Abstract
Organisms are locally adapted when members of a population have a fitness advantage in one location relative to conspecifics in other geographies. For example, across latitudinal gradients, some organisms may trade off between traits that maximize fitness components in one, but not both, of somatic maintenance or reproductive output. Latitudinal gradients in life history strategies are traditionally attributed to environmental selection on an animal's genotype, without any consideration of the possible impact of associated microorganisms (“microbiota”) on life history traits. Here, we show in Drosophila melanogaster, a key model for studying local adaptation and life history strategy, that excluding the microbiota from definitions of local adaptation is a major shortfall. First, we reveal that an isogenic fly line reared with different bacteria varies the investment in early reproduction versus somatic maintenance. Next, we show that in wild fruit flies, the abundance of these same bacteria was correlated with the latitude and life history strategy of the flies, suggesting geographic specificity of the microbiota composition. Variation in microbiota composition of locally adapted D. melanogaster could be attributed to both the wild environment and host genetic selection. Finally, by eliminating or manipulating the microbiota of fly lines collected across a latitudinal gradient, we reveal that host genotype contributes to latitude-specific life history traits independent of the microbiota and that variation in the microbiota can suppress or reverse the differences between locally adapted fly lines. Together, these findings establish the microbiota composition of a model animal as an essential consideration in local adaptation.
Keywords: acetic acid bacteria, Drosophila, lactic acid bacteria, latitude, life history, local adaptation, microbiome, microbiota
1 ∣. INTRODUCTION
Local adaptation is where individuals in a population are better suited to live in one geography than other members of the same species that live in a different location. Decades of documenting variation in organismal genotypes and phenotypes across geographic clines have established that locally adapted phenotypes result from environmental selection on an organism's genotype (Hereford, 2009; Kawecki & Ebert, 2004; Williams, 1966). The rationale for this study comes from abundant evidence that associated microorganisms (“microbiota”) substantially influence animal phenotypes and that their composition varies nonrandomly across geographic clines; thus, the microbiota should be considered in definitions of local adaptation. To date, research on interactions between microbiota and the life history strategy of the host has focused exclusively on how inter- and intraspecific variation in host life history strategies influences the microbiota (Emmett, Youngblut, Buckley, & Drinkwater, 2017; Neave et al., 2017). The reverse question—the impact of the microbiota on the life history strategy of the host—has, to our knowledge, rarely been considered (Kirschman & Milligan-Myhre, 2019; Macke, Tasiemski, Massol, Callens, & Decaestecker, 2017) and has not been investigated empirically.
Life history trade-offs have long been recognized as a widespread feature of local adaptation and have been the focus of many empirical studies (Hereford, 2009; Stearns, 1992). An animal's life history reflects its allocation of resources and time to maximize reproductive output, subject to natural selection and trade-offs along a “fast–slow” continuum (Lemaitre et al., 2015; Promislow & Harvey, 1990; Ricklefs & Wikelski, 2002). At the “fast” end, organisms develop to reproductive maturity more quickly and have high early fecundity, whereas a “slow” lifestyle favours somatic maintenance and lower initial reproduction across longer lifespan (MacArthur & Wilson, 1967; Pianka, 1970). These insights have been developed in the context of an environment-genotype centric framework, focused on geography-specific environmental selection on the organismal genotype mediated, for example, by temperature or photoperiod (Keller, Levsen, Ingvarsson, Olson, & Tiffin, 2011; Munch & Salinas, 2009). The consequent variation in genotype has been linked to various physiological and behavioural characters collectively described as the pace of life syndrome (Reale et al., 2010; Ricklefs & Wikelski, 2002).
The taxonomic identity and function of the animal microbiota can substantially influence animal life history traits (e.g., development rate, fecundity, lifespan) and their correlated physiological traits (Adair, Wilson, Bost, & Douglas, 2018; McFall-Ngai et al., 2013; Mushegian, Walser, Sullam, & Ebert, 2018; Shapira, 2017; Smith, McCoy, & Macpherson, 2007). Thus, factors that modify the microbiota composition can dramatically alter the adaptive traits of organisms in wild or laboratory settings. Relative to local adaptation, microbiota composition in some animals can vary across geographic clines such as latitude or altitude (Suzuki, Martins, & Nachman, 2019; Suzuki & Worobey, 2014). Currently, it is not clear if geographic patterns in microbiota composition are related to local adaptation of their hosts, but this idea is suggested by previous demonstrations that some animals can mediate their phenotypes by genetic control of their microbiota (Chaston, Dobson, Newell, & Douglas, 2016; Goodrich et al., 2014), and that the microbiota can drive rapid host evolution in a wild setting (Rudman et al., 2019). Thus, while the microbiota has the capacity to augment host genetic adaptations, clear demonstrations of this phenomenon in locally adapted populations are lacking.
Drosophila melanogaster is an excellent system to address the impact of the microbiota on host life history strategy because both its life history and microbiota are well studied. Considering its life history first, trade-offs and their role in local adaptation have been demonstrated, especially in relation to latitudinal clines in allele frequencies for fitness-associated traits (Lee et al., 2011; Parkash, Rajpurohit, & Ramniwas, 2008; Schmidt, Matzkin, Ippolito, & Eanes, 2005; Sgro et al., 2010; Travers, Garcia-Gonzalez, & Simmons, 2015), candidate genes (Oakeshott, Chambers, Gibson, & Willcocks, 1981; Overgaard, Kristensen, Mitchell, & Hoffmann, 2011; Paaby, Bergland, Behrman, & Schmidt, 2014; Schmidt et al., 2008s; Umina, Weeks, Kearney, McKechnie, & Hoffmann, 2005) and genomewide patterns (Bergland, Behrman, O'Brien, Schmidt, & Petrov, 2014; Bergland, Tobler, Gonzalez, Schmidt, & Petrov, 2016; Kolaczkowski, Kern, Holloway, & Begun, 2011). In particular, D. melanogaster adopt different life history strategies across a latitudinal gradient in the eastern United States. Flies at high latitudes, for example Maine, occupy the “slower,” somatic maintenance-promoting end of the fast–slow continuum (long lifespans and stress survival, high fat storage), whereas flies at low latitudes, for example Florida, invest in rapid development and early reproduction (Schmidt et al., 2005; Schmidt & Paaby, 2008; Sgro & Hoffmann, 2004). Turning to the microbiota, a growing body of research has revealed that the microbiota of D. melanogaster is of low diversity, represented by <100 species, usually dominated by acetic acid bacteria (AAB) of the family Acetobacteraceae, including Acetobacter species, or lactic acid bacteria (LAB) from the order Lactobacillales, including the genera Lactobacillus, Enterococcus and Leuconostoc (Bost et al., 2018; Corby-Harris et al., 2007; Ren, Webster, Finkel, & Tower, 2007; Staubach, Baines, Kunzel, Bik, & Petrov, 2013; Wong et al., 2015; Wong, Ng, & Douglas, 2011). As in many other animals, the D. melanogaster microbiota varies both among individual hosts and over time within an individual animal (Rogers et al., 2014; Wong, Chaston, & Douglas, 2013), and this variation is shaped by both deterministic factors, for example host genotype, among-microbe interactions, diet composition (Chaston et al., 2016; Coyte, Schluter, & Foster, 2015; Goodrich et al., 2016, 2014; Rakoff-Nahoum, Foster, & Comstock, 2016; Smith, Snowberg, Gregory Caporaso, Knight, & Bolnick, 2015) and stochastic processes of passive dispersal and ecological drift (Adair et al., 2018; Burns et al., 2016; Jeraldo et al., 2012; Venkataraman et al., 2015). The gut microbiota of D. melanogaster is also readily manipulated in the laboratory: it can be eliminated by bleach treatment; the dominant taxa are fully culturable; and microbial communities of defined composition can be administered by direct inoculation to bleach-sterilized fly eggs on a sterile diet, generating gnotobiotic flies (Koyle et al., 2016). If no bacteria are reapplied, the resultant “axenic” insects develop and reproduce with no evidence of generalized malaise (Ridley, Wong, Westmiller, & Douglas, 2012).
The basis for this study is the observation that presence and composition of the D. melanogaster microbiota affect key traits of D. melanogaster that underpin life history strategy, including development rate, lifespan and fecundity (Brummel, Ching, Seroude, Simon, & Benzer, 2004; Chaston, Newell, & Douglas, 2014; Clark et al., 2015; Deshpande et al., 2015; Newell et al., 2014; Ridley et al., 2012; Shin et al., 2011; Storelli et al., 2011; Yamada, Deshpande, Bruce, Mak, & Ja, 2015). We hypothesized that the microbiota might, therefore, influence patterns of local adaptation in D. melanogaster. We asked three questions: (a) How does the microbiota influence traits contributing to the life history strategy of their host? (b) Does the taxonomic composition of the microbiota in D. melanogaster vary with geographic location along the latitudinal cline in eastern United States? (c) What are the relative contributions of host genotype and the microbiota in shaping local adaptation of the host along this cline? Using studies of both laboratory and wild populations of D. melanogaster, we reveal that (a) the identity of associated microorganisms influences the position of the flies along the fast–slow axis; (b) relative abundances of key members of the microbiota in wild-caught flies correlate with life history traits and can be selected by their hosts; (c) local adaptation of the host genotype is independent of the microbiota; and (d) variation in the microbiota can amplify or suppress phenotypic differences between locally adapted fly populations. Together, these findings suggest that microbes are an essential consideration in understanding how a model animal displays its locally adapted traits.
2 ∣. MATERIALS AND METHODS
2.1 ∣. Fly rearing and bacterial culture conditions
Standard fly rearing conditions were at 25°C using a 12-hr light-dark cycle on a yeast–glucose (YG) diet (10% yeast, 10% glucose, 1.2% agar) containing 0.42% propionic acid and 0.04% phosphoric acid (Newell & Douglas, 2014). Fly lines are listed in Table S4. Wolbachia status of the flies was determined using the wsp691-R (5′-AAAAATTAAACGCTACTCCA-3′) and wsp81-F (5′-TGGTCCAATAAGTGATGAAGAAAC-3′) as described previously (Zhou, Rousset, & O'Neil, 1998) and is also reported in Table S4.
To control bacterial exposure to particular microbial partners and to test for the influence of individual microbes on life history traits, we reared flies under bacteria-free conditions or from bacteria-free eggs with an inoculated, defined microbiota. Fly eggs were collected from grape juice plates, dechorionated in 0.6% sodium hypochlorite for two 2.5 min washes, rinsed three times with sterile water and transferred to sterile YG diet (no acid preservative added) in a biosafety cabinet, as in our previous work (Koyle et al., 2016). Bacteria-free eggs were left unmanipulated, or, to rear flies with a defined microbiota, were inoculated with 50 μl bacterial culture that had been grown overnight and normalized in sterile phosphate-buffered saline to OD600 = 0.1. If multiple bacterial strains were used, they were first normalized to OD600 = 0.1, mixed in equal ratios and then inoculated to the flies in a 50 μl volume.
Bacterial strains (Table S5) were cultured on specific media: modified MRS medium (mMRS; 1.25% peptone, 0.75% yeast extract, 2% glucose, 0.5% sodium acetate, 0.2% dipotassium hydrogen phosphate, 0.2% triammonium citrate, 0.02% magnesium sulphate heptahydrate, 0.005% manganese sulphate tetrahydrate, 1.2% agar [Newell & Douglas, 2014]), potato medium (pot [Sigma P6685]), lysogeny broth (LB; 1% tryptone, 0.5% yeast extract, 0.5% sodium chloride) and brain–heart infusion (BHI, Sigma 53286). All strains were grown at 30°C except Escherichia coli, which was grown at 37°C. Strains grown under normoxia were shaken (liquid) or under ambient laboratory conditions (solid). Strains requiring hypoxia were grown statically (liquid) or in a sealed container flooded with CO2 (solid).
2.2 ∣. Bacterial abundance
Bacterial abundance was assessed in whole-body fly homogenates between 4 and 7 hr into the daily light cycle. Flies from each vial were anesthetized, and a pool of five flies was directly homogenized (no rinsing) in 125 μl homogenization buffer (10 mM Tris, pH 8, 1 mM EDTA, 0.1% Triton X-100 as in Chaston et al., 2014) with 125 μl Lysing Matrix D ceramic beads (MP Biomedicals 116540434) by shaking for 30–60 s at 4.0 m/s in a FastPrep-24 or 1,500 rpm for 2 min on a GenoGrinder 2010. The homogenate was plated onto mMRS medium twice, with dilution plating under normoxic or hypoxic conditions for enumeration of bacterial abundance, and a spot test under the reverse (hypoxia or normoxia) conditions to test for contamination by other microorganisms. After incubation, the colony morphologies were inspected visually to confirm strain identity. Where ≥200 CFU/fly of the expected bacterial strain were detected, the strain was deemed “present.” Differences between Acetobacter strains could usually not be determined by colony morphology, so Acetobacter contamination of other Acetobacter strains cannot be ruled out.
2.3 ∣. Development rate of the flies
Drosophila melanogaster development rate to pupariation or eclosion was determined by counting the number of pupae formed at 1, 6.5 and 10 hr into the daily light cycle. Unless otherwise noted, three separate experiments each with triplicate fly vials were performed for each treatment.
2.4 ∣. Starvation resistance (SR)
Starvation resistance was determined in pools of ten 5- to 7-day-old flies. Between 4 and 7 hr into the daily light cycle, flies with different bacterial treatments were separated by sex under light CO2-anaesthesia in a random order and then incubated in fly vials containing 5 ml 1% agarose under standard fly rearing conditions. Timing of fly mortality relative to the start of sorting was recorded 1, 5, 9, 13 and 17 hr after the start of the daily light cycle until all flies in a vial were dead. Unless otherwise noted, three separate experiments each with triplicate fly vials were performed for each treatment.
2.5 ∣. Glucose content
Glucose content was measured from homogenized pools of five 5- to 7-day-old female flies as in our previous work (Chaston et al., 2014). Briefly, the pool of flies was homogenized in homogenization buffer and analysed by the Sigma Glucose Assay kit (GAGO20-1KT) according to manufacturer instructions.
2.6 ∣. Lifespan
Drosophila melanogaster adult lifespan was measured by recording the number and sex of dead flies and transferring surviving flies to fresh sterile diet every 2–3 days until all flies in a vial were dead. For every transfer of adult flies (P generation) to fresh diet, one spent vial per week was retained for 2–3 weeks, when the offspring (F1 generation) were homogenized to check for bacterial persistence and contamination during transfer. Where ≥200 CFU/fly of an unexpected bacterial species were detected in 2 consecutive weeks, the flies were deemed contaminated from the first date contamination was detected. In the survival analysis, flies were marked as leaving the experiment alive at that time. At least three separate experiments with triplicate fly vials were performed for each treatment.
2.7 ∣. Fecundity
Drosophila melanogaster fecundity was defined as the number of F1 offspring per female that reached pupation and was measured in pools of 30–60 mixed sex flies aged 12–14 days post-egg deposition (approximately 2- to 4-day-old adults). First, 30–60 P generation D. melanogaster per vial were mono-associated with different bacterial strains. Two to four days after >90% of flies had eclosed, P generation adults were transferred to sterile YG diet between 8 and 10 hr into the daily light cycle. Eighteen hours later (between 2 and 4 hr into the daily light cycle the following day), the flies were transferred to new, sterile food. The number of F1 offspring that reached pupation in the spent vials was counted, and normalized to the number of live adult females that laid eggs in the vial. Three separate experiments with triplicate vials were performed on 3 consecutive days. If contaminating microbes were detected in emergent F1 flies, the vial was discarded.
2.8 ∣. 16S rRNA marker gene analysis
To test for microbiota composition of wild D. melanogaster, the V4 region of the 16S rRNA marker gene was amplified from whole-body homogenates of wild flies collected from two locations. In the eastern United States between July and November of 2009 through 2011 (Dataset S8), wild flies were collected to empty fly vials, and D. melanogaster were sorted from the mixed species pools (if any) within 16 hr and stored in ethanol with <100 other flies from the sample collection. DNA was extracted from triplicate pools of 5 whole-body flies by a salting out procedure (Cenis, Perez, & Fereres, 1993). Briefly, fly bodies were homogenized in enzymatic lysis buffer with 20 mg/ml lysozyme (Amresco, 0663) and disrupted using glass beads. Cells were then lysed via incubation with 10× extraction buffer and proteinase K. After incubation with 3 M sodium acetate, DNA was extracted from the pellet using 100% isopropanol, rinsed in 70% ethanol and resuspended in sterile TE buffer. From these extracts, the V4 region of the 16S rRNA gene was amplified as described previously (Kozich, Westcott, Baxter, Highlander, & Schloss, 2013). Primer sequences are listed in Datasets S8 and S9. Sequences were normalized using the SequalPrep Normalization kit (Invitrogen) and sequenced via 2 × 250 Illumina v2 chemistry on a HiSeq 2500 at the BYU DNA Sequencing Center. Sequence reads are available at NCBI (Bioproject numbers PRJNA589702, PRJNA589709). Operational taxonomic units (OTUs) were clustered and assigned to the sequencing data in QIIME 1.9.1 using UCLUST with open-reference OTU picking and the GreenGenes Core reference alignment at 97% similarity (Caporaso, Bittinger, et al., 2010; Caporaso, Kuczynski, et al., 2010; Edgar, 2010; McDonald et al., 2012; Wang, Garrity, Tiedje, & Cole, 2007). Taxonomy was assigned using the GreenGenes reference database (Price, Dehal, & Arkin, 2010; Werner et al., 2012). Wolbachia reads were filtered from the OTU table, which was rarefied to 65 reads per sample, which was still sufficient to near-saturate most samples (Figure S7). Spearman rank correlations between order-level OTU classifications and latitude were performed in r. Raw calculations and graphics are presented in Script S1. Geographic coordinates were estimated from the noted sampling locations before correlations between latitude and microbiota composition were calculated.
The microbiomes of flies from the state of Utah, USA, were analysed as above except for the following: wild flies were collected between 1991 and 1993 by Duane Jeffries and James Farmer, stored in ethanol, including at least some time at −20°C after placing in ethanol (Dataset S9). In 2019, DNA was extracted from individual male and female flies, using the salting out method above, based on which sex (sometimes both) was collected at these locations. OTUs were clustered and assigned to taxonomy using QIIME2 (Bolyen et al., 2019), including denoising with DADA2 including trimming forward reads to between bp 6 and 249 and reverse reads to between 6 and 229 based on quality scores, and rarefied to 11,025 reads per sample for subsequent analyses.
16S rRNA marker gene reads were also mined from shotgun sequencing data sets of fruit flies using standard procedures. A reference database for 16S rRNA sequences was built from the Silva 16S (90%) reference database using BWA (Li & Durbin, 2009), and reads from each data set (see accession number in Figure S3) were aligned to it using SAMTOOLS (Li, 2011; Li & Durbin, 2009). For each data set, any sequence in the Silva database that had 10 or more matches in that data set was retained for subsequent analysis. The taxonomic identity for the sequence was manually extracted from NCBI, and taxa were manually clustered as AAB, LAB, gamma-proteobacteria or other.
2.9 ∣. Host genetic selection on the microbiota
To test for host genetic selection on microbiota composition of clinally adapted fly lines, we enumerated bacterial abundance in wild D. melanogaster isofemale lines that were inoculated with a defined 5-species bacterial community as in our previous work (Koyle et al., 2016). The isofemale lines were derived from collections at Rocky Ridge Orchards in Bowdoin, ME, USA, and Spice Park in Homestead, FL, USA, in 2011–2012 (Table S4). After individual capture, progeny derived from a single female were grown in Philadelphia, PA on a 21-day generation cycle on a Bloomington-style diet (1 L H2O, 11.6 g agar, 147.5 g corn meal, 20.2 g soy flour, 34.9 g yeast, 165 ml molasses boiled and mixed together, then mixed with 1.25 L H2O. Separately, 4.9 g methyl paraben was dissolved in 99 ml ethanol and add to the diet after it was cooled). Of several dozen derived lines, five random lines from each geography were shipped from Philadelphia, PA, to Provo, UT, where they were reared for at least two generations on the YG diet. The defined community was introduced to bleach-sterilized fly eggs as an equal-ratio mixture (normalized to OD600 = 0.1) of strains DmCS_001, DmCS_002, DmCS_003, DmCS_004, DmCS_006 (Table S5). The microbial communities associated with the flies were assessed by dilution plating as described above. Visual differences in colony colour and morphology were used to distinguish Lactobacillaceae (large, white or yellow) and Acetobacter (small, tan) colonies. In a follow-up study, the same fly lines were reared with a 6-species inoculum composed of 4 Lactobacillaceae species isolated from wild Drosophila and the two previously used Acetobacter strains, which were isolated from laboratory Drosophila (strains DmW_98, DmW_103, DmW_181, DmW_196, DmCS_004, DmCS_006; Table S5). Samples were prepared, and microbiota composition was enumerated by the same methods as above.
2.10 ∣. Life history traits in wild isofemale D. melanogaster lines
Using only the Wolbachia-free wild isofemale lines from the host genetic selection experiment (Table S4), we measured the influence of the microbiota on life history traits in flies reared under different microbial treatments: bacteria free; 5- and 6-species gnotobiotic; and in mono-association with each of LAB strains DmCS_002, DmW_098, DmW_107100, and DmW_140; and AAB strains DmCS_006, DmW_043, DmW_045, DmW_12512 (Table S5). Then, development rate and SR were measured as described above. Early fecundity was also measured as described above, but only for 1- to 4-day-old adults.
2.11 ∣. Statistical analyses
To define the relationships between life history traits in mono-associated CantonS flies (Figure 1), Pearson (if normal by a Shapiro test) or Spearman rank (if not normal by a Shapiro test) correlations between mean phenotype values were calculated in r (R Core Team, 2018). All assays were performed on the same fly genotype using the same diet formulation, bacterial strains and general methods. One major bifurcation in the data is that the previously published TAG content, development rate, feeding rate and glucose content data were collected in Ithaca, NY, whereas the SR, lifespan and fecundity data were collected in Provo, UT. The previously published data were used for different purposes, and this is a nonredundant analysis of those data. Significant effects of bacterial treatment were defined using Cox proportional hazards model or a Kruskal–Wallis test.
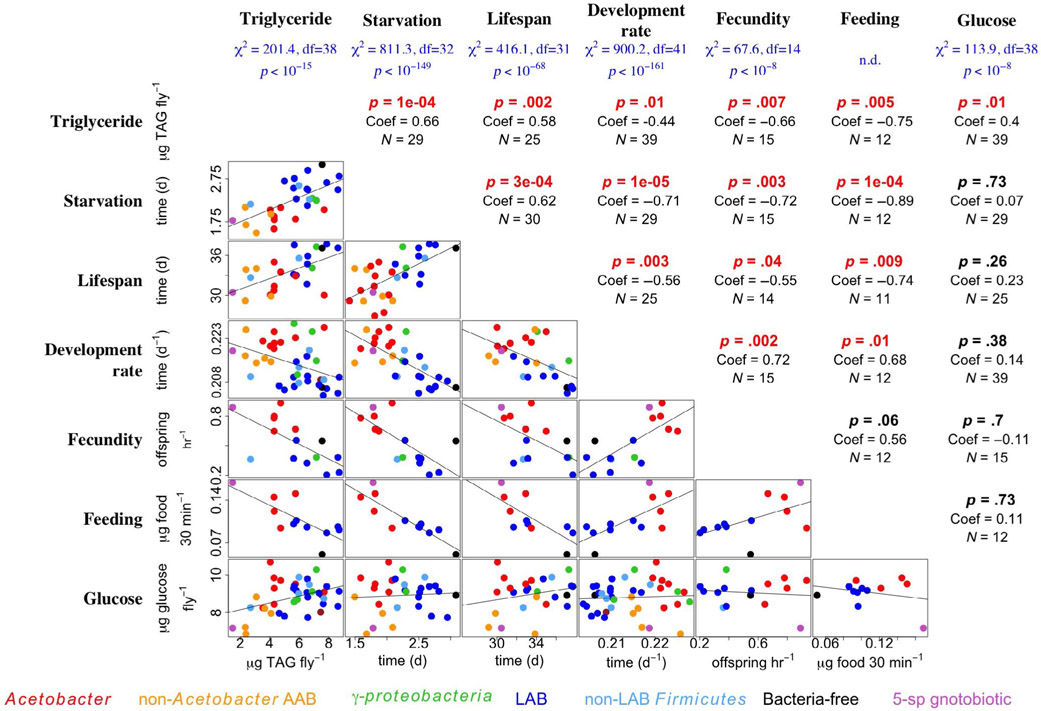
FIGURE 1.
Microbial variation influences life history patterns in a laboratory-reared isogenic fly line. Six life history traits were measured in Drosophila melanogaster that were mono-associated with different bacterial species and reared on a YG diet: whole-body triacylglyceride content (Triglyceride), survival under starvation conditions (Starvation), lifespan, the rate of development to pupariation (Development rate), number of pupariating offspring produced in the first 2–4 days after eclosion (Fecundity) and feeding rate (Feeding). Fly whole-body glucose content (Glucose), a trait that is not correlated with most other life history traits, was also measured. Significant influence of microbial treatment on the trait is shown at the top of the figure in blue under each trait. For triglyceride, fecundity and glucose, the chi-square statistic is from a Kruskal–Wallis test; for the other traits, it is from a Cox proportional hazards model. In the table portion, mean trait values conferred by different bacteria are plotted in the bottom half whereas the top half shows the results of correlation tests between traits in flies reared individually with the same microbe: p-values (p), correlation coefficients (Coef) and number of different mono-associations (N). p-values that were significant are shown in red. The data for triglyceride content, SR, development rate and feeding rate were published previously (Chaston et al., 2014; Judd et al., 2014; Newell et al., 2014)
To test for statistically significant differences in the CFU abundance data in Figure 2c and Figures S4 and S5, we used two approaches. First, we examined the raw CFU counts as a ratio of Lactobacillaceae to Acetobacter abundance in flies grouped by geographic cline or fly genotype using a generalized linear mixed-effects model with a binomial family (Bates, Maechler, Bolker, & Walker,2015; Hothorn, Bretz, & Westfall, 2008; R Core Team, 2018). If there was a cline-specific difference, then the difference in the microbiota of fly lines was tested using fly genotype as a fixed effect instead of geographic cline. We also compared the absolute abundances of the bacteria. A Shapiro test was used to confirm the CFU abundances were not distributed normally and differences between bacterial abundances in lines from ME and FL geographies were determined by a Kruskal-Wallis test. Differences in bacterial abundances by fly line were determined by a Dunn test with Benjamini–Hochberg correction after confirming a significant line effect by a Kruskal–Wallis test. All tests were performed in r (Dinno, 2017; Mangiafico, 2017; R Core Team, 2018).
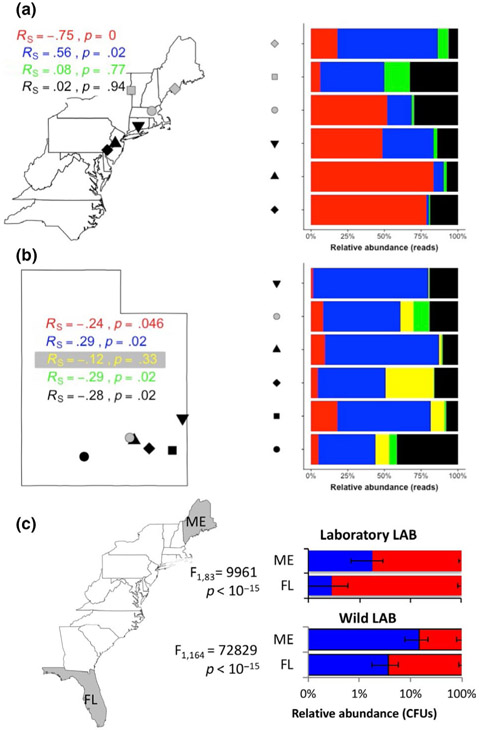
FIGURE 2.
Latitudinal variation in the microbiota of wild sampled Drosophila melanogaster. (a) Relative abundances of reads assigned to different bacterial orders in a 16S rRNA marker gene survey of D. melanogaster caught fresh in the wild in 2009. Spearman's rank correlations revealed significant positive and negative correlations between latitude and AAB (red) or LAB (blue) read abundances, respectively, but not for the Enterobacteriaceae (green) or all other bacterial reads detected (black). RS, Spearman's rho. p, p-value. N = 1–3 replicate pools of 10 flies each per geographic site. (b) Relative abundances of reads assigned to different bacterial orders in a 16S rRNA marker gene survey of fresh, wild-caught flies collected in summer, 1991–1993. Spearman's rank correlations for the AAB (red), LAB (blue), Clostridiales (yellow), Enterobacteriales (green) or all other bacterial reads detected (black). N = 6–15 individual flies per sampled location and time. (c) Relative abundance of AAB (red) and LAB (blue) in isofemale lines derived from Maine (ME) and Florida (FL) wild populations, kept in the laboratory for several years and then reared in the laboratory under gnotobiotic conditions. Flies were reared with a 5-species microbiota, including 3 LAB isolated from laboratory D. melanogaster (Laboratory LAB) or with a 6-species gnotobiotic microbiota, including 4 LAB isolated from wild D. melanogaster (Wild LAB). The statistical difference between relative LAB and AAB abundance was determined by a generalized linear mixed (GLM) effects model using a binomial family. F, F statistic of the GLM. N = 9 per treatment (triplicate vials in three separate experiments), except where vials were discarded for contamination
For Figure 3 analyses, Cox proportional hazards models were used to test for differences in development rate and SR in isofemale lines from ME and FL reared under different microbiota treatments (Therneau, 2018, 2014). Differences in fecundity were determined by a Kruskal–Wallis test. Compact letter displays identifying significant differences between treatments were defined using the multcomp package (Hothorn et al., 2008). The number of flies per vial in each condition is reported in Table S6.
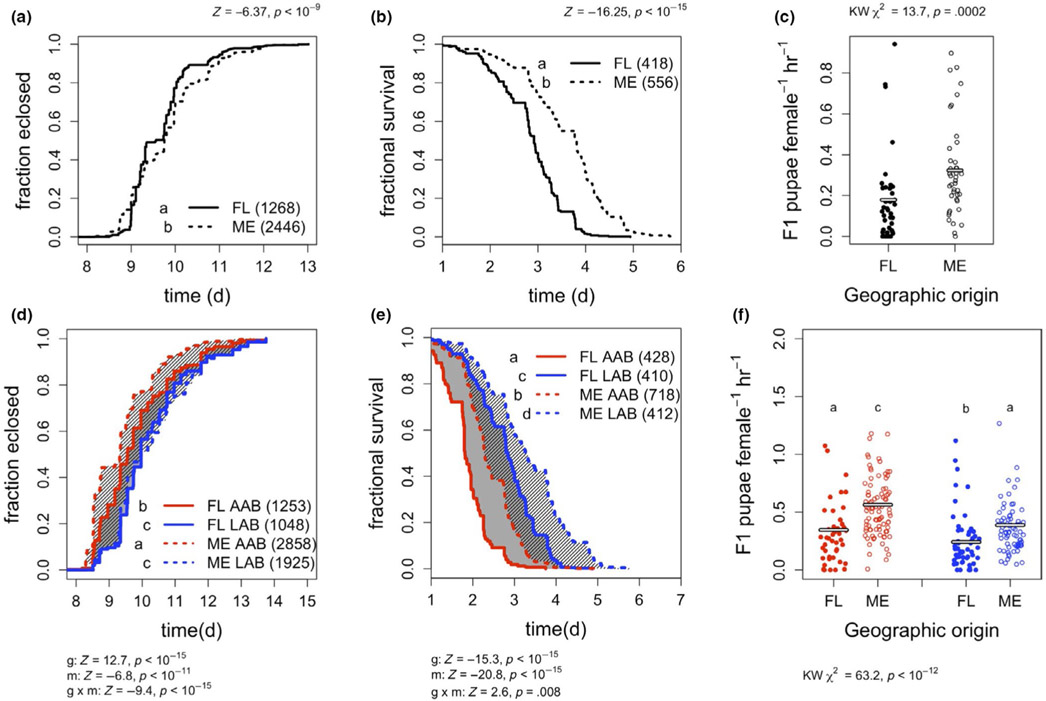
FIGURE 3.
Microbial presence and identity influence life history of wild female Drosophila melanogaster. Locally adapted ME (dashed lines) and FL (solid lines) flies reared with different bacterial treatments were tested for variation in time to eclosion (a, d), SR (b, e) and fecundity (c, f) when reared bacteria free (a–c); or in mono-association with AAB (red lines) or LAB (blue lines) strains (d–f). Solid or hatched shading represents the capacity for microbe-dependent variation in FL or ME fly phenotypes, respectively. Data were collected from triplicate vials in three separate experiments, except where vials were discarded for contamination or where low egg yields reduced the number of vials (exact N in parentheses and Table S6). Significant differences in the development or survival curves were determined by a Cox proportional hazards model (Z- and p-values are next to the genotype [g; or data in a, b], microbiota [m], or interactive [g × m] effect). Differences in fecundity were determined by a Kruskal–Wallis test, followed by a Dunn test for multiple comparisons. Different letters next to the legends represent significant differences between treatments
The code used to produce each of the figures, including many raw statistical outputs, is included in Script S1.
3 ∣. RESULTS
3.1 ∣. The microbiota influences Drosophila melanogaster life history strategy
In an evaluation of previously and newly collected data sets (Datasets S1-S7; Chaston et al., 2014; Newell et al., 2014), we noticed correlated influences of AAB and LAB on D. melanogaster life history traits. Specifically, rearing the isogenic D. melanogaster CantonS line with different bacterial strains led to two distinct outcomes. First, as was shown in the previously cited analyses, rearing the flies with different bacteria led to significant variation in the phenotypes (Figure 1, blue text at top). Second, the variation conferred by the different species manifested as a trade-off between early reproduction and somatic maintenance (Figure 1, central panels). In other words, bacteria that conferred fast development rates and high early fecundity had lower lipid (TAG) levels, lifespan and starvation resistance (SR) than strains that had low early reproduction and development rates. The correlations between phenotypes were specific to the investigated traits since SR, lifespan, fecundity, and development and feeding rates were not correlated with glucose content (Figure 1), a nutritional index that is not usually considered with other life history traits (Hoffmann & Harshman, 1999). Among all tested life history traits, the correlation coefficients were consistent with established patterns of life history trade-offs in D. melanogaster (Hoffmann & Harshman, 1999; Kalra & Parkash, 2014) although one correlation with low replication (n = 12) was not significant (Figure 1). Together, this analysis reveals that variation in the identify of D. melanogaster-associated microorganisms can shift the phenotype of an isogenic host between a fast and slow strategy.
3.2 ∣. The environment and host genetic selection contribute to geographic patterns in the D. melanogaster microbiota
To better understand the relationship between variation in the microbiota and the life history strategy adopted by an animal, we turned to the well-studied latitudinal cline in the eastern United States. Across this cline, low- and high-latitude flies, respectively, invest preferentially in fast and slow strategies (Schmidt et al., 2005; Schmidt & Paaby, 2008). We predicted that if the microbiota composition of the flies was consistent with the influence of microbes on the animal's life history strategy, D. melanogaster from low-latitude populations would bear more AAB than flies from high latitude populations, which would be dominated by LAB. Using 16S rRNA marker gene sequencing, we determined the relative abundance of AAB and LAB in wild male flies from six sites along the eastern United States coast in 2009, all collected within a 3-week period (Figure 2a, Dataset S8). Reads were clustered at the order level since LAB are an order-level designation; AAB, from the family Acetobacteraceae represented 99.97% of the Rhodospirillales reads, and Rhodospirillales reads are referred to as AAB hereafter for simplicity. Consistent with our predictions, relative AAB and LAB abundances were negatively and positively correlated with latitude, respectively (Figure 2a). When we broadened our analysis across this cline to include samples collected from the same or additional sites in two subsequent years, similar, but not identical, trends were apparent. For example, the correlation coefficients for the relationship of latitude with AAB or LAB abundance had the same directionality, but the significance of the correlation between relative LAB abundance and latitude was no longer significant (Figure S1A). These findings show heterogeneity in the spatial patterns of the D. melanogaster microbiota and may suggest that additional factors, such as season, contribute to geographic variation in the D. melanogaster microbiota. Additionally, the spatial patterning in the microbiota with latitude was not restricted to the eastern United States, as the microbiota of wild-caught flies sampled from various locations in southern Utah, USA, showed that AAB and LAB read abundances were negatively and positively correlated with latitude, respectively (Figure 2b, Figure S2, Dataset S9). Therefore, while these results do not provide a comprehensive view of spatial patterning in the microbiota of D. melanogaster, an animal distributed across the globe, they do reveal a trade-off between the abundance of the two major taxonomic groups in the D. melanogaster microbiota with latitude at two locations in the United States and match a pattern previously observed in humans (Suzuki & Worobey, 2014), where the Firmicutes, which contain the LAB, are more abundant in individuals sampled at high latitudes.
We reasoned that numerous factors, including the environment or host genotype, could contribute to the geographic patterns we detected in the microbiota of wild flies. Therefore, we first tested whether latitudinal variation in the microbiota was dependent on the wild condition of the flies, by analysing the microbiota of wild-caught D. melanogaster that were subsequently reared in the laboratory for a short period of time (<5 generations). This was done by analysing 16S reads present in D. melanogaster whole-genome shotgun sequencing projects. In D. melanogaster collected from the eastern United States or from Australia, a second geographic area showing parallel patterns of phenotypic differentiation as is observed in the eastern United States, the negative and positive correlations of AAB and LAB reads with latitude were sometimes, but not always, detected in wild-caught flies (Figure S3, Tables S1-S3). Therefore, these patterns in microbiota are not infallibly resilient to the transfer of flies from the wild to the laboratory and depend upon unknown environmental characteristics.
We also tested if host genotype contributed to variation in the microbiota composition of geographically distinct D. melanogaster populations. The gold standard to test for host genetic influence on the microbiota is to eliminate inconstant access of the flies to different sets of microorganisms (Wong et al., 2013) by rearing the flies under microbiologically defined, gnotobiotic conditions (Chaston et al., 2016). We performed these analyses with flies kept in the laboratory since their collection in ME and FL, USA, several years prior, and reared each population from birth with defined, mixed communities of AAB and LAB. Unlike the previous analyses, we measured microbiota composition using culture-dependent methods because the taxonomic identity of the community was perfectly defined, fully recoverable by culturing, and culture-dependent and culture-independent analyses of the D. melanogaster microbiota in the wild can yield similar outcomes (Rudman et al., 2019). The first microbial community comprised 5 bacterial species all derived from laboratory Drosophila. By the time these flies reached adulthood, the ratio of AAB:LAB in the ME and FL (USA) fly populations was different, even though the flies started at birth with the same ratio of the five bacteria (Figure 2c, Figure S4, Dataset S10). Because there was a low relative abundance of LAB in both fly populations, we also conducted a second experiment. The second experiment replaced the laboratory LAB with four LAB isolated from wild D. melanogaster and yielded a fourfold increase of LAB in ME flies relative to the first experiment, which was also statistically significant (Figure 2c, Figure S5; Dataset S11). The absolute AAB abundance did not differ significantly between ME- and FL-derived flies in either of the two experiments (Figures S4 and S5), suggesting a genetic effect primarily on LAB. Thus, host genetic selection on the microbiota composition can occur in locally adapted D. melanogaster adults, yielding compositional patterns consistent with the trends seen in surveys of the wild fly microbiota.
Together, the experiments using wild-caught and laboratory-reared wild flies reveal the following: (a) wild environment-dependent geographic patterns in the D. melanogaster microbiota composition; (b) host genetic selection on the microbiota that varied with geographic source of the flies and the bacteria; and (c) host genetic control of the microbiota was stronger for LAB than for other associated bacteria. Additionally, heterogeneity was apparent when samples were included from multiple years and sampling times, emphasizing that these patterns do not provide an absolute definition of the patterns in the microbiota for global D. melanogaster populations.
3.3 ∣. Microbiota composition can influence life history differences between locally adapted D. melanogaster
The wild flies captured in the eastern United States (Figure 2a) harboured a microbiota that was consistent with the life history strategy naturally adopted by those flies (Schmidt et al., 2005; Schmidt & Paaby, 2008; Sgro & Hoffmann, 2004)—that is, low-latitude flies with “fast” traits naturally bore more AAB, which promote “fast” traits, whereas high-latitude flies with slow traits had higher loads of slow-trait-conferring LAB. The correlation between latitude, ratio of AAB:LAB and life history traits of the flies raised the question whether the microbiota variation was necessary for life history variation in the locally adapted fly populations. We defined this relationship by measuring life history traits in two sets of experiments: first in bacteria-free flies, exposing the phenotypic influence of host genotype alone, and then in flies bearing individual LAB or AAB species, to determine the genotype × microbiota interactions.
The development rate, SR and early fecundity in bacteria-free treatments of flies from ME and FL were significantly different, consistent with a role for host genotype in local adaptation (Figure 3a-c, Datasets S12-S14). Relative to FL flies, ME populations displayed the established trade-off between decreased development rate and increased SR, but unexpectedly had higher early fecundity. The same trends were observed when the flies were reared under gnotobiotic conditions (Figure S6, Datasets S12-S14). Thus, bacteria-free ME flies did not display a trade-off between fecundity and development rate, suggesting that the high fecundity of ME flies could result from a condition of our laboratory experiments, such as the use of a high-nutrient diet or the relatively low abundance of LAB in gnotobiotic flies (as in Figure 2c). Overall, the results reveal that phenotypic differences between the populations are not driven exclusively by the microbiota and confirm local adaptation in the host genotype.
The second question is whether variation in microbiota composition influences the difference in phenotypes between locally adapted fly populations. When we compared the life history traits of ME and FL fly lines reared with extreme LAB:AAB ratios by using mono-association experiments, we detected that microbiota variation could reproduce, suppress or reverse the phenotypic differences found in the bacteria-free flies (Figure 3d-f, Datasets S12-S14). For example, the different fly genotypes generally showed similar phenotypic trends to the bacteria-free state when they were reared with the same microorganisms (either AAB or LAB). However, comparing traits between flies reared with different microorganisms could eliminate (fecundity in AAB-colonized FL vs. LAB-colonized ME flies) or reverse (SR and development in AAB-colonized ME vs. LAB-colonized FL flies) the difference observed between the same genotypes when bacteria free. Thus, variation in the microbiota can suppress or reinforce phenotypic differences between locally adapted populations of D. melanogaster and, therefore, the composition of the microbiota matters for flies to adopt a locally adapted life history strategy.
4 ∣. DISCUSSION
Decades of work have established that organisms adapt to their environments in response to local environmental variation (Adrion, Hahn, & Cooper, 2015; Bueno et al., 2017; Koske, 1987; Reimer et al., 2017; Savage et al., 2002). Historically, life history adaptation has been examined from the perspective of an organism's genetic adaptations to environmental circumstances that vary in different geographic locations, such as temperature, photoperiod, nutrient availability and predator pressure. There is also clear evidence that the microorganisms living within, on or near a plant or animal exert substantial influence on host traits that contribute to the life history strategy, including for latitudinal clines in microbiota composition in both animals and plants (Bueno et al., 2017; Dikongue & Segurel, 2017; Koske, 1987; Reimer et al., 2017; Savage et al., 2002; Suzuki & Worobey, 2014). Three key findings in this study extend these existing findings to conclude that geographic variation in the microbiota is associated with local adaptation in a model animal. First, the life history strategy of Drosophila melanogaster along the “fast–slow” axis can be driven by the composition of the microbiota under experimental conditions. Second, the host genetic factors driving life history variation between locally adapted natural populations include host selection of bacterial partners with congruent effects on host life history traits. Finally, variation in the microbiota composition matters to maintain genetically controlled differences in life history traits between locally adapted fly populations.
This study revealed that the bacteria can function as a rheostat to determine the fast-slow strategy adopted by the host. The molecular basis of this effect may involve bacterial production or catabolism of key metabolites. For example, bacterial production of acetic acid and other fermentation products, as well as branched-chain amino acids, can influence the activity of insulin-like/target of rapamycin (IIS/TOR) signalling (Shin et al., 2011; Storelli et al., 2011). IIS/TOR signalling has a central role in regulating cell and organismal growth, as well as female fecundity, and, consequently, life history strategies (McGaugh et al., 2015; Oldham, 2011). Drosophila melanogaster life history and survival can also be influenced by bacterial consumption of glucose (Chaston et al., 2014; Huang & Douglas, 2015) or by synthesis of methionine (Judd et al., 2018) and B vitamins (Sannino, Dobson, Edwards, Angert, & Buchon, 2018; Wong, Dobson, & Douglas, 2014), suggesting candidate bacterial functions that may mediate shifts in D. melanogaster life history strategy. Regardless of the mechanisms under question, the different bacterial functions may influence how the insect host detects its environment (especially diet) in the fruit or other ephemeral habitat that support larval development. Thus, the many Acetobacteraceae that are highly competitive in aerobic environments with high concentrations of sugars and other readily assimilated nutrients (Lievens et al., 2015; Wong et al., 2015) may represent a reliable cue for high-nutrient but ephemeral resources, favouring a “fast” phenotype of the host, while many Lactobacillales, which utilize complex carbon and nitrogen sources that are consumed more slowly (Duar et al., 2017), favour a “slow” phenotype of the host. Evidence from several studies suggest that the blend of fermentation products produced by microbial communities of different composition in the food and gut of Drosophila may represent a reliable cue for habitats of different nutritional content and persistence (Farine, Habbachi, Cortot, Roche, & Ferveur, 2017; Fischer et al., 2017; Kim, Huang, McMullen, Newell, & Douglas, 2018).
We have also obtained evidence that host genetic selection of its microbiota plays a role in shaping the fast–slow strategy adopted by D. melanogaster. Consistent with this finding, there is a strong overlap between D. melanogaster genes associated with local adaptation and host-microbe interactions. For example, of 160 previously identified genes that vary in flies across latitudinal gradients (Fabian et al., 2012), 45% have known or predicted (GWA, transcription) effects on microbiota interactions (Chaston et al., 2016; Dobson, Chaston, & Douglas, 2016; Dobson et al., 2015) relative to 31% of 13,991 total D. melanogaster genes (X2 = 7.03, p = .008). A causal role of the microbiota in these correlations is indicated by the demonstration that TAG content of D. melanogaster is regulated in part through genetic control of microbiota composition (Chaston et al., 2016). Furthermore, the genetic determinants of Acetobacter abundance in D. melanogaster include many genes that are expressed predominantly or exclusively in neurons (Chaston et al., 2016), raising the possibility that sensory functions and behavioural traits (e.g., response to microbial volatiles, diet preference and feeding rate) can mediate differences in microbiota composition. These considerations raise the possibility that an individual fly might modify its lifespan/fecundity schedule in response to altered environmental circumstances, by seeking out and filtering a different suite of microorganisms. An important topic for future research is the significance of host genetic factors in driving microbiota composition in natural populations of D. melanogaster. These host effects are more diffuse than in many associations, for example legume–rhizobia and squid–Vibrio symbioses, where exquisite specificity is dictated by defined molecular interactions (Garg & Geetanjali, 2007; Gibson, Kobayashi, & Walker, 2008; Hillman & Goodrich-Blair, 2016; McFall-Ngai, 2014) because the D. melanogaster association is an open system, continually exposed to microbes ingested in the food. Other deterministic factors, for example among-microbe interactions, as well as stochastic processes (see Section 1), may reinforce or suppress the effect of host factors on microbiota composition. Although the relative importance of these different factors is largely unknown, the demonstration of host genetic determinants of microbiota in various open associations in animals suggests that host determinants of microbiota composition contribute to the fitness of natural populations (Benson et al., 2010; Blekhman et al., 2015; Bonder et al., 2016; Davenport et al., 2015; Gomez et al., 2017; Goodrich et al., 2016, 2014; Human Microbiome Project Consortium, 2012; Rogers et al., 2014).
The strongest evidence that variation in the microbiota composition influences the phenotypic differences between locally adapted fly populations come from our findings that the microbiota can suppress or reverse genetically programmed differences in life history traits between fly lines. For example, a ME fly population that does not maintain high LAB loads could display a poorer SR phenotype than a FL fly population rich in LAB. Thus, the microbiota composition of D. melanogaster must be geography-specific to maintain the trait differences attributed to these fly lines. A major question arising from this conclusion is: How are geography-specific patterns in microbiota determined, given the established understanding that the microbiota of laboratory-reared D. melanogaster is inconstant and varies substantially with diet (Chandler, Lang, Bhatnagar, Eisen, & Kopp, 2011; Staubach et al., 2013; Wong et al., 2013)? While all the details to address this gap are unknown, our data confirm that both environmental and host genetic influences together contribute to these processes: wild flies reared in the laboratory for only a few generations failed to display the same patterns in microbiota composition as wild-caught flies, and wild flies reared in the laboratory with a defined starting set of microbes displayed key differences in their microbiota composition as adults. The environmental characters responsible for the variation could include temperature or diet (Chandler et al., 2011; Moghadam et al., 2018; Staubach et al., 2013), and at least some genetic factors that shape the microbiota composition of D. melanogaster have been described (Broderick, Buchon, & Lemaitre, 2014; Dobson et al., 2015). An additional or alternative explanation is that the characteristics described for laboratory flies may not reflect the biology of wild flies, since the interactions of Drosophila and their microbiota can vary depending if the partners are from the wild or the laboratory (Blum, Fischer, Miles, & Handelsman, 2013; Gould et al., 2018; Inamine et al., 2018; Obadia et al., 2017; Pais, Valente, Sporniak, & Teixeira, 2018; Winans et al., 2017). Therefore, we cannot rule out that characteristics defining interactions between laboratory Drosophila and their microbiota, such as inconstancy, are different for laboratory and wild flies. Regardless of the mechanism, the key importance of these findings is that models to describe local adaptation will fall short if they do not account for variation in the taxonomic and functional capacity of the microbiota.
The finding that the microbiota can mask host genetic determinants of life history traits is not without precedent. In particular, laboratory studies have revealed that the penetrance of various mutations on metabolic traits of D. melanogaster is altered, and frequently reduced, in flies colonized with microorganisms, relative to axenic flies (Dobson et al., 2015). However, these experiments have not previously been conducted relative to locally adapted populations. Further research is required to elucidate the underlying mechanisms and to establish the extent to which the impact of individual microbial taxa on host life history traits in mono-associations (as used in these experiments), or controlled multispecies associations may be displayed in the taxonomically diverse microbial communities in natural fly populations.
This study extends our understanding of natural populations by combining studies of the microbiota in natural populations of D. melanogaster and laboratory analysis of precisely controlled host–microbe combinations. For example, axenic D. melanogaster is a contrived state not likely experienced by flies in the wild, but analysis of axenic flies was essential to establish the flies' genetic contributions to life history variation. Our study further emphasizes that an exclusive focus on laboratory systems can miss important interactions because it fails to reproduce the biological context underlying evolved interactions; for example, in wild flies, the difference in abundance between wild- and laboratory-fly-isolated LAB. Similarly, Drosophila-isolated Acetobacter are discordant for key functions (uric acid utilization and motility) but not taxonomy between wild versus laboratory flies (Winans et al., 2017). These issues are of general significance for the conduct of microbiome research, with parallels coming from the evidence that the microbiota of the mouse differs between laboratory inbred strains and wild (or pet-shop) mice, with associated major differences in host physiological traits, especially relating to the immune system (Beura et al., 2016; Masopust, Sivula, & Jameson, 2017; Weldon et al., 2015).
In conclusion, the impact of the microbiota on the life history strategy of D. melanogaster is most unlikely to be a unique trait of this insect species. Microbial influence on life history traits of animals may be widespread, representing an important, but hitherto neglected, determinant of intraspecific variation, including local adaptation. We recommend that analysis of the microbiota is included as an integral part of research on life history traits and local adaptation in animals, to determine the magnitude of microbial effects in different systems and to establish the proximate and ultimate mechanisms.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Duane Jeffries and the late Dr. James Farmer for collecting wild fly samples from southern Utah, and Dr. R. Paul Evans for storing the samples and making them available to us. This work was supported by a Ruth L. Kirschstein NRSA postdoctoral fellowship (1F32GM099374-01) to P.D.N, NIH Grant R01GM095372 to A.E.D., NIH Grant R01GM100366 to P.S.S. and start-up funds from Brigham Young University to J.M.C. Some sequencing was supported by an Illumina Pilot Data Sequencing Award to J.M.C.
Funding information
NRSA postdoctoral fellowship, Grant/Award Number: 1F32GM099374-01; NIH, Grant/Award Number: R01GM095372 and R01GM100366; Brigham Young University
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
DATA AVAILABILITY STATEMENT
All fly lines and bacterial strains will be made freely available upon request to the corresponding author. Sequence data are uploaded to the short read archive (Bioproject numbers PRJNA589702, PRJNA589709). Raw phenotype data are available in Dryad (Walters et al., 2019).
REFERENCES
- Adair KL, Wilson M, Bost A, & Douglas AE (2018). Microbial community assembly in wild populations of the fruit fly Drosophila melanogaster. The ISME Journal, 12(4), 959–972. 10.1038/S41396-017-0020-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrion JR, Hahn MW, & Cooper BS (2015). Revisiting classic clines in Drosophila melanogaster in the age of genomics. Trends in Genetics, 31(8), 434–444. 10.1016/j.tig.2015.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, & Walker S (2015). Fitting linear mixed-effects models using lme4. Journal of Statistical Software, 67(1), 1–48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, … Pomp D (2010). Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proceedings of the National Academy of Sciences of the United States of America, 107(44), 18933–18938. 10.1073/pnas.1007028107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland AO, Behrman EL, O'Brien KR, Schmidt PS, & Petrov DA (2014). Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genetics, 10(11), e1004775 10.1371/journal.pgen.1004775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland AO, Tobler R, Gonzalez J, Schmidt P, & Petrov D (2016). Secondary contact and local adaptation contribute to genome-wide patterns of clinal variation in Drosophila melanogaster. Molecular Ecology, 25(5), 1157–1174. 10.1111/mec.13455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, … Masopust D (2016). Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature, 532(7600), 512–516. 10.1038/nature17655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blekhman R, Goodrich JK, Huang K, Sun QI, Bukowski R, Bell JT, … Clark AG (2015). Host genetic variation impacts microbiome composition across human body sites. Genome Biology, 16, 191 10.1186/s13059-015-0759-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum JE, Fischer CN, Miles J, & Handelsman J (2013). Frequent replenishment sustains the beneficial microbiome of Drosophila melanogaster. mBio, 4(6), e00860–13. 10.1128/mBio.00860-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, … Caporaso JG (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology, 37(8), 852–857. 10.1038/S41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, … Zhernakova A (2016). The effect of host genetics on the gut microbiome. Nature Genetics, 48(11), 1407–1412. 10.1038/ng.3663 [DOI] [PubMed] [Google Scholar]
- Bost A, Franzenburg S, Adair KL, Martinson VG, Loeb G, & Douglas AE (2018). How gut transcriptional function of Drosophila melanogaster varies with the presence and composition of the gut microbiota. Molecular Ecology, 27(8), 1848–1859. 10.1111/mec.14413 [DOI] [PubMed] [Google Scholar]
- Broderick NA, Buchon N, & Lemaitre B (2014). Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. mBio, 5(3), e01117–14. 10.1128/mBio.01117-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummel T, Ching A, Seroude L, Simon AF, & Benzer S (2004). Drosophila lifespan enhancement by exogenous bacteria. Proceedings of the National Academy of Sciences of the United States of America, 101(35), 12974–12979. 10.1073/pnas.0405207101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno CG, Moora M, Gerz M, Davison J, Öpik M, Pärtel M, … Zobel M (2017). Plant mycorrhizal status, but not type, shifts with latitude and elevation in Europe. Global Ecology and Biogeography, 26, 690–699. 10.1111/geb.12582 [DOI] [Google Scholar]
- Burns AR, Stephens WZ, Stagaman K, Wong S, Rawls JF, Guillemin K, & Bohannan BJ (2016). Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. The ISME Journal, 10(3), 655–664. 10.1038/ismej.2015.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, & Knight R (2010). PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics, 26(2), 266–267. 10.1093/bioinformatics/btp636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, … Knight R (2010). QIIME allows analysis of high-throughput community sequencing data. Nature Methods, 7(5), 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenis JL, Perez P, & Fereres A (1993). Identification of aphid (Homoptera, Aphididae) species and clones by random amplified polymorphic DNA. Annals of the Entomological Society of America, 86(5), 545–550. 10.1093/aesa/86.5.545 [DOI] [Google Scholar]
- Chandler JA, Lang JM, Bhatnagar S, Eisen JA, & Kopp A (2011). Bacterial communities of diverse Drosophila species: Ecological context of a host-microbe model system. PLoS Genetics, 7(9), e1002272 10.1371/journal.pgen.1002272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaston JM, Dobson AJ, Newell PD, & Douglas AE (2016). Host genetic control of the microbiota mediates the Drosophila nutritional phenotype. Applied and Environmental Microbiology, 82(2), 671–679. 10.1128/AEM.03301-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaston JM, Newell PD, & Douglas AE (2014). Metagenome-wide association of microbial determinants of host phenotype in Drosophila melanogaster. mBio, 5(5), e01631–14. 10.1128/mBio.01631-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RI, Salazar A, Yamada R, Fitz-Gibbon S, Morselli M, Alcaraz J, … Walker DW (2015). Distinct shifts in microbiota composition during Drosophila aging impair intestinal function and drive mortality. Cell Reports, 12(10), 1656–1667. 10.1016/j.celrep.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corby-Harris V, Pontaroli AC, Shimkets LJ, Bennetzen JL, Habel KE, & Promislow DE (2007). Geographical distribution and diversity of bacteria associated with natural populations of Drosophila melanogaster. Applied and Environmental Microbiology, 73(11), 3470–3479. 10.1128/AEM.02120-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyte KZ, Schluter J, & Foster KR (2015). The ecology of the microbiome: Networks, competition, and stability. Science, 350(6261), 663–666. 10.1126/science.aad2602 [DOI] [PubMed] [Google Scholar]
- Davenport ER, Cusanovich DA, Michelini K, Barreiro LB, Ober C, & Gilad Y (2015). Genome-wide association studies of the human gut microbiota. PLoS ONE, 10(11), e0140301 10.1371/journal.pone.0140301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande SA, Yamada R, Mak CM, Hunter B, Soto Obando A, Hoxha S, & Ja WW (2015). Acidic food pH increases palatability and consumption and extends Drosophila lifespan. The Journal of Nutrition, 145(12), 2789–2796. 10.3945/jn.115.222380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikongue E, & Segurel L (2017). Latitude as a co-driver of human gut microbial diversity? BioEssays, 39(3), 1600145 10.1002/bies.201600145 [DOI] [PubMed] [Google Scholar]
- Dinno A (2017). dunn.test: Dunn's test of multiple comparisons using rank sums (Version R package version 1.3.4.). Retrieved from https://CRAN.R-project.org/package=dunn.test [Google Scholar]
- Dobson AJ, Chaston JM, & Douglas AE (2016). The Drosophila transcriptional network is structured by microbiota. BMC Genomics, 17(1), 975 10.1186/s12864-016-3307-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson AJ, Chaston JM, Newell PD, Donahue L, Hermann SL, Sannino DR, … Douglas AE (2015). Host genetic determinants of microbiota-dependent nutrition revealed by genome-wide analysis of Drosophila melanogaster. Nature Communications, 6, 6312 10.1038/ncomms7312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duar RM, Lin XB, Zheng J, Martino ME, Grenier T, Pérez-Muñoz ME, … Walter J (2017). Lifestyles in transition: Evolution and natural history of the genus Lactobacillus. FEMS Microbiology Reviews, 41(Supp_1), S27–S48. 10.1093/femsre/fux030 [DOI] [PubMed] [Google Scholar]
- Edgar RC (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26(19), 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Emmett BD, Youngblut ND, Buckley DH, & Drinkwater LE (2017). Plant phylogeny and life history shape rhizosphere bacterial microbiome of summer annuals in an agricultural field. Frontiers in Microbiology, 8, 2414 10.3389/fmicb.2017.02414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian DK, Kapun M, Nolte V, Kofler R, Schmidt PS, Schlotterer C, & Flatt T (2012). Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Molecular Ecology, 21(19), 4748–4769. 10.1111/j.1365-294X.2012.05731.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farine JP, Habbachi W, Cortot J, Roche S, & Ferveur JF (2017). Maternally-transmitted microbiota affects odor emission and preference in Drosophila larva. Scientific Reports, 7(1), 6062 10.1038/s41598-017-04922-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer CN, Trautman EP, Crawford JM, Stabb EV, Handelsman J, & Broderick NA (2017). Metabolite exchange between microbiome members produces compounds that influence Drosophila behavior. eLife, 6, 18855 10.7554/eLife [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg N, & Manchanda G (2007). Symbiotic nitrogen fixation in legume nodules: Process and signaling: A review. Dordrecht, The Netherlands: Springer. [Google Scholar]
- Gibson KE, Kobayashi H, & Walker GC (2008). Molecular determinants of a symbiotic chronic infection. Annual Review of Genetics, 42, 413–441. 10.1146/annurev.genet.42.110807.091427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez A, Espinoza JL, Harkins DM, Leong P, Saffery R, Bockmann M, … Nelson KE (2017). Host genetic control of the oral microbiome in health and disease. Cell Host & Microbe, 22(3), 269–278.e3. 10.1016/j.chom.2017.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, … Ley RE (2016). Genetic determinants of the gut microbiome in UK twins. Cell Host & Microbe, 19(5), 731–743. 10.1016/j.chom.2016.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, … Ley RE (2014). Human genetics shape the gut microbiome. Cell, 159(4), 789–799. 10.1016/j.cell.2014.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould AL, Zhang V, Lamberti L, Jones EW, Obadia B, Korasidis N, … Ludington WB (2018). Microbiome interactions shape host fitness. Proceedings of the National Academy of Sciences of the United States of America, 115(51), e11951–e11960. 10.1073/pnas.1809349115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hereford J (2009). A quantitative survey of local adaptation and fitness trade-offs. The American Naturalist, 173(5), 579–588. 10.1086/597611 [DOI] [PubMed] [Google Scholar]
- Hillman K, & Goodrich-Blair H (2016). Are you my symbiont? Microbial polymorphic toxins and antimicrobial compounds as honest signals of beneficial symbiotic defensive traits. Current Opinion in Microbiology, 31, 184–190. 10.1016/j.mib.2016.04.010 [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, & Harshman LG (1999). Desiccation and starvation resistance in Drosophila: Patterns of variation at the species, population and intrapopulation levels. Heredity, 83(Pt 6), 637–643. 10.1046/j.1365-2540.1999.00649.x [DOI] [PubMed] [Google Scholar]
- Hothorn T, Bretz F, & Westfall P (2008). Simultaneous inference in general parametric models. Biometrical Journal, 50(3), 346–363. 10.1002/bimj.200810425 [DOI] [PubMed] [Google Scholar]
- Huang JH, & Douglas AE (2015). Consumption of dietary sugar by gut bacteria determines Drosophila lipid content. Biology Letters, 11(9), 20150469 10.1098/rsbl.2015.0469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium (2012). Structure, function and diversity of the healthy human microbiome. Nature, 486(7402), 207–214. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamine H, Ellner SP, Newell PD, Luo Y, Buchon N, & Douglas A (2018). Spatiotemporally heterogeneous population dynamics of gut bacteria inferred from fecal time series data. mBio, 9(1), e01453–17. 10.1128/mBio.01453-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeraldo P, Sipos M, Chia N, Brulc JM, Dhillon AS, Konkel ME, … Goldenfeld N (2012). Quantification of the relative roles of niche and neutral processes in structuring gastrointestinal microbiomes. Proceedings of the National Academy of Sciences of the United States of America, 109(25), 9692–9698. 10.1073/pnas.1206721109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd AM, Matthews MK, Hughes R, Veloz M, Sexton CE, & Chaston JM (2018). Bacterial methionine metabolism genes influence Drosophila melanogaster starvation resistance. Applied and Environmental Microbiology, 84(17), e00662–18. 10.1128/AEM.00662-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra B, & Parkash R (2014). Trade-off of ovarian lipids and total body lipids for fecundity and starvation resistance in tropical populations of Drosophila melanogaster. Journal of Evolutionary Biology, 27(11), 2371–2385. 10.1111/jeb.12480 [DOI] [PubMed] [Google Scholar]
- Kawecki TJ, & Ebert D (2004). Conceptual issues in local adaptation. Ecology Letters, 7(12), 1225–1241. 10.1111/j.1461-0248.2004.00684.x [DOI] [Google Scholar]
- Keller SR, Levsen N, Ingvarsson PK, Olson MS, & Tiffin P (2011). Local selection across a latitudinal gradient shapes nucleotide diversity in balsam poplar, Populus balsamifera L. Genetics, 188(4), 941–952. 10.1534/genetics.111.128041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G, Huang JH, McMullen JG 2nd , Newell PD, & Douglas AE (2018). Physiological responses of insects to microbial fermentation products: Insights from the interactions between Drosophila and acetic acid. Journal of Insect Physiology, 106(Pt 1), 13–19. 10.1016/j.jinsphys.2017.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschman LJ, & Milligan-Myhre KC (2019). The costs of living together: Immune responses to the microbiota and chronic gut inflammation. Applied and Environmental Microbiology, 85(10), e02147–18. 10.1128/AEM.02147-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowski B, Kern AD, Holloway AK, & Begun DJ (2011). Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics, 187(1), 245–260. 10.1534/genetics.110.123059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koske RE (1987). Distribution of Va mycorrhizal fungi along a latitudinal temperature-gradient. Mycologia, 79(1), 55–68. 10.2307/3807744 [DOI] [Google Scholar]
- Koyle ML, Veloz M, Judd AM, Wong AC, Newell PD, Douglas AE, & Chaston JM (2016). Rearing the fruit fly Drosophila melanogaster under axenic and gnotobiotic conditions. Journal of Visualized Experiments, 113, e54219 10.3791/54219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, & Schloss PD (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Applied and Environmental Microbiology, 79(17), 5112–5120. 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SF, Chen Y, Varan AK, Wee CW, Rako L, Axford JK, … Hoffmann AA (2011). Molecular basis of adaptive shift in body size in Drosophila melanogaster: Functional and sequence analyses of the Dca gene. Molecular Biology and Evolution, 28(8), 2393–2402. 10.1093/molbev/msr064 [DOI] [PubMed] [Google Scholar]
- Lemaitre JF, Berger V, Bonenfant C, Douhard M, Gamelon M, Plard F, & Gaillard JM (2015). Early-late life trade-offs and the evolution of ageing in the wild. Proceedings of the Royal Society B: Biological Sciences, 282(1806), 20150209 10.1098/rspb.2015.0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics, 27(21), 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, & Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25(14), 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievens B, Hallsworth JE, Pozo MI, Belgacem ZB, Stevenson A, Willems KA, & Jacquemyn H (2015). Microbiology of sugar-rich environments: Diversity, ecology and system constraints. Environmental Microbiology, 17(2), 278–298. 10.1111/1462-2920.12570 [DOI] [PubMed] [Google Scholar]
- MacArthur RH, & Wilson EO (1967). The theory of island biogeography. Princeton, NJ: Princeton University Press. [Google Scholar]
- Macke E, Tasiemski A, Massol F, Callens M, & Decaestecker E (2017). Life history and eco-evolutionary dynamics in light of the gut microbiota. Oikos, 126(4), 508–531. 10.1111/oik.03900 [DOI] [Google Scholar]
- Mangiafico S (2017). rcompanion: Functions to support extension education program evaluation (Version R package version 1.10.1.). Retrieved from https://CRAN.R-project.org/package=rcompanion [Google Scholar]
- Masopust D, Sivula CP, & Jameson SC (2017). Of mice, dirty mice, and men: Using mice to understand human immunology. Journal of Immunology, 199(2), 383–388. 10.4049/jimmunol.1700453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, … Hugenholtz P (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. The ISME Journal, 6(3), 610–618. 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai MJ (2014). The importance of microbes in animal development: Lessons from the squid-vibrio symbiosis. Annual Review of Microbiology, 68, 177–194. 10.1146/annurev-micro-091313-103654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, … Wernegreen JJ (2013). Animals in a bacterial world, a new imperative for the life sciences. Proceedings of the National Academy of Sciences of the United States of America, 110(9), 3229–3236. 10.1073/pnas.1218525110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh SE, Bronikowski AM, Kuo C-H, Reding DM, Addis EA, Flagel LE, … Schwartz TS (2015). Rapid molecular evolution across amniotes of the IIS/TOR network. Proceedings of the National Academy of Sciences of the United States of America, 112(22), 7055–7060. 10.1073/pnas.1419659112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadam NN, Thorshauge PM, Kristensen TN, de Jonge N, Bahrndorff S, Kjeldal H, & Nielsen JL (2018). Strong responses of Drosophila melanogaster microbiota to developmental temperature. Fly, 12(1), 1–12. 10.1080/19336934.2017.1394558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch SB, & Salinas S (2009). Latitudinal variation in lifespan within species is explained by the metabolic theory of ecology. Proceedings of the National Academy of Sciences of the United States of America, 106(33), 13860–13864. 10.1073/pnas.0900300106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian AA, Walser JC, Sullam KE, & Ebert D (2018). The microbiota of diapause: How host-microbe associations are formed after dormancy in an aquatic crustacean. Journal of Animal Ecology, 87(2), 400–413. 10.1111/1365-2656.12709 [DOI] [PubMed] [Google Scholar]
- Neave MJ, Rachmawati R, Xun L, Michell CT, Bourne DG, Apprill A, & Voolstra CR (2017). Differential specificity between closely related corals and abundant Endozoicomonas endosymbionts across global scales. The ISME Journal, 11(1), 186–200. 10.1038/ismej.2016.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell PD, Chaston JM, Wang Y, Winans NJ, Sannino DR, Wong ACN, … Douglas AE (2014). In vivo function and comparative genomic analyses of the Drosophila gut microbiota identify candidate symbiosis factors. Frontiers in Microbiology, 5, 576 10.3389/fmicb.2014.00576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell PD, & Douglas AE(2014). Interspecies interactions determine the impact of the gut microbiota on nutrient allocation in Drosophila melanogaster. Applied and Environmental Microbiology, 80(2), 788–796. 10.1128/AEM.02742-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakeshott JG, Chambers GK, Gibson JB, & Willcocks DA (1981). Latitudinal relationships of esterase-6 and phosphoglucomutase gene frequencies in Drosophila melanogaster. Heredity, 47(3), 385–396. 10.1038/hdy.1981.99 [DOI] [PubMed] [Google Scholar]
- Obadia B, Guvener ZT, Zhang V, Ceja-Navarro JA, Brodie EL, Ja WW, & Ludington WB (2017). Probabilistic invasion underlies natural gut microbiome stability. Current Biology, 27(13), 1999–2006. 10.1016/j.cub.2017.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S (2011). Obesity and nutrient sensing TOR pathway in flies and vertebrates: Functional conservation of genetic mechanisms. Trends in Endocrinology and Metabolism, 22(2), 45–52. 10.1016/j.tem.2010.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard J, Kristensen TN, Mitchell KA, & Hoffmann AA (2011). Thermal tolerance in widespread and tropical Drosophila species: Does phenotypic plasticity increase with latitude? The American Naturalist, 178(Suppl 1), S80–S96. 10.1086/661780 [DOI] [PubMed] [Google Scholar]
- Paaby AB, Bergland AO, Behrman EL, & Schmidt PS (2014). A highly pleiotropic amino acid polymorphism in the Drosophila insulin receptor contributes to life-history adaptation. Evolution, 68(12), 3395–3409. 10.1111/evo.12546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pais IS, Valente RS, Sporniak M, & Teixeira L (2018). Drosophila melanogaster establishes a species-specific mutualistic interaction with stable gut-colonizing bacteria. PLoS Biology, 16(7), e2005710 10.1371/journal.pbio.2005710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkash R, Rajpurohit S, & Ramniwas S (2008). Changes in body melanisation and desiccation resistance in highland vs. lowland populations of D. melanogaster. Journal of Insect Physiology, 54(6), 1050–1056. 10.1016/j.jinsphys.2008.04.008 [DOI] [PubMed] [Google Scholar]
- Pianka ER (1970). R-selection and K-selection. The American Naturalist, 104(940), 592–597. 10.1086/282697 [DOI] [Google Scholar]
- Price MN, Dehal PS, & Arkin AP (2010). FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE, 5(3), e9490 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promislow DE, & Harvey PH (1990). Living fast and dying young: A comparative analysis of life-history variation among mammals. Journal of Zoology, 220,417–437. 10.1111/j.1469-7998.1990.tb04316.x [DOI] [Google Scholar]
- R Core Team (2018). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; Retrieved from http://www.R-project.org [Google Scholar]
- Rakoff-Nahoum S, Foster KR, & Comstock LE (2016). The evolution of cooperation within the gut microbiota. Nature, 533(7602), 255–259. 10.1038/nature17626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reale D, Garant D, Humphries MM, Bergeron P, Careau V, & Montiglio PO (2010). Personality and the emergence of the pace-of-life syndrome concept at the population level. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 365(1560), 4051–4063. 10.1098/rstb.2010.0208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer JD, Herrera M, Gatins R, Roberts MB, Parkinson JE, & Berumen ML (2017). Latitudinal variation in the symbiotic dinoflagellate Symbiodinium of the common reef zoantharian Palythoa tuberculosa on the Saudi Arabian coast of the Red Sea. Journal of Biogeography, 44(3), 661–673. 10.1111/jbi.12795 [DOI] [Google Scholar]
- Ren C, Webster P, Finkel SE, & Tower J (2007). Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metabolism, 6(2), 144–152. 10.1016/j.cmet.2007.06.006 [DOI] [PubMed] [Google Scholar]
- Ricklefs RE, & Wikelski M (2002). The physiology/life-history nexus. Trends in Ecology & Evolution, 17(10), 462–468. 10.1016/S0169-5347(02)02578-8 [DOI] [Google Scholar]
- Ridley EV, Wong AC, Westmiller S, & Douglas AE (2012). Impact of the resident microbiota on the nutritional phenotype of Drosophila melanogaster. PLoS ONE, 7(5), e36765 10.1371/journal.pone.0036765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GB, Kozlowska J, Keeble J, Metcalfe K, Fao M, Dowd SE, … Bruce KD (2014). Functional divergence in gastrointestinal microbiota in physically-separated genetically identical mice. Scientific Reports, 4, 5437 10.1038/srep05437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudman SM, Greenblum S, Hughes RC, Rajpurohit S, Kiratli O, Lowder DB, … Schmidt P (2019). Microbiome composition shapes rapid genomic adaptation of Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America, 116(40), 20025–20032. 10.1073/pnas.1907787116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannino DR, Dobson AJ, Edwards K, Angert ER, & Buchon N (2018). The Drosophila melanogaster gut microbiota provisions thiamine to its host. mBio, 9(2), e00155–18. 10.1128/mBio.00155-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage AM, Goodson MS, Visram S, Trapido-Rosenthal H, Wiedenmann J, & Douglas AE (2002). Molecular diversity of symbiotic algae at the latitudinal margins of their distribution: Dinoflagellates of the genus Symbiodinium in corals and sea anemones. Marine Ecology Progress Series, 244, 17–26. 10.3354/meps244017 [DOI] [Google Scholar]
- Schmidt PS, Matzkin L, Ippolito M, & Eanes WF (2005). Geographic variation in diapause incidence, life-history traits, and climatic adaptation in Drosophila melanogaster. Evolution, 59(8), 1721–1732. 10.1554/05-115.1 [DOI] [PubMed] [Google Scholar]
- Schmidt PS, & Paaby AB (2008). Reproductive diapause and life-history clines in North American populations of Drosophila melanogaster. Evolution, 62(5), 1204–1215. 10.1111/j.1558-5646.2008.00351.x [DOI] [PubMed] [Google Scholar]
- Schmidt PS, Zhu CT, Das J, Batavia M, Yang L, & Eanes WF (2008). An amino acid polymorphism in the couch potato gene forms the basis for climatic adaptation in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America, 105(42), 16207–16211. 10.1073/pnas.0805485105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgro CM, & Hoffmann AA (2004). Genetic correlations, tradeoffs and environmental variation. Heredity, 93(3), 241–248. 10.1038/sj.hdy.6800532 [DOI] [PubMed] [Google Scholar]
- Sgro CM, Overgaard J, Kristensen TN, Mitchell KA, Cockerell FE, & Hoffmann AA (2010). A comprehensive assessment of geographic variation in heat tolerance and hardening capacity in populations of Drosophila melanogaster from eastern Australia. Journal of Evolutionary Biology, 23(11), 2484–2493. 10.1111/j.1420-9101.2010.02110.x [DOI] [PubMed] [Google Scholar]
- Shapira M (2017). Host-microbiota interactions in Caenorhabditis elegans and their significance. Current Opinion in Microbiology, 38, 142–147. 10.1016/j.mib.2017.05.012 [DOI] [PubMed] [Google Scholar]
- Shin SC, Kim S-H, You H, Kim B, Kim AC, Lee K-A, … Lee W-J (2011). Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science, 334(6056), 670–674. 10.1126/science.1212782 [DOI] [PubMed] [Google Scholar]
- Smith CC, Snowberg LK, Gregory Caporaso J, Knight R, & Bolnick DI (2015). Dietary input of microbes and host genetic variation shape among-population differences in stickleback gut microbiota. The ISME Journal, 9(11), 2515–2526. 10.1038/ismej.2015.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, McCoy KD, & Macpherson AJ (2007). Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Seminars in Immunology, 19(2), 59–69. 10.1016/j.smim.2006.10.002 [DOI] [PubMed] [Google Scholar]
- Staubach F, Baines JF, Kunzel S, Bik EM, & Petrov DA (2013). Host species and environmental effects on bacterial communities associated with Drosophila in the laboratory and in the natural environment. PLoS ONE, 8(8), e70749 10.1371/journal.pone.0070749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns SC (1992). Evolution of life histories: Theory and analysis. New York, NY: Oxford University Press. [Google Scholar]
- Storelli G, Defaye A, Erkosar B, Hols P, Royet J, & Leulier F (2011). Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metabolism, 14(3), 403–414. 10.1016/j.cmet.2011.07.012 [DOI] [PubMed] [Google Scholar]
- Suzuki TA, Martins FM, & Nachman MW (2019). Altitudinal variation of the gut microbiota in wild house mice. Molecular Ecology, 28(9), 2378–2390. 10.1111/mec.14905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki TA, & Worobey M (2014). Geographical variation of human gut microbial composition. Biology Letters, 10(2), 20131037 10.1098/rsbl.2013.1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau TM (2018). coxme: Mixed Effects Cox Models. R package version 2.2-10. Retrived from https://CRAN.Rproject.org/package=coxme [Google Scholar]
- Therneau T (2014). A package for survival analysis in S (Version 2.37-7). Retrieved from http://cran.r-project.org/package=survival [Google Scholar]
- Travers LM, Garcia-Gonzalez F, & Simmons LW (2015). Live fast die young life history in females: Evolutionary trade-off between early life mating and lifespan in female Drosophila melanogaster. Scientific Reports, 5, 15469 10.1038/srep15469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umina PA, Weeks AR, Kearney MR, McKechnie SW,& Hoffmann AA (2005). A rapid shift in a classic clinal pattern in Drosophila reflecting climate change. Science, 308(5722), 691–693. 10.1126/science.1109523 [DOI] [PubMed] [Google Scholar]
- Venkataraman A, Bassis CM, Beck JM, Young VB, Curtis JL, Huffnagle GB, & Schmidt TM (2015). Application of a neutral community model to assess structuring of the human lung microbiome. mBio, 6(1), e02284–14. 10.1128/mBio.02284-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters AW, Hughes RC, Call TB, Walker CJ, Wilcox H, Petersen SC, … Chaston JM (2019). The microbiota influences the Drosophila melanogaster life history strategy, v3. Dryad, 10.5061/dryad.ttdz08kt5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, &Cole JR (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology, 73(16), 5261–5267. 10.1128/Aem.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldon L, Abolins S, Lenzi L, Bourne C, Riley EM, & Viney M (2015). The gut microbiota of wild mice. PLoS ONE, 10(8), e0134643 10.1371/journal.pone.0134643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner JJ, Koren O, Hugenholtz P, DeSantis TZ, Walters WA, Caporaso JG, … Ley RE (2012). Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. The ISME Journal, 6(1), 94–103. 10.1038/ismej.2011.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GC (1966). Adaptation and natural selection. Princeton, NJ: Princeton University Press. [Google Scholar]
- Winans NJ, Walter A, Chouaia B, Chaston JM, Douglas AE, & Newell PD (2017). A genomic investigation of ecological differentiation between free-living and Drosophila-associated bacteria. Molecular Ecology, 26(17), 4536–4550. 10.1111/mec.14232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AC, Chaston JM, & Douglas AE (2013). The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. The ISME Journal, 7(10), 1922–1932. 10.1038/ismej.2013.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AC, Dobson AJ, & Douglas AE (2014). Gut microbiota dictates the metabolic response of Drosophila to diet. Journal of Experimental Biology, 217(Pt 11), 1894–1901. 10.1242/jeb.101725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AC, Luo Y, Jing X, Franzenburg S, Bost A, & Douglas AE (2015). The host as the driver of the microbiota in the gut and external environment of Drosophila melanogaster. Applied and Environmental Microbiology, 81(18), 6232–6240. 10.1128/AEM.01442-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CN, Ng P, & Douglas AE (2011). Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environmental Microbiology, 13(7), 1889–1900. 10.1111/j.1462-2920.2011.02511.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada R, Deshpande SA, Bruce KD, Mak EM, & Ja WW (2015). Microbes promote amino acid harvest to rescue undernutrition in Drosophila. Cell Reports, 10(6), 865–872. 10.1016/j.celrep.2015.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Rousset E, & O'Neil S (1998). Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proceedings of the Royal Society of London. Series B: Biological Sciences, 265(1395), 509–515. 10.1098/rspb.1998.0324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All fly lines and bacterial strains will be made freely available upon request to the corresponding author. Sequence data are uploaded to the short read archive (Bioproject numbers PRJNA589702, PRJNA589709). Raw phenotype data are available in Dryad (Walters et al., 2019).