Abstract
Objectives
NCOA6 is a transcription coactivator; its deletion in mice results in growth retardation and lethality between 8.5 and 12.5 dpc with defects in the placenta. However, the transcription factor(s) and the mechanism(s) involved in the function of NCOA6 in placentation have not been elucidated. Here, the roles of NCOA6 in human cytotrophoblast invasion and migration were studied.
Materials and Methods
Human placenta tissues were collected from normal pregnancies and pregnancies complicated by early‐onset severe preeclampsia (sPE). Immunofluorescence, RT‐qPCR and Western blotting were used to determine NCOA6 expression. Transwell invasion/migration assays were performed to explore whether NCOA6 knockdown affected human placenta‐derived HTR‐8/SVneo cell invasion/migration. Gelatin zymography was performed to examine the change in the gelatinolytic activities of secreted MMP2 and MMP9. Luciferase reporter assays were used to explore whether NCOA6 coactivated NF‐κB‐mediated MMP9 transcription.
Results
NCOA6 is mainly expressed in the human placental trophoblast column, as well as in the EVTs. HTR‐8/SVneo cell invasion and migration were significantly attenuated after NCOA6 knockdown, and the secretion of MMP9 was decreased due to transcriptional suppression. NCOA6 was further found to coactivate NF‐κB‐mediated MMP9 transcription. Moreover, expression of NCOA6 was impaired in placentas of patients complicated by early‐onset sPE.
Conclusions
Thus, we demonstrated that NCOA6 is important for cytotrophoblast invasion/migration, at least partially, by activating NF‐κB‐mediated MMP9 transcription; the downregulation of NCOA6 may contribute to the pathogenesis of early‐onset sPE.
Keywords: invasion and migration, MMP9, NCOA6, NF‐κB, placental trophoblast
1. INTRODUCTION
Normal placentation is indispensable for foetal development. Two major types of differentiation exist in trophoblasts during human placentation, which include trophoblast invasion and syncytialization. During invasion, cytotrophoblasts (CTBs) proliferate to form the trophoblast column (TC), from which they differentiate into the invasive extravillous trophoblasts (EVTs) that either invade into the maternal decidua/myometrium to anchor the placenta (as interstitial EVTs) or invade and remodel maternal spiral arteries to improve maternal blood flow as required for pregnancy (as endovascular EVTs). During syncytialization, mononucleated CTBs fuse to become multinucleated syncytiotrophoblasts (STBs). 1 Abnormal trophoblast differentiation and subsequent functional impairment result in pregnancy‐related diseases, including preeclampsia (PE) due to shallow interstitial EVT invasion into the maternal decidua/myometrium and incomplete remodelling of maternal vessels caused by impaired endovascular EVT invasion into blood vessels. 2 , 3 , 4
Nuclear receptor coactivator 6 (NCOA6) was first identified in human breast tumours and was originally named Amplified in breast 3 (AIB3). 5 , 6 In addition to gene amplification in breast cancer, NCOA6 was also found to be amplified and overexpressed in colon cancers and lung cancers. 7 As a transcription coactivator, NCOA6 plays multiple roles by coactivating specific transcription factors, including PPARα, 8 PPARγ, 9 AP‐1, CRE and NF‐κB. 10 Mice with Ncoa6 deletion showed growth retardation and lethality between 8.5‐12.5 days post‐conception (dpc), most likely due to developmental defects in the placenta. 9 , 11 , 12 Since both the labyrinth and spongiotrophoblast layers of the placenta in Ncoa6 −/− mice were severely affected, the essential roles of NCOA6 might be the coactivation of key transcription factors in placentation. 9 , 11 However, the transcription factor(s) and the mechanism(s) involved in the function of NCOA6 in placentation have not been elucidated.
In this study, we explored the functions of NCOA6 in human trophoblast invasion and migration. NCOA6 was mainly expressed in the TC and EVT of human placentas. NCOA6 knockdown significantly suppressed the invasion and migration of human placenta‐derived HTR‐8/SVneo cells with reduced matrix metalloproteinase 9 (MMP9) secretion. Luciferase reporter assays were further used to test whether NCOA6 contributed to NF‐κB‐mediated MMP9 transcription. In addition, the transcription of NCOA6 in placentas from patients with early‐onset sPE, showing inadequate trophoblast invasion and subsequent incomplete remodelling of maternal spiral arteries, was found to be impaired. Thus, we demonstrated that NCOA6 promotes the invasion and migration of HTR‐8/SVneo cells, at least partially, by coactivating NF‐κB‐mediated MMP9 transcription.
2. MATERIALS AND METHODS
2.1. Human placenta collection
Placental villi from the first trimester (6‐8 weeks of gestation, n = 3) were sampled from normal pregnancies after legal abortion; placental tissues from the second trimester (17‐21 weeks of gestation, n = 3) were collected after inevitable abortions that had been accidentally caused by external hurt; the third‐trimester placenta samples were obtained from normal pregnancies (normal group, between 36 and 40 weeks of gestation; n = 15) and pregnancies complicated by early‐onset sPE (early‐onset sPE group, between 33 and 37 weeks of gestation; n = 13). All tissues were sampled with informed consent at the Department of Gynecology and Obstetrics in the First Affiliated Hospital of Zhengzhou University. Individuals with early‐onset sPE were recruited as previously reported, without any other maternal complications. 13 , 14 The clinical characteristics of all the pregnant women enrolled in this study are listed (Table 1), and the protocol for sample collection was authorized by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University (2019‐KY‐288). Six small tissue blocks (~0.2 cm3 each) were collected randomly from the foetal side of each third‐trimester/term placenta to achieve uniform sampling and avoid contamination of maternal tissue, followed immediately by snap‐freezing and storage in liquid nitrogen within 30 minutes of caesarean birth.
Table 1.
Clinical characteristics of the pregnant women enrolled in this study
| Characteristics | Normal (n = 15) | Early‐onset sPE (n = 13) | P value |
|---|---|---|---|
| Maternal age (y) | 29.8 ± 2.7 | 29.8 ± 4.0 | .962 |
| Pre‐pregnancy body mass index (kg/m2) | 21.5 ± 2.0 | 22.9 ± 2.0 | .123 |
| Systolic blood pressure (mm Hg) | 114.9 ± 9.9 | 159.0 ± 18.0* | <.001 |
| Diastolic blood pressure (mm Hg) | 73.8 ± 7.9 | 99.9 ± 11.8* | <.001 |
| Proteinuria (g/24 h) | Normal/non‐detected | 4.5 ± 2.3* | <.001 |
| Primiparae (n) | 10 (66.7%) | 8 (61.5%) | NA |
| Current smoker (n) | 0 (0%) | 0 (0%) | NA |
| Han ethnicity (n) | 15 (100%) | 13 (100%) | NA |
| Female foetus (n) | 8 (53.3%) | 9 (69.2%) | NA |
| Gestational age at delivery (wk) | 38.4 ± 1.2 | 34.5 ± 1.5* | <.001 |
| Birth weight (g) | 3241.6 ± 467.8 | 2076.2 ± 385.4* | <.001 |
Values are expressed as the mean ± SD, and statistical analyses were performed by using independent‐samples t test in SPSS Statistics 17.0.
Abbreviations: NA, not analysed; sPE, severe preeclampsia.
Compared to normal pregnancy, P < .001.
2.2. Paraffin‐embedded section preparation and immunofluorescence
Paraffin‐embedded sections of placental tissue were prepared as previously reported. 15 After antigen retrieval and blocking, the serial sections were treated with the following primary antibodies overnight at 4°C: rabbit anti‐human NCOA6 serum that was generously provided by Professor Jian‐ming Xu at Baylor College of Medicine in USA and Professor Hong‐mei Wang at Institute of Zoology, CAS in China (1:500), 16 mouse anti‐human Cytokeratin 7 (CK 7, 180234, Invitrogen; 1:200), mouse anti‐human HLA‐G (sc‐21799, Santa Cruz; 1:200) and mouse anti‐human vimentin (MA5‐11883, Invitrogen; 1:200). After incubations with secondary antibodies: Alexa Fluor Plus 647‐conjugated antibody (A32733, Invitrogen) for NCOA6 and Alexa Fluor 488‐conjugated antibody (A‐11001, Invitrogen) for others, the serial sections were finally treated with DAPI (R37606, Invitrogen) in Vectashield® antifade mounting medium (Vector Laboratories). Florescence images were obtained with a LSM 780 confocal laser‐scanning microscope (ZEISS). All photographs were arranged and processed by PowerPoint (Microsoft Office Professional Plus 2010, Microsoft Corporation) and Photoshop CS6 (Adobe Systems).
2.3. Primary CTB isolation and subsequent in vitro induction of EVT
Primary CTBs were isolated as previously reported. 17 For the in vitro differentiation of primary CTBs into EVTs, 2.5 × 106 CTBs were placed into 35‐mm dishes precoated with 1 mL of liquid Matrigel (BD Biosciences) at 1 mg/mL and maintained in DMEM/F12 (Gibco) containing 10% foetal bovine serum (FBS, Gibco) for 48 hours.
2.4. Cell culture
The trophoblast cell line HTR‐8/SVneo, a generous gift from Professor C. H. Graham (Queen's University, Canada), was cultured as previously reported. 14 , 18 Plasmocin™ prophylactic (5 μg/mL; InvivoGen) was also added to the culture medium to avoid mycoplasma contamination. Mycoplasma contamination detection was performed every month by a Mycoplasma Detection Kit (Yeasen). The HTR‐8/SVneo cell line has never been listed in the Database of Cross‐Contaminated or Misidentified Cell Lines maintained by ICLAC and NCBI Biosample. 19
2.5. Plasmid construction
The pGL3‐Basic‐MMP9 promoter luciferase reporter (inserted with a 726‐bp MMP9 proximal promoter fragment) and its mutant vector M1 were both generous gifts from Professor Anthony J. Valente (University of California, USA). 20 The MMP9 promoter mutant vectors M2 and M3 were constructed with a Gibson Assembly Cloning Kit (NEB) and confirmed by complete nucleotide sequencing (Invitrogen). The pRenilla‐TK vector was a generous gift from Professor Qiang Wang (Institute of Zoology). 21 The vector for the ectopic expression of NCOA6 was kindly provided by Professor Jian‐ming Xu (Baylor College of Medicine) and Professor Hong‐mei Wang (Institute of Zoology). The vector for the ectopic expression of RELA/p65 was kindly provided by Professor Qin‐miao Sun (Institute of Zoology).
2.6. siRNA and plasmid transfections
Stealth RNAi™ siRNAs that interfere with NCOA6 (HSS118106, HSS118107 and HSS177130) and an unconjugated Med GC negative control (12935300) were both bought from Invitrogen. Plasmid and siRNA transfections in HTR‐8/SVneo cells were performed as previously reported. 14 Among the set of three Stealth RNAi™ siRNAs targeting NCOA6, two (HSS118106 and HSS177130) that significantly interfered NCOA6 expression and decreased HTR‐8/SVneo cell invasion and migration (data not shown) were pooled together to transfect HTR‐8/SVneo cells to exclude potential off‐target effects.
2.7. Transwell invasion/migration assays
Transwell invasion/migration assays were carried out as previously reported. 14 1.0 × 105 HTR‐8/SVneo cells were added without FBS to the upper compartment of Corning Costar® transwell inserts pretreated with Matrigel (60 μL of 1 mg/mL for invasion assays) or not (for migration assays). After 22 hours of culture, all of the invaded/migrated cells outside the insert membrane were stained with haematoxylin and photographed. The number of invaded/migrated cells after the knockdown of NCOA6 was normalized to that from the negative control treatment.
2.8. Cell counting kit‐8 (CCK‐8) assay
CCK‐8 incubations were carried out with CCK‐8 reagent (Dojindo Laboratories) 0, 24, 48 and 72 hours after cell‐seeding as previously reported. 14 The cell medium was renewed every day to ensure sufficient growth nutrition for plated cells. The absorbance at 450 nm of CCK‐8 incubation medium was measured by a Varioskan™ Flash Microplate Reader (Thermo Scientific).
2.9. Gelatin zymography
Gelatin zymography was carried out by using an MMP zymography assay kit (Applygen). Before the cells that invaded through the insert membrane in the invasion assays were fixed, 20 μL cell medium inside the insert was mixed with 2× SDS‐PAGE non‐reducing buffer, loaded on a gel (20 μL each) and separated via 10% SDS‐PAGE containing 1.0 mg/mL gelatin. After Buffer A rinsing, Buffer B incubation and R‐250 staining, bands representing the gelatinolytic activities of secreted MMP2 and MMP9 were recorded with a ChemiDoc MP System (Bio‐Rad).
2.10. RT‐qPCR
RT‐qPCR assays were carried out as previously reported. 14 Total RNA extracted by TRIzol reagent (Life Technologies) was applied in the RT reaction (Takara). Quantitative PCR was performed by using a QuantiNova® SYBR Green PCR Kit (QIAGEN) in a QuantStudio 5 Real‐Time PCR System (Applied Biosystems). NCOA6 expression was examined by using the corresponding glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) expression level as endogenous control. All primers used (Invitrogen) are listed (Table 2).
Table 2.
Primers used in RT‐qPCR analyses
| Genes | Primers | Sequence (5ʹ‐3ʹ) |
|---|---|---|
| NCOA6 | PCR‐F | AAAACGTGCCCAATTTGTTACAC |
| PCR‐R | CAATCTGAACGGAGAGAATCCC | |
| HLA‐G | PCR‐F | CCATCATGGGTATCGTTGCT |
| PCR‐R | GCTCCCTCCTTTTCAATCTG | |
| ITGA5 | PCR‐F | GTCGGGGGCTTCAACTTAGAC |
| PCR‐R | CCTGGCTGGCTGGTATTAGC | |
| MMP2 | PCR‐F | TACAGGATCATTGGCTACACACC |
| PCR‐R | GGTCACATCGCTCCAGACT | |
| MMP9 | PCR‐F | CAGTCCACCCTTGTGCTCTTC |
| PCR‐R | TGCCACCCGAGTGTAACCAT | |
| TIMP1 | PCR‐F | ACCACCTTATACCAGCGTTATGA |
| PCR‐R | GGTGTAGACGAACCGGATGTC | |
| TIMP2 | PCR‐F | AAGCGGTCAGTGAGAAGGAAG |
| PCR‐R | AGGGTTGCCATAAATGTCGTT | |
| GAPDH | PCR‐F | ATGGAAATCCCATCACCATCTT |
| PCR‐R | CGCCCCACTTGATTTTGG |
Abbreviations: F, Forward; R, Reverse.
2.11. TF binding site prediction
Three computational algorithms, namely, LASAGNA‐Search 2.0 (https://biogrid-lasagna.engr.uconn.edu/lasagna_search/), 22 Tfsitescan (http://www.ifti.org/cgi-bin/ifti/Tfsitescan.pl) and Gene2Promoter (http://www.genomatix.de), were utilized to search for binding sites of RELA/p65 in the MMP9 proximal promoter with the default system parameters.
2.12. Dual‐luciferase assay
Cotransfection in HTR‐8/SVneo cells was performed using a wild type (WT) or mutant luciferase vector of the MMP9 promoter, pRenilla‐TK (as the internal control), and the NCOA6 siRNAs/ectopic expression vectors of NCOA6 and RELA/p65 or corresponding negative controls. A dual‐luciferase reporter assay kit (Promega) was applied to determine the change in the luciferase activity of cells 48 hours after transfection with a Varioskan™ Flash Microplate Reader.
2.13. Western blotting
Western blotting was carried out as previously reported. 14 Primary antibodies against human NCOA6 (PA5‐52071, Invitrogen; 1:1000) and GAPDH (ab181603, Abcam, Cambridge, UK; 1:10 000) were used in an overnight incubation at 4°C with cut membranes including NCOA6 or GAPDH protein, respectively. After incubation with HRP‐conjugated antibody (ab6721, Abcam; 1:10 000), specific NCOA6 and GAPDH bands were detected with a ChemiDoc MP System.
2.14. Statistical analysis
All data are represented as the mean ± SD (standard deviation) following three and more independent experiments, and an independent‐samples t test in SPSS Statistics 17.0 was used to determine the significance of the differences between treatments and corresponding controls. Statistical significance is indicated mainly by one asterisk (*) for P < .05 or two asterisks (**) for P < .01.
3. RESULTS
3.1. NCOA6 is expressed in human placental TC and invasive EVT cells
The expression of NCOA6 in the first‐ and second‐trimester human placental tissues was first examined by immunofluorescence. The colocalization of NCOA6 and human leucocyte antigen‐G (HLA‐G, an EVT cell surface marker) 23 , 24 expression indicated that NCOA6 is mainly expressed in TC in the first‐trimester placenta and EVTs in the second‐trimester placenta (Figure 1), which implies the important roles of NCOA6 in trophoblast invasion and migration. Human primary CTBs were further isolated from placental villi with 6‐8 weeks of gestation to examine the change of NCOA6 transcription during in vitro induction of EVTs. The RNA level of NCOA6 was also significantly elevated as those of the specific EVT markers HLA‐G and integrin subunit alpha 5 (ITGA5) during the in vitro induction of primary CTBs into EVTs (Figure 2), confirming the important roles of NCOA6 in trophoblast invasion and migration.
Figure 1.
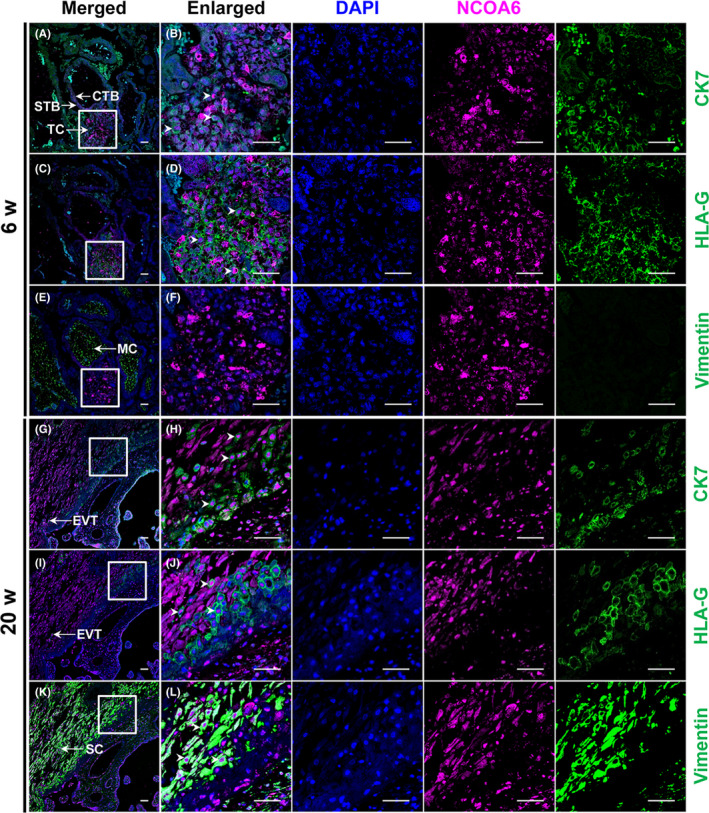
Immunofluorescence localization of NCOA6 in the first‐ and second‐trimester human placental tissues. NCOA6 staining was observed in the CTBs, STBs, TC and EVTs but not in the villous MCs of normal first‐trimester and second‐trimester placentas (A‐J). NCOA6 staining was also found in uterine SCs (K, L). In the representative images of a first‐trimester placental villus from the 6th week of gestation (A‐F), NCOA6 was highly expressed in the TC (C, D). In the representative images of a second‐trimester placenta from the 20th week of gestation (G‐L), strong expression of NCOA6 in the nucleus of EVTs was found (I, J). CK7 was used as the marker for placental trophoblasts; HLA‐G was used as the marker for EVTs; and Vimentin was used as the marker for villous MCs and uterine SCs. Scale bars: 50 μm. CTBs indicates cytotrophoblasts; EVTs, extravillous trophoblasts; HLA‐G, human leucocyte antigen‐G; MCs, mesenchymal cells; SCs, stromal cells; STBs, syncytiotrophoblasts; TC, trophoblast column
Figure 2.
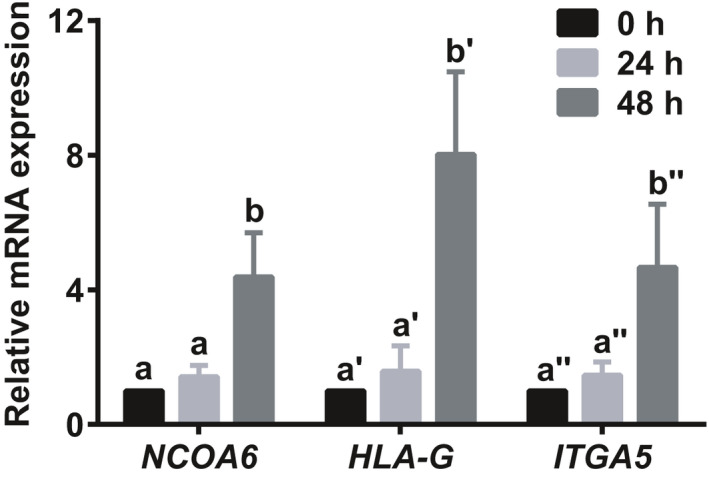
NCOA6 expression in primary CTBs isolated from the first‐trimester human placental villi was increased during in vitro EVT induction. CTBs were purified from placental villi at 6‐8 weeks of gestation by Percoll gradient and subsequently cultured in 1 mg/mL liquid Matrigel precoated plates to induce differentiation to EVTs. NCOA6 expression was measured 0, 24 and 48 hours after cell plating. The EVT markers HLA‐G and ITGA5 were used as indicators of EVT induction. The results are presented as the mean ± SD based on four independent experiments. The values with different letters are significantly different (P < .05). CTBs indicates cytotrophoblasts; EVT, extravillous trophoblast; HLA‐G, human leucocyte antigen‐G
3.2. NCOA6 knockdown attenuates the invasion and migration of HTR‐8/SVneo cells
To further explore whether NCOA6 regulates trophoblast invasion and migration, HTR‐8/SVneo cells (the most commonly applied cell model in trophoblast invasion and migration) were treated with siRNAs targeting NCOA6 before monitoring cell invasion and migration via transwell assays with/without Matrigel, respectively. HTR‐8/SVneo cell invasion and migration were both significantly attenuated after NCOA6 siRNA treatment (Figure 3A). To exclude the possibility that cell proliferation during transwell assays might have a secondary effect on the change in the invaded/migrated cell number after NCOA6 siRNA transfection, cell counting assays were introduced in parallel with transwell assays to record the changes in the number of transfected cells at 0, 24, 48 and 72 hours. NCOA6 siRNA treatment had no obvious effects on the change in the number of transfected cells (Figure 3B), suggesting that the decreased number of invaded/migrated HTR‐8/SVneo cells after NCOA6 siRNA treatment was primarily affected by impaired cell invasion and migration capacities. NCOA6 knockdown by siRNAs was confirmed by RT‐qPCR and Western blotting analyses in parallel (Figure 3C,D).
Figure 3.
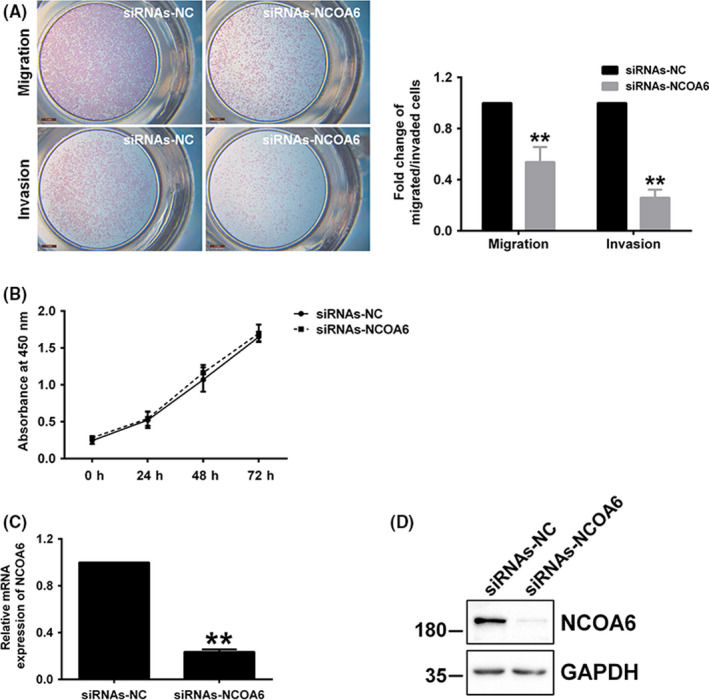
NCOA6 knockdown significantly impaired the invasion and migration of HTR‐8/SVneo cells. A, Significantly attenuated invasion and migration occurred after the transfection of NCOA6 siRNAs into HTR‐8/SVneo cells. All invaded/migrated cells outside the insert membrane were stained and photographed (left panel; scale bars: 1 mm). The fold change in cell invasion/migration capacity after treatment was estimated by counting the invaded/migrated cells with staining (right panel). B, In parallel with the transwell assays, CCK‐8 assays were carried out to determine the change in the number of cells 0, 24, 48 and 72 hours after transfection. C, D, The mRNA and protein levels of NCOA6 were examined after siRNA transfection. A representative image from a Western blot is shown, and the molecular weight markers are indicated on the left in kDa. All results are presented as the mean ± SD based on at least three independent experiments. **P < .01
3.3. NCOA6 knockdown blocks the activation of NF‐κB‐mediated MMP9 transcription
Trophoblasts degrade the extracellular matrix (ECM) when they invade and migrate into the maternal decidua and myometrium. Among the various proteinases secreted by trophoblasts, MMP2 and MMP9, belonging to the gelatinase group of the MMP family and being mainly expressed in trophoblasts of early gestation, play essential roles in the invasion and migration of trophoblasts. 25 , 26 To investigate whether MMP2 and MMP9 are involved in suppressing the invasion and migration of NCOA6‐knockdown HTR‐8/SVneo cells, gelatin zymography assays were performed to examine the changes in the gelatinolytic activities of MMP2 and MMP9. As a result, MMP9 activity was significantly decreased after NCOA6 was knocked down; however, no change in MMP2 activity was found (Figure 4A). Tissue inhibitors of metalloproteinases (TIMPs) inhibit most MMP activities in tissues with a certain degree of specificity. 26 Interestingly, in addition to the reduced level of MMP9 transcription, the transcription of TIMP1, which has been reported to preferentially bind MMP9, 27 , 28 was increased, while the transcription of TIMP2, which preferentially binds MMP2, 28 , 29 was not changed, in accordance with the MMP2 results (Figure 4B). These data suggested the important role of MMP9 but not MMP2 in regulating the invasion and migration mediated by NCOA6 in HTR‐8/SVneo cells.
Figure 4.
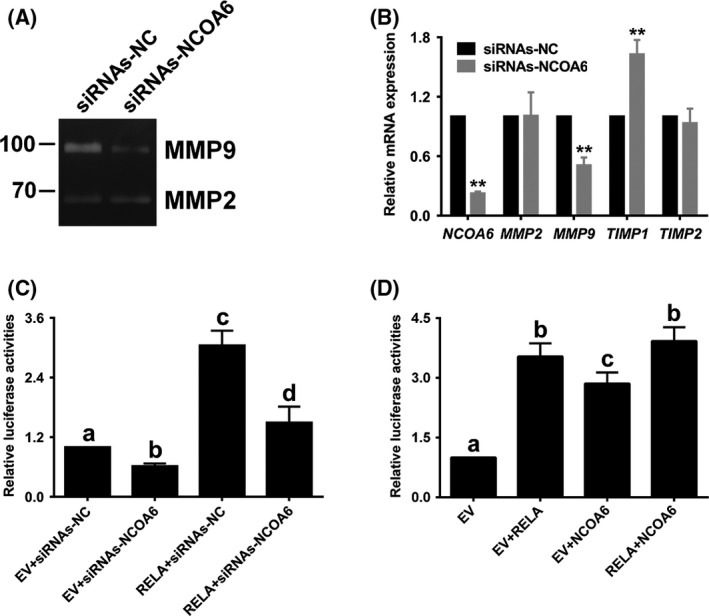
NCOA6 knockdown blocked the activation of NF‐κB‐mediated MMP9 transcription. A, The gelatinolytic activity of secreted MMP9, but not of secreted MMP2, was significantly impaired after NCOA6 was knocked down. B, The transcription of MMP9 was also significantly attenuated after NCOA6 was knocked down, while the transcription of TIMP1 was enhanced. The transcription of MMP2 and its inhibitor TIMP2 showed no change. C, Knockdown of NCOA6 not only significantly decreased the basic reporter activity of the MMP9 promoter but also blocked the increased reporter activity of the MMP9 promoter induced by RELA/p65 overexpression in luciferase assays. D, Co‐overexpression of NCOA6 and RELA/p65 did not synergistically enhance the increased reporter activity of the MMP9 promoter induced by RELA/p65 overexpression alone. All results are presented as the mean ± SD based on at least three independent experiments. **P < .01; the values with different letters are also significantly different (P < .05)
Since NCOA6 is a transcription coactivator of NF‐κB, 10 which has been reported to regulate MMP9 transcription via the binding of its subunit RELA/p65 to MMP9 promoter, 20 , 30 , 31 we further explored whether NCOA6 coactivates NF‐κB‐mediated MMP9 transcription using luciferase reporter assays. Knockdown of NCOA6 not only significantly decreased the basic reporter activity of the MMP9 promoter but also blocked increased reporter activity of the MMP9 promoter following RELA/p65 overexpression (Figure 4C). However, compared with RELA/p65 overexpression alone, the co‐overexpression of NCOA6 and RELA/p65 did not synergistically enhance the increased reporter activity of the MMP9 promoter (Figure 4D). These results further demonstrate that the coactivating role of NCOA6 is essential for NF‐κB‐mediated MMP9 transcription.
3.4. NF‐κB‐mediated transcription occurs via two binding sites in the proximal promoter of MMP9
Three computational algorithms, namely, LASAGNA‐Search 2.0, Tfsitescan and Gene2Promoter, were utilized to search for putative binding sites of NF‐κB in the MMP9 proximal promoter. Two binding sites were found: one site was located at −619 to −609 bp as reported previously, 20 and the other site was located at −331 to −322 bp (Figure 5A). These two sites were individually or both mutated, and the mutations were referred to as the M1 mutation (for site −619 to −609), the M2 mutation (for site −331 to −322) and the M3 mutation (for both sites) (Figure 5A). Only the M3 mutation in the promoter was found to completely interrupt NF‐κB‐mediated MMP9 transcription (Figure 5B), suggesting that both binding sites in the proximal promoter function in NF‐κB‐mediated MMP9 transcription. However, the knockdown of NCOA6 still significantly reduced the reporter activity of M3 (Figure 5C) with less fold change than that of WT affected by NCOA6 knockdown (Figure 4C), which suggested that in addition to NF‐κB, NCOA6 might coactivate other transcription factor(s) that bind to the proximal promoter of MMP9, for example, AP‐1, 10 , 20 during the invasion and migration of HTR‐8/SVneo cells.
Figure 5.
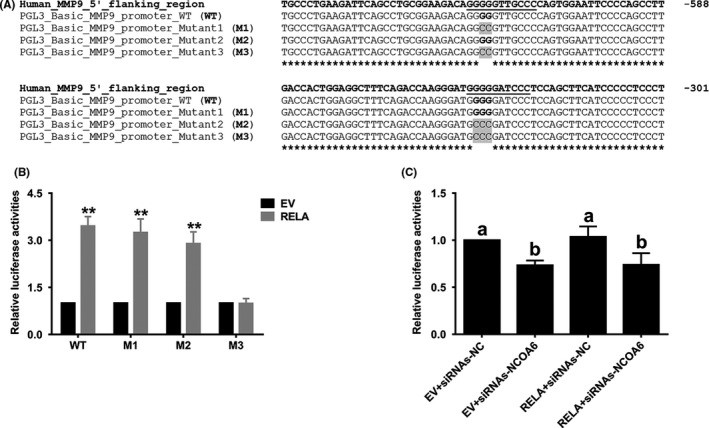
Two binding sites of NF‐κB in the MMP9 proximal promoter mediate its transcriptional regulation. A, Two putative binding sites of NF‐κB are shown in the proximal promoter of MMP9. B, Only the M3 mutant promoter, but neither M1 nor M2, could completely interrupt NF‐κB‐mediated MMP9 transcription. C, Reporter activity of M3 was still significantly decreased after NCOA6 knockdown. All presented results are expressed as the mean ± SD based on at least three independent experiments. **P < .01; the values with different letters are also significantly different (P < .01)
3.5. Transcript levels of NCOA6 were decreased in early‐onset sPE placentas
Since inadequate EVT invasion and migration primarily contribute to early‐onset PE, the transcript levels of NCOA6 in the placentas of pregnancies with early‐onset sPE were examined to evaluate whether placental transcription of NCOA6 is also altered in this diseased state. Of interest, transcript levels for NCOA6 in the early‐onset sPE placentas were significantly impaired compared with those in the paired placentas from normal pregnancies (Figure 6).
Figure 6.
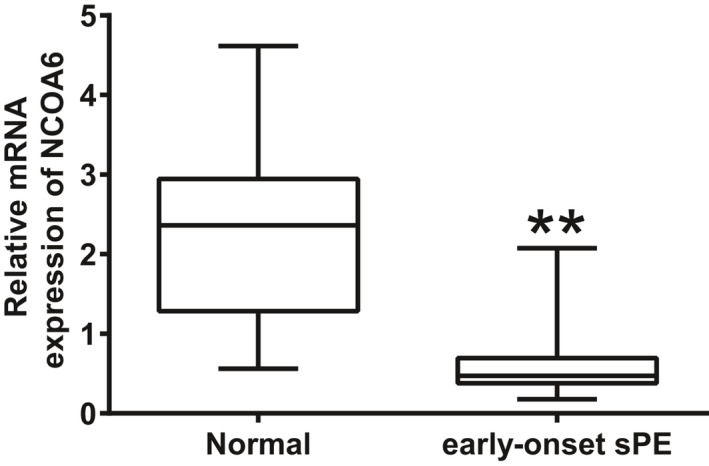
Transcription of NCOA6 was significantly impaired in early‐onset sPE placentas. Transcription of NCOA6 in early‐onset sPE placentas (n = 13) was compared with that in paired normal placentas (n = 15) by RT‐qPCR. **P < .01
4. DISCUSSION
After embryo implantation, the development of the foetus is supported by the placenta, where nutrients and oxygen are provided while waste and carbon dioxide are removed. 32 During pregnancy, the spiral arteries that ultimately supply the placenta are reconstructed by part of trophoblast cells that have invaded and migrated into the maternal decidua/myometrium and further the maternal vessels. Shallow trophoblast invasion and migration are associated with many pregnancy‐specific complications, including PE. 33
NCOA6 is a transcription coactivator that executes a variety of functions with specific transcription factors including PPARα, PPARγ, AP‐1, CRE and NF‐κB. 8 , 9 , 10 The deletion of Ncoa6 in mice resulted in developmental defects in the placenta and lethality between 8.5 and 12.5 dpc 9 , 11 , 12 ; however, the mechanisms involved remain largely unknown. In the present work, NCOA6 was found to be expressed in the TC and invasive EVTs of human placenta. In addition, the RNA level of NCOA6 was also increased as those of the specific EVT markers HLA‐G and ITGA5 during the in vitro induction of isolated primary CTBs into EVTs. These results suggested that NCOA6 is required for trophoblast invasion and migration. Due to the restricted culture days of primary trophoblasts, HTR‐8/SVneo cell line was then applied to investigate the functions of NCOA6 in trophoblast invasion and migration. HTR‐8/SVneo cell line, the most commonly used cell model in trophoblast invasion and migration, was established by stably transfecting the gene encoding simian virus 40 (SV40) large T antigen into cultured first‐trimester human trophoblasts. Apart from their extended lifespans, HTR‐8/SVneo cells possess many similar phenotypic/functional properties with the primary trophoblast cells, for example in vitro morphology, in vitro invasive abilities, and secrection of type IV collagenase. 18 As expected, the knockdown of NCOA6 significantly attenuated HTR‐8/SVneo cell invasion and migration.
Before trophoblasts invade and migrate into the maternal decidua and myometrium, the ECM, which is important for creating the cellular environments of uterine tissue, must be degraded and remodelled. MMP family is involved in tissue remodelling and angiogenesis, with 23 members identified to date. 34 Nearly all human MMPs, specifically secreted by uterine natural killer cells, decidual cells and placental trophoblasts, are present at the human fetomaternal interface 35 ; among them MMP9, but not MMP2, has been reported to exhibit reduced expression in placentas complicated by PE. 36 , 37 In addition, deficiencies in trophoblast invasion emerged soon after implantation in Mmp9‐null mouse embryos, and clinical features of PE were observed in pregnant Mmp9‐null mice with Mmp9‐null embryos. 38 These results demonstrated the crucial roles of MMP9 in trophoblast invasion during human placentation. We found in this study that the gelatinase activity of secreted MMP9, but not that of secreted MMP2, was significantly impaired after NCOA6 siRNA treatment.
NF‐κB, one of the transcription factors that can be coactivated by NCOA6, has been reported to regulate MMP9 transcription via the binding of its subunit RELA/p65 to the promoter. 20 , 30 , 31 We found in this study that NCOA6 knockdown not only significantly decreased the basic reporter activity of the MMP9 promoter but also prevented the increased reporter activity of the MMP9 promoter induced by RELA/p65 overexpression, suggesting an essential role of NCOA6 in the coactivation of NF‐κB‐mediated MMP9 transcription. However, co‐overexpression of NCOA6 and RELA/p65 did not promote increased reporter activity of the MMP9 promoter induced by RELA/p65 overexpression alone.
Two binding sites of NF‐κB on the MMP9 proximal promoter, including one (at −619 to −609 bp) reported previously, 20 were bioinformatically predicted, and both sites were validated by site mutagenesis. However, the activity of the reporter with both binding sites mutated remained significantly decreased after NCOA6 knockdown, suggested the presence of other NCOA6‐coactivated transcription factors, for example AP‐1, 10 , 20 that bind to the proximal promoter of MMP9 in HTR‐8/SVneo cells.
Since NCOA6 has essential roles in trophoblast invasion and migration and inadequate trophoblast invasion/migration and subsequent incomplete reconstruction of maternal spiral arteries are the main contributors to the early‐onset PE, 2 , 3 , 4 , 39 the transcript levels of NCOA6 were compared in placentas from pregnancies with early‐onset sPE and normal pregnancies. Notably, early‐onset sPE placentas showed significantly fewer NCOA6 transcripts than normal placentas. The transcriptomics of CTBs have been found to be dysregulated in sPE due to its diseased in vivo environment. 40 As addressed above, NCOA6 is a transcription coactivator that executes a variety of functions with many specific transcription factors. These results imply transcription coactivators including NCOA6 might play critical roles in the pathogenesis of sPE. However, one limitation exists in this section: placental tissues from normal pregnancies and pregnancies complicated by early‐onset sPE were not completely “term‐matched” (Table 1). Significant difference in genstational age might partly contribute to the decreased NCOA6 transcript levels observed in early‐onset sPE placentas.
In conclusion, our results demonstrated that NCOA6, a transcription coactivator, promotes the invasion and migration of trophoblasts, at least partially, by activating NF‐κB‐mediated MMP9 transcription. The “in vivo” functions of NCOA6 in placentation and the roles of downregulated NCOA6 in sPE development need to be investigated in the future; for example, placenta‐specific Ncoa6 knockout mice should be constructed and analysed to study the “in vivo” function of NCOA6 in placental development. Furthermore, additional researches are also needed to verify other transcription factors that are coactivated by NCOA6 during placentation and sPE development.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
AUTHOR CONTRIBUTIONS
The contributions each author made to the study are specified as follows: YPS and LW designed the study; LW, KQZ, WW, LNC and JS performed the experiments and collected the data; YPS, LW, KQZ, WW, LLH and XXJ analysed the data; LW and KQZ prepared the manuscript; YPS and LW revised the manuscript.
ACKNOWLEDGEMENTS
We honestly thank all patients enrolled in this study. We appreciate Professor Jian‐ming Xu at Baylor College of Medicine in USA and Professor Hong‐mei Wang at Institute of Zoology, CAS in China for experimental supports in plasmid and antiserum. We also appreciate Professor Aaron J. Hsueh at Stanford University in USA for his help in discussion of this study and language editing of the manuscript. The present work was supported by grants from the National Key R&D Program of China to Ying‐pu Sun (2019YFA0110900), the National Natural Science Foundation of China to Liang Wu (81971412) and the Science and Technology Research Program of Henan Province to Liang Wu (182102310138).
Wu L, Zhao K‐Q, Wang W, et al. Nuclear receptor coactivator 6 promotes HTR‐8/SVneo cell invasion and migration by activating NF‐κB‐mediated MMP9 transcription. Cell Prolif. 2020;53:e12876 10.1111/cpr.12876
Liang Wu, Kun‐qing Zhao, and Wei Wang should be considered joint first author.
DATA AVAILABILITY STATEMENT
The data that support the prediction of RELA/p65 binding sites in the MMP9 proximal promoter in this study are available from LASAGNA‐Search 2.0 (https://biogrid-lasagna.engr.uconn.edu/lasagna_search/),22 Tfsitescan (http://www.ifti.org/cgi-bin/ifti/Tfsitescan.pl) and Gene2Promoter (http://www.genomatix.de), with the default parameters.
REFERENCES
- 1. Morrish DW, Kudo Y, Caniggia I, et al. Growth Factors and Trophoblast Differentiation – Workshop Report. Placenta. 2007;28(suppl A):S121‐S124. [DOI] [PubMed] [Google Scholar]
- 2. Kadyrov M, Schmitz C, Black S, Kaufmann P, Huppertz B. Pre‐eclampsia and maternal anaemia display reduced apoptosis and opposite invasive phenotypes of extravillous trophoblast. Placenta. 2003;24:540‐548. [DOI] [PubMed] [Google Scholar]
- 3. Noris M, Perico N, Remuzzi G. Mechanisms of disease: pre‐eclampsia. Nat Clin Pract Nephrol. 2005;1:98‐114; quiz 120. [DOI] [PubMed] [Google Scholar]
- 4. Ji L, Brkic J, Liu M, Fu G, Peng C, Wang YL. Placental trophoblast cell differentiation: physiological regulation and pathological relevance to preeclampsia. Mol Aspects Med. 2013;34:981‐1023. [DOI] [PubMed] [Google Scholar]
- 5. Guan XY, Meltzer PS, Dalton WS, Trent JM. Identification of cryptic sites of DNA sequence amplification in human breast cancer by chromosome microdissection. Nat Genet. 1994;8:155‐161. [DOI] [PubMed] [Google Scholar]
- 6. Guan XY, Xu J, Anzick SL, Zhang H, Trent JM, Meltzer PS. Hybrid selection of transcribed sequences from microdissected DNA: isolation of genes within amplified region at 20q11‐q13.2 in breast cancer. Cancer Res. 1996;56:3446‐3450. [PubMed] [Google Scholar]
- 7. Lee SK, Anzick SL, Choi JE, et al. A nuclear factor, ASC‐2, as a cancer‐amplified transcriptional coactivator essential for ligand‐dependent transactivation by nuclear receptors in vivo. J Biol Chem. 1999;274:34283‐34293. [DOI] [PubMed] [Google Scholar]
- 8. Sarkar J, Qi C, Guo D, et al. Transcription coactivator PRIP, the peroxisome proliferator‐activated receptor (PPAR)‐interacting protein, is redundant for the function of nuclear receptors PParalpha and CAR, the constitutive androstane receptor, in mouse liver. Gene Expr. 2007;13:255‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Antonson P, Schuster GU, Wang L, et al. Inactivation of the nuclear receptor coactivator RAP250 in mice results in placental vascular dysfunction. Mol Cell Biol. 2003;23:1260‐1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ko L, Cardona GR, Chin WW. Thyroid hormone receptor‐binding protein, an LXXLL motif‐containing protein, functions as a general coactivator. Proc Natl Acad Sci USA. 2000;97:6212‐6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuang SQ, Liao L, Zhang H, et al. Deletion of the cancer‐amplified coactivator AIB3 results in defective placentation and embryonic lethality. J Biol Chem. 2002;277:45356‐45360. [DOI] [PubMed] [Google Scholar]
- 12. Zhu YJ, Crawford SE, Stellmach V, et al. Coactivator PRIP, the peroxisome proliferator‐activated receptor‐interacting protein, is a modulator of placental, cardiac, hepatic, and embryonic development. J Biol Chem. 2003;278:1986‐1990. [DOI] [PubMed] [Google Scholar]
- 13. Tranquilli AL, Brown MA, Zeeman GG, Dekker G, Sibai BM. The definition of severe and early‐onset preeclampsia. Statements from the International Society for the Study of Hypertension in Pregnancy (ISSHP). Pregnancy Hypertens. 2013;3:44‐47. [DOI] [PubMed] [Google Scholar]
- 14. Wu L, Song WY, Xie Y, et al. miR‐181a‐5p suppresses invasion and migration of HTR‐8/SVneo cells by directly targeting IGF2BP2. Cell Death Dis. 2018;9:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chang WL, Yang Q, Zhang H, et al. Role of placenta‐specific protein 1 in trophoblast invasion and migration. Reproduction. 2014;148:343‐352. [DOI] [PubMed] [Google Scholar]
- 16. Zhang H, Kuang SQ, Liao L, Zhou S, Xu J. Haploid inactivation of the amplified‐in‐breast cancer 3 coactivator reduces the inhibitory effect of peroxisome proliferator‐activated receptor gamma and retinoid X receptor on cell proliferation and accelerates polyoma middle‐T antigen‐induced mammary tumorigenesis in mice. Cancer Res. 2004;64:7169‐7177. [DOI] [PubMed] [Google Scholar]
- 17. Chang WL, Liu YW, Dang YL, et al. PLAC8, a new marker for human interstitial extravillous trophoblast cells, promotes their invasion and migration. Development. 2018;145:dev148932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Graham CH, Hawley TS, Hawley RG, et al. Establishment and characterization of first trimester human trophoblast cells with extended lifespan. Exp Cell Res. 1993;206:204‐211. [DOI] [PubMed] [Google Scholar]
- 19. Capes‐Davis A, Theodosopoulos G, Atkin I, et al. Check your cultures! A list of cross‐contaminated or misidentified cell lines. Int J Cancer. 2010;127:1‐8. [DOI] [PubMed] [Google Scholar]
- 20. Chandrasekar B, Mummidi S, Mahimainathan L, et al. Interleukin‐18‐induced human coronary artery smooth muscle cell migration is dependent on NF‐kappaB‐ and AP‐1‐mediated matrix metalloproteinase‐9 expression and is inhibited by atorvastatin. J Biol Chem. 2006;281:15099‐15109. [DOI] [PubMed] [Google Scholar]
- 21. Wang Q, Huang Z, Xue H, et al. MicroRNA miR‐24 inhibits erythropoiesis by targeting activin type I receptor ALK4. Blood. 2008;111:588‐595. [DOI] [PubMed] [Google Scholar]
- 22. Lee C, Huang CH. LASAGNA‐Search: an integrated web tool for transcription factor binding site search and visualization. Biotechniques. 2013;54:141‐153. [DOI] [PubMed] [Google Scholar]
- 23. Ellis SA, Palmer MS, McMichael AJ. Human trophoblast and the choriocarcinoma cell line BeWo express a truncated HLA Class I molecule. J Immunol. 1990;144:731‐735. [PubMed] [Google Scholar]
- 24. Chumbley G, King A, Holmes N, Loke YW. In situ hybridization and northern blot demonstration of HLA‐G mRNA in human trophoblast populations by locus‐specific oligonucleotide. Hum Immunol. 1993;37:17‐22. [DOI] [PubMed] [Google Scholar]
- 25. Staun‐Ram E, Goldman S, Gabarin D, Shalev E. Expression and importance of matrix metalloproteinase 2 and 9 (MMP‐2 and ‐9) in human trophoblast invasion. Reprod Biol Endocrinol. 2004;2:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cohen M, Meisser A, Bischof P. Metalloproteinases and human placental invasiveness. Placenta. 2006;27:783‐793. [DOI] [PubMed] [Google Scholar]
- 27. Strongin AY, Collier I, Bannikov G, Marmer BL, Grant GA, Goldberg GI. Mechanism of cell surface activation of 72‐kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J Biol Chem. 1995;270:5331‐5338. [DOI] [PubMed] [Google Scholar]
- 28. Olson MW, Gervasi DC, Mobashery S, Fridman R. Kinetic analysis of the binding of human matrix metalloproteinase‐2 and ‐9 to tissue inhibitor of metalloproteinase (TIMP)‐1 and TIMP‐2. J Biol Chem. 1997;272:29975‐29983. [DOI] [PubMed] [Google Scholar]
- 29. Morgunova E, Tuuttila A, Bergmann U, Tryggvason K. Structural insight into the complex formation of latent matrix metalloproteinase 2 with tissue inhibitor of metalloproteinase 2. Proc Natl Acad Sci USA. 2002;99:7414‐7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hsiao BY, Chang TK, Wu IT, Chen MY. Rad GTPase inhibits the NFkappaB pathway through interacting with RelA/p65 to impede its DNA binding and target gene transactivation. Cell Signal. 2014;26:1437‐1444. [DOI] [PubMed] [Google Scholar]
- 31. Song ZB, Ni JS, Wu P, et al. Testes‐specific protease 50 promotes cell invasion and metastasis by increasing NF‐kappaB‐dependent matrix metalloproteinase‐9 expression. Cell Death Dis. 2015;6:e1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hemberger M, Hanna CW, Dean W. Mechanisms of early placental development in mouse and humans. Nat Rev Genet. 2020;21:27‐43. [DOI] [PubMed] [Google Scholar]
- 33. Brosens I, Pijnenborg R, Vercruysse L, Romero R. The, "Great Obstetrical Syndromes" are associated with disorders of deep placentation. Am J Obstet Gynecol. 2011;204:193‐201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Visse R, Nagase H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ Res. 2003;92:827‐839. [DOI] [PubMed] [Google Scholar]
- 35. Anacker J, Segerer SE, Hagemann C, et al. Human decidua and invasive trophoblasts are rich sources of nearly all human matrix metalloproteinases. Mol Hum Reprod. 2011;17:637‐652. [DOI] [PubMed] [Google Scholar]
- 36. Shokry M, Omran OM, Hassan HI, Elsedfy GO, Hussein MR. Expression of matrix metalloproteinases 2 and 9 in human trophoblasts of normal and preeclamptic placentas: preliminary findings. Exp Mol Pathol. 2009;87:219‐225. [DOI] [PubMed] [Google Scholar]
- 37. Cohen M, Ribaux P, Epiney M, Irion O. Expression of metalloproteinases 1, 2, 7, 9, and 12 in human cytotrophoblastic cells from normal and preeclamptic placentas. Neuro Endocrinol Lett. 2012;33:406‐411. [PubMed] [Google Scholar]
- 38. Plaks V, Rinkenberger J, Dai J, et al. Matrix metalloproteinase‐9 deficiency phenocopies features of preeclampsia and intrauterine growth restriction. Proc Natl Acad Sci USA. 2013;110:11109‐11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Burton GJ, Redman CW, Roberts JM, Moffett A. Pre‐eclampsia: pathophysiology and clinical implications. BMJ. 2019;366:l2381. [DOI] [PubMed] [Google Scholar]
- 40. Zhou Y, Gormley MJ, Hunkapiller NM, et al. Reversal of gene dysregulation in cultured cytotrophoblasts reveals possible causes of preeclampsia. J Clin Invest. 2013;123:2862‐2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the prediction of RELA/p65 binding sites in the MMP9 proximal promoter in this study are available from LASAGNA‐Search 2.0 (https://biogrid-lasagna.engr.uconn.edu/lasagna_search/),22 Tfsitescan (http://www.ifti.org/cgi-bin/ifti/Tfsitescan.pl) and Gene2Promoter (http://www.genomatix.de), with the default parameters.


