Abstract
Transforming growth factor (TGF)-β is a crucial enforcer of immune homeostasis and tolerance, inhibiting the expansion and function of many components of the immune system. Perturbations in TGF-β signaling underlie inflammatory diseases and promote tumor emergence. TGF-β is also central to immune suppression within the tumor microenvironments, and recent studies have revealed roles in tumor immune evasion and poor responses to cancer immunotherapy. Here we present an overview of the complex biology of the TGF-β family and its context-dependent nature. Then focusing on cancer, we discuss the roles of TGF-β signaling in distinct immune cell types and how this knowledge is being leveraged to unleash the immune system against the tumor.
eTOC
Massague and Batlle we present an overview of the complex biology of the TGF-β family and focusing on cancer, discuss the roles of TGF-β signaling in distinct immune cell types and how this knowledge is being leveraged in the clinic.
Introduction
Correct operation of the immune system in vertebrates requires constant regulation to ensure protection against extraneous agents and tolerance of self-antigens. To achieve this critical balance, several types of regulatory components act to impose restrain on the immune system. These components include dedicated cell types such as regulatory T (Treg) cells which limit the expansion of immune effector cells, checkpoint molecules such as CTLA-4 and PD-1 which counterbalance antigen receptor signaling, and immunosuppressive cytokines (Li and Flavell, 2008) the most prominent of which is transforming growth factor β (TGF-β).
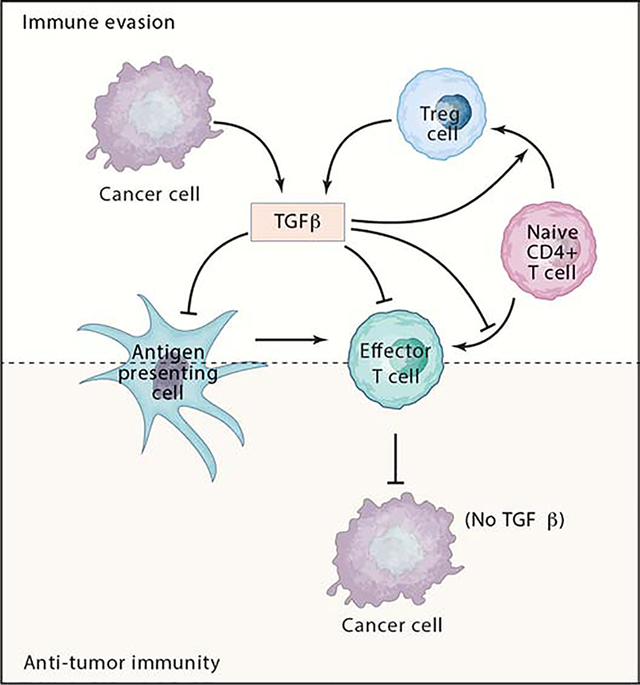
TGF-β regulates the generation and effector functions of many immune cell types (Flavell et al., 2010; Sanjabi et al., 2017). It controls adaptive immunity by directly promoting the expansion of Treg cells, and by inhibiting the generation and function of effector T cells and antigen-presenting dendritic cells (Figure 1). TGF-β similarly controls the innate immune system by inhibiting natural killer (NK) cells and regulating the complex behavior of macrophages and neutrophils, thus forming a network of negative immune regulatory inputs.
Figure 1. Key players in TGF-β suppression of tumor adaptive immunity.
Several prevalent cancer types exhibit a TGF-β-rich TME. TGF-β is produced by cancer cells and by several other cell types present in the TME including Tregs. Fibroblasts, macrophages and platelets are also main TGF-β producers in tumors (not shown). Elevated TGF-β-levels block naïve T cell differentiation towards a Th1 effector phenotype, promotes their conversion towards the Treg subset and dampens antigen presenting functions of dendritic cells.
These effects of TGF-β in immune regulation fall within a wider role of this cytokine and other members of its family in development, homeostasis and tissue regeneration. Malfunctions of this pathway cause congenital defects, fibrotic diseases, immune dysregulation and cancer. Most adult mammalian cell types respond to TGF-β with effects on cell proliferation, differentiation, adhesion, movement, metabolism, communication and death. Of particular interest here, TGF-β functions as a potent tumor suppressor by inducing growth inhibition and apoptosis in pre-malignant cells. Mutations that eliminate the TGF-β pathway or decouple it from apoptosis not only convert these cells into a full-blown malignant state but also allow them to use TGF-β to create an immune suppressive tumor microenvironment and produce additional stromal modifiers that foster tumor progression and metastasis.
The composition and function of the TGF-β signaling pathway, and the extensive role of the TGF-β family in development, homeostasis, and diseases including cancer have been reviewed in detail elsewhere (David and Massagué, 2018; Mullen and Wrana, 2017; Oshimori and Fuchs, 2012). Here, we focus on the role of TGF-β in immune regulation and its relevance to cancer. We provide an overview of the TGF-β signal transduction pathway, and summarize current knowledge about the production and mobilization of TGF-β from latent stores in the tumor microenvironment. We review how the effects of TGF-β are switched from tumor suppressive to pro-metastatic as cancers advance. We then focus on the profound effects of TGF-β on major cellular components of the adaptive and innate immune systems, and in this context we discuss recent progress in elucidating how the immunosuppressive role of TGF-β is enlisted by cancer cells to avert immune surveillance and to thwart cancer immunotherapy. We conclude with comments on the prospect of circumventing TGF-β signaling to improve the effectiveness of cancer immunotherapy.
Sources of TGF-β and regulation of its bioavailability
The thirty-two members of the TGF-β superfamily of ligands encoded in the human genome are grouped into the TGF-β and the bone morphogenetic protein (BMP) subfamilies based on sequence similarity and functional criteria (David and Massagué, 2018). The TGF-β subfamily comprises three TGF-β ligands (TGF-β1, TGF-β2 and TGF-β3), two Activins (A and B), Nodal, GDF1 (growth and differentiation factor 1), GDF3, GDF8 (also known as Myostatin), GDF9 and GDF11. The BMP subfamily includes 10 BMPs, several GDFs, and the Anti-Muellerian Hormone. Additional members encode antagonistic ligands and a few distant outliers. The three TGF-βs, and in particular TGF-β1, are the most relevant members of the family from the standpoint of immune regulation. Although we do not discuss them here, emerging evidences also suggest important roles for BMPs and other TGF-β superfamily members in the immune system (Chen and ten Dijke, 2016).
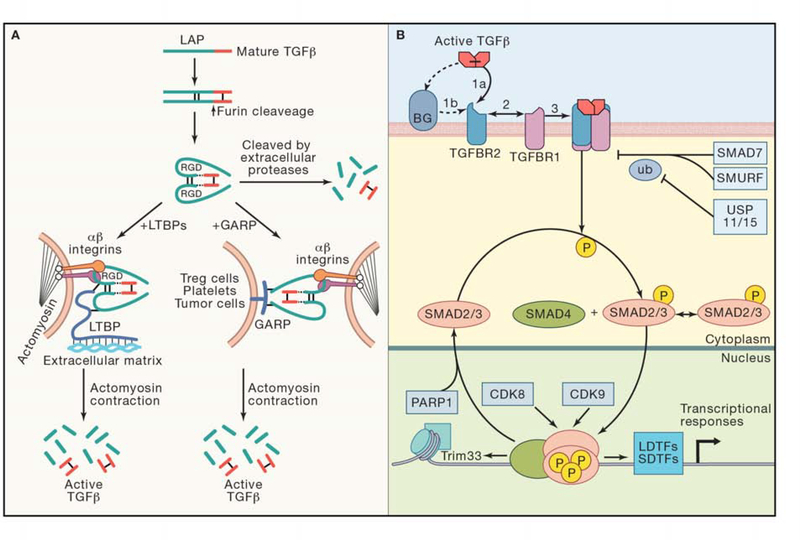
TGF-β1, TGF-β2 and TGF-β3 are synthesized as pro-hormones that include a signal sequence, a large N-terminal portion called the latency-associated peptide (LAP), and a short C-terminal segment, which corresponds to the mature active cytokine monomer (Travis and Sheppard, 2014). Upon cleavage by the protease Furin in the Golgi complex, the dimeric bioactive portion remains non-covalently associated with the disulfide-linked LAP homodimer (Figure 2A). The crystal structure of this complex revealed that LAP encircles the active TGF-β portion and hinders the relevant contact sites of the cytokine with their cognate heterotetrameric receptors (Shi et al., 2011). In many cell types, the LAP homodimer is crosslinked by disulfide bonds to a family of proteins called latent TGF-β binding proteins (LTBPs) forming the so called large latent complex (LLC) (Travis and Sheppard, 2014). After secretion, LLCs interact with Fibrillin, an extracellular matrix (ECM) protein of the elastic fibers. Mutations in the Fibrillin 1 gene cause Marfan syndrome, a human disease characterized by joint laxity, skeletal deformities, and aortic aneurysms due to excessive TGF-β signaling (Verstraeten et al., 2016). TGF-β can also be anchored to the cell surface through association with transmembrane Glycoprotein A Repetitions Predominant Protein (GARP / LRRC32) (Tran et al., 2009; Wang et al., 2012) (Figure 2A) or the related Leucine-Rich Repeat-Containing Protein 33 (LRRC33)(Qin et al., 2018). The expression of GARP is restricted to Treg cells, endothelium and platelets (Tran et al., 2009) whereas LRRC33 is expressed in macrophages and microglia (Qin et al., 2018), suggesting that these proteins regulate TGF-β signaling in restricted regions and particular contexts.
Figure 2. The TGF-β signaling pathway.
(A) Synthesis and release of active TGF-β (adapted from (Kelly et al., 2017)). In the endoplasmic reticulum, each pro-TGF-β molecule is assembled into a dimer via three interchain disulfide bonds. Following cleavage by the endoprotease Furin, the C-terminal fragment remains non-covalently associated with the disulfide-linked LAP homodimer. This molecule is termed the small latent complex. Three main mechanisms of release of active TGF-β are represented: (i) Extracellular protease cleavage of LAP domain. (ii) Tethering of small latent complex to extracellular matrix through LTBP1 and release of active TGF-beta by integrin-transmitted tension upon cell contraction. (iii) Tethering of small latent complex to GARP on the cell surface and release of active TGF-beta by integrins. (B) Signal transduction by TGF-β. The fundamental steps of ligand-induced formation of a paired-kinase receptor complex, and receptor-mediated phosphorylation of R-SMAD proteins for the formation of a trimeric receptor complex are shown. The accessory receptor proteoglycan Betaglycan (BG) presents TGF-β to the signaling receptors. In the nucleus, the activated SMAD heterotrimeric complex binds to target cis-regulatory sites as determined by interactions with lineage-determining transcription factors (LDTFs) and other signal-driven transcription factors (SDTFs). CDK8 and CDK9 phophorylate SMADs in this complex for further activation and eventual degradation. SMAD phosphatases (not shown) reverse these phosphorylation, and PARP-mediated parylation causes SMAD dissociation from DNA. The inhibitory SMAD7 recruits SMURF ubiquitin ligases to target the receptor for degradation, whereas ubiquitin specific peptidases USP11 and USP15 counterbalance this process.
The release of active TGF-β from latent complexes is a tightly regulated process, achieved through enzymatic and non-enzymatic activities present in the extracellular space. The ECM protein Thrombospondin 1 binds to a specific sequence in LAP and prevents its association with the active TGF-β molecule. Supporting this mechanism, mutations in thrombospondin 1 phenocopy several alterations present in TGF-β1 mutant mice including inflammation and epithelial hyperplasia (Crawford et al., 1998). Cleavage by extracellular serine proteases such as plasmin and cathepsin D, and several metalloproteases, including matrix metalloproteases MMP9 and MMP14, also release active TGF-β from its latent form (Travis and Sheppard, 2014) (Figure 2A).
The best understood mechanism of active TGF-β release from latent complexes involves either αvβ6 or αvβ8 integrin heterodimers, which bind to an Arg-Gly-Asp (RGD) integrin recognition motif present in the LAP domain of TGF-β1 and -β3 (Shi et al., 2011) (Figure 2A). Upon cell contraction, tension exerted by the cytoskeleton is converted by integrins into a physical force on the latent TGF-β complex that unfolds LAP and releases the active cytokine (Shi et al., 2011). This phenomenon depends critically on the actin/myosin-mediated contraction machinery (Giacomini et al., 2012), and it is facilitated by stiff substrates and by the presence of highly contractile cells such as myofibroblasts (Wipff et al., 2007). It also requires tethering of the small latent complex to the ECM via LTBP1 (Annes et al., 2004). Supporting the validity of these findings, conditional ablation of integrins β6 or β8 in Treg cells, dendritic cells, monocytes or macrophages leads to inflammatory phenotypes and loss of TGF-β-mediated immune tolerance (Kelly et al., 2017, 2018; Travis and Sheppard, 2014). GARP also acts as a chaperone that orients latent TGF-β1 for activation by integrins (Liénart et al., 2018) (Figure 2A). As discussed above, particular cell types including Tregs, platelets and macrophages expose TGF-β bound to either GARP or LRRC33 on the cell surface facilitating local TGF-β1 release and paracrine signaling (Liénart et al., 2018; Qin et al., 2018; Stockis et al., 2017; Tran et al., 2009).
In cancer cells, the release of active TGF-β1 mediated by GARP promotes TGF-β-driven epithelial-to-mesenchymal transition and immune evasion due to an expansion of tolerogenic Treg compartment (Metelli et al., 2016). Cancer cells can also evade the immune system by mobilizing active TGF-β1 through αvβ8 integrins (Takasaka et al., 2018). In some contexts, cells recruited to the tumor microenvironment (TME) act as mediators of this process. A prime example are platelets, which carry GARP-bound TGF-β on the cell surface. Mice knockout for GARP in platelets exhibit reduced TGF-β signaling in the TME and enhanced anti-tumor immune responses (Rachidi et al., 2017).
The TGF-β Signaling Pathway
The TGF-β pathway is a classic membrane–to–nucleus signaling process involving direct receptor-mediated activation of SMAD transcription factors. Activated SMAD proteins bind to multiple loci throughout the genome as dictated by partner transcription factors whose availability in a particular cellular context determines the response of this cell to TGF-β (Figure 2B). The components and operating logic of the pathway have been recently reviewed elsewhere (David and Massagué, 2018) and are only briefly summarized here.
TGF-β family members signal though paired transmembrane serine/threonine protein kinases known as the type I and type II receptors (Figure 2B). Mammalian genomes encode seven type I receptors, five type II receptors, and 8 SMAD proteins. TGFBR1 (also known as TβR-I and ALK5) and TGFBR2 (also known as TβR-II) function as TGF-β receptors. TGF-β1, TGF-β2 and TGF-β3 are the only ligands for the TGFBR1/TGFBR2 combination. SMAD2 and SMAD3 are substrates for TGF-β subfamily receptors, and SMAD1, SMAD5 and SMAD8 for BMP subfamily receptors; these are referred as receptor-regulated SMADs or R-SMADs. SMAD4 is not a receptor substrate but binds to activated R-SMADs forming heterotrimeric transcriptional complexes. SMAD6 and SMAD7 are inhibitory molecules that suppress receptor and SMAD signaling functions. SMAD7-recruited SMURF1/2 ubiquitin ligases and counteracting ubiquitin specific peptidases (USP11, USP15) regulate TGF-β receptor degradation. The induction of SMAD6 and SMAD7 expression by TGF-β and BMP family members creates negative feedback loops.
SMAD proteins consist of globular N-terminal and C-terminal domains, known as the MH1 and MH2 domains respectively, connected by a linker region. The MH1 domain in R-SMADs and SMAD4 binds DNA, and the MH2 domain binds other SMADs, cooperating transcription factors, chromatin reader and modifying factors. The components of the TGF-β–SMAD pathway are highly conserved, and the structural basis for many aspects of their interactions and function are known (Macias et al., 2015).
TGF-β released from latent complexes binds to the receptors either directly or with the assistance of the accessory co-receptor betaglycan (BG, also known as TGF-β receptor type III, TGFBR3) (Figure 2B). Betaglycan is particularly important for TGF-β2 binding. In the TGF-β-driven receptor complex, TGFBR2 phosphorylates and activates TGFBR1, which then phosphorylates SMAD2 and SMAD3 at two C-terminal serine residues. Receptor-phosphorylated SMAD2 and SMAD3 then form heterotrimeric complexes with SMAD4.
In the nucleus, activated SMAD complexes bind to hundreds of regulatory regions by interacting with other transcription factors ((Mullen et al., 2011; Trompouki et al., 2011; Wang et al., 2017), and see below)). Additional interactions with co-activators and co-repressors determine the transcriptional effect. In these complexes, R-SMADs are phosphorylated by the RNA polymerase II kinases CDK8 and CDK9, which create sites for recruitment of additional cofactors. CDK8/9-mediated phosphorylation additionally primes SMADs for phosphorylation by glycogen synthase kinase 3β (GSK3β), which marks SMADs for polyubiquitination by the HECT-domain ubiquitin ligases NEDD4L and SMURF1, leading to SMAD degradation. Alternatively, R-SMADs are dephosphorylated and dissociated from DNA for new rounds of signal transduction.
SMAD signaling variants and non-SMAD TGF-β signaling
The phenotypes of SMAD knockouts demonstrate a central role of the SMAD pathway in the effects of TGF-β in different contexts, including in the regulation of the immune system. However, variant forms of SMAD signaling and TGF-β signaling mechanisms not involving SMAD proteins should be noted. SMAD4 is essential for most but not all TGF-β family responses. Exceptions include the development of the pancreas in mice (Bardeesy et al., 2006) and the induction of the master transcription factor SOX4 in pancreatic epithelial progenitors (David et al., 2016), which require SMAD2/3 but not SMAD4. SMAD proteins also have non-transcriptional roles, a notable example being the regulation of Drosha-mediated microRNA maturation by R-SMADs (Davis et al., 2008).
SMAD-independent forms of TGF-β signaling have been described (Heldin and Moustakas, 2016; Massagué, 2012; Moustakas and Heldin, 2005). TGFBR2 directly phosphorylates the cell polarity regulator PAR6, which regulates tight junctions and cell migration (Ozdamar et al., 2005; Yi et al., 2010). TGF-β activates members of mitogen-activated protein kinase (MAPK) cascades including TGF-β-activated kinase 1 (TAK1) (Sorrentino et al., 2008), ERK (Lee et al., 2007), p38MAPK, JNK (Lee et al., 2007), and phosphatidylinositol 3-kinase (PI3K) in cell cultures (Heldin and Moustakas, 2016). The adaptor protein TRAF6 was shown to link TGF-β receptors to TAK1, p38MAPK and JNK activation (Sorrentino et al., 2008; Yamashita et al., 2008), but beyond this, the structural basis for coupling of TGF-β receptors to these various pathways remains unknown. Moreover, MAPK and PI3K pathways have their own potent agonists including mitogen receptor tyrosine kinases and cell metabolism sensors. Likewise, stress sensors regulate p38MAPK and JNK activation, whereas tumor necrosis factor (TNF), interleukin-1, and Toll-like receptors control TAK1 activation, making the role of TGF-β as a regulator of these pathways in physiology and disease quite difficult to ascertain at present.
Basis for contextual responses
Although the MH1 domain of all effector SMADs binds GC-rich, 5-bp motifs (CAGAC, GGCGC and others), the affinity of SMADs for these motifs is neither high nor specific enough to explain the selective recognition of target loci by TGF-β and BMP pathways, or the context-dependent nature of TGF-β action. Rather, activated SMAD complexes bind to hundreds of regulatory regions as dictated by context-defining transcription factors. In progenitor cells, lineage-determining transcription factors (LDTFs) play a dominant role in recruiting activated SMAD complexes (David and Massagué, 2018). This is exemplified by the interaction of the LDTF FOXH1 with Nodal-activated SMADs in mesendoderm progenitors. FOXH1 is a pioneer factor that binds to chromatin independently of TGF-β signaling and is then poised for recruitment of activated SMAD complexes (Charney et al., 2017; Chen et al., 1996). Similarly, TGF-β-activated SMADs co-bind the genome with MYOD1 in myogenic progenitors to regulate myogenic differentiation and with PU.1 in pro-B cells to regulate B cell differentiation (Mullen et al., 2011).
Several signal-driven transcription factors participate, together with LDTFs, as determinants of SMAD binding, providing integration of multiple inputs. Focusing on the immune system, TGF-β-activated SMADs cooperate with STAT5 and nuclear factor of activated T cells (NFAT) to induce FOXP3 expression in naïve CD4+ T cells, thus promoting their differentiation to a Treg phenotype (Tone et al., 2008), and with RORγ2 to induce a T helper 17 (TH17) phenotype (Martinez et al., 2010; Zhou et al., 2008). SMADs together with the transcription factor RUNX3 regulate immunoglobulin class switching in B cells (Park et al., 2005). In TGF-β-stimulated CD8+ T cells, SMADs collaborate with the transcription factor ATF1 to repress the expression of several cytolytic genes (Thomas and Massagué, 2005).
TGF-β and tumor progression
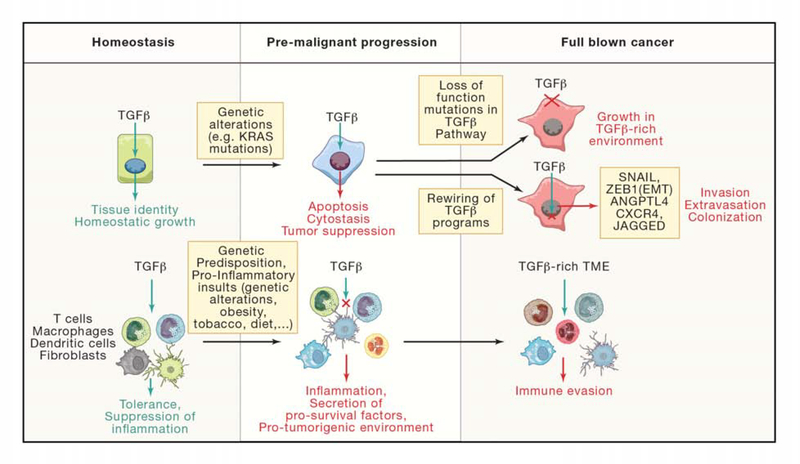
TGF-β functions as a tumor suppressor that can induce apoptosis in pre-malignant cells and inhibit proliferation in carcinoma cells. However, cancer cell clones that inactivate the TGF-β pathway or decouple it from tumor suppressive effects under this selective pressure can use TGF-β for tumor progression. In this altered context, tumor-derived TGF-β can induce tumorigenic and pro-metastatic responses in cancer cells and the stroma, including the formation of an immune suppressive tumor microenvironment (Figure 3). The basis for this dual tumor suppressive and tumor promoting role of TGF-β has been reviewed in detail elsewhere (David and Massagué, 2018; Pickup et al., 2017) and is summarized hereunder.
Figure 3. Tumor suppressor and promoting functions of TGF-β signaling. Left.
In homeostasis, TGF-β signals regulate key processes in multiple tissues including their growth, regeneration and identity. In the immune system, TGF-β instructs tolerance and suppresses inflammation. This function is particularly relevant in the gastrointestinal tract. Center. Genetic alterations can modify the output of TGF-β signals in tumor initiating cells. During the initial stages of carcinogenesis, TGF-β operates as main tumor suppressor by imposing cytostatic and apoptotic programs in tumor cells. A proinflammatory environment fosters the onset of cancer. Loss of TGF-β signals in the microenvironment contributes to exacerbate inflammation in this context. Secretion of pro-survival factors and cytokines by stromal and immune cells pushes continuous regeneration in a harsh inflammatory environment, which eventually leads to the onset of cancer. Right. During tumor progression, selective pressure promotes loss of the cytostatic and tumor suppressor function of TGF-beta in cancer cells. In general, this process occurs via two distinct mechanisms. Acquisition of loss of function mutations in TGF-beta pathway components renders tumor cells resistant to TGF-β thus enabling growth in a TGF-β-rich environment present in many advanced cancers. Alternatively, TGF-beta signals are reinterpreted in cancer cells to instruct tumor-promoting functions such as the ability to migrate and colonize foreign organs. In the TME of several prevalent tumor types, TGF-β operates as central mechanism of immune evasion.
Based on current human TCGA data sets, esophageal, gastric colorectal (CRC), and pancreatic (PDA) adenocarcinomas contain mutations or deletions in SMAD and TGF-β receptor genes in 25% to 50% of cases. Head and neck, bladder and endometrial adenocarcinomas, and cervical and lung squamous carcinomas harbor such mutations in 10% to 20% of cases. SMAD4 inactivating mutations predominate in PDA and CRC, but mutations in SMAD2, SMAD3, TGFBR1 and TGFBR2 are also frequently observed. Collectively, these loss of function mutations provide evidence for the tumor suppressor role of the TGF-β pathway in human cancer. Genetically engineered mouse models of PDA and CRC indicate that TGF-β exerts its tumor suppressive functions mostly by blocking the transition of premalignant cells to a more malignant phenotype (Figure 3). Evidence from human cancer also suggests that TGF-β pathway mutations accumulate during the malignant conversion step.
TGF-β is an archetypical growth inhibitory cytokine and inhibits cell proliferation through increased expression of the cyclin-dependent kinase (CDK) inhibitors p15INK4, p21CIP1, p27KIP1 and/or p57KIP2 (Hannon and Beach, 1994; Polyak et al., 1994; Reynisdóttir et al., 1995; Scandura et al., 2004; Seoane et al., 2004) and downregulation of MYC expression (Chen et al., 2002; Pietenpol et al., 1990). However, oncogenically transformed cells harbor strong CDK activating signals that thwart the effectiveness of TGF-β-induced cytostasis as a tumor suppressive effect. Moreover, the capacity to enter a slow-cycling state is a basic feature of stem cells, one that enables these cells to evade immune surveillance and antimitotic chemotherapy. Therefore, not only is cytostasis a milder form of tumor suppression compared to apoptosis, but cytostasis may ultimately enable disseminated tumor cells to survive for the eventual initiation of metastasis (Malladi et al., 2016).
TGF-β triggers apoptosis in pre-malignant cells that suffer oncogenic stress, in from RAS oncogenes, as documented in genetically engineered mouse models of cancer and human cancer cell lines. Notably, TGF-β–SMAD signaling causes apoptosis in pancreatic progenitors harboring KRAS mutations (Bardeesy et al., 2006). Mouse skin and mucosal epithelia showed hyperproliferation offset by TGF-β-dependent apoptosis when harboring a HRAS oncogene, mild hyperplasia when harboring TGFBR2 deletion, and overt tumor formation when both alterations were combined (Guasch et al., 2007). Thus, in these and other cancers, oncogenic mutations sensitize premalignant cells to TGF-β-induced apoptosis.
Epithelial-to-mesenchymal transitions (EMTs) are processes by which epithelial progenitor cells lose polarity, downregulate cell-cell adhesions, migrate and invade for the purpose of generating or regenerating tissues (Nieto et al., 2016). EMTs occur during development, wound healing, and in pathologies including fibrosis and cancer. EMTs are driven by master EMT regulators including SNAIL, ZEB, TWIST, which function as transcriptional repressors of epithelial genes. TGF-β is a potent inducer of EMTs but this function critically depends on inputs from the RAS-MAPK pathway (David et al., 2016; Horiguchi et al., 2009; Janda et al., 2002). However, TGF-β-induced EMT is pro-apoptotic and tumor suppressive in pre-malignant pancreatic progenitors, owing to a conflict with a pro-epithelial program that TGF-β concomitantly induces in these cells (David et al., 2016). A pro-apoptotic role of TGFβ-induced EMT is also observed in mouse mammary epithelial cells (Gal et al., 2008).
Cancer cells with an intact TGF-β pathway can avert its pro-apoptotic effect by somehow decoupling EMT from apoptosis, which in turn allows the cancer cells to use EMT for tumorigenic advantage. Beyond the contribution of TGF-β-induced EMT to tumor invasion and metastatic dissemination, the TGF-β pathway induces gene responses that support the ability of cancer cells to infiltrate and colonize specific organs (David and Massagué, 2018). Examples include the ability of hormone receptor-negative breast cancer cells in primary tumors to respond to TGF-β with induction of angiopoietin-like 4, which enhances the extravasation of these cells as they move into the circulation and lodge into capillaries in the lungs (Padua et al., 2008). Breast cancer cells to instead colonize the bone marrow respond to bone-derived TGF-β with induction of CXCR4, PTHrP, IL-11, CTGF and JAGGED1, which promote osteoclast mobilization and osteolytic metastasis (Figure 3) (Kakonen et al., 2002; Kang et al., 2003; Sethi et al., 2011; Yin et al., 1999). In addition to these effects, growing evidence points at a major role of TGF-β as a mediator of immune suppression in the tumor microenvironment.
TGF-β suppression of tumorigenic inflammation
The master role of TGF-β signaling in controlling immune tolerance and inflammatory responses was first revealed by the analysis of mice with germline null mutations in the TGF-β1 gene, which die early after birth of multiorgan inflammation reminiscent of an autoimmune disorder (Kulkarni et al., 1993; Shull et al., 1992). Subsequently, it was shown that this phenotype can be rescued by loss of function mutations in either MHC class II (Letterio et al., 1996) or β2-microglobulin genes (Kobayashi et al., 1999) implying that loss of TGF-β1 causes an unrestrained adaptive T cell response. Similarly, mice expressing a dominant-negative TGFBR2 construct under the control of the CD4 promoter (Gorelik and Flavell, 2000) or bearing T cell-specific deletion of either Tgfbr2 (Li et al., 2006; Marie et al., 2006), Tgfbr1 (Liu et al., 2008) or Tgfb1 (Li et al., 2007) by means of a CD4-Cre driver, develop enhanced T cell activation leading to a severe inflammatory disease similar to that observed in Tgfb1−/− mice. These phenotypes are consequence of CD4+ T cell activation by self-antigens present in the periphery (Robinson and Gorham, 2007). Overall, these pioneering genetic experiments demonstrated that TGF-β signaling is required for the establishment and maintenance of T cell tolerance during thymic development.
When Tgfbr2 was ablated in T cells using the dLck-Cre driver, which is only turned on after thymocyte positive selection, mice survived to adulthood and neither displayed overt T cell activation nor inflammatory disease in homeostasis (Zhang and Bevan, 2012). However, T cells in these mice exhibited enhanced proliferation and acquisition of an exacerbated effector phenotype in a lymphopenic environment, which was triggered by weak T cell receptor (TCR) stimuli (Zhang and Bevan, 2012). Similar observations were made upon deletion of Tgfbr2 in peripheral T cells of adult mice using inducible a CD4-creERT2 allele (Śledzińska et al., 2013). Therefore, in peripheral T cells, a key function of TGF-β signaling is to restrain T cell expansion and activity in response to exogenous stimuli.
The inflammatory disorders caused by genetic loss of TGF-β signaling are particularly severe in the gastrointestinal tract. TGF-β pathway mutant mice develop either spontaneous colitis or are more susceptible to experimental colitis induced by treatment with dextran sulphate sodium (DSS) (Ihara et al., 2017; Kulkarni et al., 1993; Shull et al., 1992). These phenotypes are also evident in mice with TGF-β pathway deficiencies in either T cells or dendritic cells (Gorelik and Flavell, 2000; Ihara et al., 2017; Ramalingam et al., 2012; Śledzińska et al., 2013). In agreement with these findings, multiple studies have linked dysregulation of TGF-β signaling with pathogenesis of ulcerative colitis or Crohn’s disease (Ihara et al., 2017). These inflammatory syndromes predispose to the development of cancer. Transgenic mice that overexpressed TGF-β1 in T lymphocytes under control of the CD2 promoter exhibited delayed tumor development in experimental models of azoxymethane-induced colonic tumorigenesis whereas expression of dominant negative TGFBR2 in T cells accelerated tumor progression in this model (Becker et al., 2004). This effect was linked to the modulation of IL6/STAT3 signaling in tumor cells, a cytokine input which promotes growth and survival of intestinal cancer cells in the context of inflammation (Becker et al., 2004). Similarly, deletion of Smad4 in T cells led to spontaneous formation of epithelial cancers throughout the gastrointestinal tract as a result of increased levels of pro-inflammatory cytokines IL5, IL6, IL11 and IL13 (Hahn et al., 2011; Kim et al., 2006).
TGF-β signaling also instructs a none-inflammatory program in macrophages of the intestinal epithelium, which exhibit a profound loss of response to inflammatory insults while retaining a bactericidal function (Smythies et al., 2005). This anergic phenotype is partially driven by TGF-β produced by intestinal stromal cells (Smythies et al., 2005). Mice that express dominant negative Tgfbr2 under the control of a macrophage-specific promoter display exacerbated colitis in response to the DSS treatment, an effect that correlates with increased production of proinflammatory cytokines such as IL33 (Rani et al., 2011).
Finally, fibroblast-specific Tgfbr2 knockout mice develop intraepithelial prostate neoplasias and invasive squamous cell carcinomas in the forestomach (Bhowmick et al., 2004). It was initially proposed that Hepatocyte Growth Factor (HGF) secretion by Tgfbr2 mutant fibroblasts led to hyperproliferation of adjacent epithelial cells in the affected tissues (Bhowmick et al., 2004). However, another study revealed DNA damage in epithelial cells of the forestomach due to extensive inflammation caused by loss of Tgfbr2 in fibroblasts (Achyut et al., 2013). This phenotype was enhanced by Helicobacter Pylori infection and ameliorated by anti-inflammatory drugs (Achyut et al., 2013).
Altogether, these studies highlight the pivotal role of TGF-β signaling in regulating tolerance and suppressing inflammatory reactions triggered by commensal and harmful antigens. Loss of TGF-β signaling in tissues exposed to these stimuli, such as the gastrointestinal tract, promotes exacerbated T cell activity and an unrestrained inflammatory reaction. These processes eventually result in DNA damage and increased levels of cytokines and growth factors, which foster an uncontrolled regenerative response and the onset of cancer (Figure 3).
Inhibition of Th1 helper and cytotoxic T cell responses by TGFβ
The most prominent and best-characterized T cell responses against cancers are mediated by cells of the Th1 subset. Naïve T cells cultured with TGF-β cannot differentiate into the Th1 phenotype (Sad and Mosmann, 1994). Conversely, mice lacking TGFBR2 specifically on T cells display enhanced Th1 responses (Gorelik and Flavell, 2000; Li et al., 2006; Marie et al., 2006; Śledzińska et al., 2013; Zhang and Bevan, 2012). TGF-β signaling impinges on the earliest phase of T cell activation by dampening the initial Ca2+ influx triggered T Cell Receptor (TCR) stimulation (Chen et al., 2003a). Consistent with this finding, Tgfbr2 mutant T cells exhibit increased sensitivity to TCR stimulation (Śledzińska et al., 2013). In addition, TGF-β signaling exerts an inhibitory role on T cell differentiation by silencing the expression of two Th1 master transcription factors, TBET and STAT4 (Gorelik et al., 2002; Lin et al., 2005). Blockade of STAT4 activation prevents the production of IFN-γ during the priming phase, whereas loss of TBET expression impairs IFN-γ production during re-stimulation of T cells after initial priming (Gorelik et al., 2002; Lin et al., 2005).
In addition to blocking Th1 differentiation, TGF-β inhibits T cell proliferation and effector function. Early studies revealed that during the priming phase, TGF-β silences the expression of IL-2, the cytokine that elicits subsequent CD4+ T cell proliferation (Brabletz et al., 1993). This effect is mediated by SMAD3 (McKarns et al., 2004) with SMAD4 in association with cofactor TOB1 (Tzachanis et al., 2001). A cytostatic and apoptotic response of T cells to TGF-β has also been linked to the control of several downstream cell-cycle regulators including c-Myc, p21Cip1 and p27Kip1 (Genestier et al., 1999; Wolfraim et al., 2004). As we discussed in the previous section, in vivo experiments using genetically modified mouse models revealed that the main alteration of TGFBR2-deficient peripheral CD4+ and CD8+ T cells is enhanced TCR-dependent activation upon weak antigenic stimulation (Śledzińska et al., 2013; Zhang and Bevan, 2012). These studies also demonstrated that TGFBR2-deficient T cells exhibit enhanced effector phenotypes and functions, such as expression of the receptor KLRG1 and production of granzyme-B and IFN-γ (Śledzińska et al., 2013; Zhang and Bevan, 2012). TGF-β signaling directly inhibits the cytotoxic program of CD8+ T cells (Thomas and Massagué, 2005). Mechanistically, TGF-β-stimulated SMADs together with transcription factor ATF1 repress the promoters of several genes involved the lytic function of CD8+ T cells including granzyme B and IFNγ (Figure 4).
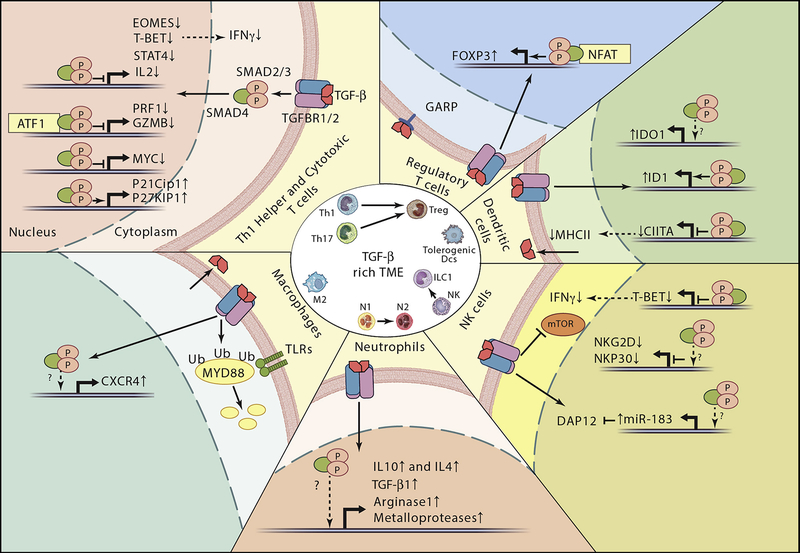
Figure 4. Immune evasion mediated by TGF-β signaling in late stage cancers.
The central circle depicts phenotypic transitions instructed by TGF-beta in the distinct immune cell types present in the TME as described in the text. The outer part summarizes the key processes regulated by TGF-beta signaling in immune cells. We depict direct transcriptional responses driven SMADs, in some cases in cooperation with cell specific transcription factors.
Echoing the above observations, widespread evidence supports a role for a TGF-β-rich TME in suppressing anti-tumor Th1 responses. Transgenic mice engineered to express dominant negative TGFBR2 in CD4+ and CD8+ T cells are resistant to tumor formation upon inoculation of syngeneic cancer cell lines, and this phenomenon is associated with a large expansion of tumor-reactive CD8+ T cells (Gorelik and Flavell, 2001). In an autochthonous genetic model of prostate cancer, TGF-β1 secreted by T cells was shown to mediate tumor evasion from adaptive immunity (Donkor et al., 2011). Authors reported that transgenic dn-TGFBR2 T cells infiltrate both the tumor beds and the tumor-draining lymph nodes, and produce high amounts of IFN-γ and GZMB. These findings suggest a role for TGF-β signaling in inhibiting tumor antigen-specific T cell priming (Donkor et al., 2011). Similarly, adoptively transferred tumor-specific CD8+ T cells modified to express a dominant-negative TGFBR2 infiltrates the tumor mass, display enhanced secretion of IFNγ and cytotoxic products leading to anti-tumor responses in models of renal cancer (Wang et al., 2010). TGF-β attenuates the effector function of antigen-specific CD8+ cells obtained from melanoma patients, a phenomenon that segregates with decreased expression of TBET (Ahmadzadeh and Rosenberg, 2005). Repression of Eomesodermin (EOMES), a homologue of TBET that is required to establish the gene program of effector cytotoxic T cells (Cruz-Guilloty et al., 2009; Pearce et al., 2003), has been also correlated with TGF-β-driven immune evasion in mouse models of melanoma (Yoon et al., 2013).
Tissue resident CD8+ memory cells T cells are maintained associated to tissues through expression of the αEβ7 integrin homodimer (also known CD103), which binds E-cadherin present in the epithelial cell junctions. TGF-β upregulates the expression of αE and β7 subunits, thus facilitating the residence of CD8+ cell in tissues (El-Asady et al., 2005; Zhang and Bevan, 2013). In addition, downregulation of the transcription factors TBET and EOMES by TGF-β is necessary to specify the CD8+CD103+ phenotype (Mackay et al., 2015). The role of CD8+CD103+ in cancer is not completely understood. Several studies have shown that a TGF-β-rich TME fosters CD8+CD103+ cells and that abundance of this T cell subset confers good prognosis (Mami-Chouaib et al., 2018). In addition, checkpoint immunotherapies reinvigorate their cytotoxic function leading to anti-tumor responses (Edwards et al., 2018; Mami-Chouaib et al., 2018). In sharp contrast, a recent study has provide evidence that TGF-β-induced CD8+CD103+ cells present in tumor beds exhibit a tolerogenic phenotype that facilitate immune evasion (Gabriely et al., 2017).
Induction of T regulatory phenotype by TGF-β signaling
Regulatory T cells (Tregs) suppress the function of effector T cells to maintain immune homeostasis and are present in low frequency in healthy tissues. However, their numbers increased in many cancers, where they enforce tolerance to tumor antigens and facilitate immune evasion. TGF-β is key to instruct the regulatory program on T cells. For example, suboptimally stimulated peripheral CD4+ T cells produce TGF-β, that in turn promotes their differentiation to the Treg stage (Strainic et al., 2013; Windhagen et al., 1995). TGF-β in combination with IL2 enforces a suppressor phenotype in ex vivo naïve CD4+ T cells by triggering expression of FOXP3, the master transcription factor of the Treg program (Chen et al., 2003b; Fantini et al., 2004). Consistent with these findings, genetic deletion of both Smad2 and Smad3 resulted in loss of FOXP3 upregulation by TGF-β (Takimoto et al., 2010). Downstream of TGF-β receptors, SMADs cooperate with NFAT transcription factor to bind a distal regulatory element in the FOXP3 locus (Tone et al., 2008) (Figure 4). TGF-β-driven induction of Tregs is counteracted by an environment rich in proinflammatory cytokines, which favors differentiation of CD4+ T cells towards an effector phenotype (Battaglia et al., 2013; Wei et al., 2007).
FoxP3 expression correlates with TGF-β levels in transcriptomic datasets of skin cutaneous melanoma and breast cancer (Ravi et al., 2018), and extensive evidence suggests that the TGFβ-rich environment characteristic of late stage cancers promotes differentiation of T cells to a Treg phenotype. High TGF-β levels secreted by cancer cells induce the Treg program on CD4+ T cells in models of pancreatic cancer (Moo-Young et al., 2009). Furthermore, TGF-β together with prostaglandin E2 trigger Th17-to-Treg transdifferentiation in tumors (Downs-Canner et al., 2017). Conversely, the inhibition of TGF-β and VEGF signaling synergistically reduces the number of Tregs and restores sensitivity to anti–PD-1 and anti–CTLA-4 treatment (Courau et al., 2016).
In addition to inducing a T regulatory program, TGF-β is utilized by Tregs to suppress anti-tumor immune responses (Chen et al., 2005; Mempel et al., 2006). As we discussed above, Tregs carry latent TGF-β1 on their surface via disulfide linkage to transmembrane protein GARP for activation by αV integrins. In B16 melanoma explants, Tregs inhibit the cytotoxic function of CD8+ T cells whereas this immunosuppressive mechanism is prevented by neutralizing antibodies against surface-bound TGF-β (Budhu et al., 2017). Indeed, expression of GARP in tumor-infiltrating Tregs characterizes an enhanced suppressive phenotype that correlates with impaired anti-tumor T cell responses (Kalathil et al., 2013).
TGF-β subversion of dendritic cell function
A central node in regulation of Th1 and Treg-mediated immune responses by TGF-β signaling are dendritic cells (DCs). DCs are the most potent antigen presenting cells and play key roles in tumor immunity. TGF-β inhibits antigen presentation capabilities of DCs in vitro by suppressing expression of MHC-II genes (Nandan and Reiner, 1997; Piskurich et al., 1998). Tgfbr2 deletion in DCs using CD11c-Cre driver causes multiorgan inflammation (Ramalingam et al., 2012). In tumor models, cancer cells instruct DCs to secrete TGF-β, which in turn promotes acquisition of a Treg phenotype by CD4+ T cells (Dumitriu et al., 2009; Ghiringhelli et al., 2005). Furthermore, the TME redirects DC differentiation towards an immature myeloid cell phenotype with potent immune suppressor functions (Papaspyridonos et al., 2015). This phenotypic switch is mediated by transcriptional regulator ID1 downstream of TGF-β signaling (Papaspyridonos et al., 2015). The tolerogenic program of DCs is in part mediated by induction of immunosuppressing molecules indoleamine 2,3-dioxygenase (IDO) and arginase by TGF-β signaling (Belladonna et al., 2008; Pallotta et al., 2011). In models of breast cancer and melanoma, TGF-β signaling mediates immune evasion by upregulating IDO in plasmacytoid DCs and the CCL22 chemokine in myeloid DCs (Hanks et al., 2013). These altered DCs facilitate Treg cell infiltration and immune suppression (Hanks et al., 2013).
TGF-β suppression of Natural Killer cells
Natural killer (NK) cells respond rapidly to virus-infected cells and tumor cells through their unique capacity to recognize stressed cells in the absence of an adaptive response, allowing a rapid immune reaction. TGF-β signaling blocks NK function at multiple levels. It silences IFN-γ and TBET expression in NK cells, thus inhibiting Th1 responses (Laouar et al., 2005; Yu et al., 2006). This mechanism is counteracted by inflammatory signals, which decrease TGFBR2 levels and suppress downstream SMAD signaling in NK cells (Yu et al., 2006). The expression of NKG2D and NKp30, two surface receptors of NK cells that mediate the recognition of stressed and malignant transformed cells, is silenced by TGF-β signaling (Castriconi et al., 2003) (Figure 4). Indeed, there is an inverse correlation between TGF-β1 and NKG2D levels in lung, colorectal carcinomas, and glioblastoma (Crane et al., 2010; Lee et al., 2004). In addition, the levels of DAP12, an adaptor of several cytotoxic receptors including NKG2D, are downregulated by TGF-β-induced miR-183 (Donatelli et al., 2014) (Figure 4).
A controversy exists regarding the role of TGF-β signaling in the ontogeny of NK cells. Early studies found increased NK cell numbers and accelerated maturation during infancy in transgenic mice that express a dominant negative TGFBR2 in NK cells under the control of the cd11c promoter (Laouar et al., 2005; Marcoe et al., 2012). In contrast, ablation of Tgfbr2 in NK cells by means of the Ncr1-cre deleter strain did not cause alterations in NK cell distribution or development. However, these mice exhibited a reduced NK-mediated anti-tumor response to lung metastasis formation (Viel et al., 2016). This study also reveal that TGF-β-mediated suppression of NK activity is due to impairment of mTOR metabolic signaling downstream of stimulatory cytokines such as IL15 (Viel et al., 2016). Finally, NK cells are plastic and can switch phenotype in response to TGF-β (Cortez et al., 2016). Analysis of tumor models revealed that a TGF-β-rich TME facilitates immune evasion by enforcing transdifferentiation of NK cells to innate lymphoid cells type 1 (ILC1) which are devoid of cytotoxic function (Gao et al., 2017) (Figure 4). This process apparently requires SMAD4-independent TGF-β signaling (Cortez et al., 2017).
Regulation of macrophage behavior by TGF-β signaling
It is well established that the TME polarizes macrophages towards a phenotype with anti-inflammatory, immune suppressive and proangiogenic functions, referred to as the M2 phenotype. TGF-β is one of the main immunosuppressive cytokines produced by tumor associated macrophages (TAMs) and, as already mentioned, subsets of human macrophages can mobilize active TGF-β through the activity of integrin αvβ8 and MMP14 (Kelly et al., 2018).
TGF-β instructs distinct programs in cells of the monocyte/macrophage lineage depending on the differentiation state and context. Early studies provided evidence that TGF-β acts as chemoattractant for monocytes to the sites of inflammation, and upregulates adhesion molecules that enable monocyte attachment to the ECM (Allen et al., 1990; Wahl et al., 1993). A similar mechanism may operate in tumors. For example, in models of breast cancer, tumor cells secrete TGF-β, which upregulates CXCR4 in monocytes, while CXCL12 secreted by perivascular fibroblasts attracts these monocytes to tumor beds (Arwert et al., 2018). Subsequently, monocytes differentiate into perivascular macrophages and facilitate tumor cell extravasation by promoting blood vessel leakiness (Arwert et al., 2018).
TGF-β signaling in macrophages also inhibits anti-inflammatory responses mediated by transcription factor NF-κB. The inhibitory SMADs, SMAD6 and SMAD7, have been implicated in this process. TGF-β promotes degradation of MYD88, an adaptor protein utilized by all Toll-like receptors (TLRs), except TLR3, to activate NF-κB signaling. SMAD6 induced polyubiquitination of MYD88 through recruitment of E3-ubiquitin ligases SMURF1/2 in TGF-β-stimulated peritoneal macrophages (Lee et al., 2011; Naiki et al., 2005) (Figure 4). SMAD6 also sequesters adaptor protein Pellino-1, which is required downstream of IL1R-TLR to promote an inflammatory response (Choi et al., 2006). Furthermore, TGF-β1 suppresses the inflammatory phenotype of macrophages by crosstalk with tumor necrosis factor signaling pathway through SMAD7, which blocks the activity of the TNF-induced kinase TAK1 (Hong et al., 2007). Although these mechanisms have not been formally investigated in the context of cancer, it is conceivable that a TGF-β-rich TME may contribute to immune evasion by dampening the inflammatory functions of macrophages. Indeed, the acquisition of a M2 phenotype in TAMs correlates with inhibition of NF-κB activity (Porta et al., 2009).
TGF-β signaling in myeloid cells of the neutrophil lineage
Neutrophils represent around 80% of all blood leukocytes and play key roles in controlling infection. Patients with various cancer types, including breast, lung and colorectal cancer, exhibit increased numbers of circulating neutrophils, often associated with poor prognosis (Gentles et al., 2015; Templeton et al., 2014). Functional analyses in models of cancer have revealed important roles of neutrophils during tumor progression (Coffelt et al., 2016). Paralleling the phenomenon of TAM polarization, tumor-associated neutrophils (TANs) adopt either an anti-tumorigenic or protumorigenic program (Fridlender et al., 2009) (Figure 4). In mice bearing mesothelioma tumors, treatment with TGF-β inhibitors enforces the anti-tumorigenic phenotype in TANs that includes cytolytic activity and elevated expression of pro-inflammatory cytokines (Fridlender et al., 2009).
A debate exists about whether TANs represent mature neutrophils or rather immature polymorphonuclear cells (Coffelt et al., 2016; Gabrilovich, 2017). It is well established that chronic infection, inflammation, or cancer induce a persistent stimulation of myelopoiesis that results in the production of cells similar to neutrophils and monocytes in morphology and phenotype yet with potent ability to suppress immune responses (Coffelt et al., 2016; Gabrilovich, 2017). These cells termed myeloid-derived suppressor cells (MDSCs) infiltrate tumors and are important players during cancer immune evasion. 80% of MDSCs display a polymorphonuclear phenotype (PMN-MDSCs) similar to that of neutrophils and can be recognized in flow cytometry analysis by the neutrophil surface marker combination CD11b+Ly6G+ (Coffelt et al., 2016; Gabrilovich, 2017). TGF-β signaling in MDSCs enhances tumor progression and metastasis in animal models. Depletion of MDSCs in a breast cancer model inhibited the therapeutic effects exerted by systemic administration of anti-TGF-β antibodies (Li et al., 2012). Indeed, mice with myeloid-specific deletion of Tgfbr2 were largely resistant to metastasis formation upon transplantation of syngeneic breast cancer, lung cancer and melanoma cell lines (Pang et al., 2013). This effect was linked to downregulation of type 2 cytokines in MDSCs and increased expression of IFNγ in CD8+ T cells leading to enhanced anti-tumor responses (Pang et al., 2013). Interestingly, in this study, primary tumor growth was not affected by loss of Tgfbr2 in myeloid cells, which suggests a specific role for TGF-β-activated MDSCs in the metastatic niche.
In contrast to the tumor promoting effects of TGF-β signaling in neutrophils and MDSCs, several studies have shown that genetic inhibition of TGF-β signaling in cancer cells results in recruitment of immature myeloid cells and MDSCs to the TME. A genetic mouse model of invasive colorectal cancer generated by inactivating Apc and Smad4 in the intestinal epithelium exhibited prominent infiltration with CCR1+ immature myeloid cells (Kitamura et al., 2007). Loss of TGF-β signaling in Apc mutant colon cancer cells led to upregulation of chemoattractant CCL9, which in turn mediates recruitment of the immature myeloid cell population (Kitamura et al., 2007). Furthermore, immature myeloid cells present in the TME expressed matrix metalloproteinases that facilitate tumor cell invasion and metastasis (Kitamura et al., 2007, 2010). In a similar study, knockout of Tgfbr2 in mammary carcinoma cells increased MDSC infiltration. In this case, the effect was mediated by upregulation of CXCL5 in Tgfbr2 deficient breast cancer cells (Yang et al., 2008). MDSCs also produced matrix metalloproteases that contributed to tumor cell invasion and metastasis in this model (Yang et al., 2008). Therefore, the anti-tumoral effects of TGF-β on cancer cells result, at least in part, from suppression of expression of myeloid cell chemoattractants.
Cancer-associated fibroblasts and TGF-β-driven immune evasion
TME of advanced stage cancers is often characterized by an abundance of fibroblasts. These cancer-associated fibroblasts (CAFs) are a heterogeneous group of cells dedicated to producing the main components of the ECM including collagens, elastins, and fibronectin, but also an array of cytokines that regulate tumor properties. CAFs represent the main TGF-β producers in many tumor types. A prime example is colorectal cancer, where CAFs secrete TGF-β1 and TGF-β3, and their abundance correlates with TGF-β pathway activity in distinct TME cell types, including T cells and macrophages (Calon et al., 2012, 2015). In addition, TGF-β activates a gene expression program in CAFs that is tightly associated with poor prognosis in CRC patients (Calon et al., 2012, 2015; Isella et al., 2015).
These findings are further strengthened by the fact that distinct molecular classifications of colorectal cancer have consistently identified a poor prognosis subtype characterized by elevated TGF-β levels and abundant CAFs (Calon et al., 2015; Guinney et al., 2015; Isella et al., 2015). The generation of genetic mouse models that develop human-like CRCs has offered insights into the interaction of the CAF-rich TGF-β-activated TME with the immune system (Tauriello et al., 2018). Elevated TGF-β levels produced by CAFs and other TME cell types exclude CD4+ and CD8+ T cells from the tumor center (Tauriello et al., 2018). T cell exclusion is associated with adverse outcome in colorectal cancer patients (Galon et al., 2006). Treatment of these mouse colorectal models with the TGFBR1 inhibitor Galunisertib triggered T cell infiltration, and upon treatment, primary tumors and metastases became susceptible to checkpoint therapies (Tauriello et al., 2018). Dual Galunisertib/PDL1 therapy induced a potent immune response, with increased TBET and IFNγ levels in CD4+ T cells and elevated granzyme B production in CD8+ cells, which eradicated overt metastatic disease (Tauriello et al., 2018). Similar therapeutic responses to dual Galunisertib/PDL1 treatment were observed in genetic mouse models of serrated colorectal cancer (Nakanishi et al., 2018).
A link between CAFs and immune evasion is further supported by another study that searched for determinants of response to checkpoint immunotherapies (Mariathasan et al., 2018). By interrogating a transcriptomic cohort of urothelial cancer, authors found that a subset of patients that did not respond to PD-L1 treatment bore tumors rich in TGF-β-activated CAF gene program. These tumors presented an immune-excluded phenotype (Mariathasan et al., 2018). This study further showed that treatment with an anti-Pan-TGF-β antibody reverted T cell exclusion and sensitized mouse tumor models to PD-L1 treatment (Mariathasan et al., 2018).
At present, the association between TGF-β-activated CAFs and cancer immune evasion remains largely correlative and therefore we can only speculate about the mechanisms utilized by CAFs to block T cell infiltration and function. CAFs secrete TGF-β, which can directly suppress several immune cell types through the mechanisms described herein. In addition, and by analogy with tissue fibrosis (Kim et al., 2018; Wipff et al., 2007), highly contractile CAFs may likely facilitate the release of active TGF-β from latent complexes stored in the TME. Alternatively (or additionally) particular genes activated by TGF-β signaling in CAFs may inhibit the migration and function of immune cells present in the tumor stroma. Supporting this hypothesis, a transcriptomic signature of ECM genes driven by TGF-β in CAFs predicts failure of PD-1 therapy (Chakravarthy et al., 2018).
TGF-β-based immunotherapies
The development of therapies based on inhibiting the TGF-β pathway has had a slow progress. Its low priority in the pharmaceutical industry pipeline is likely explained by two observations. First, the long-standing evidence supporting a tumor suppressor role for the TGF-β pathway in several contexts, which raised concerns regarding the possibility that inhibition of TGF-β signaling could worsen rather than cure cancer. Second, the finding that first generation TGFBR1 inhibitors triggered overt cardiac toxicity in experimental models (Anderton et al., 2011). Nevertheless, as we discussed throughout this review article, over the past years it has become evident that genetic ablation of TGF-β signaling pathway components in distinct immune cell types, including CD4+, CD8+ T cells, NK or dendritic cells, triggers robust anti-tumor responses in preclinical models of cancer. These studies, together with the overwhelming evidence that late-stage tumors exploit TGF-β for invasion and metastasis, have sparked the development of multiple programs aimed at blocking TGF-β signaling in cancer. Table I summarizes the main strategies, some of which are currently in clinical trials. The most extensively tested compound is Galunisertib (LY21577299), a small molecule that inhibits TGFBR1 kinase activity. Galunisertib in combination with gemcitabine has shown modest but significant therapeutic activity in a phase 2 clinical trial for pancreatic cancer (Melisi et al., 2018). Another phase 2 clinical trial demonstrated therapeutic responses in a subset of Hepatocellular carcinoma patients treated with Galunisertib as monotherapy (Faivre et al., 2014). Of note, Galunisertib displayed a safe profile across various clinical trials, without discernable cardiac toxicities (Kovacs et al., 2015).
Table I.
Therapeutic strategies to block TGF-β signaling in cancer and their progress to the clinic.
| Therapy | Target | Drug | Progress to clinic |
|---|---|---|---|
| Small molecule inhibitors | TGFBR1 kinase | Galunisertib | • Multiple completed safety and efficacy phase 1 clinical trials in HCC, glioblastoma and other tumor types. • Several on going phase 1/2 trials: - Metastatic breast cancer (GAL + Radiation therapy) - Metastatic pancreatic cancer (GAL + Durvalumab) - Advanced Hepatocellular Carcinoma (GAL + Stereotactic radiotherapy). - Metastatic Androgen Receptor Negative Triple - Negative Breast Cancer (Galunisertib and Paclitaxel) - Colorectal cancer (GAL + Capecitabine) - Rectal cancer (GAL + chemoradiation) - Advanced refractory solid tumors (GAL + Nivolumab) - Metastatic Prostate cancer (GAL + Enzalutamide) |
| Vactosertib | • Safety and efficacy phase trials in advanced stage solid tumors • Several on going phase 1/2 trials: - Metastatic Gastric Cancer (VAC + Paclitaxel) - Advanced NSCLC (VAC + Durvalumab) - Metastatic Colorectal and Gastric cancer (VAC + Pembrolizumab) - Advanced desmoid tumors (VAC + Imatinib) |
||
| LY3200882 | • Phase 1: safety and dose escalation in solid tumors | ||
| PF-06952229 | • Phase 1: safety and dose escalation in breast cancer and prostate cancer in monotherapy or combination with several drugs (palbociclib, Letrozole, Enzalutamide) | ||
| AZ12601011 AZ12799734 |
• Preclinical development (Spender et al., 2019) | ||
| Antibodies | Blocking pan-TGF- β (TGF-β1, TGF-β2, TGF-β3) | Fresolimumab | • Completed safety and efficacy phase 1 trials in renal cell carcinoma, melanoma and glioma. • On going phase 1/2 trials: - Relapsed malignant mesothelioma - Advanced renal cell carcinoma and melanoma - Early stage NSCLC (FRESO + stereotactic ablative radiotherapy). - Metastatic breast cancer (FRESO + Radiation) |
| Blocking pan-TGF-β | SAR439459 | Phase 1 (safety and dose escalation) for advanced solid tumors in monotherapy or combination with anti-PD1 antibodies. | |
| Blocking pan-TGF-β | NIS793 | Phase 1 in combination with anti-PD-1 antibodies for patients with advanced malignancies (breast, lung, HCC, CRC, Pancreatic cancer, Renal cancer) | |
| Blocking TGF-β1 & TGF-β2 specific | XPA-42-089 | Preclinical cancer models (Dodagatta-Marri et al., 2019) | |
| Chimeric antibody-TGF-β traps | CTLA4- TGFβRII | Preclinical cancer models (Ravi et al., 2018) | |
| PDL1-TGFβRII (M7824) | • Ongoing Phase 1/2: - NSCLC (compared to Pembrolizumab) - Triple negative breast cancer (M7824 + Eribulin) - Prostate cancer - Metastatic colorectal cancer - Cholangiocarcinoma and Gallbladdder |
||
| GARP | ABBV151 | Phase 1 for advanced solid tumors as monotherapy or in combination with PD-1 antibodies. | |
| αvβ8 Integrins | Preclinical cancer models (Takasaka et al., 2018) | ||
| LAP | Preclinical cancer models (Gabriely et al., 2017) | ||
| Receptor-based TGF-β traps | AVID200 | Phase 1 (safety and dose escalation) for advanced and metastatic solid tumors. | |
| Adoptive cell transfer | Autologous CD8+ T cells expressing a dnTGFR2 | Phase 1 trial in chemorefractory Epstein Barr Hodgkin lymphoma |
Preclinical studies suggest that the immunological effects of Galunisertib are strongly potentiated by combination with checkpoint inhibitors whereas TGF-β inhibition as monotherapy may exhibit limited efficacy (Holmgaard et al., 2018; Mariathasan et al., 2018; Nakanishi et al., 2018; Tauriello et al., 2018). Based on these findings, several clinical trials aimed at testing Galunisertib in combination with anti-PD1 antibodies have been recently launched. In addition, TGFBR1 small molecule inhibitors that are more potent and specific than Galunisertib have been developed and are currently being tested in patients. One of them, Vactosertib (TEW-7197), in combination with chemotherapy or antibodies against immune checkpoint molecules is in phase 1/2 clinical trials for several cancer types.
The use of antibodies that block systemic TGF-β has shown promising anti-tumor responses in multiple preclinical studies. For instance, treatment with the 1D11 anti-pan-TGF-β antibody enhances the priming of tumor-reactive CD8+ by dendritic cells upon irradiation of subcutaneous tumors (Vanpouille-Box et al., 2015). This combinatorial therapy inhibits tumor growth and metastases, and is further enhanced by treatment with PD-1 antibodies (Vanpouille-Box et al., 2015). In a similar study, radiotherapy in combination with blocking TGF-β antibodies enhances systemic anti-tumor responses as shown by analysis of non-irradiated lesions in the same mice, i.e. abscopal effects (Rodríguez-Ruiz et al., 2019). A humanized derivative of the 1D11 antibody, Fresolimumab, is being evaluated for the treatment of renal cell carcinoma, melanoma, non-small cell lung cancer, and metastatic breast cancer. Reported results from this clinical trial show that Fresolimumab in combination with focal radiotherapy increases the overall survival of patients with metastatic breast cancer (Formenti et al., 2018). This effect correlated with a large increase in the number of CD8+ central memory cell pool suggesting that the treatment triggered an immune response against the tumor (Formenti et al., 2018). Several other pan-TGF-β blocking antibodies are in phase 1 clinical trials (Table I).
There are ongoing efforts to develop TGF-β isoform-specific therapies. For example, the antibody XPA-42–089 binds TGF-β1 and TGF-β2 but not TGF-β3, and exhibit a robust synergism with PD-1 during the treatment of both genetic and mutagen-induced mouse squamous cell carcinomas (Dodagatta-Marri et al., 2019). AVID200 is a computational-designed receptor ectodomain-based trap that binds and neutralizes TGF-β1 and TGF-β3 but not TGF-β2 (O’Connor-McCourt et al., 2017). It is currently in phase 1 trials for patients with advanced solid tumors.
Another interesting approach are therapies based of anti-CTLA4 or anti-PD-L1 antibodies engineered as fusions with the extracellular domain of TGFBR2 (Ravi et al., 2018). These bi-functional molecules potentiate the efficacy of immunotherapies in mouse models of cancer by quenching TGF-β. The anti-CTLA4-TGFBR2 molecule reduced accumulation of Treg cells in immune reconstituted NSG mice bearing patient-derived melanoma and exhibited enhanced anti-tumor efficacy compared with standard anti-CTLA4 monotherapy (Ravi et al., 2018). Likewise, an anti-PDL1-TGFBR2 chimera exerted more pronounced anti-tumor responses than PD-L1 antibodies (Ravi et al., 2018). The chimeric anti-PDL1-TGFBR2 antibody is currently being tested in phase 1 clinical trials for several indications.
Blockade of TGF-β signaling also improves the outcome of immunotherapies based on adoptive cells transfer. For instance, infused CD8+ cells reactive against an autochthonous tumor model of prostate cancer mount enhanced anti-tumor responses if TGF-β signaling is prevented by transgenic expression of a dominant negative TGFBR2 (Bendle et al., 2013; Zhang et al., 2005). This strategy was recently tested in patients with chemorefractory Epstein Barr Hodgkin lymphoma (Bollard et al., 2018). The authors reported that autologous CD8+ cells directed against the Epstein Barr virus–derived tumor antigens that have been engineered to express a dominant negative TGFBR2 trigger complete responses in 4 out of 7 patients (Bollard et al., 2018). Although the patient number is small, the study suggests improved efficacy compared to a previous trial using TGF-β sensitive CD8+ T cells (Bollard et al., 2018). This strategy might be widely applicable to a wide range of adoptive cell transfer therapies.
Finally, the complex mechanism of active TGF-β release from latent complexes also offers opportunities for therapeutic intervention. Antibodies against GARP that blocked its binding to latent TGF-β prevented the formation of lung metastases generated by breast cancer cell lines implanted orthotopically (Metelli et al., 2016). Another study characterized the activity of antibodies that inhibit active TGF-β release from Treg cells through binding to a conformational epitope in GARP (Cuende et al., 2015). One of them, ABBV151, is being tested in phase 1 clinical trials for advanced solid tumors. Antibodies that bind the LAP domain of latent TGF-β elicit immune responses against tumors in models of melanoma, colorectal carcinoma, and glioblastoma (Gabriely et al., 2017). Similarly, an antibody against αvβ8 integrin that blocks the release of active TGF-β by cancer cells unleashed the immune system against tumors in preclinical models and this therapeutic effect was largely potentiated by combining anti-PD1 antibodies (Takasaka et al., 2018).
Concluding Remarks
The basic elements in the TGF-β signaling cascade were elucidated more than a decade ago. During the past few years, it has become evident that the pleiotropic nature of TGFβ responses is the result of context dependent transcriptional programs orchestrated through the interaction of SMADs with distinct tissue and cell specific transcription factors. Rewiring of transcriptional circuits in cancer cells -as result of genetic and epigenetic alterations- changes the output of TGF-β signaling from a tumor suppressing function to different tumor promoting programs that facilitate growth, invasion and metastasis. Concomitantly, TGF-β signaling operates as a major suppressor of the adaptive and innate immune responses during tumor progression. The inhibition of the immune system by TGF-β signaling leads to opposing effects depending on the context. In tissues subjected to continuous antigenic stimuli, such as the gastrointestinal tract, TGF-β limits adaptive responses and dampens inflammation. In this setting, loss of TGF-β signaling leads to a pro-inflammatory environment that facilitates the onset of cancer. In contrast, in the advanced stages of the disease, TGF-β signaling acts as an important mechanism of immune evasion in several tumor types.
The snapshots of TGF-β effects on tumor immunity covered here illustrate how distinct immune cell types present in the TME react to TGF-β in variety of experimental systems. In most cases, observations in tumor models echo the function of the TGF-β pathway in promoting tolerance and suppressing immune responses during homeostasis and infection. It remains unclear, however, how the individual cell responses described here are orchestrated in a given tumor. Do they occur in concert or sequentially along tumor progression? Which TGF-β activated immune cell types are relevant and which tumor subtypes utilize them? Addressing these questions will require cancer models that faithfully reproduce human disease. Genomic analyses, including single cell profiling, should also help understand the ecology of TGF-β-mediated immune evasion. These are particular important aspects that may contribute to rationalize the use of upcoming TGF-beta-based immunotherapies. In this regard, the observed synergism between TGF-β inhibition and checkpoint blockade perhaps reflects a hierarchy of immune suppressive events whereby TGF-β dampens the initial immune response such as the ability of T cells to infiltrate tumors and the acquisition of effector program, whereas PD1/PDL1 signaling operates at a later stage by suppressing T cell effector functions and causing exhaustion. These investigations together with the development of more potent and specific TGF-β pathway inhibitors holds promise for the treatment of prevalent tumor types that thrive in a TGF-β-rich environment.
Acknowledgments
We apologize to authors whose papers could not be cited due to space restrictions. We thank Elena Sancho for proofreading the manuscript. Work in the laboratory of Eduard Batlle is supported by the European Research Council Advanced Grant 340176, MINECO I+D Retos 2017 SAF2017-86782-R, Fundación BBVA, World Wide Cancer Research (WWCR) and Association Española de Investigation contra el Cancer (AECC). Work in the laboratory of Joan Massagué is supported by NIH grants R01-CA34610, P01-CA94060, P01-CA129243, U54-CA209975, U2C-CA233284 and P30-CA008748 and The Alan and Sandra Gerry Metastasis and Tumour Ecosystems Center at MSKCC.
Footnotes
Declaration of Interests
JM is a scientific advisor and owns company stock in Scholar Rock. EB declares no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Achyut BR, Bader DA, Robles AI, Wangsa D, Harris CC, Ried T, and Yang L (2013). Inflammation-Mediated Genetic and Epigenetic Alterations Drive Cancer Development in the Neighboring Epithelium upon Stromal Abrogation of TGF-β Signaling. PLoS Genet. 9, e1003251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadzadeh M, and Rosenberg SA (2005). TGF-beta 1 attenuates the acquisition and expression of effector function by tumor antigen-specific human memory CD8 T cells. J. Immunol. 174, 5215–5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JB, Manthey CL, Hand AR, Ohura K, Ellingsworth L, and Wahl SM (1990). Rapid onset synovial inflammation and hyperplasia induced by transforming growth factor beta. J. Exp. Med. 171, 231–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderton MJ, Mellor HR, Bell A, Sadler C, Pass M, Powell S, Steele SJ, Roberts RRA, and Heier A (2011). Induction of Heart Valve Lesions by Small-Molecule ALK5 Inhibitors. Toxicol. Pathol. 39, 916–924. [DOI] [PubMed] [Google Scholar]
- Annes JP, Chen Y, Munger JS, and Rifkin DB (2004). Integrin α V β 6 - mediated activation of latent TGF-β requires the latent TGF-β binding protein-1. J. Cell Biol. 165, 723–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arwert EN, Harney AS, Entenberg D, Wang Y, Sahai E, Pollard JW, and Condeelis JS (2018). A Unidirectional Transition from Migratory to Perivascular Macrophage Is Required for Tumor Cell Intravasation. Cell Rep. 23, 1239–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeesy N, Cheng K-H, Berger JH, Chu GC, Pahler J, Olson P, Hezel AF, Horner J, Lauwers GY, Hanahan D, et al. (2006). Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 20, 3130–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia A, Buzzonetti A, Baranello C, Fanelli M, Fossati M, Catzola V, Scambia G, and Fattorossi A (2013). Interleukin-21 (IL-21) synergizes with IL-2 to enhance T-cell receptor-induced human T-cell proliferation and counteracts IL-2/transforming growth factor-β-induced regulatory T-cell development. Immunology 139, 109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker C, Fantini MC, Schramm C, Lehr H. a, Wirtz S, Nikolaev A, Burg J, Strand S, Kiesslich R, Huber S, et al. (2004). TGF-beta suppresses tumor progression in colon cancer by inhibition of IL-6 trans-signaling. Immunity 21, 491–501. [DOI] [PubMed] [Google Scholar]
- Belladonna ML, Volpi C, Bianchi R, Vacca C, Orabona C, Pallotta MT, Boon L, Gizzi S, Fioretti MC, Grohmann U, et al. (2008). Cutting edge: Autocrine TGF-beta sustains default tolerogenesis by IDO-competent dendritic cells. J. Immunol. 181, 5194–5198. [DOI] [PubMed] [Google Scholar]
- Bendle GM, Linnemann C, Bies L, Song J-Y, and Schumacher TNM (2013). Blockade of TGF-β signaling greatly enhances the efficacy of TCR gene therapy of cancer. J. Immunol. 191, 3232–3239. [DOI] [PubMed] [Google Scholar]
- Bhowmick NA, Chytil A, Plieth D, Gorska AE, Dumont N, Shappell S, Washington MK, Neilson EG, and Moses HL (2004). TGF- Signaling in Fibroblasts Modulates the Oncogenic Potential of Adjacent Epithelia. Science (80-. ). 303, 848–851. [DOI] [PubMed] [Google Scholar]
- Bollard CM, Tripic T, Russell Cruz C, Dotti G, Gottschalk S, Torrano V, Dakhova O, Carrum G, Ramos CA, Liu H, et al. (2018). Tumor-Specific T-Cells Engineered to Overcome Tumor Immune Evasion Induce Clinical Responses in Patients With Relapsed Hodgkin Lymphoma. J Clin Oncol 36, 1128–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz T, Pfeuffer I, Schorr E, Siebelt F, Wirth T, and Serfling E (1993). Transforming growth factor beta and cyclosporin A inhibit the inducible activity of the interleukin-2 gene in T cells through a noncanonical octamer-binding site. Mol. Cell. Biol. 13, 1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhu S, Schaer DA, Li Y, Toledo-Crow R, Panageas K, Yang X, Zhong H, Houghton AN, Silverstein SC, Merghoub T, et al. (2017). Blockade of surface-bound TGF-β on regulatory T cells abrogates suppression of effector T cell function in the tumor microenvironment. Sci. Signal. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calon A, Espinet E, Palomo-Ponce S, Tauriello DVF, Iglesias M, Céspedes MV, Sevillano M, Nadal C, Jung P, Zhang XHF, et al. (2012). Dependency of Colorectal Cancer on a TGF-β-Driven Program in Stromal Cells for Metastasis Initiation. Cancer Cell 22, 571–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calon A, Lonardo E, Berenguer-llergo A, Espinet E, Hernando-momblona X, Iglesias M, Sevillano M, Palomo-ponce S, Tauriello DVF, Byrom D, et al. (2015). Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 47, 320–329. [DOI] [PubMed] [Google Scholar]
- Castriconi R, Cantoni C, Chiesa, Della M, Vitale M, Marcenaro E, Conte R, Biassoni R, Bottino C, Moretta L, and Moretta A (2003). Transforming growth factor β1 inhibits expression of NKp30 and NKG2D receptors: Consequences for the NK-mediated killing of dendritic cells. Proc. Natl. Acad. Sci. 100, 4120–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy A, Khan L, Bensler NP, Bose P, and De Carvalho DD (2018). TGF-β-associated extracellular matrix genes link cancer-associated fibroblasts to immune evasion and immunotherapy failure. Nat. Commun. 9, 4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charney RM, Forouzmand E, Cho JS, Cheung J, Paraiso KD, Yasuoka Y, Takahashi S, Taira M, Blitz IL, Xie X, et al. (2017). Foxh1 Occupies cis-Regulatory Modules Prior to Dynamic Transcription Factor Interactions Controlling the Mesendoderm Gene Program. Dev. Cell 40, 595–607.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, and ten Dijke P (2016). Immunoregulation by members of the TGFβ superfamily. Nat. Rev. Immunol. 16, 723–740. [DOI] [PubMed] [Google Scholar]
- Chen C-H, Seguin-Devaux C, Burke NA, Oriss TB, Watkins SC, Clipstone N, and Ray A (2003a). Transforming growth factor beta blocks Tec kinase phosphorylation, Ca2+ influx, and NFATc translocation causing inhibition of T cell differentiation. J. Exp. Med. 197, 1689–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-R, Kang Y, Siegel PM, and Massagué J (2002). E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell 110, 19–32. [DOI] [PubMed] [Google Scholar]
- Chen M-L, Pittet MJ, Gorelik L, Flavell RA, Weissleder R, von Boehmer H, and Khazaie K (2005). Regulatory T cells suppress tumor-specific CD8 T cell cytotoxicity through TGF- signals in vivo. Proc. Natl. Acad. Sci. 102, 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Jin W, Hardegen N, Lei K-J, Li L, Marinos N, McGrady G, and Wahl SM (2003b). Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 198, 1875–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Rubock MJ, and Whitman M (1996). A transcriptional partner for MAD proteins in TGF-beta signalling. Nature 383, 691–696. [DOI] [PubMed] [Google Scholar]
- Choi K-C, Lee YS, Lim S, Choi HK, Lee C-H, Lee E-K, Hong S, Kim I-H, Kim S-J, and Park SH (2006). Smad6 negatively regulates interleukin 1receptor–Toll-like receptor signaling through direct interaction with the adaptor Pellino-1. Nat. Immunol. 7, 1057–1065. [DOI] [PubMed] [Google Scholar]
- Coffelt SB, Wellenstein MD, and de Visser KE (2016). Neutrophils in cancer: neutral no more. Nat. Rev. Cancer 16, 431–446. [DOI] [PubMed] [Google Scholar]
- Cortez VS, Cervantes-Barragan L, Robinette ML, Bando JK, Wang Y, Geiger TL, Gilfillan S, Fuchs A, Vivier E, Sun JC, et al. (2016). Transforming Growth Factor-β Signaling Guides the Differentiation of Innate Lymphoid Cells in Salivary Glands. Immunity 44, 1127–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez VS, Ulland TK, Cervantes-Barragan L, Bando JK, Robinette ML, Wang Q, White AJ, Gilfillan S, Cella M, and Colonna M (2017). SMAD4 impedes the conversion of NK cells into ILC1-like cells by curtailing non-canonical TGF-β signaling. Nat. Immunol. 18, 995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courau T, Nehar-Belaid D, Florez L, Levacher B, Vazquez T, Brimaud F, Bellier B, and Klatzmann D (2016). TGF-β and VEGF cooperatively control the immunotolerant tumor environment and the efficacy of cancer immunotherapies. JCI Insight 1, e85974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane CA, Han SJ, Barry JJ, Ahn BJ, Lanier LL, and Parsa AT (2010). TGF- downregulates the activating receptor NKG2D on NK cells and CD8+ T cells in glioma patients. Neuro. Oncol. 12, 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford SE, Stellmach V, Murphy-Ullrich JE, Ribeiro SM, Lawler J, Hynes RO, Boivin GP, and Bouck N (1998). Thrombospondin-1 is a major activator of TGF-beta1 in vivo. Cell 93, 1159–1170. [DOI] [PubMed] [Google Scholar]
- Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG, Groner Y, and Rao A (2009). Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med. 206, 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuende J, Liénart S, Dedobbeleer O, van der Woning B, De Boeck G, Stockis J, Huygens C, Colau D, Somja J, Delvenne P, et al. (2015). Monoclonal antibodies against GARP/TGF-β1 complexes inhibit the immunosuppressive activity of human regulatory T cells in vivo. Sci. Transl. Med. 7, 284ra56. [DOI] [PubMed] [Google Scholar]
- David CJ, and Massagué J (2018). Contextual determinants of TGFβ action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol. 19, 419–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Huang Y-H, Chen M, Su J, Zou Y, Bardeesy N, Iacobuzio-Donahue CA, and Massagué J (2016). TGF-β Tumor Suppression through a Lethal EMT. Cell 164, 1015–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, and Hata A (2008). SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454, 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodagatta-Marri E, Meyer DS, Reeves MQ, Paniagua R, To MD, Binnewies M, Broz ML, Mori H, Wu D, Adoumie M, et al. (2019). α-PD-1 therapy elevates Treg/Th balance and increases tumor cell pSmad3 that are both targeted by α-TGFβ antibody to promote durable rejection and immunity in squamous cell carcinomas. J. Immunother. Cancer 7, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donatelli SS, Zhou J-M, Gilvary DL, Eksioglu EA, Chen X, Cress WD, Haura EB, Schabath MB, Coppola D, Wei S, et al. (2014). TGF-β-inducible microRNA-183 silences tumor-associated natural killer cells. Proc. Natl. Acad. Sci. U. S. A. 111, 4203–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkor MK, Sarkar A, Savage PA, Franklin RA, Johnson LK, Jungbluth AA, Allison JP, and Li MO (2011). T cell surveillance of oncogene-induced prostate cancer is impeded by T cell-derived TGF-β1 cytokine. Immunity 35, 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs-Canner S, Berkey S, Delgoffe GM, Edwards RP, Curiel T, Odunsi K, Bartlett DL, and Obermajer N (2017). Suppressive IL-17A+Foxp3+ and ex-Th17 IL-17AnegFoxp3+ Treg cells are a source of tumour-associated Treg cells. Nat. Commun. 8, 14649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitriu IE, Dunbar DR, Howie SE, Sethi T, and Gregory CD (2009). Human Dendritic Cells Produce TGF- 1 under the Influence of Lung Carcinoma Cells and Prime the Differentiation of CD4+CD25+Foxp3+ Regulatory T Cells. J. Immunol. 182, 2795–2807. [DOI] [PubMed] [Google Scholar]
- Edwards J, Wilmott JS, Madore J, Gide TN, Quek C, Tasker A, Ferguson A, Chen J, Hewavisenti R, Hersey P, et al. (2018). CD103 + Tumor-Resident CD8 + T Cells Are Associated with Improved Survival in Immunotherapy-Naïve Melanoma Patients and Expand Significantly During Anti–PD-1 Treatment. Clin. Cancer Res. 24, 3036–3045. [DOI] [PubMed] [Google Scholar]
- El-Asady R, Yuan R, Liu K, Wang D, Gress RE, Lucas PJ, Drachenberg CB, and Hadley GA (2005). TGF-{beta}-dependent CD103 expression by CD8(+) T cells promotes selective destruction of the host intestinal epithelium during graft-versus-host disease. J. Exp. Med. 201, 1647–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre SJ, Santoro A, Kelley RK, Merle P, Gane E, Douillard J-Y, Waldschmidt D, Mulcahy MF, Costentin C, Minguez B, et al. (2014). A phase 2 study of a novel transforming growth factor-beta (TGF-β1) receptor I kinase inhibitor, LY2157299 monohydrate (LY), in patients with advanced hepatocellular carcinoma (HCC). J. Clin. Oncol. 32, LBA173. [Google Scholar]
- Fantini MC, Becker C, Monteleone G, Pallone F, Galle PR, and Neurath MF (2004). Cutting Edge: TGF-β Induces a Regulatory Phenotype in CD4+CD25− T Cells through Foxp3 Induction and Down-Regulation of Smad7. J. Immunol. 172, 5149–5153. [DOI] [PubMed] [Google Scholar]
- Flavell R. a, Sanjabi S, Wrzesinski SH, and Licona-Limón P (2010). The polarization of immune cells in the tumour environment by TGFbeta. Nat. Rev. Immunol. 10, 554–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formenti SC, Lee P, Adams S, Goldberg JD, Li X, Xie MW, Ratikan JA, Felix C, Hwang L, Faull KF, et al. (2018). Focal Irradiation and Systemic TGFβ Blockade in Metastatic Breast Cancer. Clin. Cancer Res. 24, 2493–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridlender ZG, Sun J, Kim S, Kapoor V, Cheng G, Ling L, Worthen GS, and Albelda SM (2009). Polarization of Tumor-Associated Neutrophil Phenotype by TGF-β: “N1” versus “N2” TAN. Cancer Cell 16, 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriely G, da Cunha AP, Rezende RM, Kenyon B, Madi A, Vandeventer T, Skillin N, Rubino S, Garo L, Mazzola MA, et al. (2017). Targeting latency-associated peptide promotes antitumor immunity. Sci. Immunol. 2, eaaj1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich DI (2017). Myeloid-Derived Suppressor Cells. Cancer Immunol. Res. 5, 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal A, Sjöblom T, Fedorova L, Imreh S, Beug H, and Moustakas A (2008). Sustained TGF beta exposure suppresses Smad and non-Smad signalling in mammary epithelial cells, leading to EMT and inhibition of growth arrest and apoptosis. Oncogene 27, 1218–1230. [DOI] [PubMed] [Google Scholar]
- Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosolini M, Camus M, Berger A, Wind P, et al. (2006). Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313, 1960–1964. [DOI] [PubMed] [Google Scholar]
- Gao Y, Souza-Fonseca-Guimaraes F, Bald T, Ng SS, Young A, Ngiow SF, Rautela J, Straube J, Waddell N, Blake SJ, et al. (2017). Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat. Immunol. 18. [DOI] [PubMed] [Google Scholar]
- Genestier L, Kasibhatla S, Brunner T, and Green DR (1999). Transforming growth factor beta1 inhibits Fas ligand expression and subsequent activation-induced cell death in T cells via downregulation of c-Myc. J. Exp. Med. 189, 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al. (2015). The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 21, 938–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiringhelli F, Puig PE, Roux S, Parcellier A, Schmitt E, Solary E, Kroemer G, Martin F, Chauffert B, and Zitvogel L (2005). Tumor cells convert immature myeloid dendritic cells into TGF-β–secreting cells inducing CD4 + CD25 + regulatory T cell proliferation. J. Exp. Med. 202, 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomini MM, Travis MA, Kudo M, and Sheppard D (2012). Epithelial cells utilize cortical actin/myosin to activate latent TGF-β through integrin αvβ6-dependent physical force. Exp. Cell Res. 318, 716–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelik L, and Flavell RA (2000). Abrogation of TGFbeta signaling in T cells leads to spontaneous T cell differentiation and autoimmune disease. Immunity 12, 171–181. [DOI] [PubMed] [Google Scholar]
- Gorelik L, and Flavell RA (2001). Immune-mediated eradication of tumors through the blockade of transforming growth factor-β signaling in T cells. Nat. Med. 7, 1118–1122. [DOI] [PubMed] [Google Scholar]
- Gorelik L, Constant S, and Flavell RA (2002). Mechanism of transforming growth factor beta-induced inhibition of T helper type 1 differentiation. J. Exp. Med. 195, 1499–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasch G, Schober M, Pasolli HA, Conn EB, Polak L, and Fuchs E (2007). Loss of TGFβ Signaling Destabilizes Homeostasis and Promotes Squamous Cell Carcinomas in Stratified Epithelia. Cancer Cell 12, 313–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, et al. (2015). The consensus molecular subtypes of colorectal cancer. Nat. Med. 21, 1350–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn JN, Falck VG, and Jirik FR (2011). Smad4 deficiency in T cells leads to the Th17-associated development of premalignant gastroduodenal lesions in mice. J. Clin. Invest. 121, 4030–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks BA, Holtzhausen A, Evans KS, Jamieson R, Gimpel P, Campbell OM, Hector-Greene M, Sun L, Tewari A, George A, et al. (2013). Type III TGF-β receptor downregulation generates an immunotolerant tumor microenvironment. J. Clin. Invest. 123, 3925–3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ, and Beach D (1994). p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature 371, 257–261. [DOI] [PubMed] [Google Scholar]
- Heldin C-H, and Moustakas A (2016). Signaling Receptors for TGF-β Family Members. Cold Spring Harb. Perspect. Biol. 8, a022053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgaard RB, Schaer DA, Li Y, Castaneda SP, Murphy MY, Xu X, Inigo I, Dobkin J, Manro JR, Iversen PW, et al. (2018). Targeting the TGFβ pathway with galunisertib, a TGFβRI small molecule inhibitor, promotes anti-tumor immunity leading to durable, complete responses, as monotherapy and in combination with checkpoint blockade. J. Immunother. Cancer 6, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Lim S, Li AG, Lee C, Lee YS, Lee E-K, Park SH, Wang X-J, and Kim S-J (2007). Smad7 binds to the adaptors TAB2 and TAB3 to block recruitment of the kinase TAK1 to the adaptor TRAF2. Nat. Immunol. 8, 504–513. [DOI] [PubMed] [Google Scholar]
- Horiguchi K, Shirakihara T, Nakano A, Imamura T, Miyazono K, and Saitoh M (2009). Role of Ras signaling in the induction of snail by transforming growth factor-beta. J. Biol. Chem. 284, 245–253. [DOI] [PubMed] [Google Scholar]
- Ihara S, Hirata Y, and Koike K (2017). TGF-β in inflammatory bowel disease: a key regulator of immune cells, epithelium, and the intestinal microbiota. J. Gastroenterol. 52, 777–787. [DOI] [PubMed] [Google Scholar]
- Isella C, Terrasi A, Bellomo SE, Petti C, Galatola G, Muratore A, Mellano A, Senetta R, Cassenti A, Sonetto C, et al. (2015). Stromal contribution to the colorectal cancer transcriptome. Nat. Genet. 47, 312–319. [DOI] [PubMed] [Google Scholar]
- Janda E, Lehmann K, Killisch I, Jechlinger M, Herzig M, Downward J, Beug H, and Grünert S (2002). Ras and TGF[beta] cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. J. Cell Biol. 156, 299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakonen S-M, Selander KS, Chirgwin JM, Yin JJ, Burns S, Rankin WA, Grubbs BG, Dallas M, Cui Y, and Guise TA (2002). Transforming growth factor-beta stimulates parathyroid hormone-related protein and osteolytic metastases via Smad and mitogen-activated protein kinase signaling pathways. J. Biol. Chem. 277, 24571–24578. [DOI] [PubMed] [Google Scholar]
- Kalathil S, Lugade AA, Miller A, Iyer R, and Thanavala Y (2013). Higher Frequencies of GARP+CTLA-4+Foxp3+ T Regulatory Cells and Myeloid-Derived Suppressor Cells in Hepatocellular Carcinoma Patients Are Associated with Impaired T-Cell Functionality. Cancer Res. 73, 2435–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordón-Cardo C, Guise TA, and Massagué J (2003). A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3, 537–549. [DOI] [PubMed] [Google Scholar]
- Kelly A, Houston SA, Sherwood E, Casulli J, and Travis MA (2017). Regulation of Innate and Adaptive Immunity by TGFβ. Adv. Immunol. 134, 137–233. [DOI] [PubMed] [Google Scholar]
- Kelly A, Gunaltay S, McEntee CP, Shuttleworth EE, Smedley C, Houston SA, Fenton TM, Levison S, Mann ER, and Travis MA (2018). Human monocytes and macrophages regulate immune tolerance via integrin αvβ8-mediated TGFβ activation. J. Exp. Med. 215, 2725–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B-G, Li C, Qiao W, Mamura M, Kasperczak B, Anver M, Wolfraim L, Hong S, Mushinski E, Potter M, et al. (2006). Smad4 signalling in T cells is required for suppression of gastrointestinal cancer. Nature 441, 1015–1019. [DOI] [PubMed] [Google Scholar]
- Kim KK, Sheppard D, and Chapman HA (2018). TGF-β1 Signaling and Tissue Fibrosis. Cold Spring Harb. Perspect. Biol. 10, a022293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Kometani K, Hashida H, Matsunaga A, Miyoshi H, Hosogi H, Aoki M, Oshima M, Hattori M, Takabayashi A, et al. (2007). SMAD4-deficient intestinal tumors recruit CCR1+ myeloid cells that promote invasion. Nat. Genet. 39, 467–475. [DOI] [PubMed] [Google Scholar]
- Kitamura T, Fujishita T, Loetscher P, Revesz L, Hashida H, Kizaka-Kondoh S, Aoki M, and Taketo MM (2010). Inactivation of chemokine (C-C motif) receptor 1 (CCR1) suppresses colon cancer liver metastasis by blocking accumulation of immature myeloid cells in a mouse model. Proc. Natl. Acad. Sci. U. S. A. 107, 13063–13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Yoshida K, Ward JM, Letterio JJ, Longenecker G, Yaswen L, Mittleman B, Mozes E, Roberts AB, Karlsson S, et al. (1999). Beta 2-microglobulin-deficient background ameliorates lethal phenotype of the TGF-beta 1 null mouse. J. Immunol. 163, 4013–4019. [PubMed] [Google Scholar]
- Kovacs RJ, Maldonado G, Azaro A, Fernández MS, Romero FL, Sepulveda-Sánchez JM, Corretti M, Carducci M, Dolan M, Gueorguieva I, et al. (2015). Cardiac Safety of TGF-β Receptor I Kinase Inhibitor LY2157299 Monohydrate in Cancer Patients in a First-in-Human Dose Study. Cardiovasc. Toxicol. 15, 309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni AB, Huh CG, Becker D, Geiser A, Lyght M, Flanders KC, Roberts AB, Sporn MB, Ward JM, and Karlsson S (1993). Transforming growth factor beta 1 null mutation in mice causes excessive inflammatory response and early death. Proc. Natl. Acad. Sci. U. S. A. 90, 770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laouar Y, Sutterwala FS, Gorelik L, and Flavell RA (2005). Transforming growth factor-β controls T helper type 1 cell development through regulation of natural killer cell interferon-γ. Nat. Immunol. 6, 600–607. [DOI] [PubMed] [Google Scholar]
- Lee J-C, Lee K-M, Kim D-W, and Heo DS (2004). Elevated TGF-beta1 secretion and down-modulation of NKG2D underlies impaired NK cytotoxicity in cancer patients. J. Immunol. 172, 7335–7340. [DOI] [PubMed] [Google Scholar]
- Lee MK, Pardoux C, Hall MC, Lee PS, Warburton D, Qing J, Smith SM, and Derynck R (2007). TGF-beta activates Erk MAP kinase signalling through direct phosphorylation of ShcA. EMBO J. 26, 3957–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Park JS, Kim JH, Jung SM, Lee JY, Kim S-J, and Park SH (2011). Smad6-specific recruitment of Smurf E3 ligases mediates TGF-β1-induced degradation of MyD88 in TLR4 signalling. Nat. Commun. 2, 460. [DOI] [PubMed] [Google Scholar]
- Letterio JJ, Geiser AG, Kulkarni AB, Dang H, Kong L, Nakabayashi T, Mackall CL, Gress RE, and Roberts AB (1996). Autoimmunity associated with TGF-beta1-deficiency in mice is dependent on MHC class II antigen expression. J. Clin. Invest. 98, 2109–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MO, and Flavell RA (2008). Contextual regulation of inflammation: a duet by transforming growth factor-beta and interleukin-10. Immunity 28, 468–476. [DOI] [PubMed] [Google Scholar]
- Li MO, Sanjabi S, and Flavell RA (2006). Transforming Growth Factor-β Controls Development, Homeostasis, and Tolerance of T Cells by Regulatory T Cell-Dependent and -Independent Mechanisms. Immunity 25, 455–471. [DOI] [PubMed] [Google Scholar]
- Li MO, Wan YY, and Flavell RA (2007). T Cell-Produced Transforming Growth Factor-β1 Controls T Cell Tolerance and Regulates Th1- and Th17-Cell Differentiation. Immunity 26, 579–591. [DOI] [PubMed] [Google Scholar]
- Li Z, Pang Y, Gara SK, Achyut BR, Heger C, Goldsmith PK, Lonning S, and Yang L (2012). Gr-1+CD11b+ cells are responsible for tumor promoting effect of TGF-β in breast cancer progression. Int. J. Cancer 131, 2584–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liénart S, Merceron R, Vanderaa C, Lambert F, Colau D, Stockis J, Woning B, van der Haard, De H, Saunders M, Coulie PG, et al. (2018). Structural basis of latent TGF-β1 presentation and activation by GARP on human regulatory T cells. Science (80-. ). 362, 952–956. [DOI] [PubMed] [Google Scholar]
- Lin JT, Martin SL, Xia L, and Gorham JD (2005). TGF-beta 1 uses distinct mechanisms to inhibit IFN-gamma expression in CD4+ T cells at priming and at recall: differential involvement of Stat4 and T-bet. J. Immunol. 174, 5950–5958. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang P, Li J, Kulkarni AB, Perruche S, and Chen W (2008). A critical function for TGF-β signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat. Immunol. 9, 632–640. [DOI] [PubMed] [Google Scholar]
- Macias MJ, Martin-Malpartida P, and Massagué J (2015). Structural determinants of Smad function in TGF-β signaling. Trends Biochem. Sci. 40, 296–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay LK, Wynne-Jones E, Freestone D, Pellicci DG, Mielke LA, Newman DM, Braun A, Masson F, Kallies A, Belz GT, et al. (2015). T-box Transcription Factors Combine with the Cytokines TGF-β and IL-15 to Control Tissue-Resident Memory T Cell Fate. Immunity 43, 1101–1111. [DOI] [PubMed] [Google Scholar]
- Malladi S, Macalinao DG, Jin X, He L, Basnet H, Zou Y, de Stanchina E, and Massagué J (2016). Metastatic Latency and Immune Evasion through Autocrine Inhibition of WNT. Cell 165, 45–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mami-Chouaib F, Blanc C, Corgnac S, Hans S, Malenica I, Granier C, Tihy I, and Tartour E (2018). Resident memory T cells, critical components in tumor immunology. J. Immunother. Cancer 6, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcoe JP, Lim JR, Schaubert KL, Fodil-Cornu N, Matka M, McCubbrey AL, Farr AR, Vidal SM, and Laouar Y (2012). TGF-β is responsible for NK cell immaturity during ontogeny and increased susceptibility to infection during mouse infancy. Nat. Immunol. 13, 843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, Kadel III EE, Koeppen H, Astarita JL, Cubas R, et al. (2018). TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie JC, Liggitt D, and Rudensky AY (2006). Cellular Mechanisms of Fatal Early-Onset Autoimmunity in Mice with the T Cell-Specific Targeting of Transforming Growth Factor-β Receptor. Immunity 25, 441–454. [DOI] [PubMed] [Google Scholar]
- Martinez GJ, Zhang Z, Reynolds JM, Tanaka S, Chung Y, Liu T, Robertson E, Lin X, Feng X-H, and Dong C (2010). Smad2 positively regulates the generation of Th17 cells. J. Biol. Chem. 285, 29039–29043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J (2012). TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKarns SC, Schwartz RH, and Kaminski NE (2004). Smad3 Is Essential for TGF-β1 to Suppress IL-2 Production and TCR-Induced Proliferation, but Not IL-2-Induced Proliferation. J. Immunol. 172, 4275–4284. [DOI] [PubMed] [Google Scholar]
- Melisi D, Garcia-Carbonero R, Macarulla T, Pezet D, Deplanque G, Fuchs M, Trojan J, Oettle H, Kozloff M, Cleverly A, et al. (2018). Galunisertib plus gemcitabine vs. gemcitabine for first-line treatment of patients with unresectable pancreatic cancer. Br. J. Cancer 119, 1208–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mempel TR, Pittet MJ, Khazaie K, Weninger W, Weissleder R, von Boehmer H, and von Andrian UH (2006). Regulatory T Cells Reversibly Suppress Cytotoxic T Cell Function Independent of Effector Differentiation. Immunity 25, 129–141. [DOI] [PubMed] [Google Scholar]
- Metelli A, Wu BX, Fugle CW, Rachidi S, Sun S, Zhang Y, Wu J, Tomlinson S, Howe PH, Yang Y, et al. (2016). Surface Expression of TGFβ Docking Receptor GARP Promotes Oncogenesis and Immune Tolerance in Breast Cancer. Cancer Res. 76, 7106–7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moo-Young TA, Larson JW, Belt BA, Tan MC, Hawkins WG, Eberlein TJ, Goedegebuure PS, and Linehan DC (2009). Tumor-derived TGF-beta mediates conversion of CD4+Foxp3+ regulatory T cells in a murine model of pancreas cancer. J. Immunother. 32, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustakas A, and Heldin C-H (2005). Non-Smad TGF-beta signals. J. Cell Sci. 118, 3573–3584. [DOI] [PubMed] [Google Scholar]
- Mullen AC, and Wrana JL (2017). TGF-β Family Signaling in Embryonic and Somatic Stem-Cell Renewal and Differentiation. Cold Spring Harb. Perspect. Biol. 9, a022186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Lovén J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, and Young RA (2011). Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell 147, 565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiki Y, Michelsen KS, Zhang W, Chen S, Doherty TM, and Arditi M (2005). Transforming growth factor-beta differentially inhibits MyD88-dependent, but not TRAM- and TRIF-dependent, lipopolysaccharide-induced TLR4 signaling. J. Biol. Chem. 280, 5491–5495. [DOI] [PubMed] [Google Scholar]
- Nakanishi Y, Duran A, L’Hermitte A, Shelton PM, Nakanishi N, Reina-Campos M, Huang J, Soldevila F, Baaten BJG, Tauriello DVF, et al. (2018). Simultaneous Loss of Both Atypical Protein Kinase C Genes in the Intestinal Epithelium Drives Serrated Intestinal Cancer by Impairing Immunosurveillance. Immunity 49, 1132–1147.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandan D, and Reiner NE (1997). TGF-beta attenuates the class II transactivator and reveals an accessory pathway of IFN-gamma action. J. Immunol. 158, 1095–1101. [PubMed] [Google Scholar]
- Nieto MA, Huang RY-J, Jackson RA, and Thiery JP (2016). EMT: 2016. Cell 166, 21–45. [DOI] [PubMed] [Google Scholar]
- O’Connor-McCourt MD, Lenferink AE, Zwaagstra J, Sulea T, Collins C, Singh R, Durocher Y, Cantin C, and Koropatnick J (2017). Abstract 4688: AVID200: a novel computationally-designed TGF beta trap promoting anti-tumor T cell activity. Cancer Res. 77, 4688–4688. [Google Scholar]
- Oshimori N, and Fuchs E (2012). The harmonies played by TGF-β in stem cell biology. Cell Stem Cell 11, 751–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdamar B, Bose R, Barrios-Rodiles M, Wang H-R, Zhang Y, and Wrana JL (2005). Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science 307, 1603–1609. [DOI] [PubMed] [Google Scholar]
- Padua D, Zhang XH-F, Wang Q, Nadal C, Gerald WL, Gomis RR, and Massagué J (2008). TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell 133, 66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallotta MT, Orabona C, Volpi C, Vacca C, Belladonna ML, Bianchi R, Servillo G, Brunacci C, Calvitti M, Bicciato S, et al. (2011). Indoleamine 2,3-dioxygenase is a signaling protein in long-term tolerance by dendritic cells. Nat. Immunol. 12, 870–878. [DOI] [PubMed] [Google Scholar]
- Pang Y, Gara SK, Achyut BR, Li Z, Yan HH, Day C-P, Weiss JM, Trinchieri G, Morris JC, and Yang L (2013). TGF-β signaling in myeloid cells is required for tumor metastasis. Cancer Discov. 3, 936–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaspyridonos M, Matei I, Huang Y, do Rosario Andre M, Brazier-Mitouart H, Waite JC, Chan AS, Kalter J, Ramos I, Wu Q, et al. (2015). Id1 suppresses anti-tumour immune responses and promotes tumour progression by impairing myeloid cell maturation. Nat. Commun. 6, 6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-R, Seo G-Y, Choi A-J, Stavnezer J, and Kim P-H (2005). Analysis of transforming growth factor-beta1-induced Ig germ-line gamma2b transcription and its implication for IgA isotype switching. Eur. J. Immunol. 35, 946–956. [DOI] [PubMed] [Google Scholar]
- Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao C-A, et al. (2003). Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science 302, 1041–1043. [DOI] [PubMed] [Google Scholar]
- Pickup MW, Owens P, and Moses HL (2017). TGF-β, Bone Morphogenetic Protein, and Activin Signaling and the Tumor Microenvironment. Cold Spring Harb. Perspect. Biol. 9, a022285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietenpol JA, Holt JT, Stein RW, and Moses HL (1990). Transforming growth factor beta 1 suppression of c-myc gene transcription: role in inhibition of keratinocyte proliferation. Proc. Natl. Acad. Sci. U. S. A. 87, 3758–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurich JF, Wang Y, Linhoff MW, White LC, and Ting JP (1998). Identification of distinct regions of 5’ flanking DNA that mediate constitutive, IFN-gamma, STAT1, and TGF-beta-regulated expression of the class II transactivator gene. J. Immunol. 160, 233–240. [PubMed] [Google Scholar]
- Polyak K, Lee MH, Erdjument-Bromage H, Koff A, Roberts JM, Tempst P, and Massagué J (1994). Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell 78, 59–66. [DOI] [PubMed] [Google Scholar]
- Porta C, Rimoldi M, Raes G, Brys L, Ghezzi P, Di Liberto D, Dieli F, Ghisletti S, Natoli G, De Baetselier P, et al. (2009). Tolerance and M2 (alternative) macrophage polarization are related processes orchestrated by p50 nuclear factor kappaB. Proc. Natl. Acad. Sci. U. S. A. 106, 14978–14983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Garrison BS, Ma W, Wang R, Jiang A, Li J, Mistry M, Bronson RT, Santoro D, Franco C, et al. (2018). A Milieu Molecule for TGF-β Required for Microglia Function in the Nervous System. Cell 174, 156–171.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachidi S, Metelli A, Riesenberg B, Wu BX, Nelson MH, Wallace C, Paulos CM, Rubinstein MP, Garrett-Mayer E, Hennig M, et al. (2017). Platelets subvert T cell immunity against cancer via GARP-TGFβ axis. Sci. Immunol. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam R, Larmonier CB, Thurston RD, Midura-Kiela MT, Zheng SG, Ghishan FK, and Kiela PR (2012). Dendritic cell-specific disruption of TGF-β receptor II leads to altered regulatory T cell phenotype and spontaneous multiorgan autoimmunity. J. Immunol. 189, 3878–3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani R, Smulian AG, Greaves DR, Hogan SP, and Herbert DR (2011). TGF-β limits IL-33 production and promotes the resolution of colitis through regulation of macrophage function. Eur. J. Immunol. 41, 2000–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi R, Noonan KA, Pham V, Bedi R, Zhavoronkov A, Ozerov IV, Makarev E, Artemov V, A., Wysocki PT, Mehra R, et al. (2018). Bifunctional immune checkpoint-targeted antibody-ligand traps that simultaneously disable TGFβ enhance the efficacy of cancer immunotherapy. Nat. Commun. 9, 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynisdóttir I, Polyak K, Iavarone A, and Massagué J (1995). Kip/Cip and Ink4 Cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-beta. Genes Dev. 9, 1831–1845. [DOI] [PubMed] [Google Scholar]
- Robinson RT, and Gorham JD (2007). TGF-beta 1 regulates antigen-specific CD4+ T cell responses in the periphery. J. Immunol. 179, 71–79. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ruiz ME, Rodríguez I, Mayorga L, Labiano T, Barbes B, Etxeberria I, Ponz-Sarvise M, Azpilikueta A, Bolaños E, Sanmamed MF, et al. (2019). TGFβ Blockade Enhances Radiotherapy Abscopal Efficacy Effects in Combination with Anti-PD1 and Anti-CD137 Immunostimulatory Monoclonal Antibodies. Mol. Cancer Ther. 18, 621–631. [DOI] [PubMed] [Google Scholar]
- Sad S, and Mosmann TR (1994). Single IL-2-secreting precursor CD4 T cell can develop into either Th1 or Th2 cytokine secretion phenotype. J. Immunol. 153, 3514–3522. [PubMed] [Google Scholar]
- Sanjabi S, Oh SA, and Li MO (2017). Regulation of the Immune Response by TGF-β: From Conception to Autoimmunity and Infection. Cold Spring Harb. Perspect. Biol. 9, a022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandura JM, Boccuni P, Massagué J, and Nimer SD (2004). Transforming growth factor beta-induced cell cycle arrest of human hematopoietic cells requires p57KIP2 up-regulation. Proc. Natl. Acad. Sci. U. S. A. 101, 15231–15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J, Le H-V, Shen L, Anderson SA, and Massagué J (2004). Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117, 211–223. [DOI] [PubMed] [Google Scholar]
- Sethi N, Dai X, Winter CG, and Kang Y (2011). Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell 19, 192–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Zhu J, Wang R, Chen X, Mi L, Walz T, and Springer TA (2011). Latent TGF-β structure and activation. Nature 474, 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull MM, Ormsby I, Kier AB, Pawlowski S, Diebold RJ, Yin M, Allen R, Sidman C, Proetzel G, Calvin D, et al. (1992). Targeted disruption of the mouse transforming growth factor-β1 gene results in multifocal inflammatory disease. Nature 359, 693–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Śledzińska A, Hemmers S, Mair F, Gorka O, Ruland J, Fairbairn L, Nissler A, Müller W, Waisman A, Becher B, et al. (2013). TGF-β Signalling Is Required for CD4+ T Cell Homeostasis But Dispensable for Regulatory T Cell Function. PLoS Biol. 11, e1001674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythies LE, Sellers M, Clements RH, Mosteller-Barnum M, Meng G, Benjamin WH, Orenstein JM, and Smith PD (2005). Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J. Clin. Invest. 115, 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino A, Thakur N, Grimsby S, Marcusson A, von Bulow V, Schuster N, Zhang S, Heldin C-H, and Landström M (2008). The type I TGF-beta receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat. Cell Biol. 10, 1199–1207. [DOI] [PubMed] [Google Scholar]
- Spender LC, Ferguson GJ, Hughes GD, Davies BR, Goldberg FW, Herrera B, Taylor RG, Strathearn LS, Sansom OJ, Barry ST, et al. (2019). Preclinical Evaluation of AZ12601011 and AZ12799734, Inhibitors of Transforming Growth Factor β Superfamily Type 1 Receptors. Mol. Pharmacol. 95, 222–234. [DOI] [PubMed] [Google Scholar]
- Stockis J, Liénart S, Colau D, Collignon A, Nishimura SL, Sheppard D, Coulie PG, and Lucas S (2017). Blocking immunosuppression by human Tregs in vivo with antibodies targeting integrin αVβ8. Proc. Natl. Acad. Sci. U. S. A. 114, E10161–E10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strainic MG, Shevach EM, An F, Lin F, and Medof ME (2013). Absence of signaling into CD4+ cells via C3aR and C5aR enables autoinductive TGF-β1 signaling and induction of Foxp3+ regulatory T cells. Nat. Immunol. 14, 162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaka N, Seed RI, Cormier A, Bondesson AJ, Lou J, Elattma A, Ito S, Yanagisawa H, Hashimoto M, Ma R, et al. (2018a). Integrin αvβ8–expressing tumor cells evade host immunity by regulating TGF-β activation in immune cells. JCI Insight 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto T, Wakabayashi Y, Sekiya T, Inoue N, Morita R, Ichiyama K, Takahashi R, Asakawa M, Muto G, Mori T, et al. (2010). Smad2 and Smad3 Are Redundantly Essential for the TGF- -Mediated Regulation of Regulatory T Plasticity and Th1 Development. J. Immunol. 185, 842–855. [DOI] [PubMed] [Google Scholar]
- Tauriello DVF, Palomo-Ponce S, Stork D, Berenguer-Llergo A, Badia-Ramentol J, Iglesias M, Sevillano M, Ibiza S, Cañellas A, Hernando-Momblona X, et al. (2018). TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 554, 538–543. [DOI] [PubMed] [Google Scholar]
- Templeton AJ, McNamara MG, Šeruga B, Vera-Badillo FE, Aneja P, Ocaña A, Leibowitz-Amit R, Sonpavde G, Knox JJ, Tran B, et al. (2014). Prognostic Role of Neutrophil-to-Lymphocyte Ratio in Solid Tumors: A Systematic Review and Meta-Analysis. JNCI J. Natl. Cancer Inst. 106, dju124. [DOI] [PubMed] [Google Scholar]
- Thomas DA, and Massagué J (2005). TGF-β directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell 8, 369–380. [DOI] [PubMed] [Google Scholar]
- Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, and Tone M (2008). Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 9, 194–202. [DOI] [PubMed] [Google Scholar]
- Tran DQ, Andersson J, Wang R, Ramsey H, Unutmaz D, and Shevach EM (2009). GARP (LRRC32) is essential for the surface expression of latent TGF- on platelets and activated FOXP3+ regulatory T cells. Proc. Natl. Acad. Sci. 106, 13445–13450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis MA, and Sheppard D (2014). TGF-β Activation and Function in Immunity. Annu. Rev. Immunol. 32, 51–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu D-C, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, et al. (2011). Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 147, 577–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzachanis D, Freeman GJ, Hirano N, van Puijenbroek AAFL, Delfs MW, Berezovskaya A, Nadler LM, and Boussiotis VA (2001). Tob is a negative regulator of activation that is expressed in anergic and quiescent T cells. Nat. Immunol. 2, 1174–1182. [DOI] [PubMed] [Google Scholar]
- Vanpouille-Box C, Diamond JM, Pilones KA, Zavadil J, Babb JS, Formenti SC, Barcellos-Hoff MH, and Demaria S (2015). TGFβ Is a Master Regulator of Radiation Therapy-Induced Antitumor Immunity. Cancer Res. 75, 2232–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten A, Alaerts M, Van Laer L, and Loeys B (2016). Marfan Syndrome and Related Disorders: 25 Years of Gene Discovery. Hum. Mutat. 37, 524–531. [DOI] [PubMed] [Google Scholar]
- Viel S, Marçais A, Guimaraes FS-F, Loftus R, Rabilloud J, Grau M, Degouve S, Djebali S, Sanlaville A, Charrier E, et al. (2016). TGF-β inhibits the activation and functions of NK cells by repressing the mTOR pathway. Sci. Signal. 9, ra19. [DOI] [PubMed] [Google Scholar]
- Wahl SM, Allen JB, Weeks BS, Wong HL, and Klotman PE (1993). Transforming growth factor beta enhances integrin expression and type IV collagenase secretion in human monocytes. Proc. Natl. Acad. Sci. U. S. A. 90, 4577–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wen W, Yuan J, Helfand B, Li Y, Shi C, Tian F, Zheng J, Wang F, Chen L, et al. (2010). Immunotherapy for Human Renal Cell Carcinoma by Adoptive Transfer of Autologous Transforming Growth Factor -Insensitive CD8+ T Cells. Clin. Cancer Res. 16, 164–173. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zou Y, Nowotschin S, Kim SY, Li QV, Soh C-L, Su J, Zhang C, Shu W, Xi Q, et al. (2017). The p53 Family Coordinates Wnt and Nodal Inputs in Mesendodermal Differentiation of Embryonic Stem Cells. Cell Stem Cell 20, 70–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Zhu J, Dong X, Shi M, Lu C, and Springer TA (2012). GARP regulates the bioavailability and activation of TGFβ. Mol. Biol. Cell 23, 1129–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Duramad O, Perng OA, Reiner SL, Liu Y-J, and Qin FX-F (2007). Antagonistic nature of T helper 1/2 developmental programs in opposing peripheral induction of Foxp3+ regulatory T cells. Proc. Natl. Acad. Sci. 104, 18169–18174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windhagen A, Scholz C, Höllsberg P, Fukaura H, Sette A, and Hafler DA (1995). Modulation of cytokine patterns of human autoreactive T cell clones by a single amino acid substitution of their peptide ligand. Immunity 2, 373–380. [DOI] [PubMed] [Google Scholar]
- Wipff P-J, Rifkin DB, Meister J-J, and Hinz B (2007). Myofibroblast contraction activates latent TGF-β1 from the extracellular matrix. J. Cell Biol. 179, 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfraim LA, Walz TM, James Z, Fernandez T, and Letterio JJ (2004). p21Cip1 and p27Kip1 act in synergy to alter the sensitivity of naive T cells to TGF-beta-mediated G1 arrest through modulation of IL-2 responsiveness. J. Immunol. 173, 3093–3102. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Fatyol K, Jin C, Wang X, Liu Z, and Zhang YE (2008). TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol. Cell 31, 918–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Huang J, Ren X, Gorska AE, Chytil A, Aakre M, Carbone DP, Matrisian LM, Richmond A, Lin PC, et al. (2008). Abrogation of TGFβ Signaling in Mammary Carcinomas Recruits Gr-1+CD11b+ Myeloid Cells that Promote Metastasis. Cancer Cell 13, 23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JJ, Barnes AP, Hand R, Polleux F, and Ehlers MD (2010). TGF-beta signaling specifies axons during brain development. Cell 142, 144–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin JJ, Selander K, Chirgwin JM, Dallas M, Grubbs BG, Wieser R, Massagué J, Mundy GR, and Guise TA (1999). TGF-beta signaling blockade inhibits PTHrP secretion by breast cancer cells and bone metastases development. J. Clin. Invest. 103, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J-H, Jung SM, Park SH, Kato M, Yamashita T, Lee I-K, Sudo K, Nakae S, Han JS, Kim O-H, et al. (2013). Activin receptor-like kinase5 inhibition suppresses mouse melanoma by ubiquitin degradation of Smad4, thereby derepressing eomesodermin in cytotoxic T lymphocytes. EMBO Mol. Med. 5, 1720–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wei M, Becknell B, Trotta R, Liu S, Boyd Z, Jaung MS, Blaser BW, Sun J, Benson DM, et al. (2006). Pro- and Antiinflammatory Cytokine Signaling: Reciprocal Antagonism Regulates Interferon-gamma Production by Human Natural Killer Cells. Immunity 24, 575–590. [DOI] [PubMed] [Google Scholar]
- Zhang N, and Bevan MJ (2012). TGF-beta signaling to T cells inhibits autoimmunity during lymphopenia-driven proliferation. Nat Immunol 13, 667–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, and Bevan MJ (2013). Transforming Growth Factor-β Signaling Controls the Formation and Maintenance of Gut-Resident Memory T Cells by Regulating Migration and Retention. Immunity 39, 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Yang X, Pins M, Javonovic B, Kuzel T, Kim S-J, Parijs, Van L, Greenberg NM, Liu V, Guo Y, et al. (2005). Adoptive Transfer of Tumor-Reactive Transforming Growth Factor-β–Insensitive CD8 + T Cells: Eradication of Autologous Mouse Prostate Cancer. Cancer Res. 65, 1761–1769. [DOI] [PubMed] [Google Scholar]
- Zhou L, Lopes JE, Chong MMW, Ivanov II, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY, et al. (2008). TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature 453, 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]