Abstract
Sirtuin 2 (SIRT2) is a member of the sirtuin protein family. It is a Class III histone deacetylase (HDACs) and predominantly localized to the cytosol. SIRT2 deacetylates histones and a number of non-histone proteins and plays a pivotal role in various physiologic processes. Previously, SIRT2 has been considered indispensable during carcinogenesis; however, there is now a significant controversy regarding whether SIRT2 is an oncogene or a tumor suppressor. The purpose of this review is to summarize the physiological functions of SIRT2 and its mechanisms in cancer. We will focus on five malignancies (breast cancer, non-small cell lung cancer, hepatocellular carcinoma, colorectal cancer, and glioma) to describe the current status of SIRT2 research and discuss the clinical evaluation of SIRT2 expression and the use of SIRT2 inhibitors.
Keywords: sirtuins, sirtuin 2, tumor suppressor, oncogene, HDACs
Introduction
Posttranslational modifications fine tune the biological activity of many proteins (1, 2). In recent years, there has been a significant interest in the role of protein acetylation (3). Sirtuins are protein deacetylases, including a family of proteins (SIRT1–7) with homology to the silent information regulator 2 (Sir2) gene in Saccharomyces cerevisiae (4, 5). This family of proteins contains highly conserved enzymes categorized as Class III histone deacetylases (HDACs III), and their deacetylase activity is dependent on nicotinamide adenine dinucleotide (NAD) as a cofactor distinct from zinc-dependent HDACs (6, 7).
Sirtuins are heterogeneous in their subcellular locations, which reflect their broad range of biological functions. SIRT1 is mainly a nucleoprotein, although it can also be found in the cytoplasm (8). SIRT3–5 are constitutively localized to the mitochondria (9). SIRT6 and SIRT7 are also predominantly in the nucleus (10, 11). Sirtuin 2 (SIRT2) is the only sirtuin predominantly found in the cell cytoplasm (7); however, it can shuttle in and out of its primary location, using mechanisms which may be cell and tissue dependent (8, 12). Consistent with its predominant cytosolic location, SIRT2 deacetylates a number of non-histone proteins. Last decade, many new substrates and SIRT2-related proteins had been identified, such as CDK9, PGAM2, Par-3, and CDH1/CDC20, etc. (Table 1) (13–25). These results suggest that SIRT2 regulates multiple biological functions, including neurotoxicity, metabolism, mitosis regulation, genome integrity, oxidative stress, and autophagy (Table 1). Currently, there is growing evidence that abnormal expression of SIRT2 is primarily associated with two human diseases, neurologic diseases, and cancer.
TABLE 1.
The deacetylase substrates of SIRT2.
| Substrates | Site | Mechanism | Functions | References |
| Par-3 AMPAR α-syn | – K813/819/822/868 K6/10 | Active aPKC Degradation Aggregate | Neurogenesis (myelination) Neurogenesis (synaptic plasticity) Neurotoxicity | Beirowski et al. (13) Wang et al. (14) de Oliveira et al. (15) |
| FOXO1 ACLY PEPCK1 | – K540/546/554 K70/71/594 | Inhibition Degradate Stability | Metabolism (Adipogenesis) Metabolism (lipogenesis) Metabolism (Gluconeogenesis) | Jing et al. (16) Lin et al. (17) Jiang et al. (18) |
| CDH1/CDC20 PR-Set7 | K69/159, K66 K90 | Activate APC/C Chromatin localization | Mitosis regulation Mitosis regulation | Kim et al. (19) Serrano et al. (20) |
| CDK9 ATRIP | K48 K32 | Activate Activate ATR | Genome integrity Genome integrity | Zhang et al. (21) Zhang et al. (22) |
| PGAM2 G6PD | K100 K403 | Activate Activate | Oxidative stress Oxidative stress | Xu et al. (23) Wang et al. (24) |
| FOXO1 | K262/265/274 | Unbound to ATG7 | Autophagy | Zhao et al. (25) |
To date, the role of SIRT2 in malignancy has attracted widespread attention, but it is still under debate. There are two opposing viewpoints that support SIRT2 functioning as an oncogene and a tumor suppressor. Based on the existing research, this review summarizes the physiological functions of SIRT2 and its mechanisms in cancer. We will focus on five malignancies, breast cancer, non-small cell lung cancer, hepatocellular carcinoma (HCC), colorectal cancer, and glioma, in which the pattern of SIRT2 expression and its physiologic functions are controversial.
Physiologic Functions of SIRT2
SIRT2 and the Nervous System
Among all sirtuins, SIRT2 is the most highly expressed in brain tissue, particularly in the cortex, striatum, spinal cord, and postnatal hippocampus (26, 27), indicating that SIRT2 is involved in neural development. Several studies have reported that SIRT2 is crucial for myelination whether in the central nervous system (CNS) or peripheral nervous system. In the CNS, SIRT2 is mainly expressed in oligodendrocytes (OLs) and is considerably upregulated during OL differentiation and myelination (28, 29). In the peripheral nervous system, SIRT2 ablation in mouse Schwann cells (SCs) delayed myelin formation and postinjury remyelination (13, 30). Moreover, SIRT2 is involved in other developmental processes in the nervous system as SIRT2 gene knockout mice demonstrated dysfunctions of the nervous system, such as defects in differentiation of dopaminergic (DA) neurons (31), aberrant synaptic plasticity with impaired learning and memory (14), and morphological changes of mitochondria in the cortex (32).
In addition to act as a crucial regulator in neurodevelopment, SIRT2 is also associated with nervous system disorders, in particular, neurodegenerative diseases [Parkinson’s disease (PD), Alzheimer’s disease (AD), and Huntington’s disease (HD)] (33). SIRT2 expression participates in the aggregation process of proteins such as α-synuclein (α-syn) (15), huntingtin (34), as well as amyloid-β peptide (Aβ), and hyperphosphorylated tau protein (35, 36), involved in PD, HD, and AD, respectively. Mounting evidence showed that inhibition of SIRT2 function, either pharmacologically or genetically, provided neuroprotection in a variety of mice modals, suggesting that SIRT2 could be a potential therapeutic target for these diseases (37–39). The association between SIRT2 and neuromalignance will be discussed.
SIRT2 and Metabolism
The potential roles and effects of SIRT2 to maintain metabolic homeostasis have been recognized more recently. SIRT2 is the most prominently expressed sirtuin in the adipose tissue both in vivo and in culture (40), implicating its involvement in lipid metabolism, adipogenesis, lipid synthesis, and fatty acid oxidation. In mouse 3T3-L1 preadipocytes, SIRT2 deacetylates the nuclear transcription factor FOXO1, which results in nuclear retention of this protein where it represses the transcription of PPARγ (peroxisome proliferator-activated receptor γ), culminating in the inhibition of adipocyte differentiation (16). Another study showed the role of SIRT2 in the regulation of lipid synthesis. Under high-glucose conditions, SIRT2 deacetylates ATP-citrate lyase (ACLY), a lipogenic enzyme, leading to its ubiquitylation, and degradation (17). Krishnan and colleagues showed that fat cell-specific HIF-1α inactivation in obese mice causes accumulation of nuclear SIRT2, which deacetylates PGC-1α, thereby, promoting fatty acid oxidation (41).
Regarding glucose metabolism, SIRT2 regulates gluconeogenesis via deacetylating PEPCK1, which catalyzes the first rate-limiting step of gluconeogenesis. When glucose level is high, PEPCK1 acetylation increases, which promotes its interaction with the UBR5 E3 ubiquitin ligase and proteosomal degradation, thus, suppressing gluconeogenesis. In contrast, when glucose level is low, PEPCK1 is deacetylated by SIRT2, and its stabilization enhances gluconeogenesis (18).
SIRT2 and the Cell Cycle
Sirt2 colocalizes with microtubules and deacetylates α-tubulin (7). However, during the G2/M transition of the cell cycle, Sirt2 can transiently migrate to the nucleus to deacetylate histone H.4 lysine 16 (H4K16Ac) (8, 12), thereby regulating chromosomal condensation during mitosis. Moreover, cells with SIRT2 overexpression exhibit marked prolongation of the mitotic phase in vitro (12, 42). These results suggest a role for SIRT2 in regulating mitotic processes.
Investigation of Sirt2–/– mice ultimately uncovered the mechanism via which SIRT2 regulates mitosis. Sirt2–/– cells displayed widespread centrosome amplification and aneuploidy, which resulted in genetic instability and abnormal mitosis both in vitro and in vivo (19, 20). Indeed, Kim et al. reported that anaphase-promoting complex/cyclosome (APC/C), an E3 ubiquitin ligase with multiple subunits, which mediates ubiquitination of key regulators of the cell cycle, is positively regulated by SIRT2 through deacetylation of its coactivators, CDH1, and CDC20 (19). SIRT2 deficiency causes hyperacetylation of CDH1 and CDC20, impaired activity of APC/C, and hence upregulation of Aurora-A levels, which consequently lead to abnormalities in mitosis. Serrano et al. provided another possible mechanism (20). SIRT2 deacetylation of K90 residue of PR-Set7 modulates its chromatin localization. Consistently, SIRT2 depletion significantly reduced PR-Set7 chromatin levels, altering the size and number of PR-Set7 foci to decrease the overall mitotic deposition of H4K20me1.
Furthermore, other studies have shown that SIRT2 regulates genome integrity through deacetylation of CDK9 (21) or ataxia telangiectasia-mutated and Rad3-related (ATR)-interacting protein (ATRIP) (22). Overall, the crucial role of SIRT2 in mitosis regulation and genome integrity implies that its activity may have a significant effect on cancer, a disease with high genetic instability and abnormal mitosis (19).
SIRT2 and Cancer
In recent years, a growing body of evidence has proposed a role for SIRT2 in tumorigenesis. Because SIRT2 is expressed in a wide range of tissues and organs and exerts variable physiological functions, its role in cancers is complicated. Notably, SIRT2 has been described as both an oncogene and a tumor suppressor. In this review, we will discuss the divergent expression and function of SIRT2 in five malignancies: breast cancer, non-small cell lung cancer, HCC, colorectal cancer, and glioma.
Breast Cancer
Molecular Pathways Targeted by SIRT2
The role of SIRT2 in tumorigenesis has been extensively studied in breast cancer. Kim et al. observed a significant propensity of Sirt2–/– female mice to develop mammary tumors at an old age, suggesting a role for SIRT2 as a tumor suppressor (19). As discussed earlier, a possible mechanism proposed for this phenotype is Sirt2 regulation of APC/C activity through CDH1 and CDC20. SIRT2 deficiency causes increased levels of Aurora-A and, consequently, contribute to centrosome amplification, aneuploidy, genomic instability, mitotic cell death, and most importantly, spontaneous tumor formation. Serrano et al. used a skin papilloma model to further study the role of SIRT2 in tumorigenesis (20). Following DMBA/TPA treatment on the skin, Sirt2–/– mice developed larger papilloma at higher frequencies than wild-type mice. Histopathological study demonstrated that most of the papilloma developed into squamous cell carcinoma and fibrosarcoma.
Park et al. further investigated SIRT2 function using Sirt2–/– mammary tumor cell line (MMT) derived from the spontaneous mammary tumors in Sirt2–/– mice (43). They identified the M2 isoform of pyruvate kinase (PKM2) as a critical target of SIRT2. Indeed, loss of SIRT2 altered PKM2 activity and reprogrammed glycolytic metabolism in cancer cells, which was associated with increased tumorigenesis in Sirt2–/– mice.
Although genetic studies tend to suggest that SIRT2 is a tumor suppressor, pharmacological studies suggest the opposite. Jing et al. developed a potent SIRT2-specific inhibitor, TM (a thiomyristoyl lysine compound), which demonstrated a broad anticancer activity, including activity against several breast cancer cell lines. The study proposed that SIRT2 inhibition promotes NEDD4 expression, an E3 ubiquitin ligase for c-Myc, and thus causing c-Myc ubiquitination and degradation (44). Shah et al. developed another inhibitor of SIRT2 (RK-9123016), which also reduced the viability of human breast cancer cells via downregulated c-Myc expression. This drug was found to increase the acetylation level of eIF5A (eukaryotic translation initiation factor 5A), another physiological substrate of SIRT2 (45).
Similarly, studies focused on CSCs (cancer stem cells) and BLBC (basal-like breast cancer) suggest SIRT2 as an oncogene. CSCs are believed to contribute to tumor metastasis and poor prognosis. Notably, Zhao et al. demonstrated that SIRT2 protein levels were significantly increased in ALDH1+ CSCs isolated from primary human breast tumors compared with the levels in ALDH1– cells. In addition, they demonstrated NOTCH-induced SIRT2 deacetylation activity on K353 of ALDH1A1, which led to its enzymatic activation to maintain breast CSCs (46).
Basal-like breast cancer represents one of the most aggressive breast cancer subtypes characterized by increased propensity for metastasis and poor prognosis. SIRT2 has been reported to be frequently amplified and highly expressed in BLBC. Slug protein has been found to be a deacetylase target of SIRT2, and SIRT2 overexpression promoted Slug stability, thus, conferring aggressive, basal-like malignant features, and growth. By contrast, genetic depletion and pharmacological inactivation of SIRT2 in BLBC cells reversed Slug stabilization and, thus, abrogated relevant pathological features of BLBC and inhibited tumor growth (47). Another study using MDA-MB-231 cell (a BLBC cell line) demonstrated that SIRT2 silenced Arrestin domain-containing 3 (ARRDC3), a tumor suppressor, contributing to the aggressive nature of BLBC cells (48).
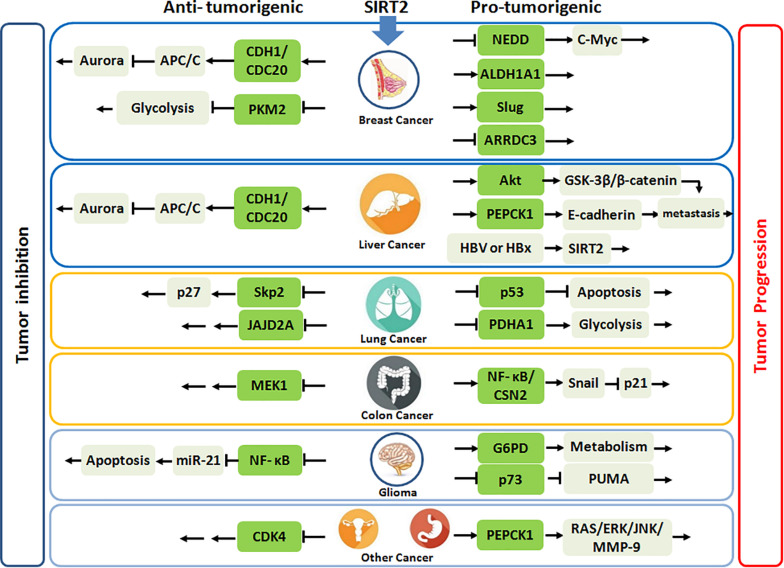
Taken together, SIRT2 may have a significant tumor suppressive role during early carcinogenesis of breast cancer, but conversely in advanced cancer, its overexpression portends more aggressive phenotype, and SIRT2 inhibition has anticancer activities. Importantly, SIRT2 functions may evolve from tumor initiation to progression with altered physiologic targets (Figure 1).
FIGURE 1.
SIRT2 plays the anti-tumorigenic or pro-tumorigenic roles in various malignances. Highlighted green frames represent substrates or partners of SIRT2.
SIRT2 Expression and the Clinical Outcome
Several studies focused on the clinical relevance of SIRT2 expression in breast carcinoma. Following the initial studies with Sirt2–/– mice, Kim et al. investigated SIRT2 expression levels in human breast cancer samples. Using a tissue array with 36 paired samples of breast cancer and adjacent normal breast tissue, they demonstrated significantly higher levels of SIRT2 in normal breast tissues compared with cancer tissues. Furthermore, lower SIRT2 expression was observed in metastatic samples suggesting that SIRT2 downregulation may be associated with more aggressive phenotype in breast cancer. This result was supported by Shi et al. who demonstrated that high expression of SIRT2 by IHC (IHC score > 3) was lower in tumor tissues compared to the normal adjacent tissues in 296 patients (49). Similarly, McGlynn et al. detected that SIRT2 transcripts were significantly lower in malignant breast tissues in comparison to non-malignant or normal breast tissue (50). These studies suggested that SIRT2 may act as a tumor suppressor in breast cancer.
Although SIRT2 expression was lower in breast cancer than in normal tissue, residual SIRT2 expression was generally observed. In Kim’s study (19), among 36 cancer tissues tested, 66.7% of the tissues maintained low or intermediate levels of SIRT2 expression. Interestingly, when McGlynn et al. investigated the correlation between SIRT2 expression levels in breast cancer and prognosis using IHC staining in 153 ER– and 392 ER+ breast cancer tissues, they observed more aggressive breast cancer phenotype with higher nuclear levels of SIRT2 (50). Indeed, in the ER– cohort with approximately 80% grade 3 tumors, they observed that high SIRT2 nuclear levels were associated with shorter time to relapse and death compared to low SIRT2 nuclear expression. Similar results also emerged in grade 3 tumors of ER+ cohort. These results highlight that SIRT2 nuclear expression is associated with poor prognosis in advanced breast cancer.
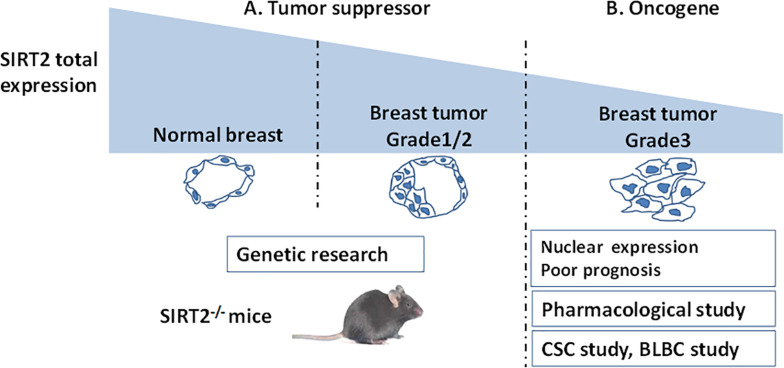
Taken together, a lower expression of SIRT2 in breast cancer compared with that in normal breast indicates that SIRT2 might act as a tumor suppressor at the initiation of tumorigenesis. However, higher SIRT2 expression in advanced tumor tissues portend poor prognosis underscoring that SIRT2 may act as an oncogene during tumor progression (Figure 2 and Table 2).
FIGURE 2.
A schema shows SIRT2’s functions evaluated by different breast cancer studies. (A) SIRT2 is highly expressed in the normal breast compared with breast tumor. This clinical result suggest that SIRT2 might act as a tumor suppressor, which is supported by Genetic research (SIRT2 Knockout). (B) However, in Grade 3 breast cancer, high levels of nuclear SIRT2 are associated with poor clinical outcome, indicating that SIRT2 might act as an oncogene in more aggressive malignancies. The studies on CSC and BLBC support this inference. Pharmacological research of SIRT2 specific inhibitor also finds the oncogenic role of SIRT2.
TABLE 2.
The clinical role of SIRT2 in five malignancies.
| Malignance | Method of study | Role of SIRT2 | References |
| Breast cancer | IHC IHC | Tumor suppressor Tumor suppressor | Kim et al. (19) Shi et al. (49) |
| qPCR IHC | Tumor suppressor Oncogene (nuclear expression) | McGlynn et al. (50) | |
| Hepatocellular carcinoma | Microarray qPCR, IHC IHC, database | Tumor suppressor Oncogene Oncogene | Kim et al. (19) Chen et al. (51) Huang et al. (52) |
| NSCLC | qPCR, WB, IHC IHC | Tumor suppressor Oncogene | Li et al. (54) Grbesa et al. (57) |
| Colorectal cancer | IHC WB, database | Tumor suppressor Oncogene | Zhang et al. (63) Hu et al. (67) |
| Glioma | qPCR, WB Northern blot | Tumor suppressor Tumor suppressor | Li et al. (70) Hiratsuka et al. (69) |
| IHC | Oncogene (nuclear expression) | Imaoka et al. (73) |
Liver Cancer
Molecular Pathways Targeted by SIRT2
Kim et al. also reported increased the development of HCC in old male Sirt2–/– mice via similar mechanisms observed for breast cancer (19). However, Chen et al. demonstrated that depletion of SIRT2 in human HCC cell lines markedly reduced cell migration with a regression of epithelial–mesenchymal transition (EMT) phenotypes (51). Ectopic SIRT2 expression in L02 cell line was found to promote cell motility and invasiveness. Notably, SIRT2 deletion increased Akt acetylation, thereby, impairing Akt/GSK-3β/β-catenin-signaling cascade to regulate EMT and cell migration. In another study, Huang et al. also found that downregulation of SIRT2 reduced migration and invasion in human HCC cells, but revealed that SIRT2 inhibition increased PEPCK1 acetylation and suppressed its downstream E-cadherin pathway (52). Interestingly, Cheng et al. revealed that hepatitis B (HBV) or HBx upregulates SIRT2 expression by targeting its promoter, which then enhanced transformation of HBV-related HCC (53). Overall, these data suggest that SIRT2 upregulation is associated with malignant transformation in the liver (Figure 1).
SIRT2 Expression and the Clinical Outcome
Kim et al. also analyzed SIRT2 expression using a microarray containing 264 human HCC samples (19). They found that many HCCs demonstrated lower levels of SIRT2 than normal liver tissue. Contrary to this finding, Chen et al. found that SIRT2 mRNA is expressed at a similar level in tumor and adjacent tissues in 45 HCC samples (51). Interestingly, SIRT2 protein was found to be expressed at a higher level in tumors (23/45) based on Western blot (WB). Overexpression of SIRT2 in primary HCC tumors was associated with increased microscopic vascular invasion and poor prognosis. Huang et al. also found that SIRT2 was significantly increased in tumor tissues than in normal adjacent tissues in tissue microarrays containing 52 HCC samples. They also queried the TCGA database, which contained clinically annotated genomic data from 286 HCC samples, and found that a higher SIRT2 level in HCC was detrimental to patient survival (52). The data from the latter two studies with both mRNA and protein detection suggest that SIRT2 expression is a negative prognostic factor (Table 2).
Lung Cancer
Molecular Pathways Targeted by SIRT2
Li et al. reported that overexpression of SIRT2 in A549 and H1299 cells was associated with delayed cell proliferation, increased apoptosis, and cell cycle arrest (54). The follow-up study revealed that overexpression of SIRT2 promoted Skp2 deacetylation and degradation, and eliminated the effect of Skp2 on p27, which resulted in an increase in p27 and suppression of NSCLC cell growth (55). Similarly Xu et al. demonstrated that SIRT2 bound to the promoter region of JMJD2A and negatively regulated JMJD2A expression, which led to the inhibition of NSCLC cell proliferation, colony formation, and tumor growth both in vitro and in vivo (56).
In contrast, Grbesa et al. reported that SIRT2 downregulation significantly decreased proliferation in NSCLC cell lines (57). Further, Hoffmann et al. identified two structurally related compounds, which selectively inhibited SIRT2, AEM1, and AEM2, and both sensitized NSCLC cells to etoposide, which damages DNA by targeting topoisomerase II. These inhibitors also increased p53 acetylation and activated p53-dependent apoptosis in NSCLC cells lines (58). Yang et al. developed N-(3-(phenoxymethylphenyl) acetamide derivatives as potent, selective SIRT2 inhibitors. Among the derivatives, 24A strongly restrained cell growth and suppressed NSCLC cell (H441) migration and invasion (59). Ma et al. combined dichloroacetic acid (DCA, a pyruvate dehydrogenase kinase inhibitor) with Sirtinol or AGK2 (SIRT2 inhibitors) to treat lung cancer cell lines and found that this combination produced a synergistic antitumor effect via activation of PDHA1 to shift metabolism from glycolysis to OXPHOS and enhance ROS generation and activation of AMPK signaling (60). Overall, there is significant discordance among studies on NSCLC, which may be related to the specific cellular contexts of NSCLC cell lines tested (Figure 1).
SIRT2 Expression and the Clinical Outcome
Li et al. evaluated SIRT2 expression in eight pairs of NSCLC and adjacent normal tissue samples by Q-PCR and WB, which demonstrated significantly lower expression of SIRT2 in tumor. This observation was reproduced in 53 paired NSCLC and normal lung tissue analyzed by microarray (54).
However, when Grbesa et al. evaluated SIRT2 protein levels in a cohort of 105 NSCLC patients using IHC, its expression, mostly confined within the cytosol, was significantly higher in primary tumors than in normal tissue. Further, patients with high levels of SIRT2 showed significantly shorter recurrence-free survival than patients with low levels (57).
These conflicting results may be due to histologic variability among NSCLC cells, which are subdivided into adenocarcinoma, squamous cell carcinoma, large-cell carcinoma, bronchoalveolar lung cancer, and mixed histologic types (e.g., adenosquamous) (61), with distinct biologic behaviors and outcome (62). Alternatively, multiple competing mechanisms may exist in NSCLC, which are preferentially elicited by SIRT2 depending on the cellular context (Table 2).
Colon Cancer
Molecular Pathways Targeted by SIRT2
Zhang et al. reported that overexpression of endogenous SIRT2 reduced migration and invasion of SW480 cells. Blocking SIRT2 expression induced the proliferation and metastatic progression of HT29 cells (63). Bajpe et al. also illustrated SIRT2 as a determinant of response to EGFR inhibitors in colon cancer (64). SIRT2 can inhibit MEK1 activation by deacetylating MEK1 and keep a check on cell proliferation. In turn, loss of SIRT2 led to increased MEK1 acetylation and its kinase activity, thus, attenuating the response to upstream inhibition of either EGFR or BRAF. These results delineate SIRT2 as a tumor suppressor in CRC.
In contrast, Cheon et al. reported that treatment of human colon cancer cells with the SIRT2-specific inhibitor, AK-1, which inactivates the NFκB/CSN2 pathway to induce proteasomal degradation of Snail, upregulated p21 to induce G1 arrest and delayed proliferation (65). Farooqi et al. presented novel lysine-based thiourea compounds as potent and selective SIRT2 inhibitors that were less hydrophobic and easier to synthesize than TM (44), which potently inhibited tumor growth in an HCT116 xenograft murine model, supporting a role for SIRT2 as a viable therapeutic target for CRC (66) (Figure 1).
SIRT2 Expression and the Clinical Outcome
Zhang et al. revealed that SIRT2 was downregulated in CRC biopsy samples (n = 31, not paired) compared with the normal adjacent tissues (n = 26). SIRT2 immunostaining was largely localized to the cytoplasm of colonic epithelial cells. Interestingly, decreased SIRT2 expression was associated with adverse clinicopathological features and poor prognosis in colon cancer (63).
In contrast, Hu et al. utilized the Oncomine database to evaluate the expression of SIRT2 in CRC and found that the level of SIRT2 was higher in CRC tissues compared to the normal tissue samples (67). This finding was verified by protein expression using WB (n = 12) and IHC (n = 46). In the CRC dataset GSE24551, SIRT2 upregulation correlated with advanced TNM stage and a lower 5-year survival rate. A similar survival outcome was observed in another online database1.
Taken together, there are potentially opposite roles of SIRT2 in CRC. The limitation of these studies is the relatively low number of clinical specimens used. Also, online databases represent mRNA expression rather than protein levels (Table 2).
Glioma
Molecular Pathways Targeted by SIRT2
Sirtuin 2 is highly expressed in brain tissue and plays a crucial role in the development of the nervous system and neurodegenerative diseases (68). In addition, SIRT2 is located at 19q13.2, a region known to be frequently deleted in human glioma (69), thereby indicating that SIRT2 may be a tumor suppressor in gliomas. Li et al. showed that Sirt2 overexpression reduced glioma cell proliferation and significantly activated proapoptotic proteins caspase 3 and Bax while inhibiting the antiapoptotic protein Bcl-2. Mechanically, Sirt2 deacetylated p65 at K310, and blocked p65 binding to the promoter region of miR-21, thus, suppressing the miR-21-modulated apoptosis pathway (70).
However, Ye et al. revealed that HSPB1 enhanced the binding between G6PD and SIRT2, which led to deacetylation and activation of G6PD, thus promoting cellular NADPH and pentose production in glioma cells and, thereby, protecting cells from oxidative and DNA damage stress (71). Further, Funato et al. demonstrated that SIRT2 deacetylated C-terminal lysine residues of p73 and inactivated its transcriptional activity, and SIRT2 inhibition in glioblastoma cell lines (GB2 or GB16) resulted in p73-mediated transactivation of PUMA and induction of apoptosis (72) (Figure 1).
SIRT2 Expression and the Clinical Outcome
Li et al. reported that SIRT2 mRNA and protein expression was downregulated in eight primary glioma samples (grade II n = 2, grade III n = 5, and grade IV n = 1) versus four normal brain tissue samples tested. Protein and mRNA levels of SIRT2 in five human glioma cell lines (T98G, U87MG, U251, A172, and CCF-STTG1) were also lower than two normal human astrocyte cell lines (NHA and HA) (70).
Using a 2D-proteomics technique, Hiratsuka et al. identified SIRT2 downregulation in glioma tissue when compared to normal adjacent tissue. Northern blot analysis also revealed that RNA expression of SIRT2 was dramatically diminished in 12 out of 17 gliomas. Ectopic expression of SIRT2 in glioma cell lines disrupted the microtubule network causing a remarkable reduction in colony formation (69). However, investigation of SIRT2 expression by IHC in samples from 23 patients with glioblastoma (grade IV), eight patients with diffuse astrocytoma (grade II), and five healthy individuals revealed that SIRT2 preferential nuclear localization was more frequent in the malignant specimens, which was positively correlated with malignant progression (73). Therefore, SIRT2 nuclear translocation may be associated with its oncogenic effects in glioma (Table 2).
Other Cancers
Dysregulated SIRT2 expression has been observed in multiple other cancers with clear clinical relevance, including renal cell carcinoma (RCC), gastric cancer, and melanoma, while SIRT2 appears downregulated in ovarian carcinoma and prostate cancer (Figure 1). Wei et al. demonstrated that SIRT2 was highly expressed in the stem-like RCC cells with adverse clinical outcome (74). In gastric cancer, SIRT2 had been found to be upregulated compared to adjacent normal tissues and also correlated with poor patient survival. Mechanically, SIRT2 altered PEPCK1 activity, and mitochondrial respiration while inducing gastric cancer cell migration and invasion by activating the RAS/ERK/JNK/MMP-9 pathway (75). SIRT2 upregulation was also found in metastatic melanoma with a predominant nuclear staining (76).
In contrast, SIRT2 was significantly downregulated in serous ovarian carcinoma (SOC) when compared with ovarian surface epithelium. Downregulated SIRT2 failed to repress CDK4 expression, eventually leading to accelerated SOC cell proliferation (77). SIRT2 loss also correlated with aggressive prostate cancer. In multiple datasets, decreased SIRT2 expression portended worse clinicopathologic outcomes (78).
Conclusion
The expression pattern of SIRT2 and its correlation with clinical outcome is variable with opposite reports (Table 2). Inconsistent results from clinical studies raise the utility of SIRT2 expression as a biomarker. To minimize variability, we suggest determining the subcellular localization of SIRT2 since nuclear or cytoplasmic expression may have different functions. Additionally, cancer subtypes should be fully considered as SIRT2 might have distinct functions in different oncopathological conditions. Because of variability involved, clinical studies evaluating SIRT2 require large sample sizes, selection of highly specific antibodies, and preferably multicenter collaboration.
The plethora of SIRT2 substrates identified reflect cell or tissue-specific functions of SIRT2, which may evolve during different phases of malignant transformation. The role of SIRT2 in cancer is thus complex with multiple competing mechanisms, and therefore, SIRT2 cannot be simply considered as an oncogene or a tumor suppressor.
Author Contributions
LZ and XR conceived and designed the review. LZ wrote the manuscript. SK reviewed and edited the manuscript. All authors read and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by grants from the National Key R&D Program (2018YFC1313400), the Natural Science Foundation of China (No. 81872166), and K08 CA194273.
References
- 1.Jensen ON. Interpreting the protein language using proteomics. Nat Rev Mol Cell Biol. (2006) 7:391–403. 10.1038/nrm1939 [DOI] [PubMed] [Google Scholar]
- 2.Westermann S, Weber K. Post-translational modifications regulate microtubule function. Nat Rev Mol Cell Biol. (2003) 4:938–47. 10.1038/nrm1260 [DOI] [PubMed] [Google Scholar]
- 3.Gil J, Ramirez-Torres A, Encarnacion-Guevara S. Lysine acetylation and cancer: a proteomics perspective. J Proteomics. (2017) 150:297–309. 10.1016/j.jprot.2016.10.003 [DOI] [PubMed] [Google Scholar]
- 4.Rine J, Strathern JN, Hicks JB, Herskowitz I. A suppressor of mating-type locus mutations in Saccharomyces cerevisiae: evidence for and identification of cryptic mating-type loci. Genetics. (1979) 93:877–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomes P, Fleming Outeiro T, Cavadas C. Emerging role of Sirtuin 2 in the regulation of mammalian metabolism. Trends Pharmacol Sci. (2015) 36:756–68. 10.1016/j.tips.2015.08.001 [DOI] [PubMed] [Google Scholar]
- 6.Lawson M, Uciechowska U, Schemies J, Rumpf T, Jung M, Sippl W. Inhibitors to understand molecular mechanisms of NAD(+)-dependent deacetylases (sirtuins). Biochim Biophys Acta. (2010) 1799:726–39. 10.1016/j.bbagrm.2010.06.003 [DOI] [PubMed] [Google Scholar]
- 7.Wilking-Busch MJ, Ndiaye MA, Liu X, Ahmad N. RNA interference-mediated knockdown of SIRT1 and/or SIRT2 in melanoma: identification of downstream targets by large-scale proteomics analysis. J Proteomics. (2018) 170:99–109. 10.1016/j.jprot.2017.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. (2007) 282:6823–32. 10.1074/jbc.m609554200 [DOI] [PubMed] [Google Scholar]
- 9.Huang JY, Hirschey MD, Shimazu T, Ho L, Verdin E. Mitochondrial sirtuins. Biochim Biophys Acta. (2010) 1804:1645–51. [DOI] [PubMed] [Google Scholar]
- 10.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. (2006) 124:315–29. 10.1016/j.cell.2005.11.044 [DOI] [PubMed] [Google Scholar]
- 11.Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. (2006) 20:1075–80. 10.1101/gad.1399706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.North BJ, Verdin E. Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS One. (2007) 2:e784. 10.1371/journal.pone.0000784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beirowski B, Gustin J, Armour SM, Yamamoto H, Viader A, North BJ, et al. Sir-two-homolog 2 (Sirt2) modulates peripheral myelination through polarity protein Par-3/atypical protein kinase C (aPKC) signaling. Proc Natl Acad Sci USA. (2011) 108:E952–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang G, Li S, Gilbert J, Gritton HJ, Wang Z, Li Z, et al. Crucial roles for SIRT2 and AMPA receptor acetylation in synaptic plasticity and memory. Cell Rep. (2017) 20:1335–47. 10.1016/j.celrep.2017.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Oliveira RM, Vicente Miranda H. The mechanism of sirtuin 2-mediated exacerbation of alpha-synuclein toxicity in models of Parkinson disease. PLoS Biol. (2017) 15:e2000374. 10.1371/journal.pbio.2000374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jing E, Gesta S, Kahn CR. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. (2007) 6:105–14. 10.1016/j.cmet.2007.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin R, Tao R, Gao X, Li T, Zhou X, Guan KL, et al. Acetylation stabilizes ATP-citrate lyase to promote lipid biosynthesis and tumor growth. Mol Cell. (2013) 51:506–18. 10.1016/j.molcel.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, et al. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell. (2011) 43:33–44. 10.1016/j.molcel.2011.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim HS, Vassilopoulos A, Wang RH, Lahusen T, Xiao Z, Xu X, et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell. (2011) 20:487–99. 10.1016/j.ccr.2011.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serrano L, Martinez-Redondo P, Marazuela-Duque A, Vazquez BN, Dooley SJ, Voigt P, et al. The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev. (2013) 27:639–53. 10.1101/gad.211342.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H, Park SH, Pantazides BG, Karpiuk O, Warren MD, Hardy CW, et al. SIRT2 directs the replication stress response through CDK9 deacetylation. Proc Natl Acad Sci USA. (2013) 110:13546–51. 10.1073/pnas.1301463110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Head PE, Daddacha W, Park SH, Li X, Pan Y, et al. ATRIP deacetylation by SIRT2 drives ATR checkpoint activation by promoting binding to RPA-ssDNA. Cell Rep. (2016) 14:1435–47. 10.1016/j.celrep.2016.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, Li F, Lv L, Li T, Zhou X, Deng CX, et al. Oxidative stress activates SIRT2 to deacetylate and stimulate phosphoglycerate mutase. Cancer Res. (2014) 74:3630–42. 10.1158/0008-5472.can-13-3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang YP, Zhou LS, Zhao YZ, Wang SW, Chen LL, Liu LX, et al. Regulation of G6PD acetylation by SIRT2 and KAT9 modulates NADPH homeostasis and cell survival during oxidative stress. EMBO J. (2014) 33:1304–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y, Wang L, Yang J, Zhang P, Ma K, Zhou J, et al. Anti-neoplastic activity of the cytosolic FoxO1 results from autophagic cell death. Autophagy. (2010) 6:988–90. 10.4161/auto.6.7.13289 [DOI] [PubMed] [Google Scholar]
- 26.Pandithage R, Lilischkis R, Harting K, Wolf A, Jedamzik B, üscher-Firzlaff JL, et al. The regulation of SIRT2 function by cyclin-dependent kinases affects cell motility. J Cell Biol. (2008) 180:915–29. 10.1083/jcb.200707126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maxwell MM, Tomkinson EM, Nobles J, Wizeman JW, Amore AM, Quinti L, et al. The Sirtuin 2 microtubule deacetylase is an abundant neuronal protein that accumulates in the aging CNS. Hum Mol Genet. (2011) 20:3986–96. 10.1093/hmg/ddr326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W, Zhang B, Tang J, Cao Q, Wu Y, Wu C, et al. Sirtuin 2, a mammalian homolog of yeast silent information regulator-2 longevity regulator, is an oligodendroglial protein that decelerates cell differentiation through deacetylating alpha-tubulin. J Neurosci. (2007) 27:2606–16. 10.1523/jneurosci.4181-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji S, Doucette JR, Nazarali AJ. Sirt2 is a novel in vivo downstream target of Nkx2.2 and enhances oligodendroglial cell differentiation. J Mol Cell Biol. (2011) 3:351–9. 10.1093/jmcb/mjr009 [DOI] [PubMed] [Google Scholar]
- 30.Nagarajan R, Le N, Mahoney H, Araki T, Milbrandt J. Deciphering peripheral nerve myelination by using Schwann cell expression profiling. Proc Natl Acad Sci USA. (2002) 99:8998–9003. 10.1073/pnas.132080999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szegõ ÉM, Gerhardt E, Outeiro TF. Sirtuin 2 enhances dopaminergic differentiation via the AKT/GSK-3β/β-catenin pathway. Neurobiol Aging. (2017) 56:7–16. 10.1016/j.neurobiolaging.2017.04.001 [DOI] [PubMed] [Google Scholar]
- 32.Liu TM, Shyh-Chang N. SIRT2 and glycolytic enzyme acetylation in pluripotent stem cells. Nat Cell Biol. (2017) 19:412–4. 10.1038/ncb3522 [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Yang J, Hong T, Chen X, Cui L. SIRT2: controversy and multiple roles in disease and physiology. Ageing Res Rev. (2019) 55:100961. 10.1016/j.arr.2019.100961 [DOI] [PubMed] [Google Scholar]
- 34.Quinti L, Casale M, Moniot S, Pais TF, Van Kanegan MJ, Kaltenbach LS, et al. SIRT2- and NRF2-Targeting Thiazole-containing compound with therapeutic activity in Huntington’s disease models. Cell Chem Biol. (2016) 23:849–61. 10.1016/j.chembiol.2016.05.015 [DOI] [PubMed] [Google Scholar]
- 35.Biella G, Fusco F, Nardo E, Bernocchi O, Colombo A, Lichtenthaler SF, et al. Sirtuin 2 inhibition improves cognitive performance and acts on amyloid-β protein precursor processing in two Alzheimer’s disease mouse models. J Alzheimers Dis. (2016) 53:1193–207. 10.3233/jad-151135 [DOI] [PubMed] [Google Scholar]
- 36.Garske AL, Smith BC, Denu JM. Linking SIRT2 to Parkinson’s disease. ACS Chem Biol. (2007) 2:529–32. 10.1021/cb700160d [DOI] [PubMed] [Google Scholar]
- 37.Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science. (2007) 317:516–9. 10.1126/science.1143780 [DOI] [PubMed] [Google Scholar]
- 38.Spires-Jones TL, Fox LM, Rozkalne A, Pitstick R, Carlson GA, Kazantsev AG. Inhibition of Sirtuin 2 with Sulfobenzoic acid derivative AK1 is non-toxic and potentially neuroprotective in a mouse model of frontotemporal dementia. Front Pharmacol. (2012) 3:42. 10.3389/fphar.2012.00042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bobrowska A, Donmez G, Weiss A, Guarente L, Bates G. SIRT2 ablation has no effect on tubulin acetylation in brain, cholesterol biosynthesis or the progression of Huntington’s disease phenotypes in vivo. PLoS One. (2012) 7:e34805. 10.1371/journal.pone.0034805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang F, Nguyen M, Qin FX, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. (2007) 6:505–14. 10.1111/j.1474-9726.2007.00304.x [DOI] [PubMed] [Google Scholar]
- 41.Krishnan J, Danzer C, Simka T, Ukropec J, Walter KM, Kumpf S, et al. Dietary obesity-associated Hif1α activation in adipocytes restricts fatty acid oxidation and energy expenditure via suppression of the Sirt2-NAD+ system. Genes Dev. (2012) 26:259–70. 10.1101/gad.180406.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dryden SC, Nahhas FA, Nowak JE, Goustin AS, Tainsky MA. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol Cell Biol. (2003) 23:3173–85. 10.1128/mcb.23.9.3173-3185.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park SH, Ozden O, Liu G, Song HY, Zhu Y, Yan Y, et al. SIRT2-mediated deacetylation and tetramerization of pyruvate kinase directs glycolysis and tumor growth. Cancer Res. (2016) 76:3802–12. 10.1158/0008-5472.can-15-2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jing H, Hu J, He B, Negron Abril YL, Stupinski J, Weiser K, et al. A SIRT2-selective inhibitor promotes c-Myc oncoprotein degradation and exhibits broad anticancer activity. Cancer Cell. (2016) 29:607. 10.1016/j.ccell.2016.03.011 [DOI] [PubMed] [Google Scholar]
- 45.Shah AA, Ito A, Nakata A, Yoshida M. Identification of a selective SIRT2 inhibitor and its anti-breast cancer activity. Biol Pharm Bull. (2016) 39:1739–42. 10.1248/bpb.b16-00520 [DOI] [PubMed] [Google Scholar]
- 46.Zhao D, Mo Y, Li MT, Zou SW, Cheng ZL, Sun YP, et al. NOTCH-induced aldehyde dehydrogenase 1A1 deacetylation promotes breast cancer stem cells. J Clin Investig. (2014) 124:5453–65. 10.1172/jci76611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou W, Ni TK, Wronski A, Glass B, Skibinski A, Beck A, et al. The SIRT2 deacetylase stabilizes slug to control malignancy of basal-like breast cancer. Cell Rep. (2016) 17:1302–17. 10.1016/j.celrep.2016.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soung YH, Pruitt K, Chung J. Epigenetic silencing of ARRDC3 expression in basal-like breast cancer cells. Sci Rep. (2014) 4:3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi P, Zhou M, Yang Y. Upregulated tumor sirtuin 2 expression correlates with reduced TNM stage and better overall survival in surgical breast cancer patients. Ir J Med Sci. (2019) 189:83–9. 10.1007/s11845-019-02071-y [DOI] [PubMed] [Google Scholar]
- 50.McGlynn LM, Zino S, MacDonald AI, Curle J, Reilly JE, Mohammed ZM, et al. SIRT2: tumour suppressor or tumour promoter in operable breast cancer? Eur J Cancer. (2014) 50:290–301. 10.1016/j.ejca.2013.10.005 [DOI] [PubMed] [Google Scholar]
- 51.Chen J, Chan AW, To KF, Chen W, Zhang Z, Ren J, et al. SIRT2 overexpression in hepatocellular carcinoma mediates epithelial to mesenchymal transition by protein kinase B/glycogen synthase kinase-3beta/beta-catenin signaling. Hepatology. (2013) 57:2287–98. 10.1002/hep.26278 [DOI] [PubMed] [Google Scholar]
- 52.Huang S, Zhao Z, Tang D, Zhou Q, Li Y, Zhou L, et al. Downregulation of SIRT2 inhibits invasion of hepatocellular carcinoma by inhibiting energy metabolism. Transl Oncol. (2017) 10:917–27. 10.1016/j.tranon.2017.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng ST, Ren JH, Cai XF, Jiang H, Chen J. HBx-elevated SIRT2 promotes HBV replication and hepatocarcinogenesis. Biochem Biophys Res Commun. (2018) 496:904–10. 10.1016/j.bbrc.2018.01.127 [DOI] [PubMed] [Google Scholar]
- 54.Li Z, Xie QR, Chen Z, Lu S, Xia W. Regulation of SIRT2 levels for human non-small cell lung cancer therapy. Lung Cancer. (2013) 82:9–15. 10.1016/j.lungcan.2013.05.013 [DOI] [PubMed] [Google Scholar]
- 55.Li Z, Huang J, Yuan H, Chen Z, Luo Q, Lu S. SIRT2 inhibits non-small cell lung cancer cell growth through impairing Skp2-mediated p27 degradation. Oncotarget. (2016) 7:18927–39. 10.18632/oncotarget.7816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu W, Jiang K, Shen M, Qian Y, Peng Y. SIRT2 suppresses non-small cell lung cancer growth by targeting JMJD2A. Biol Chem. (2015) 396:929–36. 10.1515/hsz-2014-0284 [DOI] [PubMed] [Google Scholar]
- 57.Grbesa I, Pajares MJ, Martinez-Terroba E, Agorreta J, Mikecin AM, Larrayoz M, et al. Expression of sirtuin 1 and 2 is associated with poor prognosis in non-small cell lung cancer patients. PLoS One. (2015) 10:e0124670. 10.1371/journal.pone.0124670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoffmann G, Breitenbucher F, Schuler M, Ehrenhofer-Murray AE. A novel sirtuin 2 (SIRT2) inhibitor with p53-dependent pro-apoptotic activity in non-small cell lung cancer. J Biol Chem. (2014) 289:5208–16. 10.1074/jbc.m113.487736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang LL, Wang HL, Zhong L, Yuan C, Liu SY, Yu ZJ, et al. X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells. Eur J Med Chem. (2018) 155:806–23. 10.1016/j.ejmech.2018.06.041 [DOI] [PubMed] [Google Scholar]
- 60.Ma W, Zhao X, Wang K, Liu J, Huang G. Dichloroacetic acid (DCA) synergizes with the SIRT2 inhibitor Sirtinol and AGK2 to enhance anti-tumor efficacy in non-small cell lung cancer. Cancer Biol Ther. (2018) 19:835–46. 10.1080/15384047.2018.1480281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Larsen JE, Minna JD. Molecular biology of lung cancer: clinical implications. Clin Chest Med. (2011) 32:703–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schuurbiers OC, Meijer TW, Kaanders JH, Looijen-Salamon MG, de Geus-Oei LF, van der Drift MA, et al. Glucose metabolism in NSCLC is histology-specific and diverges the prognostic potential of 18FDG-PET for adenocarcinoma and squamous cell carcinoma. J Thoracic Oncol. (2014) 9:1485–93. 10.1097/jto.0000000000000286 [DOI] [PubMed] [Google Scholar]
- 63.Zhang LL, Zhan L, Jin YD, Min ZL, Wei C, Wang Q, et al. SIRT2 mediated antitumor effects of shikonin on metastatic colorectal cancer. Eur J Pharmacol. (2017) 797:1–8. 10.1016/j.ejphar.2017.01.008 [DOI] [PubMed] [Google Scholar]
- 64.Bajpe PK, Prahallad A, Horlings H, Nagtegaal I, Beijersbergen R, Bernards R. A chromatin modifier genetic screen identifies SIRT2 as a modulator of response to targeted therapies through the regulation of MEK kinase activity. Oncogene. (2015) 34:531–6. 10.1038/onc.2013.588 [DOI] [PubMed] [Google Scholar]
- 65.Cheon MG, Kim W, Choi M, Kim JE. AK-1, a specific SIRT2 inhibitor, induces cell cycle arrest by downregulating Snail in HCT116 human colon carcinoma cells. Cancer Lett. (2015) 356:637–45. 10.1016/j.canlet.2014.10.012 [DOI] [PubMed] [Google Scholar]
- 66.Farooqi AS, Hong JY, Cao J, Lu X, Price IR, Zhao Q, et al. Novel lysine-based thioureas as mechanism-based inhibitors of Sirtuin 2 (SIRT2) with anticancer activity in a colorectal cancer murine model. J Med Chem. (2019) 62:4131–41. 10.1021/acs.jmedchem.9b00191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu F, Sun X, Li G, Wu Q, Chen Y, Yang X, et al. Inhibition of SIRT2 limits tumour angiogenesis via inactivation of the STAT3/VEGFA signalling pathway. Cell Death Dis. (2018) 10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harting K, Knoll B. SIRT2-mediated protein deacetylation: an emerging key regulator in brain physiology and pathology. Eur J Cell Biol. (2010) 89:262–9. 10.1016/j.ejcb.2009.11.006 [DOI] [PubMed] [Google Scholar]
- 69.Hiratsuka M, Inoue T, Toda T, Kimura N, Shirayoshi Y, Kamitani H, et al. Proteomics-based identification of differentially expressed genes in human gliomas: down-regulation of SIRT2 gene. Biochem Biophys Res Commun. (2003) 309:558–66. 10.1016/j.bbrc.2003.08.029 [DOI] [PubMed] [Google Scholar]
- 70.Li Y, Dai D, Lu Q, Fei M, Li M, Wu X. Sirt2 suppresses glioma cell growth through targeting NF-kappaB-miR-21 axis. Biochem Biophys Res Commun. (2013) 441:661–7. 10.1016/j.bbrc.2013.10.077 [DOI] [PubMed] [Google Scholar]
- 71.Ye H, Huang H, Cao F, Chen M, Zheng X, Zhan R. HSPB1 enhances SIRT2-mediated G6PD activation and promotes glioma cell proliferation. PLoS One. (2016) 11:e0164285. 10.1371/journal.pone.0164285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Funato K, Hayashi T, Echizen K, Negishi L, Shimizu N, Koyama-Nasu R, et al. SIRT2-mediated inactivation of p73 is required for glioblastoma tumorigenicity, EMBO Rep. (2018) 19:e45587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Imaoka N, Hiratsuka M, Osaki M, Kamitani H, Kambe A, Fukuoka J, et al. Prognostic significance of sirtuin 2 protein nuclear localization in glioma: an immunohistochemical study. Oncol Rep. (2012) 28:923–30. 10.3892/or.2012.1872 [DOI] [PubMed] [Google Scholar]
- 74.Wei R, He D, Zhang X. Role of SIRT2 in regulation of stemness of cancer stem-like cells in renal cell carcinoma. Cell Physiol Biochem. (2018) 49:2348–57. 10.1159/000493835 [DOI] [PubMed] [Google Scholar]
- 75.Li Y, Zhang M, Dorfman RG, Pan Y, Tang D, Xu L, et al. SIRT2 promotes the migration and invasion of gastric cancer through RAS/ERK/JNK/MMP-9 pathway by increasing PEPCK1-related metabolism. Neoplasia. (2018) 20:745–56. 10.1016/j.neo.2018.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilking-Busch MJ, Ndiaye MA, Huang W, Ahmad N. Expression profile of SIRT2 in human melanoma and implications for sirtuin-based chemotherapy. Cell Cycle. (2017) 16:574–7. 10.1080/15384101.2017.1288323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Du Y, Wu J, Zhang H, Li S, Sun H. Reduced expression of SIRT2 in serous ovarian carcinoma promotes cell proliferation through disinhibition of CDK4 expression. Mol Med Rep. (2017) 15:1638–46. 10.3892/mmr.2017.6183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Damodaran S, Damaschke N, Gawdzik J, Yang B, Shi C, Allen GO, et al. Dysregulation of Sirtuin 2 (SIRT2) and histone H3K18 acetylation pathways associates with adverse prostate cancer outcomes. BMC Cancer. (2017) 17:874. 10.1186/s12885-017-3853-9 [DOI] [PMC free article] [PubMed] [Google Scholar]