Abstract
The birds (class Aves) and bats (order Chiroptera, class Mammalia) are well known natural reservoirs of a diverse range of viruses, including some zoonoses. The only extant volant vertebrates, bats and birds have undergone dramatic adaptive radiations that have allowed them to occupy diverse ecological niches and colonize most of the planet. However, few studies have compared the physiology and ecology of these ecologically, and medically, important taxa. Here, we review convergent traits in the physiology, immunology, flight-related ecology of birds and bats that might enable these taxa to act as viral reservoirs and asymptomatic carriers. Many species of birds and bats are well adapted to urban environments and may host more zoonotic pathogens than species that do not colonize anthropogenic habitats. These convergent traits in birds and bats and their ecological interactions with domestic animals and humans increase the potential risk of viral spillover transmission and facilitate the emergence of novel viruses that most likely sources of zoonoses with the potential to cause global pandemics.
Keywords: Bird, Bat, Flight, Physiology, Immunology, Viral transmission
Graphical abstract
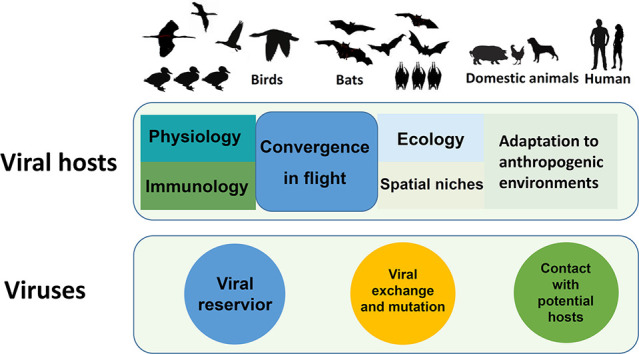
1. Introduction
The coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was characterized as a global pandemic on 11th March 2020 (World Health Organization, 2020). The COVID-19 pandemic has had a dramatic socio-economic impact due to its exceptionally rapid spread and higher number of deaths (Cash and Patel, 2020; Weiss and Murdoch, 2020), particularly among older age groups (Mahase, 2020). Epidemiological research has revealed that free-living bats are likely the native host of the SARS-CoV-2 (Zhou et al., 2020). Other recent viral epidemics are also believed to have originated from either bats or birds (Calisher et al., 2006; Chan et al., 2015; Olival et al., 2017; Nabi et al., 2020). Indeed, bats or birds are thought to host many pathogens (Morse et al., 2012; Chan et al., 2013; Hayman, 2016; Olival et al., 2017; Woolhouse and Brierley, 2018).
The relatively high number of zoonoses carried by birds and mammals is highly correlated with their diversity at the order level (Mollentze and Streicker, 2020). Birds (class Aves) and bats (order Chiroptera, class Mammalia) have both undergone dramatic adaptive radiations; there are over 10,000 bird species (Avibase; https://avibase.bsc-eoc.org/) and 1400 bat species, and both taxa have a global distribution (Wilson and Mittermeier, 2019). The high diversity of birds and bats provides an abundance of potential reservoirs for a diverse range of viruses (Calisher et al., 2006; Hayman, 2016), especially recently emerging, high-profile zoonoses (Olival et al., 2017; Woolhouse and Brierley, 2018).
Why are birds and bats reservoirs of so many zoonotic viruses (Chan et al., 2013; Chen et al., 2014; Hayman, 2016; Olival et al., 2017; Miłek and Blicharz-Domańska, 2018; Venkatesh et al., 2018; Wong et al., 2019)? One explanation lies in their many shared, convergent features, such as small body size, high population densities, close social interaction, spatial mobility, and the ability to colonize anthropogenic environments (Chan et al., 2013; Chan et al., 2015). These features predispose birds and bats to act as viral reservoirs and to transmit viruses to other vertebrates, including humans. However, few studies have compared the physiology and immunology of these ecologically, and medically, important taxa (Caviedes-Vidal et al., 2007; Mollentze and Streicker, 2020; Song et al., 2020). This paper reviews convergent traits in the physiology, immunology and flight-related ecology of birds and bats with the aim of a better understanding of why these species are such important reservoirs of viral zoonoses, and the potential risk of bat and bird viruses infecting humans.
2. Birds and bats as natural reservoirs of viruses
Wild birds are reservoirs of many emerging zoonotic viruses (Reed et al., 2003; Abulreesh et al., 2007). For example, a large variety of influenza A viruses are hosted by wild aquatic birds in the orders of Anseriformes and Charadriiformes (Olsen et al., 2006). Approximately 300 avian species have been confirmed to carry the West Nile virus (CDC, 2019) and also others gamma- and delta-CoVs have been detected in multiple avian orders on all continents (Hughes et al., 2009; Chu et al., 2011; Chamings et al., 2018). The majority of viral infections in birds are either typically of low pathogenicity or asymptomatic (Olsen et al., 2006; Kuiken, 2013; Lycett et al., 2019). However, in recent years, several birds-borne viruses, such as the highly pathogenic avian influenza virus (HPAIV) A (e.g., H5N1 and H7N9) and infectious bronchitis viruses (IBV), have caused major epidemics and mortality among humans or domestic animals (Alexander, 2007; Bui et al., 2016; Wang et al., 2020).
Bats, primates, and rodents not only have the greatest viral richness among mammals but also harbor a higher proportion of zoonotic viruses than other mammalian taxa (Olival et al., 2017; Mollentze and Streicker, 2020). Bats host a greater diversity of viruses than non-flying mammals, including the paramyxoviruses (Drexler et al., 2012), the rhabdoviruses (Rupprecht et al., 2017), the hepaciviruses, the pegiviruses (Quan et al., 2013), and the influenza A viruses (Tong et al., 2013). The updated bat-virus database indicates that 301 bat species host viruses with all known viral genomic structures and replication strategies according to the Baltimore classification system (http://www.mgc.ac.cn/DBatVir/; Chen et al., 2014; Hayman, 2016). Bats are believed to host the ancestors of all major mammalian paramyxoviruses (Drexler et al., 2012; Hayman, 2016); those hosted by non-flying mammals and birds originated from bats (Drexler et al., 2012). In the past two decades, domestic mammals and humans have contracted several viruses from bats including SARS-CoV-2 (Zhou et al., 2020), the virus that is causing the current global pandemic, but also SARS-CoV, the Middle East respiratory syndrome coronavirus (MERS-CoV), the Ebola virus, the Marburg virus, and the rabies virus (Cui et al., 2019; Hayman, 2016).
Compared with mammals, birds had significantly lower viral and zoonosis richness, but the proportion of zoonotic viruses was comparable between classes (Mollentze and Streicker, 2020). Avian CoVs are believed to be the ancestors of gamma- and delta-CoVs, whereas bat CoVs are thought to be the ancestors of the alpha- and beta-CoVs (Woo et al., 2012). Birds (particularly aquatic birds) are natural hosts of the influenza A virus (Olsen et al., 2006; Webster et al., 1992), but bats also host influenza A-like viruses (Tong et al., 2013; Zhu et al., 2013) and the conventional influenza A virus can infect bat cells (Zhou et al., 2014). The fact that they are both capable of flight means that birds and bats have coexisted within a broad range of spatial niches for over 50 Myr (Veselka et al., 2010). It is therefore unclear whether coronaviruses and influenza A virus were transmitted from birds to bats or vice versa (Brunotte et al., 2016). It is worth noting that the human influenza A virus is thought to have come from an avian ancestor, with pigs as an immediate host, approximately 100 years ago (Gammelin et al., 1990; Scholtissek, 1996).
3. Convergences in ecology, physiology and immunology between birds and bats
Because they are both endothermic vertebrates, birds and bats should be subject to similar selective pressures on flight-related morphological and physiological traits (McGuire and Guglielmo, 2009). The convergent traits of miniaturized body size, enhanced metabolic rate and antioxidant capacity, prolonged lifespan, a short but efficient digestive tract, and possessing some specific immunological features relative to non-flying mammals are thought to be the result of functional constraints on evolution imposed by the demands of powered flight (Thomas and Suthers, 1972; Norberg, 1990; Caviedes-Vidal et al., 2007; Costantini, 2008; Munshi-South and Wilkinson, 2010; Song et al., 2020).
Unlike non-flying mammals which tend to increase in size over evolutionary time (Cope's rule; Laurin, 2004), the evolutionary trend in birds and bats has been towards miniaturization; the mass of flying birds ranges from 1.5 g to 15 kg, and for bats from 1.5 g to 1.5 kg (Fig. 1 ; Norberg, 1990). In the volant groups, although the energetic costs (per unit body weight) can vary with the type of flight (e.g., sustained flapping flight, short flight, and gliding flight; Guigueno et al., 2019), in general, the energetic cost (per unit body weight) of flight is approximately comparable in birds and bats (Thomas and Suthers, 1972; Munshi-South and Wilkinson, 2010). Compared with non-flying mammals, both birds and bats have significantly higher metabolic demands for volant flight (Norberg, 1990; Guigueno et al., 2019). The enhanced metabolic demands for powered flight in birds and bats are thought to have driven the evolution of reduced cell and genome sizes (Gregory, 2001, Gregory, 2002; Organ et al., 2007). For example, bats have the smallest genome size (~1.6 to 3.54 Gb) of all mammals, and their DNA loss/gain ratio is ~4.3-fold greater than that of other mammals (Kapusta et al., 2017; Teeling et al., 2018). Similarly, birds have the smallest genome size of all vertebrates (Tiersch and Wachtel, 1991; Gregory et al., 2009). Furthermore, the reduction in DNA in both birds and bats has proceeded mostly through the deletion of large segments (>10 kb) events and gene loss (Zhang et al., 2014; Kapusta et al., 2017). The acquisition of powered flight and the evolution of smaller genome size in birds and bats is believed to have been achieved by streamlining genomic structure and reducing genomic redundancy (Teeling et al., 2018).
Fig. 1.
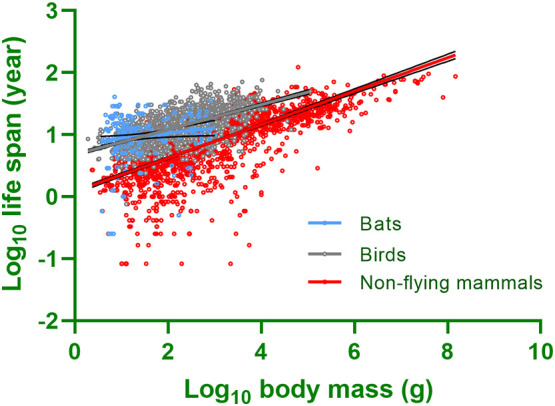
The relationships between body mass and life span in birds (n = 1660), bats (n = 260), and non-flying mammals (n = 2293). Birds (P < 0.001) and bats (P < 0.001) have significantly longer lifespans than non-flying mammals when controlled for body mass in a general linear model. Black lines around the fitted line are the 95% confidence intervals for each taxon. Body mass and lifespan data for each group were taken from Myhrvold et al. (2015).
In endotherms, body size is positively correlated with life span (Speakman, 2005). However, bats and birds have a substantially longer life span than non-flying mammals of similar body size (Fig. 1; Austad and Fischer, 1991; Prinzinger, 1993, Healy et al., 2014). On average, birds live up to four times longer than similar-sized mammals (Lindstedt and Calder, 1981; Holmes and Ottinger, 2003), and bats live 3.5 times longer than similar-sized, non-flying, placental mammals (Austad and Fischer, 1991; Wilkinson and South, 2002). The mechanisms underlying the relatively long life spans of birds and bats are associated with their enhanced capability for preventing oxidative damage to mitochondria and nuclear DNA (Costantini, 2008; Munshi-South and Wilkinson, 2010). Because of their longer life spans, widespread distributions and population connectivity, both bats and birds are exposed to a broader range of environments which may increase the probability of these taxa accumulating zoonotic pathogens over time (Figuerola and Green, 2000; Munshi-South and Wilkinson, 2010; Lucas, 2016).
In both birds and bats, the size and mass of the digestive tract have been minimized to reduce weight during the flight (Caviedes-Vidal et al., 2007). However, both birds and bats have more efficient digestive systems than non-flying mammals (Caviedes-Vidal et al., 2007). There has also been a striking convergence in the gut microbiomes of birds and bats that tends to be independent of diet or phylogeny (Song et al., 2020). Despite their smaller body size, the capacity of bats and most birds to fly allows them to easily escape from unfavorable conditions and predation (Healy et al., 2014).
Given that the immune system is highly conserved in amniotes, the basic structure and function of responses to viruses are broadly similar in mammals and birds (Schat and Kaiser, 2014; Wigley, 2017). However, birds and bats as volant groups, some specific immunological features absent in non-flying mammals have enabled them to coexist with viral pathogens (Table 1 ; Zhang et al., 2013; Schat and Kaiser, 2014) and act as natural reservoirs for emerging viruses (Brook and Dobson, 2015; Chan et al., 2013). Birds lack lymph nodes but have a specific primary lymphoid organ, the bursa of Fabricius. Birds have heterophil in their white blood cells that is the functional equivalent of mammalian neutrophil (Schat and Kaiser, 2014; Wigley, 2017). Compared to mammals, birds have different repertoires of Toll-like receptors (TLRs), inflammatory cytokines and other immune molecules (Kaiser, 2010). Genes related to innate immunity in birds are initiated immediately during antiviral responses. For example, the duck major histocompatibility complex type I (MHC-I) and interferon-induced protein with 5 tricopeptide repeats (IFIT5) are initiated in response to H5N1 virus inoculation (Vanderven et al., 2012). Avian immunoglobulin Y (IgY) is a functional counterpart of mammalian IgG and IgE, providing defense against infections. However, IgY cannot activate the complement system and promote hemagglutination inhibition (Warr et al., 1995; Zhang et al., 2017). These specific features of the avian immunological system enable birds to be tolerant of many viruses (Chan et al., 2013). Such notion has been validated by previous studies, e.g., some species can survive H5N1 virus infections and shed the virus (Sturm-Ramirez et al., 2004; Hulse-Post et al., 2005; Chen et al., 2006) and can be partially immune owing to previous exposures to the virus (Seo et al., 2002).
Table 1.
Comparison of the structure and function of the immunological systems of birds, bats, and non-flying mammals.
| Structure and function | Birds | Non-flying mammals | Bats | References |
|---|---|---|---|---|
| Lymph node | No peripheral or mesenteric lymph nodes, but have the bursa of Fabricius | Peripheral or mesenteric lymph nodes | Peripheral or mesenteric lymph nodes | Schat and Kaiser, 2014; Wigley, 2017 |
| White blood cells | Heterophil, eosinophil, basophil, lymphocyte, and monocyts | Neutrophils, eosinophil, basophil, lymphocyte, and monocyte | Neutrophils, eosinophil, basophil, lymphocyte, and monocyte | Schat and Kaiser, 2014; Wigley, 2017 |
| Immunoglobulins | Three classes: IgY, IgA, IgM | Five classes: IgG and IgE, IgA, IgM, IgD | Five classes: IgG and IgE, IgA, IgM, IgD | Schat and Kaiser, 2014; Wigley, 2017 |
| Innate or adaptive immune response | Early and quick innate antiviral immune response | Delayed innate antiviral immune response. Higher avidity and weaker association with antigens; Quick primary and secondary antibody antibody responses |
Early and quick innate antiviral immune response; Lower avidity and weaker association with antigens; Delayed, or differential, peaks of primary antibody response and a slow secondary antibody response |
Baker et al., 2010, Baker et al., 2013; Kaiser, 2010; Chan et al., 2013; Brook and Dobson, 2015; Pavlovich et al., 2018 |
Among mammalia groups, bats possess very similar virus-sensing pattern recognition receptors (PRRs) and conserved immune systems. However, bats appear to control viral replication by initiating an innate immune response earlier than non-flying mammals (Baker et al., 2013; Brook and Dobson, 2015). There are other critical differences in the adaptive immune response between bats and non-flying mammals. First, bats have a diverse antibody repertoire with relatively lower avidity and a weaker association with antigens (Baker et al., 2010). Second, bats exhibit a delayed, or differential, peak of primary antibody response and a slow secondary antibody response relative to rodents, primates, and ungulates (Baker et al., 2013; Chan et al., 2013; Pavlovich et al., 2018). In bat genomes, genes in the type I interferon family, the MHC-I, and natural killer-cell receptors, are known to be highly expanded (Zhang et al., 2013). It has recently been suggested that the low expression of C-1-tetrahydrofolate synthase in the cells and tissues of bats compared to humans is due to antiviral replication (Anderson et al., 2020). Notably, several genes, such as c-REL (a vital gene for maintaining lymphoid cell function) and the ataxia-telangiectasia mutated gene (ATM) in the DNA damage checkpoint-DNA repair pathway, were positively selected in bat ancestors (Zhang et al., 2013). Compared with non-flying mammals, these special immunological features allow bats to mount efficient immune responses against a diverse range of viruses (Banerjee et al., 2020).
4. Convergences in flight-related ecology between birds and bats
The ability to fly not only significantly reduces the risk of predation but also significantly increases the ability to colonize new niches and habitats (Norberg, 1990). This enhanced mobility also means that birds and bats transport viruses over hundreds, even thousands of kilometers during migration (Hill et al., 2012; Prosser et al., 2013). Approximately 20% of birds are migratory (Kirby et al., 2008; Newton, 2008). The migrations of billions of birds worldwide also transport viruses to stopover sites, overwintering and breeding habitats (Hill et al., 2012), although migrations can also lower infection risks by escaping from habitats where pathogen stages have accumulated and eliminating infected individuals during strenuous journey (Satterfield et al., 2018). Wild birds are associated with the dispersal of CoVs (Georgopoulou and Tsiouris, 2008; Chamings et al., 2018), the Influenza A virus (Hill et al., 2012), the Arboviruses (West Nile virus), the Usutu virus, the Newcastle disease virus, the avian pox virus, and the duck plague virus (Georgopoulou and Tsiouris, 2008; Verhagen et al., 2015; Satu et al., 2017). Fewer bats migrate (Krauel and McCracken, 2013) but some undertake migrations of over 1000 km (Plowright et al., 2015; Allocati et al., 2016). Such migrations allow bat viruses, such as CoVs, the rabies virus, the Hendra, and Nipah viruses, to spread over long distances (Calisher et al., 2006). Although migratory bird and bat species have different migration patterns, their movements, particularly during intercontinental migration, contribute to viral dissemination and also facilitate viral recombination, mutation and evolution (Bahl et al., 2013; Hill et al., 2016; Plowright et al., 2017; Lycett et al., 2019).
Given that bats and birds have evolved to compete for spatial niches such as roosting sites they may interact with each other, either directly or indirectly. The bird-bat interactions include co-occurrence in the same nest (e.g., between starling Sturnus vulgaris and noctule bats Nyctalus noctule) (Myczko et al., 2016), the predation of eggs, nestlings, or adult birds by bats (Medellín, 1988; Ibáñez et al., 2001; Perrella et al., 2020) and the predation of bats by birds (Fenton and Fleming, 1976; Camargo and Laps, 2016; Mikula et al., 2016). Perrella et al. (2020) found about 8% of bird nests were preyed by bats and 2% by reptiles. Both bats and birds species richness increases in proximity to the equator due to higher ecological productivity (Brown, 2014; De Oliveira et al., 2018). Similarly, pathogen diversity is also greater in tropical areas compared to temperate regions, and therefore, pathogen richness in birds and bats could be higher near the equator compared to temperate regions (Guernier et al., 2004). This coexistence could allow the mixing of bats and birds viruses for the generation of recombinant, novel mutant, or reassortment of RNA viruses (Chan et al., 2013; Perrella et al., 2020). Furthermore, a large number of birds and bats are gregarious with high population densities. For example, colonies of the Mexican free-tailed bat (Tadarida brasiliensis mexicana) can contain up to one million individuals per roost at an average density of about 4000 bats/m2 (McCracken and Gustin, 1991). Indirect bird-bat interactions include competition for food, and for temporal, and spatial, niches (Fenton and Fleming, 1976; Goldingay, 2009). Therefore, these convergent features of small body size, high population densities, and spatial mobility, and the bird-bat social interactions provide the opportunity for exchanging viruses, thereby facilitating the emergence of highly pathogenic, new viruses.
5. Adaptation of birds and bats to anthropogenic environments
Human activity, such as agriculture and urban development, is causing significant degradation, loss and fragmentation of bird and bat habitat (Voigt and Kingston, 2016; Walsh et al., 2017). Although the majority of the birds and bats are susceptible to anthropogenic change, some flourish in anthropogenic environments, including cities (Duchamp and Swihart, 2008; Johnson and Munshi-South, 2017) and are well adapted to urban environments by exhibiting a suite of phenotypic traits in morphology, physiology, and behavior (MacGregor-Fors et al., 2012; Magle et al., 2012; Jung and Threlfall, 2016; Afelt et al., 2018; Isaksson, 2018). Anthropogenic environments provide some human commensal species with an abundance of food and reduced numbers of parasites and predators, thereby increasing reproductive output and winter survival (Minias, 2016). The often high densities of birds and bats in anthropogenic environments facilitate viral transfer to humans (Plowright et al., 2015; Afelt et al., 2018). Domestic fowl and livestock are often also at high density in anthropogenic environments, in some countries close to captive wild game. This juxtaposition of domestic and wild animals provides an abundance of immediate hosts for bird and bat-derived viruses, many of which can become pathogenic once transmitted to humans (Chan et al., 2015). Furthermore, many birds and bats are caught and transported to markets to be sold for food, traditional medicine, ornamentation, as pets or for sport hunting (Mildenstein et al., 2016). For example, at least 167 species of bats (92 species of large-bodied fruit bats and 75 insectivorous species) are reported to be hunted in Africa, Asia, Central and South America, and across the islands of Oceania (Mildenstein et al., 2016). Similarly, 4561 bird species (45.7% of all species) are caught by humans for different purposes (Butchart, 2008). Generally, these captured bats and birds are sold and kept in close contact with humans and other taxa in overcrowded and unhygienic wet markets that have become epicenters for the mixing and transmission of viral pathogens (Chan et al., 2013; Aguirre et al., 2020). The high human population density in cities increases the risk of humans becoming infected with recombinant viruses from intermediate hosts, some of having already caused global pandemics (Sehgal, 2010). The risk of cross-species transmission depends on the spatio-temporal network connecting viral reservoirs to intermediate, and final, hosts (Hassell et al., 2017; Plowright et al., 2017). Anthropogenic environments can, therefore, be regarded as a viral nexus where bird and bat-derived viruses, a diverse range of potential intermediate hosts, and humans, all exist in close proximity. Because their primary and secondary hosts are either mildly symptomatic or asymptomatic, many bird and bat-derived viruses may spread and diversify unnoticed in anthropogenic environments (Afelt et al., 2018).
6. Conclusions
The occasional transmission of viral pathogens from asymptomatic host species to new hosts can lead to either asymptomatic infection, severe disease, or death. Birds and bats share a variety of flight-related physiological and ecological traits that predispose them to harbor, disperse, and transmit viruses. Special features of their immune systems enable them to function as asymptomatic carriers of a diverse range of viruses. Close interactions between birds and bats in the course of competition for spatial niches, further increases the probability of viral transmission, recombination, and mutation, while the migrations undertaken by many birds and bats disperse viruses over long distances. The ability of some birds and bats to flourish in anthropogenic environments increases the probability of the viral transmission to domestic animals or captive wild game, which facilitates the emergence of novel viruses pathogenic to humans. By bringing together birds, bats, domestic animals, wild game and humans, urban environments provide the ideal conditions for acquiring new viral genes, and harboring high viral burden with strains of higher transmission efficiency, thus facilitating transmission of the viruses to humans. Considering that many ongoing bird and bat-derived zoonotic viruses are probably circulating, diversifying, and spreading unnoticed, the risk of such pandemics is ongoing. More transdisciplinary and interdisciplinary investigations are warranted to unravel the complex interactions connecting bat and bird-derived viruses to immediate hosts and to humans and shed light on the origin of the current COVID-19 pandemic and reduce the risk of future pandemics.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We appreciate the valuable comments from three anonymous reviewers for the improvement of our original manuscript. This study was supported by the National Natural Science Foundation of China (NSFC, 31672292, 31971413) to D.M.L., NSFC (31770445) to Y.F.W., the biodiversity investigation, observation, and assessment program (2019-2023) of Ministry of Ecology and Environment of China to D.M.L.
CRediT authorship contribution statement
Nabi G. and Wang Y., literature search and writing the original draft; Lv L., Jiang C., Ahmad S., and Wu Y., literature search and discussion; Li D., study design and writing the final manuscript.
Editor: Daniel Wunderlin
References
- Abulreesh H.H., Goulder R., Scott G.W. Wild birds and human pathogens in the context of ringing and migration. Rining. Migr. 2007;23:193–200. [Google Scholar]
- Afelt A., Frutos R., Devaux C. Bats, coronaviruses, and deforestation: toward the emergence of novel infectious diseases? Front. Microbiol. 2018;9:702. doi: 10.3389/fmicb.2018.00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre A.A., Catherina R., Frye H., Shelley L. Illicit wildlife trade, wet markets, and COVID-19: preventing future pandemics. World Med. Health Policy. 2020 doi: 10.1002/wmh3.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander D.J. An overview of the epidemiology of avian influenza. Vaccine. 2007;25:5637–5644. doi: 10.1016/j.vaccine.2006.10.051. [DOI] [PubMed] [Google Scholar]
- Allocati N., Petrucci A., Di Giovanni P., Masulli M., Llio C.D., Laurenzi V.D. Bat–man disease transmission: zoonotic pathogens from wildlife reservoirs to human populations. Cell. Death. Discov. 2016;2:16048. doi: 10.1038/cddiscovery.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, D.E., Cui, J., Ye, Q., Huang, B., Zu, W., Gong, J., et al. (2020). Orthogonal genome-wide screenings in bat cells identify MTHFD1 as a target of broad antiviral therapy. bioRxiv. 2020.2003.2029.014209. [DOI] [PMC free article] [PubMed]
- Austad S.N., Fischer K.E. Mammalian aging, metabolism, and ecology: evidence from the bats and marsupials. J. Gerontol. 1991;46:47–53. doi: 10.1093/geronj/46.2.b47. [DOI] [PubMed] [Google Scholar]
- Bahl J., Krauss S., Kühnert D., Fourment M., Raven G., Pryor S.P., et al. Influenza A virus migration and persistence in North American wild birds. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M.L., Tachedjian M., Wang L.F. Immunoglobulin heavy chain diversity in Pteropid bats: evidence for a diverse and highly specific antigen binding repertoire. Immunogenetics. 2010;62:173–184. doi: 10.1007/s00251-010-0425-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M.L., Schountz T., Wang L.F. Antiviral immune responses of bats: a review. Zoonoses. Public. Hlth. 2013;60:104–116. doi: 10.1111/j.1863-2378.2012.01528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Baker M.L., Kulcsar K., Misra V., Plowright R., Mossman K. Novel insights into immune systems of bats. Front. Immunol. 2020;11:26. doi: 10.3389/fimmu.2020.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook C.E., Dobson A.P. Bats as ‘special’ reservoirs for emerging zoonotic pathogens. Trends Microbiol. 2015;23:172–180. doi: 10.1016/j.tim.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.H. Why are there so many species in the tropics? J. Biogeogr. 2014;41:8–22. doi: 10.1111/jbi.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunotte L., Beer M., Horie M., Schwemmle M. Chiropteran influenza viruses: flu from bats or a relic from the past? Curr. Opin. Virol. 2016;16:114–119. doi: 10.1016/j.coviro.2016.02.003. [DOI] [PubMed] [Google Scholar]
- Bui C., Bethmont A., Chughtai A.A., Gardner L., Sarkar S., Hassan S., et al. A systematic review of the comparative epidemiology of avian and human influenza a H5N1 and H7N9 – lessons and unanswered questions. Transbound. Emerg. Dis. 2016;63:602–620. doi: 10.1111/tbed.12327. [DOI] [PubMed] [Google Scholar]
- Butchart S.H.M. Red List Indices to measure the sustainability of species use and impacts of invasive alien species. Bird. Conserv. Int. 2008;18:245–262. [Google Scholar]
- Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo P.H.S.A., Laps R.R. Predation on lesser bulldog bat (Noctilio albiventris noctilionidae) by great rufous woodcreeper (Xiphocolaptes major dendrocolaptidae) J. Ornithol. 2016;128:903–912. [Google Scholar]
- Cash R., Patel V. Has COVID-19 subverted global health? Lancet. 2020;5:2020. doi: 10.1016/S0140-6736(20)31089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviedes-Vidal E., McWhorter T.J., Lavin S.R., Chediack J.G., Tracy C.R., Karasov W.H. The digestive adaptation of flying vertebrates: high intestinal paracellular absorption compensates for smaller guts. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19132–19137. doi: 10.1073/pnas.0703159104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . CDC; 2019. Species of Dead Birds in Which West Nile Virus Has Been Detected, United States, 1999–2016. (Retrieved 28 March 2019) [Google Scholar]
- Chamings A., Nelson T.M., Vibin J., Wille M., Klaassen M., Alexandersen S. Detection and characterization of coronaviruses in migratory and non-migratory Australian wild birds. Sci. Rep. 2018;8:5980. doi: 10.1038/s41598-018-24407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., To K.K.W., Tse H., Jin D.Y., Yuen K.Y. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21:544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., To K.K.W., Chen H., Yuen K.Y. Cross-species transmission and emergence of novel viruses from birds. Curr. Opin. Virol. 2015;10:63–69. doi: 10.1016/j.coviro.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Smith G.J., Li K.S., Wang J., Fan X.H., Rayner J.M., et al. Establishment of multiple sublineages of H5N1 influenza virus in Asia: implications for pandemic control. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2845–2850. doi: 10.1073/pnas.0511120103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., Liu, B., Yang, J. and Jin, Q. (2014). DBatVir: the database of bat-associated viruses. Database. 2014:bau021. [DOI] [PMC free article] [PubMed]
- Chu D.K.W., Leung C.Y.H., Gilbert M., Joyner P.H., Ng E.M., Tse T.M., et al. Avian coronavirus in wild aquatic birds. J. Virol. 2011;85:12815–12820. doi: 10.1128/JVI.05838-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini D. Oxidative stress in ecology and evolution: lessons from avian studies. Ecol. Lett. 2008;11:1238–1251. doi: 10.1111/j.1461-0248.2008.01246.x. [DOI] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Oliveira H., Oprea M., Dias R. Distributional patterns and ecological determinants of bat occurrence inside caves: a broad scale meta-analysis. Diversity. 2018;10:49. [Google Scholar]
- Drexler J.F., Corman V.M., Müller M.A., Maganga G.D., Vallo P., Binger T., et al. Bats host major mammalian paramyxoviruses. Nat. Commun. 2012;3:1–13. doi: 10.1038/ncomms1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchamp J.E., Swihart R.K. Shifts in bat community structure related to evolved traits and features of human-altered landscapes. Landsc. Ecol. 2008;23:849–860. [Google Scholar]
- Fenton M., Fleming T. Ecological interactions between bats and nocturnal birds. Biotropica. 1976;8:104–110. [Google Scholar]
- Figuerola J., Green A. Haematozoan parasites and migratory behaviour in waterfowl. Evol. Ecol. 2000;14:143–153. [Google Scholar]
- Gammelin M., Altmüller A., Reinhardt U., Mandler J., Harley V.R., Hudson P.J., et al. Phylogenetic analysis of nucleoproteins suggests that human influenza A viruses emerged from a 19th-century avian ancestor. Mol. Biol. Evol. 1990;7:194–200. doi: 10.1093/oxfordjournals.molbev.a040594. [DOI] [PubMed] [Google Scholar]
- Georgopoulou I., Tsiouris V. The potential role of migratory birds in the transmission of zoonoses. Vet. Ital. 2008;44:671–677. [PubMed] [Google Scholar]
- Goldingay R.L. Characteristics of tree hollows used by Australian birds and bats. Wildl. Res. 2009;36:394–409. [Google Scholar]
- Gregory T.R. The bigger the C-value, the larger the cell: genome size and red blood cell size in vertebrates. Blood Cells Mol. Dis. 2001;27:830–843. doi: 10.1006/bcmd.2001.0457. [DOI] [PubMed] [Google Scholar]
- Gregory T.R. A bird's-eye view of the C-value enigma: genome size, cell size, and metabolic rate in the class aves. Evolution. 2002;56:121–130. doi: 10.1111/j.0014-3820.2002.tb00854.x. [DOI] [PubMed] [Google Scholar]
- Gregory T.R., Andrews C.B., McGuire J.A., Witt C.C. The smallest avian genomes are found in hummingbirds. Proc. Biol. Sci. 2009;276:3753–3757. doi: 10.1098/rspb.2009.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guernier V., Hochberg M.E., Guégan J.F. Ecology drives the worldwide distribution of human diseases. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guigueno M.F., Shoji A., Elliott K.H., Aris-Brosou S. Flight costs in volant vertebrates: a phylogenetically-controlled meta-analysis of birds and bats. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2019;235:193–201. doi: 10.1016/j.cbpa.2019.06.003. [DOI] [PubMed] [Google Scholar]
- Hassell J.M., Begon M., Ward M.J., Fèvre E.M. Urbanization and disease emergence: dynamics at the wildlife–livestock–human interface. Trends Ecol. Evol. 2017;32:55–67. doi: 10.1016/j.tree.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman D.T.S. Bats as viral reservoirs. Annu. Rev. Virol. 2016;3:77–99. doi: 10.1146/annurev-virology-110615-042203. [DOI] [PubMed] [Google Scholar]
- Healy K., Guillerme T., Finlay S., Kane A., Kelly S.B., McClean D., et al. Ecology and mode-of-life explain lifespan variation in birds and mammals. Proc. Biol. Sci. 2014;281:20140298. doi: 10.1098/rspb.2014.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill N.J., Takekawa J.Y., Cardona C.J., Meixell B.W., Ackerman J.T., Runstadler J.A., et al. Cross-seasonal patterns of avian influenza virus in breeding and wintering migratory birds: a flyway perspective. Vector. Borne. Zoonotic. Dis. 2012;12:243–253. doi: 10.1089/vbz.2010.0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill N.J., Ma E.J., Meixell B.W., Lindberg M.S., Boyce W.M., Runstadler J.A. Transmission of influenza reflects seasonality of wild birds across the annual cycle. Ecol. Lett. 2016;19:915–925. doi: 10.1111/ele.12629. [DOI] [PubMed] [Google Scholar]
- Holmes D., Ottinger M. Birds as long-lived animal models for the study of aging. Exp. Gerontol. 2003;38:1365–1375. doi: 10.1016/j.exger.2003.10.018. [DOI] [PubMed] [Google Scholar]
- Hughes L.A., Savage C., Naylor C., Bennett M., Chantrey J., Jones R. Genetically diverse coronaviruses in wild bird populations of northern England. Emerg. Infect. Dis. 2009;15:1091–1094. doi: 10.3201/eid1507.090067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulse-Post D.J., Sturm-Ramirez K.M., Humberd J., Seiler P., Govorkova E.A., Krauss S., et al. Role of domestic ducks in the propagation and biological evolution of highly pathogenic H5N1 influenza viruses in Asia. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10682–10687. doi: 10.1073/pnas.0504662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibáñez C., Juste J., García-Mudarra J.L., Agirre-Mendi P.T. Bat predation on nocturnally migrating birds. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9700. doi: 10.1073/pnas.171140598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaksson C. In: Bird Species. Fascinating Life Sciences. Tietze D., editor. Springer; Cham: 2018. Impact of urbanization on birds. [Google Scholar]
- Johnson, M.T.J. and Munshi-South, J. (2017). Evolution of life in urban environments. Science. 358:eaam8327. [DOI] [PubMed]
- Jung K., Threlfall C.G. In: Bats in the Anthropocene: Conservation of Bats in a Changing World. Voigt C., Kingston T., editors. Springer; Cham: 2016. Urbanisation and its effects on bats-a global meta-analysis. [Google Scholar]
- Kaiser P. Advances in avian immunology-prospects for disease control: a review. Avian Pathol. 2010;39:309–324. doi: 10.1080/03079457.2010.508777. [DOI] [PubMed] [Google Scholar]
- Kapusta A., Suh A., Feschotte C. Dynamics of genome size evolution in birds and mammals. Proc. Natl. Acad. Sci. U. S. A. 2017;114:1460–1469. doi: 10.1073/pnas.1616702114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby J.S., Stattersfield A.J., Butchart S.H.M., Evans M.I., Grimmett R.F.A., Jones V.R., et al. Key conservation issues for migratory land- and waterbird species on the world's major flyways. Bird Conserv. Int. 2008;18:S49–S73. [Google Scholar]
- Krauel J.J., McCracken G.F. In: Bat Evolution, Ecology, and Conservation. Adams R., Pedersen S., editors. Springer; New York, NY: 2013. Recent advances in bat migration research. [Google Scholar]
- Kuiken T. Is low pathogenic avian influenza virus virulent for wild waterbirds? Proc. Biol. Sci. 2013;280:20130990. doi: 10.1098/rspb.2013.0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurin M. The evolution of body size, Cope's rule and the origin of amniotes. Syst. Biol. 2004;53:594–622. doi: 10.1080/10635150490445706. [DOI] [PubMed] [Google Scholar]
- Lindstedt S., Calder W. Body size, physiological time, and longevity of homeothermic animals. Q. Rev. Biol. 1981;56:1–16. [Google Scholar]
- Lucas, T.C. (2016). The Role of Population Structure and Size in Determining Bat Pathogen Richness. PhD Thesis. University College London.
- Lycett S.J., Duchatel F., Digard P. A brief history of bird flu. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2019;374:20180257. doi: 10.1098/rstb.2018.0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor-Fors I., Morales-Pérez L., Schondube J.E. In: Urban Bird Ecology and Conservation, Studies in Avian Biology (no 45) Lepczyk C.A., Warren P.S., editors. University of California Press; Berkeley: 2012. From forest to cities: effects of urbanization on tropical birds; pp. 33–38. [Google Scholar]
- Magle S.B., Hunt V.M., Vernon M., Crooks K.R. Urban wildlife research: past, present, and future. Biol. Conserv. 2012;155:23–32. [Google Scholar]
- Mahase E. Covid-19: death rate is 0.66% and increases with age, study estimates. Bmj-Brit. Med. J. 2020;369 doi: 10.1136/bmj.m1327. [DOI] [PubMed] [Google Scholar]
- McCracken G.F., Gustin M.K. Nursing behavior in Mexican free-tailed bat maternity colonies. Ethology. 1991;89:305–321. [Google Scholar]
- McGuire L.P., Guglielmo C.G. What can birds tell us about the migration physiology of bats? J. Mammal. 2009;90:1290–1297. [Google Scholar]
- Medellín R.A. Prey of Chrotopterus auritus, with notes on feeding behavior. J. Mammal. 1988;69:841–844. [Google Scholar]
- Mikula P., Morelli F., Lučan R.K., Jones D.N., Tryjanowski P. Bats as prey of diurnal birds: a global perspective. Mammal Rev. 2016;46:160–174. [Google Scholar]
- Mildenstein T., Tanshi I., Racey P.A. In: Bats in the Anthropocene: Conservation of Bats in a Changing World. Voigt C., Kingston T., editors. Springer; Cham: 2016. Exploitation of bats for bushmeat and medicine. [Google Scholar]
- Miłek J., Blicharz-Domańska K. Coronaviruses in avian species - review with focus on epidemiology and diagnosis in wild birds. J. Vet. Res. 2018;62:249–255. doi: 10.2478/jvetres-2018-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minias P. Reproduction and survival in the city: which fitness components drive urban colonization in a reed-nesting waterbird? Curr. Zool. 2016;62:79–87. doi: 10.1093/cz/zow034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollentze N., Streicker D.G. Viral zoonotic risk is homogenous among taxonomic orders of mammalian and avian reservoir hosts. Proc. Natl. Acad. Sci. U. S. A. 2020;117:9423–9430. doi: 10.1073/pnas.1919176117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse S.S., Mazet J.A.K., Woolhouse M., Parrish C.R., Carroll D., Karesh W.B., et al. Prediction and prevention of the next pandemic zoonosis. Lancet. 2012;380:1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi-South J., Wilkinson G.S. Bats and birds: exceptional longevity despite high metabolic rates. Ageing Res. Rev. 2010;9:12–19. doi: 10.1016/j.arr.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Myczko Ł., Dylewski Ł., Sparks T.H., Łochyński M., Tryjanowski P. Co-occurrence of birds and bats in natural nest-holes. Ibis. 2016;159:235–237. doi: 10.1111/ibi.12434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myhrvold N.P., Baldridge E., Chan B., Sivam D., Freeman D.L., Ernest S.K.M. An amniote life-history database to perform comparative analyses with birds, mammals, and reptiles. Ecology. 2015;96 (3109-3000) [Google Scholar]
- Nabi G., Siddique R., Ali A., Khan S. Preventing bat-born viral outbreaks in future using ecological interventions. Environ. Res. 2020;185:109460. doi: 10.1016/j.envres.2020.109460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton I. Academic Press; London and New York: 2008. The Migration Ecology of Birds. [Google Scholar]
- Norberg U.M. Springer-Verlag; Berlin Heidelberg: 1990. Vertebrate Flight: Mechanics, Physiology, Morphology, Ecology and Evolution. [Google Scholar]
- Olival K.J., Hosseini P.R., Zambrana-Torrelio C., Ross N., Bogich T.L., Daszak P. Host and viral traits predict zoonotic spillover from mammals. Nature. 2017;546:646–650. doi: 10.1038/nature22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen B., Munster V.J., Wallensten A., Waldenstrom J., Osterhaus A.D., Fouchier R.A. Global patterns of influenza a virus in wild birds. Science. 2006;312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- Organ C.L., Shedlock A.M., Meade A., Pagel M., Edwards S.V. Origin of avian genome size and structure in non-avian dinosaurs. Nature. 2007;446:180–184. doi: 10.1038/nature05621. [DOI] [PubMed] [Google Scholar]
- Pavlovich S.S., Lovett S.P., Koroleva G., Guito J.C., Arnold C.E., Nagle E.R., et al. The Egyptian rousette genome reveals unexpected features of bat antiviral immunity. Cell. 2018;173:1098–1110. doi: 10.1016/j.cell.2018.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrella D.F., Zima P.V.Q., Ribeiro-Silva L., Biagolini C.H., Jr., Carmignotto A.P., Galetti P.M., Jr., et al. Bats as predators at the nests of tropical forest birds. J. Avian Biol. 2020;51 [Google Scholar]
- Plowright R.K., Eby P., Hudson P.J., Smith I.L., Westcott D., Bryden W.L., et al. Ecological dynamics of emerging bat virus spillover. Proc. Biol. Sci. 2015;282:20142124. doi: 10.1098/rspb.2014.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright R.K., Parrish C.R., McCallum H., Hudson P.J., Ko A.I., Graham A.L., et al. Pathways to zoonotic spillover. Nat. Rev. Microbiol. 2017;15:502–510. doi: 10.1038/nrmicro.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinzinger R. Life-span in birds and the aging theory of absolute metabolic scope. Comp. Biochem. Phys. A. 1993;105:609–615. [Google Scholar]
- Prosser D.J., Nagel J., Takekawa J.Y. In: Viral Infections and Global Change. Singh S.K., editor. 2013. Animal migration and risk of spread of viral infections; pp. 151–178. [Google Scholar]
- Quan P.L., Firth C., Conte J.M., Williams S.H., Zambrana-Torrelio C.M., Anthony S.J., et al. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8194–8199. doi: 10.1073/pnas.1303037110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K.D., Meece J.K., Henkel J.S., Shukla S.K. Birds, migration and emerging zoonoses: west nile virus, lyme disease, influenza A and enteropathogens. Clin. Med. Res. 2003;1:5–12. doi: 10.3121/cmr.1.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupprecht, C., Kuzmin, I. and Meslin, F. (2017). Lyssaviruses and rabies: current conundrums, concerns, contradictions and controversies. F1000Res. 6:184-184. [DOI] [PMC free article] [PubMed]
- Satterfield D.A., Marra P.P., Sillett T.S., Altizer S. Responses of migratory species and their pathogens to supplemental feeding. Phil. Trans. R. Soc. B. 2018;373:20170094. doi: 10.1098/rstb.2017.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satu H., Lindh E., Vapalahti O., Huovilainen A. Prevalence and genetic diversity of coronaviruses in wild birds, Finland. Infect. Ecol. Epidemiol. 2017;7:1408360. doi: 10.1080/20008686.2017.1408360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schat K.A., Kaiser P. 2nd edition. Academic Press; London and New York: 2014. Avian Immunology. [Google Scholar]
- Scholtissek C. Molecular evolution of influenza viruses. Virus Genes. 1996;11:209–215. doi: 10.1007/BF01728660. [DOI] [PubMed] [Google Scholar]
- Sehgal R.N. Deforestation and avian infectious diseases. J. Exp. Biol. 2010;213:955–960. doi: 10.1242/jeb.037663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S.H., Peiris M., Webster R.G. Protective cross-reactive cellular immunity to lethal A/Goose/Guangdong/1/96-like H5N1 influenza virus is correlated with the proportion of pulmonary CD8+ T cells expressing gamma interferon. J. Virol. 2002;76:4886–4890. doi: 10.1128/JVI.76.10.4886-4890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, S.J., Sanders, J.G., Delsuc, F., Metcalf, J., Amato, K., Taylor, M.W., et al. (2020). Comparative analyses of vertebrate gut microbiomes reveal convergence between birds and bats. mBio. 11:e02901-02919. [DOI] [PMC free article] [PubMed]
- Speakman J.R. Body size, energy metabolism and lifespan. J. Exp. Biol. 2005;208:1717–1730. doi: 10.1242/jeb.01556. [DOI] [PubMed] [Google Scholar]
- Sturm-Ramirez K.M., Ellis T., Bousfield B., Bissett L., Dyrting K., Rehg J.E., et al. Reemerging H5N1 influenza viruses in Hong Kong in 2002 are highly pathogenic to ducks. J. Virol. 2004;78:4892–4901. doi: 10.1128/JVI.78.9.4892-4901.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling E.C., Vernes S.C., Dávalos L.M., Ray D.A., Gilbert M., Myers E., et al. Bat biology, genomes, and the Bat1K Project: to generate chromosome-level genomes for all living bat species. Annu. Rev. Anim. Biosci. 2018;6:23–46. doi: 10.1146/annurev-animal-022516-022811. [DOI] [PubMed] [Google Scholar]
- Thomas S.P., Suthers R.A. The physiology and energetics of bat flight. J. Exp. Biol. 1972;57:317–335. [Google Scholar]
- Tiersch T.R., Wachtel S.S. On the evolution of genome size of birds. J. Hered. 1991;82:363–368. doi: 10.1093/oxfordjournals.jhered.a111105. [DOI] [PubMed] [Google Scholar]
- Tong S., Zhu X., Li Y., Shi M., Zhang J., Bourgeois M., et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderven H.A., Petkau K., Ryan-Jean K.E.E., Aldridge J.R., Webster R.G., Magor K.E. Avian influenza rapidly induces antiviral genes in duck lung and intestine. Mol. Immunol. 2012;51:316–324. doi: 10.1016/j.molimm.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh D., Poen M.J., Bestebroer T.M., Scheuer R.D., Vuong O., Chkhaidze M., et al. Avian influenza viruses in wild birds: virus evolution in a multihost ecosystem. J. Virol. 2018;92 doi: 10.1128/JVI.00433-18. (e00433-00418) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen J.H., Herfst S., Fouchier R.A. Infectious disease. How a virus travels the world. Science. 2015;347:616–617. doi: 10.1126/science.aaa6724. [DOI] [PubMed] [Google Scholar]
- Veselka N., McErlain D.D., Holdsworth D.W., Eger J.L., Chhem R.K., Mason M.J., et al. A bony connection signals laryngeal echolocation in bats. Nature. 2010;463:939–942. doi: 10.1038/nature08737. [DOI] [PubMed] [Google Scholar]
- Voigt C.C., Kingston T. Springer; Cham: 2016. Bats in the Anthropocene: Conservation of Bats in a Changing World. [Google Scholar]
- Walsh M.G., Wiethoelter A., Haseeb M.A. The impact of human population pressure on flying fox niches and the potential consequences for Hendra virus spillover. Sci. Rep. 2017;7:8226. doi: 10.1038/s41598-017-08065-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., Zhu, W., Yang, L. and Shu, Y. (2020). The epidemiology, virology, and pathogenicity of human infections with avian influenza viruses. Cold. Spring. Harb. Perspect. Med. a038620. [DOI] [PMC free article] [PubMed]
- Warr G.W., Magor K.E., Higgins D.A. IgY: clues to the origins of modern antibodies. Immunol. Today. 1995;16:392–398. doi: 10.1016/0167-5699(95)80008-5. [DOI] [PubMed] [Google Scholar]
- Webster R.G., Bean W.J., Gorman O.T., Chambers T.M., Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss P., Murdoch D.R. Clinical course and mortality risk of severe COVID-19. Lancet. 2020;395:1014–1015. doi: 10.1016/S0140-6736(20)30633-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigley, P. (2017). Immunology of Birds. In: eLS. John Wiley and Sons, Ltd (Ed.). pp. 1-8.
- Wilkinson G.S., South J.M. Life history, ecology and longevity in bats. Aging Cell. 2002;1:124–131. doi: 10.1046/j.1474-9728.2002.00020.x. [DOI] [PubMed] [Google Scholar]
- Wilson D.E., Mittermeier R.A. Lynx Edicions; Barcelona: 2019. Handbook of the Mammals of the World. [Google Scholar]
- Wong A.C.P., Li X., Lau S.K.P., Woo P.C.Y. Global epidemiology of bat coronaviruses. Viruses. 2019;11:174. doi: 10.3390/v11020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Lam C.S.F., Lau C.C.Y., Tsang A.K.L., Lau J.H.N., et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M.E.J., Brierley L. Epidemiological characteristics of human-infective RNA viruses. Sci. Data. 2018;5:180017. doi: 10.1038/sdata.2018.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. (2020). WHO characterizes COVID-19 as a pandemic. Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen.
- Zhang G., Cowled C., Shi Z., Huang Z., Bishop-Lilly K.A., Fang X., et al. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339:456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Li C., Li Q., Li B., Larkin D.M., Lee C., et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 2014;346:1311–1320. doi: 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Calvert R.A., Sutton B.J., Doré K.A. IgY: a key isotype in antibody evolution. Biol. Rev. 2017;92:2144–2156. doi: 10.1111/brv.12325. [DOI] [PubMed] [Google Scholar]
- Zhou B., Ma J.J., Liu Q.F., Bawa B., Wang W., Shabman R.S., et al. Characterization of uncultivable bat influenza virus using a replicative synthetic virus. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Yu W., McBride R., Li Y., Chen L.M., Donis R.O., et al. Hemagglutinin homologue from H17N10 bat influenza virus exhibits divergent receptor-binding and pH-dependent fusion activities. Proc. Natl. Acad. Sci. U. S. A. 2013;110:1458–1463. doi: 10.1073/pnas.1218509110. [DOI] [PMC free article] [PubMed] [Google Scholar]


