Abstract
An acute respiratory disease caused by a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that surfaced in China in late 2019, continues to spread rapidly across the globe causing serious concerns. The coronavirus disease 2019 (COVID-19) is declared as a public health emergency worldwide by the World Health Organization (WHO). Increasing evidences have demonstrated human-to-human transmission that primarily affects the upper respiratory tract followed by lower respiratory tract damage leading to severe pneumonia. Based on the current status, the elderly population and people with prior co-morbidities are highly susceptible to serious health effects including cytokine up-regulation and acute respiratory distress syndrome (ARDS). Currently, COVID-19 research is still in the preliminary stage necessitating rigorous studies. There is no specific drug or vaccine targeting SARS-CoV-2 currently and only symptomatic treatment is being administered, but several antivirals are under active investigation. In this review, we have summarized the epidemiology, entry mechanism, immune response, and therapeutic implications, possible drug targets, their ongoing clinical trials, and put forward vital questions to offer new directions to the COVID-19 research.
Keywords: SARS-CoV-2, Immune response, SARS-CoV, MERS-CoV, Clinical trials
Graphical abstract
1. Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) provisionally named 2019-nCoV is the causative agent of the recent global pandemic COVID-19 with increasing fatality rate. It is depicted as a public health emergency of global concern by the World Health Organization (WHO). This contagion initially emerged in Wuhan city, Hubei Province, China on December 8, 2019, which caused pneumonia-like symptoms in a cluster of patients. Coronaviruses (CoVs) are members of the genus Coronaviridae, a group of pleomorphic RNA viruses that contain crown-shaped peplomers. SARS-CoV-2 possesses an 80% phylogenetic identity with severe acute respiratory syndrome coronavirus (SARS-CoV) and 50% similarity with the Middle East respiratory syndrome coronavirus (MERS-CoV) which caused global outbreaks in 2002–2003 and 2011 respectively. SARS-CoV-2 is now considered as a rapidly spreading pandemic virus that was initially transmitted from animals-to-humans and later transmitted through human-to-human [[1], [2], [3]].
At present there are no effective treatment options such as anti-viral drugs, vaccines, and monoclonal antibodies available against SARS-CoV-2. With the emergent fatalities evidenced across the globe, SARS-CoV-2 has become a major hotspot for global researchers [4]. Recently, Zhou et al., reported that the whole-genome of SARS-CoV-2 is 96% identical to bat coronavirus BatCoV RaTG13, which indicates that it is the most probable origin of SARS-CoV-2. Additionally the same group also confirmed that SARS-CoV-2 enters into human cells using angiotensin-converting enzyme 2 (ACE2) which is also the entry receptor of SARS-CoV [5]. Few reports had suggested the existence of pangolins as a possible intermediate host that warrants investigation [6]. In a clinical study with 452 COVID-19 patients, 286 were diagnosed as severely infected patients and were significantly older with a median age of 61 when compared to the remaining 166 non-severe patients with a median age of 53. This data suggested that SARS-CoV-2 is highly persistent and dangerous to elderly patients who possess weak immune system [7].
On January 20, 2020 the WHO released a situation report-1 for COVID-19 pandemic indicating 282 laboratory-confirmed cases and 6 deaths globally which has drastically increased to 21,294,845 laboratory-confirmed cases and 761,779 deaths as on August 16, 2020 [8,9]. WHO officially declared COVID-19 infection as a pandemic on March 11, 2020 [10]. Also, WHO risk assessment classified COVID-19 as a “very-high risk” global pandemic. The current treatment being administered for COVID-19 patients is only based on their symptoms and other specific therapeutic options are yet to be thoroughly investigated. High infection rates highlight the need for therapeutic strategies to develop novel therapeutics, vaccines, and other anti-viral drugs for this global pandemic threat. This review focuses on information about the SARS-CoV-2 mediated pandemic COVID-19 from the latest literature published and intends to provide comprehensive information on the topic to the readers.
2. Global statistical data and current scenario of COVID-19 pandemic
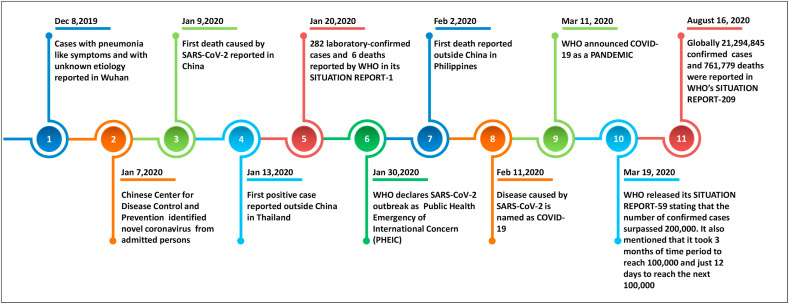
On December 8, 2019 a group of patients with pneumonia-like symptoms of unfamiliar etiology were reported from Wuhan, Hubei Province, China. Reviewing the history of these admitted persons, a notable commonality was observed. Most of the patients either worked or lived in and around the local Huanan seafood wholesale wet market. Severe acute respiratory syndrome (SARS) occurred in these patients at their early stage of pneumonia and some patients had additional complications like severe acute respiratory failure and intense acute respiratory distress syndrome (ARDS). Later, on January 7, 2020, the Chinese Center for Disease Control and Prevention detected the novel coronavirus from the throat swab sample of a hospitalized person [11]. On January 9, 2020, WHO confirmed the isolated strain from the admitted patient is a novel coronavirus and it was subsequently named as 2019-nCoV. Also, the first mortality of COVID-19 was reported on the very same day (January 9th). Apart from China, the Ministry of Public Health, Thailand witnessed and reported its first laboratory-confirmed COVID-19 case on January 13, 2020. On the same day, the Ministry of Health, Labour and Welfare, Japan and National IHR Focal Point (NFP), Republic of Korea reported their first COVID-19 case. Remarkably, both the identified cases in Thailand and Japan were imported cases from Wuhan, China [8,12].
Later, SARS-CoV-2 cases were also reported in Macau Special Administrative Region, United States of America, Hong Kong Special Administrative Region, Taipei Municipality wherein all these reported cases had a travel history to Wuhan [13]. Based on the data reported until January 26, 2020, WHO released its 6th Situation report on COVID-19 pandemic, in which 29 confirmed cases were reported worldwide in ten different countries apart from China. Out of these 29 positive cases, 26 cases had a travel history to Wuhan city of China, wherein the other 3 had no travel history to Wuhan. However, further investigation of these three particular patients revealed that one patient in Australia had direct contact with a COVID-19 positive person from Wuhan. Likewise, another person in Vietnam also had direct contact with his father, a COVID-19 positive person who recently had a travel history to Wuhan. This transfer of SARS-CoV-2 to the persons without a direct travel history to Wuhan from their close contacts and family persons who were in the vicinity of Wuhan city clearly proved that SARS-CoV-2 can be transmitted from one infected person to another healthy non-infected person leading to human-to-human transmission [14]. A timeline highlighting the key events of the COVID-19 pandemic is given in Fig. 1 .
Fig. 1.
Timeline with key events of COVID-19 pandemic.
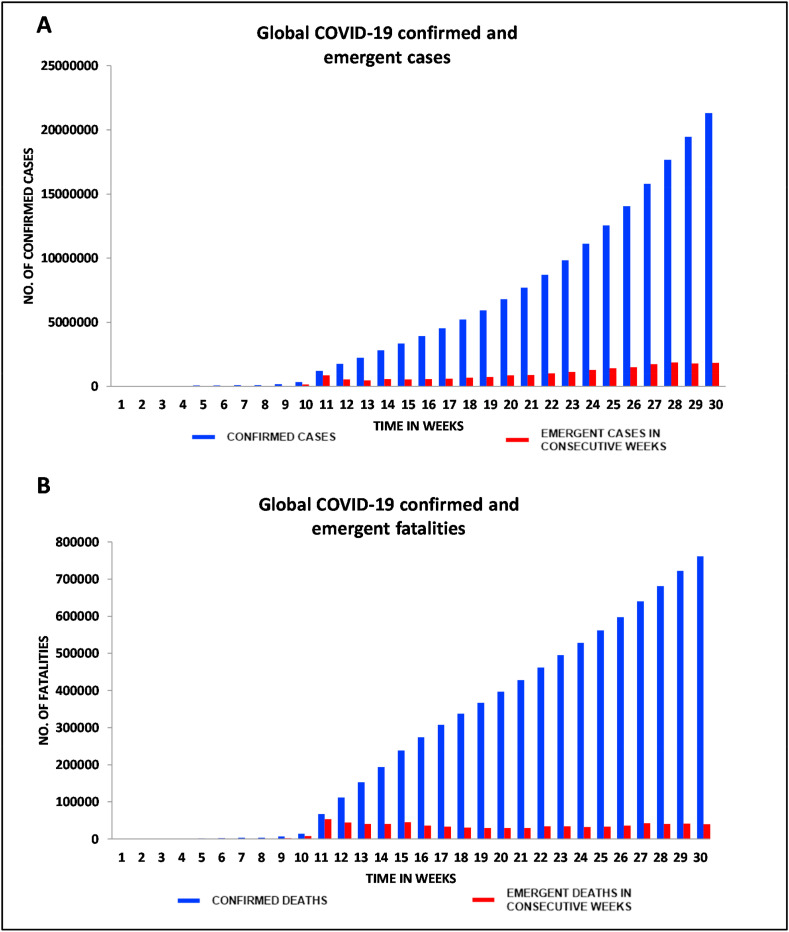
Within a duration of 210 days (30 weeks), this highly-contagious virus had caused over 761,779 deaths throughout the world and the fatalities continue to increase on a daily basis. Thus, strategic preparedness and effective public health measures are considered as the greatest need-of-the-hour to contain this global pandemic. Fig. 2 A and B are graphical representations respectively of confirmed as well as emergent COVID-19 cases and fatalities across the globe till August 16, 2020.
Fig. 2.
Graphical representation of: (A) Global COVID-19 confirmed and emergent cases, (B) Fatalities reported till August 16, 2020 (Source: WHO Situation Reports).
3. Structural and biological configuration of SARS-CoV-2
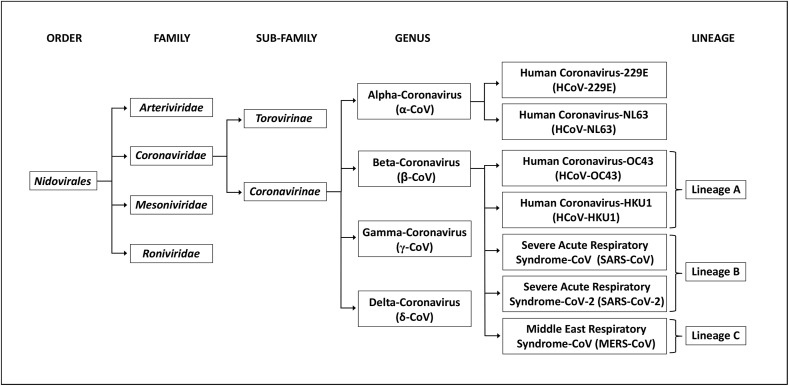
CoVs falls under the family of Coronaviridae, and the order of Nidovirales [15]. CoVs are named after their crown-shaped spikes present on their surfaces and these CoVs are classified into four main genera namely, alpha-coronavirus (α-CoV), beta-coronavirus (β-CoV), gamma-coronavirus (γ-CoV), and delta-coronavirus (δ-CoV). In the past two decades, three highly contagious and pathogenic novel CoVs emerged and caused global outbreaks. All these three CoVs namely, SARS-CoV, MERS-CoV, and the newly found SARS-CoV-2 remarkably come under the same genera of β-CoV [16,17].
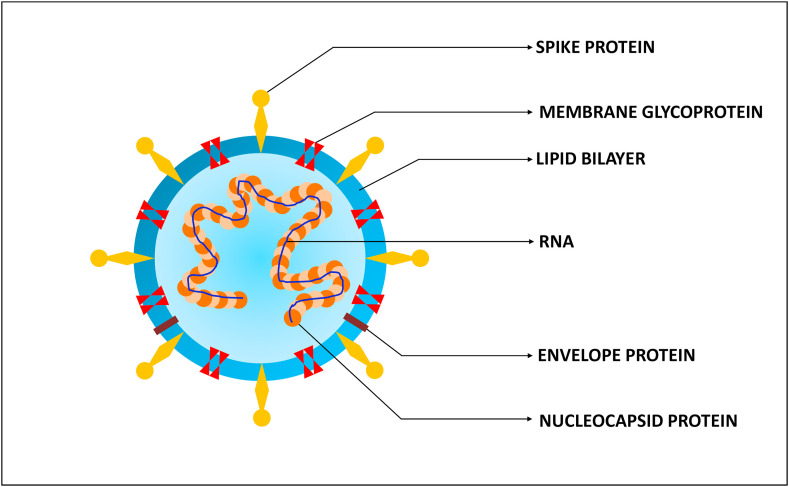
To date, seven human-affecting CoVs (HCoV) have been identified that can be segregated into two major kinds namely, high-pathogenic (SARS-CoV, MERS-CoV, and SARS-CoV-2) and low-pathogenic (HCoV 229E, NL63, OC43, and HKU1), based on their pathogenicity. CoVs are non-segmented, enveloped, positive-sense single-stranded RNA viruses which consist of spike protein (S-protein), envelope protein (E-protein), membrane glycoprotein (M-protein) and nucleocapsid protein (N-protein) in which the viral RNA is assembled. The viral RNA of CoVs with 26–32 kilo-bases length is the largest viral genome known [18,19]. This single stranded-RNA genome of SARS-CoV-2 comprises 29,891 nucleotides which encode for 9860 amino acids [20]. The structure of SARS-CoV-2 is illustrated in Fig. 3 and the schematic representation of the taxonomy of Coronaviridae (according to the International Committee on Taxonomy of Viruses) is depicted in Fig. 4 .
Fig. 3.
The structure of SARS-CoV-2.
Fig. 4.
Schematic representation of taxonomy of Coronaviridae (according to the International Committee on Taxonomy of Viruses).
4. Similarity of SARS-CoV-2 with SARS-CoV and MERS-CoV
A number of viruses from the family Coronaviridae have been identified in the past two decades and SARS-CoV-2 is the third such identified CoV after SARS-CoV and MERS-CoV. The ongoing SARS-CoV-2 outbreak was also initially transmitted from animal species just like the two previous global outbreaks caused by SARS-CoV and MERS-CoV. Evidences point out that all the three CoVs originated from bats and other different species of rodents [21,22]. Also, all these viruses, SARS-CoV, MERS-CoV, and SARS-CoV-2 caused severe respiratory diseases in humans [23,24]. Recent phylogenic analysis and full-genome sequencing showed that SARS-CoV-2 is a monophyletic group from β-CoV [25]. Kumar et al., conducted a phylogenetic analysis and reported that SARS-CoV-2 is closely associated with bat SARS-like coronaviruses. The group also conducted complete sequence alignment between SARS-CoV-2 and SARS-CoV, which showed 87.2% similarity, 76.2% identity, and 2% gaps in between their S-protein sequences. However, the team also reported that the S-protein of SARS-CoV-2 is longer when compared to bat SARS-like coronaviruses and SARS-CoV [26]. Whole-genome sequencing of SARS-CoV-2 revealed 96.2% similarity with bat-SARS-like coronaviruses (SARSr-CoV, RaTG13), 79%, and 50% similarity with SARS-CoV and MERS-CoV respectively.
Moreover, SARS-CoV-2 uses the ACE2 as entry-receptor just like SARS-CoV. Out of six crucial amino acid residues of receptor binding domain (RBD) five were different between SARS-CoV-2 and SARS-CoV. Interestingly, all the six crucial amino acid residues of SARS-CoV-2 were identical to pangolin SARS-CoV from Guangdong (GD Pangolin-CoV) and thus SARS-CoV-2 RBD region could be possibly expressed as a result of recombination in the pangolin viral strain. 3D structural analysis between SARS-CoV-2 and SARS-CoV indicated that SARS-CoV-2 has a greater binding affinity towards ACE2 when compared to SARS-CoV [27].
The protein sequence analysis reported by Zhou et al., identified that the N-protein and E-protein of SARS-CoV-2 possessed more evolutionarily conserved areas with that of SARS-CoV with a sequence identity of 89.6% and 96% respectively. However, in comparison with the S-protein of MERS-CoV, SARS-CoV-2 shares only 31.9% of sequence identity [28]. Host conditions like gender, age, and overall health strongly dictated the severity of SARS-CoV and MERS-CoV in the previous outbreaks, especially in individuals above 50 years [29]. A similar trend is also followed in the current SARS-CoV-2 outbreak wherein severe illness and high mortality rate is commonly found in elderly patients (>60 years). Initial evidences have also suggested that males may be more vulnerable to COVID-19 than females due to differences in the prevalence of smoking and their immunological responses. Males in China have a greater tendency to smoke (54%) when compared to females (2.6%). ACE2 gene expression was also found to be higher in smokers when compared to non-smokers. Few studies suggested that smoking history may play an additional role in susceptibility of smokers towards COVID-19 infection [21,22].
5. Mode of transmission, symptoms, and entry mechanism of SARS-CoV-2
5.1. Mode of transmission
At the earlier stages of the outbreak, it was considered that SARS-CoV-2 is inefficient in the case of human-to-human transmission comparing to the previous outbreaks of SARS-CoV and MERS-CoV [29]. However, the current perception on SARS-CoV-2 has witnessed a complete turn-around as the infection has been found to be highly contagious and rapidly transmissible. Several individuals were tested COVID-19 positive with just 15–50 s of exposure in public places like vegetable markets and hospitals. These patients had no history of exposure to outbreak areas and contact with symptomatic patients. Such incidents of individuals contracting SARS-CoV-2 infection in few seconds revealed the serious concern on the high infectivity and transmissibility rate unlike its predecessors SARS-CoV and MERS-CoV [30,31].
Since SARS-CoV and MERS-CoV used intermediate hosts like civets and camels respectively, SARS-CoV-2 is assumed to have a possible intermediate host for transmission. Apart from RaTG13, Pangolin-CoV extracted from dead Malayan pangolins had a nucleotide identity of 91.02% with SARS-CoV-2. This data on pangolins sufficiently proves that there is a high probability of an intermediate host existence [6]. Recently, Guangxi Customs confiscated a large consignment of pangolins smuggled into southern China in an anti-smuggling operation. Examining the frozen samples of these pangolins resulted in the identification of six complete genome sequences in the SARS-CoV-2 lineage [32]. It is noteworthy to mention that both bats and pangolins were sold live or dead either legally or illegally in the wet markets of Wuhan which could be the prime reason for the emergence of the COVID-19 pandemic. These wet-markets, which sell meats of dead and live animals, provide a conducive environment for both bacterial and viral transmission from various body fluids like blood, urine, and feces of the slaughtered animals. Moreover, poor hygiene practices followed by these wet-markets may be a key contributor to the COVID-19 pandemic [33,34].
Further, the transmission of this infection is majorly dependent on various routes of human-to-human transmission that includes direct contact with the aerial droplets released during conversation, coughing, and sneezing by infected persons. This mode of respiratory droplet transmission is considered to be the main route of viral transmission in COVID-19 infection [35]. Recent researches have suggested that the virus may also be capable of airborne transmission through aerosols (≤5 μm) of the infected persons. Even though it is impossible to confirm that transmission of SARS-CoV-2 through aerosols never occur, still there is no experimental evidence to substantiate the long-range aerosol-based transmission in COVID-19 infection [36].
5.2. Symptoms and respiratory compliance in SARS-CoV-2 infection
SARS-CoV-2 infections range from being asymptomatic to severely symptomatic. The most common symptoms that were diagnosed in the COVID-19 positive patients are fever, headache, cough, myalgia, sputum production, diarrhea, dyspnea, and pneumonia. Additional complications like ARDS, acute heart injury, and several secondary infections were found in the individuals with advanced stage of infection in hospital intensive care units [35,37].
Some patients despite being asymptomatic or with minimum symptoms have reduced pulse oximetry readings. This condition has been referred to as “Silent” or “apathetic hypoxia” [[38], [39], [40]]. These patients show relatively a normal lung compliance with lack of dyspnea but with low blood oxygen saturation levels indicating severe respiratory failure, and hence considered ‘Silent’. Such COVID-19 pneumonia patients with silent hypoxia cannot be categorized under typical ARDS definition because, patients with typical ARDS show reduced lung compliance and severe hypoxemia. Moreover the ARDS onset limit of 1 week as per Berlin criteria [41] does not apply to COVID-19 related ARDS patients as the onset time for these patients is reported to be 8–12 days [25,37,42]. Several studies have reported the clinical features of COVID-19 patients but still there exist several lacunae in our understanding. Two different phenotypes of COVID-19 pneumonia patients were proposed by Gattinoni et al., where Type L individuals exhibit normal lung compliance and low elastance (atypical ARDS) and Type H individuals show decreased respiratory compliance and high elastance fulfilling typical ARDS criteria [43].
The characteristics of COVID-19 related ARDS and typical ARDS needs to be understood fully for early diagnosis and precise treatment. As per Berlin definition, the severity of ARDS is divided into three stages based on oxygenation index (Partial pressure of arterial oxygen/Fractional inspired oxygen (PaO2/FiO2)) on positive end-expiratory pressure (PEEP) ≥5 cm H2O: mild (200 mm Hg < PaO2/FiO2 ≤300 mm Hg), moderate (100 mm Hg < PaO2/FiO2 ≤200 mm Hg), and severe (PaO2/FiO2 ≤100 mm Hg) [41]. Experts from the national health commission of China developed a standard treatment protocol for COVID-19. Based on their experience-derived classification of the oxygenation index, COVID-19-related ARDS was divided into three categories namely, (PaO2/FiO2) on PEEP≥5 cm H2O: mild (200 mm Hg ≤ PaO2/FiO2 <300 mm Hg), mild-moderate (150 mm Hg ≤ PaO2/FiO2<200 mm Hg), and moderate-severe (PaO2/FiO2 <150 mm Hg) [44].
This “silent hypoxia” may be a considered as a clinical indication to determine the status of the patients who are at increased risk of sudden decompensation leading to death. Emergency medical services (EMS) and hospital reports have suggested that COVID-19 outbreaks are associated with silent hypoxemia in the absence of dyspnea with low oxygen saturation (SpO2) levels [45]. High incidence of silent hypoxemia should be addressed with timely detection and effective clinical management strategies. Further understanding and more reliable data is awaited to evaluate the role and impact of “silent hypoxia” in the early identification and precise treatment of COVID-19 related ARDS patients.
5.3. Entry mechanism
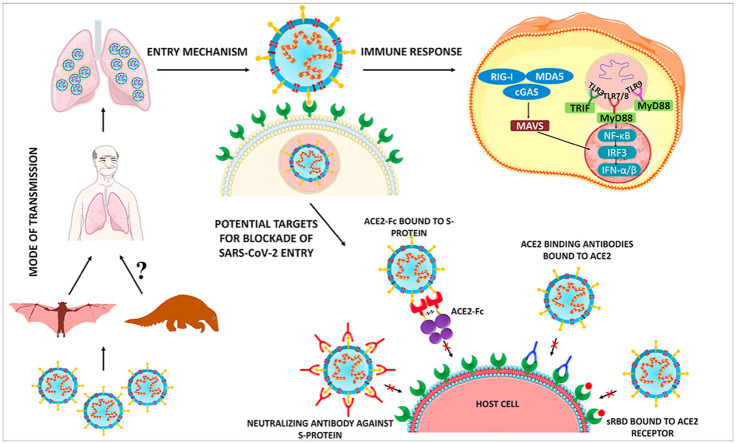
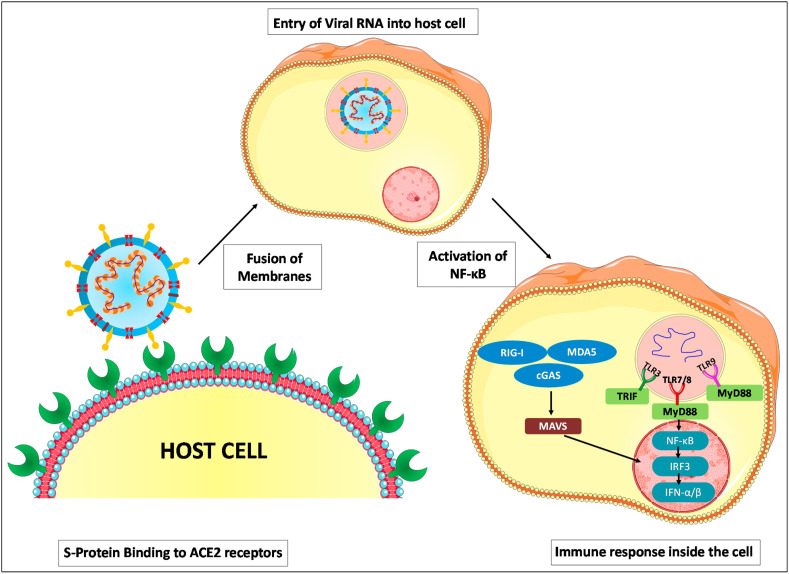
Entry of CoVs is a crucial multi-step process that includes numerous distinct subunits of S-protein that mediates the viral attachment with host-receptor, receptor engagement, protease activity and viral and host membrane fusion [46]. Zhou et al., using HeLa cells confirmed that SARS-CoV-2 and SARS-CoV share the same cell receptor namely ACE2 for their entry. Moreover, SARS-CoV-2 does not use any other conventional coronavirus entry receptors like aminopeptidase-N and dipeptidyl peptidase-4 [5]. The RBD of CoV comprises the S1 subunit of the densely glycosylated S-protein (a trimeric class I fusion protein) that facilitates viral attachment to the host receptor while the viral membrane-fusion with the host is mediated by subunit S2 of S-protein [47]. The receptor binding with S1 subunit triggers substantial structural reorganization of S-protein and also destabilizes the metastable pre-fusion conformation. This receptor engagement further leads to the shedding of S1 subunit and enables a stable post-fusion conformation of S2 subunit [48]. In case of both SARS-CoV and SARS-CoV-2, the cellular protease responsible for the S-protein priming is transmembrane serine protease 2 (TMPRSS2). Similar to SARS-CoV, SARS-CoV-2 also uses cathepsin B and L for S-protein priming [49].
The higher expression of ACE2 receptors in some patients might make them more susceptible to SARS-CoV-2 infection [50]. Since S-protein and ACE2 receptors are key mediators for the viral entry, pharmacological disruption of S-protein binding to ACE2 could be an effective treatment against SARS-CoV-2.
6. Immune response against SARS-CoV-2
Delineating the immune response against SARS-CoV-2 will aid researchers who intend to find and develop vaccines and therapeutics to handle the pandemic outbreak. However, the paucity of informative studies on the immune response against SARS-CoV-2 makes this a challenging task. Patients (82.1%) infected with SARS-CoV-2 are noted to have lymphopenia, a state of reduced level of blood lymphocytes which suggests suppression of cellular immune responses [19,35].
Even though human immune responses against SARS-CoV and MERS-CoV were elaborately investigated, studies pertaining to SARS-CoV-2 are yet to be thoroughly explored. Alessandro Sette et al., conducted a comparative analysis between B cell epitope sequences of SARS-CoV and SARS-CoV-2. Out of ten dominant regions identified, six had 90% identity, two with 80–89% and the other two regions with 69–78% identity to SARS-CoV-2. Further, T cell epitopes of SARS-CoV were mapped to SARS-CoV-2 and the same group found 12 out of 17 epitopes have a high sequence identity of ≥90%. These observations of highly conserved B and T cell epitopes of SARS-CoV and SARS-CoV-2 paves a pathway for developing vaccines against these conserved regions which could also offer a dual beneficial effect to generate cross-protective immunity against β-CoV [15].
The intricate balance between host immunity and CoV load dictates the extent of viral pathogenesis. A retrospective study to identify and correlate the role of immune cells and refractory hypoxemia in COVID-19 was conducted on three patients (mean age 74.3 ± 8.1 years) with SARS-CoV-2 infection who were admitted to Renmin Hospital of Wuhan University [51]. The patients turned to be non-responders to therapy and were reported dead at different days of hospitalization (12, 13, and 24 respectively). The research findings revealed the correlation of hypoxemia severity with decreased expressions of immune cells (CD4+ T cells, CD3+ T cells, CD19+ B cells, CD8+ T cells, and CD16 + 56+ NK cells). Such an immuno-depleted state in patients may be one of the factors for the severity of CoV infection. Also the patients had a compromised gut microbiota with severe microbial translocation resulting in secondary infections. This finding suggests that the infection mediated immuno-compromised state may lead to adverse clinical morbidities and high risk of fatal pneumonia.
Recently, a study was conducted to explore the epidemiology of the disease as well as the clinical investigation of 99 patients affected with SARS-CoV-2 admitted to Jinyintan Hospital of Wuhan [11]. This comprehensive study suggested that older adult males associated with chronic comorbidities due to their weaker immune response are more susceptible to COVID-19 infection. The study also reported that obesity and chronic co-morbidities are associated with an elevated mortality rate. The study suggests that a reduced number of lymphocytes can be a potent biomarker for the detection of severe SARS-CoV-2 infection.
Irani Thevarajan et al., detected elevated levels of follicular helper T cells (TFH cells), activated CD4+ T cells and CD8+ T cells, antibody-secreting cells (ASCs) and Immunoglobulin M (IgM) and Immunoglobulin G (IgG) against SARS-CoV-2 in the blood sample extracted from a 47-year-old infected woman before her symptomatic recovery. The patient was noted to have a peak level of ASCs of about 6.91% on day 8 after infection. Similarly, this study reported that circulating TFH cells (cTFH cells) levels in the woman was also increased on day 8 and day 9 post-infection. High-level expression of ASCs and cTFH cells in the bloodstream of the symptomatic patients were observed when compared to healthy individuals. This study presents contradicting results to those reported earlier from three fatal cases with reduced immune cells from Renmin Hospital of Wuhan University. The difference in expression levels found in both studies could probably be the reason for the different outcomes of the infected individuals. While the immune-depleted individuals could not survive the infection, the 47-year old woman with a strong immune response was found to be protected, suggesting the role of immunity status in patients to fight the pandemic.
Besides, Irani Thevarajan et al., also examined the co-expression of CD38 and HLA-DR which is an important phenotype for the CD8+ T cell activation in response to the viral infection in the same patient. They observed rapidly elevated co-expression of CD38 and HLA-DR between day 7 and day 9 of infection that decreased at day 20 of infection. The observed frequency of CD38+HLA-DR+ CD8+ T cells was comparatively higher in the affected individual than healthy individuals [52]. All three infective coronaviruses, namely, SARS-CoV, MERS-CoV, and SARS-CoV-2 induce a high level of an abnormal non-effective immune response in hosts, which is normally associated with undesirable severe lung pathology.
In most severe cases of COVID-19, the virus induces a plethora of cytokines which is identified by activated T-helper-1 (Th1) cell responses with elevated levels of Interleukins-1β, 2, 6, 7, 8 and 10 (IL-1β, IL-2, IL-6, IL-7, IL-8, and IL-10), granulocyte-colony stimulating factor (GSF), tumour necrosis factor-α (TNF-α), interferon-γ, induced protein (IP-10), monocyte chemoattractant protein-1 (MCP-1) and macrophage inflammatory-protein 1-α (MIP-1α). Such increased levels of proinflammatory cytokines may lead to pulmonary inflammation and harmful tissue damage in the heart, liver and kidney, causing multiorgan failure [37,53,54]. Abundance of cytokines is reported to be closely associated with the development and progression of ARDS in patients infected with SARS-CoV, MERS-CoV, and SARS-CoV-2 infection. The dysregulated cytokine response in severe COVID-19 cases with ARDS is also correlated with a significant increase in mortality rate especially in elderly patients [[55], [56], [57], [58]]. In addition, COVID-19 infection induced elevated secretion of Th2 (IL-4 and IL-10) and Th17 cytokines. In this line, few studies reported that IL-17 blockade in COVID-19 patients with elevated IL-17 plasma levels can be a potential therapeutic option in reducing the disease severity [37,53]. Several other strategies to reduce the inflammatory responses are under trial, like neutralizing antibodies against proinflammatory cytokines and their receptors, and small molecule inhibitors for blocking cytokine mediated immunopathology. Such approaches become crucial to minimize the mortality rate in COVID-19 patients [54].
The host immune system acts as a double-edged sword by exerting a beneficial role in controlling CoV infections and a detrimental role leading to immunopathogenesis. β-CoV has genes that encode for several structural proteins like glycosylated S-protein and non-structural proteins like RNA-dependent RNA polymerase (RdRp), papain-like protease (PLpro) and coronavirus main protease (3CLpro) which are essential for the viral life cycle. Deubiquitinase activity of PLpro on interferon regulatory factor 3 (IRF-3) and NF-κB results in immune evasion in CoV infected persons [16,59,60]. Viral RNA once released inside the cell acts as pathogen-associated molecular patterns (PAMPs), which are recognized by pattern recognition receptors (PRRs) such as toll-like receptor 3 (TLR3), TLR7/8, TLR9 present in endosomes [61,62].
Other cytosolic receptors like melanoma differentiation-associated gene 5 (MDA5), nucleotidyltransferase, cyclic GMP-AMP synthase (cGAS) and retinoic-acid inducible gene I (RIG-I) aid in detection of viral RNA and DNA present in the cytoplasm [63,64]. Further, these multiple signalling pathways employ adaptor molecules like TIR-domain-containing adaptor protein with IFN-β (TRIF) and mitochondrial antiviral-signalling protein (MAVS) to induce downstream cascade molecules that involve MyD88. MyD88 is found to activate NF-κB and IRF3 leading to the release of type I interferons (IFN-α/β) and other pro-inflammatory cytokines [65,66]. The immune response against CoV is pictorially represented in Fig. 5 . It is possible that therapeutic and prophylactic strategies to harness the host immune component could serve as a potential arsenal against COVID-19.
Fig. 5.
The immune response against CoV.
7. Existing clinical therapies and promising targets to treat COVID-19
7.1. Existing clinical therapies
There are no specific anti-viral therapeutic agents or effective vaccines for the treatment and prevention of COVID-19 respectively. In this scenario, the current management of the COVID-19 pandemic is employing supportive treatment by managing only the symptoms. These supportive cares includes oxygenation, ventilation, and fluid management. Since there are no supportive evidences to prove the stand-alone efficacy of existing antiviral drugs for SARS-CoV-2, a combinatorial therapy of low-dose systemic corticosteroids, anti-virals and respiratory supportive treatment are being followed [[67], [68]]. COVID-19 positive individuals must be kept in beds with close monitoring of their vital signs and oxygen saturation level. In addition, protective ventilation strategies are highly recommended to treat COVID-19 patients associated with ARDS. Current antivirals like lopinavir/ritonavir and remdesivir have also been suggested for treating COVID-19 positive cases [69].
Moreover, it has been reported that critically ill COVID-19 patients are often associated with systemic disorders on multiple organs like kidney, heart, and coagulation system during the early stages of disease progression. Therefore, for effective management of such patients it becomes crucial to evaluate multi-organ functionality at regular intervals along with respiratory supportive treatments [70]. Also, studies reported late manifestations of acute organ injury, such as acute kidney injury (AKI), that is found to be one of the main risk factors in COVID-19 infection [71]. Even though systemic corticosteroids are administered presently, experimental evidences for corticosteroids for their beneficial or harmful role in infected patients warrants further investigation [37]. Recently, Wang et al., has reported that convalescent plasma therapy can also be explored after detailed clinical studies [72]. Hence, there is an urgent need for the development of effective therapies, anti-viral drugs, and vaccines to suppress the increasing mortality rate and infection rate of the existing COVID-19 pandemic.
7.2. Potential targets for implementing efficacious therapeutics against SARS-CoV-2
Based on previously established literature and considerable results on SARS-CoV research, new possibilities, and several potential blocking strategies can be investigated for the blockade of SARS-CoV-2 [2].
The following potential target options for developing effective therapeutics have emerged based on previous and current experience. These include blocking of S-protein, blocking of ACE2 receptor, inhibition of type 2 serine proteases TMPRSS2, and inhibition of specific target enzymes of the virus that plays a crucial role in viral survival.
7.2.1. Blocking of S-protein
S-protein can be considered as a promising candidate for the development of therapeutics and CoV vaccine composition. It is remarkably stable in its immunogenicity and binding towards the ACE2 receptor, which makes them the crucial mediator to initiate viral entry into the host cell. A recent article has reported a real time mutation tracking study in SARS-CoV-2 coronavirus especially a mutation at the spike region called “D614G” caused by a single base change from G to A at position 23,403 of the Wuhan reference strain. Spike mutation D614G, is reported as the dominant pandemic form globally and may be associated with increased spreading and might impact transmission by its high serological reactivity, making it an urgent concern. Fourteen different mutation sites in the viral S-protein have been identified and their implications are being explored through structural modelling. The study findings suggest that the coronavirus is still evolving and such mutations could make individuals more susceptible to reinfections.
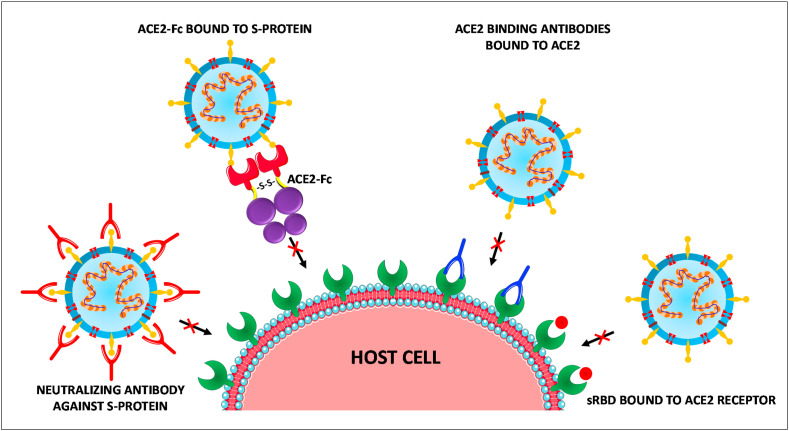
Thus, S-protein is one of the major targets for designing various antiviral therapies for CoVs. These antiviral therapies against S-protein include neutralizing antibodies against S-proteins, RBD-ACE2 blockers, S cleavage inhibitors, and S-protein inhibition. Further, blockade of S-protein can be established by human monoclonal antibodies. Such neutralizing antibodies earlier have shown a significant reduction of severity in the lung pathology of non-human primates who were infected by MERS-CoV infection [[73], [74], [75]].
7.2.2. Blocking of ACE2 receptor
Apart from targeting the inhibition of S-protein of the CoV, another efficient strategy could be blocking the ACE2 receptor so that the viral S-protein attachment and membrane fusion followed by viral entry can be prevented. This targeting of ACE2 can be achieved by administering small receptor binding domains (sRBD), a 193 amino acid-sized domain from SARS S-protein which effectively blocked the entry of SARS in vitro [76]. In vitro experiments were reported earlier involving anti-ACE2 antibodies for SARS-CoV infection, may serve as a promising strategy to block ACE2 receptors [77]. Additional in vitro experimental findings using Vero E6 cells, the human recombinant soluble ACE2 (hrsACE2) can be considered as a potential option to treat SARS-CoV-2 [78]. Phase 1 and phase 2 clinical testing has been done using hrsACE2 for the treatment of ARDS [79], however the clinical trial (NCT04287686) initiated to test the protective effect of hrsACE2 against SARS-CoV-2 infection has been withdrawn [80]. Extending the life-span of soluble circulating ACE2 receptor by converting it into an immunoadhesin form ACE2-Fc (ACE2 fused to immunoglobulin Fc domain) and making it to effectively bind to the S-protein of SARS-CoV-2 could be another promising strategy to tackle SARS-CoV-2 [2,81,82].
However, a mechanism of virus uptake that facilitates cell entry through the Fcγ receptor (FcγR) dependent manner, termed antibody-dependent enhancement (ADE) may occur when virus binds with non-neutralizing antibodies or sub-neutralizing concentrations of antibodies. Whether ADE occurs in SARS-CoV-2 infection is still controversial but such possible consequences of this alternative infection pathway contributing to disease severity must be considered while developing antibody-based therapeutics [83]. These above four different blockade methods are pictorially represented in Fig. 6 .
Fig. 6.
Potential targets for the blockade of SARS-CoV-2 entry.
ACE2 blocking-therapeutic implications: Circulating components and tissue specific components of renin–angiotensin–aldosterone system (RAAS) form an orchestrated network of both regulatory and counter-regulatory peptides. ACE2 is a vital counter-regulatory component of RAAS which degrades angiotensin II (Ang II) to angiotensin-(1–7) (Ang-(1–7)) which attenuates fibrosis, vasoconstriction and sodium retention. Pharmacological modulation of RAAS with angiotensin-converting enzyme inhibitors (ACEIs) and angiotensin receptor blockers (ARBs) are beneficial in treatment of heart failure and hypertension management. The susceptibility of SARS-CoV infection in vitro is related to the ACE2 expression. But there is no direct connection between ACE2 expression and SARS-CoV-2 infection [84]. Blocking of ACE2 is proposed in many therapeutic strategies for COVID-19. But the protective effects of ACE2 in various tissues are widely known. ACE2/Ang-(1–7) axis generally exhibits protective effects including antiapoptotic, antiproliferative, antiarrhythmic and other processes aiding cardiovascular protection [85]. Infections with SARS coronavirus is one of the multiple predisposing factors responsible for ARDS [86]. In a mice model for acute lung injury (induced by sepsis/acid aspiration), ACE2 and the angiotensin II type 2 receptor (AT2) were found to exert protective effects [87].
After viral entry, ACE2 levels decreases leading to enhanced Ang II release favouring ARDS development. Reduced ACE2 expression in lung tissues was accompanied with pulmonary edema, elevated vascular permeability in mice after SARS-CoV infection [88]. From the same study, it was found that injection of only S-protein caused acute lung injury by decreasing expression of ACE2 in lungs which was improved by administering ACEIs/ARBs. ACEIs and ARBs have different effects on Ang II. Several animal models portray mixed results in regard to the role of ACEIs on activity and levels of ACE2 in tissues [[89], [90], [91], [92]]. Animal models have also reported inconsistent findings on the effects of ARBs on ACE2 expression and activity [[92], [93], [94], [95]].
Several studies have clearly emphasized that ACEIs/ARBs possess the ability to upregulate ACE2 expression levels apart from inhibiting ACE or blocking Ang II type 1 receptor [[96], [97], [98]]. For instance, Enalapril can restore ACE2 expression levels in the rats with heart failure [99]. Similarly, olmesartan and losartan can elevate the expression of ACE2 mRNA levels in rats post myocardial-infarction (MI) [97]. ACE2 pharmacological property is not inhibited by ACEIs. Lisinopril can increase ACE2 mRNA levels in hearts of rats post MI but the activity of ACE2 in the heart of normal Lewis rats is unaltered. But, Losartan increased both mRNA levels of ACE2 and also its protein activity [98]. Although, the current investigations are focused on ACE2 mRNA levels in response to ACEIs/ARBs, its effect on human lung tissues (both mRNA and protein levels) is yet to be unraveled. There are few studies in humans assessing the effects of RAAS inhibition on ACE2 expression. Administration of intravenous ACE inhibitors in patients with coronary artery disease had no influence in Ang-(1–7) production questioning the possible effects of ACEIs on ACE2-directed Ang II metabolism [100]. Captopril administration in hypertensive patients did not affect the levels of Ang-(1–7). But, prolonged monotherapy over a period of six months increased Ang-(1–7) levels [101].
There is no clinical data regarding the effect of ACEIs and ARBs on ACE2 expression or activity in human tissues. It is difficult to conclude that ARB-induced changes in ACE2 levels mediate SARS-CoV entry even in animal models. One recent study reported that there is no increased risk for SARS-CoV-2 infection with the usage of RAAS blockers. Administration of captopril, an ACE inhibitor and telmisartan, an ARB drug has not increased the ACE2 expression in mice lung and kidney epithelia [102]. This study supports the view that in patients at risk of contracting COVID-19, the intake of RAAS blockers should be continued. Patients with diabetes, hypertension, heart disease, and chronic kidney disease with ARBs and ACEIs use are the major target groups who need to be closely monitored [103]. A recent large population-based study in Italy, concluded that the use of ACEIs and ARBs had no significant association with the risk of COVID-19 [104].
Apart from developing vaccines against COVID-19, RAAS modulation, more specifically by ACE2 and Ang II, is a vital therapeutic target. With reference to a previous study in mice models, ARB showed beneficial effects in SARS-CoV induced acute lung injury [88]. Multicenter, double-blinded placebo controlled, randomized clinical trials are in progress to study the effect of losartan, an ARB on patients requiring hospital admission (NCT04312009) and on patients who do not need hospital admission (NCT04311177). The outcome of these trials may shed more information on the therapeutic benefits of this class of drugs.
7.2.3. Inhibition of type 2 serine proteases TMPRSS2
Recent studies have confirmed that TMPRSS2 is responsible for triggering and activating the infection and entry of CoVs. Moreover, TMPRSS2 plays a crucial role in the cleavage of the SARS-CoV-2 S-protein, and thereby mediates viral entry. Thus, inhibition of this host cell protease could act as a potential anti-viral intervention against SARS-CoV-2. Further, experimental evidence has shown that camostat mesylate, a potential inhibitor of TMPRSS2 actively blocked SARS-CoV-2 infection in lung cells leading credence to this strategy [49,105,106].
7.2.4. Inhibition of specific target enzymes of virus that plays a crucial role in viral survival
Humanized nanobodies were reported to have the capacity of passing across the virus-infected cells. These trans-bodies are capable of binding or inhibiting various biological activities of viruses, which ultimately leads to the inhibition of viral replication. This approach has produced some effective progressive trans-bodies against the hepatitis C virus, dengue virus, Ebola virus, and influenza virus [74,107]. Thus, a similar approach to target the crucial components of CoVs could result in the development of an effective treatment against SARS-CoV-2 infection. The nanobodies could be raised against some of the major crucial viral proteins that are responsible for viral replication and infection of CoVs such as PLpro (an essential enzyme in CoV replication and antagonization of innate immunity of host), 3CLpro (responsible for the maturation of Nsps which is a crucial part in CoV viral life cycle), RdRp (essential for CoV transcription complex) and helicase (multifunctional protein responsible for CoV replication) [74,106]. Targeting these crucial components of CoVs could directly or indirectly affect viral replication and survivability in the host cell. The compilation of potential pharmacological targets and their investigational therapies/drug candidates are listed in Table 1 and the details of ongoing clinical trials on these investigational drugs (as experimental drug candidates) are mentioned in Table 2 .
Table 1.
Potential pharmacological targets and their investigational therapies/drug candidates.
| Pharmacological Target | Role of the target in Viral infection | Investigational drug candidate | Possible mechanism of action | References |
|---|---|---|---|---|
| ACE2/S-protein |
ACE2: Host cell receptor in which S-protein binds and initiates viral entry S-protein: Main structural protein that binds with host cell receptor; S-protein’s cleavage activation and structural integrity plays a pivotal role in virus invasion and virulence |
Umifenovir (Arbidol) | Targets S-protein/ACE2 interaction; May inhibit membrane fusion of viral envelope | [16,23,49], [[108], [109], [110], [111]], |
| RdRp | A vital enzyme responsible for CoV replication and transcription | Remdesivir, Favipiravir, Ribavirin | Inhibition of RdRp; May halt viral replication through premature termination of the viral RNA chain | [[112], [113], [114], [115]], [108,[116], [117], [118], [119]] |
| 3CLpro | Directly mediates the maturation of Non-structural proteins (Nsps) which are crucial for viral replication | Lopinavir/ritonavir | Inhibition of 3CLpro; May prevent viral replication | [120], [116,119,121], [117,122] |
| PLpro | Acts as a protease for the proteolysis of viral polyprotein to its functional units. Also responsible for antagonizing host’s innate immunity | Lopinavir | Inhibition of PLpro; May prevent viral replication | [16,[123], [124], [125], [126]], [16,106,127] |
| TMPRSS2 | Triggers the infection by priming S-protein; Facilitates S-protein’s binding to the host receptor | Camostat mesylate | Inhibition of TMPRSS2 and this inhibitory action on the enzymatic activity of TMPRSS2 may prevent CoV from entering into the cell | [16,128], [16,49,106] |
Table 2.
Ongoing clinical trials on the investigational drug candidates.
| Investigational Drug Candidate | NCT Number | Official Title | Phase |
|---|---|---|---|
| Umifenovir (Arbidol) | NCT04350684 | Efficacy and Safety of Umifenovir as an Adjuvant Therapy Compared to the Control Therapeutic Regiment of Interferon Beta 1a, Lopinavir/Ritonavir and a Single Dose of Hydroxychloroquine in Moderate to Severe COVID-19: A Randomized, Double-Blind, Placebo-Controlled, Clinical Trial | 4 |
| NCT04260594 | Randomized, Open, Multicenter Study on the Efficacy and Safety of Arbidol Hydrochloride Tablets in Treating Pneumonia in Patients Infected With Novel Coronavirus (2019-ncov) | 4 | |
| Remdesivir | NCT04292899 | A Phase 3 Randomized Study to Evaluate the Safety and Antiviral Activity of Remdesivir (GS-5734™) in Participants With Severe COVID-19 | 3 |
| NCT04292730 | A Phase 3 Randomized Study to Evaluate the Safety and Antiviral Activity of Remdesivir (GS-5734™) in Participants With Moderate COVID-19 Compared to Standard of Care Treatment | 3 | |
| NCT04252664 | A Phase 3 Randomized, Double-blind, Placebo-controlled Multicenter Study to Evaluate the Efficacy and Safety of Remdesivir in Hospitalized Adult Patients With Mild and Moderate COVID-19 | 3 | |
| NCT04409262 | A Phase III, Randomized, Double-Blind, Multicenter Study to Evaluate the Efficacy and Safety of Remdesivir Plus Tocilizumab Compared With Remdesivir Plus Placebo in Hospitalized Patients With Severe COVID-19 Pneumonia | 3 | |
| NCT04257656 | A Phase 3 Randomized, Double-blind, Placebo-controlled, Multicenter Study to Evaluate the Efficacy and Safety of Remdesivir in Hospitalized Adult Patients With Severe COVID-19 | 3 | |
| NCT04401579 | A Multicenter, Adaptive, Randomized Blinded Controlled Trial of the Safety and Efficacy of Investigational Therapeutics for the Treatment of COVID-19 in Hospitalized Adults (ACTT-II) | 3 | |
| NCT04330690 | A Multi-center, Adaptive, Randomized, Open-label, Controlled Clinical Trial of the Safety and Efficacy of Investigational Therapeutics for the Treatment of COVID-19 in Hospitalized Patients (CATCO: Canadian Treatments for COVID-19), in Conjunction With the Public Health Emergency SOLIDARITY Trial (World Health Organization) | 2 | |
| NCT04280705 | A Multicenter, Adaptive, Randomized Blinded Controlled Trial of the Safety and Efficacy of Investigational Therapeutics for the Treatment of COVID-19 in Hospitalized Adults | 3 | |
| NCT04321616 | The (Norwegian) NOR Solidarity Multicenter Trial on the Efficacy of Different Anti-viral Drugs in SARS-CoV-2 Infected Patients | 3 | |
| NCT04315948 | Multi-center, Adaptive, Randomized Trial of the Safety and Efficacy of Treatments of COVID-19 in Hospitalized Adults | 3 | |
| NCT04349410 | The Fleming [FMTVDM] Directed COVID-19 Treatment Protocol | 2, 3 | |
| Favipiravir | NCT04336904 | A Multi-center, Randomized, Double-blind, Placebo-controlled, Phase III Clinical Study Evaluating the Efficacy and Safety of Favipiravir in the Treatment of Patients With COVID-19-Moderate Type | 3 |
| NCT04358549 | Open Label, Randomized, Controlled Phase 2 Proof-of-Concept Study of the Use of Favipiravir v. Standard of Care in Hospitalized Subjects With COVID-19 | 2 | |
| NCT04359615 | Efficacy and Safety of Favipiravir Compared to the Base Therapeutic Regiment in Moderate to Severe COVID-19: A Randomized, Controlled, Double-Blind, Clinical Trial | 4 | |
| NCT04425460 | A Multi-center, Randomized, Double-blind, Placebo-controlled, Phase III Clinical Study Evaluating the Efficacy and Safety of Favipiravir in the Treatment of Adult Patients With COVID-19-Moderate Type | 3 | |
| NCT04349241 | Efficacy and Safety of Favipiravir in Management of COVID-19 | 3 | |
| NCT04402203 | Study on Safety and Efficacy of Favipiravir (Favipira) for COVID-19 Patient in Selected Hospitals of Bangladesh | 2 | |
| NCT04411433 | An Open-Label, Multicenter, Parallel-Group, Randomized, Phase III Study to Evaluate the Efficacy and Safety of Hydroxychloroquine and Favipiravir in the Treatment of Mild to Moderate COVID-19 | 3 | |
| NCT04387760 | Treatment of Covid-19 With Favipiravir Versus Hydroxychloroquine: a Randomized Comparator Trial | 3 | |
| NCT04392973 | A Trial of Favipiravir and Hydroxychloroquine Combination in Adults Hospitalized With Moderate and Severe Covid-19 | Not Applicable | |
| NCT04303299 | A 6 Week Prospective, Open Label, Randomized, in Multicenter Study of, Oseltamivir Plus Hydroxychloroquine Versus Lopinavir/Ritonavir Plus Oseltamivir Versus Darunavir/Ritonavir Plus Oseltamivir Plus Hydroxychloroquine in Mild COVID-19 AND Lopinavir/Ritonavir Plus Oseltamivir Versus Favipiravir Plus Lopinavir/Ritonavir Versus Darunavir/Ritonavir Plus Oseltamivir Plus Hydroxychloroquine Versus Favipiravir Plus Darunavir and Ritonavir Plus Hydroxychloroquine in Moderate to Critically Ill COVID-19 | 3 | |
| NCT04310228 | Favipiravir Combined With Tocilizumab in the Treatment of Corona Virus Disease 2019-A Multicenter, Randomized and Controlled Clinical Trial Study | Not Applicable | |
| NCT04376814 | The Regimen of Favipiravir Plus Hydroxychloroquine Can Accelerate Recovery of the COVID-19 Patients With Moderate Severity in Comparison to Lopinavir/Ritonavir Plus Hydroxychloroquine Regimen: an Open-label, Non-randomized Clinical Trial Study | Not Applicable | |
| NCT04333589 | The Mechanism, Clinical Outcome and Therapeutic Intervention of Corona Virus Disease 2019 Patients Whose Nucleic Acids Changed From Negative to Positive | Not Applicable | |
| NCT04373733 | A Randomized Controlled Trial of Early Intervention in Patients Hospitalized With COVID-19: Favipiravir Verses Hydroxychloroquine & Azithromycin & Zinc verses Standard Care | 3 | |
| NCT04351295 | Clinical Study Evaluating the Efficacy of Favipiravir in COVID-19 Treatment | 2 | |
| NCT04346628 | A Phase 2 Randomized, Double Blinded, Placebo Controlled Study of Oral Favipiravir Compared to Current Standard of Care in Subjects With Mild or Asymptomatic COVID-19 | 2 | |
| NCT04356495 | Outpatient Treatment of Elderly People With Symptomatic SARS-CoV-2 Infection (COVID-19): a Multi-arm, Multi-stage (MAMS) Randomized Trial to Assess the Efficacy and Safety of Several Experimental Treatments to Decrease the Risk of Hospitalization or Death (COVERAGE Trial) | 3 | |
| NCT04345419 | The Results of COVID 19 Treatment: A Real-life Experience on Patients With COVID 19 | 3 | |
| Ribavirin | NCT04356677 | An Open-Label Study to Evaluate the Safety and Efficacy of VIRAZOLE® (RIBAVIRIN FOR INHALATION SOLUTION, USP) in Hospitalized Adult Participants With Respiratory Distress Due to COVID-19 | 1 |
| NCT04392427 | Effect of a Combination of Nitazoxanide, Ribavirin and Ivermectin Plus Zinc Supplement on the Clearance of COVID-19: a Pilot Sequential Clinical Trial | 3 | |
| NCT04276688 | An Open-label Randomized Controlled Trial on Lopinavir/Ritonavir, Ribavirin and Interferon Beta 1b Combination Versus Lopinavir/Ritonavir Alone, as Treatment for 2019 Novel Coronavirus Infection | 2 | |
| Lopinavir/ritonavir | NCT04307693 | Randomized Controlled Clinical Trials of Lopinavir/Ritonavir or Hydroxychloroquine in Patients With Mild Coronavirus Disease (COVID-19) | 2 |
| NCT04330690 | A Multi-center, Adaptive, Randomized, Open-label, Controlled Clinical Trial of the Safety and Efficacy of Investigational Therapeutics for the Treatment of COVID-19 in Hospitalized Patients (CATCO: Canadian Treatments for COVID-19), in Conjunction With the Public Health Emergency SOLIDARITY Trial (World Health Organization) | 2 | |
| NCT04346147 | Prospective, Phase II, Randomized, Open-label, Parallel Group Study to Evaluate the Efficacy of Hydroxychloroquine Together With Baricitinib, Imatinib or Early Lopinavir/Ritonavir in Patients With SARS Cov 2 Pneumonia | 2 | |
| NCT04372628 | Trial of Early Therapies During Non-hospitalized Outpatient Window (TREAT NOW) for COVID-18 | 2 | |
| NCT04328285 | Chemoprophylaxis of SARS-CoV-2 Infection (COVID-19) in Exposed Healthcare Workers: A Randomized Double-blind Placebo-controlled Clinical Trial | 3 | |
| NCT04359095 | Effectiveness and Safety of Medical Treatment for SARS-CoV-2 (COVID-19) in Colombia: A Pragmatic Randomized Controlled Trial | 3 | |
| NCT04409483 | Evaluation of Additional Treatments for COVID-19: a Randomized Trial in Niger | 3 | |
| NCT04403100 | Hydroxychloroquine and Lopinavir/Ritonavir for Hospitalization and Mortality Reduction in Patients With COVID-19 and Mild Disease Symptoms: “The Hope Coalition" | 3 | |
| NCT04321174 | COVID-19 Ring-based Prevention Trial With Lopinavir/Ritonavir | 3 | |
| NCT04255017 | An Open, Prospective/Retrospective, Randomized Controlled Cohort Study to Compare the Efficacy of Three Antiviral Drugs (Arbidol Hydrochloride, Oseltamivir and Lopinavir/Ritonavir) in the Treatment of 2019-nCoV Pneumonia | 4 | |
| NCT04364022 | Efficacy of Pragmatic Same-day Ring COVID-19 Prophylaxis for Adult Individuals Exposed to SARS-CoV-2 in Switzerland: an Open-label Cluster Randomized Trial | 3 | |
| NCT04328012 | Comparison Of Therapeutics for Hospitalized Patients Infected With SARS-CoV-2 In a Pragmatic adaptive randomized Clinical Trial During the COVID-19 Pandemic (COVID MED Trial) | 2 | |
| NCT04295551 | Multicenter Clinical Study on the Efficacy and Safety of Xiyanping Injection in the Treatment of New Coronavirus Infection Pneumonia (General and Severe) | Not Applicable | |
| NCT04321993 | Treatment of Moderate to Severe Coronavirus Disease (COVID-19) in Hospitalized Patients | 2 | |
| NCT04351724 | A Multicenter, Randomized, Active Controlled, Open Label, Platform Trial on the Efficacy and Safety of Experimental Therapeutics for Patients With COVID-19 (Caused by Infection With Severe Acute Respiratory Syndrome Coronavirus-2) | 3 | |
| NCT04365582 | A Randomized Trial of Efficacy and Safety of an Early Outpatient Treatment of COVID-19 in Patients With Risk Factor for Poor Outcome: a Strategy to Prevent Hospitalization | 3 | |
| NCT04315948 | Multi-center, Adaptive, Randomized Trial of the Safety and Efficacy of Treatments of COVID-19 in Hospitalized Adults | 3 | |
| NCT04386070 | Preventing Pulmonary Complications in Surgical Patients at Risk of COVID-19 | 3 | |
| NCT04381936 | Randomized Evaluation of COVID-19 Therapy | 2 | |
| NCT02735707 | Randomized, Embedded, Multifactorial Adaptive Platform Trial for Community- Acquired Pneumonia | 4 | |
| NCT04276688 | An Open-label Randomized Controlled Trial on Lopinavir/Ritonavir, Ribavirin and Interferon Beta 1b Combination Versus Lopinavir/Ritonavir Alone, as Treatment for 2019 Novel Coronavirus Infection | 2 | |
| NCT04303299 | A 6 Week Prospective, Open Label, Randomized, in Multicenter Study of, Oseltamivir Plus Hydroxychloroquine Versus Lopinavir/Ritonavir Plus Oseltamivir Versus Darunavir/Ritonavir Plus Oseltamivir Plus Hydroxychloroquine in Mild COVID-19 AND Lopinavir/Ritonavir Plus Oseltamivir Versus Favipiravir Plus Lopinavir/Ritonavir Versus Darunavir/Ritonavir Plus Oseltamivir Plus Hydroxychloroquine Versus Favipiravir Plus Darunavir and Ritonavir Plus Hydroxychloroquine in Moderate to Critically Ill COVID-19 | 3 | |
| Camostat mesylate | NCT04353284 | The Effect of Camostat Mesylate on COVID-19 Infection in Ambulatory Patients: An Investigator-Initiated Randomized, Placebo-Controlled, Phase IIa Trial | 2 |
| NCT04321096 | The Impact of Camostat Mesilate on COVID-19 Infection: An Investigator-initiated Randomized, Placebo-controlled, Phase IIa Trial | 1 | |
| NCT04338906 | Evaluation of the Efficacy and Safety of Camostat Mesilate + Hydroxychloroquine Combination Therapy in Hospitalized Patients With Moderate COVID-19 Infection | 4 | |
| NCT04374019 | Randomized, Multi-arm Phase II Trial of Novel Agents for Treatment of High-risk COVID-19 Positive Patients | 2 |
8. Key questions on SARS-CoV-2
Aforementioned therapeutic targets and blockade mechanisms are being explored at different stages of infection but no successful outcome has yet been reported. Novel drugs, vaccines, and immunoglobulins are yet to be discovered for both prevention and treatment of COVID-19. There are still many unresolved questions on SARS-CoV-2 that warrant investigation. Some of the key questions that remain unresolved are:
It has been reported that COVID-19 can be spread from asymptomatic persons. Quantitative reverse-transcriptase–polymerase chain-reaction (qRT-PCR) assay of sputum samples from such asymptomatic patients revealed high viral-load (108/milliliter) [129]. This is a potential threat to the persons coming in contact with asymptomatic COVID-19 positive patients. Considering this scenario, the first key question that emerges is “What is the mechanism involved in the asymptomatic nature of SARS-CoV-2 infected persons?”
It has been widely reported by many researchers and clinicians that older people are more susceptible to COVID-19 disease. Moreover, older people with underlying disease conditions are considered to belong to the high-risk group. The answer for “Why elderly people are more susceptible to the COVID-19 infection?” remains to be addressed scientifically.
Even though there are considerable theories for the mechanism of the immune responses against CoVs, there is a definitive need for understanding the body’s immune response against SARS-CoV-2 infection to create effective treatment strategies. Hence the question, “What are the sequential immune responses against COVID-19?” needs to be answered through systematic studies.
As additional information on SARS-CoV2 is revealed, more questions may arise, the answers for which may lead to the development of superior therapeutics and preventive measures for this pandemic.
9. Concluding remarks
The current outbreak of COVID-19 has disseminated from China and swept around the globe at an alarming rate resulting in an exponential increase in infection rate and mortality. Currently, >25,000,000 laboratory confirmed cases and >850,000 deaths have been reported worldwide and the numbers continue to increase daily dramatically. Even though extensive quarantine and control methods are implemented by the federal governments, the spread of the infection still continues unabated. This highly contagious and highly pathogenic SARS-CoV-2 has posed a life threatening situation to the world population that has disrupted normal life and paralyzed the world economy in a matter of weeks. By far, the exact pathogenicity of COVID-19 is still unclear and needs rigorous studies. The current possible actions against COVID-19 are to cut-off transmission by social distancing, control the source of infection, and effective usage of existing anti-viral drugs. Amidst various technical challenges, we should focus on continuing to develop specific drugs and vaccines to control this pandemic.
Author contribution
Shibi Muralidar: Writing - Original Draft, Collection of data; Senthil Visaga Ambi: Conceptualization, Visualization, Supervision, Writing - Review & Editing; Saravanan Sekaran: Writing - Review & Editing; Uma Maheswari Krishnan: Writing - Review & Editing. All authors have approved the final article for submission.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgement
The authors express their gratitude to SASTRA-Deemed-to-be-University, Tamilnadu, India for infrastructure and financial support. Authors also extend their appreciation for the contribution of Biopharmaceutical research lab members, SASTRA-Deemed-to-be-University.
References
- 1.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kruse R.L. Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China, F1000Research. 2020. 9. 72. [DOI] [PMC free article] [PubMed]
- 3.Sahin A.R. Novel coronavirus (COVID-19) outbreak: a review of the current literature. Eurasian J. Med. Investig. 2019;4:1–7. doi: 10.14744/ejmo.2020.12220. 2020. [DOI] [Google Scholar]
- 4.Casadevall A., Pirofski L. The convalescent sera option for containing COVID-19. J. Clin. Invest. 2020;2 doi: 10.1172/jci138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang T., Wu Q., Zhang Z. CelPress; 2020. Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak; pp. 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gürbilek N. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. J. Chem. Inf. Model. 2013;53:1689–1699. doi: 10.1017/CBO9781107415324.004. [DOI] [Google Scholar]
- 8.World Health Organization (WHO) Novel coronavirus (2019-nCoV) situation report - 1 21 january 2020. WHO Bull. 2020:1–7. [Google Scholar]
- 9.Culp W.C. Coronavirus disease. In Pract. 2019;14 doi: 10.1213/xaa.0000000000001218. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WHO (World Health Organization) Coronavirus disease (COVID-2019) situation reports. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/https://pers.droneemprit.id/covid19/ Retrieved on 4 April, 2020, 2019. 1–19.
- 11.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boldog P., Tekeli T., Vizi Z., Dénes A., Bartha F.A., Röst G. Risk assessment of novel coronavirus COVID-19 outbreaks outside China. J. Clin. Med. 2020;9:571. doi: 10.3390/jcm9020571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.WHO Situation Report - 3, Novel Coronavirus (2019-nCoV) 2020. pp. 1–7. [Google Scholar]
- 14.WHO Situation Report - 6, Novel Coronavirus (2019-nCoV) 2020. pp. 1–8. [Google Scholar]
- 15.Alba Grifoni, Sidney John, Zhang Yun, Scheuermann Richard H., Peters Bjoern, Alessandro Sette . 2020. Candidate Targets for Immune Responses to 2019-Novel Coronavirus (nCoV): Sequence Homology- and Bioinformatic-Based Predictions, bioRxiv Preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Smoot J., Gregg A.C., Daniels A.D., Jervey S., Albaiu D. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020 doi: 10.1021/acscentsci.0c00272. acscentsci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li G., Fan Y., Lai Y., Han T., Li Z., Zhou P., Pan P., Wang W., Hu D., Liu X., Zhang Q., Wu J. Coronavirus infections and immune responses. J. Med. Virol. 2020;92:424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raoult D., Zumla A., Locatelli F., Ippolito G., Kroemer G. Coronavirus infections : Epidemiological , clinical and immunological features and hypotheses. 2020. (n.d.) 1–10. [DOI] [PMC free article] [PubMed]
- 20.Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. StatPearls; 2020. Features, Evaluation and Treatment Coronavirus (COVID-19)http://www.ncbi.nlm.nih.gov/pubmed/32150360 [PubMed] [Google Scholar]
- 21.Wenham C., Smith J., Morgan R. COVID-19: the gendered impacts of the outbreak. Lancet. 2020;395:846–848. doi: 10.1016/S0140-6736(20)30526-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rabi F.A., Al Zoubi M.S., Al-Nasser A.D., Kasasbeh G.A., Salameh D.M. Sars-cov-2 and coronavirus disease 2019: what we know so far. Pathogens. 2020;9:1–15. doi: 10.3390/pathogens9030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann M., Kleine-weber H., Krüger N., Müller M. 2020. The Novel Coronavirus 2019 (2019-nCoV) Uses the SARS-Coronavirus Receptor ACE2 and the Cellular Protease TMPRSS2 for Entry into Target Cells. [Google Scholar]
- 24.Shanmugaraj B., Malla A., Phoolcharoen W. Emergence of novel coronavirus 2019-nCoV: need for rapid vaccine and biologics development. Pathogens. 2020;9:1–10. doi: 10.3390/pathogens9020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA, J. Am. Med. Assoc. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar S., Maurya V.K., Prasad A.K., Bhatt M.L.B., Saxena S.K. Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV) VirusDisease. 2020;31:1–9. doi: 10.1007/s13337-020-00571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiaolu Tang, Changcheng Wu, Xiang Li, Yuhe Song, Xinmin Yao, Xinkai Wu, Yuange Duan, Hong Zhang, Yirong Wang, Zhaohui Qian, Jie Cui, and Jian Lu, On the origin and continuing evolution of SARS-CoV-2, Microbiology, doi: 10.1093/nsr/nwaa036/5775463. [DOI] [PMC free article] [PubMed]
- 28.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6 doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12:1–8. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han Y., Yang H. The transmission and diagnosis of 2019 novel coronavirus infection disease (COVID-19): a Chinese perspective. J. Med. Virol. 2020;92,:639–644. doi: 10.1002/jmv.25749. 0–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng Y.Y., Ma Y.T., Zhang J.Y., Xie X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 2020;17,:259–260. doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao Y., Wei W., Cheung W.Y., Li W., Li L., Leung G.M., Holmes E.C., Hu Y., Guan Y. 2020. Identifying SARS-CoV-2 Related Coronaviruses in Malayan Pangolins. [DOI] [PubMed] [Google Scholar]
- 33.Trafficked pangolins can carry coronaviruses closely related to pandemic strain, (n.d). https://www.nationalgeographic.com/animals/2020/03/pangolins-coronavirus-covid-possibility/(accessed March 31, 2020).
- 34.Malta M., Rimoin A.W., Strathdee S.A. The coronavirus 2019-nCoV epidemic: is hindsight 20/20? EClinicalMedicine. 2020;20:100289. doi: 10.1016/j.eclinm.2020.100289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu D., Wu T., Liu Q., Yang Z. The SARS-CoV-2 outbreak: what we know. Int. J. Infect. Dis. 2020;94,:44–48. doi: 10.1016/j.ijid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.A.M. Association . 2020. Airborne Transmission of SARS-CoV-2 Theoretical Considerations and Available Evidence. [DOI] [PubMed] [Google Scholar]
- 37.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ottestad W., Signe S. 2020. COVID-19 Patients with Respiratory Failure: what Can We Learn from Aviation Medicine? [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gattinoni L., Coppola S., Cressoni M., Busana M., Rossi S., Chiumello D. COVID-19 does not lead to a “typical” acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 2020;201:1299–1300. doi: 10.1164/rccm.202003-0817LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkerson R.G., Adler J.D., Shah N.G., Brown R. Silent hypoxia: a harbinger of clinical deterioration in patients with COVID-19. Am. J. Emerg. Med. 2020 doi: 10.1016/j.ajem.2020.05.044. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ranieri V.M., Rubenfeld G.D., Thompson B.T., Ferguson N.D., Caldwell E., Fan E., Camporota L., Slutsky A.S. Acute respiratory distress syndrome: the Berlin definition. JAMA, J. Am. Med. Assoc. 2012;307:2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 42.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gattinoni L., Chiumello D., Caironi P., Busana M., Romitti F., Brazzi L., Camporota L. COVID-19 pneumonia: different respiratory treatments for different phenotypes? Intensive Care Med. 2020;46:1099–1102. doi: 10.1007/s00134-020-06033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X., Ma X. Acute respiratory failure in COVID-19: is it “typical” ARDS? Crit. Care. 2020;24:198. doi: 10.1186/s13054-020-02911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friedman J., Calderon-Villarreal A., Bojorquez I., Hernandez C.V., Schriger D., Hirashima E.T. MedRxiv; Mexico: 2020. Excess Out-Of-Hospital Mortality and Declining Oxygen Saturation: the Sentinel Role of EMS Data in the COVID-19 Crisis in Tijuana; p. 2020. 05.13.20098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5 doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li F. Evidence for a common evolutionary origin of coronavirus spike protein receptor-binding subunits. J. Virol. 2012;86:2856–2858. doi: 10.1128/jvi.06882-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 84. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020:1–10. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu Zhao W.Z., Zhao Zixian, Wang Yujia, Zhou Yueqing, Ma Yu. Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. 2020. https://www.medrxiv.org/content/10.1101/2020.06.07.20121939v1 [DOI] [PMC free article] [PubMed]
- 51.Yu L., Tong Y., Shen G., Fu A., Lai Y., Zhou X., Yuan Y., Wang Y., Pan Y., Yu Z., Li Y., Liu T., Jiang H. Immunodepletion with hypoxemia: a potential high risk subtype of coronavirus disease 2019. MedRxiv. 2020:2020. doi: 10.1101/2020.03.03.20030650. 03.03.20030650. [DOI] [Google Scholar]
- 52.Breadth of concomitant immune responses prior to patient recovery : a case report of non-severe. 2019. 1. [DOI] [PMC free article] [PubMed]
- 53.Zumla A., Hui D.S., Azhar E.I., Memish Z.A., Maeurer M. Reducing mortality from 2019-nCoV: host-directed therapies should be an option. Lancet. 2020;395:e35–e36. doi: 10.1016/S0140-6736(20)30305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020;2019 doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cameron M.J., Bermejo-Martin J.F., Danesh A., Muller M.P., Kelvin D.J. Human immunopathogenesis of severe acute respiratory syndrome (SARS) Virus Res. 2008;133:13–19. doi: 10.1016/j.virusres.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reghunathan R., Jayapal M., Hsu L.Y., Chng H.H., Tai D., Leung B.P., Melendez A.J. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol. 2005;6 doi: 10.1186/1471-2172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang Y., Xu J., Zhou C., Wu Z., Zhong S., Liu J., Luo W., Chen T., Qin Q., Deng P. Characterization of cytokine/chemokine profiles of seven acute respiratory syndrome. Am. J. Respir. Crit. Care Med. 2005;171:850–857. doi: 10.1164/rccm.200407-857OC. [DOI] [PubMed] [Google Scholar]
- 59.Frieman M., Basu D., Matthews K., Taylor J., Jones G., Pickles R., Baric R., Engel D.A. Yeast based small molecule screen for inhibitors of SARS-CoV. PloS One. 2011;6:1–9. doi: 10.1371/journal.pone.0028479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frieman M., Ratia K., Johnston R.E., Mesecar A.D., Baric R.S. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-κB signaling. J. Virol. 2009;83:6689–6705. doi: 10.1128/jvi.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 62.Wu J., Chen Z.J. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 63.Yoo J.S., Kato H., Fujita T. Sensing viral invasion by RIG-I like receptors. Curr. Opin. Microbiol. 2014;20:131–138. doi: 10.1016/j.mib.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 64.Hotový I., Pullmannová A., Predanocy M., Hotový J., Řeháček V., Kups T., Spiess L. Structural and morphological investigations of TiO2 sputtered thin films. J. Electr. Eng. 2009;60:354–357. doi: 10.1126/science.1229963.Cyclic-GMP-AMP. [DOI] [Google Scholar]
- 65.Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 66.Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat. Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 67.Shanmugaraj B., Siriwattananon K., Wangkanont K., Phoolcharoen W. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19) Asian Pac. J. Allergy Immunol. 2020;38 doi: 10.12932/AP-200220-0773. 10.12932/AP-200220–0773. [DOI] [PubMed] [Google Scholar]
- 68.Cunningham A.C., Goh H.P., Koh D. Treatment of COVID-19: old tricks for new challenges. Crit. Care. 2020;24:91. doi: 10.1186/s13054-020-2818-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li T. Diagnosis and clinical management of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection: an operational recommendation of peking union medical college hospital (V2.0): Working group of 2019 novel coronavirus, peking union medical colle, Emerg. Microb. Infect. 2020;9:582–585. doi: 10.1080/22221751.2020.1735265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li T., Lu H., Zhang W. Clinical observation and management of COVID-19 patients. Emerg. Microb. Infect. 2020;9:687–690. doi: 10.1080/22221751.2020.1741327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.S.M. Devi, Comparative Genomics and Diversity of SARS-CoV-2 Suggest Potential Regional Virulence, (n.d).
- 72.Wang L., Wang Y., Ye D., Liu Q. A review of the 2019 Novel Coronavirus (COVID-19) based on current evidence. Int. J. Antimicrob. Agents. 2020:105948. doi: 10.1016/j.ijantimicag.2020.105948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Korber B., Wm F., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Ee G., Bhattacharya T., Dg P., Cm E., Silva D., Genomics S.C., Cc L., Teaching S., Nhs H., Trust F., Group C.-G., Angyal A., Brown R.L., Green L.R., Groves D.C., Johnson K.J., Keeley A.J., Benjamin B., Parsons P.J., Raza M., Rowland-jones S., Tucker R.M., Wyles M.D. 2020. Spike Mutation Pipeline Reveals the Emergence of a More Transmissible Form of SARS-CoV-2 Introduction; pp. 1–33. [Google Scholar]
- 74.Dhama K., Sharun K., Tiwari R., Dadar M., Malik Y.S., Singh K.P., Chaicumpa W. COVID-19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum. Vaccines Immunother. 2020;5515:1–7. doi: 10.1080/21645515.2020.1735227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pang J., Wang M.X., Ang I.Y.H., Tan S.H.X., Lewis R.F., Chen J.I.-P., Gutierrez R.A., Gwee S.X.W., Chua P.E.Y., Yang Q., Ng X.Y., Yap R.K., Tan H.Y., Teo Y.Y., Tan C.C., Cook A.R., Yap J.C.-H., Hsu L.Y. Potential rapid diagnostics, vaccine and therapeutics for 2019 novel coronavirus (2019-nCoV): a systematic review. J. Clin. Med. 2020;9 doi: 10.3390/jcm9030623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li W., Moore M.J., Vasllieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greeneugh T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado del Pozo C., Prosper F., Romero J.P., Wirnsberger G., Zhang H., Slutsky A.S., Conder R., Montserrat N., Mirazimi A., Penninger J.M. Inhibition of SARS-CoV-2 infections in Engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913. doi: 10.1016/j.cell.2020.04.004. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Khan A., Benthin C., Zeno B., Albertson T.E., Boyd J., Christie J.D., Hall R., Poirier G., Ronco J.J., Tidswell M., Hardes K., Powley W.M., Wright T.J., Siederer S.K., Fairman D.A., Lipson D.A., Bayliffe A.I., Lazaar A.L. A pilot clinical trial of recombinant human angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Crit. Care. 2017;21:234. doi: 10.1186/s13054-017-1823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K., Moore M.J., Tallarico A.S.C., Olurinde M., Choe H., Anderson L.J., Bellini W.J., Farzan M., Marasco W.A. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moore M.J., Dorfman T., Li W., Wong S.K., Li Y., Kuhn J.H., Coderre J., Vasilieva N., Han Z., Greenough T.C., Farzan M., Choe H. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 2004;78:10628–10635. doi: 10.1128/jvi.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Taylor A., Foo S., King N.J.C. Fc receptors in antibody- dependent enhancement of viral infections. 2015;268:340–364. doi: 10.1111/imr.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hofmann H., Geier M., Marzi A., Krumbiegel M., Peipp M., Fey G.H., Gramberg T., Pöhlmann S. Susceptibility to SARS coronavirus S protein-driven infection correlates with expression of angiotensin converting enzyme 2 and infection can be blocked by soluble receptor. Biochem. Biophys. Res. Commun. 2004;319:1216–1221. doi: 10.1016/j.bbrc.2004.05.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Santos R.A.S., Sampaio W.O., Alzamora A.C., Motta-Santos D., Alenina N., Bader M., Campagnole-Santo M.J. The ACE2/Angiotensin-(1-7)/Mas axis of the renin-angiotensin system: focus on Angiotensin-(1-7) Physiol. Rev. 2018;98:505–553. doi: 10.1152/physrev.00023.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Drosten C., Günther S., Preiser W., Van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M., Berger A., Burguière A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H.D., Osterhaus A.D.M.E., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348 doi: 10.1056/NEJMoa030747. 1967–1976. [DOI] [PubMed] [Google Scholar]
- 87.Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H., Crackower M.A., Fukamizu A., Hui C.C., Hein L., Uhlig S., Slutsky A.S., Jiang C., Penninger J.M. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hamming I., Van Goor H., Turner A.J., Rushworth C.A., Michaud A.A., Corvol P., Navis G. Differential regulation of renal angiotensin-converting enzyme (ACE) and ACE2 during ACE inhibition and dietary sodium restriction in healthy rats. Exp. Physiol. 2008;93:631–638. doi: 10.1113/expphysiol.2007.041855. [DOI] [PubMed] [Google Scholar]
- 90.Ocaranza M.P., Godoy I., Jalil J.E., Varas M., Collantes P., Pinto M., Roman M., Ramirez C., Copaja M., Diaz-Araya G., Castro P., Lavandero S. Enalapril attenuates downregulation of angiotensin-converting enzyme 2 in the late phase of ventricular dysfunction in myocardial infarcted rat. Hypertension. 2006;48:572–578. doi: 10.1161/01.HYP.0000237862.94083.45. [DOI] [PubMed] [Google Scholar]
- 91.Burchill L.J., Velkoska E., Dean R.G., Griggs K., Patel S.K., Burrell L.M. Combination renin-angiotensin system blockade and angiotensin-converting enzyme 2 in experimental myocardial infarction: implications for future therapeutic directions. Clin. Sci. 2012;123:649–658. doi: 10.1042/CS20120162. [DOI] [PubMed] [Google Scholar]
- 92.Burrell L.M., Risvanis J., Kubota E., Dean R.G., MacDonald P.S., Lu S., Tikellis C., Grant S.L., Lew R.A., Smith A.I., Cooper M.E., Johnston C.I. Myocardial infarction increases ACE2 expression in rat and humans. Eur. Heart J. 2005;26:369–375. doi: 10.1093/eurheartj/ehi114. [DOI] [PubMed] [Google Scholar]
- 93.Soler M.J., Ye M., Wysocki J., William J., Lloveras J., Batlle D. Localization of ACE2 in the renal vasculature: amplification by angiotensin II type 1 receptor blockade using telmisartan. Am. J. Physiol. Ren. Physiol. 2009;296:398–405. doi: 10.1152/ajprenal.90488.2008. [DOI] [PubMed] [Google Scholar]
- 94.Sukumaran V., Veeraveedu P.T., Gurusamy N., Lakshmanan A.P., Yamaguchi K., Ma M., Suzuki K., Nagata M., Takagi R., Kodama M., Watanabe K. Olmesartan attenuates the development of heart failure after experimental autoimmune myocarditis in rats through the modulation of ANG 1-7 mas receptor. Mol. Cell. Endocrinol. 2012;351:208–219. doi: 10.1016/j.mce.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 95.Lakshmanan A.P., Thandavarayan R.A., Watanabe K., Sari F.R., Meilei H., Giridharan V.V., Sukumaran V., Soetikno V., Arumugam S., Suzuki K., Kodama M. Modulation of AT-1R/MAPK cascade by an olmesartan treatment attenuates diabetic nephropathy in streptozotocin-induced diabetic mice. Mol. Cell. Endocrinol. 2012;348:104–111. doi: 10.1016/j.mce.2011.07.041. [DOI] [PubMed] [Google Scholar]
- 96.Karram T., Abbasi A., Keidar S., Golomb E., Hochberg I., Winaver J., Hoffman A., Abassi Z. Effects of spironolactone and eprosartan on cardiac remodeling and angiotensin-converting enzyme isoforms in rats with experimental heart failure. Am. J. Physiol. Heart Circ. Physiol. 2005;289:1351–1358. doi: 10.1152/ajpheart.01186.2004. [DOI] [PubMed] [Google Scholar]
- 97.Ishiyama Y., Gallagher P.E., Averill D.B., Tallant E.A., Brosnihan K.B., Ferrario C.M. Upregulation of angiotensin-converting enzyme 2 after myocardial infarction by blockade of angiotensin II receptors. Hypertension. 2004;43:970–976. doi: 10.1161/01.HYP.0000124667.34652.1a. [DOI] [PubMed] [Google Scholar]
- 98.Ferrario C.M., Jessup J., Chappell M.C., Averill D.B., Brosnihan K.B., Tallant E.A., Diz D.I., Gallagher P.E. Effect of angiotensin-converting enzyme inhibition and angiotensin II receptor blockers on cardiac angiotensin-converting enzyme 2. Circulation. 2005;111:2605–2610. doi: 10.1161/CIRCULATIONAHA.104.510461. [DOI] [PubMed] [Google Scholar]
- 99.Karram T., Abbasi A., Keidar S., Golomb E., Hochberg I., Winaver J., Hoffman A., Abassi Z. Effects of spironolactone and eprosartan on cardiac remodeling and angiotensin-converting enzyme isoforms in rats with experimental heart failure. Am. J. Physiol. Heart Circ. Physiol. 2005;289:1351–1358. doi: 10.1152/ajpheart.01186.2004. [DOI] [PubMed] [Google Scholar]
- 100.Campbell D.J., Zeitz C.J., Esler M.D., Horowitz J.D. Evidence against a major role for angiotensin converting enzyme-related carboxypeptidase (ACE2) in angiotensin peptide metabolism in the human coronary circulation. J. Hypertens. 2004;22:1971–1976. doi: 10.1097/00004872-200410000-00020. [DOI] [PubMed] [Google Scholar]
- 101.Luque M., Martin P., Martell N., Fernandez C., Bridget Brosnihan K., Ferrario Carlos M. Effects of captopril related to increased levels of prostacyclin and angiotensin-(1-7) in essential hypertension. J. Hypertens. 1996;14 doi: 10.1097/00004872-199606000-00017. No 6. [DOI] [PubMed] [Google Scholar]
- 102.Wysocki J., Lores E., Ye M., Soler M.J., Batlle D. Kidney and lung ACE2 expression after an ACE inhibitor or an Ang II receptor blocker: implications for COVID-19. J. Am. Soc. Nephrol. 2020;31,:1941–1943. doi: 10.1681/ASN.2020050667. ASN.2020050667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.D’Ardes D., Boccatonda A., Rossi I., Guagnano M.T., Santilli F., Cipollone F., Bucci M. COVID-19 and RAS: unravelling an unclear relationship. Int. J. Mol. Sci. 2020;21:3003. doi: 10.3390/ijms21083003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mancia G., Rea F., Ludergnani M., Apolone G., Corrao G. Renin–angiotensin–aldosterone system blockers and the risk of covid-19. N. Engl. J. Med. 2020:1–10. doi: 10.1056/nejmoa2006923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Martinez M.A. Compounds with therapeutic potential against novel respiratory 2019 coronavirus. Antimicrob. Agents Chemother. 2020:1–18. doi: 10.1128/AAC.00399-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Seesuay W., Jittavisutthikul S., Sae-Lim N., Sookrung N., Sakolvaree Y., Chaicumpa W. Human transbodies that interfere with the functions of Ebola virus VP35 protein in genome replication and transcription and innate immune antagonism article, Emerg. Microb. Infect. 2018;7 doi: 10.1038/s41426-018-0031-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. JAMA, J. Am. Med. Assoc. 2020;323 doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- 109.Kadam R.U., Wilson I.A. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc. Natl. Acad. Sci. U. S. A. 2017;114:206–214. doi: 10.1073/pnas.1617020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xia S., Liu Q., Wang Q., Sun Z., Su S., Du L., Ying T., Lu L., Jiang S. Middle East respiratory syndrome coronavirus (MERS-CoV) entry inhibitors targeting spike protein. Virus Res. 2014;194:200–210. doi: 10.1016/j.virusres.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Subissi L., Imbert I., Ferron F., Collet A., Coutard B., Decroly E., Canard B. SARS-CoV ORF1b-encoded nonstructural proteins 12-16: replicative enzymes as antiviral targets. Antivir. Res. 2014;101:122–130. doi: 10.1016/j.antiviral.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stockman L.J., Bellamy R., Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3:1525–1531. doi: 10.1371/journal.pmed.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.T Furuta Y., Komeno T., Nakamuba Polymerase activity (%) 100 μ mol/L favipiravir favipiravir-RMP control. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017;93:449–463. doi: 10.2183/pjab.93.027. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5713175/pdf/pjab-93-449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mulangu S., Dodd L.E., Davey R.T., Mbaya O.T., Proschan M., Mukadi D., Manzo M.L., Nzolo D., Oloma A.T., Ibanda A., Ali R., Coulibaly S., Levine A.C., Grais R., Diaz J., Clifford Lane H., Muyembe-Tamfum J.J., Sivahera B., Camara M., Kojan R., Walker R., Dighero-Kemp B., Cao H., Mukumbayi P., Mbala-Kingebeni P., Ahuka S., Albert S., Bonnett T., Crozier I., Duvenhage M., Proffitt C., Teitelbaum M., Moench T., Aboulhab J., Barrett K., Cahill K., Cone K., Eckes R., Hensley L., Herpin B., Higgs E., Ledgerwood J., Pierson J., Smolskis M., Sow Y., Tierney J., Sivapalasingam S., Holman W., Gettinger N., Vallée D., Nordwall J. A randomized, controlled trial of Ebola virus disease therapeutics. N. Engl. J. Med. 2019;381:2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li L., Li R., Wu Z., Yang X., Zhao M., Liu J., Chen D. Therapeutic strategies for critically ill patients with COVID-19. Ann. Intensive Care. 2020;10 doi: 10.1186/s13613-020-00661-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Barlow A., Landolf K.M., Barlow B., Yeung S.Y.A., Heavner J.J., Claassen C.W., Heavner M.S. Review of emerging pharmacotherapy for the treatment of coronavirus disease 2019. Pharmacotherapy. 2020;40(5):416–437. doi: 10.1002/phar.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wu R., Wang L., Kuo H.-C.D., Shannar A., Peter R., Chou P.J., Li S., Hudlikar R., Liu X., Liu Z., Poiani G.J., Amorosa L., Brunetti L., Kong A.-N. An update on current therapeutic drugs treating COVID-19. Curr. Pharmacol. Reports. 2020:1–15. doi: 10.1007/s40495-020-00216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tu Y.F., Chien C.S., Yarmishyn A.A., Lin Y.Y., Luo Y.H., Lin Y.T., Lai W.Y., Yang D.M., Chou S.J., Yang Y.P., Wang M.L., Chiou S.H. A review of sars-cov-2 and the ongoing clinical trials. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.H. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chu C.M., Cheng V.C.C., Hung I.F.N., Wong M.M.L., Chan K.H., Chan K.S., Kao R.Y.T., Poon L.L.M., Wong C.L.P., Guan Y., Peiris J.S.M., Yuen K.Y. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59:252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Croxtall J.D., Perry C.M. LopinavirRitonavir: a review of its use in the management of HIV-1 infection. Drugs. 2010;70:1885–1915. doi: 10.2165/11204950-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 123.Li S.W., Wang C.Y., Jou Y.J., Huang S.H., Hsiao L.H., Wan L., Lin Y.J., Kung S.H., Lin C.W. SARS coronavirus papain-like protease inhibits the TLR7 signaling pathway through removing Lys63-linked polyubiquitination of TRAF3 and TRAF6. Int. J. Mol. Sci. 2016;17:1–10. doi: 10.3390/ijms17050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yuan L., Chen Z., Song S., Wang S., Tian C., Xing G., Chen X., Xiao Z.X., He F., Zhang L. P53 degradation by a coronavirus papain-like protease suppresses type I interferon signaling. J. Biol. Chem. 2015;290:3172–3182. doi: 10.1074/jbc.M114.619890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen X., Yang X., Zheng Y., Yang Y., Xing Y., Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5:369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78:13600–13612. doi: 10.1128/jvi.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sheahan T.P., Sims A.C., Leist S.R., Schäfer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Babusis D., Clarke M.O., Spahn J.E., Bauer L., Sellers S., Porter D., Feng J.Y., Cihlar T., Jordan R., Denison M.R., Baric R.S. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11 doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., Pohlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/jvi.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rothe Camilla, Schunk Mirjam, Peter Sothmann, Bretzel Gisela, Froeschl Guenter, Wallrauch Claudia, Zimmer Thorbjörn, Thiel Verena, Janke Christian, Guggemos Wolfgang, Michael Seilmaier, Drosten Christian, Vollmar Patrick, Zwirglmaier Katrin, Zange Sabine, Roman Wölfel, Hoelscher Michael. 2020. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany; pp. 2019–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]