Abstract
Cardiac glucose uptake and oxidation are reduced in diabetes despite hyperglycemia. Mitochondrial dysfunction contributes to heart failure in diabetes. It is unclear whether these changes are adaptive or maladaptive. To directly evaluate the relationship between glucose delivery and mitochondrial dysfunction in diabetic cardiomyopathy, we generated transgenic mice with inducible cardiomyocyte-specific expression of the GLUT4. We examined mice rendered hyperglycemic following low-dose streptozotocin prior to increasing cardiomyocyte glucose uptake by transgene induction. Enhanced myocardial glucose in nondiabetic mice decreased mitochondrial ATP generation and was associated with echocardiographic evidence of diastolic dysfunction. Increasing myocardial glucose delivery after short-term diabetes onset exacerbated mitochondrial oxidative dysfunction. Transcriptomic analysis revealed that the largest changes, driven by glucose and diabetes, were in genes involved in mitochondrial function. This glucose-dependent transcriptional repression was in part mediated by O-GlcNAcylation of the transcription factor Sp1. Increased glucose uptake induced direct O-GlcNAcylation of many electron transport chain subunits and other mitochondrial proteins. These findings identify mitochondria as a major target of glucotoxicity. They also suggest that reduced glucose utilization in diabetic cardiomyopathy might defend against glucotoxicity and caution that restoring glucose delivery to the heart in the context of diabetes could accelerate mitochondrial dysfunction by disrupting protective metabolic adaptations.
Introduction
Of the numerous complications associated with diabetes, cardiovascular diseases remain the major cause of death. Both insulin-deficient (type 1) and insulin-resistant (type 2) diabetes are accompanied by a complex milieu of systemic changes including hyperlipidemia and hyperglycemia. It has long been known that despite increased availability of multiple myocardial substrates, in uncontrolled diabetes the heart shows increased fatty acid (FA) utilization and reduced glucose utilization (1). These metabolic changes acting in concert with mitochondrial dysfunction, oxidative stress, and aberrant intracellular signaling are believed to contribute to ventricular dysfunction even in the absence of coronary artery disease and hypertension (2).
Typically, the healthy heart shows flexibility in its ability to utilize ketone bodies, lactate, FA, and glucose to meet its constant energy demands. These substrates are utilized in a concentration-dependent manner, with a preference for FA. The relative balance between glucose and FA utilization is maintained via allosteric regulation by intermediates of glucose and FA metabolism (3). In diabetes, impaired insulin signaling increases circulating free FA as a result of increased lipolysis (4), which contributes to increased myocardial FA utilization. The mechanisms linking increased FA utilization, lipotoxicity, mitochondrial dysfunction, and diabetic cardiomyopathy are well studied (5). In contrast, many mechanisms including reduced cardiac GLUT4 protein levels contribute to impaired myocardial glucose utilization in diabetes (6), but the contribution of GLUT4 remains incompletely defined. Furthermore, the contribution of glucose toxicity in diabetic cardiomyopathy is less well understood, particularly in light of the relative reduction in myocardial glucose utilization in diabetes. Support for a causative role of increased glucose has been suggested by the emerging relationship between certain glucose-lowering therapies and reduction of heart failure in diabetes (7). Specifically, evidence linking glucose lowering by sodium–glucose cotransporter 2 inhibitors and the reduction of heart failure has reignited interest in the relationship between myocardial energetics and glucose availability in modulating heart failure risk in diabetes (8–10). Thus, increased understanding of mechanisms by which glucose may directly contribute to myocardial dysfunction and mitochondrial impairment, characteristic of diabetic cardiomyopathy, may further inform the complex interaction between myocardial metabolism and heart failure, particularly in the context of diabetes.
Slc2a4 (a.k.a. GLUT4) is the major mediator of myocardial glucose uptake in the contracting heart (11). Overexpression of GLUT4 in skeletal muscle, adipose tissue, and heart prevents the metabolic abnormalities characteristic of diabetic cardiomyopathy; however, it has not been clear whether these benefits are directly attributable to GLUT4 in cardiomyocytes or improved systemic metabolic homeostasis (12). Increased expression from birth of the related family member Slc2a1 (i.e., GLUT1) protected against pressure overload–induced heart failure (13). We have more recently reported that inducible GLUT1 overexpression in the context of pressure overload hypertrophy prevented mitochondrial dysfunction without rescuing contractile dysfunction (14). Conversely, loss of GLUT4 expression predisposes the heart to failure in response to either chronic pressure overload or intermittent exercise training (15). Together, these and other studies strongly support the concept that glucose utilization and GLUT expression may play an important role in the myocardial adaptation to hemodynamic stress. In contrast, constitutive increases in glucose uptake might exacerbate ventricular dysfunction in response to diet-induced obesity (16). However, it remains to be determined whether maintenance of normal rates of myocardial glucose uptake and utilization in the context of diabetes could be cardioprotective. Under conditions of sustained glucose exposure in cultured cardiomyocytes, impaired mitochondrial function has been correlated with the protein posttranslational modification of O-GlcNAcylation that may contribute to decreased mitochondrial complex activity (17,18). In addition to direct modification of mitochondrial proteins, glucose also regulates gene expression by modifying transcription factors and modulating pathways that induce epigenetic changes (19). These data support that under certain circumstances restoring glucose delivery in the context of high extracellular glucose may actually impair cellular function. Thus, increased understanding of mechanisms by which glucose may directly contribute to altered gene expression and differential protein modifications will inform mechanisms linking myocardial mitochondrial impairment to the complex interaction between myocardial metabolism and heart failure in the context of diabetes.
In the current study, we use an inducible cardiac-specific, myc-tagged GLUT4 overexpression mouse model (mG4H) that normalizes myocardial glucose uptake in the context of diabetes. This model also enabled us to define glucose-specific changes in gene expression, mitochondrial protein content, and mitochondrial bioenergetics. We observed that restoration of glucose delivery by maintaining or increasing GLUT4 expression in the context of diabetes enhances glycolytic function but exacerbates the decline in glucose oxidation, prevents the increase in FA oxidation, and accelerates the decline in mitochondrial oxidative phosphorylation (OXPHOS) capacity. These changes were associated with posttranslational modifications (i.e., O-GlcNAcylation) of mitochondrial proteins and transcription factors that contribute to changes in gene expression that would predict mitochondrial impairment. Thus, the decline in cardiac glucose utilization in the context of diabetes may play an adaptive role in reducing glucotoxicity, and circumventing this could worsen outcomes.
Research Design and Methods
Generation of Inducible Cardiomyocyte-Specific GLUT4 Overexpression in Mouse
Mice were studied in accordance with protocols approved by the Institutional Animal Care and Use Committee of the University of Utah and University of Alabama at Birmingham. To generate the inducible mycGLUT4 transgene, a c-myc epitope–tagged Rattus norvegicus GLUT4 cDNA (20) was subcloned into the pTRE2 vector (Clontech Laboratories) that contains a promoter region under regulation of tetracycline response elements and injected into oocytes from FVB/NJ mice. Offspring with germ line transmission were crossed with mice expressing αMHC-tON (21), for inducible cardiomyocyte expression. For all studies, 8- to 10-week-old double transgenic mice (mG4H) were the experimental group with αMHC-tON as controls (Con). Both male and female mice were used for a subset of studies, e.g., echocardiography and isolated working heart. As no differences between the sexes were seen, data were combined. For all other studies, male mice were used. The mice were housed at 22°C on a 12 h–12 h light-dark cycle with ad libitum access to food (8656; Harlan Teklad) and water. The transgene was induced by injection with 100 µg doxycycline (DOX) (500 µg/mL in 0.9% NaCl) (Sigma-Aldrich) and switched to 1 g/kg DOX-containing chow (F5820; Bio-Serv).
Cell Culture, Promoter Reporter Construction, and Luciferase Assays
Cell Culture
C2C12 Mus musculus muscle myoblasts were purchased from ATCC (Manassas, VA). Low passage cells (<10) were maintained at 37°C under 5% CO2 in DMEM containing 4.5 g/L glucose supplemented with 10% FBS. When cells were confluent, cell medium was changed to DMEM supplemented with 2% horse serum for myotube differentiation.
Adenoviral Expression Vectors
Adenoviral green fluorescent protein (GFP) and O-GlcNAcase (OGA) have previously been described (22).
Mammalian Expression Vectors
pCMV-Sp1 and empty control plasmids were provided by Dr. Jonathan M. Horowitz, and their construction has previously been described (23).
Reporter Constructs
The mouse Ndufa9 gene promoter was generated by PCR amplification of FVB/NJ genomic DNA followed by cloning into the pGL4.1 as previously described (24). All promoter deletions had the same 3′-primer: 5′-GAGAGCTCCAAATATCCCTTCTCTGC-3′, which has a SacI-cloning site 994 bp downstream from the transcription start site (TSS). The −2 kb, “full-length,” promoter reporter 5′-primer, 5′ACTGGCCGGTACCACAGGAAG-3′, has a KpnI-cloning site 1,855 bp upstream from the TSS. The deletions had KpnI sites for the −0.5-kb fragment 504 bp, 5′-AGGTACCGTGAACACTAA-3′, and the −0.3-kb fragment 326 bp, 5′-AGGTACCCTGCAGAGCT-3′, upstream from the TSS, respectively.
Site-Directed Mutagenesis
Site-directed mutagenesis of the candidate Sp1 response elements (REs) was performed using the QuikChangeII Kit (Agilent, Santa Clara, CA) following the manufacturer’s protocol with the following modifications. Candidate Sp1-REs were mutated as shown in Fig. 5A. The sites were mutated with the following primers: Sp1-RE1, 5′-CAGTGACTAGATGTGTGGGAGCTCGGGTGATACTCAGAGGGGA-3′/5′-TCCCCTCTGAGTATCACCCGAGCTCCCACACATCTAGTCACTG-3′; Sp1-RE2, 5′-GCGTGCGAGGGGCTTAGAGCTCGGGCGCCTCTGCAT-3′/5′-ATGCAGAGGCGCCCGAGCTCTAAGCCCCTCGCACGC-3′; and Sp1-RE3 (M3) 5′-CAGGGGAAAGGGAGCTCGGACGGCAGGAC-3′/5′-GTCCTGCCGTCCGAGCTCCCTTTCCCCTG-3′.
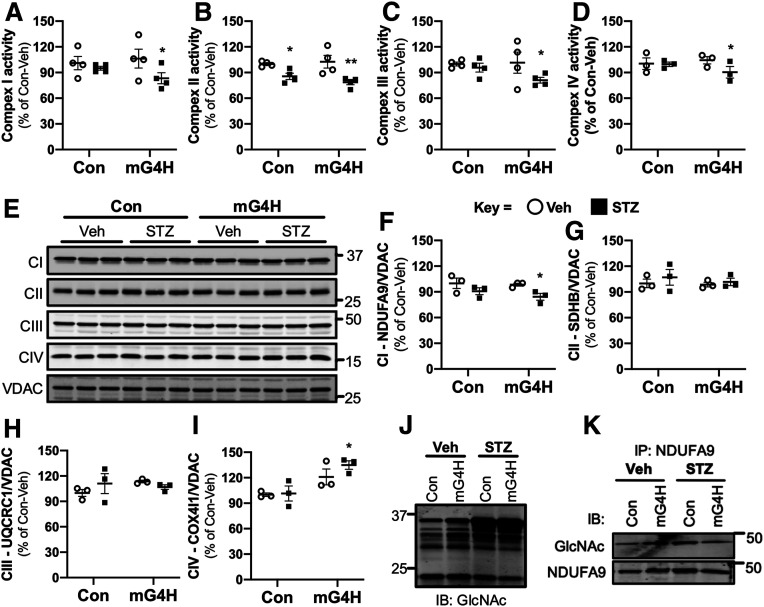
Figure 5.
Induction of cardiac GLUT4 in diabetes decreases respiratory chain complex activities in parallel with O-GlcNAc modifications of mitochondrial proteins. Specific respiratory complex activities in mitochondria isolated from Con and mG4H mice treated as in Fig. 1E and beyond. NADH dehydrogenase CI activity (A) (n = 4), succinate dehydrogenase CII activity (B) (n = 4), coenzyme Q–cytochrome c reductase CIII activity (C) (n = 4), and cytochrome c oxidase CIV activity (D) (n = 3). E: Representative subunit protein levels from CI to CIV in isolated mitochondria corrected to VDAC. (CI, NDUFA9; CII, SDHB; CIII, UQCRC1; CIV, COX5I1.) F–I: Quantification of data presented in E. J: Mitochondrial protein O-GlcNAcylation in isolated mitochondria. K: O-GlcNAcylation of NDUFA9 as assessed by immunoprecipitation (IP) followed by O-GlcNAc immunoblot (IB). Quantitative data are means ± SEM. *P < 0.05 or **P < 0.01 vs. Con-Veh.
Luciferase Assays
The constructs described above and shown in Fig. 5A were used for promoter reporter luciferase assays as previously described (24). C2C12 cells were plated at a density of 140,000 cells/mL in 12-well plates and transfected using Lipofectamine 2000 (Invitrogen, Waltham, MA) as previously described (24). Firefly luciferase reporter plasmids were cotransfected with CMV promoter–driven Renilla luciferase to control for transfection efficiency and the appropriate mammalian expression vector (e.g., pCMV-Sp1) or its corresponding empty vector. For luciferase reporter assays, cells were harvested and analyzed using Dual-Glo (Promega, Madison, WI) and were read according to the manufacturer’s protocols.
Electron Microscopy
Tissue samples were fixed and processed for histology and electron microscopy as previously described (25). Mitochondrial volume density was determined as the mitochondrion-containing fraction of a 32-by-32 grid placed on pictures of ×3,500 magnification (n = 3 hearts and 2–3 pictures/heart). Mitochondrial number was determined in identical-size pictures of ×18,000 magnification (n = 3 hearts and 2–3 pictures/heart).
Histological Analysis
Fresh hearts were fixed by immersion in 10% buffered formalin and analyzed as previously described (15). Tissue was embedded in paraffin, sectioned, and stained by the manufacturer’s protocol with Masson trichrome for visualization of fibrotic tissue on 5–10 images per group. Light microscopy was performed using a Leica DMRB inverted microscope (Leica Microsystems, Wetzlar, Germany) and captured using XnView software (XnSoft, Reims, France).
Protein Analysis by Immunoprecipitation and Western Blotting
Whole-cell, mitochondria-enriched, or nuclear-enriched protein extracts were resolved by SDS-PAGE and following transfer to polyvinylidene fluoride membranes were probed with antibodies as defined below. Detection and quantification were performed by measuring fluorescently labeled secondary antibodies using the Odyssey Infrared Imaging System and accompanying software (version 3.0; LI-COR Biosciences, Lincoln, NE). Immunoprecipitations were performed as follows. Nuclear fractions were isolated from left ventricular tissue, and protein was quantified. A total of 5% of the lysate was saved for input. Anti–O-GlcNAc antibody (5 μg) (RL2, ab2735; Abcam, Cambridge, U.K.) was conjugated with magnetic beads (cat. no. 10004D, Dynabeads; Thermo Fisher Scientific, Waltham, MA) in PBS with 0.02% Tween-20 at room temperature for 90 min. Protein lysate (1 mg) was diluted with 1 mL radioimmunoprecipitation assay buffer and added to the antibody-beads conjugate and incubated overnight at 4°C. Beads were washed five times with radioimmunoprecipitation assay buffer, and protein was eluted with Tris-Glycine SDS sample buffer (cat. no. LC2676; Invitrogen, Carlsbad, CA) containing NuPAGE sample reducing agent (NP009; Invitrogen) and blotted (immunoblot) with the anti-Sp1 antibody (07-645; EMD Millipore, Burlington, MA). Other antibodies used were for 4-aminobutyrate aminotransferase (ABAT) (ab108259; Abcam), complex IV (CIV) subunit COX4l1 (A21348; Invitrogen), GAPDH (CS-2118; Cell Signaling Technology, Danvers, MA), GlcNAc (CTD110.6, MMS-248R; Covance, Princeton, NJ), GlcNAc (RL2, ab2735; Abcam), GLUT4 (ab654; Abcam), Myc (sc-13922; Santa Cruz Biotechnology, Santa Cruz, CA), complex I (CI) subunit NDUFA9 (459100; Invitrogen), Sp1 (07-645; EMD Millipore), complex II (CII) subunit, SDHB (A21345; Invitrogen), complex III (CIII) subunit UQCRC1 (ab110252; Abcam), and VDAC (PA1-954A; Affinity BioReagents, Golden, CO).
In Vivo Cardiac Contractile Function
Echocardiography
Echocardiography was performed in a subset of the mice prior to respiration studies as previously described (26). Mice were anesthetized with isoflurane, first at an induction dose of 2%, and then transferred to a warmed platform with a nose cone at 1%–1.5% maintenance isoflurane and imaged in the left lateral decubitus position with a linear 13-MHz probe (Vivid V echocardiograph; GE Healthcare). Cardiac dimensions and function were calculated from these digital images as previously described (27). Diastolic function was assessed with the VisualSonics Vevo 2100 Imaging System (Toronto, Canada) with the mouse under isoflurane anesthesia using the same protocol as above. Spectral Doppler was used to determine transmitral early (E) and atrial (A) wave peak velocities with the ratio of E to A (E/A) calculated. Peak early (E′) annular velocities were recorded in tissue Doppler mode.
Left Ventricular Catheterization
Left ventricular catheterization was performed on an overlapping subset of animals, invasive left ventricular hemodynamic measurements were performed with a temperature-calibrated 1.4-Fr micromanometer-tipped catheter (Millar Instruments, Houston, TX) inserted through the right carotid artery in anesthetized mice and analyzed as previously described (27).
Microarray and Bioinformatics Analysis
For transcriptomic array-based analysis, left ventricular tissue was harvested from transgenic mG4H or Con mice following 4 weeks of treatment as outlined above (n = 6). RNA isolation was performed as described below. Extracted RNA was analyzed to ensure RNA quality, with RNA integrity number >7. Gene expression intensity was quantified using the Mouse GE 4x44K v2 Microarray (Agilent) by the Health Sciences Center (HSC) Core at the University of Utah as previously described (25). Significant changes in gene expression were then analyzed for functional and network gene set enrichment analysis, along with curated literature-supported candidate upstream regulators, using Ingenuity Pathway Analysis (QIAGEN, Hilden, Germany) unless otherwise specified.
Mitochondrial Purification
Hearts were freshly excised; the atria were separated from the ventricles, and the right ventricle was dissected from the left ventricle. Mitochondria were isolated from the left ventricle by differential centrifugation as previously described (28).
OXPHOS Complex Activity Assays
Activities of individual complexes of the respiratory chain were determined spectrophotometrically, corrected to mitochondrial protein levels, and presented as percent of vehicle (Veh)-treated Con. CI activity, NADH dehydrogenase, was measured as reduction of 2,6-dichloroindophenol by electrons from decylubiquinol following oxidation of NADH by CI (29); CII activity, succinate dehydrogenase, CIII activity, coenzyme Q–cytochrome c reductase, and CIV activity, cytochrome c oxidase, were measured as previously described (30).
Preparation of Isolated Cardiomyocytes and Glucose Uptake/GLUT4 Translocation Assays
Cardiac myocytes were isolated as previously described (31). Perfused hearts were digested with type I collagenase, and once the heart became translucent and soft, it was minced and myocytes were dissociated by sequential washing in buffer. Cells were pelleted by centrifugation and used for glucose transport assays. Studies commenced after waiting for 90 min to allow cardiomyocytes to attach. Cells were washed and then treated with or without insulin, 2-deoxyglucose (2DG), and 1 μCi/μL [3H]2DG (NEN Life Science Products, Boston, MA), and glucose uptake was determined as previously described (14). To measure GLUT4 translocation by exofacial exposure of myc epitope in nonpermeabilized cardiomyocytes, we performed immunofluorescence assay as previously described (32).
Respiration and ATP Measurements in Saponin-Permeabilized Cardiac Fibers
The respiratory rates of saponin-permeabilized fibers were determined using a fiber-optic oxygen sensor (Ocean Optics, Largo, FL) as previously described (33). Studies were performed with three independent substrates: 1) glutamate/malate, 2) pyruvate/malate, or 3) palmitoylcarnitine/malate. Respiratory rates are defined as follows: V0, permeabilized fibers in the presence of substrate; VADP, maximally stimulated respiration following addition of ADP; and VOligo, oligomycin uncoupled, following addition of oligomycin, which inhibits ATP synthase. ATP generation was determined with the ENLITEN ATP Assay System (Promega). ATP/O was calculated as the ratio of ATP synthesis rates and VADP.
RNA Isolation and Quantification
For each group, six individual male mice with at least one sibling pair in another group were harvested between 6:00 a.m. and 7:00 a.m., right ventricle was dissected away, and left ventricle was immediately placed in RNA, later followed by isolation, using the RNeasy Fibrous Tissue Mini Kit (QIAGEN), purification, cDNA synthesis, and labeling. Either microarray quantification (as described above) or quantitative PCR (qPCR) was performed as previously described (25). Primer pairs were designed based on GenBank accession numbers with default settings. Dissociation curves followed by agarose gel size confirmation were analyzed for all primer pairs to ensure single product amplification. Oligos for qPCR analysis were used as follows: for transforming growth factor β1 (Tfgb1) NM_011577.2 forward 5′-CGGAATACAGGGCTTTCGAT-3′, reverse 5′-TTCATGTCATGGATGGTGCC-3′; collagen type I α1 (Col1a1) NM_007743.3 forward 5′-TACAGTGGATTGCAGGGTCT-3′, reverse 5′-TCTACCATCTTTGCCAACGG-3′; transgelin (i.e., SM22α, Tagln) NM_011526.5 forward 5′-GCGACTAGTGGAGTGGATTG-3′, reverse 5′-GATCCCTCAGGATACAGGCT-3′; actin α2 (αSMA, Acta2) NM_007392.3 forward 5′-CCGCCATGTATGTGGCTATT-3′, reverse 5′-AGATAGGCACGTTGTGAGTC-3′; elastin (Eln) NM_012751.1 forward 5′-GCAGAGGAGCAAAAGCTTATTTC-3′, reverse 5′-CCAGCACAGCCAAGACATTGT-3′; ribosomal protein lateral stalk subunit P0 (36B4, Rplp0) NM_007475.5 forward 5′-ACCTCCTTCTTCCGGCTTT-3′, reverse 5′-CTCCAGTCTTTATCAGCTGC-3′; 4-aminobutyrate aminotransferase (Abat) NM_001170978.1 forward 5′-CCAGGAAGCCTTACCGGAT-3′, reverse 5′-TTGTTGAGCAGGTCTTCCCG-3′; and peptidylprolyl isomerase (Cphn, Ppia) NM_008907.2 forward 5′-AGCACTGGAGAGAAAGGATTTGG-3′, reverse 5′-TCTTCTTGCTGGTCTTGCCATT-3′.
Streptozotocin-Induced Diabetes
Mice were made diabetic using the low-dose streptozotocin (STZ) protocol as outlined by the Animal Models of Diabetic Complications Consortium. After 4–6 h of fasting, mice were injected with a stock solution of STZ (7.5 mg/mL) at 55 mg/kg body wt or an equivalent volume of sodium citrate buffer (0.1 mol/L, pH 4.5) for five consecutive days. On day 7 and weekly thereafter, blood glucose was monitored using a standard Contour glucometer (Ascensia, Basel, Switzerland). Mice were considered diabetic if their blood glucose was >200 mg/dL.
Substrate Metabolism and Contractile Function in Isolated Working Hearts
Cardiac substrate metabolism was measured in hearts isolated from Con and mG4H mice 4 weeks after STZ administration and 2 weeks of DOX treatment. Hearts were prepared and perfused using protocols previously described (15). For determination of metabolism, rates of glycolysis for exogenous glucose, glucose oxidation, and palmitate oxidation were measured over a 60-min period in working hearts, and aortic pressures and cardiac performance including coronary flow were monitored by collection of coronary sinus effluent, with the caveat that Thebesian vein drainage may be included in the measurement, as we have previously described (34).
Tissue and Serum Metabolite Levels
Blood was obtained from the submandibular facial vein following Goldenrod Animal Lancet (MEDIpoint, Inc., Mineola, NY) puncture. Serum was used for measurement of free FA (FFA) and triglyceride (TG) levels. Circulating TG and FFA concentrations were determined using kits (Wako Diagnostics, Mountain View, CA). Blood glucose was measured from tail vein puncture using the Contour glucometer. Tissue glycogen levels were measured in extract from liver, gastrocnemius, or heart as previously described (35). Results are presented as glucose released from glycogen corrected to tissue weight.
Two-Dimensional Electrophoresis Analysis and Identification of Glycosylated Mitochondrial Proteins
Two-dimensional PAGE (2D-PAGE) was performed as previously described with the following modifications (36). In brief, following protein separation, gels were stained with Pro-Q Emerald 488 Glycoprotein Gel and Blot Stain Kit (P21875; Thermo Fisher Scientific) following the manufacturer’s protocol. Quantitative differences in the protein patterns between the Con-Veh group and the three treatment groups (i.e., Con-STZ, mG4H-Veh, and mG4H-STZ) were analyzed. Spots that significantly changed in these groups were selected using the statistical module of the MELANIE III software package.
In-Gel Digestion and Mass Spectrometry
Gel spots were destained and subjected to trypsin (20 ng/μL; Promega) digest. Further extraction of peptides from the gel material was performed twice, and these solutions were collected and combined. The supernatant solutions of extracted peptides were combined and dried in a vacuum centrifuge. The samples were then subjected to liquid chromatography (LC)–tandem mass spectrometry (MS/MS) analysis by the HSC Core at the University of Utah. Specifically, peptides were analyzed using a nano–LC-MS/MS system comprised of a nano-LC pump (Eksigent, AB SCIEX, Framingham, MA) and an LTQ-FT mass spectrometer (Thermo Electron Corporation, Waltham, MA). LTQ-FT MS raw data files were processed to peak lists with BioWorks Browser 3.2 software (Thermo Electron Corporation). Resulting DTA files from each data acquisition file were merged, and the data file was searched for identified proteins against the National Center for Biotechnology Information (NCBI) database, mouse taxonomy subdatabase, using Mascot search engine (version 2.2.1; Matrix Science). Identified peptides were accepted only when the Mascot ion score value exceeded 20.
Quantification and Statistical Analysis
Data are presented as means ± SEM. Sample number (n) indicates the number of independent biological samples (individual mice or wells of cells) in each experiment. Unpaired Student t test was used to analyze comparisons between two groups. Data sets with more than two groups were analyzed by two-way ANOVA, and significance was assessed using Tukey post hoc analysis. Statistical calculations were performed using the JMP Pro 9.0 software package (SAS Institute, Cary, NC) or GraphPad Prism 8 software (GraphPad, San Diego, CA), and significance was set at P < 0.05. For array-based gene expression analysis, statistical significance was assessed with a Benjamini-Hochberg P value adjustment.
Data and Resource Availability
All data from this article are available from the corresponding authors upon request. The gene expression data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) (37) and are accessible through GEO Series accession number GSE123975 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE123975).
Results
Development of Inducible Cardiomyocyte-Specific GLUT4 Transgenic Mice
To modulate glucose uptake in the adult mouse heart, we generated double transgenic mice with DOX-inducible cardiac-specific expression of an myc-tagged GLUT4 transgene (mG4H). Western blot analysis revealed a fivefold increase in GLUT4 protein following 2 weeks of transgene induction (Fig. 1A). Increases in GLUT4 protein levels were observed as early as 2 days after introduction of DOX, reaching maximum levels at about 1 week (Supplementary Fig. 1A). Transgene induction was not observed in single transgenic animals, in double transgenic animals without DOX, or in other tissues examined (Fig. 1A, Supplementary Fig. 1A and B, and data not shown).
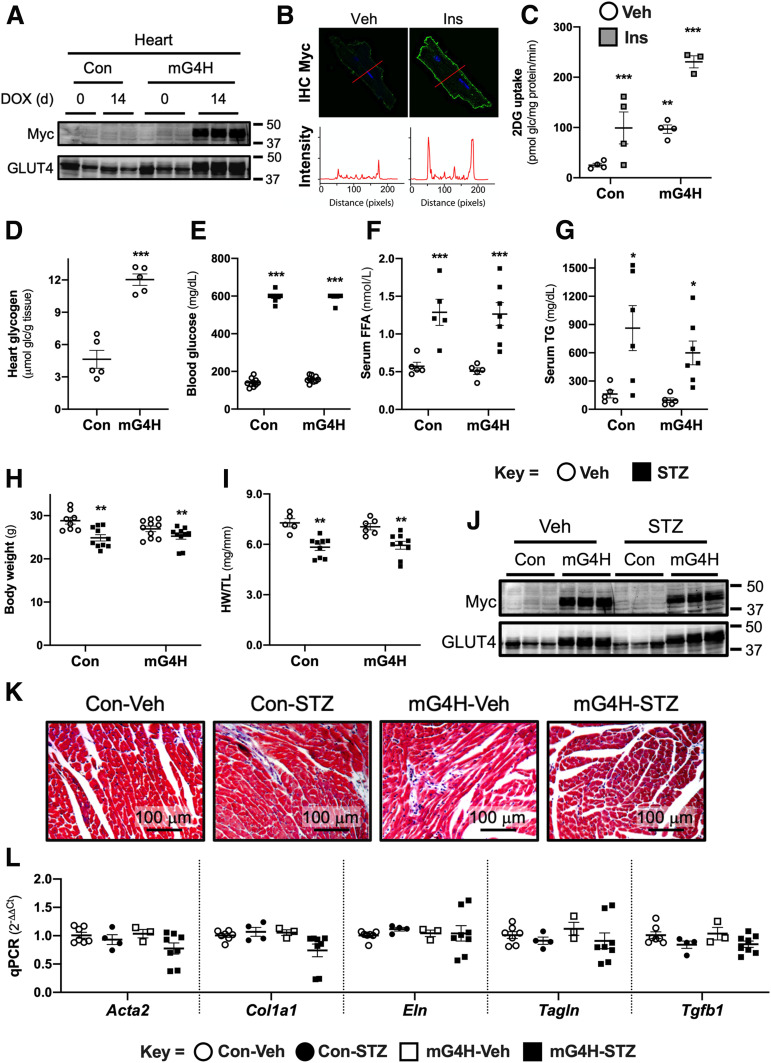
Figure 1.
mG4H mice exhibit inducible cardiomyocyte-specific expression of the mycGLUT4 transgene that enhances glucose (glc) uptake and storage in the heart without influencing systemic metabolic homeostasis or cellular structure. A: Western blot analysis of whole cell extracts for Myc-tag or GLUT4 protein from biventricular heart tissue in the hearts of single transgenic (Con) and double transgenic (mG4H) mice in the absence or presence of DOX. d, days (additional conditions and tissues described in Supplementary Fig. 1A and B). B: IHC for Myc tag of primary adult cardiomyocytes isolated from mG4H mice 2 weeks following DOX induction in the absence or presence of insulin. C: Basal and insulin-stimulated 2DG uptake in primary adult cardiomyocytes isolated from Con and mG4H mice following 2 weeks of DOX treatment (n = 3–4). Ins, 0.1 nmol/L insulin. D: Heart glycogen content of Con and mG4H mice following 2 weeks of DOX treatment (n = 5), with additional tissues in Supplementary Fig. 1C and D. E: Development of hyperglycemia following STZ treatment compared with Veh-injected mice. Serum FFAs (F) (n = 5–7) and TG (G) (n = 5–7). Body weight (H) (n = 11–14) and heart weight (HW)–to–tibia length (TL) ratio (I) (n = 5–9) at 4 weeks following Veh or STZ treatment. J: Western blot analysis as in A, with STZ treatment (n = 3). Histological examination of cardiac tissue using trichrome staining (K) and qPCR for RNA levels of fibrotic markers (n = 3–7) (L). Additional contractile function data associated with these different conditions is listed in Supplementary Tables 1 and 2. All quantitative data are means ± SEM. *P < 0.05, **P < 0.01, or ***P < 0.001 vs. Con-Veh.
Enhanced Glucose Uptake and Storage Following GLUT4 Transgene Induction
To assess transgene function, we isolated cardiac myocytes from mG4H mice following 2 weeks of DOX treatment. We performed immunofluorescence histochemistry (IHC) for detection of the exofacial myc-tag epitope of the transgene in nonpermeabilized cells. There was patchy GLUT4 protein on the sarcolemma of cardiomyocytes under basal conditions, and recruitment to the sarcolemma was markedly increased in response to insulin treatment (Fig. 1B). Using similarly prepared cells from both littermate Con and mG4H mice, we measured [3H]2DG uptake. 2DG uptake was increased 3.2-fold by transgene induction alone, which was similar to insulin-treated cells isolated from Con mice (Fig. 1C). Insulin further increased 2DG uptake by another 2.2-fold above the increased baseline uptake in cells isolated from mG4H mice (Fig. 1C). Compared with hearts of Con mice, mG4H hearts contained ∼2.6-fold more glucose stored as glycogen (Fig. 1D) but not in other tissues examined including liver and skeletal muscle (Supplementary Fig. 1C and D).
Development of Insulin-Deficient Diabetes Is Not Altered by Cardiac GLUT4 Transgene Induction
To determine the role of restoring glucose delivery to the heart under diabetic conditions, we used the low-dose STZ model of insulin-deficient diabetes. By 1 week after the initial STZ injection, all treated mice were hyperglycemic (blood glucose >200 mg/dL) compared with Veh-injected Con. At 2 weeks after the initial STZ or Veh injection, all mice were switched to DOX-containing chow to induce GLUT4 transgene expression in mG4H mice and followed an additional 2 weeks. At this 4-week time point, all STZ-treated mice had blood glucose >500 mg/dL compared with values <150 mg/dL in Veh-injected mice (Fig. 1E). Blood was collected, mice were euthanized, weights were collected, and tissue was snap-frozen for molecular analysis at this 4-week time point. Consistent with the development of diabetes, STZ-treated mice were also hyperlipidemic as shown by significantly elevated serum FFA (Fig. 1F) and TG (Fig. 1G) concentrations. As expected, STZ-induced diabetes resulted in lower body weight (Fig. 1H) and a lower heart weight–to–tibia length ratio (Fig. 1I). Cardiomyocyte-specific overexpression of GLUT4 had no effect on these parameters. Furthermore, Western blot analysis revealed that STZ treatment in turn had no significant effect on DOX-induced GLUT4 overexpression (Fig. 1J). Interestingly, GLUT4 overexpression had relatively little effect on cardiac histology (Fig. 1K) or fibrotic marker gene expression (Fig. 1L).
Induction of GLUT4 Transgene Following Short-term Hyperglycemia Did Not Alter In Vivo Systolic Cardiac Function or Exacerbate Diastolic Dysfunction
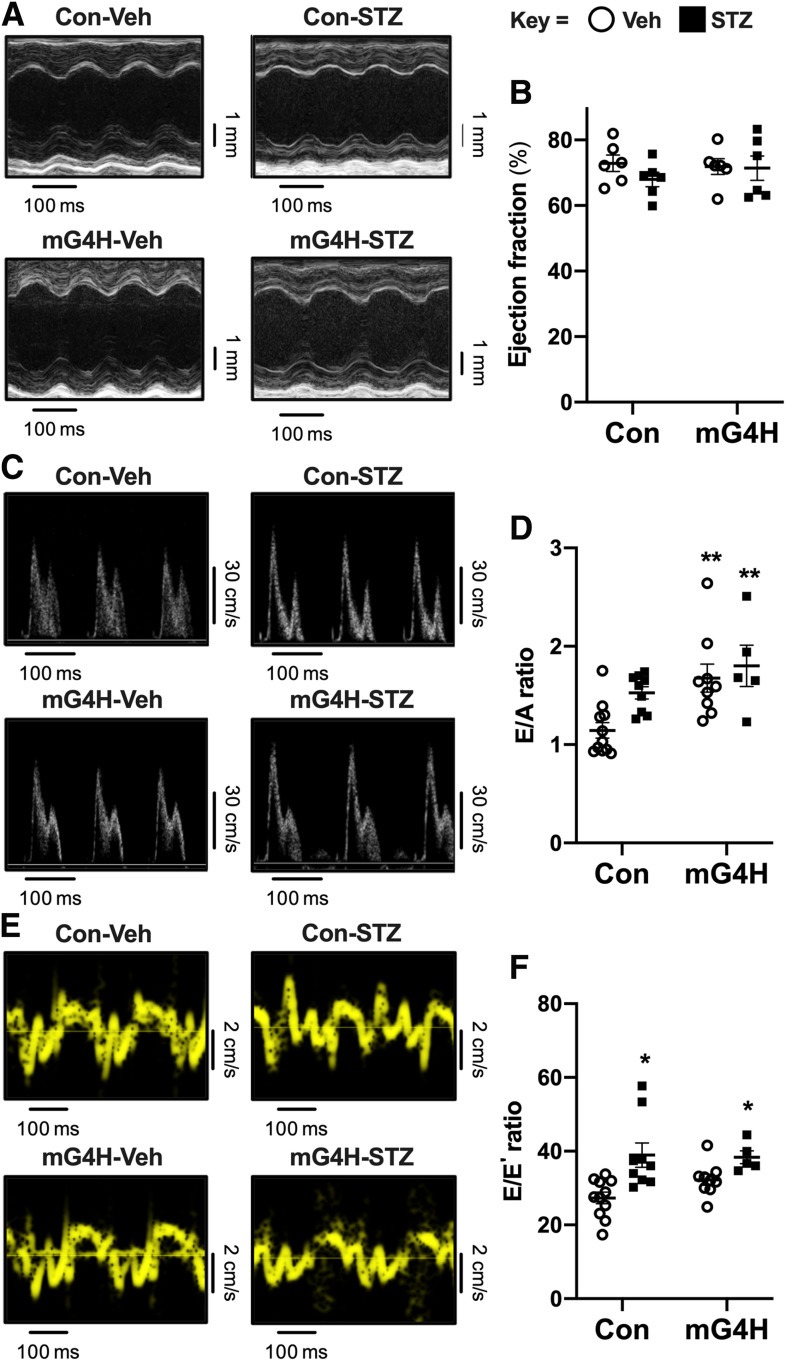
Systolic function was assessed by echocardiography (Fig. 2A). The primary difference observed was reduced cardiac output in STZ-treated animals irrespective of genotype, with a trend toward lower cardiac output in Veh-treated mG4H mice and no differences in ejection fraction (Fig. 2B and Supplementary Table 1). The change in cardiac output may reflect the smaller heart size and body weight. Invasive left ventricular catheterization revealed equivalent degrees of systolic dysfunction in STZ-treated Con and mG4H mice (Supplementary Table 2). Two independent echocardiographic measurements were used to estimate diastolic function: E/A ratio and the E/E′ ratio. GLUT4 induction was characterized by decreased A-wave amplitude leading to an increase in the E/A ratio that was independent of STZ treatment (Fig. 2C and D). In Con animals, the mean E/A ratio was higher following STZ treatment, but overlapping values in a subset of Veh-treated Con animals precluded statistical significance. Tissue Doppler imaging showed reduced E′-wave amplitude in both STZ-treated groups leading to comparable elevations in E/E′ ratios. Although the mean E/E′ ratio was higher in Veh-treated mG4H mice, values were partially overlapping in Veh-treated Con animals, precluding statistical significance (Fig. 2E and F). With results taken together, it appears that short-term STZ-induced diabetes does not impair systolic dysfunction in vivo, but evidence for some degree of diastolic dysfunction is apparent, and depending on the measurement used, transgene induction might also induce diastolic dysfunction. However, there is little evidence of synergistic exacerbation by transgene induction and diabetes.
Figure 2.
Echocardiographic analysis of in vivo cardiac dimensions and function in diabetic mice with short-term overexpression of GLUT4. Representative M-mode images (A) show preserved systolic function following STZ treatment or GLUT4 transgene induction. B: Quantification of ejection fraction from M-mode; additional parameters are reported in Supplementary Table 1. Transmitral Doppler flow reveals diastolic dysfunction in mG4H mice manifested by increased E/A ratio (n = 5–11); representative images (C) and quantification (D). Tissue Doppler imaging also identified an elevated E/E′ ratio in STZ-treated diabetic mice independent of genotype (n = 5–11); representative images (E) and quantification (F). Quantitative data are means ± SEM. *P < 0.05 or **P < 0.01 vs. Con-Veh.
Substrate Metabolism and Ex Vivo Function in Diabetes Are Altered by GLUT4 Transgene Induction
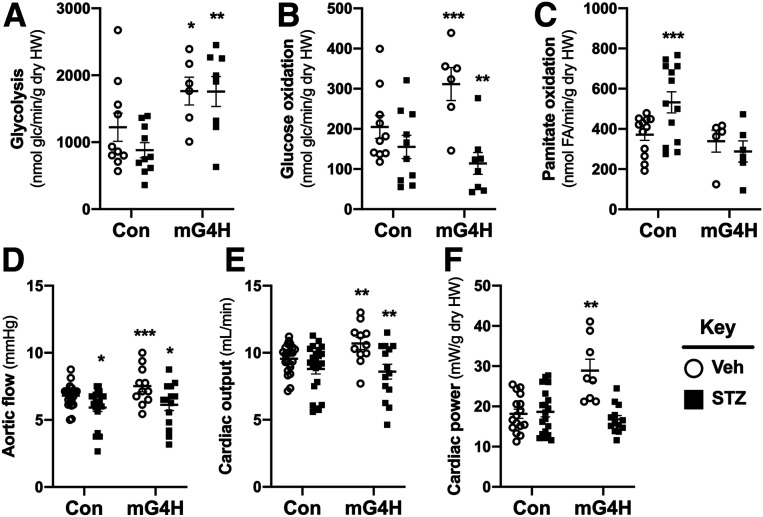
Cardiac glucose utilization by either glycolysis or oxidation was increased in perfused isolated working heart (IWH), following GLUT4 overexpression in mG4H (Fig. 3A and B). As expected, glucose utilization was decreased in hearts of Con-STZ mice. Diabetes-induced repression of glycolysis was reversed with GLUT4 overexpression (Fig. 3A), but impairment in glucose oxidation was exacerbated (Fig. 3B). The expected compensatory increase in FA utilization, as measured by palmitate oxidation in IWH, was observed in hearts of Con-STZ mice (Fig. 3C). Consistent with mitochondrial oxidative impairment as suggested by reduced glucose oxidation, palmitate oxidation failed to increase in hearts of mG4H-STZ mice (Fig. 3C). There were subtle, but statistically significant, increases in basal ex vivo contractile function in hearts of mG4H-Veh mice including aortic flow, cardiac output, and cardiac power, which were reversed in diabetic animals (Fig. 3D–F and Supplementary Table 3).
Figure 3.
Cardiac function and substrate metabolism in IWH is altered in diabetic mice with short-term overexpression of GLUT4. Glycolysis (A) (n = 6–10), glucose (glc) oxidation (B) (n = 6–10), and palmitate oxidation (C) (n = 5–13) in IWH from mice 4 weeks after STZ treatment and DOX-induced transgene expression for 2 weeks. DOX was administered 2 weeks after STZ treatment (heart weight [HW]). Ex vivo contractile function of data pooled from both glucose and palmitate metabolism studies for aortic flow (D), cardiac output (E), and cardiac power (F). Additional contractile function data associated with these perfusions are shown in Supplementary Table 3. Quantitative data are means ± SEM. *P < 0.05, **P < 0.01, or ***P < 0.001 vs. Con-Veh.
Mitochondrial Oxygen Consumption and Respiratory Coupling
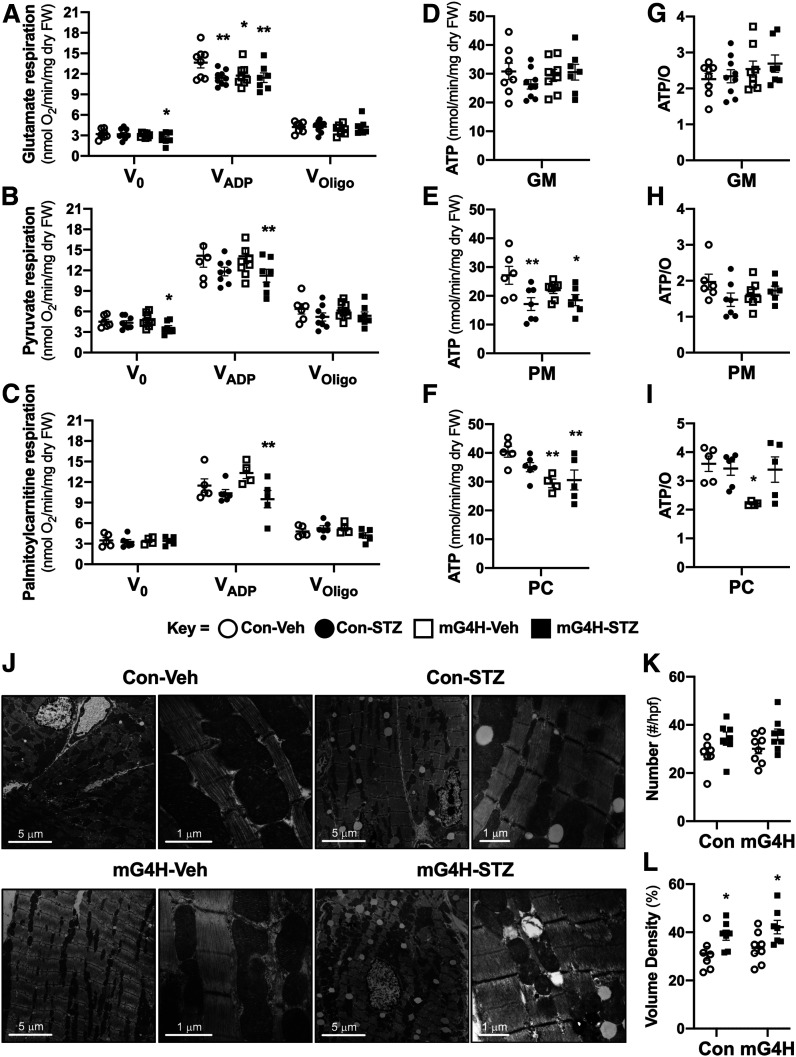
Mitochondrial respiratory function was further assessed using saponin-permeabilized cardiac fibers from hearts of Con and mG4H mice in the presence or absence of diabetes. Basal respiration (V0) was significantly reduced only in the mG4H-STZ group with glutamate or pyruvate as substrates (Fig. 4A and B). Maximal glutamate-supported ADP-stimulated mitochondrial oxygen consumption (VADP) was equivalently suppressed by diabetes in Con and mG4H mice and in animals in the mG4H-STZ group. In contrast, VADP was suppressed in pyruvate- and palmitoylcarnitine-treated samples only in the mG4H-STZ group (Fig. 4A–C). ATP synthesis was suppressed in the mG4H-STZ group with either pyruvate or palmitoylcarnitine as substrates (Fig. 4E and F) but was unchanged with glutamate (Fig. 4D). Interestingly, the only change in ATP/O ratio was in the mG4H-Veh treatment group incubated with palmitoylcarnitine (Fig. 4G–I). There were no statistically significant changes from GLUT4 overexpression (mG4H) in mitochondrial number, and a modest, but significant, increase in mitochondrial volume was observed in both STZ groups (Fig. 4J–L).
Figure 4.
Mitochondrial respiratory parameters and ATP synthesis rates are decreased by induction of GLUT4 following short-term hyperglycemia with minor changes in mitochondrial number and structure. Mitochondrial respiration in saponin-permeabilized cardiac fibers examining V0, VADP, and VOligo oxygen utilization with glutamate/malate (GM) (A) (n = 7–9), pyruvate/malate (PM) (B) (n = 6–9), or palmitoylcarnitine/malate (PC) (C) (n = 4–6) as substrates (fiber weight [FW]). ATP synthesis rates (D–F) and ATP/O ratios (G–I) in comparably treated fibers from the same mice as in A–C. J: Representative electron micrographs of cardiac tissue; left, low magnification, and right, high magnification of different heart sections. K: Mitochondrial number per high-power field (hpf) (n = 3 hearts). L: Mitochondrial volume density as a percentage (n = 3 hearts). Quantitative data are means ± SEM. *P < 0.05 or **P < 0.01 vs. Con-Veh.
Mitochondrial Respiratory Complex Activity and O-GlcNAcylation of OXPHOS Proteins and Mitochondrial Enzymes
For further elucidation of potential mechanisms for the mitochondrial respiratory defects observed, mitochondria were isolated from the four groups. In mG4H-STZ mice, there was a decrease in electron transport chain complex activity for all four electron transport chain complexes measured spectrophotometrically (Fig. 5A–D). Diabetes alone reduced CII activity in Con mice (Fig. 5B). Analysis of mitochondrial OXPHOS protein content revealed a modest but significant decrease in CI protein NDUFA9 in mG4H-STZ mice (Fig. 5E and F). There were no significant changes in CII or CIII proteins, but a significant increase in CIV protein COX4l1 was observed in the mG4H-STZ group (Fig. 5G–I). Various mechanisms have been evaluated to determine the role of glucose in regulating mitochondrial protein function, including changes in the posttranslational modification O-GlcNAcylation (38). Western blot analysis of isolated cardiac mitochondrial proteins (as in Fig. 5E) revealed a modest increase in protein O-GlcNAcylation in mG4H-Veh and a robust increase in STZ groups (Fig. 5J). To begin to identify O-GlcNAc–modified proteins, we first used a candidate approach and examined the previously identified NDUFA9 (17). There was a modest increase in protein O-GlcNAcylation of immunoprecipitated NDUFA9 by transgene induction and diabetes (Fig. 5K). We next performed targeted glycoproteomics to identify additional proteins with GlcNAc modifications. Specifically, we used isolated mitochondrial fractions (as in Fig. 5) and separated proteins by two-dimensional PAGE followed by Pro-Q Emerald 488 glycoprotein staining. Fourteen spots were then selected that showed similar staining in each of the three treatment groups relative to Con-Veh and were subjected to MS identification. All 14 proteins had one predominant peptide identified by Mascot database searches (Table 1). Furthermore, all proteins identified are known to be associated with mitochondria, and the majority have previously been shown to possess O-GlcNAcylation sites on proteins in the diabetic rat heart (39). The latter finding supports the conclusion that increasing glucose uptake alone (mG4H) may replicate certain changes in protein posttranslational regulation associated with the complex metabolic milieu that occurs in diabetes.
Table 1.
MS-identified mitochondrial proteins with differential glycosylation in Veh- and STZ-treated Con and mG4H mice
| Protein symbol | UniProt (NCBI) identifier | Description | Known protein O-GlcNAc target ref.† |
|---|---|---|---|
| ACADL | P51174 | Acyl-CoA dehydrogenase, long-chain | A |
| ATP5A1 | Q03265 | ATP synthase F1 complex, α subunit 1 | A, B, C |
| ATP5H | Q9DCX2 | ATP synthase F0 complex, subunit D | Novel |
| ATP5O | Q9DB20 | ATP synthase F1 complex, O subunit | A |
| GATD3A | Q9D172 | Glutamine amidotransferase-like class 1 domain-containing protein 3A | Novel |
| DLAT | Q8BMF4 | Dihydrolipoamide S-acetyltransferase (pyruvate dehydrogenase complex) | A, B |
| ETFB | Q9DCW4 | Electron transferring flavoprotein, β polypeptide | A |
| HADHB | Q99JY0 | Hydroxyacyl-CoA dehydrogenase trifunctional complex, β subunit | A |
| MDH2 | P08249 | Malate dehydrogenase 2, NAD | A |
| OGDH | Q60597 | Oxoglutarate (α-ketoglutarate) dehydrogenase | A, D |
| PRDX3 | P20108 | Peroxiredoxin 3 | A |
| SUCLA2 | Q9Z2I9 | Succinate-CoA ligase, ADP-forming, β subunit | A |
| UQCRC1 | Q9CZ13 | Ubiquinol–cytochrome c reductase core protein 1 | B |
| UQCRFS1 | Q9CR68 | Ubiquinol–cytochrome c reductase, Rieske iron-sulfur polypeptide 1 | A |
Expression Levels of the Mitochondrial CI Subunit Ndufa9 Is Directly Regulated by Glucose and O-GlcNAcylation
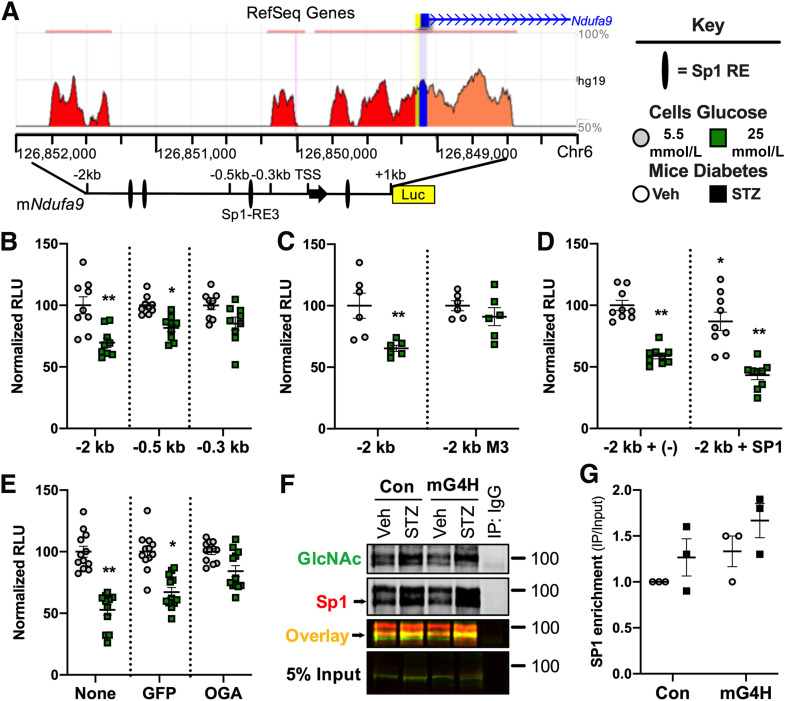
To further explore the mechanisms by which glucose availability may regulate mitochondrial oxidative capacity, we used a candidate approach to define the molecular pathways by which glucose can alter gene expression. We reasoned that given the widespread changes in mitochondrial protein O-GlcNAcylation, parallel changes could be taking place in the nucleus. We focused initially on the transcription factor Sp1, a known target of regulation by O-GlcNAcylation (40), and determined whether its regulation of mitochondrial OXPHOS gene expression could be modulated by O-GlcNAcylation in the hearts of diabetic and mG4H mice. Transcript levels of the Ndufa9 subunit of OXPHOS CI were repressed in mG4H-STZ mice (Fig. 5F), suggesting a potential role for glucose-dependent repression of gene expression. Many regions of the Ndufa9 gene are evolutionarily conserved from human to mouse (Fig. 6A) and harbor candidate Sp1 REs. We cloned a large portion of the mouse Ndufa9 gene promoter (−2 to 1 kb) into a luciferase reporter and generated two deletion mutants that removed some of these candidate REs. Following transfection of the reporter plasmids into C2C12 myoblasts, cells were differentiated into myotubes. Promoter activity was assessed, revealing it was suppressed by high-glucose treatment (25 mmol/L) in the full-length and first deletion (−0.5 kb). However, this repression was lost upon further truncation (Fig. 6B). We then sequentially mutated each of the three candidate Sp1 REs in the glucose-responsive regions and found that only one site, between the −0.5 and −0.3 kb promoter fragments, was actually required for glucose-mediated transcriptional suppression (Fig. 6C and data not shown). Lastly, we used an Sp1 overexpression plasmid to demonstrate that Sp1 was sufficient to confer transcriptional suppression and further enhanced glucose-mediated repression of promoter activity (Fig. 6D).
Figure 6.
Evidence linking O-GlcNAcylation of Sp1 to the glucose-mediated regulation of Ndufa9 gene expression. A: Conservation of the mouse and human Ndufa9 gene promoter regions (https://ecrbrowser.dcode.org) and schema of luciferase (Luc) reporter constructs. Sp1 RE identified by Transcriptional Regulatory Element Database (TRED) (purple), exon 1 of Ndufa9 (black arrow), and TSS (e.g., −2 kb is 2,000 bp from the TSS). B: Luciferase reporter assay of Ndufa9 promoter deletion series in C2C12 myotubes (−2 kb, full-length reporter; −0.5 kb, 1.5-kb deletion; or −0.3 kb, minimal promoter [as in Fig. 5A]) mapping glucose-mediated transcriptional suppression (n = 3). C: Site-directed Sp1 response element mutation M3 in the full-length −2 kb Ndufa9 promoter luciferase reporter assay defining requirement of the site for glucose-mediated transcriptional suppression (n = 3). D: Cotransfection of Sp1 expression vector and luciferase reporter assay showing Sp1 supported glucose suppression of the full-length −2-kb Ndufa9 promoter in C2C12 myotubes (n = 3). E: Adenoviral overexpression of a GFP or OGA luciferase reporter assay supports an O-GlcNAcylation mechanism of glucose-mediated transcriptional suppression of the full-length −2-kb Ndufa9 promoter in C2C12 myotubes (n = 3). F: O-GlcNAcylation of Sp1 of nuclear proteins from hearts of mice treated as in Fig. 4 and assessed by immunoprecipitation (IP) and Sp1 immunoblot. IgG-negative Con was against a pooled sample containing an equal amount of lysate from each sample group. G: Quantification of Sp1 enriched over input following O-GlcNAcylation immunoprecipitation as in Fig. 5F (n = 3); P = 0.052 genotype effect by two-way ANOVA. Quantitative data are means ± SEM. *P < 0.05 or **P < 0.01 vs. promoter, 5.5 mmol/L glucose.
To test the hypothesis that O-GlcNAcylation mediates the glucose-dependent transcriptional repression of Ndufa9 via Sp1, we repeated experiments following adenoviral overexpression of a control GFP or OGA (the enzyme that removes protein O-GlcNAcylation). OGA-mediated O-GlcNAc removal prevented glucose-mediated transcriptional repression, which was present when GFP alone was overexpressed (Fig. 6E). To determine whether a similar mechanism existed in the hearts of our mouse models, we performed protein immunoprecipitations from left ventricular nuclear lysates of mice treated as in Fig. 5, using an O-GlcNAc–specific antibody (RL2), and Sp1 was detected by using anti-Sp1 antibody (Fig. 6F). Diabetes induced a modest enrichment of cardiac Sp1 O-GlcNAcylation compared with nondiabetic mice that was enhanced in mG4H-STZ hearts (Fig. 6G). mG4H mice also showed increased glycosylation of cardiac Sp1, further supporting a glucose-specific role for O-GlcNAcylation of Sp1. Collectively, these results suggest that increased glucose availability may independently regulate Ndufa9 expression via protein O-GlcNAcylation.
Identification of Glucose-Regulated Gene Expression Profile in the Heart
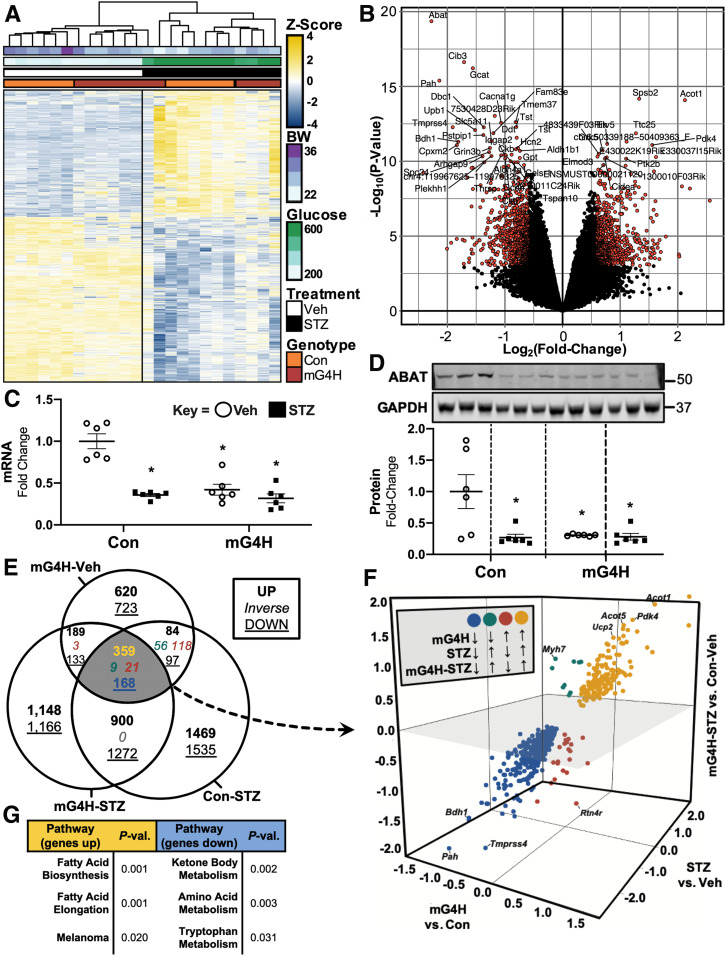
To globally identify gene expression pathways impacted by increased myocardial cardiac glucose delivery versus STZ-induced diabetes, we used microarray-based gene expression analysis. Heatmap and hierarchical clustering of differentially expressed genes (DEGs) by STZ treatment (P < 0.01) revealed a distinct clustering by genotype within the Veh-treated mice, whereas samples failed to cluster by genotype within the STZ-treated group (Fig. 7A). To examine the genes that were regulated in the mG4H-STZ group, we used a volcano plot of mG4H-STZ DEGs (Fig. 7B). This approach revealed robust changes in numerous transcripts encoding enzymes that regulate intermediary metabolism, including pyruvate dehydrogenase kinase 4 (Pdk4) (2.9-fold increase, P = 1.6 × 10−5), acyl-CoA thioesterase 1 (Acot1) (4.4-fold increase, P = 4.2 × 10−7), and β-hydroxybutyrate dehydrogenase (Bdh1) (3.5-fold decrease, P = 1.3 × 10−5). The most differentially suppressed gene, Abat (4.8-fold decrease, P = 4.3 × 10−9), was further validated via qPCR (Fig. 7C) and Western blotting (Fig. 7D), which confirmed its repression in mG4H to the same extent as observed with Con-STZ. To further identify those genes whose regulation is mediated by glucose alone, we compared differential expression based on mG4H and/or STZ treatment via Venn diagram. This analysis identified 557 DEGs between pairwise differential expression analyses of mG4H-Veh, Con-STZ, and mG4H-STZ relative to Con-Veh (Fig. 7E and Supplementary Table 4). Three-dimensional scatterplot analysis revealed that the ketone metabolic enzyme Bdh1 was among the most suppressed (6.0-fold decrease in mG4H-STZ, P = 1.2 × 10−5; 3.7-fold decrease in mG4H, P = 7.9 × 10−7; and 6.1-fold decrease in STZ, P = 6.3 × 10−7), whereas Ucp2, Acot5, Acot1, and Pdk4 were among the most highly induced (Fig. 7F). Within the minority of DEGs inversely changed by mG4H alone, the gene encoding βMHC (Myh7) was the most differentially suppressed in mG4H (3.0-fold decrease, P = 0.004) but was induced in both Con-STZ (3.1-fold increase, P = 1.7 × 10−6) and mG4H-STZ (1.9-fold increase, P = 0.004) relative to Con-Veh. Pathway analysis of the overlapping and upregulated DEGs (Fig. 7E [yellow 359]) enriched for FA metabolic pathways (Fig. 7G and Supplementary Table 5), whereas downregulated overlapping DEGs (Fig. 7E [blue 168]) enriched for ketone body and amino acid metabolic pathways (Fig. 7G and Supplementary Table 5). Together, these findings support additional metabolic signaling connections between glucose delivery and regulation of cardiac metabolism.
Figure 7.
Analysis of glucose-dependent vs. diabetes-dependent gene expression patterns in the mouse heart. A: Heatmap and hierarchical clustering of DEGs from array-based analysis of left ventricles of mice following 4 weeks of low-dose STZ or Veh treatment. B: Volcano plot of mG4H-STZ relative to Con-Veh with the top 50 DEGs labeled. Differential expression of Abat in mG4H or Con mice treated with STZ or Veh by qPCR for RNA (C) and Western blot for protein levels (n = 6) (D). E: Venn diagram comparing gene expression in mG4H-Veh, Con-STZ, and mG4H-STZ relative to Con-Veh. Genes are listed in Supplementary Table 4. F: Coexpressed genes visualized on a three-dimensional scatterplot of fold change for mG4H-Veh vs. Con-Veh, Con-STZ vs. Con-Veh, and mG4H-STZ vs. Con-Veh. G: Gene set enrichment analysis of upregulated (yellow) and downregulated (blue) coexpressed DEGs. Genes listed in Supplementary Table 5. Quantitative data are means ± SEM. *P < 0.05 vs. Con-Veh. BW, body weight; P-val., P value.
Discussion
The objective of the current study was to test the hypothesis that restoring glucose delivery to the heart in the context of diabetes may ameliorate the pathophysiology of diabetic cardiomyopathy. Prior studies revealed that overexpression of GLUT4 in muscle, heart, and adipose tissue from birth prevented the characteristic cardiac metabolic changes and contractile dysfunction in mouse models of diabetes (12,41,42). We more recently reported that loss of cardiac GLUT4 increases susceptibility to heart failure in response to either physiologic or pathologic stress (15). Together, these studies support the hypothesis that maintaining GLUT4-mediated myocardial glucose utilization may be required to prevent diabetes-related ventricular dysfunction and to maintain function following a hemodynamic stress. However, our results indicate that GLUT4 restoration in the context of diabetes not only is unable to rescue either metabolic or contractile dysfunction but also may accelerate the development of diabetic cardiomyopathy. These observations support the perspective that myocardial insulin resistance, as defined by decreased glucose uptake, may provide cardioprotection by defending against fuel overload (43). The current study provides additional molecular insights into mechanisms linking glucotoxicity with mitochondrial dysfunction. Specifically, we observed that increasing myocardial glucose uptake promoted widespread O-GlcNAcylation of core mitochondrial OXPHOS subunits and metabolic enzymes and a representative transcription factor Sp1, which repressed expression of the CI subunit Ndufa9 in a glucose- and O-GlcNAc–dependent manner. Moreover, increased myocardial glucose alone induced a transcriptional program consistent with induction of FA metabolism and repression of ketone body and amino acid metabolism.
Cardiac-specific overexpression of GLUT1 prevents contractile dysfunction and dilation following pressure overload (13), rescues cardiac dysfunction that is secondary to PPARα deficiency (44), and ameliorates ischemic injury associated with aging (45). In contrast, increased myocardial glucose uptake enhances cardiac dysfunction in diet-induced obesity (16), which supports the hypothesis that increasing myocardial glucose delivery in the context of nutrient excess becomes maladaptive.
The short-term duration of hyperglycemia in this study was not expected to induce severe ventricular dysfunction. As such, the current study focused on early mechanisms that could contribute to diabetic cardiomyopathy. We therefore focused on potential mechanisms linking increased myocardial glucose utilization and altered myocardial mitochondrial metabolism. Although not exhaustive, two important modes of regulation were explored. The first was evidence of direct regulation of enzymatic function by posttranslational O-GlcNAcylation of mitochondrial proteins. We identified enzymes and mitochondrial proteins in which a link between O-GlcNAcylation and impaired catalytic activity was previously described in diabetes (17,39). The second was transcriptional regulation, in part, through O-GlcNAcylation of transcriptional regulators (46,47). It should be noted that additional mechanisms likely exist, such as an emerging role of metabolic regulation of epigenetics and its impact on gene expression (19).
There has been resurgent interest in the relationship between cardiac ketone body utilization in heart failure (48,49). In fact, we found that one of the most heavily repressed genes by either GLUT4 expression or diabetes was Bdh1, a rate-limiting enzyme in ketone body oxidation. Of note, these changes linked to diabetes or glucotoxicity in rodent hearts are opposite to changes described in models of nondiabetic heart failure. Thus, the extent to which altered ketone metabolism might contribute to the mitochondrial and metabolic phenotypes of mG4H mice remains to be determined.
The robust changes in the regulation of Abat are intriguing, and the relationship between this change and glucose-induced metabolic and mitochondrial impairment in response to glucotoxicity in the heart remains to be clarified. Abat encodes a metabolic enzyme that catabolizes γ-aminobutyric acid (GABA) for recycling into the tricarboxylic acid cycle as succinate. Prior studies have revealed significantly elevated levels of ABAT protein in failing human dilated cardiomyopathy (1.84-fold) and following trans-aortic constriction (1.95-fold) in the mouse (49). The impact of altered ABAT on mitochondrial substrate utilization and respiratory function remains to be determined, but its pattern of expression identifies another metabolic pathway that is differentially regulated in diabetic cardiomyopathy relative to heart failure arising from other causes.
Despite its exciting and novel findings, it should be noted that the current study has some limitations that should be carefully considered. First, it has recently been shown that tetracyclines can alter mitochondrial function (50). We observed subtle effects of DOX in our pilot studies and chose to control for them by placing all mice on DOX. As such, our design should primarily reveal differences that can be attributed to increased glucose delivery in a background of DOX exposure. Second, this study focused on candidate proteins such as NDUFA9 and ABAT. We demonstrated that O-GlcNAcylation not only might directly regulate the function of NDUFA9 but also may regulate its expression via a similar modification of the Sp1 transcription factor. Although the regulation of gene expression by O-GlcNAcylation of transcription factors likely targets multiple pathways as suggested by our microarray pathway analysis, we have not definitively demonstrated that this specific mechanism mediates the global changes in gene expression that were observed. It is also unlikely that the modest change observed in NDUFA9 exclusively accounts for the mitochondrial oxidative impairment observed. Although germ line defects in NDUFA9 alone may lead to neonatal lethality (51), mice with cardiomyocyte-restricted loss of C1 (NDUFS4) are viable, exhibit preserved ventricular function in nonstressed hearts (52), and might even be protected from ischemia/reperfusion injury (53). Thus, it is likely that multiple pathways converge to limit mitochondrial oxidative capacity when glucose availability is increased. Furthermore, we chose to inducibly overexpress GLUT4, while prior studies have focused on constitutive GLUT1 and GLUT4 expression. Despite the known roles of insulin in regulating GLUT4 vesicle trafficking, and the notion that GLUT1 mediates basal levels of glucose uptake, it is now accepted that GLUT4 is the major contributor of basal glucose uptake in the beating heart, given its greater abundance and the role of myocardial contraction in regulating GLUT4 translocation to the sarcolemma. Moreover, given that GLUT4 is repressed in diabetic cardiomyopathy, we believe that increasing or maintaining GLUT4 content in the heart represents a reasonable approach for preserving myocardial glucose utilization in the context of diabetes. We used STZ-induced hyperglycemia for the current study. Although it is possible that other forms of diabetes may result in different findings, studies in GLUT1 transgenic mice following high-fat feeding–induced insulin resistance and diabetes suggest that maintaining glucose uptake in the context of increased FA delivery to the heart that occurs in type 1 and type 2 diabetes will be deleterious.
In conclusion, the current study shows that in the presence of hyperglycemia, increasing myocardial glucose utilization accelerates the decline in glucose and FA oxidative capacity in IWH, reduces mitochondrial oxygen consumption in saponin-permeabilized cardiac fibers, and impacts mitochondrial OXPHOS activity. Our data suggest that the reduction in glucose uptake in the hyperglycemic state likely plays a protective role that limits glucotoxicity. Although increasing glucose delivery to the heart increased glucose uptake, storage, and utilization and was tolerated in the absence of metabolic stress, it clearly sensitized the heart to develop impaired mitochondrial function in the context of nutrient excess. Thus, in the presence of uncontrolled hyperglycemia, normalizing or increasing myocardial glucose utilization is likely to sensitize the heart to glucotoxicity, which will exacerbate mitochondrial dysfunction and worsen diabetic cardiomyopathy.
Article Information
Acknowledgments. HSC Core services from the University of Utah were provided for the following: microarray by Brian Dalley and Qian Yang, bioinformatics by Brett Milash, and mass spectrometry and proteomics by Chad Nelson and Krishna Parsawar. Electron microscopy images were provided by Elizabeth W. Weeks of EM Laboratories, Inc., Birmingham, AL.
Funding. This work was supported by National Institutes of Health (NIH) grants R00 HL111322 and R01 HL133011 and JDRF Advanced Postdoctoral Fellowship 10-2009-267 to A.R.W. M.E.P. was supported by NIH grant F30 HL137240, O.K. was supported by NIH grant R01 GM108975, R.O.P. and M.K.B. were both supported by postdoctoral fellowships from the American Heart Association (AHA), and E.D.A. was supported by NIH grants R01 DK092065, R01 HL108379, and U01 HL087947 and is an established investigator of the AHA.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. A.R.W. and E.D.A. secured funding, conceived the study, and wrote the manuscript. All authors reviewed and edited the manuscript. A.R.W. performed mouse studies, left ventricular catheterization and analysis, and promoter cloning and analysis, as well as multiple molecular studies (e.g., RNA quantification, protein quantification, metabolite analysis). J.C.S., M.K.B., L.W., and C.A.A. performed mouse studies, collected samples, and performed Western blots. J.C.S. performed and analyzed the fiber studies. C.M.-H. performed qPCR and echocardiography data analysis and reviewed all statistical analysis of the data as well as graphing of the data. M.E.P. performed microarray analysis and qPCR. O.K. performed and analyzed the respiratory complex activities. H.S. performed and analyzed the two-dimensional PAGE. R.O.P. performed 2DG uptake and the quantification of the mitochondria as appears in the electron micrographs. M.K.B. performed immunoprecipitation studies. J.T. performed IWH perfusion and analysis. A.C.-F. performed IHC. C.D.O., W.E.B., L.J.D., and S.E.L. performed and assisted in analysis of the echocardiography data. W.H.D. provided critical reagents for the O-GlcNAc studies. A.R.W. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
See accompanying article, p. 2054.
This article contains supplementary material online at https://doi.org/10.2337/figshare.12200918.
References
- 1.Ungar I, Gilbert M, Siegel A, Blain JM, Bing RJ. Studies on myocardial metabolism. IV. Myocardial metabolism in diabetes. Am J Med 1955;18:385–396 [DOI] [PubMed] [Google Scholar]
- 2.Bugger H, Abel ED. Molecular mechanisms of diabetic cardiomyopathy. Diabetologia 2014;57:660–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Randle PJ, Garland PB, Hales CN, Newsholme EA. The glucose fatty-acid cycle. Its role in insulin sensitivity and the metabolic disturbances of diabetes mellitus. Lancet 1963;1:785–789 [DOI] [PubMed] [Google Scholar]
- 4.Sztalryd C, Kraemer FB. Regulation of hormone-sensitive lipase in streptozotocin-induced diabetic rats. Metabolism 1995;44:1391–1396 [DOI] [PubMed] [Google Scholar]
- 5.Wende AR, Abel ED. Lipotoxicity in the heart. Biochim Biophys Acta 2010;1801:311–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armoni M, Harel C, Bar-Yoseph F, Milo S, Karnieli E. Free fatty acids repress the GLUT4 gene expression in cardiac muscle via novel response elements. J Biol Chem 2005;280:34786–34795 [DOI] [PubMed] [Google Scholar]
- 7.Kenny HC, Abel ED. Heart failure in type 2 diabetes mellitus. Circ Res 2019;124:121–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiviott SD, Raz I, Bonaca MP, et al.; DECLARE–TIMI 58 Investigators . Dapagliflozin and cardiovascular outcomes in type 2 diabetes. N Engl J Med 2019;380:347–357 [DOI] [PubMed] [Google Scholar]
- 9.Ha C-M, Wende AR. The growing case for use of SGLT2i in heart failure: additional benefits of empagliflozin in a HFpEF rodent model. JACC Basic Transl Sci 2019;4:38–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connelly KA, Zhang Y, Visram A, et al. . Empagliflozin improves diastolic function in a nondiabetic rodent model of heart failure with preserved ejection fraction. JACC Basic Transl Sci 2019;4:27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abel ED. Glucose transport in the heart. Front Biosci 2004;9:201–215 [DOI] [PubMed] [Google Scholar]
- 12.Belke DD, Larsen TS, Gibbs EM, Severson DL. Altered metabolism causes cardiac dysfunction in perfused hearts from diabetic (db/db) mice. Am J Physiol Endocrinol Metab 2000;279:E1104–E1113 [DOI] [PubMed] [Google Scholar]
- 13.Liao R, Jain M, Cui L, et al. . Cardiac-specific overexpression of GLUT1 prevents the development of heart failure attributable to pressure overload in mice. Circulation 2002;106:2125–2131 [DOI] [PubMed] [Google Scholar]
- 14.Pereira RO, Wende AR, Olsen C, et al. . Inducible overexpression of GLUT1 prevents mitochondrial dysfunction and attenuates structural remodeling in pressure overload but does not prevent left ventricular dysfunction. J Am Heart Assoc 2013;2:e000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wende AR, Kim J, Holland WL, et al. . Glucose transporter 4-deficient hearts develop maladaptive hypertrophy in response to physiological or pathological stresses. Am J Physiol Heart Circ Physiol 2017;313:H1098–H1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan J, Young ME, Cui L, Lopaschuk GD, Liao R, Tian R. Increased glucose uptake and oxidation in mouse hearts prevent high fatty acid oxidation but cause cardiac dysfunction in diet-induced obesity. Circulation 2009;119:2818–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Y, Suarez J, Fricovsky E, et al. . Increased enzymatic O-GlcNAcylation of mitochondrial proteins impairs mitochondrial function in cardiac myocytes exposed to high glucose. J Biol Chem 2009;284:547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medikayala S, Piteo B, Zhao X, Edwards JG. Chronically elevated glucose compromises myocardial mitochondrial DNA integrity by alteration of mitochondrial topoisomerase function. Am J Physiol Cell Physiol 2011;300:C338–C348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wende AR. Post-translational modifications of the cardiac proteome in diabetes and heart failure. Proteomics Clin Appl 2016;10:25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanai F, Nishioka Y, Hayashi H, Kamohara S, Todaka M, Ebina Y. Direct demonstration of insulin-induced GLUT4 translocation to the surface of intact cells by insertion of a c-myc epitope into an exofacial GLUT4 domain. J Biol Chem 1993;268:14523–14526 [PubMed] [Google Scholar]
- 21.Valencik ML, McDonald JA. Codon optimization markedly improves doxycycline regulated gene expression in the mouse heart. Transgenic Res 2001;10:269–275 [DOI] [PubMed] [Google Scholar]
- 22.Clark RJ, McDonough PM, Swanson E, et al. . Diabetes and the accompanying hyperglycemia impairs cardiomyocyte calcium cycling through increased nuclear O-GlcNAcylation. J Biol Chem 2003;278:44230–44237 [DOI] [PubMed] [Google Scholar]
- 23.Udvadia AJ, Rogers KT, Higgins PD, et al. . Sp-1 binds promoter elements regulated by the RB protein and Sp-1-mediated transcription is stimulated by RB coexpression. Proc Natl Acad Sci U S A 1993;90:3265–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wende AR, Huss JM, Schaeffer PJ, Giguère V, Kelly DP. PGC-1α coactivates PDK4 gene expression via the orphan nuclear receptor ERRalpha: a mechanism for transcriptional control of muscle glucose metabolism. Mol Cell Biol 2005;25:10684–10694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wende AR, O’Neill BT, Bugger H, et al. . Enhanced cardiac Akt/protein kinase B signaling contributes to pathological cardiac hypertrophy in part by impairing mitochondrial function via transcriptional repression of mitochondrion-targeted nuclear genes. Mol Cell Biol 2015;35:831–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sena S, Rasmussen IR, Wende AR, et al. . Cardiac hypertrophy caused by peroxisome proliferator- activated receptor-γ agonist treatment occurs independently of changes in myocardial insulin signaling. Endocrinology 2007;148:6047–6053 [DOI] [PubMed] [Google Scholar]
- 27.McQueen AP, Zhang D, Hu P, et al. . Contractile dysfunction in hypertrophied hearts with deficient insulin receptor signaling: possible role of reduced capillary density. J Mol Cell Cardiol 2005;39:882–892 [DOI] [PubMed] [Google Scholar]
- 28.Tsushima K, Bugger H, Wende AR, et al. . Mitochondrial reactive oxygen species in lipotoxic hearts induce post-translational modifications of AKAP121, DRP1, and OPA1 that promote mitochondrial fission. Circ Res 2018;122:58–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janssen AJ, Trijbels FJ, Sengers RC, et al. . Spectrophotometric assay for complex I of the respiratory chain in tissue samples and cultured fibroblasts. Clin Chem 2007;53:729–734 [DOI] [PubMed] [Google Scholar]
- 30.Barrientos A. In vivo and in organello assessment of OXPHOS activities. Methods 2002;26:307–316 [DOI] [PubMed] [Google Scholar]
- 31.Belke DD, Betuing S, Tuttle MJ, et al. . Insulin signaling coordinately regulates cardiac size, metabolism, and contractile protein isoform expression. J Clin Invest 2002;109:629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Contreras-Ferrat AE, Toro B, Bravo R, et al. . An inositol 1,4,5-triphosphate (IP3)-IP3 receptor pathway is required for insulin-stimulated glucose transporter 4 translocation and glucose uptake in cardiomyocytes. Endocrinology 2010;151:4665–4677 [DOI] [PubMed] [Google Scholar]
- 33.Boudina S, Sena S, O’Neill BT, Tathireddy P, Young ME, Abel ED. Reduced mitochondrial oxidative capacity and increased mitochondrial uncoupling impair myocardial energetics in obesity. Circulation 2005;112:2686–2695 [DOI] [PubMed] [Google Scholar]
- 34.Abel ED, Kaulbach HC, Tian R, et al. . Cardiac hypertrophy with preserved contractile function after selective deletion of GLUT4 from the heart. J Clin Invest 1999;104:1703–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wende AR, Schaeffer PJ, Parker GJ, et al. . A role for the transcriptional coactivator PGC-1α in muscle refueling. J Biol Chem 2007;282:36642–36651 [DOI] [PubMed] [Google Scholar]
- 36.Schwertz H, Längin T, Platsch H, et al. . Two-dimensional analysis of myocardial protein expression following myocardial ischemia and reperfusion in rabbits. Proteomics 2002;2:988–995 [DOI] [PubMed] [Google Scholar]
- 37.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Banerjee PS, Ma J, Hart GW. Diabetes-associated dysregulation of O-GlcNAcylation in rat cardiac mitochondria. Proc Natl Acad Sci U S A 2015;112:6050–6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma J, Banerjee P, Whelan SA, et al. . Comparative proteomics reveals dysregulated mitochondrial O-GlcNAcylation in diabetic hearts. J Proteome Res 2016;15:2254–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamemura K, Hart GW. Dynamic interplay between O-glycosylation and O-phosphorylation of nucleocytoplasmic proteins: a new paradigm for metabolic control of signal transduction and transcription. Prog Nucleic Acid Res Mol Biol 2003;73:107–136 [DOI] [PubMed] [Google Scholar]
- 41.Semeniuk LM, Kryski AJ, Severson DL. Echocardiographic assessment of cardiac function in diabetic db/db and transgenic db/db-hGLUT4 mice. Am J Physiol Heart Circ Physiol 2002;283:H976–H982 [DOI] [PubMed] [Google Scholar]
- 42.Gibbs EM, Stock JL, McCoid SC, et al. . Glycemic improvement in diabetic db/db mice by overexpression of the human insulin-regulatable glucose transporter (GLUT4). J Clin Invest 1995;95:1512–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taegtmeyer H, Beauloye C, Harmancey R, Hue L. Insulin resistance protects the heart from fuel overload in dysregulated metabolic states. Am J Physiol Heart Circ Physiol 2013;305:H1693–H1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luptak I, Balschi JA, Xing Y, Leone TC, Kelly DP, Tian R. Decreased contractile and metabolic reserve in peroxisome proliferator-activated receptor-alpha-null hearts can be rescued by increasing glucose transport and utilization. Circulation 2005;112:2339–2346 [DOI] [PubMed] [Google Scholar]
- 45.Luptak I, Yan J, Cui L, Jain M, Liao R, Tian R. Long-term effects of increased glucose entry on mouse hearts during normal aging and ischemic stress. Circulation 2007;116:901–909 [DOI] [PubMed] [Google Scholar]
- 46.Krause MW, Love DC, Ghosh SK, et al. . Nutrient-driven O-GlcNAcylation at promoters impacts genome-wide RNA Pol II distribution. Front Endocrinol (Lausanne) 2018;9:521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ha C, Lim K. O-GlcNAc modification of Sp3 and Sp4 transcription factors negatively regulates their transcriptional activities. Biochem Biophys Res Commun 2015;467:341–347 [DOI] [PubMed] [Google Scholar]
- 48.Bedi KC Jr, Snyder NW, Brandimarto J, et al. . Evidence for intramyocardial disruption of lipid metabolism and increased myocardial ketone utilization in advanced human heart failure. Circulation 2016;133:706–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aubert G, Martin OJ, Horton JL, et al. . The failing heart relies on ketone bodies as a fuel. Circulation 2016;133:698–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moullan N, Mouchiroud L, Wang X, et al. . Tetracyclines disturb mitochondrial function across eukaryotic models: a call for caution in biomedical research. Cell Rep 2015;10:1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van den Bosch BJ, Gerards M, Sluiter W, et al. . Defective NDUFA9 as a novel cause of neonatally fatal complex I disease. J Med Genet 2012;49:10–15 [DOI] [PubMed] [Google Scholar]
- 52.Karamanlidis G, Lee CF, Garcia-Menendez L, et al. . Mitochondrial complex I deficiency increases protein acetylation and accelerates heart failure. Cell Metab 2013;18:239–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang H, Gong G, Wang P, et al. . Heart specific knockout of Ndufs4 ameliorates ischemia reperfusion injury. J Mol Cell Cardiol 2018;123:38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clark PM, Dweck JF, Mason DE, et al. . Direct in-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. J Am Chem Soc 2008;130:11576–11577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teo CF, Ingale S, Wolfert MA, et al. . Glycopeptide-specific monoclonal antibodies suggest new roles for O-GlcNAc. Nat Chem Biol 2010;6:338–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nandi A, Sprung R, Barma DK, et al. . Global identification of O-GlcNAc-modified proteins. Anal Chem 2006;78:452–458 [DOI] [PubMed] [Google Scholar]