Abstract
The ability to express genes ectopically in bacteria is essential for diverse academic and industrial applications. Two major considerations when utilizing regulated promoter systems for ectopic gene expression are (1) the ability to titrate gene expression by addition of an exogenous inducer and (2) the leakiness of the promoter element in the absence of the inducer. Here, we describe a modular chromosomally integrated platform for ectopic gene expression in Vibrio cholerae. We compare the broadly used promoter elements Ptac and PBAD to versions that have an additional theophylline-responsive riboswitch (Ptac-riboswitch and PBAD-riboswitch). These constructs all exhibited unimodal titratable induction of gene expression, however, max induction varied with Ptac > PBAD > PBAD-riboswitch > Ptac-riboswitch. We also developed a sensitive reporter system to quantify promoter leakiness and show that leakiness for Ptac > Ptac-riboswitch > PBAD; while the newly developed PBAD-riboswitch exhibited no detectable leakiness. We demonstrate the utility of the tightly inducible PBAD-riboswitch construct using the dynamic activity of type IV competence pili in V. cholerae as a model system. The modular chromosomally integrated toolkit for ectopic gene expression described here should be valuable for the genetic study of V. cholerae and could be adapted for use in other species.
Subject terms: Microbiology, Bacteriology
Introduction
Regulated promoter systems for ectopic gene expression have been widely used in bacterial systems. Two commonly employed system for ectopic gene expression are the isopropyl β-d-1-thiogalactopyranoside (IPTG)-inducible tac promoter (Ptac) and the arabinose inducible araBAD promoter (PBAD)1,2. Both of these systems, however, exhibit some degree of leakiness, which allows for gene expression in the absence of the inducer when cells are grown in rich LB medium. Leakiness of PBAD can be reduced, to some extent, by addition of glucose to the growth medium because this promoter is catabolite repressed3, however, addition of glucose to the growth medium can change the physiology of cells which may introduce a confounding variable for some experiments.
Riboswitches are control elements that can regulate gene expression via direct interactions between a small molecule ligand and mRNA. Synthetic riboswitches that are responsive to the small molecule theophylline have recently been developed, which allow for regulated gene expression in diverse biological systems4,5. These riboswitch elements likely fold the mRNA to occlude the ribosome binding site in the absence of theophylline. And binding of theophylline to the riboswitch alters the conformation of the mRNA to expose the ribosome binding site and allow for translation of the downstream gene. For this reason, it is important to note that these riboswitches likely have limited utility for controlling the expression of genetic elements like non-coding RNAs, which do not need to be translated to exert their effect.
Generally, plasmids are employed for ectopic gene expression. However, many commonly used plasmids are poorly maintained by Vibrio species and/or their copy number can vary relative to model systems like Escherichia coli6,7. Integration of ectopic expression constructs onto the genome can bypass these issues. For many Vibrio species (e.g. Vibrio cholerae, Vibrio natriegens, Vibrio campbellii, Vibrio vulnificus, Vibrio parahaemolyticus, and Vibrio fischeri), it is remarkably easy to integrate novel sequences into the bacterial genome by exploiting their inherent capacity to undergo horizontal gene transfer by natural transformation8–13, which can be exploited for ectopic gene expression10,13–17.
Here, we generate a chromosomally integrated modular platform for ectopic gene expression in Vibrio species based on the widely-used Ptac and PBAD promoters in conjunction with a previously described theophylline responsive riboswitch5. We demonstrate that this toolkit allows for differing levels of ectopic gene expression (i.e. max induction), and that one of these promoter constructs (PBAD-riboswitch) allows for a broad-range of titratable gene expression without detectable leakiness. We highlight the utility of this tight expression construct to study the dynamic surface appendages required for natural transformation in V. cholerae.
Results and discussion
Design of modular ectopic expression constructs
All of the ectopic expression constructs are distinct ‘cassettes’ that can be integrated at any location in the bacterial genome (Fig. 1). We accomplish this via simple splicing by overlap extension (SOE) PCR18 to stitch these expression cassettes to upstream and downstream regions of homology (see Fig. S1 for details) to generate linear PCR products that can then be integrated into the V. cholerae genome by chitin-induced natural transformation8. Once an expression construct is integrated at a genomic locus, the gene of interest to be ectopically expressed can be easily exchanged by SOE PCR and natural transformation (see Fig. S1 for details). In this study, all ectopic expression constructs are integrated at the VCA0692 locus. This is a frame-shifted gene in the N16961 reference genome19. Furthermore, the disruption of VCA0692 does not alter the fitness of V. cholerae during growth in rich medium or in environments this facultative pathogen encounters during its pathogenic life cycle20, thus, highlighting this locus as a useful “neutral” location for the integration of novel sequences. These constructs can also be integrated at other commonly used “neutral” loci in V. cholerae including the lacZ gene, the frame-shifted transposase VC1807, or within intergenic spaces between convergently transcribed genes21.
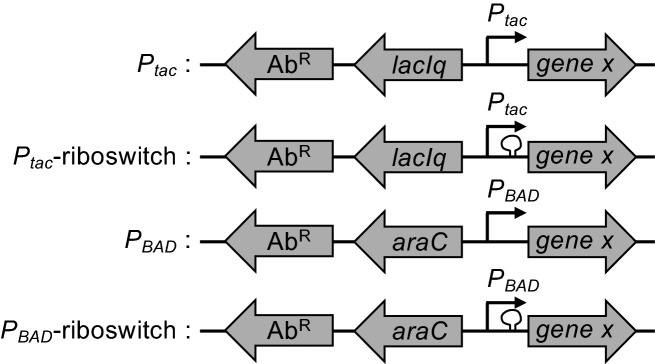
Figure 1.
Diagram of ectopic expression constructs. The four ectopic expression constructs characterized in this study are indicated. All have a linked antibiotic resistance cassette (AbR) to facilitate selection during integration into the genome by natural transformation. The gene encoding a transcription factor (lacIq or araC) and the promoter (Ptac or PBAD) required for inducible gene regulation are indicated. The presence of a theophylline-dependent riboswitch is demarcated by a loop before the gene of interest (gene x). For details on how these constructs were assembled, see Fig. S1.
The constructs for ectopic gene expression have a modular design where all have a linked antibiotic resistance marker (AbR) (Fig. 1). This AbR facilitates selection during integration into the genome by natural transformation and can be easily altered depending on the need. Linked to this AbR, there are the gene control elements. For Ptac and Ptac-riboswitch constructs, this includes the LacIq repressor and tac promoter. By contrast the PBAD and PBAD-riboswitch constructs contain AraC and the araBAD promoter. Both the Ptac and PBAD promoter constructs can be engineered to have a user-defined ribosome binding site (Fig. S1). By contrast, the two riboswitch constructs (Ptac-riboswitch and PBAD-riboswitch) contain a defined ribosome binding site within the theophylline-dependent riboswitch (riboswitch “E” in5) that is located immediately upstream of the gene of interest (Fig. 1 and Fig. S1).
Testing inducibility of ectopic expression constructs with GFP
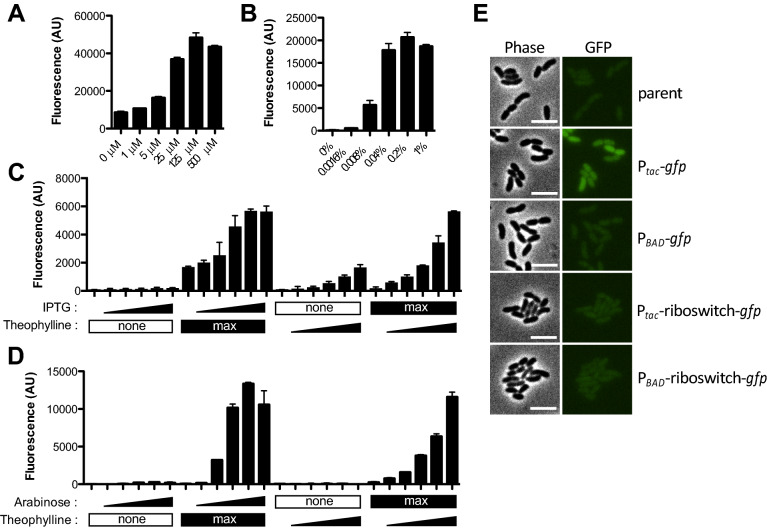
To test whether these different chromosomally integrated constructs allow for inducible gene expression and to compare the maximum level of expression they support, we generated constructs for ectopic expression of gfp22. The maximum level of gene expression varied among constructs with Ptac > PBAD > PBAD-riboswitch > Ptac-riboswitch (Fig. 2). Also, all of these constructs allowed for titratable gene expression (Fig. 2). This is particularly notable for PBAD, because this inducible system is known to have an “all-or-none” or autocatalytic gene expression phenotype in wildtype strains of E. coli23. This autocatalytic expression profile is due to high affinity transport of arabinose in E. coli, which further stimulates increased expression of arabinose transporters. Uncoupling this autoregulatory loop in E. coli can allow for titratable gene expression24. V. cholerae does not catabolize arabinose and lacks high affinity arabinose transporters. Arabinose may be transported into V. cholerae nonspecifically through one (or more) of its other carbohydrate transporters. Regardless, this low affinity transport of arabinose in V. cholerae allows for titratable gene expression from PBAD (Fig. 2), which is consistent with a number of previous studies14,25. Also, for constructs that contained the additional riboswitch control element (Ptac-riboswitch and PBAD-riboswitch), ectopic gene expression was dependent on addition of theophylline (Fig. 2), as expected5.
Figure 2.
Testing inducibility of ectopic expression constructs with GFP. (A–D) Cells harboring ectopic expression constructs driving gfp integrated at the VCA0692 locus were grown with inducer as indicated and assessed for GFP fluorescence in bulk cultures on a plate reader. (A) Cells with a Ptac-gfp construct were grown with the indicated amount of IPTG. (B) Cells with a PBAD-gfp construct were grown with the indicated amount of arabinose. (C) Cells with a Ptac-riboswitch construct were grown with increasing doses (denoted by a triangle below bars) of IPTG (from left to right: 0 µM, 1 µM, 5 µM, 25 µM, 125 µM, 500 µM) or theophylline (from left to right: 0 mM, 0.018 mM, 0.054 mM, 0.16 mM, 0.5 mM, 1.5 mM). ‘Max’ below bars denotes that cells were incubated with the highest concentration of IPTG (500 µM) or theophylline (1.5 mM) as indicated, while ’none’ indicates that none of that inducer was added. (D) Cells with a PBAD-riboswitch construct were grown with increasing doses (denoted by a triangle below bars) of arabinose (from left to right: 0%, 0.0016%, 0.008%, 0.04%, 0.2%, 1%) or theophylline (from left to right: 0 mM, 0.018 mM, 0.054 mM, 0.16 mM, 0.5 mM, 1.5 mM). ‘Max’ below bars denotes that cells were incubated with the highest concentration of arabinose (1%) or theophylline (1.5 mM) as indicated, while ’none’ indicates that none of that inducer was added. All data in (A–D) are from at least two independent biological replicates and shown as the mean ± SD. (E) Representative phase and epifluorescence images of cells with the indicated ectopic expression construct grown without any inducer added. Scale bar, 4 µM.
Testing ectopic expression constructs for the distribution of GFP fluorescence within single cells
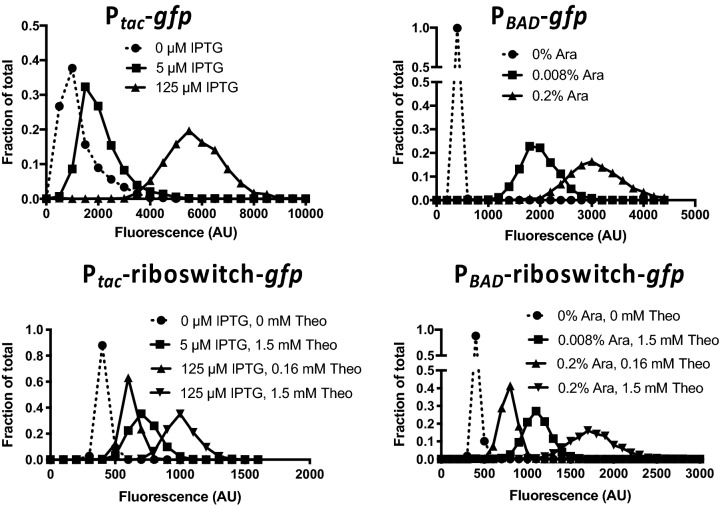
While we observed titratable gene expression above (Fig. 2), this was assessed in bulk cultures. Thus, titratable gene expression could be the result of bimodality in gene expression where cells in the population exhibit either a highly fluorescent or poorly fluorescent phenotype (similar to an ON/OFF light switch); and increased inducer simply results in a shift within the population where a higher proportion of cells exhibit the highly fluorescent phenotype. This is in contrast to titratable gene expression where the population responds uniformly to yield a unimodal distribution where increased inducer simply shifts the fluorescence intensity of the entire population (similar to a dimmer switch). Generally, for ectopic expression constructs the latter phenotype is preferred. To distinguish between these possibilities, we assessed the distribution of fluorescence among single cells within induced populations by epifluorescence microscopy. In the absence of inducer, only the Ptac construct exhibited detectable GFP fluorescence (Fig. 2A,E), which is consistent with this construct being very leaky in V. cholerae; a phenotype that is already widely appreciated. In the presence of inducer, all four constructs exhibited unimodal distributions, which supports the latter model and suggests that there is a uniform response to inducer among single cells within the population (Fig. 3).
Figure 3.
Testing ectopic expression constructs for the distribution of GFP fluorescence within single cells. Cells harboring the indicated ectopic expression constructs integrated at the VCA0692 locus were grown with inducer as indicated and assessed for GFP fluorescence in single cells via epifluorescence microscopy (Theo theophylline; Ara arabinose). The distribution of fluorescence among cells in the population is indicated on the plotted histograms. Data are from > 1,000 cells per condition tested and representative of two independent experiments.
Testing leakiness of ectopic expression constructs with Flp recombinase
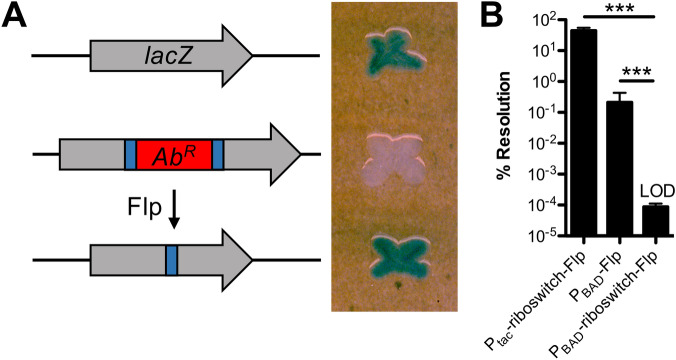
A major consideration for ectopic expression constructs is leakiness, which is defined as the basal expression of regulated genes in the absence of inducer. As mentioned above, only the Ptac construct exhibited detectable leakiness when using GFP fluorescence as a readout. This, however, is a poor readout for leaky gene expression because a substantial amount of GFP protein is required to generate an observable fluorescent readout. We sought to develop a sensitive and easily measured phenotype for leakiness from our ectopic expression constructs. To that end, we employed flippase (Flp), a highly-efficient recombinase that can mediate site-specific recombination between two Flp recombinase target (FRT) sequences26,27. Flanking FRT sequences can be engineered to leave behind an in-frame scar following Flp excision28. To generate a simple readout for Flp-mediated activity, we introduced a FRT-flanked AbR into the lacZ gene in V. cholerae (Fig. 4A). Strains with lacZ::FRT-AbR-FRT yielded a white colony phenotype on X-gal plates. Following Flp-mediated excision, however, the resulting lacZ gene (containing an in-frame 81 bp insertion) is active, and strains harboring lacZ::FRT exhibit a blue colony phenotype on X-gal plates (Fig. 4A). Thus, following Flp-mediated resolution, cells are irreversibly converted from LacZ- (white colonies) to LacZ + (blue colonies). To determine whether our expression constructs exhibited leaky expression, we generated ectopic expression constructs to drive Flp expression in strains that harbored the lacZ::FRT-AbR-FRT construct. Ptac-Flp was so leaky that all cells where we introduced this construct resolved lacZ::FRT-AbR-FRT to yield only blue colonies even in the absence of inducer. All of the other constructs (Ptac-riboswitch-Flp, PBAD-Flp, and PBAD-riboswitch-Flp) yielded only resolved blue colonies when grown in the presence of inducer. In the absence of inducer, both Ptac-riboswitch-Flp and PBAD-Flp exhibited some degree of leakiness, while PBAD-riboswitch-Flp did not exhibit any detectable leakiness in this assay (Fig. 4B). It is notable that the limit of detection of this assay is ~ 3 logs below the leakiness observed from the PBAD promoter, a construct that is traditionally considered tightly repressed in the absence of inducer2,3. This further validates our Flp-based assay as a highly sensitive approach to assess promoter leakiness.
Figure 4.
Testing leakiness of ectopic expression constructs with Flp recombinase. (A) Diagram of the approach used to test leakiness in gene expression with Flp recombinase. Wildtype V. cholerae cells have intact lacZ and form blue colonies on X-gal plates (top). Cells with lacZ::FRT-AbR-FRT have inactive LacZ and are white on X-gal plates (middle). Flp recombination resolves the FRT-AbR-FRT cassette within lacZ (making lacZ::FRT), which restores LacZ activity (bottom). (B) Single colonies of cells harboring the indicated ectopic expression constructs integrated at the VCA0692 locus and lacZ::FRT-AbR-FRT were grown in LB medium without any inducer overnight. Then, % resolution of lacZ::FRT-AbR-FRT was determined by plating for quantitative culture on X-gal plates. Percent resolution is defined as the number of blue colonies/total CFUs. Data in (B) are from 6 independent biological replicates and shown as the mean ± SD. Statistical comparisons were made by one-way ANOVA with Tukey’s post-test. LOD, limit of detection, ***P < 0.001.
Employing ectopic expression constructs to study the dynamic activity of type IV competence pili in V. cholerae
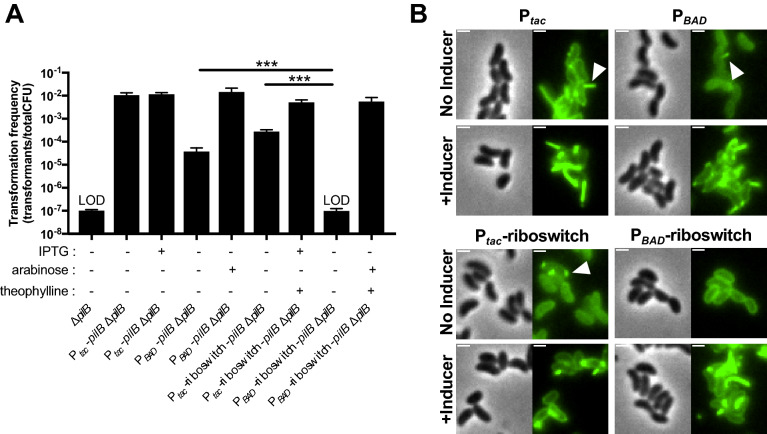
Horizontal gene transfer by natural transformation in V. cholerae is dependent on type IV competence pili29,30. These pili extend from the bacterial surface, bind to DNA in the environment, and then retract to pull DNA across the outer membrane29. This ingested DNA can then be translocated into the cytoplasm and integrated into the bacterial genome by homologous recombination. PilB is the motor ATPase that is required for extension of type IV competence pili31. To study the role of PilB in dynamic pilus activity, we sought to establish a strain that allowed for titratable and tightly regulated control of pilus extension. To that end, we generated strains where the native copy of pilB was deleted, and pilB expression was ectopically driven by our expression constructs. We then tested whether these strains were naturally transformable when grown without any inducer added. Only leaky expression of pilB would allow for natural transformation in this assay because ∆pilB mutants are not transformable (Fig. 5A). As observed using our Flp reporter readout, in the absence of inducer, Ptac, Ptac-riboswitch, and PBAD all exhibited leaky expression of pilB as evidenced by detectable natural transformation, while there was no detectable leakiness observed for PBAD-riboswitch because no transformants were obtained in this background without inducer added (Fig. 5A). Importantly, this experiment was performed in plain LB medium, thus, induction of catabolite repression (by the addition of glucose to the medium) was not necessary to prevent leaky gene expression. As expected, all strains transformed at high rates in the presence of inducer (Fig. 5A). These results suggest that leaky expression of pilB from Ptac, PBAD, and Ptac-riboswitch allow for some degree of pilus assembly even without inducer added, while pilus assembly is completely inhibited in the PBAD-riboswitch construct when no inducer is present. To test this idea further, we deleted the retraction ATPase pilT in these strains. This prevents extended pili from being easily retracted and sensitizes the direct observation of pilus assembly via a recently developed pilus labeling approach32. To determine whether leaky pilB expression allowed for pilus assembly, we assessed piliation in strains that were grown in the absence of inducer. Indeed, for Ptac, PBAD, and Ptac-riboswitch, we observed cells that contained extended pilus fibers (Fig. 5B), which is consistent with the leaky expression detected using our Flp recombinase reporter (Fig. 4B). We did not, however, observe extended pili in strains with PBAD-riboswitch when grown without inducer (Fig. 5B), which is consistent with a lack of leaky expression for this construct (Fig. 4B). As expected, all strains generated extended pili when grown with the appropriate inducer (Fig. 5B).
Figure 5.
Employing ectopic expression constructs to study the type IV competence pilus extension ATPase PilB. (A) Natural transformation of the indicated strains was tested. All ectopic expression constructs in these strains were integrated at the VCA0692 locus. IPTG (100 µM), arabinose (0.2%), and/or theophylline (1.5 mM) were added to reactions as indicated below each bar. Data are from at least 3 independent biological replicates and shown as the mean ± SD. Statistical comparisons were made by one-way ANOVA with Tukey’s post-test. LOD, limit of detection, ***P < 0.001. (B) Representative images of surface piliation. All strains harbor ∆pilB and ∆pilT mutations at the native locus and the indicated ectopic pilB expression construct. Where indicated, cells were grown with inducer as follows: 100 µM IPTG for Ptac, 0.2% arabinose for PBAD, 100 µM IPTG + 1.5 mM theophylline for Ptac-riboswitch, and 0.2% arabinose + 1.5 mM theophylline for PBAD-riboswitch. Examples of extended pili in no inducer samples are indicated by white arrows. Scale bars, 1 µm.
Together, these data indicate that only our newly generated PBAD-riboswitch-pilB construct allows for tightly regulated and titratable control of pilus biogenesis / extension in V. cholerae. This provides a valuable resource that will be critical for addressing diverse questions related to type IV pilus biology, which will be the focus of future work.
Methods
Bacterial strains and growth conditions
All strains used in this study are derivates of E7946, an El Tor isolate of V. cholerae33. See Table S1 for a complete list of strains used in this study. Strains were routinely grown at 30 °C or 37 °C in LB Miller broth and agar (BD Difco). When appropriate, media was supplemented with carbenicillin (20 µg/mL), spectinomycin (200 µg/mL), kanamycin (50 µg/mL), trimethoprim (10 µg/mL), chloramphenicol (1 µg/mL), or erythromycin (10 µg/mL).
Construction of mutant strains
All strains were generated by SOE PCR and chitin-induced natural transformation exactly as previously described8,21. See Fig. S1 for details on how the ectopic expression constructs were assembled and Table S2 for a detailed list of all primers used to generate all of the mutant constructs in this study. The araC–PBAD region for our PBAD and PBAD-riboswitch constructs was amplified from pBAD18-Kan3. The Ptac-riboswitch construct was amplified from DNA generously provided by Kim Seed15.
GFP fluorescence assays
Cells harboring ectopic expression constructs driving gfp were grown rolling at 30 °C to mid-log in LB medium. Then, inducer was added as indicated in each experiment and cells were grown for two additional hours rolling at 30 °C. Next, cells were washed and resuspended to the same optical density in instant ocean medium (7 g/L; Aquarium Systems) and fluorescence was measured on a Biotek H1M plate reader: excitation 500 nm/emission 540 nm exactly as previously described34. The parent strain lacking any ectopic expression construct was assayed alongside and used to subtract the background fluorescence of cells.
To image cells for GFP fluorescence, cultures were grown rolling at 30 °C in LB medium in the presence of the indicated inducers for 5 h to late-log. Then, cells were washed in instant ocean medium and mounted on 0.2% gelzan pads made with instant ocean medium exactly as previously described35.
Flp recombinase assays
Cells harboring ectopic expression constructs driving Flp and lacZ::FRT-SpecR-FRT were struck out onto LB + X-gal (40 µg/mL) + spectinomycin (200 µg/mL) plates. Single white colonies were picked, inoculated into plain LB medium, and grown overnight rolling at 30 °C. Then, each culture was plated quantitatively on LB + Xgal plates to determine the frequency of blue colonies within the population. Empirically, we determined that we could only reliably detect blue colonies at a rate of ~ 1 in 1,000,000 cells or 0.0001%. This equated to scoring for blue colonies on 100 µL spread plates of a dilution of 10–3 or greater. If no blue colonies were observed at 10–3, we assumed a single blue colony was present to define the limit of detection for that sample. Data are reported as the % Resolution = (CFU/mL blue colonies/CFU/mL total colonies) × 100.
Natural transformation assays
Cells harboring ectopic expression constructs driving pilB and the native pilB gene deleted were tested for rates of natural transformation. All of these strains also harbored Pconstitutive-tfoX and ∆luxO mutations, which rendered these strains constitutively competent. Strains were tested for natural transformation using chitin-independent transformation assays exactly as previously described36. The transforming DNA using in these experiments was 100 ng of a 6 kb ∆VC1807::ErmR PCR product.
Pilus labeling
All strains harbored the indicated ectopic expression construct, a cysteine amino acid substitution in the major pilin subunit PilA (PilAS56C), a deletion of the native copy of pilB (∆pilB), Pconstitutive-tfoX, a ∆luxO mutation, and a deletion of the retraction ATPase pilT (∆pilT). Strains were grown and labeled with Alexa fluor 488-maleimide exactly as previously described36. And mounted on 0.2% gelzan pads made with instant ocean medium.
Microscopy
Phase contrast and fluorescence images were collected on a Nikon Ti-2 microscope using a Plan Apo × 60 objective, a GFP filter cube, a Hamamatsu ORCAFlash 4.0 camera and Nikon NIS Elements imaging software. For Fig. 2E, the lookup tables for each phase and fluorescent image were adjusted to the same range so that they can be compared between samples. Fluorescence intensity of cells was determined using the MicrobeJ plugin37 in Fiji38. Representative images were prepared for figures using Fiji v2.0.0 (https://imagej.net/Fiji).
Graphs and statistical comparisons
All data were plotted for figures using GraphPad Prism software v8.4.3 (https://www.graphpad.com/scientific-software/prism/). The same software was used to perform the statistical comparisons indicated in the Figure legends.
Supplementary information
Acknowledgements
We gratefully acknowledge Eric Bruger, Christopher Marx, Julia van Kessel, and Johann Strnat for helpful discussions; and Kim Seed for generously providing DNA with the Ptac-riboswitch construct. This work was supported by funds from the National Science Foundation (1714949), Department of Energy (DE-SC0019436), and the National Institutes of Health (R35GM128674) to ABD.
Author contributions
All authors performed experiments and analyzed data. A.B.D. wrote the manuscript. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-72387-8.
References
- 1.de Boer HA, Comstock LJ, Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc. Natl. Acad. Sci. USA. 1983;80:21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee N, Francklyn C, Hamilton EP. Arabinose-induced binding of AraC protein to araI2 activates the araBAD operon promoter. Proc. Natl. Acad. Sci. USA. 1987;84:8814–8818. doi: 10.1073/pnas.84.24.8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch SA, Gallivan JP. A flow cytometry-based screen for synthetic riboswitches. Nucleic Acids Res. 2009;37:184–192. doi: 10.1093/nar/gkn924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Topp S, et al. Synthetic riboswitches that induce gene expression in diverse bacterial species. Appl. Environ. Microbiol. 2010;76:7881–7884. doi: 10.1128/AEM.01537-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tschirhart T, et al. Synthetic biology tools for the fast-growing marine bacterium Vibrio natriegens. ACS Synth. Biol. 2019;8:2069–2079. doi: 10.1021/acssynbio.9b00176. [DOI] [PubMed] [Google Scholar]
- 7.Delavat F, Bidault A, Pichereau V, Paillard C. Rapid and efficient protocol to introduce exogenous DNA in Vibrio harveyi and Pseudoalteromonas sp. J. Microbiol. Methods. 2018;154:1–5. doi: 10.1016/j.mimet.2018.09.022. [DOI] [PubMed] [Google Scholar]
- 8.Dalia AB. Natural cotransformation and multiplex genome editing by natural transformation (MuGENT) of Vibrio cholerae. Methods Mol. Biol. 1839;53–64:2018. doi: 10.1007/978-1-4939-8685-9_6. [DOI] [PubMed] [Google Scholar]
- 9.Simpson CA, Podicheti R, Rusch DB, Dalia AB, van Kessel JC. Diversity in natural transformation frequencies and regulation across vibrio species. mBio. 2019 doi: 10.1128/mBio.02788-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalia TN, et al. Multiplex Genome Editing by Natural Transformation (MuGENT) for Synthetic Biology in Vibrio natriegens. ACS Synth. Biol. 2017;6:1650–1655. doi: 10.1021/acssynbio.7b00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chimalapati S, et al. Natural transformation in Vibrio parahaemolyticus: a rapid method to create genetic deletions. J. Bacteriol. 2018 doi: 10.1128/JB.00032-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brooks JF, 2nd, Gyllborg MC, Kocher AA, Markey LE, Mandel MJ. TfoX-based genetic mapping identifies Vibrio fischeri strain-level differences and reveals a common lineage of laboratory strains. J. Bacteriol. 2015;197:1065–1074. doi: 10.1128/JB.02347-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Visick KL, Hodge-Hanson KM, Tischler AH, Bennett AK, Mastrodomenico V. Tools for rapid genetic engineering of Vibrio fischeri. Appl. Environ. Microbiol. 2018 doi: 10.1128/AEM.00850-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo Scrudato M, Blokesch M. The regulatory network of natural competence and transformation of Vibrio cholerae. PLoS Genet. 2012;8:e1002778. doi: 10.1371/journal.pgen.1002778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKitterick AC, Seed KD. Anti-phage islands force their target phage to directly mediate island excision and spread. Nat. Commun. 2018;9:2348. doi: 10.1038/s41467-018-04786-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borgeaud S, Blokesch M. Overexpression of the tcp gene cluster using the T7 RNA polymerase/promoter system and natural transformation-mediated genetic engineering of Vibrio cholerae. PLoS ONE. 2013;8:e53952. doi: 10.1371/journal.pone.0053952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalia AB, Lazinski DW, Camilli A. Identification of a membrane-bound transcriptional regulator that links chitin and natural competence in Vibrio cholerae. mBio. 2014;5:e01028-01013. doi: 10.1128/mBio.01028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 19.Heidelberg JF, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamp HD, Patimalla-Dipali B, Lazinski DW, Wallace-Gadsden F, Camilli A. Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS Pathog. 2013;9:e1003800. doi: 10.1371/journal.ppat.1003800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dalia AB, McDonough E, Camilli A. Multiplex genome editing by natural transformation. Proc. Natl. Acad. Sci. USA. 2014;111:8937–8942. doi: 10.1073/pnas.1406478111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 23.Siegele DA, Hu JC. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci. USA. 1997;94:8168–8172. doi: 10.1073/pnas.94.15.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khlebnikov A, Risa O, Skaug T, Carrier TA, Keasling JD. Regulatable arabinose-inducible gene expression system with consistent control in all cells of a culture. J. Bacteriol. 2000;182:7029–7034. doi: 10.1128/jb.182.24.7029-7034.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Judson N, Mekalanos JJ. TnAraOut, a transposon-based approach to identify and characterize essential bacterial genes. Nat. Biotechnol. 2000;18:740–745. doi: 10.1038/77305. [DOI] [PubMed] [Google Scholar]
- 26.Broach JR, Guarascio VR, Jayaram M. Recombination within the yeast plasmid 2mu circle is site-specific. Cell. 1982;29:227–234. doi: 10.1016/0092-8674(82)90107-6. [DOI] [PubMed] [Google Scholar]
- 27.Cox MM. The FLP protein of the yeast 2-microns plasmid: expression of a eukaryotic genetic recombination system in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1983;80:4223–4227. doi: 10.1073/pnas.80.14.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellison CK, et al. Retraction of DNA-bound type IV competence pili initiates DNA uptake during natural transformation in Vibrio cholerae. Nat. Microbiol. 2018;3:773–780. doi: 10.1038/s41564-018-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seitz P, Blokesch M. DNA-uptake machinery of naturally competent Vibrio cholerae. Proc. Natl. Acad. Sci. USA. 2013;110:17987–17992. doi: 10.1073/pnas.1315647110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ellison CK, et al. A bifunctional ATPase drives tad pilus extension and retraction. Sci. Adv. 2019;5:eaay2591. doi: 10.1126/sciadv.aay2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellison CK, Dalia TN, Dalia AB, Brun YV. Real-time microscopy and physical perturbation of bacterial pili using maleimide-conjugated molecules. Nat. Protoc. 2019;14:1803–1819. doi: 10.1038/s41596-019-0162-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller VL, DiRita VJ, Mekalanos JJ. Identification of toxS, a regulatory gene whose product enhances toxR-mediated activation of the cholera toxin promoter. J. Bacteriol. 1989;171:1288–1293. doi: 10.1128/JB.171.3.1288-1293.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dalia AB. RpoS is required for natural transformation of Vibrio cholerae through regulation of chitinases. Environ. Microbiol. 2016;18:3758–3767. doi: 10.1111/1462-2920.13302. [DOI] [PubMed] [Google Scholar]
- 35.Dalia AB, Dalia TN. Spatiotemporal analysis of DNA integration during natural transformation reveals a mode of nongenetic inheritance in bacteria. Cell. 2019;179:1499–1511. doi: 10.1016/j.cell.2019.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chlebek JL, et al. PilT and PilU are homohexameric ATPases that coordinate to retract type IVa pili. PLoS Genet. 2019;15:e1008448. doi: 10.1371/journal.pgen.1008448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ducret A, Quardokus EM, Brun YV. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 2016;1:16077. doi: 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.