Abstract
The potential role of enteric viral infections and the developing infant virome in affecting immune responses to the oral poliovirus vaccine (OPV) is unknown. Here we performed viral metagenomic sequencing on 3 serially collected stool samples from 30 Bangladeshi infants following OPV vaccination and compared findings to stool samples from 16 age-matched infants in the United States (US). In 14 Bangladeshi infants, available post-vaccination serum samples were tested for polio-neutralizing antibodies. The abundance (p = 0.006) and richness (p = 0.013) of the eukaryotic virome increased with age and were higher than seen in age-matched US infants (p < 0.001). In contrast, phage diversity metrics remained stable and were similar to those in US infants. Non-poliovirus eukaryotic virus abundance (3.68 log10 vs. 2.25 log10, p = 0.002), particularly from potential viral pathogens (2.78log10 vs. 0.83log10, p = 0.002), and richness (p = 0.016) were inversely associated with poliovirus shedding. Following vaccination, 28.6% of 14 infants tested developed neutralizing antibodies to all three Sabin types and also exhibited higher rates of poliovirus shedding (p = 0.020). No vaccine-derived poliovirus variants were detected. These results reveal an inverse association between eukaryotic virome abundance and poliovirus shedding. Overall gut virome ecology and concurrent viral infections may impact oral vaccine responsiveness in Bangladeshi infants.
Subject terms: Genomic analysis, Microbiology techniques, Sequencing, Medical research, Infectious diseases
Introduction
Oral vaccines are more effective in high-income countries than in low-income countries1. One hypothesis for this finding is potential blocking of effective immune responses by bacteria, phages and eukaryotic viruses that inhabit similar niches as live vaccine strains. Concurrent infection with non-polio enteroviruses has been previously reported to interfere with oral poliovirus vaccine (OPV) efficacy2. Concurrent administration of oral poliovirus and rotavirus vaccine has also been shown to reduce rotavirus vaccine immunogenicity3,4.
Most infections in children under five years old are caused by viruses. Our understanding of the pediatric virome, however, remains rudimentary. Metagenomic next-generation sequencing approaches have previously demonstrated that the virome in children is dynamic and that virus distributions are driven by a combination of maternal, geographic and environmental factors5–7.
In this study, we applied metagenomic virus sequencing to characterize the gastrointestinal virome in serially collected stool samples from infants from Bangladesh. All of the infants evaluated in this study completed the first two OPV vaccinations (out of 3 for the full series), with the first dose administered within the first 4 months of life. OPV consists of trivalent live-attenuated Sabin poliovirus that replicates at mucosal sites in the infant gastrointestinal tract and induces mucosal and systemic antibody production1. Here, we describe the infant virome in Bangladesh in comparison to that of US children and investigate the role of the virome on shedding of vaccine-associated poliovirus and the development of effective poliovirus neutralizing antibody responses.
Methods
Study population and sample collection
Stool samples for virome analysis were obtained from a phase I trial of probiotics in infants in Bangladesh, as previously described8. These infants all came from households of low socioeconomic status living in peri-urban slums. Infants were chosen from among the 160 babies in the trial based on the completeness of fecal sampling. Preference was given to those receiving the lowest dose of the combined lactobacillus and Bifidobacterium probiotic: five subjects were in the control arm (no probiotic), 12 were in the twice per month probiotic arm and 13 were in the weekly probiotic arm. Probiotics, consisting of Lactobacillus reuteri DSM 17,938 combined with Bifidobacterium longdum subspecies infantis 35,624, were given for one month.
Infants aged 4–12 weeks (mean age 8 weeks) were recruited from three vaccination clinics near the International Center for Diarrheal Disease Research, Bangladesh (icddr,b) in Dhaka between October 2013 and April 2014. All infants received at least the first 2 doses of the trivalent oral polio vaccination (OPV) that is administered 6, 10 and 14 weeks old; no infants were given OPV vaccine at birth. The dates of vaccination were documented by vaccination cards (> 90% of the time), or in rare instances, estimated from parents’ recollection. Other vaccines given at these timepoints included pentavalent (diphtheria, tetanus, pertussis, Haemophilus influenzae type B, and hepatitis B virus) and pneumococcal vaccines (Supplementary Table S1). Demographic and socioeconomic data were collected at enrollment. Health information for the infants in the study, including illness, gastrointestinal and respiratory symptoms and breastfeeding practices were collected at weekly intervals (Table 1).
Table 1.
Characteristics of the Bangladeshi infants in the study.
| Characteristic | Median (IQR) or N (%) |
|---|---|
| Age in weeksa | 9.9 (9.6–10.9) |
| Weight Z-scorea | − 0.76 (− 1.22 to − 0.26) |
| Height Z-scorea | − 0.45 (− 1.29 to − 0.54) |
| Head circumference Z-scorea | − 1.14 (− 1.92 to − 0.49) |
| Female | 14 (47) |
| Born by cesarean section | 7 (23) |
| Years of maternal education | 5 (4–8) |
| Household size | 5 (3–6) |
| Household monthly income | |
| < $100 | 7 (23) |
| $100–$150 | 12 (40) |
| > $150 | 11 (37) |
| Diet | |
| Exclusive breastfed | 10 (30) |
| Partial breastfed (Supplementary feeding) | 20 (60) |
| Cow’s milk | 3 (6.7) |
| Water, formula, or baby cereal | 20 (60) |
| Antibiotic exposure during study | 4 (13.3) |
| Illness symptoms during study | |
| No symptoms | 8 (26.6) |
| Fever | 12 (40) |
| Respiratory (cough, congestion) | 19 (63) |
| Gastrointestinal (vomiting, watery stool) | 3 (10) |
aAt first stool sample.
IQR interquartile range.
The study was approved by the institutional review boards at both icddr,b (Protocol ID 13,022) and Stanford University (Protocol ID 25,487) and was registered on ClinicalTrials.gov (NCT01899378). Written informed consent was provided by parents or guardians. As part of their consent, subjects agreed to unspecified “advanced tests” of their stool samples “that may help us better understand the infections that your baby may have had”’; the virome analyses described here fall in this category. All samples were anonymized prior to virome analyses. All research was performed in accordance with guidelines and regulations on human subjects research established by the IRBs at UCSF and Stanford University, the National Institutes of Health, and the World Medical Association (WMA) Declaration of Helsinki.
Viral metagenomic analysis was performed on three stool samples from 30 infants collected at 4 weeks after the first dose of OPV vaccine (prior to the second dose), 2 weeks after the second dose of OPV vaccine, and 4 weeks after the second dose of OPV vaccine (immediately prior to the third dose) (Fig. 1). Stool samples from infants prior to OPV vaccination were not available as infants had already received the first OPV vaccination at time of enrollment. For purposes of comparison, stool samples were also tested from age-matched infants in California, USA from the Stanford’s Outcome Research in Kids (STORK) cohort, a longitudinal study of the impact of the developing virome and pediatric infections on weight, growth and immune development in infants9. Specifically, we included 16 infants from the STORK cohort with available stool samples collected prior to 8 weeks of age and administration of rotavirus vaccine. The STORK study was approved by the Institutional Boards of Stanford University and the Santa Clara Valley Medical Center, and written informed consent was obtained from parents or guardians.
Figure 1.
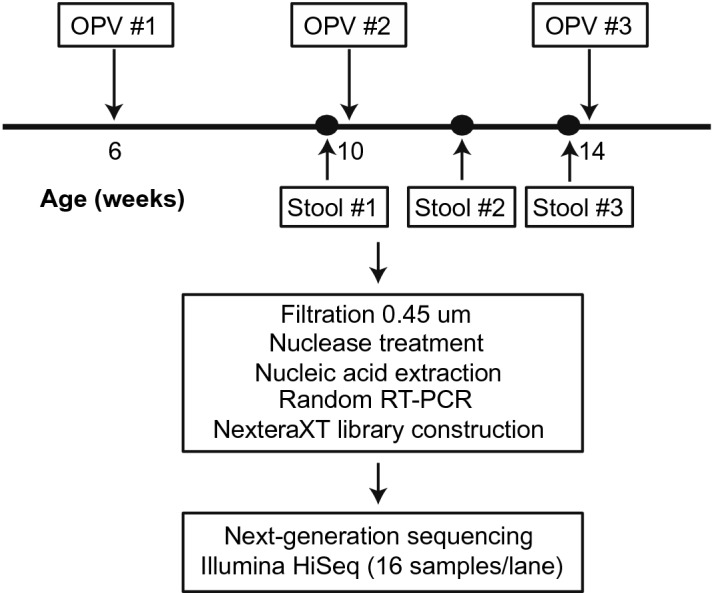
Overview of sample collection and virus metagenomic sequencing protocol. Abbreviations: OPV, oral poliovirus vaccine.
Stool samples from Bangladeshi and California infants were collected in sterile containers and processed in an identical fashion for virome analysis. For the Bangladeshi cohort, fresh stool samples collected in the field were placed on ice and then brought to the lab the same day on ice and frozen within 10 h of collection. For the California infants in the STORK cohort, fresh stool samples collected in the clinic were placed on ice packs and frozen within 24 h of collection. Frozen stool samples were stored at – 80 °C prior to processing.
Nucleic acid extraction
Nucleic acid extraction of stool samples was performed as previously described10. Stool samples were diluted 20% in phosphate buffered saline (PBS) (1,500 μl) and centrifuged for 5 min at 10,000g, followed by filtration using a 0.45 μM filter and treatment using a nuclease cocktail of TURBO DNase (Invitrogen), Baseline Zero DNase (Ambion), Benzonase (Novagen) and RNase A (Roche) for 30 min at 37 °C. This procedure digests host cell and non-protected (“naked”) viral nucleic acids, while maintaining viral RNA in particles that are protected from the action of nucleases11. Nuclease activity was then immediately inactivated by addition of guanidium-thiocyanate containing lysis buffer (Qiagen), followed by total nucleic acid extraction of 400 μl of pretreated stool using the EZ1 Virus Mini Kit v2.0 (Qiagen). Extracts were eluted in 60 μl volume.
Library preparation and viral metagenomic next-generation sequencing
Amplified cDNA was prepared using random nonamer primers attached to a primer linker sequence with 25 cycles of PCR amplification, as previously described12,13. Ultra-pure bovine serum albumin (BSA) (Ambion) was added to the reverse transcription and PCR steps to minimize PCR inhibition. Amplified cDNA was purified using AMPure XP beads (Beckman-Coulter) and quantitated using the Qubit Fluorometer and Qubit dsDNA HS Assay (Life Technologies). Purified dsDNA (2 ng) was used for NGS library generation using the Nextera XT kit (Illumina). Nextera XT libraries were purified using AMPure XP beads (Beckman Coulter) and sequenced on an Illumina HiSeq 2,500 in rapid mode using 150-bp paired-end sequencing. Samples were batched (12–16 samples per lane) and sequenced in parallel with negative controls (PBS) (one for each batch of samples) to monitor for potential reagent, laboratory, or cross-contamination.
Bioinformatics analysis
Metagenomic next-generation sequencing (mNGS) data were analyzed for viral nucleic acids using SURPI + , a bioinformatics pipeline for pathogen detection and discovery from metagenomic data14, modified to incorporate enhanced filtering and classification algorithms15. The SNAP nucleotide aligner was run using an edit distance of 16 against the NCBI NT database containing the entirety of GenBank (March 2015)16. This enabled the detection of reads with ≥ 90% identity to reference sequences in the database.
Next, RAPSearch was used to screen for divergent viruses by translated nucleotide alignment to the NCBI nonredundant (NR) protein database (March 2015)14. In accordance with a prior clinical validation study15, the pre-established criterion for viral detection by SNAP or RAPSearch was the presence of reads mapping to at least 3 nonoverlapping regions of the viral genome15. In the current study, no highly divergent viruses were detected using RAPSearch.
For quantification of viral reads8,14,17, reads were normalized according to the number of preprocessed reads (adaptor-trimmed reads with exclusion of low-quality and low-complexity sequences) and expressed in reads per million (RPM). RPM values corresponding to presumptive viral laboratory, reagent, and/or cross-contaminants in the negative control samples were subtracted from clinical stool samples in the same sequencing batch, with negative RPM values after subtraction set to 0. To ensure accurate read counts, confirmation of reads corresponding to poliovirus was performed by manual BLASTn18 analysis using a stringent threshold e-value of 10–20. Reference-based assembly of poliovirus genomes was performed using Geneious 10.2.4 software (https://www.geneious.com/download/previous-versions/, accessed February 11th, 2020)19. Phylogenetic sequence analysis of recovered whole-genome poliovirus sequences in parallel with reference poliovirus Sabin 1–3 sequences (accession numbers AY184219, AY184220, AY184221) was also performed using Geneious 10.2.4 software19. Briefly, genome sequences were aligned using MAFFT20, followed by tree construction using PHYML21 at default settings.
Although the virome is predominantly populated with phages22, fewer prokaryotic than eukaryotic reads were identified on average in samples from this study. This is likely due to the use of a normalized RPM metric as a measure of relative (but not absolute) abundance, and the lack of precise species-specific taxonomic classification and sequence representation for phages relative to eukaryotic viruses in the GenBank reference database.
Poliovirus antibody assay
Available serum samples from infants collected after the third OPV vaccination were assessed for neutralizing antibodies against human poliovirus Sabin 1–3 serotypes, as previously described23,24. Poliovirus neutralization was assessed by plaque-reduction neutralization testing (PRNT) using replication-competent poliovirus propagated in HeLa S3 cells over a period of 7 days20. Replication competent poliovirus was generated from infectious cDNA clones, as previously described20. HeLa S3 cells were seeded in 30 ml of 10% NCS DMEM/F12 48 h prior to infection with 7.5 ml of cDNA clones diluted with serum-free DMEM/F12. Virus was recovered after 3 infectious cycles (24 h) when cytopathic effect (CPE) is visible. Propagated virus was then titrated using TCID50 assay as previously described20. For poliovirus neutralization, 80 µL of serum was diluted with PBS and a series of twofold dilutions series was made (1/8 to 1/1,024 dilution) by adding 80 µL of DMEM/F12 into subsequent wells. Next, 80 μl of virus stocks diluted to 2000 TCID50/mL were added to each well. After a 90 min incubation, 100 μL of the virus and serum mix was transferred onto a plate seeded with 10,000 HeLa S3 cells/well and incubated for 7 days at 33 °C. After 7 days, plates were fixed with 2% formaldehyde and stained with 0.5% crystal violet. Neutralizing activity against each serotype was defined as 100% inhibition of infection (no plaques visualized after staining of the cell monolayer) at 1/8 dilution. The neutralizing antibody titers of the serum against human poliovirus serotype 1, 2 and 3 were determined as the highest dilution of the serum that produced 100% inhibition. Control sera for human poliovirus Sabin 1, 2, and 3 strains all demonstrated neutralizing antibodies at 1/8 dilution.
Statistical analysis
Samples were considered positive for poliovirus if reads covered at least 3 gene regions of poliovirus genome15 and read counts were > 5X that of the negative control buffer sample (the “no-template” control or NTC). Based on manual inspection of the distribution of read frequencies, high poliovirus shedding was defined as ≥ 10 RPM. Categorical data were reported as counts and percentages and continuous data as medians with ranges, and significance tested by Fisher’s Exact and Mann–Whitney tests, respectively. Diversity metrics, including the Chao Richness Score, Shannon Diversity Index and Bray–Curtis dissimilarity between samples at the genus level were calculated using R package vegan2.5–325. Principal coordinate analysis with Bray–Curtis dissimilarity was performed to visualize differences in community composition between groups based on degree of poliovirus shedding (based on the normalized RPM mapping to poliovirus in stool samples) and country of origin (Bangladesh or US), with significant differences assessed by permutation-based analysis of variance (PERMANOVA) using the Adonis function (default 1,000 permutations)26. Comparisons of virome abundance, richness and alpha diversity between groups were analyzed using either the Kruskal–Wallis rank sum test or a generalized estimating equations (GEE) model to account for the subject-structure of the longitudinal data and included age adjustment where applicable. All statistical tests were calculated as two-sided at the 0.05 significance level with virus family and genera associations adjusted for multiple comparisons using the Benjamini–Hochberg method for false discovery rate correction27. Analyses were performed using R version 3.3.3 software (RStudio version 1.1.383).
Results
Stool virome composition in Bangladeshi infants
Of 90 stool samples tested from 30 Bangladeshi infants, 87 were successfully sequenced (Table 1). Three samples failed amplification due to low nucleic acid concentration following extraction and were excluded from further analysis. Median ages of infants at the time of the first, second and third stool sampling were 9.9 (9.6–10.8), 12.0 (11.4–12.9) and 14.8 (14.4–15.8) weeks, respectively. Infants were sampled a median of 27 (24–28) days after administration of the first OPV vaccine and 9.5 (8–10) and 28.8 (27–30) days after the second OPV vaccination.
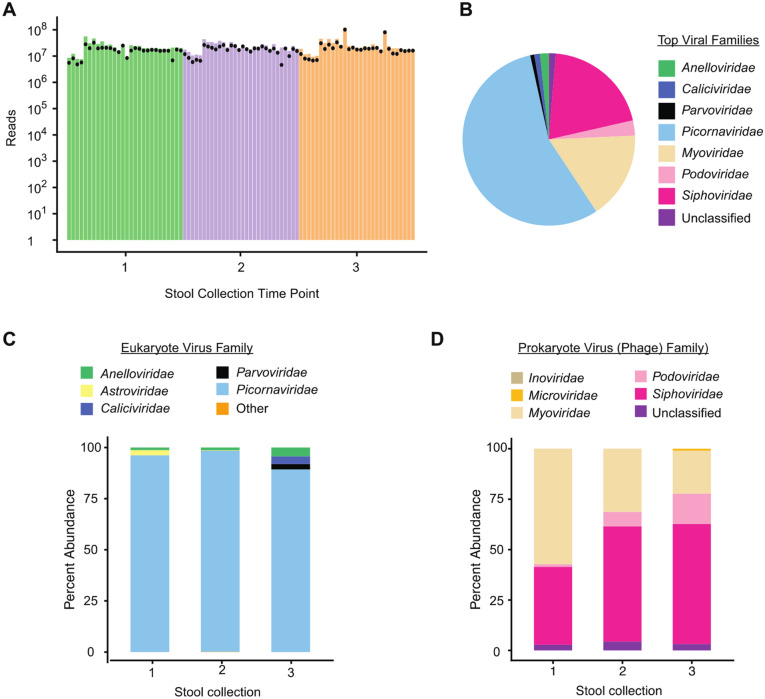
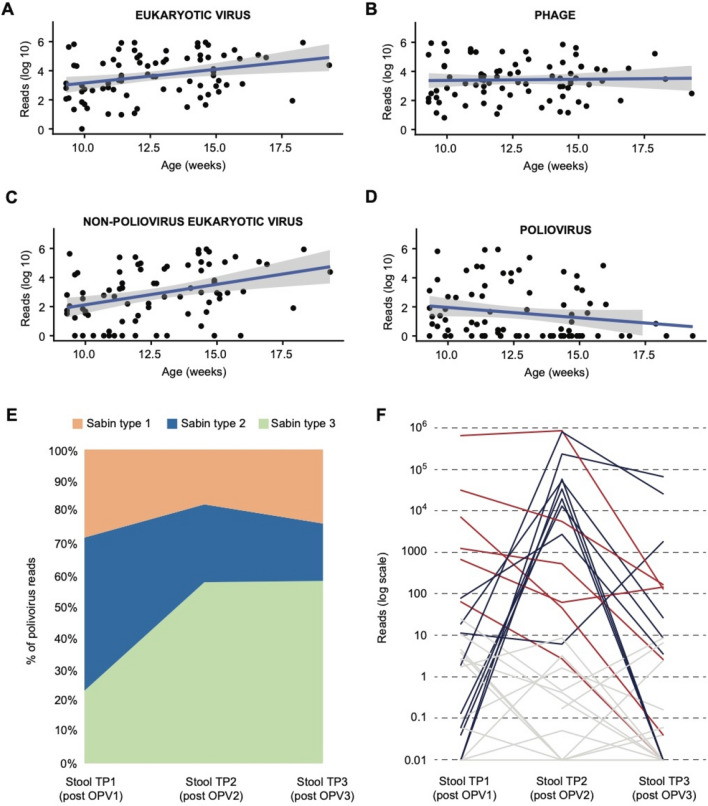
Viral metagenomic sequencing analysis of the 87 stool samples yielded a total of 1.6 billion sequence reads, with a median of 17.6 million reads per sample (14.7–20.4 million; Fig. 2). By SURPI + analysis, the mean percentages of matched viral, bacterial and human reads across all samples were 12.26%, 23.36% and 10.71% of preprocessed reads, respectively (Supplementary Table S2). An average of 23.4% of reads did not map to any reference sequence in NCBI NT. Eukaryotic viruses of the vertebrate taxa (59.4%) and prokaryotic viruses (referred hereafter as phages) (40.6%) accounted for the majority of viral reads, with less than 0.01% of reads attributed to eukaryotic plant, invertebrate, fungi and protozoa viruses. Among eukaryotic viruses, the abundance as expressed by normalized reads per million (RPM) (heretofore referred to as “abundance”) increased with infant age (Pearson’s r = 0.30, p = 0.006) whereas phage abundance remained relatively static over time (Pearson’s r = 0.03, p = 0.88; Fig. 3A,B). The increase in eukaryotic viral abundance was driven by non-polioviruses (Pearson’s r = 0.358, p < 0.001) and was not due to the number of poliovirus reads, for which there was a non-significant opposite trend towards a decline in read counts with age (Pearson’s r = 0.18, p = 0.11) (Fig. 3C,D). Richness for eukaryotic viruses increased with age as well (p = 0.013), while alpha diversity for eukaryotic viruses and phages did not differ significantly over time (p = 0.41 and p = 0.70 respectively). No significant differences in total, eukaryotic or phage richness or abundance were observed among infants who had received weekly, biweekly, or no probiotics (Supplementary Table S3).
Figure 2.
Stool Virome Composition in Bangladeshi Infants (A) Sequenced reads obtained per sample by stool collection time point. Dots reflect recovered number of reads after preprocessing raw data with trimming of adaptors and removing low-quality and low-complexity sequences. (B) Distribution of most abundant virus families reads in infant stool. (C) Percent abundance of eukaryotic vertebrate virus family by infant stool sample. (D) Percent abundance of phage family by infant stool sample.
Figure 3.
Viral abundance and poliovirus shedding. (A-D) Log-abundance of virus by infant age. (A) Eukaryote virus abundance. (B) Phage abundance (C) Non-poliovirus eukaryotic virus abundance (D) Poliovirus abundance; the regression line is plotted in blue, with the 95% confidence interval shown in gray. (E) Relative abundance of Sabin poliovirus strains at each time point following vaccination. The plot only includes data from subjects for whom samples at all 3 timepoints were available and ≥ 50 reads were detected in at least one timepoint. (F) Total poliovirus abundance at each time point. Colored lines denote infant stools with minimal viral shedding (gray), high shedding following second vaccine (blue), and high shedding following the first vaccine with gradual decline (red). Abbreviations: TP, time point, OPV, oral poliovirus vaccine.
Among eukaryotic viruses, a total of 12 families, 33 genera and 179 species were identified. Picornaviruses accounted for the vast majority of reads (93.6%), followed by anelloviruses (2.7%), caliciviruses (1.8%) and parvoviruses (1.3%; Fig. 2). The majority of picornavirus reads mapped to the Enterovirus genus (62.1%) of which more than half (52.8%) aligned to polioviruses, followed by saliviruses (18.2%), parechoviruses (16.8%) and cosaviruses (2.8%). Notably, even after excluding poliovirus reads, enteroviruses remained the most abundant viruses identified. Cardioviruses and unclassified picornaviruses accounted for < 1% of picornavirus reads. Of the detected caliciviruses, norovirus represented 69.1% and sapovirus 30.9%. Bocaviruses comprised 100% of parvoviruses found in infant stool.
All infants had picornaviruses in at least one stool sample, with 90% of infants shedding poliovirus during at least one of the sampled timepoints. Anelloviruses and circoviruses were found in 96.7% and 76.6% of infants, respectively, followed by caliciviruses (33.3%), papillomaviruses (23.3%) and astroviruses (23.3%). Other viruses, including herpesviruses, (13.3%), adenoviruses (6.7%), hepatitis B virus (3.3%) and MW polyomavirus (3.3%), were less commonly detected in infant stool.
Many of the viruses identified in stool from Bangladeshi infants, including norovirus, sapovirus, mamastrovirus, salivirus, rotavirus, bocavirus, adenovirus, cosavirus, parechovirus, or cytomegalovirus, were known or suspected pathogens associated with acute diarrheal or respiratory infection (Supplementary Table S4). The majority (86.7%) of infants shed at least one of these viruses on at least one occasion. Nearly two-thirds of infants (63.3%) had norovirus, sapovirus, astrovirus, salivirus and/or rotavirus detected in at least one sample, whereas nearly half (43.3%) had bocavirus, adenovirus, cosavirus, parechovirus and/or cytomegalovirus detected. MW polyomavirus was identified in one infant and hepatitis B virus in another. Across the sampling period, infants shed on average 1.5 (range 0–6) potential viral pathogens in their stool samples. At the time of stool collection, infants were symptomatic (defined as one or more of fever (T > 38 °C), cough, congestion, vomiting, or watery diarrhea based on a documented clinical assessment by a nurse or physician) for 34 of 87 time points (39.1%) (Supplementary Table S5). Symptomatic infants had a putative viral pathogen detected in stool approximately half of the time (18/34, 52.9%) (Supplementary Table S5). Conversely, the presence of a viral pathogen in stool was only occasionally associated with symptoms (18/50, 36%), with most identified viruses found in asymptomatic infants (32/50, 64%). Overall, there was no apparent correlation between symptoms and detection of pathogenic viruses in stool (p = 0.51, Fisher’s Exact Test). During the study period, only one infant was hospitalized; this child presented with fever and cough and had a high number of parechovirus reads (269,383 RPM, Supplementary Table S5, P167) detected in stool at the time of acute illness.
Among phages, 5 families, 30 genera and 541 species were identified. Phages in infant stool were primarily caudoviruses belonging to the siphovirus (49.3%), myovirus (40.2%) and podovirus (6.9%) families. Phage species most predominantly identified were unclassified siphoviruses, followed by phages from bacterial species largely comprising gastrointestinal flora.
Stool poliovirus shedding varies among Bangladeshi infants
Polioviruses accounted for 31.7% of eukaryotic virus reads; 63.2% (55/87) of samples were positive for poliovirus, of which 65.4% (36/55) of samples were classified as having high poliovirus reads (> 10 RPM). 70.4% of vaccinated infants shed poliovirus in stool 30 days from the first vaccine (time point 1); these infants predominantly shed Sabin type 2 and 3 polioviruses with all simultaneously shedding multiple Sabin types at roughly equal overall proportion (Table 2). At the second stool sampling (time point 2, median 10 days after second OPV vaccination), 74.1% of infants predominantly shed Sabin type 3 poliovirus (60%), with shedding of multiple Sabin types seen in 75%. Poliovirus shedding was reduced to 48% of infants by the third stool sample (time point 3, median 29 days after second vaccination). Overall, the relative proportion of poliovirus reads comprising Sabin poliovirus types 1, 2, and 3 were roughly equal at time point 1, with the relative proportion of Sabin type 3 poliovirus increasing with a concomitant decrease in Sabin type 2 poliovirus by time points 2 and 3 (Fig. 3E).
Table 2.
Poliovirus shedding characteristics among stool samples.
| Stool sample | Median days (range) after vaccine | % shedders | % of the most common poliovirus strain detected in sheddersc | ||
|---|---|---|---|---|---|
| Sabin 1 | Sabin 2 | Sabin 3 | |||
| First sample (n = 29) | 27 (24–48)a | 70.4% | 15.8% | 47.4% | 36.8% |
| Second sample (n = 30) | 9.5 (8–10)b | 74.1% | 15% | 25% | 60% |
| Third sample (n = 28) | 28.8 (27–30)b | 48.3% | 15.4% | 15.4% | 69.2% |
aAfter first poliovirus vaccine.
bAfter second poliovirus vaccine.
cDefined as the number of subjects shedding Sabin 1, 2, or 3 as the dominant poliovirus strain (strains with the greatest proportion of mapped reads) divided by the total number of subjects with poliovirus detected in stool.
The kinetics of poliovirus shedding differed among infants with three dominant patterns observed: (1) minimal or no shedding of vaccine at any time point (n = 11), (2) high shedding soon after second vaccine but not after the first (n = 13) and high shedding after first vaccine that gradually declined after second vaccine (n = 6) (Fig. 3F).
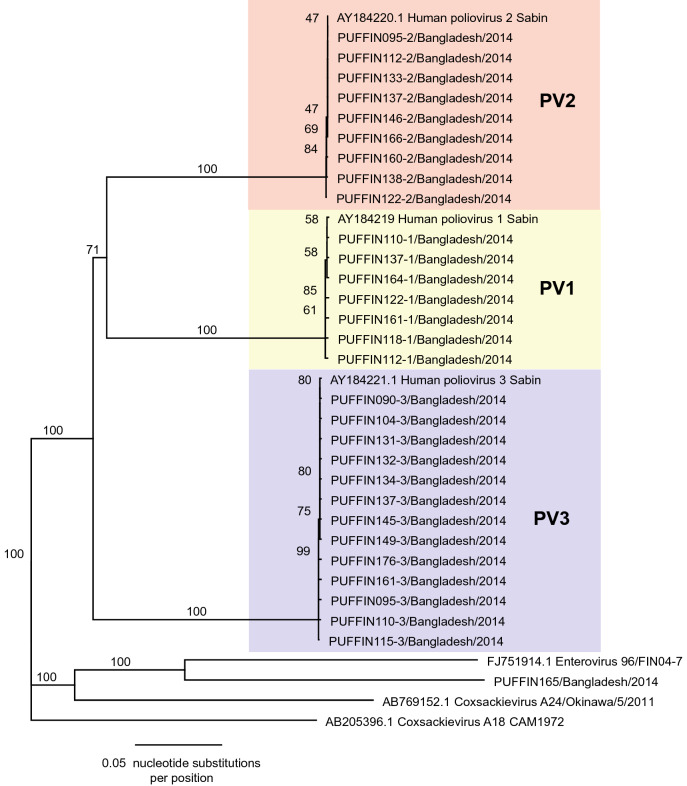
Viral metagenomic sequencing of infant stool allowed for the assembly of 29 whole genome consensus poliovirus sequences. Comparison of whole genome sequences to vaccine reference strains revealed viruses that were identical (Fig. 4). Thus, no significant vaccine-derived poliovirus variants were observed in this cohort.
Figure 4.
Phylogenetic analysis of whole genome poliovirus sequences. Comparison of 29 whole genome sequences to vaccine reference strains revealed viruses with percent pairwise identity of 100% for Sabin 1 and 3 and 100% identity for Sabin 2 viruses (differences only due to slight variations in total coverage) and no significant vaccine-derived poliovirus variants.
According to Bray–Curtis dissimilarity scores, the eukaryotic virome composition of infant stools containing poliovirus differed significantly from those that did not (p = 0.007), even after exclusion of poliovirus reads, whereas the phage virome did not differ between groups (p = 0.50). In particular, abundance (3.68 log10 vs. 2.25 log10, p = 0.002) and richness (p = 0.016) of non-poliovirus eukaryotic viruses, particularly those associated with acute respiratory or gastrointestinal illness (2.78log10 vs. 0.83log10, p = 0.002), was inversely associated with poliovirus shedding (Supplementary Table S6). Phage abundance (3.47log10 vs, 3.37log10, p = 0.50), richness (p = 0.06) and alpha diversity (p = 0.07) also did not significantly differ between groups (Supplementary Table S6). No specific virus genus or family was significantly associated with shedding of poliovirus after false discovery rate correction (Supplementary Tables S7 and S8). No associations between infant sex, maternal education, economic status, or breastfeeding status (exclusive breastfeeding or partial breastfeeding with supplementation) with poliovirus or other non-poliovirus eukaryotic viruses were observed (Supplementary Table S9).
Low neutralizing antibody response to poliovirus in Bangladeshi infants
Serum available from 14 infants, collected a median of 31 days (range: 14–34) after administration of third OPV vaccine, was tested for neutralizing antibodies against all three Sabin poliovirus types. The median infant age at time of collection was 19.6 (range 16.3–24.3) weeks old. All 14 infants had detectable neutralizing antibodies to at least one Sabin type, although only four infants (28.6%) developed antibodies to all three types (Supplementary Table S10). Infants who had neutralizing antibodies to only one Sabin type had significantly lower stool poliovirus reads during the sampled time points compared to those who developed neutralizing antibodies to all three types (0.36log10 vs. 3.72log10, p = 0.02, (Supplementary Table S11). Evaluation of virus genera, abundance, richness and diversity did not reveal significant differences among the infants among the varied serologic outcomes (Supplementary Table S12).
Stool virome composition is distinct in Bangladeshi compared to US infants
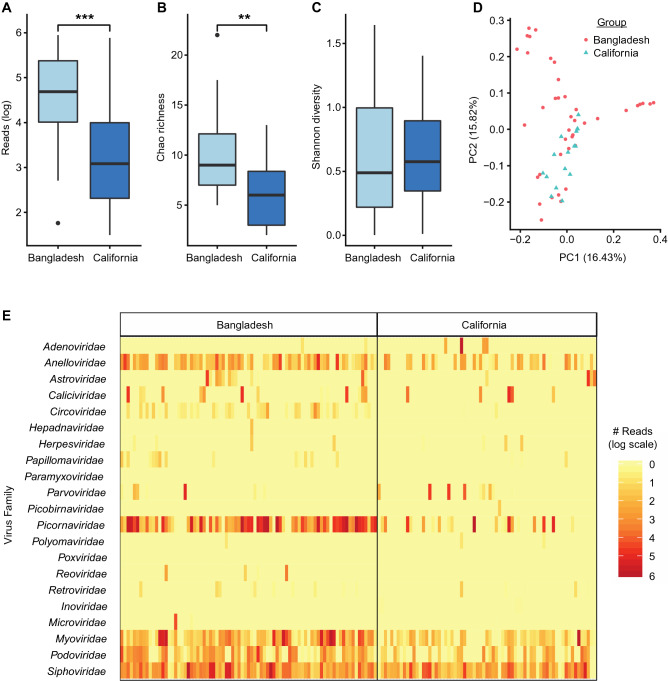
A total of 40 stool samples from the Bangladeshi study were age-matched (12–16 weeks of age) to stool samples collected from 16 California infants. Comparison of the stool samples from Bangladeshi versus California infants demonstrated marked differences in virome composition (p = 0.002 by PERMANOVA analysis; Fig. 5, Supplementary Table S13). Total virome abundance (4.69log10 vs. 3.08log10, p = 0.004) and richness (p = 0.01) were significantly higher in Bangladeshi infants, in large part due to increased abundance (4.22log10 vs. 1.51log10, p < 0.001) and richness (p = 0.003) of the eukaryotic virome. Since OPV is no longer given to US children, poliovirus shedding was exclusively seen in Bangladeshi infants. After removing polioviruses from the analysis, the abundance (3.43log10 vs. 1.51log10, p = 0.003) and richness (p = 0.005) of the non-poliovirus eukaryotic virome remained significantly higher in Bangladeshi infants. In contrast, phage abundance (3.44log10 vs. 3.08log10, p = 0.14), richness (p = 0.10) and alpha diversity (p = 0.27) did not differ significantly between Bangladeshi and US infants.
Figure 5.
Comparison of stool virome in Bangladeshi and California infants. (A) After exclusion of poliovirus reads, virome abundance and (B) richness (Chao) were significantly higher in Bangladeshi infants compared to California (USA) infants (p < 0.001). (C) Alpha diversity (Shannon) was not significantly different between groups (p = 0.27). (D) Principal coordinates analysis of Bray–Curtis dissimilarity shows co-clustering of Bangladeshi and California infants (p = 0.002 by PERMANOVA analysis). (E) Heat map showing distribution of virus families at each geographic site.
Discussion
In this study, we characterized the gut virome of Bangladeshi infants and evaluated its association with poliovirus shedding. Consistent with prevailing hypotheses2,29,30, we found that shedding of enteric viral pathogens is associated with decreased poliovirus shedding. However, unlike prior studies2,23, the observed association was not confined to nonpolio enteroviruses alone2. Bangladeshi infants with decreased or no shedding of poliovirus following vaccination exhibited a higher abundance of eukaryotic viruses overall, particularly viruses that are known causes of gastrointestinal or respiratory illness, than infants with higher rates of shedding.
Here we also found a direct correlation between the degree of poliovirus shedding and the development of neutralizing antibody responses, as previously observed31. Notably, only 28.6% (4 of 14) of infants generated neutralizing antibodies to all three Sabin types in the current study, lower than the 60% (33 of 55) seroconversion rate previously reported from India32 and nearly 100% seroconversion rate in Western populations33. Given the low numbers, the differences in observed seroconversion rate between Indian infants from the 1970’s32 or Bangladeshi infants in the current study are not statistically significant at a p-value threshold of 0.05 (p = 0.0693 by Fisher’s Exact Test). These differences are very likely multi-factorial24 and may reflect, among other possibilities, variability in environmental exposures and/or probiotic administration in our study cohort. Taken together, our results suggest that eukaryotic virome abundance may contribute to the diminished OPV seroconversion rates observed in low socioeconomic settings, such as Bangladesh. However, in the current study, we did not observe a significant association between virome metrics and OPV seroconversion, perhaps due to the low number of samples (n = 14) available for serological analysis.
Compared to similarly aged infants from the United States, the gut virome of Bangladeshi infants relative to that of US children was strikingly more abundant and richer in non-poliovirus eukaryotic viruses. This variation is likely due to a combination of factors, including differences in culture, urbanization, socioeconomic status and geography34. Although viral pathogens were commonly shed, only ~ 36% were detected in symptomatic individuals, underscoring the significant contribution of asymptomatic infection or colonization to the virome. In addition to viruses known to cause respiratory or gastrointestinal illness, nearly a quarter of Bangladeshi infants shed human papillomavirus and one infant had hepatitis B virus identified in stool, suggesting perinatal acquisition. In contrast, the phage virome in Bangladeshi and US infants were comparable in abundance, richness and diversity. Caudoviruses comprised the majority of the phage virome, consistent with prior reports of bacteriophages in breast milk and infant stool and a shift in from Caudovirales-dominated to Microviridae-dominated communities over the first 2 years of life5,35.
This study used a viral metagenomic next-generation sequencing approach to evaluate gut virome associations with poliovirus shedding and oral vaccine response. Current literature evaluating potential interference of oral poliovirus vaccine responses by enteric viruses has relied on targeted molecular or culture-based detection methods, providing only data on a limited number of individual viruses2,29,30. Our data suggest that the abundance and richness of the virome in its entirety may play a more important role in poliovirus vaccine responses than infection by any specific viral pathogen. Metagenomic virome analysis of poliovirus sequences also allowed us to document shedding of multiple poliovirus Sabin serotypes, recover 29 whole poliovirus genome sequences and determine that no vaccine-derived poliovirus variants or vaccine revertants were evident in this cohort.
This study had some limitations. First, a weakness of the study is that no samples were available for analysis before the administration of OPV, so a baseline virome could not be determined. Thus, it is possible that poliovirus replication may also alter the composition of the virome, in addition to the baseline virome potentially affecting the rates of poliovirus shedding. Second, the number of samples and time points remains limited. Testing of infants over a longer period of time and with additional time points would have enhanced power for multiple comparisons and enabled more detailed evaluation of virome dynamics. Associations between maternal, environmental and diet exposures34,36,37 would also more likely be uncovered in a larger study. In addition, variability in sampling times in the setting of an evolving virome may have decreased the power of the study to detect associations between the virome, poliovirus shedding, and OPV seroconversion. Third, potential bias by varying exposure to probiotics and selection of only an available subset (30 of 160) of Bangladeshi infants may have affected the results of the study. Of note, probiotic organisms were cultured only rarely from infant stool, even soon after administration (KJ and JP, personal communication), and it is more likely that the use of probiotics would disproportionately affect the phage rather than eukaryotic virome. Fourth, phage identification was determined using nucleotide similarity to available, taxonomically classified reference sequences in the GenBank database. This likely underestimated the true phage population, as the phage sequence database is incomplete and the vast majority of phages remain unclassified38. Finally, serum samples were also available for only 14 (47%) infants at a single time point, limiting interpretation of our study results; more work is needed to establish the impact of the virome on poliovirus vaccine serological responsiveness over time.
In summary, this study characterizes the gut virome composition in Bangladeshi infants following poliovirus vaccination. Metagenomic virus sequencing revealed dynamic exposures to eukaryotic vertebrate viruses, which were found to be inversely associated with poliovirus shedding. This finding lends support to the premise that the gut virome composition and infection from viruses other than poliovirus may contribute to oral vaccine responsiveness in infants.
Supplementary information
Author contributions
J.P. and C.Y.C. conceived and designed the experiments. A.C.G., J.B., Y.E.H–S., L.G., T.D.H., C.L. M-T.Y., and Y.A.M. performed the experiments. S.K.T., A.C.G., J.B., S.F., D.S., M-T.Y., J.P., and C.Y.C. analyzed the data. R.A., J.P., and C.Y.C. contributed reagents/materials/analysis tools. S.K.T., A.G., J.P., and C.Y.C. wrote the paper. K.J. collected stool samples from Bangladeshi infants for metagenomic analysis, was involved with project implementation, and edited the paper. All authors reviewed the manuscript and agree to its contents.
Funding
The study was funded by National Institutes of Health (NIH) grants R01501HD063142 (to JP), R01-HD008837 (to JP and CYC), the Bill and Melinda Gates Foundation Pilot Grant (to JP and CYC), the Thrasher Research Fund Early Career Award (to YEHS), Global Health Equity Scholars Fellowship (to YEHS), Stanford Child Health Research Institute Postdoctoral Grant through Lucile Packard Foundation for Children’s Health and the Stanford CTSA (UL1 RR025744) (to YEHS) and the TL1 Clinical Research Training Program of the Stanford Clinical and Translational Science Award to Spectrum (NIH TL1 TR 001084) (to YEHS). The research protocol for the Bangladeshi study was funded by Stanford University. icddr,b acknowledges with gratitude the commitment of Stanford University to its research efforts. icddr,b is also grateful to the Governments of Bangladesh, Canada, Sweden and the UK for providing core/unrestricted support.
Data availability
Reads with human sequences removed by Bowtie228 high-sensitivity local alignment to the human genome (GRChg38/hg38 build) have been deposited in the NCBI Sequence Read Archive (SRA) (PRJNA644725). Poliovirus genome sequences have been deposited in NCBI GenBank (MT957178-MT957207).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Susanna K. Tan, Andrea C. Granados, Julie Parsonnet and Charles Y. Chiu.
Supplementary information
is available for this paper at 10.1038/s41598-020-71791-4.
References
- 1.Parker EP, et al. Causes of impaired oral vaccine efficacy in developing countries. Future Microbiol. 2018;13:97–118. doi: 10.2217/fmb-2017-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parker EP, Kampmann B, Kang G, Grassly NC. Influence of enteric infections on response to oral poliovirus vaccine: a systematic review and meta-analysis. J. Infect. Dis. 2014;210:853–864. doi: 10.1093/infdis/jiu182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel M, Steele AD, Parashar UD. Influence of oral polio vaccines on performance of the monovalent and pentavalent rotavirus vaccines. Vaccine. 2012;30(Suppl 1):A30–35. doi: 10.1016/j.vaccine.2011.11.093. [DOI] [PubMed] [Google Scholar]
- 4.Emperador DM, et al. Interference of monovalent, bivalent, and trivalent oral poliovirus vaccines on monovalent rotavirus vaccine immunogenicity in rural Bangladesh. Clin. Infect. Dis. 2016;62:150–156. doi: 10.1093/cid/civ807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim ES, Wang D, Holtz LR. The bacterial microbiome and virome milestones of infant development. Trends Microbiol. 2016;24:801–810. doi: 10.1016/j.tim.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Lim ES, et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat. Med. 2015;21:1228–1234. doi: 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siqueira JD, et al. Complex virome in feces from Amerindian children in isolated Amazonian villages. Nat. Commun. 2018;9:4270. doi: 10.1038/s41467-018-06502-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoy-Schulz YE, et al. Safety and acceptability of Lactobacillus reuteri DSM 17938 and Bifidobacterium longum subspecies infantis 35624 in Bangladeshi infants: a phase I randomized clinical trial. BMC Complement Altern. Med. 2016;16:44. doi: 10.1186/s12906-016-1016-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quick J, et al. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Legoff J, et al. The eukaryotic gut virome in hematopoietic stem cell transplantation: new clues in enteric graft-versus-host disease. Nat Med. 2017;23:1080–1085. doi: 10.1038/nm.4380. [DOI] [PubMed] [Google Scholar]
- 11.Temmam S, et al. Host-associated metagenomics: a guide to generating infectious RNA viromes. PLoS ONE. 2015;10:e0139810. doi: 10.1371/journal.pone.0139810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luk KC, et al. Utility of metagenomic next-generation sequencing for characterization of HIV and human pegivirus diversity. PLoS ONE. 2015;10:e0141723. doi: 10.1371/journal.pone.0141723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reyes GR, Kim JP. Sequence-independent, single-primer amplification (SISPA) of complex DNA populations. Mol Cell Probes. 1991;5:473–481. doi: 10.1016/s0890-8508(05)80020-9. [DOI] [PubMed] [Google Scholar]
- 14.Naccache SN, et al. A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res. 2014;24:1180–1192. doi: 10.1101/gr.171934.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller S, et al. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res. 2019;29:831–842. doi: 10.1101/gr.238170.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaharia M, et al. Alignment in a SNAP: cancer diagnosis in the genomic age. Lab. Invest. 2012;92:458a–458a. doi: 10.1038/labinvest.2011.169. [DOI] [PubMed] [Google Scholar]
- 17.van Rijn, A. L. et al. The respiratory virome and exacerbations in patients with chronic obstructive pulmonary disease. PLoS One14, e0223952, 10.1371/journal.pone.0223952 (2019). [DOI] [PMC free article] [PubMed]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Kearse M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol59, 307–321. 10.1093/sysbio/syq010 (2010). [DOI] [PubMed]
- 22.Garmaeva S, et al. Studying the gut virome in the metagenomic era: challenges and perspectives. BMC Biol. 2019;17:84. doi: 10.1186/s12915-019-0704-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arita M, Iwai M, Wakita T, Shimizu H. Development of a poliovirus neutralization test with poliovirus pseudovirus for measurement of neutralizing antibody titer in human serum. Clin. Vaccine Immunol. 2011;18:1889–1894. doi: 10.1128/CVI.05225-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burrill, C. P., Strings, V. R. & Andino, R. Poliovirus: generation, quantification, propagation, purification, and storage. Curr Protoc Microbiol, Unit 15H 11, 10.1002/9780471729259.mc15h01s29 (2013). [DOI] [PMC free article] [PubMed]
- 25.25R Core Team. R: A Language and Environmental for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria (2018), https://www.R-project.org.
- 26.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 2001;26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B-Stat. Methodol. 1995;57:289–300. [Google Scholar]
- 28.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Praharaj I, et al. Influence of nonpolio enteroviruses and the bacterial gut microbiota on oral poliovirus vaccine response: a study from South India. J. Infect. Dis. 2019;219:1178–1186. doi: 10.1093/infdis/jiy568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taniuchi M, et al. Impact of enterovirus and other enteric pathogens on oral polio and rotavirus vaccine performance in Bangladeshi infants. Vaccine. 2016;34:3068–3075. doi: 10.1016/j.vaccine.2016.04.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giri S, et al. Quantity of vaccine poliovirus shed determines the titer of the serum neutralizing antibody response in indian children who received oral vaccine. J. Infect. Dis. 2018;217:1395–1398. doi: 10.1093/infdis/jix687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.John TJ. Antibody response of infants in tropics to five doses of oral polio vaccine. Br. Med. J. 1976;1:812. doi: 10.1136/bmj.1.6013.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patriarca PA, Wright PF, John TJ. Factors affecting the immunogenicity of oral poliovirus vaccine in developing countries: review. Rev. Infect. Dis. 1991;13:926–939. doi: 10.1093/clinids/13.5.926. [DOI] [PubMed] [Google Scholar]
- 34.Holtz LR, et al. Geographic variation in the eukaryotic virome of human diarrhea. Virology. 2014;468–470:556–564. doi: 10.1016/j.virol.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pannaraj PS, et al. Shared and distinct features of human milk and infant stool viromes. Front. Microbiol. 2018;9:1162. doi: 10.3389/fmicb.2018.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duranti S, et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome. 2017;5:66. doi: 10.1186/s40168-017-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altan E, et al. Enteric virome of Ethiopian children participating in a clean water intervention trial. PLoS ONE. 2018;13:e0202054. doi: 10.1371/journal.pone.0202054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chibani, C. M., Farr, A., Klama, S., Dietrich, S. & Liesegang, H. Classifying the Unclassified: A Phage Classification Method. Viruses11, 10.3390/v11020195 (2019). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Reads with human sequences removed by Bowtie228 high-sensitivity local alignment to the human genome (GRChg38/hg38 build) have been deposited in the NCBI Sequence Read Archive (SRA) (PRJNA644725). Poliovirus genome sequences have been deposited in NCBI GenBank (MT957178-MT957207).