Abstract
Certain high-risk factors related to the death of COVID-19 have been reported, however, there were few studies on a death prediction model. This study was conducted to delineate the clinical characteristics of patients with coronavirus disease 2019 (covid-19) of different degree and establish a death prediction model. In this multi-centered, retrospective, observational study, we enrolled 523 COVID-19 cases discharged before February 20, 2020 in Henan Province, China, compared clinical data, screened for high-risk fatal factors, built a death prediction model and validated the model in 429 mild cases, six fatal cases discharged after February 16, 2020 from Henan and 14 cases from Wuhan. Out of the 523 cases, 429 were mild, 78 severe survivors, 16 non-survivors. The non-survivors with median age 71 were older and had more comorbidities than the mild and severe survivors. Non-survivors had a relatively delay in hospitalization, with higher white blood cell count, neutrophil percentage, D-dimer, LDH, BNP, and PCT levels and lower proportion of eosinophils, lymphocytes and albumin. Discriminative models were constructed by using random forest with 16 non-survivors and 78 severe survivors. Age was the leading risk factors for poor prognosis, with AUC of 0.907 (95% CI 0.831–0.983). Mixed model constructed with combination of age, demographics, symptoms, and laboratory findings at admission had better performance (p = 0.021) with a generalized AUC of 0.9852 (95% CI 0.961–1). We chose 0.441 as death prediction threshold (with 0.85 sensitivity and 0.987 specificity) and validated the model in 429 mild cases, six fatal cases discharged after February 16, 2020 from Henan and 14 cases from Wuhan successfully. Mixed model can accurately predict clinical outcomes of COVID-19 patients.
Keywords: novel coronavirus pneumonia, risk factors, death prediction model, random forest, epidemiology investigation
Introduction
In late December 2019, Wuhan City, Hubei Province, China found several cases of unexplained pneumonia. On January 7, 2020, a new coronavirus was detected in the laboratory and the whole genome sequence of the virus was obtained. On January 12, 2020, the World Health Organization temporarily named this new virus 2019 novel coronavirus (2019-nCoV). On February 11, 2020, the World Health Organization announced that the same time the International Virus Classification Committee named the new coronavirus “SARS-CoV-2.” Although the lethal rate of SARS-CoV-2 is not as high as SARS and MERS, it is more infectious than other viruses including influenza virus (1–3). The range of basic regeneration number (Ro) is estimated to be 2–5 (4, 5). China has effectively controlled the epidemic by adopting strict prevention and control measures, but in areas outside China, the epidemic of novel coronavirus is still spreading. The number of infections caused by SARS-CoV-2 is large and no specific therapeutic is available yet, which is the main cause of so many deaths. SARS-CoV-2 can cause pneumonia and systemic inflammation, leading to multiple organ failure in high-risk patients. More and more studies have focused on the high-risk factors of death. Demographic factors, advanced age, combined underlying diseases, and D-dimer exceeding 1 μg/L have been confirmed as risk factors for death in adult patients (6). In the absence of vaccines and specific antiviral drugs, targeted application of supportive therapy may be beneficial to relieve symptoms and protect organ functions (7). How to quickly identify high-risk patients in the early stage of the disease and actively adopt supportive treatment to reduce mortality is an urgent problem to be solved in the clinic. Cao Bin (6) and others reported some characteristics and clinical progress of the early stage of severe and dead patients, which improved our further understanding of the characteristics of dead patients. However, there are no relevant studies on the application of models to predict COVID-19 death. Using admission characteristics and laboratory test results to establish a predictive model can calculate the probability of over-all mortality due to SARS-CoV-2, identify high-risk patients as early as possible and give support to reduce mortality as soon as possible.
In this study, we collected data of 523 discharged cases of novel coronavirus infection in Henan Province, China and compared the demographics, clinical characteristics, laboratory test, imaging between the mild, severe survivors and non-survivors. We established a death prediction model using the data upon admission of the severe survivors and non-survivors.
Methods
Study Design and Participants
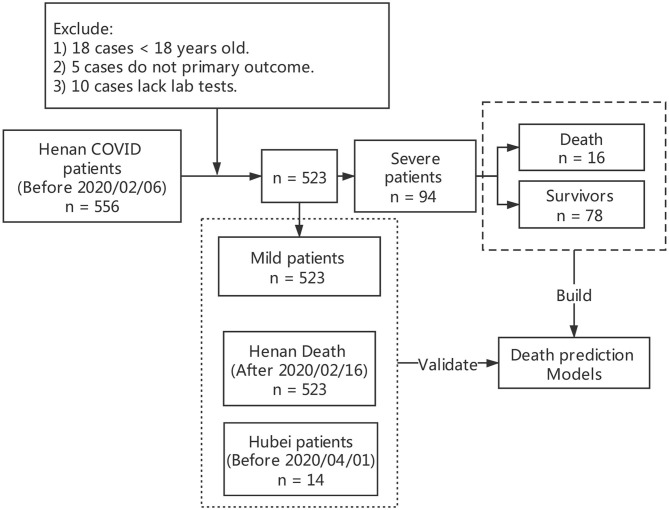
From January 22, 2020 to February 20, 2020, a total of 717 patients confirmed COVID-19 were discharged in 18 cities of Henan Province, China, of which 19 died. We designed a data collection table, including age, gender, epidemiological history, past history, clinical symptoms, laboratory examination, chest CT and recorded the treatment process and clinical outcome, and data of 556 patients with novel coronavirus pneumonia discharged before February 20, 2020 was collected. All data were checked by two physicians (AL and XM) and a third researcher (QZ) adjudicated any difference in interpretation between the two primary reviewers. For different interpretations and missing data, we contacted the doctor who filled out the form and the patient or their family members to review and supplement. Excluding 18 cases under the age of 18, 10 cases missing key information and five cases transferred to other hospitals with no end point, 523 cases were included for statistical analysis, of which 19 cases died including three fatal cases with data missing. According to the Guidance for Corona Virus Disease 2019 (6th edition) released by the National Health Commission of China, the enrolled cases were categorized as mild or severe (8). There were no deaths in the mild. According to the clinical outcome, we divided the severe into severe survivors and non-survivors. Up to April 1, there were 22 cases died of COVID-19 in Henan Province. We have managed to collect data of another six fatal cases of Henan Province and 14 cases from the Fourth People's Hospital of Wuhan to validate the predictive power of the model. The flow diagram of included patients is shown in Figure 1.
Figure 1.
Flow diagram of included patients.
Definition
The incubation period was defined as the interval between the potential earliest date of contact of the transmission source (wildlife or person of suspected or confirmed case) and the potential earliest date of symptom onset (i.e., cough, fever, fatigue, or myalgia). We excluded cases with an incubation period of <1 day or cases of continuous exposure, because those patients continued to be infected. Fever was defined as an axillary temperature of 37.3°C or higher. Lymphopenia was defined as a lymphocyte count of <1,200 per cubic millimeter. Thrombocytopenia was defined as a platelet count of <100,000 per cubic millimeter. Chest CT was divided into normal, mild, moderate and severe infections according to the range of lesions. The range of lesions <15% was mild; the range of lesions 15–40% was moderate; the range of lesions > 40% was severe.
Statistical Analysis
Statistical analyses on cohort characteristics were performed on R version 3.6.1. Participants' demographic, laboratory findings and questionnaire were summarized with a standardized statistical significance test method, categorical variables were shown as counts and percentages [n (%)], and associations were tested using a fisher' exact test. Continuous variables were shown as median (interquartile range, IQR), and differences between groups were analyzed with non-parametric test (Wilcoxon's rank-sum test). A single-sided p < 0.05 was considered statistically significant. Discriminative models were constructed by using random forest with leave-one-out cross validation, features were selected by using embedded backward selection. Missing data were filled by chose median value in relative cohort (Severe death, severe survival, and mild) for model construction and validation. Receiver operating characteristic (ROC) curve and Precision-Recall curve were visualized by using R program package “pROC” and “precrec,” respectively.
Results
Clinical Characteristics of the Study Patients According to Disease Severity and Clinical Outcome in Severe
Table 1 shows that among the 523 patients 429/523 (82.03%) were mild, 94/523 (17.97%) severe, and 16/94 (17.02%) in severe cases died of COVID-19. The median age of the 523 patients was 44.0 years (IQR 32–54), with male patients (55.26%) accounting for the majority. Severe patients were older than mild patients (50.00 [IQR 38.25–61.5] vs. 42 [IQR 31–54]), and non-survivors were older than 65 years with a median age of 71 years (IQR 67.75–80). Age difference between the cases was statistically significant.
Table 1.
Clinical characteristics of the study patients according to disease severity and clinical outcome in severe.
| Characteristics | All patients (N = 523) | Disease severity | Clinical outcome in severe | ||||
|---|---|---|---|---|---|---|---|
| Mild (N = 429) | Severe (N = 94) | p-value | Non-survivors (N = 16) | Survivors (N = 78) | p-value | ||
| Age, years | 44 (32–54) | 42 (31–54) | 50 (38.25–61.5) | 0.00004 | 71 (67.75–80) | 47 (34–54.75) | 0 |
| 18–49 | 314/523 (60.04) | 270/429 (62.94) | 44/94 (46.81) | 0.00384 | 1/16 (6.25) | 43/78 (55.13) | 0.00036 |
| 50–64 | 146/523 (27.92) | 121/429 (28.21) | 25/94 (26.6) | 0.75274 | 1/16 (6.25) | 24/78 (30.77) | 0.03505 |
| ≥65 | 46/523 (8.8) | 23/429 (5.36) | 23/94 (24.47) | 0 | 14/16 (87.5) | 9/78 (11.54) | 0 |
| Female sex | 234/523 (44.74) | 199/429 (46.39) | 35/94 (37.23) | 0.06599 | 3/16 (18.75) | 32/78 (41.03) | 0.07824 |
| Exposure to Source of Transmission Within Past 14 Days | |||||||
| Had the history of travel or residence in Wuhan and its surrounding areas, or other communities where the case of COVID-19 had been reported | 324/523 (61.95) | 272/429 (63.4) | 52/94 (55.32) | 0.08999 | 5/16 (31.25) | 47/78 (60.26) | 0.03216 |
| Had contact with Wuhan residents | 158/523 (30.21) | 132/429 (30.77) | 26/94 (27.66) | 0.32199 | 6/16 (37.5) | 20/78 (25.64) | 0.24972 |
| Cluster | 76/523 (14.53) | 66/429 (15.38) | 10/94 (10.64) | 0.15317 | 2/16 (12.5) | 8/78 (10.26) | 0.5381 |
| Had contact with patients confirmed0020COVID-19 | 136/523 (26) | 115/429 (26.81) | 21/94 (22.34) | 0.22412 | 5/16 (31.25) | 16/78 (20.51) | 0.263 |
| Not clear | 67/523 (12.81) | 48/429 (11.19) | 19/94 (20.21) | 0.01702 | 3/16 (18.75) | 16/78 (20.51) | 0.58924 |
| Comorbidity | |||||||
| COPD | 13/463 (2.81) | 6/373 (1.61) | 7/90 (7.78) | 0.00526 | 5/15 (33.33) | 2/75 (2.67) | 0.00117 |
| Asthma | 3/459 (0.65) | 2/369 (0.54) | 1/90 (1.11) | 0.48126 | 0/15 (0) | 1/75 (1.33) | 0.83333 |
| Interstitial pneumonia | 8/462 (1.73) | 7/372 (1.88) | 1/90 (1.11) | 0.51749 | 1/15 (6.67) | 0/75 (0) | 0.16667 |
| Diabetes | 42/462 (9.09) | 29/371 (7.82) | 13/91 (14.29) | 0.0475 | 5/15 (33.33) | 8/76 (10.53) | 0.03604 |
| Coronary heart disease | 24/461 (5.21) | 13/371 (3.5) | 11/90 (12.22) | 0.00237 | 5/15 (33.33) | 6/75 (8) | 0.01681 |
| Hypertension | 74/465 (15.91) | 48/375 (12.8) | 26/90 (28.89) | 0.00034 | 7/15 (46.67) | 19/75 (25.33) | 0.09088 |
| Cerebral infarction | 11/461 (2.39) | 9/371 (2.43) | 2/90 (2.22) | 0.63343 | 1/15 (6.67) | 1/75 (1.33) | 0.30712 |
| Cerebral hemorrhage | 6/459 (1.31) | 4/369 (1.08) | 2/90 (2.22) | 0.33495 | 1/15 (6.67) | 1/75 (1.33) | 0.30712 |
| Cancer | 7/230 (3.04) | 4/191 (2.09) | 3/39 (7.69) | 0.09676 | 2/10 (20) | 1/29 (3.45) | 0.15593 |
| Pregnancy | 2/469 (0.43) | 2/382 (0.52) | 0/87 (0) | 0.66309 | 0/16 (0) | 0/71 (0) | 1 |
| Digestive system disease | 11/307 (3.58) | 10/257 (3.89) | 1/50 (2) | 0.44053 | 0/10 (0) | 1/40 (2.5) | 0.8 |
| Chronic kidney disease | 5/307 (1.63) | 3/256 (1.17) | 2/51 (3.92) | 0.19413 | 0/11 (0) | 2/40 (5) | 0.61176 |
| Symptoms | |||||||
| Fever | 449/506 (88.74) | 360/412 (87.38) | 89/94 (94.68) | 0.02648 | 15/16 (93.75) | 74/78 (94.87) | 0.6154 |
| Fatigue | 190/480 (39.58) | 151/387 (39.02) | 39/93 (41.94) | 0.34364 | 10/16 (62.5) | 29/77 (37.66) | 0.06087 |
| Cough | 309/496 (62.3) | 241/403 (59.8) | 68/93 (73.12) | 0.01064 | 11/16 (68.75) | 57/77 (74.03) | 0.43858 |
| Dyspnea | 41/471 (8.7) | 18/378 (4.76) | 23/93 (24.73) | 0 | 7/16 (43.75) | 16/77 (20.78) | 0.05703 |
| Gasp | 44/468 (9.4) | 22/375 (5.87) | 22/93 (23.66) | 0 | 4/16 (25) | 18/77 (23.38) | 0.55775 |
| Chest tightness | 92/471 (19.53) | 60/378 (15.87) | 32/93 (34.41) | 0.0001 | 11/16 (68.75) | 21/77 (27.27) | 0.00236 |
| Nasal congestion | 32/467 (6.85) | 20/374 (5.35) | 12/93 (12.9) | 0.01303 | 1/16 (6.25) | 11/77 (14.29) | 0.34485 |
| Runny nose | 31/467 (6.64) | 21/374 (5.61) | 10/93 (10.75) | 0.06599 | 1/16 (6.25) | 9/77 (11.69) | 0.45525 |
| Sore throat | 65/470 (13.83) | 51/378 (13.49) | 14/92 (15.22) | 0.38807 | 2/16 (12.5) | 12/76 (15.79) | 0.54396 |
| Expectoration | 138/480 (28.75) | 107/387 (27.65) | 31/93 (33.33) | 0.16827 | 7/16 (43.75) | 24/77 (31.17) | 0.24499 |
| Anorexia | 61/473 (12.9) | 44/380 (11.58) | 17/93 (18.28) | 0.06352 | 5/16 (31.25) | 12/77 (15.58) | 0.13271 |
| Diarrhea | 25/467 (5.35) | 17/374 (4.55) | 8/93 (8.6) | 0.10097 | 2/16 (12.5) | 6/77 (7.79) | 0.41557 |
| Headache | 37/471 (7.86) | 27/378 (7.14) | 10/93 (10.75) | 0.17119 | 0/16 (0) | 10/77 (12.99) | 0.13578 |
| Dizziness | 28/470 (5.96) | 23/377 (6.1) | 5/93 (5.38) | 0.50991 | 0/16 (0) | 5/77 (6.49) | 0.38017 |
| Muscle and joint pain | 39/464 (8.41) | 26/371 (7.01) | 13/93 (13.98) | 0.02981 | 1/16 (6.25) | 12/77 (15.58) | 0.29732 |
| The Basic Vital Signs on Admission | |||||||
| Respiratory rate >24 breaths per min | 4/489 (0.82) | 3/399 (0.75) | 1/90 (1.11) | 0.02936 | 1/13 (7.69) | 0/77 (0) | 0.00554 |
| Pulse oxygen saturation <90% | 8/165 (4.85) | 0/122 (0) | 8/43 (18.6) | 0 | 5/11 (45.45) | 3/32 (9.38) | 0.00019 |
| Fever on admission, °C | 37.2 (36.7-37.9) | 37.1 (36.7-37.9) | 37.2 (36.8-38) | 0.26312 | 36.7 (36.6-37.1) | 37.3 (36.8-38) | 0.00776 |
| <37.5 | 293/496 (59.07) | 237/404 (58.66) | 56/92 (60.87) | 0.68771 | 12/15 (80) | 44/77 (57.14) | 0.09703 |
| 37.5–38.0 | 90/496 (18.15) | 78/404 (19.31) | 12/92 (13.04) | 0.15947 | 1/15 (6.67) | 11/77 (14.29) | 0.37752 |
| 38.1–39.0 | 97/496 (19.56) | 77/404 (19.06) | 20/92 (21.74) | 0.55865 | 1/15 (6.67) | 19/77 (24.68) | 0.10886 |
| >39.0 | 16/496 (3.23) | 12/404 (2.97) | 4/92 (4.35) | 0.34306 | 1/15 (6.67) | 3/77 (3.9) | 0.51568 |
| Time form illness onset to seeing a doctor, days | 2 (0–5) | 2 (0–5) | 2 (1–5) | 0.41842 | 4.5 (1.75–7) | 2 (1–4) | 0.04202 |
| Time form illness onset to hospital admission, days | 4 (2–7) | 4 (2–7) | 3 (2–7) | 0.29878 | 8 (6–10) | 3 (1–6) | 0.00047 |
| Incubation period, days | 5 (1–9) | 5 (1–9) | 4 (1–9) | 0.30537 | 5 (2–10) | 4 (1–8) | 0.10345 |
Data are median (IQR)or n/N (%). P-values were calculated by Fisher's exact test or Mann-Whitney U-test. COPD, chronic obstructive pulmonary disease.
Within 14 days before the onset, 324 (61.95%) had lived in or visited Wuhan; 158 (30.21%) had contact history with Wuhan returnees; 136 (26%) had confirmed contact history of COVID-19 cases; 76 (14.53%) occurred from familial clusters, and 67 (12.81%) had unknown contact history. Non-survivors were not much different from the mild or severe survivors in terms of epidemiological history.
Hypertension (74/465 [15.91%]), diabetes (42/462 [9.09%]), coronary heart disease (24/461 [5.21%]) were the most common comorbidities. The average number of comorbid diseases in the non-survivors was 1.94 which was significantly higher than that of the mild and severe survivors. The most common symptoms on admission were fever (449 [88.74%], cough (309 [62.3%] and fatigue (190 [39.58%]); the more common symptoms were expectoration (138 [28.75%]), chest tightness (92 [19.53%]), sore throat (65 [13.83%]), anorexia (61 [12.9%]), gasp (44 [9.4%]), and dyspnea (41 [8.7 %]). Muscle and joint pain, runny nose, diarrhea, dizziness, and headache were rare. The symptoms of fever, cough, dyspnea, gasp, chest tightness, nasal congestion, and muscle and joint pain had a higher incidence in severe cases, and the difference was significant; the incidence of chest tightness in non-survivors was higher than that in severe survivors. The patients in the non-survivors had more symptoms at the onset.
Four (0.82%) had a respiratory rate > 24 breaths/min, one of them died; 8 (4.85%) pulse oxygen saturation < 90%, all severe; median body temperature 37.2°C (IQR 36.7–37.9), 293 (59.07%) body temperature < 37.5°C, 16 (3.23%) body temperature > 39°C and 80% non-survivors body temperature < 37.5°C upon admission.
The median duration from onset of symptoms to first visit to doctor was 2 days (IQR 0–5), from onset of symptoms to first hospitalization 4 days (IQR 2–7) while 8 days (IQR 6–10) in non-survivors. The median incubation period was 5 days (IQR 1–9), with no significant difference between the cases.
Radiographic and Laboratory Findings on Admission
Table 2 shows the imaging and laboratory examination results. Of all the cases, 419 patients had detailed chest CT data on initial admission, with 17 (4.06%) being normal; 224 (53.46%) chest CT lesions < 15%; 154 (36.75%) chest CT lesions between 15 and 40%; 24 (5.73%) chest CT lesions > 40%, of which 15 were severe. In the non-survivors, 100% of patients had a chest CT lesion area of more than 15% for the first time.
Table 2.
Radiographic and laboratory findings on admission.
| Characteristics | All patients (N = 523) | Disease severity | Clinical outcome in severe | ||||
|---|---|---|---|---|---|---|---|
| Mild (N = 429) | Severe (N = 94) | p-value | Non-survivors (N = 16) | Survivors (N = 78) | p-value | ||
| Radiographic Findings | |||||||
| Chest CT | |||||||
| Normal | 17/419 (4.06) | 13/342 (3.8) | 4/77 (5.19) | 0.38245 | 0/10 (0) | 4/67 (5.97) | 0.56639 |
| Mild | 224/419 (53.46) | 202/342 (59.06) | 22/77 (28.57) | 0 | 0/10 (0) | 22/67 (32.84) | 0.02666 |
| Moderate | 154/419 (36.75) | 118/342 (34.5) | 36/77 (46.75) | 0.04398 | 6/10 (60) | 30/67 (44.78) | 0.2873 |
| Severe | 24/419 (5.73) | 9/342 (2.63) | 15/77 (19.48) | 0 | 4/10 (40) | 11/67 (16.42) | 0.09699 |
| Laboratory Findings | |||||||
| Pathogens identified | |||||||
| COVID-19 viral nucleic acid test positive on the first time | 323/495 (65.25) | 258/404 (63.86) | 65/91 (71.43) | 0.10514 | 16/16 (100) | 49/75 (65.33) | 0.00249 |
| Influenza A virus Ag+ | 8/323 (2.48) | 8/263 (3.04) | 0/60 (0) | 0.18936 | 0/7 (0) | 0/53 (0) | 1 |
| Influenza B virus Ag+ | 13/324 (4.01) | 10/264 (3.79) | 3/60 (5) | 0.44419 | 1/7 (14.29) | 2/53 (3.77) | 0.31543 |
| Mycoplasma pneumonia IgM Ab+ | 28/319 (8.78) | 22/262 (8.4) | 6/57 (10.53) | 0.3824 | 1/7 (14.29) | 5/50 (10) | 0.5621 |
| HBsAg+ | 26/357 (7.28) | 23/288 (7.99) | 3/69 (4.35) | 0.2214 | 0/11 (0) | 3/58 (5.17) | 0.58892 |
| HCV-Ab+ | 7/353 (1.98) | 7/285 (2.46) | 0/68 (0) | 0.22041 | 0/11 (0) | 0/57 (0) | 1 |
| TP-Ab+ | 7/345 (2.03) | 7/277 (2.53) | 0/68 (0) | 0.21186 | 0/11 (0) | 0/57 (0) | 1 |
| Blood routine | |||||||
| Leucocyte count, × 109 /L | 4.82 (3.585–6.225) | 4.74 (3.5025–6.0475) | 5.38 (4–7.05) | 0.00281 | 8.66 (7–12.335) | 5.12 (3.8675–5.9875) | 0.00007 |
| >10 | 24/487 (4.93) | 14/398 (3.52) | 10/89 (11.24) | 0.00537 | 5/15 (33.33) | 5/74 (6.76) | 0.01073 |
| 4–10 | 310/487 (63.66) | 252/398 (63.32) | 58/89 (65.17) | 0.74263 | 9/15 (60) | 49/74 (66.22) | 0.64496 |
| <4 | 153/487 (31.42) | 132/398 (33.17) | 21/89 (23.6) | 0.07869 | 1/15 (6.67) | 20/74 (27.03) | 0.07944 |
| Platelet count, ×109 /L | 175.5 (143–210) | 176 (143–209.5) | 168 (145–216) | 0.29403 | 153 (93.25–210.75) | 171 (147–213.5) | 0.08684 |
| <100 | 26/452 (5.75) | 19/375 (5.07) | 7/77 (9.09) | 0.13423 | 4/14 (28.57) | 3/63 (4.76) | 0.01824 |
| Absolute lymphocyte count, ×109 /L | 1.1 (0.84–1.49) | 1.12 (0.9–1.55) | 0.92 (0.595–1.225) | 0.00005 | 0.72 (0.44–0.89) | 0.985 (0.6325–1.2475) | 0.03876 |
| <0.8 | 98/448 (21.88) | 67/369 (18.16) | 31/79 (39.24) | 0.00004 | 7/13 (53.85) | 24/66 (36.36) | 0.23802 |
| Lymphocyte percentage, % | 24.5 (17.055–33.18) | 24.8 (18.575–33.625) | 19.75 (9.36–29.025) | 0.00029 | 8.34 (5.3–13.405) | 21.75 (11.225–29.775) | 0.00526 |
| <20 | 152/456 (33.33) | 112/376 (29.79) | 40/80 (50) | 0.0005 | 11/14 (78.57) | 29/66 (43.94) | 0.01858 |
| Absolute neutrophil count, ×109 /L | 3.09 (2.085–4.415) | 3.04 (2.06–4.3) | 3.73 (2.37–5.765) | 0.01557 | 6.29 (4.03–11.09) | 3.54 (2.25–4.62) | 0.01067 |
| Neutrophil percentage, % | 65.5 (56.35–74.65) | 65.4 (56.5–73.7) | 68.6 (56.6–85.1) | 0.00649 | 86.105 (80.1125–92.025) | 65.95 (55.4–77.1775) | 0.00172 |
| Absolute eosinophil count, ×109 /L | 0.01 (0–0.03) | 0.01 (0–0.03) | 0.01 (0–0.03) | 0.24402 | 0 (0–0.02) | 0.01 (0–0.03) | 0.15484 |
| <0.2 | 375/388 (96.65) | 314/324 (96.91) | 61/64(95.31) | 0.36519 | 10/11 (90.91) | 51/53 (96.23) | 0.43774 |
| Eosinophil percentage, % | 0.2 (0–0.7) | 0.2 (0–0.78) | 0.1 (0–0.4) | 0.00506 | 0 (0–0.2) | 0.1 (0–0.4) | 0.0478 |
| <0.1 | 143/405 (35.31) | 109/330 (33.03) | 34/75 (45.33) | 0.04418 | 9/15 (60) | 25/60 (41.67) | 0.20205 |
| <0.5 | 276/405 (68.15) | 217/330 (65.76) | 59/75 (78.67) | 0.03031 | 13/15 (86.67) | 46/60 (76.67) | 0.32384 |
| Hemagglutination examination | |||||||
| Fibrinogen, g/L | 3.48 (2.8–4.76) | 3.43 (2.79–4.72) | 3.607 (2.99–5.075) | 0.18678 | 3.7055 (2.685–5.035) | 3.607 (2.99–5.0825) | 0.42719 |
| >4 | 127/367 (34.6) | 97/297 (32.66) | 30/70 (42.86) | 0.10666 | 5/14 (35.71) | 25/56 (44.64) | 0.54597 |
| Prothrombin time, s | 11.9 (10.7–12.9) | 11.9 (10.77–12.9) | 11.8 (10.6–13) | 0.41355 | 12.5 (11.25–14.6) | 11.7 (10.5–12.8) | 0.06903 |
| ≥16 | 21/370 (5.68) | 17/302 (5.63) | 4/68 (5.88) | 0.56081 | 1/11 (9.09) | 3/57 (5.26) | 0.51496 |
| D-Dimer, mg/L | 0.41 (0.18–1.77225) | 0.39 (0.16–0.98) | 0.8 (0.19–4.18) | 0.04076 | 6.9 (1–32.47) | 0.46 (0.1625–1.63225) | 0.00199 |
| 0.5–1 | 57/304 (18.75) | 47/241 (19.5) | 10/63 (15.87) | 0.54445 | 2/13 (15.38) | 8/50 (16) | 0.66288 |
| ≥1 | 87/304 (28.62) | 60/241 (24.9) | 27/63 (42.86) | 0.00498 | 10/13 (76.92) | 17/50 (34) | 0.00534 |
| Liver function examination | |||||||
| Glutamic-pyruvic transaminase, U/L | 22.45 (15–35) | 22 (15–34) | 26.5 (19.375–38.825) | 0.00972 | 28 (21–41.4) | 26 (19.3–38.1) | 0.38532 |
| >40 | 86/378 (22.75) | 69/310 (22.26) | 17/68 (25) | 0.62525 | 3/11 (27.27) | 14/57 (24.56) | 0.55735 |
| Glutamic-oxalacetic transaminase, U/L | 25 (19–33) | 24 (18.8275–31.875) | 30 (20.75–38.075) | 0.00755 | 37 (32–42.25) | 27 (20–35.4) | 0.01613 |
| >40 | 55/374 (14.71) | 40/306 (13.07) | 15/68 (22.06) | 0.09858 | 5/11 (45.45) | 10/57 (17.54) | 0.05567 |
| Total bilirubin, μmol/L | 10.25 (7.575–15.35) | 10.2 (7.5–14.7) | 11.18 (8.05–17) | 0.09421 | 14.755 (8.975–17.505) | 10.9 (8.05–16.2) | 0.16849 |
| >17.1 | 61/384 (15.89) | 45/311 (14.47) | 16/73 (21.92) | 0.11717 | 4/12 (33.33) | 12/61 (19.67) | 0.24509 |
| Lactate dehydrogenase, U/L | 208 (170.69–255.445) | 202.38 (166.9–235) | 246.5 (186.75–387.795) | 0 | 437 (306.315–715.5) | 223 (177.4–343.74) | 0.00182 |
| ≥250 | 102/383 (26.63) | 64/307 (20.85) | 38/76 (50) | 0 | 9/11 (81.82) | 29/65 (44.62) | 0.02248 |
| Creatine Kinase, U/L | 69.66 (45–113) | 69 (43–106.81) | 72.5 (53.99–168.3125) | 0.0169 | 172.875 (119.75–282.84) | 67.195 (53–113.75) | 0.01175 |
| ≥200 | 36/363 (9.92) | 22/299 (7.36) | 14/64 (21.88) | 0.00042 | 4/10 (40) | 10/54 (18.52) | 0.13805 |
| Albumin, g/L | 40.4 (36.7–44) | 40.7 (37.2–44.15) | 39.9 (34–42.8) | 0.00594 | 32.8 (29.75–39.45) | 39.95 (34.975–43.175) | 0.01982 |
| <30 | 17/380 (4.47) | 8/311 (2.57) | 9/69 (13.04) | 0.00094 | 3/11 (27.27) | 6/58 (10.34) | 0.14825 |
| Renal function examination | |||||||
| Creatinine, μmol/L | 65.29 (54–78.35) | 65.29 (54–78.1) | 64.85 (54.775–79.55) | 0.47011 | 83.5 (61.38–88) | 63 (54.85–71) | 0.0379 |
| ≥133 | 5/354 (1.41) | 3/284 (1.06) | 2/70 (2.86) | 0.25749 | 2/11 (18.18) | 0/59 (0) | 0.02277 |
| Blood urea nitrogen, mmol/L | 3.94 (3.035–5.17) | 3.905 (2.9925–5.03) | 4.2 (3.2–6.9625) | 0.08176 | 5.695 (4.2075–9.99) | 3.815 (3.175–5.5725) | 0.07124 |
| >8 | 49/332 (14.76) | 35/266 (13.16) | 14/66 (21.21) | 0.09869 | 4/10 (40) | 10/56 (17.86) | 0.12549 |
| Glomerular filtration rate, ml/min/1.73 m2 | 106.39 (89.12–120.9885) | 106.49 (88.239–120.9885) | 105.0085 (89.6875–121.645) | 0.48903 | 114 (106.59–122.84) | 104.17 (88.48–118.3) | 0.35179 |
| >120 | 16/55 (29.09) | 12/39 (30.77) | 4/16 (25) | 0.46779 | 1/3 (33.33) | 3/13 (23.08) | 0.60714 |
| Arterial blood gas analysis | |||||||
| PH value | 7.435 (7.41–7.47) | 7.43 (7.409–7.46) | 7.45 (7.42625–7.48) | 0.02645 | 7.4475 (7.425–7.4675) | 7.455 (7.4285–7.48) | 0.29188 |
| >7.45 | 41/97 (42.27) | 21/61 (34.43) | 20/36 (55.56) | 0.04183 | 5/10 (50) | 15/26 (57.69) | 0.48092 |
| 7.35–7.45 | 54/97 (55.67) | 39/61 (63.93) | 15/36 (41.67) | 003294 | 4/10 (40) | 11/26 (42.31) | 0.60234 |
| <7.35 | 2/97 (2.06) | 1/61 (1.64) | 1/36 (2.78) | 0.60696 | 1/10 (10) | 0/26 (0) | 0.27778 |
| PCO2, mmHg | 35.7 (31.7–39) | 36.3 (32–39.2) | 35 (30.775–38.45) | 0.0975 | 32.5 (26.4–35.55) | 35.05 (31.85–38.55) | 0.10487 |
| <35 | 40/97 (41.24) | 23/61 (37.7) | 17/36 (47.22) | 0.35762 | 7/10 (70) | 10/26 (38.46) | 0.09239 |
| 35–45 | 51/97 (52.58) | 34/61 (55.74) | 17/36 (47.22) | 0.41712 | 2/10 (20) | 15/26 (57.69) | 0.04698 |
| >45 | 6/97 (6.19) | 4/61 (6.56) | 2/36 (5.56) | 0.60563 | 1/10 (10) | 1/26 (3.85) | 0.48413 |
| PO2, mmHg | 80.8 (67–96.6) | 86.9 (78.8–103) | 68.5 (53.8–83.65) | 0.00008 | 62.6 (49.275–86.45) | 68.5 (58.575–81.5) | 0.2567 |
| <60 | 17/97 (17.53) | 5/61 (8.2) | 12/36 (33.33) | 0.00166 | 5/10 (50) | 7/26 (26.92) | 0.17793 |
| Other examination | |||||||
| Brain natriuretic peptide, pg/mL | 117.8 (53.5625–604.9) | 90.71 (41–109) | 612.5 (256–1121) | 0 | 455.95 (314.715–666.75) | 736.4 (256–1166) | 0.22236 |
| >100 | 46/74 (62.16) | 17/41 (41.46) | 29/33 (87.88) | 0.00004 | 8/8 (100) | 21/25 (84) | 0.30914 |
| C-reactive protein, mg/L | 11.425 (4.075–28) | 10 (3.45–22.715) | 30.86 (11–73.49) | 0 | 68 (30.91–96) | 21.105 (10.6175–50.175) | 0.02518 |
| >10 | 211/372 (56.72) | 152/303 (50.17) | 59/69 (85.51) | 0 | 12/13 (92.31) | 47/56 (83.93) | 0.39436 |
| Procalcitonin, ng/mL | 0.07 (0.05–0.1525) | 0.055 (0.05–0.11) | 0.13 (0.05925–0.20475) | 0.00012 | 0.1835 (0.15625–0.3225) | 0.12 (0.05225–0.20475) | 0.04056 |
| <0.1 | 132/235 (56.17) | 116/187 (62.03) | 16/48 (33.33) | 0.00035 | 0/8 (0) | 16/40 (40) | 0.02787 |
| 0.1–0.25 | 63/235 (26.81) | 41/187 (21.93) | 22/48 (45.83) | 0.00085 | 6/8 (75) | 16/40 (40) | 0.07686 |
| 0.25–0.5 | 25/235 (10.64) | 20/187 (10.7) | 5/48 (10.42) | 0.95548 | 0/8 (0) | 5/40 (12.5) | 0.38428 |
| ≥0.5 | 15/235 (6.38) | 10/187 (5.35) | 5/48 (10.42) | 0.16861 | 2/8 (25) | 3/40 (7.5) | 0.18874 |
| Cardiac troponin T, μg/L | 0.01 (0.004–0.0895) | 0.01 (0.003–0.2) | 0.0135 (0.0075–0.04525) | 0.37652 | 0.015 (0.012–0.079) | 0.01 (0.007–0.031) | 0.1928 |
| >0.2 | 11/47 (23.4) | 9/33 (27.27) | 2/14 (14.29) | 0.28702 | 1/5 (20) | 1/9 (11.11) | 0.6044 |
| Cardiac troponin I, μg/L | 0.24 (0.055–0.4775) | 0.24 (0.1–0.4) | 0.29 (0.02–0.9) | 0.38233 | 0.045 (0.014–0.6925) | 0.4 (0.25–5.25) | 0.2669 |
| >1.5 | 6/42 (14.29) | 3/29 (10.34) | 3/13 (23.08) | 0.262 | 1/6 (16.67) | 2/7 (28.57) | 0.56294 |
Data are median (IQR)or n/N (%). P-values were calculated by Fisher's exact test or Mann-Whitney U-test. CT, computerized tomography; Ag, antigen; IgM, immunoglobulin m; Ab, antibody; HBsAg, hepatitis B surface antigen; HCV-Ab, hepatitis C virus antibody; TP-Ab, treponema pallidum antibody; PH, pondus hydrogenii; PCO2, partial pressure of carbon dioxide; PO2, partial pressure of oxygen.
In the first nucleic acid testing, 323 (65.25%) were confirmed positive for SARS-CoV-2. The leucocyte count in non-survivors (8.66 × 109/L [IQR 7–12.335]) was significantly higher than that in mild and severe survivors. Lymphocytopenia is more common in the severe than in the mild (39.24 vs. 18.16%). 96.65% of patients experienced a decrease in eosinophil count. The level of D-dimer at admission was significantly higher in severe patients (0.8 mg/L [IQR 0.19–4.18] vs. 0.39 mg/L [IQR 0.16–0.98], p = 0.04076), and the non-survivors was significantly higher than the survivors (6.9 mg/L [IQR 1–32.47] vs. 0.46 mg/L [IQR 0.1625–1.63225, p = 0.00199). The alanine aminotransferase, lactate dehydrogenase and creatine kinase in the severe were significantly higher than those in the mild, and the non-survivors was more obviously, the difference was significant. The incidence of renal impairment was higher in the non-survivors. The incidence of arterial blood gas hypoxia and respiratory alkalosis on admission in the non-survivors was higher than that in the mild and the severe survivors. Three hundred and seventy-two people were tested for C-reactive protein (CRP) upon admission. Two hundred and eleven (56.72%) had CRP > 10 mg/L. The increase rate in the severe (85.51%) was significantly higher than that in the mild (50.17%). Two hundred and thirty-five patients were tested for procalcitonin (PCT) upon admission, and 100% patients in the non-survivors had elevated PCT. Patients in non-survivors had more laboratory abnormalities than those in mild and severe.
Treatments During the Hospitalization
Two hundred and seventeen (41.49%) patients received respiratory support during hospitalization, of which 18 (4.2%) of mild patients received nasal catheter inhalation, as shown in Table 3. The respiratory support rate of the severe was higher than that of the mild, and the non-survivors all received mechanical ventilation treatment, of which six received non-invasive mechanical ventilation treatment and 11 received invasive mechanical ventilation treatment. Nine patients in the severe received ECMO treatment, and no one survived. Thirty-nine (52.7%) of the severe survivors were treated with CRRT, and only 5 (33.33%) of the non-survivors applied this technique. In terms of drug treatment, antiviral treatment was commonly used in each group. The severe had a higher proportion of antibiotics than the mild, and the non-survivors had a higher proportion of carbapenem and glycopeptide antibiotics than the survivors. One hundred and twelve (21.41%) received glucocorticoid therapy, and the non-survivors received a higher proportion of glucocorticoid therapy than the severe survivors (62.5 vs. 41.03%).
Table 3.
Treatments during the hospitalization.
| Treatments | All patients (N = 523) | Disease severity | Clinical outcome in severe | ||
|---|---|---|---|---|---|
| Mild (N = 429) | Severe (N = 94) | Non-survivors (N = 16) | Survivors (N = 78) | ||
| Oxygen support | 217/523 (41.49) | 152/429 (35.43) | 65/94 (69.15) | 15/16 (93.75) | 50/78 (64.1) |
| Oxygen inhalation through nasal catheter | 73/523 (13.96) | 18/429 (4.2) | 55/94 (58.51) | 9/16 (56.25) | 46/78 (58.97) |
| Usage time, days | 10 (6–14.5) | 10 (7–14) | 7.5 (4–14.75) | 3.5 (2–4.25) | 11 (4–15.75) |
| High-flow oxygen | 48/523 (9.18) | 0/429 (0) | 48/94 (51.06) | 10/16 (62.5) | 38/78 (48.72) |
| Usage time, days | 6 (4–11) | NA (NA-NA) | 6 (4–11) | 3.5 (2–4.75) | 8 (4.75–12.25) |
| Non-invasive mechanical ventilation | 15/523 (2.87) | 0/429 (0) | 15/94 (15.96) | 6/16 (37.5) | 9/78 (11.54) |
| Usage time, days | 6 (5–12) | NA (NA-NA) | 6 (5–12) | 4.5 (3.25–7.25) | 7 (6–13) |
| Invasive mechanical ventilation | 13/523 (2.49) | 0/429 (0) | 13/94 (13.83) | 11/16 (68.75) | 2/78 (2.56) |
| Usage time, days | 2 (1.25–7.75) | NA (NA-NA) | 2 (1.25–7.75) | 2 (1–4) | 13 (13–13) |
| ECMO | 9/523 (1.72) | 0/429 (0) | 9/94 (9.57) | 9/16 (56.25) | 0/78 (0) |
| Usage time, days | 7 (2–9) | NA (NA-NA) | 7 (2–9) | 7 (2–9) | NA (NA-NA) |
| CRRT | 44/449 (9.8) | 0/360 (0) | 44/89 (49.44) | 5/15 (33.33) | 39/74 (52.7) |
| Adsorptive | 12/44 (27.27) | 0/0 (NA) | 12/44 (27.27) | 3/5 (60) | 9/39 (23.08) |
| Usage time, times | 3 (2–3) | NA (NA-NA) | 3 (2–3) | 2 (1.5–2.5) | 3 (3–3) |
| Non-adsorptive | 32/44 (72.73) | 0/0 (NA) | 32/44 (72.73) | 2/5 (40) | 30/39 (76.92) |
| Usage time, times | 2 (2–2) | NA (NA-NA) | 2 (2–2) | NA (NA-NA) | 2 (2–2) |
| Drug Treatment | |||||
| Antiviral treatment | 497/523 (95.03) | 408/429 (95.1) | 89/94 (94.68) | 14/16 (87.5) | 75/78 (96.15) |
| Other | 174/523 (33.27) | 142/429 (33.1) | 32/94 (34.04) | 4/16 (25) | 28/78 (35.9) |
| Immunotherapy | 181/523 (34.61) | 126/429 (29.37) | 55/94 (58.51) | 13/16 (81.25) | 42/78 (53.85) |
| Methylprednisolone | 103/523 (19.69) | 61/429 (14.22) | 42/94 (44.68) | 10/16 (62.5) | 32/78 (41.03) |
| Usage time, days | 5 (3–6) | 5 (3–6) | 5 (3–6) | 5 (3–6) | 5 (3–6) |
| Prednisone | 9/523 (1.72) | 9/429 (2.1) | 0/94 (0) | 0/16 (0) | 0/78 (0) |
| Usage time, days | 6 (4–14) | 6 (4–14) | NA (NA-NA) | NA (NA-NA) | NA (NA-NA) |
| Immunoglobulin | 80/523 (15.3) | 43/429 (10.02) | 37/94 (39.36) | 12/16 (75) | 25/78 (32.05) |
| Usage time, days | 4 (3–5) | 4 (3–4.25) | 4 (2–5) | 2.5 (1.75–4.25) | 4 (3–5) |
| Thymosin α | 68/523 (13) | 50/429 (11.66) | 18/94 (19.15) | 4/16 (25) | 14/78 (17.95) |
| Usage time, days | 8.5 (6–13.75) | 8 (5–13) | 11 (8–15) | 9 (6.5–10) | 11 (8.25–15.75) |
| Antibiotics | 347/523 (66.35) | 279/429 (65.03) | 68/94 (72.34) | 12/16 (75) | 56/78 (71.79) |
| Quinolone | 266/523 (50.86) | 220/429 (51.28) | 46/94 (48.94) | 5/16 (31.25) | 41/78 (52.56) |
| Penicillin | 67/523 (12.81) | 51/429 (11.89) | 16/94 (17.02) | 3/16 (18.75) | 13/78 (16.67) |
| Cephems | 99/523 (18.93) | 72/429 (16.78) | 27/94 (28.72) | 6/16 (37.5) | 21/78 (26.92) |
| Carbapenem | 25/523 (4.78) | 11/429 (2.56) | 14/94 (14.89) | 6/16 (37.5) | 8/78 (10.26) |
| Glycopeptide | 9/523 (1.72) | 2/429 (0.47) | 7/94 (7.45) | 5/16 (31.25) | 2/78 (2.56) |
| Tetracycline | 11/523 (2.1) | 3/429 (0.7) | 8/94 (8.51) | 2/16 (12.5) | 6/78 (7.69) |
| Antifungai agents | 26/523 (4.97) | 6/429 (1.4) | 20/94 (21.28) | 10/16 (62.5) | 10/78 (12.82) |
| Fluconazole | 4/523 (0.76) | 0/429 (0) | 4/94 (4.26) | 2/16 (12.5) | 2/78 (2.56) |
| Voriconazole | 11/523 (2.1) | 1/429 (0.23) | 10/94 (10.64) | 6/16 (37.5) | 4/78 (5.13) |
| Caspofungin | 8/523 (1.53) | 0/429 (0) | 8/94 (8.51) | 5/16 (31.25) | 3/78 (3.85) |
| Anti-inflammatory treatment | 198/523 (37.86) | 148/429 (34.5) | 50/94 (53.19) | 11/16 (68.75) | 39/78 (50) |
| Acetylcysteine Effervescent Tablets | 38/523 (7.27) | 27/429 (6.29) | 11/94 (11.7) | 1/16 (6.25) | 10/78 (12.82) |
Data are median (IQR)or n/N (%). ECMO, extracorporeal membrane oxygenation; CRRT, continuous renal replacement therapy.
Death Prediction Model
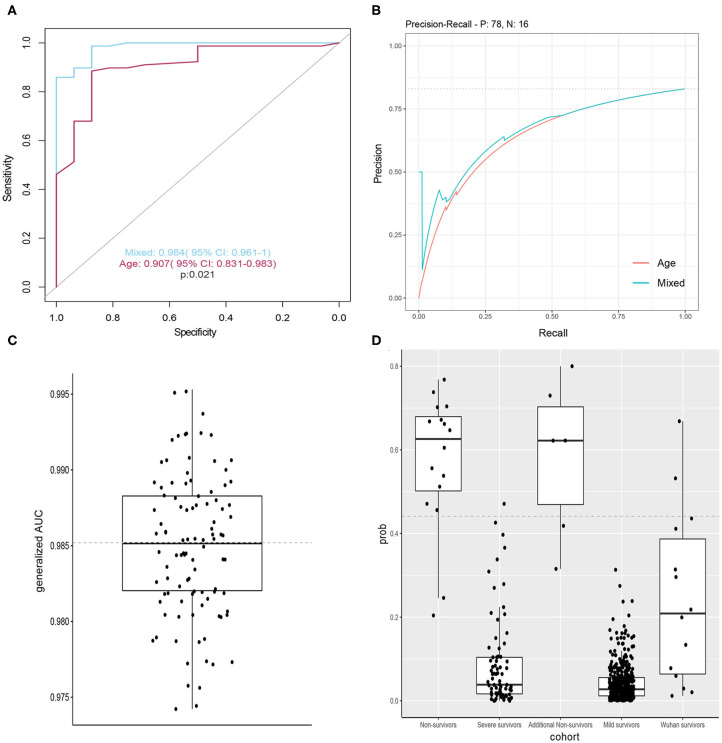
We constructed classification models to evaluate death risk for severe patients. Model performance was assessed by receiver operating characteristic (ROC) curve analysis using the area under the curve (AUC). In considering age is among leading risk factors for poor prognosis in several studies (3, 6, 7, 9–11), we firstly constructed models by using single age, which could achieve and AUC of 0.907 (95% CI 0.831–0.983) for death and alive severe COVID-19 patients. Mixed models constructed with combination of age, demographics, symptoms, and laboratory tests when firstly admitted to hospital had better performance (p = 0.021) and could achieved an AUC of 0.984 (95% CI 0.961–1) for death and alive severe COVID patients (Figures 2A,B). In considering fetal cases are with a small sample size, we randomly chose 40 samples from severe cases, then calculated the generalized AUC by using death probabilities and the median generalized AUC was 0.9852 (Figure 2C). Pulse oxygen, age, creatinine, creatine kinase, D-Dimer are the most important features (Table 4). We chose 0.441 as death prediction threshold (with 0.85 sensitivity and 0.987 specificity), then used six additional fatal cases (Henan), 429 mild cases and 14 cases (Wuhan) as independent validation cohort, and four in six death cases (0.67%) were assigned as death and majority of predicted death probabilities in the mild Henan cases and those Wuhan cases were below 0.441 (Figure 2D). Summary characteristics of six Henan additional fatal cases and 14 Wuhan cases and were outlined in Table 5.
Figure 2.
Models to predict death risk. (A) Performance of the classifiers using AUCs, significance determined by single sided AUC comparison by using bootstrap method with 10,000 permutations (boot. n = 10,000). (B) Precision-recall curves of models based on mixed features and age. (C) Distribution of generalized AUC by using bootstrap sampling (n = 100). (D) Boxplots showing distribution of death probabilities among different cohorts. horizon dashed line indicates selected threshold.
Table 4.
Importance of features in death risk prediction model.
| Feature | Mean decrease Gini |
|---|---|
| Pulse oxygen saturation <90% | 3.234 |
| Age | 3.025 |
| Creatinine | 1.907 |
| Creatine Kinase | 1.903 |
| D-Dimer | 1.787 |
| Neutrophil percentage | 1.292 |
| Lactic dehydrogenase | 1.274 |
| Leucocyte count | 1.207 |
| Albumin | 1.047 |
| Time form illness onset to hospital admission | 0.815 |
| Glutamic-oxalacetic transaminase | 0.737 |
| Neutrophil count | 0.731 |
| Lymphocyte percentage | 0.685 |
| Respiratory rate >24 breaths per min | 0.613 |
| Prothrombin time | 0.587 |
| Blood urea nitrogen | 0.562 |
| Platelet | 0.504 |
| Direct bilirubin | 0.486 |
| C-reactive protein | 0.405 |
| Incubation period | 0.388 |
| Eosinophil percentage | 0.264 |
| Temperature | 0.259 |
| Chronic respiratory disease | 0.207 |
| Chronic obstructive pulmonary disease | 0.202 |
| Chest tightness | 0.125 |
| Diabetes | 0.119 |
| Coronary heart disease | 0.116 |
| Chest CT | 0.095 |
| Cardiovascular disease | 0.072 |
| Hypertension | 0.064 |
| Dyspnea | 0.057 |
| Cluster | 0.050 |
| Expectoration | 0.031 |
CT, computerized tomography.
Table 5.
Summary characteristics of six Henan additional fatal cases and 14 Wuhan cases.
| Feature | WH (n=14) | HN (N=6) |
|---|---|---|
| Age | 63.5 (45–75.5) | 77 (65–78.5) |
| Time form illness onset to hospital admission | 10 (7–14.75) | 2.5 (1–7) |
| Breath | 21.5 (20.25–22.75) | 24.5 (23.25–25.75) |
| Cardiovascular disease | 3/14 (21.43) | 4/5 (80) |
| Diabetes | 2/14 (14.29) | 1/5 (20) |
| Coronary heart disease | 2/14 (14.29) | 1/5 (20) |
| Systolic pressure | 128 (123.25–133) | 157.5 (144.5–160) |
| Chest tightness | 10/14 (71.43) | 2/6 (33.33) |
| Pulse oxygen saturation | 92.5 (90–94.75) | 93.5 (89–95) |
| Chronic respiratory disease | 1/14 (7.14) | 2/5 (40) |
| Chronic obstructive pulmonary disease | 1/14 (7.14) | 2/5 (40) |
| Hypertension | 5/14 (35.71) | 4/5 (80) |
| Temperature | 36.75 (36.5–36.88) | 37 (36.58–37.43) |
| Sputum | 2/14 (14.29) | 4/6 (66.67) |
| Cluster | 2/14 (14.29) | 1/6 (16.67) |
| Dyspnea | 7/14 (50.00) | 1/6 (16.67) |
| Incubation period | 0 (0–0) | 8 (4–10.5) |
| White Blood Cell | 5.95 (4.74–7.09) | 8.04 (6.52–10.53) |
| Aspartate Aminotransferase | 25 (23–30) | 28.5 (18.5–50.5) |
| Lactic dehydrogenase | 175.5 (153.5–235) | 566.5 (294.25–876.25) |
| Blood urea nitrogen | 6.13 (4.32–7.27) | 10.46 (5.01–18.28) |
| Neutrophil percentage | 69.4 (62.7–79.1) | 92.35 (90.83–94.4) |
| Albumin | 32.8 (30.2–37.6) | 33.2 (29.38–35.75) |
| Creatinine | 70 (60.2–103) | 57 (42.36–67.25) |
| D-Dimer | 0.71 (0.33–0.86) | 40.31 (40.31–40.31) |
| Neutrophil count | 4.33 (3.23–5.29) | 7.33 (6.1–9.92) |
| Creatine Kinase | 66.5 (50.25–115.25) | 133.5 (66.25–197.75) |
| Platelet | 235.5 (196.25–309) | 110 (98–156.5) |
| Direct bilirubin | 4.2 (3.1–6.1) | 4.1 (3.1–13.5) |
| Lymphocyte percentage | 18.6 (12.8–28) | 5 (3.33–6.3) |
| Eosinophil percentage | 0.6 (0–1) | 0.1 (0.03–0.33) |
| C-reactive protein | 23.35 (3.95–66.53) | 55.25 (19.74–106.43) |
| Prothrombin time | 11.5 (10.7–12.2) | 14.2 (14.1–14.3) |
Data are median (IQR)or n/N (%).
Discussion
Henan Province has a large population of 95.593 million people, bordering Hubei Province, China. As of April 1, 2020, there were 1,273 people confirmed COVID-19 in Henan, which was the second most in China outside Hubei Province. We collected data of 523 confirmed COVID-19 cases who had been discharged from 18 cities in Henan Province before February 20, 2020 and conducted statistical analysis. Our data showed that the main epidemiological characteristics of novel coronavirus pneumonia in Henan Province were import and cluster, which were similar to other provinces and cities outside Hubei in China. Among the 523 cases, there were 289 males (55.26%) and 234 females (44.74%). Other reports also showed a higher percentage of males (9, 12, 13), suggesting that males were more susceptible. Our study suggested that people of all ages were generally susceptible, with people aged 18–64 accounting for 87.96%, which was consistent with the Chinese CDC report (3). In our study, there were 16 fatal cases before February 20, 2020, and 87.5% of the deaths were ≥65 years old, with a median age of 71 years, while the median age for the mild and severe survivors was 42 and 50 years, respectively. The most common comorbidities in the non-survivors were hypertension (46.67%), coronary heart disease (33.33%), diabetes (33.33%), and COPD (33.33%). The average number of comorbidities in non-survivors was 1.94. Several studies about severe novel coronavirus pneumonia in China suggested that advanced age and comorbidities were high-risk factors for COVID-19 patients to develop into severe and death (10, 13, 14). In our study, advanced age was the biggest risk factor for death, which was consistent with that. A study from Italy involving 1,043 critically ill COVID-19 cases showed similar results, but male patients accounted for a higher proportion (82%) (9).
The median incubation period of the 523 cases in Henan Province was 5 days, and there was no significant difference between mild and severe. The median time from the onset of symptoms to hospitalization in the non-survivors was 8 days, and it was significantly longer than the severe survivors, suggesting that a delay in hospitalization might be one of the factors leading to death. Fever (88.74%), cough (62.3%), fatigue (39.58%), and expectoration (28.75%) were the most common symptoms. In spite of more symptoms, 60.87% of the severe and 80% of non-survivors had a temperature below 37.5°C at the time of admission. Zhong et al.'s study on 1,099 cases of COVID-19 also found that 52% of patients did not have fever when they became ill (12). The lack of fever symptoms made it difficult to identify COVID-19 patients and could also be one of the factors that caused a delay in visiting the doctor. Another study on refractory COVID-19 also found that the refractory pneumonia cases had a significantly lower fever incidence than the common pneumonia cases, suggesting that slow or poor response to SARS-CoV-2 was more likely to cause severe illness (15).
Compared with the mild and severe survivors, the non-survivors had higher leucocyte count, neutrophil percentage, D-dimer, LDH, BNP, and PCT levels, while the proportion of eosinophils, lymphocytes and albumin were lower, which was consistent with other studies. White blood cell count, neutrophil percentage and elevated PCT suggested that the non-survivors might be hospitalized with bacterial infection. Low albumin indicated that the patient was seriously depleted and the nutritional level was poor. D-dimer elevation had been confirmed in multiple studies as a high-risk factor for severe illness and death (10, 16, 17), which was consistent with our study. Chen et al.'s study found that in the non-survivors 56% had increased leucocyte count and 91% had lymphopenia, while in the severe survivors 4% had increased leucocyte count and 47% had lymphopenia (10). Zhang et al.'s study found that most COVID-19 cases combined with lymphopenia (75.4%) and eosinophilia (52.9%), and lymphopenia and eosinophilia were associated with disease severity (17). In our study, eosinophilia generally occurred in all cases, and there was no significant difference between the non-survivors and the severe survivors, but most of the eosinophils in the severe survivors returned to normal when discharged, while that of the non-survivors continued to decrease. Liu et al. also found that eosinophilia might be an indicator of disease improvement (18).
In the non-survivors, 100% of the patients had chest CT pneumonia area > 15% at admission, which was more severe in imaging than the mild and severe survivors. In terms of respiratory support, the rate of mechanical ventilation in the non-survivors was significantly higher than that in the mild and the severe survivors, which also suggested that the lung function of the non-survivors was more seriously impaired. In the non-survivors, the percentage of invasive mechanical ventilation was 68.75%, higher than other reports from Wuhan, China, but lower than those reported by the United States (71%) and Italy (88%), and Henan Province's mortality rate was also lower than that of the United States and Italy (9, 19). In addition to the aging factor, the fatal rate difference between Italy and Henan Province could be due to the fact that the number of COVID-19 cases in Henan province was relatively smaller and the medical resources were relatively more sufficient. Nine patients were applied with extracorporeal membrane oxygenation and technology (ECMO), but no one survived. Research showed application of ECMO could reduce mortality of patients with H1N1-related ARDS and MERS-related ARDS (20, 21), but there was no large-scale clinical report on the application of ECMO in the treatment of COVID-19, and its success rate is still unclear. In Yang et al.'s report, six patients applied ECMO with only one survived (13). Only a few cases were reported with successful ECMO treatment (22, 23). The recovery of lymphocyte count was the key factor for improvement of COVID-19. The application of ECMO destroyed lymphocytes and affected the function of lymphocytes. At the same time, it could cause IL-6 increase. This could be a reason for the low success rate of ECMO treatment. How to successfully apply ECMO in the treatment of COVID-19 still requires further research. 52.7% of patients in the critical severe survivors applied CRRT technology, while 33.33% patients in the non-survivors, suggesting that CRRT could help improve the prognosis of COVID-19. The application rate of glucocorticoids in the non-survivors was significantly higher than that in the mild and the severe survivors, which was consistent with other studies. Glucocorticoids had been widely used in SARS-CoV and MERS-CoV, but studies showed that the application of glucocorticoids prolonged the clearance time of virus and the probability of mental illness was significantly increased (24). Similarly, there was no evidence that glucocorticoids were beneficial to improve the prognosis of patients with COVID-19. Whether glucocorticoids can improve the prognosis of COVID-19 still requires long-term follow-up and further research.
In our study, some independent risk factors for death were found and we firstly developed a forest tree to accurately predict clinical outcomes of patients with COVID-19 based on combination of age, demographic features, symptoms and clinical tests at admission. Old age was the most important risk factor for poor prognosis of COVID-19 patients. The mixed model conducted by forest tree performed well in predicting survival and death, with AUC of 0.984 (95% confidence interval 0.961–1) for survival and death, which is helpful for further understanding and improve clinical strategies against COVID-19. We also found the predicted value was positively correlated with the severity of COVID-19. Of the 14 confirmed cases from Wuhan, seven were mild, seven were severe, 13 were cured and discharged, and one was referred to other hospital due to critical illness. In the death prediction model based on Wuhan data, those with a predicted value >0.3 were all critically ill, and the respiratory support treatment intensity was higher than the other 10 cases. The predictive value of the case transferred to other hospitals due to critical illness was 0.673, unfortunately we failed to follow up on the clinical outcome. The death prediction model we have established has also been validated in mild and six other fatal cases in Henan Province. The prediction of death for all mild survivors was below 0.3 and 4 in six death cases (66.67%) were assigned as death.
Mild patients have rare fetal cases thus we excluded mild cases in the death prediction models. Several studies have constructed models for early identification of cases at high risk of progression to severe COVID-19 (11) or improved prognosis (25). However, fatal cases were always rapid disease progression and died in hospitals in a short time, though we have plenty of medical support in Henan province. To the best of our knowledge, this is the first death prediction model for COVID-19 established by random forest. The model can accurately predict the prognosis of patients with COVID-19. Our study provided a new method for the evaluation of disease severity. Early identification of high-risk COVID-19 cases and early supportive therapy is critical to the prognosis.
There are some limitations of our study. Firstly, this is a retrospective study. There was incomplete documentation of the history, symptoms, or laboratory findings in some cases, even after trying to feedback and recollect. Secondly, as a retrospective and observational study, although this random forest model was validated in mild cases and additional fatal cases in Henan Province and 14 cases from Wuhan and showed good predictive effects, there were few validators outside Henan Province. Thirdly, imageology lacked objective judgment standards, and the investigators' judgment was subjective, which might lead to some bias.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation. The datasets generated for this study can be found here: https://github.com/xiaoshubaba/COVID-Henan.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee from The First Affiliated Hospital of Zhengzhou University. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
QZ, AX, and ZY made substantial contributions to conception, designed the study, had full access to all of the data in the study, take responsibility for the integrity of the data, and the accuracy of the data analysis. XM and AL drafted the manuscript, critically revised the manuscript for important intellectual content, and gave final approval for the version to be published. XM, AL, MJ, QS, and XA did the data analysis. XM, AL, MJ, QS, XA, YF, HLia, JC, HLi, JL, ZR, RS, GC, YZ, MC, LX, PJ, and YW collected the data and checked the data. All authors agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are very grateful to all the investigators who collected the clinical data of this study.
Footnotes
Funding. This work was supported by Chinese National Science and Technology Major Project (2018ZX10305410); Henan Province Medical Science and Technique Project (2018020001); Henan Province postdoctoral research grant (001801005); Henan Province Key Scientific Research Projects of Higher Education Institutions (20B320032); and Beijing Medical and Health Public Welfare Foundation (YWJKJJHKYJJ-B182121-1).
References
- 1.Meo SA, Alhowikan AM, Al-Khlaiwi T, Meo IM, Halepoto DM, Iqbal M, et al. Novel coronavirus 2019-nCoV: prevalence, biological and clinical characteristics comparison with SARS-CoV and MERS-CoV. Eur Rev Med Pharmacol Sci. (2020) 24:2012–9. 10.26355/eurrev_202002_20379 [DOI] [PubMed] [Google Scholar]
- 2.Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY. Coronaviruses—drug discovery and therapeutic options. Nat Rev Drug Discov. (2016) 15:327–47. 10.1038/nrd.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72,314 cases from the Chinese Center for Disease Control and Prevention. JAMA. (2020) 323:1239–42. 10.1001/jama.2020.2648 [DOI] [PubMed] [Google Scholar]
- 4.Zhao S, Lin Q, Ran J, Musa SS, Yang G, Wang W, et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: a data-driven analysis in the early phase of the outbreak. Int J Infect Dis. (2020) 92:214–7. 10.1101/2020.01.23.916395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J. Pathogenicity and transmissibility of 2019-nCoV-A quick overview and comparison with other emerging viruses. Microbes Infect. (2020) 22:69–71. 10.1016/j.micinf.2020.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. (2020) 395:1054–62. 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss P, Murdoch DR. Clinical course and mortality risk of severe COVID-19. Lancet. (2020) 395:1014–5. 10.1016/S0140-6736(20)30633-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Health Commission of the Pople's Republic of China New Coronavirus Pneumonia Prevention and Control Program. 6th ed National Health Commission of the Pople's Republic of China; (2020) Available online at: http://www.nhc.gov.cn/yzygj/s7653p/202002/8334a8326dd94d329df351d7da8aefc2/files/b218cfeb1bc54639af227f922bf6b817 (accessed May 7, 2020). [Google Scholar]
- 9.Grasselli G, Zangrillo A, Zanella A, Antonelli M, Cabrini L, Castelli A, et al. Baseline characteristics and outcomes of 1,591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy Region, Italy. JAMA. (2020) 323:1574–81. 10.1001/jama.2020.5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen T, Wu D, Chen H, Yan W, Yang D, Chen G, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. (2020) 368:m1091. 10.1136/bmj.m1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong J, Ou J, Qiu X, Jie Y, Chen Y, Yuan L, et al. A tool to early predict severe corona virus disease 2019 (COVID-19): a multicenter study using the risk nomogram in Wuhan and Guangdong, China. Clin Infect Dis Off Publ Infect Dis Soc Am. (2020) 71:833–40. 10.1093/cid/ciaa443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. (2020) 382:1708–20. 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. (2020) 8:475–81. 10.1016/S2213-2600(20)30079-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. (2020) 180:1–11. 10.1001/jamainternmed.2020.0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mo P, Xing Y, Xiao Y, Deng L, Zhao Q, Wang H, et al. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin Infect Dis. (2020) 58:ciaa270. 10.1093/cid/ciaa270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han H, Yang L, Liu R, Liu F, Wu KL, Li J, et al. Prominent changes in blood coagulation of patients with SARS-CoV-2 infection. Clin Chem Lab Med. (2020) 58:1116–20. 10.1515/cclm-2020-0188 [DOI] [PubMed] [Google Scholar]
- 17.Zhang JJ, Dong X, Cao YY, Yuan YD, Yang YB, Yan YQ, et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. (2020) 75:1730–41. 10.1111/all.14238 [DOI] [PubMed] [Google Scholar]
- 18.Liu F, Xu A, Zhang Y, Xuan W, Yan T, Pan K, et al. Patients of COVID-19 may benefit from sustained lopinavir-combined regimen and the increase of eosinophil may predict the outcome of COVID-19 progression. Int J Infect Dis. (2020) 95:183–91. 10.1016/j.ijid.2020.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arentz M, Yim E, Klaff L, Lokhandwala S, Riedo FX, Chong M, et al. Characteristics and outcomes of 21 critically ill patients with COVID-19 in Washington State. JAMA. (2020) 323:1612–4. 10.1001/jama.2020.4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noah MA, Peek GJ, Finney SJ, Griffiths MJ, Harrison DA, Grieve R, et al. Referral to an extracorporeal membrane oxygenation center and mortality among patients with severe 2009 influenza A(H1N1). JAMA. (2011) 306:1659–68. 10.1001/jama.2011.1471 [DOI] [PubMed] [Google Scholar]
- 21.Hong X, Xiong J, Feng Z, Shi Y. Extracorporeal membrane oxygenation (ECMO): does it have a role in the treatment of severe COVID-19. Int J Infect Dis. (2020) 94:78–80. 10.1016/j.ijid.2020.03.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhan WQ, Li MD, Xu M, Lu YB. Successful treatment of COVID-19 using extracorporeal membrane oxygenation, a case report. Eur Rev Med Pharmacol Sci. (2020) 24:3385–9. 10.26355/eurrev_202003_20705 [DOI] [PubMed] [Google Scholar]
- 23.Tavazzi G, Pellegrini C, Maurelli M, Belliato M, Sciutti F, Bottazzi A, et al. Myocardial localization of coronavirus in COVID-19 cardiogenic shock. Eur J Heart Fail. (2020) 22:911–5. 10.1002/ejhf.1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russell CD, Millar JE, Baillie JK. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet. (2020) 395:473–5. 10.1016/S0140-6736(20)30317-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiaojiao Xie, Ding Shi, Mingyang Bao, Xiaoyi Hu, Wenrui Wu, Jifang Sheng, et al. A predictive nomogram for predicting improved clinical outcome probability in patients with COVID-19 in Zhejiang Province, China. Engineering. (2020). [Epub ahead of print]. 10.1016/j.eng.2020.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation. The datasets generated for this study can be found here: https://github.com/xiaoshubaba/COVID-Henan.