Fig. 1.
Volcano plot of differentially expressed genes (DEGs). The green color represents the upregulated DEGs, and blue color represents downregulated DEGs. The read color represents the hub genes which were identified from protein-protein interactions. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Table 1.
Top 25 upregulated differentially expressed genes.
| Gene Symbol | logFC | logCPM | LR | PValue | FDR |
|---|---|---|---|---|---|
| SPRR2F | 5.88 | −1.29 | 19.38 | 1.07E-05 | 0.001176 |
| CSF3 | 4.81 | 2.31 | 137.45 | 9.61E-32 | 1.14E-28 |
| PLA2G4E | 4.46 | −0.82 | 26.66 | 2.42E-07 | 3.99E-05 |
| S100A7 | 4.22 | 0.016 | 61.55 | 4.31E-15 | 1.79E-12 |
| S100A7A | 3.69 | −1.41 | 13.49 | 0.00024 | 0.016674 |
| SPRR2E | 3.57 | 1.87 | 116.79 | 3.18E-27 | 2.79E-24 |
| CXCL5 | 3.47 | 2.99 | 176.17 | 3.32E-40 | 7.15E-37 |
| MIR936 | 3.18 | −1.15 | 14.57 | 0.000135 | 0.010371 |
| CCL20 | 3.14 | 4.91 | 416.20 | 1.64E-92 | 1.29E-88 |
| CSF2 | 2.99 | 1.86 | 86.20 | 1.62E-20 | 8.73E-18 |
| SPRR2D | 2.97 | 4.75 | 229.26 | 8.61E-52 | 3.40E-48 |
| HMGN2P46 | 2.97 | −0.19 | 36.24 | 1.74E-09 | 4.21E-07 |
| CES1P1 | 2.92 | −0.22 | 33.77 | 6.20E-09 | 1.34E-06 |
| IL6 | 2.92 | 3.07 | 135.23 | 2.94E-31 | 3.17E-28 |
| TNIP3 | 2.91 | −0.97 | 11.52 | 0.000688 | 0.037079 |
| IFI27 | 2.91 | 4.57 | 156.46 | 6.71E-36 | 1.14E-32 |
| DDIT4L | 2.85 | −0.74 | 20.20 | 6.96E-06 | 0.000817 |
| IL36G | 2.73 | 4.31 | 285.04 | 5.97E-64 | 2.83E-60 |
| VNN3 | 2.65 | 0.27 | 46.05 | 1.15E-11 | 3.74E-09 |
| XAF1 | 2.52 | 2.82 | 63.81 | 1.37E-15 | 5.78E-13 |
| MX1 | 2.51 | 4.97 | 151.20 | 9.45E-35 | 1.49E-31 |
| PDZK1IP1 | 2.43 | 3.29 | 114.22 | 1.16E-26 | 9.50E-24 |
| SAA2 | 2.42 | 5.39 | 406.19 | 2.47E-90 | 1.46E-86 |
| IFI6 | 2.36 | 3.93 | 48.78 | 2.86E-12 | 1.01E-09 |
| IL8 | 2.33 | 7.43 | 196.16 | 1.43E-44 | 3.40E-41 |
Table 2.
25 top down-regulated differentially expressed genes.
| Gene Symbol | logFC | logCPM | LR | PValue | FDR |
|---|---|---|---|---|---|
| METTL7A | −1.03 | 3.45 | 33.82 | 6.03E-09 | 1.31E-06 |
| PPARGC1A | −1.03 | 2.49 | 25.44 | 4.55E-07 | 7.10E-05 |
| CCDC121 | −1.04 | 0.47 | 12.07 | 0.00051 | 0.029432 |
| ZNF718 | −1.04 | 1.47 | 11.34 | 0.000755 | 0.039905 |
| MIR221 | −1.05 | 1.85 | 15.04 | 0.000105 | 0.00854 |
| FBLN2 | −1.06 | 0.62 | 12.61 | 0.000383 | 0.023545 |
| CLDN23 | −1.06 | 0.62 | 10.93 | 0.000944 | 0.047456 |
| CYP4F3 | −1.07 | 2.25 | 14.82 | 0.000118 | 0.009293 |
| VTCN1 | −1.07 | 3.72 | 32.35 | 1.28E-08 | 2.69E-06 |
| ATG9B | −1.13 | 2.25 | 24.02 | 9.49E-07 | 0.000133 |
| ZNF488 | −1.14 | 1.79 | 25.33 | 4.82E-07 | 7.40E-05 |
| CCDC138 | −1.16 | 1.17 | 17.70 | 2.57E-05 | 0.002565 |
| LINC00672 | −1.17 | 0.88 | 18.02 | 2.19E-05 | 0.002243 |
| FAM131B | −1.18 | 0.56 | 12.96 | 0.000318 | 0.020438 |
| SEMA6D | −1.19 | 0.35 | 14.54 | 0.000137 | 0.010489 |
| CASD1 | −1.20 | 0.72 | 13.00 | 0.000311 | 0.020125 |
| POU2AF1 | −1.21 | 0.50 | 17.11 | 3.52E-05 | 0.003274 |
| NID1 | −1.23 | 2.48 | 24.31 | 8.16E-07 | 0.000117 |
| EAF2 | −1.24 | 0.10 | 11.42 | 0.000725 | 0.03882 |
| LOC284080 | −1.25 | 0.26 | 15.27 | 9.28E-05 | 0.007691 |
| CD36 | −1.28 | 0.31 | 15.96 | 6.44E-05 | 0.005551 |
| TNNI2 | −1.28 | 0.18 | 15.00 | 0.000107 | 0.008618 |
| PALM2 | −1.31 | −0.00 | 12.40 | 0.000428 | 0.025546 |
| SLC27A6 | −1.33 | 0.02 | 13.44 | 0.000246 | 0.017024 |
| LOC440028 | −1.35 | 0.46 | 16.89 | 3.95E-05 | 0.003614 |
Fig. 2.
Significant biological process (top 10) enriched by differentially expressed genes.
3.2. Molecular pathway analysis
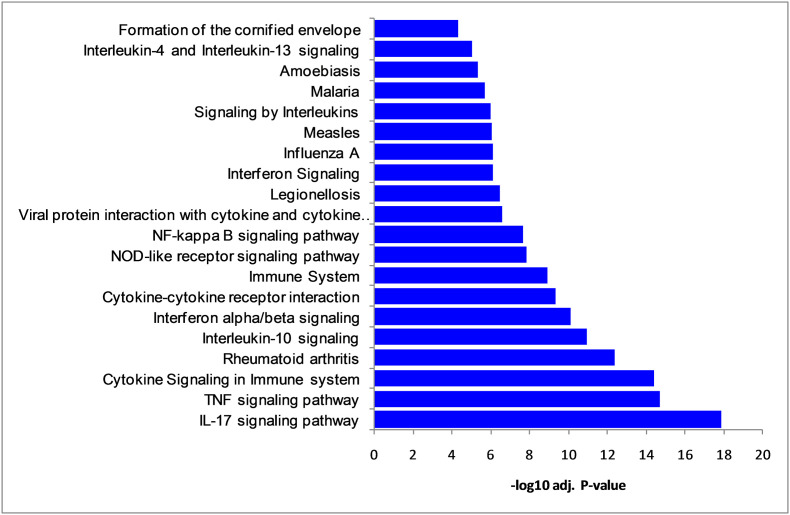
To identify potential roles of DEGs, we have performed the pathways analysis. The important pathways showed enriched pathways namely, cytokine signaling in the immune system, interleukin-10 signaling, immune system, interleukin-4, and interleukin-13 signaling, IL-17 signaling pathway, TNF signaling pathway, rheumatoid arthritis, viral protein interaction with cytokine, and influenza A (Fig. 3 and Supplementary Table S2).
Fig. 3.
Significant molecular pathways (top 20) enriched by differentially expressed genes.
3.3. Protein-protein interaction analysis to identify hub proteins
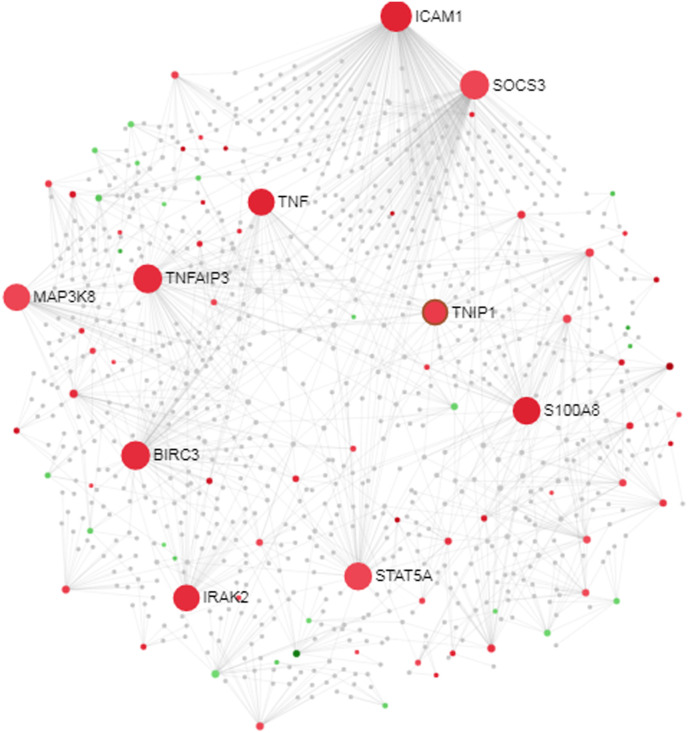
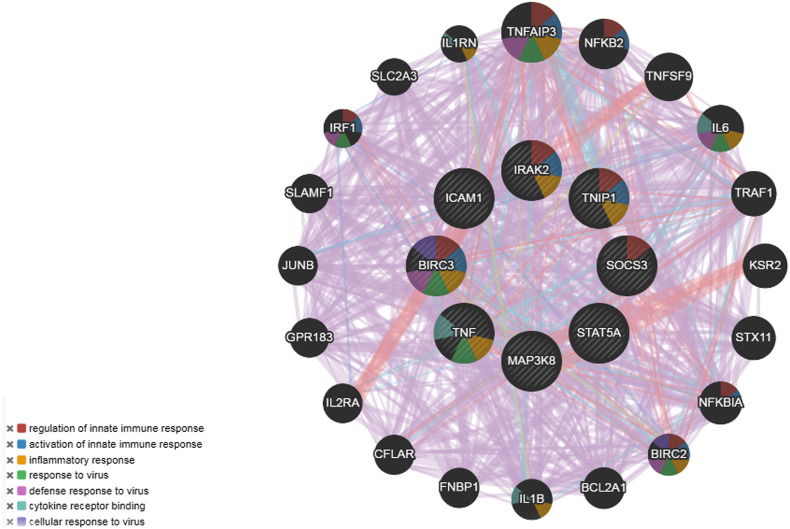
Using the PPI network analysis, we can determine the molecular pathwaysthat disturbed from the disease. For the identification of hub proteins, tissue-specific PPI sub-network of the DEGs was reconstructed (Fig. 4, Fig. 5 ). We have obtained 86 seeds with 1291 edges and 985 nodes. The topological assessment of the network revealed 10 hub genes (BIRC3, ICAM1, IRAK2, MAP3K8, S100A8, SOCS3, STAT5A, TNF, TNFAIP3, TNIP1) using degree metrics (Table 3 and Fig. 4, Fig. 5, Fig. 6 ). Fig. 6 represents hub genes and its predicted functions in GeneMania. During SARS CoV-2 infection development, these hub proteins may be of an important role in signal transduction.
Fig. 4.
Protein-Protein interaction network (Force atlas view). Red color nodes represent up-regulated genes. Gene color nodes represent down-regulated genes. Larger nodes represent the hub genes. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 5.
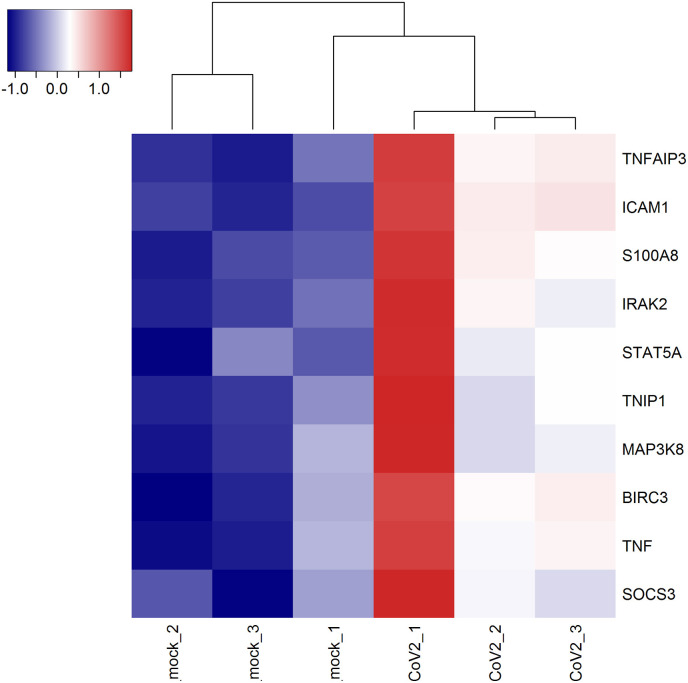
Heatmap shows the expression profiles of hub genes.
Table 3.
Hub proteins identified from protein-protein interaction network analysis of corresponding proteins encoded differentially expressed genes.
| Hub protein | Description | Regulation | Degree | Biological significance | Reference |
|---|---|---|---|---|---|
| ICAM1 | Intercellular Adhesion Molecule 1 | Up | 155 | Higher expression was demonstrated in human bronchial epithelial cells and nasopharyngeal epithelial cells in response to influenza virus infection | (Othumpangat et al., 2016; Traub et al., 2013) |
| SOCS3 | Suppressor of cytokine signaling 3 | Up | 77 | Upregulated SOCS3 was induced in lung epithelial cells infected with the Zika virus | (Liu et al., 2019; Seong et al., 2020) |
| TNFAIP3 | Tumor necrosis factor alpha-induced protein 3 | Up | 74 | Genetic variants associated with the hepatitis B virus in the Chinese population | Zhang et al. (2015) |
| BIRC3 | Baculoviral IAP Repeat Containing 3 | Up | 72 | Single nucleotide polymorphism (SNP) is associated with asthma | Roscioli et al. (2017) |
| STAT5A | Signal transducer and activator of transcription 5A | Up | 50 | STAT5 signaling plays a crucial role in human parvovirus B19 (B19V) replication | Ganaie et al. (2017) |
| S100A8 | S100 calcium binding protein A8 | Up | 49 | Released in infection-induced inflammation. | Wang et al. (2018) |
| MAP3K8 | Mitogen-Activated Protein Kinase Kinase 8 | Up | 39 | Downregulated in response to coronavirus infection | Mamoor (2020) |
| TNF | Tumor necrosis factor | Up | 36 | Higher levels expression of pro-inflammatory effector cytokines are elevated in patients with COVID-19.. | Huang et al. (2020) |
| IRAK2 | Interleukin-1 receptor-associated kinase | Up | 32 | The roles of IRAK2 and TNIP are yet to determine in COVID-19. | |
| TNIP1 | TNFAIP3 Interacting Protein 1 | Up | 31 | TNIP1 is a hub protein associated with autoimmune disease (Shamilov and Aneskievich, 2018). | (Chen et al., 2020) |
Fig. 6.
Protein-protein interaction of hub proteins obtained from Gene Mania.
3.4. Transcription factor analysis
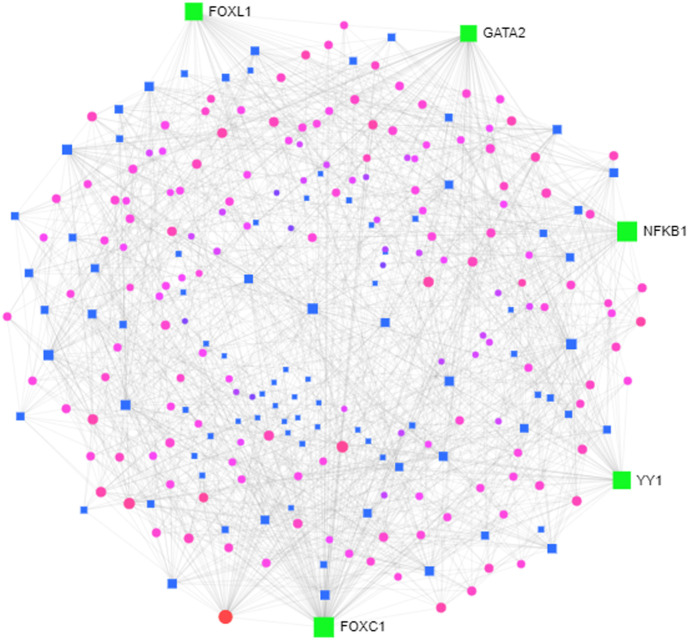
Genes are regulated at the transcriptional and post-transcription phases, gene dynamics, and expression rates are controlled by TFs and we, therefore, have developed gene-TF networks for the identification of master regulators in the DEGs (Fig. 7 ). The analyses of the network found TFs that target DEGs, namely FOXC1, GATA2, YY1, FOXL1, NFKB1.
Fig. 7.
Transcription factor gene interaction network (Force atlas view). Green color denotes hub transcription factor, blue color represents transcription factor, pink color denotes interacting genes. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. Drug repositioning based on hub proteins revealed candidate drugs
Hub proteins-based drug repositioning was carried out via L1000CDS2 tool. The search engine gave 50 drugs which have potential to mimic or reverse the COVID19 situation. We present all drugs in Supplementary Table S3 (Sheet-1). To assessing drugs that might potentially reverse the scenario of COVID19 gene expression, we selected 10 top-scored (ones with larger cosine distance value) drugs for further investigation (Table 4 ). After obtaining these 10 drugs, we searched them for more detailed information like drug category, clinical drug status and mechanism of actions through CTD (Davis et al., 2017), DrugBank and ChEBI (Degtyarenko et al., 2007). Table 4 summarizes top 10 repurposed drugs with their mechanisms of actions, approval status and associated diseases. Among these 10 drugs, 7 of them were approved whereas 3 drugs were in investigational status. The 50% of these 10 drugs were tyrosine kinase inhibitors, 30% of drugs have antibiotic activity, 20% of drugs have antifungal activity, and 80% of drugs were associated at least one type of cancer treatment. To develop docking strategies, we narrowed the results according to two criteria (i) drugs that were approved, (ii) drugs that were associated at least once with the treatment of a disease study or repurposing study. We selected top three high scored drugs that fitted above-mentioned criteria. These three drugs were radicicol, dabrafenib and AT-7519. The radicicol is an antifungal macrolactone antibiotic, derived from Diheterospora chlamydosporia and Chaetomium chiversii that is an inhibitor of protein tyrosine kinase and heat shock protein 90 (Hsp90) (Zhou et al., 2010). Also, radicicol was reported in colon cancer, prostate cancer, and ovarian cancer studies (Wu et al., 2013) as enhancer of apoptosis by inducing activation of apoptosis-related proteins (Kim et al., 2012). The dabrafenib is a selective Raf kinase inhibitor that is capable of inhibiting the kinase activity of wild-type B-Raf, B-RafV600E and c-Raf (Laquerre et al., 2009). In BRAF-mutated metastatic melanoma patients, dabrafenib reported to significantly improved progression-free survival (Hauschild et al., 2012). Also, dabrafenib plus trametinib combination targeted therapy for patients with BRAFV600E–mutated ATC was found highly promising by showing a high overall response rate (Subbiah et al., 2018). The AT-7519 is a novel small-molecule with multi-CDK inhibition activity, AT7519 showed antitumor activity in vivo for multiple myeloma resulting in prolonged survival (Santo et al., 2010).
Table 4.
Top ten scored repurposed drug candidates based on COVID-19 specific hub proteins.
| Drug name | Mechanism of Action | Approval Status | Associated Conditions | Reference |
|---|---|---|---|---|
| SYK-inhibitor | spleen tyrosine kinase inhibitor | investigational | chronic immune thrombocytopenia | Bussel et al. (2018); |
| Radicicol | HSP90 tyrosine kinase inhibitor with antifungal macrolactone antibiotic activity | approved | fungal infections, prostate cancer | Wu et al. (2013); Harashima et al., 2005 |
| Dabrafenib | BRAF inhibitor with antineoplastic activity | approved | metastatic melanoma, thyroid cancer | Hauschilde et al. (2012); Subbiah et al. (2018) |
| AT-7519 | CDK inhibitor leading to tumor regression | approved | cervical cancer, colon cancer | Xi et al. (2019) |
| Dasatinib | SRC family tyrosine kinase receptor inhibitor | approved | chronic myeloid leukemia, lung cancer | Kantarjianet al., 2010; Li et al. (2010) |
| Lovastatin | Inhibitor of HMG-CoA reductase (3-hydroxy-3-methylglutaryl-coenzyme A) with antilipemic activity | approved | cardiovascular diseases | Tobert (2003) |
| Thiostrepton | A cyclic oligopeptide and protein synthesis inhibitor, FOXM1 inhibitor with antibiotic activity | approved | colon cancer, lung cancer | Ju et al. (2015); Huang et al. (2019) |
| Linifanib | RTK inhibitor with antineoplastic activity | approved | gastric cancer, hepatocellular carcinoma | Chen et al. (2016); Cainap et al. (2015) |
| JNK–IN–5A | Selective inhibitor of JNK2 and JNK3 | Investigational | not available | Angell et al. (2007) |
| Withaferin-A | Steroidal lactone with anti-inflammatory, antiangiogenic, antifungal, and anticancer activities | Investigational | fungal infections, ovarian cancer, breast cancer | Kakar et al. (2017); Stan et al. (2008) |
3.6. Molecular docking analyses revealed in silico validated efficient repurposed drugs
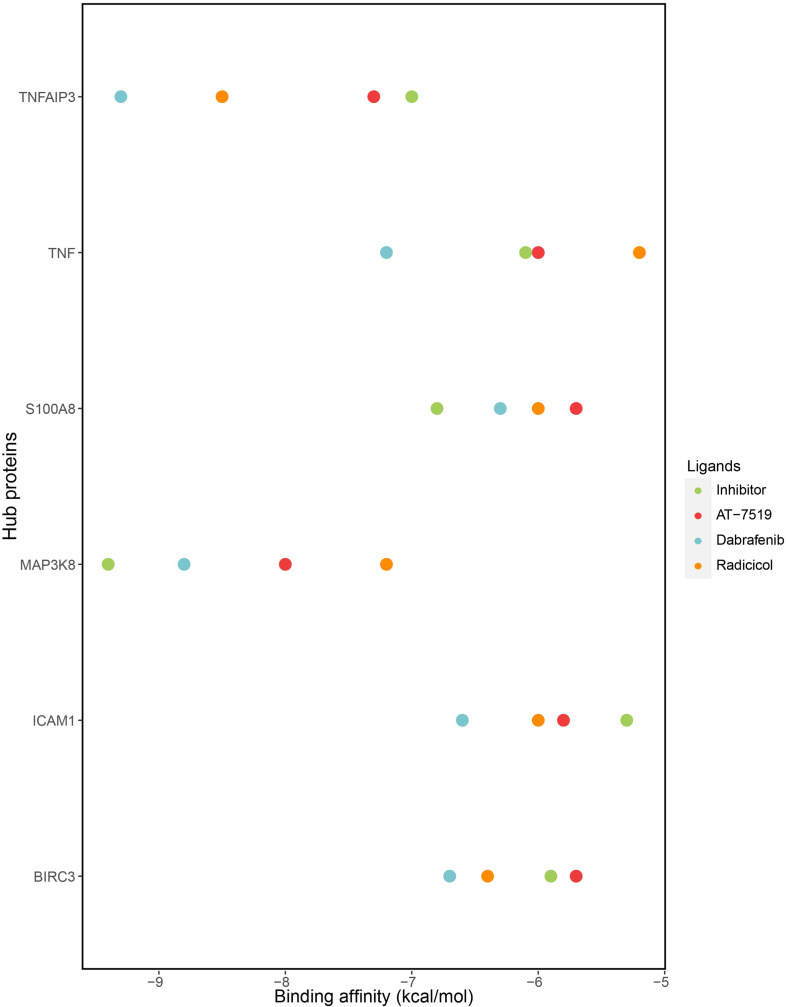
Molecular docking simulations were carried out for each six hubs, their inhibitors and three candidate drugs. The results of binding affinities were given in Fig. 8 and Supplementary Table S3 (Sheet-3). As a result, the binding affinities inhibitor of ICAM1 and candidate drugs was found −5.3 kcal/mol (A-205804), −6 kcal/mol (radicicol), −6.6 kcal/mol (dabrafenib), −5.8 kcal/mol (AT-7519). The results for TNFAIP3 were −7 kcal/mol (its inhibitor (lenalidomide)) and −8.5 kcal/mol (radicicol), −9.3 kcal/mol (dabrafenib), −7.3 (AT-7519). For BIRC3, binding affinities were −5.9 kcal/mol, −6.4 kcal/mol, −6.7 kcal/mol, −5.7 kcl/mol for its inhibitor (lenalidomide), radicicol, dabrafenib and AT-7519 kcal/mol respectively. For S100A8, its inhibitor (JQ1), radicicol, dabrafenib and AT-7519 were −6.8 kcal/mol, −6 kcal/mol, −6.3 kcal/mol, −5.7 kcal/mol respectively. For MAP3K8 -9.4 kcal/mol (its inhibitor (TPL2 kinase)), −7.2 kcal/mol (Radicicol), −8.8 kcal/mol (Dabrafenib), −8 kcal/mol (AT-7519). Finally, the results for TNF were −6.1 kcal/mol, −5.2 kcal/mol, −7.2 kcal/mol and −6 kcal/mol for its inhibitor (Thalidomide), radicicol, dabrafenib and AT-7519 respectively.
Fig. 8.
Molecular docking analysis revealed hub proteins and their binding affinities with repurposed drug candidates and their inhibitors.
4. Discussion
The latest SARS-CoV-2 infections contribute to severe pulmonary conditions and substantial morbidities and mortality complications. The prevalence of infections and mortality is still rising sharply. There are no drugs or vaccines that can be used for the curing of the disease. Few drugs for other disease are being tested for clinical trials to repurpose it in the treatment of COVID-19, one such example; FDA has given emergency approval remdesivir in COVID-19. Thus, identification of critical genes, pathways, potential candidate drugs targeting the hub genes may provide rapid identification of candidate drugs for starting repurposing processes in COVID-19. Our knowledge of host immune responses to SARS-CoV-2 has been limited, and the development of new therapeutics has become difficult. Viral infection typically induces significant changes in host transcriptome, leading to abnormal metabolism of the host cells and to an immune response that is suitable for viral replication. In this study, we analyzed lung epithelial RNA-Seq transcriptomes infected with SARS-CoV-2 compared to non-infected cells to decipher transcriptional signatures and drug targets. We detected 177 DEGs where 122 upregulated genes and 55 down-regulated DEGs.
To elucidate the roles of the identified DEGs, GO analysis revealed significant enrichment of biological process, namely, immune response, defense response, response to cytokines, cytokine-mediated signaling, and response to external stimuli. These genes are predicted to come under various categories, including defense response, immunological response. These play crucial roles in restricting viral infections. Inflammatory reactions can be a dual-edge sword in viral pneumonia. While active inflammation is essential in regional tissues to prevent infection, intensified inflammatory responses in pneumonic patients lead to the unnecessary secretion of the “cytokine storm” and lead to adverse effects such as alveolar damage and fibrosis (Huang et al., 2020; Teijaro et al., 2014). The novel SARS-CoV-2 virus has human receptor ACE2 as like previous SARS-CoV virus (Zhou et al., 2020). Systemic inflammation and cytokines storm (IL-6, IL-8, CXCL10/IP-10, CCL2/MCP-1, and CCL3/MIP-1A) are some of the important pathophysiological mechanisms (Channappanavar et al., 2014). We still know very little about the immunological mechanism in pathophysiological mechanisms of COVID-19. Recent studies demonstrated the increased expression of cytokines and chemokines in both mild and critical patients (Huang et al., 2020). Consistent with the previous finding, the pathways enrichments revealed cytokines signaling pathways, cytokine signaling in immune systems, interferon signaling, IL-4, IL-13, IL-10, and IL-17 signaling pathways, TNF signaling, cytokine-cytokine receptor interaction, viral protein interaction with cytokines and cytokines receptor were enriched highly in COVID-19. Viral diseases such as Influenza A and measles pathways were identified. An autoimmune disease, rheumatoid arthritis, was also significantly enriched. These pathways suggest the immune systems and cytokines are significantly involved in the host response to infection. These pathways show immense importance to drug repurposing opportunities in COVID-19. Throughout biological processes, PPI network analysis is a highly essential since it serves essential signal molecules. PPI analytics are worthwhile in the detection of the drug target to decipher molecules with a high number of links called hub proteins. Such core components may be a treatment target for COVID19. We detected hub genes signature (BIRC3, ICAM1, IRAK2, MAP3K8, S100A8, SOCS3, STAT5A, TNF, TNFAIP3, TNIP1) from PPI interaction network analysis. Among the hub proteins, ICAM1 has been previously demonstrated that its expression levels at the level of mRNA and protein levels increased in human bronchial epithelial cells and nasopharyngeal epithelial cells in response to influenza virus infection (Othumpangat et al., 2016). More than 90% of Rhinovirus serotypes use ICAM1 as the receptor. These results consistently support our finding that COVID-19 infection also enhanced its expression in lung epithelial cells. Another study showed anti-ICAM1 antibody could inhibit rhinovirus induced exacerbation of lung inflammation (Traub et al., 2013). Respiratory syncytial virus infection upregulates genes in the cytokine signal suppressor (SOCS) family that use a feedback loop to prevent antiviral signaling pathway, which is based on type I interferon. The upregulation of SOCS2 and SOCS3 gene was induced in lung epithelial cells infected with the Zika virus (Liu et al., 2019; Seong et al., 2020). It is thought this upregulation of SOCS in response to the Zika virus regulated viral replication, possibly via the regulation of antiviral innate immune responses. The Janus kinase 2 (JAK2)-signal transducer and activator of transcription 5 (STAT5) signaling plays a crucial role in human parvovirus B19 (B19V) replication (Ganaie et al., 2017). S100A8 and S100A9 come predominantly from immunocytes, like the neutrophils and macrophages, containing abundant S100A8/A9, which are inflammatory. One of the significant causes of S100A8/A9 release is infection-induced inflammation. After S100A8/A9 has been intensively expressed and secrete with infected bacteria, neutrophils, macrophages and monocytes, the inductive inflammatory cytokines, the reactive oxygen (ROS) species, and Nitric Oxide (NO) are modulated (Wang et al., 2018). Genetic variants of hub TNFAIP3 have been associated with the hepatitis B virus in the Chinese population (Zhang et al., 2015). MAP3K8 expression dramatically decreased within 12–24 h of coronavirus infection, both in the lungs of ferrets infected with SARS coronavirus and in primary human microvascular endothelial cells infected with MERS coronavirus. Thus, MAP3K8 may be involved in the cellular response to COVID19 infection or host cell vulnerability to coronaviruses. Hub gene TNF expression was significantly upregulated (FDR<0.05 and logFC>1) in the lung epithelial in response to COVID-19. Also, in patients needing hospitalization at intensive care units elevated IL-6 and TNF-α level has also been observed previously. This may lead to undergo clinical trials of anti-rheumatic medicines as possible therapies for this serious viral infection (Perricone et al., 2020). Upregulation of pro-inflammatory cytokines in the blood is present in patients with COVID-19, including interleukins IL-1, IL-6, TNF and interferon (α), with elevated levels of other cytokines of patients in intensive care unit (Feldmann et al., 2020; Gong et al., 2020; Huang et al., 2020). Anti-tumor necrosis factor (TNF) antibodies in serious autoimmune disorders, such as rheumatoid arthritis, inflammatory bowel disease, have been used for more than 20 years. TNF is present in blood and the tissues of COVID-19 patients and TNF is essential in virtually all acute inflammatory response and acts as an inflammatory amplifier. To avoid progression to intensive care, we recommend that an anti-TNF treatment should be tested in patients with COVID-19 on hospital admission. This justifies consideration for the possible role of anti-TNF therapy. The roles of IRAK2 and TNIP are yet to be determined in COVID-19. TNIP1 is a hub protein associated with autoimmune disease (Shamilov and Aneskievich, 2018). ACE2 is expressed on the surface cells and facilitates the viral entry into cells with interacting viral spike protein. ACE2 expression is crucial for infection of pneumonia for pneumonia infection by SARS-CoV-2. Zhang et al. studied the differential expression of ACE2 in response to Influenza A, suggesting that infection by viral influenza could be a high risk factor in COVID-19 development (Zhang and Zhang, 2020). This observation is consistent with the present study finds Influenza A pathway overrepresented. Another pathway, viral protein interaction with cytokine and cytokine receptor also denotes pathways that play a role in viral infection. This research showed that the IL-17 signaling pathway was substantially enriched. IL-17, a pro-cytokine, initiates an immune response to infections [21]. It is complicated to link RA to infection. The possible relation between viral infection and the progression of RA was examined in low numbers (Arleevskaya et al., 2017). In particular, the greater proportion of RA in the Korean study showed that parainfluenza and coronavirus were correlated [23]. The DEGs were enriched in TNF signaling pathways. Cytokine storm is characterized in accordance with our observation by COVID-19 infection [25]. Feldmann et al. suggested urgent clinical trials of anti-TNF drugs for controlling symptoms of COVID-19 (Feldmann et al., 2020). Anti-cytokine therapies are suggested to prevent pneumonia associated complications in COVID-19 (Monteleone et al., 2020). We then identified several significant TFs as the regulator of DEG.
COVID-19 hub proteins specific drug repositioning ended up with 50 repurposed candidate drugs (Supplementary File S3). Among those 50 drugs, we selected 10 of them with larger cosine distances which allow us to reverse COVID-19 expression scenario. These 10 drugs were listed in Table 4. Some of them were eliminated since they have not been approved. Docking analyses were carried out to validate our findings in silico. We employed three drugs, namely, radicicol, dabrafenib and AT-7519 to test their binding affinities to COVID-19 specific hub proteins. Among 10 hub proteins (BIRC3, ICAM1, IRAK2, MAP3K8, S100A8, SOCS3, STAT5A, TNF, TNFAIP3, and TNIP1), we chose ones that have available 3D protein structure for homo sapiens. At the end, we have three drugs with six hub proteins (BIRC3, ICAM1, MAP3K8, S100A8, TNF, and TNFAIP3). Docking analyses of these three drugs with hub proteins resulted in varied binding affinities (Fig. 8). Hub proteins were docked also their known inhibitors (LCL161, A-205804, Tpl-2 kinase, JQ1, and Thalidomide) to compare binding affinities (kcal/mol) with repurposed drugs to conclude whether docking is valid or not. To determine a docking analysis acceptable or not, we considered one main criterion for binding affinities which was approximate or lower compared to hubs’ known inhibitors. According to docking results among these three drugs, dabrafenib, radicicol, and AT-7519 demonstrated good binding affinities, respectively. Dabrafenib has showed the binding affinities that covers 83% of the hubs whereas radicicol covers 67% of the hubs, and AT-7519 covers 50% of the hubs. Therefore, dabrafenib was the best drug that was docked efficiently to target hubs.
Docking results unveiled that ICAM1 and TNFAIP3 were docked efficiently with the all three repurposed drug candidates. It can be proposed that hub and drug prediction algorithms and methods are reliable, since in silico validation of at least candidate pairs between two hubs (ICAM1 and TNFAIP3) and three drugs (radicicol, dabrafenib, AT-7519) showed good binding affinities compared to their inhibitors. BIRC3, MAP3K8, and TNF demonstrated good binding affinities with at least two out of three drugs. MAP3K8 and TNF were docked efficiently with dabrafenib and AT-7519 whereas BIRC3 showed efficient docking with radicicol and dabrafenib. Molecular docking analysis results of S100A8 ended up with higher binding affinities with these three repurposed drugs when compared to its inhibitor. Therefore, we could not mention of any docking with repurposed drugs for S100A8.
In the light of the findings, ICAM1 and TNFAIP3 were the best hubs when we consider the molecular docking results with the all repurposed candidate drugs. In addition, dabrafenib was the best repurposed drug candidate that demonstrated good binding affinity values with all COVID-specific hubs. Therefore, it can be concluded that ICAM1 and TNFAIP3 might be potential targets and dabrafenib might be a potential therapeutic for the management of the COVID-19. It was reported in a recent review that COVID-19 and episodes of thromboembolism might lead to disseminated intravascular coagulation (DIC) and display the main reason of lethality during COVID- 19 infection (Cavalli et al., 2020b). Dabrafenib were also associated with prompt resolution of DIC with use of trametinib in a melanoma patient (Chuang et al., 2019). Moreover, synergistic usage of dabrafenib and trametinib under-expressed tissue factor in BRAFv600e mutated melanoma cells consequential to inhibition of the coagulation cascade (Scatena et al., 2019). It can be hypothesized that dabrafenib might be the promising treatment option for COVID-19 via solutions of coagulation associated phenomena.
The integration of the molecular signatures and the drug knowledgehelped to find three in silico validated repurposable drugs for COVID-19. We hope that this study can help in developing and conducting clinical studies to control COVID-19 quickly at this phase. We think that in the future some of the priority therapeutics will be used to offer appropriate stronger and probably personalized care for COVID-19 in combination, as many forms of disease like viral diseases like HIV can be more effective in combinatorial therapies. However, since multifarious multi-omics data integrations are provided in this study, cautions should be made to interpret these findings. Moreover, the large cohorts of transcriptomics datasets of COVID-19 infected lungs tissues and biological evaluation of presented repurposable drugs are needed in future studies.
5. Conclusion
The present research aimed to identify molecular signatures (i.e. hub genes) and molecular pathways altered in response to SARS-CoV-2 in lung epithelial cells compared to controls. We identified 177 DEGs were 122 upregulated and 55 down-regulated DEGs altered in lung epithelial cells in response to infection SARS-CoV-2 from RNA-Seq transcriptomes. The enriched significant pathways were detected involved in immune response, influenza A, cytokines related. The integration of DEGs with tissue PPI network analysis showed the presence of ten hub genes (BIRC3, ICAM1, IRAK2, MAP3K8, S100A8, SOCS3, STAT5A, TNF, TNFAIP3, and TNIP1). TFs (FOXC1, FOXL1, GATA2, YY1, NFKB1) were also identified as potential regulators of the DEGs. The gene-drugs interactions network analysis revealed 3 in silico validated repurposable drug candidates for the treatment of COVID-19. The molecular signatures and pathways identified suggested ICAM1 and TNFAIP3 as the best hubs that responded the molecular docking results of the all repurposed candidate drugs. Dabrafenib, radidiciol and AT7519 should be evaluated with experimental studies in prior to clinical studies.
6. Data availability
All data utilized in this manuscript are available online from their respective database.
Funding
No funding received.
CRediT authorship contribution statement
Tania Islam: Data curation, Formal analysis, Writing - original draft, Formal analysis, Methodology. Md Rezanur Rahman: Conceptualization, Data curation, Formal analysis, Methodology, Writing - original draft, Writing - review & editing, Supervision. Busra Aydin: Data curation, Formal analysis, Writing - review & editing. Hande Beklen: Writing - review & editing, Data curation, Formal analysis. Kazim Yalcin Arga: Methodology, Writing - review & editing, Supervision. Md Shahjaman: Formal analysis, Methodology, Supervision, Writing - review & editing.
Declaration of competing interest
None.
Acknowledgments
We thank Professor Dr AHM Abual Islam and Mr Md Shaon Akter, MA, Department of English, Khwaja Yunus Ali University, Bangladesh for editing and checking the spelling and grammatical errors of this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejphar.2020.173594.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Differentially expressed genes in COVID-19 compared to controls
Significant molecular pathways enriched by differentially expressed genes
All repuposed drug candidates
References
- Arleevskaya M.I., Shafigullina A.Z., Filina Y.V., Lemerle J., Renaudineau Y. Associations between viral infection history symptoms, granulocyte reactive oxygen species activity, and active rheumatoid arthritis disease in untreated women at onset: results from a longitudinal cohort study of Tatarstan women. Front. Immunol. 2017;8:1725. doi: 10.3389/fimmu.2017.01725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angell R.M., Atkinson F.L., Brown M.J., Chuang T.T., Christopher J.A., Cichy-Knight M., Neu M. N-(3-Cyano-4, 5, 6, 7-tetrahydro-1-benzothien-2-yl) amides as potent, selective, inhibitors of JNK2 and JNK3. Bioorg. Med. Chem. Lett. 2007;17:1296–1301. doi: 10.1016/j.bmcl.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Basha O., Shpringer R., Argov C.M., Yeger-Lotem E. The DifferentialNet database of differential protein–protein interactions in human tissues. Nucleic Acids Res. 2018;46:D522–D526. doi: 10.1093/nar/gkx981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Battistuz T., Bhat T.N., Bluhm W.F., Bourne P.E., Burkhardt K., Fagan P. The protein data bank. Acta Crystallogr. D. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B., Liu W.-C., Moeller R., Panis M., Sachs D., Albrecht R. bioRxiv; 2020. SARS-CoV-2 Launches a Unique Transcriptional Signature from in Vitro, Ex Vivo, and in Vivo Systems. [Google Scholar]
- Bussel J., Arnold D.M., Grossbard E., Mayer J., Treliński J., Homenda W. Fostamatinib for the treatment of adult persistent and chronic immune thrombocytopenia: results of two phase 3, randomized, placebo‐controlled trials. Am. J. Hematol. 2018;93:921–930. doi: 10.1002/ajh.25125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cainap C., Qin S., Huang W.T., Chung I.J., Pan H., Cheng Y., Gorbunova V. Linifanib versus Sorafenib in patients with advanced hepatocellular carcinoma: results of a randomized phase III trial. J. Clin. Oncol. 2015;33:172. doi: 10.1200/JCO.2013.54.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campillos M., Kuhn M., Gavin A.C., Jensen L.J., Bork P. Drug target identification using side-effect similarity. Science. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- Cavalli E., Petralia M.C., Basile M.S., Bramanti A., Bramanti P., Nicoletti F. Transcriptomic analysis of COVID-19 lungs and bronchoalveolar lavage fluid samples reveals predominant B cell activation responses to infection. Int. J. Mol. Med. 2020;46:1266–1273. doi: 10.3892/ijmm.2020.4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli E., Bramanti A., Ciurleo R., Tchorbanov A.I., Giordano A., Fagone P. Entangling COVID-19 associated thrombosis into a secondary antiphospholipid antibody syndrome: diagnostic and therapeutic perspectives. Int. J. Mol. Med. 2020;46:903–912. doi: 10.3892/ijmm.2020.4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Zhao J., Perlman S. T cell-mediated immune response to respiratory coronaviruses. Immunol. Res. 2014;59:118–128. doi: 10.1007/s12026-014-8534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Guo J., Chen Z., Wang J., Liu M., Pang X. Linifanib (ABT-869) potentiates the efficacy of chemotherapeutic agents through the suppression of receptor tyrosine kinase-mediated akt/mtor signaling pathways in gastric cancer. Sci. Rep. 2016;6:1–11. doi: 10.1038/srep29382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang J., Uche A., Gupta R., Margolin K., Kim P. Fulminant disseminated intravascular coagulation as initial presentation of BRAF-mutated melanoma. Case Rep Oncol Med. 2019;2019 doi: 10.1155/2019/9246596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta bio-medica Atenei Parm. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A.P., Grondin C.J., Johnson R.J., Sciaky D., King B.L., McMorran R., Wiegers J., Wiegers T.C., Mattingly C.J. The comparative toxicogenomics database: update 2017. Nucleic Acids Res. 2017;45:D972–D978. doi: 10.1093/nar/gkw838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degtyarenko Kirill, de Matos Paula, Ennis Marcus, Hastings Janna, Zbinden Martin, McNaught Alan. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Research. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q., Reid S.P., Clark N.R., Wang Z., Fernandez N.F., Rouillard A.D., Readhead B., Tritsch S.R., Hodos R., Hafner M., Niepel M. L1000CDS 2: LINCS L1000 characteristic direction signatures search engine. NPJ Syst Biol Appl. 2016;2:16015s. doi: 10.1038/npjsba.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagone P., Ciurleo R., Lombardo S.D., Iacobello C., Palermo C.I., Shoenfeld Y., Bendtzen K., Bramanti P., Nicoletti F. Transcriptional landscape of SARS-CoV-2 infection dismantles pathogenic pathways activated by the virus, proposes unique sex-specific differences and predicts tailored therapeutic strategies. Autoimmun. Rev. 2020;19:102571. doi: 10.1016/j.autrev.2020.102571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Channappanavar R., Perlman S. Middle East respiratory syndrome: emergence of a pathogenic human coronavirus. Annu. Rev. Med. 2017;68:387–399. doi: 10.1146/annurev-med-051215-031152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann M., Maini R.N., Woody J.N., Holgate S.T., Winter G., Rowland M., Richards D., Hussell T. Trials of anti-tumour necrosis factor therapy for COVID-19 are urgently needed. Lancet. 2020;395:1407–1409. doi: 10.1016/S0140-6736(20)30858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz M., Rodriguez H., Lopes C., Zuberi K., Montojo J., Bader G.D., Morris Q. GeneMANIA update 2018. Nucleic Acids Res. 2018;46(W1):W60–W64. doi: 10.1093/nar/gky311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganaie S.S., Zou W., Xu P., Deng X., Kleiboeker S., Qiu J. Phosphorylated STAT5 directly facilitates parvovirus B19 DNA replication in human erythroid progenitors through interaction with the MCM complex. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Dong H., Xia S.Q., Huang Y.Z., Wang D., Zhao Y., Liu W., Tu S., Zhang M., Wang Q. Correlation analysis between disease severity and inflammation-related parameters in patients with COVID-19 pneumonia. MedRxiv. 2020 doi: 10.1101/2020.02.25.20025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauschild A., Grob J.J., Demidov L.V., Jouary T., Gutzmer R., Millward M. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T.H., Wu A.T., Cheng T.S., Lin K.T., Lai C.J., Hsieh H.W., Chen K.Y. In silico identification of thiostrepton as an inhibitor of cancer stem cell growth and an enhancer for chemotherapy in non–small cell lung cancer. J. Cell Mol. Med. 2019;23:8184–8195. doi: 10.1111/jcmm.14689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju S.Y., Huang C.Y., Huang W.C., Su Y. Identification of thiostrepton as a novel therapeutic agent that targets human colon cancer stem cells. Cell Death Dis. 2015;6 doi: 10.1038/cddis.2015.155. e1801-e1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakar S.S., Parte S., Kelsey Carter I.G.J., Worth C., Rameshwar P., Ratajczak M.Z. Withaferin A (WFA) inhibits tumor growth and metastasis by targeting ovarian cancer stem cells. Oncotarget. 2017;8:74494. doi: 10.18632/oncotarget.20170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian H., Shah N.P., Hochhaus A., Cortes J., Shah S., Ayala M., Nakamae H. Dasatinib versus imatinib in newly diagnosed chronic-phase chronic myeloid leukemia. N. Engl. J. Med. 2010;362:2260–2270. doi: 10.1056/NEJMoa1002315. [DOI] [PubMed] [Google Scholar]
- Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Zaslavsky L. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47:D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Lee S.A., Myung S.C., Kim W., Lee C.S. Radicicol, an inhibitor of Hsp90, enhances TRAIL-induced apoptosis in human epithelial ovarian carcinoma cells by promoting activation of apoptosis-related proteins. Mol. Cell. Biochem. 2012;359:33–43. doi: 10.1007/s11010-011-0997-9. [DOI] [PubMed] [Google Scholar]
- Kindler E., Thiel V. SARS-CoV and IFN: too little, too late. Cell Host Microbe. 2016;19:139–141. doi: 10.1016/j.chom.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laquerre S., Arnone M., Moss K., Yang J., Fisher K., Kane‐Carson L.S., Adjabeng G. 2009. Abstract B88: A Selective Raf Kinase Inhibitor Induces Cell Death and Tumor Regression of Human Cancer Cell Lines Encoding B‐RafV600E Mutation. AACR-NCI-EORTC International Conference: Molecular Targets and Cancer Therapeutics--Nov 15-19, 2009. (Boston, MA) [Google Scholar]
- Li J., Rix U., Fang B., Bai Y., Edwards A., Colinge J., Superti-Furga G. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat. Chem. Biol. 2010;6:291–299. doi: 10.1038/nchembio.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Yan R., Chen B., Pan Q., Chen Y., Hong J., Zhang L., Liu W., Wang S., Chen J.-L. Influenza virus-induced robust expression of SOCS3 contributes to excessive production of IL-6. Front. Immunol. 2019;10:1843. doi: 10.3389/fimmu.2019.01843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamoor S. 2020. MAP3K8 (COT/Tpl-2) is differentially expressed and transcriptionally repressed in models of coronavirus infection. [Google Scholar]
- Monaco G., Lee B., Xu W., Mustafah S., Hwang Y.Y., Carre C., Burdin N., Visan L., Ceccarelli M., Poidinger M. RNA-Seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep. 2019;26:1627–1640. doi: 10.1016/j.celrep.2019.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteleone G., Sarzi-Puttini P.C., Ardizzone S. Preventing COVID-19-induced pneumonia with anticytokine therapy. Lancet Rheumatol. 2020;2:e255–e256. doi: 10.1016/S2665-9913(20)30092-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelemans T., Kikkert M. Viral innate immune evasion and the pathogenesis of emerging RNA virus infections. Viruses. 2019;11:961. doi: 10.3390/v11100961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton A.H., Cardani A., Braciale T.J. Seminars in Immunopathology. Springer; 2016. The host immune response in respiratory virus infection: balancing virus clearance and immunopathology; pp. 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othumpangat S., Noti J.D., McMillen C.M., Beezhold D.H. ICAM-1 regulates the survival of influenza virus in lung epithelial cells during the early stages of infection. Virology. 2016;487:85–94. doi: 10.1016/j.virol.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perricone C., Triggianese P., Bartoloni E., Cafaro G., Bonifacio A.F., Bursi R., Perricone R., Gerli R. The anti-viral facet of anti-rheumatic drugs: lessons from COVID-19. J. Autoimmun. 2020;102468 doi: 10.1016/j.jaut.2020.102468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., Vilo J. g: profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscioli E., Hamon R., Ruffin R.E., Grant J., Hodge S., Zalewski P., Lester S. BIRC3 single nucleotide polymorphism associate with asthma susceptibility and the abundance of eosinophils and neutrophils. J. Asthma. 2017;54:116–124. doi: 10.1080/02770903.2016.1196371. [DOI] [PubMed] [Google Scholar]
- Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): A review. JAMA. 2020;323:1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- Santo L., Vallet S., Hideshima T., Cirstea D., Ikeda H., Pozzi S. AT7519, A novel small molecule multi-cyclin-dependent kinase inhibitor, induces apoptosis in multiple myeloma via GSK-3β activation and RNA polymerase II inhibition. Oncogene. 2010;29:2325–2336. doi: 10.1038/onc.2009.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong R.-K., Lee J.K., Shin O.S. Zika virus-induction of the suppressor of cytokine signaling 1/3 contributes to the modulation of viral replication. Pathogens. 2020;9:163. doi: 10.3390/pathogens9030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scatena C., Franceschi S., Franzini M., Sanguinetti C., Romiti N., Caponi L. Dabrafenib and Trametinib prolong coagulation through the inhibition of tissue factor in BRAF v600e mutated melanoma cells in vitro. Canc. Cell Int. 2019;19:223. doi: 10.1186/s12935-019-0938-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamilov R., Aneskievich B.J. TNIP1 in autoimmune diseases: regulation of toll-like receptor signaling. J. Immunol. Res. 2018;3491269 doi: 10.1155/2018/3491269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stan S.D., Hahm E.R., Warin R., Singh S.V. Withaferin A causes FOXO3a-and Bim-dependent apoptosis and inhibits growth of human breast cancer cells in vivo. Canc. Res. 2008;68:7661–7669. doi: 10.1158/0008-5472.CAN-08-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbiah V., Kreitman R.J., Wainberg Z.A., Cho J.Y., Schellens J.H., Soria J.C. Dabrafenib and trametinib treatment in patients with locally advanced or metastatic BRAF V600–mutant anaplastic thyroid cancer. J. Clin. Oncol. 2018;36:7. doi: 10.1200/JCO.2017.73.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro J.R., Walsh K.B., Rice S., Rosen H., Oldstone M.B.A. Mapping the innate signaling cascade essential for cytokine storm during influenza virus infection. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111:3799–3804. doi: 10.1073/pnas.1400593111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobert J.A. Lovastatin and beyond: the history of the HMG-CoA reductase inhibitors. Nat. Rev. Drug Discov. 2003;2:517–526. doi: 10.1038/nrd1112. [DOI] [PubMed] [Google Scholar]
- Traub S., Nikonova A., Carruthers A., Dunmore R., Vousden K.A., Gogsadze L., Hao W., Zhu Q., Bernard K., Zhu J. An anti-human ICAM-1 antibody inhibits rhinovirus-induced exacerbations of lung inflammation. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turanli B., Gulfidan G., Arga K.Y. Transcriptomic-guided drug repositioning supported by a new bioinformatics search tool: geneXpharma. OMICS. 2017;21:584–591. doi: 10.1089/omi.2017.0127. [DOI] [PubMed] [Google Scholar]
- Wang S., Song R., Wang Z., Jing Z., Wang S., Ma J. S100A8/A9 in inflammation. Front. Immunol. 2018;9:1298. doi: 10.3389/fimmu.2018.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J.A.C., Prow N.A., Schroder W.A., Ellis J.J., Cumming H.E., Gearing L.J., Poo Y.S., Taylor A., Hertzog P.J., Di Giallonardo F. RNA-Seq analysis of chikungunya virus infection and identification of granzyme A as a major promoter of arthritic inflammation. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.S., Lien G.S., Shen S.C., Yang L.Y., Chen Y.C. HSP90 inhibitors, Geldanamycin and Radicicol, enhance Fisetin-induced cytotoxicity via induction of apoptosis in human colonic cancer cells. Evid. Based Complementary Altern. Med. 2013;2013:987612. doi: 10.1155/2013/987612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi C., Wang L., Yu J., Ye H., Cao L., Gong Z. Inhibition of cyclin-dependent kinases by AT7519 is effective to overcome chemoresistance in colon and cervical cancer. Biochem. Biophys. Res. Commun. 2019;513:589–593. doi: 10.1016/j.bbrc.2019.04.014. [DOI] [PubMed] [Google Scholar]
- Zhang L., Zhang Y. 2020. Influenza Viral Infection Is a High-Risk Factor for Developing Coronavirus Disease 2019 (COVID-19).Preprints; p. 2020030307. [DOI] [Google Scholar]
- Zhang P., Li N., Zhu Q., Li F., Yang C., Zeng X., Lv Y., Zhou Z., Han Q., Liu Z. Association between TNFAIP3 nonsynonymous single-nucleotide polymorphism rs2230926 and chronic hepatitis B virus infection in a Chinese Han population. Virol. J. 2015;12:33. doi: 10.1186/s12985-015-0268-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., Soufan O., Ewald J., Hancock R.E.W., Basu N., Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47:W234–W241. doi: 10.1093/nar/gkz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Qiao K., Gao Z., Vederas J.C., Tang Y. Insights into radicicol biosynthesis via heterologous synthesis of intermediates and analogs. J. Biol. Chem. 2010;285:41412–41421. doi: 10.1074/jbc.M110.183574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differentially expressed genes in COVID-19 compared to controls
Significant molecular pathways enriched by differentially expressed genes
All repuposed drug candidates
Data Availability Statement
All data utilized in this manuscript are available online from their respective database.