Abstract
Inter‐organelle communication between closely apposed membranes is proposed at membrane contact sites (MCS). However, the regulation of MCS structure and their functional relevance in vivo remain debated. The extended synaptotagmins (Esyt) are evolutionarily conserved proteins proposed to function at MCS. However, loss of all three Esyts in yeast or mammals shows minimal phenotypes questioning the functional importance of Esyt. We report that in Drosophila photoreceptors, MCS number is regulated by PLCβ activity. Photoreceptors of a null allele of Drosophila extended synaptotagmin (dEsyt) show loss of ER‐PM MCS. Loss of dEsyt results in mislocalization of RDGB, an MCS localized lipid transfer protein, required for photoreceptor structure and function, ultimately leading to retinal degeneration. dEsyt depletion enhanced the retinal degeneration, reduced light responses and slower rates of plasma membrane PIP 2 resynthesis seen in rdgB mutants. Thus, dEsyt function and PLCβ signaling regulate ER‐PM MCS structure and lipid transfer in Drosophila photoreceptors.
Keywords: Drosophila, ER‐PM contact sites, extended synaptotagmins, lipid transfer, PIP2 signaling
Subject Categories: Membrane & Intracellular Transport
This study presents evidence for the requirement of E‐synaptotagmin (dESyt) in supporting lipid transfer activity and ER‐plasma membrane contact site structure in Drosophila photoreceptors, thereby revealing a functional role for membrane contact sites.
Introduction
One of the key attributes of eukaryotic cells is the existence of membrane bound organelles each with its own unique protein and lipid composition. The role of vesicular transport between organelles in establishing and maintaining this composition is well accepted. However, it has also been observed that in intact cells, organelle membranes are often placed in close proximity to each other without undergoing fusion as described for vesicular transport. This concept of closely apposed organelle membranes is referred to as Membrane Contact Sites (MCS) that are now defined as sites where organelle membranes are found separated by a gap of 10–30 nm. MCS were noted in the electron microscopy studies of George Palade (Palade & Porter, 1957), and subsequent studies have described MCS in several cell types (Carrasco & Meyer, 2011; Hayashi et al, 2008; Hepler et al, 1984; Suzuki & Hirosawa, 1994). The endoplasmic reticulum (ER), the largest organelle in the cell, forms an elaborate tubular network that occupies much of the interior of the cell and therefore establishes MCS with most other organelles (Gatta & Levine, 2017) including the plasma membrane (PM). A number of molecular processes are proposed to occur at MCS; these include Ca2+ homeostasis, lipid transfer between organelle membranes, and signaling interactions between proteins located at membranes on either side of the MCS [reviewed in refs. Saheki and De Camilli (2017), Balla et al (2019)]. However, the mechanisms by which MCS are established and maintained as well as their importance for cell physiology in vivo remain unclear.
Every organelle has a unique lipid composition that is a critical determinant of its function. Since lipids are hydrophobic and cannot diffuse in aqueous cytosol, lipid composition depends on the transport of lipid to and away from organelle membranes. Such intracellular lipid trafficking is mediated both by vesicular transport and by the activity of lipid transfer proteins (LTP). A major function that has been proposed for MCS is the activity of LTP at these sites, and several studies have demonstrated that LTPs can be localized to membrane contact sites [reviewed in ref. Cockcroft and Raghu (2018)]. Thus, the integrity of MCS is likely to be central to lipid homeostasis in eukaryotic cells. However, the depletion of multiple proteins localized to MCS and is thought to contribute to their structure and functions have showed limited effect on cell physiology, questioning the relevance, and importance of MCS function for lipid homeostasis in vivo.
Membrane contact sites between the PM and the ER are found in many cell types. Studies in yeast have shown that a major fraction of the ER shows a cortical localization (Pichler et al, 2001) and is confined in close proximity to the PM. The ER and PM have distinct lipid compositions; the ER is a major site of lipid biosynthesis, and multiple lipids including phosphatidylinositol are enriched in this compartment. By contrast, the PM is enriched in distinct lipid classes including phosphatidylinositol 4,5‐bisphosphate (PIP2) and phosphatidylinositol 4‐phosphate (PI4P) that are not found at the ER membrane. Remarkably, the PI4P phosphatase Sac1 is localized at the ER while its substrate PI4P is enriched at the PM. Elegant studies in yeast have proposed that Sac1 is localized to ER‐PM MCS and functions to dephosphorylate PI4P in trans (Stefan et al, 2011). Proteomic studies in yeast have identified a set of 6 proteins, referred to as tethers that interact with Sac1 and are localized to ER‐PM contact sites. These include Scs2 and Scs22 (VAP orthologs; Loewen et al, 2007), Ist2 (member of TMEM16 ion channel family; Manford et al, 2012), and the tricalbin proteins (Tcb1, Tcb2, Tcb3—Esyt orthologs; Manford et al, 2012). Depletion of all these proteins using a so‐called Δtether strain leads to the collapse of the cortical ER (cER) away from the PM and elevated PI4P levels at the PM (Manford et al, 2012). However, the Δtether strain shows remarkably mild phenotypes, with only a limited change in PM permeability described on heat stress (Omnus et al, 2016) raising the question of whether the presence of contact sites are required to support cell physiology.
Extended synaptotagmins (Esyts) are a group of ER anchored proteins belonging to the tubular lipid binding (TULIP) superfamily that comprise an N‐terminal transmembrane domain, a synaptotagmin like mitochondrial lipid binding protein domain (an SMP domain) and several acidic phospholipid binding C2 domains that bind to PM lipids (Chang et al, 2013). Mammalian cells have three independent genes that encode for Esyt: Esyt1, Esyt2 and Esyt3. Studies in mammalian cells have described localization of these proteins to ER‐PM contact sites and proposed a role for Esyt in mediating lipid transfer function (Giordano et al, 2013). It has been proposed that Ca2+ and PIP2 mediate the localization and tethering of Esyt to ER‐PM contact sites (Bian et al, 2018; Yu et al, 2016) and that this is linked to the lipid transfer activity of the SMP domain (Idevall‐hagren et al, 2015; Yu et al, 2016). Thus, two independent roles have been proposed for Esyt: one as a member of a group of proteins that function as a tether at ER‐PM contact sites and another as a lipid transfer protein via its SMP domain. Yet, the yeast Δtether strain that lacks all Esyt genes shows remarkably mild phenotypes (Omnus et al, 2016). A triple knockout of mouse Esyt1, Esyt2 and Esyt3 is viable and shows no major changes in brain morphology, synaptic protein composition, and stress response (Sclip et al, 2016). Even so, another in vitro study on Esyt2 −/− /Esyt3 −/− mouse embryonic fibroblast cells exhibit declined migratory potential and are vulnerable to stressed culture conditions (Herdman et al, 2014) questioning the role of Esyts in supporting function in vivo. Interestingly, in Drosophila, Esyt is reported to be localized to the presynaptic compartment in the neuromuscular junction and implicated in regulating neurotransmitter release and synaptic growth (Kikuma et al, 2017). Therefore, the in vivo function of this highly conserved protein family and the role of Esyt in supporting contact site functions remain unresolved.
In Drosophila photoreceptors, sensory transduction is mediated by G protein‐coupled phospholipase C mediated PIP2 turnover (Raghu et al, 2012). The biochemical reactions that constitute the PIP2 cycle in these cells are organized around an ER‐PM contact site between the sub‐microvillar cisternae (SMC), a specialized compartment of the ER and the microvillar PM (reviewed in ref. Yadav et al, 2016). Localized to this contact site is a multi‐domain protein retinal degeneration B (RDGB) that includes an N‐terminal phosphatidylinositol transfer domain (PITP) that can transfer PI and PA in vitro; binding of PI to the PITP domain is essential for the in vivo function of RDGB (Yadav et al, 2015). Loss of this function in flies lacking RDGB protein results in key in vivo phenotypes: reduced electrical response to light (reduction in ERG amplitude), delay in PIP2 resynthesis, and light‐dependent degeneration of photoreceptors (retinal degeneration) (reviewed in ref. Cockcroft et al, 2016). Thus, Drosophila photoreceptors offer an ideal in vivo model to test the mechanisms that control ER‐PM contact site structure and lipid transfer. In this study, we have generated a loss‐of‐function mutant for the only Esyt in Drosophila and studied its phenotypes in photoreceptors. We find that PLCβ‐dependent Esyt function is required to support ER‐PM contact site structure as well as the lipid transfer function of RDGB in vivo.
Results
A single gene encodes for Esyt in Drosophila
Bioinformatic analysis revealed that the Drosophila genome encodes a single ortholog of human Esyt and the tricalbin proteins of yeast (Fig 1A). The Drosophila extended synaptotagmin gene (dEsyt) is CG6643 located on the third chromosome (cytolocation 96A7‐96A7). dEsyt encodes for four different transcripts: RA, RB, RC, and RD that are identical except for a 75 bp sequence between the 2nd and 3rd C2 domain coding regions where each isoform has different sequences. Sequence comparison for all four transcripts showed that the RA CDS is the shortest, lacking 100 nucleotides at the 5′ end and the RD transcript specifically lacks 9 nucleotides which encode for 3 amino acid residues that are identical in the other three transcripts. When aligned with human Esyt1 (GI: 296317244), CG6643 shows 93% query coverage and 35% sequence identity with all four isoforms. The protein domain structure of dEsyt is conserved with that of the human and yeast orthologs (Fig 1B). For, e.g., in hEsyt proteins, a single N‐terminal SMP domain and multiple C2 domains are described. dEsyt sequence shows the presence of an SMP domain and three C2 domains. This feature along with the conserved interdomain distance makes dEsyt most similar to hEsyt2 and hEsyt3. Transcriptome analysis indicates that dEsyt is ubiquitously expressed across tissues and during all developmental stages (http://flybase.org/reports/FBgn0266758). However, qPCR analysis showed that dEsyt transcripts are enriched in the head compared to the body (Fig 1C). To determine whether dEsyt expression is enriched in the eye, we made use of so D (Roederer et al, 2005), a dominant allele of the sine oculis gene that results in a block in eye development and eliminates eye tissue. If a given gene is enriched in the eye, transcript expression is expected in wild‐type heads but will be reduced in so D heads since eye tissue, where the gene is mainly expressed is missing. For dEsyt, a comparison between transcript expression in wild‐type and so D heads shows similar levels of expression, suggesting that RNA for this gene is not enriched in the eye (Fig 1D).
Figure 1. Single gene encoding Esyt expressed in Drosophila melanogaster .
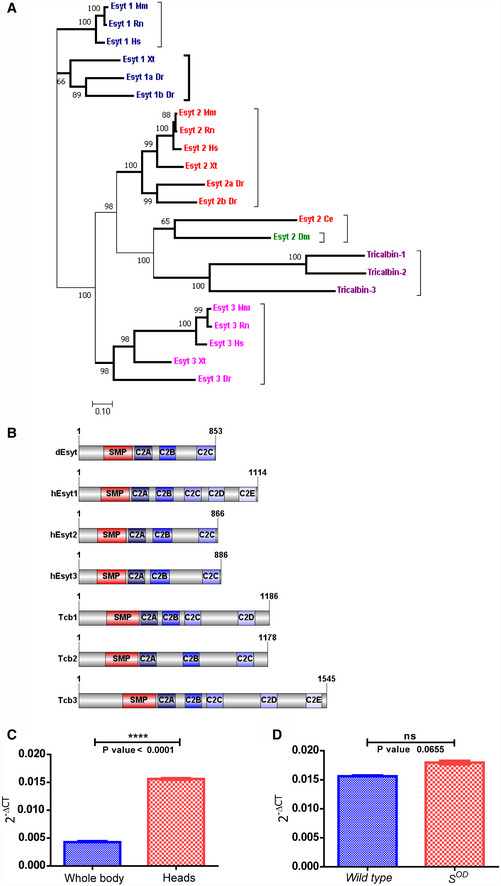
- Schematic showing the phylogeny of extended synaptotagmin in different organisms: Drosophila melanogaster (Dm), Saccharomyces cerevesiae (Tricalbin), Homo sapiens (Hs), Mus Musculus (Mm), Rattus norvegicus (Rn), Caenorhabditis elegans (Ce), Danio rerio (Dr), and Xenopus tropicalis (Xt). Bold lines—subnodes. Numbers on the nodes—bootstrap value. Colors—Esyt1, Esyt2, and Esyt3 clusters.
- Schematic of dEsyt‐PA protein domain structure aligned with mammalian Esyts and yeast tricalbins (Image generated using IBS, Illustrator for Biological sciences, version 1.0 software http://ibs.biocuckoo.org/) (Liu et al, 2015). Amino acid numbers in each protein are indicated. SMP‐SMP domain. C2A, B, C, D, and E are the various C2 domains.
- Quantitative real‐time PCR analysis showing head enrichment of dEsyt; the X‐axis indicates the wild‐type tissues from which RNA was isolated, and Y‐axis indicates the dEsyt transcript‐level expression normalized to the loading control (RP49‐ribosomal protein 49), n = 3 replicates (biological).
- Quantitative real‐time PCR analysis showing dEsyt transcript‐level expression in head RNA samples from wild‐type and s OD. Y‐axis indicates the dEsyt transcript‐level expression normalized to the loading control (RP49‐ribosomal protein 49), n = 3 replicates (biological).
dEsyt is dispensable for normal phototransduction
To examine a possible function of dEsyt, a null allele was generated using CRISPR/Cas9‐based genome editing (Kondo & Ueda, 2013; Appendix Method S1). The resulting genome edit deleted the nucleotide sequence corresponding to 488 amino acids of the dEsyt coding region; this was verified using molecular techniques (Refer to Appendix Fig S1). During larval development, dEsyt KO homozygous larvae show a delay in development and adult flies that eclose are smaller than controls (Appendix Fig S2A and B). These phenotypes can be rescued by pan larval reconstitution with dEsyt cDNA (Appendix Fig S2C and D). Although most dEsyt KO homozygotes do not eclose as adults, when grown on nutrient‐enriched medium, a small proportion is viable as homozygous adults.
To test the potential role of dEsyt in supporting lipid transfer at MCS, we studied the phenotypes of these homozygous dEsyt KO in adult photoreceptors. We measured electrical responses to light in adult flies. Electroretinograms (ERG) were recorded from wild‐type and dEsyt KO adults of matched age and eye color. We found that the absolute ERG response of dEsyt KO was smaller than that of controls (Fig 2A). However, the ERG response is an extracellular recording that scales with the size of the eye. Since dEsyt KO are smaller in size than controls, we normalized the raw ERG response of each animal to its body size (Fig 2B); this normalized ERG amplitude was not significantly different between control and dEsyt KO.
Figure 2. dEsyt is dispensable for phototransduction.
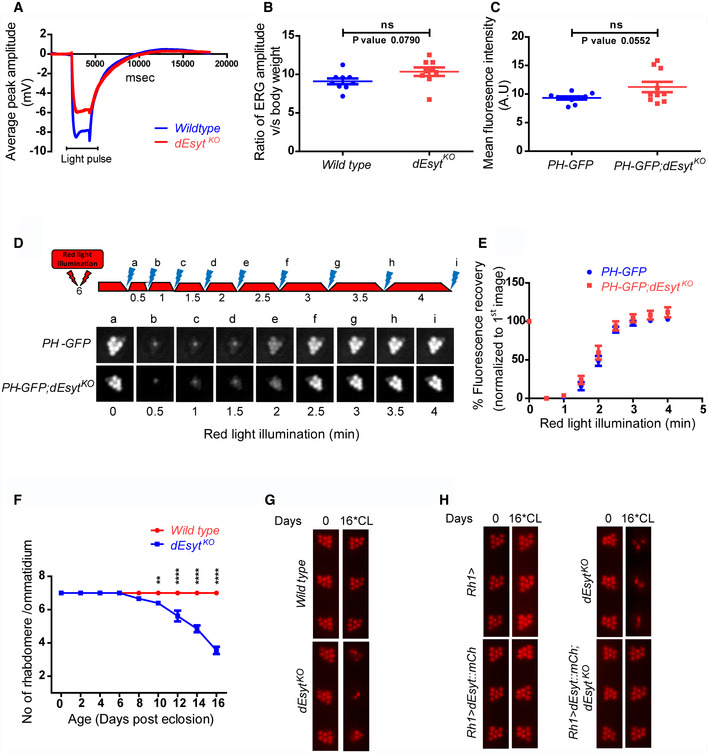
- Representative ERG trace from dark reared 0 to 1‐day‐old flies of the indicated genotype stimulated by 2‐s flash of green light. The duration of the stimulating light is shown. Y‐axis shows the amplitude of the ERG response. X‐axis shows the duration of the recording.
- Graph showing ERG amplitude normalized to body weight. Y‐axis shows the ratio of ERG amplitude of individual flies to their body weight. X‐axis indicates genotypes. Each data point represents an individual fly tested. Error bar represents s.e.m.
- Quantification of the mean fluorescence intensity of the deep pseudopupil formed by the PIP2 probe PH‐GFP in the one‐day‐old flies of the indicated genotypes (n = 10 biological replicates).
- Deep pseudopupil imaging of PIP2 levels in the microvillar membrane of photoreceptors. The fluorescence of the PH‐GFP probe is depicted. The protocol used is shown with red light illumination periods shown as red bars and flashes of blue light (a–f) used for image capture depicted. Representative deep pseudopupil images acquired at specified time points are depicted. Genotypes as indicated.
- Graph quantifying the recovery kinetics of the fluorescent pseudopupil with time, X‐axis represents the genotypes, Y‐axis represents intensity normalized to the intensity of the first image (n = 10 biological replicates). Error bars represent s.e.m.
- Quantification of the time course of retinal degeneration. 10 ommatidia from 5 separate flies of each genotype were scored and plotted. Y‐axis is the number of intact rhabdomeres/ommatidium. The maximum value possible is 7. X‐axis is the age of the flies post‐eclosion.
- Representative optical neutralization (ON) images showing rhabdomere integrity of the indicated genotypes. Rearing conditions and the age of the flies are indicated on top (*CL‐constant light).
- Representative ON images showing the rescue of rhabdomere structural integrity in dEsyt KO mutants on expressing the dEsyt::mCherry transgene. Genotypes of the flies are indicated on the left. Rearing conditions and the age of the flies are indicated on top.
Since contact sites are proposed to support lipid transfer reactions during cell signaling, we tested the requirement for dEsyt in supporting PIP2 turnover during phototransduction. We compared the level of PIP2 at the microvillar PM of photoreceptors in wild‐type and dEsyt KO flies. To monitor the changes in the PIP2 levels, the PH domain of PLCδ fused to GFP (hereafter referred to as PH‐GFP) was utilized (Yadav et al, 2015). This analysis revealed no difference in the basal levels of PIP2 between controls and dEsyt KO (Fig 2C). Further, following bright light stimulation to activate PLCβ and deplete PIP2 at the microvillar PM, we found no difference in the rate at which PIP2 levels were restored at the microvillar PM (Fig 2D and E) in dEsyt KO. Similarly, the P4M‐GFP probe was utilized to monitor the levels of PI4P which acts as the substrate for dPIP5K to produce PIP2 (Balakrishnan et al, 2018); basal PI4P levels and PI4P turnover kinetics were similar between wild‐type and dEsyt KO (Appendix Fig S3A, B and E).
Light‐dependent retinal degeneration is a common outcome when genes essential for phototransduction are disrupted (Raghu et al, 2012). To test the role if any, of dEsyt in regulating phototransduction, we monitored the integrity of rhabdomere (apical PM) structure following illumination. Although normal at eclosion, when flies were grown under constant bright illumination, normal photoreceptor ultrastructure was retained until 10 days post‐eclosion. However, photoreceptors of dEsyt KO underwent progressive retinal degeneration when observed till day 16 with substantial loss of rhabdomere integrity (Fig 2F and G). This light‐dependent degeneration exhibited by the mutant dEsyt homozygotes was rescued by reconstitution with wild‐type dEsyt transgene (Fig 2H).
Loss of dEsyt enhances rdgB 9 phenotypes
It is reported that mammalian Esyt1 enhances the activity of Nir2 by recruiting it to the ER‐PM junctions subsequent to an elevation in cytosolic calcium levels (Chang et al, 2013). We tested the function of dEsyt in supporting the activity of RDGB, the Drosophila ortholog of Nir2. For this, we generated a double‐mutant strain of dEsyt KO with rdgB 9, a hypomorphic allele of rdgB (rdgB 9 ;dEsyt KO). Using this strain, we compared the impact of dEsyt deletion on the phenotypes of the rdgB 9 single mutant.
In Drosophila, rdgB mutants show a number of key phenotypes including light‐dependent retinal degeneration, reduced electrical response to light, and altered PIP2 homeostasis at the microvillar PM (Yadav, et al. 2015). We compared the time course of retinal degeneration in rdgB 9 v rdgB 9 ;dEsyt KO and noted that dEsyt KO accelerates the time course of retinal degeneration in rdgB 9 (Fig 3Ai, ii and 3B). Over this time period, dEsyt KO itself does not show any retinal degeneration; the retinal degeneration of the dEsyt KO occurs much later after eclosion.
Figure 3. Loss of dEsyt enhances rdgB mutant phenotypes.
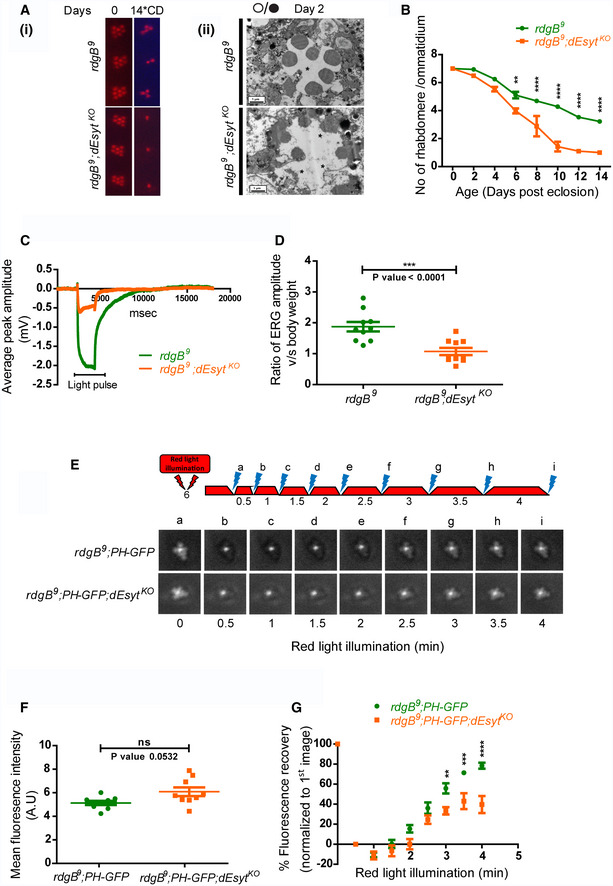
- (i) Representative optical neutralization (ON) images showing rhabdomere structure of the indicated genotypes. Rearing conditions and the age of the flies are indicated on top (*CD‐constant dark). (ii) TEM images showing a single ommatidium from the photoreceptors of rdgB 9 and rdgB 9 ;dEsyt KO mutants reared in 12‐h L/D cycle for 2 days. Scale bar: 1 μm (asterisk indicates degenerated rhabdomere).
- Quantification of the time course taken for retinal degeneration. 10 ommatidia were scored from 5 flies of each genotype and plotted.
- Representative ERG traces showing the duration of light pulse, X‐axis indicates time in msec, and Y‐axis indicates the average ERG amplitude in mV.
- Graph showing ERG amplitude of rdgB 9 and rdgB 9 ;dEsyt KO normalized to body weight. Y‐axis shows the ratio of ERG amplitude of individual flies to their body weight. X‐axis indicates genotypes. Each data point represents an individual fly tested. Error bar represents s.e.m.
- Deep pseudopupil imaging of PIP2 levels in the microvillar membrane of photoreceptors. The fluorescence of the PH‐GFP probe is depicted. The protocol used is shown with red light illumination periods shown as red bars and flashes of blue light (a–f) used for image capture depicted. Representative deep pseudopupil images acquired at specified time points are depicted. Genotypes as indicated.
- Quantification of the mean fluorescence intensity of the PIP2 probe PH‐GFP from the deep pseudopupil formed by one-day‐old flies of the indicated genotypes (n = 10 biological replicates).
- Graph quantifying the recovery kinetics of the fluorescent pseudopupil with time, X‐axis represents the genotypes, and Y‐axis represents intensity normalized to the intensity of the first image (n = 10 biological replicates).
We also studied the ERG amplitude in response to a 1 s flash of light. As previously reported (Yadav et al, 2015), rdgB 9 mutants show a reduced ERG amplitude and this was further reduced in rdgB 9 ;dEsyt KO (Fig 3C and D). Finally, we measured PIP2 levels at the microvillar PM under resting conditions. The PIP2 levels in rdgB 9 are lower than in wild type (Yadav et al, 2015), but there was no difference in resting PIP2 levels between rdgB 9 and rdgB 9 ;dEsyt KO (Fig 3F). However, when we measured the rate of recovery of microvillar PIP2 following its depletion by strong PLCβ activation, the reduced rate of PIP2 recovery in rdgB 9 was further delayed in rdgB 9 ;dEsyt KO (Fig 3E and G) and a similar observation was also made for PI4P levels (Appendix Fig S3C–E). Collectively, loss of dEsyt worsened the three key functional and biochemical outcomes in a hypomorphic allele of rdgB.
dEsyt is localized to ER‐PM contact sites in photoreceptors
To understand the mechanism by which loss of dEsyt enhances rdgB 9 phenotypes, it was crucial to decipher the localization of dEsyt. Since Esyt proteins have been described as components of MCS in both yeast and mammalian cells, we tested whether the same is also true in Drosophila photoreceptors. For this, we labeled wild‐type retinae with anti‐dEsyt serum (Kikuma et al, 2017). Single confocal images of photoreceptor cross‐sections (R1–R6) revealed a crescent‐shaped distribution of dEsyt at the MCS between the microvillar PM and the SMC (Fig 4A); this staining was absent in dEsyt KO photoreceptors demonstrating the specificity of the labeling detected. To further confirm this observation, we expressed dEsyt tagged to the fluorescent protein mCherry in photoreceptors and examined its localization using an antibody against mCherry; here too we found that dEsyt::mCherry was localized to the MCS at the base of the microvilli in photoreceptors (Fig 4B); staining was not seen in wild‐type photoreceptors not expressing dEsyt::mCherry indicating the specificity of the staining. Collectively, these findings demonstrate that dEsyt is localized at the ER‐PM MCS in photoreceptors.
Figure 4. dEsyt localizes to MCS and determines RDGB localization.
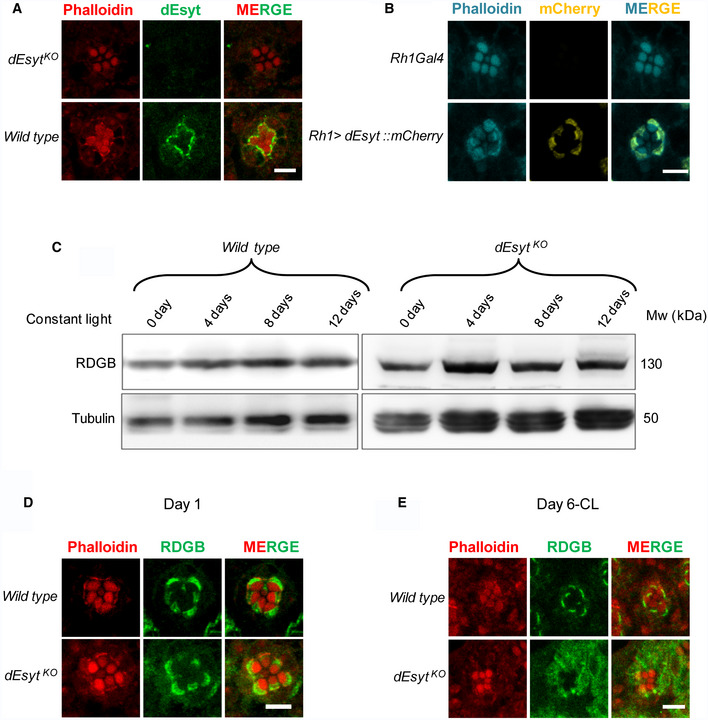
-
AConfocal images showing the localization of endogenous dEsyt protein in photoreceptors of one‐day-old dark‐reared flies probed with antibody against dEsyt. Scale bar: 5 μm. dEsyt KO shows no staining when probed with dEsyt antibody.
-
BConfocal images showing the localization of exogenously expressed dEsyt::mCherry protein expressed using the eye‐specific Rh1‐Gal4 in one-day‐old dark‐reared flies. Rh1‐Gal4 is shown as a control. A single ommatidium is shown. Scale bar: 5 μm. Phalloidin marks F‐actin staining and highlights rhabdomeres R1–R7.
-
CWestern blot from the head extracts of wild type and dEsyt KO probed with the antibody against RDGB. Rearing conditions, age of the flies, and genotype is indicated on top of the blot. Tubulin was used as the loading control.
-
D, EConfocal images showing the localization of RDGB in wild‐type and dEsyt KO photoreceptors of flies which are (D) 1‐day-old dark‐reared and (E) 6‐day-old exposed to constant illumination. For (D) and (E) RDGB visualized using an antibody against the endogenous protein. Rhabdomeres are outlined using phalloidin which marks F‐actin. Scale bar: 5 μm.
dEsyt function is required to localize RDGB to ER‐PM contact sites
One mechanism by which loss of dEsyt might enhance rdgB 9 phenotypes is by reducing the levels of the endogenous RDGB protein. We checked the levels of RDGB protein in dEsyt KO mutants exposed to bright light illumination for increasing time intervals and found that the RDGB protein levels remain unaltered even after retinal degeneration is initiated in dEsyt KO (Fig 4C). A second mechanism could be the mislocalization of the RDGB protein away from the ER‐PM MCS. In wild‐type Drosophila photoreceptors, RDGB is strictly localized to the MCS and we found that this localization was not disrupted in newly eclosed dEsyt KO flies (Fig 4D). However, when exposed to constant illumination for 6 days post‐eclosion, dEsyt KO photoreceptors showed mislocalization of RDGB away from the MCS; the protein was now diffused across the photoreceptor cell body (Fig 4E). By contrast, under the same conditions, RDGB localization to the MCS was retained in wild‐type photoreceptors (Fig 4E). Thus, dEsyt function is required for the normal localization of RDGB at the MCS in photoreceptors.
dEsyt is necessary for PLCβ‐dependent modulation of ER‐PM MCS
The most direct way of assessing MCS structure is by transmission electron microscopy (TEM). We used fixation methods that allow enhanced membrane preservation and better visualization of membranes and MCS (Matsumoto‐Suzuki et al, 1989; Fig 5A i, ii); in addition to the membranes seen, the perfectly spherical black bead‐like structures in the micrographs are pigment granules. Such images allowed us to quantify the fraction of PM at the base of the microvilli that was in contact with the SMC (hereafter referred to as MCS density) in each ultrathin section of a photoreceptor. On day 1 after eclosion, in the light‐sensitive R1‐R6 photoreceptors of dark‐reared wild‐type flies, the MCS density was ca. 50% (Fig 5A i, ii, and 5B i). As flies aged, the MCS density decreased to ca. 30% by day 14 (Fig 5A iii, iv, and 5B i). Under the same conditions, the MCS density in the UV‐sensitive R7 photoreceptor remained constant as a function of age (Fig 5B ii); this serves as an internal control for phenotypes directly linked to illumination since the light used for our illumination experiments contains visible wavelengths but not those in the ultraviolet range. We also measured the MCS density in the norpA P24 mutant allele that lacks PLCβ protein expression (Pearn et al, 1996). MCS density in norpA P24 was ca. 30% at eclosion (Fig 5C i, ii) but in contrast to wild‐type flies, norpA P24 flies did not show any reduction as a function of age; i.e., 14‐day‐old norpA P24 photoreceptors showed the same MCS density as wild‐type cells at eclosion (Fig 5D i). Thus, MCS density in photoreceptors is dependent on age and PLCβ activity.
Figure 5. dEsyt stabilizes MCS in photoreceptors.
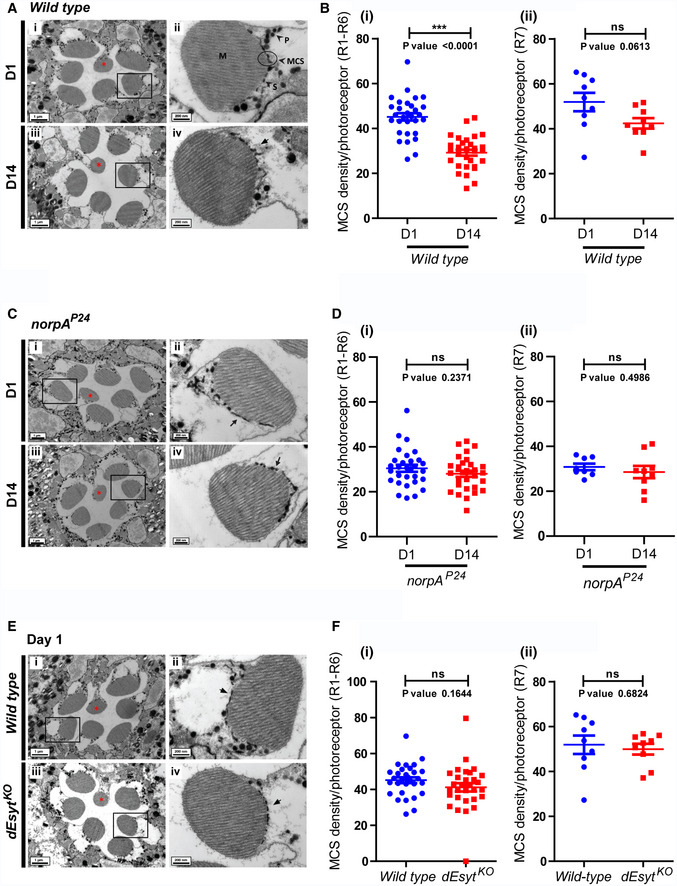
- TEM images of a single ommatidium from wild‐type photoreceptors of flies reared in dark (i) day 1 (D1) and (iii) day 14 (D14), scale bar: 1 μm (red asterisk indicates R7 photoreceptor). Magnified image showing a single photoreceptor from the same ommatidium (ii) day 1 (iv) day 14. Scale bar: 200 nm. Origin boxes for magnification shown. Arrow in (Aiv) indicates the SMC forming an MCS with the microvillar PM. M—microvillar plasma membrane, S—sub‐microvillar cisternae, MCS—membrane contact site, P—pigment granules.
- Quantification of the number of MCS per wild‐type photoreceptor of 1‐day and 14‐day‐old flies reared in dark (from Fig 6A), Y‐axis indicates the number of MCS/photoreceptor. n = 30 photoreceptors from 3 separate flies (R1–R6) for (i). n = 9 photoreceptors from 3 separate flies (R7) in (ii).
- TEM images of a single ommatidium from norpA P24 photoreceptors of flies reared in dark (i) day 1 and (iii) day 14, scale bar: 1 μm (red asterisk indicates R7 photoreceptor). Magnified image showing a single photoreceptor from the same ommatidium (ii) day 1 (iv) day 14. Scale bar: 200 nm. Arrows indicate the SMC forming an MCS with the microvillar PM.
- Quantification of the number of MCSs per photoreceptor (from Fig 6C), X‐axis indicates the genotype and age of the flies and Y‐axis indicates the number of MCS/photoreceptor. (i) n = 30 photoreceptors from 3 separate flies (R1‐R6). (ii) n = 9 photoreceptors from 3 separate flies (R7).
- TEM images of a single ommatidium from (i) wild‐type and (iii) dEsyt KO photoreceptors of 0 to 1‐day‐old flies reared in dark, scale bar: 1 μm (red asterisk indicates R7 photoreceptor). (ii, iv) Magnified image showing a single photoreceptor from the ommatidium image shown on the left. Scale bar: 200 nm. Arrows indicate the SMC forming an MCS with the microvillar PM.
- Quantification of the number of MCS per photoreceptor of wild‐type 1‐day v/s dEsyt KO 1-day‐old flies reared in dark, X‐axis indicates the genotype and age of the flies and Y‐axis indicates the number of MCS/photoreceptor. (i) n = 30 photoreceptors from 3 separate flies (R1‐R6). (ii) n = 9 photoreceptors from 3 separate flies (R7).
To assess the contribution of dEsyt to ER‐PM MCS structure, we performed TEM on dEsyt KO photoreceptors to visualize and quantify MCS density (Fig 5E iii, iv). At eclosion, MCS density was not different between wild‐type and dEsyt KO for R1‐R6 photoreceptors (Fig 5E i–iv and 5F i); a similar result was seen in UV‐sensitive R7 photoreceptors (Fig 5F ii). Thus, loss of dEsyt does not impact the establishment of ER‐PM contact sites during development in Drosophila photoreceptors. We then examined MCS density as a function of age in dEsyt KO (Fig 6A i–iv). While MCS density was reduced from ca. 50% (day 1) to 30% at day 14 in wild‐type flies (Fig 5B i), in dEsyt KO flies, the number of MCS was reduced to almost none by day 14 (Fig 6B i). By contrast, in UV‐sensitive R7 photoreceptors, MCS density was only modestly reduced in dEsyt KO (Fig 6B ii).
Figure 6. dEsyt is necessary for PLCβ‐dependent modulation of ER‐PM MCS.
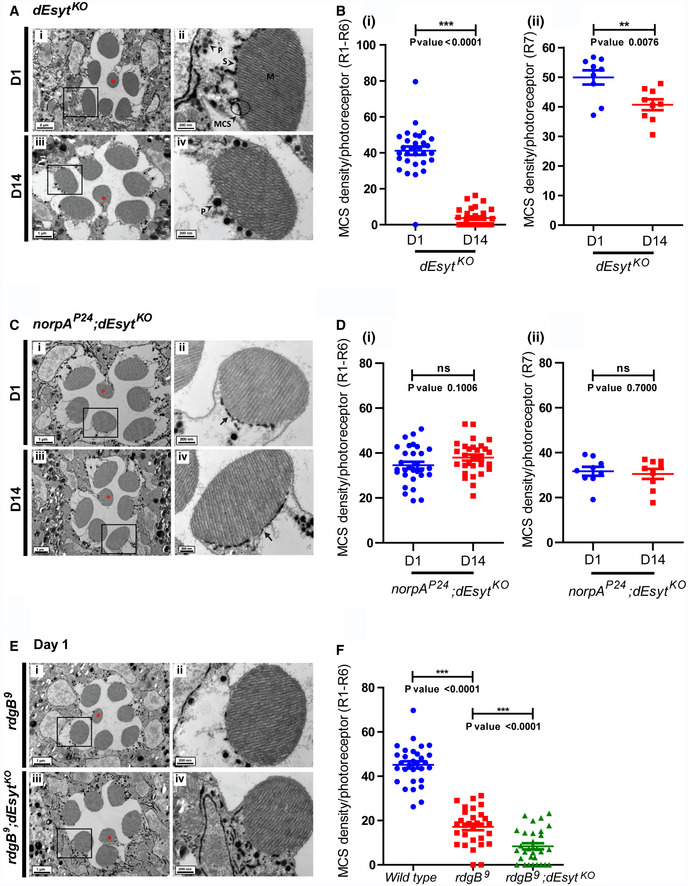
- TEM images of a single ommatidium from photoreceptors of (i) dEsyt KO‐day 1 (iii) dEsyt KO‐day 14 flies reared in dark, scale bar: 1 μm except for (Ai): 2 μm (red asterisk indicates R7 photoreceptor). (A‐ii, iv) Magnified image showing a single photoreceptor from the ommatidium image shown on the left. Scale bar: 200 nm. Boxes represent the origin boxes for magnification. M—microvillar plasma membrane, S—sub‐microvillar cisternae, MCS—membrane contact site, P—pigment granules.
- Quantification of the number of MCS per photoreceptor, X‐axis indicates the genotype and age of the flies and Y‐axis indicates the number of MCS/photoreceptor. n = 9 photoreceptors from 3 separate flies for R7.
- TEM images of a single ommatidium from photoreceptors of (i) norpA P24 ;dEsyt KO‐day 1 (iii) norpA P24 ;dEsyt KO‐day 14 flies reared in dark, scale bar: 1 μm (red asterisk indicates R7 photoreceptor). (C‐ii, iv) Magnified image showing a single photoreceptor from the ommatidium image shown on the left. Scale bar: 200 nm. Arrows indicate the SMC forming an MCS with the microvillar PM.
- Quantification of the number of MCS per photoreceptor, X‐axis indicates the genotype and age of the flies and Y‐axis indicates the number of MCS/photoreceptor. (i) n = 30 photoreceptors from 3 separate flies for R1–R6. (ii) n = 9 photoreceptors from 3 separate flies for R7.
- TEM images of a single ommatidium from photoreceptors of (i) rdgB 9‐day 1 and (iii) rdgB 9 ;dEsyt KO‐day 1 flies reared in dark, scale bar: 1 μm (red asterisk indicates R7 photoreceptor). (E‐ii, iv) Magnified image showing a single photoreceptor from the ommatidium image shown on the left. Scale bar: 200 nm.
- Quantification of the number of MCS per photoreceptor for R1‐R6 in 1-day‐old dark‐reared wild‐type (from Fig 5B i), rdgB 9 and rdgB 9 ;dEsyt KO flies. n = 30 photoreceptors from 3 separate flies.
We tested the impact of PLCβ activity on the enhanced loss of MCS in dEsyt KO. For this, we compared MCS density in dEsyt KO with that in the norpA P24 ;dEsyt KO double mutant (Fig 6C i–iv). Strikingly, the absence of basal PLCβ activity in norpA P24 ;dEsyt KO photoreceptors completely rescued MCS number in 14‐day‐old flies; this was in sharp contrast to dEsyt KO flies which at this stage typically shows very few or no MCS (Fig 6C iii, iv, and 6D i). Thus, the loss of MCS in dEsyt KO with age depends on ongoing PLCβ activity.
We also examined MCS structure in rdgB 9 photoreceptors (Fig 6E i, ii). In dark‐reared flies, just after eclosion, MCS density in R1‐R6 photoreceptors was already reduced significantly (Fig 6F i). rdgB 9 ;dEsyt KO photoreceptors also showed very few MCS in the photoreceptors (Fig 6E iii,iv); these were quantified only from photoreceptors in which there was minimal degeneration of rhabdomeres (Fig 6E iii). Further, rdgB 9 ;dEsyt KO double mutants showed substantially greater rhabdomeral degeneration than rdgB 9 (Fig 3A ii) when reared for 2 days under conditions of 12‐h L/D cycle reflecting the ultrastructural level observations seen in the optical neutralization assays (Fig 3B).
Discussion
Studies in yeast and mammalian cells have identified proteins that when overexpressed, localize to ER‐PM MCS. However, in most cases, surprisingly, loss of function of these proteins appears to not impact cellular function or in vivo physiology. Though Esyt can localize to ER‐PM MCS in cultured mammalian cells (Chang et al, 2013; Giordano et al, 2013), depletion of Esyt in cultured mammalian cells apparently does not impact cell physiology and knockout of Esyt in mice does not have discernible impact on animal physiology (Sclip et al, 2016; Tremblay & Moss, 2016) raising questions on their role in vivo. However, in this study, we found that a null mutant of the only Esyt in Drosophila showed developmental delay (data not shown), reduced adult viability and homozygotes that eclosed were smaller than controls; the reduced body size could be rescued by reconstitution with a wild‐type transgene suggesting an important role for dEsyt in growth. While the cellular and molecular basis of these phenotypes remains to be established, these data clearly demonstrate a function for Esyt in supporting animal physiology in vivo.
Importantly, we analyzed photoreceptor structure and function in dEsyt KO since this cell type critically depends on lipid transfer activity at its ER‐PM MCS for phototransduction (Yadav et al, 2016, 2018). In newly eclosed dEsyt KO flies, photoreceptor ultrastructure was unaffected and ER‐PM MCS density was not different from that in wild‐type flies. Thus, dEsyt is not required during development to establish ER‐PM MCS. We also found that dEsyt KO showed normal electrical responses to light and basal levels of PM PI4P and PIP2 as well as their turnover during PLCβ signaling was unaffected; this finding implies that dEsyt function is dispensable for PIP2 turnover (that in turn depends on PI transfer at ER‐PM MCS) during PLCβ signaling. Our observation is consistent with that reported for mammalian Esyt that the SMP domain does not exhibit specific PI transfer activity in vitro (Saheki et al, 2016; Schauder et al, 2014), that loss of Esyt function in cultured mammalian cells does not impact PIP2 turnover during PLCβ signaling, and that depletion of Esyt does not impact store‐operated calcium influx, a key output of PLCβ activation (Giordano et al, 2013).
Despite the apparent lack of a phenotype in dEsyt KO photoreceptors, we found that loss of dEsyt enhances all three key phenotypes of rdgB 9, a hypomorphic allele with ca. 5% residual RDGB protein in photoreceptors. Importantly, the impact of dEsyt KO on the reduced light response and PM PIP2 levels in rdgB 9 occurred in newly eclosed flies, prior to the onset of retinal degeneration. Conceptually, this enhancement could be due to a change in the levels of the major phototransduction proteins but this was ruled out since we found no alteration in the levels of these proteins (Rhodopsin, PLC, Gq, and TRP) in rdgB 9 ;dEsyt KO (Appendix Fig S4). Interestingly, we found that dEsyt KO did not enhance the reduced ERG amplitude resulting from the depletion of dPI4KIIIα (Appendix Fig S5), the enzyme implicated in the conversion of PI to PI4P at the PM of photoreceptors (Balakrishnan et al, 2018). The reason for this is unclear; however, it indicates that the enhancement of rdgB 9 phenotypes by dEsyt KO is specific and not seen even in cells depleted of the immediate next component of the PIP2 cycle in photoreceptors. These findings strongly indicate a key role for dEsyt in specifically supporting the function of RDGB at the ER‐PM MCS. Our data suggest that in photoreceptors with a full capacity of RDGB function, cells can dispense with the requirement for dEsyt. However, in cells with limiting RDGB function (such as rdgB 9), the requirement for dEsyt function becomes apparent. Thus, our data clearly support a role for dEsyt in supporting the lipid transfer function of RDGB at ER‐PM contact sites in vivo. Consistent with this model, we found that the dEsyt protein is localized to the base of the microvillar PM at the ER‐PM contact site where the RDGB protein is also specifically localized (Vihtelic et al, 1993; Yadav et al, 2018).
Although we found a normal density of ER‐PM MCS in dEsyt KO photoreceptors at eclosion, we noted that this changed as a function of age and illumination. Wild‐type photoreceptors show a reduction in ER‐PM MCS density with age; however, the drop in MCS density under the same conditions was much greater in dEsyt KO with virtually no MCS observed in 14 days old, constant dark‐reared flies. In addition to this, the overexpression of dEsyt specifically in the eye resulted in the recruitment of ER structures towards the apical PM with some part of the recruited structures invading the rhabdomere. This observation further supports a role dEsyt in supporting the positioning of the ER at the base of the microvilli and hence stabilizing MCS between the PM and ER at this location (Appendix Fig S6). Together, our observations imply that dEsyt function is essential to maintain the stability of ER‐PM MCS in photoreceptors. We also observed that although the RDGB protein was correctly localized at the MCS at eclosion, in dEsyt KO photoreceptors, the protein became delocalized as a function of age in the absence of dEsyt function. By day 6, the RDGB protein was substantially delocalized from the base of the microvilli in dEsyt KO, although the total amount of RDGB protein in the cell was not reduced. During this study, we also noted that the MCS density was lower in rdgB 9 ;dEsyt KO compared to rdgB 9 alone. This observation offers an explanation for the ability of dEsyt KO to enhance the physiological phenotypes of rdgB 9 on day 1 post‐eclosion; i.e, the reduced MCS density in rdgB 9 with limited lipid transfer activity is further compromised by the additional loss of MCS resulting from dEsyt KO. Taken together, these findings suggest that in the absence of dEsyt function, it is likely that RDGB (and perhaps other MCS localized proteins) become delocalized leading to disruption of its lipid transfer function. These findings lead us to a model of dEsyt function in photoreceptors where its key role is to stabilize ER‐PM MCS and facilitate the correct localization of relevant proteins at this MCS.
Despite intense interest in the function of MCS, very little is known about the mechanisms by which MCS density is tuned in response to signals. In photoreceptors, a key signal is light and this is transduced through G protein‐coupled PLCβ activity (reviewed in ref. Raghu et al, 2012). In this study, we made several observations that link MCS density to PLCβ activity: (i) In newly eclosed flies, MCS density was reduced in norpA mutants that lack PLCβ activity, (ii) in wild‐type photoreceptors, ER‐PM MCS density decreased as a function of age and this decrease was dependent on ongoing PLCβ activity, and (iii) the loss of MCS in dEsyt KO photoreceptors could be suppressed by norpA mutants. Together, these findings suggest that PLCβ activity is a key element of the mechanism by which MCS density is regulated. Interestingly, in rdgB 9, in which PM PIP2 levels are lower than wild type, MCS density was reduced. Since both PLCβ and RDGB regulate PIP2 levels, our observations suggest that MCS density in photoreceptors is modulated by PIP2 turnover at the PM. As PIP2 turnover is a key step of phototransduction in Drosophila photoreceptors, it is possible that in this cell type, illumination modulates MCS density. In turn, this will alter the biochemical activity of proteins localized at the MCS and their ability to maintain microvillar PM homeostasis. Future studies will likely reveal the molecular mechanism by which MCS density is tuned to ambient illumination and the broader question of PM homeostasis by MCS in eukaryotic cells.
Materials and Methods
Fly stocks
Flies (Drosophila melanogaster) were raised on rich medium (composition : corn flour, black jaggery, agar, dry yeast, propionic acid, methyl para hydroxy benzoate, orthophosphoric acid) at 25°C maintaining 50% relative humidity in a constant temperature laboratory incubator. The environment within the incubator was completely devoid of illumination except for the brief pulses of light to which the flies were subjected when the incubator doors were opened. The experiments were carried out in three different conditions: constant illumination, constant dark and 12‐h light–dark cycle. For experiments involving constant illumination and 12‐h light–dark cycle, incubators were maintained at 25°C with a white light source (light intensity: 8,000 lux) for the required time period. For constant dark rearing, flies within the vials were kept in tightly closed dark boxes and maintained in the constant temperature laboratory incubator. Gal4‐UAS system was used for the selective activation of the transgene spatially and temporally for targeted gene expression.
Sequence and phylogeny analysis
The protein sequences for sequence alignment were obtained from NCBI: Drosophila melanogaster: dEsyt‐PA (NP_733010.1), Homo sapiens: hEsyt1‐isoform 1 (NP_001171725.1), hEsyt2‐isoform 1 (NP_001354702.1), hEsyt3‐isoform a (NP_001309760.1), Saccharomyces cerevisiae: yeast tcb1 (NP_014729.1), yeast tcb2 (NP_014312.1), yeast tcb3 (NP_013639.1), Mus musculus: Esyt1 Mm (NP_035973.1), Esyt2 Mm (NP_083007.2), Esyt3 Mm (NP_808443.2), Rattus norvegicus: Esyt1 Rn (NP_058945.2), Esyt2 Rn (NP_001258098.1), Esyt3 [Predicted (XP_343455.4)], Xenopus tropicalis: Esyt1 Xt (XP_002934579.1), Esyt2 Xt (XP_031759913.1), Esyt3 Xt (NP_001011364.1), Danio rerio: Esyt1a Dr (XP_695611.3), Esyt1b Dr (XP_699731.6), Esyt2a Dr (XP_005171454.1), Esyt2b Dr (XP_021333611.1), Esyt3 Dr (NP_001116705.1), and Caenorhabditis elegans: Esyt2 Ce (NP_741181.1). Multiple sequence alignment of dEsyt protein sequence with the human Esyts and yeast tricalbins were obtained using MUSCLE (MUltiple Sequence Comparison by Log‐Expectation; Edgar, 2004). The phylogenetic tree reflecting the evolution of these proteins was obtained using neighbor‐joining method MEGA6 (Molecular Evolutionary Genetics Analysis, version 6.0; Tamura et al, 2013).
Optical neutralization
The flies reared under experimental conditions were immobilized by cooling on ice, carefully decapitated, and fixed on the microscope slide using a drop of colorless nail varnish. The refractive index of the cornea was neutralized using a drop of immersion oil, viewed, and imaged under the 40× oil immersion objective of Olympus BX43 microscope. The digital image acquisition and documentation were done by using CellSens software.
Scoring retinal degeneration
To obtain a quantitative index of degeneration, a total of 50 ommatidia from 5 different flies of each genotype were assessed for each time point. The single, central UV‐sensitive photoreceptor that did not show any light‐dependent retinal degeneration was used as the reference and the rest of the photoreceptors were scored. A score of 1 was given to each rhabdomere that appeared to be wild type. Thus, the control photoreceptors will have a score of 7 and the mutants undergoing degeneration will have a score from 1 to 7. The results were analyzed, and the graph was plotted using GraphPad Prism software.
Electroretinogram
Anesthetized flies were introduced into truncated 200‐μl disposable pipette tips such that the head protruded from the small opening. The fly's head was then immobilized using colorless nail varnish. Two glass microelectrodes (640786, Harvard Apparatus, Massachusetts, USA) filled with 0.8% NaCl solution were used for recordings such that the voltage changes were recorded by placing the experimental electrode on the surface of the eye and the reference electrode on the thorax. The flies were dark adapted for 5 min prior to ERG recordings followed by 2‐s flash of green light stimulus, with 10 stimuli (flashes) per recording and 15 s of recovery time between two subsequent flashes. Green light stimulus was emitted using an LED light source to within 5 mm of fly's eye through a fiber optic guide. Voltage changes were recorded using pCLAMP 10.7 and amplified using DAM50 amplifier (SYS‐DAM50, WPI, Florida, USA). Data analysis was done using Clampfit 10.7 (Molecular Devices, California, USA). Graphs were plotted using GraphPad Prism software.
Western blot
Age‐matched flies were decapitated, and the heads were homogenized in 2× Laemmli sample buffer followed by boiling at 95°C for 5 min. For rhodopsin blot, the protein samples were processed differently. The fly heads were snap‐frozen in liquid nitrogen and stored at −80°C for a period of 2 days. These head samples were homogenized in 2× Laemmli sample buffer followed by boiling at 37°C for 30 min. Protein extracts from fly heads were separated using SDS–PAGE and transferred onto nitrocellulose filter membrane [Hybond‐C Extra; (GE Healthcare, Buckinghamshire, UK)] using wet transfer apparatus (Bio‐Rad, California, USA). The membrane was blocked using 5% Blotto (sc‐2325, Santa Cruz Biotechnology, Texas, USA) in phosphate buffer saline (PBS) with 0.1% Tween 20 (Sigma‐Aldrich) (PBST) for 2 h at room temperature (RT). Primary antibody incubation was done overnight at 4°C using appropriate antibody dilutions: anti‐RDGB (lab generated), 1:1,000; anti‐dEsyt (kind gift from Dr. Dion Dickman's lab), 1:2,000; anti‐rhodopsin (4C5‐DHSB, lowa, USA), 1:250; anti‐Gq, 1:1,000; anti‐TRP (lab generated), 1:4,000; anti‐norpA, 1:1,000; and anti‐α‐tubulin‐E7c (DHSB, lowa, USA), 1:4,000. Following this, the membrane was washed in PBST and incubated with 1:10,000 dilutions of appropriate secondary antibody (Jackson ImmunoResearch Laboratories, Pennsylvania, USA) coupled to horseradish peroxidase at RT for 2 h. Three PBST washes were given, and the blots were developed with ECL (GE Healthcare) and imaged using LAS 4000 instrument (GE Healthcare).
Fluorescent pseudopupil
To monitor changes in PIP2 levels at the microvillar PM in live flies, the PIP2 biosensor PH‐PLCδ coupled to GFP driven by the transient receptor (trp) promoter of flies was used (Yadav et al, 2015). The flies were made insentient and were immobilized using the same protocol as in ERG recordings. The pseudopupil formed from the summed fluorescence of approximately 20–40 adjacent ommatidia were focused and imaged using the 10× objective of Olympus IX71 microscope. The program created using the software Micromanager captured time‐lapse images of the pseudopupil by collecting fluorescence emitted from the eye when GFP was stimulated by a 90‐ms flash of blue light. Prior to the recordings, the flies were dark adapted (resting conditions) for 6 min during which the probe binds to PIP2 and localizes to the microvillar PM. Following a flash of blue light, the PLCβ activity triggers the hydrolysis of PIP2, and thereby, the probe is displaced from the PM and this leads to the loss of the pseudopupil. The central UV‐sensitive photoreceptor is unresponsive to blue light and thereby retains the probe at the apical PM. Subsequent red light illuminations following the blue light stimulus after each time‐lapse image acquisition hastens the retrieval of the probe back to the PM. Mean fluorescence intensity indicating the basal PIP2 pools and the PIP2 recovery kinetics were calculated using ImageJ from NIH (Bethesda, MD, USA). Quantification of DPP fluorescence intensity was done by measuring the intensity values per area of the pseudopupil.
Similarly, flies expressing the P4M‐GFP probe (PI4P biosensor) were subjected to the same protocol for monitoring PI4P levels. P4M‐GFP expression was driven by an eye‐specific promoter GMR using Gal4‐UAS system.
RNA extraction and qPCR
RNA isolation was done from Drosophila heads using TRIzol reagent (Invitrogen) followed by treatment with amplification grade DNAse I (Invitrogen). cDNA synthesis was done using the Superscript II RNAse H Reverse Transcriptase (Invitrogen) and random hexamers (Applied biosystems). dEsyt primers were designed at the exon–exon junction of the dEsyt genomic region satisfying the parameters recommended for qPCR primer designing, and the run was performed in the Applied Biosystem7500 Fast Real Time PCR instrument using Ribosomal Protein 49 (RP49) primers as the control primers flanking the housekeeping gene region. Triplicates of each sample were measured to ensure the consistency of the data. The primers used for qPCR include the following:
RP49 forward: CGGATCGATATGCTAAGCTGT
RP49 reverse: GCGCTTGTTCGATCCGTA
dEsyt forward: GTTGTGGATAGTTGGCTCACCTT
dEsyt reverse: GCCTGGCTGAATCGATGAATAC
U6.1 forward: GATCCTGTGGCGGCTAC
U6.1 reverse: GAGGTGGAGTCTGGAAAGC
Immunohistochemistry
Retinae were dissected in PBS and fixed with 4% paraformaldehyde in PBS with 1 mg/ml saponin at room temperature for 30 min. Postfixation, the samples were washed in PBS with 0.3% Triton X‐100 (0.3% PBTX) followed by incubation with blocking solution (5% fetal bovine solution in PBTX) for 2 h at RT. The samples were then incubated overnight with the respective antibody [anti‐RDGB (lab generated); 1:300, dEsyt (kind gift from Dr. Dion Dickman's lab); 1:200, mCherry (Thermo Fisher Scientific‐PA5‐34974); 1:300] at 4°C. After this, samples were washed thrice with 0.3% PBTX and incubated with appropriate secondary antibody [Alexa Fluor 488 anti‐rabbit (A11034), anti‐rat (A11006) and Alexa Fluor 633 anti‐rabbit (A21070), Molecular Probes; 1:300 dilution] for 4 h at RT. Along with the secondary antibody incubation, Alexa Fluor 568–Phalloidin (Invitrogen, A12380; 1:300) was used to mark F‐actin. Samples were washed thrice with 0.3% PBTX, followed by one final wash in PBS and were mounted with 70% glycerol in 1× PBS. The whole‐mounted preparations were imaged under 60 × 1.4 NA objective, in Olympus FV 3000 microscope.
Electron microscopy
The fly heads of mentioned genotypes were cut and immersed in 2% osmium tetroxide, kept at 4°C for 1 h followed by incubation at 40°C for 4 days. Specimens were washed with distilled water, stained enbloc with uranyl acetate (0.5% in distilled water) for 3 h. After washing with distilled water, specimens were subjected to dehydration step and embedded in epon. Ultrathin sections of 60 nm were cut, and grids were subjected to poststaining with 2% uranyl acetate (in 70% ethanol) and Reynold's lead solution. Sections were imaged at 120 KV on a Tecnai G2 Spirit Bio‐TWIN (FEI) electron microscope.
Scoring MCS density
For scoring MCS density/photoreceptor cell, a total of 30 cells were taken to conduct analysis for R1‐R6 and 9 cells for R7. Using freehand line tool of ImageJ, length of MCS (μm)/the total length of the base of the rhabdomere (μm) was calculated. Fractions of MCS coverage were multiplied with 100 to show the percentage. Groups were compared using the GraphPad Prism 8 software.
Statistical analysis
Unpaired two‐tailed t‐test or ANOVA, followed by Tukey's multiple comparison test, were carried out where applicable.
Author contributions
VRN and PR designed research; VRN, SM, and BB performed research; DT generated reagents; VRN, SM, and BB analyzed data; and VRN and PR wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Review Process File
Acknowledgements
This work was supported by the Department of Atomic Energy, Government of India, under project no. 12‐R&D‐TFR‐5.04‐08002 and 12‐R&D‐TFR‐5.04‐0900, a Wellcome‐DBT India Alliance Senior Fellowship (IA/S/14/2/501540) to PR, a Wellcome‐DBT India Alliance Early Career Fellowship (IA/E/17/1/503653) to SM and the Department of Biotechnology, Government of India (BT/PRJ3748/GET/l 19/27/2015). We thank the Drosophila Facility, Central Imaging Facility and Electron Microscopy Facility at NCBS for support. We thank Dr. D. Dickman for sharing valuable reagents.
EMBO Reports (2020) 21: e50264
Data availability
No data were deposited in any public database.
References
- Balakrishnan SS, Basu U, Shinde D, Thakur R, Jaiswal M, Raghu P (2018) Regulation of PI4P levels by PI4KIIIα during G‐protein‐coupled PLC signaling in Drosophila photoreceptors. J Cell Sci 131: jcs217257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T, Kim YJ, Alvarez‐Prats A, Pemberton J (2019) Lipid dynamics at contact sites between the endoplasmic reticulum and other organelles. Annu Rev Cell Dev Biol 35: 85–109 [DOI] [PubMed] [Google Scholar]
- Bian X, Saheki Y, De Camilli P (2018) ER‐plasma membrane tethering with lipid transport. EMBO J 37: 219–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco S, Meyer T (2011) STIM proteins and the endoplasmic reticulum‐plasma membrane junctions Annu Rev Biochem 80: 973–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C‐L, Hsieh T‐S, Yang TT, Rothberg KG, Azizoglu DB, Volk E, Liao J‐C, Liou J (2013) Feedback regulation of receptor‐induced Ca2+ signaling mediated by E‐Syt1 and Nir2 at endoplasmic reticulum‐plasma membrane junctions. Cell Rep 5: 813–825 [DOI] [PubMed] [Google Scholar]
- Cockcroft S, Garner K, Yadav S, Gomez‐Espinoza E, Raghu P (2016) RdgBα reciprocally transfers PA and PI at ER‐PM contact sites to maintain PI(4,5)P2 homoeostasis during phospholipase C signalling in Drosophila photoreceptors. Biochem Soc Trans 44: 286–292 [DOI] [PubMed] [Google Scholar]
- Cockcroft S, Raghu P (2018) Phospholipid transport protein function at organelle contact sites. Curr Opin Cell Biol 53: 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE : multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta AT, Levine TP (2017) Piecing together the patchwork of contact sites. Trends Cell Biol 27: 214–229 [DOI] [PubMed] [Google Scholar]
- Giordano F, Saheki Y, Idevall‐Hagren O, Colombo SF, Pirruccello M, Milosevic I, Gracheva EO, Bagriantsev SN, Borgese N, De Camilli P (2013) PI(4,5)P2‐dependent and Ca2+‐regulated ER‐PM interactions mediated by the extended synaptotagmins. Cell 153: 1494–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Raimondi A, Toole EO, Paradise S, Collesi C, Cremona O, Ferguson SM, De Camilli P (2008) Cell‐ and stimulus‐dependent heterogeneity of synaptic vesicle endocytic recycling mechanisms revealed by studies of dynamin 1‐null neurons. Proc Natl Acad Sci USA 105: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler PK, Palevitz BA, Lancelle SA, Mccauley MM, Lichtscheidl I (1984) Cortical endoplasmic reticulum in plants. J Cell Sci 96: 355–373 [Google Scholar]
- Herdman C, Tremblay MG, Mishra PK, Moss T (2014) Loss of Extended Synaptotagmins ESyt2 and ESyt3 does not affect mouse development or viability, but in vitro cell migration and survival under stress are affected. Cell Cycle 13: 2616–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idevall‐hagren O, Lü A, Xie B, De Camilli P (2015) Triggered Ca2+ influx is required for extended membrane tethering. EMBO J 34: 2291–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuma K, Li X, Kim D, Sutter D, Dickman DK (2017) Extended synaptotagmin localizes to presynaptic ER and promotes neurotransmission and synaptic growth in Drosophila . Genetics 207: 993–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Ueda R (2013) Highly improved gene targeting by germline‐specific Cas9 expression in Drosophila . Genetics 195: 715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Xie Y, Ma J, Luo X, Nie P, Zuo Z, Lahrmann U, Zhao Q, Zheng Y, Zhao Y et al (2015) IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31: 3359–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen CJR, Young BP, Tavassoli S, Levine TP (2007) Inheritance of cortical ER in yeast is required for normal septin organization. J Cell Biol 179: 467–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manford AG, Stefan CJ, Yuan HL, MacGurn JA, Emr SD (2012) ER‐to‐plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev Cell 23: 1129–1140 [DOI] [PubMed] [Google Scholar]
- Matsumoto‐Suzuki E, Hirosawa K, Hotta Y (1989) Structure of the subrhabdomeric cisternae in the photoreceptor cells of Drosophila melanogaster. J Neurocytol 18: 87–93 [DOI] [PubMed] [Google Scholar]
- Omnus DJ, Manford AG, Bader JM, Emr SD, Stefan CJ (2016) Phosphoinositide kinase signaling controls ER‐PM cross‐talk. Mol Biol Cell 27: 1170–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade GE, Porter KR (1957) Studies on the endoplasmic reticulum: Its form and distribution in striated muscle cells. J Biophys Biochem Cytol 3: 269–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearn MT, Randall LL, Shortridge RD, Burg MG, Pak WL (1996) Molecular, biochemical, and electrophysiological characterization of Drosophila norpA mutants. J Biol Chem 271: 4937–4945 [DOI] [PubMed] [Google Scholar]
- Pichler H, Gaigg B, Hrastnik C, Achleitner G, Kohlwein SD, Daum È (2001) A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. Eur J Biochem 268: 2351–2361 [DOI] [PubMed] [Google Scholar]
- Raghu P, Yadav S, Mallampati NBN (2012) Lipid signaling in Drosophila photoreceptors. Biochim Biophys Acta 1821: 1154–1165 [DOI] [PubMed] [Google Scholar]
- Roederer K, Cozy L, Anderson J, Kumar JP (2005) Novel dominant‐negative mutation within the six domain of the conserved eye specification genesine oculis inhibits eye development in Drosophila . Dev Dyn 232: 753–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saheki Y, Bian X, Schauder CM, Sawaki Y, Surma MA, Klose C, Pincet F, Reinisch KM, De Camilli P (2016) Control of plasma membrane lipid homeostasis by the extended synaptotagmins. Nat Cell Biol 18: 504–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saheki Y, De Camilli P (2017) Endoplasmic reticulum‐plasma membrane contact sites. Annu Rev Biochem 86: 659–684 [DOI] [PubMed] [Google Scholar]
- Schauder CM, Wu X, Saheki Y, Narayanaswamy P, Torta F, Wenk MR, De Camilli P, Reinisch KM (2014) Structure of a lipid‐bound extended synaptotagmin indicates a role in lipid transfer. Nature 510: 552–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclip A, Bacaj T, Giam LR, Südhof TC (2016) Extended synaptotagmin (ESyt) triple knock‐ out mice are viable and fertile without obvious endoplasmic reticulum dysfunction. PLoS One 11: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan CJ, Manford AG, Baird D, Yamada‐Hanff J, Mao Y, Emr SD (2011) Osh proteins regulate phosphoinositide metabolism at ER‐plasma membrane contact sites. Cell 144: 389–401 [DOI] [PubMed] [Google Scholar]
- Suzuki E, Hirosawa K (1994) Immunolocalization of a Drosophila phosphatidylinositol transfer protein (rdgB) in normal and rdgA mutant photoreceptor cells with special reference to the subrhabdomeric cisternae. J Electron Microsc 189: 183–189 [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay MG, Moss T (2016) Loss of all 3 Extended Synaptotagmins does not affect normal mouse development, viability or fertility. Cell Cycle 15: 2360–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihtelic TS, Goebl M, Milligan S, O'Tousa JE, Hyde DR (1993) Localization of Drosophila retinal degeneration B, a membrane‐associated phosphatidylinositol transfer protein. J Cell Biol 122: 1013–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S, Garner K, Georgiev P, Li M, Gomez‐Espinosa E, Panda A, Mathre S, Okkenhaug H, Cockcroft S, Raghu P (2015) RDGBα, a PtdIns‐PtdOH transfer protein, regulates G‐protein coupled PtdIns(4,5)P2 signalling during Drosophila phototransduction. J Cell Sci 128: 3330–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S, Cockcroft S, Raghu P (2016) The Drosophila photoreceptor as a model system for studying signalling at membrane contact sites. Biochem Soc Trans 44: 447–451 [DOI] [PubMed] [Google Scholar]
- Yadav S, Thakur R, Georgiev P, Deivasigamani S, Krishnan H, Ratnaparkhi G, Raghu P (2018) RDGBα localization and function at a membrane contact site is regulated by FFAT/VAP interactions. J Cell Sci 131: jcs207985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Liu Y, Gulbranson DR, Paine A, Rathore SS, Shen J (2016) Extended synaptotagmins are Ca2+‐dependent lipid transfer proteins at membrane contact sites. Proc Natl Acad Sci USA 113: 4362–4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Review Process File
Data Availability Statement
No data were deposited in any public database.


