Abstract
Cardiac hypertrophy (CH) is closely related to a range of cardiovascular diseases, including heart failure and sudden cardiac death. The present study aimed to elucidate the role of long non-coding RNA (lncRNA) ZEB2 antisense RNA 1 (ZEB2-AS1) in regulating the hypertrophic process of cardiomyocytes and the potential underlying mechanism. An in vivo CH mouse model was established by performing transverse aortic constriction procedures. An in vitro CH model was established in primary cardiomyocytes isolated from mice by phenylephrine (PE) treatment. The relative protein levels of BNP, ANP and PTEN in cells with different groups (CH group and control group) were determined by western blotting. Relative expression levels of ZEB2-AS1, natriuretic peptide A (ANP) and brain natriuretic peptide (BNP) were determined in both in vivo and in vitro CH models. The regulatory effects of ZEB2-AS1/phosphatase and tensin homolog (PTEN) on cell surface area, and the relative expression levels of ANP and BNP were explored. ZEB2-AS1, ANP and BNP expression levels were increased in both in vivo and in vitro CH models compared with the sham and negative control groups, respectively. ZEB2-AS1 knockdown decreased cell surface area, and downregulated ANP and BNP expression levels in PE-treated primary cardiomyocytes. Similarly, PTEN overexpression reduced cell surface area, and downregulated ANP and BNP expression levels in PE-treated primary cardiomyocytes. Moreover, PTEN reversed the regulatory effects of ZEB2-AS1 on hypertrophic cardiomyocytes. Therefore, the present study suggested that lncRNA ZEB2-AS1 may influence the progression of CH by downregulating PTEN.
Keywords: long non-coding RNA ZEB2 antisense RNA 1, phosphatase and tensin homolog, phenylephrine, cardiac hypertrophy
Introduction
Cardiac hypertrophy (CH) is closely related to a range of cardiovascular diseases, including heart failure and sudden cardiac death (1). CH is the adaptive response to counteract stresses and maintain normal cardiac function (2). In response to external mechanical or pathological stresses, the heart undergoes cardiac remodeling, which manifests as CH by increasing the size and surface area of cardiomyocytes (3). Continuous CH usually coincides with maladaptive cardiac remodeling (4). Diverse factors are involved in the pathological progression of CH, including altered levels of noradrenaline, angiotensin II, interleukin-6 and long non-coding (lncRNA) (4); however, understanding the specific molecular mechanisms underlying CH is of significance for improving the diagnostic and therapeutic efficacies of CH.
lncRNAs are non-coding RNAs that are >200 nucleotides in length (5). lncRNAs are involved in a number of processes, such as epigenetics, cell cycle and cell differentiation (6). In addition, some studies have reported that lncRNAs are also key regulators of various cardiovascular diseases (7-9). For example: lncRNA TINCR ubiquitin domain containing inhibits CH by epigenetically silencing calcium/calmodulin dependent protein kinase II (8); lncRNA myocardial infarction-associated transcript contributed to CH by regulating toll-like receptor 4 via microRNA (miR)-93(9); and lncRNA cardiac hypertrophy-related factor (CHRF) promotes CH by targeting miR-93 to further regulate Akt3 expression levels (10). It has also been reported that lncRNA ZEB2-AS1 is a molecule involved in tumor biology (11) that promotes bladder cell proliferation and inhibits apoptosis by modulating miR-27b (12). By targeting the miR-204/high mobility group box 1 axis, ZEB2-AS1 promotes pancreatic cancer cell proliferation and invasion (13). In gastric cancer, lncRNA ZEB2-AS1 is upregulated, and affects cell proliferation and invasion via the miR-143-5p/hypoxia inducible factor 1 subunit α axis (14). However, the specific function of ZEB2-AS1 in cardiovascular diseases, especially CH, is not completely understood.
The present study constructed both in vivo and in vitro CH models by TAC procedures and PE treatment, respectively. The aim of the present study was to investigate the role of ZEB2-AS1 in CH.
Materials and methods
Experimental animals
Specific pathogen free male C57BL6 mice (age, 8 weeks; weight, 20-25 g) were selected for constructing the in vivo CH model. Mice in the CH group (n=8) were anesthetized with an intraperitoneal injection of 100 mg/kg ketamine and 5 mg/kg xylazine. Intubation was performed with a volume circulatory ventilator and a midline incision was made above the sternum. Muscles were carefully separated to expose the trachea. After trachea cannula, the second rib on the left side of the thoracic cavity was cut using surgical scissors, and both thymuses were pushed aside to expose the ascending aortic arch. A 27G needle was punctured into the ascending aorta in its natural growth direction. After ligation of the ascending aorta using a 5-0 suture, the needle was gently pulled out to perform transverse aortic constriction (TAC) and establish pressure overload-induced cardiac hypertrophy. Mice in the sham group (n=8) underwent anesthesia and exposure of the ascending aortic arch without puncture and ligation. Mice were euthanized via cervical dislocation at 6 weeks after the cardiac hypertrophy model was established. Subsequently, the heart and lung were harvested for subsequent experiments. Signs of severe pain, including abnormal movement and sound, were considered as humane endpoints requiring immediate euthanasia in the present study. The present study was approved by the Animal Ethics Committee of Nanjing Medical University Animal Center (approval no. 2016NJMU-043A-23).
Cell culture of primary cardiomyocytes
As previously described (15), primary cardiomyocytes were isolated from newborn mice (Nanjing Medical University). Briefly, five newborn mice were sacrificed by cervical dislocation. Following isolation, heart tissues were cut into small pieces and digested. After centrifugation at 4˚C and 1,050 x g for 10 min, the pellet was resuspended in DMEM (Gibco; Thermo Fisher Scientific, Inc.) and inoculated in collagen-coated plates. For 24 h, cardiomyocytes were cultured in serum-free medium at 37˚C. Subsequently, the medium was replaced with DMEM containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.). To induce an in vitro CH model, primary cardiomyocytes (3x106) were treated with 100 µM phenylephrine (PE) (Beyotime Institute of Biotechnology) for 36 h at 37˚C. Cell surface area was measured as described previously (3).
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cardiomyocytes using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and the concentration of total RNA was measured using an ultraviolet spectrophotometer (Hitachi, Ltd.). Total RNA was reverse transcribed into cDNA at 50˚C for 45 min using a PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol. qPCR was subsequently performed using the SYBR-Green Master kit (Roche Diagnostics). The reaction system volume was 25 µl in total and the cycling conditions were: Pre-denaturation at 95˚C for 5 min, denaturation at 95˚C for 30 sec, annealing at 60˚C for 45 sec, extension at 72˚C for 3 min, with 35 cycles, and then extension at 72˚C for 5 min. qPCR products were stored at 4˚C. The relative levels were quantitatively analyzed using the 2-ΔΔCq method (16). GAPDH was used as internal reference. Primer sequences were as follows: GAPDH forward, 5'-GCAAGGATACTGAGAGCAAGAG-3' and reverse, 5'-GGATGGAATTGTGAGGGAGATG-3'; ANP forward, 5'-GCCCTCATTTTGGCCATCAG-3' and reverse, 5'-TTCCCACTTGAGCAGCATTG-3'; BNP forward, 5'-TGTCCTACAGGGACCCCTTC-3' and reverse, 5'-CGCTCAGGGAACCGATTCTA-3'; PTEN forward, 5'-TGTGGTCTGCCAGCTAAAGG-3' and reverse, 5'-ACACACAGGTAACGGCTGAG-3'.
Western blotting
The cells were lysed using cell lysis buffer (cat. no. QC25-05099; Shanghai Qincheng Biological Technology Co., Ltd.). Total protein from cells was extracted using radioimmunoprecipitation assay buffer and was quantified using the bicinchoninic acid method (both reagents supplied by Beyotime Institute of Biotechnology) method. A total of 30 ug of protein was added into each lane for the electrophoresis. The extracted proteins were separated using a 10% sodium dodecyl sulphate polyacrylamide electrophoresis gel. After transfer onto polyvinylidene fluoride membranes (EMD Millipore), the protein was blocked in 5% skim milk for 2 h, incubated with primary antibodies at 4˚C overnight and secondary antibodies at 20˚C for 2 h. Bands were developed with an enhanced chemiluminescence (ECL) detection kit (GE Healthcare) and analyzed using ImageJ Software (version 1.38; National Institutes of Health). Rabbit polyclonal ANP antibody (1:1,000; cat. no. ab225844), rabbit monoclonal BNP antibody (1:2,000; cat. no. ab243440), rabbit polyclonal PTEN antibody (1:1,000; cat. no. ab170941), rabbit polyclonal GAPDH antibody (1:500; cat. no. ab37168) and secondary goat anti-rabbit (HRP) IgG antibody (1;2,000; cat. no. ab6721) were all purchased from Abcam.
Vector construction and transfection
pcDNA3.0-ZEB2-AS1 and pcDNA3.0-PTEN vectors were constructed by cloning the cDNAs of ZEB2-AS1 and PTEN into the mammalian expression vector pcDNA3.0 (Invitrogen; Thermo Fisher Scientific, Inc.). Cardiomyocytes (3x106) were transfected with pcDNA3.0-ZEB2-AS1 (100 nM), pcDNA3.0-PTEN (100 nM), pcDNA-control (NC) (100 nM), si-ZEB2-AS1 (100 nM; 5'-CAAAGGACACCTTTGGTTACCTGAA-3') or control siRNA (100 nM; all supplied by Shanghai Qincheng Biological Technology Co., Ltd.) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h post-transfection, cells were used for subsequent experiments.
Statistical analysis
Statistical analyses were conducted using SPSS software (version 18.0; SPSS Inc.). Figure editing was performed using GraphPad Prism software (version 6.0; GraphPad Software, Inc.). Data are expressed as the mean ± standard deviation. The experiments were repeated three times. Differences between two groups were analyzed using the paired Student's t-test. Comparisons among multiple groups were analyzed using one-way ANOVA followed by the Bonferroni post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
ZEB2-AS1 upregulation in CH model mice
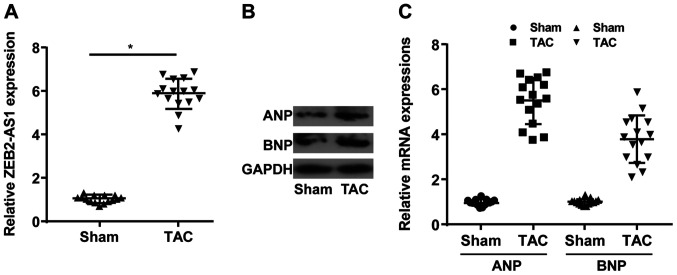
lncRNA ZEB2-AS1 expression levels in mice undergoing TAC or sham operation were determined. Compared with the sham group, the TAC group displayed higher expression levels of ZEB2-AS1 (Fig. 1A). Moreover, the relative expression levels of ANP and BNP were upregulated in the TAC group compared with the sham group (Fig. 1B and C). The results indicated that ZEB2-AS1 may be associated with CH.
Figure 1.
ZEB2-AS1 upregulation in cardiac hypertrophy model mice. (A) Relative expression level of ZEB2-AS1 in the sham and TAC groups. (B) Protein and (C) mRNA expression levels of ANP and BNP in the sham and TAC groups. ZEB2-AS1, ZEB2 antisense RNA 1; TAC, transverse aortic constriction; ANP, natriuretic peptide A; BNP, brain natriuretic peptide. *P<0.05.
ZEB2-AS1 knockdown protects against PE-induced CH
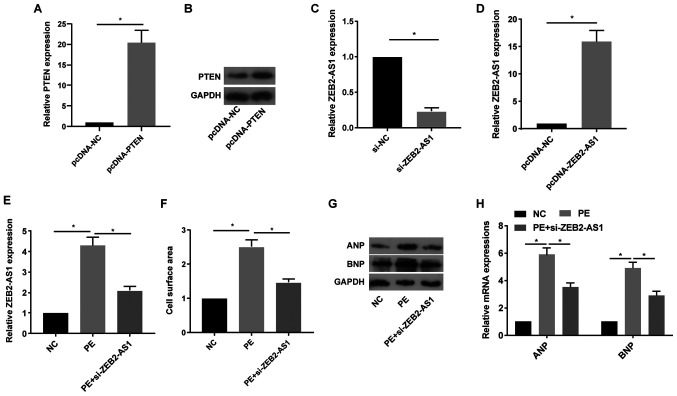
Following treatment with pcDNA-PTEN, the mRNA and protein expression levels of PTEN were notably increased in primary cardiomyocytes compared with the pcDNA-NC group (Fig. 2A and B). Following treatment with si-ZEB2-AS1 or pcDNA-ZEB2-AS1, ZEB2-AS1 expression levels were significantly decreased or increased in primary cardiomyocytes compared with the si-NC and pcDNA-NC groups, respectively (Fig. 2C and D). Primary cardiomyocytes were treated with PE (100 µM), a trigger for in vitro CH, for 36 h. PE treatment significantly upregulated ZEB2-AS1 expression levels in primary cardiomyocytes compared with the NC group; however, si-ZEB2-AS1 transfection reversed PE-mediated effects on ZEB2-AS1 expression (Fig. 2E). Compared with the NC group, cell surface area was significantly increased by PE treatment, which was reversed by ZEB2-AS1 knockdown (Fig. 2F). ANP and BNP protein and mRNA expression levels were notably increased by PE treatment compared with the NC group. By contrast, si-ZEB2-AS1 transfection decreased PE-induced ANP and BNP expression (Fig. 2G and H). The results suggested that dysregulated ZEB2-AS1 expression may serve as a vital factor leading to CH.
Figure 2.
ZEB2-AS1 knockdown protects against PE-induced cardiac hypertrophy. (A) mRNA and (B) protein expression levels of PTEN in primary cardiomyocytes transfected with pcDNA-NC or pcDNA-PTEN. ZEB2-AS1 expression levels in primary cardiomyocytes transfected with (C) si-NC, si-ZEB2-AS1, (D) pcDNA-NC or pcDNA-ZEB2-AS1. (E) ZEB2-AS1 expression levels in untreated, PE-treated (100 µM) or PE-treated + si-ZEB2-AS1-transfected primary cardiomyocytes. (F) Cell surface area in untreated, PE-treated (100 µM) or PE-treated + si-ZEB2-AS1-transfected primary cardiomyocytes. (G) Protein and (H) mRNA expression levels of ANP and BNP in untreated, PE-treated (100 µM) or PE-treated + si-ZEB2-AS1-transfected primary cardiomyocytes. ZEB2-AS1, ZEB2 antisense RNA 1; PE, phenylephrine; PTEN, phosphatase and tensin homolog; NC, negative control; si, small interfering RNA; ANP, natriuretic peptide A; BNP, brain natriuretic peptide. *P<0.05.
PTEN overexpression protects against PE-induced CH
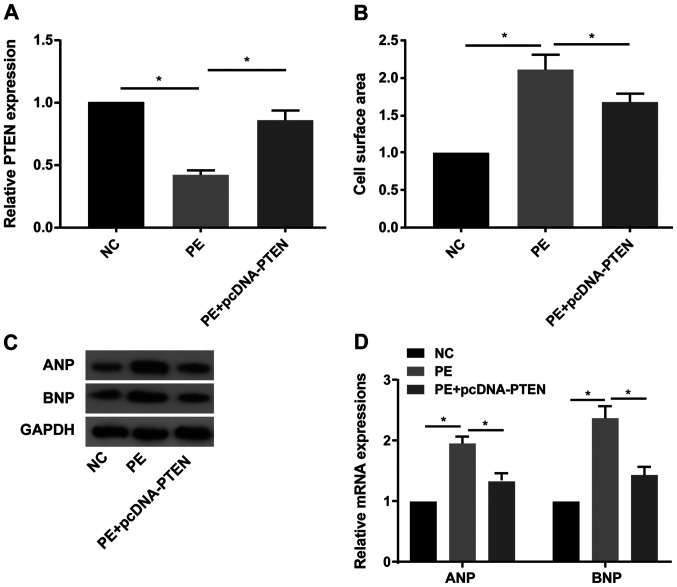
Subsequently, the involvement of PTEN in the process of CH was explored. The RT-qPCR results indicated that PTEN expression was significantly decreased in PE-treated primary cardiomyocytes compared with the NC group, and pcDNA-PTEN transfection reversed PE-mediated downregulation of PTEN expression (Fig. 3A). PTEN overexpression significantly decreased PE-mediated increased cell surface area in primary cardiomyocytes (Fig. 3B). Furthermore, PE treatment obviously increased ANP and BNP protein and mRNA expression levels compared with the NC group, whereas PE-mediated effects on ANP and BNP expression were reversed by PTEN overexpression (Fig. 3C and D).
Figure 3.
PTEN overexpression protects against PE-induced cardiac hypertrophy. (A) PTEN expression levels in untreated, PE-treated (100 µM) or PE-treated + pcDNA-PTEN-transfected primary cardiomyocytes. (B) Cell surface area in untreated, PE-treated (100 µM) or PE-treated + pcDNA-PTEN-transfected primary cardiomyocytes. (C) Protein and (D) mRNA expression levels of ANP and BNP in untreated, PE-treated (100 µM) or PE-treated + pcDNA-PTEN-transfected primary cardiomyocytes. PTEN, phosphatase and tensin homolog; PE, phenylephrine; ANP, natriuretic peptide A; BNP, brain natriuretic peptide; NC, negative control. *P<0.05.
PTEN reverses ZEB2-AS1-mediated effects on CH
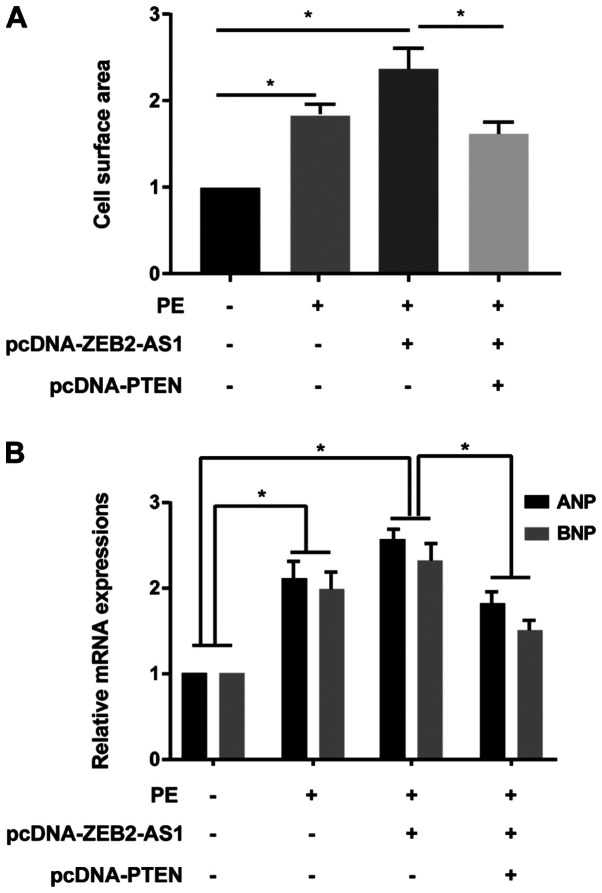
A series of rescue assays were conducted to clarify the effects of ZEB2-AS1/PTEN in CH. ZEB2-AS1 overexpression-induced enlarged cell surface area in cardiomyocytes was partially reversed by PTEN overexpression (Fig. 4A). Additionally, ZEB2-AS1 overexpression upregulated ANP and BNP expression levels in PE-treated cardiomyocytes, which were reversed by co-transfection with pcDNA-PTEN (Fig. 4B). Therefore, the results indicated that ZEB2-AS1 aggravated CH by downregulating PTEN.
Figure 4.
PTEN reverses ZEB2-AS1-mediated effects on cardiac hypertrophy. (A) Cell surface area in untreated, PE-treated (100 µM), PE-treated + pcDNA-ZEB2-AS1-transfected, and PE-treated + pcDNA-ZEB2-AS1- and pcDNA-PTEN-transfected primary cardiomyocytes. (B) ANP and BNP expression levels in untreated, PE-treated (100 µM), PE-treated + pcDNA-ZEB2-AS1-transfected, and PE-treated + pcDNA-ZEB2-AS1- and pcDNA-PTEN-transfected primary cardiomyocytes. PTEN, phosphatase and tensin homolog; ZEB2-AS1, ZEB2 antisense RNA 1; PE, phenylephrine; ANP, natriuretic peptide A; BNP, brain natriuretic peptide. *P<0.05.
Discussion
CH is a common heart disease (1-3). Pathological hypertrophy of the heart leads to a decline in cardiac function and eventually results in heart failure (17). CH is a hallmark of cardiovascular diseases and an important predictor of adverse cardiovascular outcomes, including hypertension and myocardial infarction (18). CH is initially an adaptive response to persistent overload; however, long-term progression leads to heart failure and death (19). Due to alterations to lifestyle and diet, the mortality and morbidity of cardiovascular diseases have increased annually (20). Cardiovascular diseases, such as coronary heart disease and severe heart failure, remain the leading causes of human death worldwide (21). CH is a precursor lesion and independent risk factor for coronary heart disease, heart failure, sudden cardiac death and other heart diseases (22). In particular, pathological CH leads to impaired cardiac function and is a major determinant of common heart diseases (23,24).
Persistent CH is closely related to the expression of embryonic genes, including ANP and BNP (1). The present study indicated that lncRNA ZEB2-AS1 was significantly upregulated in mice undergoing TAC procedures compared with the sham group. ZEB2-AS1 knockdown reversed PE-induced cardiomyocyte hypertrophy, including enlarged cell surface area, and upregulation of ANP and BNP expression. Therefore, it was suggested that ZEB2-AS1 may aggravate the progression of CH.
PTEN can alleviate tumor progression by antagonizing the activities of phosphorylases, such as tyrosine kinases (25). It has been reported that PTEN mutations exist in multiple types of tumors, which is considered to be the star tumor-suppressor gene after the discovery of p53 (26-33). lncRNA growth arrest-specific transcript 5 induces PTEN expression by inhibiting miR-103 in endometrial cancer cells (34). lncRNA maternally expressed 3 alters ovarian cancer cell proliferation, invasion and migration by regulating PTEN (35). lncRNA fer-1 like family member 4 (pseudogene) suppresses endometrial cancer cell proliferation by regulating PTEN expression (36). However, the role of PTEN in CH is not completely understood. The present study indicated that PTEN expression was decreased in PE-induced hypertrophic cardiomyocytes compared with the NC group. The results indicated that PTEN overexpression protected against CH, manifesting as reduced cell surface area, and downregulation of ANP and BNP expression levels compared with the PE group. Moreover, enlarged cell surface area, and upregulated ANP and BNP expression levels in ZEB2-AS1-overexpression cardiomyocytes were partially reversed by PTEN overexpression. Collectively, the results indicated that ZEB2-AS1 aggravated CH by targeting PTEN; therefore, it was suggested that lncRNA ZEB2-AS1 may influence the progression of CH by downregulating PTEN.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81670421) and the ‘Six Peak Talents’ Innovation Talent Team Project (grant no. BE2016798).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ZC and QL designed the study, performed the experiments and drafted the manuscript. ZC and LL established the animal models. QL and LL collected the data. ZC, LL and QL analyzed the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Animal Ethics Committee of Nanjing Medical University Animal Center (approval no. 2016NJMU-043A-23).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Li X, Lan Y, Wang Y, Nie M, Lu Y, Zhao E. Telmisartan suppresses cardiac hypertrophy by inhibiting cardiomyocyte apoptosis via the NFAT/ANP/BNP signaling pathway. Mol Med Rep. 2017;15:2574–2582. doi: 10.3892/mmr.2017.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Facundo H, Brainard RE, Caldas F, Lucas A. Mitochondria and cardiac hypertrophy. Adv Exp Med Biol. 2017;982:203–226. doi: 10.1007/978-3-319-55330-6_11. [DOI] [PubMed] [Google Scholar]
- 3.Tham YK, Bernardo BC, Ooi JY, Weeks KL, McMullen JR. Pathophysiology of cardiac hypertrophy and heart failure: Signaling pathways and novel therapeutic targets. Arch Toxicol. 2015;89:1401–1438. doi: 10.1007/s00204-015-1477-x. [DOI] [PubMed] [Google Scholar]
- 4.Haque ZK, Wang DZ. How cardiomyocytes sense pathophysiological stresses for cardiac remodeling. Cell Mol Life Sci. 2017;74:983–1000. doi: 10.1007/s00018-016-2373-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen S, Jiang H, Bei Y, Xiao J, Li X. Long non-coding RNAs in cardiac remodeling. Cell Physiol Biochem. 2017;41:1830–1837. doi: 10.1159/000471913. [DOI] [PubMed] [Google Scholar]
- 6.Viereck J, Kumarswamy R, Foinquinos A, Xiao K, Avramopoulos P, Kunz M, Dittrich M, Maetzig T, Zimmer K, et al. Long noncoding RNA Chast promotes cardiac remodeling. Sci Transl Med. 2016;8(326ra22) doi: 10.1126/scitranslmed.aaf1475. [DOI] [PubMed] [Google Scholar]
- 7.Zhou G, Li C, Feng J, Zhang J, Fang Y. lncRNA UCA1 is a novel regulator in cardiomyocyte hypertrophy through targeting the miR-184/HOXA9 axis. Cardiorenal Med. 2018;8:130–139. doi: 10.1159/000487204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shao M, Chen G, Lv F, Liu Y, Tian H, Tao R, Jiang R, Zhang W, Zhuo C. lncRNA TINCR attenuates cardiac hypertrophy by epigenetically silencing CaMKII. Oncotarget. 2017;8:47565–47573. doi: 10.18632/oncotarget.17735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu XH, Yuan YX, Rao SL, Wang P. lncRNA MIAT enhances cardiac hypertrophy partly through sponging miR-150. Eur Rev Med Pharmacol Sci. 2016;20:3653–3660. [PubMed] [Google Scholar]
- 10.Wo Y, Guo J, Li P, Yang H, Wo J. Long non-coding RNA CHRF facilitates cardiac hypertrophy through regulating Akt3 via miR-93. Cardiovasc Pathol. 2018;35:29–36. doi: 10.1016/j.carpath.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 12.Wu X, Yan T, Wang Z, Wu X, Cao G, Zhang C. lncRNA ZEB2-AS1 promotes bladder cancer cell proliferation and inhibits apoptosis by regulating miR-27b. Biomed Pharmacother. 2017;96:299–304. doi: 10.1016/j.biopha.2017.08.060. [DOI] [PubMed] [Google Scholar]
- 13.Gao H, Gong N, Ma Z, Miao X, Chen J, Cao Y, Zhang G. lncRNA ZEB2-AS1 promotes pancreatic cancer cell growth and invasion through regulating the miR-204/HMGB1 axis. Int J Biol Macromol. 2018;116:545–551. doi: 10.1016/j.ijbiomac.2018.05.044. [DOI] [PubMed] [Google Scholar]
- 14.Wang F, Zhu W, Yang R, Xie W, Wang D. lncRNA ZEB2-AS1 contributes to the tumorigenesis of gastric cancer via activating the Wnt/β-catenin pathway. Mol Cell Biochem. 2019;451:73–83. doi: 10.1007/s11010-018-03491-7. [DOI] [PubMed] [Google Scholar]
- 15.Wang K, Lin ZQ, Long B, Li JH, Zhou J, Li PF. Cardiac hypertrophy is positively regulated by MicroRNA miR-23a. J Biol Chem. 2012;287:589–599. doi: 10.1074/jbc.M111.266940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forlenza M, Kaiser T, Savelkoul HF, Wiegertjes GF. The use of real-time quantitative PCR for the analysis of cytokine mRNA levels. Methods Mol Biol. 2012;820:7–23. doi: 10.1007/978-1-61779-439-1_2. [DOI] [PubMed] [Google Scholar]
- 17.Creemers EE, Wilde AA, Pinto YM. Heart failure: Advances through genomics. Nat Rev Genet. 2011;12:357–362. doi: 10.1038/nrg2983. [DOI] [PubMed] [Google Scholar]
- 18.Frey N, Olson EN. Cardiac hypertrophy: The good, the bad, and the ugly. Annu Rev Physiol. 2003;65:45–79. doi: 10.1146/annurev.physiol.65.092101.142243. [DOI] [PubMed] [Google Scholar]
- 19.Braunwald E. The war against heart failure: The Lancet lecture. Lancet. 2015;385:812–824. doi: 10.1016/S0140-6736(14)61889-4. [DOI] [PubMed] [Google Scholar]
- 20.Huang Q, Cai B. Exosomes as new intercellular mediators in development and therapeutics of cardiomyocyte hypertrophy. Adv Exp Med Biol. 2017;998:91–100. doi: 10.1007/978-981-10-4397-0_6. [DOI] [PubMed] [Google Scholar]
- 21.McMullen JR, Jennings GL. Differences between pathological and physiological cardiac hypertrophy: Novel therapeutic strategies to treat heart failure. Clin Exp Pharmacol Physiol. 2007;34:255–262. doi: 10.1111/j.1440-1681.2007.04585.x. [DOI] [PubMed] [Google Scholar]
- 22.Bernardo BC, Weeks KL, Pretorius L, McMullen JR. Molecular distinction between physiological and pathological cardiac hypertrophy: Experimental findings and therapeutic strategies. Pharmacol Ther. 2010;128:191–227. doi: 10.1016/j.pharmthera.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 23.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bao Q, Zhao M, Chen L, Wang Y, Wu S, Wu W, Liu X. MicroRNA-297 promotes cardiomyocyte hypertrophy via targeting sigma-1 receptor. Life Sci. 2017;175:1–10. doi: 10.1016/j.lfs.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Malaney P, Uversky VN, Dave V. PTEN proteoforms in biology and disease. Cell Mol Life Sci. 2017;74:2783–2794. doi: 10.1007/s00018-017-2500-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wise HM, Hermida MA, Leslie NR. Prostate cancer, PI3K, PTEN and prognosis. Clin Sci (Lond) 2017;131:197–210. doi: 10.1042/CS20160026. [DOI] [PubMed] [Google Scholar]
- 27.Zhang HM, Fan TT, Li W, Li XX. Expressions and significances of TTF-1 and PTEN in early endometrial cancer. Eur Rev Med Pharmacol Sci. 2017;21:20–26. [PubMed] [Google Scholar]
- 28.Zhang R, Guo Y, Ma Z, Ma G, Xue Q, Li F, Liu L. Long non-coding RNA PTENP1 functions as a ceRNA to modulate PTEN level by decoying miR-106b and miR-93 in gastric cancer. Oncotarget. 2017;8:26079–26089. doi: 10.18632/oncotarget.15317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen W, Li HL, Liu L, Cheng JX. Expression levels of PTEN, HIF-1α, and VEGF as prognostic factors in ovarian cancer. Eur Rev Med Pharmacol Sci. 2017;21:2596–2603. [PubMed] [Google Scholar]
- 30.Ngeow J, Sesock K, Eng C. Breast cancer risk and clinical implications for germline PTEN mutation carriers. Breast Cancer Res Treat. 2017;165:1–8. doi: 10.1007/s10549-015-3665-z. [DOI] [PubMed] [Google Scholar]
- 31.Sun J, Li T, Zhao Y, Huang L, Sun H, Wu H, Jiang X. USP10 inhibits lung cancer cell growth and invasion by upregulating PTEN. Mol Cell Biochem. 2018;441:1–7. doi: 10.1007/s11010-017-3170-2. [DOI] [PubMed] [Google Scholar]
- 32.Li MF, Guan H, Zhang DD. doi: 10.4238/gmr.15028120. Effect of overexpression of PTEN on apoptosis of liver cancer cells. Genet Mol Res 15, 2016. [DOI] [PubMed] [Google Scholar]
- 33.Beg S, Siraj AK, Jehan Z, Prabakaran S, Al-Sobhi SS, Al-Dawish M, Al-Dayel F, Al-Kuraya KS. PTEN loss is associated with follicular variant of Middle Eastern papillary thyroid carcinoma. Br J Cancer. 2015;112:1938–1943. doi: 10.1038/bjc.2015.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo C, Song WQ, Sun P, Jin L, Dai HY. lncRNA-GAS5 induces PTEN expression through inhibiting miR-103 in endometrial cancer cells. J Biomed Sci. 2015;22(100) doi: 10.1186/s12929-015-0213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Xu W, He Y, Xia Q, Liu S. lncRNA MEG3 impacts proliferation, invasion, and migration of ovarian cancer cells through regulating PTEN. Inflamm Res. 2018;67:927–936. doi: 10.1007/s00011-018-1186-z. [DOI] [PubMed] [Google Scholar]
- 36.Qiao Q, Li H. lncRNA FER1L4 suppresses cancer cell proliferation and cycle by regulating PTEN expression in endometrial carcinoma. Biochem Biophys Res Commun. 2016;478:507–512. doi: 10.1016/j.bbrc.2016.06.160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.