Abstract
Human gut microbiota plays an important role in individual health. When the balance between host and gut microbiota is disrupted, changes in microbiota composition and function occur, which is referred as dysbiosis. Environmental factors as diet, proton pump inhibitors, and antibiotics can lead to a permanent dysbiotic disruption. Clarification of these imbalances was made possible by recent advances in genome sequencing methods that supported acknowledgment of the interplay between microbiome and intestinal and extraintestinal disorders. This review focuses on the microbiota impact in inflammatory bowel disease, gastric cancer, colorectal cancer, nonalcoholic fatty liver disease (NAFLD), irritable bowel syndrome (IBS), and Clostridium difficile infection (CDI). Furthermore, novel therapies are summarized. Fecal microbiota transplant (FMT) is a successful and established therapy in recurrent CDI, and its application in other dysbiosis-related diseases is attracting enormous interest. Pre- and probiotics target microbial rebalance and have positive effects mainly in NAFLD, ulcerative colitis, IBS, and CDI patients. Promising anticarcinogenic effects have also been demonstrated in animal models. The literature increasingly describes microbial changes in many dysbiotic disorders and shows what needs to be treated. However, probiotics and FMT application in clinical practice suffers from a shortage of randomized controlled trials with standardized therapy regimens to support their recommendation.
Keywords: Microbiota, Dysbiosis, Dysbiosis-associated diseases, Probiotics, Fecal microbiota transplantation
Resumo
A microbiota gastrointestinal desempenha um papel essencial na saúde humana.Quando surge um desequilibrio entre esta e o hospedeiro, ocorrem alterações na composição e fundo da microbiota que se designa de dísbiose. Fatores ambientais como a dieta, inibidores da bomba de protões e antibióticos podem induzir um estado disbiótico permanente. O esclarecimento destas desregulações foi possível graças a avanços recentes nos métodos de sequenciação genómica que, por sua vez, nos ajudaram a perceber a interação entre a microbiota e distúrbios intestinais e extra-intestinais. Esta revisão foca-se no impacto da microbiota na Doença Inflamatória Intestinal, Cancros gástrico e colorretal, Fígado Gordo Não-Alcoólico (FGNA), Síndroma do Intestino Irritável (SII) e Infeção por Clostridium difficile (ICD). Além disso, resume-se no final as estratégias terapéuticas que têm surgido. O transplante fecal é um tratamento com eficácia estabelecida na infeção recorrente por C. difficile, reunindo um crescente interesse na sua aplicado em outras doenças relacionadas com disbiose. Os pre- e probióticos promovem o reequilíbrio microbiano e têm evidência de efeitos positivos sobretudo no FGNA, Colite Ulcerosa, Sll e ICD. Em modelos animais, estes também demonstraram efeitos anti-carcinogénicos promissores. É evidente na literatura uma descrido cada vez mais pormenorizada das alterações microbianas em vários distúrbios disbióticos, oferecendo ferramentas que permitem saber o que pode ser tratado. No entanto, a aplicado de probióticos e transplante fecal na prática clínica sofre aínda de carência de ensaios clínicos randomizados, com regimes terapêuticos standard, em número que permita a sua recomendação formal.
Palavras Chave: Microbiota, Disbiose, Doenças associadas a disbiose, Probióticos, Transplante de microbiota fecal
Introduction
The gastrointestinal tract is our most colonized organ, with one hundred trillion organisms symbiotically related to humans [1]. Human microbiota refers to the entire population of microorganisms (bacteria, fungi, archaea, viruses, and protozoans) that live in the human body, with their collective genomes forming the human metagenome [2].
In a healthy individual, microbiota has important metabolic [3] and immune functions [4]. In the colon, it provides the enzymes needed to fermentation of carbohydrates, generating short-chain fatty acids (SCFA) as butyrate. SCFA provide about 70% of energy to the colonocytes, preserving the intestinal barrier and enhancing anti-inflammatory and anticancer properties [5]. Host-microbiome interplay is essential for the maintenance of a strong immune system, allowing gastrointestinal tract to remain healthy and free from pathogenic bacteria [6].
Gut microbial composition varies greatly between individuals and over different gastrointestinal segments. However, among healthy adults, it is consistently dominated by bacteria of two phyla, Bacteroidetes and Firmicutes, while Actinobacteria and Proteobacteria are less frequently found [7]. Notably, the diversity and density of bacterial species increases from proximal to distal gut.
A challenging aspect of microbiota study is the difficulty in defining an ideal eubiosis state, due to this high variability between healthy individuals. However, in most studies, certain commensal bacteria, as Roseburia, Akkermansia, Bifidobacterium, and Faecalibacterium prausnitzii seem to be associated with a healthy gut. Regardless, sometimes the balance between the host and gut microbiota is lost, and changes in its composition and function occur, which is often referred to as dysbiosis [8]. Microbial colonies live in highly competitive surroundings, fighting for the same niches and resources. Dysbiosis typically features one or more of the following characteristics: loss of commensals, excessive growth of potentially harmful organisms, and a reduction of overall diversity [9].
Changes in microbiota abundance and composition have been related to several human intestinal and extraintestinal disorders. However, there is a gap between basic scientific findings and clinical practice. In this review, we describe factors affecting microbiota, and then summarize microbiota imbalances associated with prevalent gastrointestinal diseases and potential therapies, namely inflammatory bowel disease (IBD), gastric cancer (GC), colorectal cancer (CRC), nonalcoholic fatty liver disease (NAFLD), Clostridium difficile infection (CDI), and irritable bowel syndrome (IBS).
Methods for Microbiome Analysis
Generally, gastrointestinal microorganisms are difficult to isolate in culture, making it challenging to understand the true microbial complexity. This limitation has been partially overcome over the last few decades by using DNA sequencing to define the genetic content of the microbiome.
Most techniques target the 16S ribosomal RNA (16SrRNA) gene, as it is highly conserved between different species of bacteria. While sequencing this gene, we answer the question “who is there”? revealing the relative abundance of a given bacterial taxa.
Marker gene analysis differs from metagenomic shotgun sequencing (so called whole genome shotgun sequencing − WGS) as it is based on targeting an amplicon (a target sequence or gene that is amplified) instead of trying to sequence all genes in a sample [10].
16SrRNA sequencing technique hardly approaches the deepest taxonomic levels and does not infer a relation between a bacterial species and its functional capacity. However, this technique remains cheaper, easily accessible and still acknowledges the microbial diversity and composition [11].
WGS sequencing followed by metagenomic analysis also identifies the quantity and diversity of microbial community but adds more detailed information to the taxonomic characterization of a sample. With this approach, we answer the question “what can they do,” by identifying the gene content and inferred potential functions of the microbial DNA [12]. Genomic DNA is isolated from the environment understudy, but it does not distinguish whether it comes from viable cells or not, or whether the anticipated genes are expressed and in under what circumstances [13].
The introduction of metatranscriptomic analysis has surpassed the limitations of all other gut microbiome methods. Metatranscriptomics reveals the total content of gene transcripts (tRNA) in the microbiota, answering the question “what are they doing”? [11]. As metatranscriptomic analysis complements metagenomics, we are able to know which of the genes that were marked in the metagenomic analysis are transcribed and to what extent, demonstrating functional changes that contribute to health and disease even before a compositional modification occurs [14]. Nevertheless, metatranscriptomics is even more expensive and complex than metagenomics.
The combination of all approaches may open a window to deeper understanding of potential genes and pathways that can contribute to homeostasis and disease susceptibility, so new pharmacological targets could be explored [15].
Factors Affecting Microbiota
Many studies have been conducted on factors capable of influencing the human gastrointestinal microbiome, including host genetics and environmental factors [7]. Here, we will focus on the latter, namely diet, proton pump inhibitors (PPI), and antibiotics.
Diet
Within environmental factors, diet is perhaps the most easily modifiable. Many studies tried to elucidate the link between western diet and intestinal dysbiosis as a trigger to impairment of intestinal permeability and barrier function [16]. Western diet mainly consists of animal proteins, total and saturated fats and simple sugars, rather than fruits and vegetables, and was associated with reduced bacterial colonization and an increased number of Bacteroides (mainly mucin-degrading bacteria), Firmicutes, and Proteobacteria phyla [17]. A recent study reported that a low-fiber diet resulted in microbiota consuming the mucus layer, which leads to a thinner epithelium, enhancing the chances of invasive pathogenic attack [18].
Mediterranean diet is based on high consumption of fruits, vegetables, unrefined cereals, and fish and low consumption of red meat and was associated with benefits to gut microbiota, lowering Escherichia coli load and increasing SCFA levels [19].
Gut microbiota undergoes relatively rapid changes upon exposure to different diets. David et al. [20] showed that high-fat diets consisting exclusively of animal-based foods changed the microbiome in only 2 days in both structure and function. Nonetheless, the magnitude of this effect seems modest, as dietary change does not necessarily result in permanent compositional modification at least at phylum level, since daily changes are mainly at genus and species level. In the same study, RNA sequencing analysis revealed increased expression of genes related to amino acid catabolism and the production of carcinogenic polycyclic aromatic hydrocarbons in the animal-based diet, compared with plant-based diet.
Knowing the fact that diet can promote gut dysbiosis and consequently associated diseases, it is clear that diet modification should be considered as an adjuvant therapy.
Proton Pump Inhibitors
The luminal gastric pH is normally 1.5–3.5, and PPI raise pH to values above 3.5–5.0, which can affect the composition of microbiota due to increased pH and also by directly targeting the proton pump of microorganisms.
PPI use can lead to the development of enteric infections, such as CDI [21]. Tsuda et al. [22] analyzed 18 subjects taking PPI for more than 2 years and 27 healthy volunteers who had never taken a PPI and found a significant correlation between pH value and bacterial count in all gastric fluid samples. Their data suggested that bacterial overgrowth in the stomach while using PPI may arise from a lack of killing rather than proliferation of bacteria due to low gastric acidity.
A recent study carried out in Japan also examined fecal samples of patients at three points (before and 4 and 8 weeks after starting PPI treatment) [23]. Differences in total bacterial counts at these 3 points were not significant, although the total counts of facultative anaerobes, such as Lactobacillus, Streptococcus, Staphylococcus,and Enterobacteriaceae were significantly increased after treatment [23]. Streptococci are commensals of the human oral cavity, throat, and nasal cavity and are inactivated by gastric acidity, which makes a barrier against bacterial invasion of lower gastrointestinal tract. Increased gastric pH after using PPI may be one of the reasons why many case-control studies detect Streptococcus in fecal samples, suggesting that bacterial translocation has occurred [22, 24].
PPI are widely prescribed both in out and inpatient care, in many cases without formal indication. PPI side effects are not negligible, and thus the decision-making for their use should be more careful.
Antibiotics
Antibiotics are powerful weapons against pathogenic bacteria, but can also affect commensal organisms, resulting in loss of microbial diversity, shifts in metabolic capacity, and reduced colonization resistance against invasive pathogens [25].
Within days after antibiotic treatment, profound and rapid changes in gut microbiota were described. A case-control study found that oral broad-spectrum antibiotics (ciprofloxacin, vancomycin, and metronidazole) for 7 days were associated with a loss of diversity and drastic shifts in community composition at day 9 [26]. Despite great short-term effects, there was a remarkable return towards baseline after 8–31 months of treatment. Nevertheless, increasing evidence suggests that microbiome might be at least slightly disturbed for long periods of time, some strains are lost indefinitely [27], and consequences as antibiotic resistance can persist for longer periods [28].
A relationship between the use of antibiotics in early life and the development of diseases in later childhood, as obesity and Crohn's disease (CD), has also been suggested [29, 30]. The number of antibiotic treatments and the sooner the children take them seem to be important contributing factors.
Another concern about antibiotic therapy is C. difficile colonization and infection, as the use of antibiotics are their major cause. A 2016 meta-analysis showed that clindamycin was the antibiotic most often associated with CDI in hospitalized patients, followed by carbapenems [31]. We will later focus on the effects of CDI itself on gut microbiota environment.
Potential Role of Dysbiosis in Gastrointestinal Diseases
Several highly prevalent diseases have been associated with imbalances in microbiota composition and function, including IBD, obesity, diabetes, allergy, IBS, GC, colorectal polyps and CRC, liver cirrhosis, NAFLD, neurodevelopmental disorders, cardiovascular disorders, cholesterol gallstones, diarrhea, malnutrition, and kidney disease [32]. However, it has yet to be studied if this change in microbial composition is a cause or a consequence of the disease itself. We will now give emphasis to microbiome-gastrointestinal disease associations recently described in the literature.
Colorectal Cancer
CRC is the third most common and the fourth most deadly cancer in the world [33]. Although there are also genetic components in the development of colorectal adenomas and cancer, it is a multifactorial disease as it involves various elements such as host immunity, environmental factors, and intestinal microbiota [34].
Colon bacteria counts are one million times higher than in the small intestine, and there are 12-fold more cancers there, which may in part be due to a role of colonic microbiota in carcinogenesis [35]. Previously, CRC was associated with specific bacteria such as Streptococcus gallolyticus (former S. bovis) [36], E. coli [37], Bacteroides fragilis [38], Clostridium septicum [39], and others. More recently, cancer cases have been related not only to a specific bacterium but to dysbiosis, with a decrease in diversity and community stability of gut microbiota [40].
Microbiota changes in patients with CRC are illustrated in Table 1. In various studies, an increased prevalence of Fusobacterium was shown in CRC patients [41, 42, 43] and in adenomatous tissue comparing to surrounding tissue [44], suggesting a possible role in the adenoma-cancer cascade. According to Bullman et al. [45], treatment with metronidazole highly decreases Fusobacterium load in tumor tissue and reduces colon tumor growth in mouse models. Therefore, Fusobacterium could be a stool marker for increased CRC risk.
Table 1.
Microbiota changes in gastrointestinal cancer
| First author [Ref.], year | Study group | Samples | Phylum | Genus/species |
|---|---|---|---|---|
| Colorectal cancer | ||||
| Liang [43], 2017 | 203 CRC/236 healthy controls | stool | ↑ Fusobacterium nucleatum | |
| Feng [42], 2015 | 46 CRC/57 healthy controls | stool | ↑ Fusobacterium
↑ Bacteroides ↑ Alistipes ↑ Escherichia ↑ Bilophila ↓ Ruminococcus ↓ Bifidobacterium ↓ Streptococcus |
|
| Ahn [41], 2013 | 47 CRC/94 control subjects | stool | ↑ Bacteroidetes ↓ Firmicutes |
↑ Fusobacterium
↑ Porphyromonas ↑ Atopobium ↑ Porphyromonas |
| Wang [46], 2012 | 46 CRC/56 healthy controls | stool | ↑ Proteobacteria ↓ Bacteroidetes |
↑ Escherichia/Shigella
↑ Streptococcus ↑ Peptostreptococcus ↑ Enterococcus ↓ Bacteroides ↓ Roseburia ↓ Alistipes |
| Gastric cancer | ||||
| Ferreira [62], 2018 | 54 GC/8 chronic gastritis | gastric mucosa biopsy | ↑ Actinobacteria ↑ Firmicutes ↓ Bacteroidetes ↓ Fusobacteria |
↑ Phyllobacterium
↑ Achromobacter ↑ Lactobacillus ↑ Clostridium ↑ Rhodococcus ↓ Helicobacter spp. |
| Coker [65], 2018 | 20 GC/21 superficial gastritis/23 atrophic gastritis/17 IM | gastric mucosa biopsy | ↑ Slackia exigua
↑ Parvimonas micra ↑ Dialister pneumosintes ↑ Prevotella intermedia and oris ↑ F. nucleatum ↑ Catonella morbi |
|
| Wang [63], 2016 | 103 GC/212 chronic gastritis | gastric mucosa biopsy | ↑ Lactobacillus
↑ Escherichia-Shigella ↑ Nitrospirae ↑ Burkholderia fungerum ↑ Lachospiraceae = Helicobacter pylori |
|
| Aviles-Jimenez [61], 2014 | 5 intestinal-type GC/5 non-atrophic gastritis/5 IM | gastric mucosa biopsy | ↑ Lactobacillus
↑ Lachospiraceae ↓ Porphyromonas ↓ Neisseria ↓ Streptococcus sinensis |
|
CRC, colorectal cancer; GC, gastric cancer; IM, intestinal metaplasia.
Bacterial RNA sequencing also revealed reduced levels of Firmicutes in CRC patients in both stool and tumor biopsy samples, compared to healthy individuals [41]. On the other hand, data concerning Bacteroidetes are inconsistent [41, 46].
In a recent study, Rezasoltani et al. [47] found a significant relationship between polyp size and quantity of Fusobacterium nucleatum, Streptococcus bovis, Lactobacillus spp., Roseburia spp., and Porphyromonas spp. This case-control study elucidated the role that some specific bacteria could have in adenoma development.
Studies in mouse models also suggest that dysbiosis can be a trigger to inflammation-induced CRC. One suggested mechanism involves the lack of SCFAs as the bacteria responsible for its production, like Faecalibacterium and Roseburia, are reduced in the fecal microbiota during CRC development [48]. Butyrate is known to be anticarcinogenic, as it inhibits cell proliferation, induces apoptosis of tumoral cells, and mediates T cells [49].
Gastric Cancer
GC is the fifth most common malignancy and the third leading cause of cancer-related death [33]. Around 81% of GC incidence is attributable to Helicobacter pylori, making this microorganism the most common pathogen linked to this malignancy [50].
Effective eradication of H. pylori does not entirely prevent gastric carcinogenesis [51, 52] and only 1–2% of H. pylori -positive individuals develop GC [53]. Besides, in insulin-gastrin transgenic mice, simultaneous colonization by H. pylori and a complex gastric microbiota increased the incidence of GC [54], hastening the onset of malignancy, when compared to those infected by H. pylori only [55]. These lines of evidence suggest that other commensals in gastric microbiota might contribute to cancer development.
H. pylori infection decreases gastric acidity, allowing the establishment of a new microbiota that could potentiate carcinogenesis. These bacteria overgrow in a hypoacid environment and are capable of converting nitrogen compounds to potentially carcinogenic N-nitroso compounds [56]. Numerous bacteria, such as E. coli, Haemophilus, Neisseria, Staphylococcus, Lactobacillus, Nitrospirae, and Veilonella can produce these compounds [57, 58], and some authors already demonstrated an increase in some of these bacteria in GC [59, 60, 61, 62].
GC microbial environment is illustrated in Table 1. Data concerning bacterial diversity in GC are inconsistent. Aviles-Jimenez et al. [61] reported a decreased microbial diversity from nonatrophic gastritis to intestinal metaplasia and GC. In fact, most of the studies reported a decreased microbial diversity [61, 62], though some authors described the opposite [59, 63].
Curiously, numerous studies have observed the same increasing pattern when studying Lactobacillus species in GC [61, 62, 63]. These species are used in probiotics, being beneficial to the host. However, in the stomach Lactobacillus increase lactic acid that can serve as energy for tumor cells and also promote inflammation and tumor angiogenesis [64].
Oral bacterial commensals may also have an important role in the pathogenesis and progression of gastric malignancy as many studies show increasing levels of these microorganisms in GC samples [60, 65].
Inflammatory Bowel Disease
IBD is a chronic, relapsing and remitting inflammatory disorder of the gastrointestinal tract, which consists of two major forms, CD and ulcerative colitis (UC). Despite many efforts, no exact etiology has been described, but it is thought to involve an interaction between genetic and environmental factors that can deregulate host immune response to gut microbiota.
Presently, it is accepted that intestinal dysbiosis is a potentially relevant mechanism underlying IBD pathogenesis [66]. Of note, regions with high loads of bacteria are correlated with greater incidence of CD and UC, particularly terminal ileum and colon.
Microbiota changes related to IBD patients are summarized in Table 2. Characteristic findings in IBD include reduced bacterial diversity and stability, mainly due to depleted members of Firmicutes and enriched Proteobacteria and Actinobacteria phyla [67, 68]. Dysbiosis can happen prior to the onset of intestine inflammation in genetically susceptible mouse, as proved by Glymenaki et al. [69, 70] in two different studies.
Table 2.
Microbiota changes in inflammatory bowel syndrome patients
| First author [Ref.], year | Study group | Samples | Phyla | Genus/species |
|---|---|---|---|---|
| Fuentes [75], 2017 | 29 UC/15 healthy controls | rectum biopsies | ↑ Bacteroidetes | ↑ Clostridium cluster XIVa ↓ Bacteroides fragilis and vulgatus |
| Ramakrishna [76], 2015 | 32 UC/28 CD/30 healthy controls | mucosal biopsies | ↓ Firmicutes/Bacteroidetes ratio | UC: ↑ Bacteroides ↑ Lactobacillus ↑ Escherichia coli CD: ↓ Clostridium coccoides |
| Rajca [71], 2014 | 33 CD/29 healthy controls | stool | ↓ Firmicutes | ↑ Escherichia coli
↓ Clostridium coccoides and leptum ↓ Faecalibacterium prausnitzii |
| Wang [72], 2014 | stool: 21 CD/34 UC/21 healthy controls mucosa-associated samples: 15 CD/29 UC/ 21 healthy control |
stool and mucosa-associated samples | IBD: ↓ Faecalibacterium prausnitzii UC: ↑ Bifidobacterium CD: ↑ Lactobacillus |
|
IBD, inflammatory bowel disease; CD, Crohn's disease; UC, ulcerative colitis.
The depletion of certain bacteria with the consequent loss of their protective functions has an important influence on disease. Several recent studies reported lower proportion of F. prausnitzii in patients with IBD [71, 72]. This commensal is a member of the phylum Firmicutes and a major producer of the SCFA, with important anti-inflammatory and cellular protective functions [73, 74]. In addition, a research team from France [71] found a link between CD relapse and low levels of F. prausnitzii, high C-reactive protein, and low rate of Bacteroides.
The UC dysbiosis status has been associated with increased Bacteroidetes phylum [75] and Bacteroides genus [76], more specifically with B. fragilis and vulgatus [75].
With advances in DNA sequencing, Morgan et al. [74] indicated that microbial function was consistently more perturbed (12%) than composition (2%), leading to the conclusion that microbiota composition studies may underestimate its impact on IBD pathogenesis.
Nonalcoholic Fatty Liver Disease
NAFLD encloses a spectrum of liver diseases from steatosis to nonalcoholic steatohepatitis, fibrosis, cirrhosis, and eventually hepatocellular carcinoma, in which liver fat deposition occurs. NAFLD requires the exclusion of a daily alcohol consumption of more than 30 g for men and 20 g for women. In recent years, NAFLD has emerged as one of the most common causes of liver disease worldwide [77].
The effectiveness of lifestyle changes, as modifications in diet and physical activity, have a great impact on metabolic control and liver histology [78], making clear the importance of environmental factors in this disease. However, there is a large variability in NAFLD progression that is not explained by genetics or environment.
Liver is the first organ to be exposed, through portal tract, to gut-origin metabolites, such as dietary nutrients and microbiota-related products. Due to this straight interaction between gut and liver, microbiota dysbiosis has been outlined as a major factor in the pathology of all stages of NAFLD [79]. Dysbiosis can lead to increased intestinal permeability, promoting translocation of commensal metabolites through the vascular system into the liver (endotoxemia), which directly contributes to hepatic lipid metabolism disruption and inflammatory processes in the liver [80].
Numerous investigators have been studying the microbiota composition in NAFLD patients. In a study comparing intestinal microbiome between nonobese NAFLD patients and healthy controls, the first group had more Bacteroidetes and less Firmicutes [81]. Within Firmicutes, SCFA-producing bacteria and 7α-dehydroxylating bacteria were decreased [81].
Some authors have been trying to correlate a specific bacterium with liver fibrosis in NAFLD. Duarte et al. [82] found an association with increased Lactobacillus, and Boursier et al. [79] described a correlation with Ruminococcus. A recent study [83] described NAFLD gut microbiota as dominated by Firmicutes and Bacteroidetes, followed by Proteobacteria and Actinobacteria in lower proportions. However, with fibrosis progression there is a significant increase in the abundance of Proteobacteria phylum, while that of Firmicutes decreases. Dysbiosis starts early in liver disease and largely depends on the etiological factors [84].
It is clear that microbiota varies during the course of NAFLD, and this could explain the differences found in microbiota composition. However, these studies differ in definitions used, study design, and clinical endpoints, making it inappropriate to draw general conclusions.
C. difficile Infection
C. difficile is an anaerobic, Gram-positive, sporulating, bacterial pathogen, and is recognized as the primary cause of nosocomial, antibiotic-associated diarrhea [85]. In normal circumstances, gut commensals prevent C. difficile colonization, suppressing its pathogenic activity in the colon, by spore germination and toxin production.
Human gut microbiota undergoes enormous changes throughout life; consequently, it is not surprising that CDI is more common in elderly people [86]. As well as age, many risk factors contribute to gut microbiota disruption and susceptibility to CDI, but antimicrobial therapy is the one with a major impact [87].
Generally, in CDI patients, reduced overall bacterial diversity is observed [88]. Augmented Proteobacteria and reduction of Firmicutes and Bacteroidetes phyla are the most frequent findings in these patients [89, 90].
Presently, the standard treatment for an initial nonsevere CDI episode is oral vancomycin or fidaxomicin [91]. Around a quarter of all patients with CDI will have a recurrence, with increasing rates in each subsequent episode [92]. An investigation by Seekatz et al. [93] elucidated that intestine microbiota in recurrent CDI patients and severe CDI trended towards a lower diversity community, but differences between these two groups were less pronounced when comparing CDI with healthy patients. The authors also concluded that gut microbiota recovery during treatment was more dynamic in patients without recurrence. The deeper changes in intestinal microbiota composition and the low recovery dynamics of recurrent CDI patients compared to individuals with one-time infection corroborates the fact that recurrent infection is harder to treat and has a higher reinfection rate with each new episode. Recurrent CDI management is therefore a major clinical challenge.
Irritable Bowel Syndrome
IBS is one of the most common gastrointestinal problems in clinical practice, with high morbidity and a global prevalence around 11% [94].
IBS has been classically linked to functional changes on the brain-gut axis that involve gut dysmotility, sensory-motor disfunction, and psychological stress. Recent evidence also endorses gut dysbiosis as a potential risk factor for IBS, directly causing abnormal intestinal immune activation and chronic gut inflammation [95]. The belief that dysbiosis may have a role in IBS pathogenesis is based on the fact that bacterial gastroenteritis event is the best accepted predictor of IBS development [96].
FODMAP (fermentable oligo-, di-, and monosaccharides and polyols) have been extensively associated with IBS, as they promote visceral hypersensitivity, increase gut motility, and promote dysbiotic imbalance by suppressing bacteria involved in gas consumption. Despite the beneficial effects of a low FODMAP diet on IBS symptoms, it reduces luminal concentration of Bifidobacterium and F. prausnitzii [97]. Long-term consequences of a low FODMAP diet remain unclear.
Numerous studies evidenced a different gut microbiota in IBS patients, and even intestinal symptoms were found to be positively associated with specific microorganisms. Changes in gut microbiota in IBS patients are represented in Table 3. In general, patients with IBS have decreased microbial diversity [98]. Data suggested an increased level of Firmicutes and a decrease in Bacteroidetes [99, 100]. In a 2019 systematic review involving 777 patients, the authors corroborate these microbiota changes [101]. In contrast, Wang et al. [102], presented contradictory results.
Table 3.
Microbiota changes in irritable bowel syndrome patients
| First author [Ref.], year | Study group | Samples | Phyla | Genus/species |
|---|---|---|---|---|
| Wang [102], 2019 | 20 diarrhea-predominant IBS/40 healthy controls | stool | ↑ Bacteroidetes ↓ Firmicutes |
↑ Bacteroides
↑ Prevotella ↑ Paraprevotella ↓ Faecalibacterium ↓ Lachnospiracea incertae sedis ↓ Blautia ↓ Coprococcus |
| Labus [99], 2017 | 29 IBS/23 healthy controls | stool | ↑ Firmicutes ↑ Actinobacteria ↑ Proteobacteria ↓ Bacteroidetes |
|
| Giamarellos-Bourboulis [98], 2015 | 74 IBS/163 non-healthy non-IBS/21 healthy controls | duodenal aspirate | ↑ Escherichia/Shigella
↑ Escherichia coli ↑ Klebsiella pneumoniae ↓ Acinetobacter ↓ Citrobacter ↓ Leuconostoc ↓ Lactococcus |
|
| Rajilic-Stojanovic [100], 2011 | 62 IBS/46 healthy individuals | stool | ↑ Firmicutes ↓ Bacteroidetes |
↑ Dorea
↑ Ruminococcus ↑ Clostridium ↑ Roseburia ↓ Bifidobacterium ↓ Faecalibacterium ↓ Bacteroides |
IBS, irritable bowel syndrome.
Potential pathogenic bacteria such as Ruminococcus, Clostridium, and E. coli have been reported to be increased in IBS [98, 100]. Even though Lactobacillus and Bifidobacterium have been used as probiotics, both are increased in IBS patients in numerous trials [103], raising the question about the role of these bacteria in IBS.
In several studies of dysbiosis in IBS, CD, and UC, Faecalibacterium, more specifically F. prausnitzii, were confirmed to be decreased, making this microorganism a “healthy gut marker.” F. prausnitzii has a strong anti-inflammatory effect both in vitro and in vivo, maintaining intestinal health [100, 102].
Nonetheless, results to date are inconsistent, probably due to influence of confounders as diet, phenotypic characterization of patients, and geographical environments.
Therapeutic Modulation of Gastrointestinal Microbiota
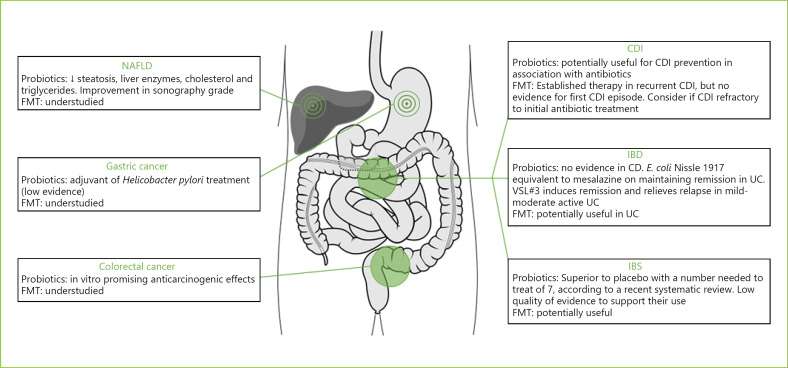
Novel therapeutic options emerged, namely probiotics, prebiotics, synbiotics, and fecal microbiota transplant (FMT) (Fig. 1). All of these aim at the restoration of microbiota composition, gut homeostasis, and physical barrier defense.
Fig. 1.
Overview of modulator therapies that target dysbiotic conditions and their current relevance. NAFLD, nonalcoholic fatty liver disease; CDI, Clostridium difficile infection; IBD, inflammatory bowel disease; IBS, irritable bowel syndrome; UC, ulcerative colitis; CD, Crohn's disease; FMT, fecal microbiota transplant.
Probiotics, Prebiotics, and Synbiotics
Probiotics are live microorganisms that confer a health benefit to the host when administered in adequate amounts [104]. Most commonly used probiotics are Bifidobacterium spp., Lactobacillus spp., Lactococcus spp., and Saccharomyces boulardii. Probiotic benefits are obtained by increasing gastrointestinal microbiota diversity and decreasing pathogenic microorganisms (Clostridium cluster XI, C. difficile, Clostridium perfringens, Enterococcus faecium,and Campylobacter) [105], in a setting of bacterial competition for nutrition and mucosal adherence [106]. Bifidobacteria, in particular, are able to inhibit potential pathogens, reduce blood ammonia levels, produce vitamins and digestive enzymes [107].
Probiotics had to overcome disbelief through the years concerning lack of evidence for its use, due to difficulties in reproducibility and standardization of doses and delivery methods. However, recent evidence suggests that probiotics may have a role in particular disease context, as discussed later.
Prebiotics are nondigestible polysaccharides and oligosaccharides, like lactulose, fructo-oligosaccharides, galacto-oligosaccharides, and inulin, that are fermented in the colon, resulting in specific changes in the composition and/or functions of gastrointestinal microbiota. Prebiotic fibers are naturally found in a common diet, mainly in vegetables such as asparagus, garlic, leeks, and onions [108]. Prebiotics selectively support intestinal growth of protective organisms, notably Bifidobacterium and Lactobacilli, and reduce intestinal permeability and metabolic endotoxemia [109]. Resistant starch is a particular form of prebiotics attracting increasing interest regarding benefits on bowel movement and inflammation on IBD and CRC [110].
Finally, synbiotics are a combination of prebiotics and probiotics, with potential benefits to the host.
The effectiveness of probiotics, prebiotics or synbiotics in clinical practice depends on the strain, dosage and context of application. These are the aspects under investigation that will now be discussed.
Colorectal and Gastric Cancer
In addition to the immunologic and metabolic benefits of pre- and probiotics on gut health, anticarcinogenic effects have been under research.
Experimental studies with probiotics in vitro have shown promising anticarcinogenic results, as they have the power to degrade carcinogenic metabolites, increase anticarcinogenic compounds (SCFA and conjugated linoleic acid), and inhibit cancer cell proliferation [111]. A symbiotic preparation with oligofructose-enriched inulin, Lactobacillus rhamnosus GC, and Bifidobacterium lactis Bb12 reduced colorectal proliferation and improved epithelial barrier function in polypectomized patients [112]. However, the evidence of clinically important benefits is still limited, and there are no human studies that show a decreased risk of adenomas/CRC.
Studies on pro- and prebiotics in GC have been focused on H. pylori eradication. Probiotics used as an adjuvant of H. pylori treatment seem to increase eradication rates, by directly inhibiting H. pylori, and decreasing first-line therapy side effects [113]. Still, the 2016 Maastricht V/Florence Consensus assumes very low evidence for probiotics' beneficial effect on H. pylori eradication [114], although they significantly decrease treatment-related antibiotic-associated diarrhea and can increase compliance.
Despite overall microbiome impact in gastrointestinal cancer, little attention has been paid to probiotics as a means to control gastric and colorectal carcinogenesis in humans.
Inflammatory Bowel Disease
In IBD, discrepant results have been observed between the power of probiotics and prebiotics in CD and UC. Multiple studies comparing probiotics and placebo in CD patients showed no clear evidence for the usefulness of probiotics in this disease [115]. However, different results were observed in UC, where adding a probiotic to conventional treatment improved induction and maintenance remission in mild to moderate active UC colitis [115]. E. coli Nissle 1917 was shown to be equivalent to mesalazine in maintaining remission in UC patients [116]. A high-potency probiotic medical food containing 8 different strains, called VSL#3®, induces remission and relieves relapse in mild to moderate active UC [117].
Other probiotics as Bifidobacterium [118] and Lactobacillus [119] seem to have beneficial effects in managing active UC. Also, supplementation with prebiotics like oligofructose-enriched inulin in active UC was associated with early reduction in fecal calprotectin [120]. Therefore, no clear recommendation on dose or strains can be given for active UC, and more research is needed.
Pouchitis is the most common complication after an ileal pouch-anal anastomosis in UC. According to European Crohn's and Colitis Organisation, there is good evidence for the usefulness of VSL#3® in preventing an initial episode of pouchitis and to maintain remission of chronic pouchitis after antibiotic treatment [121].
Nonalcoholic Fatty Liver Disease
Certain probiotics have been suggested to be useful in NAFLD, even altering biopsy liver characteristics, but no long-term benefits are yet known [122]. A trial conducted in obese children with sonography-confirmed NAFLD demonstrated that those receiving probiotics capsule (Lactobacillus acidophilus and rhamnosis, Bifidobacterium lactis and bifidum) for 12 weeks had lower liver enzymes, triglycerides, and cholesterol, also with an improvement in sonography steatosis grade compared with the placebo group [123].
The use of synbiotics as Bifidobacterium longum with fructo-oligosaccharides, when compared to lifestyle modifications, significantly reduced tumor necrosis factor-α, C-reactive protein, serum aspartate transaminase levels, insulin resistance, serum endotoxin, and biopsy-documented steatosis [124].
A recent systematic review and meta-analysis analyzed 25 clinical trials that studied the influence of pre-, pro-, and synbiotics in NAFLD patients and supports the potential use of microbial therapies in NAFLD treatment, as positive results were found for body mass index, hepatic enzymes, serum cholesterol, low-density lipoprotein cholesterol, and triglycerides [122].
C. difficile Infection
As CDI rates increase, and conventional treatment strategies become less successful, new approaches emerge. A recent Cochrane review showed that coadministration of probiotics with antibiotics was associated with a lower risk for CDI, but current guidelines do not yet recommend the routine use of preventive probiotics [125].
Prebiotic formulations as a means of treatment and prevention of recurrent CDI has still no proven clinical application [126].
Irritable Bowel Syndrome
According to World Gastroenterology Organization, some probiotic strains may reduce pain and provide global relief in IBS with a consistent reduction of abdominal bloating and flatulence in published trials [127]. A significant improvement in pain after the administration of probiotics containing Bifidobacterium breve, Bifidobacterium longum, or Lactobacillus acidophilus was also demonstrated by Ortiz-Lucas et al. [128]. Interestingly, patients who had an abnormal interleukin 10(IL-10)/IL-12 ratio, indicated as a proinflammatory state, showed normalized levels after Bifidobacterium infantis 36624 [129].
A recent meta-analysis promoted by American College of Gastroenterology studied 53 RCTs involving 5,545 IBS patients. Taken together, probiotics were statistically superior to placebo with a number needed to treat of 7, but with low quality of evidence to support their use [130]. Based on a 2016 update by British Dietetic Association, there is no good evidence to recommend a specific probiotic, and they should all be used one at a time for a minimum of 4 weeks [131]. A 2019 systematic review in IBS patients found no differences between prebiotics and placebo for symptoms and quality of life [132].
Fecal Microbiota Transplant
FMT consists in the infusion of feces from a healthy donor to the gastrointestinal tract of a recipient patient [133]. Hence, there is an opportunity to restore the complexity and diversity of the gut microbiota, overpassing the benefits of probiotics [134].
According to several recently published systematic reviews and meta-analyses [135, 136, 137], FMT has been successfully used in treating recurrent CDI, with a cure rate of roughly 90%. According to US Guidelines published in 2018, FMT is proposed in patients with at least two recurrences [138]. The European Consensus guidelines [133] recommend FMT for mild and severe recurrent CDI and suggest consideration for CDI refractory to the initial antibiotic treatment. There is still no evidence for FMT as a treatment for the first episode of CDI. In a study in Portugal, the overall cure rate of recurrent or refractory CDI with FMT was 96% (87.5% when upper endoscopy was used and 100% with colonoscopy) [139].
In a short-term follow-up, FMT seems to be safe, with the most common reported adverse effects being abdominal discomfort, diarrhea, constipation, and low-grade fever. Severe side effects seem to be uncommon and include relapse of IBD, transmission of enteric pathogens, pneumonia, infection/sepsis, and postinfectious IBS. Potential long-term effects are related to the possibility of transmission of unrecognized infections that may cause disease years later and induce chronic diseases, as reported in case reports, such as obesity, diabetes, NAFLD, asthma, autism, and others [140, 141]. To ensure safety conditions, there is a donor selection process, that includes a medical interview, as well as blood and stool tests. The European Consensus proposed clinical and analytic follow-up at least for 8 weeks after FMT [133].
Currently, due to FMT success rates in recurrent CDI, there is a great interest in evaluating FMT for diseases implicated in altered gut microbiota, in particular IBD, IBS, and idiopathic constipation. An interesting finding was that FMT was less effective in recurrent CDI patients with IBD compared with patients without IBD. In fact, 25% of the IBD patients had a flare after FMT [142].
Like pre- and probiotics in IBD, CD is less likely to respond to FMT than UC. However, a pilot study [143] demonstrated high rates of clinical improvement and remission in refractory CD patients after FMT. There are still no FMT randomized controlled trials in CD. Active UC patients under FMT therapy achieved remission in a significantly greater proportion compared to the placebo group. Those in early-stage UC reached higher success rates than those in late-stage UC. Curiously, most of the patients who achieved remission had received feces from one healthy donor [144]. Hence, FMT stands as a promising alternative for IBD requiring further investigation to support recommendation.
IBS is greatly expected to benefit from FMT potential. A 2018 randomized control trial analyzed 83 patients with IBS, 55 of whom received FMT. Sixty-five percent of patients in the FMT group and 43% in the placebo group had decreased IBS severity scores 3 months after treatment [145]. Still, conflicting results do not currently support a widespread use of this treatment [146].
FMT may theoretically be useful in NAFLD and gastrointestinal cancer since microbiota plays a role in their pathogenesis. Nonetheless, its potential benefits in these conditions have not yet been studied.
The description of dysbiosis in many nongastrointestinal disorders has raised investigation into potential FMT indication for those. Currently, FMT studies are expanding to autism, multiple sclerosis, metabolic diseases, idiopathic thrombocytopenic purpura, autoimmune diseases, allergic disorders, and other psychiatric disorders [141]. Further FMT investigation should focus on describing dosage and timing of FMT, besides patient's demographic and clinical data.
Conclusion
Recent outgrowth of microbiome genomic sequencing is revealing numerous correlations between dysbiotic imbalances and gastrointestinal diseases. However, these represent early stages of research where few causal relationships have been established. Studies in mouse models have stated that dysbiosis plays a role in the pathogenesis of inflammation-induced carcinogenesis and can happen prior to the onset of diseases.
Most Proteobacteria are considered disadvantageous, as proven by their increase in IBD and CDI, in contrast to Firmicutes that are decreased in IBD, CRC, NAFLD, and CDI patients. Even in healthy patients, factors such as diet, PPI, or antibiotics modulate gut microbiota. Long-term therapy with broad-spectrum antibiotics, Western diet, and the inhibition of gastric acidity by PPI are well-established risk factors for dysbiosis-associated diseases. However, so far there is no consensus on what constitutes a normal microbiota, and in clinical practice it is difficult to integrate the results of microbiota analysis of individual patients, namely for choosing the best way to restore normal microbiota.
Increasing evidence supports probiotics as adjuvant therapy mainly in NAFLD, CDI, UC, and IBS. The authors believe that further clinical trials of pre- and probiotics using standardized strains, doses, and duration of treatment are indispensable for placing these formulations in international guidelines. On the other hand, FMT is an established option for recurrent CDI. However, FMT application in other dysbiosis-associated diseases lacks randomized controlled trials to support recommendation.
Disclosure Statement
The authors have no conflicts of interest to declare.
References
- 1.Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010 Jul;90((3)):859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 2.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001 May;292((5519)):1115–8. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 3.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012 Jul-Aug;3((4)):289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto M, Yamaguchi R, Munakata K, Takashima K, Nishiyama M, Hioki K, et al. A microarray analysis of gnotobiotic mice indicating that microbial exposure during the neonatal period plays an essential role in immune system development. BMC Genomics. 2012 Jul;13((1)):335. doi: 10.1186/1471-2164-13-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Encarnação JC, Abrantes AM, Pires AS, Botelho MF. Revisit dietary fiber on colorectal cancer: butyrate and its role on prevention and treatment. Cancer Metastasis Rev. 2015 Sep;34((3)):465–78. doi: 10.1007/s10555-015-9578-9. [DOI] [PubMed] [Google Scholar]
- 6.Bäumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016 Jul;535((7610)):85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012 Jun;486((7402)):207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hooks KB, O'Malley MA. Dysbiosis and Its Discontents. MBio. 2017 Oct;8((5)):e01492-17. doi: 10.1128/mBio.01492-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017 Apr;17((4)):219–32. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- 10.Lepage P, Leclerc MC, Joossens M, Mondot S, Blottière HM, Raes J, et al. A metagenomic insight into our gut's microbiome. Gut. 2013 Jan;62((1)):146–58. doi: 10.1136/gutjnl-2011-301805. [DOI] [PubMed] [Google Scholar]
- 11.Rojo D, Méndez-García C, Raczkowska BA, Bargiela R, Moya A, Ferrer M, et al. Exploring the human microbiome from multiple perspectives: factors altering its composition and function. FEMS Microbiol Rev. 2017 Jul;41((4)):453–78. doi: 10.1093/femsre/fuw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wooley JC, Godzik A, Friedberg I. A primer on metagenomics. PLOS Comput Biol. 2010 Feb;6((2)):e1000667. doi: 10.1371/journal.pcbi.1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gosalbes MJ, Durbán A, Pignatelli M, Abellan JJ, Jiménez-Hernández N, Pérez-Cobas AE, et al. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS One. 2011 Mar;6((3)):e17447. doi: 10.1371/journal.pone.0017447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franzosa EA, Morgan XC, Segata N, Waldron L, Reyes J, Earl AM, et al. Relating the metatranscriptome and metagenome of the human gut. Proc Natl Acad Sci USA. 2014 Jun;111((22)):E2329–38. doi: 10.1073/pnas.1319284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bikel S, Valdez-Lara A, Cornejo-Granados F, Rico K, Canizales-Quinteros S, Soberón X, et al. Combining metagenomics, metatranscriptomics and viromics to explore novel microbial interactions: towards a systems-level understanding of human microbiome. Comput Struct Biotechnol J. 2015 Jun;13:390–401. doi: 10.1016/j.csbj.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pendyala S, Walker JM, Holt PR. A high-fat diet is associated with endotoxemia that originates from the gut. Gastroenterology. 2012;142((5)):1100–1 e2. doi: 10.1053/j.gastro.2012.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hold GL. Western lifestyle: a ‘master’ manipulator of the intestinal microbiota? Gut. 2014 Jan;63((1)):5–6. doi: 10.1136/gutjnl-2013-304969. [DOI] [PubMed] [Google Scholar]
- 18.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell. 2016;167((5)):1339–53 e21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitsou EK, Kakali A, Antonopoulou S, Mountzouris KC, Yannakoulia M, Panagiotakos DB, et al. Adherence to the Mediterranean diet is associated with the gut microbiota pattern and gastrointestinal characteristics in an adult population. Br J Nutr. 2017 Jun;117((12)):1645–55. doi: 10.1017/S0007114517001593. [DOI] [PubMed] [Google Scholar]
- 20.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014 Jan;505((7484)):559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janarthanan S, Ditah I, Adler DG, Ehrinpreis MN. Clostridium difficile-associated diarrhea and proton pump inhibitor therapy: a meta-analysis. Am J Gastroenterol. 2012 Jul;107((7)):1001–10. doi: 10.1038/ajg.2012.179. [DOI] [PubMed] [Google Scholar]
- 22.Tsuda A, Suda W, Morita H, Takanashi K, Takagi A, Koga Y, et al. Influence of Proton-Pump Inhibitors on the Luminal Microbiota in the Gastrointestinal Tract. Clin Transl Gastroenterol. 2015 Jun;6((6)):e89. doi: 10.1038/ctg.2015.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hojo M, Asahara T, Nagahara A, Takeda T, Matsumoto K, Ueyama H, et al. Gut Microbiota Composition Before and After Use of Proton Pump Inhibitors. Dig Dis Sci. 2018 Nov;63((11)):2940–9. doi: 10.1007/s10620-018-5122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takagi T, Naito Y, Inoue R, Kashiwagi S, Uchiyama K, Mizushima K, et al. The influence of long-term use of proton pump inhibitors on the gut microbiota: an age-sex-matched case-control study. J Clin Biochem Nutr. 2018 Jan;62((1)):100–5. doi: 10.3164/jcbn.17-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lange K, Buerger M, Stallmach A, Bruns T. Effects of Antibiotics on Gut Microbiota. Dig Dis. 2016;34((3)):260–8. doi: 10.1159/000443360. [DOI] [PubMed] [Google Scholar]
- 26.Haak BW, Lankelma JM, Hugenholtz F, Belzer C, de Vos WM, Wiersinga WJ. Long-term impact of oral vancomycin, ciprofloxacin and metronidazole on the gut microbiota in healthy humans. J Antimicrob Chemother. 2019 Mar;74((3)):782–6. doi: 10.1093/jac/dky471. [DOI] [PubMed] [Google Scholar]
- 27.Palleja A, Mikkelsen KH, Forslund SK, Kashani A, Allin KH, Nielsen T, et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat Microbiol. 2018 Nov;3((11)):1255–65. doi: 10.1038/s41564-018-0257-9. [DOI] [PubMed] [Google Scholar]
- 28.Jakobsson HE, Jernberg C, Andersson AF, Sjölund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010 Mar;5((3)):e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hviid A, Svanström H, Frisch M. Antibiotic use and inflammatory bowel diseases in childhood. Gut. 2011 Jan;60((1)):49–54. doi: 10.1136/gut.2010.219683. [DOI] [PubMed] [Google Scholar]
- 30.Scott FI, Horton DB, Mamtani R, Haynes K, Goldberg DS, Lee DY, et al. Administration of Antibiotics to Children Before Age 2 Years Increases Risk for Childhood Obesity. Gastroenterology. 2016;151((1)):120–9 e5. doi: 10.1053/j.gastro.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vardakas KZ, Trigkidis KK, Boukouvala E, Falagas ME. Clostridium difficile infection following systemic antibiotic administration in randomised controlled trials: a systematic review and meta-analysis. Int J Antimicrob Agents. 2016 Jul;48((1)):1–10. doi: 10.1016/j.ijantimicag.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 32.Wang WL, Xu SY, Ren ZG, Tao L, Jiang JW, Zheng SS. Application of metagenomics in the human gut microbiome. World J Gastroenterol. 2015 Jan;21((3)):803–14. doi: 10.3748/wjg.v21.i3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015 Mar;136((5)):E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 34.Castells A, Castellví-Bel S, Balaguer F. Concepts in familial colorectal cancer: where do we stand and what is the future? Gastroenterology. 2009 Aug;137((2)):404–9. doi: 10.1053/j.gastro.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 35.Proctor LM. The Human Microbiome Project in 2011 and beyond. Cell Host Microbe. 2011 Oct;10((4)):287–91. doi: 10.1016/j.chom.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Abdulamir AS, Hafidh RR, Abu Bakar F. The association of Streptococcus bovis/gallolyticus with colorectal tumors: the nature and the underlying mechanisms of its etiological role. J Exp Clin Cancer Res. 2011 Jan;30((1)):11. doi: 10.1186/1756-9966-30-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, et al. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology. 2004 Jul;127((1)):80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 38.Purcell RV, Pearson J, Aitchison A, Dixon L, Frizelle FA, Keenan JI. Colonization with enterotoxigenic Bacteroides fragilis is associated with early-stage colorectal neoplasia. PLoS One. 2017 Feb;12((2)):e0171602. doi: 10.1371/journal.pone.0171602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mirza NN, McCloud JM, Cheetham MJ. Clostridium septicum sepsis and colorectal cancer - a reminder. World J Surg Oncol. 2009 Oct;7((1)):73. doi: 10.1186/1477-7819-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhatt AP, Redinbo MR, Bultman SJ. The role of the microbiome in cancer development and therapy. CA Cancer J Clin. 2017 Jul;67((4)):326–44. doi: 10.3322/caac.21398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013 Dec;105((24)):1907–11. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015 Mar;6((1)):6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 43.Liang Q, Chiu J, Chen Y, Huang Y, Higashimori A, Fang J, et al. Fecal Bacteria Act as Novel Biomarkers for Noninvasive Diagnosis of Colorectal Cancer. Clin Cancer Res. 2017 Apr;23((8)):2061–70. doi: 10.1158/1078-0432.CCR-16-1599. [DOI] [PubMed] [Google Scholar]
- 44.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012 Feb;22((2)):292–8. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017 Dec;358((6369)):1443–8. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012 Feb;6((2)):320–9. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rezasoltani S, Asadzadeh Aghdaei H, Dabiri H, Akhavan Sepahi A, Modarressi MH, Nazemalhosseini Mojarad E. The association between fecal microbiota and different types of colorectal polyp as precursors of colorectal cancer. Microb Pathog. 2018 Nov;124:244–9. doi: 10.1016/j.micpath.2018.08.035. [DOI] [PubMed] [Google Scholar]
- 48.Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013 Aug;66((2)):462–70. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- 49.Canani RB, Costanzo MD, Leone L, Pedata M, Meli R, Calignano A. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J Gastroenterol. 2011 Mar;17((12)):1519–28. doi: 10.3748/wjg.v17.i12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plummer M, de Martel C, Vignat J, Ferlay J, Bray F, Franceschi S. Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob Health. 2016 Sep;4((9)):e609–16. doi: 10.1016/S2214-109X(16)30143-7. [DOI] [PubMed] [Google Scholar]
- 51.Wong BC, Lam SK, Wong WM, Chen JS, Zheng TT, Feng RE, et al. China Gastric Cancer Study Group Helicobacter pylori eradication to prevent gastric cancer in a high-risk region of China: a randomized controlled trial. JAMA. 2004 Jan;291((2)):187–94. doi: 10.1001/jama.291.2.187. [DOI] [PubMed] [Google Scholar]
- 52.Ma JL, Zhang L, Brown LM, Li JY, Shen L, Pan KF, et al. Fifteen-year effects of Helicobacter pylori, garlic, and vitamin treatments on gastric cancer incidence and mortality. J Natl Cancer Inst. 2012 Mar;104((6)):488–92. doi: 10.1093/jnci/djs003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Correa P, Piazuelo MB. Helicobacter pylori Infection and Gastric Adenocarcinoma. US Gastroenterol Hepatol Rev. 2011 Jun;7((1)):59–64. [PMC free article] [PubMed] [Google Scholar]
- 54.Lertpiriyapong K, Whary MT, Muthupalani S, Lofgren JL, Gamazon ER, Feng Y, et al. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut. 2014 Jan;63((1)):54–63. doi: 10.1136/gutjnl-2013-305178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lofgren JL, Whary MT, Ge Z, Muthupalani S, Taylor NS, Mobley M, et al. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology. 2011 Jan;140((1)):210–20. doi: 10.1053/j.gastro.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plottel CS, Blaser MJ. Microbiome and malignancy. Cell Host Microbe. 2011 Oct;10((4)):324–35. doi: 10.1016/j.chom.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jo HJ, Kim J, Kim N, Park JH, Nam RH, Seok YJ, et al. Analysis of Gastric Microbiota by Pyrosequencing: Minor Role of Bacteria Other Than Helicobacter pylori in the Gastric Carcinogenesis. Helicobacter. 2016 Oct;21((5)):364–74. doi: 10.1111/hel.12293. [DOI] [PubMed] [Google Scholar]
- 58.Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013 Feb;339((6120)):708–11. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Castaño-Rodríguez N, Goh KL, Fock KM, Mitchell HM, Kaakoush NO. Dysbiosis of the microbiome in gastric carcinogenesis. Sci Rep. 2017 Nov;7((1)):15957. doi: 10.1038/s41598-017-16289-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu YL, Pang W, Huang Y, Zhang Y, Zhang CJ. The Gastric Microbiome Is Perturbed in Advanced Gastric Adenocarcinoma Identified Through Shotgun Metagenomics. Front Cell Infect Microbiol. 2018 Dec;8:433. doi: 10.3389/fcimb.2018.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, Mantilla A, Torres J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci Rep. 2014 Feb;4((1)):4202. doi: 10.1038/srep04202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, Machado JC, et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. 2018 Feb;67((2)):226–36. doi: 10.1136/gutjnl-2017-314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang L, Zhou J, Xin Y, Geng C, Tian Z, Yu X, et al. Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur J Gastroenterol Hepatol. 2016 Mar;28((3)):261–6. doi: 10.1097/MEG.0000000000000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. J Clin Invest. 2013 Sep;123((9)):3685–92. doi: 10.1172/JCI69741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. 2018 Jun;67((6)):1024–32. doi: 10.1136/gutjnl-2017-314281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rutgeerts P, Goboes K, Peeters M, Hiele M, Penninckx F, Aerts R, et al. Effect of faecal stream diversion on recurrence of Crohn's disease in the neoterminal ileum. Lancet. 1991 Sep;338((8770)):771–4. doi: 10.1016/0140-6736(91)90663-a. [DOI] [PubMed] [Google Scholar]
- 67.Duvallet C, Gibbons SM, Gurry T, Irizarry RA, Alm EJ. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat Commun. 2017 Dec;8((1)):1784. doi: 10.1038/s41467-017-01973-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matsuoka K, Kanai T. The gut microbiota and inflammatory bowel disease. Semin Immunopathol. 2015 Jan;37((1)):47–55. doi: 10.1007/s00281-014-0454-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Glymenaki M, Singh G, Brass A, Warhurst G, McBain AJ, Else KJ, et al. Compositional Changes in the Gut Mucus Microbiota Precede the Onset of Colitis-Induced Inflammation. Inflamm Bowel Dis. 2017 Jun;23((6)):912–22. doi: 10.1097/MIB.0000000000001118. [DOI] [PubMed] [Google Scholar]
- 70.Glymenaki M, Barnes A, O'Hagan S, Warhurst G, McBain AJ, Wilson ID, et al. Stability in metabolic phenotypes and inferred metagenome profiles before the onset of colitis-induced inflammation. Sci Rep. 2017 Aug;7((1)):8836. doi: 10.1038/s41598-017-08732-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rajca S, Grondin V, Louis E, Vernier-Massouille G, Grimaud JC, Bouhnik Y, et al. Alterations in the intestinal microbiome (dysbiosis) as a predictor of relapse after infliximab withdrawal in Crohn's disease. Inflamm Bowel Dis. 2014 Jun;20((6)):978–86. doi: 10.1097/MIB.0000000000000036. [DOI] [PubMed] [Google Scholar]
- 72.Wang W, Chen L, Zhou R, Wang X, Song L, Huang S, et al. Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J Clin Microbiol. 2014 Feb;52((2)):398–406. doi: 10.1128/JCM.01500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008 Oct;105((43)):16731–6. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012 Apr;13((9)):R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fuentes S, Rossen NG, van der Spek MJ, Hartman JH, Huuskonen L, Korpela K, et al. Microbial shifts and signatures of long-term remission in ulcerative colitis after faecal microbiota transplantation. ISME J. 2017 Aug;11((8)):1877–89. doi: 10.1038/ismej.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kabeerdoss J, Jayakanthan P, Pugazhendhi S, Ramakrishna BS. Alterations of mucosal microbiota in the colon of patients with inflammatory bowel disease revealed by real time polymerase chain reaction amplification of 16S ribosomal ribonucleic acid. Indian J Med Res. 2015 Jul;142((1)):23–32. doi: 10.4103/0971-5916.162091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016 Jul;64((1)):73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 78.Vilar-Gomez E, Martinez-Perez Y, Calzadilla-Bertot L, Torres-Gonzalez A, Gra-Oramas B, Gonzalez-Fabian L, et al. Weight Loss Through Lifestyle Modification Significantly Reduces Features of Nonalcoholic Steatohepatitis. Gastroenterology. 2015;149((2)):367–78 e5. doi: 10.1053/j.gastro.2015.04.005. quiz e14-5. [DOI] [PubMed] [Google Scholar]
- 79.Boursier J, Mueller O, Barret M, Machado M, Fizanne L, Araujo-Perez F, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016 Mar;63((3)):764–75. doi: 10.1002/hep.28356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chassaing B, Etienne-Mesmin L, Gewirtz AT. Microbiota-liver axis in hepatic disease. Hepatology. 2014 Jan;59((1)):328–39. doi: 10.1002/hep.26494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang B, Jiang X, Cao M, Ge J, Bao Q, Tang L, et al. Altered Fecal Microbiota Correlates with Liver Biochemistry in Nonobese Patients with Non-alcoholic Fatty Liver Disease. Sci Rep. 2016 Aug;6((1)):32002. doi: 10.1038/srep32002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Duarte SM, Stefano JT, Miele L, Ponziani FR, Souza-Basqueira M, Okada LS, et al. Gut microbiome composition in lean patients with NASH is associated with liver damage independent of caloric intake: A prospective pilot study. Nutr Metab Cardiovasc Dis. 2018 Apr;28((4)):369–84. doi: 10.1016/j.numecd.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 83.Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2017;25((5)):1054–62 e5. doi: 10.1016/j.cmet.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, White MB, Monteith P, et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014 May;60((5)):940–7. doi: 10.1016/j.jhep.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Polage CR, Solnick JV, Cohen SH. Nosocomial diarrhea: evaluation and treatment of causes other than Clostridium difficile. Clin Infect Dis. 2012 Oct;55((7)):982–9. doi: 10.1093/cid/cis551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Claesson MJ, Cusack S, O'Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA. 2011 Mar;108(Suppl 1):4586–91. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Theriot CM, Koenigsknecht MJ, Carlson PE, Jr, Hatton GE, Nelson AM, Li B, et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Milani C, Ticinesi A, Gerritsen J, Nouvenne A, Lugli GA, Mancabelli L, et al. Gut microbiota composition and Clostridium difficile infection in hospitalized elderly individuals: a metagenomic study. Sci Rep. 2016 May;6((1)):25945. doi: 10.1038/srep25945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Weingarden A, González A, Vázquez-Baeza Y, Weiss S, Humphry G, Berg-Lyons D, et al. Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome. 2015 Mar;3((1)):10. doi: 10.1186/s40168-015-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fuentes S, van Nood E, Tims S, Heikamp-de Jong I, ter Braak CJ, Keller JJ, et al. Reset of a critically disturbed microbial ecosystem: faecal transplant in recurrent Clostridium difficile infection. ISME J. 2014 Aug;8((8)):1621–33. doi: 10.1038/ismej.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ramsay I, Brown NM, Enoch DA. Recent Progress for the Effective Prevention and Treatment of Recurrent Clostridium difficile Infection. Infect Dis (Auckl) 2018 Mar;11:1178633718758023. doi: 10.1177/1178633718758023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lübbert C, Zimmermann L, Borchert J, Hörner B, Mutters R, Rodloff AC. Epidemiology and Recurrence Rates of Clostridium difficile Infections in Germany: A Secondary Data Analysis. Infect Dis Ther. 2016 Dec;5((4)):545–54. doi: 10.1007/s40121-016-0135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Seekatz AM, Rao K, Santhosh K, Young VB. Dynamics of the fecal microbiome in patients with recurrent and nonrecurrent Clostridium difficile infection. Genome Med. 2016 Apr;8((1)):47. doi: 10.1186/s13073-016-0298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Canavan C, West J, Card T. The epidemiology of irritable bowel syndrome. Clin Epidemiol. 2014 Feb;6:71–80. doi: 10.2147/CLEP.S40245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bolino CM, Bercik P. Pathogenic factors involved in the development of irritable bowel syndrome: focus on a microbial role [ix.] Infect Dis Clin North Am. 2010 Dec;24((4)):961–75. doi: 10.1016/j.idc.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 96.Halvorson HA, Schlett CD, Riddle MS. Postinfectious irritable bowel syndrome—a meta-analysis. Am J Gastroenterol. 2006 Aug;101((8)):1894–9. doi: 10.1111/j.1572-0241.2006.00654.x. [DOI] [PubMed] [Google Scholar]
- 97.McIntosh K, Reed DE, Schneider T, Dang F, Keshteli AH, De Palma G, et al. FODMAPs alter symptoms and the metabolome of patients with IBS: a randomised controlled trial. Gut. 2017 Jul;66((7)):1241–51. doi: 10.1136/gutjnl-2015-311339. [DOI] [PubMed] [Google Scholar]
- 98.Giamarellos-Bourboulis E, Tang J, Pyleris E, Pistiki A, Barbatzas C, Brown J, et al. Molecular assessment of differences in the duodenal microbiome in subjects with irritable bowel syndrome. Scand J Gastroenterol. 2015;50((9)):1076–87. doi: 10.3109/00365521.2015.1027261. [DOI] [PubMed] [Google Scholar]
- 99.Labus JS, Hollister EB, Jacobs J, Kirbach K, Oezguen N, Gupta A, et al. Differences in gut microbial composition correlate with regional brain volumes in irritable bowel syndrome. Microbiome. 2017 May;5((1)):49. doi: 10.1186/s40168-017-0260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rajilić-Stojanović M, Biagi E, Heilig HG, Kajander K, Kekkonen RA, Tims S, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011 Nov;141((5)):1792–801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 101.Duan R, Zhu S, Wang B, Duan L. Alterations of Gut Microbiota in Patients With Irritable Bowel Syndrome Based on 16S rRNA-Targeted Sequencing: A Systematic Review. Clin Transl Gastroenterol. 2019 Feb;10((2)):e00012. doi: 10.14309/ctg.0000000000000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang Z, Xu CM, Liu YX, Wang XQ, Zhang L, Li M, et al. Characteristic dysbiosis of gut microbiota of Chinese patients with diarrhea-predominant irritable bowel syndrome by an insight into the pan-microbiome. Chin Med J (Engl) 2019 Apr;132((8)):889–904. doi: 10.1097/CM9.0000000000000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhuang X, Xiong L, Li L, Li M, Chen M. Alterations of gut microbiota in patients with irritable bowel syndrome: A systematic review and meta-analysis. J Gastroenterol Hepatol. 2017 Jan;32((1)):28–38. doi: 10.1111/jgh.13471. [DOI] [PubMed] [Google Scholar]
- 104.Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, et al. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017 Aug;14((8)):491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 105.Rampelli S, Candela M, Severgnini M, Biagi E, Turroni S, Roselli M, et al. A probiotics-containing biscuit modulates the intestinal microbiota in the elderly. J Nutr Health Aging. 2013 Feb;17((2)):166–72. doi: 10.1007/s12603-012-0372-x. [DOI] [PubMed] [Google Scholar]
- 106.Hemarajata P, Versalovic J. Effects of probiotics on gut microbiota: mechanisms of intestinal immunomodulation and neuromodulation. Therap Adv Gastroenterol. 2013 Jan;6((1)):39–51. doi: 10.1177/1756283X12459294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gibson GR, Wang X. Regulatory effects of bifidobacteria on the growth of other colonic bacteria. J Appl Bacteriol. 1994 Oct;77((4)):412–20. doi: 10.1111/j.1365-2672.1994.tb03443.x. [DOI] [PubMed] [Google Scholar]
- 108.Roberfroid M. Functional food concept and its application to prebiotics. Dig Liver Dis. 2002 Sep;34(Suppl 2):S105–10. doi: 10.1016/s1590-8658(02)80176-1. [DOI] [PubMed] [Google Scholar]
- 109.Vulevic J, Juric A, Tzortzis G, Gibson GR. A mixture of trans-galactooligosaccharides reduces markers of metabolic syndrome and modulates the fecal microbiota and immune function of overweight adults. J Nutr. 2013 Mar;143((3)):324–31. doi: 10.3945/jn.112.166132. [DOI] [PubMed] [Google Scholar]
- 110.Higgins JA, Brown IL. Resistant starch: a promising dietary agent for the prevention/treatment of inflammatory bowel disease and bowel cancer. Curr Opin Gastroenterol. 2013 Mar;29((2)):190–4. doi: 10.1097/MOG.0b013e32835b9aa3. [DOI] [PubMed] [Google Scholar]
- 111.Dos Reis SA, da Conceição LL, Siqueira NP, Rosa DD, da Silva LL, Peluzio MD. Review of the mechanisms of probiotic actions in the prevention of colorectal cancer. Nutr Res. 2017 Jan;37:1–19. doi: 10.1016/j.nutres.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 112.Rafter J, Bennett M, Caderni G, Clune Y, Hughes R, Karlsson PC, et al. Dietary synbiotics reduce cancer risk factors in polypectomized and colon cancer patients. Am J Clin Nutr. 2007 Feb;85((2)):488–96. doi: 10.1093/ajcn/85.2.488. [DOI] [PubMed] [Google Scholar]
- 113.Lv Z, Wang B, Zhou X, Wang F, Xie Y, Zheng H, et al. Efficacy and safety of probiotics as adjuvant agents for Helicobacter pylori infection: A meta-analysis. Exp Ther Med. 2015 Mar;9((3)):707–16. doi: 10.3892/etm.2015.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Malfertheiner P, Megraud F, O'Morain CA, Gisbert JP, Kuipers EJ, Axon AT, et al. European Helicobacter and Microbiota Study Group and Consensus panel Management of Helicobacter pylori infection-the Maastricht V/Florence Consensus Report. Gut. 2017 Jan;66((1)):6–30. doi: 10.1136/gutjnl-2016-312288. [DOI] [PubMed] [Google Scholar]
- 115.Ghouri YA, Richards DM, Rahimi EF, Krill JT, Jelinek KA, DuPont AW. Systematic review of randomized controlled trials of probiotics, prebiotics, and synbiotics in inflammatory bowel disease. Clin Exp Gastroenterol. 2014 Dec;7:473–87. doi: 10.2147/CEG.S27530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kruis W, Fric P, Pokrotnieks J, Lukás M, Fixa B, Kascák M, et al. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut. 2004 Nov;53((11)):1617–23. doi: 10.1136/gut.2003.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sood A, Midha V, Makharia GK, Ahuja V, Singal D, Goswami P, et al. The probiotic preparation, VSL#3 induces remission in patients with mild-to-moderately active ulcerative colitis. Clin Gastroenterol Hepatol. 2009;7((11)):1202–9. doi: 10.1016/j.cgh.2009.07.016. 9 e1. [DOI] [PubMed] [Google Scholar]
- 118.Kato K, Mizuno S, Umesaki Y, Ishii Y, Sugitani M, Imaoka A, et al. Randomized placebo-controlled trial assessing the effect of bifidobacteria-fermented milk on active ulcerative colitis. Aliment Pharmacol Ther. 2004 Nov;20((10)):1133–41. doi: 10.1111/j.1365-2036.2004.02268.x. [DOI] [PubMed] [Google Scholar]
- 119.Oliva S, Di Nardo G, Ferrari F, Mallardo S, Rossi P, Patrizi G, et al. Randomised clinical trial: the effectiveness of Lactobacillus reuteri ATCC 55730 rectal enema in children with active distal ulcerative colitis. Aliment Pharmacol Ther. 2012 Feb;35((3)):327–34. doi: 10.1111/j.1365-2036.2011.04939.x. [DOI] [PubMed] [Google Scholar]
- 120.Casellas F, Borruel N, Torrejón A, Varela E, Antolin M, Guarner F, et al. Oral oligofructose-enriched inulin supplementation in acute ulcerative colitis is well tolerated and associated with lowered faecal calprotectin. Aliment Pharmacol Ther. 2007 May;25((9)):1061–7. doi: 10.1111/j.1365-2036.2007.03288.x. [DOI] [PubMed] [Google Scholar]
- 121.Magro F, Gionchetti P, Eliakim R, Ardizzone S, Armuzzi A, Barreiro-de Acosta M, et al. European Crohn's and Colitis Organisation [ECCO] Third European Evidence-based Consensus on Diagnosis and Management of Ulcerative Colitis. Part 1: Definitions, Diagnosis, Extra-intestinal Manifestations, Pregnancy, Cancer Surveillance, Surgery, and Ileo-anal Pouch Disorders. J Crohn's Colitis. 2017 Jun;11((6)):649–70. doi: 10.1093/ecco-jcc/jjx008. [DOI] [PubMed] [Google Scholar]
- 122.Loman BR, Hernández-Saavedra D, An R, Rector RS. Prebiotic and probiotic treatment of nonalcoholic fatty liver disease: a systematic review and meta-analysis. Nutr Rev. 2018 Nov;76((11)):822–39. doi: 10.1093/nutrit/nuy031. [DOI] [PubMed] [Google Scholar]
- 123.Famouri F, Shariat Z, Hashemipour M, Keikha M, Kelishadi R. Effects of Probiotics on Nonalcoholic Fatty Liver Disease in Obese Children and Adolescents. J Pediatr Gastroenterol Nutr. 2017 Mar;64((3)):413–7. doi: 10.1097/MPG.0000000000001422. [DOI] [PubMed] [Google Scholar]
- 124.Malaguarnera M, Vacante M, Antic T, Giordano M, Chisari G, Acquaviva R, et al. Bifidobacterium longum with fructo-oligosaccharides in patients with non alcoholic steatohepatitis. Dig Dis Sci. 2012 Feb;57((2)):545–53. doi: 10.1007/s10620-011-1887-4. [DOI] [PubMed] [Google Scholar]
- 125.Goldenberg JZ, Yap C, Lytvyn L, Lo CK, Beardsley J, Mertz D, et al. Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. Cochrane Database Syst Rev. 2017 Dec;12:CD006095. doi: 10.1002/14651858.CD006095.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lewis S, Burmeister S, Cohen S, Brazier J, Awasthi A. Failure of dietary oligofructose to prevent antibiotic-associated diarrhoea. Aliment Pharmacol Ther. 2005 Feb;21((4)):469–77. doi: 10.1111/j.1365-2036.2005.02304.x. [DOI] [PubMed] [Google Scholar]
- 127.Guarner F, Sanders ME, Eliakiml R, Fedorak R, Gangl A, Garisch J, et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat Microbiol. 2018 Nov;3((11)):1255–65. doi: 10.1038/s41564-018-0257-9. [DOI] [PubMed] [Google Scholar]
- 128.Ortiz-Lucas M, Tobías A, Saz P, Sebastián JJ. Effect of probiotic species on irritable bowel syndrome symptoms: A bring up to date meta-analysis. Rev Esp Enferm Dig. 2013 Jan;105((1)):19–36. doi: 10.4321/s1130-01082013000100005. [DOI] [PubMed] [Google Scholar]
- 129.O'Mahony L, McCarthy J, Kelly P, Hurley G, Luo F, Chen K, et al. Lactobacillus and bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology. 2005 Mar;128((3)):541–51. doi: 10.1053/j.gastro.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 130.Ford AC, Harris LA, Lacy BE, Quigley EM, Moayyedi P. Systematic review with meta-analysis: the efficacy of prebiotics, probiotics, synbiotics and antibiotics in irritable bowel syndrome. Aliment Pharmacol Ther. 2018 Nov;48((10)):1044–60. doi: 10.1111/apt.15001. [DOI] [PubMed] [Google Scholar]
- 131.McKenzie YA, Thompson J, Gulia P, Lomer MC, IBS Dietetic Guideline Review Group on behalf of Gastroenterology Specialist Group of the British Dietetic Association British Dietetic Association systematic review of systematic reviews and evidence-based practice guidelines for the use of probiotics in the management of irritable bowel syndrome in adults (2016 update) J Hum Nutr Diet. 2016 Oct;29((5)):576–92. doi: 10.1111/jhn.12386. [DOI] [PubMed] [Google Scholar]
- 132.Wilson B, Rossi M, Dimidi E, Whelan K. Prebiotics in irritable bowel syndrome and other functional bowel disorders in adults: a systematic review and meta-analysis of randomized controlled trials. Am J Clin Nutr. 2019 Apr;109((4)):1098–111. doi: 10.1093/ajcn/nqy376. [DOI] [PubMed] [Google Scholar]
- 133.Cammarota G, Ianiro G, Tilg H, Rajilić-Stojanović M, Kump P, Satokari R, et al. European FMT Working Group European consensus conference on faecal microbiota transplantation in clinical practice. Gut. 2017 Apr;66((4)):569–80. doi: 10.1136/gutjnl-2016-313017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shahinas D, Silverman M, Sittler T, Chiu C, Kim P, Allen-Vercoe E, et al. Toward an understanding of changes in diversity associated with fecal microbiome transplantation based on 16S rRNA gene deep sequencing. MBio. 2012 Oct;3((5)):e00338–12. doi: 10.1128/mBio.00338-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kassam Z, Lee CH, Yuan Y, Hunt RH. Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am J Gastroenterol. 2013 Apr;108((4)):500–8. doi: 10.1038/ajg.2013.59. [DOI] [PubMed] [Google Scholar]
- 136.Brandt LJ, Aroniadis OC, Mellow M, Kanatzar A, Kelly C, Park T, et al. Long-term follow-up of colonoscopic fecal microbiota transplant for recurrent Clostridium difficile infection. Am J Gastroenterol. 2012 Jul;107((7)):1079–87. doi: 10.1038/ajg.2012.60. [DOI] [PubMed] [Google Scholar]
- 137.Quraishi MN, Widlak M, Bhala N, Moore D, Price M, Sharma N, et al. Systematic review with meta-analysis: the efficacy of faecal microbiota transplantation for the treatment of recurrent and refractory Clostridium difficile infection. Aliment Pharmacol Ther. 2017 Sep;46((5)):479–93. doi: 10.1111/apt.14201. [DOI] [PubMed] [Google Scholar]
- 138.McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, et al. Clinical Practice Guidelines for Clostridium difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA) Clin Infect Dis. 2018 Mar;66((7)):987–94. doi: 10.1093/cid/ciy149. [DOI] [PubMed] [Google Scholar]
- 139.Ponte A, Pinho R, Mota M, Silva J, Vieira N, Oliveira R, et al. Fecal microbiota transplantation in refractory or recurrent Clostridium difficile infection: a real-life experience in a non-academic center. Rev Esp Enferm Dig. 2018 May;110((5)):311–5. doi: 10.17235/reed.2018.5099/2017. [DOI] [PubMed] [Google Scholar]
- 140.Wang JW, Kuo CH, Kuo FC, Wang YK, Hsu WH, Yu FJ, et al. Fecal microbiota transplantation: review and update. J Formos Med Assoc. 2019 Mar;118(Suppl 1):S23–31. doi: 10.1016/j.jfma.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 141.Choi HH, Cho YS. Fecal Microbiota Transplantation: Current Applications, Effectiveness, and Future Perspectives. Clin Endosc. 2016 May;49((3)):257–65. doi: 10.5946/ce.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Khoruts A, Rank KM, Newman KM, Viskocil K, Vaughn BP, Hamilton MJ, et al. Inflammatory Bowel Disease Affects the Outcome of Fecal Microbiota Transplantation for Recurrent Clostridium difficile Infection. Clin Gastroenterol Hepatol. 2016 Oct;14((10)):1433–8. doi: 10.1016/j.cgh.2016.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cui B, Feng Q, Wang H, Wang M, Peng Z, Li P, et al. Fecal microbiota transplantation through mid-gut for refractory Crohn's disease: safety, feasibility, and efficacy trial results. J Gastroenterol Hepatol. 2015 Jan;30((1)):51–8. doi: 10.1111/jgh.12727. [DOI] [PubMed] [Google Scholar]
- 144.Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi C, et al. Fecal Microbiota Transplantation Induces Remission in Patients With Active Ulcerative Colitis in a Randomized Controlled Trial. Gastroenterology. 2015;149((1)):102–9 e6. doi: 10.1053/j.gastro.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 145.Johnsen PH, Hilpüsch F, Cavanagh JP, Leikanger IS, Kolstad C, Valle PC, et al. Faecal microbiota transplantation versus placebo for moderate-to-severe irritable bowel syndrome: a double-blind, randomised, placebo-controlled, parallel-group, single-centre trial. Lancet Gastroenterol Hepatol. 2018 Jan;3((1)):17–24. doi: 10.1016/S2468-1253(17)30338-2. [DOI] [PubMed] [Google Scholar]
- 146.Halkjær SI, Christensen AH, Lo BZ, Browne PD, Günther S, Hansen LH, et al. Faecal microbiota transplantation alters gut microbiota in patients with irritable bowel syndrome: results from a randomised, double-blind placebo-controlled study. Gut. 2018 Dec;67((12)):2107–15. doi: 10.1136/gutjnl-2018-316434. [DOI] [PubMed] [Google Scholar]